Abstract
Understanding the intricacies of lethal prostate cancer poses specific challenges due to difficulties in accurate modeling of metastasis in vivo. Here we show that NPKEYFP mice (for Nkx3.1CreERT2/+; Ptenflox/flox; KrasLSL-G12D/+; R26R-CAG-LSL-EYFP/+) develop prostate cancer with a high penetrance of metastasis to bone, thereby enabling detection and tracking of bone metastasis in vivo and ex vivo. Transcriptomic and whole-exome analyses of bone metastasis from these mice revealed distinct molecular profiles conserved between human and mouse and specific patterns of subclonal branching from the primary tumor. Integrating bulk and single-cell transcriptomic data from mouse and human datasets with functional studies in vivo unravels a unique MYC/RAS co-activation signature associated with prostate cancer metastasis. Finally, we identify a gene signature with prognostic value for time to metastasis and predictive of treatment response in human patients undergoing androgen receptor therapy across clinical cohorts, thus uncovering conserved mechanisms of metastasis with potential translational significance.
Metastatic prostate cancer, a leading cause of cancer-related death in men, is clinically manifested often following androgen deprivation therapy, which leads to metastatic castration-resistant prostate cancer (mCRPC)1–3 that is frequently accompanied by aggressive disease variants, including neuroendocrine prostate cancer (NEPC)3,4. The major site of prostate cancer metastasis is bone5–7; however, current treatments are neither curative nor do they specifically target bone metastasis1,7–9. A major challenge in studying bone metastasis is the paucity of models that recapitulate cell-intrinsic features of tumor cells and the physiological milieu of the native tumor microenvironment as occurs in vivo10. We now demonstrate that NPKEYFP mice (for Nkx3.1CreERT2/+; Ptenflox/flox; KrasLSL-G12D/+; R26R-CAG-LSL-EYFP/+) develop highly penetrant bone metastasis. Cross-species analyses of mouse bone metastasis and human prostate cancer has established co-activation of MYC and RAS pathways as a key driver and identified a gene signature, META-16, associated with time to metastasis in primary prostate cancer and response to anti-androgen treatment in metastatic disease.
Results
A highly penetrant mouse model of bone metastasis.
Previously reported NPK mice develop highly penetrant metastatic prostate cancer with disseminated tumor cells (DTCs) in bone but not overt bone metastasis11,12. Reasoning that a challenge in identifying bone metastases is their detection, we crossed NPK mice with an enhanced fluorescence reporter allele (R26R-CAG-LSL-EYFP/+)13 to generate NPKEYFP mice (Fig. 1a). These mice utilize an inducible Cre driven by the Nkx3.1 promoter (Nkx3.1CreERT2/+) to achieve temporal and spatial regulation of gene recombination of Ptenflox/flox and KrasLSL-G12D/+ specifically in luminal prostatic cells14,15, as well as activation of R26R-CAG-LSL-EYFP/+ for lineage tracing by yellow fluorescent protein (YFP), which enables fluorescent visualization of tumors and metastases (Fig. 1a).
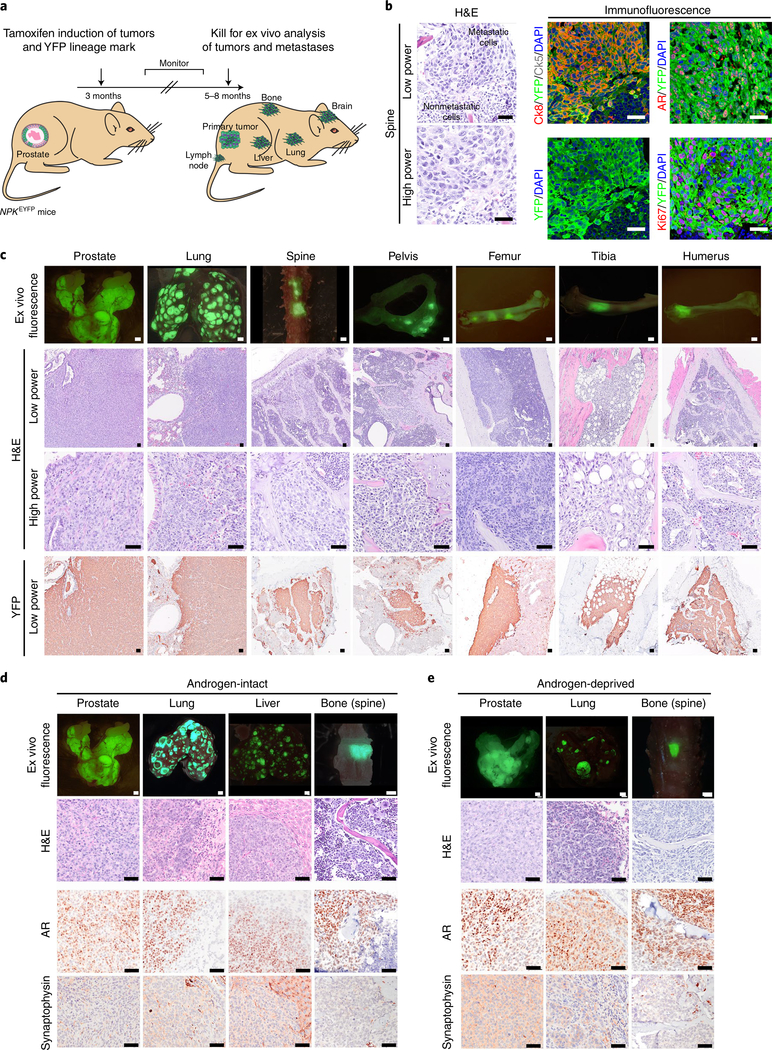
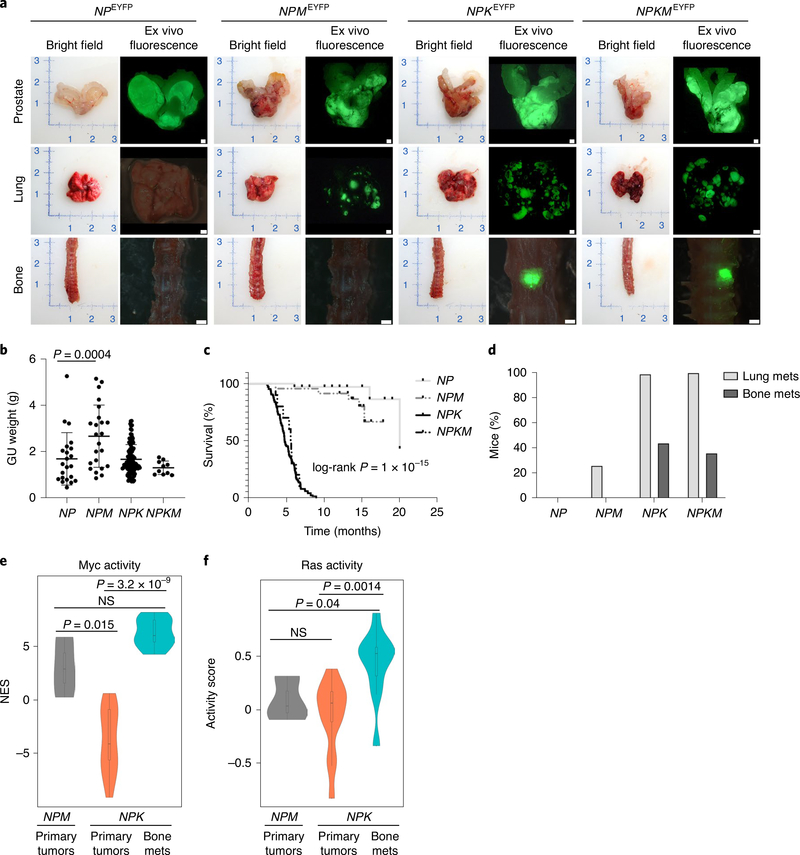
Fig. 1 |. A mouse model of highly penetrant bone metastasis.
a, Strategy. Tamoxifen delivery to NPKEYFP mice (for Nkx3.1CreERT2/+; Ptenflox/flox; KrasLSL-G12D/+; R26R-CAG-LSL-EYFP/+) at 3 months induces tumor formation and lineage marking. Tumor-induced mice are monitored for 5–8 months for development of metastases to bone as well as lymph node, lung, liver and brain. b–e, Histopathological analyses. Representative hematoxylin and eosin (H&E) (left) or confocal (right) images of bone metastases (spine) (b). DAPI, 4,6-diamidino-2-phenylindole. Coexpression of YFP with luminal cytokeratin (Ck8), basal cytokeratin (Ck5), the AR and Ki67. Representative images of prostate tumors and metastases from lung and bone (spine, pelvis, femur, tibia and humerus) showing ex vivo fluorescence, histology (H&E) and immunostaining for YFP (c). Representative images of prostate tumor and metastases from androgen-intact (d) or castrated (androgen-deprived) (e) NPKEYFP mice showing ex vivo fluorescence, histology (H&E) and immunostaining for AR or the NEPC marker, synaptophysin. Representative images from five independent mice are shown (b–e). Scale bars, 0.1 cm for ex vivo fluorescence images and 50 μm for all other images.
NPKEYFP mice develop highly penetrant metastasis as evident by ex vivo YFP fluorescence as well as YFP immunostaining (n = 106), which is not seen in control (uninduced) NPKEYFP mice (n = 3) or nonmetastatic NPEYFP mice (for Nkx3.1CreERT2/+; Ptenflox/flox; R26R-CAG-LSL-EYFP/+) (n = 35)16 (Fig. 1b,c, Extended Data Fig. 1 and Supplementary Table 1). A high percentage of NPKEYFP mice display fluorescence in bone, indicative of bone metastasis (44%; n = 47 of 106; Fig. 1c and Supplementary Table 1). In terminal mice, fluorescence was evident in spine (n = 32 of 47), pelvis (n = 18 of 47), femur (n = 22 of 47), tibia (n = 9 of 47) and humerus (n = 9 of 47) (Fig. 1c, Extended Data Fig. 1h and Supplementary Table 1), which are sites of bone metastasis in humans17. Longitudinal analyses revealed micro-metastases in bone by 3 months after tumor induction (Extended Data Fig. 1i), similar to when DTCs are first detected11. Micro-metastases occurred earlier and were more prevalent in bone than in lung (Extended Data Fig. 1j). Immunostaining revealed YFP-expressing cells in bone that express Ki67 and the luminal cytokeratin, Ck8 (Fig. 1b,c), confirming their origination from lineage-marked prostatic cells14–16.
Overall, there were few discernable differences in NPKEYFP mice that developed bone metastases (n = 47 of 106) versus those that did not (n = 59 of 106). However, those with bone metastases had a significantly augmented metastatic phenotype, with an average of >80 lung, >40 liver and at least 1 brain metastasis, whereas those without bone metastases had relatively few lung, and few if any liver or brain metastases (P < 0.0001; Extended Data Fig. 1 and Supplementary Table 1).
Similar to primary tumors and lung metastases, bone metastatic cells expressed androgen receptor (AR) protein and AR activity (based on previous works18) but negligible levels of synaptophysin, a marker of NEPC and low levels of NEPC activity (based on previous works19) (Fig. 1b–e and Extended Data Fig. 2). Notably, surgical castration did not affect median survival, incidence of bone or other metastases, or expression or activity levels of AR or NEPC when comparing castrated (n = 22) and noncastrated (n = 106) mice (Fig. 1d,e, Extended Data Fig. 2 and Supplementary Table 1). Gene set enrichment analyses (GSEA)20 comparing bone metastases with primary tumors from castrated (n = 6) and noncastrated (n = 13) mice revealed significant similarity (P < 0.001; Extended Data Fig. 2 and Supplementary Table 2).
Conservation of bone metastases in NPKEYFP with human prostate cancer.
We performed RNA sequencing (RNA-seq) on primary tumors (n = 19) and bone (n = 12), lung (n = 11), liver (n = 5), brain (n = 3) and lymph node (n = 4) metastases from 16 independent NPKEYFP mice (Supplementary Table 2). Principal-component analyses (PCA) showed that bone metastases clustered separately from primary tumors and other metastases (Fig. 2a). To evaluate conservation with human prostate cancer, we defined a mouse ‘bone metastasis signature’ comparing bone metastases (n = 12) with primary tumors (n = 19) (Supplementary Table 2). Cross-species GSEA comparing this signature with an analogous signature of human bone biopsies (n = 19) and primary tumors (n = 19) from patients with metastatic prostate cancer (Balk cohort21; Supplementary Table 3) revealed significant enrichment (Fig. 2b).
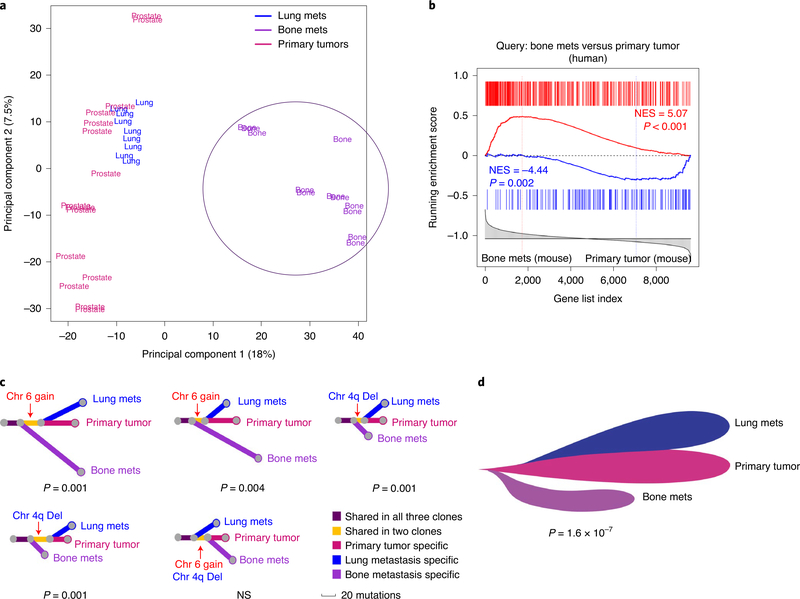
Fig. 2 |. Molecular analysis of bone metastasis from NPKEYFP mice.
a,b, Transcriptomic analyses. PCA of bulk RNA-seq of primary tumors (n = 15), lung metastases (n = 9) and bone metastases (n = 12) from androgen-intact NPKEYFP mice (Supplementary Table 2) (a). mets, metastases. The circle indicates separation of bone metastases from primary tumors and lung metastases. Conservation with human prostate cancer (b). GSEA using human bone metastasis signature based on Balk (Supplementary Table 3c) to query the reference mouse bone metastasis gene signature from NPKEYFP mice (Supplementary Table 2c). NES (normalized enrichment score) and P values were estimated using 1,000 gene permutations. c,d, Phylogenetic analysis of WES data. Evolutionary trees for matched trios of primary tumor, bone and lung metastases from five independent mice (represented by each of the trees) were constructed by WES analyses of somatic mutations (substitutions and indels) (Supplementary Table 4a) (c). The length of lines indicates the number of mutations in each branch and colors indicate the mutations unique to or shared between clones; bootstrap-derived P values for each case using 1,000 permutations are shown. Informative CNVs (gains in chromosome 6, ‘Chr 6 gain’ and deletions in chromosome 4q ‘Chr 4q Del’; Supplementary Table 4b) are shown by red arrows. Composite phylogeny tree based on consistent evolutionary patterns across all trees in c (d). The meta-analysis P value was calculated using Fisher’s method by combining bootstrap-derived P values from individual trees in c. NS, not significant.
Whole-exome sequencing (WES) of matched sets of primary tumors, bone and lung metastases, as well as control DNA (tail), from five NPKEYFP mice did not identify significant somatic mutations or alterations of tumor suppressors or oncogenes, similar to other genetically engineered mouse models (GEMMs)22–24. Nonetheless, WES enabled reconstruction of evolutionary trees for dominant clones in the primary tumor, bone and lung metastases (Fig. 2c,d and Supplementary Table 4a). In particular, phylogenetic analysis revealed that the common recent ancestor of tumor and bone metastasis preceded the common recent ancestor of tumor and lung metastasis in four of five mice (P = 1.6 × 10−7; Fig. 2c,d), suggesting that an earlier metastatic clone seeded the bone metastasis, whereas lung metastases were derived from a later clone, consistent with our finding that micro-metastases in bone arise earlier than in lung (Extended Data Fig. 1I,j). Inference of copy number variations (CNVs) from the WES data also revealed few significant gains or losses in primary tumors or metastases (Supplementary Table 4b). Nonetheless, informative CNV events reflect the history deduced by the single-nucleotide variant analyses, thus further supporting the inferred evolutionary hierarchy (Fig. 2c).
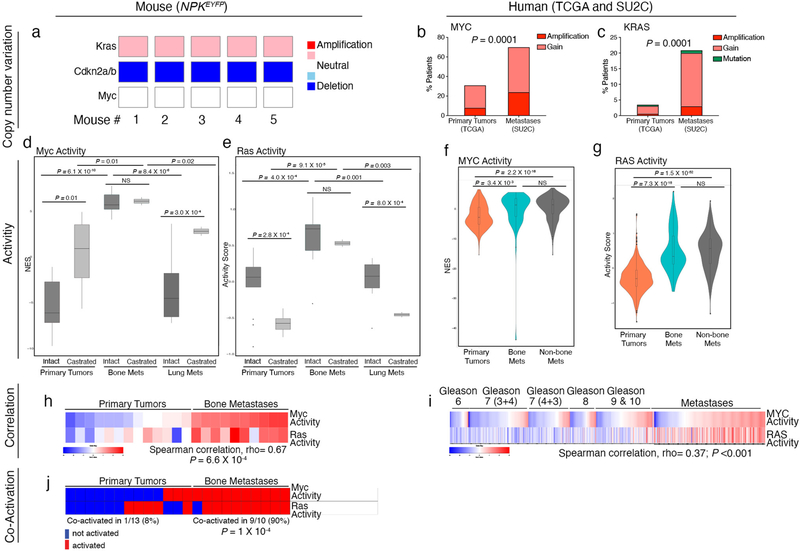
Among the few significant CNVs, Kras was amplified in tumors of all five NPKEYFP mice; analogous to other Kras-driven GEMMs22,24, this spanned the entire chromosome 6 (Extended Data Fig. 3 and Supplementary Table 4b), In human prostate cancer, KRAS copy number gains occur in 2% of primary tumors (based on the Cancer Genome Atlas (TCGA) prostate adenocarcinoma25, n = 497) but 20% of metastases (based on the Stand Up to Cancer (SU2C)26, n = 429) (Extended Data Fig. 3 and Supplementary Table 3). Thus, NPKEYFP mice model key features of human prostate cancer metastasis.
Activation of Myc pathway is cell-intrinsic to bone metastasis in NPKEYFP mice.
We performed single-cell RNA-seq on matched samples from primary tumor and bone metastases (Fig. 3 and Supplementary Table 5). As visualized using uniform manifold approximation and projection (UMAP)27, primary tumor cells (black) separated into a major (83%) and smaller (17%) group; bone cells (dark gray) separated into a major group (77%) projecting far from primary tumor cells and a smaller group (23%) projecting close to tumor cells (Fig. 3a).
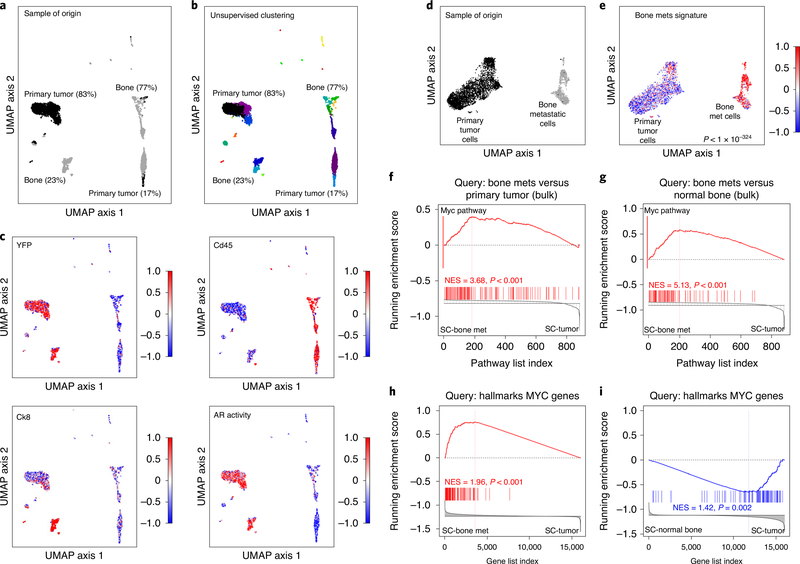
Fig. 3 |. Single-cell sequencing reveals Myc pathway activation as a cell-intrinsic feature of bone metastasis.
a–c, Single-cell RNA-seq of primary tumor and bone metastasis. UMAP visualization of matched primary tumor and bone samples from NPKEYFP mice (Supplementary Table 5a). Sample of origin; black corresponds to the primary tumor sample and dark gray to the bone sample (a). Unsupervised clustering; colors indicate distinct clusters of cells with the relative percentages of the primary tumor and bone samples indicated (b). Scaled expression (DESeq2 normalized values) of YFP, Cd45 and Ck8 expression levels and AR activity levels (based on previous work18) (c). d,e, Analysis of isolated primary tumor and bone metastatic cell clusters. Sample of origin (d). Black corresponds to the primary tumor cells and dark gray to the bone metastatic cells. Enrichment of the bone metastasis signature from bulk RNA-seq (Supplementary Table 2c) in bone metastatic versus primary tumor cells (e). P value was calculated by a two-sample two-tailed Welch t-test. f,g, Pathway-based GSEA. GSEA comparing pathways enriched in mouse bone metastasis signature (from bulk RNA-seq, Supplementary Table 6a) with those enriched in the single-cell bone metastasis signature (Supplementary Table 5c) (f). The red bar shows the location of the hallmarks MYC pathway, which is the top-most enriched pathway across the two signatures. GSEA comparing pathways enriched in a signature from the bulk RNA-seq comparing bone metastases and normal bone (Supplementary Table 6b) with those enriched in the single-cell bone metastasis signature (Supplementary Table 5c) (g). GSEA using genes from the MYC hallmarks pathway to query the single-cell bone metastasis signature (Supplementary Table 5a) h,i Gene-based GSEA. (h). GSEA using genes from MYC hallmarks pathway to query a signature based on single-cell resident nontumor bone cells versus primary tumor cells (Supplementary Table 5b) (i). NES and P values were estimated using 1,000 gene or pathway permutations, as appropriate (f–i). SC, single cell.
Unsupervised clustering revealed that the larger group of bone cells were Cd45+, whereas the smaller group were YFP+ (P < 10−324; Fig. 3b,c). This smaller group was highly enriched for Ck8 expression and AR activity (based on previous work18) (Ck8, P = 3.5 × 10−315; AR P = 2.7 × 10−134; Fig. 3c), consistent with immunostaining results (Fig. 1b,c). Thus, we infer that YFP+ (Cd45−) bone cells projecting close to the major group of primary tumor cells are metastatic cells, whereas Cd45+ (YFP−) bone cells are benign resident bone cells. In subsequent analyses, we focused on the major primary tumor cells (black) and the YFP-expressing bone metastatic cells (dark gray) (hereafter called the ‘primary tumor cells’ and ‘bone metastatic cells’, respectively; Fig. 3d).
As the ‘bone metastasis signature’ defined by bulk RNA-seq invariably includes nontumor cells, we sought to distinguish tumor cell-intrinsic features by projecting this signature on one from the single-cell bone metastatic and primary tumor cells (hereafter called the ‘single-cell bone metastasis signature’; Supplementary Table 5a). At the gene level, the bone metastasis signature was significantly enriched in the single-cell bone metastasis signature (P < 1 ×10−324; Fig. 3e). Similarly, differentially regulated pathways in the bone metastasis signature were significantly enriched with those in the single-cell bone metastasis signature (P < 0.001; Fig. 3f). Furthermore, pathway analyses using a bulk RNA signature comparing bone metastases with normal bone (Supplementary Table 2) revealed significant enrichment with the single-cell bone metastasis signature (P < 0.001; Fig. 3g). Together, these findings indicate that cell-intrinsic features of bone metastatic cells drive the bone metastasis signature.
Among leading-edge pathways enriched between the bulk and single-cell bone metastases signatures, the top-most significant was the hallmarks MYC pathway. Notably, hallmark MYC pathway genes were positively enriched in the single-cell bone metastatic signature (P < 0.001; Fig. 3h) but downregulated in a single-cell gene signature comparing the benign resident bone cells with primary tumor cells (P = 0.002; Fig. 3i and Supplementary Table 5). These analyses implicate MYC pathway activation as a principal cell-intrinsic feature of bone metastases in NPKEYFP mice.
Co-activation of MYC and RAS in prostate cancer metastasis.
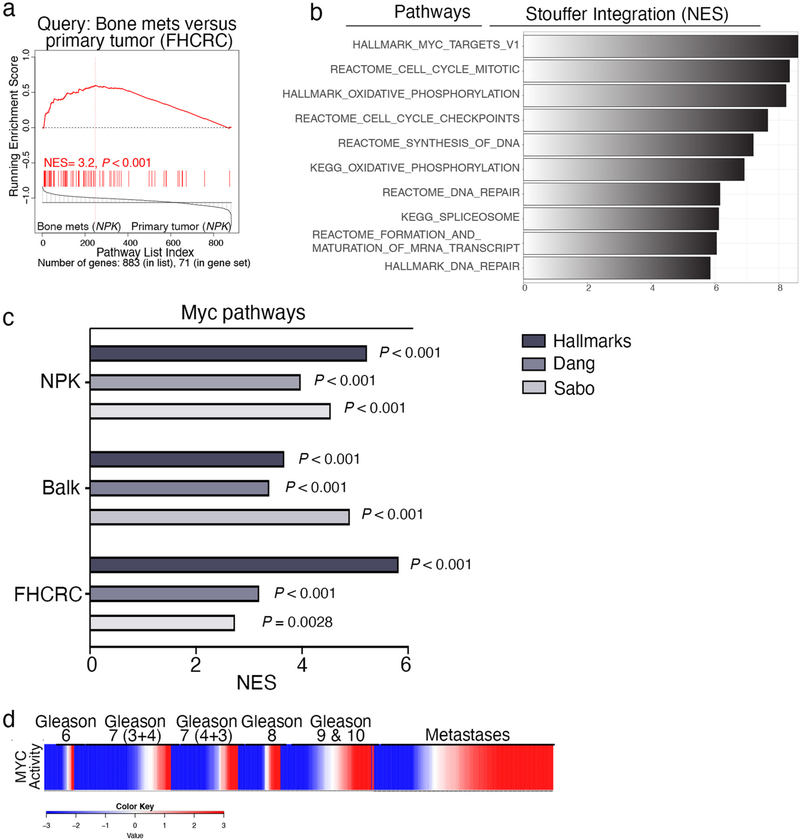
Cross-species GSEA comparing pathways enriched in the mouse single-cell bone metastasis signature with those enriched in a human signature consisting of primary tumors (n = 19) and bone biopsies (n = 19) (Balk21; Supplementary Table 3) revealed significant similarity (P < 0.001; Fig. 4a); the top-most significant was the hallmarks MYC pathway (P < 0.001; Supplementary Table 6). MYC activation was further evident by comparing pathways enriched in the mouse bone metastasis signature with the Balk signature, which consists of tumors and bone biopsies from patients living with metastatic prostate cancer21 (Supplementary Table 3c) and a second human signature consisting of primary prostate tumors (n = 14) and bone metastases (n = 20) from patients who had died of metastatic prostate cancer (FHCRC28; Supplementary Table 3d). Pathways upregulated in the Balk and FHCRC cohorts were highly enriched compared with those of the mouse bone metastasis signature (P < 0.001; Fig. 4b, Extended Data Fig. 4a and Supplementary Table 6). Stouffer integration to identify pathways significantly enriched among all three mouse and human signatures revealed the hallmarks MYC pathway as the top-most significant (Extended Data Fig. 4b and Supplementary Table 6). These mouse and human signatures were also significantly enriched with canonical MYC targets (Dang29; P < 0.001) and oncogenic MYC targets (Sabo30; P < 0.003; Extended Data Fig. 4c).
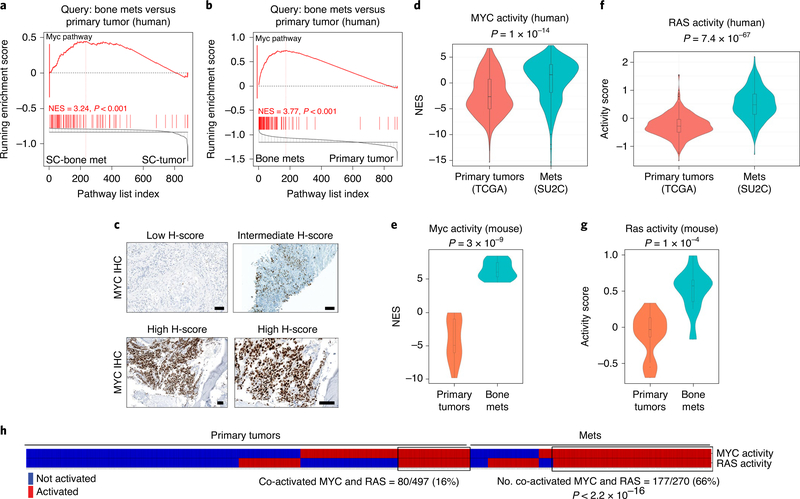
Fig. 4 |. Co-activation of MYC and RAS pathways in prostate cancer metastasis.
a,b, Cross-species pathway analysis. GSEA comparing pathways enriched in the Balk human bone metastasis signature (Supplementary Table 6c) with those in the mouse single-cell bone metastasis signature (a) (Supplementary Table 5c) or those enriched in the mouse bulk RNA bone metastasis signature (b) (Supplementary Table 6a). NES and P values were estimated using 1,000 pathway permutations. The red bar shows the hallmarks MYC pathway, which is the top-most enriched conserved pathways in both signatures. c, Representative immunohistochemical analyses of MYC expression in bone metastases, based on analysis of 34 mCRPC patient samples, including 12 with bone metastases (Supplementary Table 7). d–g, Violin plots depicting distribution of MYC and RAS pathway activation in primary tumors and metastases in human cancer and in the NPKEYFP mice. Comparison of human primary tumors (TCGA, n = 497) versus metastases (SU2C, n = 270) (Supplementary Table 3) (d,f). Comparison of primary tumors (n = 13) and bone metastases (n = 10) from the NPKEYFP mice (e,g). The distribution of the NESs (y axis) represent MYC activity levels based on single-sample GSEA (see Extended Data Fig. 4d) (d,e). The activity scores (y axis) represent RAS pathway activity levels (based on the absolute-valued average of RAS-related genes as in previous works11,33) (f,g). P values for all violin plots were estimated using two-sample one-tailed Welch t-test. In the violin plots with embedded box plots, boxes show the 25th–75th percentile, center lines show the median and whiskers show the minimum–maximum values. h, MYC and RAS co-activation in human primary tumors and metastases. Primary tumors and metastases classified as MYC- or RAS-activated are depicted in a heat map in red, whereas those without MYC- or RAS activation are represented in blue. Samples were considered MYC-activated if NES scores from single-sample GSEA using MYC hallmarks pathway were greater than the average of overall MYC activity across the cohorts. Samples were considered RAS-activated if the absolute-valued average of RAS-related genes11,33 was greater than the average of overall RAS activity across the cohorts. A black rectangle shows the samples in which MYC and RAS were co-activated. A two-tailed P value was calculated using Fisher’s exact test.
Consistent with the known upregulation of MYC in human prostate cancer31,32, immunostaining showed robust expression of MYC in human bone metastases from patients with mCRPC (n = 12; Fig. 4c and Supplementary Table 7). As observed for KRAS, copy number gains in MYC are more prevalent in human prostate cancer metastases (70% in SU2C) compared with primary tumors (31% in TCGA; Extended Data Fig. 3). Although we did not observe Myc copy number gains in NPKEYFP mice (Extended Data Fig. 3), Myc mRNA was upregulated in bone metastases relative to primary tumors and lung metastases (Supplementary Table 2).
To further investigate MYC pathway activation in human prostate cancer metastasis, we performed single-sample GSEA to estimate hallmarks MYC pathway activity levels (hereafter ‘MYC activity’) in individual cases of metastases from SU2C26 (n = 270) and primary tumors from TCGA25 (n = 497) that showed strong enrichment in metastases (Extended Data Fig. 4d). The overall distribution of MYC activity was significantly greater in metastases compared with primary tumors (P = 1 × 10−14; Fig. 4d), although MYC was not preferentially activated in bone metastasis relative to other metastatic sites (Extended Data Fig. 3).
In NPKEYFP mice, we observed strong enrichment of Myc activity in metastases relative to primary tumors (P = 3 × 10−9; Fig. 4e); however, unlike human prostate cancer, Myc activation was specific for bone metastases relative to other metastatic sites (P = 6.1 × 10−10; Extended Data Fig. 3). One major difference between the human prostate cancer and mouse cohorts is that the SU2C cohort are from mCRPC26, whereas the mouse cohort is androgen-intact. Notably, we observed robust MYC immunostaining in mCRPC (n = 34; Supplementary Table 7).
Indeed, despite strong overall similarity of their molecular profiles (Extended data Fig. 2g), Myc pathway activity was significantly upregulated in castrated versus noncastrated NPKEYFP mice in the bone metastases as well as primary tumors and other metastatic sites (P < 0.01; Extended Data Fig. 3). It is conceivable that MYC is already activated in mCRPC in SU2C, thereby obscuring activation in bone metastases. Notably, while analyses of NPKEYFP mice allows investigation in androgen-intact and -deprived contexts, we are unaware of a human cohort of bone and other metastases from patients who are castrated and noncastrated that would allow direct comparison of MYC in these contexts.
We analyzed RAS pathway activation in human and mouse metastasis based on the expression of seven genes (PTPN11, KRAS, NRAS, BRAF, RAF1, SPRY1 and SPRY2) associated with RAS/RAF signaling as described in previous works11,33 (hereafter called ‘RAS activation’). Similar to MYC activation (Fig. 4d), we observed strong enrichment of RAS activation in metastases from SU2C compared with primary tumors from TCGA (P = 7.4 × 10−67; Fig. 4f) and like MYC, RAS activation was not preferential to bone metastasis (Extended Data Fig. 3). However, in NPKEYFP mice, Ras activation was specific to bone metastases (P = 1 × 10−4; Fig. 4g and Extended Data Fig. 3).
Activation of MYC and RAS are well-correlated in human prostate cancer (Spearman correlation, rho 0.37, P < 0.001; Extended Data Fig. 3i), particularly in advanced tumors (Gleason Grades 8–10) and metastases. Case-by-case analyses revealed upregulation of MYC activity in 190 of 497 tumors from TCGA and 80 of these (16%) have co-activation of RAS, whereas 193 of 270 metastases in SU2C have upregulated MYC and 177 of these (66%) have co-activation of RAS (P < 2.2 × 10−16; Fig. 4h). In NPKEYFP mice, activation of Myc and Ras are strongly correlated (Spearman correlation, rho 0.67, P = 6.6 × 10−4; Extended Data Fig. 3h), particularly in bone. Further, co-activation of MYC and RAS occurred in only 1 of 13 (8%) primary tumors but 9 of 10 (90%) of bone metastases in NPKEYFP mice (P = 1 × 10−4; Extended Data Fig. 3j). Therefore, co-activation of MYC and RAS is significantly associated with prostate cancer metastasis and effectively modeled in NPKEYFP mice.
MYC is necessary but not sufficient for bone metastasis.
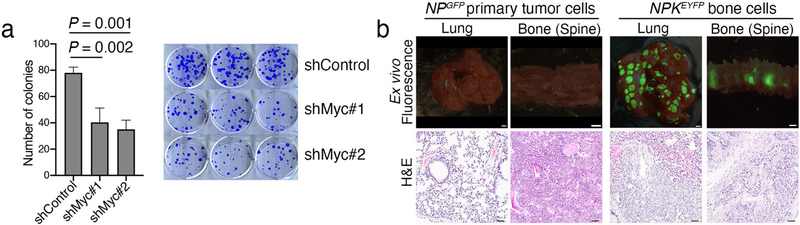
We investigated the function of MYC for bone metastasis using an in vivo allograft model derived from NPKEYFP mice (Fig. 5 and Extended Data Fig. 5). Intracardiac injection of NPKEYFP bone cells, but not control cells from nonmetastatic NP tumors16, results in metastases to bone, as well as lung and other tissues (Extended Data Fig. 5). Silencing Myc using two different short hairpin RNAs (shRNAs) (shMyc-1 or shMyc-2), but not the control shRNA (shControl), resulted in significant reduction in bone metastases, while not abrogating cellular viability (Fig. 5b,c and Extended Data Fig. 5a). Silencing Myc inhibited metastasis in each type of bone (spine, pelvis, femur, tibia and humerus), whereas lung metastases were not significantly affected (n = 8; Fig. 5c,d). While bones from mice injected with control NPKEYFP bone cells displayed YFP-marked bone metastases with robust Myc expression, bones from mice injected with Myc-silenced NPKEYFP bone cells had fewer or no YFP-marked bone metastases and low expression of Myc (Fig. 5d,e).
Fig. 5 |. Analysis of Myc function in an allograft model of bone metastasis.
a, Strategy. Cells from a bone metastasis (femur) of NPKEYFP mice were established using a procedure described previously11. The original cells were passaged in nude mouse hosts via intracardiac injection. Cells isolated from an ensuing bone metastasis, termed NPKEYFP bone cells, were used herein. b, Western blot image showing total protein extracts from NPKEYFP bone cells infected with shRNAs to silence Myc (shMyc-1, 70% inhibition; shMyc-2, 90% inhibition) or with shControl. The approximate molecular weights of markers (kDa) are indicated; actin is a control for protein loading. A representative blot is shown from two independent experiments. The uncropped western blot image is shown in Source Data Fig. 5. c, Quantification of the number of metastases in bone or lung from NPK bone cells infected with shMyc-1 or shMyc-2 or shControl and introduced into nude mouse hosts via intracardiac injection to evaluate metastasis in vivo. P values were estimated by one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons against shControl; (P < 0.05). In box plots, boxes show the 25th–75th percentile with the median and whiskers show the minimum–maximum (n = 10 mice from two independent experiments). d, Representative ex vivo imaging of n = 10 mice showing YFP fluorescence from the heart (injection site), lung and the indicated bones from nude mouse hosts following via intracardiac injection of NPKEYFP bone cells that had been infected with shControl, shMyc-1 or shMyc-2. e, Representative images (n = 3) of vertebrae showing ex vivo fluorescence, H&E or immunostaining for YFP or Myc, as indicated. Scale bars, 0.1 cm for ex vivo fluorescence images and 50 μm for all other images.
MYC is highly expressed in human PC3 cells, which were derived from a bone metastasis34 and grow in bone when implanted orthotopically35,36. Therefore, we examined MYC function for tumor growth in bone using PC3 cells engineered to express luciferase and green fluorescent protein (GFP) (herein called PC3-Luc-GFP cells) (Extended Data Fig. 6). Silencing MYC in PC3-Luc-GFP cells using two different shRNAs (shMYC-1 or shMYC-2), but not the control shRNA (shControl), inhibited tumor growth when implanted into tibia, while not completely abrogating cellular viability (P < 0.0001; Extended Data Fig. 6b–f). Tibiae of mice implanted with control PC3-Luc-GFP cells (shControl) displayed large YFP-expressing tumors, which were not observed in mice implanted with MYC-silenced cells (Extended Data Fig. 6g).
As previous studies of Myc in other prostate cancer mouse models reported no or low incidence of bone metastasis37–41, we crossed NPKEYFP mice with the Hi-MYC transgene41 to generate a series of GEMMs having activation of neither Myc nor Ras (NPEYFP), activation of either Myc (NPMEYFP) or Ras (NPKEYFP) or co-activation of Myc and Ras (NPKMEYFP) (Fig. 6). While NPMEYFP mice developed large prostate tumors and lung metastasis, they did not develop bone metastasis (n = 23; Fig. 6a–d). Further, while NPKMEYFP mice developed both lung and bone metastasis, their incidence of bone metastasis and overall survival were similar to NPKEYFP mice (n = 10; Fig. 6a–d). As above, Myc activity was significantly greater in bone metastases compared with primary tumors from NPKEYFP mice (P = 3.2 × 10−9; Figs. 6e and 4e). However, Myc activity in primary tumors of NPMEYFP mice was significantly higher than in primary tumors of the NPKEYFP mice (P = 0.015), but comparable to bone metastases (Fig. 6e). Ras pathway activity is significantly higher in NPKEYFP bone metastases relative to the primary tumors from either NPMEYFP or NPKEYFP mice (P = 0.0014; Fig. 6f). These findings show that MYC is necessary but not sufficient for bone metastasis and suggest it requires collaboration with RAS activation for bone metastasis.
Fig. 6 |. Analysis of MYC function in a new GEMM.
a–d, Comparative analyses of the tumor and metastatic phenotypes of NPEYFP (n = 35), NPMEYFP (n = 23), NPKEYFP (n = 106) and NPKMEYFP (n = 10) mice. Representative bright-field and ex vivo fluorescence images of prostate, lung and bone (spine) (a). Scale bars, 0.1 cm. Dot-plots showing tumor weights (b). GU, genitourinary. Center lines show the mean, error bars depict s.d.; P value is shown for one-way ANOVA with Dunnett’s multiple comparison test of NPMEYFP and NPEYFP mice. Kaplan–Meier curves showing overall survival (c); P value calculated using a two-tailed log-rank test. Bar graphs showing the percentage of mice with metastasis to lung and bone (d). e,f, Violin plots depicting the distribution of Myc (e) and Ras (f) pathway activity levels in primary tumors of NPMEYFP (n = 3) and NPKEYFP (n = 13) mice and bone metastases of NPKEYFP mice (n = 10). Myc activity is based on single-sample GSEA and Ras pathway activity is based on the absolute-valued average of RAS-related genes as in previous works11,33. P values were estimated using a two-sample one-tailed Welch t-test. In the violin plots with embedded box plots, boxes show the 25th–75th percentile, center lines show the median and whiskers show the minimum–maximum values.
META-16: a human gene signature prognostic for time to metastasis and treatment response.
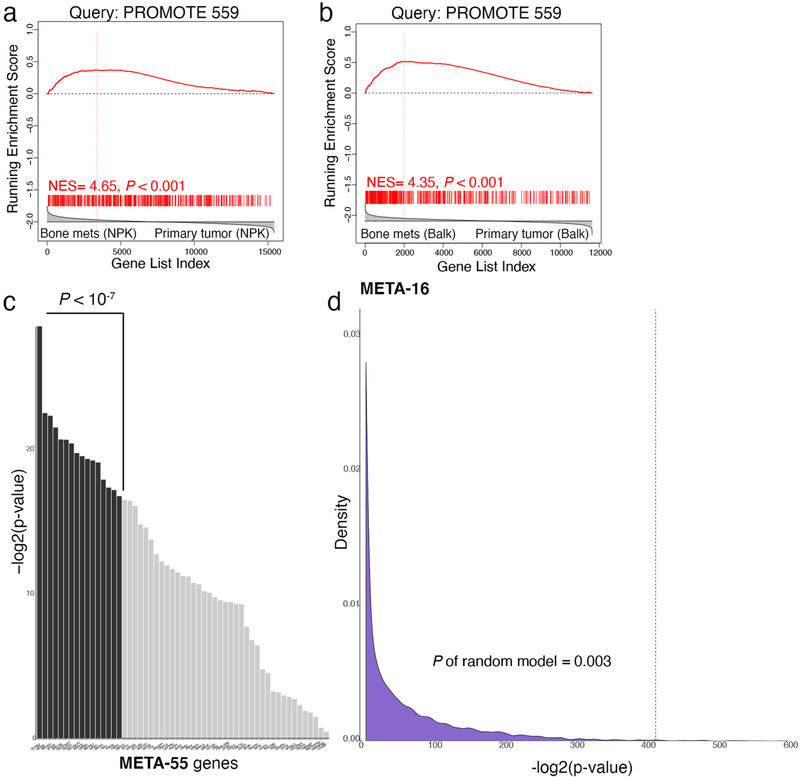
To identify a gene signature associated with co-activation of MYC and RAS, we interrogated the PROMOTE cohort (for PROstate Cancer Medically Optimized Genome-Enhanced ThErapy42), which consists of metastatic biopsies from patients with mCRPC (n = 77), the majority of which are bone (n = 55) (Fig. 7a,b and Supplementary Table 3). Genome- wide correlation in PROMOTE identified 559 genes positively correlated with MYC expression (‘PROMOTE-559’; Spearman rho > 0.5, FDR P < 0.0001; Fig. 7a,b and Supplementary Table 8); 517 of these (93%) were also correlated with RAS activation (Supplementary Table 8).
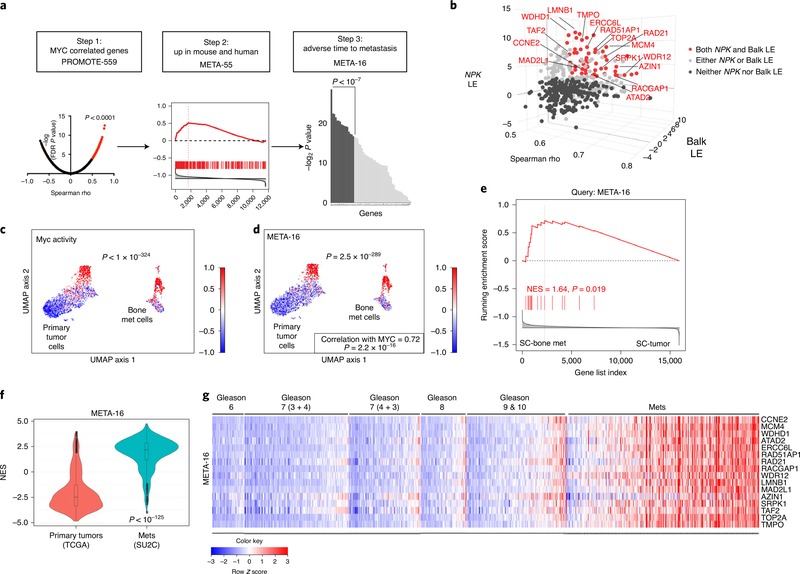
Fig. 7 |. MEtA-16 is correlated with MYC and RAS pathway activation and enriched in prostate cancer metastasis.
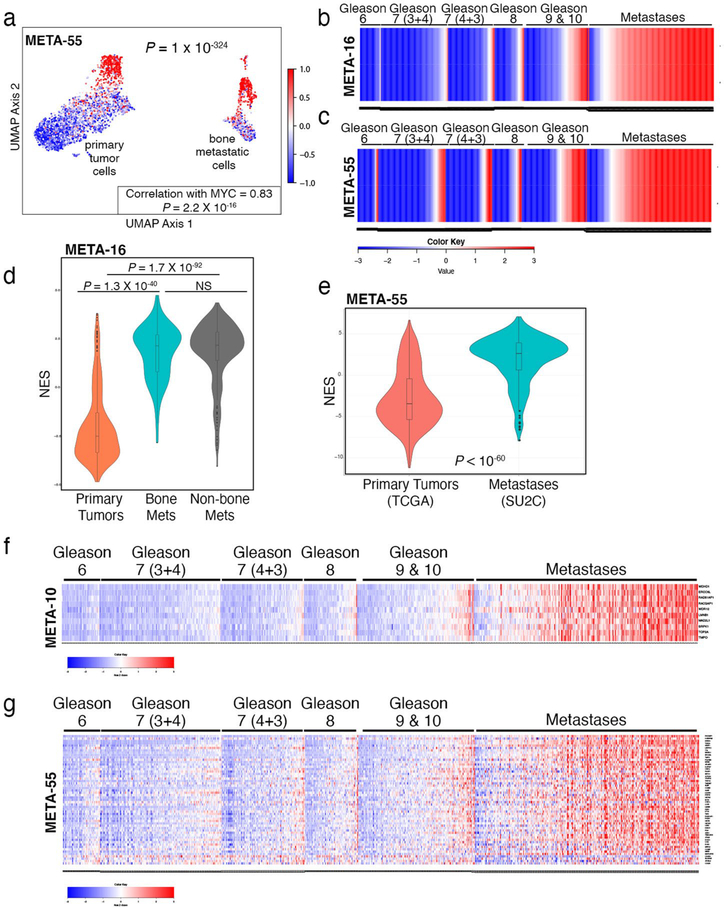
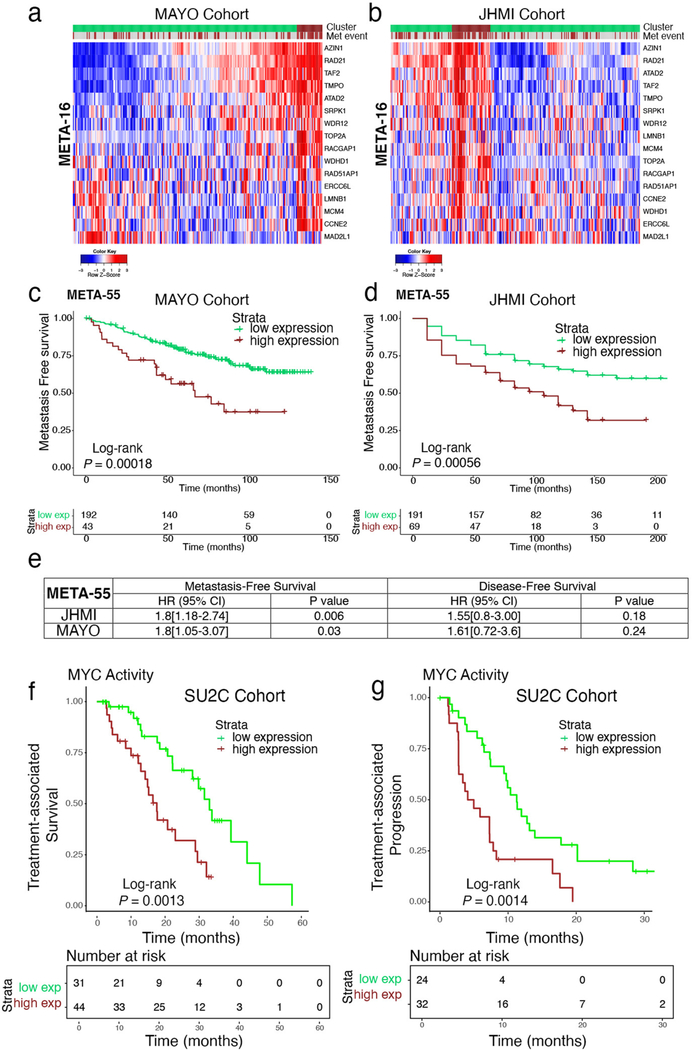
a,b, Discovery of the META-16 gene signature. Step 1, genome-wide Spearman correlation to MYC expression in PROMOTE cohort (which includes 55 bone metastases), identified 559 (PROMOTE-559) positively correlated genes (FDR P value < 0.0001, Spearman rank correlation coefficient rho plotted in the x axis in b) (a). Step 2, GSEA using PROMOTE-559 to query the mouse (NPKEYFP) and human (Balk) bone metastasis signatures (Extended Data Fig. 7a,b) (b). The leading-edge (LE) genes from mice are projected on the y axis and from humans, on the z axis (b). These analyses identified 55 genes (META-55, highlighted in red in b). Step 3, ranking of META-55 according to metastasis-free survival identified 16 genes (META-16, shown by name in b). c,d, UMAP projection of single-cell RNA-seq showing the primary tumor and bone metastatic cells (Fig. 3d). Enrichment of MYC pathway (c) and expression of META-16 (d). Scaled DESeq2 normalized values are depicted. The correlation between META-16 expression and MYC pathway activity was estimated using Spearman’s rank correlation. e, GSEA using META-16 to query single-cell bone metastasis signature (Supplementary Table 5a); NES and P values were estimated using 1,000 gene permutations. f, Violin plot depicting distribution of the NESs (y axis), which reflect activity levels of META-16 (Extended Data Fig. 8b) in primary tumors from TCGA (n = 497) compared with metastases from SU2C (n = 270) (Supplementary Table 3). P value was estimated using a two-sample one-tailed Welch t-test. In the violin plots with embedded box plots, boxes show the 25th–75th percentile, center lines show the median and whiskers show the minimum–maximum values. g, Heat map representation of individual expression levels of META-16 genes in patient samples from TCGA and SU2C cohorts. Gleason scores are shown for the primary tumors; metastases include all metastases in the SU2C cohort. Row-scaled expression values are shown (indicated by color).
Interrogation of bone metastasis signatures with PROMOTE-559 revealed significant enrichment in both the mouse (P < 0.001; Fig. 7b and Extended Data Fig. 7a) and human (P < 0.001; Fig. 7b and Extended Data Fig. 7b) signatures. Integration of leading-edge genes from the mouse (121) and human (154) signatures identified 55 genes (‘META-55’; Fig. 7b); 52 of these (95%) were also correlated with RAS activation (Supplementary Table 8).
To prioritize the META-55 genes for association with metastasis, we used a univariable Cox proportional hazards model based on metastasis-free survival for 336 patients in TCGA that had reported ‘time to metastasis’ (13 developed metastasis; Supplementary Table 3). This identified 16 genes (‘META-16’) with significant association with metastasis-free survival (P < 1 × 10−7; Extended Data Fig. 7c); all 16 (100%) are correlated with RAS activation (Supplementary Table 8). As META-16 consistently outperformed META-55, we focused on this signature in subsequent analyses but report findings for META-55 (Extended Data Figs. 7–10). Discovery of META-16 was improved by cross-species interrogation, as analyses of only human signatures identified genes of which 48.5% significantly associated to time to metastasis (P < 0.01), whereas analyses of both mouse and human signatures identified genes of which 74.5% (P < 0.01), indicating a significant improvement (P = 0.0021).
Analysis of single-cell sequencing data revealed significant enrichment of META-16 in bone metastatic versus primary tumor cells (P = 2.5 ×10−289) and strong correlation with Myc activation (Spearman correlation P = 2.2 × 10−16; Fig. 7c,d and Extended Data Fig. 8). GSEA showed enrichment of META-16 in the single-cell bone metastasis signature (P = 0.019; Fig. 7e), but not the single-cell signature based on tumor versus the benign resident bone cells.
Expression of META-16 genes was upregulated in human bone metastases relative to primary prostate tumors (P < 0.05; Extended Data Fig. 9a), whereas silencing MYC in human (PC3-Luc-GFP) or mouse (NPK bone) metastatic prostate cancer cells resulted in reducing their expression (P = 0.034 for PC3, P < 0.03 for NPK; Extended Data Fig. 9b,c). Notably, META-16 expression in SU2C is significantly higher in patients with MYC amplification (P = 0.0006) but not PTEN deletion (P = 0.11). Although META-16 includes several genes located near and potentially co-amplified with MYC on human chromosome 8q, META-16 performs equally well without these genes (P = 3.7 × 10−151; Extended Data Fig. 8).
META-16 is notably enriched across human prostate cancer metastases from various tissue sites although, as for MYC, not exclusively in bone metastases. Single-sample GSEA on each tumor from TCGA (n = 497) and each metastasis from SU2C (n = 270) showed strong enrichment of META-16, particularly in metastases (Extended Data Fig. 8). The overall distributions between the TCGA and SU2C revealed significant upregulation of META-16 in metastases compared to primary tumors (P < 10−125; Fig. 7f and Extended Data Fig. 8). Individual META-16 genes were upregulated across each metastasis versus each primary tumor (Fig. 7g and Extended Data Fig. 8). Comparing activity levels of an arbitrary, equally sized (n = 16) group of genes showed that the ability of META-16 to distinguish primary tumors from metastases was significantly nonrandom (P = 0.003; Extended Data Fig. 7d).
To ask whether META-16 is significantly associated with risk of metastasis, we used two independent prostatectomy cohorts with extensive clinical outcome data (MAYO43 and JHMI44; Fig. 8a–c, Extended Data Fig. 10 and Supplementary Table 3). Patients in MAYO (n = 235) had undergone radical prostatectomy between 2000 and 2006 with a median follow-up of 7 years; 76 patients developed metastasis43. Patients in JHMI (n = 260) had undergone radical prostatectomy between 1992 and 2010 with a median follow-up of 9 years; 99 patients developed metastasis44.
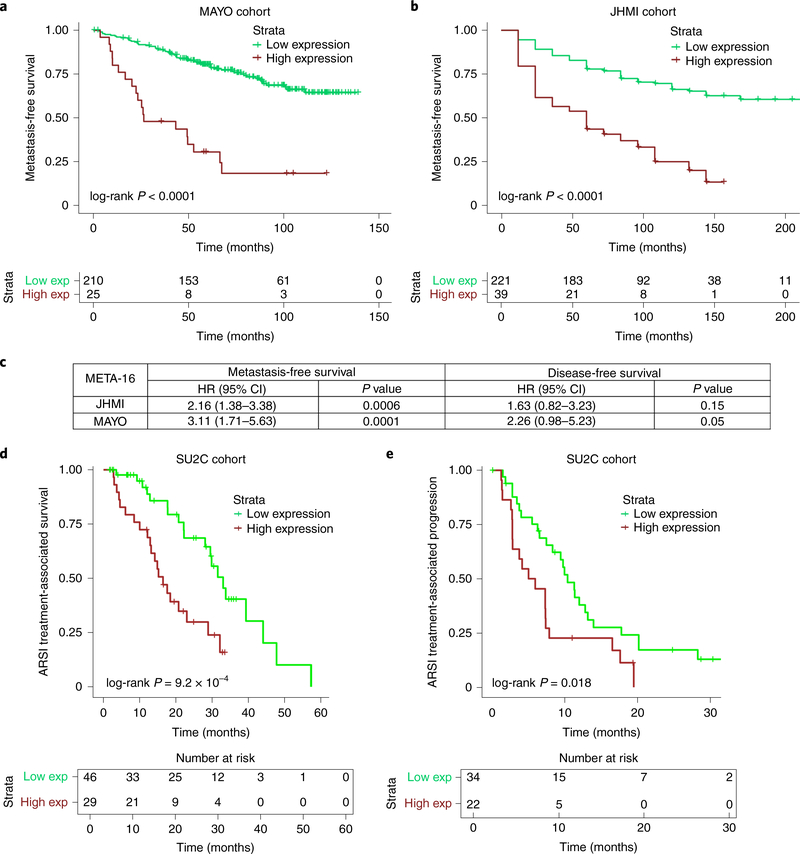
Fig. 8 |. The MEtA-16 signature is associated with metastasis-free and treatment-associated survival.
a–c, Association of META-16 with time to metastasis. Kaplan–Meier survival analyses comparing patients with low and high combined expression of META-16 in the MAYO (n = 235) and JHMI (n = 260) cohorts (Extended Data Fig. 10a,b) (a,b). P values were estimated using a log-rank test. Multivariable survival analysis of META-16 with the JHMI and MAYO cohorts (c). CI, confidence interval; HR, hazard ratio. P values were estimated from a Cox proportional hazards model. d,e, Kaplan–Meier survival analyses comparing patients from the SU2C cohort with low and high combined expression of META-16, showing treatment-associated survival (time from the start of treatment with ARSI therapy, to death or last follow-up; n = 75 patients) or treatment-associated disease progression (time on treatment with ARSIs; n = 56) as defined previously26. P values were estimated using a log-rank test.
To test the association of META-16 with metastasis-free survival45, we performed hierarchical clustering to group patients with low or high levels of combined META-16 expression (Extended Data Fig. 10a,b). In both cohorts, Kaplan–Meier survival analyses demonstrated that patients with high expression of META-16 have a shorter time to metastasis than those with low expression (P < 0.0001; Fig. 8a,b and Extended Data Fig. 10c,d). A multivariable Cox proportional hazards model, adjusted for age, pathological Gleason score/grade at diagnosis, pre-prostate-specific antigen (PSA), seminal vesicle invasion (SVI), lymph node invasion (LNI) and extra-prostatic extension, showed that the ability of META-16 to predict metastasis-free survival is not affected by those variables and is significantly associated with metastasis-free survival (MAYO, P = 0.0001; JHMI, P = 0.0006) compared to prostate-cancer-specific mortality (MAYO, P = 0.05; JHMI, P = 0.15; Fig. 8c and Extended Data Fig. 10e).
SU2C includes 75 patients with detailed clinical data regarding treatment-associated survival (time from the start of treatment with AR signaling inhibitors (ARSIs) to death or last follow-up), as well as 56 patients with detailed information about treatment-associated disease progression (time on treatment with ARSIs)26. To ask whether META-16 is associated with treatment response, we grouped these patients into low or high levels of combined META-16 expression. Subsequent Kaplan–Meier survival analyses demonstrated that patients with high META-16 expression have a shorter time to treatment-associated death (P = 9.2 × 10−4) and a shorter time to treatment-associated disease progression (P = 0.018; Fig. 8d,e). Consistent with our observations that META-16 is strongly correlated with MYC activity, when we grouped these same patients based on high versus low levels of MYC activity, we found that those with high activity had a shorter time to treatment-associated survival (P = 0.0013) as well as shorter time to treatment-associated disease progression (P = 0.0014, Extended Data Fig. 10f,g). Therefore, META-16 and MYC activity may have predictive significance for response to treatment with anti-androgens in advanced prostate cancer.
Discussion
Until now it has proven challenging to model high-efficiency bone metastasis in the context of the native tumor microenvironment during cancer evolution in the whole organism. Our description of NPKEYFP mice overcomes this limitation because these mice model lethal prostate cancer with highly penetrant bone metastasis. Indeed, our analyses of NPKEYFP mice have enabled detailed biological and molecular characterization of bone metastases as arising de novo during tumor progression in vivo in androgen-intact and androgen-deprived contexts.
Although bone metastases are only discernable in highly metastatic NPKEYFP mice, longitudinal analysis combined with phylogenetic analysis reveal that they originate early in disease progression from an early subclone of the primary tumor. We infer that bone metastases in NPKEYFP mice are seeded early, but take longer to cultivate compared with metastases to soft tissues. This parallels the scenario in human patients, wherein bone metastases are more prevalent than metastases to visceral tissues, but the latter are associated with worse clinical outcome5. As DTCs occur in bones of NPK mice early during prostate cancer progression11, we expect that analyses of circulating tumor cells in NPKEYFP mice may help to identify and molecularly characterize bone metastases early in tumor progression in future studies.
Our current findings reveal MYC activation in mCRPC and metastasis, extending previous studies that have shown the importance of MYC in prostate cancer31,32,37,39,41. Notably, MYC is upregulated in all metastases in human prostate cancer but specifically in bone metastases in NPKEYFP mice. We attribute this difference to the fact that analyses of human prostate cancer necessarily compares castration-resistant metastases and androgen-intact primary tumors25,26, as metastases from androgen-intact patients are not available. However, NPKEYFP mice enable direct comparison in androgen-intact and -deprived contexts, which is a key benefit of studying metastasis in these mice.
Notably, our findings identify co-activation of MYC and RAS as a key driver of prostate cancer metastasis. Although KRAS is rarely mutated in localized prostate cancer, it is located on the long tail of oncogenic drivers46 and gains in KRAS, as well as MYC, are more prevalent in metastatic prostate cancer than localized disease26,47. It is conceivable that RAS activation enhances the stability or localization of MYC protein in mCRPC. Our findings highlighting the importance of co-activation of RAS and MYC for prostate cancer metastasis advocate future studies to elucidate the underlying mechanisms and introduce a mouse model of bone metastases that shares key features of this subgroup of patients with lethal prostate cancer.
The translational significance of our findings is further highlighted by identification of the META-16 gene signature, which is correlated with co-activation of MYC and RAS and associated with adverse outcome for metastasis in patients with localized prostate cancer and adverse treatment response in patients with advanced disease. In this regard, META-16, may augment other prognostic signatures, such as Decipher GX, which is associated with risk of metastasis, and Prolaris CCP score, which is associated with prostate-cancer-specific survival48. We propose that further investigation of META-16, potentially in conjunction with these other signatures, is warranted to assess its potential clinical utility.
Methods
Genetically engineered mouse model of bone metastasis.
All experiments using animals were performed according to protocols approved by the Institutional Animal Care and Use Committee at Columbia University Irving Medical Center. All mice were housed in pathogen-free barrier conditions under 12-h light–dark cycles and with temperature and humidity set points at 20–25 °C and 30–70%, respectively. As our focus was prostate cancer, only male mice were used.
NPK mice11 were crossed with the Rosa-CAG-LSL-EYFP-WPRE reporter allele13 to obtain experimental NPKEYFP mice and the control (nonmetastatic) Nkx3.1CreERT2/+;Ptenflox/flox; Kras+/+; R26R-CAG-LSL-EYFP/+ (NPEYFP) mice. The Hi-MYC allele (FVB-Tg(ARR2/Pbsn-MYC41) was crossed with the NPKEYFP mice to obtain the NP-Hi-MYCEYFP (NPMEYFP) and NPK-Hi-HMYCEYFP (NPMEYFP) mice. NPK mice were maintained in our laboratory on a predominantly C57BL/6 background; the Rosa-CAG-LSL-EYFP-WPRE mice were obtained from Jackson Laboratories on a C57BL/6 background (stock no. 007903); and the Hi-MYC mice were obtained from the National Cancer Institute mouse repository on an FVB background (stock no. 01XK8). We note the significant increase in median survival of the NPKEYFP mice compared with our original report of NPK mice11 (4.7 months compared with 3.1 months, P < 0.0001). We attribute this difference to the low-level expression of the first generation YFP reporter allele in the previous NPK mice, which required that we analyzed homozygotes, whereas in the current NPKEYFP mice, we analyzed heterozygotes of the second-generation Rosa-CAG-LSL-EYFP-WPRE allele.
All studies were performed using littermates genotyped before tumor induction. Mice were induced to form tumors at 2–3 months of age by administration of tamoxifen (Sigma-Aldrich) using 100 mg kg−1 (in corn oil) once daily for four consecutive days. Control (nontumor induced) NPKEYFP mice received only vehicle (corn oil). The primary survival cohort (n = 106) were killed when their body condition score50 was <1.5 or when they experienced body weight loss ≥20% or signs of distress, such as difficulty breathing or bladder obstruction. A second survival cohort (n = 22) underwent surgical castration 1 month after tumor induction. The longitudinal cohort (n = 26) were killed at the specific time points following tumor induction as indicated.
At time of killing, YFP-positive prostatic tumors and metastases were visualized and quantified by ex vivo fluorescence using an Olympus SZX16 microscope (Ex490–500/Em510–560 filter). For accurate visualization of fluorescence, a composite image was made by superimposing a bright-field image (20% transparent) on the same area of the fluorescent image using Adobe Photoshop. Muscle and connective tissue were removed from bone before ex vivo fluorescence analyses for efficient detection of bone metastases. For histological and immunohistochemical analyses, tissues were fixed in 10% formalin (Thermo Fisher Scientific). Bones were decalcified for 3 weeks in 15% EDTA (pH 7.0) solution (Sigma-Aldrich). For isolation of genomic DNA or RNA, tumors or macrodissected metastases were snap-frozen in liquid nitrogen. For single-cell RNA-seq and establishment of cell lines, freshly dissected tissue was used directly.
Histopathological and immunohistochemical analyses were conducted on 3-μm paraffin sections as described elsewhere16. Histopathological grading of H&E-stained sections were performed blinded by two independent pathologists (A.M.D. and M.A.R.). Images were captured using an Olympus VS120 whole-slide scanning microscope. Immunofluorescence images were captured using a Leica TCS SP5 confocal microscope. Antibodies are described in Supplementary Table 9. Analysis of RNA expression was performed by quantitative reverse transcription PCR (RT–qPCR) using the QuantiTect SYBR Green PCR kit (QIAGEN)16. Primers sequences are provided in Supplementary Table 10. Western blotting was performed using total protein extracts as described16; antibodies are described in Supplementary Table 9.
RNA-seq analyses.
Transcriptomic analysis of bulk tissues was performed on primary tumors (n = 19) and matched macrodissected metastases from lung (n = 11), liver (n = 5), lymph nodes (n = 4), brain (n = 3) or bone (n = 12) from 16 independent NPKEYFP mice. RNA was prepared from snap-frozen tissues using the MagMAX-96 total RNA isolation kit (Thermo Fisher Scientific). Total RNA was enriched for mRNA using polyA pull-down; only RNA samples having between >200 ng and 1 μg and with an RNA integrity number >8 were used. Libraries were made using an Illumina TruSeq RNA prep kit v2 or TruSeq Stranded mRNA library prep kit, and sequenced using an Illumina HiSeq2500/4000 by multiplexing samples, which yields targeted number of single-end/100-bp reads for each sample, as a fraction of 180 million reads for the pooled sample. Reads were aligned to the mm9 mouse genome using TopHat (v.2.1.0)51; RNA-seq raw counts were normalized and variance stabilized using DESeq2 (v.1.36.0)52 package (Bioconductor) in R-studio 0.99.902, R v.3.3.0 (R Foundation for Statistical Computing). A list of differentially expressed genes is provided in Supplementary Table 2.
For comparison with human genes, mouse genes were mapped to their corresponding human orthologs based on the homoloGene database (NCBI). Pathway enrichment was performed using GSEA to query the Molecular Signatures Database (MSigDB), available from the Broad Institute, including the C2 (KEGG, Reactome and BioCarta) and hallmark pathway datasets53. A list of differentially expressed pathways is provided in Supplementary Table 6.
Whole-exome sequencing analyses.
WES was performed on matched trios of primary tumors, lung and bone metastases, as well as tails (as control) from five independent NPKEYFP mice. Genomic DNA was isolated from snap-frozen tissues using the DNeasy Blood & Tissue Kit (QIAGEN). Sequencing was conducted by BGI Americas Corporation using the HiSeq4000 platform and Agilent Sure Select Mouse Exon kit (50 Mb) for exome capture to produce paired-end sequenced data of up to 150-bp read length. The average sequencing depth was more than 80×, and reads were mapped to the mouse mm10 genome build using bwa (v.0.7.17)54. Substitutions and indels were called using MuTect2 (v.4.0.4)55 with default parameters; only variants with a mutant allele frequency of 5% or greater in tumors and 0% in normal tail, were included in analyses. The variant read count cutoff was 5 or more in tumor and depth was 20 or more in normal tail. A list of single-nucleotide variants is provided in Supplementary Table 4a.
CNVs were analyzed using CNVkit 0.9.5 (ref. 56) with parameters set to default. Segmentation was performed using circular binary segmentation. CNVkit uses both targeted reads and the nonspecifically captured off-target reads to infer copy number evenly across the genome, which achieves exon-level resolution in targeted regions and sufficient resolution in the larger intronic and intergenic regions to identify CNVs. To avoid fluctuations, we considered CNV events encompassing more than 30 genes. A list of CNVs is provided in Supplementary Table 4b.
Evolutionary trees were reconstructed using somatic mutations (substitutions and indels) as described elsewhere57. The number of somatic mutations specific to or shared between primary tumors, lung and bone metastases were used to build evolutionary trees, such that the lengths of the branches indicate the number of specific or shared somatic mutations in each sample. The significance was evaluated using a bootstrap test58. Within each trio (primary, lung and bone metastases), given the observed somatic mutation matrix, the mutations were randomly shuffled. An evolutionary tree was then reconstructed using this new somatic mutation matrix and the topology of the tree was compared to that of the original tree. If there are m mutations shared between primary tumor and lung metastases and n mutations between primary and bone metastases, then the tree in which m − n mutations is less than in the original tree is given a score of 0; all others are given a score of 1. This procedure of resampling and the subsequent tree reconstruction was repeated 1,000 times and the percentage of times one tree is given a score of 1 is noted as bootstrap-derived P value. Representative combined phylogeny was then constructed reflecting consistent evolutionary patterns across all trees and the meta-analysis P value was calculated using Fisher’s method through combining bootstrap-derived P value from individual trees.
Single-cell RNA-seq analyses of mouse tumors and bone metastases.
Single-cell RNA-seq was conducted on freshly dissected prostate tumor and bone metastases from NPKEYFP mice in two independent experiments using a 10X Genomics Chromium platform59. Briefly, tissues were enzymatically digested for 15 min at 37 °C in 1× collagenase/hyaluronidase, 0.5 U ml−1 dispase II and 0.1 mg ml−1 DNase 1 in DMEM-F12 medium, followed by addition of 0.025% trypsin/EDTA for 15 min (Stem Cell Technologies). Cells were resuspended in cold 10% FBS DMEM-F12, filtered through a 40-μm cell strainer and collected by centrifugation at 350g in an Eppendorf 5810R tabletop centrifuge for 5 min at 4 °C. After a 5-min incubation in cold 1× Red Blood Cell Lysis Buffer (Thermo Fisher Scientific) cells were diluted fourfold in cold PBS, centrifuged as before and resuspended in DMEM-10% FBS for cell counting and viability analysis. Cells were counted using a Countess II Automated Cell Counter (Thermo Fisher Scientific) and 10,000 cells with >70% viability were loaded into a 10X Genomics Chromium Controller for capture and barcoding following the 10X Genomics Single Cell Protocol, as described by the manufacturer (10X Genomics), with subsequent RNA-seq using Illumina NovaSeq6000. Reads were mapped to the mouse mm9 genome and processed with the CellRanger v.2.1.1 pipeline59. Data are provided in Supplementary Table 5.
Single-cell RNA-seq raw counts were normalized and variance was stabilized using DESeq2 (v.1.36.0) package (Bioconductor) in R-studio 0.99.902, R v.3.3.0. The UMAP27 dimensionality reduction technique implemented in Python was used to cluster primary and metastatic single-cell RNA-seq data. UMAP visualizations were constructed as described60; the code for visualization is available at https://github.com/simslab.
Description of human patient cohorts.
All studies using human tissue specimens were performed according to protocols approved by the Human Research Protection Office and Institutional Review Board at the respective institutions. Only male patients were involved as the focus of our study was prostate cancer. Published human patient cohorts are described in Supplementary Table 3 (refs. 21,25,26,28). These include two independent cohorts with clinical outcome data retrieved from the Decipher GRID registry (MAYO cohort, GSE62116 (ref. 43) and the JHMI cohort, GSE79957 (ref. 44)). Patients in the MAYO cohort (n = 235) had undergone radical prostatectomy between 2000 and 2006; median follow-up was 7 years with 73 patients developing metastasis43. The JHMI cohort is a case-cohort of 260 men who had undergone radical prostatectomy between 1992 and 2010 at intermediate or high risk and received no additional treatment until the time of metastasis; median follow-up was 9 years with 99 patients developing metastasis44. Both cohorts were profiled on a Human Exon 1.0 ST Array and hybridization was conducted in a Clinical Laboratory Improvement Amendments-certified laboratory facility (GenomeDx Biosciences).
The subset of SU2C patients used herein (n = 270) used polyA+ RNA isolation for transcriptomic library preparation26. Among these, clinical outcome data were available for 75 patients based on treatment-associated survival analysis (41 patients died) and 57 patients based on treatment-associated disease progression analysis (47 patients experienced disease-progression-related events). Treatment-associated survival was defined as time between start of ARSIs treatment and death or follow-up and treatment-associated progression was defined as time on ARSI treatment26.
Unpublished cohorts used anonymized human tissue specimens from Columbia University Irving Medical Center (CUIMC) or Johns Hopkins Hospital; all patients consented before inclusion. The CUIMC cohort, which was used for analysis of RNA expression, consisted of five bone metastatic resections and ten primary prostate cancer tumors (Gleason score 9) from surgical resections of patients with advanced prostate cancer that had been banked in the Molecular Pathology Shared Resource of the Herbert Irving Comprehensive Cancer Center. RNA was extracted using miRNeasy mini kit (QIAGEN) and RT–qPCR was performed using the QuantiTect SYBR Green PCR kit (QIAGEN)16.
The Johns Hopkins Hospital cohort, which was used for immunohistostaining, consisted of 34 metastatic samples, including 12 bone metastatic biopsies from patients diagnosed with advanced prostate cancer. The clinical features of the patients are summarized in Supplementary Table 7. Immunohistochemistry was performed using a rabbit monoclonal MYC antibody (Abcam)32,61. Immunostaining was quantified using an H-score system obtained by multiplying staining intensity (0, no staining; 1, weak staining; 2, moderate staining; and 3, intense staining) by the percentage (0–100) of cells showing that intensity (H-score range 0–300, with 0–100 considered low, 101–200 intermediate and 201–300 high).
Functional analyses in cell-based models.
We adapted a protocol described previously to generate prostate tumor cell lines from NPK mice11 to establish a mouse allograft model that metastasizes to bone. Briefly, bone metastases from NPKEYFP mice were visualized by ex vivo fluorescence and collected using the method described above for single-cell RNA-seq. Cells were cultured for five passages in RPMI with 10% FBS. Once established, cells were introduced via intracardiac injection into NCr nude mice (male, Taconic). Bone metastases were isolated from vertebra and the cells recultured and then introduced again into nude mice introduced via intracardiac injection. This resulted in NPKEYFP bone cells that had >90% penetrance of metastasis to bone and other tissues. The genotype of the NPKEYFP bone cells was confirmed using a commercial source (Transnetyx) and cells were tested using a multispecies mycoplasma test (Mycoplasma Detection kit, cat no. MP70114, Thermo Fisher Scientific). Cell line stocks were established at passage 5 and used for experimental assays within three passages following thawing.
For monitoring in vivo bioluminescence and ex vivo fluorescent imaging, we engineered PC3 cells to express luciferase and GFP using the pHAGE PGK-GFP-IRES-LUC-W lentiviral vector (Addgene, plasmid number 46793), herein referred to as PC3-Luc-GFP cells. Lentiviruses were produced in HEK-293 cells (ATCC), using second-generation packaging vectors (psPAX2 and pMD2.G, Addgene) as described12.
For shRNA-mediated silencing, we used two independent shRNAs based on the pLKO.1 lentiviral vector system or a nontargeting pLKO.1 lentiviral vector (SHC002) (Sigma-Aldrich). The sequences for shRNA used in this study are provided in Supplementary Table 10. Colony-formation assays were performed by plating NPKEYFP bone cells (200 cells per well) or PC3-Luc-GFP cells (1,000 cell per well) in six-well tissue culture plates. Colonies were visualized by staining with crystal violet and quantified using ImageJ software (https://imagej.nih.gov/ij/). Cell culture assays were performed in triplicate and with a minimum of two independent biological replicates.
For intracardiac metastasis assays, mouse NPKEYFP bone cells (1 × 105 cells in 100 μl of PBS) were injected percutaneously into the left heart ventricle of immunodeficient NCr nude mice (male, Taconic). Mice were killed 12–14 d after injection or sooner if their body condition score50 was <1.5 (as above). For monitoring tumor growth in bone, PC3-Luc-GFP cells (1.5 × 106 cells in 20 μl of PBS) were injected into the tibiae of male NOD-SCID mice (NOD. CB17-Prkdcscid/J, strain 001303, Jackson Laboratories) as described62. Briefly, a small longitudinal skin incision was made across the knee capsule and the tip of a scalpel was used to drill a hole into which cells (or PBS) were injected in a volume of 20 μl. Sterile surgical bone wax (QuickMedical) was used to seal the hole and the skin was closed with wound clips. Tumor growth was monitored biweekly by bioluminescence imaging using an IVIS Spectrum Optical Imaging System (PerkinElmer), following intraperitoneal injection with 150 mg kg−1 D-luciferin (PerkinElmer). Images were quantified using Living Image Software (PerkinElmer). Micro-computed tomography images were collected using a PerkinElmer Quantum FX micro-CT Imaging System.
Statistics and reproducibility.
Sample size was based on the number of available, qualified tumor samples for this study. No data were excluded in our analysis. All attempts at replication were successful for all computational work in our manuscript. Neither randomization nor blinding were relevant to our computational analysis.
Statistical analyses were performed using a two-sample two-tailed Welch t-test (for differential expression analysis), two-sample one-tailed Welch t-test (for comparison of MYC activity and META-16 activity levels between SU2C and TCGA patient cohorts), one-way ANOVA, two-way ANOVA with multiple comparison testing, chi-squared test and Fisher’s exact test as indicated in each figure legend. GraphPad Prism software (v.6.0) and R-studio 0.99.902, R v.3.3.0 were used for statistical calculations and data visualization. For all box plots, boxes show the 25th–75th percentile, center lines show the median and whiskers show the minimum–maximum values. For all bar graphs and dot-plots, means are represented and error bars show the s.d., unless specifically stated.
GSEA was performed as described previously20, where NES and P values were estimated using 1,000 gene or pathway permutations. For single-sample (single-patient) analysis, data were scaled (z scored) on a gene level, so that a set of z scores for each patient defined a ‘single-sample signature’. Subsequently, to estimate activity levels (for example, for META-16 and MYC genes) in each sample, we utilized GSEA, where we considered each single-sample signature as a reference and genes of interest as a query gene set. To compare expression levels of META-16 across different metastatic sites, we performed gene set variation analysis63, implemented as a gene set variation analysis package (Bioconductor) in R. CNVs of MYC and KRAS in the TCGA (n = 489) and SU2C (n = 429) cohorts were analyzed using cBioportal49, considering all gains as low-level gains plus amplifications.
To evaluate the nonrandom ability of candidate genes to distinguish primary tumors in the TCGA cohort from the mCRPC samples in the SU2C cohort, we selected a random (equally sized, n = 16 or n = 55) group of genes and compared their estimated activity levels between TCGA and SU2C cohorts using two-sample one-tailed Welch t-test. This random model procedure was repeated 10,000 times and two-sample one-tailed Welch t-test P values from all iterations were used to build a null model. The empirical P value was then estimated as the number of times two-sample one-tailed Welch t-test P values for a random group of 16 or 55 genes reached or outperformed our original two-sample one-tailed Welch t-test P value for the identified genes.
Cox proportional hazards model and Kaplan–Meier survival analysis were conducted with the surv and coxph functions from survcomp package (Bioconductor) or using GraphPad Prism software. Radiographical evidence of metastatic disease was the primary end point for survival analysis on human validation cohorts. Statistical significance was estimated with a Wald test and log-rank test, respectively. For Kaplan–Meier survival analysis, hierarchical consensus clustering was performed on expression levels of META-16 genes, which clustered patients into two groups: one group with high gene expression and one group with low gene expression for META-16. Time to distant metastasis from radical prostatectomy was modeled using a Cox proportional hazards model with and without adjusting for age, pathological Gleason score/grade at diagnosis, pre-PSA, SVI, LNI and extra-prostatic extension. Time from the start of the next-generation ARSI treatment to death/follow-up or time on the ARSIs were utilized for treatment-associated survival and treatment-associated progression, respectively, as defined elsewhere26.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All relevant data supporting this study are provided in this paper, and reagents will be made available upon request. RNA-seq and WES data can be found in GEO (GSE143815). Previously published genomic data, reanalyzed here, were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/), dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001141.v1.p1), TCGA data portal (https://portal.gdc.cancer.gov) and cBioPortal (https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019), as detailed in Supplementary Table 3. Databases used were HomoloGene (NCBI, https://www.ncbi.nlm.nih.gov/homologene) and hallmarks and C2 pathway gene sets from the molecular signatures database (MSigDB) 3.0 (https://www.gsea-msigdb.org/gsea/msigdb). Source data are provided with this paper.
Code availability
The code for UMAP visualization is available at https://github.com/simslab/cluster_diffex2018. The code for bootstrap analysis of evolution tree is available at https://github.com/RabadanLab.
Extended Data
Extended Data Fig. 1 |. Additional phenotypic analyses of NPKEYFP prostate tumors.
a, Representative bright field and ex vivo fluorescence images of prostate, lung, and bone (spine) from NPEYFP mice (n = 35), NPKEYFP mice without bone metastases (n = 59), and NPKEYFP mice with bone metastases (n = 47). b–j, Comparison of NPKEYFP mice with (n = 47) or without (n = 59) bone metastasis. See also Supplementary Table 1. b, Overall survival; p-value calculated using a two-tailed log-rank test. c, Bladder obstruction; p-value calculated using a two-sided Fisher’s exact test. d–g, Dot-plots showing tumor weight (d) and metastatic load (number of metastases/mouse) to lungs (e), liver (f) and brain (g). p-values were calculated by two-tailed Mann-Whitney test, center-lines show the mean and error bars depict SD. h, Distribution of metastases to specific bone types in NPKEYFP mice at the time of euthanasia. Shown is the mean with standard deviation; n = 106 mice (n = 59 without bone metastases and n = 47 with bone metastases). i, j, Longitudinal analysis of micro-metastasis in non-terminal mice dissected at the ages shown (n = 26). i, Bar graphs showing the percentage of mice with micro-metastasis at 3 months (n = 7), 4 months (n = 17) and 5 months (n = 2). j, Bar graphs showing the percentage of mice with bone or lung micro-metastases (n = 26).
Extended Data Fig. 2 |. Analyses of androgen-intact and castrated of NPKEYFP mice.
Comparison of intact (n = 106) and castrated (n = 22) NPKEYFP mice. See also Supplementary Table 1. a, Kaplan-Meier curves showing overall survival; p-value was calculated using a two-tailed log-rank test. b, c, Dot-plots showing tumor weight (b) and number of bone metastasis (c). p-values were calculated by two-tailed Mann-Whitney test, center-lines show the mean and error bars depict SD.d, Bar graphs showing the percentage of mice with metastasis to the indicated organs. e, f, Dot-plots showing relative AR activity levels (that is, NES defined based on enrichment of AR signature (based on18) in each sample) (e) and relative neuroendocrine (NE) activity (that is, NES defined based on enrichment of NEPC signature (based on19) in each sample (f) comparing intact (n = 13) or castrated (n = 6) primary tumors, intact (n = 9) or castrated (n = 2) lung metastasis and intact (n = 10) or castrated (n = 2) bone metastasis from NPKEYFP mice (Supplementary Table 2). g, Gene Set Enrichment Analyses (GSEA) comparing a bone metastasis signatures from castrated mice used as a query and bone metastasis signature from non-castrated (intact) NPKEYFP mice used as a reference (Supplementary Table 2i,j). NES (normalized enrichment score) and p-values were estimated using 1,000 gene permutations. NS, non-significant (P < 0.05).
Extended Data Fig. 3 |. Comparison of MYC and RAS in mouse and human prostate cancer.
a–c, Copy number variant (CNV) analyses of KRAS and MYC in mouse and human prostate cancer. a, Kras, Cdkn2a/b and Myc loci, inferred from whole-exome sequencing of NPKEYFP prostate tumors. Color coding reflects amplifications or deletions in five individual mice (Supplementary Table 4b). b, c, Summary of gains in MYC (b) and KRAS (c) in human prostate cancer comparing primary tumors from TCGA (n = 489) and metastases from SU2C (n = 429) using cBioportal49. P values were calculated using a Fisher’s exact test comparing samples with all gains versus no gains. d, e, Box plots depicting Myc pathway and Ras pathway activation in primary tumors and metastases from intact or castrated NPKEYFP mice (primary tumors: n = 13 intact and n = 6 castrated; lung metastases: n = 9 intact and n = 2 castrated; bone metastases: n = 10 intact and n = 2 castrated). The distribution of the activity scores (y-axis) for Myc activity is based on single-sample GSEA in panel d, and Ras activity levels is based on the absolute-valued average of RAS-related genes as in11,33 in panel e. P-values were estimated using two-sample one-tailed Welch t-test, boxes show the 25th–75th percentile with the median, and whiskers show the minimum–maximum values. f, g, Violin plots depicting the distribution of MYC and RAS pathway activation in primary tumors and metastases comparing human primary tumors (TCGA, n = 497) versus metastases (SU2C, n = 270). In panel f, the distribution of the NESs (y-axis) represent MYC activity levels based on single-sample GSEA (see Extended data Fig. 4d) In panel g, the activity scores (y-axis) represent RAS pathway activity levels based on the absolute-valued average of RAS-related genes (as in11,33). P-values were estimated using two-sample one-tailed Welch t-test. In the violin plots with embedded box plots, boxes show the 25th–75th percentile, center-lines show the median, and whiskers show the minimum–maximum values. h, i, Heatmap representation showing the correlation of MYC and RAS pathway activity in mouse (h) and human (i) prostate cancer. Panel h shows Myc and Ras pathway activity in mouse NPKEYFP primary tumors and bone metastases. Panel i shows MYC and RAS pathway activity in human primary tumors (TCGA, n = 497) and metastases (SU2C, n = 270). Gleason scores are shown for the primary tumors; metastases include all metastases in the SU2C cohort. In panels h, i, Spearman correlation rho- and p-values are shown. j, Mouse NPKEYFP primary tumors and bone metastases classified as MYC- or RAS-activated are depicted in a heatmap in red, whereas those without MYC- or RAS-activation are represented in blue. Samples were considered Myc-activated if Myc activity scores were greater than the average across the cohorts. Samples were considered Ras-activated if Ras activity scores were greater than the average across the cohorts. The percentage of cases in which Myc and Ras are co-activated are shown; two-tailed p-value was calculated using Fisher’s exact test.
Extended Data Fig. 4 |. Additional analyses of MYC activity in prostate tumors and metastases.
a, Cross-species pathway analysis. Pathway-based GSEA comparing pathways enriched in the FHCRC human bone metastasis signature (Supplementary Table 6d) with those enriched in the mouse bulk RNA bone metastasis signature (Supplementary Table 6a). NES and p-values were estimated using 1,000 pathway permutations. b, Stouffer integration of the leading-edge pathways from the GSEA comparing the mouse (Supplementary Table 6a) and the two human bone metastases signatures (Supplementary Tables 6c,d) from panel a and Fig. 4b. The x-axis shows the Stouffer integrated NES. c, Bar graphs summarizing NES scores from GSEA of bone metastasis signatures from NPKEYFP mice (Supplementary Table 2c), and the Balk and FHCRC human cohorts (Supplementary Tables 3c,d) showing enrichment of three independent MYC signatures: “Hallmarks” (human), “Dang” (human)29 and “Sabo” (mouse)30. NES and p-values were estimated using 1,000 gene permutations. d, Heatmap representation of single-sample GSEA enrichment of MYC activity based on enrichment of the Hallmarks MYC pathway in primary tumors from TCGA (n = 497) and metastases from SU2C (n = 270) (Supplementary Table 3). Gleason scores are shown for the primary tumors; metastases include all metastases in the SU2C cohort. Colors correspond to NES.
Extended Data Fig. 5 |. Additional analysis of Myc function in an allograft model of bone metastasis.
a, Bar graphs and crystal violet staining of colony formation assays of NPK bone cells two weeks after treatment with shControl or shRNAs targeting Myc (shMyc#1 and shMyc#2). b, Comparison of lung and bone from Nude mouse hosts implanted via intracardiac injection with green fluorescent protein (GFP)-tagged cells derived from primary tumors of non-metastatic NP mice (NPGFP cells, reported in;16 n = 2) or the NPKEYFP bone cells (n = 10). Shown are representative ex vivo fluorescence or H&E images. Scale bars represent 0.1 cm for the ex vivo fluorescence images and 50 μm for all other images.
Extended Data Fig. 6 |. Analyses of MYC silencing in a human tumor growth in bone.
a, Strategy. PC3 cells engineered to express luciferase and GFP (PC3-Luciferase-GFP cells). Cells were infected with a control shRNA (shControl) or shRNA to silence MYC (shMYC#1 or shMYC#2) and implanted into the tibia of NOD-SCID mouse hosts. b, Western blot image of total protein extracts. Shown are the approximate molecular weight markers (kDa); Actin is a control for protein loading. Shown is a representative blot from two independent experiments. The uncropped Western image is shown in Source data Extended Fig. 6. c. Immunostaining for MYC in tumors from mice that had been injected with cells expressing the indicated shRNAs. Scale bars represent 50 μm. d, Bar graphs and crystal violet staining of colony formation assays of PC3-Luciferase-GFP cells two weeks after treatment with the shRNAs as indicated. e, Growth curves comparing PC3-Luciferase-GFP cells infected with shRNA (n = 10/group). P-value shown for day 52 was estimated by two-way ANOVA with Sidak’s multiple comparisons against shControl. f. Representative IVIS bioluminescence imaging used for panel e. g. Representative images from the time of sacrifice of tibiae implanted with the PC3-Luciferase-GFP cells infected with shRNA (n = 10/group). Shown are ex vivo imaging of GFP fluorescence, to visualize the tumor, and corresponding micro-computed tomography (CT) images, to show areas of osteolysis as is typical of PC3 tumors in bone36. Also shown are representative H&E and immunostaining for GFP. In a and f, bars show mean and error bars the SD, (n = 3) and p-value is shown for One-way ANOVA with Dunnett’s multiple comparison test, compared to shControl. Scale bars represent 0.1 cm for the ex vivo fluorescence images and 50 μm for all other images.
Extended Data Fig. 7 |. Additional analyses of discovery of a MYC-correlated signature in prostate cancer metastasis.
a,b, GSEA using the PROMOTE-559 gene signature (Supplementary Table 8) to query the bone metastasis gene signature from the NPKEYFP mice (Supplementary Table 2c) (in a) and the human bone metastasis gene signature (Supplementary Table 3c) (in b); NESs and p-values were estimated using 1,000 gene permutations. c, Association with adverse outcome for metastasis. Each of the MEtA-55 genes was evaluated by univariable Cox proportional hazards analysis for time-to-metastasis outcome in the TCGA dataset (n = 336 with available time to follow-up, Supplementary Table 3) and ranked by the strength of the association (that is, Wald test p-value), with a cutoff at p-value<10–7 from Wald test used to identify the 16 top-genes constituting the MEtA-16 gene signature (Supplementary Table 8). d, Random model. To evaluate the probability that not any random group of 16 genes would be upregulated in the SU2C (n = 270) versus the TCGA (n = 497) cohorts, we constructed a null model using 10,000 iterations, with the x-axis showing -log2 p-value (from the two-sample one-tailed Welch t-test) between TCGA and SU2C comparisons and y-axis showing its probability density. The p-value of this random model thus represents an estimate of the number of times two-sample one-tailed Welch t-test p-values for a random 16 genes reached or outperformed two-sample one-tailed Welch t-test p-values for the MEtA-16 genes. The p-value for the analogous random model for MEtA-55 was P = 0.036.
Extended Data Fig. 8 |. Additional analyses of MEtA-55 and MEtA-16 in prostate cancer metastasis.
a, Scaled expression (DESeq2 normalized values) of MEtA-55 in single-cell UMAP projections of primary tumors and bone metastases (see Fig. 7c, d). Shown is the correlation between MEtA-55 expression at the single-cell level with MYC pathway activity (Spearman’s rank correlation rho and p-values). b, c, Heatmap representation of single-sample GSEA enrichment of the MEtA-16 (b) and MEtA-55 (c) gene signatures in primary tumors from TCGA (n = 497) and metastases from SU2C (n = 270) (Supplementary Table 3). Colors correspond to NES. d, e, Violin plots depicting the distribution of the NESs (y-axis) which reflect activity levels of MEtA-16 (d) and MEtA-55 (e) in primary tumors from TCGA (n = 497) compared with metastases from SU2C (n = 270). The p-value was estimated using two-sample one-tailed Welch t-test. In inset box-plots, boxes show the 25th–75th percentile, center-lines show the median, and whiskers show the minimum–maximum values. f, g, Heatmap representation of expression levels of MEtA genes (as indicated) in each of the individual samples from the TCGA (n = 497) and SU2C (n = 270) cohorts. Gleason scores are shown for the primary tumors; metastases include all metastases in the SU2C cohort. Shown are row-scaled expression values (color). Panel f shows the 10 genes from the MEtA-16 signature that do not co-reside with MYC on chromosome 8q, indicated as MEtA-10. Panel g shows the MEtA-55 genes.
Extended Data Fig. 9 |. Additional validation of the MEtA-16 gene signature.
a, Quantitative reverse transcriptase PCR (qRT-PCR) of MEtA-16 in the CUIMC cohort of bone metastases (n = 5) compared with high-Gleason grade primary prostate tumors (n = 10). Indicated p-values were estimated using a two-tailed Mann-Whitney test compared to the average of all primary tumors. In box plots, boxes show the 25th–75th percentile, center-lines show the median, and whiskers show the minimum–maximum values. b, c, Heatmaps showing expression levels of MEtA-16 genes determined by qRT-PCR following MYC silencing in human and mouse prostate cancer cells. b, qRT-PCR using RNA obtained from subcutaneous PC3-Luc-GFP tumors expressing shRNA against MYC (shMYC#1) or control shRNA (shControl). c, qRT-PCR using RNA obtained from NPKEYFP bone cells grown in vitro and infected with the indicated shRNAs. Scaled values represent ratios of expression compared to shControl for each gene. In b, c, p-values were estimated using z-score sums of all genes using two-tailed, unpaired t-test (b) or one-way ANOVA with Dunnett’s multiple comparisons against shControl (c).
Extended Data Fig. 10 |. Additional validation of the MEtA-55/MEtA-16 gene signatures and survival analyses.
a, b, Heatmaps of hierarchical consensus clustering analysis used to define tumors with high (brown cluster) and low (green cluster) expression of MEtA-16 in MAYO (n = 235) and JHMI (n = 260) cohorts, as indicated (Supplementary Table 3). Brown vertical bars on the second from top row represent patients that developed distant metastasis. Colors represent row-scaled expression values. c, d, Kaplan-Meier survival analyses comparing patients with low and high overall expression of MEtA-55. The p-values were estimated using a log-rank test. e, Multivariable survival analysis of the MEtA-55 gene signature in the JHMI and MAYO cohorts showing significant association with metastasis-free survival but not with prostate-cancer specific mortality (HR = hazard ratio, CI = confidence interval, p-values estimated from Coxproportional hazards model), adjusted for age, pathological Gleason score/grade at diagnosis, pre-PSA, seminal vesicle invasion (SVI), lymph node invasion (LNI), and extra-prostatic extension (EPE). f, g, Kaplan-Meier survival analyses comparing patients from SU2C cohort with the low and high MYC activity with respect to treatment-associated survival (that is, time from the start of treatment with androgen receptor signaling inhibitor (ARSi) therapy, to death or last follow-up, n = 75 patients) or treatment-associated disease progression (that is, time on treatment with ARSIs, n = 56) as defined in26. The p-values were estimated using a log-rank test.
Supplementary Material
Acknowledgements
We are grateful to D. Adams for assistance with the initial WES and E. Davicioni and T. Lotan for sharing clinical cohorts. We thank A. Aytes, R. Dalla-Favera, G. Karsenty and M. Shen for comments on the manuscript. This research was funded in part through the National Institutes of Health (NIH)/NCI Cancer Center Support Grant P30CA013696, which supported the Molecular Pathology, Flow Cytometry, Genomics and High Throughput Screening and Oncology Precision Therapeutics and Imaging Cores. The research was supported by NIH grants R01 CA193442, R01 CA173481 and R01 CA183929 (to C.A.S.), P50 CA211024 (to M.A.R.), U54CA209997 (to P.A.S. and C.A.S.), P50 CA097186 (to P.S.N.) and DOD grant W81XWH-17–1-0415 (to P.S.N.). P.S.N. was supported by Stand Up To Cancer-Prostate Cancer Foundation Prostate Cancer Dream Team Award (SU2C-AACR-DT0712). A.M. was supported by start-up funds from Rutgers School of Health Professions. J.M.A. was supported by a postdoctoral training grant from the Department of Defense Prostate Cancer Research Program (W81XWH-15–1-0185), an Irving Institute/Clinical Trials Office Pilot Award funded by the National Center for Advancing Translational Sciences, NIH (UL1TR001873) and the Dean’s Precision Medicine Research Fellowship from the Irving Institute for Clinical and Translational Research at CUIMC (UL1TR001873). S.P. was supported by New Jersey Commission on Cancer Research Pre-Doctoral Fellowship (DCHS20PPC028). M.Z. was supported by the National Center for Advancing Translational Sciences, NIH, grant number UL1TR001873. A.G. is a recipient of a Prostate Cancer Foundation Young Investigator Award. C.J.M. was supported by NIH fellowship F31CA210607. C.A.S. is supported by the TJ Martell Foundation for Leukemia, Cancer and AIDS Research and the Prostate Cancer Foundation and is an American Cancer Society Research Professor.
Competing interests
A.M.D. has sponsored research funding from Janssen R&D and consults for Cepheid. R.J.K. receives Royalties from GenomeDx. E.S.A. has served as a paid consultant/advisor to Janssen, Pfizer, Sanofi, Dendreon, Bayer, Bristol Myers Squibb, Amgen, Merck, AstraZeneca and Clovis; has received research grants to his institution from Janssen, Johnson&Johnson, Sanofi, Bristol Myers Squibb, Pfizer, AstraZeneca, Celgene, Merck, Bayer and Clovis; and is an inventor of a biomarker technology that has been licensed to QIAGEN. All other authors declare no competing interests.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s43018-020-00125-0.
Supplementary information is available for this paper at https://doi.org/10.1038/s43018-020-00125-0.
References
- 1.Sartor O & de Bono JS Metastatic prostate cancer. N. Engl. J. Med 378, 645–657 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Scher HI & Sawyers CL Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J. Clin. Oncol 23, 8253–8261 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Watson PA, Arora VK & Sawyers CL Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rickman DS, Beltran H, Demichelis F & Rubin MA Biology and evolution of poorly differentiated neuroendocrine tumors. Nat. Med 23, 1–10 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Halabi S et al. Meta-analysis evaluating the impact of site of metastasis on overall survival in men with castration-resistant prostate cancer. J. Clin. Oncol 34, 1652–1659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez RK et al. Incidence of bone metastases in patients with solid tumors: analysis of oncology electronic medical records in the United States. BMC Cancer 18, 44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logothetis C, Morris MJ, Den R & Coleman RE Current perspectives on bone metastases in castrate-resistant prostate cancer. Cancer Metastasis Rev 37, 189–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuhn P et al. Update on systemic prostate cancer therapies: management of metastatic castration-resistant prostate cancer in the era of precision oncology. Eur. Urol 75, 88–99 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Crawford ED, Higano CS, Shore ND, Hussain M & Petrylak DP Treating patients with metastatic castration resistant prostate cancer: a comprehensive review of available therapies. J. Urol 194, 1537–1547 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Arriaga JM & Abate-Shen C Genetically engineered mouse models of prostate cancer in the postgenomic era. Cold Spring Harb. Perspect. Med 9, a030528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aytes A et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc. Natl Acad. Sci. USA 110, E3506–E3515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aytes A et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat. Commun 9, 5201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ZA, Toivanen R, Bergren SK, Chambon P & Shen MM Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep 8, 1339–1346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou M et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov 7, 736–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Shen Y & Zhu S Distribution features of skeletal metastases: a comparative study between pulmonary and prostate cancers. PLoS ONE 10, e0143437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carver BS et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltran H et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanbrough M et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res 66, 2815–2825 (2006). [DOI] [PubMed] [Google Scholar]
- 22.McFadden DG et al. Mutational landscape of EGFR-, MYC- and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc. Natl Acad. Sci. USA 113, E6409–E6417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westcott PM et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 517, 489–492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller S et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 554, 62–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abida W et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Kumar A et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med 22, 369–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA & Dang CV An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4, R69 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabo A et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh CM et al. MYC and prostate cancer. Genes Cancer 1, 617–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurel B et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol 21, 1156–1167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF & Jones LW Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest. Urol 17, 16–23 (1979). [PubMed] [Google Scholar]
- 35.Wu TT et al. Establishing human prostate cancer cell xenografts in bone: induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int. J. Cancer 77, 887–894 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Corey E et al. Establishment and characterization of osseous prostate cancer models: intra-tibial injection of human prostate cancer cells. Prostate 52, 20–33 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Hubbard GK et al. Combined MYC activation and Pten loss are sufficient to create genomic instability and lethal metastatic prostate cancer. Cancer Res 76, 283–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnon C et al. Autonomic nerve development contributes to prostate cancer progression. Science 341, 1236361 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Leibold J et al. Somatic tissue engineering in mouse models reveals an actionable role for WNT pathway alterations in prostate cancer metastasis. Cancer Discov 10, 1038–1057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowak DG et al. MYC drives Pten/Trp53-deficient proliferation and metastasis due to IL-6 secretion and AKT suppression via PHLPP2. Cancer Discov 5, 636–651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellwood-Yen K et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Wang L et al. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate-prednisone. Ann. Oncol 29, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karnes RJ et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J. Urol 190, 2047–2053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross AE et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur. Urol 69, 157–165 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Beaver JA, Kluetz PG & Pazdur R Metastasis-free survival: a new end point in prostate cancer trials. N. Engl. J. Med 378, 2458–2460 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Armenia J et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung JH et al. Prospective comprehensive genomic profiling of primary and metastatic prostate tumors. JCO Precis. Oncol 10.1200/PO.18.00283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kornberg Z, Cooperberg MR, Spratt DE & Feng FY Genomic biomarkers in prostate cancer. Transl. Androl. Urol 7, 459–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Easterly ME, Foltz CJ & Paulus MJ Body condition scoring: comparing newly trained scorers and micro-computed tomography imaging. Lab. Anim. (NY) 30, 46–49 (2001). [PubMed] [Google Scholar]
- 51.Trapnell C, Pachter L & Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv https://arxiv.org/abs/1303.3997?upload=1://arxiv.org/abs/1303.3997?upload=1 (2013). [Google Scholar]
- 55.Benjamin D et al. Calling somatic SNVs and indels with Mutect2. Preprint at bioRxiv 10.1101/861054v1 (2019). [DOI]
- 56.Talevich E, Shain AH, Botton T & Bastian BC CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol 12, e1004873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oshima K et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 113, 11306–11311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsenstein J Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- 59.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levitin HM et al. De novo gene signature identification from single-cell RNA-seq with hierarchical Poisson factorization. Mol. Syst. Biol 15, e8557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trabzonlu L et al. Molecular pathology of high-grade prostatic intraepithelial neoplasia: challenges and opportunities. Cold Spring Harb. Perspect. Med 9, a030403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorek DL et al. Internalization of secreted antigen-targeted antibodies by the neonatal Fc receptor for precision imaging of the androgen receptor axis. Sci. Transl. Med 8, 367ra167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanzelmann S, Castelo R & Guinney J GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data supporting this study are provided in this paper, and reagents will be made available upon request. RNA-seq and WES data can be found in GEO (GSE143815). Previously published genomic data, reanalyzed here, were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/), dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001141.v1.p1), TCGA data portal (https://portal.gdc.cancer.gov) and cBioPortal (https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019), as detailed in Supplementary Table 3. Databases used were HomoloGene (NCBI, https://www.ncbi.nlm.nih.gov/homologene) and hallmarks and C2 pathway gene sets from the molecular signatures database (MSigDB) 3.0 (https://www.gsea-msigdb.org/gsea/msigdb). Source data are provided with this paper.