ABSTRACT
The innate immune system has numerous signal transduction pathways that lead to the production of type I interferons in response to exposure of cells to external stimuli. One of these pathways comprises RNA polymerase (Pol) III that senses common DNA viruses, such as cytomegalovirus, vaccinia, herpes simplex virus-1 and varicella zoster virus. This polymerase detects and transcribes viral genomic regions to generate AU-rich transcripts that bring to the induction of type I interferons. Remarkably, Pol III is also stimulated by foreign non-viral DNAs and expression of one of its subunits is induced by an RNA virus, the Sindbis virus. Moreover, a protein subunit of RNase P, which is known to associate with Pol III in initiation complexes, is induced by viral infection. Accordingly, alliance of the two tRNA enzymes in innate immunity merits a consideration.
KEWORDS: Pol III, RNase P, innate immune system, DNA virus, RNA virus
The innate immune system
The immune system has two divisions, the innate immune system and adaptive immune system, which together provide early and late immunity in vertebrates [1–5]. The innate immune system is a ubiquitous, primordial defense network found in vertebrates and invertebrates, including plants, insects and fungi [6]. In mammals, this system relies on macrophages, dendritic cells, neutrophils, epithelial and Natural killer cells in responding to invading pathogens and executing antigen presentation for mounting late immune response and memory by the adaptive immune system. These cells express pattern recognition receptors (PRRs) for identification of molecular features in pathogens, such as viruses and bacteria [7]. PRRs are sensors that identify two classes of molecules, pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) that represent loose components of damaged cells. In this respect, multiple context-dependent and direct molecular signatures are used to distinguish self from non-self nucleic acids of pathogenic contagions or autonomous pathologies. Common PAMPs of viral infections are viral genomic DNA and RNA that are recognized by two groups of PRRs, the Toll-like receptors (TLRs) and cytosolic PRRs [1,2,3,5,6,8–11].
TLRs are a family of receptors that sense viral and bacterial nucleic acids in endosomes of macrophages and dendritic cells and detect engulfed PAMPs in infected cells [4,9,12]. For instance, TLR3 detects dsRNA, TLR7/TLR8 recognize ssRNA, whereas TLR9 senses CpG DNA [6,13–17]. TLR3 distinguishes dsRNA that is larger than 40–50 nucleotides [18], whereas TLR9 elicits response to bacterial non-methylated CpG fragments that induce dimerization of the receptor and activation of signal transduction pathways leading to IFN production [19–21].
Cytosolic PRRs are diverse protein sensors that identify nucleic acids of viruses and bacteria in the cell cytoplasm [9,22,23]. These molecular sentinels include the retinoic acid-inducible gene (RIG)-like receptors, 2′-5′-oligoadenylate synthetase 1 (OAS1), cyclic GMP-AMP synthase (cGAS), stimulator of interferon genes (STING), additional absent in melanoma 2 (AIM2)-like receptors, DNA-dependent protein kinase (DNA-PK), as well as DEAH-box (DHX) and DEAD-box (DDX) RNA helicases [12,17,24–27]. The protein kinase RNA (PKR) is an IFN-stimulated gene and considered a PRR [28,29]. The PRR sensors are germline-encoded and conserved across evolution [30,31]. This is in contrast to somatic genetic rearrangements that produce new immunoglobulins and T-cell receptors of the adaptive immune system, endogenization of CRISPR in bacteria [32] or endogenization of retroviruses and bornaviruses in eukaryotes, including mammals [33–36]. Recent studies of insects also demonstrate the genetic acquisition of piDNAs, originating from RNA viruses, that confer protection against pathogens [37,38].
Recognition of viral DNA and RNA by cytosolic PRRs induces the production of type I IFNs via interrelated signal transduction pathways that integrate STING, TANK-binding kinase 1 (TBK1) and interferon response factor 3 (IRF3) for downstream induction of IFN-α and IFN-β [21,39,40]. Infected macrophages and dendritic cells primarily produce IFN-α, whereas fibroblasts and epithelial cells, nonimmune cells, mainly synthesize IFN-β [41]. Production of type I IFNs induces the expression of IFN-stimulated genes that finally block the dissemination of viral or bacterial infection in mammals. The aforementioned cells also respond to type I IFNs by mediating antigen presentation and producing cytokines and chemokines, e. g. tumor necrosis factors and interleukins known to act as major immune response mediators [28,42,43]. Type I IFNs also stimulate antibody production by B cells and augment the activation of T cells for adaptive immunity [41]. Viruses have evolved diverse counteracting strategies to mask their genomes, such as formation of replication complexes that shield naked viral RNA, seizing of self-identifiers of cellular RNAs (e. g. Cap-snatching) and targeting of cellular protein sensors for degradation [44–48].
Pol III, a sensor of foreign DNA
An additional but interesting cytosolic sensor of viral and bacterial DNA is RNA polymerase III (Pol III) (Figure 1) [21,49–52]. This polymerase is able to bind and transcribe AT-rich genomes of distinct viruses. The resulted 5′-pppRNA transcripts, ~70 nt in length, are recognized and bound by RIG-I that directs the signal to MAVS, TBK1 and IRF3 for downstream induction of IFN-α and IFN-β gene expression in infected cells [6,27,49,50,52–58] (Figure 1). Pol III detects the genomes of common DNA viruses, such as cytomegalovirus, vaccinia, herpes simplex virus-1 and varicella zoster virus [21,22,49,50,54,59–63]. The importance of Pol III in innate immunity is exemplified by the ability of the vaccinia virus to counteract the polymerase stimulation by its E3 protein, revealing a deep host-pathogen co-evolution [64,65]. However, the molecular mechanism by which the polymerase pinpoints its start point in viral genomes and initiates RNA synthesis remains largely unknown.
Figure 1.
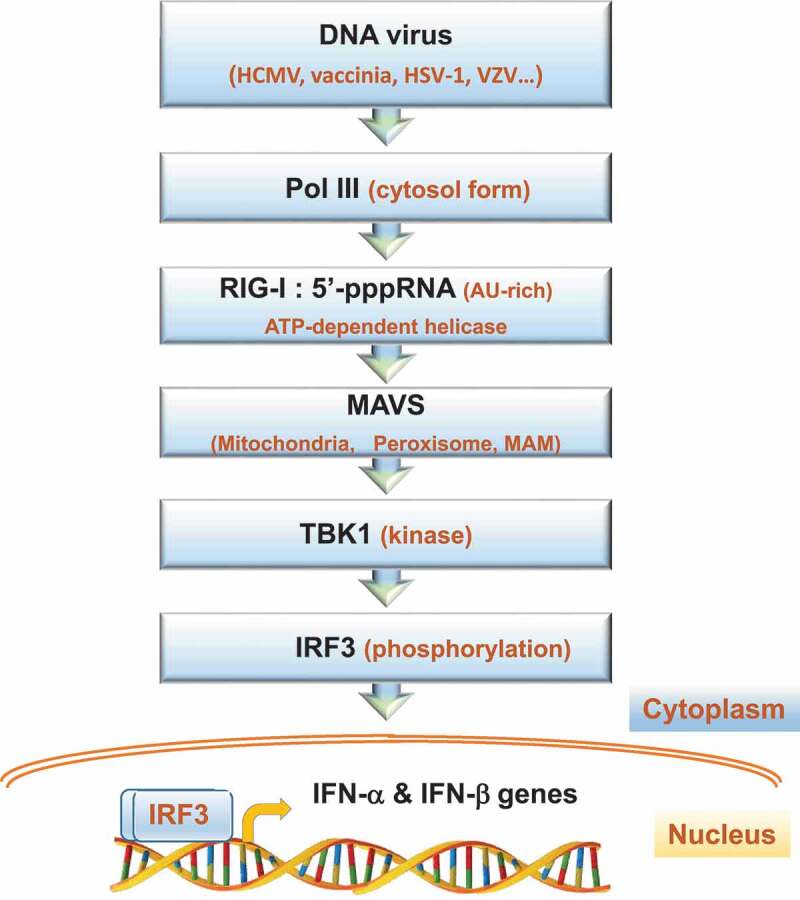
The signal transduction pathway of Pol III for induction of type I IFNs. The indicated DNA viruses with AT-rich genomes are sensed and transcribed by Pol III in the cell cytoplasm. The resulted AU-rich 5’pppRNA transcripts are then detected and bound by the RIG-I helicase. This ATP-dependent helicase requires the RING finger E3 ubiquitin ligases, Riplet and TRIM25 for K63 ubiquitination for full activation (not shown). RIG-I oligomers form a translocon complex with TRIM25 and the molecular chaperone 14-3-3ε [124] (not shown). This complex interacts with MAVS, a tail-anchored membrane protein, found on intracellular membranes, including peroxisomes and mitochondria, in addition to the mitochondrion-associated membrane (MAM), a domain of the endoplasmic reticulum. A phosphorylated and ubiquitinated form of MAVS then associates with the kinase TBK-1 and other factors to phosphorylate the IFN regulatory factor IRF3. The modified IRF3 enters the nucleus and binds its upstream regulatory element in the IFN-α and IFN-β genes for initiation of transcription (arrow)
The discovery that Pol III acts as a sensor of diverse DNA viruses raises the question if it has a general role of recognizing foreign nucleic acids in invaded cells. Previous studies show that Pol III transcribes a synthetic poly(dA-dT) template in a promoter-independent manner [66,67] and the resulted poly(A-U) transcripts trigger type I IFN induction in transfected cells [17,50,68]. The high AT content of the DNA template is critical to transcription by Pol III, probably owing to the inclination of AT-rich boxes to serve as kick-start for RNA polymerization. By contrast, non-poly(dA-dT) dsDNAs elicit type I IFN induction, but via a Pol III–independent pathway [50]. Pol III also starts transcription from synthetic circularized DNA oligonucleotides, termed coligos, in cultured cell lines and in extracts [69,70]. Transcription of a coligo template begins at a single-stranded region and it seems to take place in the cytoplasm [69,70]. In fact, the ability of Pol III to initiate transcription from ssDNA promoters is well established [71]. Moreover, the presence of linearized or circular plasmids in transfected human cell lines elicits the activity of Pol III in transcription of tRNA genes (Figure 2) [39,72]. But whether these plasmids have functional AT-rich sequences utilized as startpoints for transcription remains elusive. Together, Pol III activity is stimulated to varying extents by tiny amount of foreign DNA, viral or otherwise.
Figure 2.
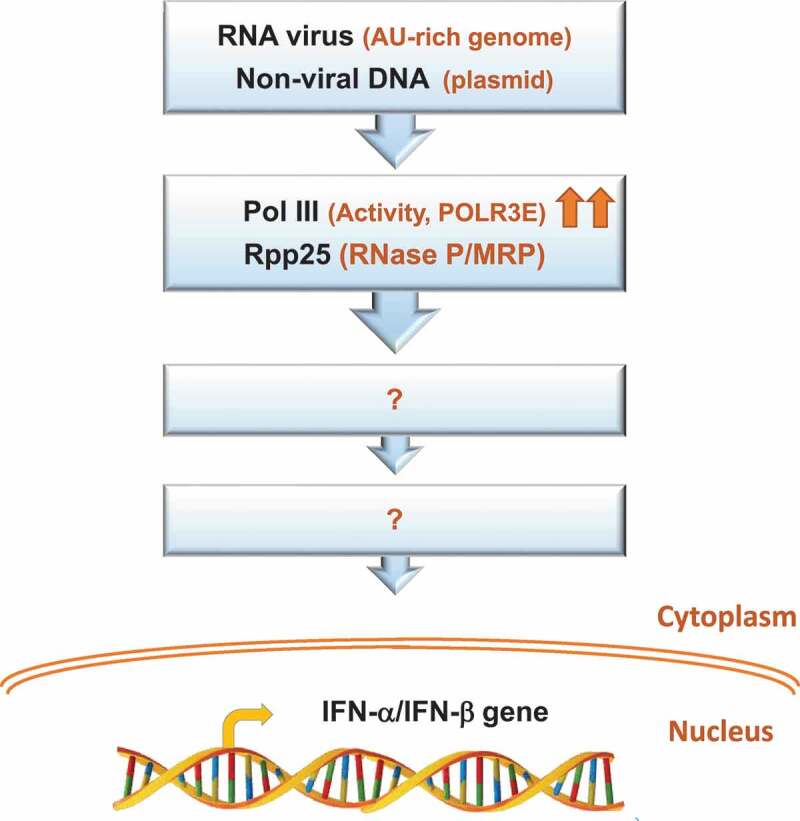
Potential foreign nucleic acids that induce human Pol III and RNase P in the innate immune system. The presence of RNA virus, whose genome is enriched with AU sequences, or non-viral DNA, such as plasmid, in the cell induces the expression of the POLR3E subunit of Pol III and its activity, as well as the expression of the protein subunit Rpp25 of RNase P/RNase MRP [72]. Arrows indicate increase in expression or activity. The downstream signal transduction pathways for induction of type I IFNs or else by these tRNA enzymes or their subunits remain unknown
Does Pol III sense RNA viruses?
As described above, many PRRs sense RNA and DNA viruses [73,74]. For instance, RIG-I senses the Sendai virus, a negative ssRNA virus that replicates in the cell cytoplasm, in addition to the vaccinia virus, a DNA virus with cytoplasmic replication cycle [27,40,49,73,75,76]. A recent study shows that infection of cultured cell lines by the Sindbis virus induces the expression of the POLR3E subunit of Pol III [72]. The finding raises the possibility that Pol III is involved in sensing a positive AU-rich ssRNA enveloped virus known to replicate its ~11.7 Kb genome in the cytoplasm (Figure 2). Preliminary results unveil that Pol III activity is central for the resistance of cells to infection by Sindbis virus, and possibly another RNA virus (Figure 2)(Mani D., unpublished data). Apparently, Pol III plays an antiviral role in cells infected with RNA viruses. This conclusion is supported by the discovery that a recessive substitution mutation in the human POLR3E gene is linked to systemic and concurrent infections with DNA and RNA viruses, including cytomegalovirus, metapneumovirus, respiratory syncytial virus, parvovirus, parainfluenza 3, and human herpesvirus 6 [72]. Fibroblasts with this rare mutation exhibit impaired induction of type I IFN and increased susceptibility to cytomegalovirus infection. The molecular mechanism underlying the mutation of POLR3E involves the assembly of malfunctioning initiation complexes of Pol III. Hence, mutated POLR3E and Pol III are linked to an innate immune deficiency condition in human [72].
RNase P and its links to the immune system
RNase P is a ubiquitous endoribonuclease that removes the 5ʹ leader of precursor tRNA [77–81]. In human cells, nuclear RNase P consists of H1 RNA and ten distinct protein subunits, termed Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40, Pop1 and Pop5 [82–86]. Cryo-electron microscopy reveals that H1 RNA, also known as RPPH1, is covered by its protein subunits arranged in three subcomplexes, Rpp20-Rpp25, Pop5-Rpp14-(Rpp30)2-Rpp40, and Rpp21-Rpp29-Rpp38 and single polypeptide Pop1. These conserved proteins [87,88] form an interlocked clamp that stabilizes the catalytic RNA in a tertiary conformation fitted for binding and cleavage of precursor tRNA substrates [89].
RNase P shares protein subunits with RNase MRP [79,83,84,88,90], a mitochondrial and rRNA processing ribonucleoprotein [86,91–93]. The RMRP gene, which codes for the RNase MRP RNA, is found to be mutated in several immunodeficiency disorders, including cartridge hair hypoplasia, Omenn syndrome, anauxetic dysplasia, kyphomelic dysplasia and metaphyseal dysplasia without hypotrichosis [94–96]. Protein subunits of the two ribonucleoproteins are recognized as Th/To autoimmune antigens [82,85,97,98]. Considering the increasing overlap in the components and functions of the adaptive and innate immune systems and their contribution to initiation and progression of autoimmune diseases [99–103], it is conceivable that RNase P, RNase MRP and/or their RNA substrates and products [104] take part in the immune system (see below). In fact, the specific subunit Rpp21 of RNase P is encoded by a gene positioned in the genetic locus of the major histocompatibility complex class I [105]. A form of this protein is fused to the ubiquitin ligase TRIM39 via transcription readthrough and alternative splicing to generate TRIM39R [106]. The hybrid polypeptide regulates type I interferon induction in response to viral infection and is genetically linked to the Behcet’s disease, a chronic inflammatory autoimmune condition characterized by eye inflammation, oral and genital sores and skin legions [107]. Of note, TRIM proteins, which possess E3 ubiquitin ligase activities, have various roles in the immune system, including antiviral innate immunity [108].
Human RNase P is evolutionary linked to viruses. Thus, screening of microbial genome databases for RNase P RNA-like genes reveals that the camelpox virus has a gene that codes for a transcript that folds into the universally conserved secondary structure of the RNase P RNA [109]. This viral gene is conserved in orthopoxviruses, including the vaccinia virus [110]. The vaccinia RNase P RNA-like transcript is detected in infected cells, but it is inactive in tRNA processing [110]. A recent study also shows that infection of cultured human cells with Sindbis virus leads to induction of the expression of the subunit Rpp25 of RNase P (Figure 2) [72]. Induction of Rpp25, which belongs to the Alba-like chromatin-binding proteins and binds the P3 domain of H1 RNA, is transient and concomitant with that of the subunit POLR3E [72].
Is RNase P linked to the sensor Pol III?
Human RNase P is implicated in transcription of small noncoding RNA genes by nuclear Pol III in cells and extracts [111–113]. Thus, targeted destruction of RNase P subunits by RNA interference inhibits transcription of tRNA and 5S rRNA genes [111,112]. This ribonucleoprotein coexists with Pol III in purified initiation complexes and inhibition of expression of its subunits demolishes the structure and function of these large complexes [112]. Of note, the coexistence of the two tRNA enzymes in initiation complexes contrasts the general view that processing of precursor tRNA occurs post-transcriptionally [114], an assessment inferred from genetic studies in the budding yeast and that show that knockout of RNase P subunits leads to accumulation of precursor tRNAs.
As a viral sensor, Pol III seems to act in the cytoplasm [49,50], whereas RNase P functions in transcription and processing of tRNA in the nucleus [105,115,116]. Nevertheless, we may speculate on possible relationship of RNase P to viral genome sensing by Pol III in the cytoplasm. Thus, it is known for a long time that a fraction of H1 RNA is localized in the cytoplasm, as local [117] and passerby to mitochondria [118–120]. Moreover, the yeast homologs of Rpp20, Rpp25 and Pop1 exist in the cytoplasm and facilitate the nuclear import of a related ribonucleoprotein, the telomerase [121–123]. Assuming that the human homologs of these three proteins are present in the cytoplasm, in which they are translated, an encounter with H1 RNA and/or Pol III is conceivable. Future studies will provide answer to whether individual subunits or new forms of RNase P act alone or together with Pol III in the innate immune system or else.
Acknowledgments
This research is supported by the Israel Science Foundation (grant #338/19) to Alexander Rouvinski and the United States-Israel Binational Science Foundation (#2015/157) and the Israel Science Foundation (grant #1205/17) to Nayef Jarrous.
Funding Statement
This work was supported by the Israel Science Foundation [1205/17]; Israel Science Foundation [338/19]; United States - Israel Binational Science Foundation [2015/157].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Lemaitre B, Nicolas E, Michaut L, et al.The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8808632 [DOI] [PubMed] [Google Scholar]
- [2].Medzhitov R, Preston-Hurlburt P, Janeway CA.. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9237759 [DOI] [PubMed] [Google Scholar]
- [3].Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9851930 [DOI] [PubMed] [Google Scholar]
- [4].Beutler BA.TLRs and innate immunity. Blood. 2009;113:1399–1407. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18757776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Akira S, Uematsu S, Takeuchi O.Pathogen recognition and innate immunity. Cell. 2006;124:783–801. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16497588 [DOI] [PubMed] [Google Scholar]
- [6].Pandey S, Kawai T, Akira S.Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol. 2014;7:a016246. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25301932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Medzhitov R, Janeway C.Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10719670 [DOI] [PubMed] [Google Scholar]
- [8].Takeda K, Kaisho T, Akira S.Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12524386 [DOI] [PubMed] [Google Scholar]
- [9].Barbalat R, Ewald SE, Mouchess ML, et al.Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21219183 [DOI] [PubMed] [Google Scholar]
- [10].Brown GD, Willment JA, Whitehead L.C-type lectins in immunity and homeostasis. Nat Rev Immunol. 2018;18:374–389. [Internet]. Available from: http://www.nature.com/articles/s41577-018-0004–8 [DOI] [PubMed] [Google Scholar]
- [11].Saxena M, Yeretssian G. NOD-like receptors: master regulators of inflammation and cancer. Front Immunol. 2014:5. [Internet]. Available from: http://journal.frontiersin.org/article/10.3389/fimmu.2014.00327/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Loo Y-M, Gale M.Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21616437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Heil F, Hemmi H, Hochrein H, et al.Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14976262 [DOI] [PubMed] [Google Scholar]
- [14].Diebold SS, Kaisho T, Hemmi H, et al.Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14976261 [DOI] [PubMed] [Google Scholar]
- [15].Alexopoulou L, Holt AC, Medzhitov R, et al.Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11607032 [DOI] [PubMed] [Google Scholar]
- [16].Hemmi H, Akira S.TLR signalling and the function of dendritic cells. Chem Immunol Allergy. 2005;86:120–135. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15976491 [DOI] [PubMed] [Google Scholar]
- [17].Spel L, Martinon F.Detection of viruses by inflammasomes. Curr Opin Virol. 2020;46:59–64. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/33176273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leonard JN, Ghirlando R, Askins J, et al.The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18172197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ohto U, Shibata T, Tanji H, et al.Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25686612 [DOI] [PubMed] [Google Scholar]
- [20].Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11130078 [DOI] [PubMed] [Google Scholar]
- [21].Briard B, Place DE, Kanneganti T-D.DNA sensing in the innate immune response. Physiology (Bethesda). 2020;35:112–124. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32027562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Luecke S, Paludan SR.Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine. 2017;98:4–14. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27751656 [DOI] [PubMed] [Google Scholar]
- [23].Ori D, Murase M, Kawai T.Cytosolic nucleic acid sensors and innate immune regulation. Int Rev Immunol. 2017;36:74–88. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28333574 [DOI] [PubMed] [Google Scholar]
- [24].Motwani M, Pesiridis S, Fitzgerald KA.DNA sensing by the cGAS–STING pathway in health and disease. Nat Rev Genet. 2019;20:657–674. [Internet]. Available from: http://www.nature.com/articles/s41576-019-0151–1 [DOI] [PubMed] [Google Scholar]
- [25].Hornung V, Hartmann R, Ablasser A, et al.OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. [Internet]. Available from: http://www.nature.com/articles/nri3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Proudfoot NJ.Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science. 2016;352:aad9926. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27284201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rehwinkel J, Gack MU.RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–551. [Internet]. Available from: http://www.nature.com/articles/s41577-020-0288–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kaempfer R.RNA sensors: novel regulators of gene expression. EMBO Rep. 2003;4:1043–1047. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14593443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gilfoy FD, Mason PW.West nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:11148–11158. [Internet]. Available from: https://jvi.asm.org/content/81/20/11148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schlee M, Hartmann G.Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–580. [Internet]. Available from: http://www.nature.com/articles/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bartok E, Hartmann G.Immune sensing mechanisms that discriminate self from altered self and foreign nucleic acids. Immunity. 2020;53:54–77. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1074761320302697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Moelling K, Broecker F, Russo G, et al. RNase H As gene modifier, driver of evolution and antiviral defense. Front Microbiol. 2017:8. [Internet]. Available from : http://journal.frontiersin.org/article/10.3389/fmicb.2017.01745/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Horie M.The biological significance of bornavirus-derived genes in mammals. Curr Opin Virol. 2017;25:1–6. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1879625717300342 [DOI] [PubMed] [Google Scholar]
- [34].Horie M, Kobayashi Y, Suzuki Y, et al.Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Philos Trans R Soc B Biol Sci. 2013;368:20120499. [Internet]. Available from: https://royalsocietypublishing.org/doi/10.1098/rstb.2012.0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Belyi VA, Levine AJ, Skalka AM.Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. [Internet]. Available from: https://dx.plos.org/10.1371/journal.ppat.1001030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Parrish NF, Tomonaga K.Endogenized viral sequences in mammals. Curr Opin Microbiol. 2016;31:176–183. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1369527416300170 [DOI] [PubMed] [Google Scholar]
- [37].Goic B, Stapleford KA, Frangeul L, et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat Commun. 2016;7:12410. [Internet]. Available from: http://www.nature.com/articles/ncomms12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Suzuki Y, Baidaliuk A, Miesen P, et al. Non-retroviral endogenous viral element limits cognate virus replication in aedes aegypti ovaries. Curr Biol. 2020;30:3495–3506.e6. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S096098222030909X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Burleigh K, Maltbaek JH, Cambier S, et al.Human DNA-PK activates a STING-independent DNA sensing pathway. Sci Immunol. 2020;5:eaba4219. [Internet]. Available from: https://immunology.sciencemag.org/lookup/doi/10.1126/sciimmunol.aba4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kato H, Sato S, Yoneyama M, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16039576 [DOI] [PubMed] [Google Scholar]
- [41].Ivashkiv LB, Donlin LT.Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24362405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jarrous N, Osman F, Kaempfer R.2-Aminopurine selectively inhibits splicing of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1996;16:2814–2822. [Internet]. Available from: http://mcb.asm.org/lookup/doi/10.1128/MCB.16.6.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Osman F, Jarrous N, Ben-Asouli Y, et al.A cis-acting element in the 3ʹ-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999;13:3280–3293. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10617576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sarkar R, Patra U, Lo M, et al.Rotavirus activates a noncanonical ATM-Chk2 branch of DNA damage response during infection to positively regulate viroplasm dynamics. Cell Microbiol. 2020;22:e13149. [Internet]. Available from:http://www.ncbi.nlm.nih.gov/pubmed/31845505 [DOI] [PubMed] [Google Scholar]
- [45].Jin H, Elliott RM.Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J Virol. 1993;67:1396–1404. [Internet]. Available from: https://jvi.asm.org/content/67/3/1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Russell AB.Cap-snatching leads to novel viral proteins. Cell. 2020;181:1450–1451. Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867420306772 [DOI] [PubMed] [Google Scholar]
- [47].Uehata T, Takeuchi O.RNA recognition and immunity—innate immune sensing and its posttranscriptional regulation mechanisms. Cells. 2020;9:1701. [Internet]. Available from: https://www.mdpi.com/2073-4409/9/7/1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hou P, Yang K, Jia P, et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021;31:62–79. [Internet]. Available from: http://www.nature.com/articles/s41422-020-0362–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chiu Y-H, Macmillan JB, Chen ZJ.RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19631370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ablasser A, Bauernfeind F, Hartmann G, et al.RIG-I-dependent sensing of poly(dA: dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19609254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].O’Neill LAJ.DNA makes RNA makes innate immunity. Cell. 2009;138:428–430. Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19665965 [DOI] [PubMed] [Google Scholar]
- [52].Unterholzner L.The interferon response to intracellular DNA: why so many receptors? Immunobiology. 2013;218:1312–1321. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23962476 [DOI] [PubMed] [Google Scholar]
- [53].Hou F, Sun L, Zheng H, et al.MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867411007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Carter-Timofte ME, Paludan SR, Mogensen TH.RNA polymerase III as a gatekeeper to prevent severe VZV infections. Trends Mol Med. 2018;24:904–915. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30115567 [DOI] [PubMed] [Google Scholar]
- [55].Yoneyama M, Kikuchi M, Natsukawa T, et al.The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15208624 [DOI] [PubMed] [Google Scholar]
- [56].Hornung V, Ellegast J, Kim S, et al. 5ʹ-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. [Internet]. Available from:http://www.ncbi.nlm.nih.gov/pubmed/17038590 [DOI] [PubMed] [Google Scholar]
- [57].Vazquez C, Horner SM, Sullivan CS.MAVS coordination of antiviral innate immunity. J Virol. 2015;89:6974–6977. [Internet]. Available from:https://pubmed.ncbi.nlm.nih.gov/25948741/ [DOI] [PMC free article] [PubMed]
- [58].Subramanian N, Natarajan K, Clatworthy MR, et al.The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867413002948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Carter-Timofte ME, Hansen AF, Mardahl M, et al. Varicella-zoster virus CNS vasculitis and RNA polymerase III gene mutation in identical twins. Neurol Neuroimmunol Neuroinflammation. 2018;5:e500. [Internet]. Available from: http://nn.neurology.org/lookup/doi/10.1212/NXI.0000000000000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Carter-Timofte ME, Hansen AF, Christiansen M, et al.Mutations in RNA Polymerase III genes and defective DNA sensing in adults with varicella-zoster virus CNS infection. Genes Immun. 2018;20:214–223. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29728610 [DOI] [PubMed] [Google Scholar]
- [61].Ahlers LRH, Bastos RG, Hiroyasu A, et al.Invertebrate iridescent virus 6, a DNA virus, stimulates a mammalian innate immune response through RIG-I-like receptors. PLoS One. 2016;11:e0166088. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27824940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Crill EK, Furr-Rogers SR, Marriott I.RIG-I is required for VSV-induced cytokine production by murine glia and acts in combination with DAI to initiate responses to HSV-1. Glia. 2015;63:2168–2180. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26146945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ogunjimi B, Zhang S-Y, Sørensen KB, et al. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J Clin Invest. 2017;127:3543–3556. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28783042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Leung DW, Basler CF, Amarasinghe GK.Molecular mechanisms of viral inhibitors of RIG-I-like receptors. Trends Microbiol. 2012;20:139–146. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0966842X11002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Valentine R, Smith GL.Inhibition of the RNA polymerase III-mediated dsDNA-sensing pathway of innate immunity by vaccinia virus protein E3. J Gen Virol. 2010;91:2221–2229. [Internet]. Available from: https://www.microbiologyresearch.org/content/journal/jgv/10.1099/vir.0.021998–0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Huet J, Riva M, Sentenac A, et al.Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985;260:15304–15310. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3905793 [PubMed] [Google Scholar]
- [67].Zaros C, Thuriaux P.Rpc25, a conserved RNA polymerase III subunit, is critical for transcription initiation. Mol Microbiol. 2005;55:104–114. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15612920 [DOI] [PubMed] [Google Scholar]
- [68].Thuillier V, Brun I, Sentenac A, et al.Mutations in the alpha-amanitin conserved domain of the largest subunit of yeast RNA polymerase III affect pausing, RNA cleavage and transcriptional transitions. Embo J. 1996;15:618–629. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8599945 [PMC free article] [PubMed] [Google Scholar]
- [69].Lama L, Seidl CI, Ryan K.New insights into the promoterless transcription of DNA coligo templates by RNA polymerase III. Transcription. 2014;5:e27913. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25764216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Seidl CI, Lama L, Ryan K.Circularized synthetic oligodeoxynucleotides serve as promoterless RNA polymerase III templates for small RNA generation in human cells. Nucleic Acids Res. 2013;41:2552–2564. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23275570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schroder O, Geiduschek EP, Kassavetis GA.A single-stranded promoter for RNA polymerase III. Proc Natl Acad Sci U S A. 2003;100:934–939. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12538860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ramanathan A, Weintraub M, Orlovetskie N, et al. A mutation in POLR3E impairs antiviral immune response and RNA polymerase III. Proc Natl Acad Sci U S A. 2020;117:22113–22121. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32843346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jensen S, Thomsen AR.Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–2910. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22258243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Brennan K, Bowie AG.Activation of host pattern recognition receptors by viruses. Curr Opin Microbiol. 2010;13:503–507. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1369527410000688 [DOI] [PubMed] [Google Scholar]
- [75].Baum A, Sachidanandam R, Garcia-Sastre A.Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci. 2010;107:16303–16308. [Internet]. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. [Internet]. Available from: http://www.nature.com/articles/nature04734 [DOI] [PubMed] [Google Scholar]
- [77].Altman S, Smith JD.Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971;233:35–39. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/4938965 [DOI] [PubMed] [Google Scholar]
- [78].Engelke DR, Fierke CA.The evolution of RNase P. RNA. 2015;21:517–518. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25780121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lygerou Z, Allmang C, Tollervey D, et al.Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8602511 [DOI] [PubMed] [Google Scholar]
- [80].Gopalan V, Jarrous N, Krasilnikov ASAS.Chance and necessity in the evolution of RNase P. RNA. 2018;24:1–5. [Internet]. Available from:https://pubmed.ncbi.nlm.nih.gov/28971852/ [DOI] [PMC free article] [PubMed]
- [81].Kikovska E, Svärd SG, Kirsebom LA.Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci U S A. 2007;104:2062–2067. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17284611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Eder PS, Kekuda R, Stolc V, et al.Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc Natl Acad Sci. 1997;94:1101–1106. [Internet]. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.94.4.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Welting TJM, Peters FMA, Hensen SMM, et al.Heterodimerization regulates RNase MRP/RNase P association, localization, and expression of Rpp20 and Rpp25. RNA. 2007;13:65–75. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17119099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lygerou Z, Mitchell P, Petfalski E, et al.The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7926742 [DOI] [PubMed] [Google Scholar]
- [85].Lygerou Z, Pluk H, van Venrooij WJ, et al.hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. Embo J. 1996;15:5936–5948. [Internet]. Available from: http://doi.wiley.com/10.1002/j.1460-2075.1996.tb00980.x [PMC free article] [PubMed] [Google Scholar]
- [86].Jarrous N, Altman S. Human ribonuclease P. Methods Enzymol. 2001;342:93–100. [DOI] [PubMed] [Google Scholar]
- [87].Tsai H-Y, Pulukkunat DK, Woznick WK, et al.Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc Natl Acad Sci U S A. 2006;103:16147–16152. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17053064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Perederina A, Li D, Lee H, et al.Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP. Nat Commun. 2020;11:3474. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32651392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wu J, Niu S, Tan M, et al. Cryo-EM structure of the human ribonuclease P holoenzyme. Cell. 2018;175:1393–1404.e11. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30454648 [DOI] [PubMed] [Google Scholar]
- [90].Chamberlain JR, Lee Y, Lane WS, et al.Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9620854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Guerrier-Takada C, Eder PS, Gopalan V, et al. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA. 2002;8:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hands-Taylor KLD, Martino L, Tata R, et al.Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res. 2010;38:4052–4066. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20215441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Khanova E, Esakova O, Perederina A, et al.Structural organizations of yeast RNase P and RNase MRP holoenzymes as revealed by UV-crosslinking studies of RNA-protein interactions. RNA. 2012;18:720–728. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22332141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mattijssen S, Welting TJM, Pruijn GJM.RNase MRP and disease. Wiley Interdiscip Rev RNA. 2010;1:102–116. [Internet]. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/wrna.9 [DOI] [PubMed] [Google Scholar]
- [95].Vakkilainen S, Taskinen M, Mäkitie O. Immunodeficiency in cartilage‐hair hypoplasia: pathogenesis, clinical course and management. Scand J Immunol. 2020:92. [Internet]. Available from: https://onlinelibrary.wiley.com/doi/10.1111/sji.12913 [DOI] [PubMed] [Google Scholar]
- [96].Martin AN, Li Y.RNase MRP RNA and human genetic diseases. Cell Res. 2007;17:219–226. [Internet]. Available from: http://www.nature.com/articles/7310120 [DOI] [PubMed] [Google Scholar]
- [97].Gold HA, Craft J, Hardin JA, et al.Antibodies in human serum that precipitate ribonuclease P. Proc Natl Acad Sci. 1988;85:5483–5487. [Internet]. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.85.15.5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jarrous N, Eder PS, Guerrier-Takada C, et al. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- [99].Zouali M, La Cava A. Editorial: innate immunity pathways in autoimmune diseases. Front Immunol. 2019:10. [Internet]. Available from : https://www.frontiersin.org/article/10.3389/fimmu.2019.01245/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Paulson JC.Innate immune response triggers lupus-like autoimmune disease. Cell. 2007;130:589–591.Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867407010318 [DOI] [PubMed] [Google Scholar]
- [101].Celhar T, Magalhães R, Fairhurst A-M.TLR7 and TLR9 in SLE: when sensing self goes wrong. Immunol Res. 2012;53:58–77. [Internet]. Available from: http://link.springer.com/10.1007/s12026-012-8270–1 [DOI] [PubMed] [Google Scholar]
- [102].Matzinger P.The danger model: a renewed sense of self. Science. 2002;296:301–305. [Internet]. Available from: https://www.sciencemag.org/lookup/doi/10.1126/science.1071059 [DOI] [PubMed] [Google Scholar]
- [103].Deerhake ME, Biswas DD, Barclay WE, et al. Pattern recognition receptors in multiple sclerosis and its animal models. Front Immunol. 2019;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Pawar K, Shigematsu M, Sharbati S, et al. Infection-induced 5′-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLOS Biol. 2020;18:e3000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jarrous N, Reiner R, Wesolowski D, et al. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7:1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Roberts JD, Chiche J-D, Kolpa EM, et al.cGMP-dependent protein kinase I interacts with TRIM39R, a novel Rpp21 domain-containing TRIM protein. Am J Physiol Cell Mol Physiol. 2007;293:L903–12. [Internet]. Available from: https://www.physiology.org/doi/10.1152/ajplung.00157.2007 [DOI] [PubMed] [Google Scholar]
- [107].Kurata R, Tajima A, Yonezawa T, et al.TRIM39R, but not TRIM39B, regulates type I interferon response. Biochem Biophys Res Commun. 2013;436:90–95. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0006291X13008607 [DOI] [PubMed] [Google Scholar]
- [108].van Gent M, Sparrer KMJ, Gack MU.TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol. 2018;5:385–405. [Internet]. Available from: https://www.annualreviews.org/doi/10.1146/annurev-virology–092917–043323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Li Y, Altman S.In search of RNase P RNA from microbial genomes. RNA. 2004;10:1533–1540. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15337843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yang L, Wesolowski D, Li Y, et al.Analysis of putative RNase P RNA from orthopoxviruses. J Mol Biol. 2005;354:529–535. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16253270 [DOI] [PubMed] [Google Scholar]
- [111].Reiner R, Ben-Asouli Y, Krilovetzky I, et al.A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 2006;20:1621–1635. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16778078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Serruya R, Orlovetskie N, Reiner R, et al. Human RNase P ribonucleoprotein is required for formation of initiation complexes of RNA polymerase III. Nucleic Acids Res. 2015;43:5442–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Jarrous N.Roles of RNase P and its subunits. Trends Genet. 2017;33:594–603. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28697848 [DOI] [PubMed] [Google Scholar]
- [114].Hummel G, Warren J, Drouard L.The multi-faceted regulation of nuclear tRNA gene transcription. IUBMB Life. 2019;71:1099–1108. [Internet]. Available from: http://doi.wiley.com/10.1002/iub.2097 [DOI] [PubMed] [Google Scholar]
- [115].Jarrous N, Wolenski JS, Wesolowski D, et al.Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol. 1999;146:559–572. [Internet]. Available from: https://rupress.org/jcb/article/146/3/559/43811/Localization-in-the-Nucleolus-and-Coiled-Bodies-of [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Bertrand E, Houser-Scott F, Kendall A, et al.Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9716399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lee B, Matera AG, Ward DC, et al.Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: a possible coordinate role in ribosome biogenesis. Proc Natl Acad Sci U S A. 1996;93:11471–11476. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8876159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Mercer TR, Neph S, Dinger ME, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867411007677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wang G, Chen H-W, Oktay Y, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867410007257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Wang G, Shimada E, Zhang J, et al.Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Sci. 2012;109:4840–4845. [Internet]. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.1116792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Garcia PD, Leach RW, Wadsworth GM, et al.Stability and nuclear localization of yeast telomerase depend on protein components of RNase P/MRP. Nat Commun. 2020;11:2173. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32358529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Garcia PD, Zakian VA. A new role for proteins subunits of RNase P: stabilization of the telomerase holoenzyme. Microb cell (Graz, Austria). 2020;7:250–254. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32904320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lemieux B, Laterreur N, Perederina A, et al.Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell. 2016;165:1171–1181. Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27156450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Lee H-C, Chathuranga K, Lee J-S.Intracellular sensing of viral genomes and viral evasion. Exp Mol Med. 2019;51:1–13. Internet]. Available from: http://www.nature.com/articles/s12276-019-0299–y [DOI] [PMC free article] [PubMed] [Google Scholar]


