Abstract
Human O-GlcNAcase (hOGA) is the unique enzyme responsible for the hydrolysis of the O-linked β-N-acetyl glucosamine (O-GlcNAc) modification, an essential protein glycosylation event that modulates the function of numerous cellular proteins in response to nutrients and stress. Here we report crystal structures of a truncated hOGA, which comprises the catalytic and stalk domains, in apo form, in complex with an inhibitor, and in complex with a glycopeptide substrate. We found that hOGA forms an unusual arm-in-arm homodimer in which the catalytic domain of one monomer is covered by the stalk domain of the sister monomer to create a substrate-binding cleft. Notably, the residues on the cleft surface afford extensive interactions with the peptide substrate in a recognition mode that is distinct from that of its bacterial homologs. These structures represent the first model of eukaryotic enzymes in the glycoside hydrolase 84 (GH84) family and provide a crucial starting point for understanding the substrate specificity of hOGA, which regulates a broad range of biological and pathological processes.
Thousands of intracellular proteins are modified by the monosaccharide O-linked β-N-acetyl glucosamine (O-GlcNAc), in a process termed O-GlcNAcylation1–3 (Fig. 1a). This nutrient-responsive modification resembles protein phosphorylation and dynamically regulates various biological processes, such as signal transduction4,5, gene expression6,7, cell cycle8,9, and proteasomal degradation10,11. Aberrant protein O-GlcNAcylation has been detected in many diseases including type 2 diabetes12,13, Alzheimer’s disease14,15, and nearly every type of cancer16–18. Unlike a large family of kinases and phosphatases that are dedicated to regulating protein phosphorylation, only a single pair of human enzymes performs the reversible O-GlcNAcylation event. This pair consists of an O-GlcNAc transferase19 (OGT) that installs the O-GlcNAc modification on serine and threonine residues and an O-GlcNAcase20 (OGA) that removes it (Fig. 1a). To date, a large number of O-GlcNAcylation sites have been mapped, but no consensus sequence can be clearly defined in the substrate proteins3,21. Hence, a central question of great interest is how OGT and OGA recognize numerous protein substrates and regulate the O-GlcNAcylation levels in a substrate- and site-specific manner. The recently reported crystal structure of human OGT and its complexes with peptide substrates provided the first insights into the molecular basis of OGT–substrate recognition8,21–23. In contrast, the structure of human OGA has long been sought by the scientific community without success, which has been a huge impediment to understanding the molecular mechanism underlying the substrate specificity of OGA.
Figure 1.
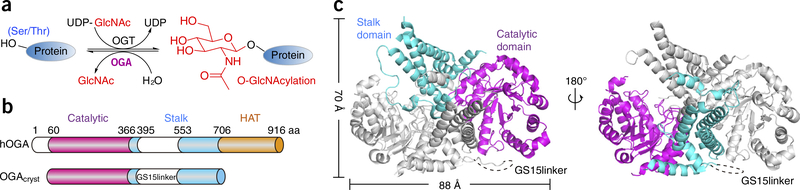
Overall structure of OGA and the construct used. (a) Schematic of the reversible O-GlcNAcylation reaction catalyzed by OGT and OGA. (b) Schematic of the domain architecture of the hOGA and OGAcryst constructs used in this study. Catalytic domain (residues 60–366), stalk domain (residues 367–706), HAT domain (residues 707–916), and low-complexity regions are highlighted in magenta, cyan, orange and white, respectively. GS15linker represents amino acid (aa) residues 401–552, which were replaced by a 15-residue glycine and serine linker in OGAcryst. (c) Ribbon representation of the OGAcryst homodimer in two different views. The OGA-α monomer is colored gray; the catalytic domain and stalk domain of OGA-β are colored magenta and cyan, respectively. GS15linker is shown as a dashed line.
Human OGA (hOGA) is a 103-kDa nucleocytoplasmic protein that consists of three distinct regions24–26 (Fig. 1b): an N-terminal catalytic domain (residues 60–366) that shows sequence similarity with glycoside hydrolases of the GH84 family and is responsible for O-GlcNAc removal, a stalk domain (residues 367–706), and a C-terminal pseudo–histone acetyltransferase (HAT) domain (residues 707–916) that possesses sequence homology to HAT enzymes but lacks the key residues for the binding of acetyl-coenzyme A (acetyl-CoA)27–29, and hence, its precise role needs to be clarified. A short isoform of hOGA (sOGA), which is encoded by an alternatively spliced transcript and is missing the HAT domain, was discovered on the surface of nascent lipid droplets30. It shows lower hydrolase activity31,32 and was suggested to modulate proteasome-mediated processes30. Although hOGA can be readily expressed and purified as a recombinant protein for biochemical assays, previous studies have not led to a crystal structure. Molecular insights into the catalytic domain of hOGA were obtained from mechanistic studies on compound analogs33–35 and from structures of its bacterial homologs—Bacteroides thetaiotaomicron hexosaminidase (BtGH84)36–38, Clostridium perfringens NagJ (CpOGA)39,40, Oceanicola granulosus glycosidase (OgOGA)24, and Thermobaculum terrenum glycoside hydrolase (TtOGA)41. Results from these studies supported the idea that hOGA performs glycoside hydrolysis with retention of the anomeric configuration via a substrate-assisted mechanism (Supplementary Fig. 1). In addition, this mechanistic and structural information has guided the rational design of a few potent and selective OGA inhibitors42,43 (for example, thiamet-G37 and GlcNAcstatin44) for biological applications.
Despite this progress, the molecular mechanism underlying the substrate specificity of hOGA is still elusive. Although the bacterial homologs had in vitro OGA activity on O-GlcNAcylated proteins from the cell lysates of human cell lines, it remains unclear whether these bacterial homologs are physiologically relevant OGAs, as no dynamic O-GlcNAcylation has been detected in bacteria24,36,37,39,41. Furthermore, although a number of bacterial GH84 proteins bear considerable similarity to the active site of hOGA, the bacterial homologs lack similarity in the stalk domain or the C-terminal HAT domain, which may also contribute to substrate binding and catalysis. Consistent with this, the N-terminal catalytic domain of hOGA alone was found to be 1,000-fold less active than the full-length protein32,45, and sOGA also has substantially lower activity than full-length hOGA31,32. Therefore, the structure of hOGA will be crucial to elucidating the essential properties of this human enzyme. Here we present the first crystal structures of a truncated hOGA, comprising the catalytic and stalk domains, by itself and in complexes with either an inhibitor or a glycopeptide. These new structures reveal a unique principle of hOGA in substrate recognition that is distinct from that of its bacterial homologs.
RESULTS
The structure of hOGA is an unusual arm-in-arm homodimer
Structural predictions illustrated that hOGA contains two low-complexity regions, one located at the N terminus, and the other inserted in the stalk domain (Fig. 1b). These two regions are predicted to be flexible and unstructured, which poses a substantial challenge to the production of diffractable protein crystals. To overcome this issue, a series of truncated constructs were prepared, and these were used to define boundaries and optimize protein expression. After an extensive number of trials to identify a truncated form of hOGA that could be used for protein crystallization, we found a construct (OGAcryst) comprising residues 60–704 of hOGA, and in which the unstructured insert residues 401–552 were replaced by a 15-residue glycine-serine (GS) linker (GS15linker) (Fig. 1b). The OGAcryst protein was stable and showed activity similar to that of full-length hOGA (Supplementary Fig. 2), suggesting that OGAcryst contained the essential functional features of hOGA. Therefore, this protein was applied for crystallization, and the structure was determined at 2.5-Å resolution (Table 1).
Table 1.
Data collection and refinement statistics
| OGAcryst (PDB 5TKE) | OGAcryst–thiamet-G (PDB 5UN9) | OGAcryst (D175N)–p53 (PDB 5UN8) | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 82.0, 96.6, 89.3 | 82.5, 96.1, 89.5 | 89.9, 95.4, 149.3 |
| α, β, γ (°) | 90.0, 115.0, 90.0 | 90.0, 115.0, 90.0 | 90.0, 96.9, 90.0 |
| Resolution (Å) | 50.0–2.5 (2.59–2.50)a | 30.0–2.5 (2.54–2.50) | 50.0–2.1 (2.18–2.14) |
| Rmerge | 8.7 (72.2) | 6.3 (58.3) | 8.2 (72.0) |
| I/σ(I) | 19.5 (2.0) | 23.0 (2.1) | 18.4 (2.0) |
| CC1/2 | 99.9 (79.1) | 99.9 (78.5) | 99.8 (78.1) |
| Completeness (%) | 100.0 (100.0) | 99.5 (95.1) | 99.9 (100.0) |
| Redundancy | 6.4 (6.2) | 5.3 (5.2) | 4.9 (4.5) |
|
Refinement Resolution (Å) |
50.0–2.5 | 30.0–2.5 | 45.0–2.1 |
| No. reflections | 44,255 | 44,310 | 139,265 |
| Rwork / Rfree | 18.2 / 23.7 | 21.0 / 26.0 | 18.4 / 22.9 |
| No. atoms Protein |
6,921 | 6,922 | 14,015 |
| Ligand/peptide | 32 | 305 | |
| Water | 396 | 238 | 1,247 |
| B factors | 43.40 | 43.38 | 30.02 |
| Protein | 43.41 | 43.49 | 29.28 |
| Ligand/peptide | 29.73 | 29.07 | |
| Water | 42.68 | 40.93 | 38.56 |
| R.m.s. deviation Bond length (Å) |
0.008 | 0.011 | 0.009 |
| Bond angle (°) | 0.970 | 1.211 | 1.016 |
Each structure was determined from one crystal.
Values in parentheses are for highest-resolution shell.
The overall structure of OGAcryst in the asymmetric unit formed an unusual arm-in-arm homodimer that resembled a butterfly (Fig. 1c). Each of the two monomers (OGA-α and OGA-β) contains a catalytic domain (residues 60–366) and a stalk domain (residues 367–400 and 553–704). The catalytic domain consisted of a classic (β/α)8-barrel (eight-stranded parallel β-sheet core mainly surrounded by eight α-helices), which formed a deep pocket for GlcNAc binding and hydrolysis (Supplementary Fig. 3). The stalk domain consisted of six α-helices: the first two (helices α12 and α13) were transitioned from the catalytic domain to the stalk region, the next three (helices α14–α16) were bundled together as a core, and the last helix (α17) was linked by a flexible loop and extended far away from the bundle core as an arm to interact with the stalk bundle of the sister monomer. Although the loop region in OGA-α was not visible due to high mobility of this region in the crystal, the corresponding loop in OGA-β displayed strong electron density and could be clearly defined. Superposition of the two monomers demonstrated that their catalytic domains were nearly identical with a root-mean-squared deviation (r.m.s. deviation) value of 0.7 Å over the Cα atoms (Supplementary Fig. 3). However, there was a slight variation between the two stalk domains (r.m.s. deviation value of 1.1 Å). As compared to the previously reported structures of the bacterial homologs BtGH84 (PDB 2CHO)36, CpOGA (PDB 2CBJ)39, OgOGA (PDB 2XSA)24, and TtOGA (PDB 5DIY)41, the conformation of hOGA was markedly different in the stalk domain, which has only 12–22% sequence similarity (Supplementary Fig. 4). The bundle core of bacterial stalk domains typically consists of four or five α-helices. In contrast, the stalk bundle of hOGA comprised only three α-helices (Supplementary Fig. 4c). Furthermore, the arm region extending from the bundle core was not found in any of the reported structures for the bacterial homologs, which represents a unique feature of hOGA. Deletion of this arm region (residues 665–704) resulted in a loss of OGAcryst stability as a recombinantly expressed protein. The stalk domain of hOGA is believed to have a pivotal role for the tight association and stabilization of the dimeric protein, and it is believed to contribute to substrate recognition (see below).
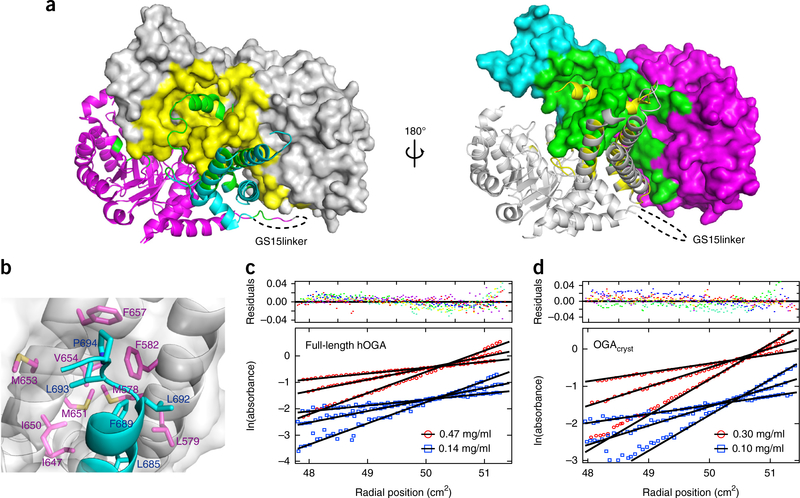
The dimerization of OGAcryst is remarkably strong, as the interface buries approximately 6,778 Å2 of the total 33,400-Å2 surface area of a monomer (20.3% of monomer surface, as calculated by the PISA server46). In addition, the residues involved in dimerization spread over the top surface of the catalytic domain, the stalk bundle, and the extended loop that links the arm region (Fig. 2a). A series of 15 residues in OGA-β made extensive hydrogen bonds with OGA-α, and another set of residues (R71, H571, D639, D675, E688, and R691) in OGA-β formed salt bridges with OGA-α residues (D639, E688, R71, K253, H571, D696, and D700, respectively) (Supplementary Fig. 5 and Supplementary Table 1). Furthermore, the side chains of another five residues (L685, F689, L692, L693, and P694) in the arm region (helix α17) contributed substantial hydrophobic interactions in the nonpolar groove that was generated by the stalk bundle of the sister monomer, such as with the M578, L579, and F582 residues in helix α14, as well as with the I647, I650, M651, M653, V654, and F657 residues in helix α16 (Fig. 2b). These massive networks of hydrogen bonds, salt bridges, and hydrophobic interactions greatly strengthen the dimerization of OGAcryst, which is expected to be critical for its activity, stability, and substrate recognition (see below).
Figure 2.
Dimerization of OGA. (a) Two different views of the dimerization interface in the OGAcryst structure. Left, OGA-α is shown in gray, and OGA-β is shown as a ribbon structure, with the catalytic and stalk domains colored magenta and cyan, respectively. Right, OGA-α is shown in the gray ribbon structure, and OGA-β is shown with the catalytic and stalk domains colored magenta and cyan, respectively. The dimerization interface of OGA-α (yellow) and OGA-β (green) are highlighted. (b) Hydrophobic interactions between the arm region of OGA-β and the nonpolar stalk bundle of OGA-α that contribute to the protein dimerization. OGA-α is shown in surface representation, and the residues involved in the hydrophobic interactions are represented in magenta sticks. OGA-β is shown as a ribbon structure, in which the hydrophobic residues are displayed in cyan sticks. (c) Sedimentation equilibrium results demonstrating that full-length hOGA is a dimer in solution (residuals fit is also shown (top); colors represent residuals for individual data sets). The data presented are the initial loading concentrations of 0.47 mg/ml (red) and 0.14 mg/ml (blue) of full-length hOGA at three different rotor speeds (4,000, 5,600 and 7,600 r.p.m.). (d) Sedimentation equilibrium results demonstrating that OGAcryst is a dimer in solution (residuals fit is also shown (top); colors represent residuals for individual data sets). The data presented are the initial loading concentrations of 0.30 mg/ml (red) and 0.10 mg/ml (blue) of OGAcryst at three different rotor speeds (6,000, 8,800 and 12,000 r.p.m.).
In the structure of OGAcryst, the GS15linker (which replaced the unstructured insert region) was pointing away from the dimerization interface (Figs. 1c and 2a), suggesting that the insert region is unlikely to participate in hOGA dimerization. We next performed analytical ultracentrifugation sedimentation equilibrium experiments to examine the association states of full-length hOGA and OGAcryst in solution. Results are presented as ln(absorbance) versus the square of the distance from the center of rotation (Fig. 2c,d). In such a plot, a single species of protein appears as a straight line with a slope proportional to the average molecular weight. The results from analyses of both proteins at varied loading concentrations and speed rates were consistent with a single species of protein. Global fitting of all of the data for each protein matched to a dimeric species, with no discernible degree of dissociation under the concentration range used in these studies (essentially 0 to >1 mg/ml). These results were consistent with the structural data and demonstrated that full-length hOGA, as well as OGAcryst, exist as stable dimers. More notably, the residues on the dimerization interface seem to be highly conserved throughout the evolution in eukaryotes but not in prokaryotes (Supplementary Note 1); therefore, the dimeric structure of hOGA may represent a prevailing model across eukaryotic species.
The structure of hOGA in complex with an inhibitor
To gain detailed insights into the active site of hOGA and to aid the future development of inhibitors with improved properties against this human enzyme, we cocrystallized OGAcryst with thiamet-G, a transition-state mimic of GlcNAc during OGA-catalyzed hydrolysis37 (Supplementary Fig. 1). This highly potent inhibitor (Ki = 21 nM for hOGA) shows 37,000-fold higher selectivity for hOGA than for human lysosomal β-hexosaminidase37. In addition, it can cross the blood–brain barrier to modulate the phosphorylation and O-GlcNAcylation levels of tau in vivo37,47,48. Thus, the crystal structure of hOGA–thiamet-G could guide the rational design of even more potent and selective inhibitors with potential therapeutic uses for the treatment of Alzheimer’s disease.
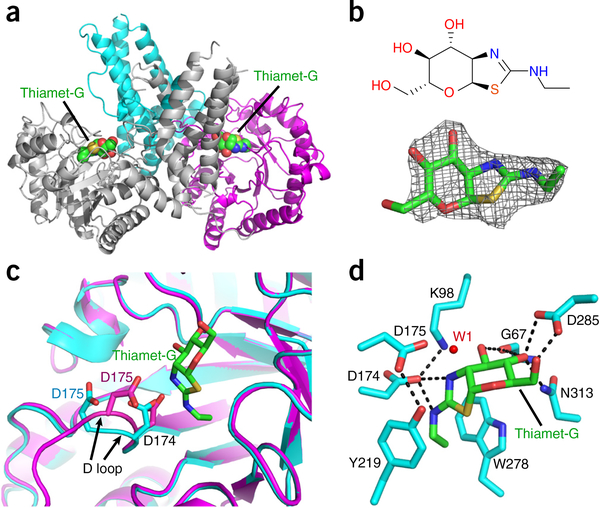
The structure of the OGAcryst–thiamet-G complex was obtained to a resolution of 2.5 Å (Fig. 3a and Table 1). The Fo – Fc difference map showed clear electron density for thiamet-G binding in the active site pocket (Fig. 3b). This inhibitor was refined at full occupancy with average B values of 29.7 Å2 (Table 1), suggesting that its binding position was well ordered in the OGA catalytic core. The overall structure of this complex was similar to the dimeric apo form of OGAcryst, but we were able to identify noticeable differences in several regions. Of note, the D loop (residues 172–182) in both monomers moved considerably toward thiamet-G after inhibitor binding (Fig. 3c). This loop was located on the top surface of the sugar-binding pocket and harbored a pair of catalytically essential aspartate residues (D174 and D175; Supplementary Fig. 6a). Previous studies with OGA mutants45,49, sugar analogs33–35, and the structures of bacterial homologs24,36–41 have produced fundamental insights into the substrate-assisted catalytic mechanism of hOGA (Supplementary Fig. 1). In the first stage, the catalytic residue D174 functions as a general base to polarize the 2-acetamido group of GlcNAc to attack the anomeric center, whereas D175 functions as a general acid to facilitate the departure of the leaving group and generate a bicyclic oxazoline intermediate45. In the second stage, deprotonated D175 activates a water molecule to attack the anomeric center of the oxazoline intermediate and promote ring opening to release the acetamido group. Because thiamet-G resembles the transition state of GlcNAc (Supplementary Fig. 1), considerable movement of the D loop in the OGAcryst–thiamet-G complex reflected tightening of the enzyme active site to stabilize the transition-state intermediate during catalysis. In particular, the aspartate side chain of D174 formed favorable electrostatic interactions with both nitrogen atoms of thiamet-G (Fig. 3d). The deprotonated state of D174 was further stabilized by the side chain amine of K98. Substitution of this lysine with alanine markedly impaired the activity of hOGA, suggesting that this residue has an essential role (Supplementary Fig. 6a). During the binding process, the other catalytic residue, D175, moved 2.8 Å closer to the anomeric center of thiamet-G. An ordered water molecule (W1 in Fig. 3d) in close proximity to the D175 side chain (2.5 Å) and the anomeric center (3.7 Å) was well positioned for the second stage of hydrolysis. Notably, the movement of D175 resulted in the formation of a new hydrogen bond with the hydroxyl group of Y219 (Fig. 3d), which is expected to stabilize the activated conformation of D175 and mediate its rapid acid and base switch during the process of glycoside hydrolysis. Indeed, a Y219F substitution dramatically reduced the catalytic efficiency of the mutant hOGA (Supplementary Fig. 6a). Consistent with the stabilized transition state, the D loop could be clearly defined in the OGAcryst–thiamet-G complex (and in the OGAcryst–p53 complex; see below), whereas the corresponding loop was highly mobile in the crystal of the apo form and could be built only in part (missing electron density for residues 176–181).
Figure 3.
Structure of OGAcryst in complex with thiamet-G. (a) Ribbon representation of the OGAcryst–thiamet-G complex, in which the thiamet-G molecules are highlighted in spheres. (b) Chemical structure (top) and Fo – Fc density map (contoured at 3σ) (bottom) of thiamet-G. (c) Superposition of OGA-α structures from apo-OGAcryst and the OGAcryst–thiamet-G complex demonstrating the marked movement of the D loop after inhibitor binding. Thiamet-G is shown in green sticks. The catalytic residues (D174 and D175) from apo OGAcryst and OGAcryst–thiamet-G are colored cyan and magenta, respectively. The movement of the D loop is highlighted using black arrows. (d) A close-up view of thiamet-G bound in the active site of OGAcryst. Hydrogen bonds are shown in dashed lines; enzyme residues are shown in cyan sticks; thiamet-G is shown in green sticks; and the catalytic water molecule is displayed as a red dot (W1).
Notably, in the OGAcryst–inhibitor complex, the pyranose ring of thiamet-G adopted a 4C1 chair conformation, supporting a late transition state that is found along the reaction pathway for oxazoline intermediate formation38. An extensive number of hydrogen bonds tightly anchored the pyranose ring of thiamet-G in the active site (Fig. 3d). For instance, the O3 hydroxyl of thiamet-G donated a hydrogen bond to the main chain carbonyl of G67, while both of the O4 and O6 hydroxyl groups were engaged in a bidentate hydrogen bond with the aspartate of D285. The essential role of this aspartate side chain was evidenced by the nearly complete loss of activity observed for the D285A mutant of hOGA (Supplementary Fig. 6a). In the meantime, the thiazoline ring of thiamet-G was tightly sandwiched by the aromatic side chains of Y219 and W278, two highly conserved residues that bordered the sugar-binding pocket of hOGA (Fig. 3d). Taken together, the OGAcryst–thiamet-G complex provides direct structural evidence to support the substrate-assisted double-displacement mechanism of hOGA. Furthermore, this study generates new insights into ancillary residues that contribute to the catalytic mechanism and will facilitate future development of improved inhibitors to target this human enzyme.
Dimeric hOGA exploits a new substrate-recognition mode
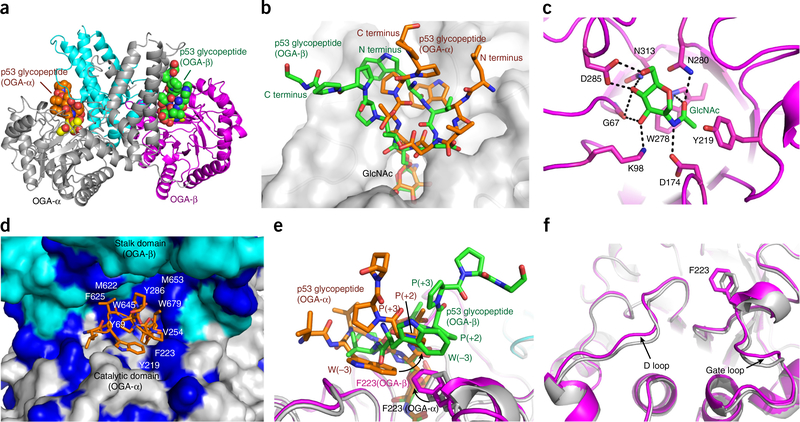
In the dimeric OGAcryst structures, the stalk domain covered the catalytic domain of the sister monomer, creating a potential substrate-binding cleft. We speculated that the residues located on the cleft surface might regulate the substrate recognition of hOGA. To investigate this, we solved the structure of OGAcryst in complex with an O-GlcNAcylated peptide substrate (Ac-QLWVDS(O-GlcNAc)TPPPG) derived from the tumor suppressor protein p53 (OGAcryst–p53 complex). To avoid O-GlcNAc hydrolysis during crystallization, a catalytically impaired mutant OGAcryst (D175N) (Supplementary Fig. 6a) that retains the ability for substrate binding24,39 was used. The structure of OGAcryst–p53 complex was determined to 2.1 Å (Fig. 4a), and we found that each asymmetric unit comprised a pair of OGAcryst dimers (Table 1). Superposition of the two OGAcryst molecules in each dimer gave r.m.s. deviation values of 0.7 Å and 1.1 Å for the catalytic domain and stalk domain, respectively, which was similar to those calculated from the structure of the apo form of OGAcryst. Notably, unambiguous electron density of the O-GlcNAcylated p53 peptide was identified in the substrate-binding cleft (Supplementary Fig. 7a). Unexpectedly, however, the peptide substrates in the two sister monomers displayed distinct binding conformations (Fig. 4b). The peptide-binding conformers were refined at full occupancy with B values of 27.6 Å2 and 30.4 Å2 for the peptides in OGA-α and OGA-β, respectively, indicating that both peptides were well ordered. This unexpected observation seems unlikely to have arisen from a random binding process of the glycopeptides during crystal soaking. In fact, we solved three OGAcryst–p53 complexes from independent soaking experiments. The glycopeptides in all of these complexes showed consistent differences in a dimer (Supplementary Fig. 7b), suggesting that the different binding conformations were strongly preferred.
Figure 4.
Structure of the OGAcryst–p53 complex. (a) Ribbon representation of OGAcryst–p53 homodimer, in which the p53 glycopeptides are highlighted in spheres. OGA-α is in gray; the catalytic domain and stalk domain of OGA-β are in magenta and cyan, respectively. (b) Superposition of the p53 glycopeptides from the two monomers of OGAcryst–p53 complex illustrates the differential peptide binding conformations, even though the GlcNAc moieties are bound nearly identically. (c) A close-up view of the GlcNAc bound in the active site of OGA. GlcNAc is displayed in green sticks, neighboring active site residues are represented in magenta sticks, and hydrogen bonds are shown in black dashed lines. (d) p53 glycopeptide (orange sticks) bound in the substrate-binding cleft (surface representation) that comprises the catalytic domain of OGA-α (gray) and the stalk domain of OGA-β (cyan). The hydrophobic residues on the inner surface of the cleft are highlighted in dark blue. (e) Superposition of the two sister monomers of the OGAcryst–p53 complex showing differential orientations of the F223 residues and their hydrophobic interactions with the corresponding p53 glycopeptide. Black arrows highlight the side chain differences of the F223 residues and the tryptophan (−3 subsite) of the peptides. (f) A close-up view of the conformational variations observed at the D loop and gate loop in the two sister monomers of the OGAcryst–p53 complex. OGA-α and OGA-β are colored in gray and magenta, respectively. The F223 residues in the two monomers are highlighted in sticks.
In spite of the apparent difference, we were able to determine a number of common features of the glycopeptides, which provided key insights into the structural elements of hOGA that contribute to substrate recognition. An evident feature was the extensive number of hydrogen bonds in the catalytic core of hOGA that tightly anchored the GlcNAc residues in a nearly identical binding conformation (r.m.s. deviation value of 0.12 Å calculated from the two sister monomers) (Fig. 4c). This observation, along with the mutation data from our study (Supplementary Fig. 6a) and previous studies36,39,45, suggests that the binding of GlcNAc is an essential element of hOGA for substrate recognition. However, the most unusual feature observed was the inner surface of the substrate-binding cleft, which comprises primarily hydrophobic residues, such as Y69, Y219, F223, and V254 from the top part of the catalytic domain, and M622, F625, W645, M653, and W679 from the stalk domain of the sister monomer (Fig. 4d). These hydrophobic residues are conserved in most eukaryotes (Supplementary Note 1), underscoring their functional importance. As an example, in the binding clefts of both sister monomers, the aromatic group of F223 made a T-shaped π-stacking interaction with the tryptophan side chain (−3 subsite) of the glycopeptide (Fig. 4e). Moreover, W679 from the stalk domain further stabilized the secondary structure of the glycopeptide via interactions with the side chain of threonine (+1 subsite) and the backbone carbonyl of the glycosylating serine (0 subsite) of the peptide (Supplementary Fig. 7c). Alanine substitution of F223 or W679 on the cleft surface markedly diminished the binding affinity of hOGA toward the glycopeptide (Supplementary Fig. 6b), emphasizing the critical contribution of these cleft surface residues for substrate recognition. More notably, substitution of the tryptophan residue (−3 subsite) of the glycopeptide to alanine substantially impaired the binding toward hOGA (Supplementary Fig. 6b). This result strongly suggests that hOGA enables recognition of specific features of substrate peptides.
In an effort to provide molecular insights into the differential binding conformations of the glycopeptides in the two sister monomers (OGA-α and OGA-β), noticeable structural variations were detected on the D loop and gate loop (Fig. 4f), which are important components of the substrate-binding cleft. Particularly, the F223 side chain in OGA-β was oriented considerably differently from that in OGA-α of the OGAcryst–p53 complex, whereas the rest of the catalytic domain remained nearly the same. Notably, the distinct orientation of F223 was only observed in OGA-β of the OGAcryst–p53 complex, but not in the apo OGAcryst or OGAcryst–thiamet-G complex. It is reasonable to infer that the conformational difference of the glycopeptides in the two monomers is associated with the unique orientation of this residue as it affords essential hydrophobic interactions with the peptide substrates (Fig. 4e). Because F223 and tryptophan (−3 subsite) offered additional hydrophobic interactions with the +2 and +3 proline residues on the other terminus of the peptide, this initial difference may subsequently induce varied binding conformation of the entire peptide. The distinct binding conformation of each glycopeptide could further be stabilized via extensive interactions with residues on the inner surface of the substrate-binding cleft (Supplementary Fig. 7c,d), revealing a new mechanism for hOGA to achieve substrate specificity. We also note that crystal packing could have potentially contributed to the stabilization of the p53 glycopeptide in OGA-β but not in OGA-α. In contrast to the p53 glycopeptide from OGA-α, which is solvent exposable and distant from the symmetric molecule (Supplementary Fig. 7e), the p53 glycopeptide from OGA-β was in proximity to the stalk domain of another OGA-β from the symmetric unit, which could potentially stabilize the differential binding conformation (Supplementary Fig. 7f).
DISCUSSION
Here we report the first crystal structures of hOGA in apo form, in complex with an inhibitor, and in complex with a glycopeptide substrate. We discovered that hOGA is an unusual arm-in-arm homodimer. Replacement of the flexible insert region in the OGAcryst construct seemed to have no significant impact on the dimerization (Fig. 2c,d), catalysis (Supplementary Fig. 2) or peptide substrate binding (Supplementary Fig. 6b), indicating that the OGAcryst protein retains essential functional properties of hOGA. Of note, the residues on the dimerization interface are highly conserved in eukaryotes but not in prokaryotes (Supplementary Note 1). Although a previous study reported that bacterial BtGH84 was crystallized as a dimer36 and another study suggested that OgOGA might have been crystallized as a domain-swapped dimer24, disorder in the latter crystal precluded a definitive determination, and in both cases the dimerization in solution was not validated. Therefore, we propose that dimerization is a featured mechanism of the eukaryotic OGAs to regulate substrate recognition.
OGA is the sole enzyme responsible for O-GlcNAc cleavage from a broad range of glycosylated human proteins; however, how OGA recognizes numerous substrates remains largely unknown, especially considering the lack of a well-defined recognition motif in the substrate proteins. As far as we know, there have been only two previous reports investigating the substrate specificity of hOGA, and they essentially reached opposite conclusions. One study found that hOGA acted on a few glycoprotein substrates with similar catalytic efficiency (kcat/Km), leading the authors to propose that hOGA recognizes only the sugar moiety but not the peptide side chains of the substrate50. In contrast, based on the crystal structure of OgOGA and on the sequence similarity between OgOGA and hOGA, another group made substitutions in a number of hOGA residues that potentially lined the top of the catalytic site and found that several of them reduced activity to protein substrates without impairing hydrolysis of the reporter substrate 4MU-NAG24. This discovery led the group to propose that a substrate-binding groove of hOGA engages in specific recognition of protein substrates.
Our OGAcryst–p53 complex provides the first direct evidence demonstrating that, in addition to the contacts with the GlcNAc moiety, there are many interactions with the peptide, including sequence-dependent side chain interactions and sequence-independent backbone interactions with the hOGA residues located on the substrate-binding cleft. Although studies with additional glycopeptide complexes are required to define generality, it is clear based on the structural and mutagenesis data that the interactions of the peptide with the cleft surface residues are key elements in substrate recognition. Because the stalk domain of hOGA showed dramatic sequence and structural variations from its bacterial homologs, it was expected that the same glycopeptide would be bound differently in the human and bacterial enzymes. Indeed, superposition of the p53 glycopeptides in the two sister monomers of the OGAcryst–p53 complex with that in the reported CpOGA–p53 complex structure (PDB 2YDR)40 revealed a distinct recognition mode for hOGA (Supplementary Fig. 8). As previously reported, the p53 glycopeptide in the CpOGA complex makes an intrapeptide hydrogen bond, but has limited side chain interactions with the enzyme40 (Supplementary Fig. 8b). In contrast, both of the glycopeptides in the OGAcryst–p53 complex made an abundant number of interactions with the cleft surface residues beyond the immediate catalytic core (Supplementary Fig. 8c,d), which could be important attributes for the substrate specificity of hOGA. Consistent with this, substitution of cleft surface residues in hOGA or of the tryptophan residue (−3 subsite) of the p53 glycopeptide recognized by hOGA indeed resulted in dramatically reduced binding. These results support the hypothesis that hOGA has an important regulatory role in O-GlcNAc biology through substrate discrimination and that it does not merely indiscriminately remove O-GlcNAc from all of its substrates.
It is conceivable that the substrate-binding cleft of hOGA may provide spatial restrictions on large protein substrates accessing the active site. For example, O-GlcNAcylated loops are favorable, whereas highly complex protein domains might not be accessible or might require considerable opening of the substrate-binding cleft for accommodation. The differential binding conformation of the p53 glycopeptide in an OGAcryst dimer was unexpected and requires further investigation. The unique orientation of the F223 residue has been observed in OGA-β of the OGAcryst–p53 complex, and its substitution markedly weakened binding of the glycopeptide, suggesting that this residue potentially affects the differential conformation of the substrate. In addition, crystal packing could be another factor that contributed to stabilizing the differential binding conformation of the glycopeptide in OGA-β. It is also possible that the two monomers of hOGA might be capable of binding and catalyzing distinct substrates or O-GlcNAcylation sites simultaneously to achieve highly efficient deglycosylation.
In summary, the hOGA structures presented here represent the first examples of eukaryotic glycoside hydrolases in the GH84 family. They provide considerable insights into the unique recognition mode of hOGA and offer a crucial starting point for future structural investigation of other regions of hOGA, such as the pseudo-HAT domain, which may participate in protein-protein interactions. Our study also provides a framework for solving the structures of additional glycopeptide or glycoprotein complexes and aids in a more complete understanding of the principles involved in hOGA regulation of a plethora of protein substrates in biology and disease.
ONLINE METHODS
Protein expression and purification.
Sequences for the full-length and truncated OGA (OGAcryst or OGA60–704-del, based on the numbering of the full-length human protein) proteins were subcloned into a modified pET24b vector or pET-SUMO vector, respectively. The plasmids were transformed into Escherichia coli strain Rosetta(DE3), and the transformants were grown at 37 °C in Luria–Bertani (LB) medium. After OD600 reached 0.8, the culture was cooled to 16 °C and induced by 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 16 h, the cells were harvested, resuspended and homogenized with an ultra-high-pressure cell disrupter (Emulsiflex-C5, Canada) at 4 °C. The supernatant was subjected to Ni–NTA column (Qiagen) for affinity purification and was subsequently eluted with buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 250 mM imidazole. The eluted full-length OGA was further purified by size-exclusion chromatography (Superdex 200 increase 10/300, GE Healthcare) in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 0.5 mM Tris(hydroxymethyl)phosphine (THP; EMD), and it was then concentrated to a concentration of 3 mg/ml for the in vitro assays. The eluted OGAcryst was digested by Sumo protease to remove the 6× His–SUMO tag and further purified by size-exclusion chromatography. The OGAcryst protein was concentrated to a concentration of 3 mg/ml for crystallization. In this construct, residues 401–552 were replaced by a 15-residue glycine–serine linker (GGGGSGGGGSGGGGS). Various OGA mutants were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent), according to the manufacturer’s instructions, using the sequence for full-length hOGA or OGAcryst as the DNA template along with the primers listed in Supplementary Table 2. The DNA sequences were verified by sequencing. Mutant proteins were expressed and purified similarly as described above.
Crystallization.
All of the crystals were generated by mixing 1 μl of protein with an equal volume of reservoir solution and equilibriation against 200 μl of reservoir solution using the hanging-drop vapor-diffusion method at 20 °C. Native OGAcryst crystals (wild-type and D175N mutant) were obtained in the reservoir solution containing 0.032 M ammonium citrate tribasic (pH 7.0), 0.02 M MES monohydrate, 0.128 M potassium thiocyanate, 0.016 M imidazole, 0.002 M zinc sulfate heptahydrate, 12.8% wt/vol polyethylene glycol 3,350, 3.2% wt/vol polyethylene glycol monomethyl ether 2,000, and 5% wt/vol polyethylene glycol monomethyl ether 550. For the inhibitor complex, OGAcryst was incubated with 2 mM thiamet-G (synthesized as reported previously37) for 1 h and co-crystallized in conditions similar to those as for the native protein. For the OGAcryst–p53 complex, the p53 glycopeptide (Ac-QLWVDS(O-GlcNAc)TPPPG) was prepared by solid-phase peptide synthesis. Native crystals (D175N mutant) were soaked for 30 min in reservoir solution containing 2 mM of glycopeptide. All of the crystals were transferred into cryoprotectant solution containing the respective mother liquor plus 10% (vol/vol) glycerol before being flash-frozen in liquid nitrogen for storage.
Data collection and structural determination.
All of the X-ray data were collected on the Life Sciences Collaborative Access Team (LS-CAT) beam lines 21-ID-G (for OGAcryst) and 21-ID-D (for the OGAcryst–thiamet-G and OGAcryst–p53 complexes) (LS-CAT, Advanced Photon Source, Argonne National Laboratory, IL, USA). The wavelength for data collection was 0.9787 Å. All data sets were processed using the HKL2000 package51. The native crystal for the complex of wild-type OGAcryst and its inhibitor belonged to the same space group of P21, containing two molecules per asymmetric unit. The crystal of OGAcryst–p53 complex also belonged to the same space group but had four molecules per asymmetric unit. The structures were solved by molecular replacement using CpOGA as a search model (PDB 2CBJ)39. Iterative model building was performed in COOT52 and further refined in PHENIX53 and CCP4 (ref. 54). Final refinement statistics are summarized in Table 1. Structural figures were drawn using the program PyMOL55. The coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 5TKE, 5UN9, and 5UN8.
Analytical ultracentrifugation sedimentation equilibrium.
Sedimentation equilibrium studies were performed using a Beckman Model XL-A Analytical Ultracentrifuge in the Biophysics Instrumentation Facility at the University of Wisconsin–Madison. Samples for the sedimentation equilibrium experiments were diluted to the desired protein concentrations in 20 mM Tris-HCl (pH 7.0), 150 mM NaCl, and 0.5 mM THP. Data were collected at 20 °C and three rotor speeds for each protein. For full-length hOGA, sedimentation equilibrium was analyzed at initial loading concentrations of 0.47 mg/ml and 0.14 mg/ml with rotor speeds of 4,000 r.p.m., 5,600 r.p.m., and 7,600 r.p.m. OGAcryst protein was analyzed at initial loading concentrations of 0.30 mg/ml and 0.095 mg/ml with rotor speeds of 6,000 r.p.m., 8,800 r.p.m., and 12,000 r.p.m. Data were analyzed at the facility by Dr. Darrell McCaslin, using approaches similar to that described by Laue56.
OGA enzymatic assays.
Steady-state kinetics of wild-type and mutant hOGA were determined using the strategy of Macauley33. The fluorogenic substrate 4-methylumbelliferyl-N-acetyl-β-D-glucosaminide (4MU-NAG; Fisher) was used in this assay. Standard reaction mixtures (25 μl) contained 2 nM of enzyme in 50 mM NaH2PO4 (pH 6.5), 100 mM NaCl, 0.1 mg/ml BSA, and various substrate concentrations (10, 50, 150, 300, 600, and 1200 μM). The reactions were incubated at 37 °C for 10 min, and then quenched by the addition of 150 μl of 200 mM glycine–NaOH (pH 10.75). The fluorescence of liberated 4-methylumbelliferone was monitored with a Microplate Fluorescence Reader (Bio-Tek), with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. The released 4-methylumbelliferone was quantified by the standard curve of 4-methylumbelliferone under the identical conditions. All of the measurements were carried out in triplicate, and the Michaelis–Menten kinetic parameters were calculated by fitting the data in GraphPad Prism (GraphPad Software, La Jolla, CA). Relative-activity assays were performed in the presence of 2 nM enzyme and 200 μM 4MU-NAG at 37 °C for 10 min in triplicate. The fluorogenic measurement was determined as mentioned above. Glycopeptide competition assays were performed in triplicate using 4MU-NAG as the reporter substrate, using multisubstrate enzyme kinetics as previously reported24,40,57. The data were analyzed by GraphPad Prism using the following equation: vi/v0 = ((1 + Km/S))/(1 + Km/S(1 + S′/Km′)) to yield Km′ (the Michaelis constant of the competition substrate p53 glycopeptide).
Data availability.
Coordinates and structural factors have been deposited in the Protein Data Bank under accession codes 5TKE, 5UN9, and 5UN8 for OGAcryst, OGAcryst–thiamet-G, and OGAcryst–p53, respectively. All other data are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Smith and L. Carlson for assistance with X-ray data collection, K. Satyshur and D. McCaslin for helpful discussions, and members of the Jiang laboratory for critical reading of the manuscript. This work was supported by start-up funds from the University of Wisconsin–Madison (J.J.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Torres CR & Hart GW Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem 259, 3308–3317 (1984). [PubMed] [Google Scholar]
- 2.Hanover JA, Krause MW & Love DC Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol 13, 312–321 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Ma J & Hart GW O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond MR & Hanover JA A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol 208, 869–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardivillé S & Hart GW Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BA & Hanover JA O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem 289, 34440–34448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BA O-GlcNAcylation at promoters, nutrient sensors, and transcriptional regulation. Biochim. Biophys. Acta 1829, 1202–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazarus MB et al. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drougat L et al. Characterization of O-GlcNAc cycling and proteomic identification of differentially O-GlcNAcylated proteins during G1/S transition. Biochim. Biophys. Acta 1820, 1839–1848 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Zachara NE & Hart GW O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell Biol. 14, 218–221 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Guinez C et al. Protein ubiquitination is modulated by O-GlcNAc glycosylation. FASEB J. 22, 2901–2911 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Vaidyanathan K & Wells L Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of, and complications arising from, type 2 diabetes. J. Biol. Chem 289, 34466–34471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J & Hart GW Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics 10, 365–380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Shan X, Yuzwa SA & Vocadlo DJ The emerging link between O-GlcNAc and Alzheimer’s disease. J. Biol. Chem 289, 34472–34481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuzwa SA & Vocadlo DJ O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Chem. Soc. Rev 43, 6839–6858 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Ferrer CM, Sodi VL & Reginato MJ O-GlcNAcylation in cancer biology: linking metabolism and signaling. J. Mol. Biol 428, 3282–3294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Z & Vosseller K Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem 289, 34457–34465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh JP, Zhang K, Wu J & Yang X O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 2 Pt A, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreppel LK, Blomberg MA & Hart GW Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem 272, 9308–9315 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Gao Y, Wells L, Comer FI, Parker GJ & Hart GW Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem 276, 9838–9845 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Pathak S et al. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol 22, 744–750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarus MB, Nam Y, Jiang J, Sliz P & Walker S Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarus MB et al. Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol 8, 966–968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schimpl M, Schüttelkopf AW, Borodkin VS & van Aalten DMF Human OGA binds substrates in a conserved peptide-recognition groove. Biochem. J 432, 1–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vocadlo DJ O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr. Opin. Chem. Biol 16, 488–497 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Alonso J, Schimpl M & van Aalten DMF O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J. Biol. Chem 289, 34433–34439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao FV et al. Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol. 3, 130021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Roth C, Turkenburg JP & Davies GJ Three-dimensional structure of a Streptomyces sviceus GNAT acetyltransferase with similarity to the C-terminal domain of the human GH84 O-GlcNAcase. Acta Crystallogr. D Biol. Crystallogr 70, 186–195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toleman C, Paterson AJ, Whisenhunt TR & Kudlow JE Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activatable O-GlcNAcase and HAT activities. J. Biol. Chem 279, 53665–53673 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Keembiyehetty CN, Krześlak A, Love DC & Hanover JA A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J. Cell Sci 124, 2851–2860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim EJ, Kang DO, Love DC & Hanover JA Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr. Res 341, 971–982 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J et al. Isoforms of human O-GlcNAcase show distinct catalytic efficiencies. Biochemistry 75, 938–943 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Macauley MS, Whitworth GE, Debowski AW, Chin D & Vocadlo DJ O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem 280, 25313–25322 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Whitworth GE et al. Analysis of PUGNAc and NAG-thiazoline as transition-state analogs for human O-GlcNAcase: mechanistic and structural insights into inhibitor selectivity and transition-state poise. J. Am. Chem. Soc 129, 635–644 (2007). [DOI] [PubMed] [Google Scholar]
- 35.He Y, Bubb AK, Stubbs KA, Gloster TM & Davies GJ Inhibition of a bacterial O-GlcNAcase homolog by lactone and lactam derivatives: structural, kinetic, and thermodynamic analyses. Amino Acids 40, 829–839 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Dennis RJ et al. Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol 13, 365–371 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Yuzwa SA et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol 4, 483–490 (2008). [DOI] [PubMed] [Google Scholar]
- 38.He Y, Macauley MS, Stubbs KA, Vocadlo DJ & Davies GJ Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J. Am. Chem. Soc 132, 1807–1809 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Rao FV et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 25, 1569–1578 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schimpl M, Borodkin VS, Gray LJ & van Aalten DMF Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem. Biol 19, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostrowski A, Gundogdu M, Ferenbach AT, Lebedev AA & van Aalten DMF Evidence for a functional O-linked N-acetylglucosamine (O-GlcNAc) system in the thermophilic bacterium Thermobaculum terrenum. J. Biol. Chem 290, 30291–30305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macauley MS & Vocadlo DJ Increasing O-GlcNAc levels: an overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 1800, 107–121 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Banerjee PS, Hart GW & Cho JW Chemical approaches to study O-GlcNAcylation. Chem. Soc. Rev 42, 4345–4357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorfmueller HC et al. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-GlcNAcylation levels. J. Am. Chem. Soc 128, 16484–16485 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cetinbas N, Macauley MS, Stubbs KA, Drapala R & Vocadlo DJ Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–3844 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Yuzwa SA et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol 8, 393–399 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Yu Y et al. Differential effects of an O-GlcNAcase inhibitor on tau phosphorylation. PLoS One 7, e35277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lameira J et al. Quantum mechanical–molecular mechanical molecular dynamics simulation of wild-type and seven mutants of CpNagJ in complex with PUGNAc. J. Phys. Chem. B 114, 7029–7036 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Shen DL, Gloster TM, Yuzwa SA & Vocadlo DJ Insights into O-linked N-acetylglucosamine ([0–9]O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem 287, 15395–15408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Adams PD et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 55.DeLano WL The PyMOL Molecular Graphics System (Delano Scientific LLC, 2002). [Google Scholar]
- 56.Laue TM Sedimentation equilibrium as a thermodynamic tool. Methods Enzymol. 259, 427–452 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Xie D, Suvorov L, Erickson JW & Gulnik AS Real-time measurements of dark-substrate catalysis. Protein Sci. 8, 2460–2464 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structural factors have been deposited in the Protein Data Bank under accession codes 5TKE, 5UN9, and 5UN8 for OGAcryst, OGAcryst–thiamet-G, and OGAcryst–p53, respectively. All other data are available from the corresponding author upon reasonable request.