Surface engineering of exosomes enhances the therapeutic efficacy of rheumatoid arthritis by macrophage reprogramming.
Abstract
Despite the remarkable advances in therapeutics for rheumatoid arthritis (RA), a large number of patients still lack effective countermeasures. Recently, the reprogramming of macrophages to an immunoregulatory phenotype has emerged as a promising therapeutic strategy for RA. Here, we report metabolically engineered exosomes that have been surface-modified for the targeted reprogramming of macrophages. Qualified exosomes were readily harvested from metabolically engineered stem cells by tangential flow filtration at a high yield while maintaining their innate immunomodulatory components. When systemically administered into mice with collagen-induced arthritis, these exosomes effectively accumulated in the inflamed joints, inducing a cascade of anti-inflammatory events via macrophage phenotype regulation. The level of therapeutic efficacy obtained with bare exosomes was achievable with the engineered exosomes of 10 times less dose. On the basis of the boosted nature to reprogram the synovial microenvironment, the engineered exosomes display considerable potential to be developed as a next-generation drug for RA.
INTRODUCTION
Rheumatoid arthritis (RA) is a debilitating and financially burdensome disease that affects up to 2.5% of the population in each country (1, 2). The RA therapeutic market worldwide was valued at US$23.82 billion in 2017 and is projected to reach US$33.96 billion by 2025 (3). The pathogenesis of RA involves the infiltration of inflammatory cells, leading to cartilage destruction and bone erosion (4). Of the cells involved in RA, M1 macrophages are known as the most prominent cells responsible for lesion formation and aggravation by releasing various types of proinflammatory cytokines such as tumor necrosis factor–α (TNF-α), interleukin-6 (IL-6), and IL-1 (5, 6). The relevant traditional therapeutic approaches have focused on suppressing the inflammation process, for instance, by inhibiting each of these cytokines or depleting M1 macrophages (7). Meanwhile, recovering the impaired resolution process in chronic inflammation by directly reversing dominant phenotype from M1 to M2 has recently gained much attention in the light of previously unidentified insights into macrophage plasticity and polarization (5). The newer approaches to fostering restorative environment have been envisioned to become therapeutic interventions closer to cure RA by ending the pathological process (8). Although mesenchymal stem cells (MSCs) have exhibited the therapeutic potential by modulating the macrophage phenotype in RA (9), their allogeneic transplantations have been limited by poor survival rates and immunogenicity in vivo (10, 11). Exosomes (EXOs), extracellular vesicles (40 to 160 nm in diameter) secreted by virtually all types of cells for intercellular communication (12), have recently emerged as cell-free therapeutic nanomedicines (13). In particular, owing to their anti-inflammatory effects, MSC-derived EXOs (MSC-EXOs) have potential to treat autoimmune diseases (14), including atopic dermatitis (15), diabetes (16), and Alzheimer’s disease (17). Also, they have shown potential to treat osteoarthritis by inducing M1-M2 macrophage polarization in the inflamed tissue (18). Although cancer-derived EXOs have the ability to affect the M1-M2 balance, they are not considered therapeutic nanomedicines due to their tumorigenicity in vivo (19, 20). Overall, for RA therapy, there is an unmet need to develop engineered MSC-EXOs, which can effectively regulate the M1-M2 balance of macrophages.
Clinical applications of EXOs have been limited because they are mostly accumulated in and removed by the liver and spleen (21, 22), resulting in short biological half-lives (<6 hours) in the body after systemic administration (23). In general, exosomal surfaces have been engineered based on genetic manipulation or chemical modification to surmount their poor in vivo distributions (24). However, the former is not applicable to the MSC-EXOs, and complicated purification steps of the latter have raised concerns about the low yield and exosomal dysfunction (24–26). Therefore, to develop safer and more effective RA therapeutics based on MSC-EXOs, an efficient surface-editing strategy that does not cost their intrinsic biological functions and/or yield is crucial. Here, we report the fine surface editing of MSC-EXOs by the metabolic glycoengineering (MGE) of adipose-derived stem cells (ADSCs) to target activated macrophages in the inflamed joints of RA (Fig. 1). These EXOs provide sounder footing as RA therapeutics by reproducing natural inflammation resolution in the lesion via M1-M2 polarization and not vice versa.
Fig. 1. Schematic illustration of DS-EXOs as a cell-free therapeutic system for RA.
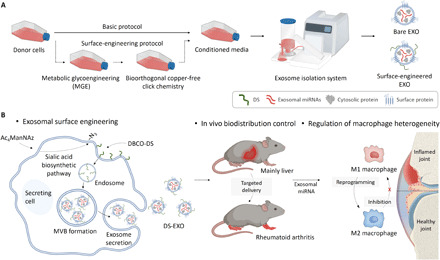
(A) The surface engineering process of MSC-EXOs by the MGE-mediated click chemistry of ADSCs. This methodology facilitated the introduction of a targeting moiety onto the MSC-EXOs without causing their dysfunction. (B) Mechanism by which the DS-EXOs reprogram macrophages. After systemic administration, the DS-EXOs reach joints inflamed by RA owing to their ability to target activated macrophages. Macrophage reprogramming in RA can be effectively promoted by the targeted delivery of the DS-EXOs. MVB, multivesicular bodies.
RESULTS
Optimization of metabolic engineering for the surface modification of MSC-EXOs
Conventional strategies to chemically modify the surface of EXOs require additional purification processes, such as ultracentrifugation (UC) and size-exclusion chromatography (SEC), which often induce exosomal dysfunction (fig. S1A). In this study, we isolated metabolically engineered EXOs using tangential flow filtration (TFF) from the conditioned medium without an additional purification step. Metabolically engineered EXOs did not exhibit substantial differences in their protein content and yield (fig. S1, B and C). Conventionally modified EXOs contained much lower amounts of proteins than bare EXOs and had poorer yields (fig. S1, D and E). When dispersed in phosphate-buffered saline (PBS) (pH 7.4), conventionally modified EXOs decreased rapidly in concentration, implying their poor stability (fig. S1F). In contrast, the concentration of metabolically engineered EXOs changed at a slower rate than that of bare EXOs. These results suggest that the MGE-based surface engineering of EXOs is a promising alternative to conventional modification approaches.
Biogenesis of metabolically engineered EXOs in ADSCs
To prepare activated macrophage-targeting MSC-EXOs, the surfaces of the ADSCs were first modified by MGE-mediated bioorthogonal copper-free click chemistry. Briefly, the azide group was exogenously introduced onto the surfaces of the ADSCs through the MGE-mediated sialic acid pathway at various concentrations of N-azidoacetylmannosamine-tetraacetylated (Ac4ManNAz) (27). Then, a dibenzocyclooctyne-conjugated dextran sulfate (DBCO-DS) was attached onto the azide-bearing ADSCs by click chemistry (fig. S2). DS, consisting of linear 1,6-linked glucopyranoses with sulfate groups, was chosen as the targeting moiety for macrophage scavenger receptor class A (SR-A) abundant at the inflamed joint of RA (28). As expected, the fluorescence signal of Cy5.5-labeled DBCO-DS on the ADSCs was enhanced in a dose-dependent manner with Ac4ManNAz (Fig. 2, A and B). Owing to their biocompatible natures, no significant toxicity was observed with up to 20 μM Ac4ManNAz and 10 μM DBCO-DS (fig. S3, A and B). These results suggest that MGE-mediated click chemistry is a safe and effective strategy for the surface modification of ADSCs.
Fig. 2. Biogenesis of DS-EXOs based on the MGE-mediated click chemistry of ADSCs.
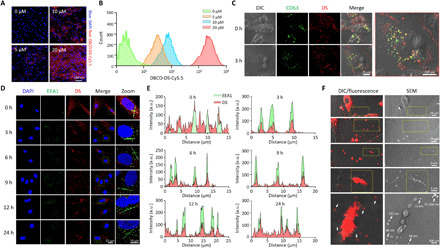
(A) Confocal microscopic images of DS-modified ADSCs at different concentrations of Ac4ManNAz. For the MGE-mediated click chemistry, ADSCs were incubated with Ac4ManNAz for 48 hours to generate azide groups on the surface and then treated with serum-free medium containing Cy5.5-labeled DBCO-DS (10 μM) for 2 hours. (B) Quantification of the DS on the ADSCs at different concentrations of Ac4ManNAz. Modified ADSCs were analyzed by flow cytometry to quantify the azide surface expression. (C) Intracellular tracking of DS in MDA-MB-231-CD63-GFP cells. The CD63-GFP cell line was used for MGE-mediated click chemistry to confirm the intracellular colocalization of CD63 and DS during biogenesis. Confocal microscopy images show CD63-GFP (green) and Cy5.5-labeled DS (red) in the MDA-MB-231-CD63-GFP cells. (D) Intracellular tracking of DS in the ADSCs. Time-dependent fluorescence images of the early endosomes and DS were obtained after the MGE-mediated click chemistry of ADSC. Confocal microscopy images show EEA1 (green) and Cy5.5-labeled DS (red) in the ADSCs. (E) Kinetic quantification of confocal imaging in (D). a.u., arbitrary units. (F) Secretion of the DS-decorated extracellular vesicles from the ADSCs. To visualize the extracellular secretion of the DS-decorated extracellular vesicles, the ADSCs were seeded onto a High Grid-500 glass-bottom μ-Dish. After the MGE-mediated click chemistry, images were separately obtained for the same spot using SEM and confocal microscopy. The dotted region in the top panel is magnified in the bottom panel.
The biogenesis of EXOs is associated with the invagination of endosomes to form multivesicular bodies, during which exosomal surface proteins (e.g., CD63 and CD9) are integrated (29). To observe the biogenesis of EXOs, CD63–green fluorescent protein (GFP)–expressing donor cells (MDA-MA-231 CD63-GFP) were treated with Cy5.5-labeled DS. After the surface modification of the donor cells based on MGE-mediated click chemistry, we confirmed the colocalization of Cy5.5-labeled DS and CD63-GFP in the cells using confocal microscopy (Fig. 2C and fig. S4). In addition, we evaluated the time-dependent localization of Cy5.5-labeled DS in the ADSCs using an endosome marker (Fig. 2, D and E). As expected, Cy5.5 fluorescence signals were initially observed in the ADSC membranes. After 3 hours of incubation, Cy5.5-labeled DS colocalized with EEA1, an early endosome marker, implying that DS was involved in the biogenesis of EXOs. We then used confocal microscopy and scanning electron microscopy (SEM) to track the DS-decorated extracellular vesicles secreted from the ADSCs; as shown in Fig. 2F, we gradually enlarged and observed the space between the cells. The fluorescence images of the DS-decorated extracellular vesicles corresponded with SEM images of particles approximately 137 nm in diameter. Overall, it was evident that the DS-decorated MSC-EXOs (DS-EXOs) were successfully secreted from the ADSCs surface-engineered by MGE-mediated click chemistry.
Characterization of DS-EXOs
To obtain a large amount of DS-EXOs without exosomal dysfunction, their isolation was carried out using TFF (Fig. 3A). TFF showed greater batch-to-batch consistency than conventional isolation approaches and provided a higher yield (30). Quantitatively, the number of DS-EXOs produced was comparable with that of bare EXOs, indicating that the MGE-mediated click chemistry did not affect the yield (Fig. 3B and table S1). Both the bare EXOs and DS-EXOs had unimodal size distributions with negatively charged surfaces (Fig. 3C and table S2). Owing to the presence of a hydrophilic DS shell, the DS-EXOs (136.3 ± 9.0 nm) were slightly larger than the bare EXOs (112.6 ± 4.3 nm). The surface of each EXO had 20,811 ± 4261 DS molecules, as confirmed by an enzyme-linked immunosorbent assay (ELISA). Both forms of EXOs were spherical in shape with strong fluorescence signals for their surface-marker proteins, including CD9 and CD63 (Fig. 3, D and E). The expression levels of CD9 and CD63 on the DS-EXOs were examined via flow cytometry and were comparable with those on the bare EXOs (Fig. 3F). Thus, surface engineering, based on MGE-mediated click chemistry, may not significantly affect the physicochemical characteristics of MSC-EXOs.
Fig. 3. Physicochemical characteristics of the DS-EXOs.
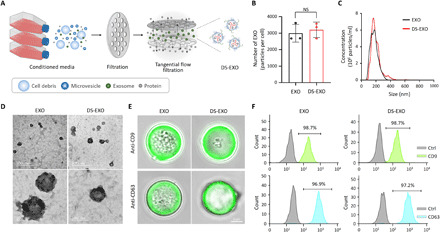
(A) Schematic illustration of the TFF-based isolation of the DS-EXOs. (B) Production yield of the bare EXOs and DS-EXOs. After surface engineering the ADSCs, the cells were incubated for 24 hours with FBS-free medium, and the DS-EXOs were isolated using TFF. The EXO number per cell was measured using an NTA. Error bars represent the SD (n = 3). NS, not significant. (C) Size distribution of the bare EXOs and DS-EXOs. The hydrodynamic size of the EXOs was measured using an NTA. (D) TEM images of the bare EXOs and DS-EXOs. Samples were dropped on a 200-mesh carbon film–coated grid and negatively stained with uranyl acetate. (E) Confocal microscopy images of exosomal surface markers on the bare EXOs and DS-EXOs. To analyze the exosomal markers, the EXOs were incubated with magnetic microbeads coated with antibody. Confocal microscopy images show anti-CD9 or anti-CD63 (green) and the microbeads. (F) Flow cytometry analysis of the exosomal surface markers on the bare EXOs and DS-EXOs.
Analysis of the microRNA expression profile in MSC-EXOs
EXOs are capable of affecting the gene expression and signaling pathways of recipient cells, principally by the transfer of microRNA (miRNA) (31). To determine whether the MGE-mediated click chemistry of the ADSCs affected their expression of exosomal miRNA, the total RNA purified from bare EXOs and DS-EXOs was used for small RNA sequencing (Fig. 4 and fig. S5A). Heatmap analysis of the top 50 significantly expressed miRNAs showed that there were no significant differences in miRNA expression between bare EXOs and DS-EXOs (Fig. 4A and table S3). Of the top four miRNAs, let-7b-5p and miR-24-3p, which are responsible for M1-M2 macrophage polarization (32), were expressed at a similar level in both bare EXOs and DS-EXOs. Next, the gene clusters and networks of the miRNA–target gene interactions were visualized using Cytoscape (Fig. 4B). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to investigate the biological processes, cellular components, molecular functions, and signaling pathways regulated by the top four miRNAs in the EXOs (Fig. 4C). GO analysis showed that the top four miRNAs are associated with “transcription” and “protein phosphorylation” in biological processes, “nucleus” and “nucleoplasm” in cellular components, and “protein binding” and “protein serine/threonine kinase activity” in molecular functions. In the KEGG analysis, the miRNAs were also found to be significantly involved in signaling pathways for M1-M2 macrophage polarization, including the “mitogen-activated protein kinase (MAPK) signaling pathway,” “neurotrophin signaling pathway,” “PI3K-Akt signaling pathway,” “Wnt signaling pathway,” “Jak-STAT signaling pathway,” and “AMPK signaling pathway” (Fig. 4C). Among a total of 278 miRNAs, only 2 had significantly different expression levels between the bare EXOs and DS-EXOs (Fig. 4D). The two down-regulated miRNAs (miR-186-5p and miR-660-5p) in the DS-EXOs are associated with the “extrinsic apoptotic signaling pathway” and “Ras signaling pathway,” which are not involved in M1-M2 macrophage polarization (Fig. 4E and fig. S7). Overall, these results suggest that the MGE-mediated click chemistry of the ADSCs did not affect exosomal miRNA expression and that MSC-EXOs contain miRNAs that promote M1-M2 macrophage polarization.
Fig. 4. miRNA profiling of the bare EXOs and DS-EXOs.
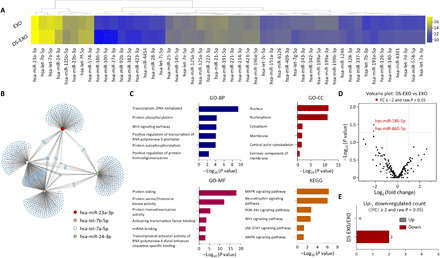
(A) Heatmap analysis of the top 50 significantly expressed miRNAs in the bare EXOs and DS-EXOs. Purified total RNA was used for the small RNA sequencing of the EXOs. (B) Schematic representation of the predicted target genes of the top four significantly expressed miRNAs enriched in the bare EXOs and DS-EXOs. The gene clusters (blue) and networks of the miRNAs were visualized using Cytoscape. (C) GO and KEGG pathway analyses of the target genes of the top four significantly expressed miRNAs in the bare EXOs and DS-EXOs. The GO terms and KEGG pathway terms enriched in the predicted target genes of the miRNAs were analyzed using DAVID Bioinformatics. (D) Volcano plot of the miRNAs in the bare EXOs and DS-EXOs. The red dots indicate the differentially expressed miRNAs between the bare EXOs and DS-EXOs. (E) Number of differentially up- and down-regulated miRNAs by fold change and P value. BP, biological processes; CC, cellular components; MF, molecular functions.
In vitro and in vivo targeted delivery of DS-EXOs
In this study, DS was chosen as the targeting moiety for surface engineering because it has a specific binding affinity for SR-A, which is overexpressed on activated macrophages (28) and is responsible for the pathogenesis of RA (fig. S7) (5). To verify the SR-A–mediated endocytosis of the DS-EXOs by the M1 macrophages, we observed the cellular uptake behavior via confocal microscopy (Fig. 5, A and B). The fluorescence signals were stronger in the cytosols of the Cy5.5-labeled DS-EXO–treated RAW264.7 cells stimulated with lipopolysaccharide and interferon gamma (LPS+ and IFN-γ+) and the bone marrow–derived macrophages (BMDMs; LPS+ and IFN-γ+) than in the Cy5.5-labeled bare EXOs at all the time points tested. Specifically, at 3 hours after incubation, the fluorescence intensities of the Cy5.5-labeled DS-EXOs in the LPS- and IFN-γ–stimulated RAW264.7 cells and BMDMs were 23.9- and 2.1-fold higher than those of the Cy5.5-labeled bare EXOs, respectively (fig. S8). Internalization of DS-EXOs into the RAW264.7 cells (LPS+ and IFN-γ+) was inhibited in the presence of free DS, implying that endocytosis of DS-EXOs was significantly dependent on interactions with SR-A (fig. S9). To maximize the in vivo therapeutic efficacy of MSC-EXOs, it is important to control their in vivo distribution after systemic administration. Therefore, we investigated the effects of surface engineering on the in vivo biodistribution of DS-EXOs after intravenous injection into mice with collagen-induced arthritis (CIA). The inflamed joints of the mice injected with the Cy5.5-labeled DS-EXOs showed stronger fluorescence signals than those of the mice treated with the Cy5.5-labeled bare EXOs (Fig. 5C). Specifically, at 1 hour after injection, the fluorescence intensity of the Cy5.5-labeled DS-EXOs was 1.52-fold higher than that of the Cy5.5-labeled bare EXOs (Fig. 5D). As expected, whole-body fluorescence imaging exhibited no significant differences in half-life between bare EXOs and DS-EXOs in CIA mice (Fig. 5, E and F). The enhanced fluorescence signal in the DS-EXO–treated joints remained visible for 24 hours (fig. S11, A and B), confirming the targeted delivery of the DS-EXOs to the inflamed joints. Ex vivo images of major organs and the inflamed joints, harvested at 24 hours after injection, showed that the number of Cy5.5-labeled DS-EXOs at the inflamed sites was significantly higher than that of Cy5.5-labeled bare EXOs (Fig. 5, G and H). We then used immunohistochemistry to examine the distribution of SR-A and DS-EXOs in the inflamed joints (fig. S11C). As expected, the fluorescence signal in the inflamed joints treated with Cy5.5-labeled DS-EXOs was stronger than that in the inflamed joints treated with Cy5.5-labeled bare EXOs. Colocalized fluorescence signals of SR-A and EXOs were detected only in the Cy5.5-labeled DS-EXO–treated joints 24 hours after injection (Fig. 5I). Thus, the enhanced RA targetability of the DS-EXOs might be attributed to the SR-A–mediated active targeting mechanism.
Fig. 5. Targeted delivery of DS-EXOs into the activated macrophages.
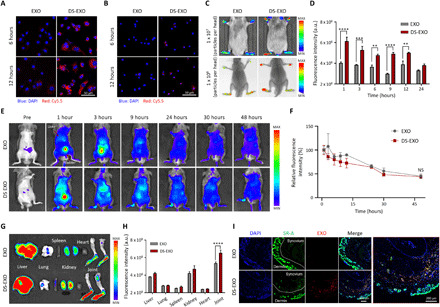
(A) Cellular uptake behavior of the Cy5.5-labeled bare EXOs and DS-EXOs in RAW264.7 cells (LPS+ and IFN-γ+). (B) Cellular uptake behaviors of the bare EXOs and DS-EXOs in BMDMs (LPS+ and IFN-γ+). Confocal microscopy images show nuclei (blue) and Cy5.5-labeled EXOs (red) in the cells (A and B). (C) In vivo fluorescence images of the Cy5.5-labeled bare EXOs and DS-EXOs in the CIA mice. The images were obtained using an IVIS Lumina III In Vivo Imaging System or SPECTRAL Lago X with an embedded x-ray. (D) Fluorescence intensity of the bare EXOs and DS-EXOs in the inflamed joints of the CIA mice as a function of time. Error bars represent the SE (n = 3). (E) Whole-body fluorescence imaging of bare EXOs and DS-EXOs in the CIA mice. After removing the hair, Cy5.5-EXOs or Cy5.5-DS-EXOs (1 × 108 particles per head) were systemically administered into the CIA mice. (F) Fluorescence intensity of bare EXOs and DS-EXOs in the whole body of the CIA mice as a function of time. Error bars represent the SE (n = 3). (G) Ex vivo organ distribution images of the bare EXOs and DS-EXOs. (H) Quantification of fluorescence intensity in the inflamed joints and major organs of the CIA mice. Error bars represent the SE (n = 3). (I) Distribution of SR-A and EXOs in the inflamed joints of the CIA mice. **P < 0.01, ***P < 0.005, and ****P < 0.001, analyzed by two-way analysis of variance (ANOVA).
In vitro macrophage polarization by DS-EXOs
To investigate the effects of the DS-EXOs on macrophage reprogramming, we examined the in vitro immunofluorescence staining images of RAW264.7 cells (Fig. 6A and fig. S12A). The DS-EXO–treated cells had much lower inducible nitric oxide synthase (iNOS) fluorescence signals (red) and much higher CD206 fluorescence signals (green) than the bare EXO-treated cells. The M1 macrophage marker in RAW264.7 cells (LPS+ and IFN-γ+) was also confirmed by flow cytometry analysis, suggesting that the expression of iNOS was much lower in the DS-EXO–treated cells than in the bare EXO-treated cells (Fig. 7, B and C). In addition, the analysis of the M2 macrophage marker in these cells indicated that the expression of CD206 was much higher in the DS-EXO–treated cells than in the bare EXO-treated cells (fig. S13). Next, to further evaluate the M1-M2 macrophage polarization caused by the DS-EXOs, we observed the in vitro immunofluorescence staining images of BMDMs (fig. S12B). As expected, the DS-EXO–treated BMDMs showed much lower iNOS fluorescence signals (red) and much higher CD206 fluorescence signals (green) than the bare EXO-treated BMDMs. These results clearly indicate that the DS-EXOs significantly improved the M1-M2 macrophage polarization efficacy in vitro. To investigate the effects of the DS-EXOs on inhibition of M2-M1 macrophage polarization, we observed the in vitro immunofluorescence staining images of RAW264.7 cells (Fig. 6D and fig. S14). To mimic the synovial environment of the inflamed joint, M2-polarized RAW264.7 cells were additionally treated with LPS and IFN-γ. It was encouraging that the DS-EXO–treated cells had much lower fluorescence signals of the M1 marker (iNOS, red) and much higher fluorescence signals of the M2 marker (CD206, green) than the bare EXO-treated cells. These results demonstrate that the DS-EXOs significantly inhibited the M2-M1 macrophage polarization in vitro.
Fig. 6. In vitro macrophage polarization by DS-EXOs.
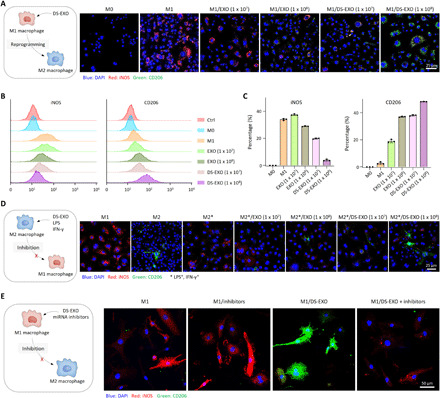
(A) Schematic illustration for M1-M2 macrophage polarization by DS-EXOs (left). The RAW264.7 cells (LPS+ and IFN-γ+) were incubated with the bare EXOs or DS-EXOs (1 × 107 or 1 × 108 particles per dish) at 37°C for 48 hours. Confocal microscopy images show iNOS (red) and CD206 (green) in cells (right). (B) Flow cytometry of the M1 and M2 macrophage markers in RAW264.7 cells (LPS+ and IFN-γ+). (C) Quantification of the expression levels of CD206 in EXO-treated RAW264.7 cells (LPS+ and IFN-γ+). Error bars represent the SD (n = 3). (D) Schematic illustration for inhibition of M2-M1 macrophage polarization by DS-EXOs (left). Confocal microscopy images show iNOS (red) and CD206 (green) in EXO-treated RAW264.7 cells (IL-4+) in the presence of LPS and IFN-γ (right). The cells were incubated with the bare EXOs or DS-EXOs (1 × 107 or 1 × 108 particles per dish) at 37°C for 48 hours. (E) Schematic illustration for inhibition of M2-M1 macrophage polarization by DS-EXOs (left). Confocal microscopy images show iNOS (red) and CD206 (green) in EXO-treated RAW264.7 cells (LPS+ and IFN-γ+) in the presence of let-7b-5p and miR-24-3p inhibitors (right).
Fig. 7. Therapeutic efficacy of the DS-EXOs.
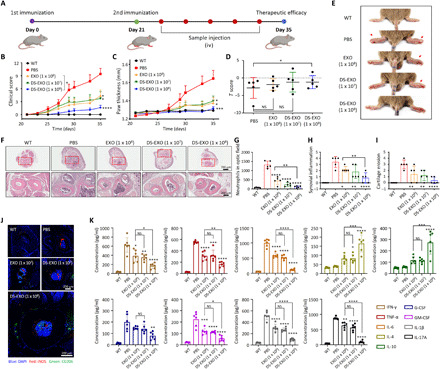
(A) Treatment regimen used for the bare EXOs and DS-EXOs. The red circles represent intravenous injections of the samples. iv, intravenous. (B and C) The severity of arthritis was determined via (B) a visual arthritis scoring system and (C) the paw thickness of the CIA mice. Error bars represent the SE (WT, n = 5; CIA, n = 10). (D) T scores of the inflamed joints in the CIA mice. On day 35 after the first immunization, the T scores of the joints were obtained. Error bars represent the SE (n = 5). (E) Representative images of the CIA mice treated with PBS, bare EXOs (1 × 108 particles per head), DS-EXOs (1 × 107 particles per head), and DS-EXOs (1 × 108 particles per head). (F) Hematoxylin and eosin–stained images of the inflamed joints in each treatment group. (G to I) Histological scores of neutrophil infiltration, synovial inflammation, and cartilage erosion. The histological scores were obtained from the images in (F). Error bars represent the SD (n = 5). (J) Regulation of macrophage heterogeneity by DS-EXOs. For immunohistochemistry, the joints were decalcified and the tissue sections were immunostained with anti-iNOS antibody Alexa Fluor 647 and anti-CD206 antibody Alexa Fluor 488. (K) Quantification of cytokines in the blood of the CIA mice. After 35 days of therapeutic monitoring, the collected blood samples were centrifuged to obtain serum. The cytokine levels in the serum samples were analyzed using ELISA. Error bars represent the SD (n = 6). *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001, analyzed by two-way ANOVA. Photo credit: Dong Gil You, Sungkyunkwan University.
According to the miRNA-sequencing data, let-7b-5p and miR-24-3p in EXOs accounted for 17% of the total miRNAs (fig. S5B). Such miRNAs are significantly associated with the “Wnt signaling pathway,” “MAPK signaling pathway,” “Hippo signaling pathway,” and “AMPK signaling pathway”, which are responsible for M1-M2 macrophage polarization (fig. S14). To verify the mechanism of M1-M2 macrophage polarization by DS-EXOs, we further obtained the in vitro immunofluorescence staining images of RAW264.7 cells by blocking the miRNAs (Fig. 6E and fig. S15). In the presence of let-7b-5p and miR-24-3p inhibitors, DS-EXO–treated cells had much higher iNOS fluorescence signals (red) and much lower CD206 fluorescence signals (green), implying that M1-M2 macrophage polarization was inhibited by blocking the miRNAs. These results suggest that the miRNAs of EXO are significantly involved in the M1-M2 macrophage polarization.
In vivo therapeutic efficacy of DS-EXOs in CIA mice
The therapeutic potential of the DS-EXOs was investigated after their systemic administration to CIA mice according to the treatment regimen presented in Fig. 7A. The clinical score and paw thickness of the inflamed joints were measured in the CIA mice for 5 weeks after the first immunization (Fig. 7, B and C); these values were found to have drastically increased in the PBS-treated group. However, the scores were lower over the entire duration of treatment in the groups treated with bare EXOs (1 × 108 particles per head) or DS-EXOs (1 × 107 particles per head) than in the control group. Although the concentration of the DS-EXOs (1 × 107 particles per head) was 10-fold lower than that of the bare EXOs (1 × 108 particles per head), they showed a similar level of therapeutic efficacy in the inflamed joints in RA. The group that received DS-EXOs at 1 × 108 particles per head had the lowest clinical scores and paw thickness. To further evaluate the therapeutic efficacy of the DS-EXOs, the T score, an indicator of bone density, was calculated for the inflamed joints in the CIA mice (Fig. 7D and fig. S16). On day 35 after the first immunization, the T scores of the PBS-, EXO (1 × 108 particles per head)–, DS-EXO (1 × 107 particles per head)–, and DS-EXO (1 × 108 particles/head)–treated CIA mice were −2.83, −1.85, −1.20, and −1.19, respectively. Only the DS-EXO (1× 108 particles per head)–treated CIA mice showed significantly lower bone erosion than the PBS-treated group. Overall, it was evident that the DS-EXOs exhibited higher therapeutic efficacy for RA than the bare EXOs, as judged by the clinical scores, paw thickness, T scores, and photographs of inflamed joints (Fig. 7, B to E). To further evaluate the therapeutic efficacy of the DS-EXOs, the joints of the CIA mice were histologically examined (Fig. 7, F to I). The mice joints in the PBS-treated group exhibited inflammatory cell infiltration, extensive synovitis and pannus formation, articular cartilage destruction, and bone erosion. However, comparatively, the mice joints in the bare EXO (1 × 108 particles per head)– and DS-EXO (1 × 107 particles per head)–treated groups showed significantly lower signs of inflammation. The mice joints in the DS-EXO (1 × 108 particles per head)–treated group had the lowest levels of cartilage erosion, neutrophil infiltration, and synovial inflammation; these levels were comparable with the wild-type (WT) levels. In addition, histological analysis of the major organs of the EXO-treated CIA mice revealed no signs of toxicity and no difference in the weight changes among the treatment groups (fig. S17). Overall, the DS-EXOs targeted to the inflamed joints showed promising therapeutic potential for RA.
Regulation of macrophage heterogeneity by DS-EXOs
Having established the feasibility of targeted delivery (Fig. 5), we next demonstrated that the systemic administration of DS-EXOs had a promising antirheumatoid effect in CIA mice (Fig. 7, B to I). Macrophages are the most dominant immune cells in the inflamed joints of RA (5), and clinically effective therapies reduce macrophage numbers in the inflamed joints (33). In particular, the polarization of M1 macrophages into the M2 phenotype, which contributes to tissue remodeling and repair through the release of anti-inflammatory cytokines (5, 34), would be a more effective treatment for RA than the total depletion of macrophages. To evaluate the effects of the DS-EXOs on macrophage heterogeneity in the CIA mice, we performed immunohistochemical analyses of the inflamed joints 5 weeks after the first immunization (Fig. 7J and fig. S18). Consistent with the therapeutic efficacy results, the DS-EXOs at 1 × 107 particles per head had an in vivo M1-M2 polarization efficacy similar to that of the bare EXOs at 1 × 108 particles per head. The CIA mice treated with 1 × 108 particles per head of DS-EXOs displayed outstanding in vivo M1-M2 polarization efficacy in their inflamed joints. Next, we evaluated the levels of proinflammatory and anti-inflammatory cytokines to confirm that the DS-EXOs drove M1-M2 macrophage polarization (Fig. 7K). Compared with the bare EXOs, the DS-EXOs targeting the activated macrophages reduced the level of IFN-γ and promoted macrophage polarization toward the M1 phenotype together with the secretion of TNF-α and IL-6. The CIA mice that received the DS-EXOs had sharply up-regulated levels of IL-4, which promotes macrophage polarization toward the M2 type, and IL-10, which is secreted by the M2 macrophages. In addition, the levels of granulocyte colony-stimulating factor (G-CSF) and granulocyte-macrophage colony-stimulating factor (GM-CSF), secreted by activated synovial fibroblasts, were significantly reduced by the DS-EXOs, implying the depletion of the M1 macrophages. DS-EXOs also down-regulated the levels of IL-1β, which is primarily secreted by activated synovial fibroblasts and activated macrophages, leading to reduced levels of IL-17A by the CD4+ T cells [T helper 17 (TH17) cells]. Overall, these results suggest that the DS-EXOs successfully reprogrammed the synovial microenvironment of the inflamed joints through the regulation of macrophage heterogeneity.
DISCUSSION
In recent years, EXOs have emerged as potential therapeutics owing to their bioactive ingredients such as mRNA, miRNA, and cytokines, which can modulate the biological activities of recipient cells (29, 35). Owing to their low immunogenicity, allogenic MSC-EXOs are available for repeated administration to patients without substantial side effects (36). The therapeutic potential of these EXOs is reflected in their application in an increasing number of clinical trials for the treatment of intractable diseases such as type I diabetes, acute ischemic stroke, and severe acute respiratory syndrome coronavirus 2–related pneumonia (ClinicalTrials.gov).
During the past three decades, MGE, a technique for adjusting cellular metabolism to modulate glycosylation, has been extensively investigated for tissue engineering, drug delivery, and immunotherapy (37). In particular, for cellular surface modification, MGE has been harnessed in combination with bioorthogonal copper-free click chemistry (27), allowing for specific chemical reactions on metabolically engineered cells (38). In this study, we prepared metabolically engineered MSC-EXOs based on MGE-mediated click chemistry using ADSCs (Fig. 1A). This approach produced a large amount of qualified DS-EXOs without exosomal dysfunction (Figs. 3B and 4A). In conventional techniques, bare EXOs isolated by TFF are used for surface engineering, thus requiring additional processes, such as UC and SEC, to remove unreacted chemicals (fig. S1). MGE-mediated click chemistry, however, allows the harvesting of the surface-engineered EXOs without additional purification steps. Notably, UC and SEC significantly reduced the protein level of the EXOs, implying that the structural integrity of conventionally surface-edited EXOs is impaired.
Although MGE-mediated click chemistry has been extensively investigated for cellular modification, there have been no reports on the biogenesis of metabolically engineered EXOs. After the MGE-mediated click chemistry of the ADSCs, DS was incorporated into the early endosomes and colocalized, suggesting that DS was effectively involved in the intrinsic biogenesis of the EXOs (Fig. 2D). In addition, the colocalization of DS and CD63-GFP in the cells further supports this hypothesis (Fig. 2C). Continued monitoring of the EXOs secreted from the metabolically engineered ADSCs demonstrated that they were released with enriched DS on their surfaces (Fig. 2F). Last, we obtained DS-EXOs to which 20,811 ± 4261 DS molecules per particle were attached; moreover, these EXOs had a stable nanostructure with a hydrodynamic size of 136.3 ± 9.0 nm.
A potential concern in using the MGE-mediated click chemistry approach to modify ADSCs may be the effect on the biological function of the ADSCs, resulting in the secretion of deformed EXOs. In the safety testing of the ADSCs, we found that DBCO-DS and Ac4ManNAz had no significant cytotoxicity (fig. S3). However, there is no previous information about the manner in which the MGE-mediated click chemistry approach affects exosomal cargos, such as mRNA, miRNA, and noncoding RNA. Although we introduced unnatural sialic acids containing an azide group onto the ADSC membranes via MGE with Ac4ManNAz, there was no significant difference in the expression levels of the top 50 miRNAs between the bare EXOs and DS-EXOs (Fig. 4A). Therefore, we demonstrated that the MGE-mediated click chemistry approach did not affect the intrinsic biological function of the surface-engineered MSC-EXOs. In addition, the top four significantly expressed miRNAs in the bare EXOs and DS-EXOs were strongly associated with signaling pathways that regulate macrophage heterogeneity (Fig. 4C). In particular, let-7b-5p and miR-24-3p, accounting for 17% of the total miRNAs in EXOs, are potent miRNAs for M1-M2 macrophage polarization (39), which was also demonstrated by KEGG analysis of their target genes (fig. S14). let-7b-5p and miR-24-3p are associated with the JAK-STAT signaling pathway to induce M2 macrophage polarization (40, 41). In the presence of the let-7b-5p and miR-24-3p inhibitors, M1-M2 macrophage polarization of DS-EXO–treated RAW264.7 cells (LPS+ and IFN-γ+) was inhibited (Fig. 6E). Overall, these results indicated that let-7b-5p and miR-24-3p of the EXOs were responsible for M1-M2 macrophage polarization via the JAK-STAT signaling pathway.
Recently, MSC-EXOs have been assessed as a potential antirheumatic medicine (42) but have not yet been tested in clinical trials (ClinicalTrials.gov); their minimal therapeutic efficacy has been primarily ascribed to their weak in vivo biodistribution (21–23). DS-EXO of this study, containing 20,811 ± 4261 DS molecules on the surface, effectively delivered therapeutic cargos (Fig. 5 and fig. S10), which are responsible for the initiation of M1-M2 macrophage polarization to reprogram the synovial microenvironment of joints inflamed by RA (Fig. 7J). This indicates that MGE-mediated click chemistry allowed for MSCs to produce surface-engineered EXOs with a sufficient amount of DS, leading to improved in vivo targetability. In addition to macrophage reprogramming, DS-EXOs exhibited the inhibitory effects on M2-M1 macrophage polarization in the RA-mimicking microenvironment (Fig. 6D). In the inflamed joints of patients with RA, proinflammatory cytokines, such as TNF-α and IL-6, are abundant, owing to the presence of M1 macrophages (5), whereas anti-inflammatory cytokines, including IL-4 and IL-10, are diminished owing to the lack of M2 macrophages (5, 43). A recent study found that, unlike the M1 macrophages, which actively contribute to inflammation, barrier-forming CX3CR1+ macrophages play a structural role and restrict the infiltration of activated immune cells (44). The barrier-forming M2 macrophages maintain their numbers through a pool of locally proliferating CX3CR1− mononuclear cells that are embedded in the synovial tissue (44). Thus, blockading and reprogramming the proinflammatory macrophages could suppress immune responses by manipulating the synovial microenvironment in the initial stages of RA, unlike currently used RA medications that do not take effect until much later in the pathogenic process. In addition, we found that the systemic administration of DS-EXOs promoted M1-M2 macrophage polarization in the bone marrow regions of joints inflamed by RA and reduced the activity of surrounding proinflammatory cells, such as M1 macrophages, activated synovial fibroblasts, and TH17 cells (Fig. 7, J and K). These findings suggest that the DS-EXOs have a strong polarization effect toward the anti-inflammatory macrophage phenotype that could allow them to be developed as a fundamental RA therapy and an alternative to TNF-α inhibitors.
In summary, to enhance targeting ability to inflamed joints, we developed DS-EXOs using the MGE-mediated click chemistry of ADSCs. This novel surface engineering strategy could be used to introduce a broad range of targeting moieties onto MSC-EXOs without causing dysfunction to their structure or functional cargos. The targeted delivery of MSC-EXOs, obtained from metabolically engineered stem cells, offers a new cell-free therapeutic system for RA, and the resulting regulation of macrophage heterogeneity can be extended to various macrophage-involving diseases, such as inflammatory bowel disease, idiopathic pulmonary fibrosis, atherosclerosis, and Alzheimer’s disease. Beyond stem cell–free therapeutics, the findings of this study have broad implications for the treatment of intractable diseases using various types of therapeutic cell–derived EXOs.
MATERIALS AND METHODS
Materials
DS sodium salt (40 kDa), thiazolyl blue tetrazolium bromide (MTT), dimethylformamide, hsa-let-7b-5p inhibitor, and hsa-miR-24-3p inhibitor were purchased from Sigma-Aldrich Co. (MO, USA). DBCO-amine and Ac4ManNAz were obtained from Click Chemistry Tools (AZ, USA), whereas sodium cyanoborohydride (NaBH3CN) was purchased from Tokyo Chemical Industry (TCI, Tokyo, Japan). Primary human ADSCs (age 38, female, 70E21-062) were obtained from Cefobio Inc. (Seoul, Korea). Water was purified using an AquaMax-Ultra water purification system from Younglin Co. (Anyang, Korea). All other chemicals were of analytical grade and used without further purification. Spectra/Por membrane was obtained from Spectrum Laboratories Inc. (CA, USA). Fetal bovine serum (FBS) and Dulbecco’s modified Eagle’s medium (DMEM) were obtained from Hyclone Laboratories Inc. (IL, USA). Dulbecco’s PBS (DPBS), antibiotic-antimycotic (AA) solution, and trypsin-EDTA were purchased from WelGENE (Gyeongsan, Korea).
Synthesis of DBCO-DS
DS (1000 mg, 0.025 mmol) and DBCO-amine (691 mg, 2.5 mmol) were dissolved in 50 ml of acetate buffer (pH 5.6). Then, NaBH3CN (157.1 mg and 2.5 mmol) was added, and the reaction mixture was stirred at 50°C for 5 days. The resulting solution was sequentially dialyzed against distilled water/methanol (1v/3v–1v/1v) for 2 days and against distilled water for another 2 days using a dialysis tube (molecular weight cut off = 3.5 kDa, Spectrum Laboratories Inc., CA, USA), followed by lyophilization. The chemical structure of the DBCO-DS was confirmed by 1H NMR analysis (500 MHz, Varian Unity INOVA, CA, USA), for which the samples were dissolved in D2O/DMSO-d6 (1v/1v) (fig. S19).
MGE of ADSCs
ADSCs were seeded at 1 × 105 and 1 × 106 cells per dish for confocal imaging and flow cytometry analysis, respectively. The cells were then incubated with various concentrations of Ac4ManNAz (0 to 20 μM) to generate azide groups on the surface through the sialic acid pathway. After 48 hours of incubation, the cells were washed twice with DPBS and treated with serum-free medium containing Cy5.5-labeled DBCO-DS (10 μM) for 2 hours. After washing the cells twice with DPBS, the cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were then observed using a confocal laser microscope (TCS SP8 HyVolution, Leica Microsystems CMS GmbH, Germany) with 405 diode (405 nm), Ar (458, 488, and 514 nm), and He-Ne (633 nm) lasers. To quantify the azide expression on the cell surface, the cells were detached from the dishes and analyzed via flow cytometry (BD FACSAria Fusion, USA) in the BIORP of Korea Basic Science Institute (KBSI).
Intracellular tracking of DBCO-DS in MDA-MB-231-CD63-GFP cells
To confirm the intracellular colocalization of CD63 and DS during EXO genesis, MDA–MB-231 cells were transfected with pCT-CD63-GFP (pCMV, Exosome/Secretory, CD63 Tetraspanin tag) plasmid (System Bioscience) to construct a stable MDA-MB-231-CD63-GFP cell line (45). CD63-GFP–transfected MDA–MB-231 cells were seeded at a density of 1 × 105 cells onto a confocal dish with RPMI 1640 medium containing 10% FBS, 1% AA, and 20 μM Ac4ManNAz for 48 hours. Subsequently, the cells were washed twice with DPBS and treated with serum-free medium containing Cy5.5-DS-DBCO (10 μM) for 2 hours. After washing twice with DPBS, the cells were incubated with serum-free medium for up to 24 hours, fixed with a 4% paraformaldehyde solution, and then observed under a confocal laser microscope.
Intracellular tracking of DBCO-DS in ADSCs
To confirm intracellular colocalization between the early endosomes and DS during the biogenesis of the EXOs, ADSCs were seeded onto a confocal dish at a density of 2 × 105 cells and incubated with DMEM containing 10% FBS, 1% AA, and 20 μM Ac4ManNAz for 48 hours. Subsequently, the cells were washed twice with DPBS and treated with serum-free medium containing Cy5.5-labeled DBCO-DS (10 μM) for 2 hours. After washing twice with DPBS, the cells were incubated with serum-free medium for up to 24 hours and fixed with a 4% paraformaldehyde solution. The cells were then incubated with 0.2% Triton X-100 in DPBS for 10 min and washed twice with DPBS. After blocking with 1% bovine serum albumin (BSA) in DPBS for 45 min, the cells were incubated with anti-EEA1 Alexa Fluor 594 antibody at 4°C for 12 hours. After washing the cells twice with DPBS, the cell nuclei were stained with DAPI to obtain confocal microscopic images.
Visualization of DS-decorated extracellular vesicle secretion
To visualize the extracellular secretion of the DS-decorated extracellular vesicles, ADSCs were seeded onto a 0.1% gelatin-coated High Grid-500 glass-bottom μ-Dish (ibidi, Gräfelfing, Germany) at a density of 5 × 104 cells with DMEM containing 10% FBS, 1% AA, and 20 μM Ac4ManNAz for 48 hours. Subsequently, the cells were washed twice with DPBS and treated with serum-free medium containing Cy5.5-labeled DBCO-DS (10 μM) for 2 hours. After washing twice with DPBS, the cells were incubated with serum-free medium for 24 hours and fixed with a 4% paraformaldehyde solution. Then, the cells were observed under a confocal laser microscope. For SEM imaging, the cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at 25°C for 1 hour and then maintained at 4°C for 12 hours. The cells were washed with 0.1 M cacodylate buffer, incubated with 1% osmium tetroxide at 25°C for 1 hour, and then washed with 0.1 M cacodylate buffer. Thereafter, the cells were dehydrated with serial concentrations of ethanol (50, 75, 95, and 100%), and the glass bottoms were detached from the dishes; this was followed by drying at 25°C for 24 hours. The dried coverslips were then mounted onto an aluminum stub with a carbon adhesive tab and sputter-coated with gold for 60 s. The samples were imaged using a JSM-7500F Field Emission SEM (JEOL, Tokyo, Japan).
Preparation of DS-EXOs
Bare EXOs and DS-EXOs were isolated by TFF. In brief, ADSCs were seeded in a T-175 flask at a density of 1 × 106 cells with DMEM containing 10% FBS and 1% AA at 37°C in a humidified 5% CO2 atmosphere and allowed to grow for 3 days. To generate azide groups on the cells, 20 μM Ac4ManNAz was added to the cell medium and incubated for 48 hours. The cells were washed twice with DPBS (pH 7.4) and incubated with 10 μM DBCO-DS for an additional 2 hours. Then, the culture medium was changed to FBS-free DMEM. After 24-hour incubation, the supernatant was collected by centrifugation at 2000g for 20 min, followed by filtration through 0.22-μm filters to remove cell debris from the medium. The filtered supernatant was sequentially concentrated using TFF with an ultrafiltration membrane filter capsule (MWCO = 300 kDa, Pall Corporation, Port Washington, NY, USA) to obtain the DS-EXOs. The DS-EXOs were kept in circulation as the retentate and concentrated in a 50-ml disposable tube to a final volume of approximately 10 ml. Bare EXOs were isolated in an identical manner using ADSCs that had not been subjected to click chemistry and MGE treatment. The bare EXOs and DS-EXOs were stored at −70°C until use. For the in vitro cell and in vivo animal experiments, the bare EXOs and DS-EXOs were labeled with Cy5.5 NHS ester dye. Bare EXOs and DS-EXOs (5 × 109) were mixed with 100 μg of Cy5.5 NHS ester dye and incubated at 4°C for 12 hours. Excess dye was removed using a PD-10 desalting column.
Characterization of DS-EXOs
The size distribution and particular concentration of the bare EXOs and DS-EXOs were measured using a nanoparticle tracking analyzer (NTA; NanoSight LM10, Malvern Instruments, Malvern, England). To evaluate the amount of DS on the exosomal surface, an ELISA was performed using a Dextran Sulfate ELISA Kit (Lifespan, UT, USA). The protein content of a single EXO was first analyzed using a Micro BCA protein assay and NTA. Then, 40 μg of DS-EXOs was dissolved in 50 μl of deionized water; the resulting solution was sonicated for 3 min to break the exosomal membrane. Afterward, ELISA proceeded according to the manufacturer’s instructions, and the absorbance at 450 nm was measured using a microplate reader (Synergy HT, BioTek Instruments, VT, USA). The number of DS on the exosomal surface was estimated using the following equation
The morphologies of bare EXOs and DS-EXOs were observed by transmission electron microscopy (TEM) (JEOL-2100F, Tokyo, Japan) at an accelerating voltage of 200 kV. For TEM sample preparation, the bare EXOs and DS-EXOs were dropped on a 200-mesh carbon-film–coated grid, negatively stained with 2% uranyl acetate for 1 min, and then washed with deionized water. To analyze the exosomal markers, bare EXOs and DS-EXOs were incubated and gently mixed with magnetic microbeads coated with antibody (ExoStep, Immunostep, Salamanca, Spain) at 25°C for 12 hours. The bead-bound EXOs were washed in buffer and placed on a magnet for 5 min before the supernatant was discarded. The bead-bound EXOs were resuspended in buffer and incubated with anti-CD9 antibody or anti-CD63 antibody at 4°C for 1 hour. After washing thrice with buffer, the antibody-attached EXOs were incubated with Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG) secondary antibody (Invitrogen, CA, USA) and washed with buffer. The fluorescent-labeled bead-bound EXOs were analyzed using a flow cytometer (Guava easyCyte, EMD Millipore, MA, USA).
Small RNA sequencing and data analysis
RNA extraction from the EXOs, library preparation, cluster generation, and sequencing were performed by Macrogen, Inc. (Seoul, Korea). Following procedures previously described in a published article (16), exosomal RNA was extracted using the Maxwell RSC miRNA Kit (Promega, WI, USA). RNA (3.672 ng/μl) from the EXOs was polyadenylated to obtain a priming sequence for an oligo (dT) primer; then, Illumina adapters were added. Complementary DNA was synthesized, followed by polymerase chain reaction amplification. Libraries for small RNA were constructed using a SMARTer smRNA-Seq Kit for Illumina (Takara Bio, Shiga, Japan). Exosomal RNA was integrated using an Agilent Technologies 2100 Bioanalyzer (Santa Clara, CA, USA). The libraries were sequenced on an Illumina HiSeq 2500 instrument (Illumina, CA, USA). Exosomal miRNA target genes were predicted using the public algorithms: miRwalk online prediction software (http://mirwalk.umm.uni-heidelberg.de/) and TargetScan 7.2 (www.targetscan.org). The GO terms and KEGG pathway terms enriched in the predicted target genes were analyzed using DAVID Bioinformatics.
Therapeutic efficacy of DS-EXO
All experiments involving live animals complied with the relevant laws and institutional guidelines of Sungkyunkwan University, and Institutional Committees approved all the experiments. The CIA model mice were prepared according to the following steps (28). Six-week-old male DBA1/J mice (Central Lab Animal Inc., Seoul, Korea) were injected intradermally at the tail with 200 μg of bovine type II collagen (2 mg/ml, Chondrex, Redmond, WA, USA) emulsified in 100 μl of Complete Freund’s Adjuvant (4 mg/ml) (46). On day 21, the mice received a booster immunization of type II collagen emulsified in Incomplete Freund’s Adjuvant. To evaluate the therapeutic efficacy of the DS-EXOs, the mice were divided into five groups. The CIA mice were administered 200 μl of PBS, bare EXO (1 × 108 particles per head), DS-EXO (1 × 107 particles per head), or DS-EXO (1 × 108 particles per head) five times (once every 3 days) by intravenous injection, starting on the day of the booster injection. The doses of EXOs were selected by considering the in vivo therapeutic potential of MSC-EXOs as reported previously (47, 48). Following procedures previously described in a published article (28), the mice were monitored every other day to measure joint inflammation, which was scored as follows: 0, normal; 1, mild swelling and erythema confined to the midfoot and ankle joint; 2, mild swelling and erythema extending from the midfoot and ankle joint; 3, moderate swelling and erythema extending from the metatarsal joints to the ankle; and 4, severe swelling and erythema encompassing the foot, ankle, and digits. The paw scores were summed for each mouse, giving a maximum possible score of 16. Paw thickness for both the right and left paws was determined using calipers. Bone density of the joints was measured using an energy x-ray absorptiometry (Lunar PIXImus DEXA, GE Medical Systems, Mississauga, ON, Canada).
For the histological analysis, the joints were decalcified for 48 hours using a 5% (v/v) formic acid solution and fixed with a 4% (v/v) buffered formalin solution. The paraffin sections of each tissue were then sliced (3 μm in thickness), stained with hematoxylin and eosin, and analyzed using a slide scanner (Aperio ScanScope CS, Leica Biosystems, Germany).
In vitro macrophage polarization
For the in vitro macrophage polarization studies, RAW264.7 cells and BMDMs were separately seeded in 35-mm confocal dishes at a density of 1 × 105 cells in RPMI 1640 medium [10% FBS, 1% AA, LPS (500 ng/ml), and IFN-γ (20 ng/ml)] at 37°C in a humidified 5% CO2 atmosphere for 24 hours. These cells were then incubated with bare EXOs or DS-EXOs (1 × 107 or 1 × 108 particles per dish) at 37°C for 48 hours. Subsequently, the cells were washed twice with DPBS and fixed with a 4% paraformaldehyde solution. The cells were then incubated with 0.2% Triton X-100 in DPBS for 10 min, washed twice with DPBS, and blocked with 1% BSA for 45 min. The cells were treated with anti-iNOS antibody Alexa Fluor 647 and anti-CD206 antibody Alexa Fluor 488 at 4°C for 12 hours. To confirm the inhibitory ability of M2-M1 polarization, RAW264.7 cells were seeded in 35-nm confocal dishes at a density of 1 × 105 cells in RPMI 1640 medium [10% FBS, 1% AA, and IL-4 (20 ng/ml)] at 37°C in a humidified 5% CO2 atmosphere for 24 hours. These cells were then incubated with bare EXOs or DS-EXOs (1 × 107 or 1 × 108 particles per dish) in the presence of LPS (500 ng/ml) and IFN-γ (20 ng/ml) at 37°C for 48 hours. Cell fixation and immunofluorescence staining were performed in an identical manner, as described earlier. To verify the effects of miRNA inhibitors on M1-M2 polarization, RAW264.7 cells (LPS+ and IFN-γ+) were incubated with bare EXOs or DS-EXOs (1 × 108 particles per dish) in the presence or absence of the let-7b-5p and miR-24-3p inhibitors (100 nM for each) at 37°C for 48 hours. Cell fixation and immunofluorescence staining were performed in an identical manner, as described earlier. The resulting cells were stained with DAPI and observed under a confocal laser microscope.
To confirm macrophage polarization using flow cytometry, RAW264.7 cells were seeded in a 100-pi dish at a density of 1 × 106 cells with RPMI 1640 medium containing 10% FBS, 1% AA, LPS (500 ng/ml), and IFN-γ (20 ng/ml) at 37°C in a humidified 5% CO2 atmosphere. After 24 hours, these cells were incubated in medium containing EXOs or DS-EXOs (1 × 107 or 1 × 108 particles per dish) at 37°C for 48 hours. Subsequently, the cells were gathered in a tube after being detached from the dishes by repetitive pipetting. The cells were washed thrice with DPBS and fixed with a 4% paraformaldehyde solution. The cells then incubated with 0.2% Triton X-100 in DPBS for 20 min and washed thrice with DPBS. After blocking with 1% BSA in DPBS for 45 min, the cells were treated with anti-iNOS antibody-PE or anti-CD206 antibody Alexa Fluor 488 at 4°C for 30 min. The control samples were treated with rat IgG2a κ isotype antibody-PE or rat IgG2a κ isotype antibody Alexa Fluor 488. Thereafter, the cells were washed with buffer and analyzed using a flow cytometer (Guava easyCyte, EMD Millipore, MA, USA).
Macrophage polarization in CIA mice
For immunohistochemistry, the joints were decalcified for 48 hours using a 5% (v/v) formic acid solution and fixed with a 4% (v/v) buffered formalin solution. The paraffin sections of each tissue were then sliced (3 μm in thickness) and immunostained with anti-iNOS antibody Alexa Fluor 647 and anti-CD206 antibody Alexa Fluor 488. Fluorescence images were obtained using a confocal laser microscope.
Cytokine analysis in CIA mice
To investigate the cytokine levels in blood, WT and CIA mice were prepared as described above. After 35 days of therapeutic monitoring, the collected blood samples were centrifuged to obtain serum. The serum samples were then analyzed using Mouse Inflammatory Cytokines Multi-Analyte ELISArray kits (Qiagen, Germany) according to the manufacturer’s instructions. The absorbance (450 nm) was measured using a microplate reader.
Statistical analysis
All values are expressed as the mean ± SEM or the mean ± SD. Statistically significant differences among the groups were analyzed using one-way or two-way analysis of variance (ANOVA). Differences with P < 0.05 were considered statistically significant. Statistical significance was assigned as *P < 0.05, **P < 0.01, ***P < 0.005, and ****P 0.001.
Acknowledgments
Funding: This research was supported by the Basic Science Research Programs (2018R1A2B3006080 and 2019R1I1A1A01063750) of the National Research Foundation (NRF), the Korea Basic Science Institute (National Research Facilities and Equipment Center, 2020R1A6C101A191), and a grant from the Ministry of Education, and Korea Health Technology R&D Project (HI20C0437) of the Ministry of Health & Welfare, Republic of Korea. Author contributions: D.G.Y. and J.H.P. designed the research and conducted the data analysis. D.G.Y., S.K., and J.H.P. wrote the manuscript. D.G.Y. and G.T.L. performed most of the experiments. W.U. and B.H.O. performed the in vivo biodistribution study. S.H.S. performed the in vitro cell study. J.L. prepared the materials. D.-G.J. and Y.W.C. conducted the data analysis. Competing interests: Y.W.C. is the chief executive officer of ExoStemTech Inc. D.-G.J. and J.H.P. are stockholders of ExoStemTech Inc. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabe0083/DC1
REFERENCES AND NOTES
- 1.Olumuyiwa-Akeredolu O.-o., Page M. J., Soma P., Pretorius E., Platelets: Emerging facilitators of cellular crosstalk in rheumatoid arthritis. Nat. Rev. Rheumatol. 15, 237–248 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Smolen J. S., Steiner G., Therapeutic strategies for rheumatoid arthritis. Nat. Rev. Drug Discov. 2, 473–488 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Fortune Business Insights (2019); www.fortunebusinessinsights.com/industry-reports/rheumatoid-arthritis-therapeutics-market-100488.
- 4.Smolen J. S., Aletaha D., Barton A., Burmester G. R., Emery P., Firestein G. S., Kavanaugh A., McInnes I. B., Solomon D. H., Strand V., Yamamoto K., Rheumatoid arthritis. Nat. Rev. Dis. Primers. 4, 18001 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Udalova I. A., Mantovani A., Feldmann M., Macrophage heterogeneity in the context of rheumatoid arthritis. Nat. Rev. Rheumatol. 12, 472–485 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M., The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Cutolo M., Sulli A., Pizzorni C., Seriolo B., Straub R., Anti-inflammatory mechanisms of methotrexate in rheumatoid arthritis. Ann. Rheum. Dis. 60, 729–735 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schett G., Neurath M. F., Resolution of chronic inflammatory disease: Universal and tissue-specific concepts. Nat. Commun. 9, 3261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin T.-H., Kim H.-S., Kang T.-W., Lee B.-C., Lee H.-Y., Kim Y.-J., Shin J.-H., Seo Y., Choi S. W., Lee S., Shin K., Seo K.-W., Kang K.-S., Human umbilical cord blood-stem cells direct macrophage polarization and block inflammasome activation to alleviate rheumatoid arthritis. Cell Death Dis. 7, e2524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freyman T., Polin G., Osman H., Crary J., Lu M.-M., Cheng L., Palasis M., Wilensky R. L., A quantitative, randomized study evaluating three methods of mesenchymal stem cell delivery following myocardial infarction. Eur. Heart J. 27, 1114–1122 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Nussbaum J., Minami E., Laflamme M. A., Virag J. A. I., Ware C. B., Masino A., Muskheli V., Pabon L., Reinecke H., Murry C. E., Transplantation of undifferentiated murine embryonic stem cells in the heart: Teratoma formation and immune response. FASEB J. 21, 1345–1357 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R., LeBleu V. S., The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andaloussi S. E. L., Mäger I., Breakefield X. O., Wood M. J. A., Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Chaput N., Théry C., Exosomes: Immune properties and potential clinical implementations. Semin. Immunopathol. 33, 419–440 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Fang Y., Zhang Y., Zhou J., Cao K., Adipose-derived mesenchymal stem cell exosomes: A novel pathway for tissues repair. Cell Tissue Bank. 20, 153–161 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Jung Y. J., Kim H. K., Cho Y., Choi J. S., Woo C. H., Lee K. S., Sul J. H., Lee C. M., Han J., Park J. H., Jo D.-G., Cho Y. W., Cell reprogramming using extracellular vesicles from differentiating stem cells into white/beige adipocytes. Sci. Adv. 6, eaay6721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Godoy M. A., Saraiva L. M., de Carvalho L. R. P., Vasconcelos-Dos-Santos A., Beiral H. J. V., Ramos A. B., de Paula Silva L. R., Leal R. B., Monteiro V. H. S., Braga C. V., de Araujo-Silva C. A., Sinis L. C., Bodart-Santos V., Kasai-Brunswick T. H., de Lima Alcantara C., Lima A. P. C. A., da Cunha-E Silva N. L., Galina A., Vieyra A., De Felice F. G., Mendez-Otero R., Ferreira S. T., Mesenchymal stem cells and cell-derived extracellular vesicles protect hippocampal neurons from oxidative stress and synapse damage induced by amyloid-β oligomers. J. Biol. Chem. 293, 1957–1975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosenza S., Ruiz M., Toupet K., Jorgensen C., Noël D., Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci. Rep. 7, 16214–16212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J. E., Dutta B., Tse S. W., Gupta N., Tan C. F., Low J. K., Yeoh K. W., Kon O. L., Tam J. P., Sze S. K., Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 38, 5158–5173 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Al-Sowayan B. S., Al-Shareeda A. T., Alrfaei B. M., Cancer stem cell-exosomes, unexposed player in tumorigenicity. Front. Pharmacol. 11, 384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiklander O. P. B., Nordin J. Z., O’Loughlkin A., Gustafsson Y., Corso G., Mäger I., Vader P., Lee Y., Sork H., Seow Y., Heldring N., Alvarez-Erviti L., Smith C. E., Blanc K. L., Macchiarini P., Jungebluth P., Wood M. J. A., Andaloussi S. E. L., Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 4, 26316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth T., Kullberg M., Malik N., Smith-Jones P., Graner M. W., Anchordoquy T. J., Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 199, 145–155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai C. P., Mardini O., Ericsson M., Prabhakar S., Maguire C. A., Chen J. W., Tannous B. A., Breakefield X. O., Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 8, 483–494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson J. J., Ejima H., Surface engineering of extracellular vesicles through chemical and biological strategies. Chem. Mater. 31, 2191–2201 (2019). [Google Scholar]
- 25.Ziaei P., Berkman C. E., Norton M. G., Review: Isolation and detection of tumor-derived extracellular vesicles. ACS Appl. Nano Mater. 1, 2004–2020 (2018). [Google Scholar]
- 26.Armstrong J. P., Holme M. N., Stevens M. M., Re-engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano 11, 69–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon H. Y., Koo H., Kim K., Kwon I. C., Molecular imaging based on metabolic glycoengineering and bioorthogonal click chemistry. Biomaterials 132, 28–36 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Heo R., You D. G., Um W., Choi K. Y., Jeon S., Park J.-S., Choi Y., Kwon S., Kim K., Kwon I. C., Jo D.-G., Kang Y. M., Park J. H., Dextran sulfate nanoparticles as a theranostic nanomedicine for rheumatoid arthritis. Biomaterials 131, 15–26 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Mathieu M., Martin-Jaular L., Lavieu G., Théry C., Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Busatto S., Vilanilam G., Ticer T., Lin W.-L., Dickson D. W., Shapiro S., Bergese P., Wolfram J., Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells 7, 273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O., Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Xu S. J., Hu H. T., Li H. L., Chang S., The role of miRNAs in immune cell development, immune cell activation, and tumor immunity: With a focus on macrophages and natural killer cells. Cells 8, 1140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haringman J. J., Gerlag D. M., Zwinderman A. H., Smeets T. J. M., Kraan M. C., Baeten D., McInnes I. B., Bresnihan B., Tak P. P., Synovial tissue macrophages: A sensitive biomarker for response to treatment in patients with rheumatoid arthritis. Ann. Rheum. Dis. 64, 834–838 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon S., Martinez F. O., Alternative activation of macrophages: Mechanism and functions. Immunity 32, 593–604 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Choi J. S., Cho W. L., Choi Y. J., Kim J. D., Park H.-A., Kim S. Y., Park J. H., Jo D.-G., Cho Y. W., Functional recovery in photo-damaged human dermal fibroblasts by human adipose-derived stem cell extracellular vesicles. J. Extracell. Vesicles 8, 1565885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiklander O. P. B., Brennan M. Á., Lötvall J., Breakefield X. O., Andaloussi S. E. L., Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 11, eaav8521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agatemor C., Buettner M. J., Ariss R., Muthiah K., Saeui C. T., Yarema K. J., Exploiting metabolic glycoengineering to advance healthcare. Nat. Rev. Chem. 3, 605–620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeForest C. A., Polizzotti B. D., Anseth K. S., Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 8, 659–664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Self-Fordham J. B., Naqvi A. R., Uttamani J. R., Kulkarni V., Nares S., MicroRNA: Dynamic regulators of macrophage polarization and plasticity. Front. Immunol. 8, 1062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong J., Xu L., Hu Y., Liu F., Yu Y., Guo H., Ni X., Huang Y., Zhao L., Wang Z., Inhibition of let-7b-5p contributes to an anti-tumorigenic macrophage phenotype through the SOCS1/STAT pathway in prostate cancer. Cancer Cell Int. 20, 470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L., Li J., Wang Q., Meng G., Lv X., Zhou H., Li W., Zhang J., The relationship between microRNAs and the STAT3-related signaling pathway in cancer. Tumor Biol. 39, 1010428317719869 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Cosenza S., Toupet K., Maumus M., Luz-Crawford P., Blanc-Brude O., Jorgensen C., Noël D., Mesenchymal stem cells-derived exosomes are more immunosuppressive than microparticles in inflammatory arthritis. Theranostics 8, 1399–1410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piccolo V., Curina A., Genua M., Ghisletti S., Simonatto M., Sabò A., Amati B., Ostuni R., Natoli G., Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 18, 530–540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Culemann S., Grüneboom A., Nicolás-Ávila J. Á., Weidner D., Lämmle K. F., Rothe T., Quintana J. A., Kirchner P., Krljanac B., Eberhardt M., Ferrazzi F., Kretzschmar E., Schicht M., Fischer K., Gelse K., Faas M., Pfeifle R., Ackermann J. A., Pachowsky M., Renner N., Simon D., Haseloff R. F., Ekici A. B., Bäuerle T., Blasig I. E., Vera J., Voehringer D., Kleyer A., Paulsen F., Schett G., Hidalgo A., Krönke G., Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 572, 670–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Im E.-J., Lee C.-H., Moon P.-G., Rangaswamy G. G., Lee B., Lee J. M., Lee J.-C., Jee J.-G., Bae J.-S., Kwon T.-K., Kang K.-W., Jeong M.-S., Lee J.-E., Jung H.-S., Ro H.-J., Jun S., Kang W., Seo S.-Y., Cho Y.-E., Song B.-J., Baek M.-C., Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat. Commun. 10, 1387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brand D. D., Latham K. A., Rosloniec E. F., Collagen-induced arthritis. Nat. Protoc. 2, 1269–1275 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Han H. S., Lee H., You D. G., Nguyen V. Q., Song D.-G., Oh B. H., Shin S., Choi J. S., Kim J. D., Pan C.-H., Jo D.-G., Cho Y. W., Choi K. Y., Park J. H., Human adipose stem cell-derived extracellular nanovesicles for treatment of chronic liver fibrosis. J. Control. Release 320, 328–336 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Woo C. H., Kim H. K., Jung G. Y., Jung Y. J., Lee K. S., Yun Y. E., Han J., Lee J., Kim W. S., Choi J. S., Yang S., Park J. H., Jo D.-G., Cho Y. W., Small extracellular vesicles from human adipose-derived stem cells attenuate cartilage degeneration. J. Extracell. Vesicles 9, 1735249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabe0083/DC1


