Abstract
Background:
Estrogens are thought to contribute to breast cancer risk through cell cycling and accelerated breast aging. We hypothesize that lifetime estrogen exposure drives early epigenetic breast aging observed in healthy women. In this study, we examined associations between hormonal factors and epigenetic aging measures in healthy breast tissues.
Methods:
We extracted DNA from breast tissue specimens from 192 healthy female donors to the Susan G. Komen Tissue Bank at the Indiana University Simon Cancer Center. Methylation experiments were performed using the Illumina EPIC 850K array platform. Age-adjusted regression models were used to examine for associations between factors related to estrogen exposure and five DNA methylation-based estimates: Grim age, Pan-tissue age, Hannum age, Phenotypic age, and Skin and Blood Clock age.
Results:
Women were aged 19–90 years, with 95 pre-menopausal, and 97 nulliparous women. The age difference (Grim age - chronologic age) was higher at earlier ages close to menarche. We found significant associations between earlier age at menarche and age-adjusted accelerations according to the Grim clock, the Skin and Blood clock, and between higher body mass index (BMI) and age-adjusted accelerations in the Grim clock, Hannum clock, Phenotypic clock, and Skin and Blood clock.
Conclusion:
Earlier age at menarche and higher BMI are associated with elevations in DNA methylation-based age estimates in healthy breast tissues, suggesting that cumulative estrogen exposure drives breast epigenetic aging.
Impact:
Epigenetic clock measures may help advance inquiry into the relationship between accelerated breast tissue aging and an elevated incidence of breast cancer in younger women.
Keywords: DNA methylation, epigenetic clock, aging, breast cancer, cancer risk, estrogen exposure, cell cycling
Introduction
Advancing age is a major risk factor for many cancers, including breast cancer. However the linear log-log relationship between cancer incidence and age does not hold for breast cancer patients (1). The concept of ‘breast tissue age’ developed by Malcolm Pike brings the age-incidence curve of breast cancer in line with other cancers, and explains key risk factors for breast cancer, including early menarche, late first full-term pregnancy, late menopause, and post-menopausal weight (1–6). In this model, breast tissues begin an accelerated aging process at menarche, and there is an initial slowing of the rate of breast aging with first full term pregnancy, followed by a gradual slowing of this rate during the perimenopausal period until the last menstrual period (1). Estrogens are thought to modulate breast cancer risk through chronic cell cycling and breast epithelial cell mitotic activity associated with the normal menstrual cycle (1,7). Estrogens and progesterone/progestins together are thought to be associated with greater cell proliferation.
We have previously shown that DNA methylation age, estimated using the pan-tissue epigenetic clock (8), is elevated in normal breast tissue compared with matched peripheral blood samples in healthy women(9). We further found that the age difference (breast age - blood age) is greatest at earlier ages closest to menarche, and that this gap closes as women approach ages close to the menopausal transition (9). These findings suggest that chronic cell cycling related to cumulative exposure to either estrogen alone or the combination of estrogens and progesterone/progestins may contribute to epigenetic age acceleration of breast tissue. We hypothesize that elevations in epigenetic age beginning shortly before menarche and ending in the post-menopausal period are related to the same risk factors associated with ‘breast tissue age’ identified in the models of Pike et al. (1), including early age at menarche, nulliparity, late first full-term pregnancy, and body mass index (BMI). In this study we examine the relationship between elevations in epigenetic age of breast tissue and self-reported measures of lifetime exposure to estrogens and progesterone. In addition to the pan tissue DNA methylation age, we examine four new additional estimates of epigenetic age: Grim Age, Hannum Age, Phenotypic Age, and Skin and Blood Age.
Materials and Methods
Study Population
We utilized specimens from the Susan G. Komen Tissue Bank (KTB) at the Indiana University Simon Cancer Center. This unique repository of breast tissue samples, donated by healthy women without a known history of breast cancer, is a resource to breast cancer researchers. Its goals are to promote an understanding of normal breast biology in order to better understand disruption occurring during breast carcinogenesis, and to promote breast cancer prevention research. Each tissue sample is richly annotated with information about the donor’s ethnicity, height, weight, family history, medical history, reproductive history, and medication use.
We requested samples from a subset of 200 healthy women donors who had fresh frozen breast tissue specimens available and did not have any history of breast cancer. We selected women from four groups: 1) pre-menopausal and nulliparous, 2) pre-menopausal and with at least 1 live birth, 3) post-menopausal and nulliparous, and 4) post-menopausal and with at least 1 live birth. This study was approved by the UCLA Institutional Review Board.
DNA extraction, bisulfite conversion, and methylation experiments
Each donor underwent six core biopsies with samples taken from the upper outer quadrant of one breast under local anesthesia. One of the six core biopsies for each donor was placed into an embedding cassette, within five minutes of procurement, and the cassettes were placed into 10% buffered formalin and stored at room temperature. The specimens were then embedded in paraffin. For the five remaining core biopsies for each donor, tissues were flash frozen in liquid nitrogen, then placed in labeled, chilled, cryovials, and stored in liquid nitrogen at −166.2 °C. 200 breast tissue samples, with 50 mg of breast tissue per sample, were shipped to the Neurogenetics Core Sequencing Laboratory at UCLA (UNGC).
DNA and RNA were extracted using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, cat # 80224). 30 mg of frozen tissue was lysed with 600 ul guanidine-isothiocyanate-containing Buffer RLT Plus in a 2.0 ml micro centrifuge tube, and homogenized using TissueLyser II (Qiagen) with 5 mm stainless steel beads. Tissue lysate was continued with the AllPrep protocol for simultaneous extraction of genomic DNA and total RNA using RNeasy Mini spin column technology. Extracted DNA was then used for bisulfite conversion and methylation analyses.
Methylation studies were performed using the Illumina Human Methylation EPIC (850K) array BeadChip (Illumina, San Diego, CA). 500 ng DNA was bisulfite-converted using the EZ-methylation kit (Zymo Research). Upon bisulfite treatment, unmethylated are converted to uracils, while methylated cytosines remain unchanged. Following bisulfite conversion, the DNA is then hybridized to the EPIC array, using site-specific probes, designed for methylated and unmethylated sites respectively. Fluorescence data from the hybridized chip were scanned on an iScan (Illumina) and analyzed to determine the level of total methylation for each interrogated locus, by calculating the ratio of the fluorescent signals from the methylated versus unmethylated sites. A well-designed selection of samples was drawn from each group (pre- or post-menopausal, nulliparous vs. at least 1 live birth). DNA methylation levels (beta-values) were determined by calculating the ratio of intensities between methylated (signal A) and un-methylated (signal B) sites. We used the “noob” normalization method implemented in the minfi R package (10,11). Specifically, the beta value was calculated from the intensity of the methylated (M corresponding to signal A) and un-methylated (U corresponding to signal B) sites, as the ratio of fluorescent signals beta = Max(M,0)/[Max(M,0) + Max(U,0) + 100]. Thus, beta values range form 0 (completely un-methylated) to 1 (completely methylated).
Epigenetic Clocks
We examined five measures of epigenetic age, estimated from weighted regression models using methylation values at selected CpGs from our bisulfite sequencing data. These measures include Grim age, based on 1030 CpGs (12), Pan-tissue age, based on the pan tissue clock comprised of 353 CpGs (8), Hannum age, based on 71 CpGs (13), Phenotypic Age, based on 513 CpGs (14), and Skin and Blood Age, based on 391 CpGs (15). All of these measures are strongly correlated with chronologic age. Pan-tissue age (8) was developed as an age estimator in multiple tissues and has been shown to be accelerated in various disease states (16–18), and predictive of mortality (19–21). In this study, we focused our analysis on Grim age, a second-generation epigenetic clock designed to be predictive of both health span and lifespan. The Grim age clock stands out in terms of its association with time to cancer and age at menopause (12,22,23). The remaining four clocks were examined to provide a robustness analysis to confirm association of epigenetic age estimates with variables we examine in this study. Supplementary Table 1 compares features of the five epigenetic clocks examined in our study. Measures of age acceleration for each variable are calculated by taking the residuals from a linear regression of each methylation age measure on chronologic age.
We used the minfi R function preprocessNoob perform quality control assessments. Data integrity were confirmed by examining the “corSampleVSgoldstandard” quality statistic for detecting outlying samples. For the Grim, Pan-tissue,Phenotypic, Skin and Blood clocks, imputation was performed using the impute.knn function based on a small subset (n=2381) of CpGs to fill in missing values. For the Hannum clock, missing values get omitted, as the coefficients in the Hannum formula are set to zero. Each of the epigenetic clock measures was normally distributed and did not require transformation.
Statistical Analysis
We examined the relationship between breast epigenetic age acceleration (dependent variable) and measures of lifetime estrogen exposure (independent variables). Four measures of age acceleration included Age Acceleration Residual (from Pan-tissue age), Hannum Age Acceleration, Phenotypic Age Acceleration, Grim Age Acceleration, and Skin and Blood Age Acceleration. All models were adjusted for chronologic age. Measures of lifetime estrogen and progesterone exposure included age at menarche, menstrual status, total menstrual years, gravidity, parity, age at first full term birth, breast feeding duration, and BMI (given the role of adipose tissue in peripheral aromatization of androgens leading to increased estrogen exposure in post-menopausal women), as well as variables related to exogenous estrogen administration including oral contraceptive use and hormone replacement therapy. Total menstrual years was defined as the age difference (current age – age at menarche) for premenopausal women, and the age difference (age at menopause - age at menarche) for postmenopausal women. In addition, we examined additional covariates related to cancer risk including tobacco smoking (current, ever, never, and pack-years), alcohol use, and BMI (measured at the time of specimen collection). BMI was examined as a continuous variable and in categories including underweight (BMI <18.5), normal (18.5 ≤ BMI < 25), overweight (25 ≤ BMI < 30), and obese (BMI ≥ 30). Because only three women were characterized as underweight, we collapsed underweight and normal individuals (N=69) into one reference category. There were balanced numbers of individuals in the underweight/normal (N=72), overweight (N=55), and obese (N=65) categories. Because the majority of women experienced menarche from age 12–14, with 32 women reporting history of early menarche from 9–11 years, and 21 women reporting history of late menarche from 15–19 years, we modeled menarche as a discrete variable. Likewise, we modeled gravidity and parity as discrete variables as these variables carry more information than their binary counterparts. We initially examined bivariate models comparing each covariate with each measure of age acceleration, and constructed multivariate models using all variables found significant in the bivariate analyses. In multivariate analyses, we excluded variables where only a small number of individuals were present in the high risk category (e.g. ethnicity and heavy tobacco smoking). In order to examine whether associations between hormonal factors and breast epigenetic age differ for pre- and post-menopausal groups, we performed additional analyses, stratified by menopausal status. We use a p-value cutoff of 0.05 to test for significance in our main analysis focused on Grim age as outcome. Because the epigenetic clocks in our robustness analysis are not independent, and a Bonferonni correction for 5 clocks would be overly conservative, we adjusted for multiple testing using a Bonferroni correction of 0.05/2.
Results
Table 1 reveals the demographic and clinical characteristics of our study sample. Women were aged 19–90 years of age, with an average age of 50.7 ± 12 years. All of the women in our sample were white, and the majority were non-Hispanic (N=183), with 9 Hispanic women. Donors aged 50 years and over were more likely to have ever smoked tobacco (36%, compared with 26% of women under 50 years), and those who smoked did so for a longer number of years on average (18 years, compared with 12 years for women under 50 years) and smoked more cigarettes per day. Women in both age groups had comparable percentages of women who currently drink any alcohol (68% young vs. 67% older women. However, women aged over 50 years who drank had more drinks per week.
Table 1.
Characteristics of the study sample
| Age < 50 years N=89 |
Age ≥ 50 years N=103 |
p-value | |
|---|---|---|---|
| Demographics | |||
| Age (years), mean (SD) | 40.8 (7.7) | 59.3 (7.1) | <0.0001 |
| Ethnicity, No. (%) | |||
| Hispanic | 3 (3) | 6 (6) | 0.64 |
| Tobacco Smoking | |||
| Ever smoked, No. (%) | 16 (26) | 37 (36) | 0.009 |
| Years smoked, mean (SD) | 12 (8.5) | 18 (12) | 0.035 |
| Current smoker | 7 (8) | 4 (4) | 0.38 |
| Cigarettes per day | |||
| 1–10 | 9 (10) | 18 (17) | 0.042 |
| 11–20 | 10 (11) | 12 (12) | |
| 21–40 | 1 (1) | 8 (8) | |
| 41–60 | 0 (0) | 2 (2) | |
| Alcohol Use | |||
| Currently drink, No. (%) | 61 (68) | 69 (67) | 0.94 |
| Drinks per week | |||
| <1 | 1 (1) | 7 (7) | 0.017 |
| 1–7 | 51 (57) | 42 (41) | |
| 14–21 | 9 (10) | 20 (19) | |
| >21 | 1 (1) | 0 | |
| Ashkenazi Jewish, No. (%) | 2 (2) | 2 (2) | 1 |
| Body mass index, mean (SD) | 28.2 (7.3) | 28.8 (6.5) | 0.57 |
| Underweight, No. (%) | 1 (1) | 2 (2) | |
| Normal, No. (%) | 44 (46) | 25 (26) | |
| Overweight, No. (%) | 28 (29) | 27 (28) | |
| Obese, No. (%) | 22 (23) | 43 (44) | |
| Gail score*, mean (SD) | 19.1 (39) | 15.5 (36) | 0.52 |
| Gynecological history | |||
| Age at menarche (years), mean (SD) | 12.79 (1.6) | 12.77 (1.6) | 0.93 |
| Postmenopausal, No. (%) | 8 (9) | 89 (86) | <0.0001 |
| Age at menopause (years, mean (SD) | 36.7 (7) | 47.4 (7) | 0.008 |
| Total menstrual years (years, mean (SD) | 27.5 (8) | 35.4 (7) | <0.0001 |
| Reproductive history | |||
| Gravidity | |||
| 0 | 40 (45) | 46 (45) | 0.26 |
| 1 | 8 (9) | 13 (13) | |
| 2 | 15 (17) | 26 (25) | |
| 3 | 14 (16) | 11 (11) | |
| 4+ | 12 (13) | 7 (7) | |
| Parity | |||
| 0 | 40 (45) | 54 (52) | 0.39 |
| 1 | 5 (6) | 12 (12) | |
| 2 | 25 (28) | 20 (19) | |
| 3 | 12 (13) | 12 (12) | |
| 4+ | 4 (4) | 5 (5) | |
| Age at first live birth, years mean (SD) | 27.3 (5.3) | 26.1 (4.8) | 0.24 |
| Breastfeeding | |||
| Ever, No. (%) | 36 (40) | 35 (34) | 0.44 |
| Total months, mean (SD) | 14 (13) | 20 (13) | 0.26 |
| Hormonal Therapy | |||
| Ever, No. (%) | 1 (1) | 44 (43) | <0.0001 |
| Birth control, | |||
| No. (%) | 22 (25) | 8 (8) | 0.0025 |
Gail score is estimated risk for breast cancer based on age, first menstrual period, age at first live birth, first-degree relatives with breast cancer, previous breast biopsy, race, and ethnicity.
Difference between epigenetic age and chronologic age
The raw age difference (Grim age - chronological age) is significantly higher with younger age (β = −0.34 per year of age, p<0.0001). This difference is also higher in pre-menopausal women (β = 5.9, p<0.0001) compared with post-menopausal women. Likewise, the age difference (Pan-tissue age - chronologic age) is higher with younger age (β = −0.33 per year of age, p<0.0001) and in pre-menopausal women (β = 5.2, p<0.0001). These findings are visualized in Figure 1.
Figure 1. Difference between estimated epigenetic age and chronological age, by age and menopausal status.
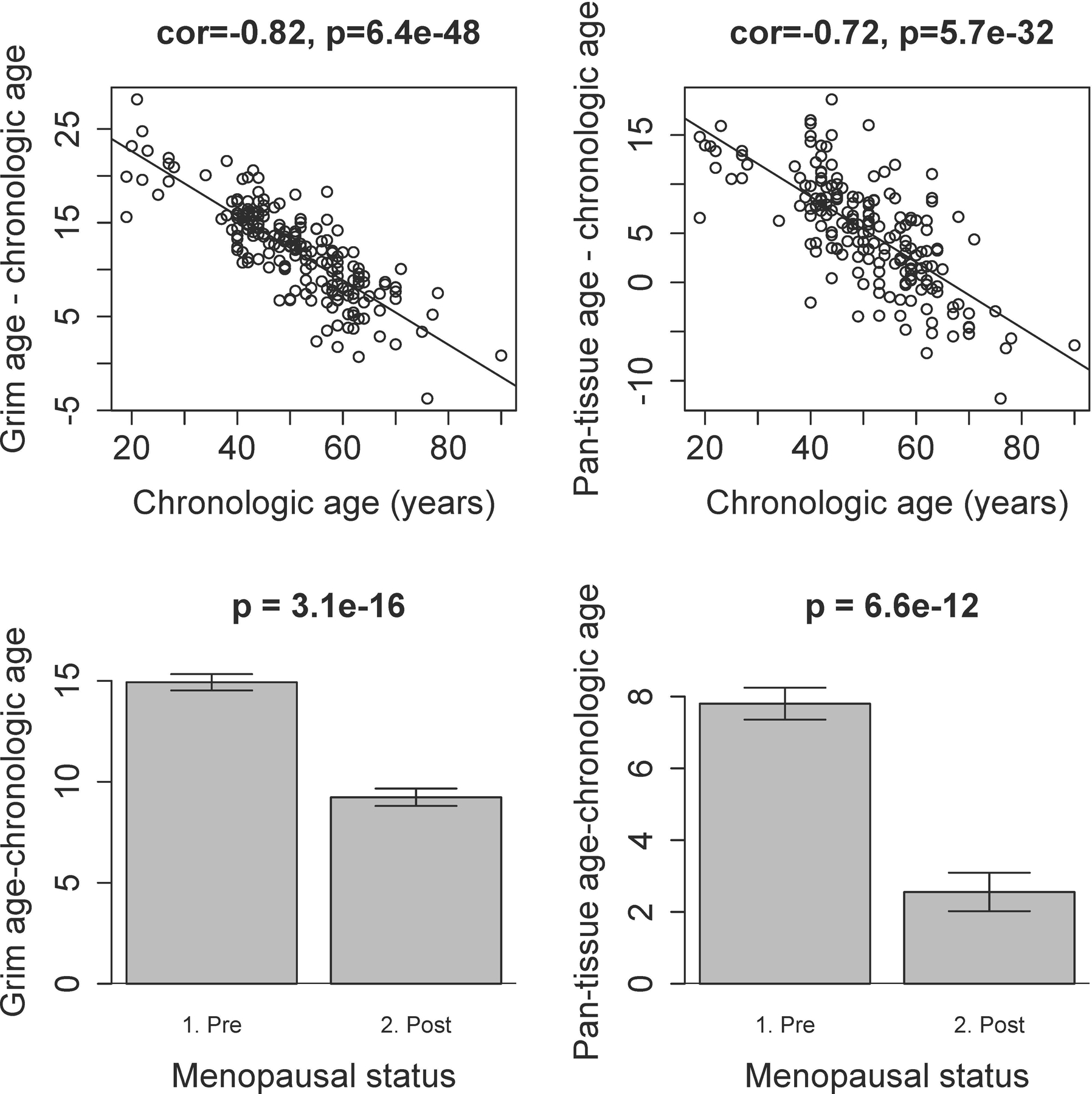
The raw age difference (estimated epigenetic age - chronological age) is significantly higher with younger age, for both Grim age (Panel A) and Pan-tissue age (Panel B). Regression lines demonstrate the linear relationship between this difference and chronologic age. The difference (estimated epigenetic age - chronologic age) is also higher in pre-menopausal women compared with post-menopausal women for both Grim age (Panel C) and Pan-tissue age (Panel D). These bar plots demonstrate the mean value (y-axis) and one standard error, with p-values from the results of a non-parametric group comparison test (Kruskal-Wallis).
Epigenetic age acceleration is associated with earlier age at menarche and higher BMI
Because measures of lifetime estrogen exposure are strongly correlated with chronologic age, we need to examine an age-adjusted measure of epigenetic acceleration. Supplementary Table 2 reveals results of our bivariate analysis examining associations between each measure of epigenetic age acceleration and each predictor variable of interest. Figure 2 shows the association of age at menarche with each of the age-adjusted measures of epigenetic age acceleration. We found that earlier age at menarche was associated with a higher degree of age-adjusted acceleration of the Grim and Skin and Blood clocks.
Figure 2. Relationship between age at menarche and age-adjusted measures of epigenetic age acceleration.
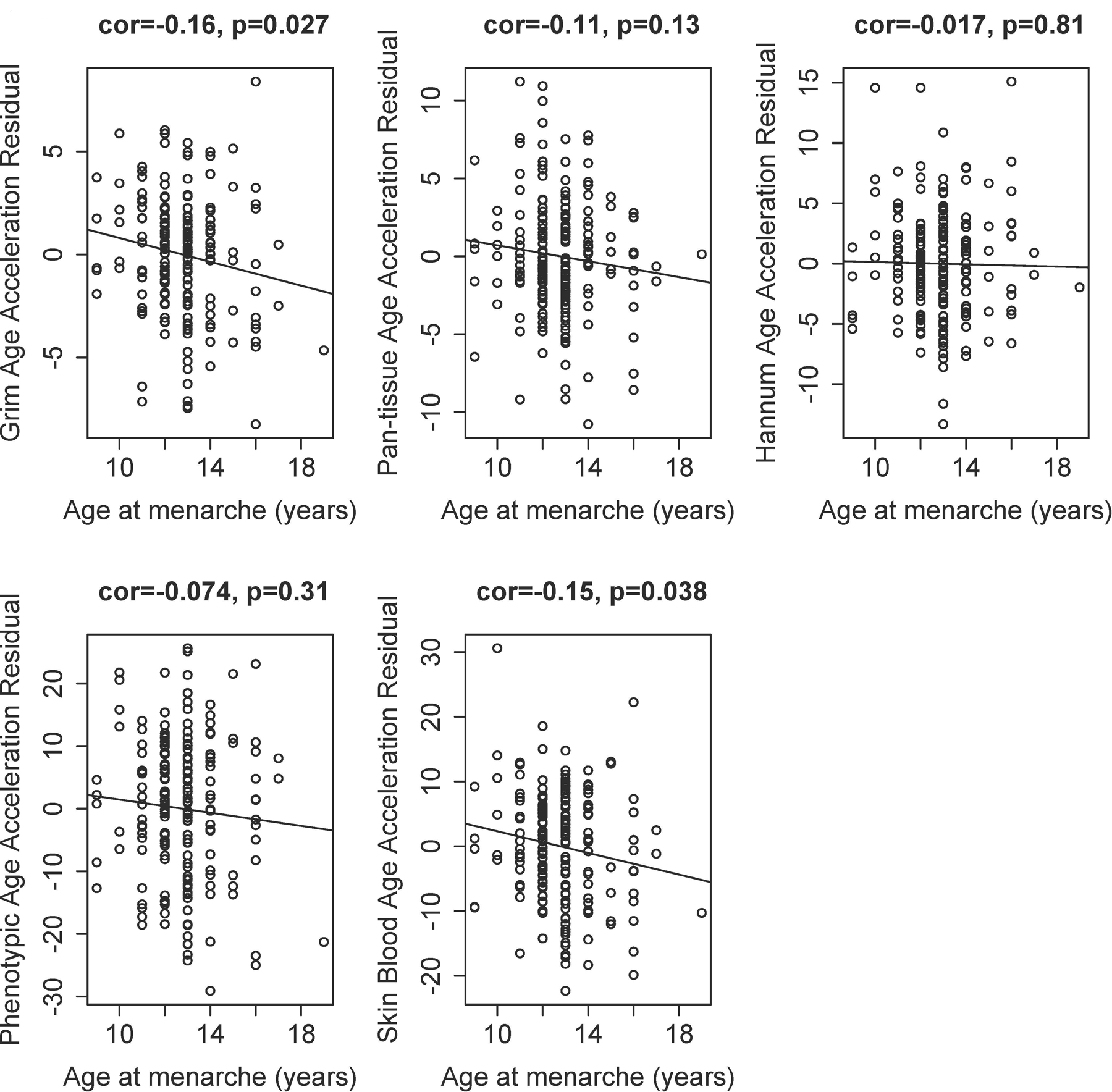
Earlier age at menarche is significantly associated with a higher degree of age-adjusted acceleration of the Grim (Panel A), and Skin and Blood (Panel E) clocks.
Elevated BMI is significantly associated with age-adjusted acceleration in Grim age, as well as Hannum, Phenotypic, and Skin and Blood clocks. Figure 3 reveals the association of BMI with each epigenetic acceleration measure. When we examine BMI as a categorical variable, we find that being overweight and obese are both significantly associated with age-adjusted acceleration in the Grim age clock (see Supplementary Table 2). In addition, being overweight is significantly associated with acceleration of the Phenotypic age clock, and obesity is significantly associated with acceleration in the Hannum, Phenotypic, and Skin and Blood clocks. Hannum and Skin and Blood age were accelerated in overweight individuals, although these findings were of borderline significance.
Figure 3. Relationship between BMI and age-adjusted measures of epigenetic age acceleration.
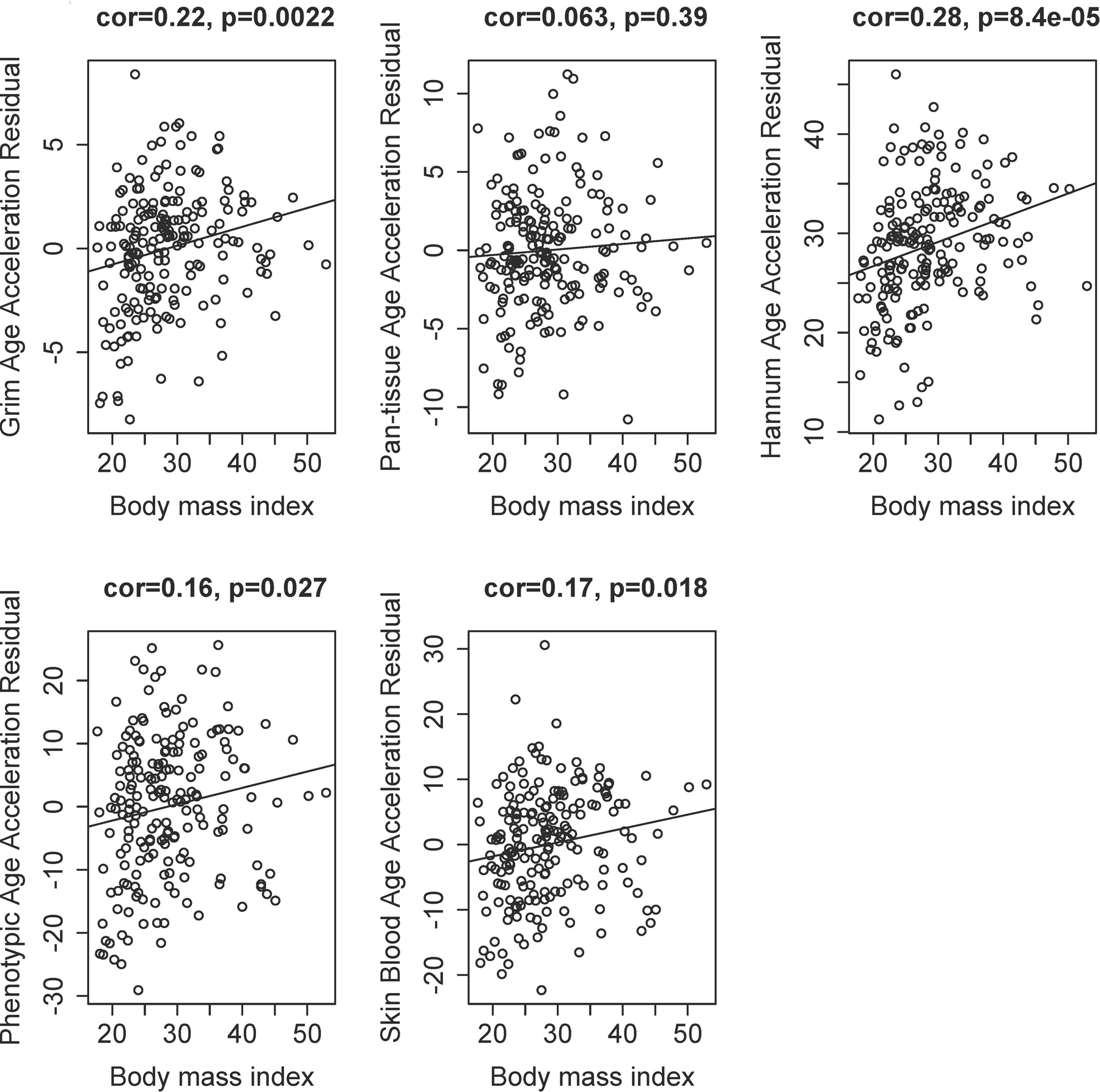
Elevated BMI is significantly associated with age-adjusted acceleration in Grim age (Panel A), Hannum (Panel C), Phenotypic (Panel D), and Skin and Blood (Panel E) clocks.
In our full sample, we did not identify significant associations between epigenetic age acceleration and gravidity and parity (see Supplementary Table 2). Exogenous estrogen was not significantly associated with elevation in age-adjusted epigenetic age acceleration measures. While women who take post-menopausal hormone replacement therapy had higher age-adjusted acceleration in both Grim age and Pan-tissue age (see Supplementary Table 2), these findings were not significant. We found that age-adjusted acceleration in Pan-tissue age was higher in nulliparous women, women with higher age at first full term birth, and earlier age at menarche, though these findings did not meet statistical significance. However, when we remove participants who undergo menopause early (age <45 years, N=27), we find that age at menarche is significantly associated with age-adjusted acceleration in Pan-tissue age (β=−0.43 for each year of age of later menarche, p=0.023).
Results of multivariate regression models examining factors associated with epigenetic age are shown in Table 2. We included three covariates for which significant associations were found with any of the measures of epigenetic age: chronologic age, BMI (categorical variable), and age at menarche. In multivariate models, being overweight or obese remained significantly associated with Grim age, as well as Hannum age, Phenotypic age, and Skin and Blood age. Women with earlier age at menarche had a higher degree of acceleration in Grim age, as well as Skin and Blood age, although in multivariate models, these associations were now of borderline significance. After adjustment for multiple comparisons, we find that being overweight or obese is significantly associated with acceleration in Grim age, while being obese is associated with acceleration in Hannum, Phenotypic, and Skin and Blood age.
Table 2.
Results of multivariate linear regression analysis* examining factors associated with breast epigenetic age acceleration.
| Grim age | Pan-tissue age | Hannum age | Phenotypic age | Skin and Blood age | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | p | β | p | β | p | β | p | β | p | |
| Age | 0.64 | <0.0001 | 0.66 | <0.0001 | 0.33 | <0.0001 | 0.20 | 0.0041 | 0.58 | <0.0001 |
| Body mass index | ||||||||||
| Overweight | 1.27 | 0.012 | 0.81 | 0.24 | 1.62 | 0.046 | 4.25 | 0.035 | 2.81 | 0.066 |
| Obese | 1.69 | 0.00065 | 0.91 | 0.17 | 2.4 | 0.0025 | 5.21 | 0.0082 | 3.74 | 0.012 |
| Age at menarche | −0.24 | 0.064 | −0.23 | 0.20 | 0.024 | 0.90 | −0.36 | 0.48 | −0.72 | 0.063 |
models adjusted for chronologic age, BMI, and age at menarche.
Differential effects of hormonal factors on breast epigenetic age acceleration based on menopausal status
Because the association of BMI with breast cancer risk differs for pre- and post-menopausal women, we examined factors associated with breast epigenetic age acceleration in separate analyses stratified by menopausal status. Supplementary Table 3 reveals the results of bivariate analyses performed in pre-menopausal and post-menopausal women separately. Table 3 reveals results of multivariate analyses, stratified by menopausal status, with models adjusted for age at menarche and BMI (categorical). We found that both having earlier age at menarche and being overweight were significantly associated with acceleration in Grim age in premenopausal women, whereas in post-menopausal women, being obese was significantly associated with acceleration in Grim age. Additionally, in pre-menopausal women, being overweight or obese was associated with acceleration in Hannum age and being overweight was associated with acceleration Phenotypic age. For post-menopausal women, obesity was associated with acceleration in Phenotypic age.
Table 3.
Results of multivariate linear regression analysis* examining factors associated with breast epigenetic age acceleration, stratified analyses based on menopausal status.
| Grim age | Pan-tissue age | Hannum age | Phenotypic age | Skin and Blood age | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre-menopausal | β | p | β | p | β | p | β | p | β | p |
| Age | 0.65 | <0.0001 | 0.73 | <0.0001 | 0.33 | <0.0001 | 0.28 | 0.052 | 0.61 | <0.0001 |
| Body mass index | ||||||||||
| Overweight | 1.49 | 0.019 | 1.07 | 0.23 | 2.35 | 0.038 | 6.44 | 0.022 | 4.08 | 0.055 |
| Obese | 1.07 | 0.13 | −0.55 | 0.57 | 2.74 | 0.030 | 3.78 | 0.22 | 3.43 | 0.15 |
| Age at menarche | −0.34 | 0.041 | −0.20 | 0.40 | −0.28 | 0.34 | −0.57 | 0.44 | −0.95 | 0.091 |
| Post-menopausal | β | p | β | p | β | p | β | p | β | p |
| Age | 0.66 | <0.0001 | 0.59 | <0.0001 | 0.35 | <0.0001 | 0.33 | 0.015 | 0.64 | <0.0001 |
| Body mass index | ||||||||||
| Overweight | 1.02 | 0.22 | 0.72 | 0.51 | 0.44 | 0.71 | 1.91 | 0.52 | 1.24 | 0.59 |
| Obese | 2.04 | 0.0071 | 1.52 | 0.12 | 1.71 | 0.11 | 6.16 | 0.024 | 3.68 | 0.078 |
| Age at menarche | −0.14 | 0.47 | −0.34 | 0.20 | 0.39 | 0.17 | −0.15 | 0.83 | −0.46 | 0.41 |
models adjusted for chronologic age, BMI, and age at menarche.
Discussion
This is the one of the first and largest studies of normal breast tissue examining epigenetic age estimates. We have confirmed that epigenetic age acceleration in healthy breast is highest at earlier ages. We have identified factors associated with the degree of epigenetic age acceleration, including earlier age at menarche, and BMI. Because age at menarche did not remain significant after adjustment for body mass index in multivariate analyses of our full population, the association between earlier age at menarche and epigenetic clock estimates is likely driven by BMI. However, in additional multivariate analyses stratified by menopausal status, both earlier age at menarche and BMI are significantly associated with acceleration in Grim age in pre-menopausal women. These findings suggest that estrogen exposure from early menarche may drive chronic cell cycling and accelerated breast aging during pre-menopause. Notably, in premenopausal women, higher BMI is inversely associated with breast cancer risk, whereas in our study we find that methylation-based aging markers are higher in premenopausal women with higher BMI. Because elevated BMI is not thought to substantially raise the estrogen levels above the level at which proliferation is further affected in premenopausal women, our findings suggest that BMI mediates increased breast cellular aging through additional mechanisms, such as inflammation. By contrast, in post-menopausal women, a major source of estrogen comes from peripheral aromatization of androgens, and higher BMI is thought to contribute substantially to estrogen exposure. Thus, a combination of increased estrogen exposure and chronic cell cycling, as well as inflammation, may contribute to accelerated breast aging in post-menopausal women.
Our finding that earlier age at menarche is associated with accelerated aging in breast is consistent with a recent study demonstrating that Pan-tissue age acceleration in peripheral blood was associated with faster pubertal development in girls (24). Recent studies have shown that comparing breast tumor tissues of very young women (age ≤ 35 years) with those of older women, the age difference (Pan-tissue age - chronologic age) is higher in breast tumor tissue from younger women with breast cancer (25). These findings, taken together with our finding that the difference (Pan-tissue age - chronologic age) is higher at younger ages, suggest that accelerated epigenetic aging may drive breast carcinogenesis in young women.
The highest degree of acceleration in breast methylation age occurs at earlier ages closer to menarche and we hypothesize that accelerated breast aging contributes to pre-menopausal breast cancer risk. While we observe in our study that the age difference (breast age -chronologic age) approaches zero as healthy women reach the age of the menopausal transition, there are a population of postmenopausal women in our study who have persistently high breast epigenetic age relative to their chronologic age. We hypothesize that these women may be at higher risk of developing post-menopausal breast cancer. Further studies with long-term follow up are needed to investigate these questions.
Our finding that elevated BMI is associated with several age-adjusted epigenetic age acceleration measures is consistent with previous studies showing obesity is associated with acceleration in Pan-tissue age in fatty liver tissue (16) and peripheral blood (26), as well as previous findings that higher BMI is associated with intrinsic and extrinsic epigenetic age acceleration (27) and Grim Age acceleration (12) in peripheral blood. Studies have shown that BMI is associated with higher breast cancer risk in postmenopausal women (28), and that epigenetic age of breast is observed in normal breast tissue of luminal breast cancer patients (29), raising the question whether accelerated epigenetic aging of breast tissue mediates this risk. Notably, the influence of BMI on postmenopausal breast cancer risk is greater in women with greater familial risk (28). A recent study demonstrated that age-adjusted breast tissue aging, a measure estimated using the Pike model, carries different associations with breast cancer risk by family history and breast density (30). Further studies are needed to investigate the link between BMI, accelerated breast aging, and breast cancer risk, particularly in women at high familial risk of breast cancer.
An important limitation of our study is the restriction of our analyses to an all white sample of women. Future studies are needed to examine the relationship between breast aging and hormonal factors in more diverse racial groups. These proposed studies are feasible as diverse groups of donors are well-represented in the Komen Tissue Bank. Cross-sectional studies are limited in their ability to examine for causal relationships between accelerated breast aging and duration of exposure to a risk factor, such as time since menarche. Prospective studies are needed to examine for longitudinal changes in accelerated breast aging associated with variables such as time since menarche. Another limitation is the lack of information on the type and formulation of hormonal medication, both for oral contraceptives and hormone replacement, as these were not always specified in patient survey responses. Different formulations have been shown to have different effects on the breast, and follow up studies are needed to examine epigenetic changes in the breast by exogenous hormone type. A further limitation is the use of biomarkers that were developed in peripheral blood. While the Pan-tissue clock was developed in multiple tissues and cell types, and has shown to be associated with chronologic age across multiple tissues including breast (8), the Hannum, Phenotypic, and Grim clocks were all developed in peripheral blood. Hannum age was developed in peripheral blood, using an elastic net regression for selection of a set of markers that were highly predictive of chronologic age (13). Phenotypic Age is estimated by formulating a weighted composite of ten clinical characteristics known to be associated with mortality and regressing that phenotypic measure on CpGs (14). Grim Age was developed by a regression of time-to-death on DNA methylation-based surrogate biomarkers of smoking pack-years and a selection of plasma proteins previously associated with mortality or moribidity (12). Skin and Blood Age was developed as an age estimator in multiple cell types, including human fibroblasts, keratinocytes, buccal cells, endothelial cells, lymphoblastoid cells, skin, blood, and saliva samples (15). All of these clocks show a significant correlation with chronologic age in breast tissue (see Supplementary Figure 1).
In our study, as demonstrated in other epigenetic studies, we note that even the most consistent associations (e.g. epigenetic age and chronologic age, epigenetic age and BMI) vary across the five epigenetic clock measures examined. In addition to being derived using different methods, each epigenetic clock captures different features of the aging process. Supplementary Table 1 describes the comparative features of each of the clocks. We focused our analysis on Grim age, as it is most strongly correlated with lifespan and healthspan, and it has been shown to be predictive of time to cancer. The other four clocks were examined as a robustness analysis to confirm whether the direction and strength of the association agreed with that of the Grim clock. For example, the Grim clock and the Phenotypic clock were designed as mortality risk predictors while the Hannum and Skin and Blood clocks were designed as predictors of chronologic age. In spite of these contrasting and complementary features, the Hannum, Phenotypic, and Skin and Blood clocks all confirmed that higher BMI was associated with accelerated breast aging in multivariate analyses.
In normal breast tissue, myeloid and lymphoid cells are present in lobules, while cytotoxic T cells, CD4 T cells, B cells, and dendritic cells are integrated in the ductal epithelium. While Grim, Phenotypic, and Hannum ages correlate with age-related changes in cell composition, correcting for estimated cell-specific composition carries the risk of eliminating informative signals. For example, Grim and Phenotypic age capture aspects of inflammation that may be relevant to breast aging and risk of carcinogenesis. Future studies are needed to examine whether Grim age and other clocks differ for cells that compose breast tissue, including ductal epithelial cells, myoepithelial cells, and immune cells within the breast.
Global methylation analyses in peripheral blood demonstrate a relationship between epigenetics and breast cancer risk. A recent study compared peripheral blood epigenetic patterns of 440 women in the EPIC-Italy with 440 unaffected women (31). Global methylation analysis revealed an epigenetic signature associated with a 5% increase in breast cancer risk for 1 year longer lifetime estrogen exposure. Pathways identified in this analysis include genes associated with cell-cell adhesion (CTTNA2), interaction with receptor tyrosine kinase signaling (GRB10), tumor suppressor genes (RPH3AL), and long non-coding RNA that binds RNA and may be involved in cancer progression (TINCR) (31). Future studies are needed to examine global methylation studies in breast to identify pathways associated with lifetime estrogen exposure.
Our findings lend support to the hypothesis that cumulative estrogen exposure is associated with elevations in breast epigenetic aging, as measured by a variety of epigenetic clocks. We further identify an association between BMI and accelerated breast epigenetic aging, which raises questions about the role of adipose and chronic inflammation in breast aging and carcinogenesis. Understanding biological pathways that underlie accelerated aging in breast tissue will aid in the development of prevention and therapeutic strategies (32). Further studies are needed to examine how gene expression patterns change with methylation patterns, whether accelerated aging is dramatically elevated in women at high risk for breast cancer, and to test whether the identification of accelerated aging signatures may play a role in earlier breast cancer detection and prevention.
Supplementary Material
Acknowledgements.
We thank survivor advocate Peggy Devine for helpful suggestions on this project, and the Komen Tissue Bank donors for participating in this study. This study was funded by a Susan G. Komen Career Catalyst Award for Basic and Translational Science CCR16380478. PAG was supported by the Breast Cancer Research Foundation (BCRF). SH was supported by NIH/NIA 1U01AG060908 - 01. In addition, PAG serves on the Scientific Advisory Board of the BCRF.
References
- 1.Pike MC, Krailo MD, Henderson BE, Casagrande JT, Hoel DG. ‘Hormonal’ risk factors, ‘breast tissue age’ and the age-incidence of breast cancer. Nature 1983;303(5920):767–70 doi 10.1038/303767a0. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein L, Pike MC, Ross RK, Judd HL, Brown JB, Henderson BE. Estrogen and sex hormone-binding globulin levels in nulliparous and parous women. J Natl Cancer Inst 1985;74(4):741–5. [PubMed] [Google Scholar]
- 3.Bernstein L, Pike MC, Ross RK, Henderson BE. Age at menarche and estrogen concentrations of adult women. Cancer Causes Control 1991;2(4):221–5 doi 10.1007/BF00052137. [DOI] [PubMed] [Google Scholar]
- 4.Kampert JB, Whittemore AS, Paffenbarger RS Jr., Combined effect of childbearing, menstrual events, and body size on age-specific breast cancer risk. Am J Epidemiol 1988;128(5):962–79 doi 10.1093/oxfordjournals.aje.a115070. [DOI] [PubMed] [Google Scholar]
- 5.Spicer DV, Pike MC. Sex steroids and breast cancer prevention. J Natl Cancer Inst Monogr 1994(16):139–47. [PubMed] [Google Scholar]
- 6.Spicer DV, Pike MC. Breast cancer prevention through modulation of endogenous hormones. Breast Cancer Res Treat 1993;28(2):179–93 doi 10.1007/BF00666430. [DOI] [PubMed] [Google Scholar]
- 7.Pike MC, Spicer DV, Dahmoush L, Press MF. Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol Rev 1993;15(1):17–35 doi 10.1093/oxfordjournals.epirev.a036102. [DOI] [PubMed] [Google Scholar]
- 8.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013;14(10):R115 doi 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sehl ME, Henry JE, Storniolo AM, Ganz PA, Horvath S. DNA methylation age is elevated in breast tissue of healthy women. Breast Cancer Res Treat 2017;164(1):209–19 doi 10.1007/s10549-017-4218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res 2013;41(7):e90 doi 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortin JP, Triche TJ Jr., Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017;33(4):558–60 doi 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11(2):303–27 doi 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49(2):359–67 doi 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10(4):573–91 doi 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY) 2018;10(7):1758–75 doi 10.18632/aging.101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A 2014;111(43):15538–43 doi 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rickabaugh TM, Baxter RM, Sehl M, Sinsheimer JS, Hultin PM, Hultin LE, et al. Acceleration of age-associated methylation patterns in HIV-1-infected adults. PLoS One 2015;10(3):e0119201 doi 10.1371/journal.pone.0119201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018;19(6):371–84 doi 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 19.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8(9):1844–65 doi 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 2015;16:25 doi 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell 2016;15(1):149–54 doi 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hillary RF, Stevenson AJ, Cox SR, McCartney DL, Harris SE, Seeboth A, et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol Psychiatry 2019. doi 10.1038/s41380-019-0616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Ploner A, Wang Y, Magnusson PK, Reynolds C, Finkel D, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife 2020;9 doi 10.7554/eLife.51507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binder AM, Corvalan C, Mericq V, Pereira A, Santos JL, Horvath S, et al. Faster ticking rate of the epigenetic clock is associated with faster pubertal development in girls. Epigenetics 2018;13(1):85–94 doi 10.1080/15592294.2017.1414127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oltra SS, Pena-Chilet M, Flower K, Martinez MT, Alonso E, Burgues O, et al. Acceleration in the DNA methylation age in breast cancer tumours from very young women. Sci Rep 2019;9(1):14991 doi 10.1038/s41598-019-51457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Wang Z, Hardy T, Huang Y, Hui Q, Crusto CA, et al. Association of Obesity with DNA Methylation Age Acceleration in African American Mothers from the InterGEN Study. Int J Mol Sci 2019;20(17) doi 10.3390/ijms20174273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9(2):419–46 doi 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopper JL, Dite GS, MacInnis RJ, Liao Y, Zeinomar N, Knight JA, et al. Age-specific breast cancer risk by body mass index and familial risk: prospective family study cohort (ProF-SC). Breast Cancer Res 2018;20(1):132 doi 10.1186/s13058-018-1056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofstatter EW, Horvath S, Dalela D, Gupta P, Chagpar AB, Wali VB, et al. Increased epigenetic age in normal breast tissue from luminal breast cancer patients. Clin Epigenetics 2018;10(1):112 doi 10.1186/s13148-018-0534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen TL, Li S, Dite GS, Aung YK, Evans CF, Trinh HN, et al. Interval breast cancer risk associations with breast density, family history and breast tissue aging. Int J Cancer 2020;147(2):375–82 doi 10.1002/ijc.32731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson A, Palli D, Masala G, Grioni S, Agnoli C, Tumino R, et al. Epigenome-wide association study for lifetime estrogen exposure identifies an epigenetic signature associated with breast cancer risk. Clin Epigenetics 2019;11(1):66 doi 10.1186/s13148-019-0664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fresques T, Zirbes A, Shalabi S, Samson S, Preto S, Stampfer MR, et al. Breast Tissue Biology Expands the Possibilities for Prevention of Age-Related Breast Cancers. Front Cell Dev Biol 2019;7:174 doi 10.3389/fcell.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


