Abstract
Background:
Chronic respiratory diseases (CRD) are a major public health problem worldwide. In the current epidemiological context, CRD have received much interest when considering their correlation with greater susceptibility to SARS-Cov-2 and severe disease (COVID-19). Increasingly more studies have investigated pathophysiological interactions between CRD and COVID-19.
Area covered:
Animal experimentation has decisively contributed to advancing our knowledge of CRD. Considering the increase in ethical restrictions in animal experimentation, researchers must focus on new experimental alternatives. Two-dimensional (2D) cell cultures have complemented animal models and significantly contributed to advancing research in the life sciences. However, 2D cell cultures have several limitations in studies of cellular interactions. Three-dimensional (3D) cell cultures represent a new and robust platform for studying complex biological processes and are a promising alternative in regenerative and translational medicine.
Expert opinion:
Three-dimensional cell cultures are obtained by combining several types of cells in integrated and self-organized systems in a 3D structure. These 3D cell culture systems represent an efficient methodological approach in studies of pathophysiology and lung therapy. More recently, complex 3D culture systems, such as lung-on-a-chip, seek to mimic the physiology of a lung in vivo through a microsystem that simulates alveolar-capillary interactions and exposure to air. The present review introduces and discusses 3D lung cultures as robust platforms for studies of the pathophysiology of CRD and COVID-19 and the mechanisms that underlie interactions between CRD and COVID-19.
Keywords: SARS-CoV-2, Organoid, Spheroid, Lung-on-a-chip, Chronic respiratory diseases
Introduction
Respiratory diseases are a major public health problem worldwide. It is worth mentioning that five (“The Big Five”) of respiratory diseases (chronic obstructive pulmonary disease—COPD, asthma, acute respiratory infections, tuberculosis, and lung cancer) are among the most common causes of morbidity and mortality worldwide [1, 2]. In addition to the high incidence and prevalence of “The Big Five,” specifically asthma and COPD have acquired greater relevance during the current epidemiological coronavirus disease (COVID-19) epidemic and received intense research interest and high media attention. Such interest is attributable to the correlation that has been established between patients who have underlying chronic respiratory diseases (CRD) and greater susceptibility to SARS-CoV-2 infection.These patients, once infected, often progress to severe acute respiratory distress syndrome (ARDS) and lung failure [3, 4].
This correlation between the higher risk of CRD patients evolving to ARDS secondary to COVID-19 has been well established since initial reports in December 2019 of new cases of pneumonia in Wuhan, China, the epicenter of the outbreak. The causal agent was identified in January 2020 as a new coronavirus [5, 6]. Among pathophysiological aspects of COVID-19, the roles of angiotensin-converting enzyme 2 (ACE2) and several other molecules have been well studied and associated with inflammation, edema, and lung hypertension, which can lead to worsening of respiratory function in COPD and asthma patients who are more susceptible to SARS-CoV-2 infection and subsequently have a higher risk of severe disease [7, 8]. However, there still exist large gaps in our knowledge of the mechanisms that underlie molecular and pathophysiological interactions between CRD and COVID-19.
In the epidemiological context of COVID-19, the present review introduces new methodologies, particularly three-dimensional (3D) lung cell cultures, as potentially promising platforms that can provide new insights into the pathogenesis of COVID-19 and advance our understanding of the mechanisms that underlie pathophysiological interactions between COVID-19 and chronic lung diseases.These new 3D cell culture platforms are also expected to serve as a valuable research tool to test new therapeutic approaches for chronic lung diseases and COVID-19.
Pathogenesis and pathophysiology of COVID-19
Novel coronavirus was first isolated and identified in the city of Wuhan, China, under the provisory name 2019-nCoV, currently designated SARS-CoV-2. SARS-CoV-2 may have originated in bats. The infectious virus in these animals shares 96% similarity to the human form of the virus, but other animals have also been considered intermediate hosts [9–12].
SARS-CoV-2 infection is mediated by the binding of viral spike proteins (S-proteins) to human cells through a two-step process that involves ACE2 and transmembrane serine protease 2 (TMPRSS2). ACE2 is a member of the renin-angiotensin system (RAS), which plays a vital role in cardiovascular and renal homeostasis. ACE2 and TMPRSS2 facilitate entry of the virus into host cells during SARS-CoV-2 infection [13].
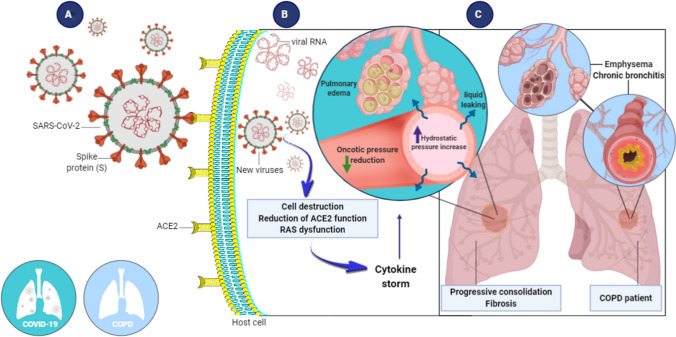
Alterations of virus structural S-proteins might be determining factors in viral infection and pathogenesis in human cells. The structural conformation of S-proteins appear to substantially aid binding to ACE2. These proteins, in turn, are highly expressed in human pulmonary cells [14–16]. The infection process occurs when S-proteins from SARS-CoV-2 bind ACE2 membrane receptors in pulmonary cells. After usually 5–14 days (i.e., the incubation period for this virus), the first COVID-19 symptoms may appear. In epithelial alveolar cells type 2 (AT2) cells, the virus recognizes ACE2 through S-proteins in their capsid (Fig. 1A). The viral envelope merges with the cellular membrane, and the virus releases its genetic material into the cell interior to finally initiate viral replication (Fig. 1B) [17–19].
Fig. 1.
Human pulmonary cell infection by SARS-CoV-2 and morphological and functional outcomes. A SARS-CoV-2 is transmitted through respiratory droplets, replicates in the respiratory tract, and infects pulmonary cells by attaching S-proteins to ACE2 membrane receptors on host cells. B Once inside host cells, new viruses are formed, resulting in a cytokine storm. Hydrostatic pressure increases in capillary walls, causing an increase in capillary permeability. C As a result, fluid leaks from the capillary into the interstitial and alveolar space, resulting in edema. In some cases, fibrosis develops. The left lung represents COVID-19. The right lung represents COPD. ACE2, angiotensin-I-converting enzyme 2; RAS, renin-angiotensin system; COPD, chronic obstructive pulmonary disease
SARS-CoV-2 infection effectively downregulates ACE2 receptors by attaching to infection-related transcription factors at ACE2 regulatory regions. Interestingly, ACE2 inhibitors and angiotensin II type 1 receptor blockers can influence ACE2 expression, and there are many common compounds and mediators that can increase the expression of ACE2 receptors, including metformin, resveratrol, vitamin B3, vitamin C, vitamin D, potassium, and nicotine. Importantly, ACE2 expression can be increased by compounds that activate or increase the expression of SIRT1 (Sirtuin 1) located next to the promotor region of the ACE2 gene. Increases in the expression or functional activation of sirtuin-1 is associated with an increase in the expression of ACE2. The prevention of COVID-19 by targeting cellular ACE2 receptors has been shown to not be a practically useful treatment strategy and can potentially increase infection, severity, or mortality [19–22].
The transmission of SARS-CoV-2 can occur by direct contact with infected people through respiratory droplets or indirectly through contact with contaminated objects and surfaces. Once the virus reaches the respiratory tract, it replicates rapidly and infects mostly type II pneumocytes and alveolar cells [23, 24]. This usually leads to cell death, which reduces ACE2 membrane receptors and triggers an inflammatory response, referred to as a cytokine storm. The cytokine storm then leads to a reduction of cell adhesion, and cytokines reach the blood circulation and evokes pulmonary arteriole dilatation through an increase in hydrostatic pressure in capillary walls and consequently an increase in capillary permeability. Liquid and exudate leakage then occurs from capillaries to the interstitium and alveoli, triggering lung edema and pneumonia. Additionally, the negative regulation of ACE2 impairs the antiinflammatory renin-angiotensin system and further exacerbates the inflammatory process [25–27]. The destruction of type II pneumocytes and increase in exudate in alveoli contribute to a surfactant reduction, and the alveolar surface undergoes an increase in superficial tension, leading to alveolar collapse and subsequent reductions of the gas exchange area and hypoxemia (Fig. 1C). In some cases, edema can result in fibrocyte accumulation in the interstitium and consequently lung fibrosis (Fig. 1C) [28, 29].
The clinical presentation of COVID-19 in humans is quite variable, with fever in 88–99%, fatigue in 38–70%, dry cough in 59–68%, anorexia in 40%, myalgias in 15–35%, dyspnea in 19–31%, and sputum production in 27–34% of cases. Muscle aches, confusion, headache, sore throat, rhinorrhea, chest pain, anosmia, conjunctivitis, and gastrointestinal disorders are also commonly described. Nearly 90% of cases present more than one symptom (e.g., fever, cough, and dyspnea). In more severe cases, low oxygenation secondary to respiratory failure and shock occur [30–32].
Notably, patients who are infected with SARS-CoV-2 and have comorbidities, such as hypertension, diabetes, cardiovascular disease, and chronic lung disease (mainly asthma and COPD), require special attention because of their higher risk of developing severe forms of COVID-19 [32]. Considering higher ACE2 expression in bronchial epithelial cells and the characteristic inflammatory process (e.g., obstructive bronchiolitis) and alveolar wall destruction (e.g., emphysema) in COPD, these patients are more susceptible to SARS-CoV-2 infection; once infected, they have a higher risk of progressing to more severe disease, including ARDS and respiratory failure [1, 3, 4, 6, 33, 34].
The main treatments for COVID-19 consist of palliative and supportive care. Several clinical trials have been conducted to repurpose or reposition existing drugs for COVID-19 [35, 36]. From a pharmacological perspective, this is a remarkably interesting approach because it reduces the usual time for drug development, which is crucial for addressing morbidity and mortality that are caused by COVID-19 [37]. Most studies have investigated the actions of remdesivir, lopinavir, ritonavir, interferon-β, ribavirin, tocilizumab, ivermectin, azithromycin, chloroquine/hydroxychloroquine, and corticosteroids [35]. A recent and controversial randomized, placebo-controlled clinical trial was performed in Brazil that evaluated whether early nitazoxanide therapy effectively accelerates symptom resolution in patients with mild COVID-19. The results of this trial showed that symptom resolution did not differ between the nitazoxanide and placebo groups after 5 days of treatment. However, early nitazoxanide therapy was safe and reduced viral load significantly after treatment with nitazoxanide (55%) in relation to placebo (45%) [38].
In November 2020, the United States Food and Drug Administration approved the emergency use of baricitinib combined with remdesivir for the treatment of COVID-19 in hospitalized adults and pediatric patients who require supplemental oxygen, invasive mechanical ventilation, or extracorporeal membrane oxygenation. The results that were obtained with the baricitinib + remdesivir combination were superior to remdesivir alone with regard to both shortening recovery time and improving clinical status in patients with COVID-19 [39].
With regard to prophylaxis and disease control, several vaccines are currently available on the market that have been shown to achieve high efficacy levels (70 to 90%) of protection against COVID-19 or against the development of more severe forms of the disease [40–44].
One current problem and cause for concern among clinicians and epidemiologists about the prophylaxis and control of COVID-19 involves new mutations of SARS-CoV-2 that have resulted in greater infectivity and consequently faster spread of the virus. These new variants of SARS-CoV-2, initially detected in South Africa and England, are worrisome in terms of the efficacy of vaccines that are currently being administered and whether they might overcome the natural immunity of individuals who are previously vaccinated or infected and lead to new waves of infection [45–47].
Vaccines can take from 3 to 4 years to reach the worldwide population. The repositioning of drugs is thus crucial for treating severe forms, symptoms, and sequelae of COVID-19. Continued follow-up of recovered COVID-19 patients is necessary. The discovery of new drugs or repositioning of existing drugs has been increasingly recognized as crucially important. Three-dimensional cell culture models are an ally in the fight against this virus and can help understand the disease and drugs that may be effective [48].
Despite undeniable advances that have been made in clinically managing patients with COVID-19 and the promising prophylactic use of vaccines, numerous doubts and concerns remain about the epidemiological context of COVID-19. There is still a lack of effective pharmacological treatments that are capable of having a significant impact on the natural course of the disease. There are also worrying epidemiological implications of SARS-CoV-2 mutations that may decrease the efficacy of current vaccines. New variants of the virus may lead to more serious clinical manifestations of COVID-19 [43, 49].
In the epidemiological context of new mutations and variants of SARS-CoV-2, it is fundamentally important to design and implement new robust and reproducible experimental models that will advance our knowledge of the pathophysiology of COVID-9 and the molecular interactions between SARS-CoV-2 and different tissues and comorbidities, such as chronic respiratory diseases that are associated with greater susceptibility to and a higher risk of progression to more serious diseases and mortality.
Research methods for lung disease and COVID-19
Much of what is currently known about human physiology and pathology has been learned through intense research using animal models. The use of such models dates back to around 500 B.C. and has allowed innumerable discoveries and advances in different areas of life sciences [50–52]. Some human diseases, however, still cannot be fully replicated in animals. Many therapies that showed promising results in rodents, for example, have failed to achieve similar results in humans. Additionally, ethical issues and guidelines have been implemented in different countries that have progressively and greatly limited the use of animals as experimental models [6, 53]. Therefore, researchers continually search for new alternatives to reduce animal use and replace experimental animal models.
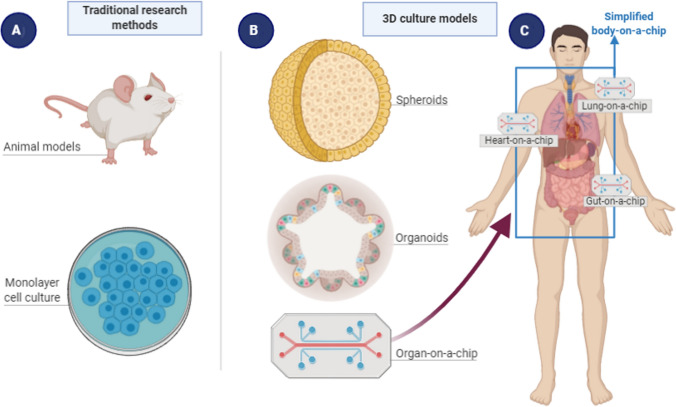
Tissue and cell culture techniques have, in parallel with and in addition to animal models, allowed great scientific advances in the biological and biomedical sciences (Fig. 2A). Classic two-dimensional (2D) cell cultures basically consist of a single cell type, with uniformity and standardization among morphological, genetic, and physiological investigations. However, this cellular uniformity of monolayer cultures is also a limiting factor in studying physiological and cellular interactions of an organ as a whole [51, 54].
Fig. 2.
Current research methods in life sciences. A Traditional methods, including animal models and 2D cell cultures. B Three-dimensional cultures, including spheroids, organoids, and organs-on-a-chip. C A more recent approach, body-on-a-chip or human-on-a-chip, with diverse organs integrated into one single circuitry
A significant advance in cell cultures in the context of regenerative and translational medicine was made in the 1970s when James Rheinwald and Howard Green reconstructed 3D tissues that derived from human progenitor cells. These researchers cultured trypsinized human skin cells that were plated together with lethally irradiated 3T3 cells. After 4 days of cultivation, human epidermal keratinocytes began to stratify in a 3D structure [55]. Afterward, cultured epithelial autografts were successfully used for wound healing in severe burns [56, 57].
Research interest in 3D cell cultures has recently gained more attention, but its history dates back to the early 20th century with the work of the French physician Alexis Carrel in 1912, who cultivated cardiomyocytes on the surface of silk threads [58]. In recent decades, especially in the last 15 years, 3D cell cultures have represented a major methodological breakthrough, enabling investigations of in vivo-like interactions from in vitro cultures. As proposed by Clevers [61], organoids can also be used to study organ development and various human pathologies “in a dish.” These 3D cell cultures today are referred to as spheroids or organoids, representing a promising alternative in regenerative and translational medicine [59–65].
The spheroid or organoid can be obtained from adult primary cells, stem cells, progenitor cells, and embryonic or induced pluripotent stem cells [59, 66]. In organoid cultures, suitable matrices are required for in vitro expansion, and hydrogels that are based on the extracellular matrix, such as Matrigel, are considered essential [67]. Considering that organoids present multiple cell lineages of a target organ, these models can be employed for numerous research purposes, including disease modeling, drug discovery, genotoxicity, and cytotoxicity [54, 60, 68–76].
With regard to lung spheroids or organoids, Tseng and colleagues from the University of Texas described in 2013 the generation of a lung spheroid model that mimics pulmonary bronchioles using a magnetic levitation technique to assemble different cell types into a single 3D culture [66]. Several research groups have already successfully generated 3D models with structures that present cell organization that is very similar to the in vivo lung [50, 66, 77–81].
More recently, further advances based on microscale fabrication technology and fluid physics have enabled the development of “microfluidic technology”. The association between these new technologies and 3D cell cultures has empowered the generation of integrated systems and the maintenance of 3D cultures in the microfluidic environment, allowing replication of the structural, functional, and environmental complexity of living organs. In this platform (Fig. 2B), different groups of 3D cells are cultivated on a microstructure (i.e., chip) and interconnected by a running fluid, resulting in an “organ-on-a-chip” structure [82, 83].
Organ-on-a-chip and lung-on-a-chip
Organs-on-a-chip have bridged microfluidic technology and living cells to investigate human and animal physiology. These chips allow contact between multicellular cultures and microfluids. The microscale technology of organs-on-a-chip is a more complex system than organoids because a microfluidic component is added. The microfluidic structure of organs-on-a-chip enables the exposure of multicellular cultures in a system that more closely mimics the physiological environment of an organ in vivo, and systemic interactions between an organ and the surrounding environment, including blood and immune cells [84–86]. These valuable research tools can then be employed to examine pharmacological effects, perform functional and toxic analyses, and assess responses to pathogens, such as bacteria and viruses [59, 66, 80, 87, 88]. The aforementioned platforms can also be applied to investigate the behavior of new mutant viruses, such as SARS-Cov-2, and their pathological (e.g., COVID-19) interactions with chronic respiratory diseases and integrated organ systems.
Interactions among diverse human tissues can also be replicated in various connected organs-on-a-chip, resulting in a body-on-a-chip or human-on-a-chip (Fig. 2C). For example, Skardal and colleagues reported integrated multi-organs-on-a-chip [89]. Their idea was to obtain a single recirculating perfusion system, comprising the liver, heart, lung, testis, colon, and brain. To date, several other authors have sought to construct integrated systems-on-a-chip, in order to replicate complete systems in a more complex and realistic model to analyze organ-organ interactions. These advances have continued to evolve to such an extent that it is expected that a model can be designed that presents nearly a full human body-on-a-chip [90].
In the specific case of the lung-on-a-chip, the 3D structure represents a microsystem that is very functionally similar to a living lung because of the possibility of replicating alveolar-capillary interactions and air exposure. The first lung-on-a-chip was proposed in 2010 by Huh and colleagues at Harvard University. These authors created a platform that simulated the vascular-alveolar structure using pulmonary epithelial cells that were exposed to air on one side and pulmonary endothelial vascular cells that were exposed to a running culture medium on the other side. Both sides were interconnected by a permeable synthetic membrane. This model presented microfluidic systems and mechanical stress, thereby simulating gas exchange [63]. The design that was developed by Stucki et al. [85] incorporated a breathing mechanism that was attached to the cultivation chambers that simulated diaphragm movements and recreated an accurate representation of the human lung.
Similar models have been used to test immune responses to pulmonary infection and the response to nanoparticles. Interestingly, periodic mechanical motions influenced the experimental data [91]. Physiologically relevant in vitro platforms that mimic human diseases and can test the effects of drugs thereon have been highlighted by several models, such as a specific alveolar model [92], an alveolar barrier in respiratory dynamics [85], intravascular thrombosis in lung alveolus [93], small airway-on-a-chip [94], and human lung airway epithelial cells and pulmonary microvascular endothelial cells [36].
Zamprogno et al. [95] proposed a new model or “the second generation” of lung-on-a-chip. In this methodological variant, the porous and stretchable polydimethylsiloxane membrane was replaced by an array of in vivo-like sized alveoli and a stretchable biological membrane. According to the authors, the proposed model allows one to mimic the functionality of the lung parenchyma at an “unprecedented level,” enabling the reproduction of different functional aspects of the alveolar environment of the lung. This model may be applied to disease modeling and drug screening.
Lung 3D cell cultures and COVID-19
Three-dimensional cell culture systems provide a consistent research tool and open new avenues for experimental tissue engineering and regenerative medicine in diverse areas of biological and biomedical sciences, including studies of tissue morphogenesis, the efficacy and toxicity of therapeutic compounds in vitro, and drug responses in personalized medicine, with prospects of replacing at least some experimental animal models [59, 61, 66, 80, 81, 96–99]. These 3D cell culture systems also enable the development of new therapeutic approaches to confronting frequent infections in patients with “The Big Five” CRD, including COVID-19 (Fig. 1).
According to a recent commentary by Hans Clevers [100], “COVID-19: organoids go viral” [100], organoids are emerging as a new platform for studying COVID-19. Although the lung stands out as the main organ that is affected by SARS-CoV-2, organoids have been applied to additionally describe the behavior of SARS-CoV-2 in the intestine, kidneys, liver, and brain [101–106].
In the context of the lungs, the 3D model of the airway epithelium that was proposed by Agostini et al. [107] represented an important advance and alternative for testing the safety and efficacy of new antiviral drugs against the Coronaviridae family. The authors structured a 3D model of the airway epithelium, provided by an air-liquid interface, thus establishing a physiological in vitro approach that resembled the in vivo pseudostratified mucociliary epithelium. This platform was used to test the inhibitory effect of remdesivir on SARS-CoV- and MERS-CoV-infected human airway epithelial cell cultures. The results demonstrated the therapeutic efficacy of remdesivir against SARS-CoV and MERS-CoV in HAE cultures at a concentration that was non-toxic for cells.
Several studies have provided robust evidence of the application of 3D cell cultures to investigate the pathophysiology and antiviral therapy of COVID-19 (Table 1). Pulmonary organoids, especially those that are related to AT2 cells, express ACE2 and are highly susceptible to infection by SARS-CoV-2 [108]. Using alveolar and basal stem cells from the human lung, Salahudeen et al. [109] developed a culture of organoids which proved to be useful for pulmonary infection diagnosis. The development and use of 3D models of the lung could be a promising strategy to elucidate the pathophysiological mechanisms that underlie COVID-19 and to screen potentially effective drugs for treatment and management of the disease [110]. Establishment of the SARS-CoV-2-infected airway organoid allowed investigations of the cytopathic effects of viral particles in different cell types by immunofluorescence, viral replication kinetics, genetic changes through complete genome sequencing and quantitative real-time polymerase chain reaction, differential transcriptomic gene expression, and cell profiles before and after viral infection using flow cytometry [111].
Table 1.
Recent evidences of 3D lung cultures as robust platforms for studying the pathogenesis of SARS-CoV-2 and testing new therapeutic approaches for COVID-19
| Approach | Main findings | Reference |
|---|---|---|
| Organoid lung model from human pluripotent stem cells infected with SARS-CoV-2 | Robust induction of chemokines and cytokines. Pre- or post-treatment with imatinib and mycophenolic acid decreased SARS-CoV-2 infection | Han et al. [108] |
| Organoid as a model of distal lung disease for COVID-19 | The basal and human alveolar epithelial type II apical-out organoids were infected by SARS-CoV-2, pointing to club cells as a new target population | Salahudeen et al. [109] |
| SARS-CoV-2 infected airway organoid | Investigation of the effects of viral particles in different cell types, viral replication kinetics and genetic changes, differential expression of genes, and cell profile before and after viral infection | Elbadawi et al. [111] |
| Organoid as a model of distal lung disease for COVID-19 | Recognition of cell functional heterogeneity and progenitor identification with a progressive proliferating human tissue | Suzuki et al. [112] |
| Organoid lung model from human pluripotent stem cells infected with SARS-CoV-2 | Evident induction of cytokines and chemokine. The treatment of the organoid with imatinib and mycophenolic acid decreased the infection by SARs-CoV-2 | Han et al. [108] |
| Bronchial organoid composed of cryopreserved human bronchial epithelial cells | After the organoid infection by SARS-CoV-2, an intracellular viral genome, progeny virus, cytotoxicity, pyknotic cells, and moderate increase of the interferon type I was observed. The treatment of organoid with camostat promoted a reduction in the viral copies | Suzuki et al. [112] |
| 3D cell culture technique for human alveolar type 2 3D cell cultures infected with SARS-CoV-2 | A rapid viral replication and modulation of the innate endogenous immune response was observed. Also, it was possible to identify an effective complete cell infection from a single viral entry | Youk et al. [113] |
| 3D cell culture technique for human alveolar type 3 3D cell cultures infected with SARS-CoV-2 | Cellular and transcriptional changes occurred, pointing to cellular tropism in viral replication and transcription as well as a consequent host cell response | Youk et al. [113] |
| Normal human distal alveolar organoids infected with SARS-CoV-2 | Pretreatment with hydroxychloroquine and remdesivir significantly reduced viral replication, however, this effect was more pronounced with remdesivir | Mulay et al. [114] |
| Lung-on-a-chip composed of human lung airway epithelium cells | Amodiaquine and toremifene significantly inhibited entry of the pseudotyped SARS-CoV-2 virus | Si et al. [36] |
Han et al. [108] described an organoid lung model that was designed from human pluripotent stem cells (hPSCs) that were infected with SARS-CoV-2. Similar to manifestation of the disease in humans, these authors applied transcriptomics and detected the induction of cytokines and chemokines and low interferon type I (IFN-I) and IFN-II signaling. They also found that treatment of the organoid with imatinib and mycophenolic acid decreased SARS-CoV-2 infection. These results indicate that this coronavirus-infected organoid can model the disease and may be a promising resource for screening new drugs to treat COVID-19.
Suzuki et al. [112] designed a bronchial organoid that was composed of cryopreserved human bronchial epithelial cells to study the effects of SARS-CoV-2 infection. The proposed organoid contained basal, club, ciliated, and goblet cells and high ACE2 and TMPRSS2 expression. After infection of the organoid with SARS-CoV-2, an intracellular viral genome, progeny virus, cytotoxicity, pyknotic cells, and a moderate increase in IFN-I were observed. Treatment of this organoid with the TMPRSS2 inhibitor camostat reduced the number of viral copies, indicating that this model can be used to study pathophysiological mechanisms and discover new and effective drugs.
Youk et al. [113] proposed a long-term 3D cell culture technique for human alveolar type 2 (hAT2) and alveolar type 2 3D cell cultures (h3ACs) and described the profile of SARS-CoV-2 infection. In h2AT cells, 3 days after infection, rapid viral replication, modulation of the innate endogenous immune response, and increases in the expression of proinflammatory and interferon-related genes were observed. By tracking acquired viral mutations, it was possible to identify effectively complete cell infection from a single virus entry. In h3ACs cells that were infected with SARS-CoV-2, cellular and transcriptional changes occurred, indicating cellular tropism in viral replication and transcription and consequently the host cell response.
The infection and replication of SARS-CoV-2 in normal human distal alveolar organoids was described by Mulay et al. [114]. The pretreatment of alveolar organoid cultures with hydroxychloroquine and remdesivir significantly reduced viral replication, but this effect was more pronounced with antiviral remdesivir treatment, indicating that pulmonary organoids may be a useful platform for drug validation.
Completing the spectrum of 3D cell cultures for studying the pathophysiology of COVID-19 and promising therapeutics, lung-on-a-chip systems have been shown to be excellent platforms for disease modeling and screening new antiviral drugs. Si et al. [36] described a microfluidic organ-on-a-chip system that was composed of the human lung airway epithelium that was cultured under an air-liquid interface with a continuous medium flow. The model contained epithelial cells that expressed high levels of ACE2 and TMPRSS2 and was used to test different drugs, including chloroquine, arbidol, toremifine clomiphene, amodiaquine, verapamil, and amiodarone, for the treatment of COVID-19. Among the tested drugs, only two, amodiaquine and toremifene, significantly inhibited entry of the pseudotyped SARS-CoV-2 virus. These results showed that human chip models are important platforms for studying the pathogenesis of SARS-CoV-2 infection and testing new therapeutic agents for COVID-19.These models will also be useful for investigating other new epidemic and pandemic viral infections that impact the human population.
Concluding remarks
Three-dimensional cell cultures, either as organoids or organs-on-a-chip, have significantly expanded different aspects of biomedical research. These new methodological approaches can mimic various important functions of different organs and tissues in vitro and constitute a more realistic approach relative to traditional 2D cell cultures. These 3D cell cultures may also be able to replace a large number of existing experimental animal models. Organoid models can complement other techniques, such as co-cultures with cells of the immune system. These new study platforms are highly likely to provide new insights into cellular interactions, pharmacological drug screening, and the pathophysiology of various diseases. In the current epidemiological scenario of COVID-19, 3D cell culture platforms have emerged as very promising for the development of novel therapeutics, especially when considering the strong likelihood of new mutations and future pathogenic variants of coronavirus and other viruses. The present review emphasized 3D cell cultures, particularly 3D organoids and lungs-on-a-chip, as robust platforms that can provide new insights into pathophysiology interactions between CRD and COVID-19. These new 3D cell culture platforms serve as valuable research tools to test new therapeutic compounds for the treatment of chronic lung diseases and COVID-19.
Acknowledgements
The authors would like to thank Prof. Ademilson Espencer Egea Soares—Department of Genetics—Ribeirão Preto Medical School—USP (Brazil) for all valuable comments and suggestions which contributed to improve the quality of the manuscript. JTRP wrote this review during the development of a project supported by the São Paulo Research Foundation—FAPESP (Proc. 2014 / 03324-4). FASC is a fellow of Coordination for the Improvement of Higher Education Personnel (CAPES—Brazil). This research was supported by grants from Coordenadoria de Pós-Graduação (COPG, UNIPAR, Brazil).
Author Contributions
JTRP suggested the theme; MJMF, MRS and FARL defined the structure of the article; JTRP, MJMF and MRS wrote introduction; MRS, GRS, FASC, FARL and MJMF wrote pathogenesis and pathophysiology of COVID-19; FASC designed the figures; FARL, MRS MJMF and JTRP wrote evolution of research methods for respiratory disease, on-a-chip methodology and conclusion. All authors contributed to the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human and animal subjects performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.GOLD (Global Strategy for Prevention, Diagnosis and Management of COPD) 2020. Global Initiat Chronic Obstr Lung Dis. 2020. Available from: http://www.goldcopd.org/. Accessed 16 Feb 2021.
- 2.WHO - World Health Organization. The top 10 causes of death (2019). https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death. Accessed 16 Feb 2021.
- 3.Halpin DMG, Faner R, Sibila O, Badia JR, Agusti A. Do chronic respiratory diseases or their treatment affect the risk of SARS-CoV-2 infection? Lancet Respir Med. 2020;8:436–438. doi: 10.1016/S2213-2600(20)30167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hui KPY, Cheung MC, Perera RAPM, Ng KC, Bui CHT, Ho JCW, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li YK, Peng S, Li LQ, Wang Q, Ping W, Zhang N, et al. Clinical and transmission characteristics of Covid-19 — a retrospective study of 25 cases from a single thoracic surgery department. Curr Med Sci. 2020;40:295–300. doi: 10.1007/s11596-020-2176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. Cell. 2020;181:271–80.e8. [DOI] [PMC free article] [PubMed]
- 14.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: Estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang D, Comish P, Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16:e1008536. doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhtar S, Benter IF, Danjuma MI, Doi SAR, Hasan SS, Habib AM. Pharmacotherapy in COVID-19 patients: a review of ACE2-raising drugs and their clinical safety. J Drug Target. 2020;28:683–699. doi: 10.1080/1061186X.2020.1797754. [DOI] [PubMed] [Google Scholar]
- 21.Cheng H, Wang Y, Wang GQ. Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. J Med Virol. 2020;92:726–730. doi: 10.1002/jmv.25785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLachlan CS. The angiotensin-converting enzyme 2 (ACE2) receptor in the prevention and treatment of COVID-19 are distinctly different paradigms. Clin Hypertens. 2020;26:14. doi: 10.1186/s40885-020-00147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO- World Health Organization, Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. 2020. Available from: https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid19-implications-for-ipc-precaution-recommendations. Accessed 15 Feb 2021.
- 25.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y, Yang H, Ji W, Wu W, Chen S, Zhang W, et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12:372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baek WK, Sohn SY, Mahgoub A, Hage R. A comprehensive review of severe acute respiratory syndrome coronavirus 2. Cureus. 2020;12:e7943. doi: 10.7759/cureus.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19) Eur Radiol. 2020;30:4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng ZJ, Shan J. 2019 Novel coronavirus: where we are and what we know. Infection. 2020;48:155–163. doi: 10.1007/s15010-020-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan. China JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Faria CA, de las Heras Kozma R, Stessuk T, Ribeiro-Paes JT. Experimental basis and new insights for cell therapy in chronic obstructive pulmonary disease. Stem Cell Rev Rep 2012; 8:1236–44. [DOI] [PubMed]
- 34.Leung JM, Yang CX, Tam A, Shaipanich T, Hackett TL, Singhera GK, et al. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J. 2020;55:2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovato ECW, Barboza LN, Wietzikoski S, de Souza ANV, Auth PA, Junior AG, et al. Repurposing drugs for the management of patients with confirmed coronavirus disease 2019 (COVID-19) Curr Pharm Des. 2021;1:115–126. doi: 10.2174/1381612826666200707121636. [DOI] [PubMed] [Google Scholar]
- 36.Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng. 2020. 10.1038/S41551-021-00718-9. [DOI] [PMC free article] [PubMed]
- 37.Mercorelli B, Palú G, Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocco PRM, Silva PL, Cruz FF, Junior MACM, Tierno PFGMM, Moura MA, et al. Early use of nitazoxanide in mild Covid-19 disease: randomized, placebo-controlled trial. Eur Respir J. 2020 doi: 10.1183/13993003.03725-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalil AC, Patterson TF, Mehta AK, Tomashek KM, Wolfe CR, Ghazaryan V, et al. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N Engl J Med. 2021;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iversen PL, Bavari S. Inactivated COVID-19 vaccines to make a global impact. Lancet Infect Dis. 2021;21:746–748. doi: 10.1016/S1473-3099(21)00020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rostad CA, Anderson EJ. Optimism and caution for an inactivated COVID-19 vaccine. Lancet Infect Dis. 2021;21:581–582. doi: 10.1016/S1473-3099(20)30988-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatatos L, Czartoski J, Wan YH, Homad L, Rubin V, Glantz H, et al. Antibodies elicited by SARS-CoV-2 infection and boosted by vaccination neutralize an emerging variant and SARS-CoV-1. medRxiv. 2021 doi: 10.1101/2021.02.05.21251182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burki T. Understanding variants ofSARS-CoV-2. Lancet. 2021;397:462. doi: 10.1016/S0140-6736(21)00298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Priesemann V, Balling R, Brinkmann MM, Ciesek S, Czypionka T, Eckerle I, et al. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet. 2021;397:469–470. doi: 10.1016/S0140-6736(21)00150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullard A. How COVID vaccines are being divvied up around the world. Nature. 2020 doi: 10.1038/d41586-020-03370-6. [DOI] [PubMed] [Google Scholar]
- 49.WHO- World Health Organization, -CoV-2 Variants. https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/. Accessed 16 Feb 2021.
- 50.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konar D, Devarasetty M, Yildiz DV, Atala A, Murphy SV. Lung-on-a-chip technologies for disease modeling and drug development. Biomed Eng Comput Biol. 2016;7:17–27. doi: 10.4137/BECB.S34252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakalem ME, Ribeiro-Paes JT. New methodologies for old problems: tridimensional gastrointestinal organoids and guts-on-a-chip. J Coloproctol. 2018;38:90–93. [Google Scholar]
- 53.Cheluvappa R, Scowen P, Eri R. Ethics of animal research in human disease remediation, its institutional teaching; and alternatives to animal experimentation. Pharmacol Res Perspect. 2017;5:e00332. doi: 10.1002/prp2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duval K, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, et al. Modeling physiological events in 2D vs 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 55.Rheinwatd JG, Green H. Seria cultivation of strains of human epidemal keratinocytes: the formation keratinizin colonies from single cell is. Cell. 1975;6:331–343. doi: 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 56.Li Z, Maitz P. Cell therapy for severe burn wound healing. Burns Trauma. 2018;28:6–13. doi: 10.1186/s41038-018-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Connor NE, Mulliken JB, Banks-Schlegel S, Kehiode O, Green H. Grafting of burns with cultured epithelium prepared from autologous epidermal cells. Lancet. 1981;1:75–78. doi: 10.1016/S0140-6736(81)90006-4. [DOI] [PubMed] [Google Scholar]
- 58.Freshney RI. Cell & molecular biology. 7. New Jersey: Wiley; 2005. Culture of animal cells: a manual of basic technique and specialized applications; p. 696. [Google Scholar]
- 59.Alhaque S, Themis M, Rashidi H. Three-dimensional cell culture: from evolution to revolution. Philos Trans R Soc Lond B Biol Sci. 2018;373:20170216. doi: 10.1098/rstb.2017.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boonekamp KE, Kretzschmar K, Wiener DJ, Asra P, Derakhshan S, Puschhof J, et al. Long-term expansion and differentiation of adult murine epidermal stem cells in 3D organoid cultures. Proc Natl Acad Sci U S A. 2019;116:14630–14638. doi: 10.1073/pnas.1715272116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 62.Doryab A, Amoabediny G, Salehi-Najafabadi A. Advances in pulmonary therapy and drug development: lung tissue engineering to lung-on-a-chip. Biotechnol Adv. 2016;34:588–596. doi: 10.1016/j.biotechadv.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Yuan Hsin H, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tseng H, Gage JA, Raphael RM, Moore RH, Killian TC, Grande-Allen KJ, et al. Assembly of a three-dimensional multitype bronchiole coculture model using magnetic levitation. Tissue Eng Part C Methods. 2013;19:665–675. doi: 10.1089/ten.tec.2012.0157. [DOI] [PubMed] [Google Scholar]
- 66.Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang M, Liu Y, Chen YG. Generation of 3D human gastrointestinal organoids: principle and applications. Cell Regen. 2020;9:6. doi: 10.1186/s13619-020-00040-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fujii M, Sato T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat Mater. 2021;20:156–169. doi: 10.1038/s41563-020-0754-0. [DOI] [PubMed] [Google Scholar]
- 69.Heidari-Khoei H, Esfandiari F, Hajari MA, Ghorbaninejad Z, Piryaei A, Baharvand H, et al. Organoid technology in female reproductive biomedicine. Reprod Biol Endocrinol. 2020;18:64. doi: 10.1186/s12958-020-00621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kabadi PK, Rodd AL, Simmons AE, Messier NJ, Hurt RH, Kane AB. A novel human 3D lung microtissue model for nanoparticle-induced cell-matrix alterations. Part Fibre Toxicol. 2019;16:15. doi: 10.1186/s12989-019-0298-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lancaster MA, Huch M. Disease modelling in human organoids. Dis Models Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cells Reports. 2017;8:1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 75.Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 76.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franzdóttir SR, Axelsson IT, Arason AJ, Baldursson Ó, Gudjonsson T, Magnusson MK. Airway branching morphogenesis in three dimensional culture. Respir Res. 2010;11:162. doi: 10.1186/1465-9921-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hegab AE, Arai D, Gao J, Kuroda A, Yasuda H, Ishii M, et al. Mimicking the niche of lung epithelial stem cells and characterization of several effectors of their in vitro behavior. Stem Cell Res. 2015;15:109–121. doi: 10.1016/j.scr.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 79.Nadkarni RR, Abed S, Draper JS. Organoids as a model system for studying human lung development and disease. Biochem Biophys Res Commun. 2016;473:675–682. doi: 10.1016/j.bbrc.2015.12.091. [DOI] [PubMed] [Google Scholar]
- 80.Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Böttinger L, Klay D, et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van der Vaart J, Clevers H. Airway organoids as models of human disease. J Intern Med. 2021;289:604–613. doi: 10.1111/joim.13075. [DOI] [PubMed] [Google Scholar]
- 82.Huh DD. A human breathing lung-on-a-chip. Ann Am Thorac Soc. 2015;12:S42–S44. doi: 10.1513/AnnalsATS.201410-442MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sosa-Hernández JE, Villalba-Rodríguez AM, Romero-Castillo KD, Aguilar-Aguila-Isaías MA, García-Reyes IE, Hernández-Antonio A, et al. Organs-on-a-chip module: a review from the development and applications perspective. Micromachines (Basel) 2018;9:536. doi: 10.3390/mi9100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramadan Q, Zourob M. Organ-on-a-chip engineering: toward bridging the gap between lab and industry. Biomicrofluidics. 2020;14:041501. doi: 10.1063/5.0011583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stucki AO, Stucki JD, Hall SR, Felder M, Mermoud Y, Schmid RA, et al. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip. 2015;15:1302–1310. doi: 10.1039/C4LC01252F. [DOI] [PubMed] [Google Scholar]
- 86.Wu Q, Liu J, Wang X, Feng L, Wu J, Zhu X, et al. Organ-on-a-chip: recent breakthroughs and future prospects. Biomed Eng Online. 2020;19:9. doi: 10.1186/s12938-020-0752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang JW, Shen YC, Lin KC, Cheng SJ, Chen SL, Chen CY, et al. Organ-on-a-chip: opportunities for assessing the toxicity of particulate matter. Front Bioeng Biotechnol. 2020;8:519. doi: 10.3389/fbioe.2020.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shrestha J, Ghadiri M, Shanmugavel M, Bazaz SR, Vasilescu S, Ding L, et al. A rapidly prototyped lung-on-a-chip model using 3D-printed molds. Organs-on-a-Chip. 2019;1:100001. doi: 10.1016/j.ooc.2020.100001. [DOI] [Google Scholar]
- 89.Skardal A, Aleman J, Forsythe S, Rajan S, Murphy S, Devarasetty M, et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication. 2020;12:025017. doi: 10.1088/1758-5090/ab6d36. [DOI] [PubMed] [Google Scholar]
- 90.Zhang B, Korolj A, Lai BFL, Radisic M. Advances in organ-on-a-chip engineering. Nat Rev Mater. 2018;3:257–278. doi: 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- 91.Azizipour N, Avazpour R, Rosenzweig DH, Sawan M, Ajji A. Evolution of biochip technology: a review from lab-on-a-chip to organ-on-a-chip. Micromachines (Basel) 2020;11:599. doi: 10.3390/mi11060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Douville NJ, Zamankhan P, Tung YC, Li R, Vaughan BL, Tai CF, et al. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip. 2011;11:609–619. doi: 10.1039/C0LC00251H. [DOI] [PubMed] [Google Scholar]
- 93.Jain A, Bansal R. Applications of regenerative medicine in organ transplantation. J Pharm Bioallied Sci. 2015;7:188–194. doi: 10.4103/0975-7406.160013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 95.Zamprogno P, Wüthrich S, Achenbach S, Stucki JD, Hobi N, Schneider-Daum N. Second - generation lung-on-a-chip array with a stretchable biological membrane. Commun Biol. 2021;4:168. doi: 10.1038/s42003-021-01695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chaicharoenaudomrung N, Kunhorm P, Noisa P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J Stem Cells. 2019;11:1065–1083. doi: 10.4252/wjsc.v11.i12.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klak M, Bryniarski T, Kowalska P, Gomolka M, Tymicki G, Kosowska K, et al. Novel strategies in artificial organ development: what is the future of medicine? Micromachines (Basel) 2020;11:646. doi: 10.3390/mi11070646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niemeyer BF, Zhao P, Tuder RM, Benam KH. Advanced microengineered lung models for translational drug discovery. SLAS Discov. 2018;23:777–789. doi: 10.1177/2472555218760217. [DOI] [PubMed] [Google Scholar]
- 99.Wang Z, Samanipour R, Kim K. Organ-on-a-chip plataforms for drug screening and tissue engineering. Biomed Eng Front Res Converg Technol. 2015;9:209–233. doi: 10.1007/978-3-319-21813-7_10. [DOI] [Google Scholar]
- 100.Clevers H. COVID-19: organoids go viral. Nat Rev Mol Cell Biol. 2020;21:355–356. doi: 10.1038/s41580-020-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mahalingam R, Dharmalingam P, Santhanam A, Kotla S, Davuluri G, Karmouty-Quintana H, et al. Single-cell RNA sequencing analysis of SARS-CoV-2 entry receptors in human organoids. J Cell Physiol. 2021;236:2950–2958. doi: 10.1002/jcp.30054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–13.e7.. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang H, Abouleila Y, Si L, Ortega-Prieto AM, Mummery CL, Ingber DE, et al. Human organs-on-chips for virology. Trends Microbiol. 2020;28:934–946. doi: 10.1016/j.tim.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang BZ, Chu H, Han S, Shuai H, Deng J, Hu YF, et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang L, Han Y, Nilsson-Payant BE, Gupta V, Wang P, Duan X, et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–36.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221-18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han Y, Yang L, Duan X, Duan F, Nilsson-Payant B, Yaron TM, et al. Identification of candidate COVID-19 therapeutics using hPSC-derived lung organoids. bioRxiv. 2020 doi: 10.1101/2020.05.05.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salahudeen AA, Choi SS, Rustagi A, Zhu J, de la O SM, Flynn RA, et al. Progenitor identification and SARS-CoV-2 infection in long-term human distal lung organoid cultures. Nature. 2020;588:670–5. [DOI] [PMC free article] [PubMed]
- 110.Shafiee A, Moradi L, Lim M, Brown J. Coronavirus disease 2019: a tissue engineering and regenerative medicine perspective. Stem Cells Transl Med. 2021;10:27–38. doi: 10.1002/sctm.20-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elbadawi M, Efferth T. Organoids of human airways to study infectivity and cytopathy of SARS-CoV-2. Lancet Respir Med. 2020;8:e55–e56. doi: 10.1016/S2213-2600(20)30238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suzuki T, Itoh Y, Sakai Y, Saito A, Okuzaki D, Motooka D, et al. Generation of human bronchial organoids for SARSCoV-2 research. bioRxiv. 2020. 10.1101/2020.05.25.115600.
- 113.Youk J, Kim T, Evans KV, Jeong YII, Hur Y, Hong SP, et al. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–19.e10. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mulay A, Konda B, Garcia G, Jr, Yao C, Beil S, Villalba JM, et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35:109055. doi: 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]