Abstract
There has been steady progress in dissecting the molecular features that define ccRCC initiation and progression. The recent publication of the TRACERx Renal papers and studies describing the interaction between tumor genomics and microenvironmental remodeling provide important new information for the field. This review summarizes common genomic and chromosomal copy number abnormalities in ccRCC, including 3p loss, and provides a mechanistic framework organizing these features into initiating events, drivers of progression and factors that confer lethality. The challenges researchers have faced developing animal models of ccRCC possessing these genomic features are described. The origins of DNA repair defects in ccRCC are discussed, with a focus on the role truncal mutations seen in ccRCC, including VHL, SET2, PBRM1 and BAP1, may play in engendering genomic instability. Molecular subtypes in ccRCC that arise from these defects are then described, placing them into clinically and therapeutically relevant categories. These findings are then placed into the context of the tumor microenvironment, with a summary of multiple studies that describe how various mutations appear to modulate immune cell populations in ccRCC tumors. These data are then used to describe opportunities for disease prevention, early detection, prognostication and treatment are described.
1. INTRODUCTION
Renal cell carcinoma (RCC) affects over 400,000 individuals worldwide per year 1. The age of diagnosis is approximately 60, and twice as many men are diagnosed as women 1. There are several subvariants of RCC, and approximately 70 percent of individuals are diagnosed with clear cell RCC (ccRCC). Although ccRCC is a disease that can be caught early and successfully treated with surgical or ablative strategies, up to a third of cases will present with or develop metastases2. This disease state is almost uniformly lethal, and represents a critical distinction when we consider the biology of ccRCC.
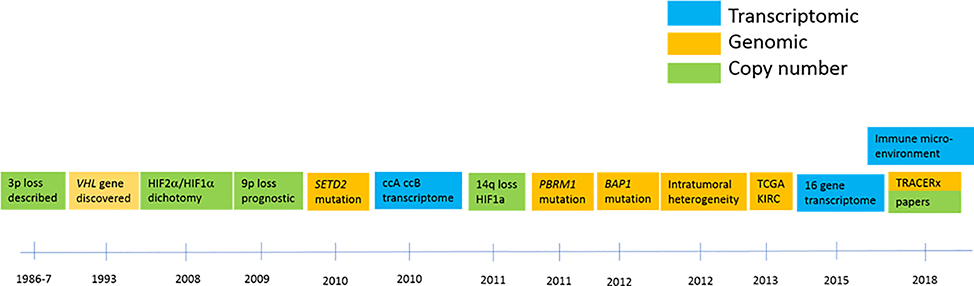
A timeline of discoveries leading to our current understanding of ccRCC ontogeny are summarized in Figure 1. These fundamental genetic and molecular features define the clear cell RCC tumors.
Figure 1: Timeline of key discoveries in ccRCC genomics.
Green boxes denote events related to copy number changes; orange boxes indicate key genomic findings, and blue boxes represent key transcriptomic contributions.
In brief, chromosomal features of three RCC cell lines were described in 1979 3 and 3p loss was identified in primary tumor samples less than a decade later 4,5. The discovery of the VHL gene in 1993 6 provided the foundation of our understanding of ccRCC biology, both in the sporadic and hereditary settings. We had to wait nearly two decades, until the widespread use of next generation sequencing, to identify additional mutations in ccRCC 7–10. Additional chromosomal copy number abnormalities were further characterized and found to be associated with prognosis 11–15. Gene expression profiles subclassified ccRCC into prognostic risk groups 16,17. From 2010 onwards a rapidly expanding series of publications describing genomic changes in ccRCC opened up a new chapter of our understanding of ccRCC genomics 7–10, and shortly thereafter studies of genomic heterogeneity provided both further insight into ccRCC clonal evolution 18–21 and raised important questions on how much tissue information is required to adequately characterize the genetic features of any one given tumor.
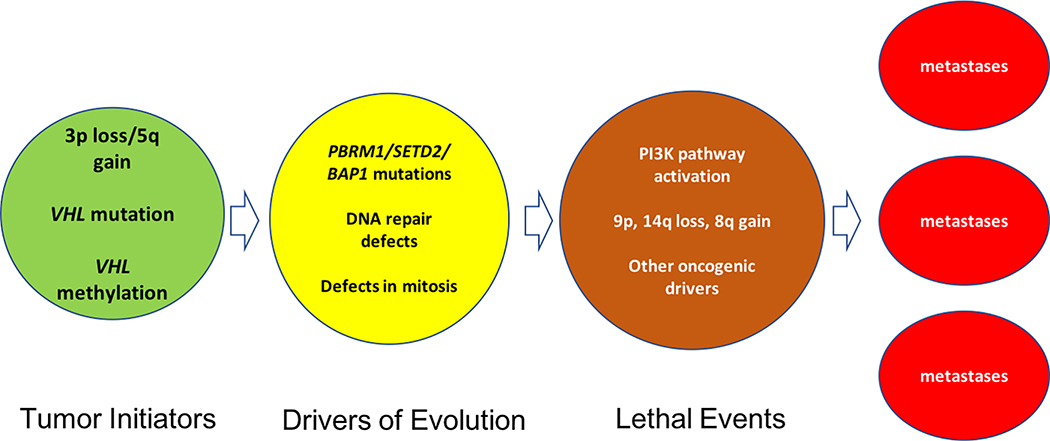
These studies raise important questions about the drivers of ccRCC ontogeny and how ccRCC evolves into a potentially lethal state. This review summarizes the consequences of VHL loss of function, the significance of 3p deletion and of additional chromosomal events on ccRCC tumor development, and mechanisms by which these changes may create the phenotypes observed in ccRCC. The review then focuses on the potential contribution of drivers of genomic instability, such as defective DNA repair, in ccRCC, an area of intense study with the potential to yield insights into the etiology and evolution of ccRCC A description of ccRCC molecular subtypes and events fostering tumor lethality then follows, and the interaction between ccRCC tumor cells and the microenvironment is discussed along with potential therapeutic implications. Figure 2 summarizes the key features of nascent and evolving ccRCC.
Figure 2: Key events in ccRCC progression.
Tumor initiators include 3p loss and VHL mutations. Evolution is driven by DNA repair defects and errors in mitosis that create additional aneuploidy. The acquisition of key chromosomal gains and losses, mutations in PI3K pathway elements and other oncogenic driver mutations confer lethal potential to intratumoral clones, and enhance the probability of developing metastases.
2. INITIATING FACTORS IN ccRCC ONTOGENY
Chromosome 3p Deletion- Initiating Factor in Nonhereditary ccRCC
The quest for additional truncal changes in RCC tumors has led to assessment of chromosomal copy number changes as a function of tumor progression and lethality 11,13. Clear cell RCC is characterized by near-universal loss of most or all of chromosome 3p 10,13,22. Polybromo 1 (PBRM1), SET domain containing 2 (SETD2) and BRCA associated protein 1 (BAP1) reside on 3p, and these genes are mutated at relatively high frequencies in ccRCC 7,9,10. Additionally, several genes on 3p may be haploinsufficient, including the mismatch repair gene MLH1 23, and SETD2 24.
Chromosome 3p loss itself was described as early as 1987 4 and the timing of 3p loss relative to subsequent genomic alterations was defined using array comparative genomic hybridization in combination with distance-based and branching-tree methodologies in 2000 15. More recently, the TRACERx Renal program prospectively collected tumor tissue samples from over 100 patients with ccRCC and collected multiple samples from each tumor to ascertain tumor ontogeny, phylogeny and intratumoral heterogeneity 19–21. These studies showed that loss of 3p was the earliest event in RCC ontogeny, likely occurring in adolescence 19,20. A study in patients with VHL disease who underwent multiple ccRCC resections demonstrated that 3p loss in synchronously arising tumors occurred via different 3p breakpoint locations, or through whole chromosome 3 loss 25, suggesting that although 3p loss itself is a necessary event for carcinogenesis, the process leading to that loss is not driven by a specific chromosomal breakpoint.
The TRACERx study demonstrated that chromothripsis is a likely driver of 3p loss, in association with 5q gain 19,20. Micronuclei form after mitosis when DNA fragments acquire a nuclear envelope, and are a caustic environment for the DNA contained within- a driver for DNA damage, mutation, and chromothripsis 58. Underlying causes of micronuclei formation include aneugens and clastrogens, the latter generally associated with formation of acentric chromosome fragments 26, which lack a centromere and fail to attach to the mitotic spindle. The presence of chromosome bridges during telophase can also lead to DNA breakage during cytokinesis and formation of micronuclei 27. Hypoxia and oxidative stress have been linked to micronucleus formation as well, 26 conditions that are potentially present in the proximal tubule. Other physical changes in the cell, including pressure, temperature, radiation, UV and ultrasound can increase micronuclei 26. It is important to bear in mind that the kidney proximal tubule and nephron is a fairly harsh environment, dictated by intense gradients of oxygen, salts, and nutrients. These factors are further misregulated in renal states including renal failure as well as in polycystic kidney disease 28. Thus, conditions around the RCC cell of origin may create a “perfect storm” that permits the development of micronuclei, chromothripsis and 3p loss.
VHL Loss in ccRCC
The loss or mutation of the von Hippel Lindau (VHL) gene is generally considered to be one of the obligate initiating steps in the development of ccRCC. Germline loss of VHL is inherited in an autosomal dominant fashion in families with VHL disease 6, and these individuals often develop multifocal, bilateral ccRCC 29,30. Somatic loss of VHL is observed in the majority of patients with sporadic clear cell RCC 31, and based on the TRACERx studies, may follow 3p loss in these patients 19.
VHL and HIF regulation
The VHL protein is an E3 ubiquitin ligase whose best understood function is the ubiquitination of the prolyl hydroxylated transcription factors hypoxia inducible factors 1 and 2 alpha (HIF1α and HIF2α) 32–34 under normoxic conditions, with their subsequent proteolytic degradation 35. HIF1α and 2α regulate transcription of a number of genes involved in angiogenesis, metabolism and chromatin remodeling 36. Even before the functional link between VHL and HIF was made, ccRCC was shown to have increased levels of vascular endothelial growth factor (VEGF) and platelet derived growth factor (PDGF) mRNA 37 and HIF mediated transcription of pro-angiogenic factors including VEGF was shown to facilitate the development of neovasculature 33.
HIF1α and HIF2α regulate overlapping gene sets, with distinct differences between the two transcription factors 38–40. HIF1α regulates a number of genes involved in enhancing glucose uptake and shifting cells towards glycolysis including pyruvate dehydrogenase kinase 41 and lactate dehydrogenase A 42, as well as mediating mitochondrial autophagy via BNIP3 43 and BNIP3L 44. Axl 45, a member of the TAM family of regulatory proteins 46,47 was found to be a HIF1α driven gene 48. AXL gene expression was associated with poor prognosis in ccRCC 48,46 and Axl protein levels were increased after sunitinib treatment 49. HIF2a has been shown to regulate the EPO, CCND1 and TGFA genes 36,39. Some evidence points towards HIF2α, but not HIF1α being the primary driver of RCC oncogenesis, with HIF2α both necessary and sufficient for the growth of VHL null cell lines 36,50. A separate set of studies in animal models implicated HIF1α 51 as the primary driver as opposed to HIF2α 52,53 in renal carcinogenesis, and questions remain on the exact role of specific HIF isoforms in the development of RCC, perhaps implying a temporal dominance in which one factor plays a larger role at various states of disease. A more recent study showed that loss of either HIF1α or HIF2α abrogated tumor formation in a Vhl/Trp53 deficient murine model 54, indicating both are required for tumor initiation in that system.
VHL and non-HIF targets
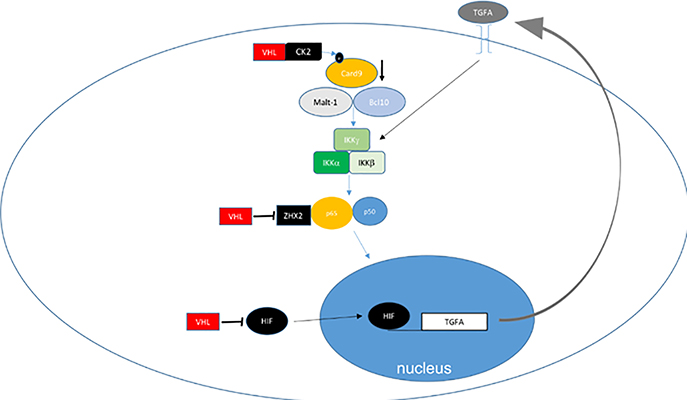
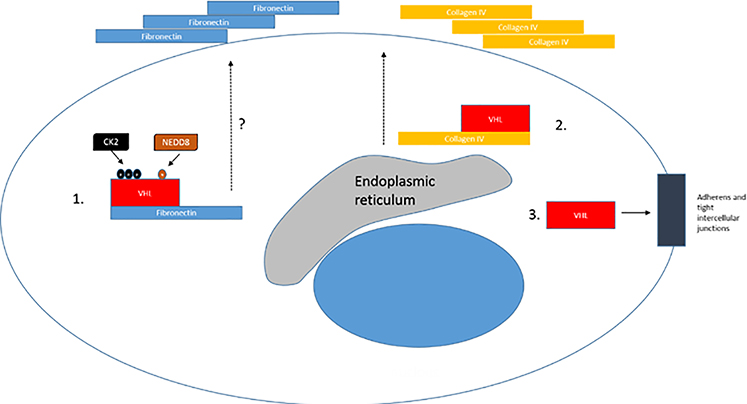
The VHL protein has been shown to regulate a number of additional molecules, pathways and processes 55. These include AKT 56, nuclear factor kappa-light-chain-enhancer of activated B cells (NK-kB) 57–59, tank-binding kinase-1 (TBK1) 60, the extracellular matrix, the primary cilium and mitosis. A few of these are depicted in Figure 3, and will be discussed briefly below. VHL is also linked to regulation of DNA repair 61–64, which is discussed in a subsequent section.
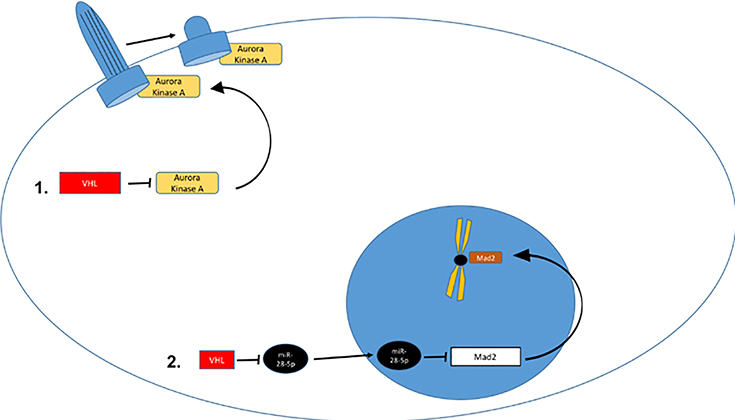
Figure 3: Non-HIF VHL Targets.
A. VHL regulates NFkB through HIF dependent and independent mechanisms. VHL loss will increase TGFA transcription which induces cell autonomous increase in NFkB activation via growth factor receptor mediated IKK inhibition. CK2 mediated CARD9 inhibition is enhanced by VHL, and ZHX2 is inhibited by VHL. B. 1. Extracellular fibronectin homeostasis is dependent on interaction with CK2-phosphorylated and neddylatedVHL and loss of VHL results in defective or deficient extracellular matrix. 2. VHL is involved in collagen IV homeostasis. 3. VHL regulates intercellular and adherens junctions. C. VHL regulates primary cilium and mitosis. 1. VHL maintains primary cilium homeostasis by blocking AURKA mediated reduction of primary ciliary stability. 2. VHL supports mitotic function by enhancing Mad2 function by downregulating Mad2 inhibitory miR-28–5p.
VHL-mediated NFkB signaling (Figure 3a):
Rettig and colleagues demonstrated that loss of VHL was associated with an upregulation of NF-kB activity 57 through HIF dependent induction of transforming growth factor alpha (TGFα). In addition to this HIF-transcription circuit, VHL was subsequently shown to serve as an adaptor to promote the inhibitory phosphorylation of the nuclear agonist Card9 by casein kinase 2 (CK2) and reduce NF-kB activation in RCC independently of HIF 58. Peri and colleagues demonstrated upregulation of NFkB and IFN signaling pathways in the absence of VHL 59. More recently, zinc fingers and homeoboxes 2 (ZHX2), an activator of NF-kB was found to be targeted and downregulated by VHL directly through a similar mechanism of ubiquitylation and proteasomal degradation 65. Thus, loss of VHL leads to coordinated stabilization of factors which can have independent activity to promote NF-kB signaling.
VHL activity related to the extracellular matrix (Figure 3b):
VHL has been reported in several lines of investigation to be involved in the regulation of intercellular junctions 66 and the extracellular matrix (ECM) 67–70. Several investigators have shown that VHL binds to fibronectin, and this step is necessary for the promotion of a correct ECM assembly in cells 68. All tumor causing VHL mutants have been reported to be defective in in fibronectin binding 71–73. This binding is regulated by CK2, as phosphorylation of VHL by CK2 at amino acids 33, 38 and 43 was found to be necessary for fibronectin binding by VHL and the development of a normal ECM 74. Mutation of serines 33, 38 and 43 was associated with eventual tumor growth in a xenograft model when compared to wildtype VHL 74. Conversely, CK2 inhibition resulted in VHL stabilization due to decreased N-terminal cleavage and proteasomal degradation, suggesting regulation by CK2 is linked to VHL protein stability 75.
VHL and microtubule stability (Figure 3c):
VHL has also been reported to enhance microtubule stability 76, an interesting observation given the more recent linkage of SETD2 to methylation of the cytoskeleton (see below). This has implications for several important microtubule structures in the cell, including the primary cilium and the mitotic spindle. Microtubules comprise the central core of the ciliary axoneme, and loss of VHL results in defects in ciliogenesis77–79. As a result, VHL disease is often classified as a “ciliopathy”, along with other diseases linked to defects of the primary cilium such as polycystic kidney disease 80. Activation of Aurora kinase A (AURKA) is known to inhibit ciliogenesis, and this kinase was recently shown to be regulated by VHL. VHL mono-, rather than poly-ubiquitinates AURKA in a PHD-independent manner, targeting AURKA for degradation, which in quiescent cells is required to maintain the primary cilium 81.
VHL and mitotic spindle function (Figure 3c):
VHL has also been linked to mitotic spindle function82–84. Krek and colleagues were the first to show that VHL loss resulted in reduced Mad2 (mitotic arrest deficient 2) levels, which was associated with spindle misorientation and loss of normal mitotic checkpoint control in an isoform-specific manner 82. The underlying molecular mechanism was identified when Mad2 translation was shown to be regulated by miR-28–5p in a VHL dependent fashion, with loss of VHL resulting in increased miR-28–5p, and decreased Mad2 translation. 84 Furthermore, ischemic injury in a kidney-targeted Vhl deficient murine model demonstrated cyst-inducing spindle misorientation and aneuploidy immediately post-injury, and longer term follow- up demonstrated the development of cysts, clear-cell type cells and dysplasia 83. Thus, VHL may play a role in regulation of both mitotic and nonmitotic cells via microtubular homeostasis.
VHL-mediated PI3K signaling:
VHL was found to downregulate AKT 56, a key component of the PI3K pathway. Upregulation of the PI3K pathway has been associated with more aggressive behavior in many cancers 85, including ccRCC 86,87. AKT phosphorylation was associated with increased tumor aggressivity and with cytoplasmic translocation and deactivation of p27 88.
VHL mutations and development of animal models for RCC
Based on the observation that germline VHL mutations in humans drove the development of pleiotropic lesions, including ccRCC, it was presumed that the loss of VHL was sufficient for tumorigenesis. However, a number of studies have shown that a Vhl knockout or mutation will not engender RCC in murine models 89–91. The first whole animal heterozygous knockout model published in 2001 did not produce a RCC phenotype 89 and subsequent efforts at creating kidney specific biallelic Vhl knockouts did not produce bona fide RCC lesions despite using a variety of kidney specific promoters 92, including a Actb-Cre 93, a proximal tubule specific Pepck-Cre 94, a pan-tubular Pax8-Cre 53,95, and a thick ascending limb specific Thp-Cre 96.
In addition, studies of renal cysts that arise in VHL patients have shown that these benign lesions have inactivated both copies of VHL97, they often do not display other characteristic hallmarks of RCC 98. These findings indicate while VHL loss is necessary for RCC formation, it is not necessarily sufficient.
Subsequent to these single-gene knockout models, knockout of Vhl was combined with other potential RCC driver genes. Several transgenic animal models employing the proximal tubule specific Ksp1.3 were used to drive a Cre recombinase and inactivate paired or multiple genes in a kidney specific manner 99,100. The combined inactivation of Pten and Vhl generated cysts100 whereas the combined inactivation of Trp53 and Vhl generated simple cysts, atypical cysts and neoplasms which demonstrated mTORC1 activation and elevated expression of Myc 99. Neither model developed tumors with metastatic potential. A further evolution of the latter model involved the combined Ksp1.3-Cre driven inactivation of Trp53, Vhl and Rb, which produced neoplastic lesions that demonstrated histological and transcriptomic features that resembled those of human ccRCC, while still not demonstrating metastatic potential 101.
More recent efforts at model creation using a polygenic approach or a focus on the recently discovered chromatin remodeling gene mutations have yielded results which are bringing us closer to producing representative model of ccRCC 102–104. These are described in greater detail in subsequent sections.
Additional Mutations in ccRCC
Several other genes, all found on chromosome 3p, are mutated at a relatively high frequency in ccRCC 7–9, and are likely associated with both convergent and divergent phenotypic characteristics in ccRCC. All are involved in some way with chromatin remodeling, and are summarized in a recent review 105.
PBRM1
PBRM1 is the most commonly mutated gene after VHL. Human PBRM1 generates a 1689 amino acid protein, consisting of six bromodomains, two bromo-associated homology domains, and a C-terminal high-mobility group 106. PBRM1 is the defining component of the switch/sucrose nonfermenting (SWI/SNF) chromatin remodeling complex known as PBAF 107. PBAF is an adenosine triphosphate (ATP)-dependent chromatin remodeler governs nucleosome structure and positioning, and gene transcription. Loss of PBRM1 and PBAF activity has potential to significantly undermine the highly organized structure of chromatin and disrupt transcription. Numerous studies have identified PBRM1 mutations in ccRCC 7,18, 108, with an incidence of 30–40 percent, although the most aggressive sarcomatoid variants tend to lack these mutations 109. PBRM1 mutations precede subsequent SETD2 mutations, suggesting these sequential mutations participate in subclonal tumor evolution19.
Several studies are beginning provide mechanistic insight in to the consequences of PBRM1/PBAF loss in ccRCC. Gao et al demonstrated that PBRM1 deficiency was associated with increased cellular growth and a HIF transcriptional signature 110. A study by Nargund et al revealed that Pbrm1 deficiency enhanced Vhl loss dependent upregulation of HIF1α and STAT3, and increased mammalian target of rapamycin (mTOR) signaling, thereby enabling the development of renal lesions in a murine transgenic model. Liao et al showed that PBRM1 and other chromatin remodeling genes lost in RCC antagonized tumor growth in an interferon-stimulated gene factor 3 dependent fashion, and loss of PBRM1 relieved this inhibition 111. Espana-Agusti and colleagues showed that Pbrm1 knockout rescued Vhl-loss induced replication stress and enhanced tumor growth in a transgenic model 63. Inactivation of Pbrm1 or Bap1 in conjunction with Vhl employing a Pax8-Cre in a murine model recapitulated the relatively low aggressivity of Pbrm1/Vhl knockout in a transgenic murine model 102. Liu et al demonstrated that PBRM1 loss downregulated interferon gamma (IFNγ) mediated signaling 112. Cai et al showed that PBRM1 acted as a lysine reader for p53, and loss of PBRM1 resulted in decreased p53 dependent CDKN1A transcriptional regulation 113. Similar to VHL, PBRM1 also plays a role in genomic stability. PBRM1 localizes to the kinetochore 114, and in yeast, the PBRM1 homolog RSC2 is essential for chromosome arm cohesions 115,116; loss of PBRM1/PBAF results in genomic instability and aneuploidy 117. Thus the potential impact of PBRM1 loss includes the augmentation of hypoxia signaling, modulation of replication stress, impairment of p53 mediated cell cycle regulation, inhibition of interferon response and genomic instability. However, in RCC, in the absence of additional mutations in SETD2, these changes appear to produce tumors that are less aggressive, with high angiogenic potential, that are immunologically cold112,118,119.
BAP1
Another 3p chromatin remodeler inactivated in RCC is the BAP1 deubiquitinase120. The BAP1 gene produces a 729 AA protein containing several functional domains: a ubiquitin C-terminal hydrolase (UCH) domain, a BRCA1-associated RING domain protein 1 (BARD1) binding domain, an HCF-C1 binding motif (HBM) and a nuclear localization signal 8. BAP1 is mutated in 10–20 percent of ccRCC 8,121. Subsequently histone 2a lysine 119 (H2AK119) was shown to be the target for this deubiquitinase 8,122, which is involved in Polycomb complex mediated gene repression. In a whole animal murine model, BAP1 also interacted with the cell cycle regulator host cell factor-1 (HCF-1), the signal transduction modulator O-linked N-acetylglucosamine transferase (OGT), and the Polycomb group proteins ASXL1 and ASXL2. OGT and HCF-1 levels were decreased by Bap1 deletion, and were associated with myeloid transformation 123. A subsequent study reported that BAP1 modulated gene expression in a cell-type specific manner, and loss of BAP1 promoted tumorigenesis in cells that did not engage an RNF2-dependent apoptotic program 124.
Similar to both VHL and PBRM1, BAP1 also plays a role in maintenance of genomic stability. BAP1 associates with the mitotic spindle regulating microspherule protein-1 (MCRS1), and loss of BAP1 resulted in increased chromosomal instability 125. The discovery of BAP1 mutations in ccRCC arose from the study of patient derived xenograft models. Brugarolas and colleagues used this model to increase relative tumor purity and to identify BAP1 as a novel tumor suppressor in ccRCC8. They then demonstrated that heterozygous Bap1 deletion coupled with homozygous Vhl loss using a SIX homeobox 2 (Six2)- Cre 126 (Six2-Cre;VhlF/F;Bap1F/+) to drive kidney specific gene inactivation could produce neoplasms in the murine kidney, 127. A follow up study employed a Pax8-Cre to generate kidney specific Vhl/Pbrm1 and Vhl/Bap1 knockouts 102. Pax8-Cre;VhlF/F;Bap1F/+ mice produced neoplastic renal lesions with nuclear pleomorphism, nucleolar prominence, atypia, mitosis and elevated phospho-S6 expression. in comparison, Pax8-Cre;VhlF/F;Pbrm1F/+ animals failed to develop renal lesions and Pax8-Cre;VhlF/F;Pbrm1F/F animals produced tumors possessing low-grade features, including moderate amounts of cytoplasm, lipid and glycogen accumulation, relative lack of nuclear atypia, and the absence of phospho-S6 staining. 102. Loss of BAP1 is strongly associated with poor risk in patients with ccRCC, even in individuals who have clinically low stage tumors 128. In patients treated with targeted therapy, presence of a BAP1 mutation was associated with worse outcome 129. A rare familial syndrome at risk for ccRCC is caused by germline mutations in BAP1 130. In the TRACERx study, BAP1 mutations were associated with high weighted genome instability index (wGII), low intratumoral heterogeneity (ITH) tumors, suggesting that the driver function of BAP1 loss is sufficient to provide ccRCC clones with a clear fitness advantage and lethal potential 19.
SETD2
SETD2 is a chromatin remodeling gene and a tumor suppressor in ccRCC 9. SETD2 mutations were first discovered through early high throughput genomic sequencing efforts 9 that preceded the Cancer Genome Atlas (TCGA) project, which also validated these findings 10. SETD2 is mutated in 10–20 percent of cases of ccRCC 9,10 SETD2 is a 2564aa long nonredundant histone H3 lysine 36 methyltransferase containing a SET domain and a C-terminal Set2 Rpb1 interacting (SRI) domain, specifically marking nucleosomes of actively transcribed genes with H3K36me3. In yeast, this mark, along with demethylation (H3K36me2) has a repressive role to prevent cryptic initiation from alternate internal transcriptional start sites 131. However, utilizing selective Set2 mutants that isolate mono, di, and trimethylation states, Rathmell and colleagues determined that the isolated loss of trimethylation, as occurs in mammalian cells with SETD2 loss, is not sufficient to promote a cryptic initiation phenotype 132. Instead, the chromatin effects of this mark appear to play major roles in governing DNA repair, as recently reviewed 133.
In addition to DNA repair, another role for SETD2 in maintenance of genomic stability is a newly discovered role in methylation of microtubules 134,135. SETD2 trimethylates α-tubulin on lysine 40 (αTubK40me3). SETD2- deficient cells demonstrated mitotic spindle defects, and in cytokinesis, and showed an increase in the formation of chromosome bridges and micronuclei. SETD2 mutated in the SET domain and the SRI domain could not methylate microtubules, which resulted in more chromosome bridges and lagging chromosomes relative to wild-type SETD2. These data indicated that in addition to the catalytic domain, a functional SRI domain was also required for αTubK40me3 134. Monoallelic SETD2 deficiency, as would be expected in cells with 3p loss, was also shown to abrogate spindle microtubule trimethylation on αTubK40, but not methylation of histones on H3K36, and this was sufficient to induce genomic instability 24. It is possible that monoallelic loss of SETD2 may further aid in the steady increase in chromosomal copy number abnormalities seen as tumors increase in size and stage 11,13. Most recently, SETD2 was found to also methylate cytoskeletal actin, and to promote polymerization of actin filaments and cell motility 136. As noted above, SETD2 mutations can arise independently of, or occur in conjunction with, PBRM1 mutations in ccRCC 19. Interestingly, functional convergence between SETD2 and PBRM1 was revealed with the discovery that PBRM1 recognizes the αTubK40me3 SETD2 mark on microtubules 137. In this study, PBRM1 was shown to localize the PBAF complex to the mitotic spindle to maintain genomic stability. Consistent with a “reader” “writer” relationship, in the TRACERx study, when both SETD2 and PBRM1 mutations co-occur, loss of PBRM1 precedes loss of SETD2. At this point in time there is no published animal tumor model of SETD2 inactivation in RCC.
3. POTENTIAL ETIOLOGY OF DNA REPAIR DEFECTS in ccRCC
The initiating events for ccRCC involve biallelic VHL loss and chromosome 3p loss 10,19,31. In addition to these events, ccRCC demonstrate an intermediate number of mutations per megabase with a relatively limited range 10,20 and a stage dependent increase in the number of chromosomal copy number alterations 13. The mechanisms driving these genomic events are slowly being elucidated, and although not part of current clinical practice, have the potential for providing new therapeutic approaches in ccRCC.
Evidence for a mismatch repair defect in ccRCC
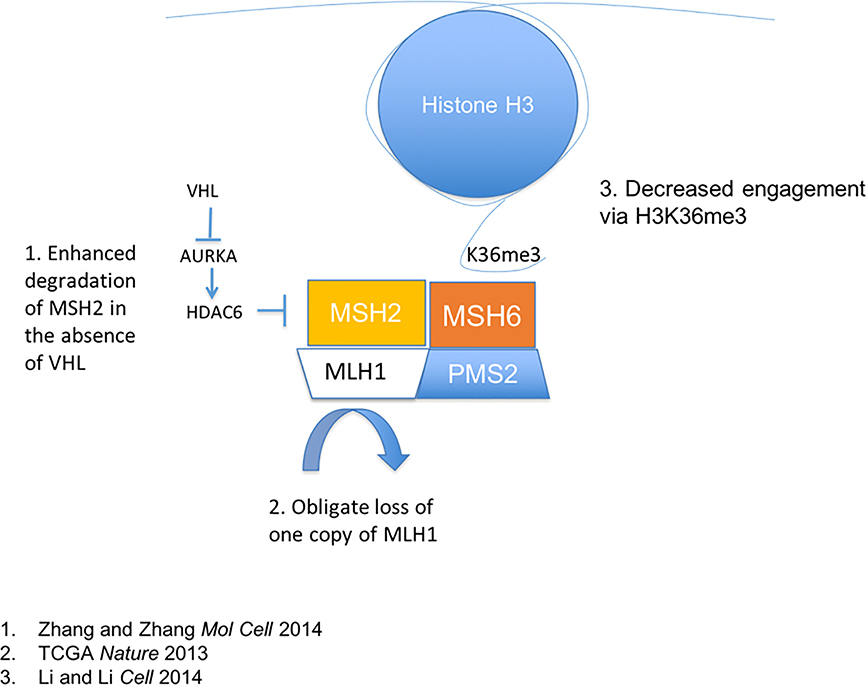
Several pathways exist involved in the repair of specific types of DNA damage, and have recently been reviewed in in the context of potential therapeutic approaches 138. Alexandrov et al analyzed mutational patterns in tumors with known genomic defects. Using an algorithm that considered nucleotides surrounding mutational sites, they identified 21 different patterns, which were associated with known germline deficiencies 139. Using this approach, ccRCC was linked to a pattern associated with aging, as well as a pattern found in tumors with mismatch repair (MMR) in a subset of individuals. There are several potential mechanisms that could lead to a mismatch repair deficiency in ccRCC (Figure 4). The MutL Homolog 1 (MLH1) gene, an essential component of the MUTL complex, is found on chromosome 3p. Evidence for MUTL haploinsufficiency has been reported in pancreatic cancer 23. Loss of VHL upregulates histone deacetylase 6 (HDAC6) levels 140. HDAC6 sequentially deacetylates and ubiquitinates MutS protein homolog 2 (MSH2) leading to MSH2 degradation 141. In addition, HDAC6 was shown to reduce sensitivity to DNA-damaging agents and cellular DNA mismatch repair activities via MSH2 downregulation 141 In addition, SETD2 mediated H3K36 trimethylation acts as an essential docking site for the MUTS/MUTL complex 142. Specifically, the hMutSα complex is directed to actively transcribed genes through interaction with the H3K36me3 mark. Thus, in the absence of H3K36me3, although MMR is intact, microsatellite instability is present, and most importantly repair may be less efficiently targeted to the most relevant areas of the genome. This interaction implies a critical activity of H3K36me3, targeting mismatch repair the active coding regions.
Figure 4: Factors influencing mismatch repair in ccRCC.
Mismatch repair mechanisms are affected by multiple axes in ccRCC. 1. Loss of VHL itself alters HDAC6 regulation via AURKA, effectively suppressing MSH2 via protein degradation. 2. MLH1 may exhibit haploinsufficiency due to 3p deletion. 3. Loss of MSH6 targeting to H3K36me3 can result in uncoupling of transcription coupled repair mechanisms for efficient targeting of DNA repair mechanisms to essential segments of the genome.
Taken together, these data point to multiple potential defects in the integrity of the MMR complex in ccRCC. However, there is a very low rate of microsatellite instability seen in RCC 143,144. Additionally, ccRCC has a relatively low absolute mutation rate, with frequencies approximately one tenth of those seen in tumors from patients with Lynch syndrome139. These findings suggest that if present, a nonclassical form of MMR defect is operative in ccRCC. Further study is needed to clearly elucidate the role of MMR defects in ccRCC mutagenesis.
Evidence for a Homologous Recombination Repair (HRR) Defect in ccRCC
The two main pathways involved in double-stranded DNA repair include nonhomologous end joining (NHEJ) which acts throughout the cell cycle and homologous recombination repair (HRR) which is operative in S phase and requires two DNA strands for error-free repair 145. Defects in HRR are associated with increased mutational frequency and cancer predisposition, classically manifested in families bearing mutations in HRR related genes, including BRCA1 146,147, BRCA2 148, PALB2 149, and ATM 150.
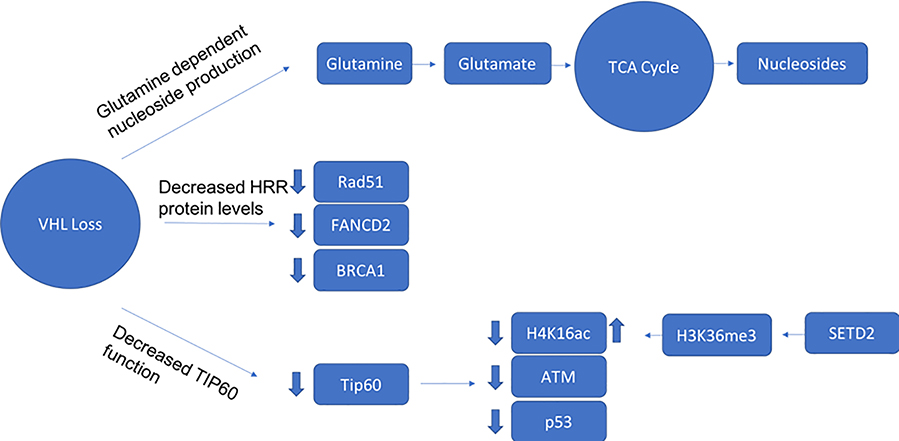
A number of groups have assessed the integrity of HRR in ccRCC (Figure 5). Metcalf and colleagues reported that suppressor of cytokine signaling 2 (SOCS2) mediated monoubiquitination of VHL at K63 61. Loss of VHL, or VHL mutations that compromise its K63-ubiquitylation attenuated the DNA-damage response (DDR), resulting in decreased HRR and persistence of DSBs 61. Espana-Agusti et al demonstrated that loss of Vhl enhanced replication stress in an animal model 63, suggesting a role in the maintenance of genomic integrity during mitosis. Glazer and colleagues demonstrated a close link between hypoxia and HRR defects, driven mainly via transcriptional regulation of key HRR components 151–153, and demonstrated that VHL loss was associated with a HRR defect 62. VHL was found to associate with, and decrease activity of KAT5/TIP60, an acetyltransferase associated with p53 activation 154. TIP60 was also found to be essential for H4K16 acetylation, which evicts 53BP1 from chromatin and favors use of HRR in DNA repair 155, and in ATM acetylation 156, which is a necessary step prior to autophosphorylation and activation. Recent work has further expanded this observation to demonstrate that VHL mediated regulation of TIP60 shifts DNA damage repair to HRR from error-prone nonhomologous end joining (NHEJ) in a HIF-independent fashion, and ATM activation was impaired in VHL deficient cells due to decreased ATM acetylation 64.
Figure 5: Factors Influencing HRR in ccRCC.
1. Loss of VHL creates glutamine dependence for nucleoside production and increased risk for nucleoside shortfall, which can create vulnerabilities during mitosis dependent DNA repair. 2. VHL loss impacts key HRR genes through a HIF dependent mechanism. 3. VHL interacts with KAT5/Tip60, which acetylates and regulates ATM, p53, and H4K16.
SETD2 is involved in regulating a number of processes associated with HRR. Most well characterized is the recruitment of DNA repair machinery to the sites of actively transcribed genes 157,158. Since H3K36me3 is preferentially placed at sites of active transcription, it provides a convenient mechanism for readers involved in DNA repair to be preferentially recruited to high priority genomic sites for repair 159. The loss of SETD2 was shown to specifically alter the efficiency of repair at these active sites. Further work demonstrated a more active role in mediating repair, via participation in engaging the p53 checkpoint, which was also discovered to be deficient in SETD2 knockout cells 158. This defect in repair is not unlike mutations in RAD51, which produce a signature of DNA defects, one hallmark of which is microdeletions due to microhomology repair mechanisms, and a reliance in the absence of SETD2 on nonhomologous repair (as is observed also with RAD51 deficiency) 160. More recently, the effect of impaired HRR to induce replication stress and mutations has been considered as a source of heterogeneity and tumor evolution in ccRCC 161. Thus, SETD2 loss may function in providing a hallmark of cancer, which is the genomic instability sufficient to generate a broad array genetic defects, and promote cancer evolution and the acquisition of deleterious events that have metastatic and lethal consequences. SETD2 was recently found to regulate H4K16 acetylation via modulation of H3K36 methylation 162, an important molecular determinant between HRR and error-prone nonhomologous end joining (NHEJ) 155. As such, it is possible the combination of SETD2 loss together with VHL deficiency potentiates the development of DNA double strand repair breaks.
A number of studies have evaluated the effects of BAP1 loss on DNA repair. Pena-Llopis et al showed that loss of BAP1 sensitized RCC cell lines to radiation and PARP inhibitors 8. BAP1 was shown to mediate poly (ADP-ribose)-dependent recruitment of the Polycomb-repressive deubiquitinase complex (PR-DUB) to sites of DNA damage, was phosphorylated by ATM, and promoted repair of DNA double-strand breaks 163. As noted above, loss of BAP1 destabilized MCRS1 and decreased mitotic stability 125. These data suggest BAP1 is regulator of DNA repair and helps maintain copy number integrity, and its loss may foster genomic instability in ccRCC.
A few significant caveats exist for the existence of a bona fide HRR defect in ccRCC. The first is that RCC is not typically sensitive to platinum and other DNA damaging agents. The second is that the Alexandrov paper does not identify ccRCC as possessing a DNA damage pattern typically seen in tumors from patients with stereotypical germline defects in HRR genes 139. The possible explanation for this discrepancy is that the signature derived from BRCA mutated tumors is sufficiently specific that it does not translate into other variants of HRR deficiency. Evidence supporting this possibility include a study that generated a homologous recombination defect (HRD) signature to identify tumors which have broad defects in HRR 164. This HRD signature identifies tumors with vulnerabilities in HRR despite not having mutations in classical HRR genes. Preliminary data suggest these signatures could help identify whether a HRD exists in ccRCC and guide therapeutic approaches 165.
4. ccRCC MOLECULAR SUBTYPES AND DRIVERS OF LETHALITY IN ccRCC
One of the key questions in ccRCC cancer evolution is whether lethality is an innate characteristic, or develops over time and through acquisition of additional genomic lesions. A number of studies in the past few years have demonstrated that both scenarios are operative 19,21.
The TRACERx study defined seven major clonal subtypes of ccRCC: VHL monodriver, PBRM1-SETD2, PBRM1-somatic copy number alteration (SCNA), PBRM1-PI3K, VHL wildtype, multiple clonal driver, and BAP1 driven 19. These subtypes are genomically distinct and are prognostically heterogeneous. A number of determinants were assessed in these subtypes, including the weighted genomic instability index (wGII), which is a measure of the fraction of the tumor genome affected by somatic copy number alterations (SCNA); percentage of cells positive for Ki67; the intratumoral heterogeneity (ITH) index, defined as the number of subclonal drivers divided by the number of clonal drivers, where drivers include all driver mutations and driver SCNAs; clonal structure; and clinicopathological parameters 19.
The VHL monodriver subcategory was largely found in small tumors, and demonstrated low ITH, wGII and Ki67 levels, and may represent temporally earlier tumors, or tumors that inherently lack the ability to develop additional lethal events. On the other end of the spectrum, the multiple clonal driver subtype showed high levels of wGII and Ki67 but relatively low levels of ITH, showing that simultaneous appearance of a small number of clonal drivers is sufficient to create RCC clones with a strong selective advantage 19.
Prognostic Implications of SCNAs in ccRCC
Several studies in addition to the TRACERx papers have shown that SCNAs are associated with poor clinical outcome 166,167,11–14. Partial or whole chromosomal losses or gains occur in chromosomes 5, 6, 7, 8, 9 and 14 at a relatively high frequency, and increase as a function of tumor size and grade 11,13.
Not only are higher overall SCNAs associated with more aggressive disease, specific chromosomal changes are linked to metastatic potential suggesting a driver function. A study of 703 ccRCC tumors published in 2009 revealed that loss of 9p was associated with poor outcome 12. An analysis of chromosomal copy number changes in 111 ccRCC patients revealed that loss of 14q together with a gain in 8q was significantly associated with poor prognosis 13,14. In the TRACERx study, 9p loss was identified as the factor most significantly associated with the formation of metastases 21, and 14q loss was significantly associated with development of metastasis prior to correction for multiple testing 21. In addition, 9p loss in metastasizing clones was a significant predictor of OS after correction for known clinical variables 21. Assessment of potential tumor suppressor or pro-oncogenic genes on these chromosomal regions identifies several candidates. Chromosome 14q contains HIF1A, which has been shown to act antagonistically to HIF2a in ccRCC progression 168. Chromosome 9p encodes CDKN2A, which generates p16/INK4A, a critical G1 cell cycle regulatory protein. Chromosome 8q harbors the MYC gene. Copy-number alterations of these genes may be responsible for engendering a selective advantage for tumor cells, or there may be additional genes on these chromosomes whose loss or gain cooperatively enhance cell fitness 21. The challenge for the field is to understand the mechanism underpinning SCNA, with the goal of either blocking its development, or targeting specific vulnerabilities in cells possessing these SCNAs.
ITH in ccRCC
ITH is a major point of concern in the molecular characterization of ccRCC. Clinically relevant ITH is generated by mutations in key genes and by driver SCNAs 19. The TRACERx Renal team defined the ITH index as the number of subclonal drivers over the number of clonal drivers, where drivers included all driver mutations and driver SCNAs 19. Future work will permit a more nuanced weighting of these driver events. Lower levels of ITH are seen at both ends of the tumor lethality spectrum. Tumors harboring only VHL mutations as well as tumors with multiple driver mutations have low ITH. In the latter case, the clonal genomic events likely confer a significant selective advantage permitting these cells to be the primary clone generating the tumor mass and impart aggressive tumor behavior.
Genotypic divergence is likely limited by constraints in cellular viability and fitness, and this may lead to phenotypic convergence and a manageable number of targetable pathways 169. Further work needs to be performed to define the phenotype to be targeted and to come up with tailored therapies that address these specific tumor features.
Sarcomatoid RCC
A special comment concerns aggressive subsets of ccRCC. Sarcomatoid variation is a histologic designation assigned to highly aggressive tumors with underlying ccRCC features. Sircar et al performed transcriptomic and genomic analyses on sarcomatoid RCC. They found a significantly lower rate of 3p mutation overall, and an enrichment of PTEN, TP53, and RELN compared with ccRCC 109. This pattern held up even when the tumors were regionally sampled in areas harboring sarcomatoid patterns as compared to regions with more traditional histology. Similar findings were observed by others 170,171. These findings suggest that sarcomatoid ccRCC may have fundamental differences in its early molecular pathogenesis when compared to non-sarcomatoid ccRCC. Interestingly, the overall mutational burden is not broadly different between sarcomatoid and nonsarcomatoid ccRCC 170,171, suggesting either similar mechanisms of mutation or a cell-type specific threshold for mutational burden that is similar between the two entities.
5. INTERACTION BETWEEN ccRCC AND THE TUMOR MICROENVIRONMENT
Multiple groups have assessed the diversity of intratumoral immune cells in ccRCC (Table 1) 119,172–177,112. One of the key questions is whether there are identifiable interactions between tumor genomic features and the microenvironment that can inform therapeutic decisions (Figure 6). A number of studies have been performed assessing these interactions, and are summarized in Table 1.
Table 1:
Tumor immune microenvironmental studies in RCC
| Paper/First Author | Tissue | Study Goal | Analytes and analytical tools | Key Findings |
|---|---|---|---|---|
| Chevrier | -Primary ccRCC | Define ccRCC macrophage and T-cell subtypes | Single cell mass cytometry; single cell RNAseq | -Described 17 tumor-associated macrophage- and 22 T-cell phenotypes -Identified prognostically significant macrophage subcategories |
| Wang | -Primary ccRCC -Tumorgrafts |
Characterize RCC TME | Bulk and single cell RNAseq, IHC, WES | -Identified novel immune/stromal transcripts -Defined an inflamed pan-RCC subtype |
| Hakimi | -Primary ccRCC | Create clinically significant ccRCC subtypes using RNA | RNA microarray, WES | -Defined four biologically distinct clusters, with specific angiogenic and immune features - Linked subtypes to clinical outcome and treatment response |
| McDermott | -Primary ccRCC | Assess response to immunotherapy, and identify genomic and microenvironmental response determinants | Bulk RNAseq, WES | -Combined immunotherapy plus anti-VEGF therapy showed trend towards improved outcome vs TKI -Response to IO therapy higher in patients whose tumor transcriptomes showed T-cell infiltrates -Presence of myeloid population attenuated IO therapy response; addition of anti-VEGF therapy rescued IO effect |
| Miao | -Primary ccRCC | Link genomic status to immune response | WES | -Showed PBRM1 deficiency was associated with improved response to IO therapy |
| Clark | -Primary ccRCC | Use proteomic and phosphoproteomic data to subclassify ccRCC | Bulk RNAseq, WES, reversed phase liquid chromatography, immobilized metal affinity chromatography | -Identified genomically unstable ccRCC subtype -Mapped metabolic changes in Krebs cycle and OXPHOS pathways -Defined CD8+ inflamed, CD8- inflamed, metabolic immune desert and VEGF immune desert subtypes |
| Liu | -Primary ccRCC -Isogenic Renca cell lines |
Assess impact of PBRM1 loss in immune response | WES, RNAseq, multispectral IHC | -Revealed Pbrm1 KO tumors less immunogenic in a Renca murine model, and in human tissue -PBRM1 deficiency reduced Ifgr2 transcription |
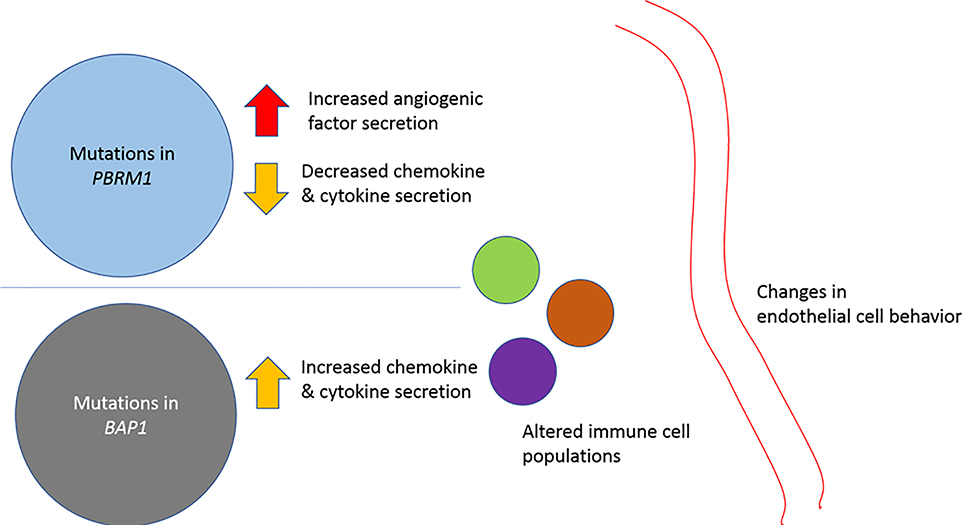
Figure 6: Alterations in the immune microenvironment.
Tumor cell genomic features may influence cell surface receptor expression as well as synthesis and secretion of immunomodulatory chemokines, cytokines and proangiogenic factors. PBRM1 mutations have been shown to increase angiogenesis, and decrease chemokine and cytokine production, whereas tumors bearing BAP1 mutations are associated with an inflamed microenvironment.
Chevrier and colleagues performed mass cytometric high-dimensional single cell analysis of ccRCC samples from 73 individuals with varying stages of disease with a focus on T-cell and tumor associated macrophages (TAM) 172. This analysis demonstrated diverse populations of immune and macrophage populations, with significant intra- and inter- cell population and patient heterogeneity. Specific patterns of T-cell and macrophage co-expression were reported, with exhausted/regulatory T-cells associated with a specific TAM subgroup expressing high levels of HLA-DR, CD68, and CD64, as well as CD204 and CD38. Other TAM subgroups expressed both pro- (CD163, CD204, CD206) - and antitumoral (CD169) TAM markers, although all of these subgroups were associated with more advanced (as defined as higher than stage 1) tumors, prevalence of additional TAM subgroups not specifically associated with a T-cell subpopulation were associated with worse progression free survival. This study illustrates the tremendous diversity of T-cell and TAM populations, and the challenges in identifying driver populations and therapeutic strategies. This study did link these cell populations to tumor genomic features. Doing so will help our understanding of the interplay between tumor-cell specific features and the immune microenvironment.
Wang et al used a comparison between uninvolved kidney, bulk tumor and tumorgraft transcriptomes to identify stromal and immune contributions to the overall expressed gene set in ccRCC and in other RCC tumor subtypes. Employing an in-house algorithm based on a Bayesian hierarchical model they named DisHet, they identified an immune/stromal gene set (eTME) specific to ccRCC, with 2080 genes expressed 3-fold higher and 904 genes expressed 20-fold or higher in the immune/stroma component than in the tumor 175. Using the eTME gene sets, the authors found that 65, 22 and 9 percent of immune signature genes in the Immunome178, ESTIMATE179 and Winslow180 sets were not abundantly expressed in the ccRCC immune/stromal compartment. They applied the eTME gene set to TCGA RCC datasets, including those representing ccRCC, papillary and chromophobe RCC. They identified an eTME high subgroup, which was associated with increased immune cell and complement transcripts, and a noninflamed group. There was a relatively low prevalence of inflamed tumors in type I papillary RCC. In ccRCC, the inflamed group was enriched for BAP1 mutations, and, using an annotated institution-specific patient cohort, demonstrated the inflamed subgroup manifested IMDC poor-risk features, including anemia and thrombocytosis, and worse prognosis 181.
A study assessing the immune microenvironment in 409 patients treated with sunitinib or pazopanib applied unsupervised consensus nonnegative matrix factorization (cNMF) clustering on expression microarray data and identified four biologically distinct clusters119. These clusters demonstrated varying response rates and OS. Cluster 4 was associated with worse OS, IMDC poor risk categorization, elevated PD-L1 levels and BAP1 and TP53 mutations, and anticorrelated with PBRM1 mutations. Tumors harboring PBRM1 mutations were associated with higher angiogenesis gene expression, and BAP1 mutated tumors with lower angiogenesis gene expression. Pathway analysis demonstrated elevated inflammatory signatures, including IFN gamma response in Cluster 4, and the highest immune score using the ESTIMATE179 algorithm. Higher macrophage infiltration was associated with worse response to antiangiogenic therapy and with worse OS. Scoring based on angiogenesis (Angio) and macrophage (Macrophage) status demonstrated that patients in this antiangiogenic therapy treated group that had AngioloMacrophagehi tumors demonstrated the worse outcomes compared to the AngiohiMacrophagelo group.
McDermott et al reported on the clinical outcomes of 301 patients with advanced ccRCC treated with the programmed death-ligand 1 (PD-L1) inhibitor atezolizumab, the combination of atezolizumab plus the anti VEGF antibody bevacizumab or the small molecule VEGFR inhibitor sunitinib. They also performed hypothesis generating analyses on the genomic and transcriptomic tumor features in the primary tumors from these patients182. These analyses identified angiogenesis, T-effector, and myeloid signatures, which were used to group patients into clinically relevant categories. The angiogenesis high subgroup was more likely to respond to sunitinib therapy whereas the T-effector high group was more likely to respond to the combination of atezolizumab plus bevacizumab. Further subdivision demonstrated that in the absence of a significant myeloid population, there was a trend towards improved response to atezolizumab monotherapy relative to sunitinib, a trend that was reversed if the myeloid population was present 182 An additional analysis demonstrated that PBRM1 mutations were significantly associated with response to sunitinib, and not with response to either atezolizumab or the combination of atezolizumab plus bevacizumab 182.
Clark et al performed a multiparametric analysis of 103 treatment naïve ccRCC and 84 matched uninvolved tissue samples, including genomic, transcriptomic, proteomic and phosphoproteomic assays 118. The authors identified a subset of ccRCC with chromosome-level genomic instability potentially associated with worse prognosis. At a protein level, upregulation of glycolysis and corresponding downregulation of oxidative phosphorylation (OXPHOS) was noted, with the changes in OXPHOS observed at a protein, but not at an mRNA level, with later stage tumors once again showing increased OXPHOS. Immune classification divided tumors into CD8+ immune inflamed, CD8- immune inflamed, metabolic immune desert, and VEGF immune desert tumors. Notably, CD8+ immune inflamed tumors were enriched for BAP1 mutations, whereas the VEGF immune desert tumors had a higher proportion of PBRM1 mutations.
Liu et al recently developed an isogenic murine Renca based model assessing the impact of PBRM1 deficiency on immune response 112. Transcriptomic mapping of immune gene expression in murine tumors closely paralleled that seen in the KIRC TCGA dataset. They found that loss of PBRM1 in both animal models and in human samples decreased immune infiltration, and Pbrm1 knockout tumors were more resistant to anti-PD-1 antibody. They determined that loss of PBRM1 in both murine and human tumors resulting in lower IFN gamma mediated transcription of the chemoattractive chemokines, which was at least partially due to loss of PBRM1 mediated upregulation of interferon receptor gamma 2 (IFNGR2) gene expression.
In contrast to these prior studies, an assessment of primary tumor genomics from 63 metastatic ccRCC patients treated with checkpoint blocking antibodies indicated that PBRM1 mutated tumors were more likely to respond to checkpoint antibody therapy 176. A follow up study by the same group demonstrated similar outcomes 183. The results of these studies may be due to patient selection or to other confounding factors including prior therapies.
6. TREATMENT IMPLICATIONS
Prevention and Early Detection
At this point in time the mechanism of 3p loss in renal precursor cells has not been formally defined. A better understanding of the etiology of this driver of ccRCC tumor initiation could lead to prevention strategies. Due to the relatively low incidence of ccRCC at a population level, this may pose challenges, but would be of greater use in high-risk populations, like those with germline losses in the VHL gene.
Early detection may be possible through either blood or urine-based detection systems with sufficient sensitivity to detect a small amount of genomic material harboring 3p loss. It may be possible to develop circulating tumor DNA (ctDNA) assays that capture specific SCNAs which identify these changes. If such a technology were successfully developed, it would encourage the development of a treatment strategy that selectively targets cells harboring 3p loss, thereby eradicating the foundational clones responsible for ccRCC development.
Identifying lethal versus nonlethal primary tumors
In the nonmetastatic setting, identifying patients who are at higher risk of developing metastatic disease would help to develop targeted adjuvant therapy strategies. A number of nomograms and algorithms have previously been generated which use a combination of clinicopathological data to risk-stratify patients 184–186. Applying our broader understanding of the impact tumor genomic features on patient outcome may help refine these algorithms. As an example, the seven TRACERx groups were divided into three broad groups of clinical context: linear evolution which include the VHL monodriver patients; branched evolution, which include the PBRM1-SCNA, PBRM1-SETD2 and the PBRM1-PI3K groups; and punctuated evolution which include the BAP-driven, multiple clonal driver and the VHL wildtype groups 21. Tumors with linear evolution patterns are less likely to metastasize, are more likely to be cured after nephrectomy, and are unlikely to need adjuvant therapy. The patients with branched evolution are potentially associated with intermediate prognosis. Surgical removal of the primary tumor would reduce the pool of potential metastatic subclones in these patients, and the genomic changes seen in these tumors may provide opportunities for neoadjuvant or adjuvant therapy development. The tumors with punctuated evolution/multiple clonal drivers are likely high risk for metastatic disease, and if metastatic, for progression. These individuals could be considered for adjuvant therapy trials, and if discovered with synchronous metastases, are most likely to benefit from upfront systemic therapy as opposed to cytoreductive nephrectomy.
The further evolution and validation of molecular subclassifiers as proposed by the TRACERx project will improve prognostication of ccRCC. As these tumor subtypes become more robustly defined, the measurement of circulating tumor DNA (ctDNA) could potentially provide an aggregate readout of tumor genomic features, and identify clones associated with tumor aggressivity and metastatic potential 187,188. As a whole this field is in a state of evolution, with significant unanswered questions on how best to develop clinically impactful ctDNA platforms 189. As of now, there are very few reported studies assessing ctDNA in ccRCC 190, and despite its clear potential utility, additional work is required to bring this approach into routine practice for patients with ccRCC.
Targeting genomically and metabolically defined vulnerabilities in ccRCC- tumor cell-centric approach
The process of developing a molecularly defined taxonomy of ccRCC provides a parallel opportunity to develop subtype-tailored therapeutic options (Table 2). At this point in time the treatment of ccRCC has not reached the granularity seen in other tumor types like breast and lung carcinomas191,192. Nonetheless, an understanding of ccRCC biology has already yielded significant advances in treatment, and further work in the next few years will undoubtedly continue this trend.
Table 2:
Opportunities for intervention in ccRCC.
| Tumor alteration | Therapeutic Opportunities | Knowledge Gap |
|---|---|---|
| VHL/3p loss | Prevention Early detection | Initiating factors for 3p loss Sensitive assays for early clonal evolution |
| Mutations in chromatin remodeling genes | Synthetic lethal strategies | Mechanism(s) of oncogenesis |
| Upregulation of oncogenic pathways | Targeted agents | Key drivers of pathway upregulation/mechanisms of resistance |
| Modulation of immune microenvironment | Tailored intervention(s) to modulate immune response | Mechanism of microenvironmental alteration(s), genotype/phenotype association |
The near-obligate loss of VHL in these tumors has spurred efforts to develop synthetic lethal targeting strategies193 and to rescue function of point-mutated VHL protein75,194,195. Metabolic consequences of VHL loss have been elegantly defined by Iliopoulos and colleagues196 and this research has led to the testing of glutaminase inhibitors in ccRCC (NCT03428217, NCT03163667). Upregulated HIF35 also provides a fairly unique target for ccRCC, and HIF2a inhibitors are showing promising results in preclinical and clinical studies 197,198. Ongoing trials are testing HIF2a inhibitors both in combination with tyrosine kinase inhibitor (TKIs) in advanced ccRCC (NCT03634540) and as monotherapy in patients with hereditary VHL disease (NCT03401788). Preliminary data from these studies show that the small molecule HIF2a inhibitor MK6482 is active both in advanced ccRCC 199 and in VHL disease related ccRCC 200.
The mechanistic consequences of BAP1, SETD2 and PBRM1 mutations on ccRCC are under active investigation. While prognostic differences exist between tumors harboring one or another of these mutations, distinct and targetable molecular features of these subtypes remain a work in progress.
Loss of BAP1 was shown to increase enhancer of zeste homolog 2 (EZH2) levels in a mesothelioma model 201, and to increase sensitivity to EZH2 inhibition. Similar efforts in uveal melanoma appeared less promising 202. An EZH2 inhibitor, tazemetostat, is currently in clinical trials. At this point in time the applicability of this approach to ccRCC harboring BAP1 mutations is not known.
Preclinical studies showed that a SETD2 deficiency increased sensitivity to WEE1 inhibitors in cell line experiments via nucleotide depletion, due to decreased expression and increased degradation of a ribonuclease reductase subunit, RRM2203. The WEE1 inhibitor adavosertib (AZD1775, MK-1775) demonstrated preclinical efficacy in H3K36me3 deficient tumor xenografts 203. A number of clinical trials are now testing this agent138, including a study focused on SETD2 deficient tumors (NCT03284385). Additionally, a preclinical study has shown that synthetic lethality exists between SETD2 deficiency and PI3Kβ inhibition in a ccRCC model 204.
Agents that target TORC1 signaling are approved for treatment of patients with advanced RCC, but have shown modest efficacy205,206, as have agents that target AKT 207. More recently, the PI3K pathway was found to modulate elements of HRR, and inhibition of PI3K pathway signaling could sensitize various tumor subtypes to PARP inhibition 208,209. In a similar manner, the RCC 786–0 cell line, known to have elevated PI3K pathway signaling, and relatively resistant to PARP inhibition, could be sensitized through concomitant blockade of PI3K pathway signaling 164. These observations provide a potential new direction in modulating ccRCC signaling in a way to render it more susceptible to DNA repair targeted therapy.
Integrating treatment of tumor cells and microenvironment
A broader understanding the proangiogenic consequences of VHL mutation in ccRCC spawned a panoply of agents targeting either vascular endothelial growth factors210,211 or their receptors212–217 in ccRCC. These agents moved the treatment of ccRCC from relatively toxic and inconsistently effective cytokines firmly to predominantly orally bioavailable agents with consistent efficacy and manageable side effects. In hereditary VHL disease, use of these agents has resulted in a salutary effect in patients harboring ccRCC 218,219, as well as showing some benefit in other affected organ systems.
As the various phenotypic and genotypic subvariants of ccRCC become defined, it is essential to understand how their specific molecular features influence the tumor immune microenvironment, and how this information can be used to add precision to therapy selection. As an example, in sarcomatoid RCC, programmed death- ligand 1 (PD-L1) is upregulated relative to nonsarcomatoid ccRCC, and an increased number of T-cells is present in the tumor microenvironment 220,221. Emerging data suggest that patients with sarcomatoid RCC demonstrate superior response to checkpoint antibody therapy222, with larger confirmatory analyses being performed (NCT02420821).
Recent studies have assessed correlations between specific molecular signatures, the tumor microenvironment and clinical outcome. Multiple studies have showed that PBRM1 mutation was associated with upregulated angiogenesis and downregulated immune cell infiltration. Congruent with the preclinical data summarized above 103,110, data from the RECORD-3 study indicated that patients with PBRM1 mutated tumors responded well to antiangiogenic therapy 129. Results from the IMmotion 150 study demonstrated that patients with PBRM1 mutated tumors responded better to antiangiogenic agents than those with wildtype tumors, and the directionality was reversed with PD-L1 blocking therapy- patients with PBRM1 mutated tumors showed a trend towards worse outcome after atezolizumab treatment177. This is in contradistinction to data reported by a different group, who indicated that PBRM1 mutations were associated with a better response to checkpoint antibody therapy 176,183. Clearly more work needs to be done to provide clarity on how this commonly mutated gene impacts immune response in ccRCC.
BAP1 mutated tumors demonstrate a more inflamed immune microenvironment 118,175, suggesting that some form of immune-targeting strategy may benefit these patients as well. Less is known about the impact of SETD2 mutations on the immune microenvironment and treatment response, with ongoing efforts to better characterize their effect.
The expression of Axl on both ccRCC tumor cells49 and on immune suppressive cell populations in the tumor microenvironment 223,224 provide additional opportunities for targeted, integrated therapeutic approaches. Clinical trials testing this hypothesis are underway using the combination of cabozantinib and nivolumab (NCT03141177), and there are clear opportunities to test agents that more precisely target TAM family members in combination with immunotherapy.
Can we target or circumvent the “engines” of tumor heterogeneity?
Tumor heterogeneity may influence the ability to fully characterize the genomic drivers of ccRCC 19, and can make it more difficult to choose molecularly targeted agents against key driver mutations. If sufficiently sensitive, ctDNA may help in identifying key drivers of tumor biology and in therapy selection.
Similarly, tumor heterogeneity may decrease the impact of targeted therapy in the treatment of ccRCC, especially if the molecules being targeted are fairly downstream from the driver events. By targeting the drivers of heterogeneity themselves, it may be possible to decrease or prevent further clonal and subclonal evolution, and the very presence of this aberrant programming may confer specific vulnerabilities to the tumor cell.
Immunotherapy response may actually be enhanced by tumor heterogeneity 169. The diverse repertoire of tumor neoantigens may enhance immune response by providing a larger number of opportunities for cytotoxic T-cells to interact with and kill tumor cells225. MSI high tumors are particularly responsive to checkpoint antibody therapy 226,227. Despite the intermediate level of tumor heterogeneity observed in ccRCC 139 ccRCC is quite responsive to checkpoint inhibitory antibodies228,229, and this response appears unrelated to qualitative characteristics of the ccRCC mutational repertoire, including insertions and deletions, and associated frameshift mutational burden 177. The reasons why ccRCC responds to immunotherapy are not fully elucidated, and future research investigating the STING pathway230 and other mechanisms of immunogenicity may provide clues.
7. CONCLUDING STATEMENTS
In the past decade our understanding of the determinants of ccRCC ontogeny and the drivers of heterogeneity and lethality has reached the point where are beginning to develop cogent mechanistic pathways outlining tumor ontogeny and progression and are creating clinically relevant subcategories of ccRCC that may benefit from subtype-specific therapeutic interventions. Robust ongoing work to elucidate the impact driver gene mutations have on ccRCC biology will help create the necessary framework to begin developing more targeted therapeutic approaches for patients with ccRCC. As our understanding of the molecular biology of ccRCC expands, there is a parallel evolution of diagnostic, imaging and therapeutic tools that will enable us to translate these advances into substantive improvements in the prevention, early detection, and treatment of ccRCC.
KEY POINTS.
Chromosome 3p loss is an almost universal finding in both hereditary and sporadic ccRCC
The near ubiquitous loss of a second copy of VHL appears to provide a selective advantage for cells, as well as enabling defects in DNA repair and an increase in genomic instability.
Secondarily mutated genes in ccRCC, including PBRM1, SETD2 and BAP1, as well as copy number changes in chromosomes 9p and 14q are associated with prognostically important molecular and phenotypic characteristics that can be used to create specific subgroups.
Tumor genomic features are associated with distinct immune phenotypes. As an example, PBRM1 mutations are associated with decreased T-cell infiltration.
Efforts are underway to link genomic features to specific therapeutic strategies and agents for patients with ccRCC
Bibliography
- 1.Bray F et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424, doi: 10.3322/caac.21492 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Jonasch E, Gao J & Rathmell WK Renal cell carcinoma. Bmj 349, g4797, doi: 10.1136/bmj.g4797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagemeijer A, Hoehn W & Smit EM Cytogenetic analysis of human renal carcinoma cell lines of common origin (NC 65). Cancer Res 39, 4662–4667 (1979). [PubMed] [Google Scholar]
- 4.Szucs S, Muller-Brechlin R, DeRiese W & Kovacs G Deletion 3p: the only chromosome loss in a primary renal cell carcinoma. Cancer genetics and cytogenetics 26, 369–373 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Yoshida MA et al. Rearrangement of chromosome 3 in renal cell carcinoma. Cancer genetics and cytogenetics 19, 351–354 (1986). [DOI] [PubMed] [Google Scholar]
- 6.Latif F et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260, 1317–1320. (1993). [DOI] [PubMed] [Google Scholar]
- 7.Varela I et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542, doi:nature09639 [pii] 10.1038/nature09639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena-Llopis S et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 44, 751–759, doi: 10.1038/ng.2323 ng.2323 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgliesh GL et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363, doi: 10.1038/nature08672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Network CGAR Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49, doi: 10.1038/nature12222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klatte T et al. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J Clin Oncol 27, 746–753, doi:JCO.2007.15.8345 [pii] 10.1200/JCO.2007.15.8345 (2009). [DOI] [PubMed] [Google Scholar]
- 12.La Rochelle J et al. Chromosome 9p deletions identify an aggressive phenotype of clear cell renal cell carcinoma. Cancer 116, 4696–4702, doi: 10.1002/cncr.25279 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Monzon FA et al. Chromosome 14q loss defines a molecular subtype of clear-cell renal cell carcinoma associated with poor prognosis. Mod Pathol 24, 1470–1479, doi: 10.1038/modpathol.2011.107 modpathol2011107 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen C et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer Discov 1, 222–235, doi: 10.1158/2159-8290.CD-11-0098 2159–8290.CD-11–0098 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang F et al. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res 60, 6503–6509 (2000). [PubMed] [Google Scholar]
- 16.Brannon AR et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer 1, 152–163, doi: 10.1177/1947601909359929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rini B et al. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: development and validation studies. Lancet Oncol 16, 676–685, doi: 10.1016/S1470-2045(15)70167-1 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366, 883–892, doi: 10.1056/NEJMoa1113205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turajlic S et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell 173, 595–610 e511, doi: 10.1016/j.cell.2018.03.043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell TJ et al. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell 173, 611–623 e617, doi: 10.1016/j.cell.2018.02.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turajlic S et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 173, 581–594 e512, doi: 10.1016/j.cell.2018.03.057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monzon FA et al. Whole genome SNP arrays as a potential diagnostic tool for the detection of characteristic chromosomal aberrations in renal epithelial tumors. Mod Pathol 21, 599–608, doi:modpathol200820 [pii] 10.1038/modpathol.2008.20 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Wang L et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res 22, 208–219, doi: 10.1101/gr.123109.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang YC et al. SETD2 Haploinsufficiency for Microtubule Methylation Is an Early Driver of Genomic Instability in Renal Cell Carcinoma. Cancer Res 78, 3135–3146, doi: 10.1158/0008-5472.CAN-17-3460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fei SS et al. Patient-specific factors influence somatic variation patterns in von Hippel-Lindau disease renal tumours. Nature communications 7, 11588, doi: 10.1038/ncomms11588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalsbeek D & Golsteyn RM G2/M-Phase Checkpoint Adaptation and Micronuclei Formation as Mechanisms That Contribute to Genomic Instability in Human Cells. Int J Mol Sci 18, doi: 10.3390/ijms18112344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podrimaj-Bytyqi A et al. The frequencies of micronuclei, nucleoplasmic bridges and nuclear buds as biomarkers of genomic instability in patients with urothelial cell carcinoma. Sci Rep 8, 17873, doi: 10.1038/s41598-018-35903-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DB, Huang E & Ward HJ Tight junction biology and kidney dysfunction. Am J Physiol Renal Physiol 290, F20–34, doi: 10.1152/ajprenal.00052.2005 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Lonser RR et al. von Hippel-Lindau disease. Lancet 361, 2059–2067 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Ho TH & Jonasch E Genetic kidney cancer syndromes. Journal of the National Comprehensive Cancer Network : JNCCN 12, 1347–1355 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Nickerson ML et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res 14, 4726–4734 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxwell PH et al. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A 94, 8104–8109 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flamme I, Krieg M & Plate KH Up-regulation of vascular endothelial growth factor in stromal cells of hemangioblastomas is correlated with up-regulation of the transcription factor HRF/HIF-2alpha. Am J Pathol 153, 25–29 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieg M et al. Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene 19, 5435–5443, doi: 10.1038/sj.onc.1203938 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Maxwell PH et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275. (1999). [DOI] [PubMed] [Google Scholar]
- 36.Keith B, Johnson RS & Simon MC HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12, 9–22, doi: 10.1038/nrc3183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi A et al. Markedly increased amounts of messenger RNAs for vascular endothelial growth factor and placenta growth factor in renal cell carcinoma associated with angiogenesis. Cancer Res 54, 4233–4237 (1994). [PubMed] [Google Scholar]
- 38.Hu CJ, Wang LY, Chodosh LA, Keith B & Simon MC Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol 23, 9361–9374 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raval RR et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol 25, 5675–5686 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll VA & Ashcroft M Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res 66, 6264–6270, doi: 10.1158/0008-5472.CAN-05-2519 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Kim JW, Tchernyshyov I, Semenza GL & Dang CV HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell metabolism 3, 177–185, doi: 10.1016/j.cmet.2006.02.002 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Semenza GL et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem 271, 32529–32537 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Zhang H et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283, 10892–10903, doi: 10.1074/jbc.M800102200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Bellot G et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29, 2570–2581, doi: 10.1128/MCB.00166-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Bryan JP et al. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol 11, 5016–5031 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemke G Biology of the TAM receptors. Cold Spring Harbor perspectives in biology 5, a009076, doi: 10.1101/cshperspect.a009076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai C, Gore M & Lemke G Structure, expression, and activity of Tyro 3, a neural adhesion-related receptor tyrosine kinase. Oncogene 9, 2567–2578 (1994). [PubMed] [Google Scholar]
- 48.Rankin EB et al. Direct regulation of GAS6/AXL signaling by HIF promotes renal metastasis through SRC and MET. Proc Natl Acad Sci U S A 111, 13373–13378, doi: 10.1073/pnas.1404848111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou L et al. Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma. Oncogene, doi: 10.1038/onc.2015.343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rankin EB et al. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol Cell Biol 25, 3163–3172, doi: 10.1128/MCB.25.8.3163-3172.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu L, Wang G, Shevchuk MM, Nanus DM & Gudas LJ Generation of a mouse model of Von Hippel-Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1alpha. Cancer Res 71, 6848–6856, doi: 10.1158/0008-5472.CAN-11-1745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu L, Wang G, Shevchuk MM, Nanus DM & Gudas LJ Activation of HIF2alpha in kidney proximal tubule cells causes abnormal glycogen deposition but not tumorigenesis. Cancer Res 73, 2916–2925, doi: 10.1158/0008-5472.CAN-12-3983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schietke RE et al. Renal tubular HIF-2alpha expression requires VHL inactivation and causes fibrosis and cysts. PLoS One 7, e31034, doi: 10.1371/journal.pone.0031034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schonenberger D et al. Formation of Renal Cysts and Tumors in Vhl/Trp53-Deficient Mice Requires HIF1alpha and HIF2alpha. Cancer Res 76, 2025–2036, doi: 10.1158/0008-5472.CAN-15-1859 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Zhang J & Zhang Q VHL and Hypoxia Signaling: Beyond HIF in Cancer. Biomedicines 6, doi: 10.3390/biomedicines6010035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo J et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science 353, 929–932, doi: 10.1126/science.aad5755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An J & Rettig MB Mechanism of von Hippel-Lindau protein-mediated suppression of nuclear factor kappa B activity. Mol Cell Biol 25, 7546–7556, doi:25/17/7546 [pii] 10.1128/MCB.25.17.7546-7556.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang H et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell 28, 15–27, doi: 10.1016/j.molcel.2007.09.010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peri S, Devarajan K, Yang DH, Knudson AG & Balachandran S Meta-analysis identifies NF-kappaB as a therapeutic target in renal cancer. PLoS One 8, e76746, doi: 10.1371/journal.pone.0076746 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu L et al. TBK1 Is a Synthetic Lethal Target in Cancer with VHL Loss. Cancer Discov 10, 460–475, doi: 10.1158/2159-8290.CD-19-0837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metcalf JL et al. K63-ubiquitylation of VHL by SOCS1 mediates DNA double-strand break repair. Oncogene 33, 1055–1065, doi: 10.1038/onc.2013.22 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Scanlon SE, Hegan DC, Sulkowski PL & Glazer PM Suppression of homology-dependent DNA double-strand break repair induces PARP inhibitor sensitivity in VHL-deficient human renal cell carcinoma. Oncotarget 9, 4647–4660, doi: 10.18632/oncotarget.23470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Espana-Agusti J, Warren A, Chew SK, Adams DJ & Matakidou A Loss of PBRM1 rescues VHL dependent replication stress to promote renal carcinogenesis. Nature communications 8, 2026, doi: 10.1038/s41467-017-02245-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou L et al. Tip60 dependent DNA homologous recombination repair is impaired in VHL-deficient clear cell renal cell carcinoma. AACR, 1364, doi:DOI: 10.1158/1538-7445.AM2018-1364 (2018). [DOI] [Google Scholar]
- 65.Zhang J et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 361, 290–295, doi: 10.1126/science.aap8411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calzada MJ et al. von Hippel-Lindau tumor suppressor protein regulates the assembly of intercellular junctions in renal cancer cells through hypoxia-inducible factor-independent mechanisms. Cancer Res 66, 1553–1560 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Stickle NH et al. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol 24, 3251–3261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohh M et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell 1, 959–968. (1998). [DOI] [PubMed] [Google Scholar]
- 69.Kurban G et al. Collagen matrix assembly is driven by the interaction of von Hippel-Lindau tumor suppressor protein with hydroxylated collagen IV alpha 2. Oncogene 27, 1004–1012, doi: 10.1038/sj.onc.1210709 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Kurban G, Hudon V, Duplan E, Ohh M & Pause A Characterization of a von Hippel Lindau pathway involved in extracellular matrix remodeling, cell invasion, and angiogenesis. Cancer Res 66, 1313–1319 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Clifford SC & Maher ER Von Hippel-Lindau disease: clinical and molecular perspectives. Advances in cancer research 82, 85–105 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Hoffman MA et al. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet 10, 1019–1027 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Tang N, Mack F, Haase VH, Simon MC & Johnson RS pVHL function is essential for endothelial extracellular matrix deposition. Mol Cell Biol 26, 2519–2530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lolkema MP et al. Tumor suppression by the von Hippel-Lindau protein requires phosphorylation of the acidic domain. J Biol Chem 280, 22205–22211, doi:M503220200 [pii] 10.1074/jbc.M503220200 (2005). [DOI] [PubMed] [Google Scholar]
- 75.German P et al. Phosphorylation-dependent cleavage regulates von Hippel Lindau proteostasis and function. Oncogene, doi: 10.1038/onc.2016.40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hergovich A, Lisztwan J, Barry R, Ballschmieter P & Krek W Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol 5, 64–70 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Schermer B et al. The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J Cell Biol 175, 547–554, doi:jcb.200605092 [pii] 10.1083/jcb.200605092 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esteban MA, Harten SK, Tran MG & Maxwell PH Formation of primary cilia in the renal epithelium is regulated by the von Hippel-Lindau tumor suppressor protein. J Am Soc Nephrol 17, 1801–1806, doi:ASN.2006020181 [pii] 10.1681/ASN.2006020181 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Lutz MS & Burk RD Primary cilium formation requires von hippel-lindau gene function in renal-derived cells. Cancer Res 66, 6903–6907, doi: 10.1158/0008-5472.CAN-06-0501 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Devlin LA & Sayer JA Renal ciliopathies. Curr Opin Genet Dev 56, 49–60, doi: 10.1016/j.gde.2019.07.005 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Hasanov E et al. Ubiquitination and regulation of AURKA identifies a hypoxia-independent E3 ligase activity of VHL. Oncogene 36, 3450–3463, doi: 10.1038/onc.2016.495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thoma CR et al. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol 11, 994–1001 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Hell MP, Duda M, Weber TC, Moch H & Krek W Tumor suppressor VHL functions in the control of mitotic fidelity. Cancer Res 74, 2422–2431, doi: 10.1158/0008-5472.CAN-13-2040 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Hell MP et al. miR-28–5p promotes chromosomal instability in VHL-associated cancers by inhibiting Mad2 translation. Cancer Res 74, 2432–2443, doi: 10.1158/0008-5472.CAN-13-2041 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Janku F, Yap TA & Meric-Bernstam F Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 15, 273–291, doi: 10.1038/nrclinonc.2018.28 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Horiguchi A, Oya M, Uchida A, Marumo K & Murai M Elevated Akt activation and its impact on clinicopathological features of renal cell carcinoma. J Urol 169, 710–713 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Sourbier C et al. The phosphoinositide 3-kinase/Akt pathway: a new target in human renal cell carcinoma therapy. Cancer Res 66, 5130–5142 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Kim J et al. Cytoplasmic sequestration of p27 via AKT phosphorylation in renal cell carcinoma. Clin Cancer Res 15, 81–90, doi:15/1/81 [pii] 10.1158/1078-0432.CCR-08-0170 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haase VH, Glickman JN, Socolovsky M & Jaenisch R Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A 98, 1583–1588, doi: 10.1073/pnas.98.4.1583 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hickey MM, Lam JC, Bezman NA, Rathmell WK & Simon MC von Hippel-Lindau mutation in mice recapitulates Chuvash polycythemia via hypoxia-inducible factor-2alpha signaling and splenic erythropoiesis. J Clin Invest 117, 3879–3889, doi: 10.1172/JCI32614 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee CM et al. VHL Type 2B gene mutation moderates HIF dosage in vitro and in vivo. Oncogene 28, 1694–1705, doi:onc200912 [pii] 10.1038/onc.2009.12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frew IJ & Moch H A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu Rev Pathol 10, 263–289, doi: 10.1146/annurev-pathol-012414-040306 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Ma W et al. Hepatic vascular tumors, angiectasis in multiple organs, and impaired spermatogenesis in mice with conditional inactivation of the VHL gene. Cancer Res 63, 5320–5328 (2003). [PubMed] [Google Scholar]
- 94.Rankin EB, Tomaszewski JE & Haase VH Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res 66, 2576–2583, doi: 10.1158/0008-5472.CAN-05-3241 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mathia S et al. Action of hypoxia-inducible factor in liver and kidney from mice with Pax8-rtTA-based deletion of von Hippel-Lindau protein. Acta Physiol (Oxf) 207, 565–576, doi: 10.1111/apha.12058 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Schley G et al. Hypoxia-inducible transcription factors stabilization in the thick ascending limb protects against ischemic acute kidney injury. J Am Soc Nephrol 22, 2004–2015, doi: 10.1681/ASN.2010121249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lubensky IA et al. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am J Pathol 149, 2089–2094 (1996). [PMC free article] [PubMed] [Google Scholar]
- 98.Paraf F et al. Renal lesions in von Hippel-Lindau disease: immunohistochemical expression of nephron differentiation molecules, adhesion molecules and apoptosis proteins. Histopathology 36, 457–465, doi: 10.1046/j.1365-2559.2000.00857.x (2000). [DOI] [PubMed] [Google Scholar]
- 99.Albers J et al. Combined mutation of Vhl and Trp53 causes renal cysts and tumours in mice. EMBO molecular medicine 5, 949–964, doi: 10.1002/emmm.201202231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frew IJ et al. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. Embo J 27, 1747–1757, doi:emboj200896 [pii] 10.1038/emboj.2008.96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harlander S et al. Combined mutation in Vhl, Trp53 and Rb1 causes clear cell renal cell carcinoma in mice. Nat Med 23, 869–877, doi: 10.1038/nm.4343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gu YF et al. Modeling Renal Cell Carcinoma in Mice: Bap1 and Pbrm1 Inactivation Drive Tumor Grade. Cancer Discov 7, 900–917, doi: 10.1158/2159-8290.CD-17-0292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nargund AM et al. The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma. Cell reports 18, 2893–2906, doi: 10.1016/j.celrep.2017.02.074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee H et al. BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget 7, 19134–19146, doi: 10.18632/oncotarget.8102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Cubas AA & Rathmell WK Epigenetic modifiers: activities in renal cell carcinoma. Nat Rev Urol 15, 599–614, doi: 10.1038/s41585-018-0052-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thompson M Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie 91, 309–319, doi: 10.1016/j.biochi.2008.10.019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pulice JL & Kadoch C Composition and Function of Mammalian SWI/SNF Chromatin Remodeling Complexes in Human Disease. Cold Spring Harb Symp Quant Biol 81, 53–60, doi: 10.1101/sqb.2016.81.031021 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Kim SH et al. The prognostic value of BAP1, PBRM1, pS6, PTEN, TGase2, PD-L1, CA9, PSMA, and Ki-67 tissue markers in localized renal cell carcinoma: A retrospective study of tissue microarrays using immunohistochemistry. PLoS One 12, e0179610, doi: 10.1371/journal.pone.0179610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Z et al. Sarcomatoid Renal Cell Carcinoma Has a Distinct Molecular Pathogenesis, Driver Mutation Profile, and Transcriptional Landscape. Clin Cancer Res 23, 6686–6696, doi: 10.1158/1078-0432.CCR-17-1057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao W, Li W, Xiao T, Liu XS & Kaelin WG Jr. Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL−/− clear cell renal carcinoma. Proc Natl Acad Sci U S A 114, 1027–1032, doi: 10.1073/pnas.1619726114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liao L et al. Multiple tumor suppressors regulate a HIF-dependent negative feedback loop via ISGF3 in human clear cell renal cancer. Elife 7, doi: 10.7554/eLife.37925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu XD et al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nature communications 11, 2135, doi: 10.1038/s41467-020-15959-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cai W et al. PBRM1 acts as a p53 lysine-acetylation reader to suppress renal tumor growth. Nature communications 10, 5800, doi: 10.1038/s41467-019-13608-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xue Y et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A 97, 13015–13020, doi: 10.1073/pnas.240208597 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang J, Hsu JM & Laurent BC The RSC nucleosome-remodeling complex is required for Cohesin’s association with chromosome arms. Mol Cell 13, 739–750, doi: 10.1016/s1097-2765(04)00103-0 (2004). [DOI] [PubMed] [Google Scholar]
- 116.Baetz KK, Krogan NJ, Emili A, Greenblatt J & Hieter P The ctf13–30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol Cell Biol 24, 1232–1244, doi: 10.1128/mcb.24.3.1232-1244.2003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brownlee PM, Chambers AL, Cloney R, Bianchi A & Downs JA BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell reports 6, 973–981, doi: 10.1016/j.celrep.2014.02.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Clark DJ et al. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 179, 964–983 e931, doi: 10.1016/j.cell.2019.10.007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hakimi AA et al. Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear cell Renal Cell Cancer - Data from a Randomized Phase III Trial. Cancer Discov, doi: 10.1158/2159-8290.CD-18-0957 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jensen DE et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16, 1097–1112, doi: 10.1038/sj.onc.1201861 (1998). [DOI] [PubMed] [Google Scholar]
- 121.Kapur P et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 14, 159–167, doi: 10.1016/S1470-2045(12)70584-3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scheuermann JC et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247, doi: 10.1038/nature08966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dey A et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546, doi: 10.1126/science.1221711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cejas P et al. Enhancer signatures stratify and predict outcomes of non-functional pancreatic neuroendocrine tumors. Nat Med, doi: 10.1038/s41591-019-0493-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peng J et al. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett 369, 167–174, doi: 10.1016/j.canlet.2015.08.013 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Kobayashi A et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181, doi: 10.1016/j.stem.2008.05.020 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang SS et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci U S A 111, 16538–16543, doi: 10.1073/pnas.1414789111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Joseph RW et al. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low-risk clear cell renal cell carcinoma. Cancer 120, 1059–1067, doi: 10.1002/cncr.28521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsieh JJ et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. Eur Urol 71, 405–414, doi: 10.1016/j.eururo.2016.10.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Farley MN et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res 11, 1061–1071, doi: 10.1158/1541-7786.MCR-13-0111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lickwar CR et al. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4, e4886, doi: 10.1371/journal.pone.0004886 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hacker KE et al. Structure/Function Analysis of Recurrent Mutations in SETD2 Protein Reveals a Critical and Conserved Role for a SET Domain Residue in Maintaining Protein Stability and Histone H3 Lys-36 Trimethylation. J Biol Chem 291, 21283–21295, doi: 10.1074/jbc.M116.739375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fahey CC & Davis IJ SETting the Stage for Cancer Development: SETD2 and the Consequences of Lost Methylation. Cold Spring Harb Perspect Med 7, doi: 10.1101/cshperspect.a026468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park IY et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 166, 950–962, doi: 10.1016/j.cell.2016.07.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Giaccia AJ A New Chromatin-Cytoskeleton Link in Cancer. Mol Cancer Res 14, 1173–1175, doi: 10.1158/1541-7786.MCR-16-0250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seervai RNH et al. SETD2 is an actin lysine methyltransferase. bioRxiv, doi: 10.1101/2020.04.13.034629 (2020). [DOI] [Google Scholar]
- 137.Karki M et al. A Cytoskeletal Function for PBRM1 Reading Methylated Microtubules. bioRxiv, doi: 10.1101/2020.04.21.053942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pilie PG, Tang C, Mills GB & Yap TA State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol, doi: 10.1038/s41571-018-0114-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421, doi: 10.1038/nature12477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dere R, Perkins AL, Bawa-Khalfe T, Jonasch D & Walker CL beta-Catenin Links von Hippel-Lindau to Aurora Kinase A and Loss of Primary Cilia in Renal Cell Carcinoma. J Am Soc Nephrol, doi: 10.1681/ASN.2013090984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang M et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol Cell 55, 31–46, doi: 10.1016/j.molcel.2014.04.028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li F et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 153, 590–600, doi: 10.1016/j.cell.2013.03.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Naqvi RA et al. Hypermethylation analysis of mismatch repair genes (hmlh1 and hmsh2) in locally advanced breast cancers in Indian women. Hum Pathol 39, 672–680, doi: 10.1016/j.humpath.2007.09.011 (2008). [DOI] [PubMed] [Google Scholar]
- 144.Altavilla G, Fassan M, Busatto G, Orsolan M & Giacomelli L Microsatellite instability and hMLH1 and hMSH2 expression in renal tumors. Oncol Rep 24, 927–932 (2010). [DOI] [PubMed] [Google Scholar]
- 145.Jasin M & Rothstein R Repair of strand breaks by homologous recombination. Cold Spring Harbor perspectives in biology 5, a012740, doi: 10.1101/cshperspect.a012740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hall JM et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689 (1990). [DOI] [PubMed] [Google Scholar]
- 147.Hall JM et al. Closing in on a breast cancer gene on chromosome 17q. Am J Hum Genet 50, 1235–1242 (1992). [PMC free article] [PubMed] [Google Scholar]
- 148.Wooster R et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789–792, doi: 10.1038/378789a0 (1995). [DOI] [PubMed] [Google Scholar]
- 149.Antoniou AC et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 371, 497–506, doi: 10.1056/NEJMoa1400382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Swift M, Morrell D, Massey RB & Chase CL Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med 325, 1831–1836, doi: 10.1056/NEJM199112263252602 (1991). [DOI] [PubMed] [Google Scholar]
- 151.Bindra RS et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol 24, 8504–8518, doi: 10.1128/MCB.24.19.8504-8518.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bindra RS et al. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res 65, 11597–11604, doi: 10.1158/0008-5472.CAN-05-2119 (2005). [DOI] [PubMed] [Google Scholar]
- 153.Scanlon SE & Glazer PM Hypoxic stress facilitates acute activation and chronic downregulation of fanconi anemia proteins. Mol Cancer Res 12, 1016–1028, doi: 10.1158/1541-7786.MCR-13-0628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roe JS et al. Phosphorylation of von Hippel-Lindau protein by checkpoint kinase 2 regulates p53 transactivation. Cell Cycle 10, 3920–3928, doi:18096 [pii] 10.4161/cc.10.22.18096 (2011). [DOI] [PubMed] [Google Scholar]
- 155.Tang J et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20, 317–325, doi: 10.1038/nsmb.2499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sun Y, Jiang X, Chen S, Fernandes N & Price BD A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 102, 13182–13187, doi: 10.1073/pnas.0504211102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pfister SX et al. SETD2-Dependent Histone H3K36 Trimethylation Is Required for Homologous Recombination Repair and Genome Stability. Cell reports 7, 2006–2018, doi: 10.1016/j.celrep.2014.05.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Carvalho S et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 3, e02482, doi: 10.7554/eLife.02482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Aymard F et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21, 366–374, doi: 10.1038/nsmb.2796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pfister SX et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 7, 2006–2018, doi: 10.1016/j.celrep.2014.05.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kanu N et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34, 5699–5708, doi: 10.1038/onc.2015.24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li L & Wang Y Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem 292, 11951–11959, doi: 10.1074/jbc.M117.788224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ismail IH et al. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res 74, 4282–4294, doi: 10.1158/0008-5472.CAN-13-3109 (2014). [DOI] [PubMed] [Google Scholar]
- 164.Peng G et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nature communications 5, 3361, doi: 10.1038/ncomms4361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pilie PG et al. Homologous repair deficiency in VHL-mutated clear cell renal cell carcinoma. J Clin Oncol 36, Suppl. 6S Abstract 585 (2018). [Google Scholar]
- 166.Carter SL, Eklund AC, Kohane IS, Harris LN & Szallasi Z A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 38, 1043–1048, doi: 10.1038/ng1861 (2006). [DOI] [PubMed] [Google Scholar]
- 167.McGranahan N, Burrell RA, Endesfelder D, Novelli MR & Swanton C Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep 13, 528–538, doi: 10.1038/embor.2012.61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gordan JD et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell 14, 435–446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jamal-Hanjani M, Quezada SA, Larkin J & Swanton C Translational implications of tumor heterogeneity. Clin Cancer Res 21, 1258–1266, doi: 10.1158/1078-0432.CCR-14-1429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Malouf GG et al. Genomic Characterization of Renal Cell Carcinoma with Sarcomatoid Dedifferentiation Pinpoints Recurrent Genomic Alterations. Eur Urol 70, 348–357, doi: 10.1016/j.eururo.2016.01.051 (2016). [DOI] [PubMed] [Google Scholar]
- 171.Bi M et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proc Natl Acad Sci U S A 113, 2170–2175, doi: 10.1073/pnas.1525735113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chevrier S et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 169, 736–749 e718, doi: 10.1016/j.cell.2017.04.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Senbabaoglu Y et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 17, 231, doi: 10.1186/s13059-016-1092-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ko JS et al. Sunitinib mediates reversal of myeloid-derived suppressor cell accumulation in renal cell carcinoma patients. Clin Cancer Res 15, 2148–2157, doi: 10.1158/1078-0432.CCR-08-1332 (2009). [DOI] [PubMed] [Google Scholar]
- 175.Wang T et al. An Empirical Approach Leveraging Tumorgrafts to Dissect the Tumor Microenvironment in Renal Cell Carcinoma Identifies Missing Link to Prognostic Inflammatory Factors. Cancer Discov 8, 1142–1155, doi: 10.1158/2159-8290.CD-17-1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Miao D et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806, doi: 10.1126/science.aan5951 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McDermott DF et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 24, 749–757, doi: 10.1038/s41591-018-0053-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bindea G et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795, doi: 10.1016/j.immuni.2013.10.003 (2013). [DOI] [PubMed] [Google Scholar]
- 179.Yoshihara K et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications 4, 2612, doi: 10.1038/ncomms3612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Winslow S, Leandersson K, Edsjo A & Larsson C Prognostic stromal gene signatures in breast cancer. Breast cancer research : BCR 17, 23, doi: 10.1186/s13058-015-0530-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heng DY et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol 27, 5794–5799 (2009). [DOI] [PubMed] [Google Scholar]
- 182.Seung SK, Shu HK, McDermott MW, Sneed PK & Larson DA Stereotactic radiosurgery for malignant melanoma to the brain. Surg Clin North Am 76, 1399–1411 (1996). [DOI] [PubMed] [Google Scholar]
- 183.Braun DA et al. Clinical Validation of PBRM1 Alterations as a Marker of Immune Checkpoint Inhibitor Response in Renal Cell Carcinoma. JAMA Oncol, doi: 10.1001/jamaoncol.2019.3158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zisman A et al. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol 19, 1649–1657 (2001). [DOI] [PubMed] [Google Scholar]
- 185.Frank I et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol 168, 2395–2400 (2002). [DOI] [PubMed] [Google Scholar]
- 186.Kattan MW, Reuter V, Motzer RJ, Katz J & Russo P A postoperative prognostic nomogram for renal cell carcinoma. J Urol 166, 63–67. (2001). [PubMed] [Google Scholar]
- 187.Bettegowda C et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6, 224ra224, doi: 10.1126/scitranslmed.3007094 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cohen JD et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930, doi: 10.1126/science.aar3247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Merker JD et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. Arch Pathol Lab Med 142, 1242–1253, doi: 10.5858/arpa.2018-0901-SA (2018). [DOI] [PubMed] [Google Scholar]
- 190.Pal SK et al. Evolution of Circulating Tumor DNA Profile from First-line to Subsequent Therapy in Metastatic Renal Cell Carcinoma. Eur Urol 72, 557–564, doi: 10.1016/j.eururo.2017.03.046 (2017). [DOI] [PubMed] [Google Scholar]
- 191.Rouzier R et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 11, 5678–5685, doi: 10.1158/1078-0432.CCR-04-2421 (2005). [DOI] [PubMed] [Google Scholar]
- 192.Travis WD et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 10, 1243–1260, doi: 10.1097/JTO.0000000000000630 (2015). [DOI] [PubMed] [Google Scholar]
- 193.Turcotte S et al. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell 14, 90–102, doi: 10.1016/j.ccr.2008.06.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ding Z et al. Genetic and pharmacological strategies to refunctionalize the von Hippel Lindau R167Q mutant protein. Cancer Res 74, 3127–3136, doi: 10.1158/0008-5472.CAN-13-3213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ding Z et al. Agents That Stabilize Mutated von Hippel-Lindau (VHL) Protein: Results of a High-Throughput Screen to Identify Compounds That Modulate VHL Proteostasis. J Biomol Screen 17, 572–580, doi:1087057112436557 [pii] 10.1177/1087057112436557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gameiro PA et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell metabolism 17, 372–385, doi: 10.1016/j.cmet.2013.02.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Courtney KD et al. Phase I Dose-Escalation Trial of PT2385, a First-in-Class Hypoxia-Inducible Factor-2alpha Antagonist in Patients With Previously Treated Advanced Clear Cell Renal Cell Carcinoma. J Clin Oncol 36, 867–874, doi: 10.1200/JCO.2017.74.2627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen W et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117, doi: 10.1038/nature19796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jonasch E et al. An open-label phase II study to evaluate PT2977 for the treatment of von Hippel-Lindau disease-associated renal cell carcinoma. Journal of Clinical Oncology 37, TPS680 (2019). [Google Scholar]
- 200.Jonasch E et al. Phase II study of the oral HIF-2α inhibitor MK-6482 for Von Hippel-Lindau disease–associated renal cell carcinoma. Journal of Clinical Oncology 38, 5003–5003, doi: 10.1200/JCO.2020.38.15_suppl.5003 (2020). [DOI] [Google Scholar]
- 201.LaFave LM et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 21, 1344–1349, doi: 10.1038/nm.3947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Schoumacher M et al. Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status. Nat Med 22, 577–578, doi: 10.1038/nm.4098 (2016). [DOI] [PubMed] [Google Scholar]
- 203.Pfister SX et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell 28, 557–568, doi: 10.1016/j.ccell.2015.09.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Terzo EA et al. SETD2 loss sensitizes cells to PI3Kbeta and AKT inhibition. Oncotarget 10, 647–659, doi: 10.18632/oncotarget.26567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Motzer RJ et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet 372, 449–456 (2008). [DOI] [PubMed] [Google Scholar]
- 206.Hudes G et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med 356, 2271–2281 (2007). [DOI] [PubMed] [Google Scholar]
- 207.Jonasch E et al. A randomized phase 2 study of MK-2206 versus everolimus in refractory renal cell carcinoma. Ann Oncol 28, 804–808, doi: 10.1093/annonc/mdw676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ibrahim YH et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov 2, 1036–1047, doi: 10.1158/2159-8290.CD-11-0348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Juvekar A et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov 2, 1048–1063, doi: 10.1158/2159-8290.CD-11-0336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Escudier B et al. A randomized, controlled, double-blind phase III study (AVOREN) of bevacizumab/interferon-α2a vs placebo/interferon- α2a as first-line therapy in metastatic renal cell carcinoma. Journal of Clinical Oncology 25, 3 (2007).17194900 [Google Scholar]
- 211.Rini BI et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol 26, 5422–5428 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rini BI et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet 378, 1931–1939, doi:S0140–6736(11)61613–9 [pii] 10.1016/S0140-6736(11)61613-9 (2012). [DOI] [PubMed] [Google Scholar]
- 213.Motzer RJ et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356, 115–124 (2007). [DOI] [PubMed] [Google Scholar]
- 214.Choueiri TK et al. Cabozantinib versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 373, 1814–1823, doi: 10.1056/NEJMoa1510016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Escudier B et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med 356, 125–134 (2007). [DOI] [PubMed] [Google Scholar]
- 216.Sternberg CN et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol 28, 1061–1068 (2010). [DOI] [PubMed] [Google Scholar]
- 217.Motzer RJ et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med 369, 722–731, doi: 10.1056/NEJMoa1303989 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Jonasch E et al. Phase II Study of Two Weeks on, One Week off Sunitinib Scheduling in Patients With Metastatic Renal Cell Carcinoma. J Clin Oncol 36, 1588–1593, doi: 10.1200/JCO.2017.77.1485 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jonasch E et al. Pilot trial of sunitinib therapy in patients with von Hippel-Lindau disease. Ann Oncol 22, 2661–2666, doi: 10.1093/annonc/mdr011mdr011 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kawakami F et al. Programmed cell death ligand 1 and tumor-infiltrating lymphocyte status in patients with renal cell carcinoma and sarcomatoid dedifferentiation. Cancer 123, 4823–4831, doi: 10.1002/cncr.30937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Joseph RW et al. PD-1 and PD-L1 Expression in Renal Cell Carcinoma with Sarcomatoid Differentiation. Cancer Immunol Res 3, 1303–1307, doi: 10.1158/2326-6066.CIR-15-0150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Raychaudhuri R et al. Immune Check Point Inhibition in Sarcomatoid Renal Cell Carcinoma: A New Treatment Paradigm. Clin Genitourin Cancer 15, e897–e901, doi: 10.1016/j.clgc.2017.05.018 (2017). [DOI] [PubMed] [Google Scholar]
- 223.Paolino M et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507, 508–512, doi: 10.1038/nature12998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Paolino M & Penninger JM The Role of TAM Family Receptors in Immune Cell Function: Implications for Cancer Therapy. Cancers (Basel) 8, doi: 10.3390/cancers8100097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Heemskerk B, Kvistborg P & Schumacher TN The cancer antigenome. EMBO J 32, 194–203, doi: 10.1038/emboj.2012.333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Le DT et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372, 2509–2520, doi: 10.1056/NEJMoa1500596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413, doi: 10.1126/science.aan6733 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Motzer RJ et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 373, 1803–1813, doi: 10.1056/NEJMoa1510665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Motzer RJ et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N Engl J Med 378, 1277–1290, doi: 10.1056/NEJMoa1712126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Woo SR, Corrales L & Gajewski TF The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol 36, 250–256, doi: 10.1016/j.it.2015.02.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]