Abstract
Purpose:
Despite the prognostic importance of immune infiltrate in colorectal cancer (CRC), immunotherapy has demonstrated limited clinical activity in refractory metastatic proficient mismatch repair (pMMR) CRC. This study explores combining anti-CTLA-4 and an anti-PDL-1 therapy in the preoperative management of resectable CRC liver metastases with the intent to improve immune responses in this disease setting.
Patients and Methods:
Patients with resectable CRC liver-only metastases received 1 dose of tremelimumab and durvalumab preoperatively followed by single-agent durvalumab postoperatively. Primary objectives were to determine feasibility and safety.
Results:
A total of 24 patients were enrolled between 11/2016-11/2019. 23 patients received treatment [21 pMMR and 2 deficient mismatch repair (dMMR)] and subsequently 17 (74%; 95%CI: 53-88%) underwent surgical resection. Grade 3/4 treatment-related immune toxicity and postoperative grade 3/4 toxicity were seen in 5/23 (22%; 95%CI: 10-44%) and 2/17 (12%; 95%CI: 2-38%) patients. The median RFS was 9.7 (95%CI: 8.1-17.8) months and OS was 24.5 (95%CI: 16.5-28.4) months. Four patients demonstrated complete pathological response, two dMMR patients and two POLE mutation patients. Pre- and post-tumor tissue analysis by flow cytometry, immunofluorescence, and RNA sequencing revealed similar levels of T cell infiltration, but did demonstrate evidence of CD8+ and CD4+ activation post treatment. An increase in B-cell transcriptome signature and B cell density was present in post-treatment samples from patients with prolonged RFS.
Conclusions:
This study demonstrates the safety of neoadjuvant combination tremelimumab and durvalumab prior to CRC liver resection. Evidence for T and B cell activation following this therapy was seen in pMMR mCRC.
Keywords: Colorectal cancer, pMMR, dMMR, immune checkpoint therapy, tumor-infiltrating lymphocytes, liver resection
Background:
Colorectal cancer is the second most common cancer in the United States with an estimated 147,950 new cases and 53,200 deaths in 2020 (1). Approximately one-third of these patients will develop liver metastases within 3 years of initial diagnosis (2). Surgery is potentially curative in the 15-20% of patients who meet criteria for resection (2–5). For surgical candidates, complete surgical resection is associated with a 20-50% overall survival rate at 5 years (6–8). Unfortunately, the majority of resected patients ultimately recur, and data demonstrating the overall survival benefit from perioperative systemic chemotherapy is limited (9, 10). To date, the role of immunotherapy in the perioperative setting in metastatic CRC has not been investigated.
The tumor microenvironment represents a complex collection of cell types that modulate tumor development. As a result, even highly immunogenic tumors can have a suppressed immune environment depending on the cells that makeup the microenvironment (11,12). Tumor-infiltrating lymphocytes (TIL) have the capacity to control the growth of many types of cancers and are emerging as an important biomarker in predicting the efficacy and outcome of treatment (13). Blockade of immune-checkpoint inhibitors such as cytotoxic T-lymphocyte associated antigen 4 (CTLA-4), programmed cell death 1 (PD-1), and programmed cell death ligand 1 (PD-L1) can exhibit clinical activity in a wide range of tumor types. PD-1 based therapy has become the standard of care for patients with mismatch repair deficient (dMMR) metastatic CRC (14, 15). However, patients with mismatch repair proficient (pMMR) mCRC have shown minimal responses to immune checkpoint monotherapy. The combination of CTLA-4 inhibition (tremelimumab) with anti-PD-L1 inhibition (durvalumab) compared to best supportive care was recently examined in the Canadian Cancer Trials Group CO.26 Study, which demonstrated an improvement in overall survival in refractory metastatic colorectal cancer, although no difference in response rate or progression-free survival was found (16). In a separate study, durvalumab and tremelimumab was demonstrated to be safe when combined with liver directed yttrium-90 resin microsphere-based radioembolization (17).
To better understand the mechanisms of immune resistance in pMMR CRC, we initiated a pilot clinical trial combining CTLA-4 inhibition (tremelimumab) with anti-PD-L1 inhibition (durvalumab) in the perioperative resection setting where the suppressive impact of the tumor microenvironment would be addressed with surgical resection and the acquisition of tumor tissue for immune characterization would be possible. In addition, adjuvant durvalumab was allowed post-operatively to evaluate the additional impact of PD-L1 based therapy, given the high risk of residual microscopic disease following CRC liver resection.
Methods:
Study design:
This study was an open-label, single center pilot trial assessing the safety and feasibility of adding neoadjuvant tremelimumab 75 mg IV flat dose and durvalumab 1500 mg IV flat dose given pre-operatively for 1 cycle prior to CRC liver metastases resection. Post-operative therapy was at the discretion of the treating physician, and patients were eligible to receive durvalumab 1500 mg IV every 4 weeks for 4 cycles. Liver resection was scheduled approximately 4 to 8 weeks after durvalumab/tremelimumab infusion.
The primary endpoints were feasibility and safety assessed by the rate of on-trial surgical resection of liver metastases, post-operative toxicity graded by the Clavien-Dindo classification (18), and treatment related toxicity graded by CTCAE v5. The combination was defined as feasible if at least 80% of patients could undergo resection or if between 60% and 80% could undergo resection with a positive toxicity and efficacy profile. Secondary endpoints included the translational evaluation of various immune-relevant factors, pre-operative response rate by RECIST v1.1, overall survival (OS) and relapse-free survival (RFS).
The trial was conducted in accordance with the Declaration of Helsinki. The protocol (ClinicalTrials.gov identifier: NCT02754856) was approved by the Institutional Review Board at University of Texas MD Anderson Cancer Center (Houston, TX), and written informed consent was obtained for all patients before performing study-related procedures.
Eligibility criteria:
Eligible patients were required to have histologically confirmed colorectal adenocarcinoma with liver-only metastases deemed resectable by a liver surgeon (resectability may involve the use of ablative techniques to some but not all liver metastases), measurable disease per RECIST v1.1, Eastern Cooperative Oncology Group (ECOG) Performance Status ≤1, normal organ and marrow function, any number of prior lines of systemic chemotherapy, and known MMR status. In addition, patients with an intact primary tumor that was planned for surgical resection were eligible.
Translational Analysis:
Flow cytometry staining of fresh tumor samples:
When feasible, tumors were subdivided for fresh flow cytometry analysis. Cells were first stained for surface antigens and a live/dead dye followed by fixation and permeabilized for intracellular staining as previously described (19). The antibody resources and dilutions used are listed in Supplemental Table S2. Gating was determined by using the fluorescence minus one approach. Samples were acquired using a BD Fortessa X20 and analyzed using FlowJo v 10.0.7. The gating strategy for T-cell and myeloid cell phenotyping is depicted in Supplementary Fig S4D and S4E.
RNA sequencing analysis:
We performed RNA sequencing on RNA extracted from fresh frozen tumor tissue. The sequencing reads were aligned to the UCSC GRCh37 assembly of the human genome with TopHat2 (20), and the read counts were resolved using HTSeq (21). The average coverage per base is ~118. We next applied DESeq2 to calculate differential expression between tissues groups of interest and transform count data for downstream analysis with variance stabilizing transformations (VST) (22). Genes with an adjusted p-value of < 0.05 were determined as differentially expressed genes (DEGs). We plotted heatmaps for DEGs and performed hierarchical clustering utilizing pheatmap, an R package (23). To further analyze the biological differences at a pathway level, we performed gene set enrichment analysis (GSEA) using the default settings of the GSEA software (24,25). Multiple pre-collected gene sets from the Molecular Signatures Database (MSigDB) were also selected, and included hallmark gene sets, C2 curated gene sets, and C7 immunologic gene sets (25).
Multiplex Immunofluorescence (mIF) and Immunohistochemistry (IHC) analyses:
The mIF analysis was conducted by a pathologist in five intratumoral areas using 660μm×500μm (0.33mm2) region of interest (ROI) at ×20 magnification to cover a total intratumoral area of 1.65mm2. In cases where five ROIs did not cover 1.65mm2 of intratumoral area, additional ROIs were included in the analysis. The final results were expressed as the average cell densities in any given area by mm2 (cells/mm2) (26). The mIF panels utilized are present in Supplementary Table S2. IHC staining and scoring of PD-L1, CD20 and CD73 are included in the supplementary material.
Microbial DNA isolation and 16S rRNA gene sequencing:
Pretreatment fecal baseline samples were collected from CRC patients. In brief, genomic DNA was isolated using QIAamp DNA stool mini kit (Qiagen), according to the manufacturer’s protocol, modified to include an intensive bead-beating lysis step. The V4 region of 16S rRNA gene was amplified by PCR from 10 ng of each of extracted and purified genomic DNA using 515 forward and 806 reverse primer pairs (27). The amplicon pool was purified with QIAquick gel extraction kit (Qiagen) and sequenced on the Illumina Miseq sequencer platform using 2 × 250 bp paired-end protocol. After sequencing, paired-end reads were de-multiplexed by QIIME and then merged and dereplicated for chimeras using VSEARCH. UNOISE 3 command algorithm was used to perform denoising of reads (28). Operational taxonomic units (OUTs) were classified using Mothur method with the Silva database version 138. For differential taxa-based univariate analysis, abundant microbiome taxa at species, genus, family, class, and order levels were analyzed using Mann-Whitney U-test after logit transformation. The detailed computational pipeline of analysis has been previously described (29). For exploratory analyses, p values have not been adjusted for multiple comparisons. Three patients analyzed for microbiome had antibiotics in the preceding six months. We did not observe any obvious or dramatic derangements in the fecal microbiome composition of patients treated with antibiotics previously, though numbers were small.
Statistical Analysis:
The planned study sample size was 25 patients, enabling a Bayesian 95% credible interval of surgery received to be (0.62, 0.92), assuming that the proportion of patients successfully getting to surgery is 80%. This utilized a beta-binomial model with a non-informative prior distribution of Beta (1.2, 0.8). Patients underwent interim analyses for futility, regimen toxicity, and post-surgical complications based on a Bayesian sequential monitoring design (30,31). The trial was able to continue by these rules, but was closed prior to completion of full enrollment due to programmatic reasons. Estimates of successful surgery rates are provided from the posterior distribution due to the multiple Bayesian interim analyses throughout the trial. OS and RFS were calculated and plotted by Kaplan-Meier methods (32). Relationships between baseline characteristics, immune markers, and outcomes (response, OS, or RFS) were explored with logistic or Cox models as previously described (33). Individual immune markers between pre and post treatment samples were compared with unpaired Student’s t-test. For the analysis across flow cytometry, IHC and RNA sequencing with efficacy, patients were stratified by relapse-free survival (RFS) into long RFS of >1.5 years or short RFS ≤1.5 years.
Results:
Patient Characteristics and Safety
A total of 24 mCRC patients were enrolled from 11/2016 to 9/2020 and 23 patients received trial treatment and are evaluable, Figure 1A. One patient withdrew from the study prior to study treatment and is not included in the study results. The median study follow-up is 2.3 years. Table 1A lists the baseline characteristics of the evaluable patients. Eighteen patients (78%) received preoperative chemotherapy with a median of 3.8 weeks from last chemotherapy dose to durvalumab and tremelimumab treatment. Of the 23 patients, 20 underwent surgical exploration (87%; 95%CI: 67-96%) and 17 underwent surgical resection (74%; 95%CI: 53-88%), Table 1B. The Bayesian posterior distribution estimate for the proportion of patients undergoing surgery with 95% credible interval (ci) is 85% (69%, 96%), while for patients undergoing complete surgical resection was 73% (54%, 88%). Immune-related toxicity did not prevent surgical intervention in any patients. The reasons for not undergoing surgical exploration were progression of previously noted sub-centimeter lung nodules in all three patients. Of the three patients who underwent surgical exploration but not resection, one patient had chemotherapy induced liver toxicity (this patient had received previous FOLFOXIRI chemotherapy) and two patients demonstrated more extensive liver metastatic disease than appreciated on imaging. Two of these patients subsequently went on to definitive liver treatment after addition interval therapy (liver resection in one patient and combined radiation/microwave ablation in one patient).
Figure 1: Overall Study Design and Clinical Data.

Figure 1A depicts the study design with treatment regime. Figure 1B shows assessment of pre-surgery RESIST response (dMMR patients are identified with # symbol). Tumor viability and response assessment shown in Figure 1C and Overall survival is shown in figure 1D. Relapse-free survival shown in Figure 1E.
Table 1A:
Baseline characteristics of the study patients (N=23)
| Patient and disease characteristics | Number |
|---|---|
| Age, median (range), years | 56 (28-69) |
| Female Sex | 11 (48%) |
| ECOG PS | |
| 0 | 14 (61%) |
| 1 | 9 (39%) |
| Primary tumor location | |
| Right | 1 (4%) |
| Left | 13 (57%) |
| Transverse | 2 (9%) |
| Rectal | 7 (30%) |
| Histological grade | |
| Moderately differentiated | 22 (96%) |
| Primary tumor stage, ≥T3 | 19 (83%) |
| Node positive primary | 13 (57%) |
| Synchronous metastases | 20 (87%) |
| Number of prior chemo lines, median (range) | 1 (1-3) |
| Extrahepatic disease | 4 (17%) |
| Pre-operative chemotherapy | |
| YesA | 18 (78%) |
| No | 5 (22%) |
| Pre-operative chemotherapy duration, median (range), months | 2 (1.5-6) |
| Pre-operative CEA, ng/mL, median (range) | 3 (0.9-65.7) |
| Tumor mutation | |
| POLE mutationB | 2 (9%) |
| BRAF mutation | 1 (4%) |
| KRAS mutation | 12 (52%) |
| TP53 mutation | 14 (61%) |
| APC mutation | 11 (48%) |
| MMR status | |
| pMMR | 21 (91%) |
| dMMR | 2 (9%) |
| Primary Tumor CMS (N=15) | |
| CMS1 | 0 (0%) |
| CMS2 | 7 (32%) |
| CMS3 | 4 (18%) |
| CMS4 | 4 (18%) |
Oxaliplatin-based in 12 (52%), irinotecan-based in 4 (17%) oxaliplatin + irinotecan based in 2 (9%); addition of bevacizumab in 15 (65%).
both POLE mutation patients were pMMR.
Table 1B:
Surgical outcomes of the patients in the study (N=20)
| Patient and disease characteristics | Number |
|---|---|
| Surgery | 20/23 (87%) |
| Type of surgery | |
| Major hepatectomy | 6 (30%) |
| Surgical margin | |
| R0 | 15 |
| R1 | 1 |
| R2 | 1 |
| Number of liver resections | |
| Median (range) | 1 (1-6) |
| Hepatic tumor size (cm) | |
| Median (range) | 2 (0.9-5.8) |
| Number of hepatic tumors | |
| Median (range) | 2 (1-10) |
| Pre surgery response | |
| PR | 3 (15%) |
| SD | 15 (75%) |
| PD | 2 (10%) |
| RFS (months) | |
| Median (range) | 9.7 (1.3-28.0) |
| OS (months) | |
| Median (range) | 24.5 (1.3-45.6) |
| Histopathological response (viability %) | |
| Median (range) | 30 (0-80) |
| Post-surgery complications | 8/20 (40%) |
| Anemia (grade 2) | 1 |
| Colon perforation (grade 4) | 1 |
| Upper respiratory infection (grade 2) | 1 |
| NGT placement (grade 1) | 1 |
| Pelvic anastomosis leak (grade 3) | 1 |
| Surgical site infection | |
| Grade 1 | 1 |
| Grade 2 | 2 |
A major hepatectomy, defined as ≥4 liver segments, was performed in 6 of the 17 resected patients, 30%. The median time from durvalumab/tremelimumab to surgical resection was 30 days (range 17 to 69 days). Sixteen of the 17 resected patients received adjuvant durvalumab in the post-operative setting, while one patient received adjuvant FOLFOX due to physician decision.
A total of 6 post-surgical complications occurred in 8 of 20 patients (40%), with 2 complications rated grade 3 or 4. A grade 3 anastomotic leak complication occurred in a patient who underwent concurrent ultra-low anterior resection of his rectal primary and a grade 4 colonic perforation occurred at another patient’s primary tumor location. In the eight patients with ≤28days between durvalumab/tremelimumab and surgical resection, two patients (25%) experienced a post-operative complication.
Durvalumab and/or tremelimumab related toxicities are presented in Supplementary Table S1. Five grade 3 or 4 treatment related adverse events occurred: fatigue, AST elevation, lipase elevation, oral mucositis, and thromboembolic event.
Clinical Efficacy
Pre-surgical radiographic response per RECIST v1.1 was stable disease in 15 (65%) patients, partial response in 3 (13%) patients and progressive disease in 5 (22%) patients (Fig. 1B, waterfall plot). Percent tumor cellularity in the 17 resected patients is shown in Fig. 1C. Four patients, 2 of whom were dMMR, demonstrated a complete pathological response with the presence of residual mucin in all cases. Of the two pMMR cases with complete pathological response, both had POLE P286R mutations with a tumor mutation burden of 61 per Foundation One assay in one patient and 42 mutations on the 138 gene Oncomine panel. Overall survival and relapse-free survival are shown in Fig. 1D–E and Table 1B. As 5 out of 17 patients did not receive pre-study systemic chemotherapy, we evaluated the various efficacy outcomes, percent tumor cellularity, RFS and OS, between these two groups and no significant differences were seen (Supplementary Figs S1A–S1C). Imaging and markers of the pMMR patients with POLE mutation are shown in the Supplementary Fig S2.
Immune profiling the tumor microenvironment following CTLA-4 and PD-L1 inhibition
Pre-treatment mandatory tumor biopsy and post-treatment surgery samples were collected along with pre-treatment microbiome samples (Fig. 1A). A total of 21 pre-treatment biopsies were collected but only 10 biopsies demonstrated >10% malignant cells. Reasons for inadequate biopsies were: normal liver in 4, necrotic tumor only in 2, and minimal tumor amount in 5. Post-treatment surgical resection samples were collected in 13 patients with 11 demonstrating >10% malignant cells and 2 demonstrating pathological complete response, no evidence of tumor cells in the samples. In total 6 paired (pre- and post-surgery) samples were obtained but one post-surgery sample was a pathological complete response with acellular mucin (pMMR patient) and thus 5 pairs with tumor tissue were available for analyses. Due to the known differences between pMMR and dMMR CRC, the two dMMR were excluded from all analyses except for when directly compared to pMMR samples.
To assess the impact of the combination of durvalumab/tremelimumab on the tumor immune infiltrates, we performed immunoprofiling using flow cytometry, IHC, mIF, and RNA sequencing of pre-treated and post-treated pMMR tumor samples.
Flow cytometry assessment of TIL activation and inhibitory receptors in pre versus post treatment samples found the intratumoral cytotoxic CD8+ T cell populations exhibited a significant increase in Lag3+ expression following treatment (p=0.017, Fig. 2A). While limited by sample size and sample heterogeneity, there were trends suggesting potential increases in PD-1+ (p=0.117), ICOS+ (p=0.284), and Tim3+ (p=0.121) expression by CD8+ TIL (Fig. 2A). No markers were significantly decreased post-treatment. When comparing post-treatment CD4+ T cell subsets (Fig. 2B), no significant changes were observed within CD4+ TIL subsets, except for a trend with increased ICOS+ (p=0.111) levels. Treatment effects on other immune cell populations are shown in Fig. 2C and demonstrate similar levels of various myeloid and dendritic subtypes.
Figure 2: Immune Infiltrate and Gene Expression Response to Therapy.
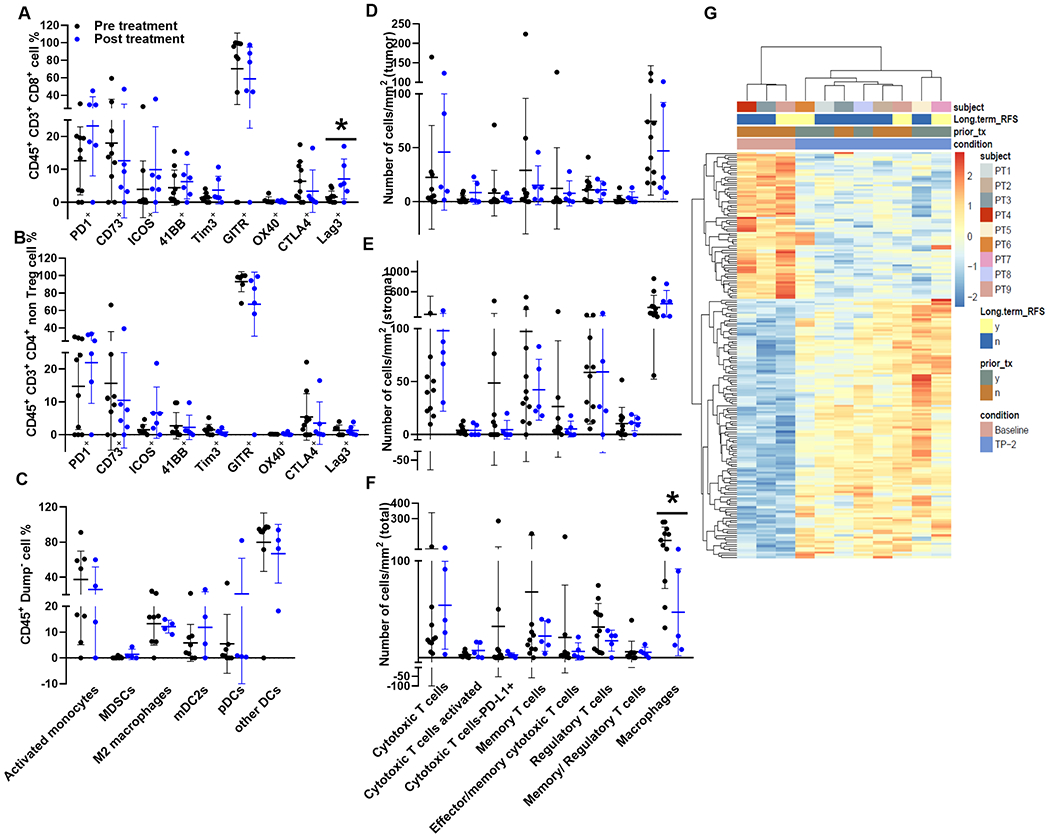
All analysis were done only on pMMR tumors. Panel 2A shows the intratumoral CD8+ immune cell subsets found within pre- and post-treatment (black vs blue) tumors that were analyzed by flow cytometry. Panel 2B illustrates comparable flow cytometry profiles for intratumoral CD4+ non-T-reg immune cell subsets. Panel 2C shows different subsets flow cytometry profiles of myeloid cells found in pre- and post-treatment samples. Immunofluorescence staining of FFPE tissue slices to identify and quantify immune cell infiltrates in the tumor (2D), stroma (2E), and total (2F) compartments based on cells/mm2 pre- and post- treatment. Panel 2G shows RNA sequencing analysis of differentially expressed genes between pre-and post-treatment.
Immune cell distribution and infiltrates were analyzed by mIF to evaluate tumor (Fig. 2D), stroma (Fig. 2E), and total (Fig. 2F) compartments between baseline and post treatment samples. These findings are represented as cells/mm2 and reflect similarities with flow cytometric analyses. Intratumor distribution illustrated in Fig. 2D demonstrates a trend towards an increase in activated cytotoxic T cells (CD8+ GZB+, p=0.094) and no significant decrease in macrophages (p=0.431). Similar trends in immune changes were also observed in the stromal region (Fig. 2E). The combined tumor and stromal region (Fig. 2F) show trends towards an increase in activated cytotoxic T cells (CD8+ GZB+, p=0.099) and decreases in both T regulatory cells (p=0.239) and macrophages (p=0.018) following treatment.
In addition, RNA sequencing analysis identified 188 differentially expressed genes (DEG) that were significantly (adjusted p<0.05) different between baseline and post treated pMMR samples (Fig. 2G, Supplementary. Table S3). Gene set enrichment analysis against hallmark gene sets, C2 curated chemical and genetic perturbations gene sets, and C7 immunologic signatures of treated versus baseline samples was performed. Supplementary. Table S3 lists modulation of gene sets involved in the T cell, myeloid cell, cytokine, and other immune related pathways between treated vs baseline samples. An upregulation in the inflammatory response genes and in signaling pathways such as JAK/STAT3 and STAT5 observed in the Hallmark gene sets along with the down modulation of FOXP3 targets in CD4 T cells in the C2 gene sets supports the activation of T cells. In addition, a downregulation of pro-inflammatory monocyte genes in C7 gene and an upregulation of DNA damage response in the C2 gene sets were also observed.
Treatment induced immune changes in paired tumor samples
Given the immune changes observed above across the cohort, we next focused on an exploratory analysis of samples that were paired pre and post- surgery from pMMR patients (n=6). Of the 6 paired (pre- and post-surgery) tumor samples, 5 pairs were analyzed using IHC and immunofluorescence, while 3 pairs passed the quality control metrics to allow flow cytometry sub-gating of CD8 and CD4 TIL subsets, and myeloid lineage stratifications. Flow analysis demonstrated non-statistically significant changes, with a decrease in CD8+ T cell percentage, yet trends toward an increase in the percentages of CD8+ ICOS+, CD8+ 41BB+, CD8+ Tim3+, CD8+ Lag3+, and CD8+ PD-1+ cells (Fig. 3A–3F). Similar results were observed by immunofluorescence staining with non-statistically significant increases in activated cytotoxic T cells (CD8+ GZB+), CD8+ PD-L1+ cells, and effector memory cells in post treated samples, panels 3M-3P.
Figure 3: Immune Infiltrate Changes in Response to Therapy.

Immune changes in paired pre vs. post treatment pMMR tumor samples are shown. Panel 3A shows CD8+ T cell percentage changes along with ICOS+ (3B), 41BB+ (3C), Lag3+ (3D), Tim3+ (3E), and PD-1+ (3F) CD8 T cells analyzed by flow cytometry. Panel 3G shows the percentage of T regulatory cells analyzed by flow cytometry along with CD4+ non-T regs (3H), CD4+ PD-1+ non T regs (3I), and CD4+ ICOS+ non T regs (3J) analyzed by flow cytometry. Panel 3K demonstrates treatment induced changes occurring in activated monocytes (3K) and M2 macrophages (3L) analyzed by flow cytometry. Additional CD8+ T cell (3M) and subsets include activated cytotoxic T cells (CD8+ GZB+) (3N), CD8+ PD-L1+ cells (3O), and effector memory cells (3P) analyzed by immunofluorescence staining. Panels 3Q shows T regulatory and panel 3R shows memory T regulatory cells analyzed by immunofluorescence. Panel 3S shows CD20+ B cell staining and Panel 3T shows CD73 tumoral staining by IHC.
Trends in CD4+ T cells were also observed with increases in CD4+ T regs by both flow cytometry (panel 3G) and immunofluorescence (panel 3Q). Decreases in the myeloid population were seen with non-statistically significant decreases in both activated monocytes (panel 3K) and M2 macrophages (panel 3L) by flow cytometry. Interestingly, B cell densities increased in 3 out of 5 samples post treatment (panel 3S).
Immune markers and disease relapse
To identify potential factors that may associate with disease relapse, pMMR tumors were stratified based upon recurrence and assessed for differences in immune populations and gene expression profiles as well as microbiome profiles at baseline and surgery. At baseline, patients with shorter RFS trended towards lower frequencies of CD8+ PD-1+ TIL compared to those patients with longer RFS (RFS short vs long, p=0.079, Fig. 4A). Other markers of activation or inhibition on CD8+ or CD4+ T cells were not observed to be different at baseline (Fig. 4A and B). Overall, myeloid populations were not differentially present at baseline between samples with longer RFS and shorter RFS (Fig. 4C).
Figure 4: Immune Infiltrate and Microbiome Changes Associated with Relapse.

All analyses were done only on pMMR tumors. Subsets of infiltrating immune cell change relative to baseline samples stratified by RFS including CD8+ (4A), CD4+ non T-regs (4B), and myeloid cells (4C). Heatmap in panel D is shows the differences in the microbiome taxa that were significant between short vs long RFS baseline samples. In an exploratory analysis, p values have not been corrected for multiple comparisons. Subsets of infiltrating immune cell change relative to post treatment samples stratified by RFS including CD8+ (4E) and CD4+ non T-regs (4F). A rank order list based on p-values of differentially expressed genes (DEG) identified by RNA sequencing between long vs short RFS in post treatment samples is shown in panel 4G. Differences observed in B cell numbers/nm2 in short vs long RFS pretreated samples are shown in panel 4H and post treated samples in panel 4I.
When comparing surgical cases post treatment based upon RFS, although CD8+ TIL percentage itself did not change (Supplementary Fig. S5A, p=0.662), we observed a significant difference in the frequency of Tim3 expression on CD8+ TIL in longer RFS cases (RFS short vs long, p=0.013 Fig. 4E). Expression of other activation and inhibitory receptors on CD4 or CD8 TIL was not different based upon RFS (Fig. 4E and F). Supervised DEG analyses between long and short RFS demonstrated a dramatic B cell immunoglobulin predominance in the post-treatment samples of the long RFS patients (Fig. 4G and Supplementary Table 4). This B cell response was characterized by a robust expression of immunoglobulin genes, including IGHG1, IGHG2, IGHG3, and IGHD genes. CD20 staining by IHC demonstrated similar findings with a trend toward an increase in B cell density in post-treatment samples with longer RFS (p=0.062, panel 4I). Interestingly, in the analysis of pre-treatment supervised DEG there was no evidence for an enrichment of a B-cell response in the long RFS cohort, Supplementary Table 4, and no increase in CD20 density in pre-treatment long RFS patients (4H, p=0.308). In addition, higher tumoral CD73 expression (≥10% versus <10%) post treatment correlated with improved RFS (S5F, p=0.002). CD20+ lymphoid aggregate densities within the intratumoral compartment were also included in the analysis (Supplementary Fig S5G).
Microbiome analysis
The genus level differences in microbiome composition stratified by patients who had short term RFS and those that had long term RFS benefit is shown in Supplementary Fig. S5C and Supplementary Table 5. In an exploratory analysis (Fig. 4D) the abundance of certain microbiome species were correlated with disease RFS with an enrichment of Murimonas intestine (p=0. 038), and a diminution of species Blautia caecimuris (p=0037), Blautia hominis (p=0.019), Enterococcus malodoratus (p=0.006), Anaerostipes caccae (p=0.023), Fusobacterium simiae (p=0.008), Lactobacillus salivarius (p=0.039), Scardovia wiggsiae (p=0.039), Maihella massiliensis (p=0.037), Actinomyces viscosus (p=0.039), Dialister propionicifaciens (p=0.039) and genus Lachnoclostridium (p=0.013), Scardovia (p=0.039), Desulfovibrionaceae unclass (p=0.037) in patients with shorter RFS.
Immune profiles of dMMR and pMMR CRC
To assess the baseline immune characteristics in pMMR and dMMR tumors, we evaluated tumor immune markers using flow cytometry and compared 2 dMMR samples with pMMR samples in Fig. 5. The expression pattern of activation and inhibitory receptors within the CD8+ (Fig. 5A) and CD4+ non-T-regs (Fig. 5B), and frequencies of TIL (Fig. 5C) and myeloid populations (Fig. 5D) in baseline pMMR vs dMMR samples are also shown. Significantly higher expression of Tim3+ (p=0.013) and surface CTLA4+ (p=0.012) was observed in the CD8+ TIL subset in baseline dMMR tumors compared to pMMR tumors suggesting the presence of a greater number of activated T cells in dMMR patients pre-treatment. However, there was no significant difference in the frequency of TIL or myeloid subsets between dMMR and pMMR tumors. Overall, more cases are needed to verify these findings. Given that the 2 dMMR patients demonstrated complete pathological responses to inhibition of the CTLA-4 and PD-1/PD-L1 axes, it suggests that this combination may be functionally important given the higher expression of CTLA-4 observed in these patient samples.
Figure 5: Treatment Related Immune Response Profile Comparisons by MMR Subsets.
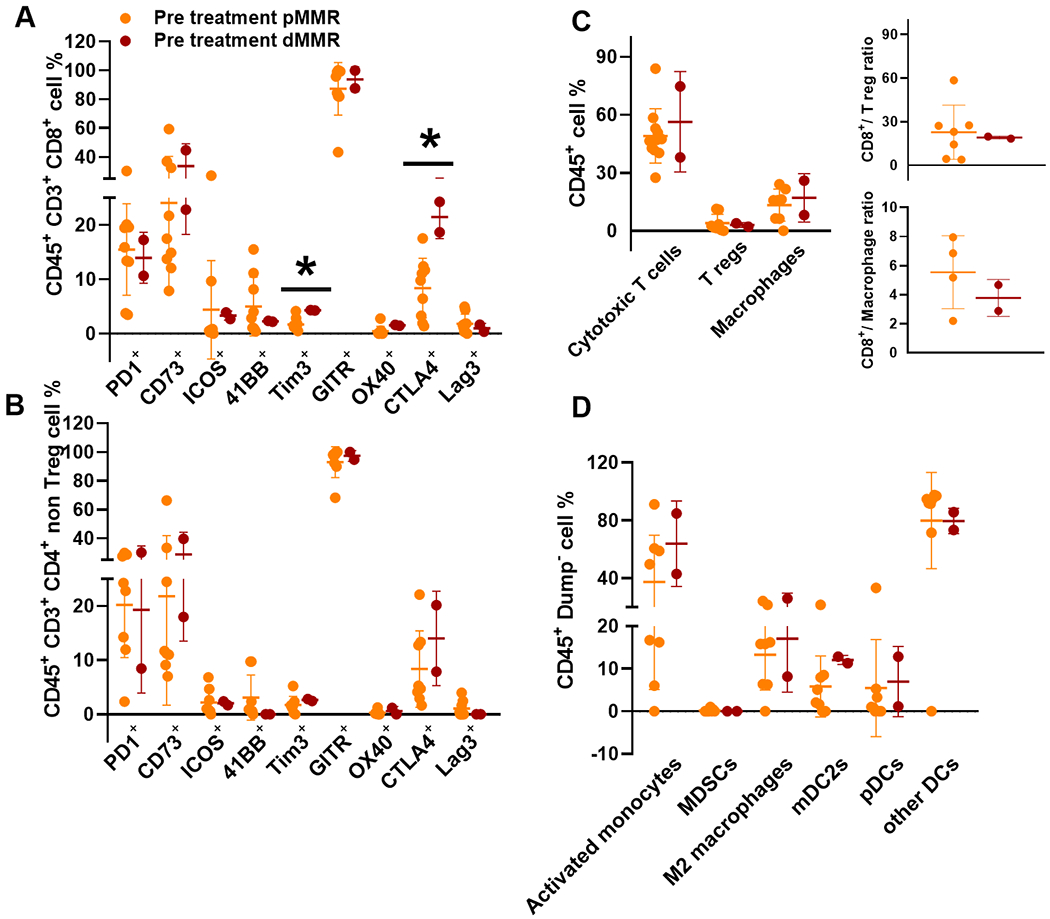
Baseline samples of pMMR (orange) vs dMMR (red) with subsets of CD45+ CD3+ CD8+ (5A), CD45+ CD3+ CD4+ non T-regs (5B), and myeloid cells (5C) that were analyzed by flow cytometry. Inserts in panel 5C show the ratios of cytotoxic T cells and T regulatory cells (top) and cytotoxic T cells and macrophages (bottom). Panel D shows the differences in the myeloid panel analyzed by flow cytometry.
Discussion:
In this 23 patient trial, neoadjuvant use of durvalumab and tremelimumab prior to CRC liver resection was well tolerated and feasible with 73% of patients undergoing surgical resection. In addition, we show the rapid activity of combined CTLA-4 and PD-L1 therapy in dMMR CRC with both dMMR patients demonstrating complete pathological responses after one dose of therapy. Within pMMR CRC we show that treatment with durvalumab and tremelimumab was able to produce modest T-cell activation and a post-treatment B cell signature was found to correlate with long-term relapse-free survival benefit.
The Bayesian posterior distribution estimate for the proportion of patients undergoing surgery of 73% in conjuncture with the overall safety demonstrated that perioperative durvalumab and tremelimumab was feasible, meeting the primary endpoint of this study. The perioperative surgical resection setting provides the potential for robust tumor acquisition from surgical resection. However, our results also demonstrate the challenges with pre-treatment biopsies in the perioperative setting as over half of our pre-treatment biopsies were inadequate for analysis. This may reflect this study’s selection of resectable liver metastases, where metastatic tumor size maybe smaller, as reflected by the median size of resected tumors being 2cm. In this trial we focused upon quantifying T and B cell percentages, phenotyping T cell subpopulations, and evaluating the changes in their distribution with combination treatment which was achieved by comparing the results obtained by flow cytometry, transcriptome, and immunofluorescence multiplex analysis. Due to the known unique nature of dMMR CRC, these two patients were excluded from all analyses, except for the direct comparison between dMMR and pMMR CRC samples. Overall, we observed a general concordance of results across approaches with an increase in cytotoxic T cell activation and B cell enrichment, in pMMR CRC following treatment with anti-PD-L1 and anti-CTLA-4 combination therapy.
Assessment of immune markers in post treated specimens showed consistent increases in CD8+ T cell activation, and a decrease in the percentages of macrophages in tumors reflecting a dynamic interplay between the immunosupportive and immunosuppressive cell types in mediating anti-tumor immune responses. Lag3+ in particular was markedly amplified in CD8+ T cells following treatment. In other studies, antibody blockade of LAG3 increased proliferation and effector cytokine production of intra-tumoral T cells isolated from pMMR metastatic CRC, suggesting a potential role for combination immune-checkpoint inhibition in pMMR (34).
RNA sequencing analysis showing increases in CD86 (an antigen presenting cell-specific marker) and CD69 (an early activation marker) gene expression, is suggestive of a response to anti-CTLA-4 therapy in these patients that mediates an upregulation in co-stimulatory signals that are necessary for T cell activation and survival (Supplementary Table 3). However, the lack of significant increases in the expression of co-inhibitory receptors CTLA-4 and PD-1 in patients after 4 weeks of treatment suggests that these T cells have not reached exhaustion and may respond to continuation of therapies that target CTLA-4 and PD-1/PDL-1 (35). In addition, enrichment in interferon gamma signature, JAK/STAT signaling, and inflammatory response genes was observed in post treated tumors suggesting a potential immune activation mechanism.
Despite these immunological changes, the median RFS of 9.7 months and median OS of 24.5 months do not appear improved over the expected outcomes for resected colorectal cancer liver metastases (9,10). Recently, in the randomized CO.26 clinical trial of durvalumab/tremelimumab versus best supportive care in refractory metastatic CRC, the response rate of durvalumab/tremelimumab was 1% and median PFS was 1.8 months, which was similar to the best supportive care arm of 1.9 months (p=0.97) (16). However, an improvement in OS (6.6m vs. 4.1m, p=0.07) was seen for durvalumab/tremelimumab. In addition the NICHE trial investigating nivolumab and ipilimumab prior to the resection of localized primary colon adenocarcinomas demonstrated a 27% pathological complete response in 20 pMMR patients (36). Whether this signal of activity relates to the earlier non-metastatic stage of patients is not known. In the context of this study, our findings demonstrating T cell activation suggest that additional factors potentially related to the immunosuppressive microenvironment or the liver or insufficient T cell recruitment may be contributors to the limited clinical activity of this combination in pMMR CRC. A recent study combining the use of durvalumab/tremelimumab following Yttrium-90 resin microsphere-based radioembolization also demonstrated the limited effects of radiation on the infiltration of TILs and tumor microenvironment in pMMR, although there was a transient increase of cyclin dependent kinase inhibitor 1A (CDKN1A: p21CIP1) and TNF receptor superfamily member 10c (TNFRSF10C: TRAILR3) expression (17).
The B cell activation signature seen in post treated tumor samples with longer RFS suggests a role of B cells in promoting response, leading to important biomarkers for therapy and contributing a novel finding in CRC. The existing data from literature captures a very limited role of tumor associated B cells within the tumor microenvironment, with few studies suggesting a positive correlation of B cell signature with improved outcome. Along with the production of tumor-specific antibodies and cytokines some of the other functions attributed to B cell presence within the tumor include presentation of B cell receptor-cognate antigens to T cells, or their role in enhancing antigen capture and presentation by dendritic cells (37,38). A recent study linking driver mutations and B-cell response showed that abundance and a high proportion of IgG1 isotype were associated with improved overall survival for KRAS mutant but not KRAS wildtype lung adenocarcinoma. Here IGH transcript upregulation was reported to be associated with longer survival in melanoma and lung adenocarcinoma (39). A recent phase II trial of neoadjuvant immune checkpoint blockade in patients with resectable melanoma, also reports an increased B cell infiltration contributing to response to therapy in patients (40). Our results similarly show that patients with longer RFS have increased IGH levels (Supplementary Table 4).
Another contributing factor in modulating the host inflammatory response and influencing the outcome of cancer therapy is the gut microbiome and dysbiosis in the colon (41,42). Gut microbiota can modulate intestinal immunity, increase inflammation and the risk of CRC. In addition, gut microbiome has been associated with response to PD-1 and CTLA-4 blockade (43,44). Analysis of pre-treatment microbiome profiles in these patients revealed that the enrichment of species Murimonas intestini correlates with shorter RFS. Similar to reports in the literature that have identified Lachnoclostridium as a marker for non-invasive diagnosis of CRC, we observed an abundance in this genus in our patient cohort (45,46). Our results show that the abundance of Blautia species and Anaerostipes caccae is associated with longer RFS. Blautia was previously reported to be associated with pMMR mCRC (47). Baseline enrichment in Blautia has also been reported to be associated with longer PFS (44). Anaerostipes caccae, a butyrate producer, can convert lactate to butyrate. Butyrate is known to function in the suppression of inflammation and cancer (48).
The main limitations of our study relates to the small number of patients and even smaller patient numbers that were able to be fully analyzed for immune phenotyping. Given these findings and the design of this study for feasibility and safety as the primary endpoint, the efficacy and translational analyses are hypothesis generating and require further confirmation in additional datasets. In addition, this study did not complete enrollment and the 95% confidence intervals for surgical resection are broad, suggesting that further efforts to confirm the feasibility of immunotherapy window studies prior to CRC liver metastases resection are needed. This study did not investigate the role of repeated dosing of durvalumab and tremelimumab, as only one dose of each agent was given prior to surgical resection.
In conclusion the use of anti–CTLA-4 and anti-PD-L1 in metastatic CRC demonstrated immune activation with regard to both T cell and B cells, though robust clinical activity was only seen in dMMR patients. However, the finding of a post-treatment B cell signature suggests the importance of improved understanding of the B cell context within the immunotherapy treatment space. Finally, this study demonstrates the safety of a perioperative immunotherapy approach in mCRC and supports further efforts to utilize this disease space in order to make both clinical and translational improvements in CRC.
Supplementary Material
Statement of translational relevance:
This study evaluates the feasibility and effectiveness of durvalumab and tremelimumab combination immunotherapy in colorectal cancer patients with resectable liver metastases. Treatment was associated with an expected toxicity profile and no apparent increase in post-operative complications. Neoadjuvant durvalumab and tremelimumab demonstrated no increase in the number of T-cells but evidence of T-cell activation was observed in mismatch repair proficient metastatic colorectal cancer patients. In addition, a post-treatment tumoral B-cell signature was associated with patients that had prolonged relapse-free survival. In summary, this study demonstrates the safety of a perioperative immunotherapy approach in metastatic colorectal cancer (CRC). Further efforts to utilize this treatment space are needed in order to make both clinical and translational improvements in CRC.
Acknowledgments
Financial support: This work was supported through a Medimmune/Astra-Zeneca MD Anderson Cancer Center alliance, by the National Cancer Institute through both the Cancer Center Support Grant P30CA16672 (Institutional Tissue Bank (ITB) and Research Histology Core Laboratory (RHCL)), and SPORE Grant P50CA221707, and supported by the generous philanthropic contributions to The University of Texas MD Anderson Cancer Center Moon Shots Program™. Adaptive Patient-Oriented Longitudinal Learning and Optimization (APOLLO) Moonshot Program, Strategic Alliances and the Translational Molecular Pathology-Immunoprofiling lab (TMP-IL) at the Department Translational Molecular Pathology, the University of Texas MD Anderson Cancer Center and the National Cancer Institute (NCI) Cooperative Agreement U24CA224285 (to the MD Anderson Cancer Center CIMAC).
Author’s disclosures of competing interests:
Overman (Research funding: Medimmune/Astra-Zeneca, BMS, Merck, Takeda, and Apexigen. Consulting: Merck, Astra-Zeneca, Takeda, Pfizer, Array, Gritstone, and Promega).
Haymaker (advisory board for Briacell).
Tidwell (salary support provided to department by Galera Pharmaceuticals).
Kopetz (Research funding: Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, Daiichi Sankyo. Consulting: Roche, Genentech, EMD Serono, Merck, Karyopharm Therapeutics, Amal Therapeutics, Navire Pharma, Symphogen, Holy Stone, Biocartis, Amgen, Novartis, Lilly, Boehringer Ingelheim, Boston Biomedical, AstraZeneca/MedImmune, Bayer Health, Pierre Fabre, Redx Pharma, Ipsen, Daiichi Sankyo, Natera, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata).
All other authors declare no competing interests.
References
- 1.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA: a cancer journal for clinicians 2020. doi 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Adam R, Vinet E. Regional treatment of metastasis: surgery of colorectal liver metastases. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2004;15 Suppl 4:iv103–6 doi 10.1093/annonc/mdh912. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians 2005;55(2):74–108. [DOI] [PubMed] [Google Scholar]
- 4.Leporrier J, Maurel J, Chiche L, Bara S, Segol P, Launoy G. A population-based study of the incidence, management and prognosis of hepatic metastases from colorectal cancer. The British journal of surgery 2006;93(4):465–74 doi 10.1002/bjs.5278. [DOI] [PubMed] [Google Scholar]
- 5.Ismaili N Treatment of colorectal liver metastases. World J Surg Oncol 2011;9:154 doi 10.1186/1477-7819-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choti MA, Sitzmann JV, Tiburi MF, Sumetchotimetha W, Rangsin R, Schulick RD, et al. Trends in long-term survival following liver resection for hepatic colorectal metastases. Annals of surgery 2002;235(6):759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdalla EK, Adam R, Bilchik AJ, Jaeck D, Vauthey JN, Mahvi D. Improving resectability of hepatic colorectal metastases: expert consensus statement. Annals of surgical oncology 2006;13(10):1271–80 doi 10.1245/s10434-006-9045-5. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher DJ, Zheng J, Capanu M, Haviland D, Paty P, Dematteo RP, et al. Response to neoadjuvant chemotherapy does not predict overall survival for patients with synchronous colorectal hepatic metastases. Annals of surgical oncology 2009;16(7):1844–51 doi 10.1245/s10434-009-0348-1. [DOI] [PubMed] [Google Scholar]
- 9.Nordlinger B, Guiguet M, Vaillant JC, Balladur P, Boudjema K, Bachellier P, et al. Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Association Francaise de Chirurgie. Cancer 1996;77(7):1254–62. [PubMed] [Google Scholar]
- 10.Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. The Lancet Oncology 2013;14(12):1208–15 doi 10.1016/s1470-2045(13)70447-9. [DOI] [PubMed] [Google Scholar]
- 11.Shields JD, Kourtis IC, Tomei AA, Roberts JM, Swartz MA. Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21. Science (New York, NY) 2010;328(5979):749–52 doi 10.1126/science.1185837. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev 2018;32(19-20):1267–84 doi 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. British journal of cancer 2011;105(1):93–103 doi 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(8):773–9 doi 10.1200/jco.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 15.Katsiampoura A, Raghav K, Jiang ZQ, Menter DG, Varkaris A, Morelli MP, et al. Modeling of Patient-Derived Xenografts in Colorectal Cancer. Molecular cancer therapeutics 2017;16(7):1435–42 doi 10.1158/1535-7163.mct-16-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen EX, Jonker DJ, Loree JM, Kennecke HF, Berry SR, Couture F, et al. Effect of Combined Immune Checkpoint Inhibition vs Best Supportive Care Alone in Patients With Advanced Colorectal Cancer: The Canadian Cancer Trials Group CO.26 Study. JAMA oncology 2020;6(6):1–8 doi 10.1001/jamaoncol.2020.0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Park J, Ouyang C, Longmate JA, Tajon M, Chao J, et al. A Pilot Feasibility Study of Yttrium-90 Liver Radioembolization Followed by Durvalumab and Tremelimumab in Patients with Microsatellite Stable Colorectal Cancer Liver Metastases. Oncologist 2020;25(5):382–e776 doi 10.1634/theoncologist.2019-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dindo D, Demartines N, Clavien PA. Classification of surgical complications: a new proposal with evaluation in a cohort of 6336 patients and results of a survey. Annals of surgery 2004;240(2):205–13 doi 10.1097/01.sla.0000133083.54934.ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radvanyi LG, Bernatchez C, Zhang M, Fox PS, Miller P, Chacon J, et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2012;18(24):6758–70 doi 10.1158/1078-0432.CCR-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14(4):R36 doi 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9 doi 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolde R, Vilo J. GOsummaries: an R Package for Visual Functional Annotation of Experimental Data. F1000Research 2015;4:574 doi 10.12688/f1000research.6925.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34(3):267–73 doi 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget M-A, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Scientific reports 2017;7(1):13380- doi 10.1038/s41598-017-13942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal 2012;6(8):1621–4 doi 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv; 2016. [Google Scholar]
- 29.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med 2018;24(12):1804–8 doi 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thall PF, Simon RM, Estey EH. Bayesian sequential monitoring designs for single-arm clinical trials with multiple outcomes. Stat Med 1995;14(4):357–79. [DOI] [PubMed] [Google Scholar]
- 31.Thall PF, Sung HG. Some extensions and applications of a Bayesian strategy for monitoring multiple outcomes in clinical trials. Stat Med 1998;17(14):1563–80 doi . [DOI] [PubMed] [Google Scholar]
- 32.Kaplan EL, Meier P. Nonparametric estimator from incomplete observations. . J Am Stat Assoc 1958;53:457–81. [Google Scholar]
- 33.Cox DR. Regression Models and Life-Tables. J R Stat Soc B 1972;34(2):187-+. [Google Scholar]
- 34.Zhou G, Noordam L, Sprengers D, Doukas M, Boor PPC, van Beek AA, et al. Blockade of LAG3 enhances responses of tumor-infiltrating T cells in mismatch repair-proficient liver metastases of colorectal cancer. Oncoimmunology 2018;7(7):e1448332 doi 10.1080/2162402X.2018.1448332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016;44(5):989–1004 doi 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalabi M, Fanchi LF, Dijkstra KK, Van den Berg JG, Aalbers AG, Sikorska K, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nature Medicine 2020;26(4):566–76 doi 10.1038/s41591-020-0805-8. [DOI] [PubMed] [Google Scholar]
- 37.Germain C, Gnjatic S, Tamzalit F, Knockaert S, Remark R, Goc J, et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. American journal of respiratory and critical care medicine 2014;189(7):832–44 doi 10.1164/rccm.201309-1611OC. [DOI] [PubMed] [Google Scholar]
- 38.Rivera A, Chen CC, Ron N, Dougherty JP, Ron Y. Role of B cells as antigen-presenting cells in vivo revisited: antigen-specific B cells are essential for T cell expansion in lymph nodes and for systemic T cell responses to low antigen concentrations. International immunology 2001;13(12):1583–93 doi 10.1093/intimm/13.12.1583. [DOI] [PubMed] [Google Scholar]
- 39.Isaeva OI, Sharonov GV, Serebrovskaya EO, Turchaninova MA, Zaretsky AR, Shugay M, et al. Intratumoral immunoglobulin isotypes predict survival in lung adenocarcinoma subtypes. Journal for immunotherapy of cancer 2019;7(1):279 doi 10.1186/s40425-019-0747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med 2018;24(11):1649–54 doi 10.1038/s41591-018-0197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergsten E, Mestivier D, Amiot A, DeAngelis N, Khazaie K, Sobhani I. Immune tolerance to colon cancer is mediated by colon dysbiosis: Human results and experimental In Vivo validation. Journal of Clinical Oncology 2020;38(15_suppl):e16062–e doi 10.1200/JCO.2020.38.15_suppl.e16062. [DOI] [Google Scholar]
- 42.Toker J, Arora R, Wargo JA. The Microbiome in Immuno-oncology. Advances in experimental medicine and biology 2020;1244:325–34 doi 10.1007/978-3-030-41008-7_19. [DOI] [PubMed] [Google Scholar]
- 43.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science (New York, NY) 2018;359(6371):97–103 doi 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2019;30(12):2012 doi 10.1093/annonc/mdz224. [DOI] [PubMed] [Google Scholar]
- 45.Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2019. doi 10.1136/gutjnl-2019-318532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangifesta M, Mancabelli L, Milani C, Gaiani F, de’Angelis N, de’Angelis GL, et al. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci Rep 2018;8(1):13974 doi 10.1038/s41598-018-32413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hale VL, Jeraldo P, Chen J, Mundy M, Yao J, Priya S, et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome medicine 2018;10(1):78 doi 10.1186/s13073-018-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwiertz A, Hold GL, Duncan SH, Gruhl B, Collins MD, Lawson PA, et al. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Systematic and applied microbiology 2002;25(1):46–51 doi 10.1078/0723-2020-00096. [DOI] [PubMed] [Google Scholar]
- 49.Posch F, Silina K, Leibl S, Mündlein A, Moch H, Siebenhüner A, et al. Maturation of tertiary lymphoid structures and recurrence of stage II and III colorectal cancer. Oncoimmunology 2018;7(2):e1378844 doi 10.1080/2162402x.2017.1378844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vijayan D, Barkauskas DS, Stannard K, Sult E, Buonpane R, Takeda K, et al. Selective activation of anti-CD73 mechanisms in control of primary tumors and metastases. Oncoimmunology 2017;6(5):e1312044 doi 10.1080/2162402x.2017.1312044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21(11):1350–6 doi 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


