Abstract
Triple-negative breast cancer (TNBC) is an aggressive subtype, with a peak recurrence rate within the first few years post diagnosis. Few targeted therapies are available to treat this breast cancer (BC) subtype, defined by the lack of estrogen and progesterone receptors (ER and PR) and without amplification of human epidermal growth factor receptor 2 (HER2). While cell cycle cyclin dependent kinase (CDK) 4/6 inhibitors are approved for treatment of ER-positive BC, they have not proven effective as monotherapy in patients with TNBC. The androgen receptor (AR) has emerged as a therapeutic target in a subset of TNBC and with significant clinical benefit observed in multiple trials. The purpose of this study was to investigate the pre-clinical activity of the CDK4/6 inhibitor abemaciclib in combination with an agent that targets both androgen biosynthesis and AR activity, seviteronel, using TNBC cell lines expressing high AR, cell line xenografts and an AR positive, androgen-responsive TNBC patient-derived xenograft (PDX). Single cell RNA-sequencing demonstrated heterogeneity in AR levels, even in a highly AR+ cell line, and identified cell cycle pathway activation in ARHigh versus ARLow expressing cells. Combination treatment with the cell cycle CDK4/6 inhibitor abemaciclib and seviteronel showed synergy in an AR+ TNBC model compared to each drug alone.
INTRODUCTION
Breast cancer (BC) is the most common cancer among women in the United States, and approximately 1 in 8 women will be diagnosed with breast cancer within their lifetime [1]. Breast cancers can be divided into three main subtypes: estrogen receptor alpha (ERα)-positive, human epidermal growth factor receptor 2 (HER2)-positive or triple-negative (TNBC); and treatment regimens are determined based on the subtype at diagnosis [2]. TNBCs, which account for approximately 15–20% of diagnosed BC cases [2], lack expression of ERα and progesterone receptor (PR) and do not have amplified HER2. TNBC is notoriously difficult to treat, particularly once chemoresistant metastatic disease develops, and few targeted therapies are available for the majority of patients. Additionally, TNBC is the most aggressive BC subtype with a high rate of metastases in the first three years and a low 5-year survival rate [3]. Thus, development of additional therapeutic strategies for TNBC is an important avenue of study.
The androgen receptor (AR) has emerged as a potential therapeutic target in BC. More than 80% of all BCs and up to 50% of TNBCs overexpress AR [4–9]. In TNBC, AR supports cancer stem cell-like properties including anchorage-independent survival, mammosphere formation and tumor initiation [10]. AR activation also promotes cell survival through regulation of the cell cycle [11]. Androgen withdrawal results in G1 arrest [12], and AR-dependent gene expression is highest during G1 and decreases throughout the cell cycle [13]. In fact, AR+ TNBC cells are highly sensitive to cyclin dependent kinase (CDK) 4/6 inhibition due to their dependence on these proteins to progress through the cell cycle [14]. Combining standard of care chemotherapy with AR antagonists and/or CDK4/6 inhibitors may prove effective in this patient population and this strategy is being employed in clinical trials with older generation drugs. Given the recent advancements in both AR antagonist and CDK4/6 inhibitor drug design, newer generation inhibitors may prove even more effective.
Anti-androgens have been under investigation in BC for over a decade [15, 16]. First generation anti-androgens, such as bicalutamide (Bic), function as competitive inhibitors that block AR transcriptional activity. The second-generation anti-androgen enzalutamide (Enza) is a competitive inhibitor that blocks nuclear localization of AR, and consequently has less partial agonist activity than first generation anti-androgens [17, 18]. Both of these AR antagonists are well tolerated in patients and have shown beneficial clinical activity in TNBC, with subsets of patients showing prolonged stable disease [19–21].
In order to maximize response rates of AR+ TNBC to anti-androgens, studies are ongoing to determine the best diagnostic cutoff levels of AR expression, identify gene expression signatures predictive of AR dependency [22], and evaluate newer generation AR targeted therapies that block AR activity through the inhibition of androgen biosynthesis. One such therapy is abiraterone acetate, a CYP17 hydroxylase inhibitor that blocks androgen synthesis and has shown promise in the clinical setting against AR+ TNBC [23]. However, a consequence of inhibiting CYP17 hydroxylase is the interruption of cortisol production, which necessitates supplementation with the corticosteroid prednisone [23, 24]. Seviteronel (Sevi; INO-464) is a dual CYP17 lyase inhibitor and AR antagonist (Supplemental Fig. 1), with preclinical activity in multiple tumor types including castration-resistant prostate cancer (CRPC), ER+ BC, TNBC and glioblastoma (GBM) [25–30]. It has undergone clinical investigation in CRPC, BC [31, 32] and GBM (NCT03600467). Since Sevi preferentially inhibits CYP17 lyase over hydroxylase, there is less impact on cortisol production compared to hydroxylase inhibitors such as abiraterone [33]. Thus, Sevi’s dual mechanism of action of CYP17 lyase inhibition and AR antagonism combines the actions of Enza and abiraterone, potentially providing increased efficacy in treating AR+ TNBCs. The purpose of this study was to investigate the pre-clinical activity of Sevi in AR+ TNBC and further explore the efficacy of targeting both androgen biosynthesis and AR activity in combination with CDK4/6 inhibition [34].
MATERIALS AND METHODS
Cell culture and reagents.
Human TNBC cell lines were cultured in 5% carbon dioxide (CO2). Luminal AR MDA-MB-453 cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD) and maintained in DMEM with 10% FBS. SUM159PT cells, expressing moderate levels of AR [35], were obtained in 2013 from the University of Colorado Cancer Center (UCCC) Tissue Culture Core (Aurora, CO) and maintained in Ham’s/F-12 with 5% FBS, 1% HEPES, 1 μg/mL hydrocortisone and 5 μg/mL insulin. Only cells of under ten passages were used in this study. All cell lines were routinely tested for mycoplasma contamination, and human cell lines were authenticated in 2017 by short tandem repeat analysis in the UCCC Tissue Culture Core. The androgen dihydrotestosterone (DHT; Sigma-Aldrich Corporation, St. Louis, MO) used for in vitro experiments was diluted in ethanol (EtOH). The AR and CYP17 lyase inhibitor seviteronel (Sevi) was provided by Innocrin Pharmaceuticals, Inc. (Durham, NC). The CDK4/6 inhibitor Abemaciclib (Abem) was purchased from Selleck Chemical LLC. (Houston, TX). All drugs were diluted in dimethyl sulfoxide (DMSO).
Drug sensitivity assays.
Cells were plated in 96-well plates in quadruplicate or quintuplicate, and treated with either DMSO or increasing concentrations of Sevi or Abem. After 5 days of drug treatment cells were fixed with 10% formalin, stained with 0.1% crystal violet dye, and dye was then solubilized with 10% acetic acid. Absorbance was measured at 570 nm. Data are presented as percent cell growth and normalized to the mean absorbance of DMSO-treated cells. Synergy was calculated with CalcuSyn Software (Biosoft Inc.) using mean percent growth inhibition values from crystal violet cell viability assays. CalcuSyn software uses the Median Effect method to determine synergy where a combination index (CI) < 0.9 indicates synergy, CI = 0.9–1.1 indicates additivity, and CI > 1.1 indicates antagonism [36].
In vivo preclinical models.
All in vivo experiments were performed in accordance with NIH Guidelines of Care and Use of Laboratory Animals. Mice were euthanized by CO2 inhalation followed by cervical dislocation. Tissue was immediately frozen whole in liquid nitrogen for RNA analysis or fixed in 10% buffered formalin for histological analysis. Sevi experiments were limited to 4 weeks since rodents experience induction metabolism in which Sevi is degraded by activated liver enzymes following 4 weeks of treatment [37]. Xenograft studies. 1×106 MDA-MB-453 cells were bilaterally, orthotopically injected into the mammary fat pads of cycling female nu/nu mice. There was no hormone supplementation in this experiment. When tumors reached a size of approximately 100 mm3, mice were randomized and matched into treatment groups (n=5–12 mice per group). Varying doses of Sevi and/or Abem were administered by oral gavage daily for 4 weeks. Tumors were measured weekly by caliper. Patient-derived xenograft (PDX) studies. HCI-009 is a TNBC PDX originally developed in the laboratory of Dr. Alana Welm [38] and propagated in immunodeficient NOD/SCID/gamma (NSG) mice. In the DHT experiment (n=8 mice per group), cycling female NSG mice were implanted with cellulose control or slow release 8mg DHT pellets, prepared in Dow Corning silastic tubing (Fisher Scientific, Hampton, NH). PDX tumor portions were then bilaterally, orthotopically injected into the mammary fat pads, and tumors were measured weekly by caliper for 9 weeks. In the Sevi experiment, PDX tumor portions were bilaterally, orthotopically injected into the mammary fat pads of cycling female NSG mice. When tumors reached a size of approximately 100 mm3, mice were randomized and matched into treatment groups (n=4–5 mice per group). 150mg/kg/day Sevi was administered daily by oral gavage for 2 weeks. Tumors were measured weekly by caliper.
Histology.
For the analysis of MDA-MB-453 cultured cells, cells were fixed in 10% buffered formalin and pelleted in Histogel from ThermoFisher Scientific Inc. (Waltham, MA). The UC Denver Tissue Biobanking and Processing Core performed all tissue and cell processing and paraffin embedding. Five-micrometer sections of formalin-fixed paraffin-embedded (FFPE) samples were used for analyses. Immunohistochemistry (IHC). Sections were deparaffinized in a series of xylenes and ethanols, and antigens were heat-retrieved in either 10mM citrate buffer pH 6.0 or 10mM Tris, 1mM EDTA pH 9.0. Antibodies used included the following: rabbit monoclonal antibody specific for AR (#200R clone SP107, Cell Marque), rabbit polyclonal antibody specific for FKBP5 (#8245, Cell Signaling Technology), rabbit polyclonal antibody specific for PSA (#A0562, Agilent Technologies Inc., Santa Clara, CA), goat polyclonal antibody specific for GDF15 (#AF957, R&D Systems) and rabbit polyclonal antibody specific for CHI3L1 (#ab77528, Abcam). Tris-buffered saline with 0.05% Tween 20 was used for all washes. AR, FKBP5, PSA, and CHI3L1 antibodies were detected with EnVision-HRP anti-rabbit polymer (#K4003, Agilent), while the GDF15 antibody was detected with a biotinylated donkey anti-goat secondary antibody (#705–065-147, Jackson ImmunoResearch Laboratories, Inc.) followed by streptavidin-HRP (#P0397, Agilent). Representative images were taken using a BX40 microscope (Olympus, Center Valley, PA) with a SPOT Insight Mosaic 4.2 camera and software (Diagnostic Instruments, Inc., Sterling Heights, MI). Expression levels were scored visually by an experienced histotechnician for the entire sample and presented as an IHC score, calculated by multiplying the average staining intensity by the percentage of positive cells. Tumor necrosis. Mammary tumor sections were H&E stained and analyzed for necrosis by quantifying by a board-certified veterinary pathologist. Data was presented as the percentage of each tumor that was composed of necrotic dead cells.
Bioinformatics.
Bulk RNA-sequencing.
RNA isolated from frozen HCI-009 PDX mammary tumors was used to prepare Illumina HiSeq libraries according to manufacturer’s instructions for the TruSeq Stranded RNA kit (Illumina Inc, San Diego, CA). The mRNA template libraries were then sequenced as single pass 50bp reads on the Illumina HiSeq4000 platform at the University of Colorado’s Genomics and Sequencing Core Facility. Derived sequences were analyzed by applying a custom computational pipeline consisting of the open-source gSNAP, Cufflinks, and R for sequence alignment and ascertainment of differential gene expression [39]. In short, reads generated were mapped to the human genome (GRCh38) by gSNAP [40], expression (FPKM) derived by Cufflinks [41], and differential expression analyzed with ANOVA in R. Differentially expressed genes (FDR < 0.05 for DHT and p < 0.05 for Sevi, due to a lack of statistical power) with a minimum expression ratio of 1.15 were used for downstream analyses. These data are available in the Gene Expression Omnibus (GEO) database as GSE152246 for the DHT experiment (Supplemental Fig. 2A) and GSE152318 for the Sevi experiment (Supplemental Fig. 2B).
Single cell RNA-sequencing.
500,000 MDA-MB-453 cells were plated in two T25 flasks. After 48 hours cells were trypsinized, washed and prepared for scRNAseq analysis via the 10X Genomics platform and Illumina NovSeq 6000 platforms [42] at the University of Colorado’s Genomics and Sequencing Core Facility using the 10X Genomics Sample Preparation Protocol. 3000 cells were sequenced with a minimum read-depth of 75,000 reads/cell. Read mapping and expression quantification was performed using a combination of the 10X Cellranger pipeline and custom analytic scripts. Briefly, single-cell reads were mapped to the human genome (GRCh38) and assigned to genes using the standard CellRanger pipeline. The R packages Monocle [43–45] and Seurat [46] were used for differential expression. The cell cycle heatmap, using 10 cell aggregates, was generated using the Morpheus Software Package. These data are available in the GEO database as GSE152315. Downstream RNAseq analyses were performed using Ingenuity Pathway Analysis (IPA, Qiagen, Hilden, Germany) and gene set enrichment analyses (GSEA) [47, 48]. Overlap between significantly differentially expressed genes from the MDA-MB-453 scRNAseq and HCI-009 Sevi experiments were compared using the web application BioVenn [49].
Statistical analysis.
Statistically significant differences (p values < 0.05) were calculated using the GraphPad Prism 7.0 statistical program. Single variable comparisons were made with two-tailed unpaired t-tests. A repeated measure two-way ANOVA was used for in vivo tumor data analysis.
RESULTS
Seviteronel effectively limits AR activity in TNBC.
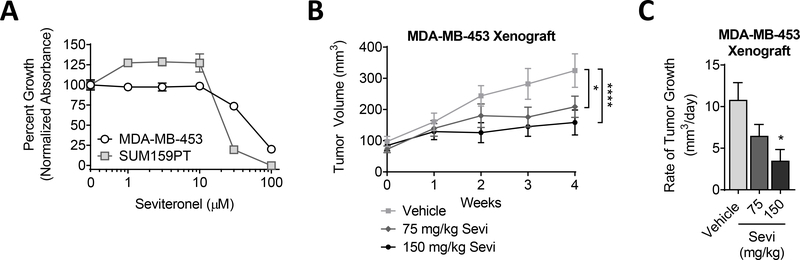
Sevi has been shown to target AR transcriptional activity in prostate and BC with preclinical activity similar to previously approved AR-targeted therapies [25–27, 50]. ChIP qPCR indicated that Sevi significantly blocks DHT-induced binding of AR to the promoters of aquaporin 3 (AQP3), transmembrane serine protease 2 (TMPRSS2), and fatty acid synthase (FASN) [50]. Similarly, Sevi blocked the DHT-induced transcription of multiple AR-target genes [50]. In both cases, Sevi inhibited AR activity with an efficiency similar to that observed with the second-generation AR-antagonist Enza [50]. Moreover, a recent publication found that Sevi sensitizes TNBC cells to standard radiation [51]. To determine the effect of Sevi on TNBC proliferation, AR+ MDA-MB-453 cells were treated with increasing concentrations of Sevi, resulting in the inhibition of cell proliferation at concentrations greater than 10μM (Fig. 1A). This was further confirmed in the TNBC cell line SUM159PT (Fig. 1A), which express moderate levels of AR [10]. Given the ability of Sevi to inhibit androgen synthesis in addition to inhibiting AR directly [50], as compared to other AR antagonists, we examined its effectiveness against TNBC tumor growth in vivo using a xenograft model. Normal cycling female mice were used as a more clinically relevant model of female patients with low levels of androgen synthesis. Sevi significantly inhibited tumor volume and growth rate in a dose-dependent manner (Fig. 1B–C). Together these data indicate that Sevi effectively targets the pro-proliferative activities of AR in TNBC cell lines in culture and in vivo.
Figure 1. TNBC sensitivity to Sevi in a preclinical models.
(A) TNBC cell lines were cultured in full serum media. Cell viability was determined by crystal violet assay after a 5-day exposure to DMSO (0) or increasing concentrations of Sevi. Data was normalized to the mean absorbance of DMSO-treated cells. Mean ± standard deviation. (B) MDA-MB-453 cells were injected into the mammary fat pads of cycling female nu/nu mice. Supplemental DHT (dihydrotestosterone) was not given in this experiment. Sevi was administered daily. Tumor volume was measured by calipers and (C) the rate of tumor growth per day over four weeks was calculated. Mean ± standard error of the mean; * p<0.05, **** p<0.0001.
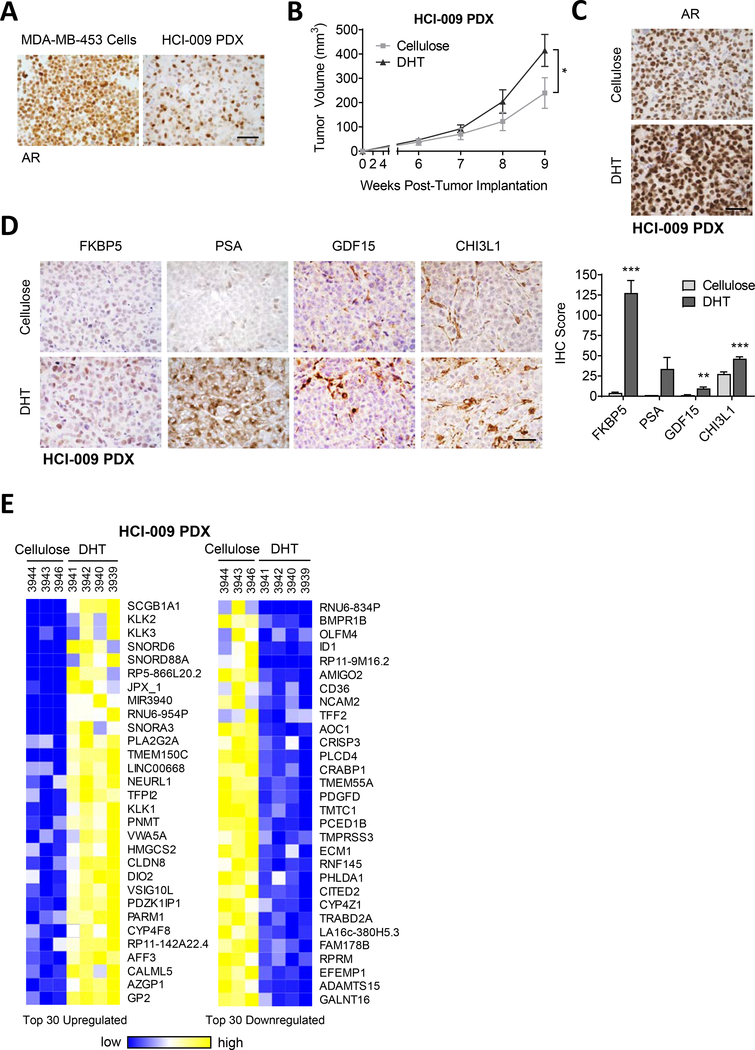
Patient-derived xenografts (PDXs) are preclinical models that more closely mimic patient tumor heterogeneity and clinical responses to therapeutic agents than traditional cultured cell lines [52]. HCI-009 is an AR+ TNBC PDX with a distinctly heterogeneous AR staining pattern (Fig. 2A). In mice supplemented with AR agonist dihydrotestosterone (DHT) HCI-009 PDX tumor growth was significantly increased (Fig. 2B) and AR protein levels were upregulated (Fig. 2C), indicating that this PDX model is androgen-responsive. To evaluate the transcriptional activity of AR in these tumors, known AR-regulated genes were examined for upregulation at the protein level by immunohistochemistry (IHC). Expression of the AR-regulated proteins FKBP prolyl isomerase 5 (FKBP5), prostate specific antigen (PSA), growth differentiation factor 15 (GDF15) and chitinase 3-like 1 (CHI3L1) was significantly upregulated in HCI-009 PDX tumors from mice supplemented with DHT (Fig. 2D). Similarly, RNA-sequencing (RNAseq) of HCI-009 tumors from mice supplemented with DHT (Supplemental Fig. 2A) showed upregulation of known AR-regulated genes, such as KLK3, the gene that encodes PSA (Fig. 2E, Supplemental Table 1) and many others such as KLK1 (kallikrein 1), KLK2, AZGP1 (zinc-alpha-2-glycoprotein 1), PARM1 (prostate androgen-regulated mucin-like protein 1), SCGB1A1 (secretoglobin family 1A member 1), PLA2G2A (phospholipase A2 group IIA), CLDN8 (claudin 8), and HMGCS2 (3-hydroxy-3-methylglutaryl-coA synthase 2).
Figure 2. Androgen responsive TNBC PDX model.
(A) TNBC PDX HCI-009 was propagated in NSG mice. Tumor tissue was stained for AR by IHC. AR staining in MDA-MB-453 cells is shown for comparison. (B-E) HCI-009 PDX tumor portions were injected into the mammary fat pads of female NSG mice supplemented with cellulose, as a control, or DHT (dihydrotestosterone; 8mg/pellet). (B) Tumor volume was measured with calipers. (C) AR was stained by IHC. (D) IHC staining of known AR-regulated genes. Quantification of staining is presented as an IHC score, combining cell positivity and staining intensity. (E) Shown are the top 30 up- and downregulated genes identified by RNAseq in HCI-009 PDX tumors from mice treated with DHT. Mean ± standard error of the mean; * p<0.05, ** p<0.01, *** p<0.001; scale bars = 50μm.
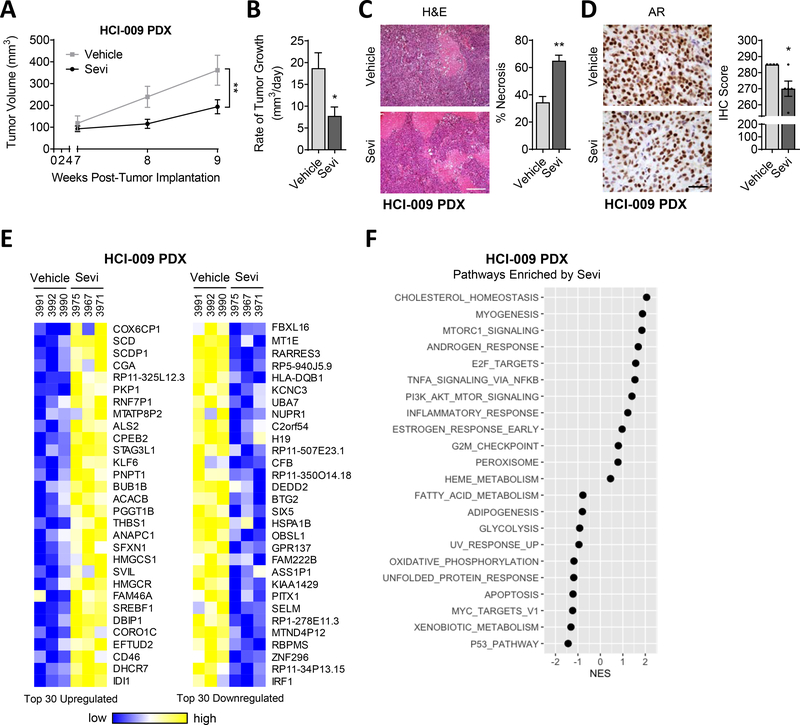
After confirming that HCI-009 PDX tumors have active AR and are growth stimulated by exogenous androgen, we tested responsiveness to Sevi in mice that did not have exogenous androgen supplementation. Mice with HCI-009 PDX mammary tumors were treated with 150mg/kg/day Sevi, and tumor volume and rate of growth were significantly decreased by Sevi treatment (Fig. 3A–B). Sevi increased tumor necrosis (Fig. 3C) and significantly decreased AR protein levels (Fig. 3D). RNAseq showed significant changes in gene expression following Sevi treatment, inducing a distinctive Sevi gene signature that included known AR-regulated genes (Fig. 3E, Supplemental Fig. 2B, Supplemental Table 2). Pathway analysis performed on RNAseq data further demonstrated that while Sevi affects several biological pathways, the Androgen Response pathway (normalized enrichment score, NES=1.69) was one of the top altered pathways (Fig. 3F). Overall, these data support the function of Sevi as a potent inhibitor of AR and tumor growth in this AR+ TNBC PDX.
Figure 3. AR-positive TNBC PDX sensitivity to AR-targeted therapy.
HCI-009 PDX tumor portions were injected into the mammary fat pads of female NSG mice. Mice were treated daily with vehicle or 150mg/kg/day Sevi (seviteronel). (A) Tumor volume was measured with calipers and (B) the rate of tumor growth per day over two weeks was calculated. (C) The percentage of tumor necrosis and (D) AR positive cells per tumor were quantified; representative images of H&E staining, scale bars = 200μm and 50μm, respectively. (E) Shown are the top 30 up- and downregulated genes and (F) most highly correlated biological pathways identified by Gene Set Enrichment (GSEA) in RNAseq data from HCI-009 PDX tumors from mice treated with Sevi. Mean ± standard error of the mean; * p<0.05, ** p<0.01.
AR is a positive regulator of the cell cycle
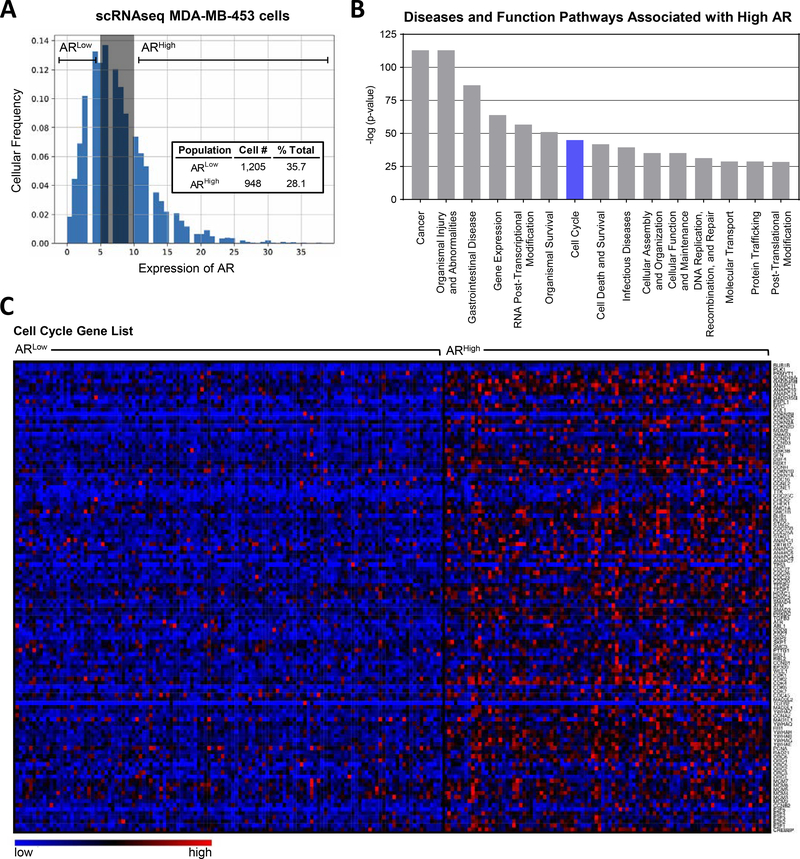
While MDA-MB-453 cells are highly AR+, IHC demonstrated heterogeneity in cell-specific AR levels within this cell line (Fig. 2A). To examine gene expression associated with varying levels of AR, single-cell RNAseq (scRNAseq) was performed on MDA-MB-453 cells cultured in 10% serum containing endogenous levels of androgens (Supplemental Fig. 3A). MDA-MD-453 cells were divided into ARHigh (1,205 cells) and ARLow (948 cells) populations based on the degree of AR mRNA expression (Fig. 4A, Supplemental Tables 3 and 4 for gene lists). Pathway analysis on genes associated with ARHigh versus ARLow levels indicated that the Cell Cycle (z=4.12, p=1.13E-45) was one of the top pathways associated with high AR expression (Fig. 4B, Supplemental Fig. 4B). A large percentage (54%) of cell cycle related genes were upregulated in high AR expressing MDA-MB-453 cells (Fig. 4C, Supplemental Table 5). Interestingly, when the genes associated with ARHigh MDA-MB-453 cells (Fig. 3) were compared to the genes altered in HCI-009 PDX tumors treated with Sevi approximately 65% of genes altered by Sevi overlap with the MDA-MB-453 ARHigh related gene signature (Supplemental Fig. 4, Supplemental Table 6), providing further evidence to support the AR-directed effects of Sevi in TNBC, although non-AR-directed Sevi effects cannot be ruled out.
Figure 4. The association between high AR expression and cell cycle pathway activation.
scRNAseq was performed on untreated MDA-MB-453 cells cultured in full serum media. Cells were divided into groups based on their expression of AR, ARLow and ARHigh. (A) Histogram of the distribution of AR expression in single MDA-MB-453 cells including AR level group determination. (B) Ingenuity pathway analysis was performed on genes differentially expressed between ARLow and ARHigh groups. Shown are the top 15 most significantly altered disease and function pathways. (C) Differential expression of genes involved in cell cycle indicating cell cycle activation associated with high AR expression, shown as 10 cell aggregates.
Combination targeting of AR and CDK4/6 is effective in AR+ TNBC
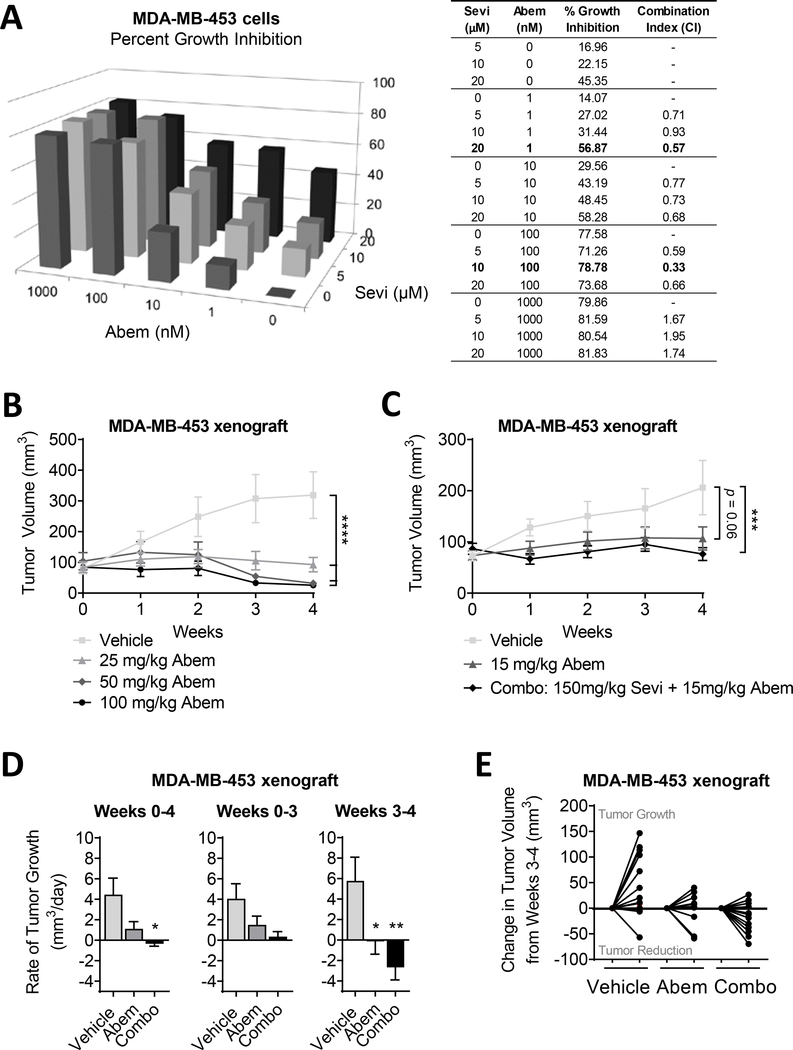
Based on the link between AR levels and cell cycle activation in TNBC cells (Fig. 4), we aimed to evaluate dual targeting of AR and the cell cycle as a potential treatment strategy for AR+ TNBC. A previous investigation of combination therapy, using different early generation inhibitors, showed some promise clinically (NCT02605486; pablociclib with bicalutamide in AR+ metastatic BC patients) [53–55]. We tested combination treatment with Sevi and the CDK4/6 inhibitor abemaciclib (Abem, [56]) in a preclinical TNBC model. In MDA-MB-453 cells dual treatment showed synergy at multiple concentration combinations, analyzed by the Chou Talalay method [36], indicated by combination indexes less than 0.9 (Fig. 5A). While Abem is currently FDA approved for use in ER-positive breast cancer patients [57], our pre-clinical analysis showed that it is effective in inhibiting MDA-MB-453 tumor growth as well (Fig. 5B). These data led us to examine how dual treatment with Sevi and Abem affected MDA-MB-453 tumor growth in vivo. Combination treatment significantly inhibited tumor growth when compared to vehicle alone and was more effective than Abem treatment alone (Fig. 5C). Interestingly, the anti-tumor effects of combined treatment predominantly occurred during the final week of treatment (weeks 3–4) where the combination treatment led to tumor shrinkage indicated by an average negative rate of growth. Indeed 8 out of 12 (67%) tumors shrank during the last week of treatment in the combination of Sevi plus Abem group as compared to just 3 out of 12 (25%) that shrank with Abem alone. (Fig. 5D–E). Moreover, the combination treatment was well tolerated with no negative effects on mouse weight gain (Supplemental Fig. 5). These data suggest that the combination of AR target therapies with cell cycle inhibitors may prove to be an effective treatment for patients with AR+ TNBC.
Figure 5. Combination treatment of TNBC cells with an anti-androgen and cell cycle inhibitor.
(A) MDA-MB-453 cells were cultured in full serum media. Cell viability was determined by crystal violet assay after a 5-day exposure to DMSO (0) or increasing concentrations of Sevi (seviteronel) and Abem (abemaciclib; CDK4/6 inhibitor). Data was normalized to the mean absorbance of DMSO-treated cells and presented as mean percent growth inhibition. CalcuSyn software was used to calculate the degree of synergy between combination treatments, represented as the combination index. Combination indexes < 0.9 indicate synergy. (B) MDA-MB-453 cells were bilaterally injected into the mammary fat pads of cycling female nu/nu mice. Abem was administered daily. Tumor volume was measured with calipers. (C-E) MDA-MB-453 cells were injected into the mammary fat pads of cycling female nu/nu mice. Abem and/or Sevi were administered daily. (C) Tumor volume was measured with calipers and (D) the rate of tumor growth per day over four weeks was calculated. Rate of tumor growth was broken down to illustrate the differences in tumor growth over the first 3 weeks of treatment compared to the final week alone. (E) Change in tumor volume, either growth or reduction, during the final week of treatment is depicted for each individual mouse. Mean ± standard error of the mean; * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
DISCUSSION
AR signaling has emerged as an effective targetable pathway in a subset of TNBCs, and first- and second-generation anti-androgens such as bicalutamide and enzalutamide have shown promising clinical activity in patients with AR+ TNBC. Sevi is unique in its ability to directly antagonize AR as well as inhibit the androgen-synthesizing enzyme CYP17 lyase, with less interference on corticosteroid production than earlier generations of anti-androgens [27]. Similar to what has previously been reported in prostate cancer [58], Sevi is as effective as Enza at inhibiting AR activity in TNBC cells [50]. Due to its mechanistic differences from earlier generation AR anti-androgens, Sevi’s effectiveness was predicted to be particularly pronounced in vivo because of its ability to decrease ligand availability [25, 50]. Our data demonstrate that Sevi significantly inhibited an AR+ TNBC cell line xenograft in a dose-dependent manner (Fig. 1). Our data is the first evidence that Sevi is able to inhibit proliferation of a AR+ TNBC PDX tumor model as well (Fig. 3). Thus, Sevi shows promising anti-tumor activity in both cell line and PDX preclinical models of TNBC.
While the activity of Sevi as an inhibitor of both AR activity and ligand synthesis has been demonstrated [50, 58], there are likely other mechanisms of action contributing to its effect on TNBC growth. Our data indicate that a proportion of the anti-tumor effects of Sevi are AR-directed, as shown by the “androgen response pathway” being one of the top biological pathways associated with Sevi treatment and tumor shrinkage in vivo (Fig. 3F) and the large overlap between genes altered by Sevi treatment and upregulated in ARHigh MDA-MB-453 cells (Supplemental Fig. 4). Pathway analysis (Fig. 3F) also indicates that cholesterol homeostasis, myogenesis, and MTORC1 (mammalian target of rapamycin complex 1) signaling are also highly altered by Sevi treatment. The influence of Sevi on these pathways has yet to be investigated but may be linked to its role as a dual CYP17A1 and AR inhibitor since the CYP17A1 enzyme catalyzes multiple reactions involved in the synthesis of steroids and other lipids, including cholesterol [59]. AR inhibition has previously been shown to activate mTOR signaling in hepatocellular carcinoma [60], and our lab identified a feedback loop between AR and mTOR signaling in breast cancer [61], providing a potential link between Sevi and mTOR pathway activation. Finally, androgens are known to regulate myogenic differentiation [62, 63]; thus, Sevi likely has dramatic effects on myogenesis as well. Future studies are needed to confirm the anti-tumor contribution of these and other pathways affected by Sevi.
In addition to targeting AR directly and inhibiting ligand synthesis, targeting additional points within the AR signaling cascade may be therapeutically beneficial since clinical data from prostate cancer show that acquired resistance to AR targeted therapies can develop over time [64]. While this has yet to be shown for BC, being mindful of acquired resistance from the outset could lead to more effective combination therapies for patients prior to their clinical need. Our data indicate that blocking the cell cycle may be an effective cooperative therapeutic approach to add to anti-androgen therapy in AR+ TNBC (Fig. 4).
During the cell cycle, CDK4/6 binds to cyclin D to mediate phosphorylation and deactivation of Retinoblastoma (Rb) protein, which allows for G0/G1 to S phase progression through the cell cycle. ERα-positive BCs often overexpress cyclin D, promoting cell cycle activation, making CDK4/6 targeted therapies an attractive therapeutic option [65], and the FDA (Food and Drug Administration) approved multiple CDK4/6 inhibitors for use in the treatment of ERα-positive BC, including palbociclib, ribociclib and Abem between 2015–2017 [66]. While a portion of TNBCs have RB1 mutations and/or loss, making CDK4/6 inhibition impractical, AR and Rb expression are positively associated and AR promotes cyclin D-CDK4/6 activation [66–68], suggesting the AR+ TNBCs may benefit from CDK4/6 blockade. In fact, our data indicate that not only do AR+ TNBC xenografts respond well to the CDK4/6 inhibitor Abem, but that the combination of Abem and Sevi was more effective than Abem alone (Fig. 5). The synergistic effect is likely due to the interconnectedness and known overlap between the AR signaling and cell cycle pathways [34, 67]. While CDK4/6 inhibitors are not currently approved for use in TNBC, our pre-clinical data suggest that this may be an effective treatment strategy in AR+ TNBC, especially when combined with AR targeting therapies.
Supplementary Material
Implications.
While cell cycle inhibitors are FDA-approved for use in ER-positive breast cancer, our studies suggest that they may also be effective in AR+ TNBC, perhaps combined with AR targeted agents.
Acknowledgments.
The authors acknowledge the shared resources of the University of Colorado Cancer Center NCI Support Grant (P30CA046934), particularly the Genomics Shared Resource and the Office of Laboratory Animal Resources. The authors would also like to thank Dr. Jim Lambert for his assistance with cell line preparations for in vivo xenograft experiments. Financial support included NIH NRSA T32 CA190216-01A1 (J. Christenson), DOD CTRA W81SWH-13-1-0091/90 (A. Elias and J. Richer), and NIH R01CA187733 (J. Richer).
Footnotes
Conflict of Interest Statement.
Joel Eisner was an employee and shareholder of Innocrin Pharmaceuticals, Inc. All other authors have no conflicts of interest to disclose.
REFERENCES
- 1.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438–51. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LAG, Cronin KA. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst 2014;106:dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429–34. [DOI] [PubMed] [Google Scholar]
- 4.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod Pathol 2011;24:924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safarpour D, Pakneshan S, Tavassoli FA. Androgen receptor (AR) expression in 400 breast carcinomas: is routine AR assessment justified? Am J Cancer Res 2014;4:353–68. [PMC free article] [PubMed] [Google Scholar]
- 6.Qi J-P, Yang Y-L, Zhu H, Wang J, Jia Y, Liu N et al. Expression of the androgen receptor and its correlation with molecular subtypes in 980 chinese breast cancer patients. Breast Cancer (Auckl) 2012;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mrklić I, Pogorelić Z, Capkun V, Tomić S. Expression of androgen receptors in triple negative breast carcinomas. Acta Histochem 2013;115:344–48. [DOI] [PubMed] [Google Scholar]
- 8.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F et al. A refined molecular taxonomy of breast cancer. Oncogene 2012;31:1196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNamara KM, Yoda T, Takagi K, Miki Y, Suzuki T, Sasano H. Androgen receptor in triple negative breast cancer. J Steroid Biochem Mol Biol 2013;133:66–76. [DOI] [PubMed] [Google Scholar]
- 10.Barton VN, Christenson JL, Gordon MA, Greene LI, Rogers TJ, Butterfield K et al. Androgen Receptor Supports an Anchorage-Independent, Cancer Stem Cell-like Population in Triple-Negative Breast Cancer. Cancer Res 2017;77:3455–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nuclear receptor signaling 2008;6:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. The Journal of biological chemistry 1998;273:20213–22. [DOI] [PubMed] [Google Scholar]
- 13.Koryakina Y, Knudsen KE, Gioeli D. Cell-cycle-dependent regulation of androgen receptor function. Endocrine-related cancer 2015;22:249–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asghar US, Barr AR, Cutts R, Beaney M, Babina I, Sampath D et al. Single-Cell Dynamics Determines Response to CDK4/6 Inhibition in Triple-Negative Breast Cancer. Clin Cancer Res 2017;23:5561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christenson JL, Trepel JB, Ali HY, Lee S, Eisner JR, Baskin-Bey ES et al. Harnessing a Different Dependency: How to Identify and Target Androgen Receptor-Positive Versus Quadruple-Negative Breast Cancer. Horm Cancer 2018;9:82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannelli P, Di Donato M, Galasso G, Di Zazzo E, Bilancio A, Migliaccio A. The Androgen Receptor in Breast Cancer. Frontiers in endocrinology 2018;9:492–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kono M, Fujii T, Lim B, Karuturi MS, Tripathy D, Ueno NT. Androgen Receptor Function and Androgen Receptor-Targeted Therapies in Breast Cancer: A Review. JAMA Oncol 2017;3:1266–73. [DOI] [PubMed] [Google Scholar]
- 18.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science (New York, NY) 2009;324:787–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res 2013;19:5505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castan JC, Schmid P, Awada A, Uppal H, Tudor I, Blaney M et al. Stage 1 results from MDV3100–11: A 2-stage study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in advanced AR+ triple-negative breast cancer (TNBC). Annals of Oncology 2015;26:iii6. [Google Scholar]
- 21.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O’Shaughnessy J et al. Enzalutamide for the Treatment of Androgen Receptor–Expressing Triple-Negative Breast Cancer. Journal of Clinical Oncology 2018;36:884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker J, Peterson A, Tudor I, Hoffman J, Uppal H: A novel biomarker to predict sensitivity to enzalutamide (ENZA) in TNBC. In: American Society of Clinical Oncology. Chicago, IL; 2015. [Google Scholar]
- 23.Bonnefoi H, Grellety T, Tredan O, Saghatchian M, Dalenc F, Mailliez A et al. A phase II trial of abiraterone acetate plus prednisone in patients with triple-negative androgen receptor positive locally advanced or metastatic breast cancer (UCBG 12–1). Annals of Oncology 2016;27:812–18. [DOI] [PubMed] [Google Scholar]
- 24.Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. New England Journal of Medicine 2017;377:352–60. [DOI] [PubMed] [Google Scholar]
- 25.Norris JD, Ellison SJ, Baker JG, Stagg DB, Wardell SE, Park S et al. Androgen receptor antagonism drives cytochrome P450 17A1 inhibitor efficacy in prostate cancer. J Clin Invest 2017;127:2326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maity SN, Titus MA, Gyftaki R, Wu G, Lu J-F, Ramachandran S et al. Targeting of CYP17A1 Lyase by VT-464 Inhibits Adrenal and Intratumoral Androgen Biosynthesis and Tumor Growth of Castration Resistant Prostate Cancer. Sci Rep 2016;6:35354–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toren PJ, Kim S, Pham S, Mangalji A, Adomat H, Guns EST et al. Anticancer activity of a novel selective CYP17A1 inhibitor in preclinical models of castrate-resistant prostate cancer. Mol Cancer Ther 2015;14:59–69. [DOI] [PubMed] [Google Scholar]
- 28.Gordon MA, D’Amato NC, Gu H, Babbs B, Wulfkuhle J, Petricoin EF et al. Synergy between Androgen Receptor Antagonism and Inhibition of mTOR and HER2 in Breast Cancer. Mol Cancer Ther 2017;16:1389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahl D, Sun H, Werner C, Dresser J, Wilder-Romans K, Baskin-Bey ES et al. : Targeting androgen signaling in glioblastoma (GBM) using seviteronel (SEVI), a CYP17 lyase and androgen receptor (AR) inhibitor, alone and in combination with radiation (RT). In: Society for NeuroOncology. New Orleans, LA; 2018. [Google Scholar]
- 30.Speers C, Chandler B, Zhao S, Liu M, Wilder-Romans K, Olsen E et al. : Radiosensitization of androgen receptor (AR) positive triple-negative breast cancer (TNBC) cells using seviteronel (SEVI), a selective CYP17 lyase and AR inhibitor. In: American Society of Clinical Oncology. Chicago, IL; 2017. [Google Scholar]
- 31.Gupta S, Nordquist LT, Fleming MT, Berry WR, Zhang J, Ervin SL et al. Phase I Study of Seviteronel, a Selective CYP17 Lyase and Androgen Receptor Inhibitor, in Men with Castration-Resistant Prostate Cancer. Clin Cancer Res 2018;24:5225–32. [DOI] [PubMed] [Google Scholar]
- 32.Bardia A, Gucalp A, DaCosta N, Gabrail N, Danso M, Ali H et al. Phase 1 study of seviteronel, a selective CYP17 lyase and androgen receptor inhibitor, in women with estrogen receptor-positive or triple-negative breast cancer. Breast cancer research and treatment 2018;171:111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisner JR, Abbott DH, Bird IM, Rafferty SW, Moore W. Assessment of steroid hormones upstream of p450c17 (CYP17 in chemically castrate male rhesus monkeys following treatment with the CYP17 inhibitors VT-464 and abiraterone acetate (AA. Endocr Rev 2012; [Google Scholar]
- 34.Patel JM, Goss A, Garber JE, Torous V, Richardson ET, Haviland MJ et al. Retinoblastoma protein expression and its predictors in triple-negative breast cancer. NPJ breast cancer 2020;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christenson JL, Butterfield KT, Spoelstra NS, Norris JD, Josan JS, Pollock JA et al. MMTV-PyMT and Derived Met-1 Mouse Mammary Tumor Cells as Models for Studying the Role of the Androgen Receptor in Triple-Negative Breast Cancer Progression. Horm Cancer 2017;8:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 37.Toren PJ, Kim S, Pham S, Mangalji A, Adomat H, Guns ES et al. Anticancer activity of a novel selective CYP17A1 inhibitor in preclinical models of castrate-resistant prostate cancer. Mol Cancer Ther 2015;14:59–69. [DOI] [PubMed] [Google Scholar]
- 38.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature medicine 2011;17:1514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baird NL, Bowlin JL, Cohrs RJ, Gilden D, Jones KL. Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts. Journal of virology 2014;88:5877–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics (Oxford, England) 2010;26:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R et al. Massively parallel digital transcriptional profiling of single cells. Nature communications 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 2014;32:381–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nature methods 2017;14:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nature methods 2017;14:979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 49.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC genomics 2008;9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellison S, Norris J, Wardell S, Eisner J, Hoekstra W, Stagg D et al. Abstract P3–14-04: Effects of the dual selective CYP17 lyase inhibitor and androgen receptor (AR) antagonist, VT-464, on AR+ and ER+ tumor models in vitro and in vivo. Cancer Res 2016;76:P3–14-04–P3–14-04. [Google Scholar]
- 51.Michmerhuizen AR, Chandler B, Olsen E, Wilder-Romans K, Moubadder L, Liu M et al. Seviteronel, a Novel CYP17 Lyase Inhibitor and Androgen Receptor Antagonist, Radiosensitizes AR-Positive Triple Negative Breast Cancer Cells. Frontiers in endocrinology 2020;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung J, Seol HS, Chang S. The Generation and Application of Patient-Derived Xenograft Model for Cancer Research. Cancer research and treatment : official journal of Korean Cancer Association 2018;50:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gucalp A, Edelweiss M, Patil S, Gounder M, Feigin K, Corben A et al. Abstract P3–11-04: Phase I/II trial of palbociclib in combination with bicalutamide for the treatment of androgen receptor (AR)+ metastatic breast cancer (MBC). Cancer Res 2018;78:P3–11-04–P3–11-04. [Google Scholar]
- 54.Liu C-Y, Lau K-Y, Hsu C-C, Chen J-L, Lee C-H, Huang T-T et al. Combination of palbociclib with enzalutamide shows in vitro activity in RB proficient and androgen receptor positive triple negative breast cancer cells. PLoS One 2017;12:e0189007–e07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gucalp A, Boyle LA, Alano T, Arumov A, Gounder MM, Patil S et al. Phase II trial of bicalutamide in combination with palbociclib for the treatment of androgen receptor (+) metastatic breast cancer. Journal of Clinical Oncology 2020;38:1017–17. [Google Scholar]
- 56.Martin JM, Goldstein LJ. Profile of abemaciclib and its potential in the treatment of breast cancer. Onco Targets Ther 2018;11:5253–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sledge GW Jr, Toi M, Neven P, Sohn J, Inoue K, Pivot X et al. The Effect of Abemaciclib Plus Fulvestrant on Overall Survival in Hormone Receptor–Positive, ERBB2-Negative Breast Cancer That Progressed on Endocrine Therapy—MONARCH 2: A Randomized Clinical Trial. JAMA Oncol 2020;6:116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norris JD, Ellison SJ, Baker JG, Stagg DB, Wardell SE, Park S et al. Androgen receptor antagonism drives cytochrome P450 17A1 inhibitor efficacy in prostate cancer. J Clin Invest 2017;127:2326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uno Y, Hosaka S, Yamazaki H. Identification and analysis of CYP7A1, CYP17A1, CYP20A1, CYP27A1 and CYP51A1 in cynomolgus macaques. The Journal of veterinary medical science 2014;76:1647–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Li XX, Yang Y, Zhang Y, Wang HY, Zheng XFS. Significance and mechanism of androgen receptor overexpression and androgen receptor/mechanistic target of rapamycin cross-talk in hepatocellular carcinoma. Hepatology (Baltimore, Md) 2018;67:2271–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon MA, D’Amato NC, Gu H, Babbs B, Wulfkuhle J, Petricoin EF et al. Synergy between Androgen Receptor Antagonism and Inhibition of mTOR and HER2 in Breast Cancer. Mol Cancer Ther 2017;16:1389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee DK. Androgen receptor enhances myogenin expression and accelerates differentiation. Biochemical and biophysical research communications 2002;294:408–13. [DOI] [PubMed] [Google Scholar]
- 63.Rana K, Lee NK, Zajac JD, MacLean HE. Expression of androgen receptor target genes in skeletal muscle. Asian journal of andrology 2014;16:675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nature reviews Cancer 2015;15:701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah M, Nunes MR, Stearns V. CDK4/6 Inhibitors: Game Changers in the Management of Hormone Receptor–Positive Advanced Breast Cancer? Oncology (Williston Park, NY) 2018;32:216–22. [PMC free article] [PubMed] [Google Scholar]
- 66.Matutino A, Amaro C, Verma S. CDK4/6 inhibitors in breast cancer: beyond hormone receptor-positive HER2-negative disease. Therapeutic advances in medical oncology 2018;10:1758835918818346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michmerhuizen AR, Spratt DE, Pierce LJ, Speers CW. ARe we there yet? Understanding androgen receptor signaling in breast cancer. NPJ breast cancer 2020;6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pernas S, Tolaney SM, Winer EP, Goel S. CDK4/6 inhibition in breast cancer: current practice and future directions. Therapeutic advances in medical oncology 2018;10:1758835918786451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.