Live imaging of chromatin in an intact organism reveals a novel mode of mesoscale chromatin organization at nuclear periphery.
Abstract
The three-dimensional organization of chromatin contributes to transcriptional control, but information about native chromatin distribution is limited. Imaging chromatin in live Drosophila larvae, with preserved nuclear volume, revealed that active and repressed chromatin separates from the nuclear interior and forms a peripheral layer underneath the nuclear lamina. This is in contrast to the current view that chromatin distributes throughout the nucleus. Furthermore, peripheral chromatin organization was observed in distinct Drosophila tissues, as well as in live human effector T lymphocytes and neutrophils. Lamin A/C up-regulation resulted in chromatin collapse toward the nuclear center and correlated with a significant reduction in the levels of active chromatin. Physical modeling suggests that binding of lamina-associated domains combined with chromatin self-attractive interactions recapitulate the experimental chromatin distribution profiles. Together, our findings reveal a novel mode of mesoscale organization of peripheral chromatin sensitive to lamina composition, which is evolutionary conserved.
INTRODUCTION
While three-dimensional (3D) organization of the genome has been directly linked to gene regulation, information regarding nuclear-scale 3D chromatin organization under native physiological conditions is limited. Recent advances in imaging, sequencing, and modeling approaches have greatly enhanced our understanding of 3D genome organization, bridging the gap between single-cell spatial information and genome-wide linear sequence interactions (1). However, the absence of imaging data on the global 3D chromatin organization within the native tissue environment limits the interpretation of current data.
The genome is packed within the nucleus in a nonrandom manner and organized in a hierarchical manner, with several characteristic length scales. The basic nucleosome unit consists of DNA wrapped around a core octamer of histone proteins H2A, H2B, H3, and H4. Nucleosomes are organized into 10-nm “beads on a string” fiber that, together with additional proteins, defines the chromatin. While the textbook view suggests hierarchical higher-order folding of chromatin fibers (10, 30, 100 nm, and higher), recent advanced imaging under more physiological conditions challenged this view, describing a more complex and heterogeneous chromatin organization into clutches and domains of nucleosomes of varying sizes and densities (1–4). On the nuclear scale, a range of imaging approaches, as well as Hi-C analysis, suggests that chromatin of an interphase nucleus is partitioned into distinct chromosomal territories several micrometers long, as was documented in various mammalian and Drosophila cells (5–8). A functional model for global chromatin organization suggests that chromatin territories occupied by the chromosomes are pervaded by a channel network devoid of chromatin, termed interchromatin compartments (9, 10).
Several lines of evidence suggest a nonuniform distribution of chromatin as a function of the radial distance from the center of the nucleus. Interphase chromosomes and specific gene loci within chromosomes have preferential radial positions, with gene rich chromosomes positioned toward the nucleus interior (7, 11). However, a general view of chromatin architecture in nondividing mature cells in live tissues is still missing. The nuclear lamina, a thick meshwork of intermediate filaments associated with the inner nuclear membrane, is a major regulator of chromatin architecture, as it tethers mostly dense heterochromatin at specific sequences termed lamina-associated domains (LADs). Consistently, genome-wide DNA adenine methyl-transferase identification (DamID) analysis performed with lamin B or lamin A under culture conditions identified LADs mostly as gene poor and transcriptionally silenced sequences (12). Specifically, variations in lamin A/C levels, induced by either its up- or down-regulation, or as observed in laminopathy-associated mutations in lamin A/C, drive heterochromatin detachment from the nuclear lamina (13, 14). This was accompanied by changes in chromatin condensation that correlated with alterations in gene activation/repression in mammals (15, 16), Caenorhabditis elegans (17), and Drosophila cells (18, 19). Together, these findings suggested that LADs are specifically sensitive to the levels of lamin A/C at the nuclear lamina.
Chromatin is estimated to fill about 15 to 60% of the nuclear volume (7, 9) and shares the nucleoplasm with other dense, membrane-less organelles such as the nucleolus and nuclear speckles. Furthermore, imaging of fixed nuclei, live cells in culture, or cells isolated from their native tissue environment suggests that chromatin is distributed throughout the entire nuclear space. However, the lack of information about nuclei within their intrinsic tissue environment, where cells and nuclei adopt a specific 3D morphology, obscures this view in nondividing, differentiated cells. Chromatin organization can change in space and time in a physiological, adaptive manner, where nuclear morphology is an important contributor that reflects the balance between cytoplasmic forces acting on the nucleus and the collective mechanical resistance of the lamina and the chromatin (20–22). In addition, nuclear morphology and chromatin organization can be altered because of experimental methodologies such as reduction in cell and nucleus volume due to fixation, change in osmolality, or as a result of culture on surfaces of various stiffness conditions (23–25). Recent study that analyzed live telomere dynamics in the liver in vivo reported significantly different dynamics from that observed in cultured cells (26). It is therefore critical to reveal the structural principles of chromatin within its tissue intrinsic physiological conditions (27).
Here, we combined a custom-made device placed on top of a confocal microscope stage (28), with genetic labeling of the chromatin and the nuclear lamina, to characterize in vivo nuclear scale 3D chromatin organization in fully differentiated muscles of live, intact Drosophila larva. Imaging of live 3D chromatin organization was reported for various cell lines (2, 29, 30), as well as in the isolated Drosophila larva salivary gland (31). We present here the first 3D analysis of global chromatin organization of fully differentiated nuclei, with preserved physiologic environment, within a live, intact organism. Our live imaging demonstrates significantly different nuclear dimensions and higher nuclear volume compared to fixed preparations (32, 33). The preserved shape and volume of the live nuclei enabled visualization of a novel, nuclear mesoscale mode of global chromatin organization, where both active and nonactive chromatin regions are distributed at the periphery of the muscle nucleus, forming a substantial region that is devoid of chromatin in the nucleus interior. We detected comparable peripheral chromatin organization in other tissues of live Drosophila larva, as well as in live human effector T cells and neutrophils, two types of fully differentiated immune cells. This peripheral chromatin architecture was sensitive to the levels of lamin A/C, because overexpression (OE) of lamin A/C resulted in chromatin condensation toward the center of the nucleus. This shift in global chromatin distribution was accompanied by a reduction in nuclear histone 3 lysine 9 acetylated (H3K9ac) density. We further present simulation results of a polymer model, demonstrating that a balance between chromatin association with the lamina and effective chromatin self-attraction can explain the global peripheral chromatin distribution that we observed experimentally. The model predicts that reduced LAD attachment to the lamina leads to chromatin collapse toward the center, consistent with the experimental observation. Our results reveal a novel mode of nuclear organization in fully differentiated live cells sensitive to nuclear volume and lamina composition.
RESULTS
Live imaging of 3D chromatin distribution in intact Drosophila larval muscles reveals novel, nuclear-scale chromatin organization
The current view of higher-order chromatin organization depicts chromatin as distributed throughout the entire nuclear volume (7). So far, most of the studies describing 3D chromatin organization were conducted either on cultured cells or on cells in fixed tissue preparations (3, 8, 19, 21, 34). To study 3D chromatin organization under physiological conditions in a live organism, we have developed a setup to visualize nuclei and chromatin in live, intact Drosophila larva. This setup is based on a minimally constrained device designed for imaging stationary or contractile muscles and their nuclei in an intact Drosophila larva, preserving tissue and nuclear native environment (28). Figure 1A shows a 3D view of a single muscle nucleus, cut through the middle, of live, intact third instar Drosophila larva. The nuclear envelope was labeled with nesprin/klar–GFP (green fluorescent protein) driven by a muscle-specific driver (Mef2-Gal4), and chromatin was labeled by expression of His2B-mRFP (histone H2B–red fluorescent protein) controlled by its endogenous promoter, a common live mark for total chromatin (2, 8, 29, 30).
Fig. 1. Peripheral chromatin organization in live Drosophila larva muscle nuclei, not detected in fixed nuclei.
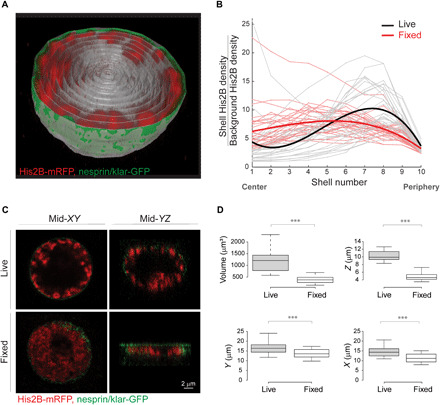
(A) 3D view of a single live muscle nucleus cut through the middle. Chromatin is labeled with His2B-mRFP (red) and nuclear envelope with nesprin/klar-GFP (green). For quantification of radial chromatin distribution, the segmented nucleus is divided into 10 concentric 3D radial shells (gray). (B) His2B fluorescent signal density for each shell, normalized by the background signal density, is plotted from center to periphery. The resulting radial chromatin distribution profile demonstrates peripheral trend in live nuclei (individual nuclei in gray, fitted average in bold black, n = 27), which is significantly different from the more uniform radial chromatin distribution profile in fixed preparations (individual nuclei in light red, fitted average in bold red, n = 30; P < 0.001). (C) Mid-confocal XY plane of a representative nucleus from live, intact larva shows that chromatin is organized at the periphery of the nucleus (top left), whereas in a representative nucleus from a fixed preparation, chromatin is expanded into most of the nuclear space (bottom left). Mid-YZ plane of the live nucleus demonstrates preserved ellipsoid shape and peripheral chromatin (top right), whereas the mid-YZ plane from the fixed nucleus reveals substantial flattening and altered chromatin organization (bottom right). (D) Significant reduction (***P < 0.001) in nuclear volume, as well as in the short, middle, and long nuclear dimensions in fixed preparations (n = 30) compared to live (n = 27).
3D visualization of the live muscle nuclei revealed a peripheral distribution of the chromatin, with a substantial region in the interior of the nucleus that was devoid of chromatin (movie S1). To quantify chromatin distribution along the radial direction, we segmented each nucleus in 3D and divided into 10 concentric shells (Fig. 1A, gray traces). Chromatin density for each shell was calculated from the sum of His2B-mRFP fluorescence intensity, divided by the shell volume. The background His2B-mRFP density was calculated from an additional shell outside of the nuclear envelope. For each nucleus, we generated a radial chromatin distribution profile, by plotting the chromatin fluorescent density, normalized by the background fluorescent density, from the center to the periphery of the nucleus (Fig. 1B). Note that shell 10 was defined as the outer nuclear envelope. To study the robustness of the phenomena, we analyzed the radial chromatin distribution profiles of nuclei from at least three different, randomly chosen muscles along the larva length and from five different larvae (gray traces in Fig. 1B, n = 27). Mean radial chromatin distribution demonstrates a robust peripheral profile for the live nuclei (bold black in Fig. 1B, fitted with third-order polynomial, linear mixed-effect model). Note that a few individual live nuclei also have high chromatin density in the center, in addition to the high peripheral chromatin density. Examination of these few nuclei revealed a chromatin branch extended from one side of the nucleus to the other, but still most of the nucleus center was devoid of chromatin, as demonstrated in fig. S1.
In contrast to the peripheral chromatin distribution that we observed in muscles of live larvae, previous studies from our group and of others, performed on muscle nuclei from fixed Drosophila larvae, have depicted chromatin that is fairly uniformly distributed throughout the entire nuclear volume (32, 33). To investigate this discrepancy, we compared chromatin distribution in muscle nuclei visualized in the live larvae, with that observed in fixed larval preparations, using identical genotype and method of chromatin labeling and similar imaging setup and image analysis. Notably, the radial chromatin distribution profile of fixed nuclei (Fig. 1B, individual nuclei in light red, n = 30, fitted average in bold red) was significantly different from that of the live larvae (black, P < 0.001), indicating a relatively homogenous chromatin distribution throughout the nuclear volume (Fig. 1C).
To reveal possible explanations for the marked difference in global chromatin organization between live and fixed nuclei, we measured nuclear dimensions and volume of live and fixed nuclei. Figure 1C shows a single midsection in the XY plane (left) and a single midsection in the XZ plane (right) of a confocal Z-stack taken from live (top) or fixed (bottom) nuclei of comparable size. In the XY plane, chromatin distribution of the live nucleus appeared peripheral, whereas in the fixed nucleus, the chromatin distributed throughout most of the nuclear volume, excluding only the region occupied by the nucleolus. Whereas the live nucleus preserved its volume and ellipsoidal shape, the fixed nucleus exhibited substantial flattening, forming a disk-like shape (33), as depicted in the XZ plane. Figure 1D summarizes the reduction in nuclear volume and nuclear dimensions upon fixation. On average, there was a 3.1-fold decrease in nuclear volume from 1183.1 μm3 in live nuclei to 380.8 μm3 in fixed nuclei (P < 0.001), mostly due to a 2.1-fold reduction in the Z-axis diameter, from an average of 10.3 μm in live nuclei to 4.9 μm in fixed nuclei (P < 0.001). Note that the Z-axis dimension is perpendicular to the muscle fiber axis. There was also a smaller but significant 1.3-fold decrease in the X and Y diameters of fixed nuclei, from 14.4 and 16.3 μm in live nuclei to 11.2 and 13.7 μm in the fixed nuclei, respectively (P < 0.001). We excluded a possible effect of submerging the larvae in water before imaging, because similar peripheral chromatin organization was observed in live larvae imaged immediately following their collection from the vials (movies S2 and S3).
It has been reported that tissue fixation can cause cellular and nuclear volume reduction due to dehydration of the sample (24, 35). We, therefore, speculate that Drosophila larval muscle nuclei appeared flat in fixed preparations due to dehydration. The spherical shape of the nuclei observed in the live setup allowed us to distinguish between the chromatin-containing compartment localized at the periphery of the nucleus and the chromatin-devoid compartment in the interior of the nucleus.
To further support the observation of peripheral chromatin organization in the live, intact setup, we used histone H2A variant (His2Av)–GFP as an additional, commonly used marker for total chromatin labeling (31). Figure 2A shows a mid-XY plane of a nucleus from live larval muscle, colabeled with His2Av-GFP (A) and His2B-mRFP (A′). The merged view (A″) indicates high colocalization of both histone tags, providing additional support to the hypothesis that the observed peripheral chromatin distribution is independent of the labeling type.
Fig. 2. Peripheral chromatin organization observed with His2Av and analysis of chromatin and nucleolus volumes relative to nuclear volume.
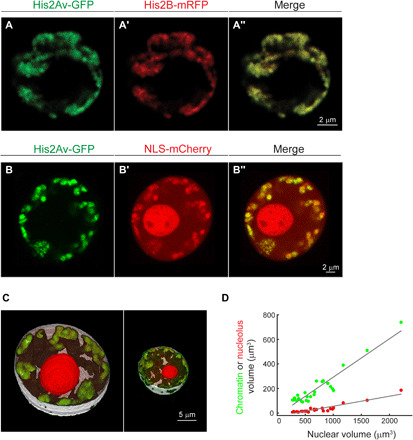
(A, A′, and A″) Colabeling of a live muscle nucleus with two independent histone labels, His2Av-GFP (A) and His2B-mRFP (A′), shows high colocalization (A″), supporting peripheral chromatin organization. (B, B′, and B″) Live muscle nucleus colabeled with His2Av-GFP (B) and NLS-mCherry (B′). (C) Representative 3D segmentation of the nuclear volume (white), chromatin volume (green), and nucleolar volume (red) in large or small nuclei. (D) Linear scaling between chromatin (green) and nucleolar (red) volumes with the corresponding nuclear volume (R2 = 0.89 and R2 = 0.86, respectively, linear mixed-effect model). Chromatin and the nucleolus occupy 31 and 8% of the nuclear volume, respectively, leaving 61% remaining nucleoplasm space.
The nucleolus is a membrane-less, phase-separated organelle within the nucleus. To address whether a large nucleolus compartment excludes chromatin from the central region of the nucleus to the periphery, we expressed mCherry fused to nuclear localization signal (NLS) in muscles, together with His2Av-GFP (Fig. 2, B and B′, respectively). The NLS-mCherry labeled the entire nucleoplasm with a denser accumulation inside the nucleolus (verified by colabeling with fibrillarin; fig. S2). The merged view in Fig. 2B″ demonstrates a large chromatin-devoid region in the nucleus interior that did not overlap the nucleolus domain. We further quantified the nucleolus and chromatin volumes as a fraction of the total nuclear volume of at least three different, randomly chosen muscles, along the larval length, from seven different larvae (n = 25). Figure 2C shows representative image of 3D segmentation of the total nuclear volume (white), chromatin volume (green), and nucleolus volume (red), in either a large (left) or small (right) muscle nucleus. Note that Drosophila muscle nuclei are polyploid, with higher DNA content in larger nuclei (33). Figure 2D shows the dependency of either chromatin or nucleolar volumes on the entire nuclear volume, demonstrating that both structures scale linearly with the nuclear volume. Our measurements indicated that chromatin occupies on average 31% of the nucleus (R2 = 0.89, linear mixed-effect model), the nucleolus occupies on average 8% of the nucleus volume (R2 = 0.86, linear mixed-effect model), and both structures are barely contacting each other. Collectively, these data suggest that the chromatin in live Drosophila larva muscle nuclei is not squeezed passively to the periphery by the nucleolus.
Compartmentalization into peripheral chromatin layer is observed in other larval tissues and in live human leukocytes
To test the generalization of our observations, we examined 3D chromatin distribution in other tissues of the live, intact Drosophila larva, as well as in primary human leukocytes. Figure 3A demonstrates a representative nucleus from the Drosophila larva epidermal layer, labeled with His2B-mRFP. Despite the flat nuclear morphology typical to the epidermal cells, peripheral organization of the chromatin was detectable from a middle section of all three imaging planes, when imaged within the live, intact larva. Similarly, peripheral chromatin distribution was observed in the salivary gland nuclei labeled with His2Av-GFP (Fig. 3B), where the high polyploidy results in large nuclei and thick chromatin fibers (31). Radial chromatin distribution profiles for the epidermis and salivary gland nuclei are shown in Fig. 3 (A′ and B′, respectively). Similar to the muscle nuclei, we imaged the epidermis and salivary gland nuclei in the live setup, in larvae labeled with NLS-mCherry and His2Av-GFP, to visualize the nucleolus with respect to the peripheral chromatin. In both tissues, the nucleolus does not occupy the entire chromatin devoid region in the center of the nucleus (fig. S3). Although nuclei of the Drosophila muscle, epidermis, and salivary gland tissues are widely studied, we report here a high-resolution analysis of the native 3D chromatin organization that reveals consistent peripheral chromatin distribution in these three tissues that cannot be explained by passive chromatin pushing to the periphery by the nucleolus.
Fig. 3. Peripheral chromatin distribution in additional larval tissues and in non-Drosophila models.
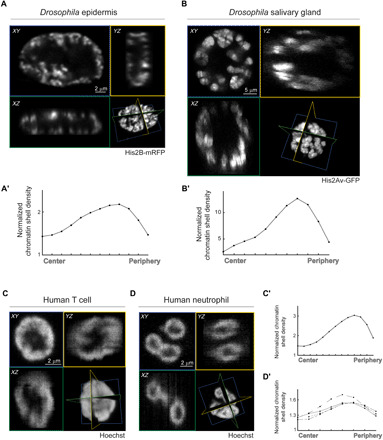
Mid-XY (blue), -XZ (green), and -YZ (yellow) sections of Drosophila larva epidermal nucleus labeled with His2B-mRFP (A), salivary gland nucleus labeled with His2Av-GFP (B), human T cell nucleus (C), and human neutrophil nucleus (D), labeled with Hoechst. (A′ to D′) His2B or Hoechst fluorescent signal density for each shell, normalized by the background signal density, is plotted from center to periphery for each of the depicted nuclei. For the human neutrophile, radial profile is presented separately for each lobe.
Further, to rule out the possibility that peripheral chromatin distribution is a unique phenomenon of the differentiated Drosophila larval tissues, we analyzed live nuclei of freshly isolated blood-derived human neutrophils and cultured effector T lymphocytes, two types of fully differentiated leukocytes. Both types of immune cells were labeled with the nuclear dye Hoechst under conditions validated to retain the viability, motility, and nuclear dynamics (36). The native environment and nuclear shape of the blood circulating T cells and neutrophils are well preserved when imaged live in suspension. The global chromatin organization pattern of the T cells was reported to be modified with developmental stages, to support functional requirements such as transmigration and altered gene expression (37). Here, we demonstrate that effector T cells exhibited typical spherical shape with peripheral distribution of labeled DNA (Fig. 3, C and C′). The peripheral chromatin organization could not result from large nucleoli, which were previously described to only fill about 8 to 10% of the nucleus size (38). Furthermore, we demonstrate that live neutrophils freshly isolated from human blood, exhibiting typical multilobulated nuclear shape, also depicted peripheral arrangement of their labeled DNA, preserved within each lobe (Fig. 3, D and D′). Notably, neutrophils nucleoli have extremely small volume and are localized to the periphery close to the nuclear envelope (39). In summary, these findings suggest that compartmentalization of the nucleus into peripheral chromatin layer is not a tissue-specific or organism-specific phenomenon.
Chromatin localized at the nuclear periphery contains active chromatin regions
A nonrandom higher-order chromatin organization is a hallmark of the interphase nucleus 3D structure (9, 13). With respect to the radial distribution, it is well established that the nuclear periphery is enriched with condensed heterochromatic regions of the genome and that LADs, representing DNA sequences that interact with the nuclear lamina, are largely transcriptionally repressive (11, 12, 40). The nucleus interior, on the other hand, has been reported to contain gene-rich chromatin, which is often actively transcribed (7). To rule out the possibility that the peripherally organized chromatin observed in our study represents mostly dense heterochromatin, while more open, active chromatin in the nucleus interior was undetected by our imaging approach, we colabeled nuclei with a live tag for active chromatin. Sato et al. (41) have developed a fluorescently labeled modification-specific antibody (mintbody), by fusing enhanced GFP (EGFP) to a single-chain variable fragment antibody with high specificity to H3K9ac, which allows in vivo tracking of active chromatin domains with minimal interference to cell function. Muscle nuclei of live Drosophila larvae colabeled with H3K9ac-EGFP and His2B-mRFP were imaged as described above, to identify the active chromatin regions with respect to the peripherally distributed chromatin compartment (mid confocal planes are shown in Fig. 4, A and A′, respectively). In contrast to the His2B-mRFP signal, which shows stronger fluorescence within dense chromatin regions, the H3K9ac-EGFP is expected to exhibit higher signal intensity within the less dense, open chromatin regions. The merged image in Fig. 4A″ (and movie S4) shows that active chromatin regions were distributed in the periphery of the nucleus, with a spatial pattern similar to that of total chromatin. The dashed boxed region of Fig. 4A″ is enlarged in Fig. 4 (B and B″) and highlights the inverse pattern of both labels, with respect to signal intensity. The filled white arrow points to the large, repressed chromocenter of the nucleus with a strong His2B-mRFP signal and lacks the H3K9ac-EGFP signal of the active chromatin. The empty white arrow points to one of the active regions with diffuse chromatin indicated by weak His2B-mRFP fluorescence but strong H3K9ac-EGFP signal intensity.
Fig. 4. Live labeling of active chromatin regions shows spatial overlap with His2B-mRFP–labeled chromatin compartment at the nuclear periphery.
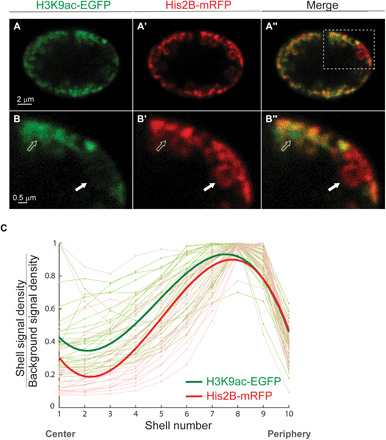
(A) H3K9ac-EGFP mintbody expressed in the live larva muscle nucleus demonstrates peripheral distribution of the euchromatin, similar to the His2B-mRFP spatial distribution of the total chromatin. (A′) Merged image of active and total chromatin (A″); boxed area is enlarged in the bottom (B, B′, and B″). Solid white arrow points to a dense heterochromatin with strong His2B-mRFP signal (B′) and weak H3K9ac-EGFP (B). Hollow arrow points to an active region with strong H3K9ac-EGFP signal and diffused chromatin emphasized by weak His2B-mRFP. These regions coexist in the same radial layer but at different positions perpendicular to the radial direction. (C) H3K9ac (light green) and His2B (light red) signal density for each shell, normalized by background signal density, is plotted from center to periphery. Each light line represents a single nucleus, with signal normalized to maximum value (n = 27). Bold lines represent the fitted average, with peak signal at the periphery of the nucleus for both active and overall chromatin.
Figure 4C compares the 3D radial fluorescent signal distribution profiles for the active, H3K9ac (light green), and total chromatin, His2B (light red), in individual nuclei (n = 27). Each radial profile was scaled to its maximum value to simplify the comparison of the profile shapes. The fitted average radial distribution profiles indicated peripheral distribution of the active chromatin regions (bold green), similar to that of total chromatin (bold red). Statistical analysis comparing the shape descriptors of the profiles showed no significant differences between the radial distributions of the active relative to the total chromatin. The lack of a strong active chromatin signal in the nucleus interior reduces the possibility of undetectable, decondensed chromatin, which could possibly be the case when imaged solely by His2B-mRFP labeling. Moreover, our results suggest that the active euchromatin is not separated from the heterochromatin along the radial direction, i.e., localized internally to the heterochromatin, but rather forms hubs of varying sizes along the circumference of the nuclear lamina, within the peripheral chromatin layer. This is in agreement with the observations in nuclei of fixed eight-cell bovine embryo demonstrating no radial segregation between active and repressed regions in the peripherally organized chromatin (10) and with a recent study showing that active chromatin forms spatially segregated clusters, mostly excluded from larger dense, heterochromatic clusters (34).
Lamin C OE disrupts chromatin localization at the nuclear periphery
Drosophila is the only known invertebrate model expressing both types of the human lamins (i.e., A/C and B types), along with genome representation of additional nuclear envelope components such as the LINC (linker of nucleoskeleton and cytoskeleton) complex, LEM (LAP2, emerin, MAN1) domain, and BAF (barrier to autointegration factor) proteins. Drosophila lamin C, which is the homolog of mammalian A/C type lamin, displays tissue-specific expression levels in fully differentiated cells (42, 43). Mutations in lamin A/C have been associated with aberrant chromatin organization and global detachment of chromatin from the nuclear lamina in a wide range of model organisms and tissues (15, 18, 19). Therefore, we used the genetic advantages of the Drosophila to study the effect of OE of lamin C on the peripheral chromatin localization that we observed in live larva muscles. Toward that end, lamin C–GFP was expressed in muscles using the Mef2-Gal4 muscle driver in combination with the ubiquitously expressed His2B-mRFP, allowing live imaging of the nuclear envelope and chromatin (Fig. 5A). We did not observe in lamin C–GFP OE larvae altered development or apparent viability phenotypes. Figure 5A (and movie S5) shows a 3D view of a single muscle nucleus with lamin C OE, cut through the middle, in our live, intact setup. The chromatin detached from the nuclear lamina and was condensed toward the center. The nucleus in Fig. 5A is shown at a single mid-XY plane in Fig. 5B (nucleus 1), along with two additional nuclei (nuclei 2 and 3), that represent variable phenotypes of global chromatin distribution obtained following lamin C–GFP OE. This phenotypic variability was presumably caused by differential levels of the Mef2-Gal4 driver in different muscles. Significantly, a general trend of disruption to the peripheral chromatin localization and a collapse of chromatin toward the nucleus interior are demonstrated in Fig. 5C showing radial His2B-mRFP density profiles for the control nuclei (gray; n = 27) and nuclei with lamin C–GFP OE (light red; n = 23). Each radial profile was scaled to its maximum value to aid with shape comparison. Despite the variability in radial chromatin distribution within the lamin C–GFP OE group, the fitted average radial density profile (bold red) demonstrates a substantial shift in chromatin density from the periphery toward the center, compared with control nuclei (bold black). Statistical analysis comparing the shape descriptors of control and lamin C–GFP OE profiles showed significant differences in their radial chromatin distribution (P < 0.001).
Fig. 5. Lamin A/C OE in the Drosophila larva muscle disrupts peripheral chromatin localization, driving chromatin condensation toward the center of the nucleus.
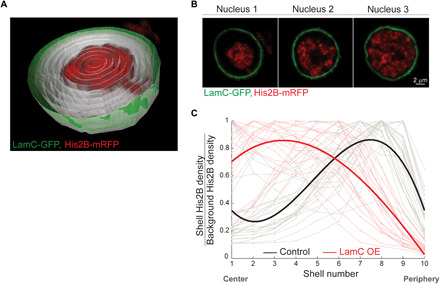
(A) 3D view of a live nucleus, overexpressing lamin C, cut through the middle. Chromatin is labeled with His2B-mRFP and nuclear envelope with lamin C–GFP. The segmented nucleus is divided into 10 concentric 3D radial shells (gray) for quantification of radial chromatin distribution. (B) Variable phenotypes of chromatin distribution in lamin C OE nuclei from different muscles. (C) His2B signal density for each shell, normalized by background signal density, is plotted from center to periphery. Each individual line was normalized to its maximum value. The resulting radial chromatin density profile demonstrates the trend toward central localization in the live lamin C OE group (individual nuclei in light red and fitted average in bold red; n = 23), compared with the peripheral trend in live control nuclei (individual nuclei in gray and fitted average in bold black; n = 27).
Together, our results demonstrate that elevated lamin C levels in the fully differentiated larva muscle caused detachment of chromatin from the lamina and loss of peripheral organization, in agreement with previous studies demonstrating that both up- and down-regulation of lamin C levels change the 3D chromatin organization (14, 18). Moreover, the observed central chromatin in the live lamin C OE further supports the ability of our imaging setup to detect chromatin in the interior of the nucleus, when it is present.
Epigenetic consequences of altered global chromatin organization upon lamin A/C OE
How structural changes in 3D chromatin organization regulate nuclear function is an exciting open question. Specifically, up-regulation and down-regulation of lamin A/C have been linked to transcriptional regulation via alteration of chromatin-lamina tethering (16, 18). We investigated the functional consequences of altered global 3D chromatin organization in the lamin C–up-regulated nuclei, by quantitative immunofluorescence measurements of H3K9ac and H3K9me3 (histone H3 lysine 9 tri-methylated) epigenetic marks that have been implicated in regulation of transcription. We quantified the levels of active and repressive epigenetic marks, DNA content, and nuclear volume from muscles 6 and 7 in the fixed preparation of Drosophila third instar larvae. When lamin C–GFP was driven fully to the muscle with Mef2-Gal4 driver line, we identified DNA leakage from the nucleus. We therefore performed a milder lamin C up-regulation in the muscle, only during the second and third instar larva stage, using temporal induction of the Mef2-Gal4 (in combination with Tub-gal80ts), where cytoplasmic DNA levels were comparable with the control group, as observed by Hoechst labeling in the fixed preparations (Fig. 6, A and B). Disruption of peripheral chromatin organization with temporal lamin C OE was confirmed by live imaging (fig. S4). Immunofluorescence analysis revealed significantly reduced levels of the active, H3K9ac chromatin mark relative to control (Fig. 6A) and preserved levels of the repressed, H3K9me3 chromatin mark (Fig. 6B), where the Hoechst levels were unchanged between the groups. H3K9ac density for each nucleus was calculated from the total nuclear H3K9ac fluorescence intensity, divided by nuclear volume. This volume normalization was performed to account for the large variation in nuclear volumes in the lamin C OE group observed between and within muscles (Fig. 6, C and D). Quantification of the total nuclear fluorescence intensity versus nuclear volume demonstrates 56% reduction of H3K9ac density in the temporal lamin C–GFP OE nuclei (Fig. 6C, red; n = 288) compared to control (black; n = 281), confirmed by mixed linear model analysis (P < 0.001). Similar analysis for H3K9me3 density showed no significant difference (P = 0.42) between the lamin C OE (Fig. 6D, red; n = 181) and control groups (black; n = 171). Mean DNA content and nuclear volumes were not significantly different between the control and lamin C OE groups (P > 0.1). We further verified that OE of a fluorescent label per se in the muscle nuclei has no effect on the H3K9ac fluorescence by quantitative immunofluorescence of H3K9ac in Mef2-Gal4/+ versus Mef2-Gal4 < NLS-mCherry groups (fig. S5). Overall, our findings suggest that a peripheral to central shift in chromatin distribution, caused by conditional and temporal up-regulation of lamin C, is associated with reduced levels of H3K9ac, an active chromatin mark, and support the link between 3D mesoscale genome structure and transcriptional regulation.
Fig. 6. Temporal lamin A/C OE associates with decreased density of the active H3K9ac epigenetic mark.
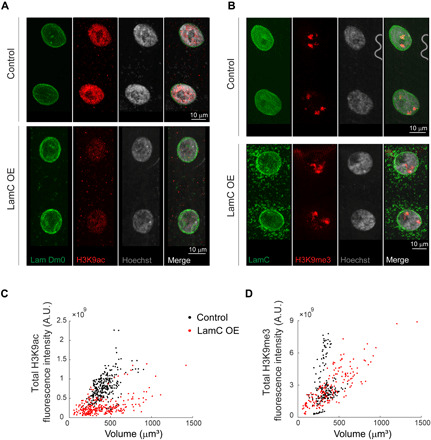
Muscle nuclei labeled with lamin Dm0, H3K9ac, and Hoechst show reduced intensity of the active mark in the temporal lamin C OE group (A). Muscle nuclei labeled with lamin C, H3K9me3, and Hoechst show preserved intensity of the repressive mark between the control and temporal lamin C OE groups (B). Quantification of the total nuclear fluorescence intensity versus nuclear volume demonstrates reduced H3K9ac density (slope; P < 0.001) in the temporal Lamin C OE group (red; n = 288) versus control (black; n = 281) (C) and preserved H3K9me3 density (slope; P = 0.42) between temporal lamin C OE (red, n = 181) and control groups (black; n = 171) (D). Nuclei for each group were pulled from 5 to 6 different larvae and 8 to 10 different muscles in each larva. A.U., arbitrary units.
A computational chromatin polymer model to describe the shift from peripheral to central chromatin organization
Computational modeling is a powerful tool to study spatial genome organization and has the potential to suggest mechanistic insights and governing principles of the 3D chromatin organization, based on physical and biological processes and experimental data (44). To gain mechanistic insights into the observed shift in nuclear-scale chromatin distribution from the periphery to the interior, we performed simulations of a polymer-based chromatin model.
The computational model approximates chromatin as a semiflexible, bead-spring polymer (4, 45–47) and shown schematically in Fig. 7A. Each spherical bead has a diameter of 10 nm that accounts for 600 base pairs (bp) of DNA and coarse grains over three nucleosomes (45). Neighboring beads along the chain are connected via springs (not depicted in the figure), and each bead interacts with any other bead in the chain with a repulsive, Lennard-Jones potential (4, 45) that is repulsive at very short range of about one bead diameter and can be attractive in the range of a few bead diameters. We simulated two cases: a case of self-attraction, where the interaction is attractive in the range of one to about two bead diameters, and a case of no self-attraction, where the potential is zero for particle separations of more than one bead diameter (48). The former case represents generically “self-attracting” chromatin, without the need to specify the small molecular actors, whereas the latter case results in an “excluded volume random walk” of chromatin in an aqueous solvent. This bead-spring model behaves as a flexible polymer on large length scales. To account for the local rigidity in the polymer, we added angular interactions between beads (bending energies) so that our model polymer has a persistence length of 1.2 kb, consistent with a previous estimate of the persistence length for the interphase chromosome as 1 to 2 kb (49). The overall polymer length included 37,333 beads, representing the section (22.4 Mb) of Drosophila chromosome X (ChrX) for which the LAD and non-LAD regions are known. The nucleus was modeled as a hollow spherical shell with a thickness of 10 nm (representing the thickness of the nuclear lamina) and beads that comprise this shell account for the nuclear lamina layer, which interacts attractively with LADs of the chromatin (15). The volume fraction of chromatin (and its strongly bound aqueous/small protein molecules) in the nucleus was set to 0.3 based on our experimental observation and the literature (50, 51).
Fig. 7. Simulations of 3D chromatin polymer model suggest governing principles for global chromatin organization and its dependence on LAD–nuclear lamina interactions.
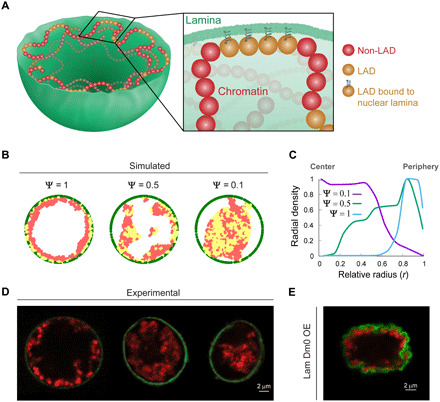
(A) Chromatin is described by a semiflexible, bead-spring polymer model that is confined to a sphere, with non-LAD (red) and LAD (yellow) chromatin beads. Yellow LAD beads bound to the lamina are represented with a spring. (B) Simulated results of chromatin concentration maps (LAD in yellow and non-LAD in red) confined within the nuclear lamina (green) for decreasing (left to right) fraction of LADs bound to nuclear lamina (ψ). Equatorial plane of the spherical model nucleus is shown. (C) Mean simulated radial chromatin density profiles describe a shift in chromatin distribution, from periphery to the center, with ψ decrease from 1 to 0.5 to 0.1. (D) Mid-XY planes of experimental live larva muscle nuclei show similar trend in chromatin shift from peripheral distribution (left; control), to more central chromatin distribution in lamin C OE (middle and left; intermediate and strong phenotype, respectively). (E) Experimental OE of lamin Dm0–GFP in larval muscle does not show chromatin collapse phenotype and preserves peripheral chromatin organization despite substantial deformations in the nuclear lamina.
The polymer comprises two types of beads: lamin-associating and nonlamin-associating (red and yellow in Fig. 7A, respectively). When a lamin-associating bead is within a distance of 25 nm of a lamin bead, a strong bond is formed (represented by a spring in Fig. 7A). The LAD locations were based on experimentally sequenced data on Drosophila ChrX, which also suggests that about 48% of the chromatin consists of LADs (52). While we have used numerical values for relevant variables such as bead size, chain length, etc., the general trends shown below did not depend on these details and were mostly sensitive to the values of chromatin volume fraction and the fraction of LAD domains interacting with the lamina, as discussed below.
Simulations were performed with coarse-grained molecular dynamics using LAMMPS software (53). When the simulation begins, all the chromatin beads are distributed randomly in the interior of the lamin confined sphere (movie S6). To model changes in the interactions of LADs with the nuclear lamina due to genetic changes of lamin proteins, as well as different fractions of LADs that may be applicable to chromosomes in other organisms, we defined a parameter ψ as the fraction of LAD beads in chromatin that can form bonds with the nuclear lamina beads. Experimental observations upon lamin C OE suggest weaker chromatin–nuclear lamina interactions. We thus allowed for the possibility that in some systems, not all LADs form bonds with the nuclear lamina, by considering general values of 0 < ψ ≤ 1.
Figure 7B shows typical simulated results of the chromatin concentration map (yellow for LAD and red for non-LAD beads), confined within the nuclear lamina (green), in the equatorial plane of the spherical model nucleus. The fraction of bound LADs, ψ, decreases from left to right, from ψ = 1 representing all estimated 48% LADs in ChrX as bound to ψ = 0.5 and ψ = 0.1. Decreased fraction of bound LADs resulted in a shift in the peak average radial chromatin density profile, from the periphery toward the center (Fig. 7C). In parallel, Fig. 7D shows the experimental mid-XY planes of nuclei from control larva muscle (left), lamin C OE with intermediate phenotype (middle), and lamin C OE with strong phenotype (right). The simulated trends in chromatin concentration profiles and their central shift with decreased LAD binding closely resemble the experimental results. Notably, experimental OE of lamin Dm0–GFP (lamin B–type homolog) in the larval muscles, in contrast to lamin C OE, preserves peripheral chromatin organization (Fig. 7E) and does not result in chromatin detachment from the lamina and collapse to the center of the nucleus, supporting the dominant-negative effect of lamin C on lamina-LAD interactions.
While the simulated results are not designed to mimic the details of the experiments, the ability to achieve peripheral chromatin organization at equilibrium, with reasonable biophysical parameters, suggests possible governing principles for 3D nuclear-scale chromatin organization in live nuclei related to the competition between lamin attraction, confinement and polymer entropy, and self-attraction of the chromatin. Moreover, the qualitative agreement of the simulations shown in Fig. 7 with the experiments suggests that self-attraction must be included in the model. If the chromatin interactions were purely repulsive (excluding the volume of beads that are close in 3D space), then the chromatin would uniformly fill the nucleus for volume fractions of 0.3 (fig. S6). We further performed a set of simulations for the full length of ChrX (40 Mbp). Because experimental LAD data are available only for the largely euchromatic 22.4-Mbp region simulated above, we accounted for two cases of the remaining, mostly heterochromatic region: (i) All the remaining sequence is LAD. (ii) None of it is LAD. We found peripheral organization of chromatin for case 1 (fig. S7). Therefore, if the remaining region includes a significant amount of LAD domains, then we still expect peripheral organization. The peripheral chromatin distribution in live control nuclei might therefore arise from strong chromatin interaction with the nuclear lamina, coupled with high chromatin-chromatin affinity (Fig. 7A). When LAD–nuclear lamina interactions are decreased, the balance is shifted, and the strong chromatin-chromatin interaction gradually drives chromatin toward the center of the nucleus.
DISCUSSION
Robust chromatin organization within the nucleus has been linked to the control of gene transcription (44), yet information about the 3D chromatin distribution under physiological conditions in a living organism is limited. Here, we have revealed the 3D distribution of chromatin in muscle nuclei of live Drosophila larvae at high resolution. Our analysis demonstrates a novel mode of chromatin organization at the nuclear mesoscale of fully differentiated cells, wherein a high density of chromatin, including both active and repressed regions, is distributed at the nuclear periphery. Chromatin density decreases sharply as a function of distance to the center of nucleus, giving rise to a chromatin-devoid region at the center of the nucleus. We show that this novel mode of nuclear-scale chromatin architecture was detectable only in unfixed tissue within a live organism and preservation of nuclear volume was critical for that. We show that the peripheral chromatin organization was sensitive to lamin A/C levels and contributed to maintaining adequate levels of the active epigenetic mark H3K9ac. Model simulations show that a peripheral chromatin organization was obtained when LADs and chromatin attractive interactions dominate the entropic tendency to distribute the chromatin uniformly throughout the nucleus. Peripheral chromatin organization was observed not only in larval muscles but also in other live Drosophila larval tissues, as well as in live human effector T lymphocytes and neutrophils, suggesting an evolutionary conserved mode of chromatin organization at the nuclear periphery.
A key experimental feature that allowed us to view nuclear partitioning was preservation of nuclear volume. Previous global chromatin organization studies were performed primarily on fixed preparation (6, 34) or on live flat cultured cells (2, 29, 30), which might obscure the native spatial chromatin distribution. Specifically, prevailing fixation methods may cause cell and nuclear volume changes (8, 24), often at the Z axis (54), while the dimensions in the imaging X-Y plane are less affected, possibly due to robust nuclear-cytoskeletal interactions in this plane. Furthermore, cells grown on rigid surfaces are relatively spread and flat, with 40 to 50% less water content relative to cells grown on soft matrices (24, 25). As nuclear and cytoplasmic volumes were shown to be interconnected and adapt to external signals, through tight control of their mutual water content (23, 25, 55), preservation of nuclear volume within the tissue (as in our experimental system) appears to be critical. This was also deduced from our model simulations where nuclear volume was found to be directly linked to chromatin mesoscale organization. Significantly, changes in nuclear volume and shape were shown to have critical functional effects such as altered chromatin organization (22), gene expression (56, 57), and DNA synthesis (55).
Although a few previous studies already reported specific cases of peripheral chromatin distribution, it was considered as a transient stage that correlated with the differentiation state of the cell, for example, in the transition from 8- to 20-cell stages of in vitro fertilized bovine embryos (10), T cell differentiation (37), or during myeloid cell differentiation (58). The reported alternations in chromatin organization were accompanied by volume changes; however, the link between these two parameters was not explored. Our data suggest that chromatin peripheral organization is common to a wide range of differentiated tissues. Furthermore, chromatin to nucleus volume fraction is a major regulator of chromatin mesoscale organization, as indicated by our experimental and model results. Drosophila myonuclei are polyploid, where DNA copy number was shown to correlate with nuclear size (33). Here, we demonstrate a constant chromatin/nucleus volume ratio (0.31) for a wide range of muscle nuclei volumes suggesting that it is a tightly regulated feature. A constant chromatin volume fraction in polyploid nuclei is not unique to larval myonuclei and was also found in Drosophila embryos (59) as well as in plants (60) and fish (61).
A recent study challenged the prevailing view that repressed and active chromatin compartments partition radially within the nuclear volume, i.e., repressed chromatin predominates at the nuclear periphery and active chromatin prevails near the nuclear center (34). In agreement with this report, we did not observe a clear radial separation between active and repressed chromatin distributions and also detected active chromatin at the nuclear peripheral compartment.
The nuclear lamina controls radial chromatin organization by its dynamic tethering of chromatin to the nuclear periphery through LADs (13, 14). Because the nucleus is thought to be a mechanosensitive organelle (62–64) in which the close proximity of the chromatin to the nuclear envelope might sensitize the chromatin to both mechanical inputs and changes in the nuclear lamina composition, a peripheral chromatin organization might couple between mechanical inputs and transcriptional regulation. This is especially relevant for fully differentiated nondividing cells, in which the program of gene transcription has been well established; however, because our analysis did not include undifferentiated cells, it remains unclear whether chromatin peripheral organization also takes place in all cell types. In addition, the global chromatin detachment from the periphery and its condensation toward the center that was observed upon elevated lamin C levels agrees with previous reports on reduced chromatin-lamina tethering following up- or down-regulation of lamin C, as well as with observations in cells with laminopathy-associated mutations. These chromatin structural changes were accompanied by chromatin decondensation and were linked to transcriptional regulation (15–19). Our data support a link between 3D chromatin distribution and transcriptional regulation as we observed a significant reduction in the active, H3K9ac levels in the nuclei overexpressing lamin C–GFP. The centrally located, condensed chromatin might prompt reduced availability to enzymes (such as acetyltransferase), leading to a net decrease in acetylated H3K9, predicted to induce reduction in gene transcription. Chromatin regions that are already densely packed (such as repressed chromatin) might be less affected from the collapse of chromatin to the center as observed with the preserved H3K9me3 levels in nuclei overexpressing lamin C–GFP.
The simulations of chromatin distribution dynamics involved only a small number of parameters extracted from the literature, including the percentage of LADs per chromosome, the tendency of chromatin to effectively self-attract (48) and the extent of nuclear confinement. The similarity between the simulation and the experimental results suggests that these are among the most critical factors in determination of the partitioning of chromatin on the nuclear scale. It further suggests that a balance between chromatin-lamina interactions through LADs, entropy, and chromatin self-association may represent the major driving forces for the nuclear partitioning observed experimentally. Our physical model suggests that the experimentally observed mesoscale chromatin collapse from the periphery to the nucleus interior upon lamin C OE might be driven by reduced lamina-LAD interactions. It has been shown that both lamin A/C and B types compete with one another on similar LAD-binding sites (40, 65). Although lamin B contacts with LAD exclusively at the nuclear periphery because its localization is restricted to the nuclear membrane due to its farnesylation, lamin A/C was shown to interact with LADs at the nuclear periphery and at the nucleoplasm. Here, we demonstrate that in contrast to chromatin collapse following lamin C OE, lamin Dm0 (lamin B homolog) OE did not result in chromatin detachment from the lamina and the peripheral chromatin organization was preserved. This might be due to the nucleoplasmic localization of lamin C or due to distinct association of each of the lamin types with other nuclear membrane components affecting the recruitment and preservation of chromatin-lamina interactions at the periphery.
In summary, our study reveals a novel mode of nuclear mesoscale chromatin organization in fully differentiated cells in which chromatin density is high at the nuclear periphery and undetectable in the nuclear center, creating an effectively central chromatin-devoid region. Simulation of chromatin organization based on LAD binding and chromatin self-attraction recapitulated the experimental observations, supported that preservation of nuclear volume is critical and predicted that changing each of these parameters may disrupt peripheral chromatin density profile. Experimental OE of lamin C disrupted the peripheral chromatin organization, supporting the basic assumptions of the model, and had functional consequence of reduced active epigenetic mark.
MATERIALS AND METHODS
Fly stocks and handling
The following fly stocks were used: ubi-H2B-mRFP/CyO; Dr/TM6B (66), CyO/Sp; UAS-laminC-GFP (43), UASp-mintbody-EGFP; Pr,Dr/TM3,Sb,Ser (41), His2Av-GFP (FBst0005941), GAL4-Mef2.R (FBst0027390), UAS-mCherry-NLS (FBst0038424), UAS-klar-GFP (67), arm-gal4 (FBst0001560), Sallimus (SLS)–GFP (obtained from B. Bullard, Department of Biology, University of York), Hand-Gal4 (68), Tub-Gal80ts; Mef2-Gal4 (obtained from F. Schnorrer IBDM, Marseille), and UAS-lam-GFP (FBst0007377). All crosses were carried and maintained at 25°, 18°, or 29°C and raised on cornmeal agar.
The minimal constraint device for imaging live intact Drosophila larvae
For imaging live nuclei in their intrinsic environment, a minimal constraint device for Drosophila larvae was designed in our laboratory, to be placed on top of a confocal microscope stage, as previously described (28). Briefly, the center of the device is a thin plastic bar positioned in a larger frame, with a groove crossing the frame and bar from side to side. The larva is placed in the groove, and glass capillaries (Drummond Scientific Company, PA, USA, catalog no. 1-000-0010, outside diameter 0.026″ and inside diameter 0.0079″), which are aligned with the larval body, are glued to its head and tail (Gorilla super glue, Gorilla Glue Company, Ohio, USA). The device base with a high-precision coverslip glass in it (catalog no. 0107222, thickness no. 1.5H, Pauk Marienfeld GmbH & Co. KG) is placed on top of the larva in the bar, and all the parts of the system are inverted together as a unit. To keep the larva moist during the experiment, it is enclosed with alginate hydrogel made by polymerizing a solution of 4% alginate (catalog no. 180947, Sigma-Aldrich) dissolved in 0.9% saline (NaCl; catalog no. 0277, J.T.Baker) with a solution of 0.8 M CaCl2·2H2O (catalog no. 2382.0.0500, Merck).
Live preparations
Third instar, wandering larvae were selected for imaging. For stationary, 3D imaging, before placement of the larvae in the device, it was immersed in water for ~4 hours to decrease its movement (larval movement could be restored by exposure to air). For each larva, at least three nuclei were imaged from randomly chosen muscles along the entire larval body. Before and after obtaining full Z-stacks of the nucleus, a single mid-XY plane and mid-YZ planes were taken to verify that the nucleus did not deform or move during image acquisition. To verify peripheral chromatin organization in dynamic larvae that were not immersed in water before imaging, we performed time-lapse imaging (spinning disk confocal) on wandering larvae imaged immediately following their collection. Dynamic larval contractions in this case did not allow 3D imaging; nevertheless, peripheral chromatin organization was observed with 2D time lapse in multiple muscle nuclei when the imaging plane is in the middle of the nucleus (movie S2) and when manually moving through the height of the nuclei (movie S3).
Fixed preparations
For comparison of chromatin organization between live and fixed samples, the same crosses were used, with no antibody staining of the fixed samples. Third instar, wandering larvae were selected and dissected in phosphate-buffered saline (PBS), as previously described (67). Paraformaldehyde (4% from 16% stock of electron microscopy grade; Electron Microscopy Sciences, 15710) was used for fixation. Specimens were fixed for 20 min, washed several times in PBS, and mounted in Shandon Immu-Mount (Thermo Fisher Scientific).
For quantitative immunofluorescence experiments, temporal expression of UAS-laminC-GFP was performed using a combination of Mef2-Gal4, tubGal80ts drivers as follows: Embryo collection was performed at 25°C for 6 hours, then embryos were transferred to permissive temperature of 18°C up to first instar larval stage, and then larvae were transferred to restrictive temperature of 29°C up to early third instar stage. Control and mutant larvae were staged and grown in parallel time intervals and dissected and fixed as described above. Fixation and antibody staining of both groups were performed at the same tube marking one group by head excision. Imaging and analysis were consistently performed on muscles 6 and 7.
Antibodies and synthetic dyes
The following primary antibodies were used: mouse anti–lamin C (Developmental Studies Hybridoma Bank, no. LC28.26-c), monoclonal anti–lamin Dm0 (provided by Y. Gruenbaum, Hebrew University of Jerusalem, Jerusalem, Israel), rabbit anti-H3K9ac (Abcam, AB4441), and rabbit anti-H3K9me3 (Abcam, AB176916). The following conjugated secondary antibodies were used: Alexa Fluor 555 goat anti-rabbit (Renium, #A27039) and Alexa Fluor 647 goat anti-mouse (Renium, #A21235). Hoechst 33342 (1 μg/ml; Sigma-Aldrich) was used for labeling DNA.
Human cells
Human neutrophils and T cells were isolated from citrate–anticoagulated whole blood of healthy donors by dextran sedimentation and density separation over Ficoll-Paque Plus (Merck, GE17-1440-03), as described (69). Neutrophils were stored in Ca2+- and Mg2+-free medium to minimize premature activation and assayed up to 3 hours after purification. For generation of T effectors, isolated lymphocytes were seeded on plates coated with anti-CD3 (BioLegend, catalog no. 317302) and anti-CD28 (BioLegend, catalog no. 302933), activated for 48 hours, and then cultured with interleukin-2 (PeproTech, catalog no. 200-02), as previously described (69). T effectors were assayed on days 5 to 6 after original isolation. All in vitro experiments using human leukocytes were approved by the Institutional Review Board of the Rambam Medical Center, in accordance with the provisions of the Declaration of Helsinki.
For imaging, Ibidi chamber slides (μ-Slide VI 0.4, Ibidi) were coated overnight with 0.01% poly-l-lysine solution (Sigma-Aldrich, catalog no. P4832). Human neutrophils or effector T cells were labeled with 20 μM Hoechst 33342 (Thermo Fisher Scientific, catalog no. 62249) at 37°C for 5 min, washed three times with PBS, and resuspended in a solution of cold collagen I (Corning, catalog no. 354236) and Ca2+- and Mg2+-containing binding medium as described (70). The different leukocytes were introduced into an Ibidi chamber, sedimented at 50g for 3 min at 4°C, and subsequently incubated at 37°C for 30 min to allow collagen polymerization.
Microscopy and image acquisition
Confocal imaging of the live and fixed preparations genetically expressing fluorescent markers was performed using an inverted Leica SP8 STED3× microscope, equipped with internal Hybrid detectors and acousto-optical tunable filter (Leica Microsystems CMS GmbH, Germany) and a white light laser excitation laser. Spectral analysis of His2B-RFP and klar-GFP channels revealed their maximal emission to be at excitation wavelengths of 586 and 478 nm, respectively. RFP emission signal was collected at the range of 597 to 699 nm, and GFP emission signal was collected in the range of 488 to 559 nm. All nuclei were imaged with a HC PL APO 86×/1.20 water STED white objective, a numerical aperture of 1.2, at a scan speed of 400 Hz and a pinhole of 0.8 A.U. Z-stacks were acquired using the galvo stage, with 0.308-μm intervals. Bit depth was 12, and to enhance image quality, field of view and laser intensity were adjusted separately for each nucleus sampled. The acquired images were visualized during experiments using LAS-X software (Leica Application Suite X, Leica Microsystems CMS GmbH).
Imaging of human leukocytes and Drosophila epidermis cells and dynamic muscle contraction were performed using a Dragonfly spinning disk confocal system (Andor Technology PLC) connected to an inverted Leica Dmi8 microscope (Leica Microsystems CMS GmbH). The signals were detected by an sCMOS Zyla (Andor) 2048X2048 camera, and bit depth was 12. Human leukocyte images were acquired with a 100×/1.47 total internal reflection fluorescence oil objective, and Hoechst dye was excited with a 405-nm laser line. Drosophila epidermal nuclei expressing His2B-mRFP images were acquired with a 63×/1.3 glycerol objective and excited with a 561-nm laser line. Dynamic contractions of Drosophila muscle labeled with His2B-mRFP and SLS-GFP were acquired with 63×/1.3 glycerol objective, excited with a 561- and 488-nm laser lines for 20-ms exposure time, and detected simultaneously with two sCMOS Zyla cameras.
Images for quantitative epigenetic immunofluorescence were acquired at 23°C on confocal microscopes Zeiss LSM 800 with a Zeiss C-Apochromat 40×/1.20 W Korr M27 lens and an Immersol W 2010 immersion medium. The samples were embedded with a coverslip high precision of 1.5H ± 5 μm (Marienfeld-Superior, Lauda-Königshofen, Germany) and acquired using Zen 2.3 software (blue edition).
Image processing and data analysis
For live imaging, only nuclei that did not move or deform during image acquisition were included. Arivis Vision4D 3.1.2-3.4 was used for image visualization and analysis. Nuclei labeled for nuclear membrane (control and lamin C–GFP OE groups) were segmented in 3D with a dedicated pipeline based on the “blob finder” operation. For nuclei colabeled with His2B-mRFP and H3K9ac-mintbody-EGFP, a separate pipeline was used for segmentation, based on automatic “otsu” threshold operation, with segment modification to close a 3D nucleus object. All segmented nuclei were further divided into 10 concentric 3D shells (Fig. 1A), using a dedicated Python script embedded within the Arivis Vision4D environment. Cytoplasmic background intensity was calculated from additional shell generated outside of the segmented nucleus using the “dilate” segment modification operation. Nuclei and corresponding shell segment volumes, dimensions, and channel intensities were then exported to MATLAB R2019b (MathWorks) to generate radial distribution profiles. Chromatin density for each shell was calculated by summing the His2B fluorescence intensity within the shell and dividing it by shell volume. To compare radial profiles of nuclei from different muscles and larvae, the absolute His2B density values were divided by His2B density in the cytoplasmic shell. Quantification of chromatin and nucleolar volumes from the muscle nuclei labeled with His2Av-GFP and NLS-mCherry was performed with Arivis Vision4D (V4D) 3.4, combining two dedicated pipelines. First, the His2Av-GFP channel was denoised and background-subtracted, and then chromatin was segmented in 3D using the automatic “Li” threshold. Further, the entire nucleus was segmented in 3D from the NLS-mCherry channel using the automatic Li threshold. Last, to segment the nucleolus, the chromatin mask was applied on the NLS-mCherry channel, and it was further background-corrected to detect only the stronger nucleolar signal within the nucleus and segmented in 3D with the automatic otsu threshold.
Quantitative immunofluorescence analysis was performed with dedicated pipeline in Arivis V4D 3.3. Multiple nuclei per stack were automatically segmented in 3D, using automatic otsu threshold operation on the nuclear envelope channel. Nuclear boundaries were defined by the inner nuclear envelope to avoid introducing error due variation in the thickness of the nuclear lamina. Nuclear volumes and total fluorescent intensities of the epigenetic marks and the DNA were calculated and exported to MATLAB R2019b for pulled analysis.
Statistics
To compare radial chromatin distribution between groups, each radial distribution profile of an individual nucleus was divided by its maximum value. The nuclei were fitted with a third-order polynomial, using a linear mixed-effects model fit by Restricted Maximum Liklihood (REML), accounting for group as a fixed effect and for nucleus and larva as random effects. Analyses were performed using the packages “lme4” and “lmerTest” in R v.3.5.1. Quantitative immunofluorescence parameters from control and temporal lamin C OE nuclei were compared using a mixed linear model, with genotype as a fixed effect and with larva and muscles as random effects. Statistics were performed in R v.4.0.2 using the package lmerTest v.3.1-2. Linear fits to the chromatin and nucleolar volumes as function of the nuclear volume were generated using linear mixed-effects model fit, and Rm2 (marginal) is reported for each line.
Modeling
More details on the modeling approach are currently submitted for publication elsewhere (G.B., D.A. -P., D.L., T. V., and S.S).
Acknowledgments
We thank K. Hiroshi (Osaka University, Japan), K. Furukawa (Niigata University, Japan), and the Bloomington Stock Center for providing fly lines. We are grateful to D. Deviri and O. Cohen from the Department of Chemical and Biological Physics, Weizmann Institute of Science for discussions and support. The images in this paper were acquired at the Advanced Optical Imaging Unit, de Picciotto-Lesser Cell Observatory unit, at the Moross Integrated Cancer Center Life Science Core Facilities, Weizmann Institute of Science. We thank M. Shemesh and Y. Addadi for advising on image acquisition, O. Golani for advising on imaging analysis, and R. Rotkopf for statistical analysis, all from the Life Sciences Core Facility, Weizmann Institute. Special thanks to M. Abbate from Arivis V4D support team for incorporating custom Python script for 3D radial shells. We are grateful to G. Ankaoua and B. Pasmantirer from the Physics Core Facilities at Weizmann Institute for helping in the design of the live imaging device. We thank D. Antes (DAntes Scientific Illustration) for the schematic model illustration. Funding: This study was supported by grants from “The French Muscular Dystrophy Association (AFM-Téléthon)” grant no. 22339, NSF-BSF (BSF grant no. 2016738), Israel Science Foundation (ISF) grant no. 750/17 awarded to T.V., Weizmann Krenter-Katz Interdisciplinary Research at the Interfaces of Life and Exact Sciences awarded to T.V. and S.S., and Tandem Call Weizmann–PIC3i Curie 2019-2021. R.A.’s research was supported by ISF grant no. 791/17. Author contributions: D.A.P., D.L., and T.V. conceptualized the experiments. G.B. and S.S. conceptualized the theoretical model. R.A. conceptualized the live leukocytes experiments. D.A.P. and D.L. performed the experiments and image acquisition and analysis. F.R. conducted the live leukocytes experiments. A.R. performed fixed immunofluorescence experiments. G.B. performed model computer simulations and analysis. G.B. and S.S. reviewed and edited the manuscripts. D.A.P., D.L., and T.V. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabf6251/DC1
REFERENCES AND NOTES
- 1.Lakadamyali M., Cosma M. P., Visualizing the genome in high resolution challenges our textbook understanding. Nat. Methods 17, 371–379 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Ricci M. A., Manzo C., García-Parajo M. F., Lakadamyali M., Cosma M. P., Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Ou H. D., Phan S., Deerinck T. J., Thor A., Ellisman M. H., O’Shea C. C., ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajpai G., Padinhateeri R., Irregular chromatin: Packing density, fiber width, and occurrence of heterogeneous clusters. Biophys. J. 118, 207–218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cremer T., Cremer C., Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292–301 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Rosin L. F., Nguyen S. C., Joyce E. F., Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLOS Genet. 14, e1007393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dekker J., Misteli T., Long-range chromatin interactions. Cold Spring Harb. Perspect. Biol. 7, a019356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeshima K., Tamura S., Hansen J. C., Itoh Y., Fluid-like chromatin: Toward understanding the real chromatin organization present in the cell. Curr. Opin. Cell Biol. 64, 77–89 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Cremer T., Cremer M., Hübner B., Strickfaden H., Smeets D., Popken J., Sterr M., Markaki Y., Rippe K., Cremer C., The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 589, 2931–2943 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Popken J., Brero A., Koehler D., Schmid V. J., Strauss A., Wuensch A., Guengoer T., Graf A., Krebs S., Blum H., Zakhartchenko V., Wolf E., Cremer T., Reprogramming of fibroblast nuclei in cloned bovine embryos involves major structural remodeling with both striking similarities and differences to nuclear phenotypes of in vitro fertilized embryos. Nucleus 5, 555–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar Y., Sengupta D., Bickmore W. A., Recent advances in the spatial organization of the mammalian genome. J. Biosci. 45, 18 (2020). [PubMed] [Google Scholar]
- 12.van Steensel B., Belmont A. S., Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bickmore W. A., The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14, 67–84 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Briand N., Collas P., Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 21, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solovei I., Wang A. S., Thanisch K., Schmidt C. S., Krebs S., Zwerger M., Cohen T. V., Devys D., Foisner R., Peichl L., Herrmann H., Blum H., Engelkamp D., Stewart C. L., Leonhardt H., Joffe B., LBR and Lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Zheng X., Hu J., Yue S., Kristiani L., Kim M., Sauria M., Taylor J., Kim Y., Zheng Y., Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell 71, 802–815.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harr J. C., Schmid C. D., Muñoz-Jiménez C., Romero-Bueno R., Kalck V., Gonzalez-Sandoval A., Hauer M. H., Padeken J., Askjaer P., Mattout A., Gasser S. M., Loss of an H3K9me anchor rescues laminopathy-linked changes in nuclear organization and muscle function in an Emery-Dreifuss muscular dystrophy model. Genes Dev. 34, 560–579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurudatta B. V., Shashidhara L. S., Parnaik V. K., Lamin C and chromatin organization in Drosophila. J. Genet. 89, 37–49 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Ulianov S. V., Doronin S. A., Khrameeva E. E., Kos P. I., Luzhin A. V., Starikov S. S., Galitsyna A. A., Nenasheva V. V., Ilyin A. A., Flyamer I. M., Mikhaleva E. A., Logacheva M. D., Gelfand M. S., Chertovich A. V., Gavrilov A. A., Razin S. V., Shevelyov Y. Y., Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 10, 1176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens A. D., Banigan E. J., Marko J. F., Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 58, 76–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crosetto N., Bienko M., Radial organization in the mammalian nucleus. Front. Genet. 11, 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhler C., Shivashankar G. V., Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 18, 717–727 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Irianto J., Swift J., Martins R. P., McPhail G. D., Knight M. M., Discher D. E., Lee D. A., Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophys. J. 104, 759–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie K., Yang Y., Jiang H., Controlling cellular volume via mechanical and physical properties of substrate. Biophys. J. 114, 675–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo M., Pegoraro A. F., Mao A., Zhou E. H., Arany P. R., Han Y., Burnette D. T., Jensen M. H., Kasza K. E., Moore J. R., Mackintosh F. C., Fredberg J. J., Mooney D. J., Lippincott-Schwartz J., Weitz D. A., Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. U.S.A. 114, E8618–E8627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan J., Lu G., Hong Y., Hu Q., Mai X., Guo J., Si X., Wang F., Zhang Y., Live imaging and tracking of genome regions in CRISPR/dCas9 knock-in mice. Genome Biol. 19, 192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaban H. A., Seeber A., Monitoring the spatio-temporal organization and dynamics of the genome. Nucleic Acids Res. 48, 3423–3434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorber D., Rotkopf R., Volk T., A minimal constraint device for imaging nuclei in live Drosophila contractile larval muscles reveals novel nuclear mechanical dynamics. Lab Chip 20, 2100–2112 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Nozaki T., Imai R., Tanbo M., Nagashima R., Tamura S., Tani T., Joti Y., Tomita M., Hibino K., Kanemaki M. T., Wendt K. S., Okada Y., Nagai T., Maeshima K., Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67, 282–293.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Sherrard A., Bishop P., Panagi M., Villagomez M. B., Alibhai D., Kaidi A., Streamlined histone-based fluorescence lifetime imaging microscopy (FLIM) for studying chromatin organisation. Biol. Open. 7, bio031476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasulo B., Deuring R., Murawska M., Gause M., Dorighi K. M., Schaaf C. A., Dorsett D., Brehm A., Tamkun J. W., The Drosophila MI-2 chromatin-remodeling factor regulates higher-order chromatin structure and cohesin dynamics in vivo. PLOS Genet. 8, e1002878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S., Stoops E., CP U., Markus B., Reuveny A., Ordan E., Volk T., Mechanotransduction via the LINC complex regulates DNA replication in myonuclei. J. Cell Biol. 217, 2005–2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Windner S. E., Manhart A., Brown A., Mogilner A., Baylies M. K., Nuclear scaling is coordinated among individual nuclei in multinucleated muscle fibers. Dev. Cell 49, 48–62.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J., Ma H., Jin J., Uttam S., Fu R., Huang Y., Liu Y., Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells. Cell Rep. 24, 873–882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Almassalha L. M., Chandler J. E., Zhou X., Stypula-Cyrus Y. E., Hujsak K. A., Roth E. W., Bleher R., Subramanian H., Szleifer I., Dravid V. P., Backman V., The effects of chemical fixation on the cellular nanostructure. Exp. Cell Res. 358, 253–259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barzilai S., Yadav S. K., Morrell S., Roncato F., Klein E., Stoler-Barak L., Golani O., Feigelson S. W., Zemel A., Nourshargh S., Alon R., Leukocytes breach endothelial barriers by insertion of nuclear lobes and disassembly of endothelial actin filaments. Cell Rep. 18, 685–699 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Gupta S., Marcel N., Talwar S., Garg M., R I., Perumalsamy L. R., Sarin A., Shivashankar G. V., Developmental heterogeneity in DNA packaging patterns influences T-cell activation and transmigration. PLOS ONE 7, e43718 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popp W., Wachtler F., Changes in nucleolar structure, number and size in cellular activation and inactivation: Observations in human phytohaemagglutinin-treated lymphocytes. Cell Tissue Res. 234, 377–388 (1983). [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y., Gong K., Denholtz M., Chandra V., Kamps M. P., Alber F., Murre C., Comprehensive characterization of neutrophil genome topology. Genes Dev. 31, 141–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S. S., Janssen H., Amendola M., Nolen L. D., Bickmore W. A., van Steensel B., Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178–192 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Sato Y., Mukai M., Ueda J., Muraki M., Stasevich T. J., Horikoshi N., Kujirai T., Kita H., Kimura T., Hira S., Okada Y., Hayashi-Takanaka Y., Obuse C., Kurumizaka H., Kawahara A., Yamagata K., Nozaki N., Kimura H., Genetically encoded system to track histone modification in vivo. Sci. Rep. 3, 2436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pałka M., Tomczak A., Grabowska K., Machowska M., Piekarowicz K., Rzepecka D., Rzepecki R., Laminopathies: What can humans learn from fruit flies. Cell. Mol. Biol. Lett. 23, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchino R., Nonaka Y.-k., Horigome T., Sugiyama S., Furukawa K., Loss of Drosophila A-type lamin C initially causes tendon abnormality including disintegration of cytoskeleton and nuclear lamina in muscular defects. Dev. Biol. 373, 216–227 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Dekker J., Belmont A. S., Guttman M., Leshyk V. O., Lis J. T., Lomvardas S., Mirny L. A., O’Shea C. C., Park P. J., Ren B., Ritland Politz J. C., Shendure J., Zhong S.; 4D Nucleome Network , The 4D nucleome project. Nature 549, 219–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B. R., Mirny L. A., Dekker J., Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bajpai G., Jain I., Inamdar M. M., Das D., Padinhateeri R., Binding of DNA-bending non-histone proteins destabilizes regular 30-nm chromatin structure. PLOS Comput. Biol. 13, e100536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z., Wippo C. J., Wal M., Ward E., Korber P., Pugh B. F., A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332, 977–980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson B. A., Doolittle L. K., Schneider M. W. G., Jensen L. E., Gamarra N., Henry L., Gerlich D. W., Redding S., Rosen M. K., Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marko J. F., Siggia E. D., Polymer models of meiotic and mitotic chromosomes. Mol. Biol. Cell 8, 2217–2231 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hochstrasser M., Sedat J. W., Three-dimensional organization of Drosophila melanogaster interphase nuclei. I. Tissue-specific aspects of polytene nuclear architecture. J. Cell Biol. 104, 1455–1470 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinney N. A., Sharakhov I. V., Onufriev A. V., Investigation of the chromosome regions with significant affinity for the nuclear envelope in fruit fly—A model-based approach. PLOS ONE. 9, e91943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho J. W. K., Jung Y. L., Liu T., Alver B. H., Lee S., Ikegami K., Sohn K.-A., Minoda A., Tolstorukov M. Y., Appert A., Parker S. C. J., Gu T., Kundaje A., Riddle N. C., Bishop E., Egelhofer T. A., Hu S. S., Alekseyenko A. A., Rechtsteiner A., Asker D., Belsky J. A., Bowman S. K., Chen Q. B., Chen R. A.-J., Day D. S., Dong Y., Dose A. C., Duan X., Epstein C. B., Ercan S., Feingold E. A., Ferrari F., Garrigues J. M., Gehlenborg N., Good P. J., Haseley P., He D., Herrmann M., Hoffman M. M., Jeffers T. E., Kharchenko P. V., Kolasinska-Zwierz P., Kotwaliwale C. V., Kumar N., Langley S. A., Larschan E. N., Latorre I., Libbrecht M. W., Lin X., Park R., Pazin M. J., Pham H. N., Plachetka A., Qin B., Schwartz Y. B., Shoresh N., Stempor P., Vielle A., Wang C., Whittle C. M., Xue H., Kingston R. E., Kim J. H., Bernstein B. E., Dernburg A. F., Pirrotta V., Kuroda M. I., Noble W. S., Tullius T. D., Kellis M., MacAlpine D. M., Strome S., Elgin S. C. R., Liu X. S., Lieb J. D., Ahringer J., Karpen G. H., Park P. J., Comparative analysis of metazoan chromatin organization. Nature 512, 449–452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plimpton S., Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 117, 1–19 (1995). [Google Scholar]
- 54.J. B. Pawley, Handbook Of Biological Confocal Microscopy (Springer, ed. 3, 2006). [Google Scholar]
- 55.Versaevel M., Grevesse T., Gabriele S., Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 3, 671 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Jain N., Iyer K. V., Kumar A., Shivashankar G. V., Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. U.S.A. 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas C. H., Collier J. H., Sfeir C. S., Healy K. E., Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. U.S.A. 99, 1972–1977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hübner B., Lomiento M., Mammoli F., Illner D., Markaki Y., Ferrari S., Cremer M., Cremer T., Remodeling of nuclear landscapes during human myelopoietic cell differentiation maintains co-aligned active and inactive nuclear compartments. Epigenetics Chromatin 8, 47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puah W. C., Chinta R., Wasser M., Quantitative microscopy uncovers ploidy changes during mitosis in live Drosophila embryos and their effect on nuclear size. Biol. Open. 6, 390–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jovtchev G., Schubert V., Meister A., Barow M., Schubert I., Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114, 77–82 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Fiske J. A., Van Eenennaam J. P., Todgham A. E., Young S. P., Holem-Bell C. E., Goodbla A. M., Schreier A. D., A comparison of methods for determining ploidy in white sturgeon (Acipenser transmontanus). Aquaculture 507, 435–442 (2019). [Google Scholar]
- 62.Wang N., Tytell J. D., Ingber D. E., Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Cho S., Irianto J., Discher D. E., Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 216, 305–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirby T. J., Lammerding J., Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 20, 373–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kind J., van Steensel B., Stochastic genome-nuclear lamina interactions. Nucleus 5, 124–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bosveld F., Ainslie A., Bellaïche Y., Sequential activities of Dynein, Mud and Asp in centrosome-spindle coupling maintain centrosome number upon mitosis. J. Cell Sci. 130, 3557–3567 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Elhanany-Tamir H., Yu Y. V., Shnayder M., Jain A., Welte M., Volk T., Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 198, 833–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Albrecht S., Wang S., Holz A., Bergter A., Paululat A., The ADAM metalloprotease Kuzbanian is crucial for proper heart formation in Drosophila melanogaster. Mech. Dev. 123, 372–387 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Shulman Z., Alon R., Chapter 14 real-time in vitro assays for studying the role of chemokines in lymphocyte transendothelial migration under physiologic flow conditions. Methods Enzymol. 461, 311–332 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Yadav S. K., Feigelson S. W., Roncato F., Antman-Passig M., Shefi O., Lammerding J., Alon R., Frontline Science: Elevated nuclear lamin A is permissive for granulocyte transendothelial migration but not for motility through collagen I barriers. J. Leukoc. Biol. 104, 239–251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabf6251/DC1


