Abstract
Expanding the chemical diversity of metal complexes provides a robust platform to generate functional bioactive reagents. To access an excellent repository of metal-based compounds for probe/drug discovery, we capitalized on the rich chemistry of gold to create organometallic gold(iii) compounds by ligand tuning. We obtained novel organogold(iii) compounds bearing a 1,2-bis(diphenylphosphino)benzene ligand, providing structural diversity with optimal physiological stability. Biological evaluation of the lead compound AuPhos-89 demonstrates mitochondrial complex I-mediated alteration of the mitochondrial electron transport chain (ETC) to drive respiration and diminish cellular energy in the form of adenosine triphosphate (ATP). Mechanism-of-action efforts, RNA-Seq, quantitative proteomics, and NCI-60 screening reveal a highly potent anticancer agent that modulates mitochondrial ETC. AuPhos-89 inhibits the tumor growth of metastatic triple negative breast cancer and represents a new strategy to study the modulation of mitochondrial respiration for the treatment of aggressive cancer and other disease states where mitochondria play a pivotal role in the pathobiology.
Expanding the chemical diversity of metal complexes provides a robust platform to generate functional bioactive reagents.
Introduction
Platinum drugs, e.g. cisplatin, remain the first-line treatment option for many cancer types.1,2 The induction of drug resistance and toxic side effects from platinum therapy has prompted the generation of new metal-based anticancer agents.3–6 Metal complexes with different mechanisms of action in the biological context hold great promise. The mode of action of metal complexes is usually by covalent modification of biomolecules,7–9 non-covalent interaction with target proteins,10–12 redox activation by biomolecules,13–15 and photosensitizer action.16 This has propelled the broad application of metal-based compounds in the treatment of diseases such as microbial infection, rheumatoid arthritis, diabetes, and cancer.17 Alternative metals used in recent metal-based drug discovery include ruthenium,18,19 rhenium,20,21 osmium,22,23 and gold.24–27 Additionally, developing metal-based anticancer compounds with a known mechanism of action, well differentiated from cisplatin is essential to overcome cisplatin resistance and improve therapeutic performance.
Gold compounds are resurging as an important class of therapeutics across a broad range of pathophysiology, but major challenges in gold-based bioinorganic chemistry remain, including: (i) the kinetic lability of gold(iii) compounds under physiological conditions; (ii) the lack of a rational design of gold-based drugs; (iii) affinity for thiol-rich proteins and enzymes;28,29 (iv) incomplete understanding of gold reagent localization in living systems; and (v) difficulty in accessing stable high-valent gold(iii) compounds. Significant progress in understanding the structure and reactivity of selected gold(iii) complexes has provided new impetus for the use of gold compounds.30–36 Previous and ongoing contributions to surmount challenges associated with physiologically relevant gold compounds as anticancer libraries through innovative chemistries37–39 continue to expand via the development of new scaffolds including gold-porphyrin,9,40–44 gold-phosphine,45–51 and cyclometalated gold variations.52–57 Additionally, our synthetic efforts have led to the generation of [C^N]-cyclometalated gold(iii) compounds, which present strong sigma-donation of electrons to stabilize the gold center.58,59 Furthermore, we expanded the repertoire of gold-based agents with the synthesis of distorted gold constructs bearing chiral phosphine ligands.60
Accumulating evidence shows that aggressive tumors including triple negative breast cancer (TNBC, the model system for this report)61–65 have strong dependence on both glycolytic and oxidative pathways. Thus, a key promising strategy is targeting mitochondrial metabolism for cancer therapy.66 Rosner et al. showed that combining BACH1 targeting with oxidative phosphorylation electron transport chain (OXPHOS) inhibition by metformin is an effective TNBC therapy.67 The lack of potency of metformin has prompted several groups to develop superior inhibitors of OXPHOS as therapeutic agents for several diseases including cancer.68–73 Parada et al. demonstrated the use of a small-molecule, Gboxin, as an inhibitor of OXPHOS to target glioblastoma.74 Their work revealed that aggressive tumors rely on mitochondrial respiration for their energy needs, which can be exploited for therapeutic gains.
Here, we have synthesized and characterized new organogold(III) compounds supported by a diphenylphosphine benzene ligand. These compounds are stable toward biological thiols and demonstrate enhanced mitochondrial oxygen consumption rate and inducing proton leak in TNBC. Strikingly, in a comparative screening of a reference set of 60 cancer cell lines compiled by the National Cancer Institute (NCI-60) these compounds induce lethality across all tumor types. Whole-cell transcriptomics reveal a broader mechanism of action in addition to the dominant oxidative phosphorylation target supported by quantitative proteomic studies in response to the gold compound, AuPhos-89. We tested the efficacy of the lead compound, AuPhos-89 in a metastatic 4T1 TNBC mouse model and found significant tumor inhibition. Together, these studies demonstrate the translational potential of gold(iii) compounds in anticancer drug development.
Results and discussion
Anticancer compound design, synthesis, and characterization
The organogold(iii) bearing bisphosphine ligands, herein referred to as the AuPhos class of compounds, were synthesized as part of our research efforts to generate diverse gold compounds for biological evaluation. We rationalized that stabilizing the gold(iii) center by efficient ligand tuning will influence biological reactivity and function. To minimize premature deactivation and degradation of anticancer agents, in vivo, it is imperative to enhance their kinetic stability. The use of strongly donating phosphorus containing ligands coupled with (C^N)-cyclometalation provides a unique balance between compound stability and efficacy. Optimizing ligands around the gold center decorated with (C^N)-cyclometalation has been hampered by the rapid reductive elimination induced by donor ligands to form C(sp2)–X bonds. To resolve this, we utilized bisphosphine coupled with mild reaction conditions which minimizes reductive elimination but gives rise to the target compound. We selected diphenylphosphine benzene as the donor ligand of choice and reacted it with a series of cyclometalated gold(iii) starting materials for structure activity relationship (SAR) studies. Briefly, treatment of HAuCl4 with the respective benzyl/benzoyl pyridine ligands in refluxing water affords the cyclometalated compounds. Ligand substitution reactions with bisphosphine ligands in chloroform at room temperature overnight resulted in organometallic gold(iii) bisphosphine compounds, AuPhos (Fig. 1). It is worth noting that the products of the reaction scheme in Fig. 1A were verified by structural insights obtained by X-ray crystallography data (vide infra). Thus, the Au(iii)-bisphosphine studied in this work likely existed as organgold(iii)-bisphosphine compounds without Au–N (sp2) coordination or with Au–N (sp2) coordination as in AuPhos-83. Complexes AuPhos-81–AuPhos-89 were characterized by 1H NMR, 31P NMR, 13C NMR spectroscopy (Fig. S1–S24†), and high-resolution mass spectrometry (Fig. S25–S32†). The purity of the compounds was confirmed by elemental analysis. The 31P NMR spectra of these compounds show two characteristic peaks in the downfield region of >50 ppm. The resonances show a doublet, which is as a result of cis-P–P coupling. The spectroscopic data obtained were validated by HRMS (ESI) for all compounds, showing a characteristic [M–Cl]+ molecular ion peak. The ionization pattern and isotopic distribution confirm the potential coordination of one chloride ligand to the gold center.
Fig. 1. Chemical structures of AuPhos compounds. (A) Synthetic scheme to access organogold(iii) compounds investigated in this study. (B)–(E) Crystal structures of AuPhos-81, 82, 84, and 83. Outer-sphere solvent molecules are omitted for clarity. Thermal ellipsoids are shown at the 50% probability level. In (D), only one representative molecule from the asymmetric unit is shown.
The AuPhos complexes display moisture and air stability. To study the biological stability of AuPhos complexes, reactivity with glutathione (L-GSH), a representative intracellular antioxidant, was evaluated using HPLC in aqueous solution (Fig. S33†). Interestingly, the chromatogram of AuPhos-89 in the reaction showed little change over the estimated 24 h period. Similarly, stability studies of AuPhos-89 using 1H-NMR spectroscopy were performed (Fig. S34†). The proton resonances corresponding to AuPhos-89 or L-GSH in the mixed reaction solution of AuPhos-89 and L-GSH in DMSO-d6 were minimally altered after 24 h. Changes in proton resonances at 0 and 1 h seem to be due to the slow solubility of L-GSH in DMSO solvent. Together, these experiments confirm the stability of AuPhos-89 in the biological environment.
Single crystals of AuPhos-81, 82, and 84 were grown by vapor diffusion of ether into a solution of concentrated gold complex in DMF at 4 °C or room temperature, and AuPhos-83 was crystallized using acetone/THF. The single crystals were analyzed by X-ray diffraction to determine the molecular structures. The crystal structures solved for AuPhos-81, 82 and 84 reveal a four-coordinate structure with the carbon from the arylpyridine bonded to the gold atom, chloride and diphosphine coordinated to the gold atom in a square-planar geometry. Based on the crystal structure of AuPhos-83, a cyclometalated compound is obtained, where the arylpyridine ring is bonded through the carbon and nitrogen atoms to the gold. The Au–N bond length is 2.096(4), indicative of a strong bond. The organogold character is defined by the existence of the Au–C bond with a bond length within the range of 2.072(8)–2.0819(19) for AuPhos-81, 82 and 84. In contrast, the Au–C bond for AuPhos-83 is slightly longer at 2.100(5). Another interesting feature of the crystal structure of AuPhos-83 is the potential axial interaction of the cationic Au center and chloride ligand. Whereas the observation is found in the solid state with a relatively longer bond length of 2.8790(11), it presents opportunities to investigate the type of interaction or bonding in solution and whether Au(iii) systems can accommodate a fifth coordination. The use of the 1,2-bis(diphenylphosphino)benzene (DPPB) ligand provides a rigid backbone for gold chelation. Table 1 shows selected interatomic bond distances and angles of the crystal structure of AuPhos-81, 82, 83, and 84 as displayed in Fig. 1. Further analysis of the P1–Au1–Cl1 angles of AuPhos-81, 82, and 84 shows a close to linear angle of ∼173–178°. Overall, the structures of organogold(iii)-bisphosphine complexes have been elucidated.
Selected interatomic distances (Å) and angles (°) from the crystal structures shown in Fig. 1 below.
| Bond/angle | Gold complexes | ||||
|---|---|---|---|---|---|
| AuPhos-81 | AuPhos-82 | AuPhos-83 | AuPhos-84 | ||
| Au1–N1 | — | — | 2.096(4) | — | — |
| Au1–C1 | 2.0819(19) | 2.074(3) | 2.100(5) | 2.079(8) | 2.072(8) |
| Au1–P1 | 2.2808(5) | 2.2843(7) | 2.3396(12) | 2.289(2) | 2.283(2) |
| Au1–P2 | 2.3602(5) | 2.3451(7) | 2.3406(12) | 2.341(2) | 2.344(2) |
| Au1–Cl1 | 2.3387(5) | 2.3354(6) | 2.8790(11) | 2.342(2) | 2.341(2) |
| P1–C24/C25 | 1.808(2) | 1.811(3) | 1.797(4) | 1.807(8) | 1.804(8) |
| P1–C12/C13 | 1.8180(19) | 1.815(3) | 1.810(5) | 1.806(9) | 1.814(9) |
| P2–C36/C37 | 1.808(2) | 1.799(3) | 1.801(5) | 1.800(8) | 1.808(8) |
| C1–Au1–P1 | 91.10(6) | 93.46(7) | 96.22(14) | 88.7(2) | 178.63(7) |
| N1–Au1–P2 | — | — | 96.46(13) | — | — |
| C1–Au1–P2 | 172.04(6) | 175.92(7) | 175.16(15) | 175.3(2) | 175.5(2) |
| P1–Au1–P2 | 84.459(17) | 82.91(2) | 82.76(4) | 86.55(8) | 86.57(8) |
| N1–Au1–Cl1 | — | — | 96.05(14) | — | — |
| C1–Au1–Cl1 | 89.76(6) | 90.44(7) | 97.65(14) | 90.5(2) | 90.5(2) |
| P1–Au1–Cl1 | 176.814(18) | 173.47(2) | 88.58(4) | 178.91(8) | 178.63(7) |
| P2–Au1–Cl1 | 94.331(17) | 93.01(2) | 87.07(4) | 94.15(8) | 93.79(8) |
NCI-60 screening
Encouraged by the anti-proliferative potency of the compounds (Table S1 and Fig. S35–S39†), we sought to expand the cell lines used and possibly gain insight into the mechanism of action (MOA). Comparative profiling and recent advances in omics technology have expanded the toolkit for target identification and mechanism of action of bioactive molecules. A phenotypic comparison of compounds with known mechanisms can reveal the MOA of compounds with unknown mechanisms, particularly, metal-based compounds. The use of comparative profiling approaches relies on correlating phenotypes obtained with an unknown compound to those within a set of reference compounds with known MOA. It is inferred that compounds with similar MOA will exhibit similar phenotypes. In 1990, the National Cancer Institute compiled a reference set of 60 cancer cell lines (NCI-60) and profiled the sensitivity of each cell line to a large panel of compounds.75,76 This has yielded large-scale datasets that are rich resources for the MOA of unknown compounds and target identification. We were interested in assessing the comparative profiles of three of our AuPhos compounds (AuPhos-83, 84, and 89) in the NCI-60 screening service. Following a single dose screening at 10 μM, the Developmental Therapeutic Program prioritized the compounds for 5-point dose response assays in the NCI-60 panel. The results reveal that AuPhos-83, 84 and 89 display profound lethality across all cancer types including breast cancer with GI50 in the range of 150 nM–700 nM. All three compounds (AuPhos-83, 84, and 89) showed excellent results across the NCI-60 panel with unbiased lethal effects, especially in central nervous system (CNS) cancer, melanoma, and renal cancers. Surprisingly, the lethal effect on liquid tumors such as leukemia was relatively diminished compared to the solid tumors in the NCI-60 panel. Thus, we prioritized the compound with superior cell growth inhibitory ability across all cell lines (i.e.AuPhos-89) for further biological experiments. These results are shown in the ESI (Fig. S40–S42†) and summarized in Fig. 2.
Fig. 2. Summary of the NCI-60 human tumor cell line screen. AuPhos 83, 84, and 89 show superior activity across the panel of cell lines tested including breast. Numbers in parentheses represent the number of cell lines tested for each indication. The breast panel includes TNBC (MDA-MB-231, BT549, Hs578T, MDA-468) and luminal cell lines (MCF7, T47D).
Additionally, the COMPARE algorithm, which is a pattern recognition algorithm, can be used to suggest a putative mechanism or the uniquely distinct mechanism of a given agent. Pearson (PCC) and Spearman correlations (SC) are used to estimate the degree of similarity of a test compound, in this case AuPhos, to compounds with known mechanisms in the NCI-60 database. We used an FDA-approved and investigational compound library from the Cancer Chemotherapy National Service Center (NSC) for comparison. The rationale for this compound set was the well-characterized MOA of approved drugs. We found that the AuPhos compounds screened in the NCI-60 panel had no reliable correlation with the drug set used. The filtering conditions utilized GI50, LC50, and TGI derived from the NCI-60 results obtained for AuPhos. Taken together, the comparative profiling of the novel class of gold compounds revealed a different mechanism of action from known drugs, which presents new opportunities for efficacious drugs.
Differential gene expression and biological pathway analysis
Based on the exceptional potency of the AuPhos class of compounds and different mechanism of action as derived from the NCI-60 screening, we employed a systems biology approach to assist in our quest to unravel the mechanism of action or validate findings from comparative profiling. We used RNA sequencing to determine differential gene expression in MDA-MB-231 cells in response to treatment with 1 μM of AuPhos-89 for 12 h. Briefly, MDA-MB-231 cells were exposed to AuPhos-89 and following the isolation of highly pure RNA, RNA-sequencing was performed for both control- and AuPhos-treated cells with ∼30 million 50-bp paired-end reads generated per sample using the Illumina Hi-seq. Sequence reads were mapped to the hg19 (GRCh37) human genome. We found 922 DEG with 83 upregulated and 94 downregulated genes in response to AuPhos (Fig. 3a). Kyoto encyclopaedia of genes and genomes (KEGG)77–79 (Fig. 3b) pathway analysis uncovered potential processes perturbed by AuPhos-89. The pathway analysis software employed is an extensive library database capable of integrating chemical and biological pathway perturbation processes and is well suited for drug development studies. Differentially expressed genes that contributed to the top five pathways in response to AuPhos-89 included rheumatoid arthritis, oxidative phosphorylation, Parkinson's disease, cytokine–cytokine receptor interaction, and NOD-like receptor signaling. Interestingly, these processes are related to inflammation, which is chiefly regulated by the mitochondria and linked to metabolism. Several differentially expressed genes were related to the electron transport chain, the driver of oxidative phosphorylation. Thus, we hypothesized that AuPhos-89 induces cell death by modulating oxidative phosphorylation and redox pathways in breast cancer cells. Recent findings suggest OXPHOS as a viable therapeutic target in cancer including breast cancer. This led us to investigate the effect of AuPhos-89 on mitochondrial metabolism.
Fig. 3. Whole-cell transcriptomics. (a) Representative heat map of DEGs in response to AuPhos-89. (b) KEGG analysis plots outlining varying pathways perturbed upon treatment with AuPhos-89. MDA-MB-231 cells were treated with AuPhos-89 (1 μM for 12 h) and pure RNA isolated for sequencing. Data are representative of two independent replicates.
Cellular and mitochondrial uptake of AuPhos
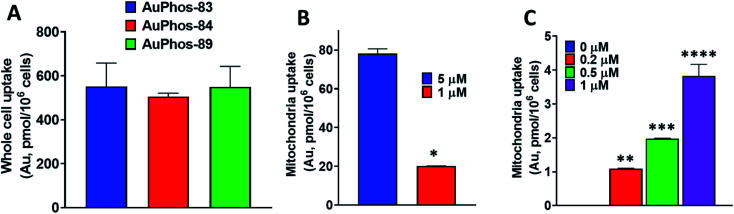
In our efforts to deepen the insight into the mechanism of action of AuPhos, we sought to examine the intracellular accumulation. Cell permeability is an essential physicochemical property of drug-like molecules or chemical probes. The ability of compounds to cross the cell membrane to induce cytotoxic effects or engage their targets contributes to their efficacy. To study the intracellular uptake for AuPhos, we performed whole-cell uptake experiments in OVCAR8 cells using AuPhos-83, 84 and 89 and found that an appreciable amount of compound was detected in cells after 15 h (Fig. S43†). Compound uptake was confirmed by measuring gold accumulation using graphite furnace atomic absorption spectroscopy (GF-AAS). Given that the preliminary targets identified via NCI-60 screening and RNA-seq were localized in the mitochondria, quantification of AuPhos in mitochondria is imperative. We further assessed compound uptake in mitochondria organelle fractions of MDA-MB-231 cells after incubation with AuPhos-89 (Fig. 4). This was to confirm mitochondria localization given the dominant oxidative phosphorylation pathway identified. The gold content measured by GF-AAS showed that the gold compound taken up in the mitochondria of cancer cells is significantly higher (>5-fold) than normal cell mitochondria (Fig. 4B and C). This could be attributed to the high mitochondrial membrane potential in cancer cells compared to normal cells. Additionally, gold uptake by the mitochondria of MDA-MB-231 compared to NCM460 demonstrates the selective accumulation of the compound for cancerous cells in comparison to normal cells. The high degree of cancer cell selectivity is driven by the upregulation of vitamin and fatty acid transporters in cancer cells. In addition, the lipophilic cationic character of AuPhos facilitates mitochondrial uptake. Together, these findings suggest that AuPhos-89 is taken up into intracellular locations of cancer cells, specifically into the mitochondria.
Fig. 4. Cellular and mitochondrial uptake study. (A) Whole-cell uptake results of representative AuPhos compounds in MDA-MB-231 cells. (B) Mitochondrial accumulation of AuPhos-89 in breast cancer epithelial cells. (C) Mitochondrial accumulation of AuPhos-89 in normal colon epithelial cells (NCM 460). Data are a mean of three independent replicates. For (B), unpaired t-test, *P < 0.0001, for (C), ordinary one-way ANOVA, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Role of AuPhos in enhancing mitochondrial respiration
The direct interaction and effect of AuPhos-89 on mitochondria was investigated to assess mitochondrial bioenergetics. A characteristic hallmark of mitochondrial uncoupling activity is the increase of mitochondrial oxygen consumption rate (OCR), even in the presence of FoF1 ATP synthase inhibitors including oligomycin. Using isolated mitochondria from the liver of C57BL/6J mice, AuPhos-89 at a concentration of 1 μM increased oxygen consumption in the presence of oligomycin (Fig. 5A) as determined using a Seahorse XF analyzer. This phenomenon was dose-dependent up to about 10 μM. Moreover, the induction of OCR was independent of Ca2+ (Fig. 5B). Furthermore, at the same concentrations, AuPhos-89 acutely induced proton leak in live MDA-MB-231 cells (Fig. 5D) with diminished ATP production (Fig. 5E). Assessment of the mitochondrial membrane potential (MMP) indicated that there was depolarization of the MMP of MDA-MB-231 cells using the TMRE (Fig. 5F) or JC-1 assay at short compound exposure periods (Fig. 5G). Our results support the mitochondrial OXPHOS as a potential target of AuPhos-89.
Fig. 5. Effect of AuPhos on bioenergetics. (A) OCR mediated by complex I (State IV) from the effect of AuPhos-89 on isolated liver mitochondria in the presence of oligomycin. (B) OCR mediated by complex I (State IV) from the effect of AuPhos-89 on isolated liver mitochondria in the presence of oligomycin and Ca2+. (C) Schematic diagram of the mitochondrial respiration experiment. (D) Proton leak extrapolated from the mito-stress test of MDA-MB-231 cells treated with AuPhos-89, (E) ATP production extrapolated from the mito-stress test of AuPhos-89 treated MDA-MB-231 cells. (F) Changes in the membrane potential of MDA-MB-231 cells depending on the concentration of AuPhos-89 using TMRE assay and FCCP as control. (G) Mitochondrial membrane potential measured by FACS of MDA-MB-231 cells, control (left), AuPhos-89-treated (middle) using JC-1 assay. Ordinary one-way or two-way ANOVA, *P < 0.01 and ****P < 0.0001.
Cellular responses evoked by AuPhos
Analysis of cell population in the different phases of the cell cycle revealed no significant changes to the cell cycle, indicating that the compound does not stall cells in these phases under the treatment condition used (Fig. 6A). The DNA binding agent, cisplatin, is known to induce S and G2/M cell cycle arrest in a number of cell lines, supporting its mechanism of DNA cross-linking. When MDA-MB-231 cells were exposed to AuPhos-89 at 1 μM for 15 h, a significant population (∼40%) of cells were in early to late apoptosis as detected by FACS (Fig. S43A†). As expected, the control treated with hydrogen peroxide showed significant apoptosis (Fig. S44B†). The results suggest that AuPhos induces significant apoptosis as a mode of cell death in breast cancer cells. Furthermore, to determine if AuPhos-89 induces apoptosis through the caspase-mediated apoptotic pathway, we performed western blotting to assess the induction of caspases and cleaved PARP (Fig. S44C†). We found that AuPhos-89 activates cleaved caspases, PARP and the cellular energy regulator, AMPK (Fig. 6B), a hallmark of apoptosis. We investigated global changes in proteins using quantitative proteomics after 12 h of exposing MDA-MB-231 cells to AuPhos-89 at 1 μM. In this tandem mass tag (TMT)-based quantification, protein was extracted from cryo-preserved cell pellets and following tryptic digestion, TMT-labeling was performed. After fractionation, tandem LC-MS/MS was conducted and database search against HUMAN protein database was carried out. Proteins of relative quantification were divided into two categories. A quantitative ratio over 1.5 was considered upregulation while a quantitative ratio less than 1/1.5 was considered as downregulation. We found a total of 163 differentially expressed proteins, 75 of which were upregulated and 88 downregulated (Fig. 6C, ESI table†). Additionally, protein-related mitochondrial metabolism including ADP-ribosylation 4, glutathione-S-transferase, thioredoxin, and enzymes in the glucose pathway was upregulated to compensate for energy deprivation by mitochondrial stress. Overall, the proteomics study supports the other profiling studies described in this work and clearly lays out a systematic pipeline to study new metal-based agents and elucidate their mode of action in an unbiased way.
Fig. 6. Cellular responses to AuPhos-89. (A) Cell cycle distribution by PI staining: br graph representing the percentage of cells in G1, G2, and S. MDA-MB-231 cell bars represent an average of 3 measurements. Error bars represent means ± SD. (B) Time-dependent activation of AMPKα and of MDA-MB-231 treated with AuPhos-89 (1 μM) and analyzed by immunoblotting. (C) TMT-labelled quantitative proteomics. Differentially expressed protein data; only Log2 FC values more than 1 and less than −1 were extracted and displayed. Proteins with high values are shown in the table. MDA-MB-231 cells were treated with AuPhos-89 (1 μM, 12 h).
In vivo anticancer potential of AuPhos-89
Despite the augmented in vitro potency and the novel mechanistic activities reported with other experimental metal-based anticancer agents, many of these compounds have been limited by lack of preclinical evaluation in animal models.
We evaluated the relative in vivo antitumor efficacy of AuPhos-89 as compared to vehicle control. A murine 4T1 cell-line syngeneic of the TNBC model was employed, and AuPhos-89 was administered by 3 weekly IV injections at a dose equivalent of 10 mg kg−1 (n = 5 mice per treatment group). A separate group of mice was administered equal volumes of vehicle control at the same time points and via the analogous route. During the first three days after tumor cell implantation and prior to treatment, all mice exhibited equivalent rates of tumor growth. After the first injection, the growth rate of the AuPhos-89 treated group decreased significantly, and this trend was maintained until the end of the experiment (Fig. 7A).
Fig. 7. Antitumor effect induced by AuPhos-89. (A) Impact of AuPhos-89 on the tumor volume of 4T1 (1 million cells inoculated, n = 5). Unpaired t-test, *P < 0.05. (B) Weight of mice (n = 3) following intravenous administration of AuPhos-89 and observed over 19 days. (C) Tissue biodistribution of AuPhos-89 in mice as determined by GF-AAS, which measures gold content. The compound was administered by intravenous injection and at indicated time points, tissues were collected after mice (n = 3) were euthanized. (D) Hematoxylin and eosin (H&E) staining indicates reduced cellularity and proliferation in tumors treated with AuPhos-89. Liver metastasis is observed in control mice with no palpable metastatic lesions in treated mice. (E) Comparative in vivo efficacy study of AuPhos-89 and cisplatin. Impact of AuPhos-89 and cisplatin on the tumor volume of 4T1 (two million cells inoculated, n = 5). Ordinary one-way ANOVA test, *P < 0.05. (F) Weight of mice following intraperitoneal administration of AuPhos-89 and cisplatin.
We conducted a separate experiment to compare the antitumor efficacy of the gold compound, AuPhos-89, with that of the FDA approved platinum drug, cisplatin, which is used to treat TNBC. AuPhos-89 (10 mg kg−1) was injected by the intraperitoneal route, and cisplatin was administered at a low concentration of 3 mg kg−1via intraperitoneal injection due to the toxicity of the drug. Compared with the control group, significant tumor inhibition was observed in the two treatment groups. The weight was kept constant in all the control groups and the experimental group (Fig. 7).
Tissue biodistribution of therapeutic agents can shed light on drug accumulation and clearance in various organs within test animals. Biodistribution contributes to validating a drug candidate for clinical use. To determine the in vivo biodistribution profile and to validate the effective delivery of the gold compound, we injected AuPhos-89 intravenously at 10 mg kg−1 into female BALB/c mice. After respective time points of 1 h and 24 h, mice were euthanized. Organs including heart, lung, liver, spleen, kidney, and tumor were excised and the gold concentration was analyzed by GF-AAS. Furthermore, the relative quantification of Au levels (as measured by GF-AAS) in the different organs confirmed the uptake of AuPhos-89 that had been introduced into separate mice via IV administration (Fig. 7B). These data give preliminary indication that AuPhos-89 may travel in circulation to reach various organs. Further studies to determine metabolized or degraded gold compounds in circulation using detailed LC-MS/MS are underway.
Together, the data suggest that AuPhos-89 induces significant tumor inhibition in 4T1 tumor bearing mice. Importantly, TNBC is an aggressive form of breast cancer with limited treatment options. Thus, therapeutic agents with different mechanisms of action could provide therapeutic benefit to patients. At the end of the observation period, mice in the AuPhos-89 treatment arm did not exhibit significant change in their body weights, and histological tissue evaluation after H&E staining of excised tumor tissue shows high cellularity in the treated group compared to vehicle control (Fig. 7D). Additionally, signs of liver metastasis were observed in control tissue, suggesting that AuPhos-89 can inhibit liver metastasis associated with 4T1 tumors. Overall, AuPhos shows promise as a potent anticancer agent in vivo.
Conclusion
This report details the synthesis of novel gold(iii)-bisphosphines that modulate mitochondrial respiration as anticancer agents in vitro and in vivo. Compounds that disrupt mitochondria with good pharmacokinetics and a wide therapeutic window remain an unmet need. Thus, a gold-based platform as a mitochondria respiration modulator holds promise as a useful therapeutic agent in a variety of diseases including cancer. The developed organometallic gold(iii) compounds are relatively stable in solution even in the presence of physiologically relevant concentrations of glutathione, a major biological thiol. These complexes are potent across a wide range of cancer cell lines with significant lethality in the NCI-60 cell line with no cross-resistance. The compounds display high cellular uptake in cancer cells of >100 pmol per million cells. We used a systems biology approach to gain insight into the mechanism of action of the prioritized gold agent, AuPhos-89. The global effects of AuPhos on MDA-MB-231 cells identified biological processes related to oxidative phosphorylation, rheumatoid arthritis, Parkinsons disease, cytokine–cytokine receptor interaction, and NOD-like receptor signalling as prominent pathways by KEGG analysis. Using bioenergetics to assess mouse liver mitochondria and liver cancer cells, it was confirmed that AuPhos stimulates OCR leading to oxidative stress and, consequently, cell death via apoptosis. Therapeutic indices including the maximum tolerated dose, tissue biodistribution and efficacy reveal that AuPhos is a promising gold(iii) anticancer agent.
Author contributions
S. O. performed the Western Blot and animal study. H. V. and J. H. K. accomplished mitochondria metabolism assay. H. V., P. G. S., J. H. K., and S. G. A. analyzed the mitochondria metabolism data. S. P. solved the X-ray structures of compounds. J. H. K. performed the synthesis and all characterization, stability assay and reactivity with GSH, cell viability assay, cellular/mitochondria uptake assay, differentially expressed gene/protein experiments, JC-1 assay, cell cycle, apoptosis, and animal experiments. J. H. K. and S. G. A. designed the project, analyzed the results, prepared figures, and wrote the manuscript.
Funding sources
We are grateful to the University of Kentucky for funding. The authors acknowledge the support of the Center for Pharmaceutical Research and Innovation (NIH P20 GM130456). The work was also supported by NIH R01NS112693-01A1 (PGS).
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
Our study was performed in compliance with the NIH guidelines (NIH Publication No. 85-23 Rev. 1985) for the care and use of laboratory animals and all experimental procedures were monitored and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Kentucky (USA). We thank the UK NMR Center supported by NSF (CHE-997738) and the UK X-ray facility supported by the MRI program from NSF (CHE-1625732). For the flow cytometry analysis, we thank Dr Greg Bauman and the UK Flow Cytometry & Immune Monitoring core facility supported in part by the Office of the Vice President for Research, the Markey Cancer Center, and an NCI Center core support grant (P30 CA177558) to the University of Kentucky Markey Cancer Center. We thank Dr Tomoko Sengoku and Mr Michael Alstott for the redox metabolism analysis and their program supported by the Redox Metabolism Shared Resource Facility of the University of Kentucky Markey Cancer Center (P30 CA177558). We sincerely thank Dr Wendy Katz and Ms Megan A. Peterson for helping with H&E staining, and their research program supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P30 GM127211. We also thank Dr Thomas Lee and the University of Colorado Boulder, College of Arts and Sciences, Mass Spectrometry Facility, for analyzing HRMS samples. We thank Dr Neeraj Kapur for providing the NCM460 cell. We also thank Ms Sailajah Gukathasan for helping with the LC-MS and Dr Steven Van Lanen of the UK College of Pharmacy, who allowed the use of the LC-MS.
Electronic supplementary information (ESI) available: All synthetic procedures, chemical experiments, biological experiments and characterization data. CCDC 2034162 (AuPhos-81), 2034159 (AuPhos-82), 2034161 (AuPhos-83), 2034160 (AuPhos-84). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1sc01418h
References
- Sirohi B. Ashley S. Norton A. Popat S. Hughes S. Papadopoulos P. Priest K. O'Brien M. Early Response to Platinum-Based First-Line Chemotherapy in Non-small Cell Lung Cancer May Predict Survival. J. Thorac. Oncol. 2007;2(8):735–740. doi: 10.1097/JTO.0b013e31811f3a7d. [DOI] [PubMed] [Google Scholar]
- Li M. Zhang Q. Fu P. Li P. Peng A. Zhang G. Song X. Tan M. Li X. Liu Y. Wu Y. Fan S. Wang C. Pemetrexed plus platinum as the first-line treatment option for advanced non-small cell lung cancer: a meta-analysis of randomized controlled trials. PLoS One. 2012;7(5):e37229. doi: 10.1371/journal.pone.0037229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabec V. Kasparkova J. Modifications of DNA by platinum complexes. Relation to resistance of tumors to platinum antitumor drugs. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. Drug Resist. Updates. 2005;8(3):131–146. doi: 10.1016/j.drup.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Siddik Z. H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- McWhinney S. R. Goldberg R. M. McLeod H. L. Platinum neurotoxicity pharmacogenetics. Mol. Cancer Ther. 2009;8(1):10–16. doi: 10.1158/1535-7163.MCT-08-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott I. Gust R. Non platinum metal complexes as anti-cancer drugs. Arch. Pharm. 2007;340(3):117–126. doi: 10.1002/ardp.200600151. [DOI] [PubMed] [Google Scholar]
- Jung Y. Lippard S. J. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007;107(5):1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- Zou T. Lum C. T. Lok C.-N. Zhang J.-J. Che C.-M. Chemical biology of anticancer gold(III) and gold(I) complexes. Chem. Soc. Rev. 2015;44(24):8786–8801. doi: 10.1039/c5cs00132c. [DOI] [PubMed] [Google Scholar]
- Tong K.-C. Lok C.-N. Wan P.-K. Hu D. Fung Y. M. E. Chang X.-Y. Huang S. Jiang H. Che C.-M. An anticancer gold(III)-activated porphyrin scaffold that covalently modifies protein cysteine thiols. Proc. Natl. Acad. Sci. U. S. A. 2020;117(3):1321–1329. doi: 10.1073/pnas.1915202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimoska J. Feng L. Harms K. Yi C. Kissil J. Marmorstein R. Meggers E. Targeting large kinase active site with rigid, bulky octahedral ruthenium complexes. J. Am. Chem. Soc. 2008;130(47):15764–15765. doi: 10.1021/ja805555a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggers E. Targeting proteins with metal complexes. Chem. Commun. 2009:1001–1010. doi: 10.1039/b813568a. [DOI] [PubMed] [Google Scholar]
- Dorr M. Meggers E. Metal complexes as structural templates for targeting proteins. Curr. Opin. Chem. Biol. 2014;19:76–81. doi: 10.1016/j.cbpa.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Zhang P. Sadler P. J. Redox-Active Metal Complexes for Anticancer Therapy. Eur. J. Inorg. Chem. 2017;2017(12):1541–1548. [Google Scholar]
- Graf N. Lippard S. J. Redox activation of metal-based prodrugs as a strategy for drug delivery. Adv. Drug Delivery Rev. 2012;64(11):993–1004. doi: 10.1016/j.addr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth U. Kowol C. R. Keppler B. K. Hartinger C. G. Berger W. Heffeter P. Anticancer activity of metal complexes: involvement of redox processes. Antioxid. Redox Signaling. 2011;15(4):1085–1127. doi: 10.1089/ars.2010.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer N. J. Salassa L. Sadler P. J. Photoactivated chemotherapy (PACT): the potential of excited-state d-block metals in medicine. Dalton Trans. 2009:10690–10701. doi: 10.1039/b917753a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjos K. D. Orvig C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014;114(8):4540–4563. doi: 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- Antonarakis E. S. Emadi A. Ruthenium-based chemotherapeutics: are they ready for prime time? Cancer Chemother. Pharmacol. 2010;66(1):1–9. doi: 10.1007/s00280-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker-Lakhai J. M. van den Bongard D. Pluim D. Beijnen J. H. Schellens J. H. M. A phase I and pharmacological study with imidazolium-trans-DMSO-imidazole-tetrachlororuthenate, a novel ruthenium anticancer agent. Clin. Cancer Res. 2004;10(11):3717–3727. doi: 10.1158/1078-0432.CCR-03-0746. [DOI] [PubMed] [Google Scholar]
- Knopf K. M. Murphy B. L. MacMillan S. N. Baskin J. M. Barr M. P. Boros E. Wilson J. J. In vitro anticancer activity and in vivo biodistribution of rhenium(I) tricarbonyl aqua complexes. J. Am. Chem. Soc. 2017;139(40):14302–14314. doi: 10.1021/jacs.7b08640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam K. Awuah S. G. Bruno P. M. Johnstone T. C. Wang F. Lin W. Zheng Y.-R. Page J. E. Hemann M. T. Lippard S. J. Necroptosis-Inducing Rhenium(V) Oxo Complexes. J. Am. Chem. Soc. 2015;137(8):2967–2974. doi: 10.1021/ja511978y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn J. M. Romero-Canelón I. Munro A. F. Fu Y. Pizarro A. M. Garnett M. J. McDermott U. Carragher N. O. Sadler P. J. Potent organo-osmium compound shifts metabolism in epithelial ovarian cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112(29):E3800–E3805. doi: 10.1073/pnas.1500925112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam K. Johnstone T. C. Bruno P. M. Lin W. Hemann M. T. Lippard S. J. Bidentate Ligands on Osmium(VI) Nitrido Complexes Control Intracellular Targeting and Cell Death Pathways. J. Am. Chem. Soc. 2013;135(38):14060–14063. doi: 10.1021/ja4075375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens R. T. Parkin S. Awuah S. G. Cancer cell-selective modulation of mitochondrial respiration and metabolism by potent organogold(III) dithiocarbamates. Chem. Sci. 2020;11:10465–10482. doi: 10.1039/d0sc03628e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gukathasan S. Parkin S. Awuah S. G. Cyclometalated Gold(III) Complexes Bearing DACH Ligands. Inorg. Chem. 2019;58(14):9326–9340. doi: 10.1021/acs.inorgchem.9b01031. [DOI] [PubMed] [Google Scholar]
- Kim J. H. Reeder E. Parkin S. Awuah S. G. Gold(I/III)-Phosphine Complexes as Potent Antiproliferative Agents. Sci. Rep. 2019;9(1):12335. doi: 10.1038/s41598-019-48584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott I. On the medicinal chemistry of gold complexes as anticancer drugs. Coord. Chem. Rev. 2009;253(11):1670–1681. [Google Scholar]
- Anvarhusein I. Sadler P. J. A carbon-13 nuclear magnetic resonance study of thiol-exchange reactions of gold(I) thiomalate (“Myocrisin”) including applications to cysteine derivatives. J. Chem. Soc., Dalton Trans. 1982:135–141. [Google Scholar]
- Zou T. Lum C. T. Lok C. N. Zhang J. J. Che C. M. Chemical biology of anticancer gold(III) and gold(I) complexes. Chem. Soc. Rev. 2015;44(24):8786–8801. doi: 10.1039/c5cs00132c. [DOI] [PubMed] [Google Scholar]
- Preiß S. Förster C. Otto S. Bauer M. Müller P. Hinderberger D. Hashemi Haeri H. Carella L. Heinze K. Structure and reactivity of a mononuclear gold(II) complex. Nat. Chem. 2017;9:1249. doi: 10.1038/nchem.2836. [DOI] [PubMed] [Google Scholar]
- Wu C.-Y. Horibe T. Jacobsen C. B. Toste F. D. Stable gold(III) catalysts by oxidative addition of a carbon-carbon bond. Nature. 2015;517(7535):449–454. doi: 10.1038/nature14104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand B. Spreckelmeyer S. Bodio E. Cocco F. Picquet M. Richard P. Le Gendre P. Orvig C. Cinellu M. A. Casini A. Exploring the potential of gold(III) cyclometallated compounds as cytotoxic agents: variations on the C^N theme. Dalton Trans. 2015;44(26):11911–11918. doi: 10.1039/c5dt01023c. [DOI] [PubMed] [Google Scholar]
- Carboni S. Zucca A. Stoccoro S. Maiore L. Arca M. Ortu F. Artner C. Keppler B. K. Meier-Menches S. M. Casini A. Cinellu M. A. New Variations on the Theme of Gold(III) C(wedge)N(wedge)N Cyclometalated Complexes as Anticancer Agents: Synthesis and Biological Characterization. Inorg. Chem. 2018;57(23):14852–14865. doi: 10.1021/acs.inorgchem.8b02604. [DOI] [PubMed] [Google Scholar]
- Marcon G. Carotti S. Coronnello M. Messori L. Mini E. Orioli P. Mazzei T. Cinellu M. A. Minghetti G. Gold(III) complexes with bipyridyl ligands: solution chemistry, cytotoxicity, and DNA binding properties. J. Med. Chem. 2002;45(8):1672–1677. doi: 10.1021/jm010997w. [DOI] [PubMed] [Google Scholar]
- Massai L. Cirri D. Michelucci E. Bartoli G. Guerri A. Cinellu M. A. Cocco F. Gabbiani C. Messori L. Organogold(III) compounds as experimental anticancer agents: chemical and biological profiles. BioMetals. 2016;29(5):863–872. doi: 10.1007/s10534-016-9957-x. [DOI] [PubMed] [Google Scholar]
- Messori L. Marcon G. Cinellu M. A. Coronnello M. Mini E. Gabbiani C. Orioli P. Solution chemistry and cytotoxic properties of novel organogold(III) compounds. Bioorg. Med. Chem. 2004;12(23):6039–6043. doi: 10.1016/j.bmc.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Mertens R. T. Parkin S. R. Awuah S. G. Synthesis and crystal structure of 1,3-bis-(4-hy-droxy-phen-yl)-1H-imidazole-3-ium chloride. Acta Crystallogr., Sect. E: Crystallogr. Commun. 2019;75(Pt 9):1311–1315. doi: 10.1107/S2056989019011058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H. Mertens R. T. Agarwal A. Parkin S. Berger G. Awuah S. G. Direct intramolecular carbon(sp(2))-nitrogen(sp(2)) reductive elimination from gold(III) Dalton Trans. 2019;48(18):6273–6282. doi: 10.1039/c8dt05155k. [DOI] [PubMed] [Google Scholar]
- Mertens R. T. Kim J. H. Jennings W. C. Parkin S. Awuah S. G. Revisiting the reactivity of tetrachloroauric acid with N,N-bidentate ligands: structural and spectroscopic insights. Dalton Trans. 2019;48(6):2093–2099. doi: 10.1039/c8dt04960b. [DOI] [PubMed] [Google Scholar]
- Hu D. Liu Y. Lai Y.-T. Tong K.-C. Fung Y.-M. Lok C.-N. Che C.-M. Anticancer Gold(III) Porphyrins Target Mitochondrial Chaperone Hsp60. Angew. Chem., Int. Ed. 2016;55(4):1387–1391. doi: 10.1002/anie.201509612. [DOI] [PubMed] [Google Scholar]
- Che C.-M. Sun R. W.-Y. Yu W.-Y. Ko C.-B. Zhu N. Sun H. Gold(III) porphyrins as a new class of anticancer drugs: cytotoxicity, DNA binding and induction of apoptosis in human cervix epitheloid cancer cells. Chem. Commun. 2003:1718–1719. doi: 10.1039/b303294a. [DOI] [PubMed] [Google Scholar]
- Lammer A. D. Cook M. E. Sessler J. L. Synthesis and anti-cancer activities of a water soluble gold(III) porphyrin. J. Porphyrins Phthalocyanines. 2015;19(1–3):398–403. doi: 10.1142/S1088424615500236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong K.-C. Hu D. Wan P.-K. Lok C.-N. Che C.-M. Anticancer Gold(III) Compounds with Porphyrin or N-heterocyclic Carbene Ligands. Front. Chem. 2020;8(919):587207. doi: 10.3389/fchem.2020.587207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubia I. Nguyen C. Diring S. Ali L. M. A. Larue L. Aoun R. Frochot C. Gary-Bobo M. Kobeissi M. Odobel F. Synthesis and Anticancer Activity of Gold Porphyrin Linked to Malonate Diamine Platinum Complexes. Inorg. Chem. 2019;58(18):12395–12406. doi: 10.1021/acs.inorgchem.9b01981. [DOI] [PubMed] [Google Scholar]
- Serratrice M. Cinellu M. A. Maiore L. Pilo M. Zucca A. Gabbiani C. Guerri A. Landini I. Nobili S. Mini E. Messori L. Synthesis, Structural Characterization, Solution Behavior, and in Vitro Antiproliferative Properties of a Series of Gold Complexes with 2-(2′-Pyridyl)benzimidazole as Ligand: Comparisons of Gold(III) versus Gold(I) and Mononuclear versus Binuclear Derivatives. Inorg. Chem. 2012;51(5):3161–3171. doi: 10.1021/ic202639t. [DOI] [PubMed] [Google Scholar]
- Stenger-Smith J. R. Mascharak P. K. Gold Drugs with {Au(PPh3)}+ Moiety: Advantages and Medicinal Applications. ChemMedChem. 2020;15(22):2136–2145. doi: 10.1002/cmdc.202000608. [DOI] [PubMed] [Google Scholar]
- Srinivasa Reddy T. Privér S. H. Rao V. V. Mirzadeh N. Bhargava S. K. Gold(I) and gold(III) phosphine complexes: synthesis, anticancer activities towards 2D and 3D cancer models, and apoptosis inducing properties. Dalton Trans. 2018;47(43):15312–15323. doi: 10.1039/c8dt01724g. [DOI] [PubMed] [Google Scholar]
- Cinellu M. A. Zucca A. Stoccoro S. Minghetti G. Manassero M. Sansoni M. Synthesis and characterization of gold(III) adducts and cyclometallated derivatives with 2-substituted pyridines. Crystal structure of [Au{NC5H4(CMe2C6H4)-2}Cl2] J. Chem. Soc., Dalton Trans. 1995:2865–2872. [Google Scholar]
- Berners-Price S. J. Mirabelli C. K. Johnson R. K. Mattern M. R. McCabe F. L. Faucette L. F. Sung C. M. Mong S. M. Sadler P. J. Crooke S. T. In vivo antitumor activity and in vitro cytotoxic properties of bis[1,2-bis(diphenylphosphino)ethane]gold(I) chloride. Cancer Res. 1986;46(11):5486–5493. [PubMed] [Google Scholar]
- Rackham O. Nichols S. J. Leedman P. J. Berners-Price S. J. Filipovska A. A gold(I) phosphine complex selectively induces apoptosis in breast cancer cells: implications for anticancer therapeutics targeted to mitochondria. Biochem. Pharmacol. 2007;74(7):992–1002. doi: 10.1016/j.bcp.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Berners-Price S. J. Girard G. R. Hill D. T. Sutton B. M. Jarrett P. S. Faucette L. F. Johnson R. K. Mirabelli C. K. Sadler P. J. Cytotoxicity and antitumor activity of some tetrahedral bis(diphosphino)gold(I) chelates. J. Med. Chem. 1990;33(5):1386–1392. doi: 10.1021/jm00167a017. [DOI] [PubMed] [Google Scholar]
- Buckley R. G. Elsome A. M. Fricker S. P. Henderson G. R. Theobald B. R. C. Parish R. V. Howe B. P. Kelland L. R. Antitumor Properties of Some 2-[(Dimethylamino)methyl]phenylgold(III) Complexes. J. Med. Chem. 1996;39(26):5208–5214. doi: 10.1021/jm9601563. [DOI] [PubMed] [Google Scholar]
- Liu H.-Q. Cheung T.-C. Peng S.-M. Che C.-M. Novel luminescent cyclometaiated and terpyridine gold(III) complexes and DNA binding studies. J. Chem. Soc., Chem. Commun. 1995;17:1787–1788. [Google Scholar]
- Carboni S. Zucca A. Stoccoro S. Maiore L. Arca M. Ortu F. Artner C. Keppler B. K. Meier-Menches S. M. Casini A. Cinellu M. A. New Variations on the Theme of Gold(III) C∧N∧N Cyclometalated Complexes as Anticancer Agents: Synthesis and Biological Characterization. Inorg. Chem. 2018;57(23):14852–14865. doi: 10.1021/acs.inorgchem.8b02604. [DOI] [PubMed] [Google Scholar]
- Frik M. Fernández-Gallardo J. Gonzalo O. Mangas-Sanjuan V. González-Alvarez M. Serrano del Valle A. Hu C. González-Alvarez I. Bermejo M. Marzo I. Contel M. Cyclometalated Iminophosphorane Gold(III) and Platinum(II) Complexes. A Highly Permeable Cationic Platinum(II) Compound with Promising Anticancer Properties. J. Med. Chem. 2015;58(15):5825–5841. doi: 10.1021/acs.jmedchem.5b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S. K. Zou T. Cao B. Lee P. Y. Fung Y. M. Hu D. Lok C. N. Che C. M. Cyclometalated Gold(III) Complexes Containing N-Heterocyclic Carbene Ligands Engage Multiple Anti-Cancer Molecular Targets. Angew. Chem. 2017;56(14):3892–3896. doi: 10.1002/anie.201612583. [DOI] [PubMed] [Google Scholar]
- Massai L. Cirri D. Michelucci E. Bartoli G. Guerri A. Cinellu M. A. Cocco F. Gabbiani C. Messori L. Organogold(III) compounds as experimental anticancer agents: chemical and biological profiles. BioMetals. 2016;29(5):863–872. doi: 10.1007/s10534-016-9957-x. [DOI] [PubMed] [Google Scholar]
- Kumar R. Nevado C. Cyclometalated Gold(III) Complexes: Synthesis, Reactivity, and Physicochemical Properties. Angew. Chem., Int. Ed. 2017;56(8):1994–2015. doi: 10.1002/anie.201607225. [DOI] [PubMed] [Google Scholar]
- Gukathasan S. Parkin S. Awuah S. G. Cyclometalated Gold(III) Complexes Bearing DACH Ligands. Inorg. Chem. 2019;58(14):9326–9340. doi: 10.1021/acs.inorgchem.9b01031. [DOI] [PubMed] [Google Scholar]
- Dennis E. K. Kim J. H. Parkin S. Awuah S. G. Garneau-Tsodikova S. Distorted Gold(I)-Phosphine Complexes as Antifungal Agents. J. Med. Chem. 2020;63(5):2455–2469. doi: 10.1021/acs.jmedchem.9b01436. [DOI] [PubMed] [Google Scholar]
- Jia D. Lu M. Jung K. H. Park J. H. Yu L. Onuchic J. N. Kaipparettu B. A. Levine H. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 2019;116(9):3909–3918. doi: 10.1073/pnas.1816391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H. Vithayathil S. Kumar S. Sung P.-L. Dobrolecki L. E. Putluri V. Bhat V. B. Bhowmik S. K. Gupta V. Arora K. Wu D. Tsouko E. Zhang Y. Maity S. Donti T. R. Graham B. H. Frigo D. E. Coarfa C. Yotnda P. Putluri N. Sreekumar A. Lewis M. T. Creighton C. J. Wong L.-J. C. Kaipparettu B. A. Fatty Acid Oxidation-Driven Src Links Mitochondrial Energy Reprogramming and Oncogenic Properties in Triple-Negative Breast Cancer. Cell Rep. 2016;14(9):2154–2165. doi: 10.1016/j.celrep.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu V. S. O'Connell J. T. Gonzalez Herrera K. N. Wikman H. Pantel K. Haigis M. C. de Carvalho F. M. Damascena A. Domingos Chinen L. T. Rocha R. M. Asara J. M. Kalluri R. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porporato P. E. Payen V. L. Pérez-Escuredo J. De Saedeleer C. J. Danhier P. Copetti T. Dhup S. Tardy M. Vazeille T. Bouzin C. Feron O. Michiels C. Gallez B. Sonveaux P. A Mitochondrial Switch Promotes Tumor Metastasis. Cell Rep. 2014;8(3):754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- Camarda R. Zhou A. Y. Kohnz R. A. Balakrishnan S. Mahieu C. Anderton B. Eyob H. Kajimura S. Tward A. Krings G. Nomura D. K. Goga A. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 2016;22(4):427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg S. E. Chandel N. S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11(1):9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Yesilkanal A. E. Wynne J. P. Frankenberger C. Liu J. Yan J. Elbaz M. Rabe D. C. Rustandy F. D. Tiwari P. Grossman E. A. Hart P. C. Kang C. Sanderson S. M. Andrade J. Nomura D. K. Bonini M. G. Locasale J. W. Rosner M. R. Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature. 2019;568(7751):254–258. doi: 10.1038/s41586-019-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby G. Donnelly L. A. Donnan P. T. Alessi D. R. Morris A. D. Evans J. M. M. New users of metformin are at low risk of incident cancer. A cohort study among people with type 2 diabetes. Diabetes Care. 2009;32(9):1620–1625. doi: 10.2337/dc08-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton W. W. Weinberg S. E. Hamanaka R. B. Soberanes S. Sullivan L. B. Anso E. Glasauer A. Dufour E. Mutlu G. M. Budigner G. R. S. Chandel N. S. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib A. Mathew G. Reczek C. R. Watrud K. Ambrico A. Herzka T. Salas I. C. Lee M. F. El-Amine N. Zheng W. Di Francesco M. E. Marszalek J. R. Pappin D. J. Chandel N. S. Trotman L. C. Mitochondrial Complex I Inhibitors Expose a Vulnerability for Selective Killing of Pten-Null Cells. Cell Rep. 2018;23(1):58–67. doi: 10.1016/j.celrep.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J. R. Sun Y. Protopopova M. Gera S. Bandi M. Bristow C. McAfoos T. Morlacchi P. Ackroyd J. Agip A.-N. A. Al-Atrash G. Asara J. Bardenhagen J. Carrillo C. C. Carroll C. Chang E. Ciurea S. Cross J. B. Czako B. Deem A. Daver N. de Groot J. F. Dong J.-W. Feng N. Gao G. Gay J. Do M. G. Greer J. Giuliani V. Han J. Han L. Henry V. K. Hirst J. Huang S. Jiang Y. Kang Z. Khor T. Konoplev S. Lin Y.-H. Liu G. Lodi A. Lofton T. Ma H. Mahendra M. Matre P. Mullinax R. Peoples M. Petrocchi A. Rodriguez-Canale J. Serreli R. Shi T. Smith M. Tabe Y. Theroff J. Tiziani S. Xu Q. Zhang Q. Muller F. DePinho R. A. Toniatti C. Draetta G. F. Heffernan T. P. Konopleva M. Jones P. Di Francesco M. E. Marszalek J. R. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018;24(7):1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- Liu X. Romero I. L. Litchfield L. M. Lengyel E. Locasale J. W. Metformin Targets Central Carbon Metabolism and Reveals Mitochondrial Requirements in Human Cancers. Cell Metab. 2016;24(5):728–739. doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord S. R. Cheng W.-C. Liu D. Gaude E. Haider S. Metcalf T. Patel N. Teoh E. J. Gleeson F. Bradley K. Wigfield S. Zois C. McGowan D. R. Ah-See M.-L. Thompson A. M. Sharma A. Bidaut L. Pollak M. Roy P. G. Karpe F. James T. English R. Adams R. F. Campo L. Ayers L. Snell C. Roxanis I. Frezza C. Fenwick J. D. Buffa F. M. Harris A. L. Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer. Cell Metab. 2018;28(5):679–688. doi: 10.1016/j.cmet.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Lim S. K. Liang Q. Iyer S. V. Wang H. Y. Wang Z. Xie X. Sun D. Chen Y. J. Tabar V. Gutin P. Williams N. De Brabander J. K. Parada L. F. Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature. 2019;567(7748):341–346. doi: 10.1038/s41586-019-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull K. D. Shoemaker R. H. Hodes L. Monks A. Scudiero D. A. Rubinstein L. Plowman J. Boyd M. R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989;81(14):1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Shoemaker R. H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Sato Y. Furumichi M. Morishima K. Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2018;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.