Abstract
Axonal degeneration contributes to the pathogenesis of many neurodegenerative disorders, motivating efforts to dissect the mechanism of pathological axon loss in order to develop therapies for axonal preservation. SARM1 is a particularly attractive therapeutic target, as it is an inducible NAD+ cleaving enzyme that is required for axon loss in multiple mouse models of traumatic and degenerative neurological disease. However, it is essential to establish whether SARM1 triggers axon degeneration in human neurons before proceeding with the development of SARM1-directed therapeutics. Here we combine genome engineering with the production of human stem cell-derived neurons to test the role of human SARM1 in traumatic and neurotoxic axon degeneration. We have generated two independent SARM1 knockout human iPSC lines that do not express SARM1 protein upon differentiation into neurons. We have developed a modified sensory neuron differentiation protocol that generates human sensory neurons with high yield and purity. We find that SARM1 is required for axon degeneration in response to both physical trauma and in a cellular model of chemotherapy-induced peripheral neuropathy. Finally, we identify cADPR as a biomarker of SARM1 enzyme activity in both healthy and injured human sensory neurons. These findings are consistent with prior molecular and cellular studies in mouse neurons, and highlight the therapeutic potential of SARM1 inhibition for the prevention and treatment of human neurological disease.
Keywords: neurodegeneration, TIR domain, sarmoptosis, NADase, axon degeneration
Introduction
Axon degeneration (AxD) is a regulated process of axonal self-destruction that is an early and contributing event in many neurological disorders including peripheral neuropathies, Parkinson’s disease, and traumatic brain injury1–3. The molecular pathway underlying pathological axon degeneration is rapidly being deciphered4,5, with SARM1 (sterile α and TIR motif containing 1) recognized as a central executioner of this degenerative process6,7
SARM1 is required for axon loss in mouse models of traumatic nerve injury 6,7 chemotherapy-induced peripheral neuropathy8–10, traumatic brain injury1, and glaucoma11. In addition, SARM1-dependent neuronal cell death, termed Sarmoptosis, occurs in response to mitochondrial dysfunction12,13, as well as in mouse models of retinitis pigmentosa14 and Leber’s congenital amaurosis15. This wide-ranging role for SARM1 in promoting neurodegeneration identified SARM1 as an important therapeutic target and motivated efforts to define the SARM1 mechanism of action.
SARM1 is a multidomain protein consisting of an autoinhibitory N-terminal domain, tandem SAM domains that mediate multimerization, and a C-terminal TIR (Toll-like interleukin 1 receptor) domain6. The SARM1 TIR domain is the founding member of a new class of NAD+ cleaving enzymes16 that are evolutionarily conserved in bacteria17 and plants18,19. In healthy axons, SARM1 is predominantly in the ‘off’ state20, but upon injury the autoinhibition mediated by the N-terminal domain is relieved and the TIR NADase is activated6,21. Activation of the TIR domain is sufficient22 and TIR NADase activity is necessary16 for axon degeneration. The discovery that SARM1 is an enzyme that cleaves NAD+ identifies it as a druggable target whose inhibition may treat a wide variety of neurodegenerative diseases23.
The SARM1 protein is highly conserved between human and mouse (94% identity) suggesting that functional conservation of the human protein is likely. Here we test that assumption in human neurons. We have developed SARM1 knockout induced pluripotent stem cells (hiPSCs) using CRISPR-mediated gene editing and developed a modified one-step protocol24 for generating human sensory neurons with high yield and purity. We demonstrate that loss of SARM1 blocks axonal degeneration in iPSC-derived human sensory neurons after both traumatic injury and in a cellular model of vincristine-induced peripheral neuropathy. Moreover, SARM1 KO prevents depletion of axonal NAD+ after injury. Finally, we identify the SARM1 enzymatic product cADPR as a biomarker of SARM1 activity in both healthy and injured human axons. These results demonstrate the essential role of SARM1 in pathological axon degeneration in human peripheral sensory neurons and support the nomination of SARM1 as a therapeutic candidate for the prevention and treatment of neurodegenerative diseases.
Materials and methods
iPSC cell culture.
BJ (CRL-2522) fibroblasts from ATCC were reprogrammed to establish the BJFF.6 iPSC line using the non-integrating Sendai virus by Genome Engineering and iPSC Center (GEiC) at Washington University in St. Louis. iPSC cells were maintained on Matrigel (Corning) coated plates in Stem-Flex medium (ThermoFisher). Matrigel coating was performed by diluting the concentrated matrix in ice-cold DMEM/F12 medium, transferring the diluted solution to plates, and allowing the solution to coat the plates for at least 1h at RT. The human iPSCs were passaged by washing once with DPBS, followed by dissociation into small clumps using ReLeSR passaging solution (STEMCELL Technologies). The cells were collected and diluted (from 1:4 to 1:8) according to the initial density of the culture for passaging. iPSC cultures were maintained at 37°C in a humidified incubator with 5% CO2.
Generation of SARM1 knockout iPSC.
The human BJFF.6 iPSC cells were used to generate SARM1-deficient iPSCs using CRIPSR/Cas9 technology. The hPSCs were dissociated into single cells with 0.75X TrypLE-select reagent. Approximately 1 to 1.5 × 106 hPSCs were washed in DPBS and resuspended in P3 primary buffer with 1 μg gRNA (5’- AAACCAGGCGTTTCAGGCTC −3’) expression construct (MLM3636 was a gift from Keith Joung; Addgene plasmid # 43860), 1.5 μg Cas9 vectors (p3s-Cas9HC was a gift from Jin-soo Kin; Addgene plasmid #43945) and 0.5 ug GFP expression construct and then electroporated using a 4D-Nucleofector (Lonza) using CA-137 program. Following nucleofection, cells were plated in a 6-well plate with warm medium supplemented with ROCKi for 24 hrs. The next day, the medium was replaced with fresh medium without ROCKi. After growth for 1–2 days, the nucleofected cells were sorted into 96-well plates with one cell per well as previously described25. Single cell clones were screened for SARM1 disruption via Cas9 editing using targeted NGS analysis26.
Sensory neuron differentiation.
The sensory neuron differentiation was performed as previously described with slight modifications24 to improve differentiation efficiency. Human iPSCs were dissociated with 0.75X TrpLE-select, and 30,000 cells/well were seeded onto a Matrigel-coated 6-well plate in Stem-Flex medium supplemented with 1X RevitaCell (Thermo Fisher). The next day the medium was replaced with differentiation medium (DMEM/F12 supplemented with 5% KnockOut Serum Replacement (KOSR) (Thermo Fisher), 1% penicillin/streptomycin, 0.3 μM LDN-193189, 2 μM A83–01, 6 μM CHIR99021, 2 μM RO4929097, 3 μM SU5402 and 0.3 μM Retinoic acid) for 10 days. It is critical to perform total medium change every 2 days. On day 10, the medium was switched to sensory neuron maturation medium [neurobasal plus medium supplemented with 1X B-27 plus, 10 ng/ml neurotrophin 3 (NT3), 20 ng/ml brain-derived neurotrophic factor (BDNF), 20 ng/ml nerve growth factor (NGF), and 20 ng/ml glial cell line-derived growth factor (GDNF)) with 2 μM 5-Fluoro-2’-deoxyuridine (FdU)] for 5 days. On average, 2.5 million sensory neurons per well of 6-well plate could be expected on day 15. Continued culture in maturation process in maturation medium plus Culture One supplement (Thermo Fisher) for additional 5 days (day 15–20 of the differentiation protocol). On day 28, matured sensory neurons were ready for experiments. For spot cultures, early differentiated neurons (day 15), are dissociated and pipetted into a small area and then cultured in maturation medium plus Culture One supplement as above and continue the differentiation process for total 28 days.
Immunofluorescence and immunoblotting assays.
For immunostaining, cells were rinsed once with phosphate-buffer saline (PBS), fixed in 4% paraformaldehyde for 15 mins at room temperature (RT), and treated with blocking buffer (PBS containing 2% bovine serum albumin and 0.1% Triton X-100) for 1 hour at RT. Cells were then incubated with primary antibodies overnight at 4 °C. Next day, the cells were rinsed with PBS three times and incubated with Alexa 488- or Alex 568-conjugated secondary antibodies (Thermo Fisher) for 1 hour at RT. Nuclei were counterstained with 4,6-diamidino-2-Phenylindole (DAPI). Images of the stained cells were captured using the Nikon fluorescence microscope and CCD camera. Antibody against SARM1 (Cell Signaling Technology, D2M5I, cat# 13022) and HRP-conjugated secondary antibody (Jackson ImmunoResearch) were used for Western blotting.
Flow cytometry.
iPSC-derived sensory neurons were dissociated and harvested at day 17 or 28 after initiation of neuronal differentiation using TrypLE-Select. Cyto-Fast Fix/Perm buffer set (BioLegened) was used for intracellular staining. The dissociated cells were fixed in Cyto-Fast Fix/Perm buffer for 20 min at RT, then washed twice with 1X Cyto-Fast Perm Wash solution. After washing, the cells were incubated with appropriate antibodies (10ng/μl) for 30 min at RT. Lastly, cells were washed and resuspended in cell staining buffer. Flow cytometry was performed using a Sony SH800S cell sorter and the data was analyzed using FlowJo software.
Axotomy and vincristine induced axon degeneration.
Human iPSCs derived sensory neurons (15 days after initiation of differentiation protocol described above) were dissociated with TrypLE-Select for 3 min at 37 °C. The cell suspensions at a density of 1×107 cells/ml were prepared in the maturation medium and 1.2 ul (96 well) or 10 ul (24 well) of cell suspension was placed in the middle of culture wells coated with Poly-D-Lysine (PDL) and laminin. After the neurons were attached, maturation medium was gently added and neurons were cultured for an additional 13 days to allow axonal extension. At 28 days after differentiation axons were injured either by transection with a microsurgical blade or by addition of vincristine. The imaging fields were predetermined in a unbiased manner and continuously imaged using an IncuCyte S3 (Sartorius) for 48 hours after axon injury.
Quantification of axon degeneration
Axonal images were captured using an IncuCyte S3 (Sartorius) and analyzed using the trainable Weka segmentation plugin provided in ImageJ (version 2.0.0-rc-69/1.52n, NIH) 27. Axonal degeneration was measured as the increase of vacuolated axons (Figure S1). To quantify the extent of axon vacuolation, the Weka segmentation tool was trained to identify the area containing 1) healthy axon, 2) vacuolated axon, and 3) no cellular component (Figure S1). An example of phase contrast and corresponding Weka segmented images, both before and after axonal injury are shown in Figure S1A. The vacuolation index (Vi) was determined by the ratio of the number of pixels corresponding to area of vacuolated axons divided by pixels corresponding to all axon space (summation of healthy and unhealthy vacuolated axons). The vacuolation index (Vi) increases with time after axonal injury (Figure S1C). Axon degeneration was determined by the ratio of vacuolation index (Vi) at the time after axon injury (24 hr or 48 hr) compared with time at the injury (0 hr) obtained from the same field of the well and expressed as axon degeneration index.
Axonal metabolite extraction and measurement by LC-MS/MS.
For axon metabolite measurements, spot cultures of iPSC-derived sensory neurons were prepared in 24-well plates. The iPSC-derived neuron cell bodies and axons were separated using a microsurgical blade under the microscope, the plate was placed on ice, culture medium was replaced with ice-cold saline (0.9% NaCl), and the cell bodies were removed by pipetting. Metabolites were extracted from the remaining axons by incubating them with ice-cold 1:1 mixture of MeOH and water (160 μl per well) for 10 min. The MeOH/water solution containing axon metabolites was transferred to a microfuge tube and extracted with chloroform (30 μl per sample) to extract lipids and proteins. The aqueous phase (140 μl) was lyophilized and stored at −20°C until analysis. On the day of analysis, metabolites were dissolved by adding 15 μl of 5 mM ammonium formate in water and vortexing. The samples were centrifuged (12,000 × g, 10 min, 4 °C) and clear supernatants were transferred the sample plate for LC-MS/MS analysis of metabolites as described previously28.
Lentiviral constructs and infection
Lentiviruses were produced in HEK293T cells (RRID: CVCL_0063) as previously described29. Cells were seeded at a density of 1 × 106 cells per 35 mm well the day before transfection. FCIV lentivirus constructs harboring cDNAs encoding Venus fluorescent protein, human SARM1, or dominant negative human SARM1 (SARM1(K193R, H685A (each 400 ng) were co-transfected with VSV-G (400 ng) and pSPAX2 (1.2 μg) using X-tremeGene (Roche). The virus containing medium was collected 3–5 days after transfection and lentivirus particles were concentrated from the cleared culture supernatant using Lenti-X concentrator (Clontech). Lentivirus stocks at final concentration of 1–10 × 106 infectious particles/ml were stored at −80°C.
Statistical analysis.
Sample number (n) was defined as the number of cell culture wells that were independently manipulated and measured. No statistical evaluations were performed to predetermine sample sizes, but our sample sizes are similar to those generally used in the field. Data comparisons were performed using Mann-Whitney U test or Kruskal-Wallis test using R (RRID:SCR_002394). P values were reported for each comparison in corresponding figure legends. For multiple comparisons, pairwise Wilcox comparison with Bonferroni adjusted p-value was used. Statistical significance was noted as * in each graph with indicated P values in the corresponding legends. The box height of each graph represents the first (bottom) and third (top) quartiles, and the median is indicated as a horizontal line in the box, and the top error bar represents the smaller of 1.5 times interquartile range or maximum data point. The bottom error bar represents 1.5 times interquartile range or minimum data point. All data points are shown as scatter plots over the box plot.
Results and Discussion
Generation of SARM1 knockout human induced pluripotent stem cells
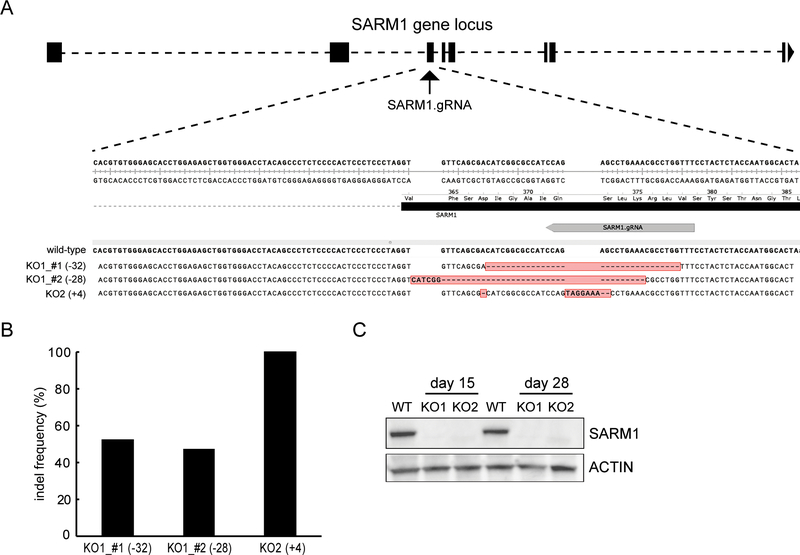
To investigate the function of SARM1 in human neurons, we generated SARM1 knockout iPSC lines using CRISPR/Cas9 gene editing. A guide RNA (gRNA) was designed to target the third exon of the human SARM1 gene locus (Figure 1A). After the CRISPR/Cas9 editing, single cell clones were screened using targeted Next Generation Sequencing (NGS). Two independent genetic knock-out clones were identified from the screen. Clone one has a thirty-two nucleotide deletion in one SARM1 allele and a twenty-eight nucleotide deletion in the other allele, while clone two has an insertion of four nucleotides in each SARM1 allele (Figure 1A). NGS analysis identified the indels and their allele frequency in the two knockout (KO) clones (Figure 1B). The genomic aberrations in exon3 of the SARM1 gene in both clones results in a downstream premature stop codon, potentially leading to nonsense-mediated mRNA decay (NMD) and the complete loss of SARM1 protein. To confirm that these SARM1 edited cells do not express SARM1 protein, wild-type and SARM1 KO clones were differentiated into sensory neurons as described below. Protein lysates were prepared from neurons harvested after 15 or 28 days after initiation of differentiation protocol and subjected to Western blot analysis using an antibody targeting around Pro324 of SARM1. We find that SARM1 is well expressed in neurons derived from wild type iPSCs. However, SARM1 is undetectable in neurons derived from either of the SARM1 KO clones we tested (Figure 1C), demonstrating that the engineered SARM1 mutation effectively disrupted SARM1 expression. While we focus here on the function of SARM1 in human sensory neurons, the development of SARM1 KO iPSCs will enable functional analysis of SARM1 in many types of human iPSC-derived cell types.
Figure 1. CRIPSR-mediated SARM1 knock-out in human iPSCs.
(A) Schematic representation of the genomic locus of hSARM1 with exons highlighted in black. The gRNA with TGG PAM is shown in gray arrow. Indels in the SARM1 knockout lines KO1 and KO2 are shown in red and aligned with the wild-type sequence (+: inserted based; -: deleted bases). (B) NGS-based targeted deep sequencing indicated the indel frequency in both knock-out (KO) clones. (C) Western blotting shows loss of SARM1 expression in both KO1 and KO2 clones) at day 15 and day 28 after initation of the sensory neuron differentiation protocol. ACTIN was used as loading control.
Development of an efficient one-step sensory neuron differentiation protocol from iPSCs.
The role of SARM1 in the mechanism of axon degeneration has been most clearly defined in mouse sensory neurons. To assess the function of human SARM1, we wished to test its function in human sensory neurons derived from iPSCs. A number of protocols exist for generating human sensory neurons from iPSCs24,30–32. By modifying a previously published method24, we established an efficient one-step sensory neuron differentiation protocol from human iPSCs that does not require mouse embryonic fibroblast (MEF) conditioned media and demonstrates excellent viability of the differentiated neurons (Figure 2A and movie S1). In this one-step protocol, human iPSCs are treated concurrently with 5 small molecule inhibitors (lDN-193189: BMP signaling inhibitor, A83–01: ALK-5 inhibitor, CHIR99021: GSK3 inhibitor, RO4929097: Gamma-secretase inhibitor, and SU5402: FGFR inhibitor) plus 5% KnockOut Serum Replacement (KOSR). They showed progressive changes in morphology, from compact cell bodies characteristic of iPSCs, to the extension of multiple processes that in time formed interconnecting networks as expected from neurons (Movie S1). During these optimization experiments, we found that a 5% KOSR concentration and 3,000 cells/cm2 seeding density provided both high levels of neuronal differentiation and minimal non-neuronal cell contamination. To eliminate any residual dividing non-neuronal cells, we added 5-Fluoro-2’-deoxyuridine (FdU)33 early in the differentiation protocol (day 10). Finally, we took advantage of the commercially available supplement, CultureOne, to improve neuronal differentiation and maturation.
Figure 2. One-step protocol for the robust differentiation of iPSCs to sensory neurons.
(A) A schematic workflow outlining the high efficiency sensory neuron differentiation protocol. Morphological changes in the progression from iPSCs to sensory neurons were observed throughout the differentiation process. (B) Neuronal lineage marker expression by immunostaining using antibodies for TUJ1, neurofilament, BRN3A and ISL1. (C) Representative dot plot of two-color flow cytometric data showing population of iPSC-derived sensory neurons at day 17 and day 28 after initiation of differentiation protocol: Q1, high expression in peripherin; Q2, high expression in both BRN3A and peripherin; Q3, high expression in BRN3A; Q4, negative for both BRN3A and peripherin. Note the preponderance of neurons produced are Peripherin+, BRN3A+ sensory neurons. (D) Representative pictures of iPSC-derived sensory neuron spot cultures showing axonal outgrowth. FITC, fluorescein isothiocyanate; PE, phycoerythrin. Nuclei were counterstained with DAPI. Scale bar, 100 μm.
To assess sensory neuron differentiation, we performed immunocytochemistry on neurons 28 days post-differentiation using a variety of specific markers. We found that the putative sensory neurons produced by our differentiation protocol expressed markers of neuronal cytoskeleton, TUJ1 (TUBB3) and neurofilament, as well as canonical markers of sensory neurons, BRN3A (POU4F1) and ISL1 (Islet1) (Figure 2B). Moreover, flow cytometric analysis showed that ~90% of the population was doubly positive for the sensory neuron markers peripherin and BRN3A at day 28 post differentiation (Figure 2C and table S1). Hence, this modified one step differentiation protocol yields excellent purity of sensory neurons. Finally, we wished to plate the neurons in spot cultures, as this format allows for optimal analysis of axon degeneration and for collection of axon-only lysates for protein34 or metabolite28 analysis. For the human iPSC-derived sensory neurons, day 15 differentiated neurons were plated as spot cultures. The extensive axonal growth away from the cell bodies is sufficient for the subsequent mechanistic analysis of axon degeneration (Figure 2D and Movie S2 and S3). While our focus here is on the mechanism of axon degeneration, the development of this simple and efficient differentiation protocol for human sensory neurons will enable high throughput phenotypic screens and promote the study of their biology.
SARM1 promotes severed axon degeneration in human sensory neurons.
To test the role of SARM1 in human neurons, we first established an axon degeneration assay. Sensory neurons derived from parental wildtype control or SARM1 KO iPSCs were pipetted into a small area (spot culture) and axons were allowed to grow for 13 days, at which point axons were severed with a scalpel. The wild type transected axons formed blebs at 6 hours post-axotomy and fragmented within 24 hours after injury (Figure 3A and Movie S4). To quantify axon degeneration, we developed an automated image analysis pipeline that assesses axon degeneration. We previously developed an algorithm to quantify axon degeneration for brightfield images35,36, however, the phase contrast images from the IncuCyte S3 system used in the current analysis requires additional image processing steps due to the low contrast of the cellular structure and artifacts associated with the phase-contrast optical system37. We therefore developed a novel image analysis method based on axon vacuolization using ImageJ and Weka segmentation plugin (see Methods and Figure S1). From analysis of the images and the use of this new algorithm, it is clear that the profile of axon degeneration in human iPSC-derived sensory neurons is strikingly similar to that observed in mouse DRG sensory neurons6.
Figure 3. SARM1 is required for injury-induced axon degeneration.
(A) Representative images of sensory neuron (SN) axons at 24 hours after axotomy. Axotomy-induced axon degeneration was observed in human wild-type (WT) axons while axons from SARM1 knockout (KO) sensory neurons did not degenerate after injury. Scale bar, 100μm. (B) Axonal degeneration 24 hours post-axotomy was quantified. WT axons degenerate after axotomy. SARM1 KO axons remained intact post injury. Expression of human SARM1 restores normal injury-induced axon degeneration in SARM1 knockout (KO) SNs. Kruskal-Wallis test, H=163.9 and p < 2.2 × 10−16, showed significant difference among groups (sample numbers are indicated on top of bars). Pairwise Wilcox comparison with Bonferroni adjusted p-value was used to calculate significant differences. *p<2 × 10−16 denotes significant difference compared with wild-type control axons. # denotes no significant difference compared with wild-type control axons. **p<1 × 10−7 denotes significant difference compared with SARM1 KO control axons. (C) Axotomy-induced axon degeneration is blocked by dominant negative SARM1 (DNSARM1) but not EGFP (control) in wild-type SNs (sample numbers are indicated on top of bars). *p<1 × 10−5 denotes significant difference compared with wild-type control axons with Mann-Whitney U test. (D,E) Axonal NAD+ and cADPR was quantified at 0 and 16 hr post axotomy using LC-MS/MS. NAD+ depletion and cADPR increase were observed in severed axons from wildtype neurons, but no changes were observed in axons from SARM1 KO neurons. For NAD+, Kruskal-Wallis test, H=29.3 and p = 2.0 × 10−5, showed significant difference among groups (n = 12). For cADPR, Kruskal-Wallis test, H=49.6 and p = 1.7 × 10−9, showed significant difference among groups (n = 12). Pairwise Wilcox comparison with Bonferroni adjusted p-value was used to calculate significant differences. *p<1 × 10−5 denotes significant difference from metabolite levels of wild-type axons at 0 hour post axotomy. # denotes no significant differences compared with 0 hour post axotomy of each group.
We next investigated the axon degeneration process in human sensory neurons derived from the two independent SARM1 KO iPSC lines. In contrast to the wild type neurons whose axons degenerate rapidly, axons from SARM1-deficient neurons derived from either iPSC SARM1 KO line showed no signs of degeneration for up to 48 hours (Figure 3A, B (control, red dots), and Movie S5).
To further investigate the utility of these human sensory neurons for axon degeneration therapeutic studies, we performed experiments wherein lentivirus was used to express SARM1 in these neurons. First, we re-introduced SARM1 into wildtype control and SARM1 KO human sensory neurons by lentivirus infection in an attempt to rescue the blockade in axon degeneration caused by lack of SARM1 in the neurons derived from the SARM1 KO engineered iPSCs. In both wildtype and SARM1 KO neurons expressing SARM1, injured axons degenerated within 24 hours (Figure 3B (SARM1, green dots)). This successful rescue demonstrates that the defect in axon degeneration in the SARM1 KO neurons is due to the specific absence of SARM1 in the Cas9 edited iPSCs.
Next, we tested a dominant negative version of SARM1 that potently blocks axon degeneration in mouse neurons both in vitro and in vivo9. This SARM1 derivative has two mutations (K193A and H685A) that prevent it from being activated by injury. This SARM1 mutant is a strong gene therapy candidate for conditions where axon degeneration is an important component of the disorder. Its potential therapeutic utility will depend on its ability to prevent axon degeneration in human neurons, so we tested its activity in human iPSC-derived sensory neurons. Lentivirus expressing SARM1(K193A, H685A) was used to infect wildtype human sensory neurons and axons were severed. We found that axon degeneration was blocked to a similar extent as that observed in the SARM1 KO derived sensory neurons (Figure 3C). Together these findings demonstrate that SARM1 mediates axon degeneration in human sensory neurons, and that a gene therapeutic targeting SARM1 that was initially validated using mice is also effective in preventing axon degeneration in human neurons.
SARM1 is an injury-activated NADase in mouse. Once activated, SARM1 cleaves NAD+ and generates cADPR, which serves as a selective biomarker of SARM1 activity20. Here we explored the metabolic consequences of SARM1 activation in human neurons. Because SARM1 is activated locally, all metabolite measurements are made from axon-only lysates. In wild-type neurons, there is a significant decrease in the level of axonal NAD+ 16 hours after injury. In contrast, in neurons derived from either SARM1 KO clone there is no significant change in NAD+ levels 16 hours after injury (Figure 3D). These findings demonstrate that SARM1-dependent NAD+ depletion occurs in injured human neurons.
In exploring the mechanism of SARM1-mediated axon degeneration and NAD+ depletion, we identified cADPR as an important biomarker of SARM1 activity in mouse neurons. To determine whether cADPR was generated by SARM1 in human neurons, and could thus serves as a biomarker of SARM1 activity in humans, we performed a series of metabolite assays. First, we examined the levels of cADPR in uninjured axons. While the levels of cADPR are low in uninjured axons from wild type neurons, they are even lower, and indeed are near the limit of detection, in axons from SARM1 KO neurons (Figure 3E). These findings are consistent with a low, basal level of SARM1 activity in healthy human axons. Upon axonal injury there is a dramatic increase in cADPR levels in axons from wild type neurons, consistent with the activation of SARM1 NADase activity. In contrast, there is no change in axonal cADPR levels in injured SARM1 KO neurons (Figure 3E). These findings show that SARM1 is the major source of cADPR in both uninjured and injured axons, and therefore identify cADPR as a useful biomarker of SARM1 activity in human neurons. Together, these findings demonstrate the conservation of both metabolic and prodegenerative functions of SARM1 in human and mouse neurons.
SARM1 promotes axon degeneration in a human cellular model of chemotherapy-induced peripheral neuropathy.
Chemotherapy-induced peripheral neuropathy (CIPN) is the major dose-limiting side effect of many common chemotherapeutic agents, and there is therefore great interest in developing preventative treatments for CIPN38,39. SARM1 is the central executioner of axon degeneration in in vitro and in vivo mouse models of CIPN8,10,39. Here we developed a human in vitro model of CIPN to assay the role of SARM1 in driving axon loss in response to chemotherapeutic agents. Vincristine is often used to treat leukemias and lymphomas as well as other tumors. Its use is commonly associated with painful neuropathy, a neurotoxicity that has been modeled in rodent neurons8. We treated iPSC-derived human sensory neurons with 0.5 nM, and 5 nM vincristine or vehicle (DMSO) for 48 hours and assessed axonal health (Figure 4A, B and Movie S6). Axon degeneration was observed in wildtype neurons within 48 hours after exposure to 5 nM vincristine. In contrast, axons from sensory neurons derived from the human SARM1 KO iPSC lines were protected from vincristine-induced axon degeneration at 5 nM vincristine (Figure 4A, B and Movie S7). Hence, the absence of SARM1 protects axons from vincristine-induced axon degeneration in human neurons, highlighting the potential utility of SARM1 inhibition for the prevention of CIPN11,39,40.
Figure 4. Human neurons lacking SARM1 are resistant to vincristine-mediated axon degeneration.
(A) Representative images of sensory neuron axons at 48 hr post DMSO or vincristine treatments (5 nM). Scale bar, 100μm. (B) Axon degeneration post 48 hr DMSO or vincristine treatment (0.5, and 5 nM) was quantified. After vincristine treatment, significant axonal degeneration was observed in wild-type. This degeneration was delayed in SARM1 knockout axons. Kruskal-Wallis test, H=160.8 and p = 2.2 × 10−16, showed significant difference among groups (sample numbers are indicated on top of bars). Pairwise Wilcox comparison with Bonferroni adjusted p-value was used to calculate significant differences. *p < 5 × 10−4 denotes significant difference from the DMSO treated wild-type axons at a same dose.
Conclusion
There is great interest in targeting SARM1 for the prevention and treatment of neurodegenerative diseases because SARM1 is required for axon and/or cell body loss in many mouse models of neurodegeneration and, as an NAD+ cleavage enzyme, it is a likely druggable target. However, prior to this study, there was no evidence that SARM1 promotes human neurodegeneration. Here we generate SARM1 KO human iPSCs, develop a modified one-step protocol for the efficient generation of human sensory neurons, and demonstrate that the metabolic and prodegenerative role of SARM1 is conserved in human neurons. These findings support the potential utility of SARM1-directed therapeutics for the prevention and treatment of human neurodegenerative diseases.
Supplementary Material
Supplemental Figure 1 The quantification of axon degeneration. (A) Example of phase contrast images of the same field of axons (left column) and corresponding Weka segmented images (right column) at 0, 9, and 24 hours post axotomy. Segmented images were generated using Weka segmentation plugin in ImageJ. Briefly, the representative structures corresponding to the healthy axon, vacuolated axon, and no cellular component were assigned manually in the Weka segmentation model. This model was used to train the Weka segmentation tool to determine these structures using a series of image segmentation methods. The Weka segmentation model was further trained by modifying the parameters for segmentation methods until the best correlation between automated segmentation and segmentation defined by observers was obtained. The resultant segmented images were displayed with color coded structures including the healthy axon (indicated by red pixels), vacuolated axon (indicated by green pixels), and no cellular component (indicated by yellow pixels). After the optimization, a series of axon images was analyzed automatically using ImageJ script that run the trained Weka segmentation plugin. Examples of segmented images are shown in the right column. (B) The definition of the color code in the Weka segmented image. (C) Injury-induced change of vacuolation index (Vi) determined by ratio of vacuolated vs. total axon areas (the number of green pixels / the summation of the number of green and red pixels) of the region shown in (A). Axons were severed at 0 hours. The Vi gradually increased over time and plateaued around 20 hr after injury. Axon degeneration was determined by the ratio of Vi at the time after axon injury (24 hr or 48 hr) compared with time at the injury (0 hr) obtained from the same field of the well and expressed as axon degeneration index. Scale bar, 50 μm.
Highlights.
Human sensory neurons are derived from SARM1 null iPSC with high yield and purity
SARM1 promotes axon degeneration in human sensory neurons
cADPR is a biomarker of SARM1 enzyme activity in human sensory neurons
Acknowledgements
This work was supported by funds from the National Institutes of Health grants (R01CA219866 and RO1NS087632 to A.D. and J.M. and RF1AG013730 to J.M.).
We thank Kelli Simburger, Tim Fahrner, and Alicia Neiner for technical support. We thank Mahoto Sasaki for generating the ImageJ script analyzing axon degeneration. We thank members of the Milbrandt and DiAntonio labs for helpful discussions. We thank Amber Neilson and Yong Miao for technical advices. We thank the Siteman Flow Cytometry Core which provided single-cell sorting services. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA091842.
Footnotes
Declaration of Competing Interest
A.D and J.M. co-founders of, consultants to, and shareholders in Disarm Therapeutics, a wholly owned subsidiary of Eli Lilly & Co. Y.S. is a consultant to Disarm Therapeutics.
References
- 1.Johnson VE, Stewart W & Smith DH Axonal pathology in traumatic brain injury. Experimental neurology 246, 35–43, doi: 10.1016/j.expneurol.2012.01.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cashman CR & Hoke A Mechanisms of distal axonal degeneration in peripheral neuropathies. Neuroscience letters 596, 33–50, doi: 10.1016/j.neulet.2015.01.048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tagliaferro P & Burke RE Retrograde Axonal Degeneration in Parkinson Disease. Journal of Parkinson’s disease 6, 1–15, doi: 10.3233/jpd-150769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figley MD & DiAntonio A The SARM1 axon degeneration pathway: control of the NAD(+) metabolome regulates axon survival in health and disease. Current opinion in neurobiology 63, 59–66, doi: 10.1016/j.conb.2020.02.012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman MP & Höke A Programmed axon degeneration: from mouse to mechanism to medicine. Nature reviews. Neuroscience 21, 183–196, doi: 10.1038/s41583-020-0269-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdts J, Summers DW, Sasaki Y, DiAntonio A & Milbrandt J Sarm1-mediated axon degeneration requires both SAM and TIR interactions. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 13569–13580, doi: 10.1523/jneurosci.1197-13.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osterloh JM et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science (New York, N.Y.) 337, 481–484, doi: 10.1126/science.1223899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisler S et al. Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain : a journal of neurology 139, 3092–3108, doi: 10.1093/brain/aww251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geisler S et al. Gene therapy targeting SARM1 blocks pathological axon degeneration in mice. The Journal of experimental medicine 216, 294–303, doi: 10.1084/jem.20181040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turkiew E, Falconer D, Reed N & Höke A Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. Journal of the peripheral nervous system : JPNS 22, 162–171, doi: 10.1111/jns.12219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauss R, Bosanac T, Devraj R, Engber T & Hughes RO Axons Matter: The Promise of Treating Neurodegenerative Disorders by Targeting SARM1-Mediated Axonal Degeneration. Trends in pharmacological sciences 41, 281–293, doi: 10.1016/j.tips.2020.01.006 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Loreto A et al. Mitochondrial impairment activates the Wallerian pathway through depletion of NMNAT2 leading to SARM1-dependent axon degeneration. Neurobiology of disease 134, 104678, doi: 10.1016/j.nbd.2019.104678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers DW, DiAntonio A & Milbrandt J Mitochondrial dysfunction induces Sarm1-dependent cell death in sensory neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 9338–9350, doi: 10.1523/jneurosci.0877-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozaki E et al. SARM1 deficiency promotes rod and cone photoreceptor cell survival in a model of retinal degeneration. Life science alliance 3, doi: 10.26508/lsa.201900618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki Y et al. SARM1 depletion rescues NMNAT1-dependent photoreceptor cell death and retinal degeneration. eLife 9, doi: 10.7554/eLife.62027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Essuman K et al. The SARM1 Toll/Interleukin-1 Receptor Domain Possesses Intrinsic NAD(+) Cleavage Activity that Promotes Pathological Axonal Degeneration. Neuron 93, 1334–1343.e1335, doi: 10.1016/j.neuron.2017.02.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essuman K et al. TIR Domain Proteins Are an Ancient Family of NAD(+)-Consuming Enzymes. Current biology : CB 28, 421–-430.e424., doi: 10.1016/j.cub.2017.12.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horsefield S et al. NAD(+) cleavage activity by animal and plant TIR domains in cell death pathways. Science (New York, N.Y.) 365, 793–799, doi: 10.1126/science.aax1911 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Wan L et al. TIR domains of plant immune receptors are NAD(+)-cleaving enzymes that promote cell death. Science (New York, N.Y.) 365, 799–803, doi: 10.1126/science.aax1771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki Y et al. cADPR is a gene dosage-sensitive biomarker of SARM1 activity in healthy, compromised, and degenerating axons. Experimental neurology 329, 113252, doi: 10.1016/j.expneurol.2020.113252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers DW, Gibson DA, DiAntonio A & Milbrandt J SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proceedings of the National Academy of Sciences of the United States of America 113, E6271–e6280, doi: 10.1073/pnas.1601506113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A & Milbrandt J SARM1 activation triggers axon degeneration locally via NAD(+) destruction. Science (New York, N.Y.) 348, 453–457, doi: 10.1126/science.1258366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiAntonio A Axon degeneration: mechanistic insights lead to therapeutic opportunities for the prevention and treatment of peripheral neuropathy. Pain 160 Suppl 1, S17–s22, doi: 10.1097/j.pain.0000000000001528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai S, Han L, Ao Q, Chan YS & Shum DK Human Induced Pluripotent Cell-Derived Sensory Neurons for Fate Commitment of Bone Marrow-Derived Schwann Cells: Implications for Remyelination Therapy. Stem cells translational medicine 6, 369–381, doi: 10.5966/sctm.2015-0424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YH & Pruett-Miller SM Improving single-cell cloning workflow for gene editing in human pluripotent stem cells. Stem cell research 31, 186–192, doi: 10.1016/j.scr.2018.08.003 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Sentmanat MF, Peters ST, Florian CP, Connelly JP & Pruett-Miller SM A Survey of Validation Strategies for CRISPR-Cas9 Editing. Scientific reports 8, 888, doi: 10.1038/s41598-018-19441-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arganda-Carreras I et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426, doi: 10.1093/bioinformatics/btx180 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Sasaki Y, Nakagawa T, Mao X, DiAntonio A & Milbrandt J NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD(+) depletion. eLife 5, doi: 10.7554/eLife.19749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Araki T, Sasaki Y & Milbrandt J Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science (New York, N.Y.) 305, 1010–1013, doi: 10.1126/science.1098014 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Chambers SM et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nature biotechnology 30, 715–720, doi: 10.1038/nbt.2249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young GT et al. Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Molecular therapy : the journal of the American Society of Gene Therapy 22, 1530–1543, doi: 10.1038/mt.2014.86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartzentruber J et al. Molecular and functional variation in iPSC-derived sensory neurons. Nature genetics 50, 54–61, doi: 10.1038/s41588-017-0005-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui CW, Zhang Y & Herrup K Non-Neuronal Cells Are Required to Mediate the Effects of Neuroinflammation: Results from a Neuron-Enriched Culture System. PloS one 11, e0147134, doi: 10.1371/journal.pone.0147134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin JE et al. SCG10 is a JNK target in the axonal degeneration pathway. Proceedings of the National Academy of Sciences of the United States of America 109, E3696–3705, doi: 10.1073/pnas.1216204109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki Y, Vohra BP, Lund FE & Milbrandt J Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. The Journal of neuroscience : the official journal of the Society for Neuroscience 29, 5525–5535, doi: 10.1523/jneurosci.5469-08.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerdts J, Sasaki Y, Vohra B, Marasa J & Milbrandt J Image-based screening identifies novel roles for IkappaB kinase and glycogen synthase kinase 3 in axonal degeneration. The Journal of biological chemistry 286, 28011–28018, doi: 10.1074/jbc.M111.250472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaccard N, Szita N & Griffin LD Segmentation of phase contrast microscopy images based on multi-scale local Basic Image Features histograms. Comput Methods Biomech Biomed Eng Imaging Vis 5, 359–367, doi: 10.1080/21681163.2015.1016243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukuda Y, Li Y & Segal RA A Mechanistic Understanding of Axon Degeneration in Chemotherapy-Induced Peripheral Neuropathy. Front Neurosci 11, 481, doi: 10.3389/fnins.2017.00481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geisler S et al. Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight 4, doi: 10.1172/jci.insight.129920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loring HS & Thompson PR Emergence of SARM1 as a Potential Therapeutic Target for Wallerian-type Diseases. Cell Chem Biol 27, 1–13, doi: 10.1016/j.chembiol.2019.11.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 The quantification of axon degeneration. (A) Example of phase contrast images of the same field of axons (left column) and corresponding Weka segmented images (right column) at 0, 9, and 24 hours post axotomy. Segmented images were generated using Weka segmentation plugin in ImageJ. Briefly, the representative structures corresponding to the healthy axon, vacuolated axon, and no cellular component were assigned manually in the Weka segmentation model. This model was used to train the Weka segmentation tool to determine these structures using a series of image segmentation methods. The Weka segmentation model was further trained by modifying the parameters for segmentation methods until the best correlation between automated segmentation and segmentation defined by observers was obtained. The resultant segmented images were displayed with color coded structures including the healthy axon (indicated by red pixels), vacuolated axon (indicated by green pixels), and no cellular component (indicated by yellow pixels). After the optimization, a series of axon images was analyzed automatically using ImageJ script that run the trained Weka segmentation plugin. Examples of segmented images are shown in the right column. (B) The definition of the color code in the Weka segmented image. (C) Injury-induced change of vacuolation index (Vi) determined by ratio of vacuolated vs. total axon areas (the number of green pixels / the summation of the number of green and red pixels) of the region shown in (A). Axons were severed at 0 hours. The Vi gradually increased over time and plateaued around 20 hr after injury. Axon degeneration was determined by the ratio of Vi at the time after axon injury (24 hr or 48 hr) compared with time at the injury (0 hr) obtained from the same field of the well and expressed as axon degeneration index. Scale bar, 50 μm.