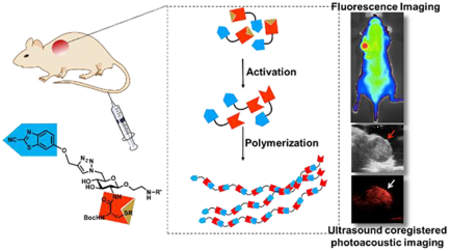
Abstract
“Smart” biomaterials that are responsive to physiological or biochemical stimuli have found many biomedical applications for tissue engineering, therapeutics and molecular imaging. In this work we describe in situ polymerization of activatable biorthogonal small molecules in response to reducing environment change in vivo. We designed a carbohydrate linker- and cyanobenzothiazole-cysteine condensation reaction-based small molecule scaffold that can undergo rapid condensation reaction upon physiochemical changes (such as a reducing environment) to form polymers (pseudopolysaccharide). The fluorescent and photoacoustic properties of fluorophore-tagged condensation scaffold before and after the transformation have been examined with a dual-modality optical imaging method. These results confirmed the in situ polymerization of this probe after both local and systemic administration in living mice.
Graphical Abstract
Introduction
Biomaterials that can undergo in situ transformation with exposure to specific biomarkers or environmental cues in living subjects can potentially lead to new biomedical applications.1 For example, preformed polymers that can cross-link or solidify in vivo after injected locally when exposed to light,2–3 temperature,4 pH changes,5 enzymes,6–7 or presence of specific ions8 can provide better tissue-material contact and can be used for facilitating local transplantation of medical scaffolds,9 tissue engineering,10–11 drug delivery12–13 and imaging.14 Compared to polymeric materials, small molecules possess properties of efficient tissue distribution, deep penetration, and fast systemic clearance in normal tissues. However, small molecules also suffer from poor retention compare to polymeric material. Thus in situ self-assembly of small molecules into large macromolecules or polymeric materials at target sites can take advantage of the efficient distribution and clearance of small molecules as well as the enhanced retention of macromolecules and offer unique opportunities in designing new materials for diagnostics and therapeutics.15–17
One of the strategies for assembling small molecules into macromolecules inside complex biological environment is through covalent bonding enabled by bioorthogonal reactions.18 These reactions such as ketone/hydroxylamine condensation,19–22 copper free azide-alkyne cycloaddition,23–26 Staudinger ligation27–29 and tetrazine-trans-cyclooctene cycloaddition,30–36 have been applied to site-directed protein labeling,37 studying metabolic pathways24, 38–39 and engineering polymeric biomaterials.40–42 Their applications in “smart” biomaterials that are responsive to physiological or biochemical stimuli are limited due to the fact that most of the chemical moieties used in these reactions cannot be biochemically masked. Recently there have been some reports on using light or enzymes to modulate bioorthogonal reactions,43–52 but not yet demonstrated in vivo. For the past few years, we have been studying the reaction between aminothiols and aromatic nitriles53–56 and its applications for in vivo molecular imaging.57–63 One of the most attractive features of this reaction is that its reactivity can be modulated by pH, reducing environment or enzyme activities by masking the aminothiol with different responsive moieties. One of the fastest reactions we’ve tested so far is between the cyanobenzothiazole (CBT) group and free cysteine, which has a second-order rate constant of 9.19 M−1s−1.53 We and others have used this reaction for protein labeling,64–67 formation of hydrogels,53, 68–69 fluorescence lifetime imaging70 and in vivo imaging of protease activity.53, 57–58 This condensation reaction therefore holds great potential for constructing “smart” molecules that can go through in situ transformation into large macromolecules or polymeric materials when encounters specific biological stimuli.
Our previously reported CBT conjugates tend to form cyclic dimers or oligomers after condensation instead of forming linear polymers due to the flexible peptide linkers within the scaffolds.53, 57 We hypothesized that a rigid linker in the scaffold would promote the elongation of the linear polymers over cyclization and provide an attractive strategy for in situ formation of hydrogels from small molecules (Fig. 1). The carbohydrate moieties are used extensively in nature to form rigid polymers, such as cellulose. The equatorial conformation of glucose in cellulose provides rigidity for the linear polysaccharides. Here we report a carbohydrate-based small molecule scaffold that can undergo CBT-cysteine condensation to form polymers under reducing conditions. We used a dual-modality optical imaging method to measure the fluorescence and photoacoustic properties (conversion of non-ionizing radiation to pressure/ultrasound waves)71–73 of new scaffold based fluorescent probes before and after the polymerization and demonstrated the in situ formation of polymers in living mice.
Figure 1. Structures of molecular probes used in the study and their activation to polymers.
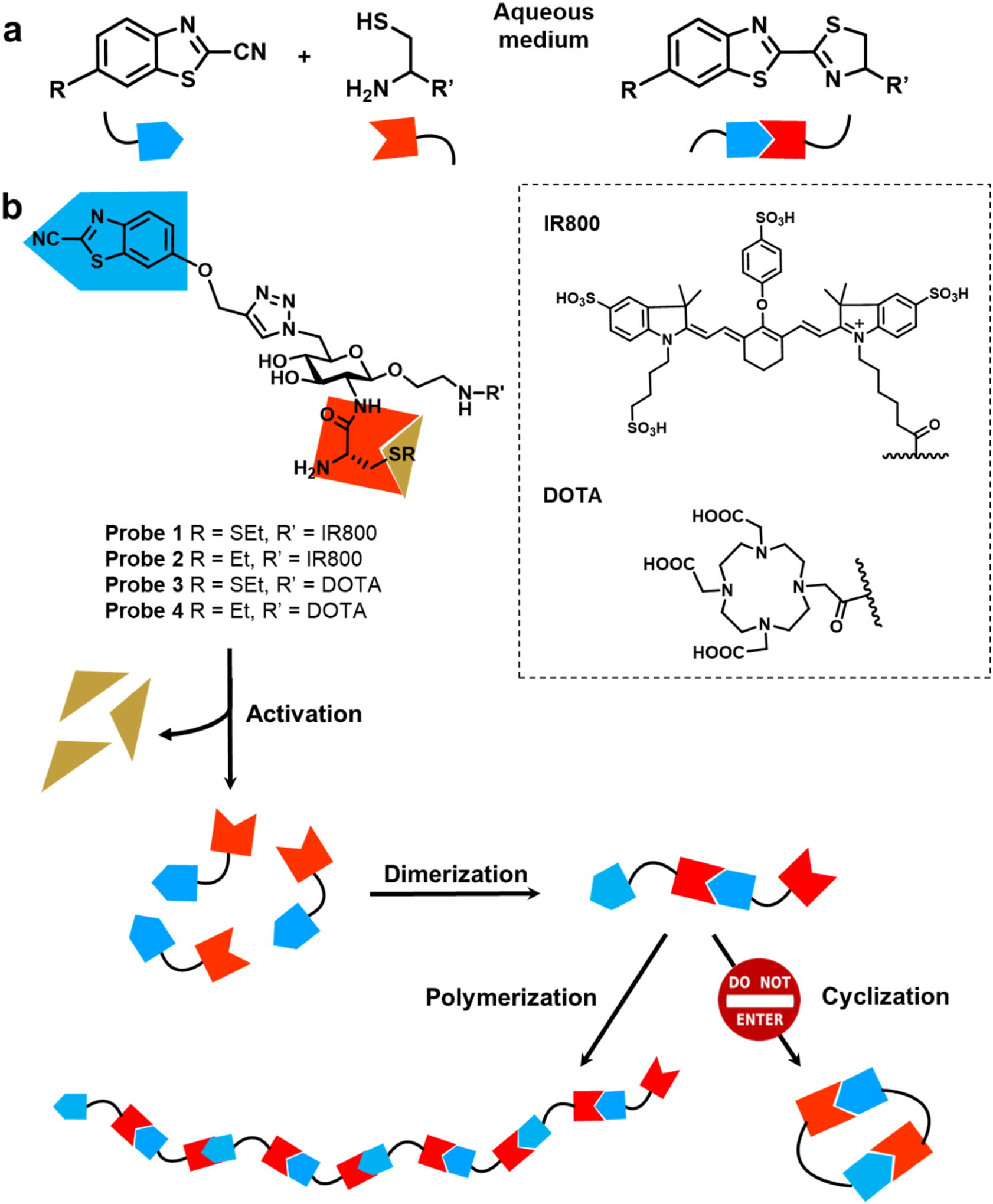
a. Condensation reaction between cyanobenzothiazole and cysteine (a.k.a. CBT Condensation). b. Probes 1 and 3 undergo polymerization upon reductive activation. Control probes 2 and 4 remain as monomers under the same conditions.
Results and discussion
Design and synthesis of polymerizable molecular scaffold.
To evaluate the polymerization reaction, we synthesized two sets of probes with D-glucosamine as the backbone to facilitate the polymerization (Fig. 1). The CBT is linked to C-6 on the glucosamine via click reaction and cysteine is linked to the C-2 amine via a peptide linker. The fluorescent probes 1 and 2 contained an IR800 dye as an imaging moiety to detect their fluorescence and photoacoustic properties. IR800 dye was selected for its suitable excitation/emission wavelength (774 nm/789 nm) for in vivo imaging, its large extinction coefficient (240,000 M−1 cm−1), and its good water solubility. The other set of probes 3 and 4 contained a 2,2’,2’’-(10-(2-((2,5-dioxopyrrolidin-1-yl)oxy)-2-oxoethyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (DOTA) moiety for its water solubility and its potential to chelate Gd3+ for magnetic resonance imaging (MRI) or radioactive metal ions such as Cu-64 or Ga-68 for positron emission tomography (PET) imaging in the future. Probes 1 and 3, containing a disulfide linkage on the cysteine, can be activated by reducing environment to generate the free N-terminal cysteine group that then reacts with CBT group on the other molecule to initiate the polymerization (Fig. 1). The control probes 2 and 4 cannot expose the thiol group in reducing environment and should remain as small molecules.
A convergent synthetic scheme was developed to obtain all probes 1-4 (Supporting Information Fig. S1). The synthesis started from free D-glucosamine, and an azido group was linked to the C-1 position to lock its chair conformation. A cysteine group was then conjugated to the amino group, and the CBT group was introduced at the C-6 position via a copper-catalyzed azide-alkyne cycloaddition (CuAAC). Both IR800 and DOTA were introduced via their NHS esters.
Characterization of polymerization reactions using SEC-MALS.
The polymerization reactions were monitored by the size exclusion chromatography (SEC) coupled with multiangle light scattering (MALS) to analyze the sizes, mass and relative distribution of the resulting polymers (Fig. 2, Supporting Fig. S2). The activated or un-activated probe 3 was injected onto a WTC-010S5 column, and the elution profile was monitored by light scattering at 658 nm and differential refractometry. The triggered polymerization was very efficient in the presence of GSH (10 mM): the majority (> 60%) of probe 3 (at a concentration of 10 mM) was converted to polymers at a size of around 40 kDa (~40 repeating units, Supporting Fig. S2 peak 1) and hydrodynamic radius of 3 nm (Supporting Fig. S2) in 10 minutes, together with monomer 3, dimer (Supporting Fig. S2 peak 4, S5), oligomers (Supporting Fig. S2 peak 3, S6) and a second portion of polymers of around 20 kDa (Supporting Fig. S2 peak 2, S4) as detected by matrix-assisted laser desorption/ionization mass spectroscopy (MALDI-MS). To evaluate the polymerization at low concentrations, probe 3 was activated at 1, 10 and 100 μM by GSH (10 mM), and the polymerization profiles were monitored from the initial 10 minutes to 8 hours after incubation in reaction buffer (Fig. 2). Formation of polymers was concentration-dependent, with larger polymer species formed at higher concentrations (Fig. 2a, 2b). At 100 μM, nearly 80% of probe 3 was converted to polymers at around 25 kDa (~25 repeating units) within the first 1 hour of reaction (Fig. 2a, 2b, 2e). At concentrations of 10 μM or even 1 μM, around 40% of probe 3 formed polymer species at around 25 kDa within 1 hour (Fig. 2a, 2b, 2c, 2d). These in vitro results demonstrated the efficiency of the polymerization reactions at various concentrations. To our delight, this polymerization can proceed efficiently at as low as a few micro molar concentrations, making it attractive for applications in living systems.
Figure 2. Characterization of polymerization.
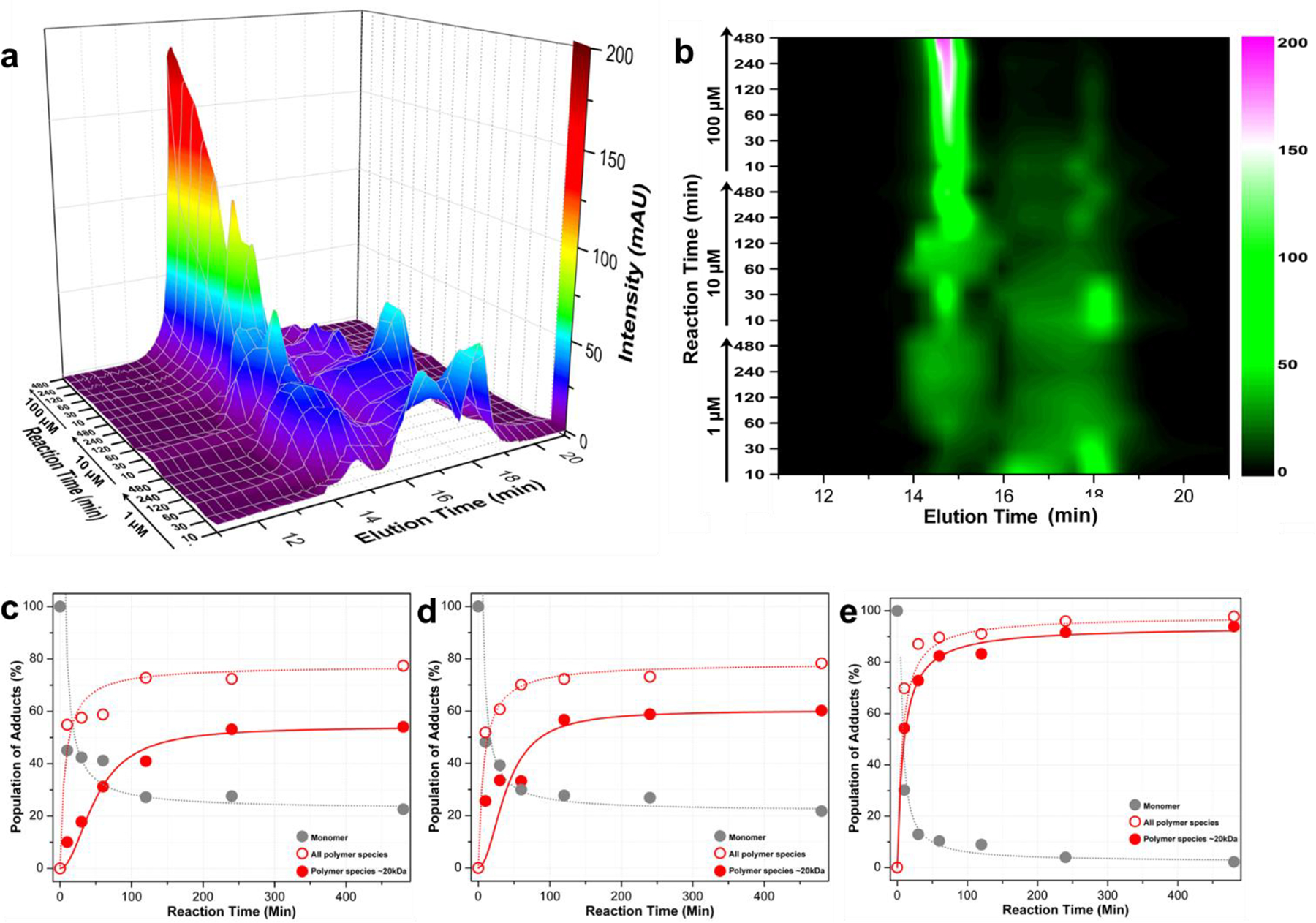
a. Comparison of elution profiles of the polymerization reaction of probe 3 at various concentrations (1, 10, 100 μM, as indicated) after different reactions times (10, 30, 60, 120, 240, 480 min, as indicated). The molecular weights of peaks at TE = 14.8 min and 17.7 min are 25 kDa and 1 kDa respectively (Supporting Fig. S4); the peak at 16.5 min contains trimers and higher-order oligomers as detected by MALDI-MS (Supporting Fig. S6). b. 2D intensity contour image of the elution profiles at indicated concentrations and reaction time points. c, d, e. Products population of the reaction at 1, 10, and 100 μM vs. time, respectively.
Optical properties of small molecule probes upon in situ polymerization.
We evaluated the optical properties of probes 1 and 2 before and after activation by glutathione (Fig. 3, Supporting Fig. S7). Glutathione plays a key role in regulating the redox potential in mammalian cells and its concentration can range from several μM in plasma to several mM in cells and tumor tissues.74–80 Our group and others have demonstrated that the CBT condensation reaction reactions can proceed under physiological conditions where glutathione is present in cells as well as in living animals.53, 58, 70, 81–83 Incubation of probe 1 (100 μM) in PBS buffer containing reducing agent glutathione (GSH, 10 mM) or tris(2-carboxyethyl)phosphine (TCEP, 10 mM) at pH 7.4 resulted in a fluorescence signal drop by around 70% within 30 min (Fig. 3a, 3c) likely due to the close proximity of IR800 dye and self-quenching after polymerization; in comparison, probe 2 under the same conditions or probe 1 without incubation with GSH or TCEP barely showed any changes in fluorescence signals within two hours (Fig. 3a, 3c). Electrophoretic gels of the GSH-treated probe 1 and 2 were stained using silver nitrate (Fig. 3b) and scanned using Maestro hyperspectral fluorescence imaging system (PerkinElmer, 635±25 nm excitation filter, 675 nm long-pass emission filter, image acquisition from 670 to 900 nm). From the silver staining, control probe 2 showed single bands at all concentrations (6, 0.6, and 0.06 nmol per 6 μL loaded) tested; while probe 1 (6 nmol per 6 μL loaded) showed smeared bands at a position similar to that of the 25–40 kDa fraction from SEC (Fig. 3b, left), and probe 1 at lower concentrations (0.6 nmol per 6 μL loaded) produced slightly smaller polymer species (Fig. 3b, left). Interestingly, in comparison to the fluorescence intensity of probe 2 at the test concentrations, probe 1 only gave a major fluorescence band of the dimer (detected by MALDI-MS after gel extraction), however the larger polymer species remained dark due to self-quenching of IR800 dye after polymerization occurred (Fig. 3b, right).
Figure 3. Characterization of polymerization using fluorescence and photoacoustic signals.
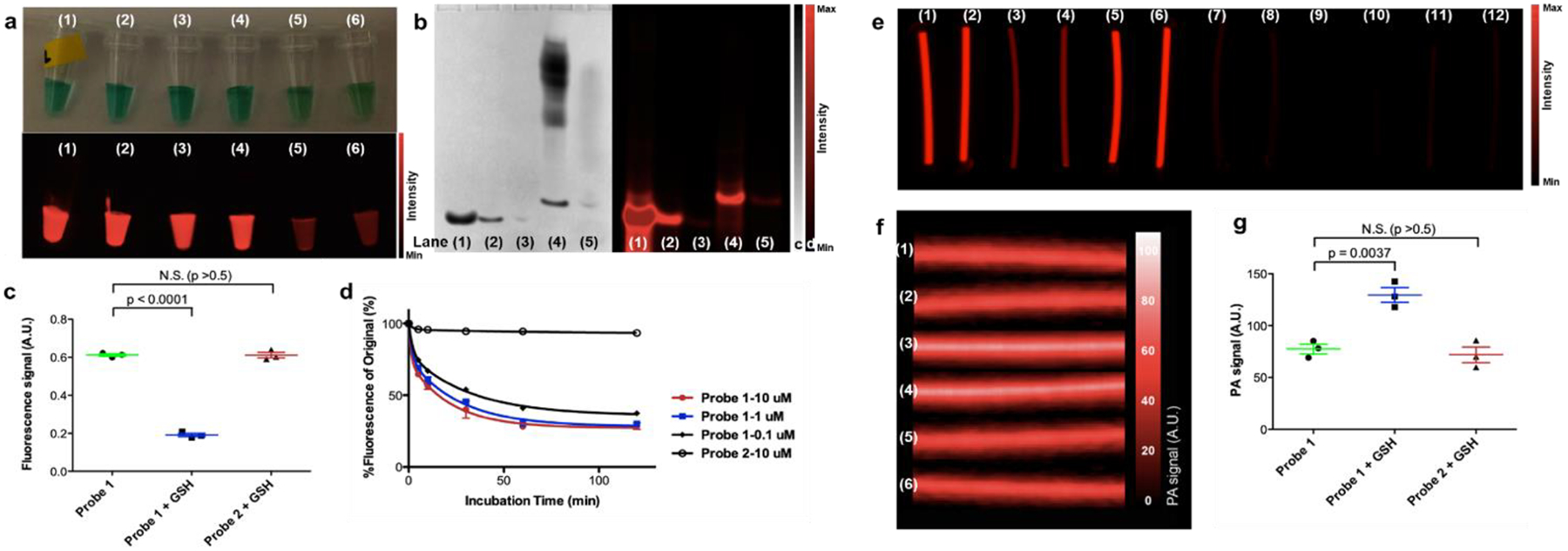
a. White light (upper) and fluorescence (lower) images of (1) probe 2 (100 μM) in PBS buffer at pH 7.4, (2) probe 2 (100 μM) in PBS buffer containing TCEP (10 mM) at pH 7.4, (3) probe 2 (100 μM) in PBS buffer containing GSH (10 mM) at pH 7.4, (4) probe 1 (100 μM) in PBS buffer at pH 7.4, (5) probe 1 (100 μM) in PBS buffer containing TCEP (10 mM) at pH 7.4, (6) probe 1 (100 μM) in PBS buffer containing GSH (10 mM) at pH 7.4. b. Silver nitrate staining (left) and fluorescence image (right) of gel electrophoresis (25% acrylamide gel) of (1) probe 2 (6 nmol), (2) probe 2 (0.6 nmol), (3) probe 2 (0.06 nmol), (4) probe 1 (6 nmol), and (5) probe 1 (0.6 nmol) in loading buffer containing GSH (10 mM) at pH 7.4. c. Quantification of fluorescence signals of probe 1 (100 μM) in PBS buffer at pH 7.4, probe 1 (100 μM) with GSH (10 mM) at pH 7.4, and probe 2 (100 μM) with GSH (10 mM) at pH 7.4 after 30 min incubation. d. Fluorescence changes of probe 1 at 0.1, 1, 10 μM and probe 2 at 10 μM over two hours at pH 7.4. e. Fluorescence images of sealed PE tubes of (1) and (2) probe 1 (100 μM) in PBS buffer at pH 7.4, (3) and (4) probe 1 (100 μM) in PBS buffer containing GSH (10 mM) at pH 7.4, (5) and (6) probe 2 (100 μM) in PBS buffer containing GSH (10 mM) at pH 7.4. (7) and (8) probe 1 (100 μM) in PBS buffer at pH 7.4, (9) and (10) probe 1 (100 μM) in PBS buffer containing GSH (10 mM) at pH 7.4, (11) and (12) probe 2 (100 μM) in PBS buffer containing GSH (10 mM) at pH 7.4. f. Processed photoacoustic images (3D maximization) of the same sample tubes (1)-(6) in e. g. Quantification of photoacoustic signals of probe 1 (100 μM) in PBS buffer at pH 7.4, probe 1 (100 μM) with GSH (10 mM) at pH 7.4, and probe 2 (100 μM) with GSH (10 mM) at pH 7.4. Standard deviations are shown by error bars.
Photoacoustic (PA) imaging detects the acoustic signals generated during thermoelastic expansion due to the photoabsorption. Therefore, PA imaging offers deeper tissue penetration and higher spatial resolution than fluorescence imaging.71–73 PA signals were measured using a Vevo LAZR instrument (Visualsonics, 2100 High-Resolution Imaging System). Probe 1 and 2 (each at 100 μM) with or without GSH (10 mM) were sealed in polyethylene tubes and buried in agarose gel, and fluorescence imaging were performed 30 min after the reaction started, immediately before acquiring photoacoustic signals (Fig. 3e, 3f). While GSH-incubated probe 1 showed reduced fluorescence signal compared with probe 2 (Fig. 4e), its PA signal increased to nearly 200% of the signal from probe 2 or probe 1 alone (Fig. 3f, 3g), suggesting the enhanced PA effects accompanying reduced fluorescence signal upon the formation of polymers.
Figure 4. Fluorescence and photoacoustic imaging of polymerizable probe in living mice.
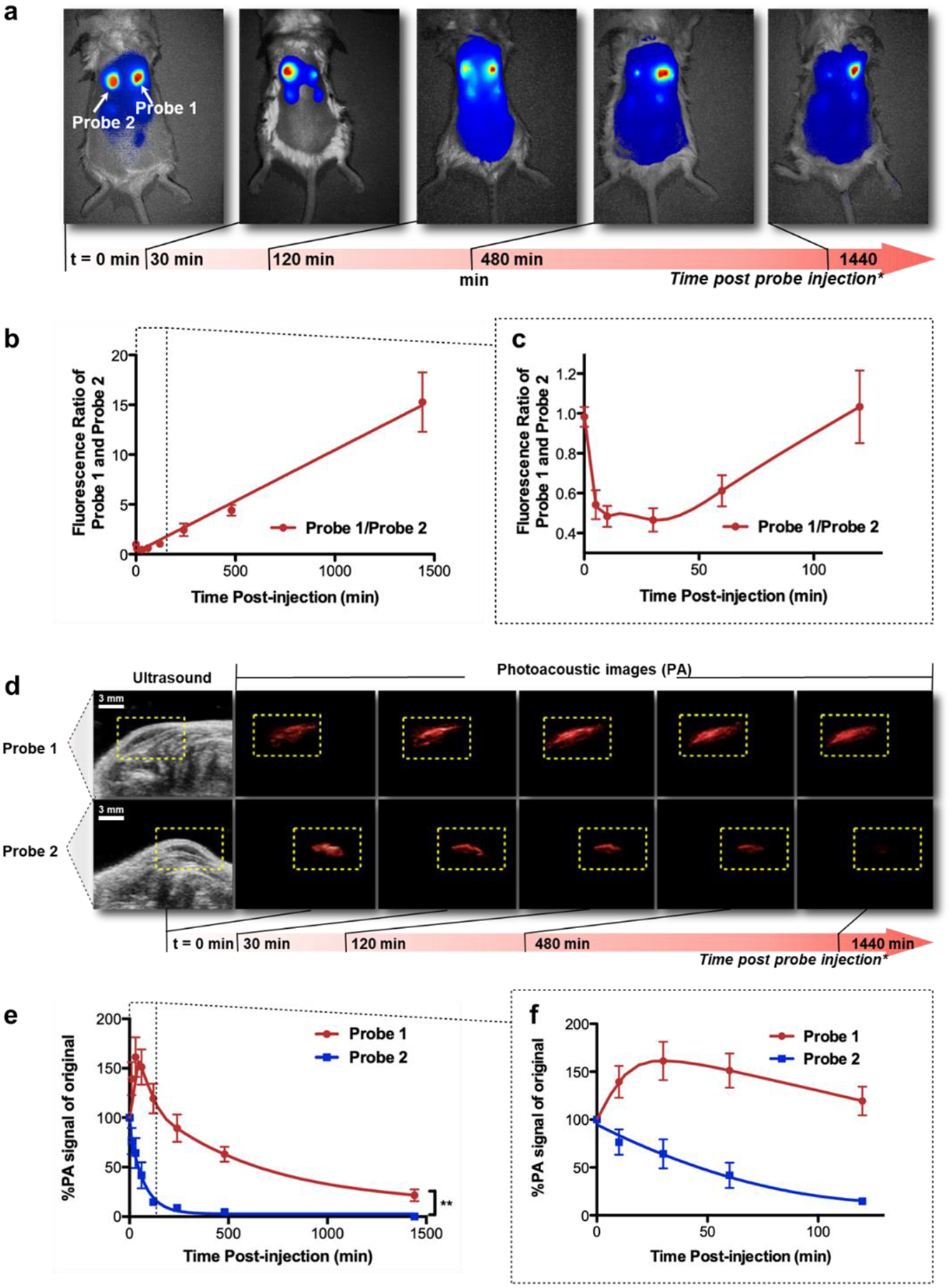
a. Fluorescence imaging of subcutaneously injected probe 1 (right spot, 1 nmol) and 2 (left spot, 1 nmol) in living mice in 24 hours after injection of the probe. Area of injection was pre-treated with GSH. The images are presented at different intensity scales to show the contrast of the two reaction spots. b. Ratio of fluorescence intensities of probe 1 and 2 vs. time. Red symbols and line, fluorescence ratio of probe 1 and 2. c. First two hours of graph b. d. Co-registered ultrasound and photoacoustic images of subcutaneously injected probe 1 (top, 1 nmol) and 2 (bottom, 1 nmol) in living mice in 24 hours after injection of the probe. Area of injection was pre-treated with GSH. White scale bar in the ultrasound windows is 3 mm. The yellow dash-lined rectangular indicates the area of interest for photoacoustic signals. e. Changes of photoacoustic intensities of probe 1 and 2 vs. time. Blue symbols and line, probe 1; red symbols and line, probe 2. f. The first two hours of graph b. Standard deviations are shown by error bars. **, P<0.0001.
Imaging polymerization in living animals.
We next evaluated the polymerizable probe with both fluorescence and PA imaging in a model system in living mice. The back of BALB/c mice was shaved, and a solution of GSH was injected subcutaneously on the furless back to form two spots of reducing environment (Fig. 4a). Probe 1 (1 nmol in 30 μL of PBS buffer) and probe 2 (1 nmol in 30 μL of PBS buffer) were then injected to each of the two reducing sockets. As expected, the two probes initially displayed the same fluorescence intensity, but the fluorescence of probe 1 decreased drastically in comparison to probe 2 within the first 30 minutes after injection (Supporting Fig. S8). Interestingly while the absolute fluorescence signal decreased in both spots due to the probe diffusion from the inoculation site, probe 1 appeared brighter than probe 2 afterwards even with self-quenching: the intensity ratio of probe 1 and 2 started to escalate, and reached around 15 after 24 hours (Fig. 4b, 4c), indicating the prolonged retention of probe 1 upon activation.
PA imaging (co-registered with ultrasound imaging) was similarly performed using the same model system, and each spot was monitored for 24 hours after probe injection (Fig. 4d). Different from fluorescence imaging, which showed drastic fluorescence decrease of probe 1 in the initial 30 min, PA signal of the same spot increased 30 min after probe 1 was injected (Supporting Fig. S9). Comparing to probe 2, which showed gradual decrease in PA signal over time, probe 1 showed initial PA enhancement in the first hour after injection, followed by a slow decline phase (Fig. 4e, 4f). Together with the fluorescence imaging results, the initial decrease in fluorescence and increase in PA signal resembled the trend in the in vitro experiments (Fig. 3), and probe 1 was retained longer in tissue than probe 2, suggesting the high probability of polymer formation.
Imaging of polymerization in tumor models.
The ideal polymerizable small molecules when injected systemically should be activated to form macromolecular structures at the target site (such as a tumor) in the living subjects, while diffusing away in other non-targeted tissues to provide better imaging contrast. To assess whether our molecular scaffold can be activated at desired sites after intravenous injection, we evaluated the probes 1 and 2 in mice xenografted tumor models. The ability to trigger the polymerization at desired sites was evaluated in female nude mice bearing subcutaneous U87MG tumors that were pre-treated with intratumoral injection of GSH or saline. Probe 1 or 2 (20 nmol in 200 μL saline) was administered to tumor-bearing mice via tail vein injection, and longitudinal fluorescence and PA imaging of these mice were performed (Fig. 5, Supporting Fig. S10). Since both probe 1 and 2 are not cell permeable, the polymerization will occur outside tumor cells, depending on the intratumoral delivery of GSH. We hypothesized that formed polymers would diffuse away slower than the free probe itself. The whole animal fluorescence imaging showed the fluorescence of probe 1 accumulated in GSH-pretreated tumor during the first one hour after probe injection and declined slowly over the monitored time course of 24 hours (Fig. 5, Supporting Fig. S10). Probe 1 administered to the mice bearing saline-pretreated tumor or probe 2 administered to the mice bearing GSH-pretreated tumor produced much faster fluorescence signal decay after injection, suggesting that probe 1 are 2 are poorly retained in tumor specifically (Fig. 5b, 5c, Supporting Fig. S10).
Figure 5. Fluorescence and photoacoustic imaging of polymerizable probe in tumor models.
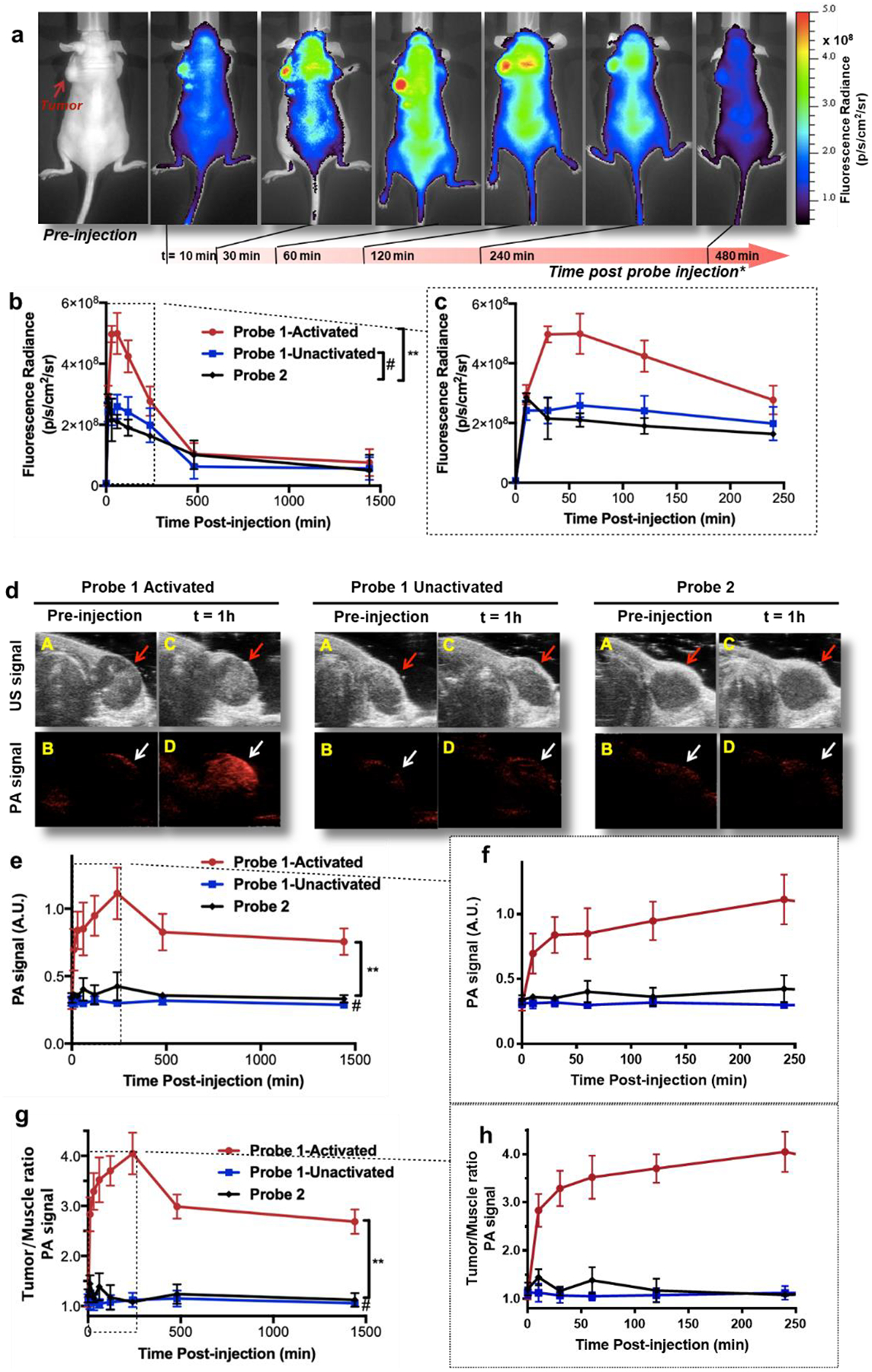
a. Longitudinal fluorescence imaging of tumor-bearing mice with i.v. injected probe 1 (20 nmol) in 8 hours. Tumor (indicated by red arrow) was pre-treated with GSH to activate probe 1. b. Comparison of the average tumor fluorescence radiance intensity of mice groups injected with probe 1 (20 nmol, GSH treatment, red circle, n = 5; 20 nmol, saline treatment, blue square, n = 5) or probe 2 (20 nmol, GSH treatment, black diamond, n = 5) vs. time (24 hours). Standard deviations are shown by error bars. c. First four hours of graph b. d. PA and US co-registered tumor images of mice with i.v. injected probe 1 and probe 2 (each at 20 nmol) before and one hour after injection. Tumor was pre-treated with intratumoral injection of GSH. Anatomical position of the tumor is indicated by red (US images) or white (PA images) arrows. A &B, US (A) and PA (B) images of target area before injection. C & D, US (C) and PA (D) images of target area one hour after i.v. injection of probe. e. Comparison of the average tumor PA intensity of mice groups injected with probe 1-activated (20 nmol, GSH treatment, red circle, n = 5), probe 1-unactivated (20 nmol, saline treatment, blue square, n = 5), and probe 2 (20 nmol, GSH treatment, black diamond, n = 5) vs. time. f. First four hours of graph e. g. Tumor to muscle ratio of the PA signal of the same mice groups in e. h. First four hours of graph g. All error bars indicate standard deviation. **, P<0.0001; #, not significant.
PA imaging of each mouse at each specific time point was performed immediately after the fluorescence scanning (Fig. 5d). PA signal in saline-pretreated tumors with probe 1 administration or GSH-pretreated tumors with probe 2 administration appeared close to the background level during the monitored time course (Fig. 5d, middle and right, Fig. 5e, 5f); in comparison, the GSH-pretreated tumors with probe 1 administration gave strikingly high PA signals in the entire time, peaking at 4 hours after probe injection (Fig. 5d, left, Fig. 5e, 5f). Importantly, the PA signal was nearly evenly distributed throughout the tumor region. While examining the results of the activated probe 1 in both of the PA imaging (Fig. 5d) and fluorescence imaging (Fig. 5a), tumor PA signal peaked (at 4 hours post probe injection) later than its fluorescence signal (at 30 min to 1 hour post probe injection). Also, PA signal in tumor remained high (nearly 50% of its peak value) even after 24 hours (Fig. 5e), but its corresponding fluorescence signal dropped to around the same level as two other control conditions after 4 hours (Fig. 5b, 5c). Together with the in vitro results, remaining PA signal in tissue without fluorescence can be attributed to the formation of polymers, which lose the fluorescence via self-quenching.
Conclusions
Our previous experience with the CBT condensation reactions often led to early cyclization and self-stacking to form nanoparticles. Molecular scaffolds that can prevent the formation of self-aggregates can further extend the application of the scaffold and chemistry to other applications such as therapeutics delivery and tissue engineering. In this work, we explored a carbohydrate scaffold for its water solubility and structural rigidity from the chair conformation to promote linear polymerization. Our results have shown that linear polymerization can occur even at a few micromolar concentration with this new rigid carbohydrate scaffold. To address the challenge of the detection of polymerization in living animals, we developed a dual-modality optical method using fluorescence and PA imaging. With this imaging method, we were able to demonstrate the in situ polymer formation with reductive activation in living mice. Future efforts may extend this system to other stimuli such as enzyme activation in tumors.
Experimental Section
Probe synthesis.
Chemical syntheses and characterizations of probes 1-4 and intermediates are provided in Supporting Information.
SEC-MALS measurements.
Size exclusion chromatography (SEC) coupled with multiangle light scattering (MALS) was performed by injecting 150 μg of activated or unactivated probe 3 or 4 onto a WTC-010S5 column (Wyatt Technology) previously equilibrated in PBS buffer at a flow rate of 0.5 mL/min. The elution profile was monitored by light scattering at 658 nm (HELEOS system, Wyatt Technology) and differential refractometry (Optilab system, Wyatt Technology). UV Absorbance was not used due to saturation of the detector at the concentrations above the detection limit of light scattering. Data analyses were carried out using ASTRA 6.0 software (Wyatt Technology). The refractive index increment (dn/dc) value of 0.163 mLμg−1 was assumed based on literature,84 as experimental determination was hindered by limited solubility of samples. Calculated molecular weights were further confirmed by MALDI-TOF mass spectrometry. SEC traces of polymerization reactions of different concentrations at different time points were plotted using Origin Lab 8.5 as 3D stacking and 2D intensity graphs. Different species (monomer, polymer species around 25 kDa alone or with other oligomers) were quantified using integration of area under curve (AUC) of the differential refractometry data and plotted against time using Origin Lab 8.5.
Measurement of photoacoustic effects of in vitro samples.
PA imaging was performed using a LAZRTight™ imaging enclosure coupled Vevo LAZR PA imaging system (VisualSonics), equipped with a MS-250 linear array transducer (21 MHz, 70% 6 dB two-way bandwidth, 256 elements) to detect ultrasound signals, and a tunable Nd:YAG laser system (OPOTEK, 680–950 nm, 20 Hz repetition rate, 5 ns pulse width, 50 mJ pulse peak energy) to generate optical pulses. For in vitro measurements, the probe solutions were injected into one-inch long polyethylene (PE100) tubing (Becton Dickinson, i.d. = 0.86 mm, o.d. = 1.52 mm), and the open ends of the tubes were heat-sealed. The tubes were then inserted horizontally into a freshly made semi-hardened agarose gel (1%) and the surface of the gel was coated with even layer of agarose solution (1%, ~3 mm thick). PA imaging was then performed after the gel completely solidified. PA signals were acquired using a 21 Hz transducer at 40 dB PA gain in 3D mode at a wavelength of 774 nm (absorbance max of IR800 dye) and with an acquisition rate of 5 frames/second. The 3D image of each sample tube was processed using loaded Visualsonics Vevo software package, and analyzed using Image J software (Fig. 3f, 3g).
Fluorescence imaging of polymerizable probes in living mice.
Female BALB/c mice (4 months old, Charles River Inc.) were anesthetized by isoflurane, and fur was removed to expose the back skin (Fig. 4a). In the following day, the mice were anesthetized and positioned on the heating platform inside a Maestro hyperspectral fluorescent imaging system (PerkinElmer). GSH (20 mM in 20 μL of PBS buffer) was injected subcutaneously under the back skin for each testing spot, where probe 1 or 2 (each of 1 nmol in 30 μL of PBS buffer) were then injected 5 min later. Fluorescence imaging was performed immediately using the Maestro scanner (635±25 nm excitation filter, 675 nm long-pass emission filter, images acquisition from 670 to 900 nm). IR800 fluorescence was deconvolved from tissue autofluorescence (Nuance v.3.0.1.2, PerkinElmer), and fluorescence intensity was quantified by region of interest (ROI) analysis (Fig. 4b, 4c).
PA imaging of polymerizable probes in living mice.
Female BALB/c mice were anesthetized and positioned on the heating platform inside the LAZRTight™ imaging enclosure of the Vevo LAZR imaging system. GSH (20 mM in 20 μL of PBS buffer) was injected subcutaneously under the furless back skin for each testing spot, where probe 1 or 2 (each of 1 nmol in 30 μL of matrigel) were then injected 5 min later. A thin layer of ultrasound scanning gel (Clear Image Singles) was applied to cover scanning area. Ultrasound co-registered PA imaging was performed immediately using a 21 MHz transducer at 100% power, 40 dB PA gain, and excitation wavelength of 774 nm (Fig. 4d). PA images were processed using Image J software, and PA intensity was quantified after subtraction of background signals (Fig. 4e, 4f).
Fluorescence imaging of polymerizable probe in tumor models.
To initiate tumors in female nu/nu mice (8-week-old, Charles River Inc.), 5-million U87MG cells suspended in saline (50 μL, original culturing medium was replaced immediately before implantation) were injected subcutaneously at the left shoulder of the mouse. Tumors were grown until reaching the largest dimension at 0.5 to 0.7 cm (~14 to 21 days). The mice were anesthetized, and the tumors of two mice groups (n=5) were treated with intratumoral injections of GSH (20 mM, pH 7.4) and a third group received saline (50 μL). Probe 1 or 2 (20 nmol in 200 μL saline) was administered 5 minutes later to the two mice groups with GSH treatment via tail vein, and probe 1 (20 nmol in 200 μL saline) was also administered to the third mice group with saline treatment. Longitudinal whole animal fluorescence was monitored over 24 hours using an IVIS spectrum fluorescence imaging system (PerkinElmer, Fig. 5a, Supporting Fig. S10). Tumor fluorescence radiance was quantified by region of interest (ROI) measurement using IVIS Living Image 4.2 software (Fig. 5b, 5c).
PA imaging of polymerizable probe in tumor models.
Immediately after the fluorescence imaging, the tumor-bearing mouse was moved to the heating platform inside the LAZRTight™ imaging enclosure of the Vevo LAZR imaging system for PA imaging. A generous layer of ultrasound scanning gel (Clear Image Singles) was applied to cover the tumor region. Ultrasound co-registered PA imaging was performed using a 21 MHz transducer at 100% power, 40 dB PA gain, and excitation wavelength of 774 nm (Fig. 5d). Photoacoustic images were processed using the Visualsonics imaging system software package, and PA intensity was quantified by region of interest (ROI) measurement (Fig. 5e, 5f).
Supplementary Material
ACKNOWLEDGMENT
We thank Prof. Axel T. Brunger (Stanford Molecular & Cellular Physiology) for use of the SEC-MALS instrument, Dr. Sean O’Leary (Joseph Puglisi lab) and Dr. Kimberly Brewer (Brian Rutt lab) for initial characterization of the probes, Mr. Ken Lau and Dr. Mark Stolowitz (Proteomics Core Facility, Stanford Canary Center) for MALDI-MS analysis, Drs. Adam J. Shuhendler (Rao lab) and Lihong Bu (Zhen Cheng lab) at Stanford School of Medicine for their kind assistance in animal work, Deju Ye (Rao lab) and Leonid Pereyaslavets (Michael Levitt lab) for helpful discussions.
Funding Sources
This work was supported by the Stanford University National Cancer Institute (NCI) Centers of Cancer Nanotechnology Excellence U54CA199075 and R01 GM128089 (to JR) and NIGMS R35GM124963 (to LC).
Footnotes
Conflict of Interests
Dr. Gambhir served on the board of directors of Endra Inc. and as a consultant for Visualsonics Inc.; both manufacturers of PA imaging equipment for small animals.
Supporting Information.
Supporting figures, experimental procedures and chemical characterizations (HRMS, 1H and 13C NMR spectra). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Anderson DG; Burdick JA; Langer R Smart biomaterials. Science 2004, 305 (5692), 1923–1924. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen KT; West JL Photopolymerizable hydrogels for tissue engineering applications. Biomaterials 2002, 23 (22), 4307–4314. [DOI] [PubMed] [Google Scholar]
- 3.Ruskowitz ER; DeForest CA Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nature Rev. Mater 2018, 3 (2), 1–17. [Google Scholar]
- 4.Klouda L; Mikos AG Thermoresponsive hydrogels in biomedical applications. Eur. J. Pharm. Biopharm 2008, 68 (1), 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizwan M; Yahya R; Hassan A; Yar M; Azzahari A; Selvanathan V; Sonsudin F; Abouloula C pH-sensitive hydrogels in drug delivery: brief history, properties, swelling, and release mechanism, material selection and applications. Polymers 2017, 9 (12), 137–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreira Teixeira LS; Feijen J; van Blitterswijk CA; Dijkstra PJ; Karperien M Enzyme-catalyzed crosslinkable hydrogels: emerging strategies for tissue engineering. Biomaterials 2012, 33 (5), 1281–1290. [DOI] [PubMed] [Google Scholar]
- 7.Hu J; Zhang G; Liu S Enzyme-responsive polymeric assemblies, nanoparticles and hydrogels. Chem. Soc. Rev 2012, 41 (18), 5933–5949. [DOI] [PubMed] [Google Scholar]
- 8.Munarin F; Guerreiro SG; Grellier MA; Tanzi MC; Barbosa MA; Petrini P; Granja PL Pectin-based injectable biomaterials for bone tissue engineering. Biomacromolecules 2011, 12 (3), 568–577. [DOI] [PubMed] [Google Scholar]
- 9.Ko DY; Shinde UP; Yeon B; Jeong B Recent progress of in situ formed gels for biomedical applications. Prog. Polym. Sci 2013, 38 (3–4), 672–701. [Google Scholar]
- 10.Sivashanmugam A; Arun Kumar R; Vishnu Priya M; Nair SV; Jayakumar R An overview of injectable polymeric hydrogels for tissue engineering. Eur. Polym. J 2015, 72, 543–565. [Google Scholar]
- 11.Liu M; Zeng X; Ma C; Yi H; Ali Z; Mou X; Li S; Deng Y; He N Injectable hydrogels for cartilage and bone tissue engineering. Bone Res 2017, 5 (1), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoare TR; Kohane DS Hydrogels in drug delivery: progress and challenges. Polymer 2008, 49 (8), 1993–2007. [Google Scholar]
- 13.Li J; Mooney DJ Designing hydrogels for controlled drug delivery. Nat. Rev. Mater 2016, 1 (12), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei K; Ma Q; Yu L; Ding J Functional biomedical hydrogels for in vivo imaging. J. Mater. Chem. B 2016, 4 (48), 7793–7812. [DOI] [PubMed] [Google Scholar]
- 15.Whitesides GM Self-assembly at all scales. Science 2002, 295 (5564), 2418–2421. [DOI] [PubMed] [Google Scholar]
- 16.Whitesides GM; Mathias JP; Seto CT Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science 1991, 254 (5036), 1312–1319. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol 2003, 21 (10), 1171–1178. [DOI] [PubMed] [Google Scholar]
- 18.Cui L; Rao J Chemical methodology for labelling and bioconjugation. In The Chemistry of Molecular Imaging; Long N, Wong W-K, Eds.; Wiley, 2014, pp25–53. [Google Scholar]
- 19.Brustad EM; Lemke EA; Schultz PG; Deniz AA A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J. Am. Chem. Soc 2008, 130 (52), 17664–17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen I; Howarth M; Lin W; Ting AY Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods 2005, 2 (2), 99–104. [DOI] [PubMed] [Google Scholar]
- 21.Mahal LK; Yarema KJ; Bertozzi CR Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 1997, 276 (5315), 1125–1128. [DOI] [PubMed] [Google Scholar]
- 22.Rideout D Self-assembling cytotoxins. Science 1986, 233 (4763), 561–563. [DOI] [PubMed] [Google Scholar]
- 23.Agard NJ; Prescher JA; Bertozzi CR A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc 2004, 126 (46), 15046–15047. [DOI] [PubMed] [Google Scholar]
- 24.Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR Copper-free click chemistry for dynamic in vivo imaging. Proc. Nat. Acad. Sci. U. S. A 2007, 104 (43), 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ning X; Guo J; Wolfert MA; Boons GJ Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem. Int. Ed 2008, 47 (12), 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikic I; Plass T; Schraidt O; Szymanski J; Briggs JA; Schultz C; Lemke EA Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed 2014, 53 (8), 2245–2249. [DOI] [PubMed] [Google Scholar]
- 27.Song W; Wang Y; Qu J; Madden MM; Lin Q A photoinducible 1,3-dipolar cycloaddition reaction for rapid, selective modification of tetrazole-containing proteins. Angew. Chem. Int. Ed 2008, 47 (15), 2832–2835. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z; Pan Y; Wang Z; Wang J; Lin Q Genetically encoded cyclopropene directs rapid, photoclick-chemistry-mediated protein labeling in mammalian cells. Angew. Chem. Int. Ed 2012, 51 (42), 10600–10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song W; Wang Y; Qu J; Lin Q Selective functionalization of a genetically encoded alkene-containing protein via “photoclick chemistry” in bacterial cells. J. Am. Chem. Soc 2008, 130 (30), 9654–9655. [DOI] [PubMed] [Google Scholar]
- 30.Blackman ML; Royzen M; Fox JM Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc 2008, 130 (41), 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devaraj NK; Weissleder R; Hilderbrand SA Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjugate Chem 2008, 19 (12), 2297–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang K; Davis L; Torres-Kolbus J; Chou C; Deiters A; Chin JW Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem 2012, 4 (4), 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. J. Am. Chem. Soc 2012, 134 (25), 10317–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plass T; Milles S; Koehler C; Szymanski J; Mueller R; Wiessler M; Schultz C; Lemke EA Amino acids for Diels-Alder reactions in living cells. Angew. Chem. Int. Ed 2012, 51 (17), 4166–4170. [DOI] [PubMed] [Google Scholar]
- 35.Seitchik JL; Peeler JC; Taylor MT; Blackman ML; Rhoads TW; Cooley RB; Refakis C; Fox JM; Mehl RA Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. J. Am. Chem. Soc 2012, 134 (6), 2898–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J; Seckute J; Cole CM; Devaraj NK Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem. Int. Ed 2012, 51 (30), 7476–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKay CS; Finn MG Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem. Biol 2014, 21 (9), 1075–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxon E; Bertozzi CR Cell surface engineering by a modified Staudinger reaction. Science 2000, 287 (5460), 2007–2010. [DOI] [PubMed] [Google Scholar]
- 39.Prescher JA; Dube DH; Bertozzi CR Chemical remodelling of cell surfaces in living animals. Nature 2004, 430 (7002), 873–877. [DOI] [PubMed] [Google Scholar]
- 40.Desai RM; Koshy ST; Hilderbrand SA; Mooney DJ; Joshi NS Versatile click alginate hydrogels crosslinked via tetrazine-norbornene chemistry. Biomaterials 2015, 50, 30–37. [DOI] [PubMed] [Google Scholar]
- 41.Truong VX; Ablett MP; Richardson SM; Hoyland JA; Dove AP Simultaneous orthogonal dual-click approach to tough, in-situ forming hydrogels for cell encapsulation. J. Am. Chem. Soc 2015, 137 (4), 1618–1622. [DOI] [PubMed] [Google Scholar]
- 42.Liu S; Dicker KT; Jia X Modular and orthogonal synthesis of hybrid polymers and networks. Chem. Comm 2015, 51 (25), 5218–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An P; Lewandowski TM; Erbay TG; Liu P; Lin Q Sterically shielded, stabilized nitrile imine for rapid bioorthogonal protein labeling in live cells. J. Am. Chem. Soc 2018, 140 (14), 4860–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J; Kong H; Huang L; Cheng B; Qin K; Zheng M; Yan Z; Zhang Y Visible light-initiated bioorthogonal photoclick cycloaddition. J. Am. Chem. Soc 2018, 140 (44), 14542–14546. [DOI] [PubMed] [Google Scholar]
- 45.McNitt CD; Cheng H; Ullrich S; Popik VV; Bjerknes M Multiphoton activation of photo-strain-promoted azide alkyne cycloaddition “Click” reagents enables in situ labeling with submicrometer resolution. J. Am. Chem. Soc 2017, 139 (40), 14029–14032. [DOI] [PubMed] [Google Scholar]
- 46.Nainar S; Kubota M; McNitt C; Tran C; Popik VV; Spitale RC Temporal labeling of nascent RNA using photoclick chemistry in live cells. J. Am. Chem. Soc 2017, 139 (24), 8090–8093. [DOI] [PubMed] [Google Scholar]
- 47.Poloukhtine AA; Mbua NE; Wolfert MA; Boons G-J; Popik VV Selective labeling of living cells by a photo-triggered Click reaction. J. Am. Chem. Soc 2009, 131 (43), 15769–15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah L; Laughlin ST; Carrico IS Light-activated Staudinger–Bertozzi ligation within living animals. J. Am. Chem. Soc 2016, 138 (16), 5186–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida S; Hatakeyama Y; Johmoto K; Uekusa H; Hosoya T Transient protection of strained alkynes from Click reaction via complexation with copper. J. Am. Chem. Soc 2014, 136 (39), 13590–13593. [DOI] [PubMed] [Google Scholar]
- 50.Kumar P; Laughlin ST Modular activatable bioorthogonal reagents. In Chemical and Synthetic Biology Approaches To Understand Cellular Functions – Part B, 2019, 153–182. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H; Trout WS; Liu S; Andrade GA; Hudson DA; Scinto SL; Dicker KT; Li Y; Lazouski N; Rosenthal J; Thorpe C; Jia X; Fox JM Rapid bioorthogonal chemistry turn-on through enzymatic or long wavelength photocatalytic activation of tetrazine ligation. J. Am. Chem. Soc 2016, 138 (18), 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang T; Kumar P; Huang W; Kao WS; Thompson AO; Camarda FM; Laughlin ST Modular enzyme‐ and light‐based activation of cyclopropene–tetrazine ligation. ChemBioChem 2019, 20 (17), 2222–2226. [DOI] [PubMed] [Google Scholar]
- 53.Liang G; Ren H; Rao J A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat. Chem 2010, 2 (1), 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye D; Liang G; Ma ML; Rao J Controlling intracellular macrocyclization for the imaging of protease activity. Angew. Chem. Int. Ed 2011, 50 (10), 2275–2279. [DOI] [PubMed] [Google Scholar]
- 55.Ye D; Shuhendler AJ; Cui L; Tong L; Tee SS; Tikhomirov G; Felsher DW; Rao J Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat. Chem 2014, 6 (6), 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z; Chen M; Cheng Y; Kowada T; Xie J; Zheng X; Rao J Exploring the condensation reaction between aromatic nitriles and aminothiols to optimize in situ nanoparticle formation for the imaging of proteases and glycosidases in cells. Angew. Chem. Int. Ed 2020, 59 (8), 3272–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang G; Ronald J; Chen Y; Ye D; Pandit P; Ma ML; Rutt B; Rao J Controlled self-assembling of gadolinium nanoparticles as smart molecular magnetic resonance imaging contrast agents. Angew. Chem. Int. Ed 2011, 50 (28), 6283–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dragulescu-Andrasi A; Kothapalli SR; Tikhomirov GA; Rao J; Gambhir SS Activatable oligomerizable imaging agents for photoacoustic imaging of furin-like activity in living subjects. J. Am. Chem. Soc 2013, 135 (30), 11015–11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen B; Jeon J; Palner M; Ye D; Shuhendler A; Chin FT; Rao J Positron emission tomography imaging of drug-induced tumor apoptosis with a caspase-triggered nanoaggregation probe. Angew. Chem. Int. Ed 2013, 52 (40), 10511–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye D; Pandit P; Kempen P; Lin J; Xiong L; Sinclair R; Rutt B; Rao J Redox-triggered self-assembly of gadolinium-based MRI probes for sensing reducing environment. Bioconjugate Chem 2014, 25 (8), 1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palner M; Shen B; Jeon J; Lin J; Chin FT; Rao J Preclinical kinetic analysis of the caspase-3/7 PET tracer 18F-C-SNAT: quantifying the changes in blood flow and tumor retention after chemotherapy. J. Nucl. Med 2015, 56 (9), 1415–1421. [DOI] [PubMed] [Google Scholar]
- 62.Witney TH; Hoehne A; Reeves RE; Ilovich O; Namavari M; Shen B; Chin FT; Rao J; Gambhir SS A systematic comparison of 18F-C-SNAT to established radiotracer imaging agents for the detection of tumor response to treatment. Clin. Cancer Res 2015, 21 (17), 3896–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z; Chen M; Zhou K; Rao J Pre‐targeted imaging of protease activity through in situ assembly of nanoparticles. Angew. Chem. Int. Ed 2020, 59 (20), 7864–7870. [DOI] [PubMed] [Google Scholar]
- 64.Ren H; Xiao F; Zhan K; Kim YP; Xie H; Xia Z; Rao J A biocompatible condensation reaction for the labeling of terminal cysteine residues on proteins. Angew. Chem. Int. Ed 2009, 48 (51), 9658–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen DP; Elliott T; Holt M; Muir TW; Chin JW Genetically encoded 1,2-aminothiols facilitate rapid and site-specific protein labeling via a bio-orthogonal cyanobenzothiazole condensation. J. Am. Chem. Soc 2011, 133 (30), 11418–11421. [DOI] [PubMed] [Google Scholar]
- 66.Ramil CP; An P; Yu Z; Lin Q Sequence-specific 2-cyanobenzothiazole ligation. J. Am. Chem. Soc 2016, 138 (17), 5499–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui L; Rao J 2-Cyanobenzothiazole (CBT) condensation for site-specific labeling of proteins at the terminal cysteine residues. I In Site-specific protien labeling; Gautier A, Hinner M, Eds; Methods Mol. Biol 2015, 1266, 81–92. [DOI] [PubMed] [Google Scholar]
- 68.Liu S; Tang A; Xie M; Zhao Y; Jiang J; Liang G Oligomeric hydrogels self-assembled from reduction-controlled condensation. Angew. Chem. Int. Ed 2015, 54 (12), 3639–3642. [DOI] [PubMed] [Google Scholar]
- 69.Zheng Z; Chen P; Xie M; Wu C; Luo Y; Wang W; Jiang J; Liang G Cell environment-differentiated self-assembly of nanofibers. J. Am. Chem. Soc 2016, 138 (35), 11128–11131. [DOI] [PubMed] [Google Scholar]
- 70.Schleyer KA; Datko BD; Burnside B; Cui C; Ma X; Grey JK; Cui L Responsive fluorophore aggregation provides spectral contrast for fluorescence lifetime imaging. ChemBioChem 2020, 21(14), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu M; Wang LV Photoacoustic imaging in biomedicine. Rev. Sci. Instrum 2006, 77 (4), 1–22. [Google Scholar]
- 72.Zackrisson S; van de Ven SMWY; Gambhir SS Light in and sound out: emerging translational strategies for photoacoustic imaging. Cancer Research 2014, 74 (4), 979–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang J; Pu K Activatible molecular probes for second near-infrared fluorescence, chemiluminescence, and photoacoustic imaging. Angew. Chem. Int. Ed 2020, 59(29), 11717–11731. [DOI] [PubMed] [Google Scholar]
- 74.Hassan SSM; Rechnitz GA Determination of glutathione and glutathione reductase with a silver sulfide membrane electrode. Anal. Chem 2002, 54 (12), 1972–1976. [Google Scholar]
- 75.Hwang C; Sinskey A; Lodish H Oxidized redox state of glutathione in the endoplasmic reticulum. Science 1992, 257 (5076), 1496–1502. [DOI] [PubMed] [Google Scholar]
- 76.Jones DP; Carlson JL; Samiec PS; Sternberg P; Mody VC; Reed RL; Brown LAS Glutathione measurement in human plasma. Clin. Chim. Acta 1998, 275 (2), 175–184. [DOI] [PubMed] [Google Scholar]
- 77.Park S; Imlay JA High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol 2003, 185 (6), 1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stipanuk MH; Dominy JE; Lee J-I; Coloso RM Mammalian Cysteine Metabolism: New Insights into Regulation of Cysteine Metabolism. J. Nutr 2006, 136 (6), 1652S–1659S. [DOI] [PubMed] [Google Scholar]
- 79.Gamcsik MP; Kasibhatla MS; Teeter SD; Colvin OM Glutathione levels in human tumors. Biomarkers 2012, 17 (8), 671–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forman HJ; Zhang H; Rinna A Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Aspects Med 2009, 30 (1–2), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan Y; Wang L; Du W; Ding Z; Zhang J; Han T; An L; Zhang H; Liang G Intracellular self-assembly of taxol Nanoparticles for overcoming multidrug resistance. Angew. Chem. Int. Ed 2015, 54 (33), 9700–9704. [DOI] [PubMed] [Google Scholar]
- 82.Ai X; Ho CJH; Aw J; Attia ABE; Mu J; Wang Y; Wang X; Wang Y; Liu X; Chen H; Gao M; Chen X; Yeow EKL; Liu G; Olivo M; Xing B In vivo covalent cross-linking of photon-converted rare-earth nanostructures for tumour localization and theranostics. Nat. Commun 2016, 7, 10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hai Z; Ni Y; Saimi D; Yang H; Tong H; Zhong K; Liang G γ-Glutamyltranspeptidase-triggered intracellular gadolinium nanoparticle formation enhances the T2-weighted MR contrast of tumor. Nano Lett. 2019, 19 (4), 2428–2433. [DOI] [PubMed] [Google Scholar]
- 84.Gilbert R Size-separation characterization of starch and glycogen for biosynthesis–structure–property relationships. Anal. Bioanal. Chem 2011, 399 (4), 1425–1438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


