Abstract
The worldwide spread of COVID-19 highlights the urgent need for an efficient approach to rapidly develop therapeutics and prophylactics against SARS-CoV-2. Extracellular vesicle(EVs) are recognized and endocytosed by tissue cells via specific interactions between surface membrane proteins, where after they deliver their molecular cargo. This provides the potential to modify membrane proteins at EV surfaces as a promising means for specific tissue targeting and drug delivery. In this study, we describe a VSVG viral pseudotyping-based approach to load EV membranes with the receptor-binding domain (RBD) of the viral spike protein, the key domain in SARS-CoV-2 attachment, fusion and cellular entry. The RBD-tagged EVs can specifically recognize ACE2 receptor on the surface of target cells, which is required for the RBD-tagged EVs cellular uptake and targeting. Further, using the hACE2 transgenic mouse model, we show the RBD-tagged EVs accumulate specifically in the target tissues that highly express ACE2. Finally, we demonstrate that the RBD-tagged EVs that encapsulate siRNAs against SARS-CoV-2 pseudovirus can specifically target lung tissues and suppress the pseudovirus infection in vivo. Together, our work presents a safe and effective engineered EV system for in vivo targeted delivery of potential antiviral agents into specific tissues which as a therapeutic potential against SARS-CoV-2 infection.
Keywords: SARS-CoV-2, Extracellular vesicles, Receptor binding domain (RBD), ACE2
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), becomes a major health concern all over the world as the rate of mortality has been rising each day [1,2]. It was observed that there was an urgent need for developing effective therapies or prophylactics to prevent SARS-CoV-2 infection. SARS-CoV-2 virus targets lungs via its binding to the angiotensin-converting enzyme 2 (ACE2) receptors, which are highly expressed at the surface of type 2 alveolar epithelial cells in the lungs [3]. In addition, ACE2 is also expressed on other tissues, including the heart, kidney and small intestine [[3], [4], [5]]. Virus entry into cells depends on the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein (S) that specifically recognizes ACE2 [6,7]. SARS-CoV-2 S protein has been experimentally shown to bind ACE2 on host cells with significantly higher affinity than SARS-CoV [8,9]. It is known that the blockage of the RBD and the ACE2 association site, is a major potential strategy for developing vaccines, neutralizing antibodies and small molecule drugs to against COVID-19 [6,7].
Extracellular vesicle (EVs) are natural transport nano-vesicles secreted by numerous cell types [10]. It is clear that EVs have the capacity to deliver specific cargo of functional biomolecules, such as nucleic acids (including plasmid DNA and siRNA), proteins and chemotherapeutic drugs, to the recipient cells, release cargo, and mediate many physiological or pathological processes [[10], [11], [12]]. It is widely recognized that EVs are promising nanocarriers for clinical application based on their nanoscale for penetration into deep tissues, long-term circulation, and their naturally biocompatible characteristics [13,14]. However, it has been shown that the majority of intravenous injected EVs are absorbed within the liver [15,16]. Therefore, if EVs are candidates for the systemic delivery of therapeutic compounds and exploited as the targeted therapeutic particles, it will be necessary to deliver exogenous cargoes to specific tissues or cell types in vivo. Targeted EVs can be obtained by displaying targeting molecules, such as tissue-specific peptides or antibody fragments recognizing target antigens, on the outer surface of EVs [17,18].
The vesicular stomatitis virus(VSV)-G protein is frequently used for viral pseudotyping, owing to its broad tropism and high efficacy in transduction. Prior research has shown that by fusing a variety of reporter proteins with VSVG, including green fluorescent protein(GFP), red fluorecent protein(RFP), and luciferase, could be successfully incorporated into EVs membranes as a means of tracking and detection [19,20]. In this study, we exploited a pseudotyping-based approach to load EV membranes with SARS-CoV-2 spike protein RBD through engineering the VSVG. We showed that the RBD-tagged EVs required the ACE2 receptor for cellular uptake and were able to specifically trend to target cells or tissues that highly express ACE2. Further, we showed that siRNAs encapsulated in the RBD-tagged EVs can effectively delivered into lung tissues and inhibit SARS-CoV-2 infection, suggesting this RBD-tagged EV-mediated delivery of antiviral agents into target tissues may serve as a promising therapeutic platform for treatment of SARS-CoV-2 infection.
2. Materials and methods
2.1. RBD-VSVG fusion vector construction
To display SARS-CoV-2 RBD domain on the outer membrane of EVs, the ectodomain sequence of VSVG was replaced by codon-optimized RBD sequence. Fusion sequence was configured from 5′ to 3′ as following: the signal peptide, the RBD domain, stem sequence, transmembrane domain and cytoplasmic domain. The fusion sequence was synthesized from the GENEWIZ company and then cloned downstream of the CMV promoter with EcoRI and XbaI restriction sites into pcDNA3.1 vector. We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV210161) [21].
2.2. Transmission and scanning electron microscope
Purified EVs resuspended in Phosphate Buffered Saline(PBS) were dropped onto a sheet of Parafilm and a 200-mesh formvar coated copper grids were floated at room temperature allowed to adsorb for 10 min, followed by wash with PBS twice and fixed with 2% paraformaldehyde. After rinsing with PBS twice, the grids were negatively stained with 2% Uranyl acetate for 2 min and observed using a transmission electron microscope(Hitachi HT7700 TEM, Tokyo, Japan) or scanning electron microscope(Hitachi Regulus SU8230, Tokyo, Japan).
2.3. Nanoparticle-tracking analysis
Control or modified EVs were carried out using an NS300 machine (Malvern Instruments, Malvern, UK). In a typical analysis, 1 mL of the 1:1000 diluted EVs was used for EV visualization by laser-light scattering, using a conventional optical microscope aligned normally to the beam axis which collects light scattered from every particle in the field of view. and three videos of 30 s each were recorded. Data analysis was performed by NTA software, and the results are presented graphically to show particle size and distribution.
2.4. EVs purification
HEK-293T cells were transfected with S-FLAG verctor, RBD-VSVG vector, CD9-GFP vector or empty vector using Lipofectamine 3000 when reached 70%–80% confluence. At 6 h after transfection, cells were switched to DMEM supplemented with 10% EV-depleted FBS(System Biosciences lnc) for the production of EVs. After 48 h, the cell supernatants was collected and centrifuged at 10,000g for 30 min, then subjected to filter twice through 0.22 μm membranes(Millpore). The EV pellets were prepared from filtered supernatants by ultracentrifugation at 100,000 g for 70 min at 4 °C (Beckman Ti70 rotor). Resuspend the pellets in phosphate buffered saline (PBS) and repeat centrifuged 70 min at 100,000g at 4 °C to remove soluble serum and secreted proteins. The purified EV pellets were resuspended in fresh PBS(0.22 μm filtration).
2.5. Western blot
Cells, purified EVs or tissues were lysed with RIPA buffer (Santa Cruz,USA) and cleared lysate was collected by centrifugation for protein separation on SDS-polyacrylamide gel(10%), followed by transfer onto PVDF membranes(Millipore), blocked with 5% nonfat dry milk in TBST and reacted with primary antibodies recognizing FLAG M2 tag(Sigma,1:2000), SARS-CoV-2 RBD(Sinobiological,1:500), CD9(Abcam,1:2000), Alix(Cell Signaling Technology,1:1000), GM130(Abcam,1:1000), Calnexin(Abcam,1:1000), hACE2(Abcam,# ab108209,only react with human species,1:2000) and GAPDH(Santa Cruz,1:2000). Blots were incubated with HRP-conjugated goat anti-mouse or rabbit IgG (Southern Biotech,1:5000) for 1 h at room temperature before imaging.
2.6. Immunoprecipitation of EVs
Purified RBD-tagged or control EVs resuspended in PBS(60 μg of total EV protein each sample, in a 200 μl volume) were incubated with 5 μg rabbit-anti-RBD antibody(Sinobiological) at 4 °C overnight. Magnetic protein A/G beads(Millipore) were washed three times in PBS and blocked for 1 h in 1.5%BSA diluted in PBS. Beads were washed twice in PBS then immune-captured purified EVs were added to the beads and incubated for 2 h at 4 °C. After three washes in PBS, beads were incubated with 1× loading sample buffer at 95 °C for 5 min and then used for western blot analysis.
2.7. Cell cultures
HEK-293T, Caco-2, A549, U251, A498, HT-29 and HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). The THP-1 cell line was cultured in RPMI-1640 containing 5 mM l-glutamine and 10% FBS. All cell lines were obtained from American Type Culture Collection (ATCC, USA). Human pluripotent stem cells (hPSCs) were purchased from CellAPY Company(CA4002106, Beijing, China) and maintained in serum-free mTeSR1 medium (STEMCELL Technologies, Canada). All cells were incubated in a 37 °C humidified atmosphere of 5% CO2.
2.8. Human intestinal organoids
Human intestinal organoids are derived from induced pluripotent stem cells (iPSC) through a three-stage differentiation process(definitive endoderm, mid−/hindgut, and small intestine) using stage-specific, specialized media as previously described [22]. Human-iPSC-derived cardiomyocytes(iPSC-CMs) using a previously published protocol [23] and cultured on Cardiomyocyte Maintenance Medium(STEMCELL Technologies, Canada).
2.9. EV protein and RNA concentration measurement
Purified RBD-tagged or control EVs resuspended in PBS. 1 × 108 of each EV sample was extracted in 100 μl RIPA buffer(Santa Cruz, USA) with freshly added protease inhibitor(Roche, USA), and then cleared by centrifugation at 15,000 g, for 10 min at 4 °C. Total protein concentration of each EV lysate was determined using the Bicinchoninic Acid(BCA) Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instructions with BSA as the standard. For total RNA concentration, 1 × 108 of each EV sample dissolved in TRIzol reagent(Life Technologies) and performed according to manufacturer's specifications.
2.10. Cytokine assay
Mouse serum concentrations of IL-6, TNF-α and IFN-β were obtained from eBioscience Elisa Kit (Thermo Scientific, USA) according to manufacturer's suggested instructions.
2.11. ACE2 binding assay based on ELISA
96-well ELISA plates(Thermo Fisher Scientific) were coated with 100 μl/well of 500 ng/ml reconstitute human ACE2 protein(Sinobiological, #10108-H08H-B) at 4 °C overnight. Plates were then washed 3 times with PBST and blocked by 1% BSA for 30 min at room temperature. 100ul/well(1 μg/mL or 10 μg/mL diluted EVs that determined by BCA protein assay) was added to the wells and incubated for 2 h at 37 °C. After incubation, the wells are washed six times with PBST to remove unbound material and 100 μl/well of diluted detection CD9 antibody solution (1:10000, Sigma, #SAB4503606) was added to the wells and incubated for 60 min at room temperature. Plates were then washed, 100 μl/well of diluted HRP anti-rabbit(Southern Biotech) solution was added to the wells and incubated for 30 min at room temperature. TMB solution(Abcam) is added and then stopped by addition of Stop Solution(Abcam) completing any colour change from blue to yellow. Signal was measured at 450 nm on Thermo Scientific Varioskan™ LUX.
2.12. Production and titration of SARS-CoV-2 pseudovirions
SARS-CoV-2 pseudovirions were produced by co-transfection 293T cells with psPAX2, pLenti-GFP, and plasmids encoding codon-optimized SARS-CoV-2 S vector(Sinobiological) by using Lipofectamine 3000(Life Technologies, USA) according to the manufacturer's instructions. The supernatants were harvested at 48, 72 h post transfection, centrifuged at 2000 ×g for 10 min to remove cell debris and passed through 0.45 μm filter(Millipore). For viral titers, we used flow cytometry to determine the number of viral particles per ml as previously described [24].
2.13. Animals
Transgenic mice that express human ACE2(hACE2 mouse) were obtained from Nanjing Biomedical Research Institute of Nanjing University (NBRI). Transgenic mice were generated by microinjection of the mouse ACE2 promoter driving the human ACE2 coding sequence into the pronuclei of fertilized ova from C57BL/6JGpt. The human ACE2 integrated was identified by PCR using the primer: hACE2-F: 5’-AAGTGGCTCCTCTCTTACACTCTGG-3’;hACE2-R:5’-CAGACCATTTGTCCCCAGCAT-3′, The expected size of amplicons was 1508 bp. All animal experimental protocols were approved by the Soochow University Animal Care Committee and research protocols were conducted in accordance with the animal behavioral guidelines, using approved protocols from the institutional animal care committee (#202006A600).
2.14. Intravital imaging of EVs
EVs at a total protein concentration were determined using the BCA protein assay kit (Pierce, USA). Purified EVs suspended in PBS were added with 1,1-Dioctadecyl-3,3,3,3-tetramethylindodicarbocyanine (DiD, purchased from MCE) to 1 μM and incubated for 15 min, followed by an additional wash to remove the free dye. DiD-labeled EVs were resuspended in PBS and injected intravenously into 6-week-old female hACE2 mice(C57BL/6) with 150 μg EV per mice. The red fluorescence of the whole body was acquired by IVIS spectrum (Caliper Life Sciences). The hACE2 mice were sacrificed at 24 h, 48 h,72 h and 96 h post injection, organs harvested and the red fluorescence images was quantified in various organs. Radiant efficiency was measured using Living Image 3.1 software (Caliper Life Sciences).
2.15. EV siRNA loading and suppression of SARS-CoV-2 pseudovirions
For siRNA encapsulated in EVs, mix 150 μg of synthetic GFP siRNA(stock in RNase-free water, Seq: 5′-AAGCTGACCCTGAAGTTCAT-3′) with 150 μg of either RBD-tagged or control EVs(stock in PBS) in the microcentrifuge tube. Make up the volume to at 200 μL with citric acid buffer(18.6 mM citric acid, 29.4 mM disodium phosphate, pH = 4.4) and transfer the mixture to 0.4 cm electroporation cuvette. Electroporations were performed by Bio-Rad Gene Pulser electroporator with capacitance extender set at 400 V and 125 μF. Then purified EVs by ultracentrifugation and removed unencapsulated siRNA at 100,000 g for 70 min at 4 °C. Encapsulated siRNA of EVs were resuspended in phosphate buffered saline (PBS) and injected into the tail veins of 6-week-old female hACE2 mice(150 μg EV per mice) after infecting with SARS-CoV-2 pseudovirions though intranasal route(MOI = 50). The mice were sacrificed 48 h later and lung tissues were excised and fixed with 4% neutral buffered formaline, dehydrated and embedded in paraffin. Paraffin-embedded tissues were sectioned at a thickness of 4 μm for immunofluorescence according to standard procedures. Nuclear was visualized by staining with DAPI (Sigma) and examined under fluorescence microscopy and images were analyzed using an Nikon A1 confocal microscope(Tokyo, Japan). The GFP fluorescence was quantified with ImageJ (National Institutes of Health, USA), and analyzed for statistical significance using Graphpad Prism 8.
2.16. RNA extraction, reverse transcription and real-time PCR analysis
Encapsulation of siRNA in EVs was analyzed by real-time quantitative reverse transcription PCR. After electroporation and purified by ultracentrifugation, RNA of EV samples or naked siRNA were isolated using TRIzol reagent(TaKaRa) according to manufacturer's instructions. A total of 250 fmol of synthetic Caenorhabditis elegans miRNA cel-miR-39 (RiBoBio, Guangzhou, China) was added to each sample as an internal control. Reverse transcription of samples using PrimeScript RT Master Mix for RT-PCR (TaKaRa) by specific reverse stemloop primers (RiBoBio, Guangzhou, China). Real-time PCR performed using ABI SYBR Green Master Mix (Life Technologies) on ABI QuantStudio 6 Detection System. All reactions were carried out using 2-ΔΔCt method.
2.17. Statistical analysis
All numerical data are presented as mean ± SEM using Graphpad Prism 8.0 software (GraphPad Software Inc., San Diego CA). Comparison of two groups were analyzed by two-tailed Student's t-test. P value of 0.05 and below was considered significant: *p < 0.05, **p < 0.01, ***p < 0.001.
3. Results
3.1. Design and construction of SARS-CoV-2 S protein RBD-tagged EVs
First, to analyze whether the spike (S) glycoprotein of the SARS-CoV-2 could be incorporated into EVs, cell lysates of 293T cells transiently transfected with different amounts of FLAG-tagged SARS-CoV-2 S gene expression plasmids were analyzed by Western blot using anti-FLAG antibody. As shown in Fig. 1A, the S proteins were readily detected in the whole cell lysates of transfected cells with a molecular weight of approximately 180 kDa, but was not detected in the purified EV fractions from the transfected 293T cells. Purified EVs were further characterized for the presence of conventional markers for exsosomes, namely, CD9 and Alix, and non-EV markers GM130 (Golgi matrix marker) calnexin (endoplasmic reticulum marker) and calnexin (endoplasmic reticulum marker) followed the ‘Minimal information for studies of extracellular vesicles 2018’ (MISEV2018) guidelines [25](Fig. 1A). To confer EVs targeting capabilities, we incorporated the RBD of S protein into the membrane of EVs. While the ectodomain of VSVG coding region was replaced by the RBD of S protein, the signal peptide (SP), transmembrane helix and cytoplasmic domain of VSVG were left intact in this fusion protein to ensure the fusion proteins were properly anchored with the correct topography on EV membranes (Fig. 1B). By domain swapping, the expression of RBD was robustly detected in transfected 293T cells-derived EVs (EV-RBD) as indicated by the Western blot analysis with RBD antibody, compared to that in EVs from VSVG-transfected(exo-VSVG) or non-transfected cells(EV-NC) (Fig. 1C). CD9 and Alix, served as the protein markers for the presence of EVs. To explore the fusion protein was properly anchored with the correct topography on EV membrane, we used immunological pull-down assay from suspensions with anti-RBD antibody followed by magnetic beads (Fig. 1D). As shown in Fig. 1E, the RBD protein was only detected in EV-RBD fraction, confirming the RBD epitope is localized at the external EV surface. The presence of CD9 and Alix in EVs from pull-down assay confirmed they were EVs. Together, our results demonstrated that the modified EVs with RBD tag was successfully localized to the external EV membrane.
Fig. 1.
Construction of SARS-CoV-2 S protein RBD-tagged EVs. (A) Detection of SARS-CoV-2 Spike protein in cell lysates or EV pellets by western blot. 293T cells were transfected with plasmid expressing the FLAG-tagged Spike protein. EVs pelleted from the media were analyzed by western blot for the presence of the Spike protein using the FLAG M2 antibody. Purified EVs were characterized for EV markers CD9 and Alix, and non-EV markers calnexin (endoplasmic reticulum marker) and GM130 (Golgi matrix marker). (B) Schematic illustration of RBD-VSVG fusion-loaded EVs. The VSVG ectodomain was replaced by the RBD domain of SARS-CoV-2 (right, red), followed by a transmembrane helix and a cytoplasmic tail of VSVG (right, green). (C) Western blot analysis of EVs purified from 293T cells transfected with or without RBD-VSVG-vector for 48 h. CD9 or Alix proteins served as markers for the presence of EVs.(D, E) Schematic presentation of affinity pull-down of EVs by anti-RBD coupled magnetic beads. (D)EVs isolated from supernatants of transfected or non-transfected 293T cells were subjected to anti-RBD pull-down assay, then total protein of EVs was analyzed by western blot (E). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Characterization of physical properties of SARS-CoV-2 S protein RBD-tagged EVs
Next, we took the ratio of particles per μg of protein as a measure of purity of EVs from supernatant under our culture conditions refer to the MISEV2018 as well as previous study [26]. Culture supernatant from RBD-transfected or non-transfected 239T cells gave the ratio of approximately 4.96 × 109 or 4.27 × 109 particles per μg protein (P/μg), respectively. As a background, the detection for cell free supernatant within 10% EV-deleted FBS represented a ratio of 4.69 × 105 P/μg(Fig. 2A). To investigate if the incorporation of S protein RBD into the membrane of EV could affect the EV overall physical properties, the size distribution of the RBD-tagged EVs was examined by Nanoparticle Tracking Analysis (NTA). As shown in Fig. 2B, the average diameters of modified EVs (EV-RBD) integrated with RBD were similar to that of non-modified EVs (EV-NC), with peaks of 121.6 nm for EV-NC and 128.1 nm for EV-RBD. Further, similar results were also obtained with the analysis by electron microscopy, showing that the EV-RBD and EV-NC displayed expected morphology of small, round vesicles with a clearly discernible lipid bilayer (Fig. 2C). Finally, the concentrations of total proteins and RNA were measured from an equal amount of EVs(~1 × 108 for each sample), showing there were no significant differences in the EV contents of proteins and RNA between EV-RBD and EV-NC EVs (Fig. 2D and E). Together, these results demonstrated that incorporation of EVs with SARS-CoV-2 S protein RBD did not cause alterations of the physical properties and the cargo amounts of the EVs.
Fig. 2.
Characterization of physical properties of SARS-CoV-2 S protein RBD-tagged EVs. (A) The ratio of particles to protein for each sample is shown. (B)Histogram showing the size distribution of the purified EVs as analyzed by NTA. (C) TEM or SEM images of RBD-tagged or control EVs purified from 293T cells. (D and E) EV protein and RNA concentration measurement. EV protein or RNA was extracted from 1 × 108 EVs and protein concentration was measured by BCA protein quantification (D) and RNA concentration was determined by NanoDrop (E). Data are shown as mean ± SEM of four independent experiments, each experiment performs triplicate.
3.3. ACE2-dependent cellular uptake and targeting of SARS-CoV-2 S protein RBD-tagged EVs
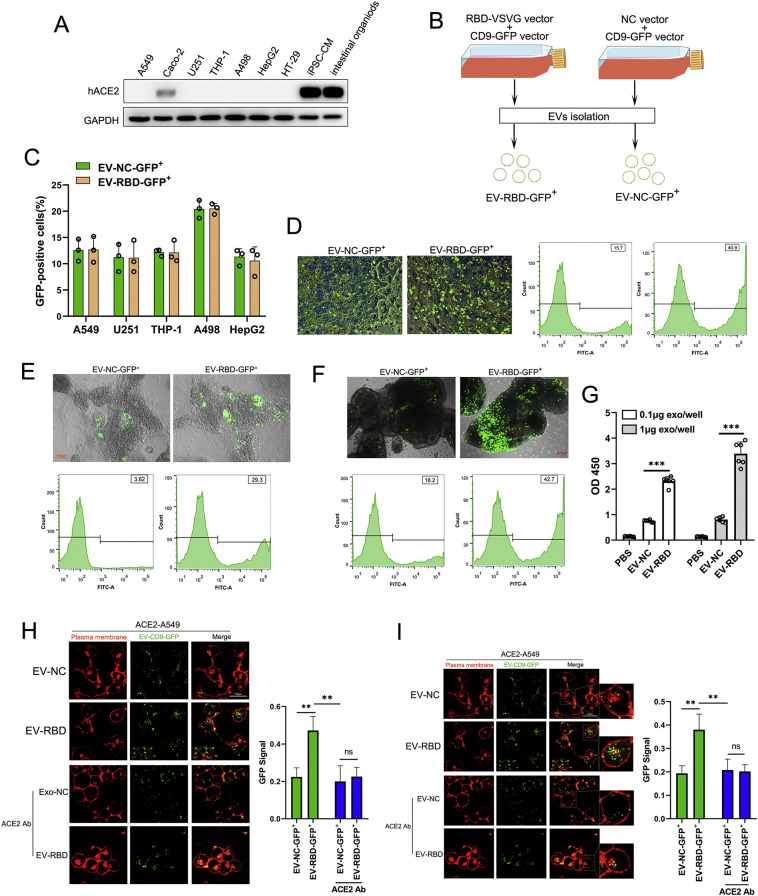
The binding of the RBD of S protein to ACE2 receptor at the surface of target cells is required for the viral cellular entry during SARS-CoV-2 infection [27]. We thus assessed whether RBD-tagged EVs are able to specifically target those cells or tissues that highly express ACE2 receptors. First, the expression level of ACE2 was examined in a number of different human tissue-derived cell lines including A549 (lung tissue), Caco-2(colon tissue), U251(neuroglia), THP-1(monocyte), A498(kidney tissue), HepG2 (liver tissue) and HT-29(colon tissue) cells. Whole cell lysates from these cell lines were extracted and subject to Western blot analysis with antibody against ACE2. As shown in Fig. 3A, there was no obvious ACE2 detected, whereas as a positive control, ACE2 was highly expressed in Caco-2 cells, human iPS cells-derived cardiomyocytes (iPS-CM) and intestinal organoids.
Fig. 3.
ACE2 is required for the cellular uptake and targeting of SARS-CoV-2 S protein RBD-tagged EVs. (A)Western blot analysis of human ACE2 was performed with cell lysates from various cell lines or organoid as where indicated using antibody against ACE2. (B) Schematic illustration of RBD-GFP-loaded EVs. Cultured cells were transfected with RBD-VSVG vector or empty vector (NC-vector) in combination with CD9-GFP vector. 48 h post transfection, the EV fraction that was labeled with GFP was collected(EV-RBD-GFP+/EV-NC-GFP+). (C) Flow cytometric analysis of GFP ratio in different cell lines after incubating with 1 × 109 of EV-RBD-GFP+ or EV-NC-GFP+ for 6 h. Different symbols correspond to independent experiments. Data are shown as mean ± SEM of three independent experiments, each individual experiment performs triplicate. (D, E and F) Representative images of Caco-2 cells (D), iPS cells-derived cardiomyocytes (E) and intestinal organoids (F) after application of 1 × 109 EV-RBD-GFP+ or EV-NC-GFP+ for 6 h. The GFP ratio was determined by flow cytometric analysis. (G) The binding affinity for RBD-tagged EVs with ACE2 protein was quantified by ELISA. Different concentration of EVs were incubated and followed by the anti-CD9 as the detector antibody. Each concentration was repeated with six wells. Data are shown as mean ± SEM of three independent experiments. (H and I) Representative images of ACE2-A549 cells showed an earlier plasma membrane distribution of EV-RBD-GFP+ or EV-NC-GFP+ incubation for 1 h (H) and 6 h (I) with or without pre-treated with ACE2 antibody. Fluorescence intensity of GFP was analyzed by ImageJ software were averaged from six different fields for each. Data are shown as mean ± SEM of three independent experiments. **p < 0.01 ***p < 0.001.ns: no significant.
In order to track the RBD-tagged EVs, we co-transfected 293T cells with the CD9-GFP fusion vector (CD9-GFP vector), RBD-VSVG fusion vector (RBD-VSVG vector) or empty vector(NC vector) as schematically illustrated in Fig. 3B. 48 h post transfection, the purified EV fraction that labeled with GFP was collected (EV-RBD-GFP+/EV-NC-GFP+) and was then incubated with different human tissue-derived cell lines or organoids for 24 h. Flow cytometric analysis showed that there was no significant difference of GFP ratio between cells inoculated with EV-RBD-GFP+ and cells inoculated with EV-NC-GFP+ (Fig. 3C). Of note, interestingly, the intensity of GFP was significantly higher with cells incubated with EV-RBD-GFP+ compared to that with cells inoculated with EV-NC-GFP+ incubation in Caco-2 cells(Fig. 3D), iPS-CM(Fig. 3E) and intestinal organoids(Fig. 3F), suggesting RBD-tagged EVs specifically target cells with high ACE2 expression. Furthermore, We also test the binding affinity for RBD-tagged EVs with ACE2 protein by enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 3G, EV-RBD have remarkable higher binding affinity with ACE2 protein compared to EV-NC or PBS, and this affinity could dependent on the dose of EVs.
To further access whether the RBD-tagged EVs require the ACE2 receptor for cellular uptake and targeting, we constructed stable ACE2-expressing A549 cells using lentiviral system (ACE2-A549 cells) and was then incubated with an equal amount(~1 × 109) of EV-RBD-GFP+ or EV-NC-GFP+. Fluorescence microscopy images showed a greater number of punctated GFP were attached onto plasma membranes of ACE2-A549 cells when incubated with EV-RBD-GFP+ compared to that with EV-NC-GFP+(Fig. 3H). To further confirm this specific binding, ACE2 specific-antibody(ACE2 Ab) was pre-treated ACE2-A549 cells for 2 h and then incubated with EV-RBD-GFP+ or EV-NC-GFP+. We observed that ACE2 Ab inhibited RBD-mediated binding with high efficiency in ACE2-A549 cells(Fig. 3H). Moreover, with the increasing time of incubation after 6 h, the number of GFP puncta accumulated at intracellular regions of ACE2-A549 cells were becoming much stronger in the presence of EV-RBD-GFP+ compared to EV-NC-GFP+ (Fig. 3I), supporting the evidence that EVs uptake through endocytic pathways rather than plasma membrane fusion. Block membrane of ACE2 by specific antibody showed an inhibitory enter of EV-RBD-GFP+ while no inhibitory effect for EV-NC-GFP+(Fig. 3I). Taken together, our results demonstrated that the RBD-tagged EVs could recognize specific ACE2 protein on the surface of recipient cells and facilitate attachment and internalization.
3.4. SARS-CoV-2 S protein RBD-tagged EVs specifically target tissues in vivo
Next, we investigated the potential for RBD-tagged EVs mediated systemic targets in vivo. To better characterize the tissue distribution, we used the transgenic human ACE2 mice (hACE2 mice) for evaluating. Western blot analysis demonstrated that human ACE2 protein was high expression in heart, lung, liver, kidney and small intestine with specific anti-human ACE2 antibody (Fig. 4A). Mice received an equal number of DiD labeled EV-RBD or EV-NC by intravenous injection, followed by fluorescence imaging in a 24-h interval. In vivo fluorescence imaging showed there were different signal distribution between EV-RBD and EV-NC (red fluorescence) from 24 h to 96 h post injection(hpi) (Fig. 4B). Ex vivo whole-organ fluorescence imaging demonstrated that DiD labeled EV-RBD were accumulated primarily in heart, lung and kidney at 24 h post injection, while no signal was detected in those tissues when injected with labeled EV-NC. Of note, the accumulated fluorescence signals of the labeled EV-RBD were decayed in heart and kidney at 72 h and 96 hpi, respectively (Fig. 4C vii and viii, Fig. 4D), but retained in lung tissues over 96 hpi (Fig. 4C viii, Fig. 4D). In contrast, upon injection of DiD labeled EV-NC, it only yielded fluorescence signals in the liver and intestine in the period of 24–96 hpi (Fig. 4C i-iv, Fig. 4D). To further determine if RBD-tagged EVs can induce immune responses in vivo, we assessed the serum concentration of IL-6, TNF-α and IFN-β in mice injected with EV-RBD or EV-NC as a mock control by ELISA. As shown in Fig. 4E-G, there were no significant changes in all cytokines assessed between EV-RBD and mock injection groups, indicating the immunologically inert profile of the EVs. Together, these observations suggested that RBD-tagged EVs were able to specifically target tissues of heart, lung and kidney, the main target organs of SARS-CoV-2 viruses.
Fig. 4.
SARS-CoV-2 S protein RBD-tagged EVs specifically target tissues in vivo. (A) Western blot analysis of tissue distribution of hACE2 in humanized mice using antibody against human ACE2. (B,C,D) After intravenous injection of DiD-labeled EV-RBD or EV-NC in hACE2 mice(n = 4), the red-fluorescence of whole animal was detected at the indicated time points by IVIS spectrum (B) Representative IVIS images of various organs were acquired at the indicated time points through intravenous injection of EVs(C). Radiant efficiency was measured using Living Image software. Four mice were sacrificed in each time-point for tissue collection(D). (E, F, G) Serum cytokines concentrations of IL-6 (E), TNF-α (F) and IFN-β (G) were measured by ELISA. Serum samples were from hACE2 mice after intravenous injection of EV-RBD or EV-NC at 24 h. Each mouse serum was repeated with four wells. All data expressed as mean ± SEM of four independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Delivery of siRNAs as a model system to identify inhibition of SARS-CoV-2 pseudovirus infection in vivo
To confirm the therapeutic potential of RBD-tagged EVs, we next investigated delivery of antiviral agents against SARS-CoV-2 genome. Given the SARS-CoV-2 is a highly infective and pathogenic native virus, we used an HIV-based lentiviral system to produce viral particles pseudotyped with SARS-CoV-2 Spike, which can infect permissive cells or tissues that express the SARS-CoV-2 receptor ACE2. As the SARS-CoV-2 pseudovirus contained the lentiviral backbone that can encode the reporter protein GFP, it was named as SARS-CoV-2-GFP. We investigated the possibility of loading RBD-tagged EVs with specific siRNA of GFP using electroporation methods adapted for nanoscale applications in vitro. To detect the electroporated efficiency of siRNA loading, EVs were purified after electroporation and the amount of siRNA in the EVs was assayed by real-time PCR. It was quantified that approximately 40% of the siRNA was loaded into the EV-NC or EV-RBD after electroporation(Fig. 5A). Moreover, we also found that EV-containing siRNA-GFP could resist benzonase degradation while naked siRNA was digested, suggesting electroporated siRNA was indeed encapsulated inside the EVs(Fig. 5A). The ACE2-A549 cell line was infected with SARS-CoV-2-GFP for 24 h(MOI = 2), and then incubated for 24 h with GFP siRNA alone(Naked si-GFP), unmodified or modified EVs electroporated with the siRNA(EV-NC-si-GFP and EV-RBD-si-GFP, respectively)(Fig. 5B). High delivery efficiency was confirmed by fluorescence microscopy, showing GFP protein knockdown was achieved with EV-RBD compared to EV-NC, suggesting that the modified constructs successfully endowed EVs with cell targeting capabilities(Fig. 5C).
Fig. 5.
Delivery of siRNAs as a model system to identify inhibition of SARS-CoV-2 pseudovirus infection in vivo. (A) The efficiency of siRNA loading in EVs after electroporation as measured by real-time PCR. Each sample contained mixture of 150 μg siRNA-GFP and 150 μg EVs or 150 μg naked siRNA. After electroporation, purified EVs by ultracentrifugation to remove unencapsulated siRNA and each EV pellet treated with or without Benzonase. All electroporation samples were prepared in triplicate and each RNA isolate was analyzed in duplicate. (B) Scheme of the possibility of loading unmodified or modified EVs with specific siRNA into target cells. (C) Representative images of pseudotyped SARS-CoV-2-GFP-infected ACE2-A549 cells after incubation with different origin of EVs. The ACE2-A549 cells was infected with SARS-CoV-2-GFP for 24 h(MOI = 2), and then incubated for 24 h with naked si-GFP, EV-NC or EV-RBD electroporated with the siRNA, respectively. The GFP ratio was determined by flow cytometric analysis. Data are shown as mean ± SEM of three independent experiments in triplicates. (D) hACE2 mice(n = 4) were inoculated intranasally with pseudotyped SARS-CoV-2-GFP at a multiplicity of infection (MOI) of 50 per 50 μl inoculum volume per mouse. At 24 h post-infection,150 μg RBD-tagged EVs(EV-RBD-si-GFP) or control EVs (EV-NC-si-GFP) loaded with 150 μg GFP siRNA were injected into tail veins, and all mice were sacrificed at 48 h post injection for lung tissues collection.(E) Immunofluorescence staining of mouse lung paraffin sections for pseudotyped SARS-CoV-2-GFP (green, white arrows) and DAPI(blue). (F) Each hACE2 mouse(n = 4) injected with 150 μg RBD-tagged EVs encapsulated GFP siRNA or control EVs in 24 h and later inoculated intranasally with pseudotyped SARS-CoV-2-GFP at MOI of 50. All mice were sacrificed at 48 h post infection for lung tissues collection (G) Immunofluorescence staining analysis for pseudotyped SARS-CoV-2-GFP infected cells in lung paraffin sections (green, white arrows). Fluorescence intensity of GFP in the regions were analyzed through ImageJ software. Data are shown as mean ± SEM of four independent experiments in triplicates.**p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To investigated the therapeutic potential of RBD-tagged EVs in vivo, hACE2 mice were inoculated intranasally with SARS-CoV-2-GFP at a multiplicity of infection (MOI) of 50 per mouse. At 24 h post-infection, the RBD-tagged EVs encapsulated GFP-siRNA (EV-RBD-si-GFP) or control EVs(EV-NC-si-GFP) were injected into tail veins. At 48 h post injection, immunostaining of mouse lung section was examined(Fig. 5D). As shown in Fig. 5E, the virus load as indicated by the GFP signals of SARS-CoV-2 were significantly lower(decreased by ~65%) in the lung from mice with injection of EV-RBD-si-GFP than that from mice with injection of EV-NC-si-GFP or naked si-GFP, suggesting RBD-tagged EVs encapsulated siRNAs as a potential means of tissue targeting for inhibiting SARS-CoV-2 infection. Furthermore, hACE2 mice were pre-injected with EV-RBD-si-GFP, EV-NC-si-GFP or naked si-GFP, followed by infection with SARS-CoV-2-GFP pseudovirus at 24 hpi. All mice were sacrificed after 48 h of infection, and lung tissues were collected(Fig. 5F). As showed in Fig. 5G, injection of EV-RBD-si-GFP resulted in a significant decrease (fell by~50%)in GFP signals detected in lung regions compared to the that with injection of EV-NC-si-GFP or naked si-GFP, indicating RBD-tagged EVs with specific siRNA against SARS-CoV-2 pseudovirus infection may have alternative approaches. Together, our experiments implied that RBD-tagged EVs-mediated delivery of antiviral agents into target tissues may as a promising vehicle for treatment of SARS-CoV-2 infection.
4. Discussion
Since December 2019, a novel coronavirus SARS-CoV-2 has emerged and rapidly spread throughout the world, resulting in a global public health emergency [1,2] . As EVs progress toward the clinic for treatment of a vast array of diseases, it is important to develop efficient, tissue-specific and non-immunogenic delivery technologies to exploit as the targeted therapeutic particles [28]. In general, EVs are recognized and endocytosed by tissue cells via specific interactions between surface membrane proteins, where after they deliver their molecular cargo [10,17,29]. This property provides the potential to modify the membrane proteins in EV surface as a promising means of tissue targeting.
In this study, we described a VSVG pseudotyping approach to load EV membranes with SARS-CoV-2 RBD proteins, the key domain in viral attachment, fusion and entry. VSVG was incorporated into the viral envelope during the budding of microdomains at the plasma membrane, the same domains involved in the formation of endosomes and EVs [19,30]. Previous study revealed that the VSVG without the ectodomain could integrate into the EV membrane [19]. This led us to propose the possibility of engineering of EVs with specific membrane-bound proteins by expressing gene-encoding VSVG fusion proteins in cells. Thus, we incorporated the RBD of S protein on the surface of EVs replaced ectodomain of VSVG coding region and fusion protein was confirmed anchored with the correct topography on EV membrane (RBD-tagged EVs) without affecting the physical properties of the modified EVs. As expected, the RBD-tagged EVs required the ACE2 receptor for cellular uptake and would be able to specifically trend to target cells or tissues that highly express ACE2 receptors. Several research groups have exploited the methods to display target peptides on the external EVs/exosomes surface by fusing Lamp2b, an EV membrane protein [17,31]. We also have tried to incorporate the RBD protein on the surface of EVs by fusing Lamp2b at the extra-EV N terminus, but we found that the RBD protein was unable to express effectively on the EV membrane(data not shown). One possibility may be RBD fused with Lamp2b could affect the protein refolding of Lamp2b resulted in failing membrane localization. In addition, previous study confirmed that viral glycoproteins could directly express on the surface membrane of cells lead to incorporation into EVs released from transfected cells, such as VSVG and HIV-1 gp120(env) protein [30,32]. However, we did not detect that the spike glycoprotein(S) of the SARS-CoV-2 could incorporated into EVs from S expression vector-transfected 293T cells. Similar results were also obtained from S proteins of SARS-CoV [33], another highly fatal coronavirus, suggesting the native S protein of coronavirus incorporated into EVs particles through transfection of expression vectors were inefficient.
In vivo fluorescence imaging demonstrated that RBD-tagged EVs could accumulate specifically in heart, lung and kidney through using the hACE2 transgenic mouse model. An advantage of the use of DiD dye in our study was that at longer excitation wavelengths the tissue auto-fluorescence background levels were low, reducing the signal-to-noise ratio. The DiD-labeled EVs was irreversible and has a long-term stability in vivo without affect basic physiological properties [[34], [35], [36]]. The transgenic mouse model expressing human ACE2 has been successfully evaluated the infectivity and pathological changes following SARS-CoV-2 infection [37,38]. Recent study observed that SARS-CoV-2 could use human, bat, or civet ACE2 as a cellular receptor but not the mouse ACE2 [39]. Therefore, it appeared that hACE2 transgenic mouse would be a reasonable choice for evaluating delivery of RBD-tagged EVs in vivo. Notably, although several pioneering investigations have confirmed that both SARS-CoV-2 and SARS-CoV utilized the same cellular receptor hACE2 to enter cells, the crystal structure of the SARS-CoV-2 S protein RBD binding to hACE2 had an estimated 10- to 20-fold higher affinity toward hACE2 than SARS-CoV has [4,27]. It could be speculated that SARS-CoV-2 RBD-tagged EVs may have a more compact conformation with binding to ACE2-expressed cells or tissues. Interestingly, negative-staining electron microscopy of SARS-CoV-2 virions showed average diameters of ~100 nm membrane-enclosed particles [40], which have a similar size to that of RBD-tagged EVs with a diameter of ~120 nm in our study. Both EVs and coronavirus shared the intracellular vesicle trafficking resulted in similarities between EVs and coronavirus envelopes in lipid composition [41]. These similarities suggested that as a coronavirus-like particles, the RBD-tagged EVs could be used as a safety tool to explore the pathogenesis and primary tissue tropism of SARS-CoV-2 virus, or even as a vaccine candidate. A study from Choudhury et al indicated that the Spike protein of SARS-CoV-2 has the potential to interact with certain members of human TLR4 and most likely to be involved in SARS-CoV-2 to induce inflammatory responses [42], however, there was no direct evidence that the RBD interaction with TLR4. In addition, recent study showed that SARS-CoV-2 spike protein S1 subunit activated TLR4 signaling to induce pro-inflammatory responses in murine and human macrophages [43]. Although SARS-CoV-2 S1 contains the RBD that specifically recognizes ACE2 as its receptor, no data supported the RBD-ACE2 binding contributed to mediating pro-inflammatory responses in macrophages. Importantly, S1 has no or low affinity for murine ACE2, suggesting the ACE2 may not be the main contributor to S1-mediated pro-inflammatory responses in macrophages. These findings reinforced safety to utilize RBD-tagged EVs for therapeutics or prophylactics.
Several reports have demonstrated engineering EVs has the capacity to deliver therapeutic compounds to specific tissues or cell types. For example, Alvarez-Erviti et al showed that loading modified EVs/EVs with rabies viral glycoprotein(RVG), a peptide that binds the acetylcholine receptor, could deliver GAPDH or BACE1 siRNA specifically to brain regions, resulting in specific genes knockdown [17]. In another study, Haney et al have utilized EV-based delivery system for a potent antioxidant, catalase, to treat Parkinson's disease (PD) with non-cytotoxic effects [44]. In our study, we introduced the RBD-tagged EVs encapsulated GFP siRNAs as a model agent means of lung tissue targeting for inhibiting pseudotyped SARS-CoV-2 infection without immune stimulation. Although there were some limitations, it could be able to conceivable that deliver the authentic therapeutic agents of COVID-19 using RBD-tagged EVs have the effect and function. Further verification should be needed. All these data highlighted the possibilities of engineering of EVs are a promising system for agents delivery with better safety profiles than those of viral vectors, cationic lipids, or polymer based particles. Of note, although electroporation of siRNA into EVs was less efficient which reported by several studies [17,[45], [46], [47]] and the loading efficiency of siRNA into EVs in our case was around 40%, electroporated method was still the most extensive approach. Our and others groups data [17,45] confirmed that siRNA encapsulated inside the EVs by electroporation was functional in vitro and in vitro, suggesting the low efficiency of siRNA loading has little influence on potential target-function.
Although the inflammation of the lung is the primary symptom of patients with SARS-CoV-2 infection because of an abundant expression of ACE2, many data revealed that the ACE2 was expressed in a wide variety of human tissues, especially in heart, kidney, small intestine and testis with the highest expression levels, which indicated an intrinsic susceptibility to SARS-CoV-2 infection [5,48]. Indeed, the virus-related cardiac injury in patients with SARS-CoV-2 infection were common for observation [2,49]. In addition, the acute kidney injury (AKI) occurrence has been often reported in patients with SARS-CoV-2 infection [50,51]. Our results showed RBD-tagged EVs would be able to specifically trend to heart and kidney tissues, implying this specific targeting of EVs may as a promising vehicle to deliver therapeutic antibodies or specific suppress drugs for treatment of SARS-CoV-2-offensive multi-organ. Nevertheless, the potential therapeutic interventions for RBD-tagged EVs were still needed to be evaluated using authentic SARS-CoV-2 in vivo. Remarkably, several studies reported that using EVs/EVs derived from Mesenchymal Stem Cells (MSCs) to treat COVID-19 pneumonia have shed some light on a potential cure for the COVID-19 infected patients [[52], [53], [54]], it may provide a potential to engineer EVs with RBD protein tag using MSCs to achieve better therapeutic effect for SARS-CoV-2 infection. Recent studies demonstrate that ACE2 alone, or in combination with TMPRSS2, could be present on the surface of EVs result in better efficacy as a decoy to capture SARS-CoV-2 S protein-containing lentivirus. Combined with our research, these studies pointed out that well-designed EV therapeutics against COVID-19 may be feasible to prevent initial infection or further internal dissemination of the virus [55,56].
5. Conclusions
In the present study, we exploited a pseudotyping-based approach to load EV membranes with SARS-CoV-2 spike protein RBD through engineering the VSVG. Using the pseudotyped SARS-CoV-2 model, we demonstrate that the RBD-tagged EVs that encapsulate siRNAs against SARS-CoV-2 pseudovirus can specifically target lung tissues and suppress the pseudovirus infection in vivo without immune stimulation. Importantly, our results showed RBD-tagged EVs would be able to specifically trend to multiple organs including heart and kidney, implying this specific targeting of EVs may as a promising vehicle to deliver antiviral agents for treatment of SARS-CoV-2-offensive multi-organ. Our work presents a safe and effective engineered EV system and may serve as a vehicle to deliver therapeutic antibodies or specific suppress drugs for treatment of SARS-CoV-2-offensive multi-organ. Nevertheless, the potential therapeutic interventions for RBD-tagged EVs were still needed to be evaluated using authentic SARS-CoV-2.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grants [31670898, 31970844 and 31900679], the National Science and Technology Key Project (2018ZX10731301-004-003), Jiangsu Provincial Innovative Research Team Project, the Priority Academic Program Development of Jiangsu Higher Education Institutions and the China Postdoctoral Science Foundation under Grants [2018M640521 and 2020T130090ZX].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2021.05.049.
Appendix A. Supplementary data
Supplementary material
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (Lond. Engl.) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. (e1019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benítez-Cardoza C.G., Vique-Sánchez J.L. Potential inhibitors of the interaction between ACE2 and SARS-CoV-2 (RBD), to develop a drug. Life Sci. 2020;256:117970. doi: 10.1016/j.lfs.2020.117970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. (e899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv Z., Deng Y.Q., Ye Q., Cao L., Sun C.Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., Chen Q., Wang N., Nie J., Cui Z., Zhu D., Shaw N., Li X.F., Li Q., Xie L., Wang Y., Rao Z., Qin C.F., Wang X. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 11.Vlassov A.V., Magdaleno S., Setterquist R., Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Kanada M., Bachmann M.H., Hardy J.W., Frimannson D.O., Bronsart L., Wang A., Sylvester M.D., Schmidt T.L., Kaspar R.L., Butte M.J., Matin A.C., Contag C.H. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1433–E1442. doi: 10.1073/pnas.1418401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan X., Sansanaphongpricha K., Myers I., Chen H., Yuan H., Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samir E.L.A., Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y., Nishikawa M., Shinotsuka H., Matsui Y., Ohara S., Imai T., Takakura Y. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 2013;165:77–84. doi: 10.1016/j.jbiotec.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Bala S., Csak T., Momen-Heravi F., Lippai D., Kodys K., Catalano D., Satishchandran A., Ambros V., Szabo G. Biodistribution and function of extracellular miRNA-155 in mice. Sci. Rep. 2015;5:10721. doi: 10.1038/srep10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 18.Vandergriff A., Huang K., Shen D., Hu S., Hensley M.T., Caranasos T.G., Qian L., Cheng K. Targeting regenerative exosomes to myocardial infarction using cardiac homing peptide. Theranostics. 2018;8:1869–1878. doi: 10.7150/thno.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer C., Losacco J., Stickney Z., Li L., Marriott G., Lu B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int. J. Nanomedicine. 2017;12:3153–3170. doi: 10.2147/IJN.S133430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., Hong Y., Nam G.H., Chung J.H., Koh E., Kim I.S. Virus-mimetic fusogenic exosomes for direct delivery of integral membrane proteins to target cell membranes. Adv. Mater. 2017;29(13) doi: 10.1002/adma.201605604. [DOI] [PubMed] [Google Scholar]
- 21.Van Deun J., Mestdagh P., Agostinis P., Akay Ö., Anand S., Anckaert J., Martinez Z.A., Baetens T., Beghein E., Bertier L., Berx G., Boere J., Boukouris S., Bremer M., Buschmann D., Byrd J.B., Casert C., Cheng L., Cmoch A., Daveloose D., De Smedt E., Demirsoy S., Depoorter V., Dhondt B., Driedonks T.A., Dudek A., Elsharawy A., Floris I., Foers A.D., Gärtner K., Garg A.D., Geeurickx E., Gettemans J., Ghazavi F., Giebel B., Kormelink T.G., Hancock G., Helsmoortel H., Hill A.F., Hyenne V., Kalra H., Kim D., Kowal J., Kraemer S., Leidinger P., Leonelli C., Liang Y., Lippens L., Liu S., Cicero A. Lo, Martin S., Mathivanan S., Mathiyalagan P., Matusek T., Milani G., Monguió-Tortajada M., Mus L.M., Muth D.C., Németh A., Nolte-'t Hoen E.N., O'Driscoll L., Palmulli R., Pfaffl M.W., Primdal-Bengtson B., Romano E., Rousseau Q., Sahoo S., Sampaio N., Samuel M., Scicluna B., Soen B., Steels A., Swinnen J.V., Takatalo M., Thaminy S., Théry C., Tulkens J., Van Audenhove I., van der Grein S., Van Goethem A., van Herwijnen M.J., Van Niel G., Van Roy N., Van Vliet A.R., Vandamme N., Vanhauwaert S., Vergauwen G., Verweij F., Wallaert A., Wauben M., Witwer K.W., Zonneveld M.I., De Wever O., Vandesompele J., Hendrix A. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017;14:228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 22.Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., Shroyer N.F., Wells J.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breckwoldt K., Letuffe-Breniere D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S., Shibamiya A., Prondzynski M., Mearini G., Schade D., Fuchs S., Neuber C., Kramer E., Saleem U., Schulze M.L., Rodriguez M.L., Eschenhagen T., Hansen A. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- 24.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., Pettie D., King N.P., Balazs A.B., Bloom J.D. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12 doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., Ayre D.C., Bach J.M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N.N., Baxter A.A., Bebawy M., Beckham C., Zavec A. Bedina, Benmoussa A., Berardi A.C., Bergese P., Bielska E., Blenkiron C., Bobis-Wozowicz S., Boilard E., Boireau W., Bongiovanni A., Borràs F.E., Bosch S., Boulanger C.M., Breakefield X., Breglio A.M., Brennan M., Brigstock D.R., Brisson A., Broekman M.L., Bromberg J.F., Bryl-Górecka P., Buch S., Buck A.H., Burger D., Busatto S., Buschmann D., Bussolati B., Buzás E.I., Byrd J.B., Camussi G., Carter D.R., Caruso S., Chamley L.W., Chang Y.T., Chen C., Chen S., Cheng L., Chin A.R., Clayton A., Clerici S.P., Cocks A., Cocucci E., Coffey R.J., Cordeiro-da-Silva A., Couch Y., Coumans F.A., Coyle B., Crescitelli R., Criado M.F., D'Souza-Schorey C., Das S., Chaudhuri A. Datta, de Candia P., De Santana E.F., De Wever O., Del Portillo H.A., Demaret T., Deville S., Devitt A., Dhondt B., Di Vizio D., Dieterich L.C., Dolo V., Rubio A.P. Dominguez, Dominici M., Dourado M.R., Driedonks T.A., Duarte F.V., Duncan H.M., Eichenberger R.M., Ekström K., El Andaloussi S., Elie-Caille C., Erdbrügger U., Falcón-Pérez J.M., Fatima F., Fish J.E., Flores-Bellver M., Försönits A., Frelet-Barrand A., Fricke F., Fuhrmann G., Gabrielsson S., Gámez-Valero A., Gardiner C., Gärtner K., Gaudin R., Gho Y.S., Giebel B., Gilbert C., Gimona M., Giusti I., Goberdhan D.C., Görgens A., Gorski S.M., Greening D.W., Gross J.C., Gualerzi A., Gupta G.N., Gustafson D., Handberg A., Haraszti R.A., Harrison P., Hegyesi H., Hendrix A., Hill A.F., Hochberg F.H., Hoffmann K.F., Holder B., Holthofer H., Hosseinkhani B., Hu G., Huang Y., Huber V., Hunt S., Ibrahim A.G., Ikezu T., Inal J.M., Isin M., Ivanova A., Jackson H.K., Jacobsen S., Jay S.M., Jayachandran M., Jenster G., Jiang L., Johnson S.M., Jones J.C., Jong A., Jovanovic-Talisman T., Jung S., Kalluri R., Kano S.I., Kaur S., Kawamura Y., Keller E.T., Khamari D., Khomyakova E., Khvorova A., Kierulf P., Kim K.P., Kislinger T., Klingeborn M., Klinke D.J., 2nd, Kornek M., Kosanović M.M., Kovács F.A., Krämer-Albers E.M., Krasemann S., Krause M., Kurochkin I.V., Kusuma G.D., Kuypers S., Laitinen S., Langevin S.M., Languino L.R., Lannigan J., Lässer C., Laurent L.C., Lavieu G., Lázaro-Ibáñez E., Le Lay S., Lee M.S., Lee Y.X.F., Lemos D.S., Lenassi M., Leszczynska A., Li I.T., Liao K., Libregts S.F., Ligeti E., Lim R., Lim S.K., Linē A., Linnemannstöns K., Llorente A., Lombard C.A., Lorenowicz M.J., Lörincz M.A., Lötvall J., Lovett J., Lowry M.C., Loyer X., Lu Q., Lukomska B., Lunavat T.R., Maas S.L., Malhi H., Marcilla A., Mariani J., Mariscal J., Martens-Uzunova E.S., Martin-Jaular L., Martinez M.C., Martins V.R., Mathieu M., Mathivanan S., Maugeri M., McGinnis L.K., McVey M.J., Meckes D.G., Jr., Meehan K.L., Mertens I., Minciacchi V.R., Möller A., Jørgensen M. Møller, Morales-Kastresana A., Morhayim J., Mullier F., Muraca M., Musante L., Mussack V., Muth D.C., Myburgh K.H., Najrana T., Nawaz M., Nazarenko I., Nejsum P., Neri C., Neri T., Nieuwland R., Nimrichter L., Nolan J.P., Nolte't Hoen E.N., Hooten N. Noren, O'Driscoll L., O'Grady T., O'Loghlen A., Ochiya T., Olivier M., Ortiz A., Ortiz L.A., Osteikoetxea X., Østergaard O., Ostrowski M., Park J., Pegtel D.M., Peinado H., Perut F., Pfaffl M.W., Phinney D.G., Pieters B.C., Pink R.C., Pisetsky D.S., von Strandmann E. Pogge, Polakovicova I., Poon I.K., Powell B.H., Prada I., Pulliam L., Quesenberry P., Radeghieri A., Raffai R.L., Raimondo S., Rak J., Ramirez M.I., Raposo G., Rayyan M.S., Regev-Rudzki N., Ricklefs F.L., Robbins P.D., Roberts D.D., Rodrigues S.C., Rohde E., Rome S., Rouschop K.M., Rughetti A., Russell A.E., Saá P., Sahoo S., Salas-Huenuleo E., Sánchez C., Saugstad J.A., Saul M.J., Schiffelers R.M., Schneider R., Schøyen T.H., Scott A., Shahaj E., Sharma S., Shatnyeva O., Shekari F., Shelke G.V., Shetty A.K., Shiba K., Siljander P.R., Silva A.M., Skowronek A., Snyder O.L., 2nd, Soares R.P., Sódar B.W., Soekmadji C., Sotillo J., Stahl P.D., Stoorvogel W., Stott S.L., Strasser E.F., Swift S., Tahara H., Tewari M., Timms K., Tiwari S., Tixeira R., Tkach M., Toh W.S., Tomasini R., Torrecilhas A.C., Tosar J.P., Toxavidis V., Urbanelli L., Vader P., van Balkom B.W., van der Grein S.G., Van Deun J., van Herwijnen M.J., Van Keuren-Jensen K., van Niel G., van Royen M.E., van Wijnen A.J., Vasconcelos M.H., Vechetti I.J., Jr., Veit T.D., Vella L.J., Velot É., Verweij F.J., Vestad B., Viñas J.L., Visnovitz T., Vukman K.V., Wahlgren J., Watson D.C., Wauben M.H., Weaver A., Webber J.P., Weber V., Wehman A.M., Weiss D.J., Welsh J.A., Wendt S., Wheelock A.M., Wiener Z., Witte L., Wolfram J., Xagorari A., Xander P., Xu J., Yan X., Yáñez-Mó M., Yin H., Yuana Y., Zappulli V., Zarubova J., Žėkas V., Zhang J.Y., Zhao Z., Zheng L., Zheutlin A.R., Zickler A.M., Zimmermann P., Zivkovic A.M., Zocco D., Zuba-Surma E.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracellular Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. (1535750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webber J., Clayton A. How pure are your vesicles? J. Extracellular Vesicles. 2013;2 doi: 10.3402/jev.v2i0.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y., El Andaloussi S., Wood M.J. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 2012;21:R125–R134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 29.Stickney Z., Losacco J., McDevitt S., Zhang Z., Lu B. Development of exosome surface display technology in living human cells. Biochem. Biophys. Res. Commun. 2016;472:53–59. doi: 10.1016/j.bbrc.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 30.Mangeot P.E., Dollet S., Girard M., Ciancia C., Joly S., Peschanski M., Lotteau V. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Therapy. 2011;19:1656–1666. doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mentkowski K.I., Lang J.K. Exosomes engineered to express a cardiomyocyte binding peptide demonstrate improved cardiac retention in vivo. Sci. Rep. 2019;9:10041. doi: 10.1038/s41598-019-46407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nanjundappa R.H., Wang R., Xie Y., Umeshappa C.S., Chibbar R., Wei Y., Liu Q., Xiang J. GP120-specific exosome-targeted T cell-based vaccine capable of stimulating DC- and CD4(+) T-independent CTL responses. Vaccine. 2011;29:3538–3547. doi: 10.1016/j.vaccine.2011.02.095. [DOI] [PubMed] [Google Scholar]
- 33.Kuate S., Cinatl J., Doerr H.W., Uberla K. Exosomal vaccines containing the S protein of the SARS coronavirus induce high levels of neutralizing antibodies. Virology. 2007;362:26–37. doi: 10.1016/j.virol.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrari A., Hannouche D., Oudina K., Bourguignon M., Meunier A., Sedel L., Petite H. In vivo tracking of bone marrow fibroblasts with fluorescent carbocyanine dye. J. Biomed. Mater. Res. 2001;56:361–367. doi: 10.1002/1097-4636(20010905)56:3<361::aid-jbm1104>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 35.Heinrich L., Freyria A.M., Melin M., Tourneur Y., Maksoud R., Bernengo J.C., Hartmann D.J. Confocal laser scanning microscopy using dialkylcarbocyanine dyes for cell tracing in hard and soft biomaterials. J Biomed Mater Res B Appl Biomater. 2007;81:153–161. doi: 10.1002/jbm.b.30648. [DOI] [PubMed] [Google Scholar]
- 36.Asahara T., Lin M., Kumazawa Y., Takeo K., Akamine T., Nishimura Y., Kayahara T., Yamamoto T. Long-term observation on the changes of somatotopy in the facial nucleus after nerve suture in the cat: morphological studies using retrograde labeling. Brain Res. Bull. 1999;49:195–202. doi: 10.1016/s0361-9230(99)00041-6. [DOI] [PubMed] [Google Scholar]
- 37.Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., Wang Q., Min J., Wang X., Zhang W., Li B., Zhang H.J., Baric R.S., Zhou P., Yang X.L., Shi Z.L. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting Enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. (e58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cleary S.J., Pitchford S.C., Amison R.T., Carrington R., Robaina Cabrera C.L., Magnen M., Looney M.R., Gray E., Page C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., Cheng L., Shi D., Lu X., Lei J., Crispin M., Shi Y., Li L., Li S. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e713. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolte-'t Hoen E., Cremer T., Gallo R.C., Margolis L.B. Extracellular vesicles and viruses: are they close relatives? Proc. Natl. Acad. Sci. U. S. A. 2016;113:9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirato K., Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., Patel T., Piroyan A., Sokolsky M., Kabanov A.V., Batrakova E.V. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J. Control. Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Andaloussi S., Lee Y., Lakhal-Littleton S., Li J., Seow Y., Gardiner C., Alvarez-Erviti L., Sargent I.L., Wood M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 46.Kooijmans S.A.A., Stremersch S., Braeckmans K., de Smedt S.C., Hendrix A., Wood M.J.A., Schiffelers R.M., Raemdonck K., Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release. 2013;172:229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 47.Wahlgren J., De L.K.T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116:1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet (Lond. Engl.) 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fanelli V., Fiorentino M., Cantaluppi V., Gesualdo L., Stallone G., Ronco C., Castellano G. Acute kidney injury in SARS-CoV-2 infected patients. Crit. Care (Lond. Engl.) 2020;24:155. doi: 10.1186/s13054-020-02872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghav A., Khan Z.A., Upadhayay V.K., Tripathi P., Gautam K.A., Mishra B.K., Ahmad J., Jeong G.B. Mesenchymal stem cell-derived exosomes exhibit promising potential for treating SARS-CoV-2-infected patients. Cells. 2021;10 doi: 10.3390/cells10030587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchiya A., Takeuchi S., Iwasawa T., Kumagai M., Sato T., Motegi S., Ishii Y., Koseki Y., Tomiyoshi K., Natsui K., Takeda N., Yoshida Y., Yamazaki F., Kojima Y., Watanabe Y., Kimura N., Tominaga K., Kamimura H., Takamura M., Terai S. Therapeutic potential of mesenchymal stem cells and their exosomes in severe novel coronavirus disease 2019 (COVID-19) cases. Inflamm. Regen. 2020;40:14. doi: 10.1186/s41232-020-00121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akbari A., Rezaie J. Potential therapeutic application of mesenchymal stem cell-derived exosomes in SARS-CoV-2 pneumonia. Stem Cell Res Ther. 2020;11:356. doi: 10.1186/s13287-020-01866-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cocozza F., Névo N., Piovesana E., Lahaye X., Buchrieser J., Schwartz O., Manel N., Tkach M., Théry C., Martin-Jaular L. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 spike protein-containing virus. J. Extracellular Vesicles. 2020;10 doi: 10.1002/jev2.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q., Jeppesen D.K., Higginbotham J.N., Franklin J.L., Crowe J.E., Jr., Coffey R.J. Angiotensin-converting Enzyme 2-containing small extracellular vesicles and exomeres bind the severe acute respiratory syndrome coronavirus 2 spike protein. Gastroenterology. 2021;160:958–961. doi: 10.1053/j.gastro.2020.09.042. (e953) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material