Abstract
The ongoing coronavirus disease 19s pandemic has yet again demonstrated the importance of the human-animal interface in the emergence of zoonotic diseases, and in particular the role of wildlife and livestock species as potential hosts and virus reservoirs. As most diseases emerge out of the human-animal interface, a better understanding of the specific drivers and mechanisms involved is crucial to prepare for future disease outbreaks. Interactions between wildlife and livestock systems contribute to the emergence of zoonotic diseases, especially in the face of globalization, habitat fragmentation and destruction and climate change. As several groups of viruses and bacteria are more likely to emerge, we focus on pathogenic viruses of the Bunyavirales, Coronaviridae, Flaviviridae, Orthomyxoviridae, and Paramyxoviridae, as well as bacterial species including Mycobacterium sp., Brucella sp., Bacillus anthracis and Coxiella burnetii. Noteworthy, it was difficult to predict the drivers of disease emergence in the past, even for well-known pathogens. Thus, an improved surveillance in hotspot areas and the availability of fast, effective, and adaptable control measures would definitely contribute to preparedness. We here propose strategies to mitigate the risk of emergence and/or re-emergence of prioritized pathogens to prevent future epidemics.
Keywords: Bacteria, Emergence drivers, Infectious diseases, Viruses, Zoonoses
Implications
The emergence of severe acute respiratory syndrome coronavirus 2 and the resulting pandemic of coronavirus disease 19 reminds us the importance of a suitable monitoring of human-animal interfaces to prevent future pandemics we could face. Livestock species are in close contact with humans, are often involved in (re)emergences, and then deserve to be carefully considered when assessing zoonotic disease emergence. Determining at least partly the next potential bacterial and viral candidates for zoonotic emergence and identifying the main drivers can help to outline new measures to decrease the risk. We discuss here all these aspects about the main already known potential candidates for emergence and we propose strategies to mitigate the risk of (re)emergence and prevent future epidemics.
Introduction
More than 60% of all human infectious diseases, described between 1940 and 2004, originate from animals (Jones et al., 2008). A broad variety of animals including species of livestock, wildlife, pet animals but also laboratory animals and captive animals are sources of pathogens. Livestock can be defined as mammals that are kept on a farm such as small and large ruminants (Bos taurus, Capra aegagrus hircus and Ovis aries) and pigs (Sus scrofa). Poultry includes chickens (Gallus gallus), ducks and geese (several species in the Anatidae family). With the largest definition of the word livestock, less common species like alpaca (Vicugna pacos), bison (Bison bison), elk (Cervus canadensis), camel and dromedary (Camelus sp), guineafowl (Numida meleagris), mink (Neovison vison), and ostrich (Struthio camelus) should also be considered as sources of pathogens. Livestock and poultry provide many goods and services to human populations such as milk, meat, eggs, hides, feathers, fibres and manure, which all pose a risk for human pathogen exposure. In this review, nonavian livestock and poultry will be considered together under the term livestock (FAO definition), which are farm domestic animals raised for subsistence or local sales, thereby mingle with other species, or supply international markets with large numbers following long distance transports. Wildlife includes a wide range of species that not only are relevant because of their direct interaction, but also because of their use as bushmeat for human and animal consumption. Conversely, pet animals will not be discussed in this review. Amongst the pathogens causing emerging infectious diseases, viruses are naturally very well-represented. This is not surprising, since 76% of viruses affecting humans are zoonotic while for bacteria a percentage of 50 has been described (Taylor et al., 2001, Jones et al., 2008). According to the World Organization for Animal health (OIE), an emerging disease is “a new infection resulting from the evolution or change of an existing pathogen or parasite resulting in a change of host range, vector, pathogenicity or strain; or the occurrence of a previously unrecognized infection or disease”. A re-emerging disease is “an already known disease that either shifts its geographical setting or expands its host range, or significantly increases its prevalence”. Interestingly, amongst animals, mammals have been identified as the main reservoir (around 80%) of human infectious agents and ungulates, probably because they are also major food sources, are the mammalian taxon sharing the highest number of pathogens with humans (Cleaveland et al., 2001, Taylor et al., 2001, Woolhouse and Gowtage-Sequeria, 2005, Jones et al., 2008). Compared to ungulates, birds are second in line although less prone to transmit infectious diseases to humans. Since ungulates and birds are massively represented amongst livestock species, these observations are particularly relevant for the current review.
Various livestock farming systems are used for domestic mammal and bird species with a huge variety in animal density worldwide (Derner et al., 2017, Gilbert et al., 2018). The systems can be intensive or extensive with a maximization of the production in the first system. With an increased demand for animal products due to a fast growing human population, intensive livestock systems developed massively in Western countries, Asia and South America (Duru and Therond, 2015, FAO, 2020). The intensive farming system has particularly been applied to pigs, dairy cattle, and poultry. When ruminant livestock (cattle, sheep, camelids and goats) are compared to monogastric species such as pigs, turkeys and chickens, a difference can be identified in terms of production system (FAO, 2020). Ruminant livestock systems are more dependent on the land and available space while chicken and pig production systems depend more on consumer demand and the level of financial investment (FAO, 2020). In the 1980s, a progressive awareness about the negative impact of productive systems – associated with intensive farming systems – on biodiversity and climate change, but also on animal production quality, animal welfare, human health, and depletion of fossil and water resources emerged in many countries (Duru and Therond, 2015). Alteration of the biodiversity and climate change were shown to drive the (re)emergence of infectious diseases (Zell, 2004, Keesing et al., 2010). Thus, organic practices promoting traditional methods without synthetic inputs and pesticides were developed in Western countries and were used as alternatives to the conventional practices. Indeed, conventional practices were accompanied by mechanization, simplification and standardization of production modes, a decreased diversity of crops varieties and livestock breeds, and the creation of uniform landscapes (Derner et al., 2017). Then, new integrated farming systems emerged (Veysset et al., 2014, Martin et al., 2016). These systems based on ecological principles combine organic and conventional practices (Duru and Therond, 2015).
Regarding disease emergence, different models have been developed to describe the process through which zoonoses could emerge (Morse et al., 2012). Morse et al originally described the process in two steps, the first being the introduction into a new species – human for instance – and the second being the establishment/dissemination (Morse, 1995). Then, Wolfe and collaborators developed a five-stage model of pathogen adaptation to humans (Wolfe et al., 2007). In stage 1, the pathogen is unable to infect humans while in stage 5 it is causing exclusively a human disease. In 2012, Morse and collaborators proposed an alternative model explaining the emergence in three steps (Morse et al., 2012). In that model, the stage 1 corresponds to the pre-emergence state, in which microorganisms are being transmitted between their natural animal reservoirs. Modifications of the surrounding environment, the ecological niches and others impact the animal populations and affect the dynamics of transmission, increasing the risk of pathogen spillover to other wildlife or livestock species (Daszak et al., 2006). Stage 2 represents the first spillover of the pathogen from wildlife or livestock to humans. At this stage there is still no, or limited human-to-human transmission (Ebola virus or H5N1 influenza virus) (Morse et al., 2012). Stage 3 corresponds to the full epidemic emergence, with sustained person-to-person transmission and a large-scale or worldwide spread of the pathogen and associated disease. Examples of stage 3 are the emergence of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) in 2002, pandemic H1N1 influenza virus in 2009, and SARS-CoV-2 in 2019.
Here, we selected pathogens likely to (re)emerge in the future at the interface of wildlife-, livestock-, and human-systems, based on the current knowledge, and discussed drivers of emergence and strategies to mitigate associated risks. We particularly developed the chapters dealing with viruses and to some extent bacteria and deliberately excluded fungi, parasites and prions. Indeed, viruses and bacteria are more likely to cause significant epidemics even if some exceptions exist such as malaria, a parasitic disease most probably of zoonotic origin.
Drivers of Emergence and Hotspots
Many factors can be involved in the emergence of zoonotic diseases (Walsh et al., 2020). Increasing populations of humans and animal species are often associated with a rise in the circulation of infectious disease agents and therefore are obvious factors in the risk of emerging zoonotic threats. The interactions between pathogens and their hosts – including the reservoir considered as a multi-host system – are closely associated with the environment they are in, and these are changing at an increasing pace. It must be underlined that the (re)emergence of zoonoses is always a multifactorial process. This process can involve, amongst others, modifications in farming and trading practices, human behaviour, animal vector distribution and in the genetics of the microorganisms and their hosts. Furthermore, the different drivers can play various and distinct roles in the emergence of multiple viruses (Cutler et al., 2010, Wang and Crameri, 2014).
It is also of high importance to evaluate and understand the impacts of these changes on the interactions between the pathogenic microorganisms and their hosts and between the host and other animal species, including livestock, wildlife and humans. These interactions are at the core of zoonoses (re)emergence, understanding these drivers and impacts will allow the development of mitigation strategies and enable an effective and timely response. Typically, when the first infected human (index case) transmits the infectious agent to more than one other human, an infection can cause an epidemic in a human population. Immunological studies investigating quantitative and qualitative differences in the host-virus equilibrium in animal reservoirs can help us to elucidate why some viruses are more hazardous than others. On a more local level, elevated transmission risk is found at food places with a high human to animal contact frequency, such as wet markets and slaughterhouses, which are essential for the daily food supply for billions of consumers.
In many regions of the world, we see changing farming practices or changing farm management. This includes the modernization of farming, particularly in the developing world, and the intensification of farming ongoing in the Western world for more cost-effective production. Besides intensification of farming practices also habitat clearance for cropping and grazing can result in alteration of biodiversity and promote (re)emergence of infectious diseases (Keesing et al., 2010). These agricultural drivers are significant and have a number of effects, including mixing diverse wildlife species together and pushing wildlife and livestock into overlapping environments, thus facilitating the transfer of novel agents into naive and susceptible species (Greger, 2007).
Global air travel easily leads to a rapid and intercontinental spread of a pathogen, for example in the case of SARS-CoV in 2003 and recently very clearly seen for SARS-CoV-2. Reservoir hosts and vectors can spread pathogens more rapidly due to international movement or through trade (Morse et al., 2012). Intensified encroachment into areas of virus endemicity increases the number of diseases attributable to vector-borne pathogens and it may also increase the number of infections coming from wildlife.
Bushmeat consumption is still a significant traditional practice and a growing food source in many countries. When the live animals are moved to markets where diverse species are in close contact, more trade in bushmeat can definitely increase the risk of pathogen transmission (Greatorex et al., 2016). It is known that the initial transmission of the SARS-CoV, from a chiroptera reservoir to the amplifying hosts (including masked palm civet, Paguma larvata), was a consequence of this type of farming and trading activities. Also, in 2019, a wildlife market may have facilitated SARS-CoV-2 transmission from the animal reservoir to the human population (Li et al., 2020a, Andersen et al., 2020).
Climate change and weather changes affect vector, reservoir and pathogen life cycles since these are influenced by multiple and complex processes in their environment (Zinsstag et al., 2018). Modifications of the climate and the habitat can have significant impact on the insect vector distribution. For instance, a pathogen previously limited to a specific area can move to another area where naive populations of animals and humans are present. Heavy rainfalls or extended drought periods can occur due to climate change, and may increase the dispersion of mosquitoes. In more urban areas, water storage facilities and swimming pools as well as the trade of used tires can increase the dispersion of mosquitoes. Zoonotic viruses such as West Nile virus (WNV) and Rift Valley fever virus (RVFV) show an increased distribution in many countries with the colonization of new habitats by their associated vectors. In the north, climate change can also increase the release of some pathogens from frozen soils, see Bacillus anthracis in bacteria section.
Different animal species, especially rodents and bats, can carry a multitude of pathogens with public and veterinary health implications. Several species have the potential to rapidly reach high population numbers, which may create unpredictable situations of high pathogen transmission risks. Rodent populations are heavily affected by environmental changes, including urbanization and climate change.
Wildlife can be considered as the main reservoir for many emerging zoonotic diseases (for an interesting review see (Yon et al., 2019)), and often also the dispersing factor. For one group of pathogens, the actual transmission of the pathogen is rare and human-to-human transmission maintains the circulation of the pathogen in the population, whereas for other pathogens, direct or vector-mediated transmission is the usual source of human infection. While viruses can jump from wild to domesticated animals without any human intervention, trade and animal transports also play a role in the spread of wildlife zoonoses (Bengis et al., 2004). Regions or activities where humans frequently interact with wildlife (hunting-bushmeat, wet markets, deforestation areas, bird migration routes) are the risk hotspots for animal-to-human transmission of zoonotic agents. Transmission risk between wildlife, livestock and humans is further increased by the massive loss of wildlife habitat and changes in land use. Today, more than 77% of the land (without Antarctica) and 87% of the ocean has been altered by the direct effects of human activities (Allan et al., 2017, Watson et al., 2018). Alterations of the last high biodiversity areas in Africa, Asia, Central and South America can significantly increase interactions between livestock and wildlife species and promote (re)emergence events.
Besides wildlife, pets, and livestock, urban fauna can act as reservoirs of zoonotic diseases. It is estimated that over 60% of western families own a pet. Many urban areas are experiencing an increase in the population of stray and semi-domestic dogs. Livestock is one of the fastest-growing agricultural subsectors in developing countries, driven by the rapidly increasing demand for livestock products due to population growth, urbanization and improved incomes (FAO, 2020).
Risk Assessment of Emergence
Risk assessment aims at integrating available knowledge to evaluate the probability and consequences of an emergence in a population. Determining the scale and localization of the population of interest for a given risk assessment is pivotal. Indeed, the occurrence and impact of an emergence can vary from local to global. Moreover, the interactions between wildlife and domestic animals vary largely from a region of the world to the other, as well as contacts between animal and human populations. For instance, in Europe, a lot of effort has been put into limiting the interactions between wildlife and livestock over the past 60 years, in order to avoid emergence of diseases in farm animals and to increase production performance. New trends in farm management, however, tend to provide more outdoor access and thus the potential for increased disease transmission. Risk assessment helps to estimate whether the conditions are present in countries or regions, so that steps can be taken to minimize transmission between animals but also to humans. Health authorities can utilize such risk assessments strategies to mitigate the risk.
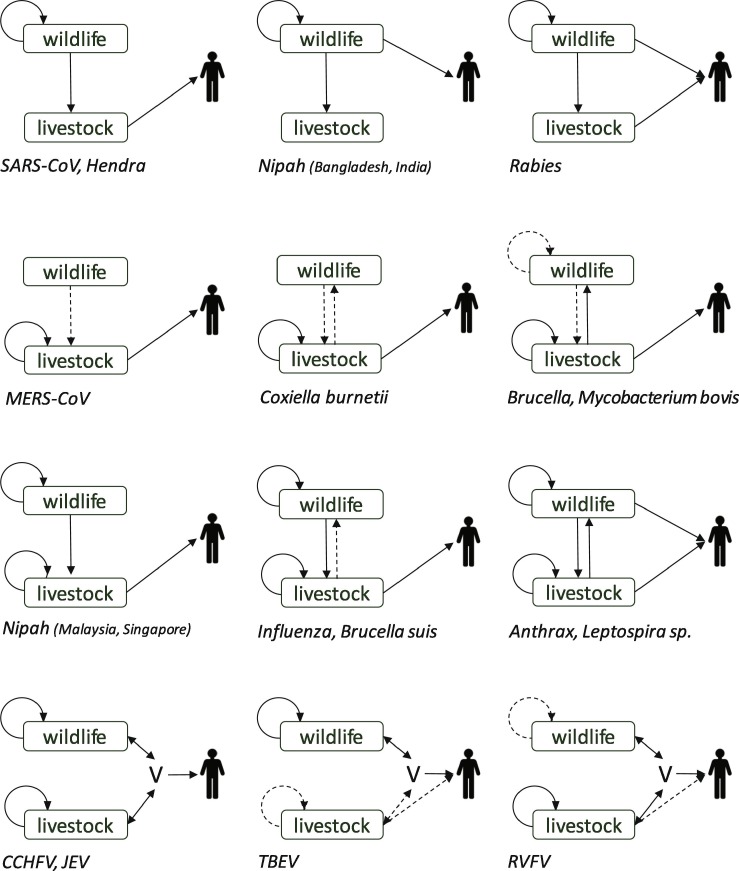
Risk assessment follows a methodology recognized and shared by different authors and institutions. It consists of estimating the probability of the different events that contribute to the emergence of a hazard: first, the probability of hazard emission by the source, and second, the probability of exposure of the at-risk population to this hazard. The risk assessment then consists of taking these probabilities into account, while estimating the consequences of emergence. The probabilities of emission and exposure are mainly a combination of several conditional probabilities, which are related to the different stages allowing the emergence to occur. In order to identify these stages, the use of an event diagram is recommended. It describes the risks pathways, which are the series of events required to occur so that the hazard under consideration results in the unwanted outcome specified (Saegerman et al., 2018). At the interface of wildlife and livestock systems, the issues for the risk assessment are (i) the wildlife surveillance, especially the determination of the health status of wild animals for the hazards under consideration and (ii) the extent of direct or indirect contacts between farm animals and wildlife. If risk assessment is possible at the interface, it helps health authorities to have a better idea of this risk and adopt management options proportionate to the risk. Depending on the hazard, many different pathways have been identified for zoonotic diseases at the wildlife-livestock interface, resulting in exposure of human populations (Fig. 1 ). In a few cases, human exposure is directly related to sources in the wildlife, but most often livestock play an important role either because close contact with farm animals is more likely to occur than with wild animals, or because livestock in turn can amplify the pathogen reservoir. Transmission between the two animal compartments can be occasional spillover, or repeated in time. Finally, the probability of an emergence in humans is largely increased when inter-human transmission of the pathogen occurs. Risk assessment methods can be qualitative (Panel EFSA AHAW, 2006, Jori et al., 2009, Dufour et al., 2011) or use quantitative, deterministic or stochastic models (Jori and Etter, 2016), depending on available data.
Fig. 1.
Diversity of the interfaces between wildlife and livestock resulting in human exposure to zoonotic pathogens. Solid and dotted arrows represent the main and secondary mechanisms for amplification and transmission in populations. V: vectors. For some pathogens, transmission pathways involve the environment (e.g. in the case of anthrax, the interface is the soil contaminated by dead animals). SARS-CoV: Severe Acute Respiratory Syndrome coronavirus; MERS-CoV: Middle East Respiratory Syndrome coronavirus; CCHFV: Crimean-Congo haemorrhagic fever virus; JEV: Japanese encephalitis virus; TBEV: tick-borne encephalitis virus; RVFV: Rift Valley fever virus.
When focusing on the risk assessment at the wildlife-livestock interface, a main issue is data availability and access, in order to estimate the different stage probabilities in the event diagram. Unlike a disease specific to farm animals, for which a number of data are often available, specific difficulties result from understanding and precise description of the wildlife compartment. First, species present and animal densities have to be known. Then, estimating the hazard emission probability by wildlife requires a good knowledge of the pathogens in the wildlife and the environment. Moreover, the diagnosis of an infection can be very complex in wildlife (challenging capture, frequent coinfections and lack of adapted tools amongst others) compromising further actions. When the presence of pathogens is known, surveillance of wildlife remains difficult and quantitative data stay scarce. Prevalence surveys (Varela-Castro et al., 2017) and long-term and risk-based wildlife health surveillance are decisive. Monitoring of the health situation cannot go without a good knowledge of the ecology of wild species and the modelling of their population dynamics, which directly influence the development of an outbreak in this compartment. Next, estimating livestock exposure probability, for the hazard under consideration, requires a good view of the interactions between wildlife and livestock in the studied area. Livestock exposure probability depends on contacts between wildlife and livestock. Specific studies to characterize the wildlife-livestock interface use different methods such as questionnaires to stakeholders (Meunier et al., 2017), telemetric monitoring of wildlife (Triguero-Ocaña et al., 2020), or rely on data analysis by modelling (Barasona et al., 2014) or risk factor identification (Sichewo et al., 2020). A systematic review has highlighted the wide variation and lack of consensus in the definitions of direct and indirect contacts between wild and farm animals (Bacigalupo et al., 2020). The authors proposed a generic unified framework for defining contacts, sufficiently flexible to be applied to most wildlife and livestock species, and adapted to each specific pathogen (for non-vector-borne diseases). This can support future data collection to describe precisely these interactions. In the case of a vector born pathogen, an additional step is needed to evaluate the vector population dynamics.
Finally, after having completed a risk assessment, the potential measures for control and their expected result can be evaluated. Indeed, the respective roles of wildlife, livestock and their interactions in emission, amplification of the pathogen sources and exposure of humans can be identified and weighed. Targeting risk mitigation for the main mechanisms which enable emergence depends widely on the pathogen as each pathosystem is specific (Fig. 1). For a given pathogen, the interactions which result in human exposure can vary in time or between regions, as was evidenced for influenza viruses, for rabies virus or for the Nipah virus. Therefore, risk assessment has to be adapted to each region, and regularly updated. Moreover, multidisciplinary collaboration is necessary, implying wildlife specialists, as well as veterinary and medical professionals. An even wider approach including environmental disciplines also may be indicated, demonstrating the need for a One Health approach elaborated below. Capacity building to perform risk assessment for disease emergence at the interface between wildlife, livestock and humans and to mitigate risk has been promoted by international agencies (World Health Organization, 2019).
Prevention and control of emergence
Old wisdom of infectious disease prevention and control
Successful infection needs the establishment of an infection chain, which includes infectious agent, transmission and host, with the interactions between an infectious agent, routes of transmission, and host factors determining the spectrum of signs and symptoms (Detels et al., 2015). Correspondingly, the measures and tools to interrupt the infection chain are effective and applicable for prevention and control of infectious diseases, including (re)emerging zoonoses. These measures and tools encompass the elimination or restriction of infectious agents, interference with the transmission routes, and identification and protection of susceptible hosts using various approaches including vaccination. Specifically, the infectious agents identified in hosts can be restricted or eliminated by isolation of the infected hosts and by chemical therapy; under certain cases, the agents will be eliminated by culling and safe disposal of the infected animals, whereas those identified in the environment are eliminated by physical or chemical disinfection and sterilization methods. The transmission routes for infectious diseases can be interfered, interrupted or blocked via improved and better hygiene and sanitation systems. For vector-borne pathogens, the vectors are controlled by chemical, environmental and biological ways. The susceptible hosts can be protected from pathogens by quarantine, immunization, human behaviour changes and other intervention measures. Among the various measures and tools, the surveillance is the most important for recognition, evaluation and control of infectious diseases (Detels et al., 2015).
One Health approach applied to prevention and control of zoonotic diseases
Zoonoses involve the mutual interactions of pathogens, humans, animals, and environment; further, some zoonotic agents like highly pathogenic A influenza virus, SARS-CoV and SARS-CoV-2 were able to spread globally and cause pandemics. Therefore, a multi-sectoral approach with interdisciplinary, collaborative strategy is required for the effective control measures and optimal health of people, animals and environment, which reflects the One Health concept or One Health approach (Rahman et al., 2020). One Health was defined by different organizations with similarity: The American Veterinary Association sets One Health as “an integrative effort of multiple disciplines working locally, nationally and globally to attain optimal health for people, animals and the environment” (AVMA – One Health Initiative Task Force, 2008). The Food and Agricultural Organization gives the definition as “a collaborative, international, multidisciplinary mechanism to address threats and reduce risks of detrimental infectious diseases at the human–animal–ecosystem interface” (FAO, 2012), whereas the One Health Initiative defines One Health as “a worldwide strategy for expanding interdisciplinary collaborations and communications in all aspects of health care for humans, animals and the environment” (One Health Initiative, 2012). During the last decade, the concept of One Health has become the international standard for zoonotic disease control (van Herten et al., 2019). However, the concept is still ambiguous and functions as a “boundary object” to leave room for interpretation and facilitate cooperation, so that the equal health of humans, animals and the environment can be possibly improved (van Herten et al., 2019). Certainly, the implementation of One Health concept depends on multiple organizations and various governments to promote and coordinate the cooperation, and fund inter-sectoral activities. Thanks to the efforts of multiple organizations, the One Health approach has become more operational, with an array of One Health tools available, among which twelve commonly implemented One Health tools are used for different countries in strengthening One Health systems (Pelican et al., 2019). Animals as the key component of the One Health concept play a vital role in security, economic and social well-being of humanity. Targeting the ‘risk at source’ in animal populations is a vital strategy in reducing the risks of emerging zoonoses. As such, by collaborating mostly with public health, food safety, and environmental authorities, veterinary authority should follow the performance of veterinary services pathway, which was proposed by the OIE under the One Health concept to prevent and control zoonoses (Stratton et al., 2019).
Prevention and control of zoonotic diseases at the interface of livestock systems
Livestock has a major economic role in the development of many countries. Because of increased interactions (outdoor access and high animal densities for instance), livestock are important source of zoonotic microorganisms and frequently involved in their spread to humans. External as well as internal measures of biosecurity, adapted to the different pathogens, are absolutely needed to limit the risks of transmission to humans. Different measures are available including, amongst others, the cleaning and sterilization of the environment, the culling of infected animals, animal vaccination, and restriction of animal and people movements (Layton et al., 2017). Culling of infected animals is a very effective measure. However, mass culling is expensive because of the loss of animals and the costs associated with waste management. In addition, a major ethical issue exists for culling healthy livestock animals. Preventive immunization of livestock is a very cost-effective measure if the vaccine is available. For instance, regarding human brucellosis for which there is no effective vaccine, the prevention is based on the education of the persons in contact with animals and on the disease control in animal populations. Eradication of brucellosis in ruminants could be achieved based on a combination of slaughter of seropositive adult animals and the vaccination of young animals (Ganter, 2015). Vaccinating livestock against vectors in combination with biological control of the vectors such as mosquito and tick which transmit certain pathogens (this strategy already existed for tick) represented promising preventive measures (Díaz-Martín et al., 2015).
The great incidence of zoonotic diseases in livestock are related to a number of factors including animal production system, increases in global travel, trade and urbanization, human behaviour, vector habitat change, wildlife reservoir, climate change etc. The pathogen needs to overcome several barriers to successfully reach livestock and humans. In addition to conventional preventive approaches such as the vaccination of wildlife species, treatments, disinfection and chemical control, ecological interventions in a One Health approach can be proposed (Sokolow et al., 2019). These interventions allow, amongst others, to control the density, distribution and infectiousness of the wildlife hosts as well as the survival and spread of the zoonotic agent in the environment and the risks of contacts with the spillover host. It has been recommended, for instance, not to plant fruit trees that attract bats, too close to pig pens in the context of Nipah virus control. Similarly, to reduce transmission from bats to horses of Hendra virus, it has been suggested to block the horse overnight access to trees in pastures. Ecological interventions need considerations of the economic, social and political factors to achieve success in managing spillover from wildlife. Globally, synergistic ecological interventions between countries such as China and US are essential for control of global emerging zoonosis spread from wildlife reservoirs (Smiley Evans et al., 2020).
Actually, all zoonotic pathogens from wildlife or other vertebrate reservoir must overcome a hierarchical series of barriers to cause spillover in humans, therefore, understanding how these barriers are intrinsically linked, and how they interact in space and time, will substantially improve our ability to predict or prevent spillover events (Plowright et al., 2017). In this regard, the coronavirus disease 19s (COVID-19) pandemic has its wildlife origin and understanding its spillover mechanism from wildlife will teach us lots of lessons (Morens and Fauci, 2020).
Candidates for the next zoonotic emergence
In this section, diverse and distinguishing examples of candidates for (re)emergence amongst viruses and bacteria are presented (see Table 1 for the main zoonotic agents and role of the different wild and livestock species and Table 2 for examples of viral orders that could lead to the next zoonotic emergences).
Table 1.
Main zoonotic agents and role of the different wild and livestock species.
| Microbial agents | Role of wildlife in epidemiological cycle - main species | Livestock |
Human | ||||
|---|---|---|---|---|---|---|---|
| Main species affected | Species found infected sometimes | Role (in zoonotic context) | Developing disease (?) (Y/N) | Notifiable disease (Y/N) | Human diseases | ||
| Zoonotic avian influenza virus | Reservoir | Poultry | / | Spillover host | Y = avian influenza | Y (if H5 or H7) | ‘Avian’ influenza |
| Influenza viruses infecting swine (swine viruses and reassortants) | Reservoir of “parental” avian influenza viruses | Swine | / | ‘Mixing vessel’ | Y (swine influenza) or N | N | Influenza |
| Wesselsbron virus | Reservoir? (wild rodents, wild fowl) Accidental hosts? | Sheep, goats | Cattle, camels, pigs, donkeys, horses, swine | Reservoir? Spillover hosts? | Y (sheep, goats) N (others) | N | Wesselsbron disease |
| Rift valley fever virus | Wild ruminants and camels are susceptible to disease and act as amplifying hosts | Sheep, goat, cattle, camelids, humans | Giraffe, kudu, warthogs, buffalo, gazelle, springbuck, waterbuck, antelope, wildebeest, impala, rhinoceros, alpaca, dog, cat, bat | Amplifying host, transmission via mosquitoes to animals and humans, and transmission to humans during slaughtering of infected animals | Y | Y | Mild to severe complications including haemorrhagic fever, encephalitis, temporal or permanent blindness |
| Crimean-Congo haemorrhagic fever virus | Hedgehogs, hares, birds, sheep, goats, cattle | Humans | Giraffe, camelids, rhinoceros, buffalo, kudu, horse, donkey, ostrich, dog | Amplifying hosts, transmission via ticks to animals and humans and nosocomial transmission to humans | N | N | Mild to severe complications including haemorrhagic fever |
| Huaiyangshan banyangvirus | Sheep, goat, cattle | Humans | Dog, pig, chicken | Amplifying hosts, transmission via ticks to animals and humans and nosocomial transmission to humans | N | N | Mild to severe complications including multiple organ failure, thrombocytopenia, leucopenia |
| Mycobacterium bovis | Part of a multihost reservoir (badger, wild boar, red deer…) | Cattle | Other domestic ruminants (bisons…) | Unique reservoir /main part of a multihost reservoir | Y = bovine tuberculosis | Y | Tuberculosis |
| Brucella melitensis | Secondary reservoir (ibex) | Small ruminants, cattle | Pigs, horses | Reservoir | Y = ruminant brucellosis | Y | Brucellosis |
| Brucella suis | Main reservoir | Swine | Domestic ruminants | Reservoir (enzootic countries) Spillover host (free countries) | Y = swine brucellosis | Y | Brucellosis |
| Coxiella burnetii | Reservoir (multihost) | Ruminants | / | Part of the multihost reservoir | Y = Q fever | N | Q fever |
| Bacillus anthracis | Contamination of the soil by dead animals | Sheep, goats, cattle | Swine, horses | Contamination of the soil and of humans by dead animals | Y = anthrax | Y | Anthrax |
Y = Yes; N = No; H = hemagglutinin.
Table 2.
Examples of viral orders that could lead to the next zoonotic emergences.
| Orders | Selected families | Examples of previous/current emergences |
|---|---|---|
|
Bunyavirales (enveloped segmented negative-strand RNA viruses) |
Nairoviridae Phenuiviridae |
|
|
Nidovirales (enveloped positive-strand RNA viruses) |
Coronaviridae |
|
| Arteriviridae |
|
|
|
Amarillovirales (enveloped positive-strand RNA viruses) |
Flaviviridae |
|
|
Articulavirales (enveloped segmented negative-strand RNA viruses) |
Orthomyxoviridae |
|
|
Mononegavirales (enveloped negative-strand RNA viruses) |
Paramyxoviridae |
|
| Others: | ||
| Chitovirales (enveloped DNA) | Poxviridae |
|
| Herpesvirales (enveloped double-stranded DNA)… |
Herpesviridae |
|
Viruses
Members of the Bunyavirales order
Recently, the World Health Organization (WHO) has acknowledged the threats posed by several viruses belonging to Bunyavirales order in the face of insufficient or absent countermeasures (Bernasconi et al., 2020). Crimean-Congo haemorrhagic fever virus (CCHFV) is endemic to Southern Europe, Africa, the Middle-East, Southern Asia, and Western China, making it the most widely distributed tick-borne virus affecting humans. The virus is maintained in a transmission cycle involving ixodid (hard) ticks and a variety of wild- and domesticated animals. The tick species most associated with CCHFV transmission is the two-host tick Hyalomma marginatum which is known as a “hunting tick” (Gargili et al., 2017). CCHFV can be transmitted by all developmental stages of H. marginatum with larvae and nymphs feeding on small animals such as hedgehogs, hares, and ground feeding birds, and adults targeting larger animals such as sheep, cattle, and humans. CCHFV infection of animals remains unapparent, while human infections may result in a severe, life-threatening disease. Symptomatic infections of humans initiate with sudden onset of flu-like symptoms. Patients progressing to the haemorrhagic syndrome develop a petechial rash, followed by haemorrhage of the conjunctiva and other mucus membranes that may exacerbate with development of large cutaneous ecchymoses, haematuria and bleeding from the gastrointestinal tract. Fatal cases are associated with multi-organ failure and shock. Whereas some small outbreaks have suggested much higher numbers, the average case-fatality rate of CCHF is estimated at 5% (Bente et al., 2013). Most humans affected are farmers, veterinarians, and abattoir workers. Infections of farmers and veterinarians are generally attributed to tick bites, whereas infections of abattoir workers are believed to result from exposure to contaminated blood. Social and cultural practices, such as the ritual slaughtering of ruminants during the Hajj and Eid-al-Adha, are epidemiologically linked to CCHF outbreaks (Sorvillo et al., 2020).
Another example of a tick-borne virus that circulates at the human-livestock-wildlife interface is Huaiyangshan banyangvirus (BHAV), formally known as Severe Fever with Thrombocytopenia Syndrome virus (SFTSV) (Maslow et al., 2019). The primary vector of BHAV is the Asian longhorned tick, Haemaphysalis longicornis, which transmits the virus to sheep, goats, cattle, pigs, dogs, chickens, and humans. Regarding cattle, a particular role in the transmission to humans, directly or indirectly, has been suggested (Xing et al., 2017). Animal infection is generally asymptomatic, whereas humans may present with thrombocytopenia, leukopenia, and multi-organ failure (Liu et al., 2014). After its first detection in rural areas of Hubei and Henan provinces in China in 2009 (Yu et al., 2011), the virus was detected in Korea (Kim et al., 2018) and Japan (Takahashi et al., 2014). The average case-fatality ratios reported by these countries vary from 5 to 16% in China to 23% in Korea and 27% in Japan (Yun et al., 2020).
Interestingly, a virus related to BHAV, named Heartland virus (HTLV), was detected in 2009, in Missouri (US), in farmers presenting with low white-blood-cell and platelet counts (McMullan et al., 2012). HTLV is believed to be transmitted by the “lone star” tick, Amblyomma americanum, and serological testing has suggested that raccoons and white-tailed deer may function as amplifying hosts (Brault et al., 2018). Although outbreaks caused by tick-borne Phenuiviridae are currently sporadic, the recent (re)emergence of these viruses calls for further assessment of their potential future impact on human health.
The member of the Bunyavirales order with clearly the most significant impact on both animal and human health is Rift valley fever virus (RVFV), a phlebovirus that is transmitted by mosquitoes (Lumley et al., 2017). Wild- and domesticated ruminants, camelids and humans are susceptible to disease, primarily resulting from hepatic necrosis following extensive replication of the virus in the liver. RVFV is endemic to Africa and the Arabian Peninsula, where large outbreaks occur after interepidemic periods that may last for decades. Globalization, climate change, and the world-wide distribution of potential mosquito vectors explain the risk of future incursions into currently unaffected areas (Wright et al., 2019). Sheep are the most susceptible to RVFV, with susceptibility being highest at young age. New-born lambs generally succumb to the infection as a consequence of extensive liver necrosis, whereas mortality rates among adult sheep may also be substantial. A characteristic feature of RVFV outbreaks are abortion storms, in which all pregnant ewes in a flock may abort their foetuses. Goats, cattle and camelids are less susceptible than sheep, although significant morbidity, new-born fatalities and abortions also occur in these species. Humans may become infected through mosquito bites, although most human cases are attributed to contact with contaminated animal tissues during slaughter. Most human cases present with a self-resolving flu-like syndrome without serious consequences, whereas a substantial number of patients develop temporal or permanent vision loss resulting from retinal lesions. A small percentage of patients (1–2%) develop neurological disorders or haemorrhagic icterus (Ikegami and Makino, 2011).
Human exposure to these arthropod-borne viruses can be prevented by avoiding mosquito and tick bites and nosocomial infections can be prevented by proper sanitary practices. The increasing acknowledgement of human health risks posed by Bunyavirales order members has stimulated novel initiatives to develop vaccines, antibodies and antiviral therapies, as well as novel animal models (Garrison et al., 2019, Maslow et al., 2019). Importantly, the coalition for epidemic preparedness innovations is currently supporting the development of Rift valley fever vaccines (Gouglas et al., 2019, Petrova et al., 2020), providing new hope for the prevention and control of future epidemics. The availability of stockpiled vaccines for immediate employment in the case of an emergence is of major importance in the prevention and control of an outbreak.
Coronaviridae
At the time of COVID-19′s pandemic, the emergence capacity of coronaviruses was already well-known. The SARS-CoV-2 is the third emerging coronavirus causing a health crisis in humans during the 21st century. The first emerging zoonotic coronavirus was SARS-CoV in 2003 in China (Guan et al., 2003). This virus, which uses horseshoe bats (Rhinolophus sp.) as reservoir (Li et al., 2005), crossed the species barrier to humans by infecting a small mammal, the palm civet (Paguma larvata) (Tu et al., 2004). The civet being a delicacy in China is bred in farms and sold on markets. The handling of infected animals or contaminated animal products on farms and markets may have facilitated passage of the virus to humans. This first major outbreak due to a zoonotic coronavirus spread to more than 30 countries on several continents, but the number of deaths remained relatively low (774) and no further cases were reported since 2004 (WHO, 2004). A second coronavirus that emerged in this century is the Middle East Respiratory Syndrome coronavirus (MERS-CoV). MERS-CoV was for the first detected in a human case in 2012 and has thus far caused over 2 500 cases with a fatality ratio of 35%. Whereas MERS-CoV is believed to find its origin in bats, transmission to humans predominantly occurs through infected dromedary camels. The exact role of dromedaries and camels in the epidemiological cycle of MERS-CoV is getting better and better understood and a recent review nicely describes the current knowledge regarding the origin of the virus (Bleibtreu et al., 2020). The study of dromedary camel serum collections, including sera collected before 1983, showed that a camel virus closely related to MERS-CoV was already widespread (>80% seropositivity rate) in the East African countries many years ago (Müller et al., 2014). This epidemic has remained more geographically circumscribed but cases are still reported every year (WHO, 2019). In 2019, SARS-CoV-2 emerged, possibly finding its origin in Rhinolophidae as suggested by the RaTG13-CoV virus isolated from these bats and having approximately 96% homology with SARS-CoV-2 (Zhou et al., 2020). By analogy with SARS-CoV and MERS-CoV, an intermediate host is being suspected but currently its clear identification remains unsuccessful.
Human coronaviruses may have emerged from animals earlier. For instance, the coronavirus OC43 that causes mild colds in humans may have emerged from bovine coronavirus, which is closely related and is responsible for diarrhoea in cattle. This zoonotic emergence could have caused what has been described by authors (Vijgen et al., 2005) as a human epidemic ascribed to influenza spreading around the world in 1889–1890. The other human coronaviruses, i.e. NL63, HKU1 and 229E, all responsible for colds in humans, are suspected to have emerged from wildlife (bats or rodents) via intermediate domesticated animals (cattle, dromedary, alpaca) (Corman et al., 2018).
Coronaviruses share with other RNA viruses several mechanisms that may help them to cross the species barriers. Amongst these mechanisms there is, for instance, the absence of corrective activity of the viral RNA dependent RNA polymerase enabling a high mutation rate in RNA viruses. However, some factors more specifically related to coronaviruses explain these numerous successful emergences. Among RNA viruses infecting humans, coronaviruses are those with the longest genome (about 30 kb). This length enables them to withstand large deletions and deleterious mutations during replication and their ability for inter- and intra-specific recombinations allows their variability and plasticity (Woo et al., 2009). Thus, within an infected host, the viral population is characterized by many variants, making spillover infection and adaptation to a new host more likely. In addition, the ability of coronaviruses to infect numerous animal species favours spillover events and stimulates emergence of novel variants resulting from exchange of genetic material between related viruses.
Among emerging coronaviruses in humans, the present or ancestral reservoir of many of them (SARS-CoV, SARS-CoV-2, MERS-CoV, NL63 and 229E) is bats and for the others (OC43 and HKU1), the supposed reservoir is rodents. It is unclear whether bat-borne viruses are more likely to emerge than viruses associated with other species and/or whether it is the high number of bat species that makes them more often the reservoir of emerging pathogens (Luis et al., 2013). Indeed, bats are, with rodents, the mammals with the largest number of species distributed in many environments around the world (Burgin et al., 2018).
Flaviviridae
Viruses of the genus Flavivirus are among the most important representatives of zoonotic arboviruses due to their worldwide distribution and significant number of human infections, amounting to 400 million cases per year (Holbrook, 2017). Their emergence is particularly influenced by anthropogenic environmental changes such as land use and climate change which are strongly impacting ecotones favouring changes in the virus’ ecology (Despommier et al., 2006). Zoonotic flaviviruses include tick-borne flaviviruses, with tick-borne encephalitis virus (TBEV) as the most important representative, and the large family of mosquito-borne flaviviruses (MBF). In this review, we describe a selection of MBFs that is far from being comprehensive with the aim to highlight important candidates for emergence and to point to the extraordinary versatility in host tropism and ecology of some flaviviruses.
The yellow fever complex comprises at least nine viruses, of which six have been found to cause disease in humans. Yellow fever virus (YFV) is endemic to tropical Africa and South America (Monath and Vasconcelos, 2015). It is transmitted by Aedes spp. in Africa and Haemagogus spp. in South America. An important virus threatening ruminants is Wesselsbron virus (WESSV) which was first isolated in 1955 in South Africa from a lamb (Weiss et al., 1956). There have been numerous reports of infections of other ruminants (Mushi et al., 1998), and ostriches have been also shown to be infected (Allwright et al., 1995). Importantly, virus isolation from a black rat in Senegal indicated spread in Africa (Diagne et al., 2017). WESSV has been reported in over 30 human cases causing fever, headache, muscle and joint pain. Interestingly, serological surveys estimated seropositivity at the level of 20–35% in the countries of Southern Africa (Weyer et al., 2013).
The Spondweni virus complex consists of Zika virus (ZIKV) and Spondweni virus (SPOV), both transmitted by Aedes mosquitoes (Haddow and Woodall, 2016). ZIKV was responsible for a recent epidemic that was associated with high occurrence of microcephaly in infants born of infected mothers (Mlakar et al., 2016). The evidence of ZIKV to circulate in nonhuman primates, as described for YFV, appears to be limited (Moreira-Soto and de Carneiro, 2018). Serological studies in Kenya demonstrated prevalence of ZIKV and closely related SPOV in cattle, goats and sheep, notably closer to irrigated areas (Johnson et al., 1977). Recent report from China using the plaque reduction neutralization test showed anti-ZIKV antibody prevalence in sheep at 6.67% (n = 30/2) (Li et al., 2019). SPOV is widely distributed in Africa causing febrile illness in humans (Wolfe et al., 1982).
The Kokobera virus (KOKV) complex consists of two viruses: KOKV and Stratford virus (Simmonds et al., 2017). KOKV, which was first isolated in 1960 in Queensland (Doherty et al., 1964), is enzootic to Australia and Papua New Guinea and appears to be transmitted by Culex and Aedes mosquitoes (Doherty et al., 1964, Doherty et al., 1979). Disease symptoms in humans are fever, arthralgia and lethargy (Mein et al., 1998). Interestingly, serological studies have shown the presence of antibodies against KOKV in macropods, cattle (Doherty et al., 1964, Doherty et al., 1971) and horses (Prow et al., 2013).
The Ntaya virus complex consists of five virus species. Amongst them, Bagaza virus (BAGV) (Fernández-Pinero et al., 2014) is transmitted by Culex spp., but vector competence has also been demonstrated experimentally in Aedes aegypti (Sudeep et al., 2013). Its geographic distribution was long thought to be limited to Africa. However, BAGV was associated with an outbreak of encephalitis in humans in 1996 in India, at least based on serological findings (Bondre et al., 2009). This flavivirus is strongly associated with birds. Outbreaks were reported in 2010 in Spain affecting several birds including red-legged partridges, ring-necked pheasants (Agüero et al., 2011), and common wood pigeons in which increased mortality was observed (Gamino et al., 2012). Over-wintering of this virus in Spain and the possibility of direct transmission make BAGV a serious threat to the European bird population (Llorente et al., 2013). Ilheus virus (ILHV) has been isolated from Aedes spp., and Psorophora mosquitoes in 1944 near Ilheus, Brazil (Laemmert and Hughes, 1947), and geographic distribution has remained confined to South America. It has been isolated from several species of wild birds (Catenacci et al., 2018) but serological studies indicated possible transmission to several mammals including primates, sloths, horses (Iversson et al., 1993) and humans, associated with febrile disease (Venegas et al., 2012).
The Japanese encephalitis virus complex consists of eight virus species, all typically but not only transmitted by Culex ssp., and often infecting bird species. West Nile virus (WNV) is widespread in Africa, Asia, Europe, Australia, and the Americas (Hubálek et al., 2014), and has been detected in more than 150 bird species (van der Meulen et al., 2005). It also infects a wide range of wild and domestic mammals and amphibians (Klenk et al., 2004, Jeffrey, 2013). In the human population, since its emergence in the USA in 1999, the virus has resulted in over 51 000 reported cases, over 25 000 reported neuroinvasive cases, almost 2 400 deaths (Centers for Disease Control and Prevention, 2020). In horses, over 27 000 cases were reported in the USA since 1999 (Animal and Plant Health Inspection Service, US Department of Agriculture, 2020). Both humans and horses are dead-end hosts unable to transmit the pathogen back to mosquitoes and infection can result in subclinical or mild disease or severe encephalitis and deaths (Colpitts et al., 2012).
Japanese encephalitis virus (JEV) is considered one of the major threats for zoonotic emergence in Europe, Africa and the Americas where it would hit a naïve vertebrate population. This is because JEV has so far only established endemicity in Southern Asia (Rosen, 1986). Despite this, JEV represents the most common cause of severe viral encephalitis in the human population (Turtle and Solomon, 2018). Birds, especially wading birds like herons and egrets, are described as most important vertebrate hosts, with pigs acting as virus amplifiers in the environment. In pigs, the virus causes severe reproductive losses through infertility, abortions, mummification of foetuses, and encephalitis in piglets (Mansfield et al., 2017). The observation of mosquito-free direct transmission of JEV between pigs is of concern for areas with intensive pig farming (Ricklin et al., 2016). Horses sporadically develop encephalitis with up to 5% mortality ratio (Hubálek et al., 2014).
Usutu virus (USUV) is another important member of the JEV serocomplex and was first isolated in 1959 in South Africa. Since then it has been detected in other countries throughout the African continent, and in 1996 in Europe (Gaibani and Rossini, 2017). USUV was directly isolated from mosquitoes, birds and bats, but serological studies indicate also infection of horses (Ashraf et al., 2015). Based on a serological survey, USUV was found to circulate in red deer (0.1–0.2%) in Spain, in some cases with indication of co-infection with WNV (García-Bocanegra et al., 2016). Recent retrospective serological study in southwestern and southeastern France showed USUV occurrence in 1% of roe deers (Bournez et al., 2019). It is highly pathogenic to certain birds of prey and passerines. Clinical signs of disease include apathy, incoordination, encephalitis, carditis, hepato- and splenomegaly, with high mortality rates in blackbirds (Hubálek et al., 2014, Ashraf et al., 2015). Human infection is possible and usually associated with mild clinical signs, although a total of 47 cases of neuroinvasive infections have been described so far in Europe (Clé et al., 2019).
Orthomyxoviridae
Viruses from the family Orthomyxoviridae are always very strong candidates for (re)emergence. All orthomyxovirus pandemics so far in human history were caused by Alphainfluenzavirus (1918–1919, 1957–1958, 1968–1969, 1977, and 2009) due to the emergence of new viruses after various reassortment events (Wright et al., 2013). Alphainfluenzavirus isolates are divided into different subtypes based on hemagglutinin (HA) and neuraminidase (NA) antigens. Aquatic birds are the natural reservoirs of Alphainfluenzavirus and 18 HA and 11N subtypes are currently identified, most of them isolated from wild aquatic birds (waterfowl and shore birds) (Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD), 2019). Potentially, 198 different Alphainfluenzavirus subtype combinations are possible but only 131 have been identified. Alphainfluenzaviruses are also able to infect various mammal species including swine, horse, ferret, dog, cat, bat, marine mammals, and humans. New strains of Alphainfluenzavirus emerge frequently as a consequence of two main mechanisms: antigenic drift (mutations resulting from the absence of correction activity of the RNA dependent viral RNA polymerase) which lead to variation within a subtype and antigenic shift (genetic reassortment) which generates novel subtypes. Alphainfluenzavirus strains are commonly species-specific; however, there are many examples of interspecies transmission. For instance, direct passages of avian H5 and H7 subtypes have been documented from birds to humans with most often poultry acting as spillover hosts facilitating the transmission to human hosts from the wild bird reservoir. In 2009, H1N1pdm09 (2009 influenza outbreak) emerged after multiple reassortment events between avian, swine, and human strains (Dawood et al., 2009). At multiple occasions, outbreaks initiated in Southeast Asia probably facilitated by the large human populations living there and traditional farming practices where ducks, humans, and pigs living in close proximity.
Regarding (re)emergence, special attention should be made to pigs, domestic and wild birds and mustelids. Indeed, these animals are clearly all susceptible to Alphainfluenzavirus with aquatic birds such as waterfowl and shore birds as the main reservoir of influenza virus subtypes and pigs known as being a mixing vessel (Ma et al., 2008), since they are at least receptive to infections with both avian and mammalian influenza viruses. However, for many years, the molecular basis of this peculiar susceptibility of pigs remains largely unknown. Recently, an interesting study showed that the swine host factor ANP32A, unlike swine ANP32B or other mammalian ANP32A or B, presented stronger supporting activity to avian viral polymerase offering a molecular basis for the mixing vessel role of the species (Zhang et al., 2020). This discovery is of particular importance since pig to human as well as human to pig transmissions have been identified (Chastagner et al., 2019) increasing the risk of emergence when humans are also in contact with domestic birds, especially when pigs and birds have outdoor access and potential contacts with wildlife.
Paramyxoviridae
Paramyxoviruses have been involved in several (re)emergence events during the past 30 years (Clayton, 2017). A famous example is measles virus, which is believed to be a descendant of the cattle-infecting Rinderpest virus that crossed the species barrier in the 11th or the 12th century (Furuse et al., 2010), occasioning more infections in the last years compared to before as a result of a lower vaccination coverage in human populations. Recent emerging infectious disease events have been caused by viruses belonging to two of the four subfamilies of the Paramyxoviridae; henipavirus and rubulavirus.
On a property in Hendra, Australia in 1994, 21 horses presented a severe respiratory disease leading to the death of 14 of them. Two weeks later, a trainer and a stable-hand also became ill. The trainer died a few days later of pneumonia. The causative virus was isolated from a dead horse and named Hendra virus. Since this first outbreak, several outbreaks were reported in Australia in horses, sometime in association with human cases. No direct human-to-human transmission has been reported. Retrospective studies carried out after outbreaks have provided the opportunity to isolate the virus from fruit bats (Pteropus sp.) (Tulsiani et al., 2011).
Later in 1998, a second henipavirus emerged from fruit bats to pigs and men. A severe outbreak of encephalitis was observed in Kampung Sungai Nipah in Malaysia, causing death of one hundred people. All infected persons had been in contact with sick pigs (Luby, 2013). The introduction of the Nipah virus on the affected pig farms did not appear to be a recent event as suggested the levels of seroprevalence at the index farm (Thibault et al., 2017). The spillover of the virus from bats (Pteropus sp.) to pigs occurred many years ago. Since this first emergence, frequent outbreaks of the Nipah virus disease have been reported in Malaysia, in Bangladesh, in India and Philippines. The disease in these different regions is due to distinct introductions by two different viral lineages (Li et al., 2020b). The first lineage was associated with disease in Malaysia and Singapore. Outbreaks were characterized by a transmission mainly from pigs, the intermediate and amplifying host, and a disease with a case-fatality ratio of approximately 40% (Wang and Anderson, 2019). The second lineage was associated with disease in Bangladesh, India and Philippines. Most of the transmission events were human-to-human and directly from bats via contaminated fruits. The case-fatality rate was higher, about 70–90% (Wang and Anderson, 2019). The genetic diversity of this lineage was higher than the lineage responsible for the outbreaks in Malaysia and Singapore, suggesting numerous spillover events from bats to humans.
The emerging paramyxovirus from the second subfamily of rubulavirus is less studied than the henipaviruses as human cases are less serious and less numerous. Menangle virus was identified in 1997 in Menangle near Sydney in Australia from a piggery with pigs presenting with flu-like disease and gestation disorders. Two piggery workers fell ill during the outbreak and developed neutralizing antibodies (Chant et al., 1998). Bats of the genus Pteropus were suspected of being the reservoir as a retrospective study highlighted neutralizing antibodies in several bat species in Australia (Philbey et al., 2008). A similar study, subsequently carried out in Malaysia in order to find the origin of Nipah virus, isolated a novel rubulavirus very close to Menangle virus (Chua et al., 2001). Due to this proximity, an experimental challenge of pigs was performed to test the pathogenicity of this new virus named Tioman virus. Pigs were productively infected, suggesting that they may act as an intermediate host for humans (Yaiw et al., 2008).
This review of emerging paramyxovirus suggests that this only concerns Asia. However, other potentially emerging paramyxoviruses have been identified in other regions of the world. In 2012, a wildlife biologist developed a severe febrile illness attributed to infection by a rubula-like virus in Uganda (Albariño et al., 2014). Infection with a paramyxovirus related to Nipah virus was highlighted by a serological study in people who reported butchering bats for bushmeat in Cameroon (Pernet et al., 2014). The latter study suggested that spillover events involving paramyxoviruses occurred regularly, but that their detection is undertaken only when clinical events are clearly identified.
The high potential for emergence of paramyxoviruses is explained by several factors, some of which are shared with other emerging pathogens and others being more specific (Thibault et al., 2017). Specifically, spillover events involving paramyxoviruses seem to be facilitated by evolutionarily conserved host proteins being used as receptors. Each spillover is accompanied by an accumulation of genetic mutations induced by the selective forces imposed by the new host. Interestingly, paramyxoviruses adapted to bats use a mechanism to bypass IFIT1 effector activity that functions at least partially in humans and artiodactyls, an order which includes pigs and ruminants (Thibault et al., 2017).
Other viruses
Besides the virus families presented above, potential candidates for future (re)emergence can also be found in other families. For instance, members of the Poxviridae are able to cross species barriers, the most concerning at this stage being the monkeypox virus (MPVX) (Bohelay and Duong, 2019). Another example is vaccinia virus (VACV), able to cross the species barrier from cows (spillover host) to humans. VACV as well as cowpox virus (CPXV) and MPXV have been recently responsible for several outbreaks of exanthematic diseases around the world – in humans as well as in bovines, equids and other animals – and are considered emergent zoonotic viral diseases (Essbauer et al., 2010, Oliveira et al., 2017). Other viruses are strongly associated with their host species after a long co-evolution process (i.e. herpesviruses – (Thiry et al., 2005, Thiry et al., 2006)). The risk of new disease emerging from these examples appears far less elevated based on current knowledge. Then, we cannot exclude new emergences from members of quickly evolving viral families such as Arteriviridae. Indeed, the porcine reproductive and respiratory syndrome virus (PRRSV), for instance, is evolving particularly rapidly − 5.14 × 10–3 nucleotide substitutions/site/year (Yu et al., 2020) – and is probably originating from another arterivirus, the mouse lactate dehydrogenase-elevating virus (Plagemann, 2003). Its successful adaptation to the African Green monkey kidney cell line MA-104 and its derivatives, such as MARC145 cells (Xie et al., 2019), should warn the scientific community about its potential to further cross new species barriers in the future.
Bacteria
Bacteria are less frequently recognized than viruses as agents of zoonotic diseases emerging at the interface of wildlife and livestock. Nevertheless, the capacity of some already known zoonotic bacteria that also affect domestic ruminants, to (re)emerge from unexpected or already identified wild reservoirs is well recognized. The most spectacular illustration is the re-emergence of bovine tuberculosis in several European countries. On the other hand, the emergence of still unknown zoonotic bacteria is considered as a very unusual event, and there is no significant example involving wildlife-livestock interface.
Bovine tuberculosis
Evolution of bovine tuberculosis infection in domestic and wild animals
The situation in England perfectly illustrates the re-emergence of a wildlife reservoir for a bovine zoonotic agent (Mycobacterium bovis, very close to M. tuberculosis, the agent of human tuberculosis). This was completely unexpected, as, by the early 1970s, the apparent annual incidence of bovine tuberculosis had declined sharply to 0.05%. When sporadic contaminations of cattle (infection of cattle or contamination of cattle carcass/meat) occurred that were difficult to explain, the possible emergence of a wild reservoir was considered. The responsibility of infected badgers (Meles meles) was unambiguously confirmed, among other more “classical” factors. Today, the situation has even worsened. In 2019, the annual prevalence of infected herds was 9.4% in England and 17.9% in the high-risk area of the southwest and centre-west (Department for Environment Food and Rural Affairs – DEFRA, 2020).
In continental Europe, wildlife infection is now described both in countries with high and low prevalences, like Spain (2.81% bovine tuberculosis herd prevalence in 2015) (Ciaravino et al., 2018) and France (France is now bovine tuberculosis-free. To be classified as officially tuberculosis-free according to the European Union, a country must fulfil the following criterion: >99.9% of its cattle herds must be bovine tuberculosis-free for at least six consecutive years), respectively. In Spain, the emergence of wild bovine tuberculosis was first observed in wild boars, in the Mediterranean Spanish region (herd prevalence reached 17.2% in Andalusia in 2015), whereas the most affected regions in France were Côte d’Or and south of New-Aquitaine. This unfavourable situation still prevails in New-Aquitaine. In both countries, several wild species are infected (Santos et al., 2020), including wild boars (Sus scrofa) (Massei et al., 2015), but also red deer (Cervus elaphus) (Vicente et al., 2006) and/or badgers. Red foxes (Vulpes vulpes) have recently been incriminated (Michelet et al., 2018). In Spain, noncattle domestic species (small ruminants and even pigs) are heavily infected too (Muñoz-Mendoza et al., 2016).
Relationships between tuberculosis infection of wild and domestic animals
The emergence of cases in wildlife was particularly observed in regions where the prevalence of bovine tuberculosis had apparently recently increased. Isolates from infected cattle and wild animals harboured the same genetic profiles. This confirms local transmission of M. bovis from cattle to autochthonous wild species. Further intra-species transmission has been associated with the increase of certain wild species populations. For example, in the UK, badgers became protected in 1970, which likely allowed badger populations to reach a critical threshold compatible with the constitution of a new reservoir. In continental Europe, the difference with the UK is that M. bovis apparently circulates within different multi-host ecosystems, adding further complexity to the epidemiological situation.
Infection prevalence in wild hosts can even exceed that of infected cattle, like in Britain and Spain, according to Santos et al. (2020). This underlines the need to address the dynamics of infection at a multi-host scale. However, at least in some French regions, the decrease of bovine infection through vigorous control measures was followed by a decrease of wildlife incidence, suggesting that cattle still play a major role in the maintenance of this multi-host transmission system (ANSES, 2019).
Impact on human infection
A retrospective cohort study of human M. bovis cases carried out in the UK during 2002–2014 (Davidson et al., 2017) showed that the incidence remained low. However, a slight but significant increase was observed (from 0.03 and 0.06 annual cases/100 000), with a decrease of the median age of autochthonous patients (from 71 years in 2002 to 53 years in 2014). The strongest risk factors among these patients was an agricultural or animal-related occupation and a history of unpasteurized milk consumption for the majority of them. In other European countries where wildlife infection is observed, the relationship between wildlife/cattle infection and human M. bovis infection is not documented to date.
Brucellosis and human infection
Brucellosis is a severe zoonotic disease characterized by acute septicemia. It can lead to chronic osteoarthritis or orchitis/epididymitis. Two main situations can be distinguished in the context of wildlife-livestock-human interfaces:
Human infections by Brucella melitensis or Brucella abortus
An interesting example is the occurrence in 2012–13 in France of two human cases due to B. melitensis biovar 1, including one clinical case. Both had eaten raw milk cheese from a farm in Haute-Savoie. The investigations revealed that cows on this farm were infected with the same strain as the humans. This was quite surprising, as France was brucellosis-free since 2005, and the farm since 1999. It turned out that these cows had become infected in summer pastures by an ibex reservoir (Bargy massif), initially contaminated by domestic ruminants that had excreted bacteria into the environment. For the first time, a wild reservoir of B. melitensis was present in France, which led to human cases via the contamination of cattle (Mailles et al., 2016).
Human infections by Brucella suis
Brucella suis is present worldwide, with three main biovars affecting swine clinically. Biovars 1 and 3 are far more virulent for humans, while biovar 2 is by far the most frequent in Europe. Wild reservoirs include primarily wild boars but also hares (and less frequently roe deer, like in Germany). B. suis 1 has also been isolated from possums, armadillos and sheep in Argentina and from dogs in the USA and Australia, and also feral swine in the Americas, whereas B. suis biovar 3 has also been isolated from horses in Croatia.
If the transmission of B. suis to humans via swine is considered as anecdotic in Europe, in other parts of the world, swine are more frequently infected, as biovars 1 and/or 3 involve a multihost transmission system, with both wild and domestic hosts, including ruminants in several countries, and also feral swine (in the USA, seroprevalence rates range from 18 to 53%). In these countries, the processing of swine carcasses is associated with a high risk of infection of human workers in slaughterhouses (Olsen and Tatum, 2017). When cattle are involved, raw milk represents a significant source of B. suis infection, as cows are generally not showing any clinical signs but can shed high levels of bacteria, even more frequently than B. abortus, as historically demonstrated (Jordan et al., 1943).
Q fever and human infection
Q fever is by essence a multi-host zoonotic pathogen, involving both wild (in particular small mammals) and domestic reservoirs. The extreme majority of human cases is linked to infected domestic animals. The difficulty relates here to the possibility or not to incriminate wild animals in the genesis of human cases, as domestic animals also represent a reservoir, which can be considered as sufficient to maintain the pathogen in a long-lasting manner. However, in particular when human patients are infected by grazing animals, which is frequently the case, the role of an initial wildlife source (via the environment as Coxiella burnetii pseudo-spores are very resistant, or less frequently via tick bites) is credible, in addition to other sources.
As a whole, the role of wildlife is obvious, but is not necessarily required in the context of a given episode of zoonotic transmission from given domestic livestock. The most spectacular outbreak ever seen in the world (more than 4 000 reported clinical cases between 2007 and 2011 in the Netherlands with an estimation of at least ten times more asymptomatic infections) illustrates the capacity of livestock to be a strong and perennial source of C. burnetii for humans, as dairy goats and, to a lesser extent, dairy sheep bred in mega-farms up to 7 500 animals, were identified as the exclusive source for humans. Conversely, recent human cases involved a particularly virulent strain, MST 17, which emerged in French Guiana in a very unexpected wild reservoir, the three-toed sloth (Bradypus tridactylus), which can also be raised as a pet. Interestingly, C. burnetii has not been described yet in domestic ruminants in French Guiana.
Other considerations regarding bacterial emergence
Several bacterial (re)emergences have been linked to climate change (Wu et al., 2016, El-Sayed and Kamel, 2020) and vector-borne zoonoses are already a reality, however, very few of them involve livestock as an interface with wild reservoirs. In contrast to viruses, bacteria can survive in complex ecosystems including soil, wild animals (dead of the disease and/or burrowing animals) and domestic animals (dead of the disease and/or grazing) from which they can (re)emerge. For example, certain regions in Russia were threatened by the emergence of anthrax, as, between 1897 and 1925, frequent outbreaks of anthrax caused the death of 1.5 million deer and affected hundreds of humans in certain areas of the Russian North, where many historical cattle burial grounds are present. An outbreak occurred in 2016, after a 75-year break, with the death of >200 000 reindeers and one child, in addition to 20 persons hospitalized (Hueffer et al., 2020). This deadly episode has been associated with the acceleration of permafrost thawing as spore viability has been estimated at about 105 years in Siberian permafrost. As about one million wild reindeers and 1.2 million domestic reindeers live nowadays in the Russian North, both can contribute to increased risks of human anthrax in this area, by amplifying domestic cases in a first stage.
Thus, the original source of human infection is not always clear in the case of bacteria, as, for the majority of them, both wild and domestic animal species can behave as reservoirs (Mycobacterium bovis, Coxiella burnetii, Brucella suis, Bacillus anthracis and even Brucella melitensis as shown recently) and/or spillover hosts, allowing the amplification of the pathogen. The separation between a wild reservoir and a domestic reservoir is not always pertinent, as in some cases, they both contribute to maintain the domestic reservoir and to generate a complex ecosystem for the persistence and the potential increase of the zoonotic risk. A priori, livestock species seem more appropriate for playing a role as spillover host, as they live in close proximity with humans. However, with the adoption of wild animals as pets, the risk of zoonotic infections remains a reality as seen for Coxiella burnetii recently (see before).
In conclusion, (re)emergence of zoonotic diseases is of major importance in our attempts to maintain good health for human populations. To date, (re)emergences have never been anticipated adequately causing major crises worldwide and reinforcing the need for a better knowledge of the emergence drivers for the various microbial candidates in current and future hotspots. Which virus family will be associated with the next emergence? Where is it going to occur? Is it going to involve wildlife and/or livestock? Many families of viruses and some bacterial species are good candidates. In the current review we presented potential candidates for future emerging disease outbreaks. We also explained the main drivers of emergence and the measures we should collectively put in place to reduce the risk of an effective emergence in a One Health context. Then, a risk analysis was discussed to orientate our future strategies to mitigate the risk of (re)emergence. Improving our preparation for the next emergences is one of the big challenges we have to face in the next decades.
Ethics approval
Not applicable.
Data and model availability statement
Not applicable.
Author ORCIDs
FM: https://orcid.org/0000-0002-0353-4871.
CD: https://orcid.org/0000-0001-6056-6442.
CF: https://orcid.org/0000-0002-7079-0955.
VG: https://orcid.org/0000-0001-8229-1611.
NH: https://orcid.org/0000-0001-7079-8319.
JK: https://orcid.org/0000-0002-0329-0176.
ML: https://orcid.org/0000-0002-4751-918X.
EML: https://orcid.org/0000-0002-3231-398X.
AS: https://orcid.org/0000-0001-8292-4634.
PJWS: https://orcid.org/0000-0001-9790-2438.
Author contributions
FM: coordinates and supervises the review work, writing and editing; CD: risk assessment of emergence, revision and editing; CF: risk assessment of emergence, Fig. 1, revision and editing; VG: revision and editing; NH: bacterium section, Table 1, revision and editing; JK: Bunyavirales, revision and editing; ML: Flavivirus, revision and editing; EML: Coronavirus, Paramyxovirus and editing; AS: Flavivirus and editing; PJWS: Bunyavirales, revision and editing; WHMvdP: drivers of emergence and hotspots, revision and editing; JZ: prevention and control of emergence, revision and editing. All authors revised and agreed to the published version of the manuscript.
Declaration of interest
The authors declare no conflict of interest
Acknowledgments
Acknowledgements
None.
Financial support statement
The work of ML was supported by the European Union’s Horizon 2020 research and innovation program under Grant No. 734548-ZIKAlliance. Besides, this research received no specific grant from any funding agency, commercial or non-for-profit section.
References
- Agüero M., Fernández-Pinero J., Buitrago D., Sánchez A., Elizalde M., San M.E., Villalba R., Llorente F., Jiménez-Clavero M.A. Bagaza virus in partridges and pheasants, Spain, 2010. Emerging Infectious Diseases. 2011;17:1498–1501. doi: 10.3201/eid1708.110077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albariño C.G., Foltzer M., Towner J.S., Rowe L.A., Campbell S., Jaramillo C.M., Bird B.H., Reeder D.M., Vodzak M.E., Rota P., Metcalfe M.G., Spiropoulou C.F., Knust B., Vincent J.P., Frace M.A., Nichol S.T., Rollin P.E., Ströher U. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerging Infectious Diseases. 2014;20:211–216. doi: 10.3201/eid2002.131620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J.R., Venter O., Watson J.E.M. Temporally inter-comparable maps of terrestrial wilderness and the Last of the Wild. Scientific Data. 2017;4 doi: 10.1038/sdata.2017.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwright D.M., Geyer A., Burger W.P., Williams R., Gerdes G.H., Barnard B.J. Isolation of Wesselsbron virus from ostriches. The Veterinary Record. 1995;136:99. doi: 10.1136/vr.136.4.99. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nature Medicine. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Animal and Plant Health Inspection Service, US Department of Agriculture 2020. West Nile Virus Maps- States with Equine Cases. Retrieved on 27 February 2021, from https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/equine/wnv/west-nile-virus.
- ANSES, 2019. Avis et rapport d’expertise collective révisé relatif à la gestion de la tuberculose bovine et des blaireaux/198 (Saisine n° 2016-SA-0200). Retrieved on 4 October 2020 from https://www.anses.fr/fr/system/files/SABA2016SA0200Ra.pdf.
- Ashraf U., Ye J., Ruan X., Wan S., Zhu B., Cao S. Usutu virus: an emerging flavivirus in Europe. Viruses. 2015;7:219–238. doi: 10.3390/v7010219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVMA - One Health Initiative Task Force, 2008. One Health: A New Professional Imperative. Retrieved on 15 October 2020 from https://www.avma.org/sites/default/files/resources/onehealth_final.pdf.
- Bacigalupo S.A., Dixon L.K., Gubbins S., Kucharski A.J., Drewe J.A. Towards a unified generic framework to define and observe contacts between livestock and wildlife: a systematic review. PeerJ. 2020;8 doi: 10.7717/peerj.10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barasona J.A., Latham M.C., Acevedo P., Armenteros J.A., Latham A.D.M., Gortazar C., Carro F., Soriguer R.C., Vicente J. Spatiotemporal interactions between wild boar and cattle: implications for cross-species disease transmission. Veterinary Research. 2014;45:122. doi: 10.1186/s13567-014-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengis R.G., Leighton F.A., Fischer J.R., Artois M., Mörner T., Tate C.M. The role of wildlife in emerging and re-emerging zoonoses. Revue Scientifique et Technique OIE. 2004;23:497–511. [PubMed] [Google Scholar]
- Bente D.A., Forrester N.L., Watts D.M., McAuley A.J., Whitehouse C.A., Bray M. Crimean-Congo hemorrhagic fever: History, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Research. 2013;100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Bernasconi V., Kristiansen P.A., Whelan M., Román R.G., Bettis A., Yimer S.A., Gurry C., Andersen S.R., Yeskey D., Mandi H., Kumar A., Holst J., Clark C., Cramer J.P., Røttingen J.-A., Hatchett R., Saville M., Norheim G. Developing vaccines against epidemic-prone emerging infectious diseases. Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz. 2020;63:65–73. doi: 10.1007/s00103-019-03061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleibtreu A., Bertine M., Bertin C., Houhou-Fidouh N., Visseaux B. Focus on Middle East respiratory syndrome coronavirus (MERS-CoV) Médecine et Maladies Infectieuses. 2020;50:243–251. doi: 10.1016/j.medmal.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohelay G., Duong T.-A. Infections humaines à poxvirus. Annales de Dermatologie et de Vénéréologie. 2019;146:387–398. doi: 10.1016/j.annder.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondre V.P., Sapkal G.N., Yergolkar P.N., Fulmani P.V., Sankararaman V., Ayachit V.M., Mishra A.C., Gore M.M. Genetic characterization of Bagaza virus (BAGV) isolated in India and evidence of anti-BAGV antibodies in sera collected from encephalitis patients. Journal of General Virology. 2009;90:2644–2649. doi: 10.1099/vir.0.012336-0. [DOI] [PubMed] [Google Scholar]
- Bournez L., Umhang G., Faure E., Boucher J.-M., Boué F., Jourdain E., Sarasa M., Llorente F., Jiménez-Clavero M.A., Moutailler S., Lacour S.A., Lecollinet S., Beck C. Exposure of Wild Ungulates to the Usutu and Tick-Borne Encephalitis Viruses in France in 2009–2014: Evidence of Undetected Flavivirus Circulation a Decade Ago. Viruses. 2019;12:10. doi: 10.3390/v12010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A.C., Savage H.M., Duggal N.K., Eisen R.J., Staples J.E. Heartland Virus Epidemiology, Vector Association, and Disease Potential. Viruses. 2018;10:498. doi: 10.3390/v10090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin C.J., Colella J.P., Kahn P.L., Upham N.S. How many species of mammals are there? Journal of Mammalogy. 2018;99:1–14. [Google Scholar]
- Catenacci L.S., Ferreira M., Martins L.C., De Vleeschouwer K.M., Cassano C.R., Oliveira L.C., Canale G., Deem S.L., Tello J.S., Parker P., Vasconcelos P.F.C., Travassos da Rosa E.S. Surveillance of Arboviruses in Primates and Sloths in the Atlantic Forest, Bahia, Brazil. EcoHealth. 2018;15:777–791. doi: 10.1007/s10393-018-1361-2. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2020. Final Cumulative Maps & Data for 1999–2019 from 2020-11-24. Retrieved on 27 February 2021, from https://www.cdc.gov/westnile/statsmaps/cumMapsData.html.
- Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD), 2019. Types of Influenza Viruses. Retrieved on 2 November 2020 from https://www.cdc.gov/flu/about/viruses/types.htm.
- Chant K., Chan R., Smith M., Dwyer D.E., Kirkland P. Probable human infection with a newly described virus in the family Paramyxoviridae. The NSW Expert Group. Emerging Infectious Diseases. 1998;4:273–275. doi: 10.3201/eid0402.980215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastagner A., Enouf V., Peroz D., Hervé S., Lucas P., Quéguiner S., Gorin S., Beven V., Behillil S., Leneveu P., Garin E., Blanchard Y., van der Werf S., Simon G. Bidirectional Human-Swine Transmission of Seasonal Influenza A(H1N1)pdm09 Virus in Pig Herd, France, 2018. Emerging Infectious Diseases. 2019;25:1940–1943. doi: 10.3201/eid2510.190068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Wang L.F., Lam S.K., Crameri G., Yu M., Wise T., Boyle D., Hyatt A.D., Eaton B.T. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology. 2001;283:215–229. doi: 10.1006/viro.2000.0882. [DOI] [PubMed] [Google Scholar]
- Ciaravino G., García-Saenz A., Cabras S., Allepuz A., Casal J., García-Bocanegra I., De Koeijer A., Gubbins S., Sáez J.L., Cano-Terriza D., Napp S. Assessing the variability in transmission of bovine tuberculosis within Spanish cattle herds. Epidemics. 2018;23:110–120. doi: 10.1016/j.epidem.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Clayton B.A. Nipah virus: transmission of a zoonotic paramyxovirus. Current Opinion in Virology. 2017;22:97–104. doi: 10.1016/j.coviro.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Clé M., Beck C., Salinas S., Lecollinet S., Gutierrez S., Van de Perre P., Baldet T., Foulongne V., Simonin Y. Usutu virus: A new threat? Epidemiology and Infection. 2019;147 doi: 10.1017/S0950268819001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S., Laurenson M., Taylor L. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts T.M., Conway M.J., Montgomery R.R., Fikrig E. West Nile Virus: biology, transmission, and human infection. Clinical Microbiology Reviews. 2012;25:635–648. doi: 10.1128/CMR.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Advances in Virus Research. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.J., Fooks A.R., van der Poel W.H.M. Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerging Infectious Diseases. 2010;16:1–7. doi: 10.3201/eid1601.081467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P., Plowright R.K., Epstein J.H., Pulliam J., Rahman S.A., Field H.E., Jamaluddin A., Sharifah S.H., Smith C.S., Olival K.J., Luby S., Halpin K., Hyatt A.D., Cunningham A.A. The emergence of Nipah and Hendra virus: pathogen dynamics across a wildlife-livestock-human continuum. In: Collinge S.K., Ray C., editors. Disease Ecology: Community Structure and Pathogen Dynamics. Oxford University Press; Cary, NC, USA: 2006. pp. 227–247. [Google Scholar]
- Davidson J.A., Loutet M.G., O’Connor C., Kearns C., Smith R.M.M., Lalor M.K., Thomas H.L., Abubakar I., Zenner D. Epidemiology of Mycobacterium bovis Disease in Humans in England, Wales, and Northern Ireland, 2002–2014. Emerging Infectious Diseases. 2017;23:377–386. doi: 10.3201/eid2303.161408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J., Gubareva L.V., Xu X., Bridges C.B., Uyeki T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. New England Journal of Medicine. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Department for Environment Food and Rural Affairs – DEFRA, 2020. Quarterly publication of National Statistics on the incidence and prevalence of tuberculosis (TB) in Cattle in Great Britain – to end June 2020. Retrieved on 7 September 2020 from https://www.gov.uk/government/collections/bovine-tb.
- Derner J., Hunt L., Filho K., Ritten J., Capper J., Han G. Livestock Production Systems. In: Briske D.D., editor. Rangeland Systems: Processes, Management and Challenges. Springer Open; Cham, Switzerland: 2017. pp. 347–372. [Google Scholar]
- Despommier D., Ellis B.R., Wilcox B.A. The role of ecotones in emerging infectious diseases. EcoHealth. 2006;3:281–289. [Google Scholar]
- Detels R., Gulliford M., Abdool Karim Q., Chuan Tan C. 6th ed. Oxford University Press; Oxford, UK: 2015. Oxford Textbook of Global Public Health. [Google Scholar]
- Diagne M.M., Faye M., Faye O., Sow A., Balique F., Sembène M., Granjon L., Handschumacher P., Faye O., Diallo M., Sall A.A. Emergence of Wesselsbron virus among black rat and humans in Eastern Senegal in 2013. One Health (Amsterdam, Netherlands) 2017;3:23–28. doi: 10.1016/j.onehlt.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Martín V., Manzano-Román R., Obolo-Mvoulouga P., Oleaga A., Pérez-Sánchez R. Development of vaccines against Ornithodoros soft ticks: An update. Ticks and Tick-Borne Diseases. 2015;6:211–220. doi: 10.1016/j.ttbdis.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Doherty R.L., Carley J.G., Gorman B.M. Studies of arthropod-borne virus infections in Queensland. IV. Further serological investigations of antibodies to group B Arboviruses in man and animals. The Australian Journal of Experimental Biology and Medical Science. 1964;42:149–164. doi: 10.1038/icb.1964.16. [DOI] [PubMed] [Google Scholar]
- Doherty R.L., Carley J.G., Kay B.H., Filippich C., Marks E.N., Frazier C.L. Isolation of virus strains from mosquitoes collected in Queensland, 1972–1976. The Australian Journal of Experimental Biology and Medical Science. 1979;57:509–520. doi: 10.1038/icb.1979.52. [DOI] [PubMed] [Google Scholar]
- Doherty R.L., Standfast H.A., Domrow R., Wetters E.J., Whitehead R.H., Carley J.G. Studies of the epidemiology of arthropod-borne virus infections at Mitchell River Mission, Cape York Peninsula, North Queensland. IV. Arbovirus infections of mosquitoes and mammals, 1967–1969. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1971;65:504–513. doi: 10.1016/0035-9203(71)90161-1. [DOI] [PubMed] [Google Scholar]
- Dufour B., Plée L, Moutou F., Boisseleau D., Chartier C., Durand B., Ganiere J.P., Guillotin J., Lancelot R., Saegerman C., Thebault A., Hattenberger A.M. and Toma B. 2011. A qualitative risk assessment methodology for scientific expert panels. Revue scientifique et technique OIE 30, 673–81. [DOI] [PubMed]
- Duru M., Therond O. Livestock system sustainability and resilience in intensive production zones: which form of ecological modernization? Regional Environmental Change. 2015;15:1651–1665. [Google Scholar]
- El-Sayed A., Kamel M. Climatic changes and their role in emergence and re-emergence of diseases. Environmental Science and Pollution Research International. 2020;27:22336–22352. doi: 10.1007/s11356-020-08896-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essbauer S., Pfeffer M., Meyer H. Zoonotic poxviruses. Veterinary Microbiology. 2010;140:229–236. doi: 10.1016/j.vetmic.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO, 2012. Thoughts of FAO on ‘One Health’. Retrieved on 5 October 2020 from http://www.fao.org/ag/againfo/home/en/news_archive/2010_one-health.html.
- FAO, 2020. Livestock systems. Retrieved on 6 October 2020 from http://www.fao.org/livestock-systems/en/.
- Fernández-Pinero J., Davidson I., Elizalde M., Perk S., Khinich Y., Jiménez-Clavero M.A. Bagaza virus and Israel turkey meningoencephalomyelitis virus are a single virus species. The Journal of General Virology. 2014;95:883–887. doi: 10.1099/vir.0.061465-0. [DOI] [PubMed] [Google Scholar]
- Furuse Y., Suzuki A., Oshitani H. Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virology Journal. 2010;7:52. doi: 10.1186/1743-422X-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani P., Rossini G. An overview of Usutu virus. Microbes and Infection. 2017;19:382–387. doi: 10.1016/j.micinf.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Gamino V., Gutiérrez-Guzmán A.-V., Fernández-de-Mera I.G., Ortíz J.-A., Durán-Martín M., de la Fuente J., Gortázar C., Höfle U. Natural Bagaza virus infection in game birds in southern Spain. Veterinary Research. 2012;43:65. doi: 10.1186/1297-9716-43-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter M. Zoonotic risks from small ruminants. Veterinary Microbiology. 2015;181:53–65. doi: 10.1016/j.vetmic.2015.07.015. [DOI] [PubMed] [Google Scholar]
- García-Bocanegra I., Paniagua J., Gutiérrez-Guzmán A.V., Lecollinet S., Boadella M., Arenas-Montes A., Cano-Terriza D., Lowenski S., Gortázar C., Höfle U. Spatio-temporal trends and risk factors affecting West Nile virus and related flavivirus exposure in Spanish wild ruminants. BMC Veterinary Research. 2016;12:249. doi: 10.1186/s12917-016-0876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargili A., Estrada-Peña A., Spengler J.R., Lukashev A., Nuttall P.A., Bente D.A. The role of ticks in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus: A review of published field and laboratory studies. Antiviral Research. 2017;144:93–119. doi: 10.1016/j.antiviral.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison A.R., Smith D.R., Golden J.W. Animal Models for Crimean-Congo Hemorrhagic Fever Human Disease. Viruses. 2019;11:590. doi: 10.3390/v11070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M., Nicolas G., Cinardi G., Van Boeckel T.P., Vanwambeke S.O., Wint G.R.W., Robinson T.P. Global distribution data for cattle, buffaloes, horses, sheep, goats, pigs, chickens and ducks in 2010. Scientific Data. 2018;5 doi: 10.1038/sdata.2018.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouglas D., Christodoulou M., Plotkin S.A., Hatchett R. CEPI: Driving Progress Toward Epidemic Preparedness and Response. Epidemiologic Reviews. 2019;41:28–33. doi: 10.1093/epirev/mxz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greatorex Z.F., Olson S.H., Singhalath S., Silithammavong S., Khammavong K., Fine A.E., Weisman W., Douangngeun B., Theppangna W., Keatts L., Gilbert M., Karesh W.B., Hansel T., Zimicki S., O’Rourke K., Joly D.O., Mazet J.A.K. Wildlife Trade and Human Health in Lao PDR: An Assessment of the Zoonotic Disease Risk in Markets. PloS One. 2016;11 doi: 10.1371/journal.pone.0150666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger M. The human/animal interface: emergence and resurgence of zoonotic infectious diseases. Critical Reviews in Microbiology. 2007;33:243–299. doi: 10.1080/10408410701647594. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science (New York, NY) 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haddow A.D., Woodall J.P. Distinguishing between Zika and Spondweni viruses. Bulletin of the World Health Organization. 2016;94 doi: 10.2471/BLT.16.181503. 711–711A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook M.R. Historical Perspectives on Flavivirus Research. Viruses. 2017;9:97. doi: 10.3390/v9050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubálek Z., Rudolf I., Nowotny N. Arboviruses pathogenic for domestic and wild animals. Advances in Virus Research. 2014;89:201–275. doi: 10.1016/B978-0-12-800172-1.00005-7. [DOI] [PubMed] [Google Scholar]
- Hueffer K., Drown D., Romanovsky V., Hennessy T. Factors Contributing to Anthrax Outbreaks in the Circumpolar North. EcoHealth. 2020;17:174–180. doi: 10.1007/s10393-020-01474-z. [DOI] [PubMed] [Google Scholar]
- Ikegami T., Makino S. The Pathogenesis of Rift Valley Fever. Viruses. 2011;3:493–519. doi: 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversson L.B., Silva R.A., da Rosa A.P., Barros V.L. Circulation of eastern equine encephalitis, western equine encephalitis, Ilhéus, Maguari and Tacaiuma viruses in equines of the Brazilian Pantanal, South America. Revista Do Instituto De Medicina Tropical De Sao Paulo. 1993;35:355–359. doi: 10.1590/s0036-46651993000400009. [DOI] [PubMed] [Google Scholar]
- Jeffrey Root J. West Nile virus associations in wild mammals: a synthesis. Archives of Virology. 2013;158:735–752. doi: 10.1007/s00705-012-1516-3. [DOI] [PubMed] [Google Scholar]
- Johnson B.K., Chanas A.C., Shockley P., Squires E.J., Gardner P., Wallace C., Simpson D.I., Bowen E.T., Platt G.S., Way H., Parsons J., Grainger W.E. Arbovirus isolations from, and serological studies on, wild and domestic vertebrates from Kano Plain, Kenya. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1977;71:512–517. doi: 10.1016/0035-9203(77)90146-8. [DOI] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan C.F., Borts I.H., Harris D.M., Jennings J.R. Brucellosis: Consideration of Its Epidemiology, Diagnosis and Control. American Journal of Public Health and the Nation’s Health. 1943;33:773–779. doi: 10.2105/ajph.33.7.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jori F., Etter E. Transmission of foot and mouth disease at the wildlife/livestock interface of the Kruger National Park, South Africa: Can the risk be mitigated? Preventive Veterinary Medicine. 2016;126:19–29. doi: 10.1016/j.prevetmed.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Jori F., Vosloo W., Du Plessis B., Bengis R., Brahmbhatt D., Gummow B., Thomson G.R. A qualitative risk assessment of factors contributing to foot and mouth disease outbreaks in cattle along the western boundary of the Kruger National Park. Revue Scientifique et Technique (International Office of Epizootics) 2009;28:917–931. doi: 10.20506/rst.28.3.1932. [DOI] [PubMed] [Google Scholar]
- Keesing F., Belden L.K., Daszak P., Dobson A., Harvell C.D., Holt R.D., Hudson P., Jolles A., Jones K.E., Mitchell C.E., Myers S.S., Bogich T., Ostfeld R.S. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.R., Yun Y., Bae S.G., Park D., Kim S., Lee J.M., Cho N.-H., Kim Y.S., Lee K.H. Severe Fever with Thrombocytopenia Syndrome Virus Infection, South Korea, 2010. Emerging Infectious Diseases. 2018;24:2103–2105. doi: 10.3201/eid2411.170756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk K., Snow J., Morgan K., Bowen R., Stephens M., Foster F., Gordy P., Beckett S., Komar N., Gubler D., Bunning M. Alligators as West Nile virus amplifiers. Emerging Infectious Diseases. 2004;10:2150–2155. doi: 10.3201/eid1012.040264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmert, H.W., Hughes, T.P., 1947. The virus of Ilhéus encephalitis; isolation, serological specificity and transmission. Journal of Immunology (Baltimore, Md.: 1950) 55, 61–67. [PubMed]
- Layton D.S., Choudhary A., Bean A.G.D. Breaking the chain of zoonoses through biosecurity in livestock. Vaccine. 2017;35:5967–5973. doi: 10.1016/j.vaccine.2017.07.110. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. The New England Journal of Medicine. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.-F. Bats are natural reservoirs of SARS-like coronaviruses. Science (New York, NY) 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li K., Yan S., Wang N., He W., Guan H., He C., Wang Z., Lu M., He W., Ye R., Veit M., Su S. Emergence and adaptive evolution of Nipah virus. Transboundary and Emerging Diseases. 2020;67:121–132. doi: 10.1111/tbed.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhou J.Z., Zhou L., Fu S.H., Tian Z.Z., Wang Q., Shao N., Li D., He Y., Lei W.W., Tang G.P., Liang G.D., Wang D.M., Zhang Y.P., Wang H.Y. Serological Survey of Zika Virus in Humans and Animals in Dejiang Prefecture, Guizhou Province, China. Biomedical and Environmental Sciences: BES. 2019;32:875–880. doi: 10.3967/bes2019.108. [DOI] [PubMed] [Google Scholar]
- Liu Q., He B., Huang S.-Y., Wei F., Zhu X.-Q. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. The Lancet Infectious Diseases. 2014;14:763–772. doi: 10.1016/S1473-3099(14)70718-2. [DOI] [PubMed] [Google Scholar]
- Llorente F., Pérez-Ramírez E., Fernández-Pinero J., Soriguer R., Figuerola J., Jiménez-Clavero M.A. Flaviviruses in game birds, southern Spain, 2011–2012. Emerging Infectious Diseases. 2013;19:1023–1025. doi: 10.3201/eid1906.130122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S.P. The pandemic potential of Nipah virus. Antiviral Research. 2013;100:38–43. doi: 10.1016/j.antiviral.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T.S., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R.C., Mills J.N., Timonin M.E., Willis C.K.R., Cunningham A.A., Fooks A.R., Rupprecht C.E., Wood J.L.N., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proceedings. Biological Sciences. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley S., Horton D.L., Hernandez-Triana L.L.M., Johnson N., Fooks A.R., Hewson R. Rift Valley fever virus: strategies for maintenance, survival and vertical transmission in mosquitoes. Journal of General Virology. 2017;98:875–887. doi: 10.1099/jgv.0.000765. [DOI] [PubMed] [Google Scholar]
- Ma W., Kahn R.E., Richt J.A. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. Journal of Molecular and Genetic Medicine: An International Journal of Biomedical Research. 2008;3:158–166. [PMC free article] [PubMed] [Google Scholar]
- Mailles A., Garin-Bastuji B., Lavigne J.P., Jay M., Sotto A., Maurin M., Pelloux I., O’Callaghan D., Mick V., Vaillant V., De Valk H. Human brucellosis in France in the 21st century: Results from national surveillance 2004–2013. Médecine et Maladies Infectieuses. 2016;46:411–418. doi: 10.1016/j.medmal.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Mansfield K.L., Hernández-Triana L.M., Banyard A.C., Fooks A.R., Johnson N. Japanese encephalitis virus infection, diagnosis and control in domestic animals. Veterinary Microbiology. 2017;201:85–92. doi: 10.1016/j.vetmic.2017.01.014. [DOI] [PubMed] [Google Scholar]
- Martin G., Moraine M., Ryschawy J., Magne M.-A., Asai M., Sarthou J.-P., Duru M., Therond O. Crop–livestock integration beyond the farm level: a review. Agronomy for Sustainable Development. 2016;36:53. [Google Scholar]
- Maslow J.N., Kwon J.J., Mikota S.K., Spruill S., Cho Y., Jeong M. Severe fever and thrombocytopenia syndrome virus infection: Considerations for vaccine evaluation of a rare disease. Human Vaccines & Immunotherapeutics. 2019;15:2249–2257. doi: 10.1080/21645515.2019.1633875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massei G., Kindberg J., Licoppe A., Gačić D., Šprem N., Kamler J., Baubet E., Hohmann U., Monaco A., Ozoliņš J., Cellina S., Podgórski T., Fonseca C., Markov N., Pokorny B., Rosell C., Náhlik A. Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Management Science. 2015;71:492–500. doi: 10.1002/ps.3965. [DOI] [PubMed] [Google Scholar]
- McMullan L.K., Folk S.M., Kelly A.J., MacNeil A., Goldsmith C.S., Metcalfe M.G., Batten B.C., Albariño C.G., Zaki S.R., Rollin P.E., Nicholson W.L., Nichol S.T. A New Phlebovirus Associated with Severe Febrile Illness in Missouri. New England Journal of Medicine. 2012;367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- Mein J., O’Grady K.A., Whelan P., Merianos A. Dengue or Kokobera? A case report from the top end of the Northern Territory. Communicable Diseases Intelligence. 1998;22:105–107. doi: 10.33321/cdi.1998.22.20. [DOI] [PubMed] [Google Scholar]
- Meunier N.V., Sebulime P., White R.G., Kock R. Wildlife-livestock interactions and risk areas for cross-species spread of bovine tuberculosis. The Onderstepoort Journal of Veterinary Research. 2017;84:e1–e10. doi: 10.4102/ojvr.v84i1.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L., De Cruz K., Hénault S., Tambosco J., Richomme C., Réveillaud É., Gares H., Moyen J.-L., Boschiroli M.L. Mycobacterium bovis Infection of Red Fox, France. Emerging Infectious Diseases. 2018;24:1150–1153. doi: 10.3201/eid2406.180094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V., Vizjak A., Pižem J., Petrovec M., Avšič Županc T. Zika Virus Associated with Microcephaly. The New England Journal of Medicine. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Monath T.P., Vasconcelos P.F.C. Yellow fever. Journal of Clinical Virology: The Official Publication of the Pan American Society for Clinical Virology. 2015;64:160–173. doi: 10.1016/j.jcv.2014.08.030. [DOI] [PubMed] [Google Scholar]
- Moreira-Soto, A., Carneiro, I. de O., Fischer, C., Feldmann, M., Kümmerer, B.M., Silva, N.S., Santos, U.G., Souza, B.F. de C.D., Liborio, F. de A., Valença-Montenegro, M.M., Laroque, P. de O., da Fontoura, F.R., Oliveira, A.V.D., Drosten, C., de Lamballerie, X., Franke, C.R., Drexler, J.F., 2018. Limited Evidence for Infection of Urban and Peri-urban Nonhuman Primates with Zika and Chikungunya Viruses in Brazil. mSphere 3, e00523-17. [DOI] [PMC free article] [PubMed]
- Morens D.M., Fauci A.S. Emerging Pandemic Diseases: How We Got to COVID-19. Cell. 2020;182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S. Factors in the emergence of infectious diseases. Emerging Infectious Diseases. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S., Mazet J.A.K., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet (London, England) 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.-J., Lattwein E., Hilali M., Musa B.E., Bornstein S., Drosten C. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerging Infectious Diseases. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Mendoza M., Romero B., Del Cerro A., Gortázar C., García-Marín J.F., Menéndez S., Mourelo J., de Juan L., Sáez J.L., Delahay R.J., Balseiro A. Sheep as a Potential Source of Bovine TB: Epidemiology, Pathology and Evaluation of Diagnostic Techniques. Transboundary and Emerging Diseases. 2016;63:635–646. doi: 10.1111/tbed.12325. [DOI] [PubMed] [Google Scholar]
- Mushi E.Z., Binta M.G., Raborokgwe M. Wesselsbron disease virus associated with abortions in goats in Botswana. Journal of Veterinary Diagnostic Investigation. 1998;10:191. doi: 10.1177/104063879801000216. [DOI] [PubMed] [Google Scholar]
- Oliveira G.P., Rodrigues R.A.L., Lima M.T., Drumond B.P., Abrahão J.S. Poxvirus Host Range Genes and Virus-Host Spectrum: A Critical Review. Viruses. 2017;9:331. doi: 10.3390/v9110331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S.C., Tatum F.M. Swine brucellosis: current perspectives. Veterinary Medicine (Auckland, NZ) 2017;8:1–12. doi: 10.2147/VMRR.S91360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Health Initiative, 2012. The One Health concept. Retrieved on 6 October 2020 from https://onehealthinitiative.com/about/.
- Panel EFSA AHAW Opinion of the Scientific Panel Animal Health and Welfare (AHAW) related with the Migratory Birds and their Possible Role in the Spread of Highly Pathogenic Avian Influenza. EFSA Journal. 2006;4:357. doi: 10.2903/j.efsa.2006.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelican K., Salyer S.J., Barton Behravesh C., Belot G., Carron M., Caya F., De La Rocque S., Errecaborde K.M., Lamielle G., Latronico F., Macy K.W., Mouille B., Mumford E., Shadomy S., Sinclair J.R., Dutcher T. Synergising tools for capacity assessment and One Health operationalisation. Revue Scientifique et Technique. 2019;38:71–89. doi: 10.20506/rst.38.1.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet O., Schneider B.S., Beaty S.M., LeBreton M., Yun T.E., Park A., Zachariah T.T., Bowden T.A., Hitchens P., Ramirez C.M., Daszak P., Mazet J., Freiberg A.N., Wolfe N.D., Lee B. Evidence for henipavirus spillover into human populations in Africa. Nature Communications. 2014;5:5342. doi: 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V., Kristiansen P., Norheim G., Yimer S.A. Rift valley fever: diagnostic challenges and investment needs for vaccine development. BMJ Global Health. 2020;5 doi: 10.1136/bmjgh-2020-002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philbey A.W., Kirkland P.D., Ross A.D., Field H.E., Srivastava M., Davis R.J., Love R.J. Infection with Menangle virus in flying foxes (Pteropus spp.) in Australia. Australian Veterinary Journal. 2008;86:449–454. doi: 10.1111/j.1751-0813.2008.00361.x. [DOI] [PubMed] [Google Scholar]
- Plagemann P.G.W. Porcine Reproductive and Respiratory Syndrome Virus: Origin Hypothesis. Emerging Infectious Diseases. 2003;9:903–908. doi: 10.3201/eid0908.030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., Lloyd-Smith J.O. Pathways to zoonotic spillover. Nature Reviews. Microbiology. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prow N.A., Tan C.S.E., Wang W., Hobson-Peters J., Kidd L., Barton A., Wright J., Hall R.A., Bielefeldt-Ohmann H. Natural exposure of horses to mosquito-borne flaviviruses in south-east Queensland, Australia. International Journal of Environmental Research and Public Health. 2013;10:4432–4443. doi: 10.3390/ijerph10094432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.T., Sobur M.A., Islam M.S., Ievy S., Hossain M.J., El Zowalaty M.E., Rahman A.T., Ashour H.M. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms. 2020;8:1405. doi: 10.3390/microorganisms8091405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin M.E., García-Nicolás O., Brechbühl D., Python S., Zumkehr B., Nougairede A., Charrel R.N., Posthaus H., Oevermann A., Summerfield A. Vector-free transmission and persistence of Japanese encephalitis virus in pigs. Nature Communications. 2016;7:10832. doi: 10.1038/ncomms10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen L. The natural history of Japanese encephalitis virus. Annual Review of Microbiology. 1986;40:395–414. doi: 10.1146/annurev.mi.40.100186.002143. [DOI] [PubMed] [Google Scholar]
- Saegerman C., Bertagnoli S., Meyer G., Ganière J.-P., Caufour P., Clercq K.D., Jacquiet P., Fournié G., Hautefeuille C., Etore F., Casal J. Risk of introduction of lumpy skin disease in France by the import of vectors in animal trucks. PLOS ONE. 2018;13 doi: 10.1371/journal.pone.0198506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N., Richomme C., Nunes T., Vicente J., Alves P.C., de la Fuente J., Correia-Neves M., Boschiroli M.-L., Delahay R., Gortázar C. Quantification of the Animal Tuberculosis Multi-Host Community Offers Insights for Control. Pathogens (Basel, Switzerland) 2020;9:421. doi: 10.3390/pathogens9060421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichewo P.R., Kelen C.V., Thys S., Michel A.L. Risk practices for bovine tuberculosis transmission to cattle and livestock farming communities living at wildlife-livestock-human interface in northern KwaZulu Natal, South Africa. PLOS Neglected Tropical Diseases. 2020;14 doi: 10.1371/journal.pntd.0007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Becher P., Bukh J., Gould E.A., Meyers G., Monath T., Muerhoff S., Pletnev A., Rico-Hesse R., Smith D.B., Stapleton J.T., Ictv Report Consortium null ICTV Virus Taxonomy Profile: Flaviviridae. The Journal of General Virology. 2017;98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley Evans T., Shi Z., Boots M., Liu W., Olival K.J., Xiao X., Vandewoude S., Brown H., Chen J.-L., Civitello D.J., Escobar L., Grohn Y., Li H., Lips K., Liu Q., Lu J., Martínez-López B., Shi J., Shi X., Xu B., Yuan L., Zhu G., Getz W.M. Synergistic China-US Ecological Research is Essential for Global Emerging Infectious Disease Preparedness. EcoHealth. 2020;17:160–173. doi: 10.1007/s10393-020-01471-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolow S.H., Nova N., Pepin K.M., Peel A.J., Pulliam J.R.C., Manlove K., Cross P.C., Becker D.J., Plowright R.K., McCallum H., De Leo G.A. Ecological interventions to prevent and manage zoonotic pathogen spillover. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2019;374:20180342. doi: 10.1098/rstb.2018.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorvillo T.E., Rodriguez S.E., Hudson P., Carey M., Rodriguez L.L., Spiropoulou C.F., Bird B.H., Spengler J.R., Bente D.A. Towards a Sustainable One Health Approach to Crimean-Congo Hemorrhagic Fever Prevention: Focus Areas and Gaps in Knowledge. Tropical Medicine and Infectious Disease. 2020;5:113. doi: 10.3390/tropicalmed5030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton J., Tagliaro E., Weaver J., Sherman D.M., Carron M., Di Giacinto A., Sharandak V., Caya F. Performance of Veterinary Services Pathway evolution and One Health aspects. Revue Scientifique et Technique. 2019;38:291–302. doi: 10.20506/rst.38.1.2961. [DOI] [PubMed] [Google Scholar]
- Sudeep A.B., Bondre V.P., Mavale M.S., Ghodke Y.S., George R.P., Aher R.V., Gokhale M.D. Preliminary findings on Bagaza virus (Flavivirus: Flaviviridae) growth kinetics, transmission potential & transovarial transmission in three species of mosquitoes. The Indian Journal of Medical Research. 2013;138:257–261. [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., Kamei T., Honda M., Ninomiya D., Sakai T., Senba T., Kaneyuki S., Sakaguchi S., Satoh A., Hosokawa T., Kawabe Y., Kurihara S., Izumikawa K., Kohno S., Azuma T., Suemori K., Yasukawa M., Mizutani T., Omatsu T., Katayama Y., Miyahara M., Ijuin M., Doi K., Okuda M., Umeki K., Saito T., Fukushima K., Nakajima K., Yoshikawa T., Tani H., Fukushi S., Fukuma A., Ogata M., Shimojima M., Nakajima N., Nagata N., Katano H., Fukumoto H., Sato Y., Hasegawa H., Yamagishi T., Oishi K., Kurane I., Morikawa S., Saijo M. The First Identification and Retrospective Study of Severe Fever With Thrombocytopenia Syndrome in Japan. The Journal of Infectious Diseases. 2014;209:816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philosophical Transactions of the Royal Society B: Biological Sciences. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault P.A., Watkinson R.E., Moreira-Soto A., Drexler J.F., Lee B. Zoonotic Potential of Emerging Paramyxoviruses: Knowns and Unknowns. Advances in Virus Research. 2017;98:1–55. doi: 10.1016/bs.aivir.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry E., Meurens F., Muylkens B., McVoy M., Gogev S., Thiry J., Vanderplasschen A., Epstein A., Keil G., Schynts F. Recombination in alphaherpesviruses. Reviews in Medical Virology. 2005;15:89–103. doi: 10.1002/rmv.451. [DOI] [PubMed] [Google Scholar]
- Thiry E., Muylkens B., Meurens F., Gogev S., Thiry J., Vanderplasschen A., Schynts F. Recombination in the alphaherpesvirus bovine herpesvirus 1. Veterinary Microbiology. 2006;113:171–177. doi: 10.1016/j.vetmic.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Triguero-Ocaña R., Laguna E., Jiménez-Ruiz S., Fernández-López J., García-Bocanegra I., Barasona J.Á., Risalde M.Á., Montoro V., Vicente J., Acevedo P. The wildlife-livestock interface on extensive free-ranging pig farms in central Spain during the “montanera” period. Transboundary and Emerging Diseases. 2020 doi: 10.1111/tbed.13854. (in press) [DOI] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., Yu Y., Eaton B.T., Xuan H., Wang L.-F. Antibodies to SARS coronavirus in civets. Emerging Infectious Diseases. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulsiani S., Graham G., Moore P., Jansen C., Van Den Hurk A., Moore F., Simmons R., Craig S. Emerging tropical diseases in Australia. Part 5. Hendra virus. Annals of Tropical Medicine and Parasitology. 2011;105:1–11. doi: 10.1179/136485911X12899838413547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle L., Solomon T. Japanese encephalitis - the prospects for new treatments. Nature Reviews. Neurology. 2018;14:298–313. doi: 10.1038/nrneurol.2018.30. [DOI] [PubMed] [Google Scholar]
- van der Meulen K.M., Pensaert M.B., Nauwynck H.J. West Nile virus in the vertebrate world. Archives of Virology. 2005;150:637–657. doi: 10.1007/s00705-004-0463-z. [DOI] [PubMed] [Google Scholar]
- van Herten J., Bovenkerk B., Verweij M. One Health as a moral dilemma: Towards a socially responsible zoonotic disease control. Zoonoses and Public Health. 2019;66:26–34. doi: 10.1111/zph.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Castro L., Lara-Vergara J., Ortega N., Salinas J., Colom-Cadena A., Lavín S., Tizzani P., Velarde R., Serrano E., Mentaberre G. Endemic caseous lymphadenitis in a wild Caprinae population. The Veterinary Record. 2017;180:405. doi: 10.1136/vr.103925. [DOI] [PubMed] [Google Scholar]
- Venegas E.A., Aguilar P.V., Cruz C., Guevara C., Kochel T.J., Vargas J., Halsey E.S. Ilheus virus infection in human, Bolivia. Emerging Infectious Diseases. 2012;18:516–518. doi: 10.3201/eid1803.111486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veysset P., Lherm M., Bébin D., Roulenc M. Mixed crop–livestock farming systems: a sustainable way to produce beef? Commercial farms results, questions and perspectives. Animal. 2014;8:1218–1228. doi: 10.1017/S1751731114000378. [DOI] [PubMed] [Google Scholar]
- Vicente J., Höfle U., Garrido J.M., Fernández-De-Mera I.G., Juste R., Barral M., Gortazar C. Wild boar and red deer display high prevalences of tuberculosis-like lesions in Spain. Veterinary Research. 2006;37:107–119. doi: 10.1051/vetres:2005044. [DOI] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.-M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. Journal of Virology. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M.G., Sawleshwarkar S., Hossain S., Mor S.M. Whence the next pandemic? The intersecting global geography of the animal-human interface, poor health systems and air transit centrality reveals conduits for high-impact spillover. One Health. 2020;11 doi: 10.1016/j.onehlt.2020.100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F., Anderson D.E. Viruses in bats and potential spillover to animals and humans. Current Opinion in Virology. 2019;34:79–89. doi: 10.1016/j.coviro.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Crameri G. Emerging zoonotic viral diseases. Revue scientifique et technique OIE. 2014;33:569–581. doi: 10.20506/rst.33.2.2311. [DOI] [PubMed] [Google Scholar]
- Watson J.E.M., Venter O., Lee J., Jones K.R., Robinson J.G., Possingham H.P., Allan J.R. Protect the last of the wild. Nature. 2018;563:27–30. doi: 10.1038/d41586-018-07183-6. [DOI] [PubMed] [Google Scholar]
- Weiss K.E., Haig D.A., Alexander R.A. Wesselsbron virus - a virus not previously described, associated with abortion in domestic animals. Onderstepoort Journal of Veterinary Research. 1956;27:183–195. [Google Scholar]
- Weyer J., Thomas J., Leman P.A., Grobbelaar A.A., Kemp A., Paweska J.T. Human cases of Wesselsbron disease, South Africa 2010–2011. Vector Borne and Zoonotic Diseases (Larchmont, NY) 2013;13:330–336. doi: 10.1089/vbz.2012.1181. [DOI] [PubMed] [Google Scholar]
- WHO, 2019. MERS Situation Update. Retrieved on 2 November 2020 from https://applications.emro.who.int/docs/EMROPub-MERS-SEP-2019-EN.pdf?ua=1&ua=1.
- WHO, 2004. Severe acute respiratory syndrome (SARS). Retrieved on 5 November 2020 from https://www.who.int/csr/don/archive/disease/severe_acute_respiratory_syndrome/en/.
- Wolfe M.S., Calisher C.H., McGuire K. Spondweni virus infection in a foreign resident of Upper Volta. Lancet (London, England) 1982;2:1306–1308. doi: 10.1016/s0140-6736(82)91511-2. [DOI] [PubMed] [Google Scholar]
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.-Y. Coronavirus diversity, phylogeny and interspecies jumping. Experimental Biology and Medicine (Maywood, NJ) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E.J., Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerging Infectious Diseases. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, Food and Agriculture Organization of the United Nations and World Organisation for Animal Health, 2019. Taking a multisectoral, one health approach: a tripartite guide to addressing zoonotic diseases in countries. Retrieved on 16 October 2020 from https://www.oie.int/fileadmin/Home/eng/Media_Center/docs/EN_TripartiteZoonosesGuide_webversion.pdf.
- Wright D., Kortekaas J., Bowden T.A., Warimwe G.M. Rift Valley fever: biology and epidemiology. Journal of General Virology. 2019;100:1187–1199. doi: 10.1099/jgv.0.001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.F., Neumann G., Kawaoka Y. Orthomyxoviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1146–1243. [Google Scholar]
- Wu X., Lu Y., Zhou S., Chen L., Xu B. Impact of climate change on human infectious diseases: Empirical evidence and human adaptation. Environment International. 2016;86:14–23. doi: 10.1016/j.envint.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Xie J., Trus I., Oh D., Kvisgaard L.K., Rappe J.C.F., Ruggli N., Vanderheijden N., Larsen L.E., Lefèvre F., Nauwynck H.J. A Triple Amino Acid Substitution at Position 88/94/95 in Glycoprotein GP2a of Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV1) Is Responsible for Adaptation to MARC-145 Cells. Viruses. 2019;11:36. doi: 10.3390/v11010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X., Guan X., Liu L., Xu J., Li G., Zhan J., Liu G., Jiang X., Shen X., Jiang Y., Wu Y., Zhang H., Huang J., Ding F., Sha S., Liu M., Zhan F. A Case-control Study of Risk Sources for Severe Fever with Thrombocytopenia Syndrome in Hubei Province, China. International Journal of Infectious Diseases. 2017;55:86–91. doi: 10.1016/j.ijid.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Yaiw K.C., Bingham J., Crameri G., Mungall B., Hyatt A., Yu M., Eaton B., Shamala D., Wang L.-F., Thong Wong K. Tioman virus, a paramyxovirus of bat origin, causes mild disease in pigs and has a predilection for lymphoid tissues. Journal of Virology. 2008;82:565–568. doi: 10.1128/JVI.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon L., Duff J.P., Ågren E.O., Erdélyi K., Ferroglio E., Godfroid J., Hars J., Hestvik G., Horton D., Kuiken T., Lavazza A., Markowska-Daniel I., Martel A., Neimanis A., Pasmans F., Price S.J., Ruiz-Fons F., Ryser-Degiorgis M.-P., Widén F., Gavier-Widén D. Recent changes in infectious diseases in European wildlife. Journal of Wildlife Diseases. 2019;55:3–43. doi: 10.7589/2017-07-172. [DOI] [PubMed] [Google Scholar]
- Yu X.-J., Liang M.-F., Zhang S.-Y., Liu Y., Li J.-D., Sun Y.-L., Zhang L., Zhang Q.-F., Popov V.L., Li C., Qu J., Li Q., Zhang Y.-P., Hai R., Wu W., Wang Q., Zhan F.-X., Wang X.-J., Kan B., Wang S.-W., Wan K.-L., Jing H.-Q., Lu J.-X., Yin W.-W., Zhou H., Guan X.-H., Liu J.-F., Bi Z.-Q., Liu G.-H., Ren J., Wang H., Zhao Z., Song J.-D., He J.-R., Wan T., Zhang J.-S., Fu X.-P., Sun L.-N., Dong X.-P., Feng Z.-J., Yang W.-Z., Hong T., Zhang Y., Walker D.H., Wang Y., Li D.-X. Fever with Thrombocytopenia Associated with a Novel Bunyavirus in China. The New England Journal of Medicine. 2011;364:1523. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yan Y., Shi M., Liu H.-Z., Zhang H.-L., Yang Y.-B., Huang X.-Y., Gauger P.C., Zhang J., Zhang Y.-H., Tong G.-Z., Tian Z.-J., Chen J.-J., Cai X.-H., Liu D., Li G., An T.-Q. Phylogenetics, Genomic Recombination, and NSP2 Polymorphic Patterns of Porcine Reproductive and Respiratory Syndrome Virus in China and the United States in 2014–2018. Journal of Virology. 2020;94:e01813–e1819. doi: 10.1128/JVI.01813-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.-M., Park S.-J., Kim Y.-I., Park S.-W., Yu M.-A., Kwon H.-I., Kim E.-H., Yu K.-M., Jeong H.W., Ryou J., Lee W.-J., Jee Y., Lee J.-Y., Choi Y.K. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (SFTSV) in South Korea. JCI Insight. 2020;5 doi: 10.1172/jci.insight.129531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R. Global climate change and the emergence/re-emergence of infectious diseases. International Journal of Medical Microbiology Supplements. 2004;293:16–26. doi: 10.1016/s1433-1128(04)80005-6. [DOI] [PubMed] [Google Scholar]
- Zhang H., Li H., Wang W., Wang Y., Han G.-Z., Chen H., Wang X. A unique feature of swine ANP32A provides susceptibility to avian influenza virus infection in pigs. PLOS Pathogens. 2020;16 doi: 10.1371/journal.ppat.1008330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Current Biology. 2020;30:2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsstag J., Crump L., Schelling E., Hattendorf J., Maidane Y.O., Ali K.O., Muhummed A., Umer A.A., Aliyi F., Nooh F., Abdikadir M.I., Ali S.M., Hartinger S., Mäusezahl D., de White M.B.G., Cordon-Rosales C., Castillo D.A., McCracken J., Abakar F., Cercamondi C., Emmenegger S., Maier E., Karanja S., Bolon I., de Castañeda R.R., Bonfoh B., Tschopp R., Probst-Hensch N., Cissé G. Vol. 365. 2018. Climate change and One Health; p. fny085. (FEMS Microbiology Letters). [DOI] [PMC free article] [PubMed] [Google Scholar]