Abstract
Background and aim
A novel coronavirus, called the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been found to cause COVID-19 in humans and some other mammals. The nonstructural protein 16 (NSP16) of SARS-CoV-2 plays a significant part in the replication of viruses and suppresses the ability of innate immune system to detect the virus. Therefore, inhibiting NSP16 can be a secure path towards identifying a potent medication against SARS-CoV-2. Tea (Camellia sinensis) polyphenols have been reported to exhibit potential treatment options against various viral diseases.
Methods
We conducted molecular docking and structural dynamics studies with a set of 65 Tea bioactive compounds to illustrate their ability to inhibit NSP16 of SARS-CoV-2. Moreover, post-simulations end state thermodynamic free energy calculations were estimated to strengthen our results.
Results and conclusion
Six bioactive tea molecules showed better docking scores than the standard molecule sinefungin. These results were further validated by MD simulations, where Theaflavin compound demonstrated lower binding free energy in comparison to the standard molecule sinefungin. The compound theaflavin could be considered as a novel lead compound for further evaluation by in-vitro and in-vivo studies.
Keywords: SARS-CoV-2, COVID-19, NSP16, MM-PBSA, Methyltransferase, MD simulations
Graphical abstract
Abbreviations
- CHARMm
Chemistry at Harvard Macromolecular Mechanics
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- GROMACS
GROningen MAchine for Chemical Simulations
- MD
Molecular Dynamics
- MM-PBSA
Molecular Mechanics Poisson Boltzmann Surface Area
- NSP16
Non-Structural Protein 16
- RMSD
Root Mean Square Deviation
- RMSF
Root Mean Square Fluctuation
- SASA
Solvent accessible surface area
1. Introduction
Coronaviruses (CoVs) caused a significant explosion of fatal human pneumonia leading to a worldwide pandemic. CoVs comprise four structural proteins: envelope protein, spike protein, nucleocapsid, and membrane protein. A dynamic RNA replication system, consisting of at least 16 nonstructural viral proteins,1 is used to reproduce and transcribe the CoV RNA genome. Previous studies have shown that MERS-CoV and SARS-CoV non structural protein 16 (NSP16) catalyze the methylation by activating the methyltransferase (MTase) activities of the first nucleotide transcribed at the ribose 2′-O (2′-O-Me) position.2, 3, 4 NSP16 forms a heterodimer with its NSP10 co-factor and activates 2′-O-MTase action. By complementing the 2′-O-MTase action, NSP16 transforms the virus's genetic material to make it seem more familiar with human RNA.5 The 2′-O-Me virus cap RNAs defend themselves from 5′ to 3′ exoribonucleases breakdown, assures effective transmission, and supports to prevent recognition by the human innate immune system.5 Previous in-vivo and in-vitro studies5, 6, 7 have illustrated the influence of 2′-O-MTase action of NSP16 for CoV pathogenesis and infection. The NSP16 2′-O-MTase activity absence results in critical attenuation for SARS-CoV, defined by reduced replication of the virus, decreased weight deficit, and fewer mice respiration dysfunction.8 Thus, the blocking of NSP16 2′-O-MTase actions will limit replication of the virus and empower the human innate immune system to recognize it.
The NSP16 residues are highly conserved across the CoV family, proposing related structural domains and functional actions.5 The structure of Nsp16 has all 301 residues. The structure of NSP16 includes a 2-O-MTase analytic action core formed of a Rossmann-like β-sheet fold shaped by eleven α-helices, seven β-strands, and loops. The NSP16 binding site is coordinated by the residues Phe6947, Asp6912, Leu6898, Cys6913, Met6929, Gly6871, Asp6897, Asn6899, Asp6928, Tyr6845, Asn6841, and Gly6871.9
Natural plant products may be a valuable source of novel drugs to fight with COVID-19 pandemic.10,11 Indeed, various phytoconstituents, including polyphenols and flavonoids, have shown the ability to prevent SARS-CoV-2 replication and infection to mitigate the clinical consequences of the infection.12,13 In this context, the tea plant (Camellia sinensis), rich in micro-nutrients, polyphenols, and vitamins in tea mixture was considered as a source for bioactive molecules in this study. Tea is a traditional and most consumed drink globally due to its economic viability and easy availability and can play an essential role in nutritional immunity.14 The tea polyphenols were reported to exhibit antiviral activities against various viruses, especially positive-sense single-stranded RNA viruses.15 Bioactive tea compounds manifest antiviral activity against a broad spectrum of human viruses, including Dengue, Chikungunya, Zika, herpes simplex virus, HIV, hepatitis B, hepatitis C, and influenza.16 Recent in-silico and experimental studies documented potent antiviral activities of bioactive tea compounds against multiple SARS-CoV-2 viral proteins.17, 18, 19, 20, 21, 22, 23, 24
The present study was aimed to recognize lead compounds that may serve as a barrier against the activity of NSP16 of SARS-CoV-2. The objectives of the study were to rank and compare the binding affinity of the selected tea bioactive compounds with standard molecule sinefungin. Secondly, to validate the molecular docking results and in-depth analysis of protein-ligand complexes by performing robust, long-term (100 ns) molecular dynamics (MD) simulations. Lastly, to identify the most potent inhibitor by calculating the thermodynamic binding free energy by Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) analysis.
2. Material and methods
2.1. Datasets
NSP16 protein (PDB ID 6W4H) was selected for study based on the best resolution of 1.80 Å compared to other NSP16 crystallographic structures. Sinefungin (standard molecule/inhibitor)9,25 and a library of 65 bioactive compounds from tea (Camellia sinensis) were prepared for in-silico study as depicted in Table S1.17,26,27 The protein crystal structure preparation was conveyed by the Discovery studio protocols “prepare protein”.28 The ligand geometry of each molecule was optimized with minimization protocols (DFT) of Gaussian16.29
2.2. Molecular docking
The molecular docking was conducted in Discovery Studio version 2018 to determine the binding poses of the selected compounds with the NSP16 protein. CDOCKER, a docking system based on Chemistry at Harvard Macromolecular Mechanics Energy (CHARMm)30 was used to perform the docking procedure. The receptor was kept rigid while the ligands were allowed to flexible during the docking. The water molecules from the protein were eliminated, because the fixed-water molecules may change the ligand-receptor complex rendering. The NSP16 binding pocket allocated as the spaces inside a radius of 10 Å from the middle of the standard compound, while the other docking parameter values were retained default. The CHARMm force field with the interaction strength of each complex was measured. The distinct conformational poses of each compound generated and analyzed based on the CDOCKER interaction energy. The number of starting random conformations and the number of rotated ligand orientations to refine for each of the conformations for 1000 dynamics steps were set to ten. Moreover, for annealing refinement, the number of heating steps were 2000 while the number of cooling sets were set to 5000. The distance to consider Pi-cation, Pi-Pi, and Pi-alkyl interactions was set to 5 Å, 6 Å, and 5.5 Å respectively.
2.3. Molecular dynamics simulations
Molecular Dynamics (MD) simulations studies were conducted to determine the stability and flexibility of the NSP16 complexes. All simulations were performed using the GROMOS96 43a1 force field available in GROMACS 4.6.7.31 Ligand topology files were produced with the server of PRODRG.32 The processed protein complexes were solvated in a cubic box of edge length 10 nm along with simple point-charge (SPC) water models. Sufficient amounts of ions were appended to sustain the system neutrality. To exclude the clashes within atoms of the system, energy minimization estimations were made with the convergence pattern of 1000 kJ/mol/nm. Particle mesh Ewald (PME) method was employed to supervise the long-range interaction electrostatics33 A cutoff radius of 9 Å was accepted for both Coulombic and van der Waals interactions. Equilibration was finished in two-stages. In the first phase, the solvent and ion molecules were put unrestrained while in the next step, the restraint weight from the protein and protein-ligand complexes was steadily decreased, in the NPT ensemble. All hydrogen bonds were retained constrained, adopting a Linear Constraint Solver (LINCS) algorithm.34 The system's pressure and temperature were held at 300 K and 1 atm, sequentially, using Berendsen's temperature and Parrinello-Rahman pressure coupling, respectively.35 The simulation production was begun from the velocity and coordinates collected after the end step of the equilibration run. All the systems were simulated for 100 ns.
2.4. MM-PBSA calculation
The Molecular Mechanics Poisson Boltzmann Surface Area (MM-PBSA) technique was utilized to calculate the binding energy of the protein-ligand complexes. MMPBSA is a combined energy system, described by the Van der Waal, Solvent accessible surface area (SASA), electrostatic, and binding free energy of the system. In MM-PBSA, the polar solvation energy is computed, adopting the linear association to the SASA. The g_mmpbsa package of GROMACS was implemented to ascertain the diverse components of the free binding energy of the complexes.36
3. Results
3.1. Molecular docking analysis
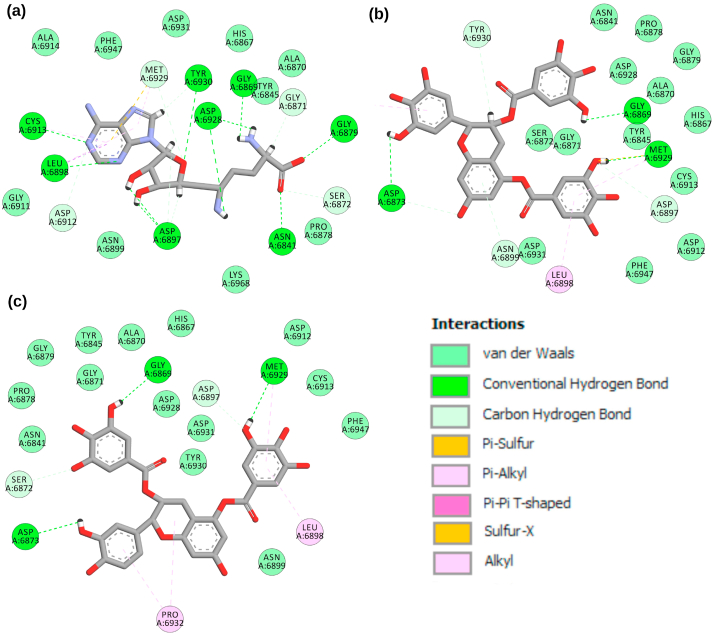
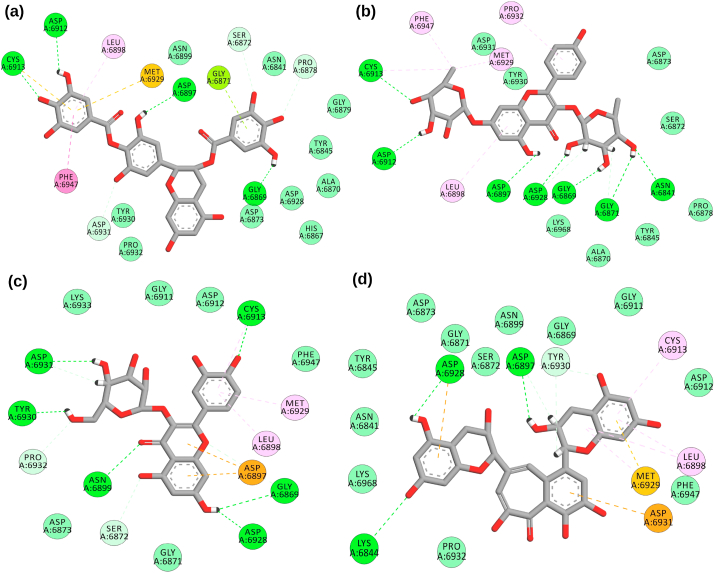
A library of 65 bioactive tea compounds was docked with SARS-CoV-2 NSP16. The compounds were docked in the binding site of the already published inhibitor sinefungin. The robustness of docking protocol was endorsed by comparing the co-crystallized sinefungin (PDB ID 6WKQ) with docked pose of sinefungin on the same protein. The root mean square deviation (RMSD) between both structures was 0.0001 Å over 1758 to 1758 atoms (Fig. S1). We selected the top six high-scoring bioactive molecules from all the docked compounds compared to sinefungin with valid -CDOCKER scores as tabulated in Table S1. The docking scores of the top selected molecules along with sinefungin were shown in Table 1. The active site of the NSP16 conferred hydrogen bonds with conserved residues Gly6869, Met6929, Asp6873, Tyr6930, Asp6928, Gly6879, Asn6841, Asp6897, Cys6913, Leu6898, Asp6912, Gly6871, Asp6930, Asn6899, and Lys6844. While hydrophobic interaction with residues Leu6898, Met6929, Gly6871, Pro6878, Asp6931, Phe6947, Pro6932, Tyr6930, Cys6913, Asp6897, Ser6872, Asp6912, and Asn6899. Fig. 1, Fig. 2 depicted the molecular interactions of the sorted compounds in the binding pocket of the NSP16.
Table 1.
Selected compounds based on CDOCKER interaction energy.
| Compounds | -CDOCKER interaction energy |
|---|---|
| Sinefungin | 76.83 |
| Epigallocatechin-3,5-di-O-gallate | 62.22 |
| Epicatechin-3,5-di-O-gallate | 60.24 |
| Epigallocatechin-3,4-di-O-gallate | 60.09 |
| Kaempferitrin | 59.29 |
| Isoquercetin | 58.47 |
| Theaflavin | 57.82 |
Fig. 1.
2-D interactions of SARS-CoV-2 NSP16 protein with standard and bioactives (a) sinefungin (b) epigallocatechin-3,5-di-O-gallate and (c) epicatechin-3,5-di-O-gallate.
Fig. 2.
2-D interactions of SARS-CoV-2 NSP16 protein with standard and bioactives (a) epigallocatechin-3,4-di-O-gallate (b) kaempferitrin, (c) isoquercetin and (d) theaflavin.
3.2. Structural stability at global and local levels
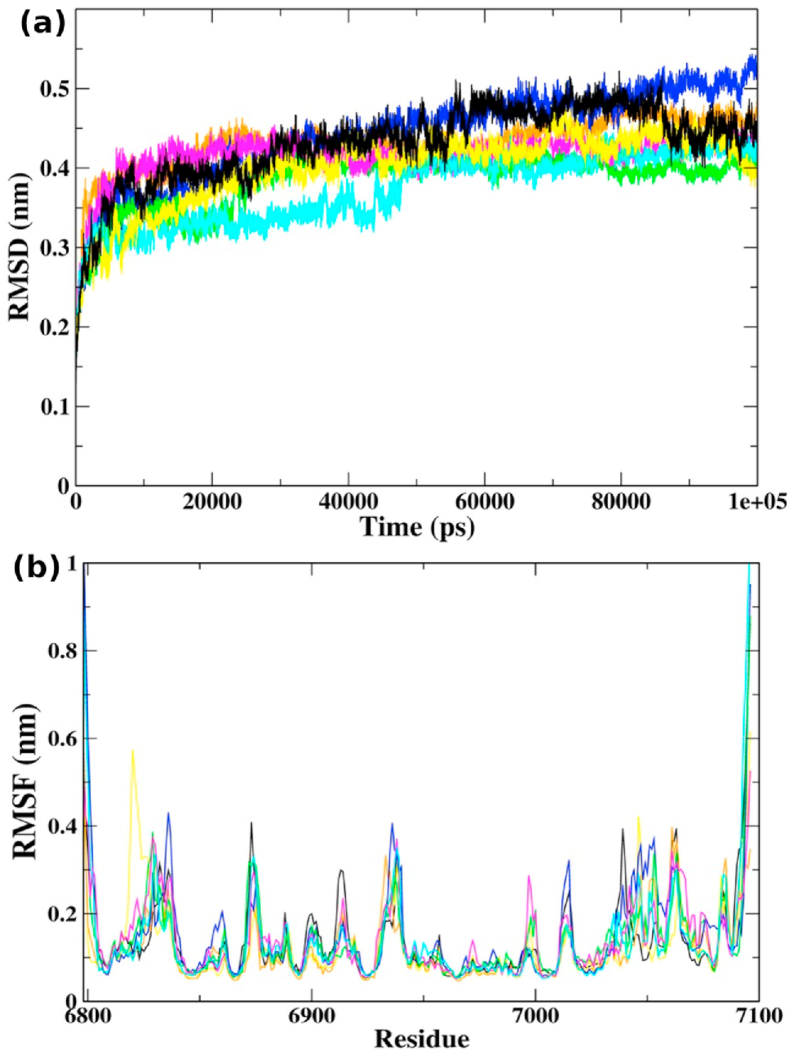
To gain insight into the dynamic behavior of the compounds at the active site of 2′-O-MTase, 100 ns MD simulations were executed for seven selected complexes (top hits and sinefungin, Table 1). To quantify the structural stability of protein-ligand complexes, root mean square deviation (RMSD) of backbone C-α atoms was measured (Fig. 3a). NSP16 complexes with theaflavin, kaempferitrin, isoquercetin, epicatechin-3,5-di-O-gallate, and epigallocatechin-3,5-di-O-gallate, pointed average RMSD values between 0.2 and 0.45 nm. At the same time, sinefungin and epigallocatechin-3,4-di-O-gallate in complex with NSP16 recorded higher fluctuations till 0.5 nm. Less fluctuations in RMSD trajectories and lower average values (<0.5 nm) indicated stable protein-ligand complexes, suitable for further analyses. Moreover, the root mean square fluctuation (RMSF) was estimated to perceive structural stability at local levels (Fig. 3b). The average RMSF values lied between 0.1 and 0.45 nm, and with moderate fluctuations, the RMSF trajectories all the seven complexes were pretty similar. However, epicatechin-3,5-di-O-gallate showed abrupt fluctuations in RMSF values around residues 6820 to 6840. All the residues belonging to the binding site showed low RMSF values.
Fig. 3.
(a) RMSD of backbone C α-atoms of NSP16 complexes, (b) RMSF for the backbone C α-atoms of NSP16 complexes with sinefungin (black), epigallocatechin-3,5-di-O-gallate (orange), epicatechin-3,5-di-O-gallate (yellow), epigallocatechin-3,4-di-O-gallate (blue),kaempferitrin (magenta), isoquercetin (green), and theaflavin (cyan).
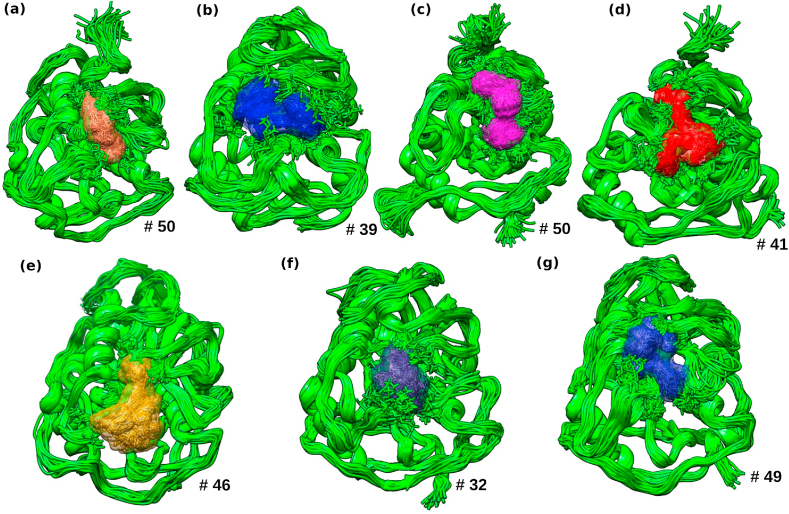
3.3. RMSD clustering analysis
Further, we estimated other structural parameters, including the RMSD clustering analysis based on the underlying dynamics approach. The clustering was performed to investigate the dynamics of molecular interactions of selected compounds with NSP16 protein, as shown in Fig. 4. In NSP16 complexes, sinefungin formed 50 clusters with an average RMSD of 0.17 nm. The bioactive compounds epigallocatechin-3,5-di-O-gallate, epigallocatechin-3,4-di-O-gallate, and isoquercetin formed 41, 39, 32 clusters with an average 0.16 nm RMSD. While compounds theaflavin, kaempferitrin, and epicatechin-3,5-di-O-gallate formed 49, 46, 50 clusters with an average RMSD of 0.17 and 0.15 and 0.19 nm. The chosen bioactive molecules covered greater space of the binding site as compared to sinefungin. These outcomes illustrated that the chosen bioactive compounds were enduring and relatively comparable to sinefungin in RMSD clustering interpretation.
Fig. 4.
Pictorial representation of conformational flexibility using cluster analysis for NSP16 in complex with (a) sinefungin, (b) epigallocatechin-3,5-di-O-gallat, (c) epicatechin-3,5-di-O-gallate, (d) epigallocatechin-3,4-di-O-gallate, (e) kaempferitrin, (f) isoquercetin, and (g) theaflavin.
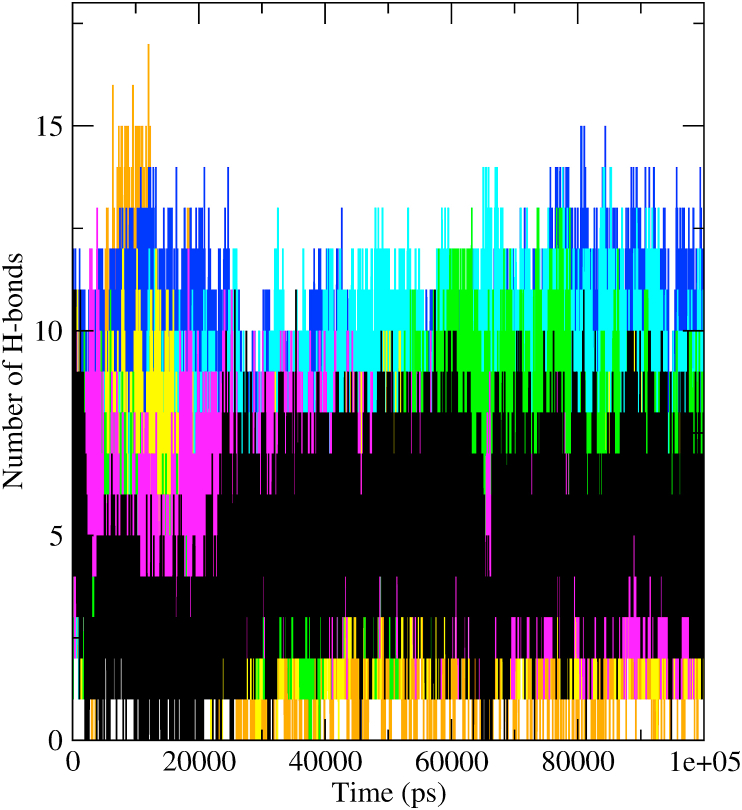
3.4. Analysis of inter-molecular hydrogen bonds
Additionally, our MD simulation analysis reported an average of five to seven hydrogen bonds for sinefungin during the entire simulation run. In comparison, all the six selected bioactive compounds of tea formed an average number of eight to twelve hydrogen bonds inside the active site of NSP16 of SARS-CoV-2. These outcomes manifested that all the complexes were adequately equilibrated and structurally abiding (Fig. 5). Moreover, we also obtained the scripts of all the selected complexes at different time intervals (5, 25, 45, 65, 85, 100 ns) to perceive the position and stability of compounds inside the binding pocket of the NSP16. It also allowed us to reaffirm the number of hydrogen bonds between protein and ligands. All the bioactive tea compounds formed a higher number of hydrogen bonds inside the binding pocket throughout the simulation than sinefungin. The number of hydrogen bonds acted as a driving efficiency for the solid binding of the ligand to its receptor. The common residues participating in hydrogen bond formations were Phe6947, Asp6912, Leu6898, Cys6913, Met6929, Gly6871, Asp6897, Asn6899, Asp6928, Tyr6845, Asn6841, and Gly6871. Also, all the selected molecules were present inside the binding site throughout the simulations (Fig. S2).
Fig. 5.
Hydrogen bond profiles of the NSP16 complexes with compounds sinefungin (black), epigallocatechin-3,5-di-O-gallate (orange), epicatechin-3,5-di-O-gallate (yellow), epigallocatechin-3,4-di-O-gallate (blue), kaempferitrin (magenta), isoquercetin (green), and theaflavin (cyan).
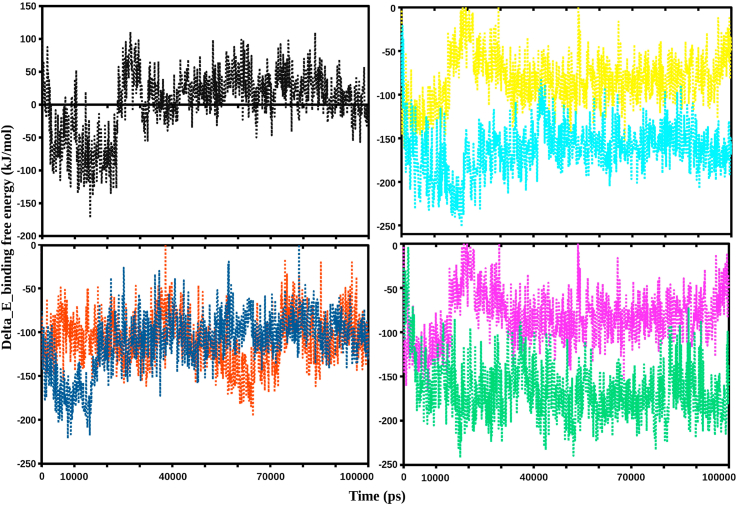
3.5. Analysis of thermodynamic binding energy
Another significant indicator that accounts for the potential affinity of a ligand with its target is the binding free energy computed by using the MM-PBSA approach. The binding free energies were measured for all conformations stored in the 0–100 ns trajectories during the simulations. The binding free energy outcomes showed that the ΔE free binding energy (kJ/mol) was higher for sinefungin, whereas lower for the other six selected bioactive compounds of tea. Lower the binding free energy, greater is the affinity between ligand and protein. The binding free energy of sinefungin recorded fluctuation after ∼22 ns and remained higher till the end of simulation, while selected bioactive compounds displayed stable trajectories with lower binding free energies (Fig. 6). In addition, the binding free energy was disintegrated into individual components, as tabulated in Table 2. On comparing the components, the polar solvation and Van der Waal energy showed unfavorable contributions, resulting in a decline in the binding free energy between sinefungin and NSP16. The favorable contribution of electrostatic energy was lower than the Van der Waal energy towards the total binding free energy. However, the binding free energy outcomes represented more favorable contributions for all the six tea compounds as compared to sinefungin.
Fig. 6.
Graphical representation of the Delta_E_Binding free energy kJ/mol showing sinefungin (black), epigallocatechin-3,5-di-O-gallate (yellow), epicatechin-3,5-di-O-gallate (cyan), kaempferitrin (blue), isoquercetin (orange), and epigallocatechin-3,4-di-O-gallate (pink), theaflavin (ocean green).
Table 2.
Binding free energy calculations of selected complexes using MM-PBSA.
| Compounds in complex with NSP16 | ΔE binding(kJ/mol) | ΔE polar solvation(kJ/mol) | SASA (kJ/mol) | ΔE Electrostatic (kJ/mol) | ΔE Van der Waal (kJ/mol) |
|---|---|---|---|---|---|
| Sinefungin | 3.723 | 372.618 | −17.442 | −170.772 | −180.682 |
| Theaflavin | −165.063 | 269.194 | −22.777 | −155.910 | −255.569 |
| Kaempferitrin | −110.230 | 215.255 | −18.984 | −115.877 | −190.624 |
| Isoquercetin | −108.973 | 222.321 | −18.693 | −117.182 | −195.419 |
| Epigallocatechin-3,5-di-O-gallate | −163.136 | 225.488 | −22.502 | −100.135 | −265.987 |
| Epigallocatechin-3,4-di-O-gallate | −143.084 | 325.824 | −24.432 | −177.909 | −266.567 |
| Epicatechin-3,5-di-O-gallate | −82.136 | 335.779 | −24.002 | −127.733 | −266.180 |
4. Discussion
The capability to effectively evade the immune detection and depress human immune responses significantly raises the virus transmission ability. This feature of SARS-CoV-2 to bypass identification and activation of the host immune response requires the RNA cap altering enzyme 2′-O-MTase, which gives the ability to viral mRNA to hide from the host cell.37,38 Several different classes of natural molecules isolated from many plants have been shown to have antiviral activity against CoV infection.10,39,40 Many studies have reported the computational screening of natural molecules for various SARS-CoV-2 targets, such as the main protease, papain-like protease, spike protein, and RNA-dependent RNA polymerase.41, 42, 43, 44, 45 In order to find the potential compounds against a target SARS-CoV-2 NSP16, we employed structural bioinformatics approaches to rank and compare bioactive tea compounds in comparison to sinefungin.
Molecular docking investigation is a wide-ranging technique that typically uncovers the binding interactions of protein-ligand complexes.46, 47, 48 The low RMSD value of superimposition between docked pose (generated by re-docking sinefungin on NSP16 PDB ID: 6WKQ) and co-crystallographic structure affirmed that the CDOCKER was notably firm for regenerating the binding poses between ligands and NSP16 of SARS-CoV-2. Our docking protocol showed six bioactive tea molecules (epigallocatechin-3,5-di-O-gallate, epigallocatechin-3,4-di-O-gallate, isoquercetin, theaflavin, kaempferitrin, and epicatechin-3,5-di-O-gallate) with high docking scores to be considered for drug development. Protein-ligand inter-communications are usually highlighted by hydrogen bonds and hydrophobic interactions, which perform a vital role in predicting the binding affinity of ligands with proteins.49,50 The active site of the NSP16 showed hydrogen bonding with the same residues, including Phe6947, Asp6912, Leu6898, Cys6913, Met6929, Gly6871, Asp6897, Asn6899, Asp6928, Tyr6845, Asn6841, and Gly6871. Similar results with different molecules were shown in different computational and experimental studies.9,51,52
Molecular docking, although very effective, provides static poses of protein-ligand interactions. In nature, these interactions are highly dynamic and limits the use of docking studies to preliminary screening only.53,54 Hence, the seven selected protein ligand complexes were subjected to MD simulations that can offer essential time-related insights into individual atomic movements and can also be used to address distinctive features of a simulated system.55, 56, 57 Based on experimental and theoretical data, MD simulations describes the fundamental relations between structure and its essential dynamics, enabling the exploration of the conformational energy environment open to biological macromolecules.58 The RMSD is a popular method used to evaluate the structural stability of protein structures via MD simulations.41,59 The stable trajectories along with low RMSD values generated during the entire simulation in the RMSD interpretation of MD simulations intimated the propriety of the simulation run. Also, the low fluctuations in RMSF values also indicated high structural stability of protein-ligand complexes. Potential inhibitors of NSP16 were screened by computational studies also showed low RMSD and RMSF values.51,60
The ensemble clustering analysis of simulated NSP16 complexes is a definitive and successful way to examine the structural stability of the protein systems.61 For the last 5ns of the simulation phase, the MD trajectories of all the selected complexes were obtained and subjected to cluster interpretation. The analysis furnished a notable amount of clusters for a protein-ligand complex considered for the flexibility of the protein, following their low average RMSD values deemed for the conformational stability of the complex.
Furthermore, we obtained the number of hydrogen bond between selected compounds and NSP16 protein to illustrate the impact of structural variations on inter-molecular interactions. The bioactive compounds of tea were firmly adhering to the binding site of NSP16 by making a higher number of hydrogen bonds than sinefungin. These findings were further verified by collecting NSP16 complexes trajectories at various time intervals and by visualizing the stability of selected compounds in the NSP16 active site. These results complemented our hydrogen bond analysis and showed hydrogen bonds throughout the simulation with different residues of the binding pocket. These results also suggested that all the selected molecules stayed inside the binding site till the end of simulation.
Moreover, an adequate and stable process of measuring the binding free energy (MM-PBSA) was adopted to compare the affinity of bioactive compounds of tea and sinefungin to NSP16 of SARS-CoV-2. The free binding energy calculation is an essential parameter of evaluation in drug discovery.62,63 Theaflavin showed most favorable binding energy among all the selected molecules, including sinefungin. The Van der Waal, Electrostatic, and SASA energies were favorable contributors to the total binding free energy for theaflavin. These results are however preliminary, and require validation by in-vivo and in-vitro studies. To conclude, our studies implemented the framework to further examine these lead candidates experimentally against SARS-CoV-2 NSP16.
5. Conclusion
In this study, we examined 65 bioactive compounds of the tea to provide a lead compound against the NSP16 of SARS-CoV-2. These molecules were compared to standard inhibitor sinefungin. Molecular docking results suggested six potential compounds for MD simulations studies. The stable trajectories and low RMSD and RMSF values revealed structural stability of selected protein-ligand complexes. Further, these results were complemented by hydrogen bond analysis, RMSD clustering, and analysis of molecular interactions at different time-intervals. Theaflavin conferred the higher binding free energy among all the selected compounds. Hence, this investigation states theaflavin as a new persuasive inhibitor of the SARS-CoV-2 NSP16 than sinefungin. However, its inhibitory potential against NSP16 of SARS-CoV-2 needs to be examined by suitable in-vivo and in-vitro analyses.
Author contribution
RP conceived of and designed the study. RS, VKB and JS analyzed and interpreted the data. RS, VKB, RP and SK critically revised it for important intellectual content. All authors gave final approval of the version to be published.
Consent for publication
All the authors have read and approved the manuscript in all respects for publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We gratefully acknowledge the CSIR-Institute of Himalayan Bioresource Technology, Palampur for providing the facilities to carry out this work. This manuscript represents CSIR-IHBT Communication No. 4678.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtcme.2021.05.005.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Jiang Y, Liu L, Manning M, Bonahoom M, Lotvola A, Yang Z-Q. Repurposing Therapeutics to Identify Novel Inhibitors Targeting 2’-O-Ribose Methyltransferase Nsp16 of SARS-CoV-2. ChemRxiv. Published online 2020. doi:10.26434/chemrxiv.12252965.
- 2.Chen Y., Su C., Ke M., et al. Biochemical and structural insights into the mechanisms of sars coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decroly E., Debarnot C., Ferron F., et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7(5) doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aouadi W., Blanjoie A., Vasseur J.-J., Debart F., Canard B., Decroly E. Binding of the methyl donor S-Adenosyl-l-Methionine to Middle East respiratory syndrome coronavirus 2′-O-methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J Virol. 2017;91(5) doi: 10.1128/jvi.02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menachery V.D., Debbink K., Baric R.S. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subissi L., Imbert I., Ferron F., et al. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antivir Res. 2014;101(1):122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menachery V.D., Yount B.L., Josset L., et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-O-methyltransferase activity. J Virol. 2014;88(8):4251–4264. doi: 10.1128/jvi.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosas-Lemus M., Minasov G., Shuvalova L., et al. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci Signal. 2020;13(651) doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuzimoto A.D., Isidoro C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J Tradit Compl Med. 2020;10(4):405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omrani M., Keshavarz M., Nejad Ebrahimi S., et al. Potential natural products against respiratory viruses: a perspective to develop anti-COVID-19 medicines. Front Pharmacol. 2021;11:2115. doi: 10.3389/fphar.2020.586993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey A., Khan M.K., Hamurcu M., Gezgin S. Natural plant products: a less focused aspect for the COVID-19 viral outbreak. Front Plant Sci. 2020;11:1356. doi: 10.3389/fpls.2020.568890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasansuklab A., Theerasri A., Rangsinth P., Sillapachaiyaporn C., Chuchawankul S., Tencomnao T. Anti-COVID-19 drug candidates: a review on potential biological activities of natural products in the management of new coronavirus infection. J Tradit Compl Med. 2021;11(2):144–157. doi: 10.1016/j.jtcme.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury P., Barooah A.K. Tea bioactive modulate innate immunity: in perception to COVID-19 pandemic. Front Immunol. 2020;11:1. doi: 10.3389/fimmu.2020.590716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mhatre S., Srivastava T., Naik S., Patravale V. Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: a review. Phytomedicine. 2021;85:153286. doi: 10.1016/j.phymed.2020.153286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan N., Mukhtar H. Tea and health: studies in humans. Curr Pharmaceut Des. 2013;19(34):6141–6147. doi: 10.2174/1381612811319340008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. Published online May 13, 2020:1-10. doi:10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed]
- 18.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput Biol Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Depicting the inhibitory potential of polyphenols from Isatis indigotica root against the main protease of SARS CoV-2 using computational approaches. J Biomol Struct Dyn. Published online December 9, 2020:1-12. doi:10.1080/07391102.2020.1858164. [DOI] [PMC free article] [PubMed]
- 20.Sharma J, Kumar Bhardwaj V, Singh R, Rajendran V, Purohit R, Kumar S. An in-silico evaluation of different bioactive molecules of Tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. Published online 2020:128933. doi:10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed]
- 21.Ohgitani E., Shin-Ya M., Ichitani M., et al. Significant inactivation of SARS-CoV-2 by a green tea catechin, a catechin-derivative and galloylated theaflavins in vitro. bioRxiv. 2020;12 doi: 10.1101/2020.12.04.412098. Published online December 6, 2020. 04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du A., Zheng R., Disoma C., et al. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int J Biol Macromol. 2021;176:1–12. doi: 10.1016/j.ijbiomac.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Bioactive molecules of Tea as potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2. Front Med. 2021;8:645. doi: 10.3389/FMED.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umashankar V., Deshpande S.H., Hegde H.V., Singh I., Chattopadhyay D. Phytochemical moieties from Indian traditional medicine for targeting dual hotspots on SARS-CoV-2 spike protein: an integrative in-silico approach. Front Med. 2021;8 doi: 10.3389/fmed.2021.672629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai M., Fukui Y., Asami S., et al. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J Agric Food Chem. 2005;53(11):4593–4598. doi: 10.1021/jf047814+. [DOI] [PubMed] [Google Scholar]
- 27.Namal Senanayake SPJ Green tea extract: Chemistry, antioxidant properties and food applications - a review. J Funct Foods. 2013;5(4):1529–1541. doi: 10.1016/j.jff.2013.08.011. [DOI] [Google Scholar]
- 28.Studio D. Dassault Systemes BIOVIA, Discovery Studio Modelling Environment, Release 4.5. Accelrys Softw Inc. Published online 2015:98-104.
- 29.Zheng J., Frisch M.J. Efficient geometry minimization and transition structure optimization using interpolated potential energy surfaces and iteratively updated hessians. J Chem Theor Comput. 2017;13(12):6424–6432. doi: 10.1021/acs.jctc.7b00719. [DOI] [PubMed] [Google Scholar]
- 30.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- 31.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 32.Schüttelkopf A.W., Van Aalten D.M.F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect D Biol Crystallogr. 2004;60(8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 33.Abraham M.J., Gready J.E. Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J Comput Chem. 2011;32(9):2031–2040. doi: 10.1002/jcc.21773. [DOI] [PubMed] [Google Scholar]
- 34.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear Constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 35.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91(1-3):43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 36.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 37.Encinar J.A., Menendez J.A. Potential drugs targeting early innate immune evasion of SARS-coronavirus 2 via 2’-O-Methylation of Viral RNA. Viruses. 2020;12(5):525. doi: 10.3390/v12050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panyod S., Ho C.T., Sheen L.Y. Dietary therapy and herbal medicine for COVID-19 prevention: a review and perspective. J Tradit Compl Med. 2020;10(4):420–427. doi: 10.1016/j.jtcme.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elgazar A.A., Knany H.R., Ali M.S. Insights on the molecular mechanism of anti-inflammatory effect of formula from Islamic traditional medicine: an in-silico study. J Tradit Compl Med. 2019;9(4):353–363. doi: 10.1016/j.jtcme.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaki AA, Ashour A, Elhady SS, Darwish KM, Al-Karmalawy AA. Calendulaglycoside A showing potential activity against SARS-CoV-2 main protease: molecular docking, molecular dynamics, and SAR studies. J Tradit Compl Med. Published online May 17, 2021. doi:10.1016/j.jtcme.2021.05.001. [DOI] [PMC free article] [PubMed]
- 42.Rahman F., Tabrez S., Ali R., Alqahtani A.S., Ahmed M.Z., Rub A. Molecular docking analysis of rutin reveals possible inhibition of SARS-CoV-2 vital proteins. J Tradit Compl Med. 2021;11(2):173–179. doi: 10.1016/j.jtcme.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rangsinth P., Sillapachaiyaporn C., Nilkhet S., Tencomnao T., Ung A.T., Chuchawankul S. Mushroom-derived bioactive compounds potentially serve as the inhibitors of SARS-CoV-2 main protease: an in silico approach. J Tradit Compl Med. 2021;11(2):158–172. doi: 10.1016/j.jtcme.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vardhan S, Sahoo SK. Virtual screening by targeting proteolytic sites of furin and TMPRSS2 to propose potential compounds obstructing the entry of SARS-CoV-2 virus into human host cells. J Tradit Compl Med. Published online April 12, 2021. doi:10.1016/j.jtcme.2021.04.001. [DOI] [PMC free article] [PubMed]
- 45.Parashar A, Shukla A, Sharma A, Behl T, Goswami D, Mehta V. Reckoning γ-Glutamyl-S-allylcysteine as a potential Main protease (M pro ) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation. Drug Dev Ind Pharm. Published online May 26, 2021:1-32. doi:10.1080/03639045.2021.1934857. [DOI] [PMC free article] [PubMed]
- 46.Zaki A.A., Al-Karmalawy A.A., El-Amier Y.A., Ashour A. Molecular docking reveals the potential of: Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J Chem. 2020;44(39):16752–16758. doi: 10.1039/d0nj03611k. [DOI] [Google Scholar]
- 47.Samra R.M., Soliman A.F., Zaki A.A., et al. Bioassay-guided isolation of a new cytotoxic ceramide from Cyperus rotundus L. South Afr J Bot. 2021;139:210–216. doi: 10.1016/j.sajb.2021.02.007. [DOI] [Google Scholar]
- 48.Singh R, Bhardwaj VK, Sharma J, Das P, Purohit R. Identification of selective cyclin-dependent kinase 2 inhibitor from the library of pyrrolone-fused benzosuberene compounds: an in silico exploration. J Biomol Struct Dyn. Published online 2021. doi:10.1080/07391102.2021.1900918. [DOI] [PubMed]
- 49.Wang G., Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med Chem. 2016;8(14) doi: 10.4155/fmc-2016-0143. [DOI] [PubMed] [Google Scholar]
- 50.Lavecchia A., Giovanni C. Virtual screening strategies in drug discovery: a critical review. Curr Med Chem. 2013;20(23):2839–2860. doi: 10.2174/09298673113209990001. [DOI] [PubMed] [Google Scholar]
- 51.Liang J., Pitsillou E., Burbury L., Hung A., Karagiannis T.C. In silico investigation of potential small molecule inhibitors of the SARS-CoV-2 nsp10-nsp16 methyltransferase complex. Chem Phys Lett. 2021;774 doi: 10.1016/j.cplett.2021.138618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tazikeh-Lemeski E, Moradi S, Raoufi R, Shahlaei M, Janlou MAM, Zolghadri S. Targeting SARS-COV-2 non-structural protein 16: a virtual drug repurposing study. J Biomol Struct Dyn. Published online 2020. doi:10.1080/07391102.2020.1779133. [DOI] [PMC free article] [PubMed]
- 53.Durrant J.D., McCammon J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9 doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front Pharmacol. 2018;9:923. doi: 10.3389/fphar.2018.00923. AUG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karplus M., Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci U S A. 2005;102(19):6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar Bhardwaj V., Purohit R. Taming the ringmaster of the genome (PCNA): phytomolecules for anticancer therapy against a potential non-oncogenic target. J Mol Liq. 2021;337 doi: 10.1016/j.molliq.2021.116437. [DOI] [Google Scholar]
- 57.Bhardwaj V.K., Purohit R. Computer simulation to identify selective inhibitor for human phosphodiesterase10A. J Mol Liq. 2021;328 doi: 10.1016/j.molliq.2021.115419. [DOI] [Google Scholar]
- 58.Hansson T., Oostenbrink C., Van Gunsteren W.F. Molecular dynamics simulations. Curr Opin Struct Biol. 2002;12(2):190–196. doi: 10.1016/S0959-440X(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 59.Elmaaty A.A., Alnajjar R., Hamed M.I.A., Khattab M., Khalifa M.M., Al-Karmalawy A.A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: theoretical study. RSC Adv. 2021;11(17):10027–10042. doi: 10.1039/d0ra10674g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El Hassab M.A., Ibrahim T.M., Al-Rashood S.T., Alharbi A., Eskandrani R.O., Eldehna W.M. In silico identification of novel SARS-COV-2 2′-O-methyltransferase (nsp16) inhibitors: structure-based virtual screening, molecular dynamics simulation and MM-PBSA approaches. J Enzym Inhib Med Chem. 2021;36(1):727–736. doi: 10.1080/14756366.2021.1885396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhardwaj V.K., Purohit R. A new insight into protein-protein interactions and the effect of conformational alterations in PCNA. Int J Biol Macromol. 2020;148:999–1009. doi: 10.1016/j.ijbiomac.2020.01.212. [DOI] [PubMed] [Google Scholar]
- 62.Huang K., Luo S., Cong Y., Zhong S., Zhang J.Z.H., Duan L. An accurate free energy estimator: based on MM/PBSA combined with interaction entropy for protein-ligand binding affinity. Nanoscale. 2020;12(19):10737–10750. doi: 10.1039/c9nr10638c. [DOI] [PubMed] [Google Scholar]
- 63.Wang C., Greene D., Xiao L., Qi R., Luo R. Recent developments and applications of the MMPBSA method. Front Mol Biosci. 2018;4:87. doi: 10.3389/fmolb.2017.00087. JAN. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.