ABSTRACT
Genetic screens are designed to target individual genes for the practical reason of establishing a clear association between a mutant phenotype and a single genetic locus. This allows for a developmental or physiological role to be assigned to the wild-type gene. We previously observed that the concurrent loss of Pax6 and Polycomb epigenetic repressors in Drosophila leads the eye to transform into a wing. This fate change is not seen when either factor is disrupted separately. An implication of this finding is that standard screens may miss the roles that combinations of genes play in development. Here, we show that this phenomenon is not limited to Pax6 and Polycomb but rather applies more generally. We demonstrate that in the Drosophila eye-antennal disc, the simultaneous downregulation of Pax6 with either the NURF nucleosome remodeling complex or the Pointed transcription factor transforms the head epidermis into an antenna. This is a previously unidentified fate change that is also not observed with the loss of individual genes. We propose that the use of multi-gene knockdowns is an essential tool for unraveling the complexity of development.
KEY WORDS: Pax6, ISWI, E(bx), NURF, Pointed, Antenna, Head epidermis, Drosophila
Summary: The simultaneous reduction of Pax6 with either the NURF nucleosome remodeling complex or the transcription factor Pointed reveals roles for these factors that cannot be discerned from removing each one individually.
INTRODUCTION
Fruit flies, nematodes and zebrafish are powerful model organisms for studying development, in part because they are amenable to having their genomes interrogated through large-scale unbiased genetic screens or targeted molecular screens. Irrespective of the type of screen, the overarching goal is to establish a clear association between a mutant phenotype and the loss of a single genetic locus. As such, screens are designed to reveal the developmental and/or physiological outcome of disrupting a single gene. An unintended consequence is that phenotypes that result from the removal of combinations of genes are neither searched for nor frequently recovered. As a result, the more complex and/or cryptic roles that genes play in development are often overlooked.
One apposite example involves Eyeless (Ey), the Drosophila homolog of vertebrate Pax6, which historically has been thought to play a rather limited role in the eye-antennal disc of the fruit fly, Drosophila melanogaster. The eye-antennal disc is an epithelial sac gives that rise to most external adult head structures, including the compound eyes, ocelli, antennae, maxillary palps, and head epidermis. It was long assumed that the only function of Ey within the disc is to specify and pattern the eye field. This is because aborted retinal development is the only phenotype observed when Ey is removed from the disc (Hoge, 1915; Morgan, 1929; Quiring et al., 1994; Baker et al., 2018). For very similar reasons, the other Pax6 homolog, Twin of Eyeless (Toy), was thought to participate only in the specification of the compound eyes and ocelli (Czerny et al., 1999; Blanco et al., 2010; Brockmann et al., 2011).
The view that these Pax6 genes play limited roles in the eye-antennal disc has been recently challenged by our demonstrations that Ey and Toy also function to control growth of the entire eye-antennal disc and to prevent the developing eye from being reprogrammed into a wing (Zhu et al., 2017; 2018; Palliyil et al., 2018). These functions were discovered under circumstances in which multiple genes are simultaneously removed from the disc. For example, a role for these transcription factors in promoting growth of the entire disc was revealed only when we removed both Pax6 genes simultaneously or in combination with the zinc-finger transcription factor Teashirt (Tsh) (Zhu et al., 2017; Palliyil et al., 2018). Similarly, the unexpected finding that Ey and Toy are required to prevent the eye from transforming into a wing is only seen when either Pax6 gene is simultaneously downregulated alongside members of the Polycomb Group (PcG) of epigenetic repressors (Zhu et al., 2018). Prior to our study of the PcG factors, this breathtaking transformation of the eye into a wing had been observed in several additional instances in which multiple genes are simultaneously removed from the developing eye field. In these cases, flies harboring loss-of-function mutations in ophthalmoptera (opht) also contained mutations in the ey, eyes reduced (eyr), Deformed (Dfd) or loboid (ld) genes (Goldschmidt and Lederman-Klein, 1958; Edwards and Gardner, 1966; Ouweneel, 1970).
In this paper, we use multi-gene knockdowns to unearth new roles for Pax6 within the eye-antennal disc. We have discovered that when Toy is simultaneously removed with either the nucleosome remodeling factor (NURF) complex or the Ets transcription factor Pointed (Pnt), the region of the disc that normally gives rise to the dorsal head epidermis and ocelli is inappropriately transformed into a fully formed second antenna. The pharate adult flies have four complete antennae instead of the normal complement of two. This dramatic change in organ fate and number is not observed when either Toy, NURF or Pnt are removed individually. Instead, the only defect seen in single-gene knockdowns is a reduction in the number of ocelli and surrounding mechanosensory bristles. Our study reveals a new and unexpected role for Toy in tissue fate decisions outside of the retina. Because this striking change in the identity of the ocelli/head epidermis is only revealed when Toy is removed in combination with other regulatory factors, these findings provide support for our contention that the use of multi-gene knockdowns is a powerful method for revealing hidden or cryptic roles for even the most extensively studied genes.
RESULTS
A single antennal field is maintained by Pax6 and chromatin-modifying enzymes
The eye-antennal discs of Drosophila give rise to most adult head structures, including the compound eyes, ocelli, antennae, maxillary palps, and surrounding head epidermis (Fig. 1A,B; Haynie and Bryant, 1986). The discs are first set aside during embryogenesis when cells from several different embryonic head segments come together to form each of the two epithelial sheets (Ferris, 1950; Cohen, 1993; Jurgens and Hartenstein, 1993; Younossi-Hartenstein et al., 1993). During the three larval instars, the eye-antennal discs undergo expansive growth, are subdivided into individual territories and undergo pattern formation (Krafka, 1924; Chen, 1929; Ready et al., 1976; Campos-Ortega and Hofbauer, 1977; Madhavan and Schneiderman, 1977; Haynie and Bryant, 1986). Later in pupal development, the two eye-antennal discs surround the brain lobes and fuse with each other as well as to a pair of labial discs, which give rise to the proboscis (Birmingham, 1942; Bodenstein, 1950; Milner and Haynie, 1979; Milner et al., 1984; Fristrom and Fristrom, 1993). Together, the four imaginal discs produce an intact adult head.
Fig. 1.
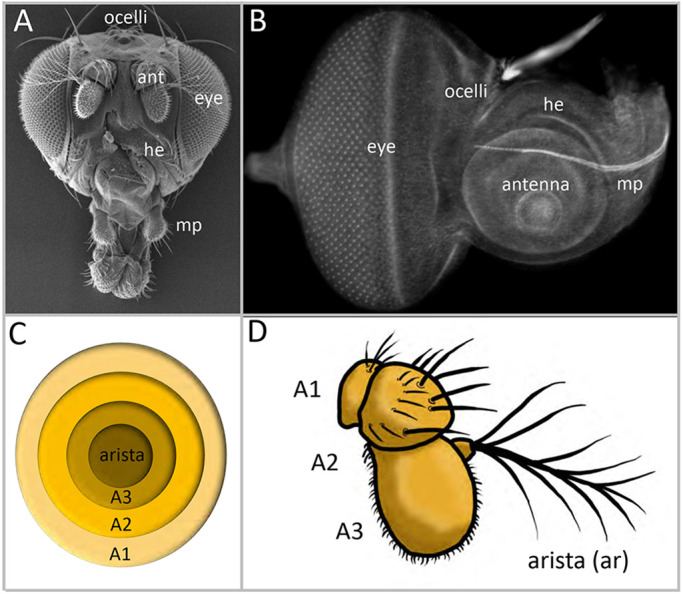
Organization of the eye-antennal disc of Drosophila. (A) SEM image of the Drosophila head. (B) Light microscopic image of the developing eye-antennal disc. ant, antenna; he, head epidermis; mp, maxillary palp. (C,D) Schematics of the developing and antennal adult showing the A1, A2, A3 and aristal segments.
We began this study by using RNA interference (RNAi) to reduce expression levels of 45 chromatin-modifying proteins that participate in the activation of transcription. These proteins add methyl and/or acetyl groups to histone core proteins or remodel nucleosomes during gene activation (Fig. S1). Whenever possible, we targeted individual genes with multiple RNAi lines as a means to eliminate false positives that result from off-target effects (Table S1). The expression of each RNAi line was placed under the control of the dorsal eye (DE-GAL4) and eyeless GAL4 (ey-GAL4) drivers. The DE-GAL4 line is an insertion of GAL4 within the mirror (mirr) locus (Morrison and Halder, 2010). The ey-GAL4 was generated by fusing the first intron of the ey gene (which contains an eye-specific enhancer) to GAL4 (Hauck et al., 1999). Both lines are initially expressed broadly throughout the entire disc, but by the second larval instar ey-GAL4 expression becomes restricted to the eye field whereas DE-GAL4 is confined to the dorsal half of the eye disc (Hauck et al., 1999; Kumar and Moses, 2001; Morrison and Halder, 2010; Palliyil et al., 2018).
We found that targeting individual chromatin-modifying genes with RNAi constructs results in a multitude of fate, patterning and growth defects (Fig. 2, Table S2). For example, targeting some genes, such as moira (mor), eliminates the eye-antennal discs themselves and leaves pharate adult flies headless (Fig. 2A-D, Table S2). Reducing the expression of other members, such as will die slowly (wds), interferes with global development of the disc and this leads to an overall deformation of the adult head (Fig. 2E,F, Table S2). We also observed cases in which only selected tissues were affected. For instance, targeting trithorax related (trr) resulted in an adult fly in which the head is relatively normal but the eye is smaller and roughened (Fig. 2G,H, Table S2). One of the more intriguing phenotypes is the presence of a second, fully formed ectopic antenna. A schematic of the developing and adult antennae showing the A1, A2, A3 and aristal segments is provided in Fig. 1C,D. The ectopic antenna presented itself when expression of several genes, such as Brahma associated protein 60kd (Bap60), Brahma associated protein 111kd (Bap111), osa, trithorax (trx), absent small homeotic discs 1 (ash1), Nipped-B and Caf1-55, were targeted (Fig. 2I,J, Table S2). Only one ectopic antenna was produced per disc.
Fig. 2.
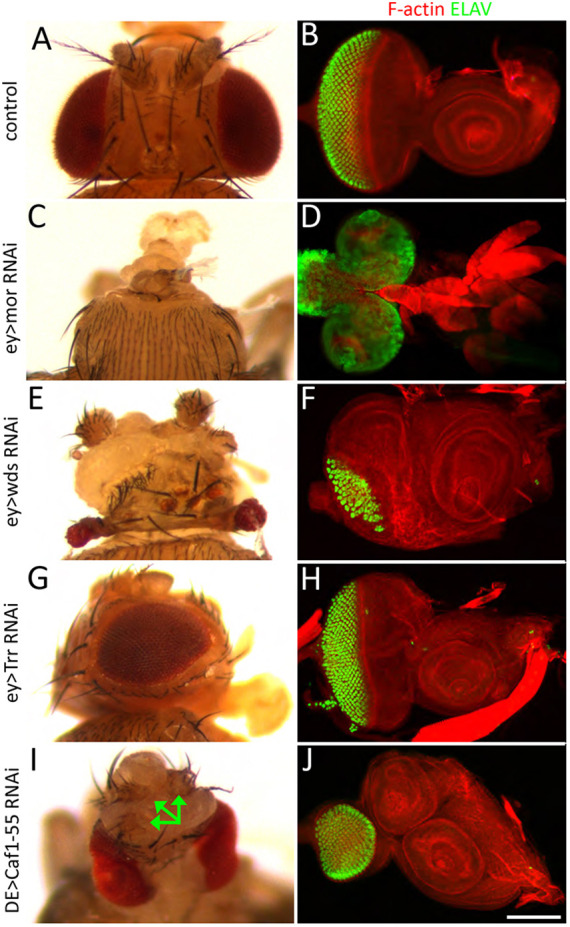
The development of the eye-antennal disc is regulated by chromatin-modifying proteins. (A,C,E,G,I) Adult heads. (B,D,F,H,J) Third instar eye-antennal discs stained for F-actin (red) and ELAV (green). (A,B) A pair of wild-type eye-antennal discs give rise to the external surface of the adult head. (C-J) Knockdown of chromatin-modifying proteins result in a variety of defects, including complete loss of the head (C,D), severe head defects (E,F), small rough eyes (G,H) and antennal duplications (I,J). The arrows in I point to three of the four antennae that are formed when Caf1-55 is knocked down. Scale bar: 50 µm.
Although reductions in the seven aforementioned chromatin-modifying genes increased antennal numbers (7/45=15.5%), the individual loss of 38 other genes (38/45=84.5%) had little to no effect on antennal fate (Table S2). The differential effects on antennal fate is reminiscent of the earlier screen that we conducted of PcG mutants. Although reductions in Polycomb (Pc) itself resulted in an eye-to-wing transformation, targeting the remaining 15 other PcG factors individually did not alter the fate of the eye to a discernable degree (Zhu et al., 2018). We therefore investigated whether the maintenance of a single antennal field similarly requires both Pax6 and the chromatin-modifying enzymes that we selected for this study. Of the 38 instances in which antennal development was unaffected by the knockdown of an individual gene, we observed 19 cases (19/38=50%) in which a second antenna emerged when the factor in question was simultaneously targeted with either Ey or Toy (Table S2). This suggests that Pax6 and chromatin-modifying factors are both required to suppress formation of extra antennal organs. A major difference between this study and our analysis of PcG members is that the eye-to-wing transformation occurs irrespective of which Pax6 gene is lost (Zhu et al., 2018), whereas the formation of a second antenna is biased towards the loss of Toy (Table S2). For example, 14 chromatin-modifying genes showed a strong bias towards requiring Toy (14/19=73.6%) whereas only two genes specifically required the simultaneous loss of Ey (2/19=10.5%). Three chromatin-modifying genes (3/19=15.7%) showed no bias toward requiring a particular Pax6 gene as antennal duplications were triggered when either Pax6 gene was lost (Table S2).
NURF and Toy are required to maintain a single antennal field
For the remainder of this study, we focus on the NURF chromatin-remodeling complex as it is known to physically interact with insulator proteins, members of the general transcription factor machinery, and tissue-specific transcription factors (Xiao et al., 2001; Song et al., 2009; Kugler and Nagel, 2010; Kwon et al., 2016; Qiu et al., 2015). We thought this might be relevant to our study because we were interested in determining whether Toy interacted directly or indirectly with chromatin-modifying factors. We specifically focused on two NURF complex components, Imitation SWI (ISWI) and Enhancer of bithorax [E(bx)]. ISWI, in addition to being a component of NURF, is a member of several other chromatin-remodeling complexes and is important for nucleosome sliding (Corona and Tamkun, 2004). E(bx), by contrast, is found only within NURF and is the DNA- and histone-binding component of the complex (Badenhorst et al., 2002; Kwon et al., 2016). Here, we show that RNAi targeting of either factor simultaneously with Toy gives identical mutant phenotypes and that both genes are required during the same developmental window. As such, we propose that our findings go beyond these two individual proteins and instead provide insights into the role that NURF itself plays in generating the correct number of individual tissues/organs within the eye-antennal disc.
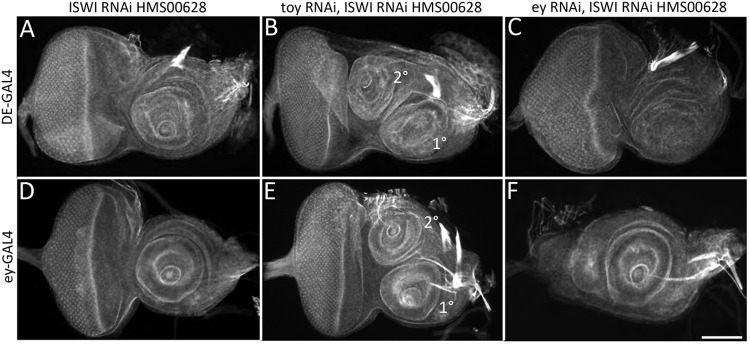
We first used quantitative real time PCR (qRT-PCR) and determined that both ISWI and E(bx) are expressed robustly within the eye-antennal disc (Fig. S2A). Interfering with either ISWI expression (with two different RNAi lines) or activity (with a dominant-negative protein; Deuring et al., 2000) resulted in the induction of a second antenna – but only when Toy was simultaneously knocked down (Fig. 3A,B,D,E, Fig. S2B-E). As was the case for most genes that we tested, the concomitant targeting of Ey with ISWI did not produce the secondary antenna (Fig. 3C,F). Identical results were observed when E(bx) was targeted along with either Ey or Toy genes (Fig. S3A-E). We note that loss of Toy and E(bx) within the ey-GAL4 domain does induce a second antenna. This may be due to the relative weakness of this driver compared with DE-GAL4. Ey and Toy have distinct spatial distribution patterns (see below) and this could account for the observed bias in requiring Toy to repress ectopic antennal formation (Jacobsson et al., 2009; Quiring et al., 1994; Czerny et al., 1999). The two Pax6 proteins also bind to somewhat dissimilar consensus sequences (Czerny et al., 1999; Nimi et al., 2002; Punzo et al., 2002) and any predicted differences in their molecular targets could be another contributing factor.
Fig. 3.
The simultaneous loss of Toy and ISWI transforms the head epidermis into an antenna. (A,D) The number of antennae within the eye-antennal disc is unchanged when ISWI alone is removed. (B,E) A fully formed second antenna is found in discs in which ISWI and Toy are simultaneously knocked down. (C,F) This effect appears to be specific to Toy, as the simultaneous loss of Ey and ISWI does not appear to affect antennal number. Scale bar: 50 µm.
Several lines of evidence suggest that Toy and NURF are likely working on distinct target genes and do not directly collaborate with or regulate each other. First, the loss of Toy, ISWI or E(bx) individually has no effect on antennal numbers (Fig. 3A,D, Fig. S2B,D, Fig. S3A,C; Zhu et al., 2018) making is unlikely that these factors are regulating a common target. This is further supported by the fact that the pnt locus, which we have identified as being relevant for the antennal duplication phenotype (see below), is bound by E(bx) (Kwon et al., 2016) but lacks intact Toy consensus binding sites. Second, it is doubtful that NURF is regulating toy because the removal of either ISWI or E(bx) from the dorsal half of the eye field did not affect toy expression (Fig. S4A-D) and because the toy locus is not bound by E(bx) (Kwon et al., 2016). Third, as consensus binding sites for Toy are absent from the ISWI and E(bx) loci it is unlikely that Toy regulates either NURF complex member. In total, these findings suggest that Toy and NURF are not regulating each other and are not directly cooperating with each other, nor are they controlling the same downstream gene target(s). It is more likely that Toy and NURF are regulating independent target genes.
Although Toy is required to prevent the formation of a second antenna, other members of the retinal determination (RD) network do not appear to be needed in a similar manner. As we have seen above, the simultaneous knockdown of ISWI and Ey did not yield a second ectopic antenna (Fig. 3C,F). We also did not see the formation of additional antennae when ISWI was knocked down in sine oculis (so) and eyes absent (eya) mutant discs (Fig. 4A). The induction of ectopic antennae is therefore not caused by or dependent upon the collapse of the entire RD network but is instead a result of specifically disrupting toy expression levels. Interestingly, toy expression is maintained at robust levels in ey, so and eya mutant discs (Czerny et al., 1999; Weasner et al., 2016; Baker et al., 2018); therefore, we propose that its continued presence in these mutants is sufficient to suppress formation of the second antenna even when NURF activity is inhibited.
Fig. 4.
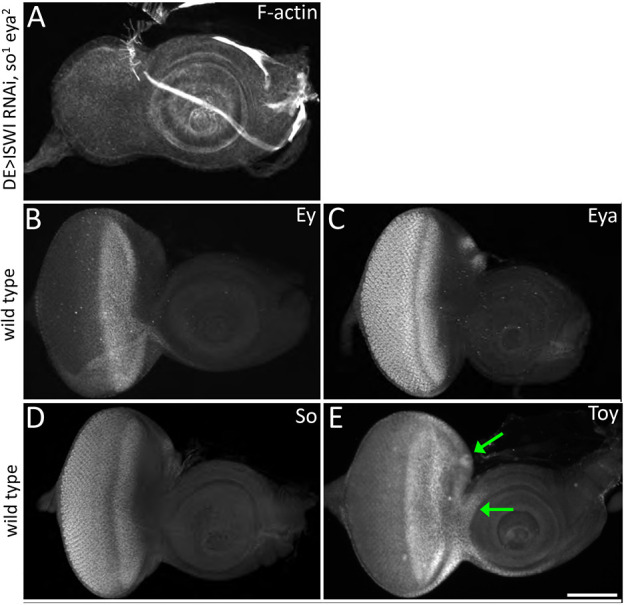
Expression of patterns of retinal determination genes within the eye-antennal disc. (A) Removal of ISWI along with the RD network genes So and Eya does not induce the formation of a second antenna. (B-E) Toy is expressed within the ocellar and anterior head epidermis domains (arrows in E). Scale bar: 50 µm.
The expression patterns of the four aforementioned RD genes provide further insight into the specificity of the genetic interactions between Toy and NURF and the physical origin of the second antennal field. The ey, so and eya genes are expressed in stripes of varying widths ahead of the morphogenetic furrow but none of the expression patterns extends to or beyond the border of the eye and antennal fields (Fig. 4B-D) (Bonini et al., 1993; Cheyette et al., 1994; Quiring et al., 1994). In contrast, toy expression extends into the ocellar portion of the eye field and into the head epidermis region of the antennal field (Fig. 4E) (Jacobsson et al., 2009). The extended distribution of Toy beyond the domains occupied by the other RD genes suggests that the second antenna might arise from the ocellar and/or head epidermis regions.
We next analyzed the development of the ectopic antennal field (refer to Fig. 1C,D for a description of the antennal field) by using a suite of transcriptional reporters and antibodies. The patterns of these reporters within the endogenous and ectopic antennae were compared with each other. The dpp-lacZ transcriptional reporter and antibodies against Wingless (Wg) protein revealed that the dorsal/ventral (D/V) axis is established correctly (Fig. 5A-D). Similarly, antibodies against the active version of Cubitus Interruptus (CiA) revealed that anterior/posterior (A/P) axis also develops properly (Fig. 5E,F). Lastly, antibodies against the transcription factors Cut, Lim1, Distalless (Dll), Dachshund (Dac) and Aristaless (Al) indicated that the A1, A2, A3 and aristal segments are patterned accurately along the proximal/distal (P/D) axis (Fig. 5G-P). Not surprisingly, the Toy/ISWI double knockdown animals had four structurally normal antennae instead of the normal complement of two (Fig. S5A). Using the same set of molecular markers, we observed that the ectopic antennae in Toy/E(bx) double knockdowns also have normally formed D/V, A/P and P/D axes (Fig. S5B-I).
Fig. 5.
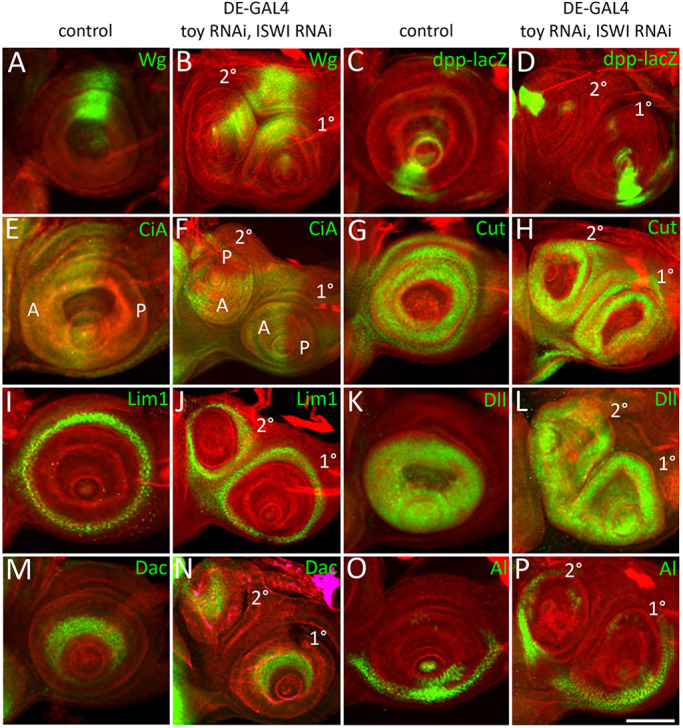
Axis and segment formation are normal in the ectopic antenna. (A-P) Expression of wingless (A,B), decapentaplegic (C,D) and the active version of cubitus interruptus (E,F) indicate that the D/V and A/P compartments of the ectopic antenna all form correctly. Distribution of the transcription factors Cut (G,H), Lim1 (I,J), Distal-less (K,L), Dachshund (M,N) and Aristaless (O,P) also shows that the A1, A2, A3, and aristal segments also develop properly along the P/D axis. Scale bar: 50 µm.
Toy/NURF are required during the second larval instar to maintain a single antennal segment
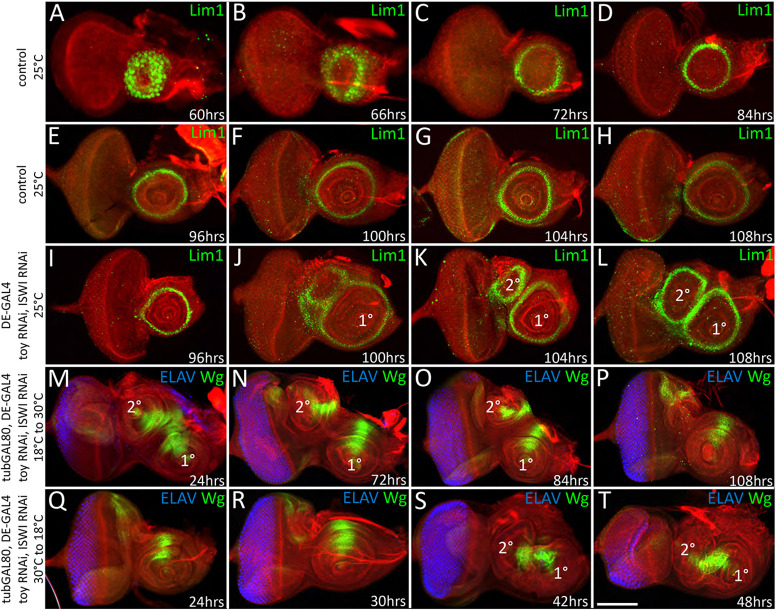
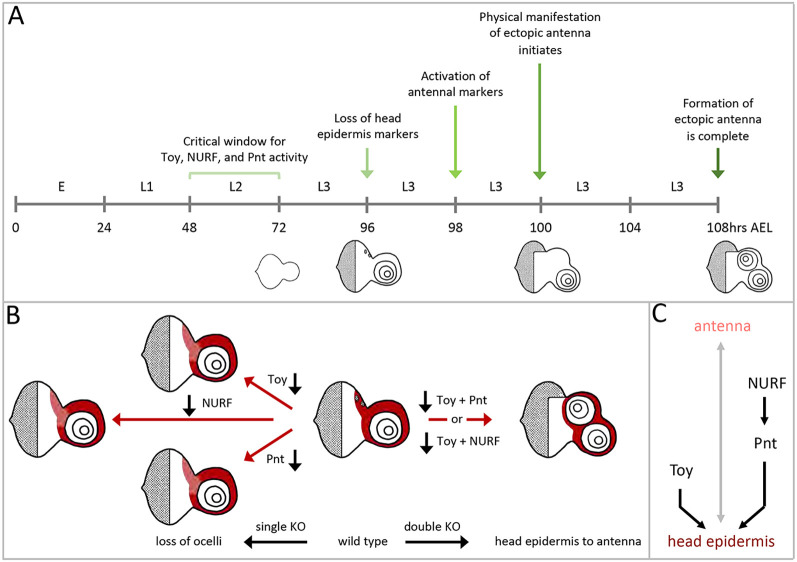
We used expression of the homeobox transcription factor Lim1, which marks the A1 antennal segment, to trace the temporal development of endogenous and ectopic antennae in control and Toy/ISWI double knockdown larvae. In control discs, Lim1 expression appeared as a near solid disc at 60 h after egg lay (AEL) (Fig. 6A). These cells will give rise to all antennal segments. By 72 h AEL, Lim1 expression was extinguished from the inner segments and remained just within the outer A1 segment (Fig. 6B,C). This mature Lim1 pattern persisted throughout the remainder of larval development (Fig. 6D-H). In Toy/ISWI knockdown discs, Lim1 expression within the endogenous antenna initiated and evolved with the same temporal/spatial dynamics as it does in control larvae. The presence of a single Lim1 ring within the endogenous antenna was seen as late as 96 h AEL (Fig. 6I). However, at 100 h AEL (middle of the third larval instar) ectopic Lim1 expression and tissue overgrowth were both observed in the dorsal head epidermis (Fig. 6J, Fig. S6). Over the next 8 h, this area was then patterned into a fully fledged second antenna (Fig. 6K,L, Fig. S6).
Fig. 6.
Toy and ISWI are required during the second larval instar to suppress formation of the second antenna. (A-H) Lim1 expression reaches its final pattern at 72 h AEL and remains just within the A1 segment for the rest of larval development. (I) In Toy/ISWI mutants a single Lim1 ring is seen as late as 96 h AEL. (J-L) The second antenna begins to develop at 100 h AEL and is complete by 108 h AEL. (M-O) Sustained reductions of Toy/ISWI that begin during embryogenesis or the first two larval instars all lead to the formation of a second ectopic antenna. (P) However, if the targeting of Toy/ISWI begins at the L2/L3 transition (or later), then a second antennal field does not form. (M) Embryogenesis (24 h at 18°C). (N) First/second larval transition (72 h at 18°C). (O) Mid/late second instar (84 h at 18°C). (P) Second/third larval transition (108 h at 18°C). (Q,R) If Toy/ISWI activity is restored during embryogenesis or the first larval instar then a second antenna is not produced. (S,T) However, if Toy/ISWI are restored only after the L1/L2 transition, then an ectopic antenna emerges. (Q) Embryonic/first instar transition (24 h at 30°C). (R) Mid first instar (30 h at 30°C). (S) First/second instar transition (42 h at 30°C). (T) Mid second instar (48 h at 30°C). Scale bar: 50 µm.
The ectopic antenna within Toy/E(bx) knockdown discs develops along a similar timeline. In this instance, we followed wg expression, which is transcribed in the dorsal quadrant of the antenna. Similar to Lim1, ectopic wg expression was initiated in the dorsal head epidermis at approximately 96 h AEL (Fig. S7A). By 100 h, we observed significant overgrowth within that region (Fig. S7B), and by 108 h AEL a complete second antenna had been produced (Fig. S7C,D). The identical temporal dynamics of antennal development in Toy/ISWI and Toy/E(bx) double knockdown discs suggests that Toy and NURF are required to ensure the production of a single antennal field in each eye-antennal disc.
We next set out to determine when Toy and the NURF complex are required to prevent ectopic antennal formation. To do this, we used the TARGET system (McGuire et al., 2003) and incorporated a temperature-sensitive GAL80 protein (GAL80ts) into our genetic backgrounds to control the timing of Toy/ISWI RNAi and Toy/E(bx) RNAi expression. At the permissive temperature of 18°C, GAL80ts is capable of inhibiting GAL4 activation of UAS-RNAi constructs. If flies are kept at 18°C throughout development, then the eye-antennal discs and adult heads are completely normal. In contrast, at the non-permissive temperature of 30°C, GAL80ts is non-functional thereby allowing GAL4 to drive expression of UAS-RNAi lines. Holding flies at this temperature leads to the generation of ectopic antennae. By toggling between these temperatures, we controlled the timing of RNAi expression and determined the critical window for Toy/NURF activity in suppressing the formation of a second antenna.
We began by holding DE>toy RNAi, ISWI RNAi embryos/larvae at 18°C (no RNAi expression) for varying periods of time before shifting flies to 30°C (RNAi activation) for the remainder of larval development. Sustained reductions of Toy/ISWI that began during embryogenesis or the first two larval instars all led to the formation of a second ectopic antenna (Fig. 6M-O). However, if the targeting of Toy/ISWI began at the L2/L3 transition (or later), then a second antennal field did not form (Fig. 6P). From this experiment, we conclude that the temporal endpoint for Toy/ISWI activity is the end of the second larval instar. We then did the reciprocal experiment in which we held embryos/larvae at 30°C for varying periods of time before stepping down to 18°C for the remainder of larval development. If Toy/ISWI activity was restored during embryogenesis or the first larval instar then a second antenna was not produced (Fig. 6Q,R). However, if Toy/ISWI were restored only after the L1/L2 transition, then an ectopic antenna emerged (Fig. 6S,T). This indicates that the beginning of the Toy/ISWI window is the start of the second larval instar. The same approach was used to find the critical window for Toy/E(bx) activity, which we found is also required during the second larval instar (Fig. S7E-L). In total, these findings indicate that the formation of a second antenna must be suppressed by Toy/NURF during the second larval instar. If Toy/NURF are removed during this period then an ectopic antenna will emerge from the head epidermis/ocellar region during the mid-third larval instar stage.
Toy/NURF mediates the head epidermis/antenna fate choice
2We then set out to determine the developmental mechanism by which the ectopic antenna arises. The second antenna could result from an early splitting of the nascent antennal field into two smaller domains. If this is the underlying mechanism then we would expect to see two equally sized antennae developing side by side from the earliest stages of development. However, Toy/ISWI and Toy/E(bx)double knockdown discs exhibited only a single antennal field until well into the third larval instar and the ectopic antenna started out at a much smaller size compared with the normal antenna (Fig. 6I-L, Fig. S7A-D). Thus, a splitting of the original antennal field into two daughter fields is an unlikely mechanism.
Another possibility is that the knockdown of Toy/NURF triggers cell death, which, in turn, induces the formation of a blastema at the genetic injury site and elicits tissue regeneration. Although a wounded imaginal disc is often accurately repaired, the fate of the regenerating tissue can, in some instances, be incorrectly specified. This is the mechanism by which the fate of imaginal discs is altered after fragmentation and transplantation (Hadorn, 1965; 1968; 1978). It is also the underlying cause for the duplication of imaginal discs after irradiation (Postlethwait and Schneiderman, 1971a, 1971b, 1971c, 1973; Verghese and Su, 2017). An ectopic antenna could thus arise if removal of Toy/NURF induces a cell death-based wound within the head epidermis that is incorrectly healed during tissue regeneration.
We initially thought that the incorrect resolution of a blastema might indeed play a role because a TUNEL assay revealed the presence of cell death at 96 h AEL within the region of the Toy/ISWI knockdown disc that will eventually give rise to the second antenna (Fig. S8A-D). To test whether this is indeed the underlying mechanism, we blocked cell death by (1) the overexpression of DIAP1 and P35, two potent inhibitors of apoptosis; (2) the expression of an RNAi construct that downregulates the cell death gene eiger; (3) the expression of a microRNA that targets the mRNA of the pro-apoptotic genes hid, grim and reaper; and (4) the expression of an RNAi construct that reduces expression of basket (bsk), the ligand for the cJun N-terminal Kinase (JNK) signaling pathway. In all cases, the inhibition of apoptosis did not affect the formation of the second antenna within Toy/ISWI knockdown discs (Fig. S8E-I). Because we also observed a slight increase in TUNEL staining in Toy/E(bx) knockdown discs (Fig. S9A,B), we repeated these experiments in the Toy/E(bx) knockdown discs and blocking apoptosis via the above methods did not lead to a suppression of the ectopic antenna (Fig. S9C-G). The only exception is that the percentage of animals with antennal duplications dropped slightly when the UAS-P35 construct was introduced into the DE>toy RNAi, E(bx) RNAi strain. As such, in total we propose that apoptosis does not contribute to the production of the ectopic antenna in Toy/NURF double knockdowns.
We also did not see evidence that cell proliferation is enhanced nor that the tissue is regenerating. The former conclusion is based on a lack of elevated levels or localized patterns of cell proliferation using pH3 staining of mitotic cells (Fig. S10A-G). Furthermore, blocking JNK signaling, which in addition to inducing cell death also plays a role in promoting proliferation, did not suppress the formation of the ectopic antenna (Fig. S8I, Fig. S9G). We were also able to rule out tissue regeneration because the blastema markers BRVB-GFP and AP1-RFP (Harris et al., 2016) were not activated consistently in the Toy/ISWI and Toy/E(bx) knockdown discs (Fig. S11A-L). Although the BRVB-GFP marker was activated at very low levels in the Toy/ISWI knockdown discs, it was not expressed in the Toy/E(bx) knockdown discs (Fig. S11A,B,E,F,I,J). Furthermore, the AP1-RFP marker was not hyperactivated in either knockdown background (Fig. S11C,D,G,H,K,L). Lastly, the loss of Toy/NURF did not induce a severe developmental delay (Fig. S12), which often characterizes situations in which the imaginal discs undergo extensive tissue regeneration (Smith-Bolton et al., 2009). Together, these results suggest that although the loss of Toy/NURF does indeed yield minor amounts cell death and activation of some damage-related gene targets, these cellular events are not the underlying cause of the formation of the ectopic antenna.
In contrast, we uncovered evidence that the dorsal head epidermis undergoes a fate change and is transformed into an ectopic antenna. DE>Toy/ISWI RNAi and DE>Toy/E(bx) RNAi knockdown adults both had a deep fissure on the dorsal side of the head between the two compound eyes. This fissure was accompanied by the loss of the head epidermis (Fig. 7A,D, Fig. S13A). DE-GAL4 is an insertion within the mirror (mirr) locus and is normally expressed in the dorsal half of the eye and in the dorsal head epidermis (Fig. S4B; Morrison and Halder, 2010). If this tissue is, in fact, being converted into a second antenna then the expression of the driver should be lost. As expected, if Toy is removed on its own, then there is no effect on the expression of the DE-GAL4 driver. This is consistent with the loss of Toy inducing very mild defects within the ocellar and dorsal head epidermis regions of the adult head (Zhu et al., 2018). However, although the driver was still active in this region at 96 h AEL when Toy and ISWI were simultaneously removed, it was shut off by 120 h AEL (Fig. 7B,C,E,F, Fig. S13B,C). This suggests that the dorsal head epidermis and ocellar domains had changed their fate and were now developing into antennae.
Fig. 7.
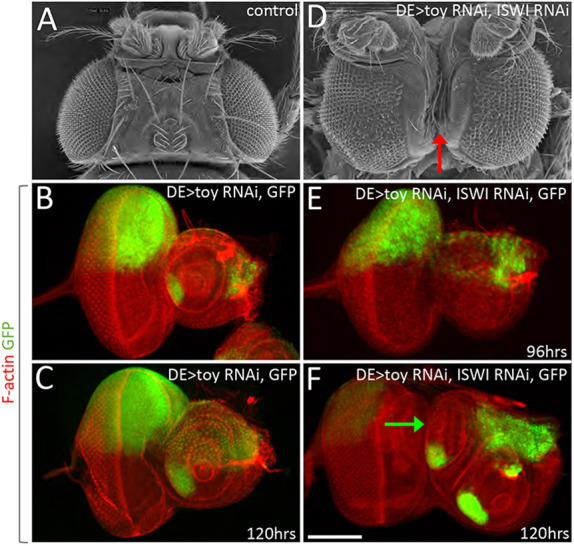
The dorsal head epidermis is lost in response to reductions of Toy/ISWI levels. (A,D) SEM images of wild-type and Toy/ISWI double-knockdown flies. A deep fissure (arrow in D) is seen along the dorsal surface of the adult head between the two compound eyes when Toy/ISWI are reduced. This region normally consists of head epidermal tissue (B,C) Expression pattern of the DE-GAL4 driver. Toy/ISWI are reduced within the dorsal eye, head epidermis, and ocellar region at both 96 and 120 h AEL. (E,F) Although expression of the DE-GAL4 is maintained at 96 h AEL in Toy/ISWI knockdowns, it is lost at 120 h AEL (arrow in F). The loss of DE-GAL4 (which is an insertion in the mirror locus) suggests that the ocellar and head epidermis domains are losing their primary fate. Scale bar: 50 µm.
To characterize further a potential fate transformation, we analyzed the pattern of the homeobox transcription factor Orthodenticle (Otd; also known as ocelliless, oc), which is also expressed in and required for the specification of the ocelli and dorsal head epidermis (Fig. 8A) (Finkelstein et al., 1990; Royet and Finkelstein, 1995). In Toy/NURF double-knockdown discs, otd expression was reduced at 96 h AEL, which again suggests that the head epidermis is losing its primary fate (Fig. 8B, Fig. S14A). As development continues, otd expression was re-activated in cells that separate the two antennae from each other. We suggest that that these cells are being re-specified as head epidermis (Fig. 8C-E, Fig. S14B). We also analyzed wg and decapentaplegic (dpp) expression, which in control discs were found within the dorsal and ventral domains of the antenna, respectively (Fig. 8F,K). In the absence of Toy/NURF, both genes were ectopically activated within the dorsal head epidermis. These patterns were ultimately juxtaposed to each other and this, in turn, triggered the establishment of a new antennal axis (Fig. 8G-J,L-O, Fig. S7A-D, Fig. S14C). In total, we observed a loss of multiple genes that normally promote a head epidermis fate and the concomitant ectopic activation of several genes that are required for antennal development.
Fig. 8.
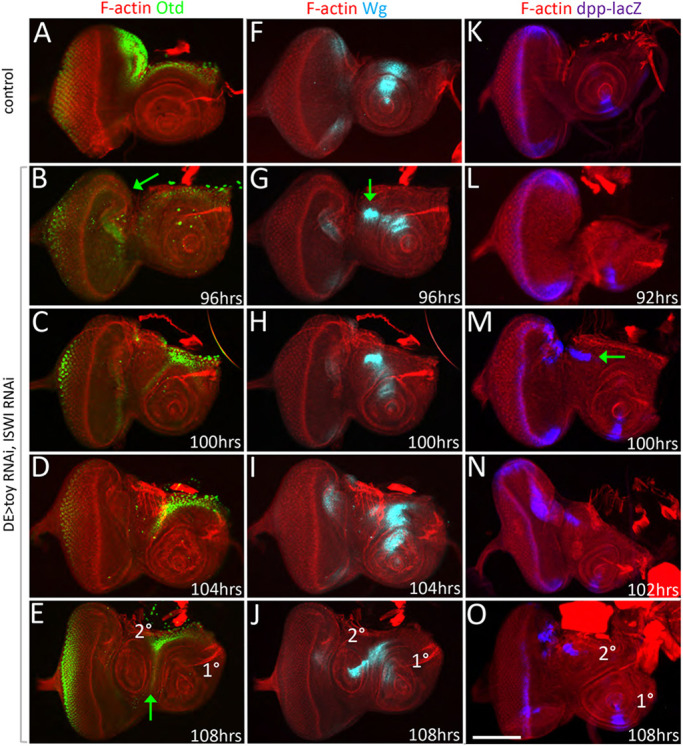
Toy and ISWI maintain the fate of the ocellar and head epidermis domains. (A) otd is expressed within the ocellar and dorsal epidermis domains of wild-type eye-antennal discs. (B-E) otd is first lost within the head epidermis region (arrow in B) at 96 h AEL but is subsequently re-activated in cells between the two antennal fields by 108 h (arrow in E). These cells will form new head epidermis tissue. (F,K) wg and dpp are expressed in the dorsal and ventral domains, respectively, of the wild-type antennal disc. (G-J) During development of the second antenna, wg expression begins to be activated starting at 96 h AEL (arrow in G). (L-O) Expression of dpp follows and is activated in regions that will give rise to the second antenna at 100 h AEL (arrow in M). The juxtaposition of the wg and dpp domains triggers the formation of the second antenna as it does in the endogenous antenna. Scale bar: 50 µm.
The Ets transcription factor Pointed is a target of the NURF complex
Because NURF participates in the activation of thousands of loci, we searched for direct gene targets that are relevant for specifying the fate of the head epidermis and for preventing it from being reprogrammed into an antenna. We consulted Flybase (http://flybase.org/), the Janelia FlyLight Project (https://www.janelia.org/project-team/flylight) to identify 400 genes that are annotated to be involved in head epidermis formation. We then used Flybase annotations to restrict this list to transcription factors and signaling pathways. RNAi targeting lines for 177 genes on this more restricted list are available from the Bloomington Drosophila Stock Center (Table S3). We combined the DE>toy RNAi line with each of these RNAi constructs in an effort to identify dual knockdown combinations that would mimic the Toy/ISWI and Toy/E(bx) mutant phenotypes. We reasoned that if knocking down a head epidermis gene (in conjunction with toy) induced an ectopic antenna, then that gene might be a target of the NURF complex. Eighteen double-knockdown combinations yielded antennal duplications at various frequencies. To narrow down the list of possible candidates, we bioinformatically analyzed ChIP-seq data on E(bx) binding in wild-type larval hemocytes and MNase-seq data of genome accessibility in E(bx) mutants from Kwon et al. (2016). Of the 18 genes that could be playing a role with Toy in the head epidermis/antennal fate choice, E(bx) appears to bind to only one locus, that of the Ets transcription factor pointed (pnt). We therefore reasoned that pnt may be a direct target of the NURF complex. We should note that as the binding of E(bx) to the pnt locus was observed in larval hemocytes (Kwon et al., 2016), it is still possible that E(bx) indirectly regulates Pnt in the eye-antennal disc.
In order to determine whether Toy is also regulating the pnt locus, we searched for the presence of Toy-binding sites (Punzo et al., 2002) within the locus itself and 50 kb of intergenic DNA flanking both ends of the gene. We failed to find any intact consensus sites within the pnt locus itself. The nearest potential sites contain several mismatches and are actually closest to the CG4374, ATPsynCF6 and CG4467 genes. Thus, it is highly unlikely that pnt is being co-regulated directly by Toy and NURF. We also determined that toy expression appears normal in pnt knockdown discs (Fig. S4E), which suggests that Toy is not regulated by Pnt.
Pnt is one of several transcription factors that are modulated by and transcribed in response to signaling from the Sevenless (Sev) and Epidermal Growth Factor Receptor (EGFR) pathways (Brunner et al., 1994; O'Neill et al., 1994; Gabay et al., 1996). EGFR signaling is required for ocellar development (Amin et al., 1999; Amin, 2003), which is consistent with our finding that the loss of pnt also blocks ocellar formation. The pnt locus encodes two isoforms: PntP1 and PntP2. Upon phosphorylation by Ras/MAPK, PntP2 is able to induce expression of the constitutively active form, PntP1 (Brunner et al., 1994; O'Neill et al., 1994). In the developing eye, both isoforms are required for the specification of photoreceptor and cone cells (O'Neill et al., 1994; Treier et al., 1995; Flores et al., 2000; Rogers et al., 2005).
An enhancer-GAL4 line from the Janelia FlyLight collection (containing a genomic fragment of the pnt locus) drives reporter expression within the ocellar/head epidermis region of the eye disc (Jin et al., 2016). We examined the expression of a Pnt-GFP BAC construct that was generated by the modERN (model organism Encyclopedia of Regulatory Networks) consortium (Kudron et al., 2018) and confirmed that pnt is transcribed in this region of the disc (Fig. 9A). Whereas removal of pnt itself affected ocelli and bristle development without inducing the head epidermis-to-antennal transformation (Fig. S15A,C), Toy/Pnt double knockdowns induced antennal duplications at 100% penetrance (Fig. 9B-E; Table S3).
Fig. 9.
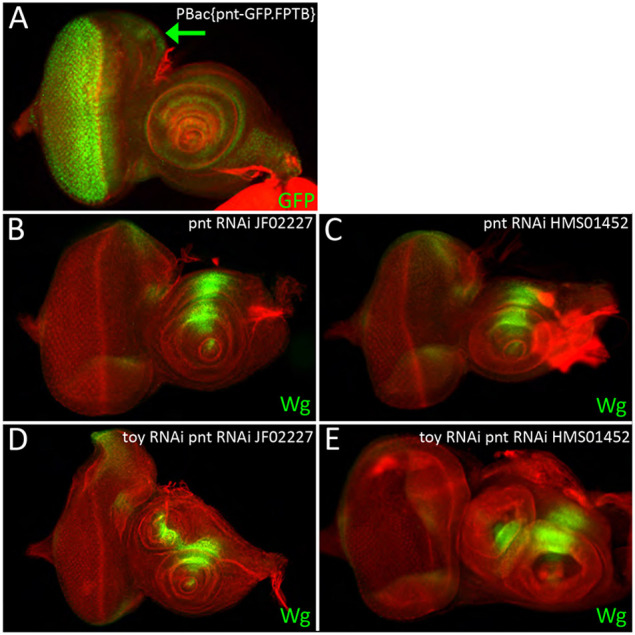
Toy and Pointed are required to prevent formation of an ectopic antenna. (A) The pnt gene is transcribed within the region of the eye-antennal disc that gives rise to the second antenna. (arrow). (B,C) Knockdown of pnt expression alone does not alter the number of antennae that are produced within the eye-antennal disc. (D,E) However, if Pnt and Toy are knocked down simultaneously, then a second ectopic antenna forms in place of the head epidermis. This appears identical to when Toy and NURF are both knocked down. Anterior is to the right. Scale bar: 50 µm.
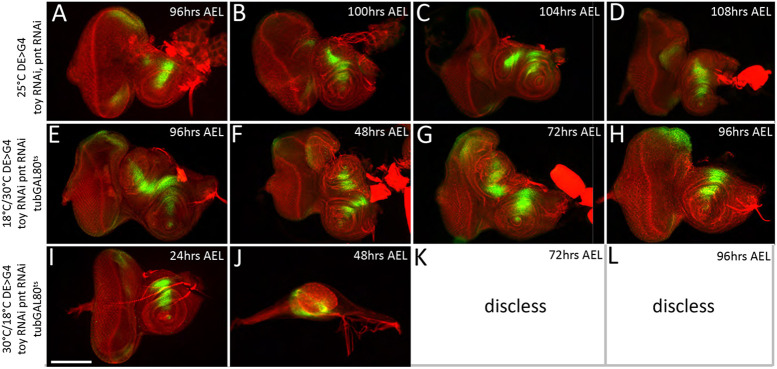
A temporal analysis of Toy/Pnt double knockdown discs revealed that the ectopic antenna arises from the same location within the disc and within the same developmental window as both Toy/ISWI and Toy/E(bx) knockdown discs – the middle of the third larval instar (Fig. 10A-D, Fig. 6I-L, Fig. S7A-D). Similarly, we determined that Toy/Pnt functions to block the transformation during the second larval instar (Fig. 10E-L), which is the same critical window for Toy/ISWI and Toy/E(bx) (Fig. 6M-T, Fig. S7E-L). The identical temporal dynamics of ectopic antennal formation and the coincident timing of Toy/Pnt, Toy/ISWI and Toy/E(bx) activity support the suggestion that pnt is indeed a downstream target of the NURF complex.
Fig. 10.
Toy/Pnt are required during the second larval instar to suppress formation of the second antenna. (A-D) In Toy/Pnt knockdown discs, the second antenna initiates its development during the middle of the third larval instar. As with Toy/NURF double knockdown discs, wg expression is activated within the dorsal head epidermis at 96 h AEL. (E-L) Controlling the expression of the pnt RNAi lines during development indicates that Toy/Pnt are required during the second larval instar stage. Toy/ISWI and Toy/E(bx) are required during the same developmental window. Red, F-actin; green, Wingless. Scale bar: 50 µm.
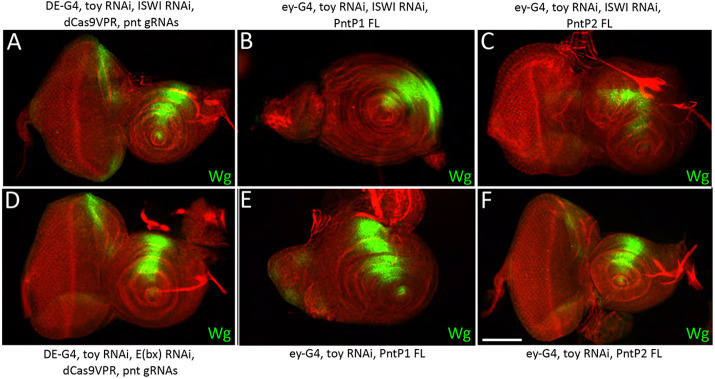
We then attempted to suppress the formation of the ectopic antenna by using an in vivo Cas9 activation method (Ewen-Campen et al., 2017) to overexpress Pnt in the Toy/ISWI and Toy/E(bx) double-knockdown discs. In this method, the DE-GAL4 driver directs expression of a catalytically dead version of Cas9 (dCas9) that has been fused to an optimized tripartite activation domain (VP64-p65-Rta: called UAS-dCas9-VPR). The dCas9-VPR is brought to its target promoter by a synthetic guide RNA (sgRNA). This method has several advantages over traditional UAS-GAL4 overexpression assays including, expression of the target gene closer to physiologically relevant levels. Overexpressing Pnt using this approach completely suppressed the head epidermis-to-antenna transformation in Toy/ISWI and Toy/E(bx) double knockdowns (Fig. 11A,D). In order to determine which of the two Pnt isoforms is maintaining the fate of the head epidermis, we forcibly expressed each isoform independently within Toy/ISWI double-knockdown discs using the traditional UAS/GAL4 system. We found that expression of PntP1 is sufficient to suppress the formation of the second antenna (Fig. 11B). We note that the eye portion of the disc was poorly formed when PntP1 was overexpressed. This is likely due to the fact that UAS/GAL4-based gain-of-function approaches invariably lead to very high levels of expression. This was confirmed when we overexpressed PntP1 in control discs and observed that the eye was similarly distorted but the antenna remained intact (Fig. 11E). In contrast, the expression of PntP2 had minimal effects on the eye-antennal disc when expressed by itself and it also failed to rescue the Toy/ISWI phenotype (Fig. 11C,F). This may be because PntP2 activity requires phosphorylation by MAPK. In contrast, PntP1 activity is not directly dependent upon MAPK activity. The rescue of Toy/ISWI by PntP1 further indicates and the binding of E(bx) to the pnt locus suggests that it lies downstream of NURF.
Fig. 11.
Expression of pnt suppresses the formation of the antenna that results from Toy/ISWI knockdown. (A) The formation of the second antenna (induced from knocking down Toy/ISWI) is completely suppressed by the expression of pnt. The locus encodes two isoforms, PntP1 and PntP2. (D) The formation of the second antenna [induced from knocking down Toy and E(bx)] is completely suppressed by the expression of pnt. (B,E) Expression of the PntP1 isoform alone using the UAS/GAL4 system suppresses ectopic antenna formation. Note that eye development is severely impaired by the ectopic expression of PntP1. (C,F) In contrast, the expression of PntP2 fails to suppress the formation of the second antenna. Scale bar: 50 µm.
DISCUSSION
The specification and patterning of tissues and organs are essential properties of metazoan development. Failure to execute either of these two programs correctly is often disastrous and is the leading cause of numerous congenital disorders. To gain a molecular understanding of both development and disease, forward genetic screens are often employed as a means to identify the full repertoire of factors that comprise the gene regulatory networks (GRNs) that underly specification and patterning. Genetic screens employ a variety of agents, including chemical mutagens, irradiation, transposable elements, morpholinos, RNAi and CRISPR approaches. Irrespective of which agent is used, considerable effort is placed, at some stage, on guaranteeing that each mutant strain contains one, and only one, disrupted gene. This ensures that a mutant phenotype can be linked to the loss of a single gene. Factors that appear to regulate the same developmental process (by displaying a common mutant phenotype) are then grouped together into a GRN. For instance, membership in the RD GRN of Drosophila is contingent upon loss-of-function mutants having severely reduced or absent compound eyes and with overexpression yielding ectopic eyes.
In this paper, we have demonstrated that tissue fates are also influenced by combinations of independently acting transcription factors and epigenetic regulators. We show that Toy, a Pax6 transcription factor, and chromatin-modifying proteins regulate independent targets that are then required together to maintain the fate of the ocellar/head epidermis within the eye-antennal disc. We focused on the NURF ATP-dependent nucleosome remodeling complex and demonstrate that the simultaneous loss of Toy/NURF results in transformation of the head epidermis into an antenna. Surprisingly, this change in fate is not observed when either Toy or the NURF complex are removed individually. We went on to identify Pointed, an Ets transcription factor, as being a potential direct target of the NURF complex within the head epidermis. As with Toy/NURF double knockdowns, the combined loss of Toy/Pnt induces the transformation of the head epidermis into an antenna. The critical window for both Toy/NURF and Toy/Pnt activity appear to coincide during the second instar. Likewise, the timing of ectopic antennal development, which takes place in the mid third larval instar, are nearly identical in Toy/NURF and Toy/Pnt knockdown discs (Fig. 12A). As E(bx) binds to an enhancer within the pnt locus, it is possible that pnt is a direct target of the NURF complex.
Fig. 12.
Combinatorial code of transcriptional networks reveals the complexity of development. (A) A schematic outlining the summary of events that take place when Toy is removed simultaneously with either NURF or Pnt. (B) Schematic illustrating how the removal of multiple transcriptional networks reveals developmental choices that are not predicted by the knockdown of individual factors. (C) Our data indicates that a hidden step in the development of the eye-antennal disc is the choice between the fate of the head epidermis and antenna. We suggest that Toy and NURF work on independent gene targets to promote the fate of the head epidermis. NURF appears to directly regulate pnt but the targets of Toy are yet to be identified.
The removal of Toy, NURF and Pnt individually disrupts specification of the ocelli and/or the surrounding head epidermis. Based on the individual loss-of-function phenotypes, these factors would be classified as being part of a GRN for either ocellar or head epidermis development. As such, there is little reason for them to be removed simultaneously and their roles in the decision to mediate the decision between head epidermis and antennal fate would have remained permanently hidden (Fig. 12B,C).
Similar observations regarding the intersection of multiple transcriptional regulators during development have been made within the neighboring eye field. Its fate is established by the RD transcriptional network, which includes the Ey and Toy transcription factors. The eye is lost and replaced with head epidermis in loss-of-function mutants. However, the eye field can be further reprogrammed and forced into adopting a wing fate if mutations in either ey, eyes reduced (eyr), Deformed (Dfd) or loboid (ld) are combined with those in ophthalmoptera (opht) (Goldschmidt and Lederman-Klein, 1958; Edwards and Gardner, 1966; Kobel, 1968; Ouweneel, 1970). Another path for redirecting the eye towards a wing fate involves simultaneously removing members of the PcG family of epigenetic regulators with either Ey or Toy (Zhu et al., 2018). A common theme that runs through these genetic combinations is that the eye-to-wing fate switch does not occur with the individual loss of any of these components. Furthermore, there is no clear connection between the mutant phenotypes of individual genes. For instance, mutations in eyr, opht, and most PcG genes affect the overall growth and patterning of the eye whereas ocellar development is disrupted in toy mutants. Similarly, the eye is transformed into head epidermis in ey mutants and an antenna in ld mutants. Because these individual phenotypes are so disparate in nature, there would be little reason to combine these loss-of-function mutants together. However, the creation of the above double-mutant combinations is the only path to revealing that the choice between an eye and wing fate is an important decision that must be made in imaginal disc development.
The results in this paper, along with those describing the eye-to-wing transformation, reveal that many fate decisions require multiple independently acting transcriptional regulators and these often elude detection by traditional genetic screens. We propose that identification of the full palette of decisions that are made by a cell or tissue will require an approach that makes use of removing or knocking down multiple factors simultaneously.
MATERIALS AND METHODS
Stocks
The following fly stocks were used in the main and supplementary figures of this study: (1) ey-GAL4 [Bloomington Drosophila Stock Center, (BDSC), BL8221]; (2) DE-GAL4 (Georg Halder, Katholic University, Leuven, Belgium); (3) eya2 (Nancy Bonini, University of Pennsylvania, Philadelphia, PA, USA); (4) so1 (Larry Zipursky, University of California, Los Angeles, CA, USA); (5) UAS-toy RNAi (BDSC, BL33679); (6) UAS-ey RNAi (BDSC, BL32486); (7) UAS-ISWI RNAi (BDSC, BL32845, BL31111); (8) UAS-ISWIDN (Brian Calvi, Indiana University, Bloomington, IN, USA; Deuring et al., 2000); (9) UAS-E(bx) RNAi (BDSC, BL33658); (10) UAS-eiger RNAi (BDSC, BL55276); (11) UAS-bsk RNAi (BDSC, BL32977, BL53310, BL57035); (12) UAS-pnt RNAi (BDSC, BL31936, BL35038); (13) UAS-GFP (BDSC, BL4775); (14) UAS-p35 (Gerald Rubin, HHMI Janelia Research Campus, Ashburn, VA, USA, BL5072); (14) UAS-DIAP1 (Bruce Hay, California Institute of Technology, Pasadena, CA, USA); (16) UAS-miR for hid/grim/rpr (Iswar Hariharan, University of California, Berkeley, CA, USA); (17) UAS-pntP1 (BDSC, BL869); (18) UAS-pntP2 (BDSC, BL399); (19) P{y[+t7.7]=Mae-UAS.6.11}pnt[LA00739] (BDSC, BL22190); (20) dpp-lacZ (Kevin Moses, Emory University, Atlanta, GA, USA); (21) PBac{y[+mDint2]w[+mC]=pnt-GFP.FPTB}VK00037 (BDSC, BL42680); (22) tub-GAL80ts10 (BDSC, BL7108); (23) w; BVRB-GFP (Iswar Hariharan, University of California, Berkeley, CA, USA); (24) w1118 (Kevin Moses, Emory University, Atlanta, GA, USA); (25) AP1 w[*]; P[y(+t7.7) w(+mC)=TRE-DsRedT4]attP40 (BDSC, BL59011); (26) y1 sc* v1 sev21; P[y(+t7.7) v(+1.8)=TOE.GS05176]attP40 (BDSC, BL82744); (27) y w*; P[y(+t7.7) w(+mC)=UAS-3xFLAG.dCas9.VPR]attP40 (BDSC, BL66561). BDSC stock numbers of RNAi lines that were used in the genetic screens of chromatin-modifying proteins and head capsule genes are listed in Tables S1 and S2, respectively.
All crosses were maintained at 25°C unless stated otherwise and all experiments were conducted on standard media, except those involving developmental time courses, which were conducted on molasses-rich media spiked with yeast. Each experiment was performed at least three times and at least 30 discs and 30 adults of the correct genotype were scored. The full genotypes for each main and supplementary figure panel are provided in Table S4.
Antibodies
The following primary antibodies were used in this study: (1) mouse anti-Ey [1:250, anti-eyeless, Developmental Studies Hybridoma Bank (DSHB)]; (2) mouse anti-Eya (1:5, eya10H6, DSHB); (3) mouse anti-Dac (1:5, mAbdac1-1, DSHB); (4) mouse anti-Wg (1:800, 4D4, DSHB); (5) mouse anti-β-galactosidase (1:250, Z3781, Promega); (6) chicken anti-β-Galactosidase (1:800, ab9361, Abcam); (7) guinea pig anti-Otd (1:750, Tiffany Cook, Wayne State University, Detroit, MI, USA); (8) mouse anti-Ct (1:100, 2B10, DSHB); (9) mouse anti-Dll (1:500, Diana Duncan, Washington University, St. Louis, MO, USA); (10) mouse anti-Al (1:50, Gerard Campbell, University of Pittsburgh, PA, USA); (11) mouse anti-Ss (1:100, Ian Duncan, Washington University, St. Louis, MO, USA); (12) mouse anti-Dcp1 (1:100, Asp216, Cell Signaling Technologies); (13) rat anti-CiACT (1:50, Robert Holmgren, Northwestern University, Evanston, IL, USA); (14) rabbit anti-Lim1 (1:800, Tetsuya Kojima, University of Tokyo, Japan); (15) rat anti-ELAV (1:100, 7E8A10, DSHB); (16) rabbit anti-pH3 (1:20,000, ab80612, Abcam); (17) rabbit anti-GFP (1:1000, A-11122, Invitrogen). The following secondary antibodies are from Jackson Laboratories and were each used at a concentration of 1:100: (1) AffiniPure donkey anti-rat IgG (712-005-153); (2) AffiniPure donkey anti-mouse IgG (715-005-151); (3) AffiniPure donkey anti-rabbit IgG (711-005-152); (4) AffiniPure donkey anti-guinea pig IgG (706-005-148); (5) AffiniPure donkey anti-chicken IgG (703-005-155). Alexa Fluor 594 Phalloidin from Thermo Fisher Scientific (A12381) was used at a concentration of 1:20 to detect F-actin.
Microscopy
Imaginal discs were prepared, viewed on a Zeiss Axioplan II compound fluorescence microscope, and photographed as described by Spratford and Kumar (2014). Adult samples were prepared, viewed with a scanning electron microscope or a Zeiss Discovery microscope, and photographed as described by Palliyil et al. (2018).
GAL80 control of RNAi induction
GAL80ts, DE-GAL4, UAS-toy RNAi, UAS-ISWI RNAi and GAL80ts, DE-GAL4, UAS-toy RNAi, UAS-E(bx) RNAi embryos were collected at 25°C for 2 h and were then incubated at either 18°C or 30°C for defined periods of time before being shifted to the alternate temperature for the remainder of development. Some larvae were then dissected at the late wandering third instar stage whereas others were allowed to mature into pharate adults.
ChIP-seq and MNase-seq data analysis
NURF ChIP-seq and MNase-seq experimental and control Bam files that are associated with Kwon et al., 2016 were downloaded from the European Nucleotide Archive (ENA). For the ChIP-seq files, peaks were called using MACS (v2.2.7.1) (Gaspar, 2018 preprint) with custom settings ‘-s 50 –q 0.1 –keep-dup ‘all’ -g ‘dm’’. Peaks were then annotated to the nearest gene (from the BDGP R5 dm3 genome annotation) using the annotatePeak function from the R ChIPseeker (v1.24.0) (Yu et al., 2015) library with settings ‘tssRegion=c(−3000, 1000), level=‘gene’’. deepTools (v3.4.3) (Ramírez et al., 2016) bamCoverage was used to create CPM normalized bigwig files with settings ‘-bs 10 –normalizeUsing ‘CPM’ -e 200’. Input and NURF experimental data sets can be found at https://www.ebi.ac.uk/ena/data/view/ERX1488474 and https://www.ebi.ac.uk/ena/data/view/ERX1488191. For the MNase-seq analysis, The R library EdgeR (v3.30.3) (McCarthy et al., 2012) was used to find differential MNase signal based on the total reads in the −250 to +250 promoter region for each gene. For visualization, replicates were first merged using Samtools (v1.10) (Li et al., 2009), and then SES normalized bigwigs were produced with deepTools bamCompare with settings ‘--scaleFactorsMethod ‘SES’ -operation ‘log2’ -bs 1’. Wild-type and knockdown data sets can be found at https://www.ebi.ac.uk/ena/data/view/ERX1375378, https://www.ebi.ac.uk/ena/data/view/ERX1375379, https://www.ebi.ac.uk/ena/data/view/ERX1375380 and https://www.ebi.ac.uk/ena/data/view/ERX1375381.
Toy-binding site analysis
We searched for Toy-binding sites (Punzo et al., 2002) using a custom R script that is available upon request. Briefly, the Biostrings (v2.56.0) (Pages et al., 2020) function ‘vmatchPattern’ was used to search the D. melanogaster dm6 genome from the BSgenome.Dmelanogaster.UCSC.dm6 (v1.4.1) library for motif matches with up to three mismatches. The coordinates of motif matches were then annotated to the nearest genomic features with the ChIPseeker (v1.24.0) (Yu et al., 2015) function ‘annotatePeak’ with promoter regions defined as ±2.5 kb, and using the TxDb.Dmelanogaster.UCSC.dm6.ensGene (v3.11.0) and org.Dm.eg.db (3.11.4) libraries (Carlson, 2019). Genomic coordinates for the pnt locus were next retrieved using the GenomicsFeatures (v1.40.0) ‘genes’ function (Lawrence et al., 2013) on the TxDb.Dmelanogaster.UCSC.dm6.ensGene library and re-sized to include 50 kb upstream and downstream of the locus. Motif matches intersecting by at least one base with the expanded pnt locus were retained.
Quantitative PCR (qPCR) of gene expression
We dissected discs from third instar larvae as previously described. RNA was extracted from approximately 60 unfixed discs per sample using the Qiagen RNeasy Plus Mini Kit (Qiagen, 74104). We then created cDNA libraries from 100 ng of RNA using the Invitrogen SuperScript III First Strand Synthesis Supermix kit (Thermo Fisher Scientific, 11752250). We then prepared samples for qPCR analysis using the LightCycler 480 SYBR Green I Master kit (Roche, 04707516001) and ran the reactions on a Roche LightCycler 96 machine. qPCR was run on three independently generated cDNA libraries per genotype with technical triplicates and all appropriate controls. Resulting data were analyzed using Microsoft Excel and GraphPad Prism 9 software. We determined relative expression values for the Toy eye selector gene, the ISWI and E(bx) NURF complex genes, and Vestigial (Vg), which is a wing selector gene. Toy primers used were: forward 5′-CCAGAGGCACGTATTCAGGTTTGG-3′, reverse 5′-TTATTTGCCGTGCTGGTTCGA-3′. Vg primers used were: forward 5′-TGCCCCGAAGTTATGTACGG-3′, reverse 5′-TGGTTGAACCTCTCATACTGGTA-3′. ISWI primers used were: forward 5′-AAGAGTCCCACGAAGCCTAAG-3′, reverse 5′-GTGAGGCATCGAAGCGAAAGA-3′. E(bx) primers used were: forward 5′-TCTGGAAGGAAAGGGTACGAG-3′, reverse 5′-CTGGGAGTAAGCAGCATATCATC-3′.
Quantification of cell proliferation
Imaginal discs were dissected, stained and imaged as previous described. All discs were imaged at a predetermined exposure time and focal plane. Representative (those on the predetermined focal plane) pH3-positive nuclei were then marked in the Fiji software package using a standard threshold value and counted using the Fiji analyze particles function. This representative number of pH3-positive nuclei was then divided by the area of the disc, producing a pH3-positive nuclei/disc area ratio. Datasets of n=15 discs were collected per genotype. Datasets collected from DE-GAL4, toy RNAi, ISWI RNAi and DE-GAL4, toy RNAi, E(bx) RNAi discs were then compared with DE-GAL4 control discs at the same time point using a two-tailed unpaired t-test in GraphPad Prism 9 software.
Supplementary Material
Acknowledgements
We would like to thank Walter Gehring (deceased), Georg Halder (Katholic University, Leuven, Belgium), Larry Zipursky (University of California, Los Angeles, CA, USA), Nancy Bonini (University of Pennsylvania, Philadelphia, PA, USA), Ken Cadigan (University of Michigan, Ann Arbor, MI, USA), Iswar Hariharan (University of California, Berkeley, CA, USA), Kevin Moses (Emory University, Atlanta, GA, USA), Brian Calvi (Indiana University, Bloomington, IN, USA), Jason Tennessen (Indiana University, Bloomington, IN, USA), Rudi Turner (deceased), Robert Holmgren (Northwestern University, Evanston, IL, USA), the Bloomington Drosophila Stock Center, the Developmental Studies Hybridoma Bank, the IU Bloomington Electron Microscopy Center, and Flybase for fly strains, antibodies, equipment, technical support with scanning electron microscopy and information on Drosophila chromatin-modifying enzymes.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.P.K.; Methodology: A.J.O., B.P.W.; Formal analysis: A.J.O., R.P.; Investigation: A.J.O., G.M.T., B.M.W.; Data curation: B.M.W.; Writing - original draft: J.P.K.; Writing - review & editing: A.J.O., G.M.T., B.M.W., J.P.K.; Supervision: J.P.K.; Project administration: J.P.K.; Funding acquisition: J.P.K.
Funding
The work was supported by the Robert C. Briggs Fellowship in Developmental Biology to A.J.O. as well as grants from the National Eye Institute (NEI) (R01 EY014863, R01 EY030847) and funds from the Department of Biology, College of Arts and Sciences, and the Office of the Vice Provost for Research at Indiana University to J.P.K. Deposited in PMC for release after 12 months.
References
- Amin, A. (2003). EGFR and wingless signaling pathways interact to specify the ocellar pattern in Drosophila. Acta Histochem. 105, 285-293. 10.1078/0065-1281-00729 [DOI] [PubMed] [Google Scholar]
- Amin, A., Li, Y. and Finkelstein, R. (1999). Hedgehog activates the EGF receptor pathway during Drosophila head development. Development 126, 2623-2630. 10.1242/dev.126.12.2623 [DOI] [PubMed] [Google Scholar]
- Badenhorst, P., Voas, M., Rebay, I. and Wu, C. (2002). Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16, 3186-3198. 10.1101/gad.1032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, L. R., Weasner, B. M., Nagel, A., Neuman, S. D., Bashirullah, A. and Kumar, J. P. (2018). Eyeless/Pax6 initiates eye formation non-autonomously from the peripodial epithelium. Development 145, dev163329. 10.1242/dev.163329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham, L. (1942). Boundaries of differentiation of cephalic imaginal discs in Drosophila. J. Exp. Zool. 91, 345-363. 10.1002/jez.1400910303 [DOI] [Google Scholar]
- Blanco, J., Pauli, T., Seimiya, M., Udolph, G. and Gehring, W. J. (2010). Genetic interactions of eyes absent, twin of eyeless and orthodenticle regulate sine oculis expression during ocellar development in Drosophila. Dev. Biol. 344, 1088-1099. 10.1016/j.ydbio.2010.05.494 [DOI] [PubMed] [Google Scholar]
- Bodenstein, D. (1950). The postembryonic development of Drosophila. In Biology od Drosophila (ed. Demerec M.). Cold Spring Harbor Laboratory Press, New York, pp. 275-367. [Google Scholar]
- Bonini, N. M., Leiserson, W. M. and Benzer, S. (1993). The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379-395. 10.1016/0092-8674(93)90115-7 [DOI] [PubMed] [Google Scholar]
- Brockmann, A., Dominguez-Cejudo, M. A., Amore, G. and Casares, F. (2011). Regulation of ocellar specification and size by twin of eyeless and homothorax. Dev. Dyn. 240, 75-85. 10.1002/dvdy.22494 [DOI] [PubMed] [Google Scholar]
- Brunner, D., Ducker, K., Oellers, N., Hafen, E., Scholz, H. and Klambt, C. (1994). The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370, 386-389. 10.1038/370386a0 [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J. A. and Hofbauer, A. (1977). Cell clones and pattern formation: on the lineage of photoreceptor cells in the compound eye of Drosophila. Wilhelm Roux's Archives of Developmental Biology 181, 227-245. 10.1007/BF00848423 [DOI] [PubMed] [Google Scholar]
- Carlson, M. (2019). org.Dm.eg.db: Genome wide annotation for Fly. R package version 3.8.2.
- Chen, T.-Y. (1929). On the development of imaginal buds in normal and mutant Drosophila melanogaster. Journal of Morphology and Physiology 47, 135-199. 10.1002/jmor.1050470105 [DOI] [Google Scholar]
- Cheyette, B. N., Green, P. J., Martin, K., Garren, H., Hartenstein, V. and Zipursky, S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977-996. 10.1016/0896-6273(94)90308-5 [DOI] [PubMed] [Google Scholar]
- Cohen, S. M. (1993). Imaginal disc develoment. In The development of Drosophila melanogaster. (ed. Bate M. and Martinez Arias M.). Cold Spring Harbor Press, New York, pp. 787-841. [Google Scholar]
- Corona, D. F. V. and Tamkun, J. W. (2004). Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochemica et Biophysica Acta 1677, 113-119. 10.1016/j.bbaexp.2003.09.018 [DOI] [PubMed] [Google Scholar]
- Czerny, T., Halder, G., Kloter, U., Souabni, A., Gehring, W. J. and Busslinger, M. (1999). Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3, 297-307. 10.1016/S1097-2765(00)80457-8 [DOI] [PubMed] [Google Scholar]
- Deuring, R., Fanti, L., Armstrong, J. A., Sarte, M., Papoulas, O., Prestel, M., Daubresse, G., Verardo, M., Moseley, S. L., Berloco, M.et al. (2000). The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5, 355-365. 10.1016/S1097-2765(00)80430-X [DOI] [PubMed] [Google Scholar]
- Edwards, J. W. and Gardner, E. J. (1966). GA menetics of the eyes-reduced mutant of Drosophila melanogaster, with special reference to homoeosis and eyelessness. Genetics 53, 785-798. 10.1093/genetics/53.4.785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen, B., Yang-Zhou, D., Fernandes, V. R., González, D. P., Liu, L.-P., Tao, R., Ren, X., Sun, J., Hu, Y., Zirin, J.et al. (2017). Optimized strategy for in vivo Cas9-activation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 114, 9409-9414. 10.1073/pnas.1707635114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, G. F. (1950). External morphology of the adult. In Biology of Drosophila (ed. Demerec M.). Cold Spring Harbor Laboratory Press, New York, pp. 368-419. [Google Scholar]
- Finkelstein, R., Smouse, D., Capaci, T. M., Spradling, A. C. and Perrimon, N. (1990). The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev. 4, 1516-1527. 10.1101/gad.4.9.1516 [DOI] [PubMed] [Google Scholar]
- Flores, G. V., Duan, H., Yan, H., Nagaraj, R., Fu, W., Zou, Y., Noll, M. and Banerjee, U. (2000). Combinatorial signaling in the specification of unique cell fates. Cell 103, 75-85. 10.1016/S0092-8674(00)00106-9 [DOI] [PubMed] [Google Scholar]
- Fristrom, D. and Fristrom, J. W. (1993). The metamorphic development of the adult epidermis. In The development of Drosophla melanogaster (ed. Bate M. and Martinez-Arias A.). Cold Spring Harbor Press, New York, pp. 467-516. [Google Scholar]
- Gabay, L., Scholz, H., Golembo, M., Klaes, A., Shilo, B. Z. and Klambt, C. (1996). EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 122, 3355-3362. [DOI] [PubMed] [Google Scholar]
- Gaspar, J. M. (2018). Improved peak-calling with MACS2. bioRxiv, 10.1101/496521 [DOI] [Google Scholar]
- Goldschmidt, E. and Lederman-Klein, A. (1958). Reoccurrence of a forgotten homeotic mutant in Drosophila. J. Hered. 49, 262-266. 10.1093/oxfordjournals.jhered.a106822 [DOI] [Google Scholar]
- Hadorn, E. (1965). Problems of determination and transdetermination. Brookhaven Symp. Biol. 18, 148-161. [Google Scholar]
- Hadorn, E. (1968). Transdetermination in cells. Sci. Amer. 219, 110-172. 10.1038/scientificamerican1168-110 [DOI] [PubMed] [Google Scholar]
- Hadorn, E. (1978). Transdetermination. In The Genetics and Biology of Drosphila, vol 2c (ed. Ashburner M. and Wright). Academic Press, New York, pp. 555-617. [Google Scholar]
- Harris, R. E., Setiawan, L., Saul, J. and Hariharan, I. K. (2016). Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, B., Gehring, W. J. and Walldorf, U. (1999). Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. USA96, 564-569. 10.1073/pnas.96.2.564 [DOI] [PMC free article] [PubMed]
- Haynie, J. L. and Bryant, P. J. (1986). Development of the eye-antennal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 237, 293-308. 10.1002/jez.1402370302 [DOI] [PubMed] [Google Scholar]
- Hoge, M. A. (1915). Another gene in the fourth chromosome of Drosophila. Am. Nat. 49, 47-49. 10.1086/279455 [DOI] [Google Scholar]
- Jacobsson, L., Kronhamn, J. and Rasmuson-Lestander, A. (2009). The Drosophila Pax6 paralogs have different functions in head development but can partially substitute for each other. Mol. Genet. Genomics 282, 217-231. 10.1007/s00438-009-0458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., Aibar, S., Ge, Z., Chen, R., Aerts, S. and Mardon, G. (2016). Identification of novel direct targets of Drosophila Sine oculis and Eyes absent by integration of genome-wide data sets. Dev. Biol. 415, 157-167. 10.1016/j.ydbio.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens, J. and Hartenstein, V. (1993). The terminal regions of the body pattern. In The development of Drosophila melanogaster (ed. Bate M. and Martinez-Arias M.). Cold Spring Harbor Press, New York, pp. 687-746. [Google Scholar]
- Kobel, H. R. (1968). Homeotic wing formation through the loboid allele “ophthalmoptera” (ld-oph) in Drosophila melanogaster. Genetica 39, 329-344. 10.1007/BF02324472 [DOI] [PubMed] [Google Scholar]
- Krafka, J. (1924). Development of the compound eye of Drosophila melanogaster and its Bar-eyed mutant. Biological Bulletin 47, 143-149. 10.2307/1536493 [DOI] [Google Scholar]
- Kudron, M. M., Victorsen, A., Gevirtzman, L., Hillier, L. W., Fisher, W. W., Vafeados, D., Kirkey, M., Hammonds, A. S., Gersch, J., Ammouri, H.et al. (2018). The ModERN Resource: genome-wide binding profiles for hundreds of Drosophila and Caenorhabditis elegans transcription factors. Genetics208, 937-949. 10.1534/genetics.117.300657 [DOI]
- Kugler, S. J. and Nagel, A. C. (2010). A novel Pzg-NURF complex regulates Notch target gene activity. Mol. Biol. Cell 21, 3443-3448. 10.1091/mbc.e10-03-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, J. P. and Moses, K. (2001). EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell 104, 687-697. 10.1016/S0092-8674(01)00265-3 [DOI] [PubMed] [Google Scholar]
- Kwon, S. Y., Grisan, V., Jang, B., Herbert, J. and Badenhorst, P. (2016). Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators. PLoS Genet. 12, e1005969. 10.1371/journal.pgen.1005969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M., Huber, W., Pages, H., Aboyoun, P., Carlson, M., Gentleman, R., Morgan, M. T. and Carey, V. J. (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G. and Durbin, R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan, M. M. and Schneiderman, H. A. (1977). Histological analysis of the dynamics of growth of imaginal discs and histobloast nests during the larval development of Drosophila melanogaster. Willhelm Roux's of Developmental Biology 183, 269-305. 10.1007/BF00848459 [DOI] [PubMed] [Google Scholar]
- McCarthy, D. J., Chen, Y. and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288-4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire, S. E., Le, P. T., Osborn, A. J., Matsumoto, K. and Davis, R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765-1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Milner, M. J. and Haynie, J. L. (1979). Fusion of Drosophila eye-antennal imaginal discs during differentiation in vitro. Willhelm Roux's Archives of Developmental Biology 185, 363-370. 10.1007/BF00848522 [DOI] [PubMed] [Google Scholar]
- Milner, M. J., Bleasby, A. J. and Pyott, A. (1984). Cell interactions during the fusion in vitro of Drosophila eye-antennal imaginal discs. Wilhelm Roux's Archives of Developmental Biology 193, 406-413. 10.1007/BF00848232 [DOI] [PubMed] [Google Scholar]
- Morgan, T. H. (1929). Variability in eyeless. Publication of the Carnegie Institute 399, 139-169. [Google Scholar]
- Morrison, C. M. and Halder, G. (2010). Characterization of a dorsal-eye Gal4 Line in Drosophila. Genesis 48, 3-7. 10.1002/dvg.20608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi, T., Clements, J., Gehring, W. J. and Callaerts, P. (2002). Dominant-negative form of the Pax6 homolog eyeless for tissue-specific loss-of-function studies in the developing eye and brain in Drosophila. Genesis34, 74-75. 10.1002/gene.10140 [DOI] [PubMed]
- O'Neill, E. M., Rebay, I., Tjian, R. and Rubin, G. M. (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137-147. 10.1016/0092-8674(94)90580-0 [DOI] [PubMed] [Google Scholar]
- Ouweneel, W. J. (1970). Genetic analysis of loboid-ophthalmoptera, a homoeotic strain in Drosophila melanogaster. Genetica 41, 1-20. 10.1007/BF00958890 [DOI] [PubMed] [Google Scholar]
- Pages, H., Aboyoun, P., Gentleman, R. and DebRoy, S. (2020). Biostrings: Efficient manipulation of biological strings. R package version 2.56.0.
- Palliyil, S., Zhu, J., Baker, L. R., Neuman, S. D., Bashirullah, A. and Kumar, J. P. (2018). Allocation of distinct organ fates from a precursor field requires a shift in expression and function of gene regulatory networks. PLoS Genet. 14, e1007185. 10.1371/journal.pgen.1007185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait, J. H. and Schneiderman, H. A. (1971a). A clonal analysis of development in Drosophila melanogaster: morphogenesis, determiantion and growth of the wild type antenna. Dev. Biol. 24, 477-519. 10.1016/0012-1606(71)90061-3 [DOI] [PubMed] [Google Scholar]
- Postlethwait, J. H. and Schneiderman, H. A. (1971b). Pattern formation and determination in the antenna of the homoeotic mutant Antennapedia of Drosophila melanogaster. Dev. Biol. 25, 606-640. 10.1016/0012-1606(71)90008-X [DOI] [PubMed] [Google Scholar]
- Postlethwait, J. H., Poodry, C. A. and Schneiderman, H. A. (1971c). Cellular dynamics of pattern duplication in imaginal discs of drosophila melanogaster. Dev. Biol. 26, 125-132. 10.1016/0012-1606(71)90112-6 [DOI] [PubMed] [Google Scholar]
- Postlethwait, J. H. and Schneiderman, H. A. (1973). Pattern formation in imaginal discs of Drosophila melanogaster after irradiation of embryos and young larvae. Dev. Biol.32, 345-360. 10.1016/0012-1606(73)90246-7 [DOI] [PubMed]
- Punzo, C., Seimiya, M., Flister, S., Gehring, W. J. and Plaza, S. (2002). Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development 129, 625-634. [DOI] [PubMed] [Google Scholar]
- Qiu, Z., Song, C., Malakouti, N., Murray, D., Hariz, A., Zimmerman, M., Gygax, D., Alhazmi, A. and Landry, J. W. (2015). Functional interactions between NURF and Ctcf regulate gene expression. Mol. Cell. Biol. 35, 224-237. 10.1128/MCB.00553-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring, R., Walldorf, U., Kloter, U. and Gehring, W. J. (1994). Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265, 785-789. 10.1126/science.7914031 [DOI] [PubMed] [Google Scholar]
- Ramírez, F., Ryan, D. P., Grüning, B., Bhardwaj, V., Kilpert, F., Richter, A. S., Heyne, S., Dundar, F. and Manke, T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160-W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready, D. F., Hanson, T. E. and Benzer, S. (1976). Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53, 217-240. 10.1016/0012-1606(76)90225-6 [DOI] [PubMed] [Google Scholar]
- Rogers, E. M., Brennan, C. A., Mortimer, N. T., Cook, S., Morris, A. R. and Moses, K. (2005). Pointed regulates an eye-specific transcriptional enhancer in the Drosophila hedgehog gene, which is required for the movement of the morphogenetic furrow. Development 132, 4833-4843. 10.1242/dev.02061 [DOI] [PubMed] [Google Scholar]
- Royet, J. and Finkelstein, R. (1995). Pattern formation in Drosophila head development: the role of the orthodenticle homeobox gene. Development 121, 3561-3572. [DOI] [PubMed] [Google Scholar]
- Smith-Bolton, R. K., Worley, M. I., Kanda, H. and Hariharan, I. K. (2009). Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell 16, 797-809. 10.1016/j.devcel.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, H., Spichiger-Haeusermann, C. and Basler, K. (2009). The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep. 10, 1140-1146. 10.1038/embor.2009.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratford, C. M. and Kumar, J. P. (2014). Dissection and immunostaining of imaginal discs from Drosophila melanogaster. J Vis Exp, 91, 51792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier, M., Bohmann, D. and Mlodzik, M. (1995). JUN cooperates with the ETS domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell 83, 753-760. 10.1016/0092-8674(95)90188-4 [DOI] [PubMed] [Google Scholar]
- Verghese, S. and Su, T. T. (2017). STAT, Wingless, and Nurf-38 determine the accuracy of regeneration after radiation damage in Drosophila. PLoS Genet. 13, e1007055. 10.1371/journal.pgen.1007055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner, B. M., Weasner, B. P., Neuman, S. D., Bashirullah, A. and Kumar, J. P. (2016). Retinal Expression of the Drosophila eyes absent Gene Is Controlled by Several Cooperatively Acting Cis-regulatory Elements. PLoS Genet. 12, e1006462. 10.1371/journal.pgen.1006462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H., Sandaltzopoulos, R., Wang, H.-M., Hamiche, A., Ranallo, R., Lee, K.-M., Fu, D. and Wu, C. (2001). Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8, 531-543. 10.1016/S1097-2765(01)00345-8 [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein, U., Tepass, U. and Hartenstein, V. (1993). Embryonic origin of the imaginal discs of the head of Drosophila melanogaster. Roux Arch Dev Biol 203, 60-73. 10.1007/BF00539891 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang, L.-G. and He, Q.-Y. (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382-2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zhu, J., Palliyil, S., Ran, C. and Kumar, J. P. (2017). Drosophila Pax6 promotes development of the entire eye-antennal disc, thereby ensuring proper adult head formation. Proc. Natl. Acad. Sci. USA 114, 5846-5853. 10.1073/pnas.1610614114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Ordway, A., Weber, L., Buddika, K. and Kumar, J. P. (2018). Polycomb group (Pc-G) proteins and Pax6 cooperate to inhibit in vivo reprogramming of the developing Drosophila eye. Development 145, dev160754. 10.1242/dev.160754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.