Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer-related death in the United States. Despite the high prevalence of Kras mutations in pancreatic cancer patients, murine models expressing the oncogenic mutant Kras (Krasmut) in mature pancreatic cells develop PDAC at a low frequency. Independent of cell of origin, a second genetic hit (loss of tumor suppressor TP53 or PTEN) is important for development of PDAC in mice. We hypothesized ectopic expression and elevated levels of oncogenic mutant Kras would promote PanIN arising in pancreatic ducts. To test our hypothesis, the significance of elevating levels of K-Ras and Ras activity has been explored by expression of a CAG driven LGSL-KrasG12V allele (cKras) in pancreatic ducts, which promotes ectopic Kras expression. We predicted expression of cKras in pancreatic ducts would generate neoplasia and PDAC. To test our hypothesis, we employed tamoxifen dependent CreERT2 mediated recombination. Hnf1b:CreERT2;KrasG12V (cKrasHnf1b/+) mice received 1 (Low), 5 (Mod) or 10 (High) mg per 20 g body weight to recombine cKras in low (cKrasLow), moderate (cKrasMod), and high (cKrasHigh) percentages of pancreatic ducts. Our histologic analysis revealed poorly differentiated aggressive tumors in cKrasHigh mice. cKrasMod mice had grades of Pancreatic Intraepithelial Neoplasia (PanIN), recapitulating early and advanced PanIN observed in human PDAC. Proteomics analysis revealed significant differences in PTEN/AKT and MAPK pathways between wild type, cKrasLow, cKrasMod, and cKrasHigh mice. In conclusion, in this study, we provide evidence that ectopic expression of oncogenic mutant K-Ras in pancreatic ducts generates early and late PanIN as well as PDAC. This Ras rheostat model provides evidence that AKT signaling is an important early driver of invasive ductal derived PDAC.
Subject terms: Cancer models, Mechanisms of disease
Summary
In this study, the authors provide evidence that ectopic expression of oncogenic mutant Kras in pancreatic ducts generates early and late (PanIN) and pancreatic ductal adenocarcinoma (PDAC) . They characterized this Ras rheostat model which reveals elevated Kras mutation frequency and loss of PTEN are important drivers of PanIN and invasive ductal derived PDAC.
Introduction
Cell of origin differences have been shown to promote molecular heterogeneity in a number of malignancies [1, 2]. Pancreatic ductal adenocarcinoma (PDAC) displays remarkable epithelial and stromal heterogeneity and both genetic and pathologic heterogeneity have been described indicating multiple cells of origin may contribute to this aggressive malignancy [3–13]. PDAC is aggressive and diagnosis before cancer cells have locally invaded or metastasized is uncommon. Understanding the biology of how exocrine pancreatic cells transform to cancer is important for development of targeted therapeutic approaches for prevention of aggressive disease. PDAC is preceded by precursor lesions postulated to arise from pancreatic exocrine epithelium. Two major precursor lesions include Pancreatic Intraepithelial Neoplasia (PanIN) and Intraductal Papillary Mucinous Neoplasia (IPMN). PanIN are most frequently observed in resected PDAC and are thought to precede development of invasive adenocarcinoma [14, 15]. A well-established autochthonous mouse model has shown PanIN arise in the setting of physiologic expression of KrasG12D in Pdx1 (or Ptf1a/p48) expressing multipotent pancreatic progenitors. In this model, at 9 months of age, mice display the full histological spectrum of early and advanced PanIN lesions [16]. In contrast, adult pancreatic exocrine cells are refractory to oncogenic Krasmut induced invasive carcinoma when KrasG12D is expressed under endogenous promoter elements unless there is induced inflammation or concomitant loss of a tumor suppressor gene [17–22].
Somatic mutation in codon 12 of KRAS is the most common point mutation in PDAC [23]. Although KRAS is mutated in more than 93% of sequenced human PDAC and is described as the initiating genetic event for cancer formation, studies using genetically engineered mouse models reveal that physiologic expression of oncogenic Krasmut alone can promote PanIN, but it is rarely sufficient for tumor development in pancreatic exocrine cells [4, 17, 18, 24–27]. Mouse models have shown oncogenic Krasmut can promote senescence in healthy cells, which is overcome by mutations in or genetic deletion of tumor suppressor genes [28–32]. These models are informative regarding development of oncogenic Krasmut driven malignancies as KRAS`mutations occur, even in the absence of neoplastic pancreata [33].
In this study, our goal was to determine if expression of cKras would promote IPMN, PanIN, or PDAC in ductal pancreatic epithelium. We wanted to test the distinct pathologies this allele could promote, as pancreatic ducts have been thought to be the cellular origin of PDAC due to the predominant ductal and glandular histology of PDAC[14, 34]. Furthermore, recent analysis of somatic variants in human PDAC and precursor lesions has shown a multi-step progression in which advanced PanIN are a single neoplasm that colonize the ductal system [35]. Despite this presumption, early studies using genetic mouse models indicated acinar cells are a possible cellular origin [36–38]. CreER mediated expression of oncogenic Krasmut in mature, adult acinar cells led to PanIN lesions that transformed to PDAC when oncogenic Krasmut was expressed in combination with chronic inflammation, high fat diet, or mutant tumor suppressor genes [17, 21, 39, 40]. Recently, pancreatic duct ligation was used to model chronic obstructive pancreatitis in the context of cell of origin. In this model, expression of oncogenic Krasmut and pancreatic duct ligation resulted in PDAC in ductal, but not acinar cells [41]. In addition, recent studies have shown that mutations in or genetic loss of Trp53 or PTEN results in ductal or acinar derived PDAC [4, 17, 22, 41, 42]. These models are important as human PDAC subtypes, which may arise from divergent cells of origin, have been characterized and indicate differences in prognosis based on histologic, genetic, and RNA sequencing data [6, 8, 23, 43, 44].
The overall hypothesis guiding this work is mouse pancreatic ductal cells, which are otherwise refractory to transformation under conditions of physiologic oncogenic Krasmut expression, will become morphologically distinct PanIN or IPMN when expressing cKras. We predict if cKras is able to generate neoplastic and invasive PDAC that this model will provide important context and a new model to study molecular requirements for ductal derived PDAC. As previously reported [19], cKras is engineered following a ubiquitously expressed CAG promoter blocked by proximal insertion of a loxP-green fluorescent protein (GFP)-STOP-loxP cassette, which allows for lineage tracing. In this model, intraperitoneal injection of tamoxifen promotes CreER dependent recombination of loxP sites flanking a STOP cassette and GFP allele; which results in expression of cKras allele and loss of GFP [19, 45]. As we have previously worked with the Hnf1b:CreER allele, in initial experiments, 6–8 week old mice were given 10 mg of tamoxifen. We observed very aggressive lethality in these mice and thus titrated our tamoxifen dose, which led to generation of this Ras rheostat model of ductal derived PanIN and PDAC. We predict this model is relevant to what has been shown in sequencing of human PanIN, wherein while >90% of sequenced PanIN have mutations in KRAS, quantification of mean KRAS mutations per PanIN significantly increases in progression from early to advanced PanIN, indicating increasing numbers of pancreatic ductal cells with oncogenic KRAS is a method by which cells elevate K-Ras oncogenic signaling pathways important for transformation [33].
Results
Tamoxifen dosage directly correlates with percentage of recombined pancreatic ducts
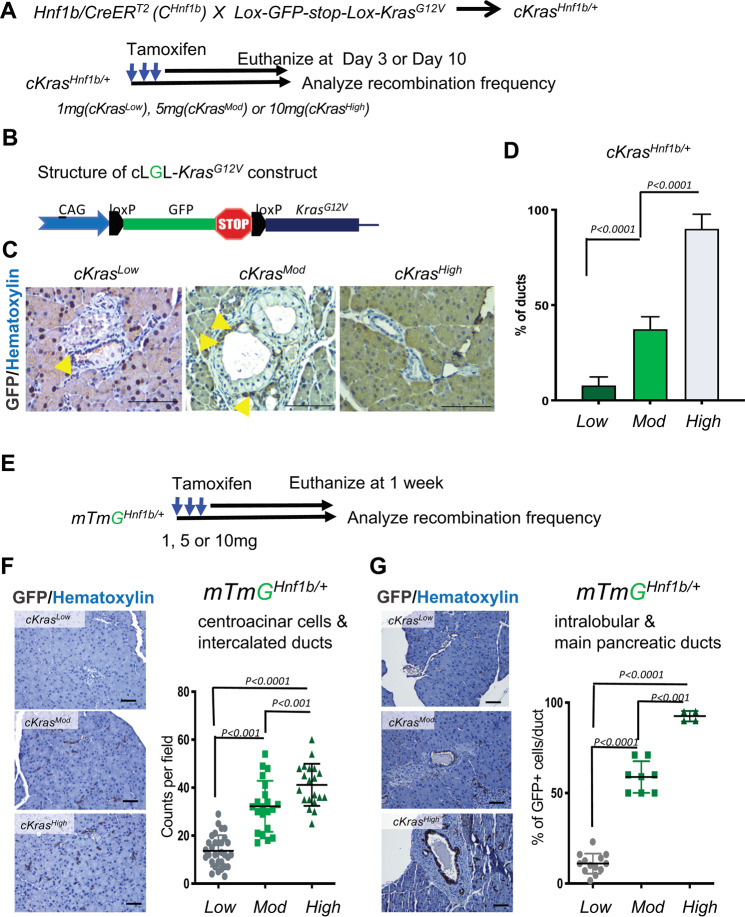
We examined tamoxifen dependent recombination of cKras allele. Figure 1a shows a schematic for generation of cKrasHnf1b/+ mice and Fig. 1b is a graphic of the inducible cKras allele. When allelic recombination of loxP sites occurs, recombined cells lose GFP. Thus, initial methodology used to study occurrence of recombination in pancreatic ducts was loss of GFP. Paraffin embedded sections from mice euthanized at both early and late time points were used for analysis. Immunohistochemistry (IHC) staining for GFP was used to detect the presence or absence of recombination. For all experiments, mice were administered tamoxifen at an age of 6–8 weeks. We observed a significant increase in recombination (loss of GFP) in ductal cells as a function of tamoxifen dose. Non-recombined GFP+ cells (Fig. 1c, yellow arrows) are present in cKrasLow and cKrasMod pancreatic sections. In cKrasHigh mice, tissue sections from early time points were evaluated as these mice rapidly developed invasive PDAC, which compromised our lineage tracing methods (Supplementary Fig. 1). We quantified recombined epithelium (number of GFP- ducts/total number of ducts) for all models, which revealed a significant increase in recombination comparing cKrasLow (7.8 ± 4.5%) to cKrasMod (37.3 ± 6.5%) and cKrasMod to cKrasHigh (90 ± 7.62%) (Fig. 1d). As a second method, we used the loxP-mTdTomato-loxP-mGFP (RosamTmG) [46] reporter allele crossed to Hnf1b:CreERT2 animals to study recombination (gain of GFP expression) in pancreatic ducts as a function of tamoxifen dose (mTmGHnf1b/+) (Fig. 1e). These mice were euthanized 1 week after they were given tamoxifen. We used immunohistochemistry and stained mTmGHnf1b/+Low, mTmGHnf1b/+Mod, and mTmGHnf1b/+High murine pancreatic sections with GFP and quantified recombined ducts (Fig. 1f, g). We observed higher numbers of GFP + ducts which indicated increased recombination in centroacinar and intercalated ducts in mTmGHnf1b/+High mice with an average count of 41/field compared to mTmGHnf1b/+Low (average count of 13/field) or mTmGHnf1b/+Mod (average count of 32/field) (Fig. 1f). mTmGHnf1b/+Mod mice also had significantly higher numbers of recombined centroacinar cells and intercalated ducts than mTmGHnf1b/+Low mice (Fig. 1f). When analyzing recombination in main pancreatic and intralobular ducts, we observed a significant increase in mTmGHnf1b/+Mod (58% ducts/field) and mTmGHnf1b/+High (92% of ducts/field) mice compared to mTmGHnf1b/+Low mice (11% ducts/field) (Fig. 1g).
Fig. 1. Tamoxifen dosage directly correlates with recombination frequency in pancreatic ducts.
a Schematic showing the experimental strategy to lineage trace Hnf1b+ duct cells in the pancreas of cKrasHnf1b/+ mice for analysis of tamoxifen mediated CreERT2 recombination. At an age of 6–8 weeks, mice received 1 mg (cKrasLow), 5 mg (cKrasMod), or 10 mg (cKrasHigh) of tamoxifen and were analyzed at both early and late timepoints. b Structure of the cLGL-KrasG12V ectopic expression transgene. cKras was engineered following a human CMV and chicken-β-actin chimeric promoter (CAG) and blocked by the proximal insertion of a loxP-green fluorescent protein (GFP)-Stop-loxP cassette (c) Lineage tracing shows that PDAC precursors (PanIN and micropapillary lesions) and associated PDAC arise in pancreatic ducts of mice. Recombination in ducts depends on tamoxifen dosage (yellow arrows show nonrecombined ducts in cKrasLow and cKrasMod at end point and cKrasHigh mice at day3 post tamoxifen). d Quantification of percentage of ductal cells recombined in cKrasLow, cKrasMod, and cKrasHigh mice. We observed a significant increase in recombination comparing cKrasLow (7.8 ± 4.5%) to cKrasMod (37.3 ± 6.5%) and cKrasMod to cKrasHigh (90 ± 7.62%). e Schematic showing the experimental strategy to lineage trace Hnf1b+ duct cells in the pancreas of mTmGHnf1b/+ for analyzing tamoxifen mediated Cre recombination. Mice received 1, 5, and 10 mg of tamoxifen and were analyzed at 1 week. f and g IHC staining for GFP and quantification of recombined centroacinar and intercalated ducts. GFP immunolabeling significantly increased in centroacinar and intercalated ducts in mTmGHnf1b/+ High mice with an average count of 41/field compared to mTmGHnf1b/+ Low (13/field) or mTmGHnf1b/+ Mod (32/field) mice. mTmGHnf1b/+ Mod mice had significantly higher numbers of recombined centroacinar cells and intercalated ducts than mTmGHnf1b/+ Low mice (P < 0.001). g When analyzing recombination in main pancreatic and intralobular ducts, we observed a significant increase in mTmGHnf1b/+ Mod (58% ducts/field) and mTmGHnf1b/+ High (92% of ducts/field) mice compared to mTmGHnf1b/+ Low mice (11% ducts/field). Statistical analysis was performed using a Two-way Anova in Prism GraphPad software.
cKras recombination in higher percentages of ductal cells directly affects survival
Endogenous expression of oncogenic Kras alone has a low penetrance of generating PanIN and PDAC in mature adult pancreatic ducts [17, 24]; thus, we examined if ectopic expression cKras would transform ductal cells. cKrasHigh mice aged 6–8 weeks were administered 10 mg of tamoxifen. (Fig. 2a) and within 10–14 days, we noted severe weight loss and lethality in 100% of injected animals (Fig. 2b, c). As such a drastic effect on survival was observed, we analyzed pancreatic head, body, and tail histology on paraffin embedded sections which revealed invasive PDAC throughout the entire pancreatic parenchyma (Supplementary Fig. 1 & 2).
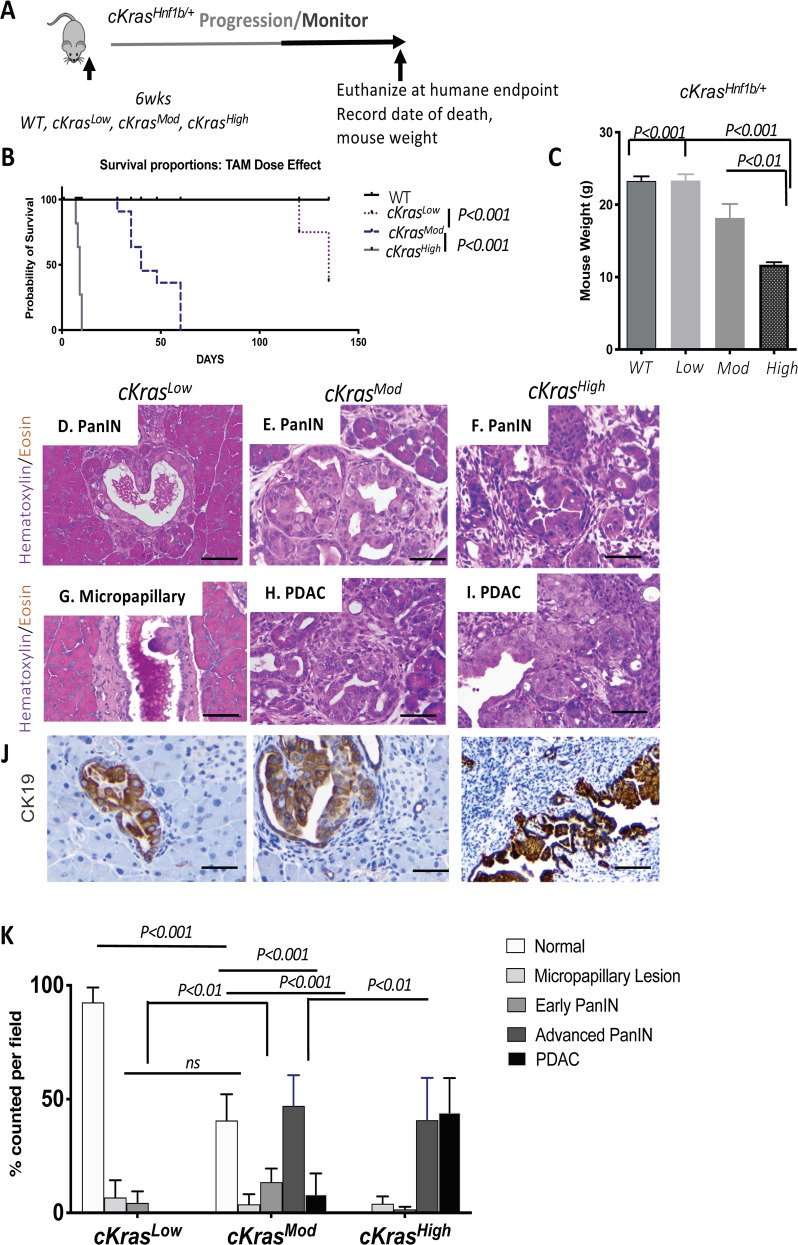
Fig. 2. Pathologic outcomes of increasing the percentage of pancreatic ductal cells expressing cKras.
a Schematic of experimental plan to study the effect of titrating tamoxifen given to mice to study recombination efficiency and effects on pancreatic histology. b Increased dosage of tamoxifen significantly decreases survival of cKrasHnf1b/+ mice. cKrasHigh mice survive an average of 10 days, cKrasMod mice survive an average of 42 days and 50% of cKrasLow mice are still alive 138 days after administered tamoxifen. The other 50% had to be euthanized. c cKrasHigh and cKrasMod have a significant reduction in body weight at time of death (P < 0.001) compared to littermate control weight and cKrasLow mice at time of death. Statistical analysis was evaluated using a student’s t test. d–i Representative H&E images and CK19 IHC of pancreas from cKrasLow, cKrasMod and cKrasHigh mice. d cKrasLow mice show limited early PanIN and (g) Micropapillary lesions. (e) cKrasMod mice have early and advanced PanIN and (h) PDAC whereas cKrasHigh mice have predominantly advanced PanIN (f) and PDAC(I). (j) CK19 labeling of ductal lesions in cKrasLow, cKrasMod and cKrasHigh mice. k Quantification of normal ducts, micropapillary lesions, early PanIN, advanced PanIN, and PDAC as a consequence of tamoxifen dosage. Lesions were counted and percentages were calculated using total ducts as the denominator. A two-way Anova using Prism Graphpad software evaluated statistical significance. An n = 8 mice per tamoxifen dosage was used for pathologic analysis. Scale bars are 50 µM.
We hypothesized varying levels of tamoxifen would be proportional to changes in histologic outcomes and survival. To test our hypothesis, cKrasLow and cKrasMod mice were injected at the age of 6–8 weeks (Fig. 2a). Mice were monitored for evidence of disease progression. cKrasMod mice survived on average 6 weeks and 50% of cKrasLow mice remained healthy and were euthanized at 16 weeks for histologic analysis (Fig. 2b). Upon necropsy and histologic examination of euthanized cKrasLow mice, we observed adenoma formation in the lungs (Supplementary Fig. 2). In the lungs of cKrasMod mice, we observed adenoma and adenocarcinoma and in cKrasHigh mice, we observed severe inflammation in interstitial spaces and alveolar lining but no adenoma or adenocarcinoma (Supplementary Fig. 2B). These results are similar to pathologies we recently published [47]. We measured mouse body weight at time of necropsy and noted significant weight loss at time of death in cKrasMod and cKrasHigh mice compared to cKrasLow mice and littermate controls that received no tamoxifen (Fig. 2c).
Levels of cKras directly modulate early and advanced PanIN and PDAC
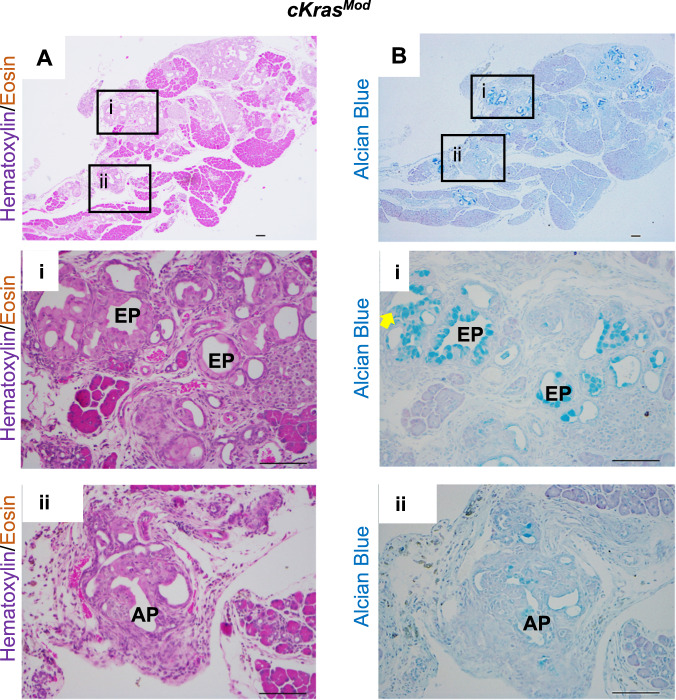
To characterize the histology of cKras mice, we used H&E staining, cytokeratin 19 (CK19) immunolabeling, and alcian blue to quantify the occurrence and abundance of early and advanced PanIN lesions (Figs. 2d and 3). For quantification, we counted the number of each type of ductal lesion and divided this number by the total number of ducts per field. For cKrasLow mice, 92.5 ± 6.5% of pancreas had normal ductal histology with evidence of proliferative ductal epithelium (Supplementary Fig. 3A and Fig. 4B), low abundance intra ductal micropapillary lesions (Fig. 2g, 5.6 ± 9.1%) and early PanIN (4.37 ± 0.5, Fig. 2d). In cKrasMod mice, we observed early and advanced PanIN and PDAC (Fig. 2e, h). We observed 38.6 ± 11.4% normal ducts and early PanIN occupied 13.5 ± 6% of pancreatic ductal tissue. Advanced PanIN occupied on average 41 ± 13.5% of ductal epithelium and PDAC was observed in 8.1 ± 9.23% of ducts. All cKrasHigh pancreata (15/15) had no remaining normal ducts and early PanIN were infrequently observed (3.25 ± 2.4%) as these mice had predominantly advanced PanIN (Figs 2f, 40.75 ± 18.59%) and PDAC (Fig. 2i, 44.1 ± 15.1%). To determine if cKrasMod mice had early or advanced PanIN, we stained with H&E and alcian blue, which confirmed mucinous PanIN (Fig. 3a, b). To confirm the absence of early PanIN in cKrasHigh mice, we analyzed alcian blue staining in pancreata of mice at earlier time points (days 5 and day 7 post tamoxifen injection), which revealed only advanced PanIN lesions (Supplementary Fig. 3A, B). As elevated Ras signaling has been shown to regulate senescence in ductal epithelium, we stained our sections for p21 and PCNA, which revealed no expression of p21 in ducts and abundant staining for PCNA (Supplementary Fig. 4). These data are interesting as we previously observed expression of p21 in pancreatic ducts expressing KrasG12D under regulation of endogenous promoter elements [17].
Fig. 3. cKrasMod expression in murine pancreata results in full spectrum of PanIN and PDAC.
a H&E images of early (i) and advanced (ii) PanIN lesions arising in pancreatic ducts of cKrasMod mice. Early PanIN (EP) have columnar epithelium and mucin and papillary outgrowths (ii) advanced PanIN (AP) occasionally have loss of basement membrane. b Serial section panel is Alcian blue staining of tissue shown in (a). Scale bars are 50 µM.
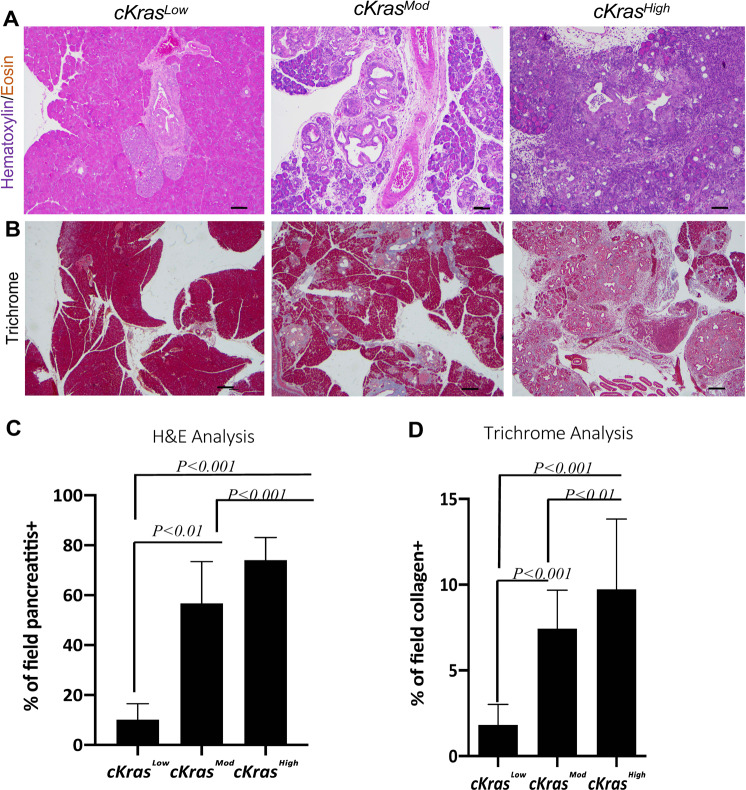
Histologic evaluation of cKrasMod and cKrasHigh pancreata revealed pancreatitis and stromal composition near areas of PanIN and PDAC arising in ductal epithelium (Fig. 4). To determine if significant differences in pancreatitis and stroma were evident, we calculated percentage of pancreatitis or collagen + fibrostroma in five mice per dosage in cKrasMod and cKrasHigh sections compared to cKrasLow sections (Fig. 4a, b). To accurately determine pancreatitis and stromal composition, we used ImageJ software and color deconvolution plugin to analyze percentage of pancreatitis (or collagen+) area divided by total area of each section (Supplementary Fig. 5 and Fig. 4C, D). Ductal neoplasia in cKrasMod and cKrasHigh mice was associated with significantly increased pancreatitis and collagen positive fibrostroma (Fig. 4c, d). In these sections, we also observed infiltration of CD68 + macrophage and CD45 + lymphocytes, indicating in this model, infiltrating immune cells are also a component of stroma (Supplementary Fig. 4).
Fig. 4. Pancreatitis and fibrostroma are elevated in cKrasMod and cKrasHigh pancreata.
a Representative H&E images of cKrasLow, cKrasMod, and cKrasHigh mice showing increasing pancreatitis surrounding the ductal lesions with increased Kras expression. Increased lymphocyte infiltration can be noted. b Trichrome staining of pancreas from cKrasLow, cKrasMod, and cKrasHigh showing increased fibrillar collagen. c and d quantification of pancreatitis and fibrillar collagen per field in cKrasLow, cKrasMod, and cKrasHigh mice. 5 fields per slide were selected for quantification and 5 mice were analyzed per tamoxifen dose. Analysis of pancreatitis area and collagen+ stroma were completed using imageJ software with color deconvolution plugin (Supplementary Fig. 5). Statistical analysis was done using a one-way Anova and Prism Graphpad software.
Increased recombination frequency correlates with total Ras in pancreatic tissue
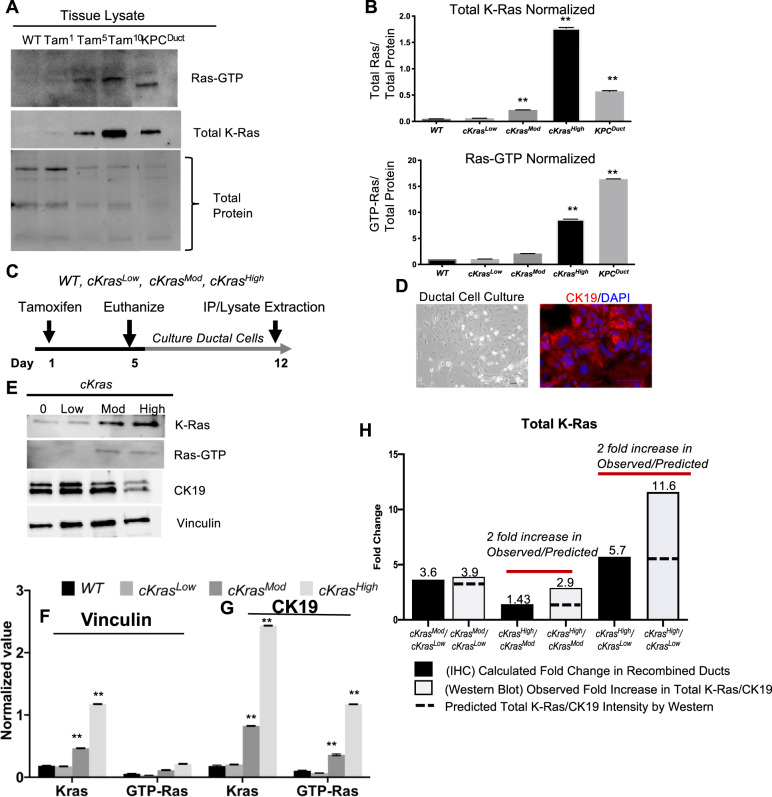
As we observed such drastic differences in histology when comparing cKrasLow, cKrasMod, and cKrasHigh mice, we wanted to examine levels of total K-Ras and Ras-GTP in this ductal cell of origin PanIN and PDAC system. We performed a Ras pull down assay to measure levels of Ras activity in pancreata from cKrasLow, cKrasMod, and cKrasHigh mice and directly compared the levels of Ras-GTP to a previous ductal cell of origin model we published by expressing KrasG12D and Trp53R175H (KPCDuct) [17]. Western blot analysis and normalization to total protein in these pancreata indicated significantly higher levels of Total K-Ras and Ras-GTP in cKrasMod, cKrasHigh, and KPCDuct tissue compared to cKrasLow (Fig. 5a, b). These data are similar to previous publications showing loss of Trp53 or gain of function mutations in Trp53 can elevate Ras-GTP and promote aggressive PDAC in mice [19, 48].
Fig. 5. Tamoxifen dose is directly related to total Ras levels and signaling.
a Lysates from wild-type pancreas (Lane1), cKrasLow, cKrasMod, and cKrasHigh murine pancreas (lanes 2,3,4) were affinity precipitated with Raf RBD agarose and subjected to immunoblot analysis with anti-Ras antibody. b Quantification of Total Ras protein and RAS-GTP to total protein confirmed a significant increase in Ras expression and GTP bound Ras with increased tamoxifen dosage. c Experimental plan to establish ductal cell culture from cKrasHnf1b/+ mice. d Brightfield image of primary pancreatic duct culture and adjacent panel showing CK19 IF labeling confirming ducts. e Western blot analysis of lysates from primary ducts cultured from WT, cKrasLow, cKrasMod, and cKrasHigh murine pancreas four days after tamoxifen administration. f Normalization of western blot image. Normalization of protein expression was done in triplicate using vinculin as the control for ductal cell culture expression analysis. g Normalization of total K-Ras and GTP bound Ras using CK19 as epithelial marker control for duct cells indicates increased K-Ras expression per cells with increased tamoxifen dose. h Comparison of K-Ras expression predicted vs calculated based on IHC and western analysis indicating increased K-Ras expression per cell with increased tamoxifen dosage. Statistical analysis was done using a Two-way Anova in Prism GraphPad.
In vitro duct cultures reveal cKrasMod and cKrasHigh ducts have significantly higher GTP bound Ras
To further study effects of increased cKras on cell autonomous signaling, we isolated and cultured ductal cells from wild type (WT), cKrasLow, cKrasMod, and cKrasHigh mice four days after mice were given tamoxifen (Fig. 5c, d). Cell lysates were prepared after 1 week in culture and subjected to Ras-GTP pull down. Western analysis confirmed a significant increase in Ras-GTP (Ras activity) in cKrasMod and cKrasHigh mice similar to what is observed with whole tissue lysates from these mice pancreata (Fig. 5e, f). We also analyzed total K-Ras levels and observed a significant increase in total K-Ras levels in cKrasHigh versus cKrasMod mice. While we had shown increased Ras-GTP as a function of increased percentage of recombined cells, we wanted to determine if this allele also increased the abundance of K-Ras per cell in our model. We probed our cell cultures with CK19 and normalized Ras-GTP and K-Ras to CK19 (Fig. 5g). This normalization indicated a highly significant increase in K-Ras and Ras-GTP in cKrasMod and cKrasHigh cultured ducts. To determine if the fold increase in Total K-Ras was due to increased recombination or increased total K-Ras per cell, we calculated the observed fold increase in recombined ducts analyzed in Fig. 1 using IHC (Fig. 5h). Using data from Fig. 1, we calculated an overall average of 3.6-fold increase in recombined ducts in cKrasMod/cKrasLow pancreata. When comparing cKrasHigh/cKrasMod, we observed a 1.43-fold change in overall recombined ducts (Fig. 5h). When comparing cKrasHigh/cKrasLow, we observed a 5.7-fold change in overall recombined ducts (Fig. 5h). The fold increases by IHC were used to determine a predicted fold increase by western blot. We then calculated the fold increase observed by western blot using normalized values from total K-Ras/CK19. Using this method, we calculated a 3.9-fold increase in observed fold increase in cKrasMod/cKrasLow ducts. When comparing cKrasHigh/cKrasMod, we calculated a 2.9-fold increase in total K-Ras (Fig. 5h) and cKrasHigh/cKrasLow quantification indicated an 11.6-fold increase in total K-Ras. We compared the observed fold increase by western blot to the predicted fold increase due to recombination analyzed using IHC. When comparing cKrasMod/cKrasLow, the observed and predicted fold increase were not significant (3.6 versus 3.9). However, when we compared the predicted and observed fold increase in cKrasHigh/cKrasMod (2.9 versus 1.43) and cKrasHigh/cKrasLow (11.6 versus 5.7), we observed two-fold increases in observed/predicted fold increase indicating expression of cKras increases the abundance of Total K-Ras per cell (Fig. 5h) in cKrasHigh pancreatic ducts.
Functional proteomics reveal important pathways expressed in ductal derived PanIN and PDAC
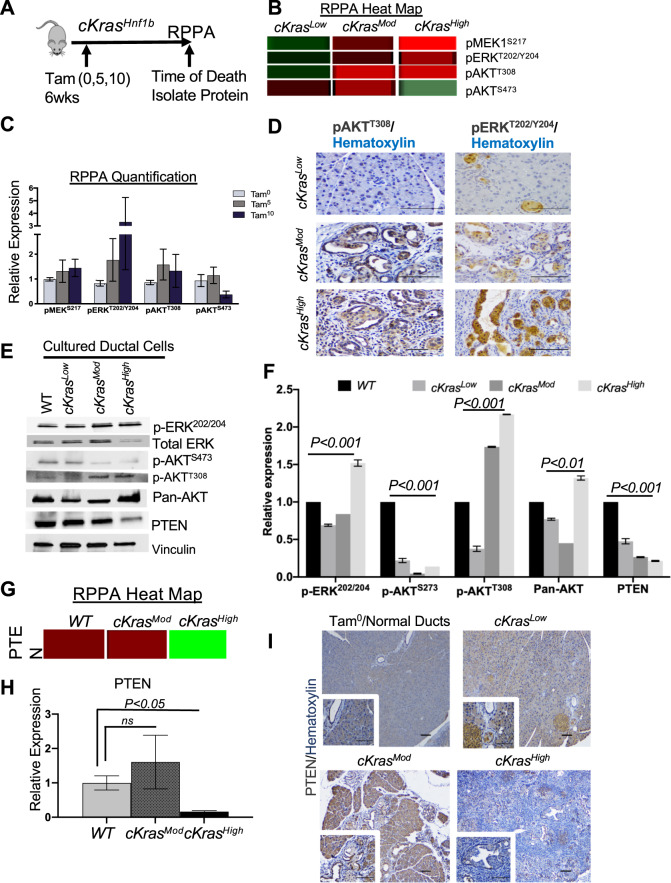
We employed a large-scale functional proteomics platform using Reverse Phase Protein Array (RPPA) at MDAnderson Cancer Center. This functional proteomics approach characterizes protein expression and post translational modifications for over 400 proteins. Three samples each of WT, cKrasMod, and cKrasHigh pancreata were analyzed (Fig. 6). Figure 6a, b and c shows a representative diagram of the experiment (Fig. 6a) and heat map analyses of proteins with high expression (red) versus low expression (green) in analyzed representative samples. Proteomic analysis yielded data regarding elevated MAPK and PI3K/PTEN/AKT pathways in transforming ducts, as pMEKS127, pERKT202/Y204, and pAKTT308 were elevated in cKrasMod and cKrasHigh pancreata relative to WT animals (Fig. 6b, c). Using our in vitro model, we analyzed downstream signaling in WT, cKrasLow, cKrasMod, and cKrasHigh cultured ducts. Similar to what we observed in RPPA samples, we observed elevated pERK202/204 and elevated pAKTT308 but observed a decrease in pAKTS473 in cKrasMod and cKrasHigh ducts compared to cKrasLow cultured ducts (Fig. 6e, f).
Fig. 6. Proteomic analysis of ductal PDAC reveals loss of PTEN.
a Schematic and (b) RPPA heat map data showing increased pMEKS127, pERKT202/Y204, pAKTT308, and decreased expression of pAKTS273 in lysates from WT, cKrasMod, and cKrasHigh pancreata. c Quantification of RPPA relative expression data. d IHC analysis of pAKTT308 and pERKT202/Y204 in WT, cKrasMod and cKrasHigh pancreata confirms RPPA data that pAKTT308 and pERKT202/Y204 are significantly increased in cKrasMod and cKrasHigh pancreata relative to cKrasLow. e Western blot analysis of downstream signaling pathways in WT, cKrasLow, cKrasMod, and cKrasHigh cultured pancreatic ducts. f Quantification of western blot data normalized to vinculin. g Heatmap and (h) quantification for PTEN relative expression in WT, cKrasMod, and cKrasHigh mice. (i) IHC staining for PTEN of pancreata from cKrasLow, cKrasMod, and cKrasHigh mice (Zoomed in ×40 magnification images in inset). PTEN expression in normal ducts can be seen in cKrasLow, cKrasMod, and cKrasHigh mice. Scale bars are 50 µM.
Our RPPA data revealed a highly significant decrease in PTEN expression in cKrasHigh tissue compared to WT (Fig. 6f, g and h). As PTEN has previously been shown to be highly expressed in pancreatic ducts and centroacinar cells and inhibits IPMN (49, 50), we probed for PTEN expression, which confirmed high levels of PTEN in cKras0 and cKrasLow ducts; however, we observed a significant reduction in PTEN in cKrasMod and cKrasHigh PanIN and PDAC (Fig. 6i). These results were interesting as PTEN has been previously shown to inhibit pAKTT308, indicating cell autonomous signaling mechanisms are present in cultured ducts with differences in levels of total K-Ras [49, 50]. To determine if PTEN reduction was an early event in ductal cell transformation, we probed for PTEN in our duct culture system which confirmed a significant reduction in PTEN in cKrasMod and cKrasHigh cultured ducts (Fig. 6e–g).
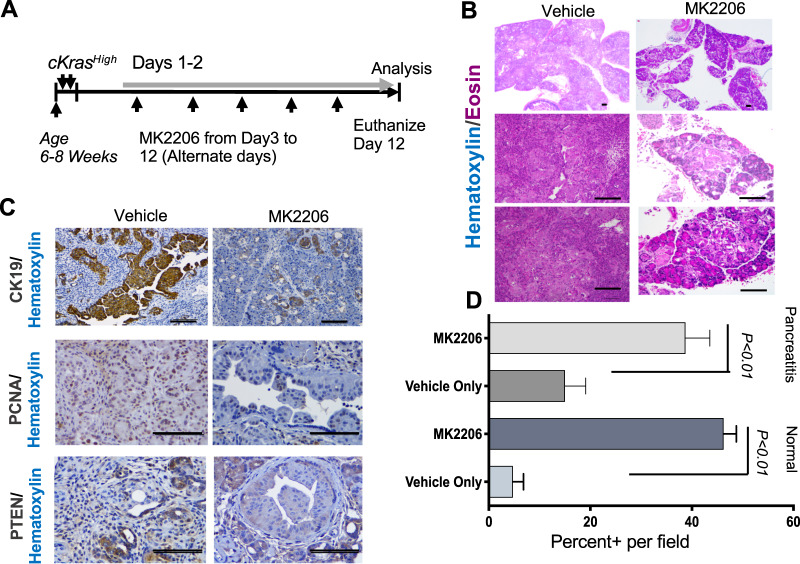
AKT inhibitor MK-2206 reduces ductal derived PDAC in cKrasHigh mice
The role of PI3K/AKT signaling as a potent driver of PDAC has been studied in transgenic mice [50]. As we observed reduced PTEN and elevated pAKTT308 in cKrasMod and cKrasHigh tissue and primary ductal cultures, we sought to determine if signaling through pAKTT308 is required for aggressive ductal derived PDAC. Using MK2206 AKT inhibitor, we treated cKrasHigh mice every other day and euthanized at day 12 to compare with vehicle treated mice (Fig. 7a). Histologic analysis revealed a significant increase in normal pancreatic area and reduced expression of CK19 in MK2206 treated mice compared to vehicle treated mice (Fig. 7b–d). While we did observe a significant reduction in PDAC (Fig. 7d), we observed sustained pancreatitis in MK2206 treated mice (Fig. 7d). In areas where we observed advanced PanIN, we observed reduced expression of PCNA (Fig. 7c). Notably, resistant neoplastic epithelium had sustained PTEN loss, indicating these lesions were resistant to AKT inhibition and loss of PTEN is important for cuboidal ductal cell to PanIN and PDAC transition. (Fig. 7c).
Fig. 7. Kras signaling through AKT is important for ductal derived PDAC.
a Schematic of experimental setup to study the role of AKT in cKrasHigh mice. b Representative H&E sections (×4 and ×20 magnification) show a significant reduction in PDAC in cKrasHigh pancreata from mice treated with MK2206 relative to vehicle control (n = 5 mice analyzed per treatment). c CK19, PCNA, and PTEN IHC in vehicle control versus MK2206 treated mice. We observe a reduction in IHC staining for CK19 and PCNA. In resistant advanced PanIN, we observe sustained loss of PTEN. d Quantification of normal and pancreatitis percent positive per ×20 field quantified (n = 5 per mouse). We observe a significant increase in normal pancreas in MK2206 treated mice. While we do not observe PDAC in MK2206 mice, we do observe significant pancreatitis, indicating sustained inflammation even in the setting of MK2206 treatment. Scale bars are 50 μM. Statistical analysis was performed using a student’s test, Prism GraphPad software.
Discussion
PDAC remains a highly lethal malignancy. Studying cellular origins of cancer and understanding mechanisms and pathways that drive aggressive tumor formation are important for devising targeted early and late therapeutics for patients. In the context of ductal adenocarcinoma of the pancreas, recent mouse models have shown that pancreatic ductal cells can serve as the cell of origin of PDAC when Kras is mutated and, from the onset of oncogenic Krasmut expression, Trp53, Fbw7, or Pten are genetically deleted, or a gain of function mutant of Trp53 is expressed, using Hnf1b:CreERT2, Sox9:CreER, or Ck19:CreER driver lines [4, 17, 22, 41, 42]. As adult pancreatic epithelial cells are refractory to invasive PDAC formation when expressing physiologic levels of Kras without loss of tumor suppressor gene and previous manuscripts have shown that cKras can produce PDAC from acinar cells [19, 51], we employed the cKras allele to study PanIN, IPMN, or and PDAC arising in adult pancreatic ductal cells.
In our genetic system, we have shown we can recombine mutant cKras as a function of tamoxifen dosage [40]. Recent sequencing data have shown in human PanIN that when PanIN1, −2, and −3 are sequenced, the percentage of mutant Kras positive cells significantly increases when comparing early and advanced PanIN [39]. In this context, we have now characterized a model highly relevant to what is observed in human PanIN, where we increase the percentage of recombined ducts as a consequence of tamoxifen dosage. We have used sophisticated lineage tracing to define the percentage of recombined ductal cells in a dose dependent manner, mimicking what is observed in human PanIN. Notably, in human PanIN, Trp53 mutations are not detected in PanIN-1 or −2 lesions; only in PanIN-3. In this new model, we observe PDAC arising in the setting of early and advanced PanIN in cKrasMod mice. cKrasMod displayed the entire spectrum of PanIN including early PanIN lesions with abundant mucin. Existing ductal cell of origin mouse models have reported PDAC formation primarily through advanced PanIN with limited early PanIN as well as atypical flat lesions (Table 1). In cKrasHigh mice, PDAC developed mainly through advanced PanIN. Neoplastic ducts also had invasive cells invading into apical duct lumens, which aggravated tumor progression.
Table 1.
Genetic engineered mouse models of PDAC arising in mature pancreatic duct cells.
| Gene promoter | Tumor suppressor loss/mutation | Pancreas histology | Reference |
|---|---|---|---|
| CK19CreERT; LSL-KrasG12D | No | Mostly normal pancreas with scattered PanIN lesions | Kevin C Ray, et al. PLoS One, 2011. |
| Hnf1bCreERT2; KrasG12D, TP53R172H/R172H | Yes | Development of invasive PDAC through High grade PanIN lesions | Bailey JM, et al., Oncogene, 2016. 35(32): p. 4282–8 |
| Sox9CreER; KrasLSL-12D; Trp53flox/flox | Yes | High Grade PanIN lesions and PDAC | Lee AYL, et al., Gut. 2018 |
| Ck19-CreER; Kras LSL-G12D/wt; Fbw7f/f | Yes | Development of invasive PDAC through non mucinous tubular lesions. | Ferreira RMM, et al., Cell Reports. 2017 |
| Sox9CreERT2; KrasG12D; Ptenflox/flox; R26RYFP | Yes | Pancreatobilliary type IPMN and invasive PDAC | Kopp JL, et al., Gastroenterology. 2018 |
| CK19CreERT2; LSL-KrasG12D; R26REYFP | |||
| Sox9CreERT2; LSL-KrasG12D; R26REYFP | |||
| with pancreatic duct ligation (chronic pancreatitis) | No | PDAC development through flat epithelial precursors. Few PanIN lesions observed. | Shi C, et al. CMGH, 2019. |
We also found that as a higher number of cells expressed mutant cKras there was an overall increase PanIN, PDAC, fibrostroma, and inflammation. In the context of ductal cells as origin of PDAC, a recent study presented PDAC arising in the setting of duct obstruction after pancreatic duct ligation which led to neoplasia and PDAC from duct cells expressing Krasmut from endogenous locus [41]. In our model, we show development of pancreatitis which increases as we enhance the abundance of intrapancreatic ductal neoplasia. It can be noted that, especially in our cKrasHigh model, significant duct obstruction may be present from advanced PanIN, which promotes widespread pancreatitis.
We report proteomics analysis performed on pancreatic tissue obtained from WT, cKrasMod, and cKrasHigh mice. RPPA analysis revealed a spectrum of differentially activated MAPK and PI3K. Similar to previous reports, we observed ductal epithelial cells lacked expression of p21 and were highly proliferative (Supplementary Fig. 4). In this model we see loss of tumor suppressor PTEN, a well-established tumor suppressor in human cancers. The role of PTEN in pancreatic ductal malignancy has been studied in recent years. Notably, loss of Pten and expression of Kras promotes an IPMN-PDAC phenotype in Sox9 expressing ductal epithelium [52]. In our model, we observed PanIN-PDAC and did not observe IPMN, indicating potential divergent mechanisms of ductal cell transformation in the context of PTEN gene expression loss as an initiating event or occurring spontaneously during tumorigenesis. Future studies using this model will aid in understanding how elevated levels of K-Ras in murine PDAC decreases PTEN levels [49, 52–54]. Using tissue lysates and our in vitro cell cultre platform, we analysed AKT phosphorylation. We observed increased phosphorylation of AKT at Threonine site at amino acid 308 (pAKTT308) whereas pAKTS473 was decreased. AKT is known to be fully active when phosphorylated at both these sites. However, recent emerging data has shown pAKTT308 is a more potent driver and marker of aggressive disease in non –small cell lung cancer and pAKTT308, rather than pAKTS473, correlates with AKT downstrem effectors [55]. In our model, while we have not completely interrogated downstrem pathways of AKT, we did employ a pan AKT inhibior (MK2206), which significanlty reduced aggressive PDAC in cKrasHigh mice. Existing neoplastic ducts were seen to be less proliferative, but remained associated with pancreatitis. Our results and previous work [56] show the importance of studying both AKT phosphoryaltion sites and its effectors in patient tumor samples for better prognosis and to develop targetted therapies. In summary, our mouse model provides an important new system to study how levels of total K-Ras and Ras activity drive intraductal lesions including PanIN and PDAC. This system will be very relevant for studying novel therapeutic strategies in the context of Ras signaling and this model provides an important Ras rheostat tool to study not only underlying mechanisms of PDAC arising in pancreatic ductal epithelium, but also better understanding how to calibrate therapeutics.
Methods
Transgenic mouse models
Hnf1bCreERT2 mice were purchased form Jackson Laboratories. Transgenic mice with CAG-lox-GFP-stop-lox-KrasG12V were received from Craig Logsdon, MD Anderson Cancer Center, Houston, TX [19]. Strains of Hnf1bCreERT2 mice were crossed with cLGL-KRASG12V to generate cKrasHnf1b/+ mice and obtain mutant cKras expression in adult pancreatic ductal cells. These mice express GFP in whole body and lose GFP after Cre mediated recombination. Mice were genotyped by PCR or Transnetyx. Mice were injected with tamoxifen at an age of 6–8 weeks and an n = 10–12 mice was evaluated for each tamoxifen dose. To achieve different levels of mutant cKras in ductal cells, mice were injected with 1 mg tamoxifen (Sigma, T5648) subcutaneously one day (1 mg dose), 5 mg tamoxifen for 1 day (5 mg dose), and 5 mg tamoxifen for 2 consecutive days (10 mg dose). All pancreatic pathologies in genetically engineered mice and humans were classified by a pathologist as has been classified and described previously [57, 58]. PanIN were classified on a two-tier grade based on cognate human classification system [59]. All animal studies were approved by the Animal Care and Use Committees at University of Texas Health Science Center at Houston.
Ras activity pull down assay
Ras activity in the cKrasHnf1b/+ mice pancreas samples was performed using the Active Ras pull down and detection kit (Thermofisher Scientific). Up to 30 mg of tissue (fresh or frozen at −80 °C) was washed with 1X cold PBS, cut into smaller pieces and homogenized in 1 ml lysis buffer (Cell Signaling Technology, 9803 S) containing protease inhibitor cocktail (Roche, 4693159001). Homogenized tissue was placed on ice for 30 min followed by sonication for 2 min with 10 s on/off cycle. The lysate was centrifuged for 20 min at 10,000 × g at 4 °C in a micro centrifuge. The pellet was discarded, and the lysate was used for protein estimation by BCA method. Aliquots of lysates were saved for further quantification of total Ras or protein loading controls by immunoblotting and a 500 μg equivalent of lysate protein were incubated for 45 min at 4 °C, with beads coated with Raf1-RBD provided with the kit. Beads were then washed 3 times with ice-cold lysis buffer, and bound protein was eluted for 15 min with Laemmli sample buffer that had been preheated to 95 °C and analyzed by immunoblotting for Ras following Western blot analysis.
Western blot analysis
Tissue extracts were prepared using cell lysis buffer (Cell signaling #9803) with protease inhibitor cocktail tablets (Cell signaling #5871). Tissue extracts were prepared using cell lysis buffer (Cell signaling #9803) with protease inhibitor cocktail tablets (Cell signaling #5871). Protein concentrations were determined by BCA method. A 20 µg of protein was analyzed using SDS page. The proteins were transferred by semi dry method on to the nitrocellulose membrane using Trans blot Turbo Transfer (Bio-Rad). Membranes were blocked using 5% skimmed milk and then incubated with primary antibodies overnight at 4 °C. Next day the membrane was washed 4 times with TBST buffer and incubation with the respective HRP-conjugated secondary antibody used at 1:5000 for 1 h. Membranes were washed and developed using ClarityTM Western ECL Substrate (Bio-Rad #1705061). Primary antibodies used in this study are described in Supplementary Table 1.
Primary pancreatic duct culture
Pancreatic ducts were cultured as defined previously [60, 61] from cKras0 (WT), cKrasLow, cKrasMod, and cKrasHigh mice four days after tamoxifen was administered. Briefly, Pancreas was collected, minced to 1 mm pieces and digested for 30 min at 37 °C in digestive solution (0.1% soybean trypsin inhibitor and 0.1% Collagenase). Cells were filtered through 40 µm filter, washed additional two times with culture medium and plated on collagen coated plates in complete medium DMEM/F12 (Life Technologies 11330-032) 500 mL, Penicillin‐streptomycin (100×; Life Technologies 15140-122) 5 mL, 1×Nu-serum IV (BD Biosciences 355104) 25 mL, 5% Bovine pituitary extract (3 mg/mL; BD Biosciences 354123) 4.2 mL, 25 μg/mL ITS + Premix (BD Biosciences 354352) 2.5 mL, Epidermal growth factor (100 μg/mL; BD Biosciences 354001) 100 μL, 20 ng/mL Cholera toxin (1 mg/mL; Sigma-Aldrich C8052) 50 μL, 100 ng/mL3,3′,5-Triiodo-L-thyronine (50 μM; Sigma-Aldrich T2877)50 μL, 5 nM Dexamethasone (100 mM; Sigma-AldrichD1756) 5 μL, 1 μM D-Glucose (Sigma-Aldrich G5400) 2.5 g 4.7 mg/mL, Nicotinamide (Sigma-Aldrich N3376) 0.66 g 1.22 mg/mL and Soybean trypsin inhibitor (type I; Sigma-Aldrich T6522) 50 mg 0.1 mg/mL. The cultures grew to confluency in one week and fibroblast contamination was reduced using differential trypsinization method. Cell lysate were collected and equal amounts of protein were subjected to Western analysis.
Immunofluorescence and immunohistochemistry
Tissues were fixed in 4% paraformaldehyde, processed according to standard protocols and embedded in paraffin. The unstained sections were baked at 60 °C for 30 min. The sections were deparaffinized with Histoclear and rehydrated. Antigen retrieval was performed using heat-mediated microwave methods and an antigen unmasking solution (Vector Laboratories, H-3300) was used for all antibodies. All sections were blocked in 10% FBS in PBST and primary antibodies were incubated overnight at 4 °C. Secondary antibodies were used at 1:500 and incubated at room Temperature for 2 h for IF and 30 min for IHC. For IF, slides were stained with IHC-Tek Dapi counterstain solution (IHC World, IW-1404) and mounted in fluorescence mounting medium (Dako, S3023). For IHC, Vectastain Elite ABC kit (Vector Laboratories, PK-6100) and DAB Peroxidase (HRP) Substrate kit (Vector Laboratories, SK-4100) were used. Primary antibodies used in this study are described in Supplementary Table 1.
Functional proteomics/RPPA analysis
Differentially tamoxifen dosed cKrasHnf1b/+ mice were monitored after tamoxfien injection and were euthanized at their humane timepoint. Pancreas tissue was collected an lysed using lysis buffer 1% Triton X-100, 50 mM HEPES, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1mMEGTA, 100 mM NaF, 10 mM Na pyrophosphate, 1 mM Na3VO4, 10% glycerol, containing freshly added protease and phosphatase inhibitors (Sigma Aldrich,St. Louis, MO) and protein concentration was measured by bicinchoninic acid (BCA) method. Protein concentrations of samples were adjusted to 1 mg/ml with lysis buffer. Cell lystes were serially diluted twofold for 5 dilutions (from undiluted to 1:16 dilution) and arrayed on nitrocellulose-coated slides in an 11 × 11 format. Samples were probed with antibodies by tyramide-based signal amplification approach and visualized by DAB colorimetric reaction. The slides were analyzed and protein expression quantitated with the use of Array-Pro Analyzer. All the data points were normalized for protein loading and transformed to linear value, designated as “Normalized Linear”.
MK-2206 treatment
Six to eight-week-old cKrasHn1b/+ mice were administered 5 mg of tamoxifen on days 1 and 2 (total 10 mg dose). MK-2206 (Selleckchem) was given on alternate days starting on Day 3 through Day 12. MK-2206 was administered using intraperitoneal injection at a dosage of 120 mg/kg.
ImageJ analysis
Area of pancreatitis was measured using imageJ (http://imagej.nih.gov/ij/) software. Five randomly chosen fields of H&Es of each pancreatic section were selected for quantification. Pancreatitis was calculated as percentage of total pancreatic area. Color deconvolution plugin was used to isolate pancreatitis versus normal pancreatic acinar cell areas. 5 fields per slide were selected for quantification and 5 mice were analyzed per tamoxifen dose. For Collagen fiber quantification, Masson’s trichrome stained pancreatic sections were used. The color deconvolution plugin of ImageJ software was used to quantify the collagen area in five randomly chosen fields of each pancreatic section as has been previously described [62]. Fibrosis was calculated as percentage of total pancreatic area. In total, 5 fields per slide were selected for quantification and 5 mice were analyzed per tamoxifen dose.
Supplementary information
Acknowledgments
Funding
JMB-L is supported by an AACR Pathway to Leadership Award, in part by PHS grant P30DK056338 and a grant from the National Pancreas Foundation. The RPPA Core facility at MDAnderson is funded by CA16672. AM is supported by the Khalifa bin Zayed Foundation, the MD Anderson Cancer Moonshot program, and NCI U24 CA224020, NCI R01CA218004, and NCI R01CA220236.
Author contributions
KS, MP Acquisition of data; KS, MP, SG, NT, AM, FM, CDL, JTC, JMB-L Analysis and interpretation of data. JMB-L Study concept and design. KS and JMB-L Drafting of manuscript. BDC, AM, JMB-L Critical revision of the manuscript. JMB-L Obtained funding.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The original online version of this article was revised: Due to a retrospective Open Access order.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/2/2021
A Correction to this paper has been published: 10.1038/s41374-021-00615-4
Supplementary information
The online version of this article (10.1038/s41374-020-00490-5) contains supplementary material, which is available to authorized users.
References
- 1.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–22. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 3.Kuboki Y, Fischer CG, Beleva Guthrie V, Huang W, Yu J, Chianchiano P, et al. Single-cell sequencing defines genetic heterogeneity in pancreatic cancer precursor lesions. J Pathol. 2019;247:347–56. doi: 10.1002/path.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AYL, Duboi CL, Sarai K, Zarei S, Schaeffer DF, Sander M, et al. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut. 2019;68:487–98. doi: 10.1136/gutjnl-2017-314426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–96. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Electronic address aadhe, cancer genome atlas research n. integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic. Cancer. Cancer Discov. 2016;6:166–75. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddipati R, Stanger BZ. Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov. 2015;5:1086–97. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim MS, Zhong Y, Yachida S, Rajeshkumar NV, Abel ML, Marimuthu A, et al. Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol Cell Proteomics. 2014;13:2803–11. doi: 10.1074/mcp.M114.038547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koay EJ, Baio FE, Ondari A, Truty MJ, Cristini V, Thomas RM, et al. Intra-tumoral heterogeneity of gemcitabine delivery and mass transport in human pancreatic cancer. Phys Biol. 2014;11:065002. doi: 10.1088/1478-3975/11/6/065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penchev VR, Rasheed ZA, Maitra A, Matsui W. Heterogeneity and targeting of pancreatic cancer stem cells. Clin Cancer Res. 2012;18:4277–84. doi: 10.1158/1078-0432.CCR-11-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic. Cancer Cell. 2019;178:160–75. doi: 10.1016/j.cell.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hruban RH, Wilentz RE, Maitra A. Identification and analysis of precursors to invasive pancreatic cancer. Methods Mol Med. 2005;103:1–13. doi: 10.1385/1-59259-780-7:001. [DOI] [PubMed] [Google Scholar]
- 15.Hruban RH, Maitra A, Goggins M. Update on pancreatic intraepithelial neoplasia. Int J Clin Exp Pathol. 2008;1:306–16. [PMC free article] [PubMed] [Google Scholar]
- 16.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 17.Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016;35:4282–8. doi: 10.1038/onc.2015.441. [DOI] [PubMed] [Google Scholar]
- 18.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Ji B, Tsou L, Wang H, Gaiser S, Chang DZ, Daniluk J, et al. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137:1072–82. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murtaugh LC. Pathogenesis of pancreatic cancer: lessons from animal models. Toxicol Pathol. 2014;42:217–28. doi: 10.1177/0192623313508250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–39. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray KC, Bell KM, Yan J, Gu G, Chung CH, Washington MK, et al. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS ONE. 2011;6:e16786. doi: 10.1371/journal.pone.0016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 24.Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–50. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L, McFaul C, Howes N, Leslie J, Lancaster G, Wong T, et al. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology. 2005;128:2124–30. doi: 10.1053/j.gastro.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Lu X, Xu T, Qian J, Wen X, Wu D. Detecting K-ras and p53 gene mutation from stool and pancreatic juice for diagnosis of early pancreatic cancer. Chin Med J. 2002;115:1632–6. [PubMed] [Google Scholar]
- 27.Parsons BL, Meng F. K-RAS mutation in the screening, prognosis and treatment of cancer. Biomark Med. 2009;3:757–69. doi: 10.2217/bmm.09.95. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 29.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–51. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 32.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 33.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–3.e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo TP, Hruban RH, Leach SD, Wilentz RE, Sohn TA, Kern SE, et al. Pancreatic cancer. Curr Probl Cancer. 2002;26:176–275. doi: 10.1067/mcn.2002.129579. [DOI] [PubMed] [Google Scholar]
- 35.Makohon-Moore AP, Matsukuma K, Zhang M, Reiter JG, Gerold JM, Jiao Y, et al. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature. 2018;561:201–5. doi: 10.1038/s41586-018-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De La OJP, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci USA. 2008;105:18907–12. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murtaugh LC, Leach SD. A case of mistaken identity? Nonductal origins of pancreatic “ductal” cancers. Cancer Cell. 2007;11:211–3. doi: 10.1016/j.ccr.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci USA. 2008;105:18913–8. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philip B, Roland CL, Daniluk J, Liu Y, Chatterjee D, Gomez SB, et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2013;145:1449–58. doi: 10.1053/j.gastro.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi C, Pan FC, Kim JA, Washington MK, Padmanabhan C, Meyer CT, et al. Differential cell susceptibilities to Kras. Cell Mol Gastroenterol Hepatol. 2019;8:579–94. doi: 10.1016/j.jcmgh.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira RMM, Sancho R, Messal HA, Nye E, Spencer-Dene B, Stone RK, et al. Duct- and acinar-derived pancreatic ductal adenocarcinomas show distinct tumor progression and marker expression. Cell Rep. 2017;21:966–78. doi: 10.1016/j.celrep.2017.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–85. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–18. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 46.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 47.Singh K, Pruski MA, Polireddy K, Jones NC, Chen Q, Yao J, et al. Mst1/2 kinases restrain transformation in a novel transgenic model of Ras driven non-small cell lung cancer. Oncogene. 2020;39:1152–64. doi: 10.1038/s41388-019-1031-z. [DOI] [PubMed] [Google Scholar]
- 48.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 49.Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–95. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 50.Maitra A, Hruban RH. A new mouse model of pancreatic cancer: PTEN gets its Akt together. Cancer Cell. 2005;8:171–2. doi: 10.1016/j.ccr.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Logsdon CD, Lu W. The significance of ras activity in pancreatic cancer initiation. Int J Biol Sci. 2016;12:338–46. doi: 10.7150/ijbs.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kopp JL, Dubois CL, Schaeffer DF, Samani A, Taghizadeh F, Cowan RW, et al. Loss of pten and activation of Kras synergistically induce formation of intraductal papillary mucinous neoplasia from pancreatic ductal cells in mice. Gastroenterology. 2018;154:1509–23. doi: 10.1053/j.gastro.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, et al. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol. 2000;157:1097–103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ying H, Elpek KG, Vinjamoori A, Zimmerman SM, Chu GC, Yan H, et al. PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer Discov. 2011;1:158–69. doi: 10.1158/2159-8290.CD-11-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincent EE, Elder DJ, Thomas EC, Phillips L, Morgan C, Pawade J, et al. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer. 2011;104:1755–61. doi: 10.1038/bjc.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vadlakonda L, Dash A, Pasupuleti M, Anil Kumar K, Reddanna P. The Paradox of Akt-mTOR Interactions. Front Oncol. 2013;3:165. doi: 10.3389/fonc.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hruban RH, Rustgi AK, Brentnall TA, Tempero MA, Wright CV, Tuveson DA. Pancreatic cancer in mice and man: the Penn Workshop 2004. Cancer Res. 2006;66:14–7. doi: 10.1158/0008-5472.CAN-05-3914. [DOI] [PubMed] [Google Scholar]
- 58.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 59.Basturk O, Yang Z, Tang LH, Hruban RH, Adsay V, McCall CM, et al. The high-grade (WHO G3) pancreatic neuroendocrine tumor category is morphologically and biologically heterogenous and includes both well differentiated and poorly differentiated neoplasms. Am J Surg Pathol. 2015;39:683–90. doi: 10.1097/PAS.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichert M, Takano S, Heeg S, Bakir B, Botta GP, Rustgi AK. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc. 2013;8:1354–65. doi: 10.1038/nprot.2013.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reichert M, Rhim AD, Rustgi AK. Culturing primary mouse pancreatic ductal cells. Cold Spring Harb Protoc. 2015;2015:558–61. doi: 10.1101/pdb.prot078279. [DOI] [PubMed] [Google Scholar]
- 62.Chen A. A convenient method for quantifying collagen fibers in atherosclerotic lesions by ImageJ software. Int J Clin Exp Me. 2017;10:14904–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.