Abstract
Congenital heart defects (CHDs) affecting the cardiac outflow tract (OFT) constitute a significant cause of morbidity and mortality. The OFT develops from migratory cell populations which include the cardiac neural crest cells (cNCCs) and secondary heart field (SHF) derived myocardium and endocardium. The related transcription factors HAND1 and HAND2 have been implicated in human CHDs involving the OFT. Although Hand1 is expressed within the OFT, Hand1 NCC-specific conditional knockout mice (H1CKOs) are viable. Here we show that these H1CKOs present a low penetrance of OFT phenotypes, whereas SHF-specific Hand1 ablation does not reveal any cardiac phenotypes. Further, HAND1 and HAND2 appear functionally redundant within the cNCCs, as a reduction/ablation of Hand2 on an NCC-specific H1CKO background causes pronounced OFT defects. Double conditional Hand1 and Hand2 NCC knockouts exhibit persistent truncus arteriosus (PTA) with 100% penetrance. NCC lineage-tracing and Sema3c in situ mRNA expression reveal that Sema3c-expressing cells are mis-localized, resulting in a malformed septal bridge within the OFTs of H1CKO;H2CKO embryos. Interestingly, Hand1 and Hand2 also genetically interact within the SHF, as SHF H1CKOs on a heterozygous Hand2 background exhibit Ventricular Septal Defects (VSDs) with incomplete penetrance. Previously, we identified a BMP, HAND2, and GATA-dependent Hand1 OFT enhancer sufficient to drive reporter gene expression within the nascent OFT and aorta. Using these transcription inputs as a probe, we identify a novel Hand2 OFT enhancer, suggesting that a conserved BMP-GATA dependent mechanism transcriptionally regulates both HAND factors. These findings support the hypothesis that HAND factors interpret BMP signaling within the cNCCs to cooperatively coordinate OFT morphogenesis.
1. Introduction
Congenital heart defects (CHDs) afflict roughly 1% of newborns according to the National Heart Lung and Blood Institute, and ultimately affect the quality of life of more than 1 million adults in the United States. (Cheng et al., 2012a) Of these affected individuals, approximately 25% require invasive treatment within the first year of life. (Mozaffarian et al., 2014) Owing to significant advances in cardiothoracic medicine, infant mortality rates from CHDs have fallen drastically in the past half-century. Unfortunately, CHDs occur with such frequency that they remain a significant cause of morbidity and mortality in both children and adults. Further progress in the prevention and treatment of CHDs hinges upon a complete understanding of their etiology.
A significant portion of CHDs can be attributed to dysfunction of the main developmental progenitors of the cardiac outflow tract (OFT), cardiac neural crest cells (cNCCs) and of the second heart field (SHF). (Mozaffarian et al., 2014) cNCCs and SHF cells are both migratory cell populations that originate outside of the primary heart-forming field. NCC subpopulations migrate from the dorsal neural tube throughout the developing embryo, whereupon they differentiate into distinct tissue types. Within the OFT, the cardiac NCCs, differentiate into smooth muscle and connective tissue to contribute to the aorta, pulmonary artery, and nascent ventricular septum. The SHF, a subpopulation of splanchnic mesoderm located anterior to the cardiac crescent, migrates into the arterial pole to contribute endocardial and myocardial cells to the growing heart. (Barnes et al., 2010b ; Harrison et al., 2005; Nakajima, 2010; Zaffran and Kelly, 2012) Within the OFT, the cNCCs coordinate both with a specific SHF subpopulation, known as the myocardial cuff (MC), and with adjacent endocardium, to divide the OFT into two separate blood vessels, and thereby sequester systemic and pulmonary circulation. Although a number of studies have identified genetic and environmental factors that affect OFT remodeling, and consequently cause CHDs, a failure to understand the local signaling cues and transcriptional mechanisms that regulate gene expression within the cNCCs after these cells have migrated into the OFT, the pleiotropic effects of crucial factors that have confounded studies of their tissue- and stage-specific cellular functions, and functional redundancy among mammalian genes that likewise mask their critical functions have hindered understanding of the transcriptional and extracellular signaling mechanisms that coordinately control cardiac NCC and MC morphogenesis.
The bHLH transcription factors HAND1 and HAND2 exhibit similar functional properties and have been proposed to perform redundant functions during development. (Barbosa et al., 2007; George and Firulli, 2019; McFadden et al., 2005; Vincentz et al., 2017) The two factors share dimer partners and bind to either of two consensus elements termed E-boxes (CANNTG) and D-boxes (CGNNTG). (Firulli et al., 2007; George and Firulli, 2019; Hollenberg et al., 1995) Studies in both chick (Srivastava et al., 1995) and mouse (McFadden et al., 2005) indicate that HAND1 and HAND2 may functionally compensate for each other’s absence during early cardiogenesis. Hand1 deletion within NCCs reveals gene dosage dependent craniofacial phenotypes in Hand1 conditional knockouts that are also on a Hand2 heterozygote background; however, cardiac defects were not investigated, and if present, are not severe enough to cause lethality (Barbosa et al., 2007).
HAND1 mutations have been observed in human patients with Tetralogy of Fallot (TOF; (Wang et al., 2017b; Wang et al., 2011a), with Double Outlet Right Ventricle (DORV; (Li et al., 2017), and with Ventricular Septal Defects (VSDs; (Cheng et al., 2012b), as well as, most recently, cardiac conduction system functional variation (Evans1 et al., 2016; Firulli et al., 2019; Sotoodehnia et al., 2010; van Setten et al., 2019; Vincentz et al., 2019). HAND1 haploinsufficiency likely contributes to CHDs associated with 5q33 chromosomal deletions (Starkovich et al., 2016). Studies of children with CHDs such as TOF, pulmonary stenosis, and VSDs with DORV, have identified multiple HAND2 missense mutations (Shen et al., 2010). Additionally, defects including VSDs, pulmonary atresia, coarctation of the aorta, and TOF, are associated with genomic duplications or deletions of chromosome 4q33, to which HAND2 maps (Borochowitz et al., 1997; Byatt et al., 1997). Mouse genetic models indicate that the OFT CHDs associated with the human chromosomal disorder partial trisomy distal 4q (4q+) are caused by overexpression of HAND2 (Tamura et al., 2013); however, the variable penetrance of CHDs associated with 4q+ suggest that unknown HAND2 genetic modifiers influence the presentation of this disease. Increased HAND2 mRNA expression is associated with TOF, whereas increased HAND1 mRNA expression is associated with hypertrophic obstructive cardiomyopathy (Ritter et al., 1999). Collectively, these findings are consistent with a mechanism whereby either too much or too little HAND factor function is detrimental to cardiac growth and morphogenesis.
As steady state expression levels of HAND1 and HAND2 appear to dictate their cellular function (Barbosa et al., 2007; Firulli et al., 2005; McFadden et al., 2005), unraveling the mechanisms by which Hand1 and Hand2 expression is regulated within these tissues, and, in turn, HAND factor dependent downstream targets, is necessary to understand their roles in heart development. To date, a Hand2 right ventricle (RV)/OFT enhancer, directly regulated by GATA factors, that drives gene expression within the right ventricle (RV) and OFT myocardium has been identified (McFadden et al., 2000). Two separate Hand1 transcriptional enhancers, one specific for the OFT and MC (Vincentz et al., 2016), and the other specific for the LV are also dependent on GATA cis-elements for their transcriptional activity (Vincentz et al., 2019; Vincentz et al., 2017). Indeed, a single nucleotide polymorphism (SNP) within one of two evolutionarily conserved GATA cis-elements located within the LV enhancer is associated with altered cardiac conduction in humans and human phenotypes are modeled in genetically engineered mice (Vincentz et al., 2019). Collectively, these data support the hypothesis that both HAND1 and HAND2 play essential roles in cardiogenesis.
In this study, we explore the molecular pathways and mechanism(s) by which HAND1 and HAND2 regulate proper development of the cNCC and MC components of the OFT. Hand1 NCC conditional knockouts (H1CKONCC) exhibit a low penetrance of OFT phenotypes. Additionally, removing a single or both Hand2 alleles within the NCC on a H1CKONCC background result in more serious and highly penetrant OFT defects that include persistent truncus arteriosus (PTA). SHF conditional Hand1 knockout (H1CKOSHF) and Hand2 reveal genetic interactions within OFT myocardium. Like Hand1 (George and Firulli, 2019; Vincentz et al., 2016), a novel Hand2 OFT transcriptional enhancer includes transcriptional inputs for HAND2, GATA and SMAD1/5. Together these findings expand our understanding of how coordinate developmental events in distinct tissue populations (the cNCCs and MC) affect proper morphogenesis of the OFT and provide insight into the etiology of the CHDs defects associated with HAND factor mutations.
2. Materials and Methods.
Experimental Mice
Genotyping of the Hand1tm1Eno/AbfiJ, (Firulli et al., 1998) Hand1f, (McFadden et al., 2005) Hand2neo, (Srivastava et al., 1997) Hand2f, (Morikawa and Cserjesi, 2008) Smad4tm2.1Cxd/J (Yang et al., 2002) Wnt1-Cre, (Vincentz et al., 2013) Tg(Mef2c-cre)2Blk, (Verzi et al., 2005) and Gt(ROSA)26Sortm1Sor (Soriano, 1999) and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (genotyped as per (Soriano, 1999)) alleles have been previously described. The mm1284 sequence was amplified from mouse genomic DNA (Clontech) and cloned into the hsp68-lacZ vector (Kothary et al., 1989; Visel et al., 2007). Pronuclear injection into fertilized eggs, implantation into pseudo pregnant mothers, and embryo dissection at embryonic day 11.5 (E11.5) was performed as described previously (Attanasio et al., 2013). Yolk sac DNA was used to assess the presence of the various alleles in embryos. Tail DNA was used to assess the presence of the various alleles in neonatal mice. All animal maintenance and procedures were performed in accordance with the Indiana University School of Medicine and Lawrence Berkeley National Laboratory Animal Care and use committees.
Embryo harvesting in situ hybridization & Xgal staining
Following dissection in cold 1X PBS, embryos of the indicated age were fixed in 4% paraformaldehyde at 4°C. Embryos were washed three times with wash buffer (2mM MgCl2; 0.01% deoxycholate; 0.02% NP-40; 100mM phosphate buffer, pH 7.3) for 30 min., then stained for 24 hrs. at room temperature with freshly made X-gal staining solution (0.8mg/mL X-gal; 4mM potassium ferrocyanide; 4mM potassium ferricyanide; 20mM Tris, pH 7.5 in wash buffer). The following day, embryos were rinsed three times in 1X PBS, then post-fixed in 4% paraformaldehyde. X-gal staining was performed as previously described (Barnes et al., 2010a). Nuclear Fast Red staining was performed as per manufacturer’s instructions.
Histology
Embryos (E10.5 and E16.5) were fixed in 4% paraformaldehyde, dehydrated, embedded, sectioned, and Hematoxylin and Eosin (H&E) or Sirus red & Fast Green stained as described. (Firulli et al., 2014; Firulli et al., 2019). All data was collected on a Leica DM5000 B compound wide field florescent microscope.
in situ hybridization was performed as described. (Vincentz et al., 2013; Vincentz et al., 2019) Antisense digoxygenin-labeled Sema3c and Hand2 riboprobes were synthesized using T7, T3 or SP6 polymerases (Promega) and DIG-Labeling Mix (Roche) using a linearized plasmid template.
Bioinformatics
Histone marks and variants, chromatin accessibility, and transcription factor / chromatin regulator binding within 100 Kb of the Hand2 transcription start site was examined using the Toolkit for Cistrome Data Browser (http://dbtoolkit.cistrome.org). SMAD1/5, HAND2, and GATA4 tracks were loaded onto the UCSC mm10 genome browser and one region of ChIP-seq binding co-occupancy was identified. A DNA fragment containing this region of co-occupancy was then cloned into a lacZ reporter construct and used to generate transient transgenic embryos via previously reported methodology (Visel et al., 2007). VISTA Enhancer Browser-a database of tissue-specific human enhancers (Visel et al., 2007).
3. Results
A critical Hand factor genetic complement is crucial for morphogenesis of the developing OFT.
Hand1 is expressed in the developing OFT within both the cardiac NCC and MC, (Vincentz et al., 2011) Vincentz et al., 2016). Mutations in human HAND1 are associated with OFT CHDs. (Wang et al., 2017a; Wang et al., 2011b) Surprisingly, Hand1fx/fx;Wnt1-Cre(+) NCC-specific conditional knockouts (CKOs) have been reported to be viable and fertile. (Barbosa et al., 2007) This discrepancy indicates that current genetic understanding of HAND1 function within the OFT, based upon animal models, is incomplete. Moreover, evidence shows that Hand2 expression shows a slight increase within Hand1 systemic knockout embryos (Morikawa and Cserjesi, 2004) To explore the possibility that Hand1 CKOs exhibits cardiac phenotypes that could be compensated by HAND2, we first tested whether Hand1 function is required for OFT development within the developing MC and/or cNCCs.
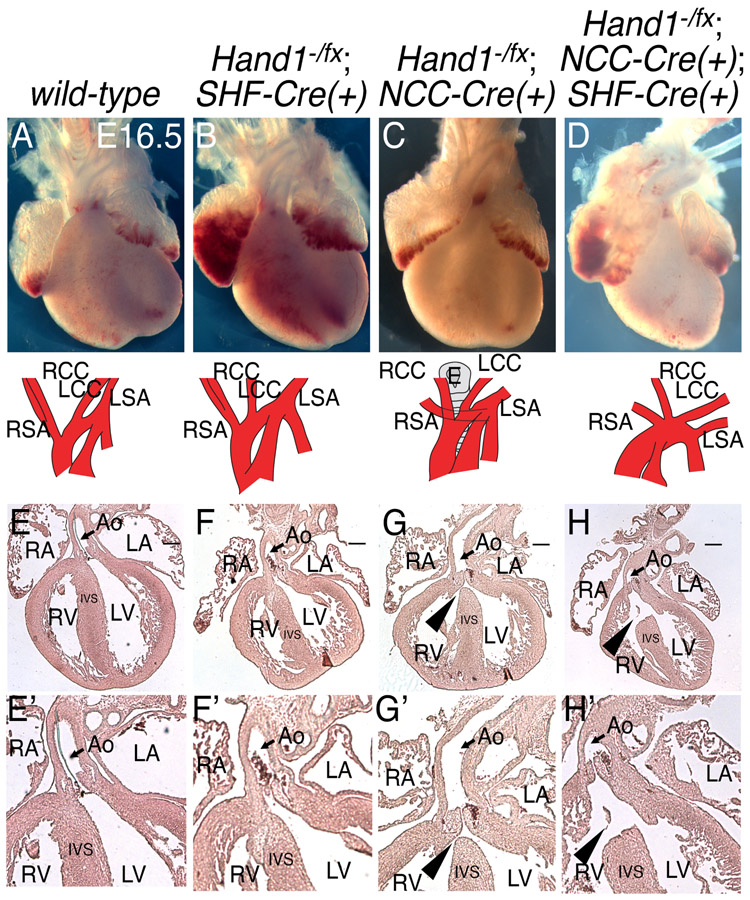
To test whether HAND1 function is necessary within the MC, Mef2c-AHF-Cre(+) mice (Verzi et al., 2005) were employed to ablate Hand1 function specifically within SHF derivatives (Fig. 1). Hand1fx/+;Mef2c-AHF-Cre(+) mice were bred with Hand1+/− mice to generate Hand1−/fx;Mef2c-AHF-Cre(+) embryos. Phenotypic analyses of E16.5 Hand1−/fx;Mef2c-AHF-Cre(+) CKOs revealed no OFT abnormalities within H1CKOsSHF (Hand1fx/fx;Mef2c-AHF-Cre(+)); Fig. 1B, F, and F’; n=5 all normal) compared with controls (Hand1+/+;Mef2c-AHF-Cre(+);Wnt1-Cre(+)); Fig. 1A, E, and E’; n=6 all normal). We next looked carefully at E16.5 hearts from H1CKOSNCC (Fig. 1C, G, and G’ n=9). One in three NCC-specific H1CKOsNCC (Hand1fx/−;Wnt1-Cre(+)) displayed membranous VSDs (Fig. 1C, G, and G’; n=3/9), and one displayed aberrant origin of the right subclavian artery (AORSA; Fig. 1C; 1/9) whereas 6 out of 9 H1CKONCC hearts appeared phenotypically normal. To test for tissue interactions of Hand1 within both SHF and NCC, we generated H1CKONCC/SHF E16.5 embryos (Fig.1D, H, and H’ n=4). We observed no increase in OFT defect severity or penetrance compared with what is observed in H1CKONCC hearts. These findings are summarized in Table 1. This data supports that Hand1 loss-of-function within NCCs can cause OFT phenotypes with incomplete penetrance consistent with those observed in human patients with HAND1 mutations, and that deletion of Hand1 within the SHF presents no obvious cardiac phenotypes.
Figure 1. Hand1 neural crest cell loss-of-function leads to a low penetrance of ventricular septal defects.
Whole mount preparations (A-D) and hematoxylin and eosin-stained (E-H) E16.5 hearts and aortic arches in which SHF- (B and F), NCC- (C and G), or both SHF and NCC-specific (D and H) Hand1 has been conditionally ablated reveals rare aortic arch patterning defects (C), and a low penetrance of VSDs (G and H, arrowheads) in NCC Hand1 conditional knockouts. Aortic arches are schematized in A-D. Scale bars = 250 mm. Arrowhead, VSD; Asterisk, PTA; Ao, aorta; LA, left atrium; LCC, left common carotid; LSA, left subclavian artery; LV, left ventricle; RA, right atrium; RCC, right common carotid; RSA, right subclavian artery; RV, right ventricle. (E’-H’) Higher magnification images of E-H to better show structures.
Table 1.
Congenital heart defects in OFT Hand1 mutants.
| Genotype | n | AoRSA | membranous VSD |
ASD | Phenotypically Normal |
|---|---|---|---|---|---|
| Hand1+/+; Wnt1-Cre(+); Mef2c-AHF-Cre(+) | 6 | 0 (0%) | 0 (0%) | 0 (0%) | 6 (100%) |
| Hand1−/fx; Mef2c-AHF-Cre(+) | 5 | 0 (0%) | 0 (0%) | 0 (0%) | 5 (100%) |
| Hand1−/fx; Wnt1-Cre(+) | 9 | 1 (11.1%) | 3 (33.3%) | 0 (0%) | 6 (66.6%) |
| Hand1−/fx; Wnt1-Cre(+); Mef2c-AHF-Cre(+) | 4 | 0 (0%) | 2 (50.0%) | 1 (25.0%) | 2 (50.0%) |
AoRSA, aberrant origin of the right subclavian artery; VSD, ventricular septal defect; ASD, atrial septal defect
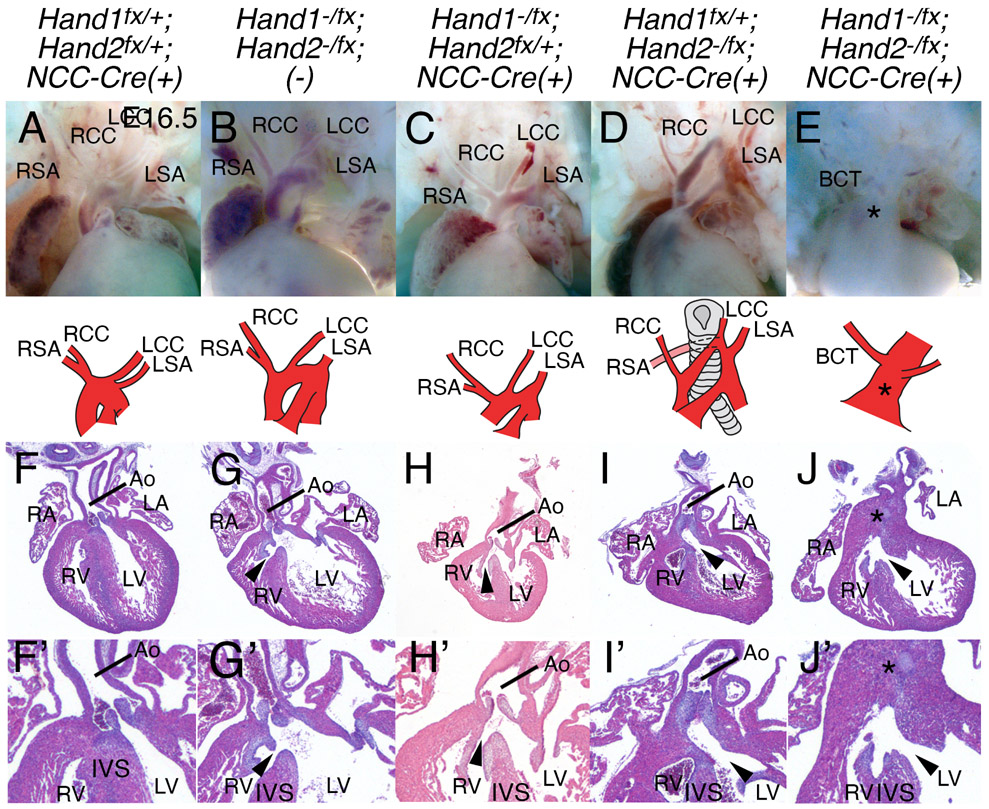
To investigate any HAND factor functional redundancy within the NCC, we next looked at hearts from combinations of H1CKONCC and H2CKONCC (Fig. 2). Hearts from double heterozygotes (Hand1fx/+;Hand2fx/+;Wnt1-Cre(+)) embryos (n=7) show normal development at E16.5 (Fig. 2A, F and F’). In contrast, double heterozygous Hand1fx/−;Hand2fx/− embryos (n=7) all present with DORV and associated VSD (Fig. 2B, G, and G’, black arrowheads). This difference between the double heterozygous genotypes likely reflects hypomorphic expression from one or both of the Hand conditional alleles. (McFadden et al., 2005; Morikawa et al., 2007) Embryos that contain a single Hand2 allele, (n=9) show a spectrum of defects that include DORV with associated VSD (5/9), dilated/hypertrophic LV (1/9), bicuspid aortic valve (1/9) and three embryos showed normal phenotype (Fig. 2C, H, and H’). E16.5 embryos that contain only a single Hand1 allele (n=9) exhibit DORV with associated VSD (9/9), interrupted aortic arch type B (3/9), and/or dilated/hypertrophic LV (4/9) (Fig. 2D, I, and I’). Finally analysis of H1CKONCC;H2CKONCC E16.5 hearts (n=3) showed 100% penetrance of persistent truncus arteriosus (Fig 2E, J, and J’, asterisk). Results are fully summarized in Table 2.
Figure 2. Hand1 and Hand2 genetically interact in the cardiac NCCs.
Whole mount preparations (A-E) and hematoxylin and eosin-stained (F-J) E16.5 hearts and aortic arches in which NCC-specific Hand1 and/or Hand2 has been conditionally ablated. Aortic arches are schematized in A-E. Arrowhead, VSD; Asterisk, PTA; Ao, aorta; BCT, brachiocephalic trunk; LA, left atrium; LCC, left common carotid; LSA, left subclavian artery; LV, left ventricle; RA, right atrium; RCC, right common carotid; RSA, right subclavian artery; RV, right ventricle. (F’-J’) higher magnification images of F-J to better show structures.
Table 2.
OFT defects in HAND factor mutants.
| Genotype | n | PTA + VSD |
IAA-B | AoRSA | DORV + VSD |
Dilated and/or hypertrophic LV |
Bicuspid Ao and PT valves |
Phenotypically Normal |
|---|---|---|---|---|---|---|---|---|
| Hand1+/fx; Hand2+/fx; Wnt1-Cre(+) | 7 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 7 (100%) |
| Hand1fx/−; Hand2fx/−; (−) | 7 | 0 (0%) | 0 (0%) | 0 (0%) | 7 (100%) | 7 (100%) | 0 (0%) | 0 (0%) |
| Hand1fx/−; Hand2fx/−; Wnt1-Cre(+) | 9 | 0 (0%) | 0 (0%) | 0 (0%) | 5 (55.6%) | 1 (11.1%) | 1* (11.1%) | 3 (33.3%) |
| Hand1+/fx; Hand2fx/−; Wnt1-Cre(+) | 9 | 0 (0%) | 3 (33.3%) | 6 (66.7%) | 9 (100%) | 4 (44.4%) | 0 (0%) | 0 (0%) |
| Hand1fx/−; Hand2fx/−; Wnt1-Cre(+) | 3 | 3 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (50%) | 0 (0%) | 0 (0%) |
AoRSA, aberrant origin of the right subclavian artery; DORV, double outlet right ventricle; IAA-B, interrupted aortic arch type-B; PTA, persistent truncus arteriosus; VSD, ventricular septal defect.
cNCC localization within the OFT is compromised within HAND loss-of-function hearts
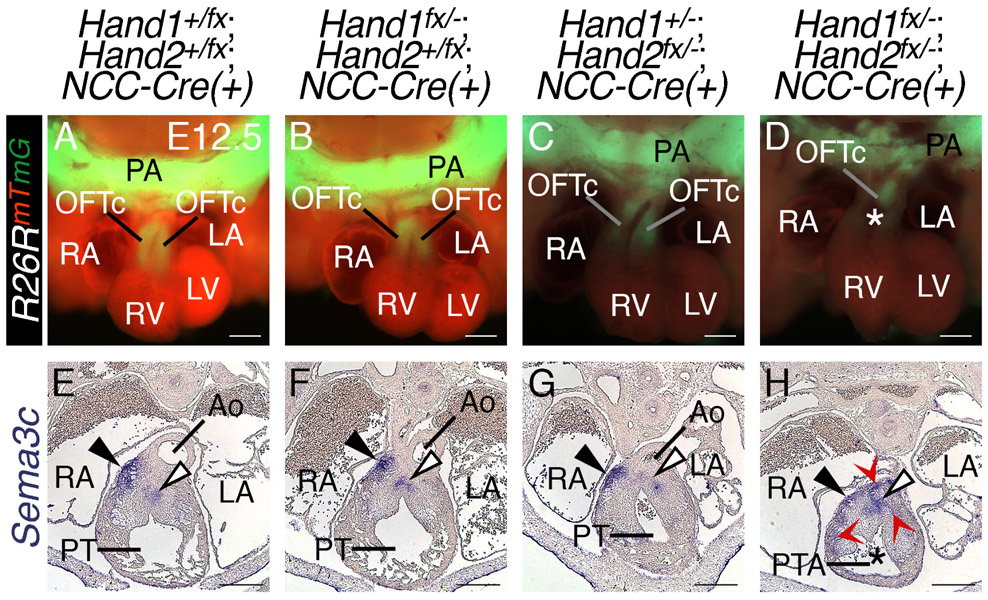
OFT defects are often correlated with diminished NCC migration into the OFT. To completely characterize the phenotypes seen in Hand1;Hand2 double NCC CKOs, we performed lineage trace analyses to monitor cNCC migration to the OFT in these double CKOs utilizing the R26RmT/mG allele, which switches from membrane-localized RFP tdTomato to a membrane-localized EGFP following Cre recombination. This allele enables visualization of NCC invasion of the OFT cushions at E12.5 (Fig. 3A-D). In contrast to Hand1+/fx;Hand2+/fx;Wnt1-Cre(+) controls (Fig. 3A) and Hand1fx/−;Hand2+/fx;Wnt1-Cre(+) or Hand1+/fx;Hand2fx/−;Wnt1-Cre(+) mutants (Fig. 3B and C), wherein two OFT cushions (OFTc) are clearly defined, the OFT cushions of H1CKONCC;H2CKONCC hearts are clearly dysmorphic and cannot be individually identified, as would be expected in a PTA (Fig. 3D; n=4/4).
Figure 3. Hand1 and Hand2 regulate cardiac NCCs localization in the developing OFT.
Wnt1-Cre;R26Rmtmg lineage mapping Control (A), H1CKO, Hand2 heterozygous (B and C) and H1CKO;H2CKO E12.5 day embryos (D). two outflow tract cushions (OFTc) are visible in control and H1CKO; H2 heterozygous hearts; however, only a single cushion is visible in the double knockout hearts. The class 3 semaphorin SEMA3C promotes septal bridge formation (white arrowhead). Sema3c in situ hybridization in Control (E) and H1CKO; H2 heterozygous hearts display Sema3c expression consistent with controls (F and G). Hand1fx/−;Hand2fx/−;Wnt1-Cre(+) mutants; however, exhibit mis-localized Sema3c-expressing cNCCs (H, red arrow heads). As Sema3c marks the septal bridge, this structure appears malformed within double CKOs. All double CKOs present with persistent truncus arteriosus (PTA, n=5/5). pharyngeal arch, PA; Myocardial cuff, denoted by black arrowhead; aorta, Ao; pulmonary trunk, PT; left atrium, LA; right atrium, RA; left ventricle, LV; right ventricle, RV; white asterisk, PTA
Although previous studies have revealed that NCC-specific Hand2 CKOs display aortic arch defects, VSDs, and DORV (Holler et al., 2010), it is unclear whether impaired cNCC contribution to the OFT underlies these defects. Coordinate with cNCC migration into the OFT, the endothelial lining of the OFT undergoes endothelial-to-mesenchymal transition to form mesenchymal cells that colonize the OFT cushions (Sugishita et al., 2004). This process is critical for the growth of the OFT cushions, which fuse to form the septal bridge (Fig. 3E-H, shown at the white arrowheads) a precursor to the mature aortico-pulmonary septum. A signaling cue that is critical for the process of aortico-pulmonary septation is the class 3 semaphorin SEMA3C (Brown et al., 2001). SEMA3C signals through the transmembrane receptor Neuropilin 1 (NRP1; (Schwarz and Ruhrberg, 2010) and loss of Sema3c expression results in interruption of the aortic arch accompanied by PTA, thus regulating development and migration of cNCCs (Feiner et al., 2001). Cardiac NCC-derived SEMA3C signaling promotes septal bridge formation through regulation of endothelial-to-mesenchymal transition (endoMT) and, indirectly, cardiac NCC relocalization (Plein et al., 2015). Sema3c ISH within H1CKONCC;H2CKONCC mutants shows that Sema3c-expressing cells are mis-localized (asterisks), and suggests that the septal bridge is malformed within H1CKONCC;H2CKONCC mutant OFTs (Fig. 3D and H, arrowhead; n=5/5). Thus, the Sema3c-positive population of cardiac NCCs migrate to the OFT in these mutants, but interaction of the NCCs with the endocardium is likely disrupted such that endoMT and cNCC localization is abnormal. Conditional ablation of Hand2 using the Hand1eGFP-Cre knock-in allele (one copy of Hand1), which recombines within both post-migratory cardiac NCCs and the MC, results in incompletely penetrant PTA (Barnes et al., 2011). ISH revealed that Sema3c expression is similarly mis-localized in Hand1Cre;Hand2 CKOs (Sup. Fig 1).
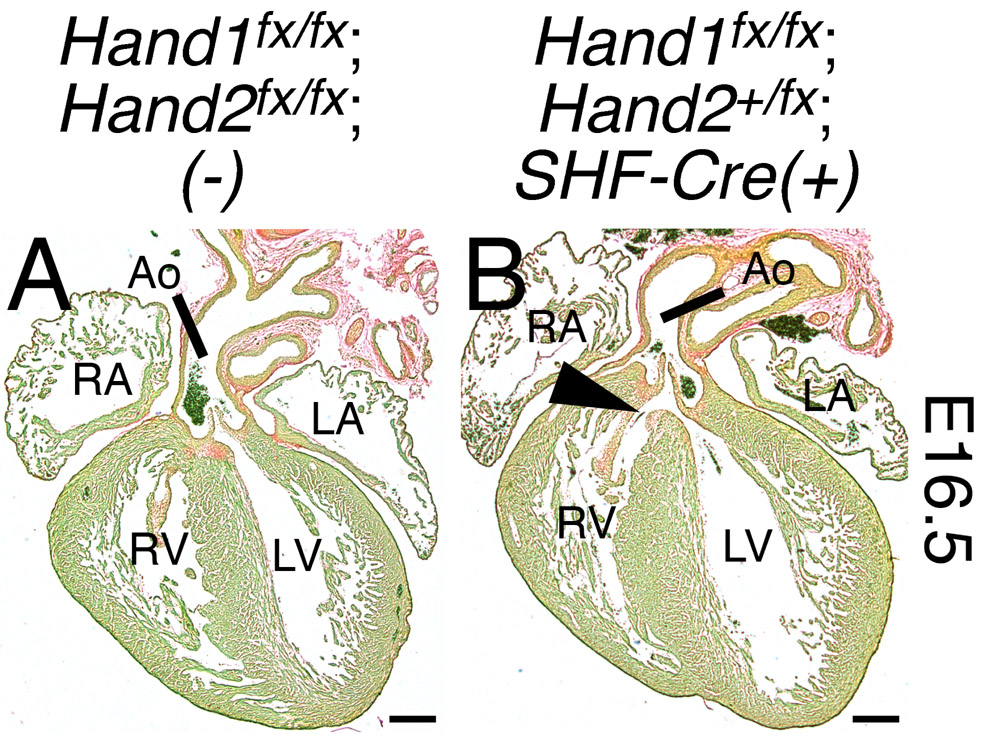
We next examined embryos in which SHF-specific Hand1 function has been conditionally ablated and Hand2 function has been reduced by half (Hand1fx/fx;Hand2fx/+;Mef2c-AHF-Cre(+)). Analyses of these embryos revealed a low penetrance of VSDs (Fig. 4, n=3/12; summarized in Table 3) and dilated coronary vessels within the ventricular free wall and septum. These coronary phenotypes are consistent with previous reports that Hand2 is necessary for coronary vasculogenesis (Vandusen et al., 2014), but have not been reported with HAND1-associated clinical heart phenotypes. Cumulatively, these findings confirm that OFT morphogenesis cannot tolerate a collective reduction in HAND factor function beyond a critical threshold.
Figure 4. Hand1;Hand2 genetic interaction in the second heart field.
E16.5 hearts in which Hand1 has been ablated and Hand2 function has been reduced by half within the SHF (Hand1fx/fx;Hand2fx/+;Mef2c-AHF-Cre(+)) display a low penetrance of VSDs (B, arrowhead; summarized in Table 3). Coronal sections have been stained with Sirius Red and Fast Green. Scale bars = 250 μm. Ao, aorta; LA, left atrium; RA, right atrium; LV, left ventricle; RV, right ventricle.
Table 3.
OFT defects in HAND factor SHF mutants.
| Genotype | n | membranous VSD |
Phenotypically Normal |
|---|---|---|---|
| Hand1fx/fx; Hand2+/fx; (−) | 3 | 0 (0%) | 3 (100%) |
| Hand1+/fx; Hand2+/fx;SHF-Cre(+) | 8 | 0 (0%) | 8 (100%) |
| Hand1fx/fx;Hand2+/fx;SHF-Cre(+) | 12 | 3 (25%) | 9 (75%) |
VSD, ventricular septal defect.
A conserved gene regulatory network regulates Hand factor expression in cardiac NCCs.
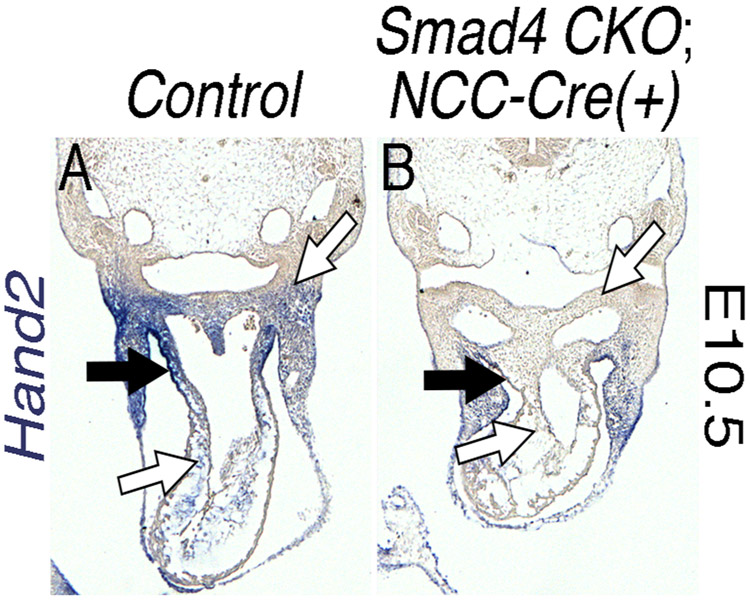
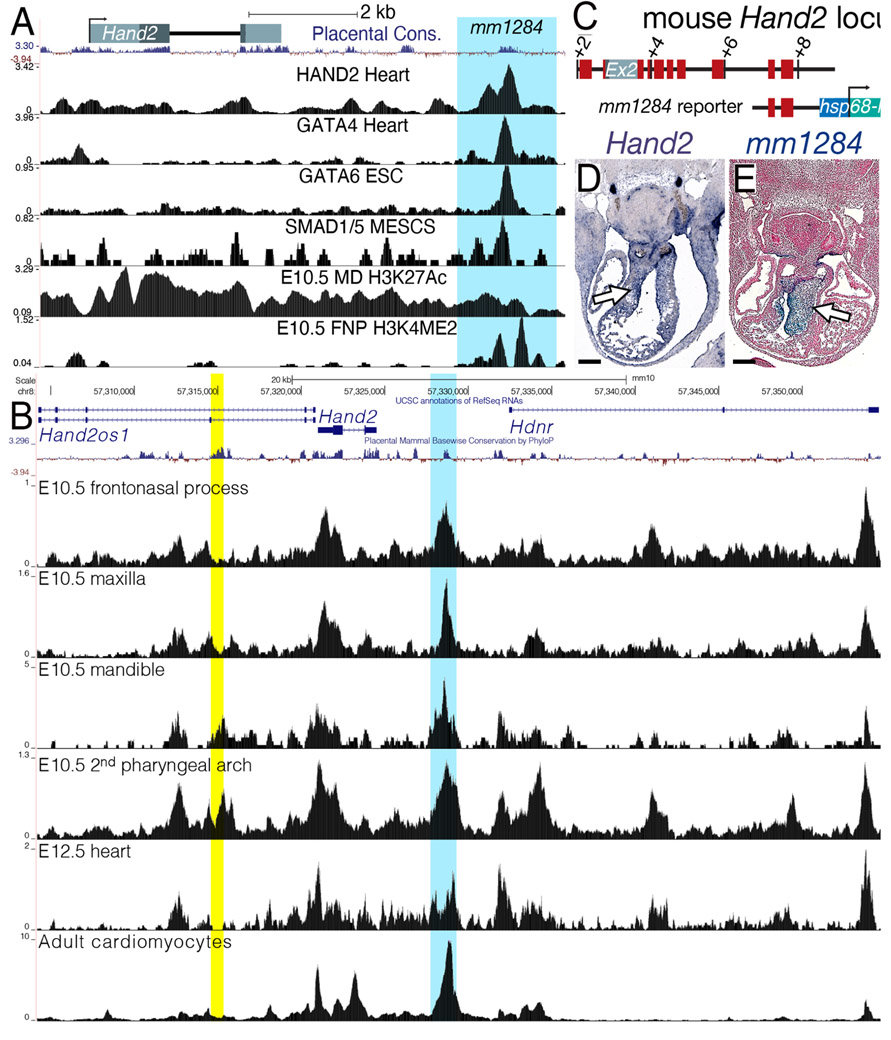
Identification and validation of tissue-specific enhancers is integral to establishing the gene regulatory networks that orchestrate OFT morphogenesis. One approach to isolate such enhancers is to identify conserved regions of DNA which developmentally important transcription factors co-occupy. Our previous findings demonstrate that the Hand1PA/OFT enhancer is activated by SMADs, HAND2, and GATA transcriptional inputs (Vincentz et al., 2016). BMP signaling is also established as being transcriptionally upstream of Hand2 within the developing OFT (Holler et al., 2010; Liu et al., 2004) and reduced Hand2 expression observed from in situ hybridization in Smad4fx/fx; Wnt1-Cre(+) embryos further supports these findings (Fig. 5). Previous studies have identified a Hand2 NCC enhancer sufficient to drive gene expression within the cranial NCCs, but not the cardiac NCCs (Charite et al., 2001). Hand2 RV expression is GATA-dependent (McFadden et al., 2000). ChIP-seq datasets for HAND2 within the embryonic heart, (Laurent et al., 2017) GATA4 (from both heart and HL-1 cells; (He et al., 2014; He et al., 2011), GATA6 (mouse embryonic stem cells; (Wamaitha et al., 2015), and SMAD1/5 (mouse embryonic stem cells; Morikawa et al., 2016) are publicly available. As both Hand1 and Hand2 are well established to respond to BMP/TGFβ transcriptional regulation (Bonilla-Claudio et al., 2012; Howard et al., 2000; Vincentz et al., 2016; Wu and Howard, 2002), HAND factor, and/or GATA factor dependent expression in NCCs (Bonilla-Claudio et al., 2012; Vincentz et al., 2016), we used this regulatory relationship to search for novel Hand2 enhancers that would express within the cNCC of the OFT. We next identified, using the publicly available ChIP-seq datasets, peaks of enrichment for HAND2/SMAD/GATA factor binding that occur within regions marked by histones associated with transcriptionally accessible chromatin, as revealed by ChIP-seq of these chromatin marks in NCCs (Minoux et al., 2017) an evolutionarily conserved Hand2 enhancer that contained conserved cis-elements that matched HAND2/SMAD/GATA factor consensus sequences.
Figure 5. Hand2 expression is lost in both the cardiac neural crest cells and the myocardial cuff of Smad4 conditional knockouts.
In situ hybridization shows that Hand2 expression is lost in both the cNCCs (B, white arrows) and MC (B, black arrows) of NCC-specific (Wnt1-Cre) Smad4 CKOs (n=3/3).
We first employed Cistrome (http://cistrome.org/db/#/) to load the aforementioned ChIP-seq tracks onto the UCSC Genome Browser (http://genome.ucsc.edu). We hypothesized that the SMAD/HAND2/GATA transcriptional synergy that drives Hand1PA/OFT enhancer expression (Vincentz et al., 2016) would also drive a Hand2 autoregulatory mechanism within these tissues. Peaks of enrichment for HAND2, GATA4/6, and SMAD1/5 DNA occupancy overlap within a region, evolutionarily conserved among placentals, located 3’ to the second exon of Hand2 (Fig. 6A). This sequence also displays histone marks that indicate active transcription (Fig. 6A). We next examined ATAC-seq from E10.5 frontonasal process, maxilla, mandible, and 2nd pharyngeal arch (Minoux et al., 2017) E12.5 hearts (Zhou et al., 2017) and adult cardiomyocytes (Monroe et al., 2019) We observed a region of accessible chromatin 3' to Hand2 and 5' to the Hand2 downstream lncRNA, Hdnr. This peak of enrichment appeared in both NCC-derived and cardiac cells. (Yellow shading highlights the mandibular arch enhancer. (Charite et al., 2001)
Figure 6. Identification of an endocardial cushion-specific Hand2 enhancer.
A) Schematic of the mouse Hand2 locus. Peaks of enrichment for HAND2, GATA4/6, and SMAD1/5 overlap with a region, evolutionarily conserved among placentals, that displays histone marks for active transcription in NCCs (H3K4ME2 in the E10.5 fronto-nasal processes (FNP) and H3K27Ac in the mandibular pharyngeal arch (MD). This mm1284 sequence is highlighted in blue. B) ATAC-seq from E10.5 frontonasal process, maxilla, mandible, and 2nd pharyngeal arch (Minoux et al., 2017), E12.5 hearts (Zhou et al., 2017), and adult cardiomyocytes (Monroe et al., 2019) (Yellow shading highlights the Hand2 mandibular arch enhancer. (Charite et al., 2001) C) Schematic of the mm1284+hsp68-lacZ reporter construct. Red blocks denote regions of significant sequence conservation D) Hand2 section in situ hybridization showing expression in the E11.5 OFT cushions (white arrow). E) Section of an X-gal stained E11.5 mm1284+hsp68-lacZ transgenic embryo showing expression in the OFT cushions (white arrow). n = 5/7. Scale bars = 250 μm.
This enhancer, denoted mm1284 (Fig. 6C), has been tested in vivo using a transient transgenic enhancer assay, and the results have been posted to the publicly available VISTA Cardiac Enhancer Browser (http://portal.nersc.gov/dna/RD/heart/index.html). Fitting our hypothesis, this enhancer’s expression overlaps with endogenous Hand2 expression within the OFT (Fig. 6D, white arrow). Reporter expression within the OFT is observed within the endocardial cushions (Fig. 6E, white arrow) in 5 of 7 transgene-positive embryos. Phylogenetic footprinting of this enhancer identified E-box, D-box, SMAD, and GATA cis-elements that exhibit conservation among mammals (Supp. Fig 2). Additionally, human ChIP-seq data sets for GATA4 in iPS-derived cardiomyocytes reveal a peak of enrichment that co-localizes with this conserved enhancer (Ang et al., 2016). These findings support that integration of mutant phenotype, gene expression, and DNA occupancy data can be used effectively to identify novel enhancers that are active specifically in the developing cNCCs.
4. Discussion
OFT defects in newborns constitute a significant cause of morbidity and mortality and are amongst the most highly encountered congenital defects in humans. Here we show that loss-of-function models of Hand1 and Hand2 indicate that these two highly related bHLH transcription factors genetically interact within both the cNCC and SHF. The data reveal OFT morphogenesis cannot tolerate a collective reduction of HAND factor function beyond a critical threshold. This critical threshold of HAND protein reflects both direct DNA occupancy on enhancer sequences of regulated genes as well as the reiteration of other bHLH factors that when unpartnered could exhibit deleterious gain of function effects such as what is observed in Saethre-Chotzen syndrome (Firulli et al., 2005). Given systemic knockout (Srivastava et al., 1997), as well as the NCC- (Holler et al., 2010) and SHF- (Tsuchihashi et al., 2011) conditional ablations of Hand2 are sufficient to disrupt OFT morphogenesis with high penetrance, it is clear that HAND2 plays a more dominant role in this process. This could reflect actual direct transcriptional target efficiencies or simply reflect variation in expression levels. Although SHF Hand1 ablation did not reveal any observable cardiac phenotypes alone (Fig. 1), NCC ablation of Hand1 did reveal a low penetrance of OFT phenotype that include membranous VSDs and AORSA (Fig. 1C and G) revealing that less than a full complement of HAND factors can be deleterious to OFT morphogenesis. Hand1 deletion within both the NCC and MC did not reveal any increase in phenotypic penetrance suggesting that HAND1 is not playing a significant role regulating the gene regulatory networks involved in modulating the cell-cell interactions of these OFT cell types. Interestingly, the observed H1CKONCC OFT defects are not severe enough to be lethal, as H1CKONCC are encountered at expected mendelian frequencies similar to as was reported (Barbosa et al., 2007). This is consistent with GWAS data showing disease numerous HAND1 associated SNPs in humans linked to cardiac defects (Cheng et al., 2012b; Evans1 et al., 2016; Firulli et al., 2019; Li et al., 2017; Sotoodehnia et al., 2010; van Setten et al., 2019; Vincentz et al., 2019; Wang et al., 2017b; Wang et al., 2011a).
Complete loss of HAND factors within the NCC results in a full PTA with 100% penetrance (Fig. 2E and J) and indeed, when Hand2 is knocked out using Hand1eGFP-Cre knock-in allele which recombines within both post-migratory cNCCs and MC, it also results in PTA at partial penetrance (Barnes et al., 2011). Within H1CKONCC;H2CKONCC mutants, the Sema3c-positive population of cNCCs migrate to the OFT, but interaction between these cNCCs and endocardium is disrupted such that endothelial-to-mesenchymal transition and cNCC localization is abnormal.
Although H1CKOsSHF OFTs are normal (Fig. 1), we do observe CHDs when Hand1 is ablated within SHF derivatives on a Hand2 heterozygous gene dosage (Fig. 4). These results suggest that reduction of SHF expressed Hand2 sensitizes OFT morphogenesis to a loss of HAND1 function. Isolated membranous VSDs are observed in SHF-specific H1CKOsSHF;Hand2 het mutants (Fig. 4), revealing a critical SHF HAND factor threshold in addition to the threshold established within the NCC (Fig. 2). The Mef2c-AHF-Cre driver recombines within both the myocardium and the endocardium of the OFT and right ventricle (Verzi et al., 2005). Hand2fx/fx;Mef2c-AHF-Cre(+) CKOs present with PTA (Tsuchihashi et al., 2011). Although conditional ablation of Hand2 within the endocardium is embryonic lethal, these H2CKOs do not exhibit OFT phenotypes (Vandusen et al., 2014). Hand1 lineage does not mark the endocardium (Barnes et al., 2010a), and therefore, we conclude that HAND factor function within the endocardium is unlikely to be critical for OFT morphogenesis with the caveat that HAND2 loss-of function within both the NCC and endocardium collectively was not investigated.
The Hand1PA/OFT enhancer is directly regulated by SMAD/HAND2/GATA transcriptional inputs (Vincentz et al., 2016). Utilizing co-occupancy data of these factors within the Hand2 locus, we predicted a Hand2 cNCC enhancer that exhibits cNCC cis-regulatory activity consistent with Hand2 mRNA expression (Fig. 6). The finding that both Hand genes are likely co-regulated by a trio of transcriptional inputs that allows cNCC co-expression is perhaps not surprising but is confirmation that HAND1 and HAND2 lie directly downstream within the BMP and GATA gene regulatory networks of the OFT. Previous findings show that Hand1 cranial NCC expression is HAND2, BMP, and GATA dependent whereas all Hand2 NCC expression is independent of HAND1 but completely dependent on BMP signaling and GATA (Bonilla-Claudio et al., 2012; Howard et al., 2000; McFadden et al., 2000; Vincentz et al., 2016; Wu and Howard, 2002). This suggests subtle differences in NCC gene regulatory networks where in cranial populations BMP and GATA upregulate HAND2, which is followed by BMP, GATA and HAND2 directly regulating HAND1. In cNCC Hand1 expression is not HAND2 dependent indicating that post-migratory NCC display overlapping but subtly unique transcriptional networks that likely convey the specific transcriptional information for normal development. Collectively, these findings provide evidence that HAND factors are a component of these overlapping yet unique transcriptional inputs that interpret BMP-mediated extracellular signaling cues and GATA transcriptional cues to effect aorticopulmonary septogenesis and OFT morphogenesis within the cNCC.
5. Conclusion
This study helps to resolve why mutations in human HAND1, which are associated with CHDs affecting the OFT, are not directly phenocopied by mutations in mouse. Hand1. SHF loss-of function, on its own, does not cause OFT defects; however, Hand1 NCC deletion does present with low penetrance CHDs. Hand1 loss-of function phenotypes are enhanced by reduction of Hand2 gene dosage in both SHF and NCC cell populations and NCC transcriptional enhancers of both Hand1 and Hand2 have similar transcriptional inputs and through comparison of cranial and cardiac NCC regulation display unique features that help define the difference in the post-migratory fates of these distinct cell populations.
Supplementary Material
Supplemental Figure 1
In situ hybridization showing E12.5 expression of Sema3C in Control and Hand1eGFP-Cre/+; Hand2f/f mutants (H2CKOs). Arrowheads show the position of the myocardial cuff. White arrowheads show the position of the septal bridge and reduced area of Sema3C expression.
Supplemental Figure 2. Nucleotide identity alignment of the Hand2 cNCC enhancer. Conserved DLX / HOX binding sites are grey shaded; GATA cis-elements are green shaded; E-Boxes are light blue shaded; ATF/CREB cis-elements pink shaded; SMAD elements brown shaded; dark blue shade inverted D-box.
Highlights.
HAND1 and HAND2 genetically interact within the cardiac neural crest
HAND1 and HAND2 double mutants reveal a mis-organization of cardiac neural crest
In silico prediction of a Hand2 cardiac neural crest enhancer based on an identified Hand1 enhancer shows conservation in upstream transcriptional regulation inputs.
Acknowledgments
We thank Danny Carney for technical assistance. We thank all the researchers that shared reagents. We thank the Riley Heart Research Center for helpful discussions.
Funding
Infrastructural support at the Herman B Wells Center for Pediatric Research, Riley Children’s Foundation, and the Carrolton Buehl McCulloch Chair of Pediatrics. This work is supported by the NIH P01. HL134599-03, R01 HL145060-02 (ABF). MO is supported by Swiss National Science Foundation grant PCEFP3_186993. The research conducted at the E.O. Lawrence Berkeley National Laboratory was supported by a National Institutes of Health grant R01HG003988 (to L.A.P.) and performed under Department of Energy Contract DE-AC02- 05CH11231, University of California.
Footnotes
Availability of data and materials
All data generated or analyzed in this study are included in this manuscript.
Declaration of competing interest
Authors have no conflict of interest regarding this work.
Ethics statement:
Animal work performed at Indiana School of Medicine (IUSM) was approved by the IUSM Animal Care and Use Committee (IACUC) under protocol 11326. Animal work performed at Lawrence Berkeley National Laboratory (LBNL) was reviewed and approved by the LBNL Animal Welfare Committee. IUSM mice were housed in room IB31. LBNL mice used for this study were housed at the Animal Care Facility (ACF) at LBNL. All mice were monitored daily for food and water intake, and animals were inspected weekly. LBNL mice were also inspected by the Chair of the Animal Welfare and Research Committee and the head of the animal facility in consultation with the veterinary staff on a weekly basis. The IUSM facilities and LBNL ACF is accredited by the American Association for the Accreditation of Laboratory Animal Care (AAALAC). LBNL transgenic mouse assays were performed in Mus musculus FVB background mice.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, Tippens ND, Li M, Narasimha A, Radzinsky E, Moon-Grady AJ, Yu H, Pruitt BL, Snyder MP, Srivastava D, 2016. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 167, 1734–1749 e1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrímsson B, Pennacchio LA, Visel A, 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. . Science, 6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H, 2007. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Developmental Biology 310, 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli B, Conway SJ, Vincentz JW, Firulli AB, 2010a. Analysis of the Hand1 Cell Lineage Reveals Novel Contributions to Cardiovascular,Neural Crest, Extra-Embryonic, and Lateral Mesoderm Derivatives. Dev. Dyn 239, 3086–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, Conway SJ, Vincentz JW, Firulli AB, 2010b. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Developmental dynamics : an official publication of the American Association of Anatomists 239, 3086–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, VanDusen NJ, Morikawa Y, Conway SJ, Cserjesi P, Vincentz JW, Firulli AB, 2011. Hand2 loss-of-function in Hand1-expressing cells Reveals Distinct Roles In Epicardial And Coronary Vessel Development. Circ Res. 108, 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF, 2012. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development 139, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borochowitz Z, Shalev SA, Yehudai I, Bar-el H, Dar H, Tirosh E, 1997. Deletion (4)(q33 --> qter): a case report and review of the literature. J Child Neurol 12, 335–337. [DOI] [PubMed] [Google Scholar]
- Brown CB, Feiner L, Lu MM, Li J, Ma X, Webber AL, Jia L, Raper JA, Epstein JA, 2001. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development 128, 3071–3080. [DOI] [PubMed] [Google Scholar]
- Byatt SA, Baker E, Richards RI, Roberts C, Smith A, 1997. Unbalanced t(4;11)(q32;q23) in a 34-year-old man with manifestations of distal monosomy 11q and trisomy 4q syndromes. Am J Med Genet 70, 357–360. [PubMed] [Google Scholar]
- Charite J, McFadden DG, Merlo G, Levi G, Clouthier DE, Yanagisawa M, Richardson JA, Olson EN, 2001. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes & development 15, 3039–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Lib L, Li Z, Liu M, Yan J, Wang B, Ma X, 2012a. Two novel HAND1 mutations in Chinese patients with ventricular septal defect. Clinica chimica acta; international journal of clinical chemistry 413, 675–677. [DOI] [PubMed] [Google Scholar]
- Evans1 DS, Avery CL, Mike A.Nalls, Guo Li4, Barnard J, Smith EN, Tanaka T, Butler AM, Buxbaum SG, Alonso A, Arking DE, Berenson GS, Bis JC, Buyske S, Carty CL, Chen W, Chung MK, Cummings SR, Deo R, Eaton CB, Fox ER, Heckbert SR, Heiss G, Hindorff LA, Hsueh W-C, Isaacs A, Jamshidi Y, Kerr KF, Liu F, Liu Y, Lohman KK, Magnani JW, Maher JF, Mehra R, Meng YA, Musani SK, Newton-Cheh C, North KE, Psaty BM, Redline S, Rotter JI, Schnabel RB, Schork NJ, Shohet RV, Singleton AB, Smith JD, Soliman EZ, Srinivasan SR, Herman A Taylor J, Wagoner DRV, Wilson JG, Young T, Zhang Z-M, Zonderman AB, Evans MK, Ferrucci L, Murray SS, Tranah GJ, Whitsel EA, Reiner AP, Sotoodehnia N, 2016. Fine-mapping, Novel Loci Identification, and SNP Association Transferability in a Genome-Wide Association Study of QRS Duration in African Americans. Hum Mol Genet 0, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner L, Webber AL, Brown CB, Lu MM, Jia L, Feinstein P, Mombaerts P, Epstein JA, Raper1 JA, 2001. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruptionTargeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development 128, 3061–3070. [DOI] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN, 1998. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nature genetics 18, 266–270. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Fuchs RK, Vincentz JW, Clouthier DE, Firulli AB, 2014. Hand1 phosphoregulation within the distal arch neural crest is essential for craniofacial morphogenesis. Development 141, 3050–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, George RM, Harkin J, Toolan KP, Gao H, Liu Y, Zhang W, Field LJ, Liu Y, Shou W, Payne RM, Rubart-von der Lohe M, Firulli AB, 2019. HAND1 loss-of-function within the embryonic myocardium reveals survivable congenital cardiac defects and adult heart failure. Cardiovasc Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Virshup DE, Conway SJ, Cserjesi P, Laufer E, Firulli AB, 2005. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat. Genet 37, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Redick BA, Conway SJ, Firulli AB, 2007. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. Journal of Biological Chemistry 282, 27536–27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RM, Firulli AB, 2019. Hand Factors in Cardiac Development. Anat Rec (Hoboken) 302, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JR, Huang YF, Wilson KA, Kelly PL, Adams DJ, Gronowicz GA, Clark SH, 2005. Col1a1 promoter-targeted expression of p20 CCAAT enhancer-binding protein beta (C/EBPbeta), a truncated C/EBPbeta isoform, causes osteopenia in transgenic mice. J Biol Chem 280, 8117–8124. [DOI] [PubMed] [Google Scholar]
- He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA, Pu WT, 2014. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun 5, 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT, 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A 108, 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H, 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler KL, Hendershot TJ, Troy SE, Vincentz JW, Firulli AB, Howard MJ, 2010. Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Developmental Biology 341, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H, 2000. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 127, 4073–4081. [DOI] [PubMed] [Google Scholar]
- Kothary RK, Allen ND, Surani MA, 1989. Transgenes as molecular probes of mammalian developmental genetics. Oxf Surv Eukaryot Genes, 145–178. [PubMed] [Google Scholar]
- Laurent F, Girdziusaite A, Gamart J, Barozzi I, Osterwalder M, Akiyama JA, Lincoln J, Lopez-Rios J, Visel A, Zuniga A, Zeller R, 2017. HAND2 Target Gene Regulatory Networks Control Atrioventricular Canal and Cardiac Valve Development. Cell Rep 19, 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang J, Liu XY, Liu H, Shi HY, Yang XX, Li N, Li YJ, Huang RT, Xue S, Qiu XB, Yang YQ, 2017. HAND1 loss-of-function mutation contributes to congenital double outlet right ventricle. Int J Mol Med 39, 711–718. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF, 2004. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc Natl Acad Sci U S A 101, 4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxan G, D'Amato G, MacGrogan D, de la Pompa JL, 2016. Endocardial Notch Signaling in Cardiac Development and Disease. Circ Res 118, e1–e18. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN, 2005. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189–201. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN, 2000. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development 127, 5331–5341. [DOI] [PubMed] [Google Scholar]
- Minoux M, Holwerda S, Vitobello A, Kitazawa T, Kohler H, Stadler MB, Rijli FM, 2017. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science 355, eaal2913. [DOI] [PubMed] [Google Scholar]
- Monroe TO, Hill MC, Morikawa Y, Leach JP, Heallen T, Cao S, Krijger PHL, de Laat W, Wehrens XHT, Rodney GG, Martin JF, 2019. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev Cell 48, 765–779 e767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Mizutani A, Kawasaki N, Holmborn K, Sundqvist A, Tsutsumi S, Watabe T, Aburatani H, Heldin CH, Miyazono K, 2016. BMP Sustains Embryonic Stem Cell Self-Renewal through Distinct Functions of Different Kruppel-like Factors. Stem Cell Reports 6, 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P, 2008. Cardiac Neural Crest Expression of Hand2 Regulates Outflow and Second Heart Field Development. Circ. Res. 103, 1422–1429. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, D'Autreaux F, Gershon MD, Cserjesi P, 2007. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Developmental Biology 307, 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres J, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB, 2014. Heart Disease and Stroke Statistics-2015 Update: A Report From the American Heart Association. Circulation. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, 2010. Second lineage of heart forming region provides new understanding of conotruncal heart defects. Congenital anomalies 50, 8–14. [DOI] [PubMed] [Google Scholar]
- Plein A, Calmont A, Fantin A, Denti L, Anderson NA, Scambler PJ, Ruhrberg C, 2015. Neural crest-derived SEMA3C activates endothelial NRP1 for cardiac outflow tract septation. J Clin Invest 125, 2661–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter O, Haase H, Schulte HD, Lange PE, Morano I, 1999. Remodeling of the hypertrophied human myocardium by cardiac bHLH transcription factors. J Cell Biochem 74, 551–561. [DOI] [PubMed] [Google Scholar]
- Schwarz Q, Ruhrberg C, 2010. Neuropilin, you gotta let me know: should I stay or should I go? Cell adhesion & migration 4, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Li XF, Shen AD, Wang Q, Liu CX, Guo YJ, Song ZJ, Li ZZ, 2010. Transcription factor HAND2 mutations in sporadic Chinese patients with congenital heart disease. Chin Med J (Engl) 123, 1623–1627. [PubMed] [Google Scholar]
- Soriano P, 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, van der Harst P, Muller M, Eijgelsheim M, Alonso A, Hicks AA, Padmanabhan S, Hayward C, Smith AV, Polasek O, Giovannone S, Fu J, Magnani JW, Marciante KD, Pfeufer A, Gharib SA, Teumer A, Li M, Bis JC, Rivadeneira F, Aspelund T, Kottgen A, Johnson T, Rice K, Sie MP, Wang YA, Klopp N, Fuchsberger C, Wild SH, Mateo Leach I, Estrada K, Volker U, Wright AF, Asselbergs FW, Qu J, Chakravarti A, Sinner MF, Kors JA, Petersmann A, Harris TB, Soliman EZ, Munroe PB, Psaty BM, Oostra BA, Cupples LA, Perz S, de Boer RA, Uitterlinden AG, Volzke H, Spector TD, Liu FY, Boerwinkle E, Dominiczak AF, Rotter JI, van Herpen G, Levy D, Wichmann HE, van Gilst WH, Witteman JC, Kroemer HK, Kao WH, Heckbert SR, Meitinger T, Hofman A, Campbell H, Folsom AR, van Veldhuisen DJ, Schwienbacher C, O'Donnell CJ, Volpato CB, Caulfield MJ, Connell JM, Launer L, Lu X, Franke L, Fehrmann RS, te Meerman G, Groen HJ, Weersma RK, van den Berg LH, Wijmenga C, Ophoff RA, Navis G, Rudan I, Snieder H, Wilson JF, Pramstaller PP, Siscovick DS, Wang TJ, Gudnason V, van Duijn CM, Felix SB, Fishman GI, Jamshidi Y, Stricker BH, Samani NJ, Kaab S, Arking DE, 2010. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet 42, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN, 1995. A subclass of bHLH proteins required for cardiac morphogenesis. Science 270, 1995–1999. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN, 1997. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nature genetics 16, 154–160. [DOI] [PubMed] [Google Scholar]
- Starkovich M, Lalani SR, Mercer CL, Scott DA, 2016. Chromosome 5q33 deletions associated with congenital heart defects. Am J Med Genet A 170, 3338–3342. [DOI] [PubMed] [Google Scholar]
- Sugishita Y, Watanabe M, Fisher SA, 2004. The development of the embryonic outflow tract provides novel insights into cardiac differentiation and remodeling. Trends Cardiovasc Med 14, 235–241. [DOI] [PubMed] [Google Scholar]
- Tamura M, Hosoya M, Fujita M, Iida T, Amano T, Maeno A, Kataoka T, Otsuka T, Tanaka S, Tomizawa S, Shiroishi T, 2013. Overdosage of Hand2 causes limb and heart defects in the human chromosomal disorder partial trisomy distal 4q. Hum Mol Genet 22, 2471–2481. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi T, Maeda J, Shin CH, Ivey KN, Black BL, Olson EN, Yamagishi H, Srivastava D, 2011. Hand2 function in second heart field progenitors is essential for cardiogenesis. Developmental Biology 351, 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Setten J, Verweij N, Mbarek H, Niemeijer MN, Trompet S, Arking DE, Brody JA, Gandin I, Grarup N, Hall LM, Hemerich D, Lyytikainen LP, Mei H, Muller-Nurasyid M, Prins BP, Robino A, Smith AV, Warren HR, Asselbergs FW, Boomsma DI, Caulfield MJ, Eijgelsheim M, Ford I, Hansen T, Harris TB, Heckbert SR, Hottenga JJ, Iorio A, Kors JA, Linneberg A, MacFarlane PW, Meitinger T, Nelson CP, Raitakari OT, Silva Aldana CT, Sinagra G, Sinner M, Soliman EZ, Stoll M, Uitterlinden A, van Duijn CM, Waldenberger M, Alonso A, Gasparini P, Gudnason V, Jamshidi Y, Kaab S, Kanters JK, Lehtimaki T, Munroe PB, Peters A, Samani NJ, Sotoodehnia N, Ulivi S, Wilson JG, de Geus EJC, Jukema JW, Stricker B, van der Harst P, de Bakker PIW, Isaacs A, 2019. Genome-wide association meta-analysis of 30,000 samples identifies seven novel loci for quantitative ECG traits. Eur J Hum Genet 27, 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandusen NJ, Vincentz JW, Firulli BA, Howard MJ, Rubart M, Firulli AB, 2014. Loss of Hand2 in a population of Periostin lineage cells results in pronounced bradycardia and neonatal death. Dev Biol 388, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL, 2005. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Developmental Biology 287, 134–145. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Firulli AB, 2011. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol. 91, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Casasnovas JJ, Barnes RM, Que J, Clouthier DE, Wang J, Firulli AB, 2016. Exclusion of Dlx5/6 expression from the distal-most mandibular arches enables BMP-mediated specification of the distal cap. Proc Natl Acad Sci U S A 113 7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Firulli BA, Lin A, Spicer DB, Howard MJ, Firulli AB, 2013. Twist1 controls a cell specification switch governing cell fate decisions within the cardiac neural crest. PLoS Genet. 9, e1003405 1003401–1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Firulli BA, Toolan KP, Arking DE, Sotoodehnia N, Wan J, Chen PS, de Gier-de Vries C, Christoffels VM, Rubart-von der Lohe M, Firulli AB, 2019. Variation in a Left Ventricle-Specific Hand1 Enhancer Impairs GATA Transcription Factor Binding and Disrupts Conduction System Development and Function. Circ Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Toolan KP, Zhang W, Firulli AB, 2017. Hand factor ablation causes defective left ventricular chamber development and compromised adult cardiac function. PLoS Genet 13, e1006922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA, 2007. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35, D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamaitha SE, del Valle I, Cho LT, Wei Y, Fogarty NM, Blakeley P, Sherwood RI, Ji H, Niakan KK, 2015. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev 29, 1239–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou B, Kong X, 2017a. Zhong Nan Da Xue Xue Bao Yi Xue Ban 42, 1383–1388. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu XQ, Guo YH, Gu JY, Xu JH, Li YJ, Li N, Yang XX, Yang YQ, 2017b. HAND1 Loss-of-Function Mutation Causes Tetralogy of Fallot. Pediatr Cardiol 38, 547–557. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu Y, Chen H, Yin M, Yu T, Fu Q, 2011a. Investigation of somatic NKX2-5, GATA4 and HAND1 mutations in patients with tetralogy of Fallot. Pathology 43, 322–326. [DOI] [PubMed] [Google Scholar]
- Wang Z, Huang G, Yan Q, Wang L, Zhu J, Lu Y, Li P, Cheng H, Ma MJ, Walker BF, Allen PW, 2011b. Inflammatory monomorphic undifferentiated sarcoma with distinct clinical and pathological features: a 'new' entity? Pathology 43, 48–53. [DOI] [PubMed] [Google Scholar]
- Wu X, Howard MJ, 2002. Transcripts encoding HAND genes are differentially expressed and regulated by BMP4 and GDNF in developing avian gut. Gene Expression 10, 279–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li C, Herrera PL, Deng CX, 2002. Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–81. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Kelly RG, 2012. New developments in the second heart field. Differentiation 84, 17–24. [DOI] [PubMed] [Google Scholar]
- Zhou P, Gu F, Zhang L, Akerberg BN, Ma Q, Li K, He A, Lin Z, Stevens SM, Zhou B, Pu WT, 2017. Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
In situ hybridization showing E12.5 expression of Sema3C in Control and Hand1eGFP-Cre/+; Hand2f/f mutants (H2CKOs). Arrowheads show the position of the myocardial cuff. White arrowheads show the position of the septal bridge and reduced area of Sema3C expression.
Supplemental Figure 2. Nucleotide identity alignment of the Hand2 cNCC enhancer. Conserved DLX / HOX binding sites are grey shaded; GATA cis-elements are green shaded; E-Boxes are light blue shaded; ATF/CREB cis-elements pink shaded; SMAD elements brown shaded; dark blue shade inverted D-box.