Abstract
Cell death and immune response are at the core of life. In past decades, the endoplasmic reticulum (ER) protein STING1 (also known as STING or TMEM173) was found to play a fundamental role in the production of type I interferons (IFNs) and pro-inflammatory cytokines in response to DNA derived from invading microbial pathogens or damaged hosts by activating multiple transcription factors. In addition to this well-known function in infection, inflammation, and immunity, emerging evidence suggests that the STING1-dependent signaling network is implicated in health and disease by regulating autophagic degradation or various cell death modalities (e.g., apoptosis, necroptosis, pyroptosis, ferroptosis, mitotic cell death, and immunogenic cell death [ICD]). Here, we outline the latest advances in our understanding of the regulating mechanisms and signaling pathways of STING1 in autophagy and cell death, which may shed light on new targets for therapeutic interventions.
Subject terms: Cell biology, Immunology
Introduction
Pathogen-associated molecular patterns (PAMPs) derived from microorganisms and damage-associated molecular patterns (DAMPs) produced by host cells are recognized by pattern recognition receptors (PRRs), which play a fundamental role in innate immunity during infection and tissue damage1,2. Major PAMPs include microbial nucleic acids (DNA and RNA) and membrane components (e.g., lipopolysaccharide [LPS]), whereas the host DNA is an important DAMPs. Dysregulation of DNA-sensing pathways is implicated in various diseases, such as autoimmune diseases and cancer. In 2008 and 2009, stimulator of interferon response cGAMP interactor 1 (STING1, also known as STING, TMEM173, MITA, or MPYS) was identified by multiple groups as a key adapter in DNA-mediated innate immunity3–6. In 2013, Dr. Chen’s group ultimately identified that cyclic GMP-AMP synthase (CGAS) is a direct cytosolic DNA sensor that elicits robust innate immune responses through STING17. These studies have established a new DNA recognition pathway in the innate immune system8,9.
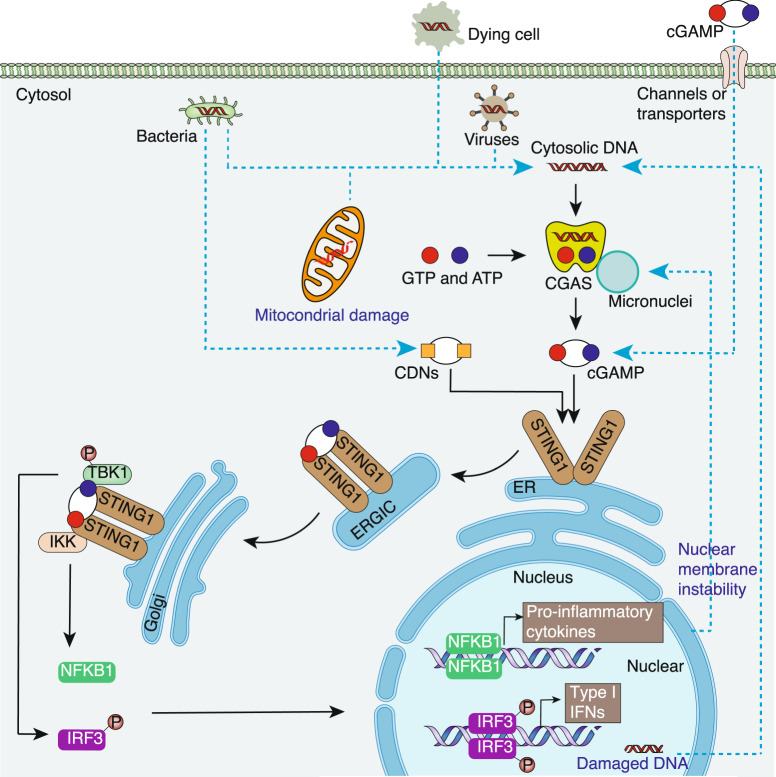
STING1 is an evolutionarily conserved transmembrane protein that localizes to the endoplasmic reticulum (ER) membrane in immune and non-immune cells. As an adapter protein, STING1 can be directly activated by bacterial cyclic dinucleotides (CDNs, such as cyclic-di-GMP and cyclic-di-AMP) or the second messenger cyclic guanosine monophosphate-adenosine monophosphate (cGAMP)10,11, a process that is involved in inflammation and immune response by producing type I interferons (IFNs) and pro-inflammatory cytokines3,8. The cGAMP is produced from CGAS, which detects and binds DNA substrates from invading pathogens (e.g., DNA viruses, retroviruses, and bacteria) or damaged hosts (including mitochondrial DNA [mtDNA] and nuclear DNA [nDNA]) during various stresses12 (Fig. 1). In addition, cGAMP or host DNA can be transferred between cells and activate STING1 through channels and transporters13–15, dying cell debris, or DNA-containing extracellular vesicles16. In addition to cytoplasmic CGAS, plasma membrane receptors (such as epidermal growth factor receptor [EGFR] and ALK receptor tyrosine kinase [ALK]) can also activate STING1 in response to exogenous cGAMP or CDNs in immune cells17. Overall, these findings indicate that the activation of STING1 can be induced by both CGAS-dependent and -independent pathways.
Fig. 1.
The CGAS-STING1 pathway. A critical cytosolic DNA sensor, CGAS elicits robust innate immune responses through the production of 2′3′-cGAMP, which binds and activates STING1 in response to DNA from pathogens or hosts. In addition, bacteria-produced CDNs can directly activate STING1 in a CGAS-independent manner. Activated STING1 exits the ER membrane and travels to Golgi via the ERGIC, leading to the production of type I IFNs by activating the TBK1-IRF3 pathway. STING1 also activates NFKB1-dependent pro-inflammatory cytokine expression. CDNs cytosolic cyclic dinucleotides, cGAMP cyclic GMP-AMP, CGAS cyclic GMP-AMP synthase, ER endoplasmic reticulum, ERGIC endoplasmic reticulum-Golgi intermediate compartment, IFN interferon, IKK IκB kinase, IRF3 interferon regulatory factor 3, NFKB1 nuclear factor kappa B subunit 1, STING1 stimulator of interferon response cGAMP interactor 1, TBK1 TANK binding kinase 1
After activation, STING1 on the ER undergoes oligomerization18, leaving the ER through the chromosome X open reading frame 56 (CxORF56) and coat protein complex II (COPII)19–21, and finally translocating to the Golgi apparatus through the endoplasmic reticulum-Golgi intermediate compartment (ERGIC)22 (Fig. 1). In the Golgi, palmitoylated STING1 recruits TANK binding kinase 1 (TBK1)23,24, and TBK1 further transphosphorylates the C-terminal tail of STING1 to recruit interferon regulatory factor 3 (IRF3) for phosphorylation25. Phosphorylated IRF3 translocates to the nucleus and triggers the expression of immune stimulated genes (ISGs) and type I IFNs, resulting in the activation and migration of immune cells (including dendritic cells [DCs], T cells, and natural killer [NK] cells) to the target cells26. Alternatively, STING1 also activates nuclear factor kappa B subunit 1 (NFKB1)-driven inflammatory cytokine (e.g., tumor necrosis factor [TNF] and interleukin 6 [IL6]) production. As a negative feedback mechanism, the degradation of cGAMP, CGAS, or STING1 at various levels can limit type I IFN responses27,28. Functionally, the activation of this classical STING1 pathway bridges innate and adaptive immunity in response to PAMPs or DAMPs. Consequently, an insufficient or excessive activation of the STING1 pathway is implicated in various pathological conditions, such as tumorigenesis, infection, disseminated intravascular coagulation, autoimmune conditions, and tissue damage3,5,6,9,17,29–34.
Beyond the canonical role of STING1 in mediating cytokine production, growing evidence highlights the emerging role of STING1 in regulating autophagy and cell death. In this review, we focus on new discoveries about the regulation mechanisms and outcomes of STING1 in autophagy and cell death, which provide another framework to understand the biological function of STING1 in health and disease.
STING1 in autophagy
Autophagy is a degradation process that can be classified into three types: macroautophagy, microautophagy, and chaperone-mediated autophagy35. Macroautophagy (hereafter referred to as autophagy) is the most studied form of autophagy and regulates homeostasis by either promoting survival or inducing cell death36–38. Autophagy is also closely related to inflammation and immune response39. Here, we not only outline the basic process of autophagy, but also discuss the interaction between STING1 activation and autophagy induction.
Membrane dynamics during autophagy
The process of autophagy involves the formation of membrane structures, especially phagophores, autophagosomes, and autolysosomes38. This dynamic membrane process can be roughly divided into five consecutive phases: initiation, nucleation, expansion, fusion, degradation, and recycling, and is controlled by autophagy-related (ATG) family proteins, kinases, and lipid metabolism36,37,40,41. The initiation step is triggered by inhibiting mechanistic target of rapamycin kinase (MTOR) or activating AMP-activated protein kinase (AMPK), leading to the assembly and activation of the ULK complex (including unc-51-like autophagy activating kinase 1/2 [ULK1/2, orthologs of yeast Atg1], ATG13, ATG101, and RB1 inducible coiled-coil 1 [RB1CC1/FIP200]) and subsequent the class III phosphatidylinositol 3-kinase (PtdIns3K) complex (containing PIK3C3 [phosphatidylinositol 3-kinase catalytic subunit type 3, an ortholog of yeast Vps34], beclin 1 [BECN1, a mammalian homolog of yeast Vps30/Atg6], ATG14L [autophagy-related protein 14-like protein], and PIK3R4 [phosphoinositide 3-kinase regulatory subunit 4, a mammalian homolog of yeast Vps15]). The PtdIns3K complex-mediated production of phosphatidylinositol 3-phosphate (PI3P) leads to the recruitment of PI3P-binding ATG proteins, WD repeat domain phosphoinositide-interacting (WIPI) proteins, and ATG9-containing vesicles, thereby supporting phagophore nucleation (mostly at the ER). In addition to regulating autophagy, BECN1 is also a multifunctional protein in cell death through different binding partners42,43. To promote the expansion and closure of the phagophore membrane, two ubiquitin-like conjugation pathways are needed to mediate the conjugation of microtubule-associated protein 1 light chain 3 (MAP1LC3, an ortholog of yeast Atg8) and phosphatidylethanolamine (PE). This process is called MAP1LC3 lipidation, which is controlled by many ATGs. The E1-like enzyme ATG7, the E2-like enzyme ATG3, and the cysteine protease ATG4 lead to the formation of MAP1LC3-II. The ATG7 and the E2-like enzyme ATG10 catalyze the formation of the ATG12-ATG5-ATG16L1 complex, which binds to WIPI and functions as an E3-like ligase to mediate the lipidation of MAP1LC3-II. Lipidated MAP1LC3-II also enables the assembly of phagophores with autophagic cargo receptors (e.g., sequestosome 1 [SQSTM1/p62]) to engulf cytoplasmic materials. The closure of the phagophore results in the formation of double-membrane structures, namely autophagosomes. Autophagosomes finally fuse with lysosomes to produce autolysosomes, where the cargoes are degraded44. The subsequent degradation of autophagic cargoes by lysosomal hydrolases leads to the release of nutrients and materials back to the cytoplasm for reuse. Overall, from yeasts to plants and animals, membrane dynamics during autophagy are highly conserved.
STING1-mediated autophagy prevents pathogen infection
Since it was first documented that ATG9a and STING1 co-localized in vesicles and restricted dsDNA-induced IFN production in mouse embryonic fibroblasts (MEFs)45, accumulating evidence has enabled us to understand how STING1 plays context-dependent roles in the induction and regulation of autophagy under different stresses. In general, pathogenic DNA can activate STING1 and subsequent autophagy to restrict pathogen infection by removing bacteria and viruses46–48, as described below.
During Mycobacterium tuberculosis infection, STING1 is activated by CGAS, triggering ubiquitin-mediated selective autophagy (namely xenophagy) by SQSTM1, ATG5, and TBK1 in macrophages to eliminate M. tuberculosis11,47–50. In contrast, c-di-AMP induces STING1-dependent autophagy via MTOR inactivation in macrophages during Gram-positive bacterial infection51, whereas the deletion of ULK1, RB1CC1, and ATG14L can limit MAP1LC3 lipidation during this process. Similar to bacterial infection, during herpes simplex virus 1 (HSV-1) infection, cytosolic viral DNA triggers STING1-dependent autophagy in bone marrow-derived DCs46. Further studies using Sting1 mutant mice suggest that STING1-mediated autophagy (but not STING1-dependent IFN production) is responsible for anti–HSV-1 responses52, although HSV may also counter this STING1-mediated autophagy induction in vivo53. This autophagy-dependent antiviral function is also observed in human rhinovirus (HRV)-infected HeLa cells after treatment with the STING1 agonist dimeric amido-benzimidazole (diABZI)54, although it is unclear whether CGAS is required for this process.
STING1-mediated autophagy during infection is also related to NF-κB signaling. For example, NF-κB-mediated STING1 expression triggers autophagy in the adult fly brain against Zika virus (ZIKV) infection55. Moreover, NF-κB mediates the expression of DNA damage-regulated autophagy modulator (DRAM1), thereby inducing selective autophagy in zebrafish and human macrophages during M. tuberculosis infection, and this process is positively regulated by STING156. STING1 can also recognize and degrade the contents of lytic Chlamydiae trachomatis in HeLa cells through a selective autophagy pathway57. Moreover, the antibacterial protein IL-26 secreted by Th17 cells can induce STING1-dependent autophagy to eliminate invasive Mycobacterium leprae in THP-1 cells58.
Together, STING1-dependent autophagic clearance of invading pathogens may be an important host defense mechanism against infection. However, the interplay between STING1-dependent xenophagy and cytokine production remains uncertain.
STING1-mediated autophagy limits tumor growth and transformation
In addition to promoting survival, excessive autophagy may also lead to cell death, known as autophagy-dependent cell death (ADCD)59. Although ADCD is also a type of regulated cell death (RCD) according to the recommendation of the Nomenclature Committee on Cell Death60, it relies on components of autophagic machinery and takes place in the context of apoptosis61, necroptosis62–64, and ferroptosis65–69. Two studies on pancreatic cancer cells have linked DNA damage, STING1 activation, and autophagy-dependent ferroptosis. The nucleoside analog zalcitabine (an antiviral drug) induces oxidative mtDNA damage and the release of mtDNA into cytosol, resulting in the activation of the CGAS-STING1 pathway, which in turn induces autophagy-dependent ferroptosis and suppresses pancreatic tumor growth in mice70. In addition to mtDNA, the release of nDNA to cytosol caused by nuclear cathepsin B (CTSB)-mediated genomic DNA damage activates STING1-dependent autophagy and subsequent glutathione peroxidase 4 (GPX4) degradation, leading to ferroptotic cell death in human pancreatic cancer cells71. These findings provide an example of how lysosomal proteins can drive STING1 activation for autophagy-dependent ferroptosis.
Replicative crisis is a senescence-independent process, characterized by chromosomal instability72. It functions as a potent tumor suppressor against oncogenic transformation and tumorigenesis and culminates in extensive cell death, which is modulated by telomeric damage signals72,73. When replicative crisis occurs, increased telomere DNA damage also initiates CGAS-STING1-dependent ADCD to limit genome instability in human mammary epithelial cells and IMR90E6E7 cells74. These results demonstrate tumor suppressive roles for STING1-dependent ADCD during transformation. Accordingly, Burkholderia pseudomallei-induced cell fusion triggers mitotic events and subsequent micronuclei formation, which leads to CGAS-STING1 activation and ultimately STING1-dependent ADCD in macrophages75, suggesting that ADCD acts as a natural defense against cellular transformation and unnatural cellular fusions.
Overall, these in vitro and in vivo data indicate that STING1 activation induced by host DNA damage can trigger ADCD to remove cancer cells or other stressed cells. It will be interesting to see whether the activation of STING1-dependent autophagy plays a direct role in degrading DNA or micronucleus in damaged cells, a process called DNAphagy.
The mechanism of STING1 regulates the autophagy pathway
The autophagy mediated by STING1 is a selective process that requires specific signals and regulators in a context-dependent manner (Fig. 2). After binding to cGAMP, STING1 leaves the ER and is transported to the ERGIC, which serves as a membrane source for autophagosome biogenesis21. STING1-mediated autophagosome formation is generally independent of the activation of TBK1 and IRF321,22,52,76 as well as ULK1, BECN1, and PIK3C321,76. Moreover, although ATG9a is a regulator of STING1 trafficking45 and nucleation of autophagy77, it is dispensable for STING1-dependent autophagy76. These findings suggest that STING1 initiates a noncanonical form of autophagy driving autophagosome formation. Subsequent studies have highlighted that WD repeat domain, phosphoinositide-interacting 2 (WIPI2), ATG5, and ATG7 are required for STING1-induced autophagosome formation21, whereas RAB7A promotes the transport of STING1 to lysosomes through autophagosomes and endosomes21,27. Finally, STING1 can be self-degraded by autophagy machinery, which requires the use of the TBK1 signal and autophagy receptor SQSTM1 in MEFs and THP-1 cells78, while SQSTM1 and TBK1 are required for STING1- and ubiquitin-mediated selective autophagy targeting M. tuberculosis in macrophages48. Alternatively, since TBK1 is not required for STING1-induced autophagy in HeLa cells, STING1 can function as an autophagy receptor itself that directly binds with MAP1LC3 via its LIR motifs in a SQSTM1-independent manner76. Consistently, the lack of ATG5 or WIPI2 abolishes SQSTM1 degradation (but not STING1 degradation) in cGAMP-treated BJ cells21. In addition, extracellular SQSTM1 is an inflammatory mediator in mice with bacterial infections, and the lysosome-dependent release of SQSTM1 induced by LPS in macrophages is initiated by STING1-TBK1–mediated phosphorylation of SQSTM1 on Ser 40331. These findings confirm that STING1 is not only an autophagy substrate, but also a modulator of autophagy during infection, which shapes host defense response, coupled to signals via SQSTM1 release.
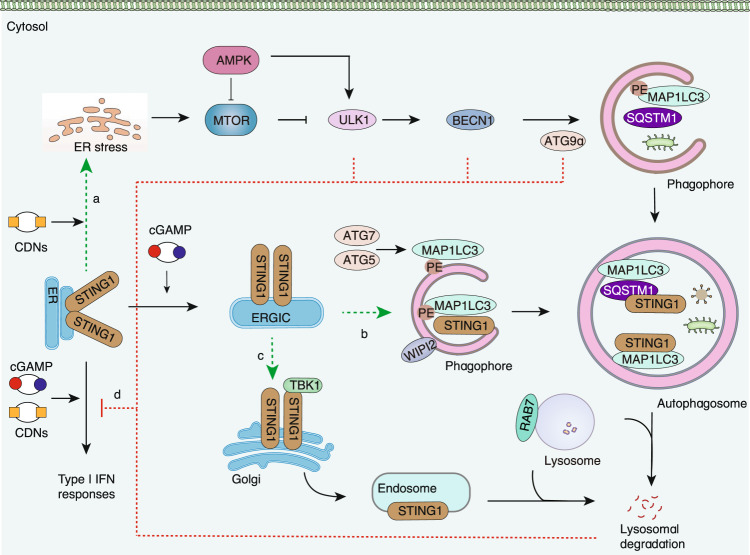
Fig. 2.
Crosstalk between STING1 and autophagy. a The activation of STING1 by bacterial CDNs triggers autophagy through ER stress or the MTOR-ULK1-BECN1 pathway. b cGAMP binds to STING1, causing it to be translocated from the ER to the ERGIC and Golgi. The ERGIC serves as a membrane source for the recruitment and lipidation of MAP1LC3 through an ATG5-, ATG7-, and WIPI2-dependent mechanism, leading to autophagosome formation. Autophagosomes engulfing DNA and pathogens target STING1 in an SQSTM1-dependent or -independent manner. c Activated STING1 can translocate to endosomes through the trans-Golgi network. Both the endosomes and autophagosomes fused with lysosomes require RAB7A GTPase. d STING1-mediated IFN response is also negatively regulated by autophagic degradation or ATGs (including ULK1, BECN1, and ATG9a). AMPK AMP-activated protein kinase, ATG autophagy-related, BECN1 beclin 1, CDNs cytosolic cyclic dinucleotides, cGAMP cyclic GMP-AMP, ER endoplasmic reticulum, ERGIC endoplasmic reticulum-Golgi intermediate compartment, IFN interferon, MAP1LC3 microtubule-associated protein 1 light chain 3, MTOR mechanistic target of rapamycin kinase, PE phosphatidylethanolamine, PIK3C3 phosphatidylinositol 3-kinase catalytic subunit type 3, RAB7A, member RAS oncogene family, SQSTM1 sequestosome 1, STING1 stimulator of interferon response cGAMP interactor 1, TBK1 TANK binding kinase 1, ULK1 unc-51–like autophagy activating kinase 1, WIPI2 WD repeat domain phosphoinositide-interacting 2
In other cases, a BECN1-related PtdIns3K complex may contribute to STING1-dependent ADCD in HeLa cells during Chlamydiae infection57. Bacterial c-di-AMP activates STING1 and triggers ER stress, leading to MTOR inactivation and subsequent ER-phagy in macrophages51. Considering that STING1 is widely involved in ER calcium homeostasis79 and the unfolded protein response80, it is possible that STING1 activation may also trigger autophagy through ER stress and the MTOR-BECN1 pathway (Fig. 2).
Moreover, the activation of STING1 by cGAMP also induces V-ATPase-dependent MAP1LC3 lipidation to single-membrane perinuclear vesicles through ATG16L181, suggesting an additional function of STING1 in membrane dynamics. In line with this, STING1 was observed to increase PI3P production and ER membrane curvature and to cluster at ER curvature-rich regions after cGAMP stimulation19. HRV replication also requires that STING1 is expressed on phosphatidylinositol 4-phosphate (PI4P)-enriched replication organelles82. Given that the IFN-dependent C-terminal tail region is not required for STING1 to induce autophagy21,52, it is possible that STING1 contributes to a wide range of cellular functions via membrane dynamics and ER exit trafficking in an IFN-independent manner. Of note, the activation of STING1 by lipotoxicity induced by saturated fatty acids may inhibit hepatocyte autophagy, which is related to the increase of SQSTM1 phosphorylation and oxidative stress83. The precise role of STING1 in autophagy regulation should be carefully evaluated under different stress conditions.
The mechanism of autophagy modulating the STING1 pathway
The function of STING1 in immunity is also dually regulated by autophagic degradation or components of autophagy machinery. On one hand, autophagy restricts the activation of the STING1 pathway through multiple mechanisms (Fig. 2). First, autophagy can remove radiation-caused cytosolic mtDNA accumulation in breast cancer cells84. Second, AMPK and ULK1 mediate the phosphorylation of STING1 on S366, leading to the degradation of STING1 and inhibition of IFN production85,86. Consistently, the inactivation of ULK1 enhances the STING1-mediated innate immune response in keratinocytes under UV treatment87. Third, the autophagic degradation of STING1 in lysosomes by V-ATPase diminishes STING1 activation21,27. In contrast, blocking STING1 degradation by the V-ATPase inhibitor bafilomycin A enhances STING1-mediated immune signaling and antitumor response. Fourth, BECN1 is a negative regulator of STING1, which partially affects the phosphorylation of STING1 by binding to STING188,89. Given that BECN1 and STING1 can bind to ER Ca2+ channel inositol 1,4,5-trisphosphate receptors (ITPRs/InsP3Rs) to promote the release of Ca2+ from the ER29,79,90, they may form a complex on the ER that controls autophagosome formation by ITPR-mediated Ca2+ signaling. Fifth, the pharmacological or genetic inhibition of ATG9a augments STING1-dependent IFN production45,91. On the other hand, a few studies show that the knockdown of PIK3C3 inhibits STING1 trafficking and activation85. Thus, it remains a challenge to distinguish the autophagy-dependent and -independent roles of ATG in regulating the activation of the STING1 pathway.
STING1 in cell death
RCD is an active process involving tightly structured signal transduction cascades, molecularly defined effector mechanisms, and membrane repair machineries92–97. It has many forms, such as but not limited to apoptosis, necroptosis, pyroptosis, ferroptosis, ADCD, and immunogenic cell death (ICD)98, and plays a vital role in various physiological or pathological processes, including tissue regeneration, infection, immunity, and tumorigenesis99. Such a homeostatic function not only reflects the elimination of damaged or aged cells, but also the ability of dying cells to expose or release DAMPs that activate immune responses100–103. Recent evidence indicates that STING1 not only mediates cell death, but also plays a role in recognizing and amplifying the immune response induced by dying cells.
STING1 in apoptosis
Apoptosis is usually a type of immune-silent RCD because it has a limited ability to release contents and can be quickly cleared by phagocytes (a process also known as efferocytosis). The activation of apoptosis generally includes two distinct pathways: the death receptor (or extrinsic) and the mitochondrial (or intrinsic) pathways. In many cases, caspases (cysteine-dependent proteases), such as caspase 3 (CASP3), CASP6, and CASP7, are effectors of apoptosis by mediating the cleavage of structural proteins104,105. The mitochondrial apoptosis pathway is initiated by various stresses, leading to increased mitochondrial outer membrane permeability (MOMP) and the release of mitochondrial proteins, such as cytochrome C, somatic (CYCS). Cytosolic CYCS binds to apoptotic peptidase activating factor 1 (APAF1), leading to the activation of CASP9 and subsequent executioner caspases CASP3 and CASP7, and ultimately causing apoptosis60,106. The interaction of anti-apoptotic or pro-apoptotic BCL2 family members orchestrates MOMP and has become a therapeutic target in clinical trials107. The death receptors include Fas receptors, TNF receptors, and TNF-related apoptosis-inducing ligand (TRAIL) receptors, which can recruit the adapter protein Fas associated via death domain (FADD) to active CASP8 or CASP10, and ultimately lead to apoptosis by activating CASP3 or CASP7108. In contrast, the mechanism and function of caspase-independent apoptosis remains poorly understood.
The activation of STING1 during apoptosis was first reported in 2014 by two groups. They found that mitochondrial apoptosis mediated by BCL2-associated X, apoptosis regulator (BAX), and BCL2 antagonist/killer 1 (BAK1) leads to mtDNA release and subsequently the activation of the CGAS-STING1 pathway in CASP9-deficient cells, which contributes to the inflammatory response109,110. Further studies confirm that BAX/BAK1-mediated MOMP is required for mtDNA-dependent STING1 activation111,112, while active CASP3 can cleave CGAS or IRF3 to prevent excessive IFN production113. Consistently, the depletion of transcription factor A, mitochondrial (TFAM, a mtDNA binding protein essential for genome maintenance) also induces mtDNA-mediated inflammation by activating STING1 in renal tubule cells114 (Fig. 3). Moreover, STING1 facilitates apoptotic DNA-induced pro-inflammatory gene expression in hematopoietic cells or apoptosis-derived membrane vesicle (AdMV)-induced type I IFN expression in THP-1 cells115,116, respectively. In addition, apoptotic tumor cells activate the STING1 pathway in tumor-associated macrophages (TAMs), thereby enhancing antitumor immune responses117. These studies from different groups suggest that apoptotic cells can activate STING1 to produce inflammation or an immune response.
Fig. 3.
STING1 in apoptosis, mitotic cell death, and necroptosis. a In response to mtDNA, nDNA, or ER stress, STING1 activates IRF3. IRF3 can increase BAX/BAK1-mediated MOMP by forming an IRF3-BAX complex or inhibiting BCL2L1, leading to caspase activation and ultimately apoptosis. In turn, increased apoptosis double-regulates the STING1-IRF3 pathway by releasing mtDNA or activating CASP3-mediated cleavage of CGAS or IRF3. b, STING1 triggers necroptosis by cooperating with downstream type I IFN-mediated MLKL expression and TNF-induced phosphorylation of RIPK1 and RIPK3. APAF1 apoptotic peptidase activating factor 1, BAK1 BCL2 antagonist/killer 1, BAX BCL2-associated X, apoptosis regulator, BCL2L1 BCL2-like 1, CASP caspase, cGAMP cyclic GMP-AMP, CGAS cyclic GMP-AMP synthase, CYCS cytochrome C, ER endoplasmic reticulum, FADD Fas associated via death domain, IFN interferon, IMM inner mitochondrial membrane, IRF3 interferon regulatory factor 3, MLKL mixed-lineage kinase domain-like pseudokinase, MOMP mitochondrial outer membrane permeability, NFKB1 nuclear factor kappa B subunit 1, RIPK1 receptor interacting serine/threonine kinase 1, RIPK3 receptor interacting serine/threonine kinase 3, STING1 stimulator of interferon response cGAMP interactor 1, TBK1 TANK binding kinase 1, TNF tumor necrosis factor
The activation of STING1 also determines the sensitivity to apoptosis in a context-dependent manner. For example, STING1 agonists can trigger apoptosis in malignant B cells118, neuroblastoma cells119, and cancerous T cells120, but not in MEFs, bone marrow-derived DCs, and bone marrow-derived macrophages (BMDMs)120. In a sepsis mouse model, elevated mtDNA induces STING1-dependent apoptosis in intestinal epithelial cells121 (Fig. 3). Interestingly, both STING1 deficiency and activation promote the polarization of TAMs into a pro-inflammatory subtype, thereby inducing apoptosis in gastric cancer cells through the IL6R-JAK-IL24 pathway122. These data also suggest a role of STING1 signaling in mediating cell–cell communication in the tumor microenvironment.
The mechanism of STING1-mediated apoptosis is mainly related to ER stress based on several findings. First, STING1-dependent apoptosis occurs simultaneously with increases in spliced X-box binding protein 1 (XBP1, a marker of ER stress) in T cells123, whereas the genetic or chemical inhibition of spliced XBP1 impairs STING1-mediated apoptosis in B cells118. Second, the constitutive activation of STING1 by gain-of-function mutation enhances the sensitivity of T cells to apoptosis through ER-related calcium signaling80. Third, Mycobacterium bovis infection in RAW.247 cells or ethanol treatment in hepatocytes triggers ER stress and subsequent STING1-dependent apoptosis124,125 (Fig. 3). Structurally, STING1 activates ER stress through an evolutionarily conserved motif within CDN binding domain, which is also crucial for autophagy induction76. In contrast, the notch intracellular signaling domain (NICD) interacts with the CDN binding domain of STING1, thereby inhibiting STING1-dependent apoptosis in CD4+ T cells126. These results establish a link between STING1, ER stress, and apoptosis, although it is unclear whether STING1 regulates the activation of CASP12, which is an initiated caspase in ER stress-induced apoptosis.
In addition to ER signaling, STING1-mediated apoptosis may require the activation of the IRF3 pathway in some conditions. These conditions include free fatty acid-induced L-O2 cells127, retrovirus-infected primary human monocytes128, Mycobacterium bovis-infected RAW.247 cells124, LPS-induced cardiomyocytes129, and ethanol-treated hepatocytes125 (Fig. 3). Mechanistically, STING1-mediated phosphorylation of IRF3 triggers the formation of the IRF3-BAX complex, which leads to BAX activation, CYCS release, and apoptosis. These findings raise concerns about the mitochondrial function of IRF3 in innate immunity.
Taken together, the STING1-IRF3 pathway engages in both apoptosis-derived immune response and apoptosis induction, and this pathway can be regulated by apoptotic caspases, while the interplay and feedback mechanism between STING1, ER stress, and apoptosis is definitely complex.
STING1 in necroptosis
Necroptosis is a form of regulated necrosis in which caspase is inhibited. It can be triggered by the activation of death receptors or PRRs, such as toll-like receptor 3 (TLR3), TLR4, and Z-DNA binding protein 1 (ZBP1/DAI). The core regulator of necroptosis is involved in the formation of necrosome, which is composed of three core components: receptor interacting serine/threonine kinase 1 (RIPK1), RIPK3, and mixed-lineage kinase domain-like pseudokinase (MLKL). After being activated by RIPK1 and RIPK3, phosphorylated MLKL oligomerizes and destroys the plasma membrane, leading to necroptotic cell death130.
The activation of STING1 is related to necroptotic stimulation. In sporadic aortic aneurysm and dissection tissue, increased stress in smooth muscle cells causes the release of mtDNA and nDNA into the cytosol, resulting in the activation of the CGAS-STING1 pathway and necroptosis131. A deficiency of STING1, TBK1, or IRF3 reduces, whereas the overexpression of STING1 and TBK1 increases, the phosphorylation of RIPK3 and MLKL, indicating that the STING1-IRF3-RIPK3-MLKL pathway drives necroptosis by a phosphorylation mechanism. Given that IFNB is required for RIPK3 activation and necroptosis in macrophages132, STING1 may also contribute to necroptosis by IFN production. Indeed, the constitutive activation of the CGAS-STING1 pathway instead of ZBP1 in macrophages elevates MLKL expression and necroptosis through IFNB133. In addition, BCL2 binding component 3 (BBC3/PUMA)-induced mtDNA release activates STING1-dependent necroptosis by elevating the expression of RIPK3 and MLKL in HT29 colon cancer cells and MEFs134 (Fig. 3). These data also indicate that MLKL is an expression of STING1-dependent ISGs, providing an additional transcription-mediated modulation mechanism of necroptosis.
However, the overexpression of MLKL alone could not restore necroptosis in IFN receptor-deficient macrophages, implying other STING1-dependent downstream products may mediate MLKL phosphorylation for necroptosis133. In agreement with this hypothesis, TNF production is also involved in STING1-mediated necroptosis based on four independent studies (Fig. 3). First, anti-TNF neutralizing antibody inhibits murine gammaherpesvirus-68 (MHV68)-induced necroptosis in the fibrosarcoma L929 cell line in a STING1-dependent manner135. Second, the activation of CGAS-STING1 in vivo and in vitro can lead to RIPK3-mediated necroptosis, which requires STING1-dependent type I IFN and TNF production136. Third, IL-22 induces STING1-dependent type I IFN and TNF expression in mouse small intestine organoids, driving necrosis137. Fourth, mtDNA-mediated STING1 signaling triggers necroptosis through synergistic IFN and TNF signaling in primary intestinal epithelial cells138. In sum, these data indicate that STING1 signaling triggers necroptosis through at least two mechanisms, inducing MLKL expression or MLKL phosphorylation.
STING1 in pyroptosis
Pyroptosis is a caspase-dependent RCD driven by pore formation protein gasdermin D (GSDMD) or gasdermin E (GSDME/DFNA5). During inflammasome activation or other stresses, GSDMD can be cleaved by CASP1, CASP11 (also known as CASP4 or CASP5 in humans) or CASP8 to produce N-terminal fragment of GSDMD (GSDMD-N)139. In contrast, the production of GSDME-N is mediated by CASP3140. After oligomerization, GSDMD-N or GSDME-N forms pores in the plasma membrane, leading to pyroptotic cell death. Inflammasomes are divided into two categories: canonical and noncanonical. Canonical inflammasome complexes are assembled in response to signaling from cytosolic PRRs, such as NLR family pyrin domain containing 3 (NLRP3) and absent in melanoma 2 (AIM2). These PRRs sense PAMPs or DAMPs to recruit the adapter protein PYD and CARD domain containing (PYCARD/ASC), leading to CASP1 activation and subsequent pyroptosis141. Noncanonical inflammasome is mainly triggered by cytoplasmic LPS-induced CASP11 or CASP4/5 activation, resulting in the production of GSDMD-N and pyroptosis142. As a feedforward control, the GSDMD pores allow potassium efflux, promoting the activation of NLRP3 inflammasome, which then causes the release of IL1 family cytokines or DAMPs.
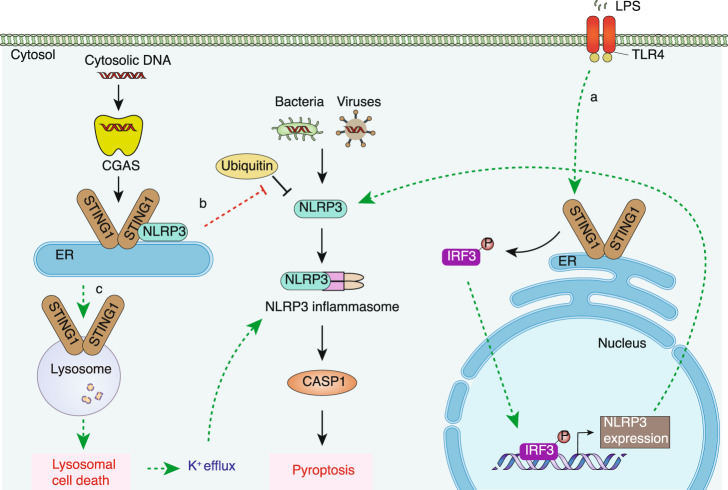
STING1 promotes cGAMP-induced NLRP3 inflammasome activation in macrophages143, cardiomyocytes, and mice129 following LPS challenge. In addition, Mycobacterium abscessus infection in murine macrophages triggers mitochondrial oxidative stress and leads to the activation of the CGAS-STING1 pathway and NLRP3 inflammasome activation144, while the inhibition of STING1 suppresses NLRP3 activation in mtDNA-stimulated BMDMs from aged mice145. In terms of mechanisms, three pathways may contribute to STING1-dependent inflammasome activation and pyroptosis (Fig. 4). First, the transcriptional upregulation of NLRP3 requires the production of STING1-mediated IRF3 in a TLR4-dependent manner129. Second, STING1 can directly bind and promote NLRP3 localization in the ER, thereby inhibiting NLRP3 ubiquitin degradation146. Third, in response to cytosolic DNA in human myeloid cells, the CGAS-STING1 pathway instead of AIM2 activates potassium efflux-mediated NLRP3 inflammasome via lysosomal STING1-mediated lysosomal cell death147. Consistent with this, G10 (an agonist of STING1) induces potassium efflux and NLRP3 activation in porcine cells148. Conversely, the inhibition of NLRP3 inflammasomes increases STING1-dependent IFN production148, while the activation of NLRP3 or AIM2 in murine macrophages leads to the inactivation of the STING1 pathway149,150, suggesting a yet unknown mechanism to balance the activation of STING1 and inflammasomes in response to infection.
Fig. 4.
STING1 in pyroptosis. STING1 activates NLRP3 and subsequent pyroptosis via three pathways: a LPS directly triggers STING1-IRF3 activation via TLR4 and activated IRF3 increases the expression of NLRP3; b STING1 interacts with NLRP3 and prevents ubiquitin-mediated degradation of NLRP3; c STING1 induces lysosomal cell death, causing potassium efflux to activate an NLRP3 inflammasome. CASP caspase, CGAS cyclic GMP-AMP synthase, ER endoplasmic reticulum, IRF3 interferon regulatory factor 3, LPS lipopolysaccharide, NLRP3 NLR family pyrin domain containing 3, STING1 stimulator of interferon response cGAMP interactor 1, TLR4 toll-like receptor 4
STING1 in ferroptosis
Ferroptosis is a type of regulated necrosis151, which is characterized by iron-dependent lipid peroxidation and was first described as a mutant RAS-dependent cancer cell death152. In addition to cancer153, ferroptotic injury is related to various tissue injuries and infections154–156. Ferroptosis can be induced by destroying certain antioxidant systems, especially the SLC7A11-glutathione (GSH)-GPX4 axis152. Ferroptosis may be a type of ADCD because several key anti-ferroptosis regulators157, such as ferritin65, aryl hydrocarbon receptor nuclear translocator-like (ARNTL)158, GPX4159, solute carrier family 40 member 1 (SLC40A1, best known as ferroportin)68, and lipid droplets160, can be eliminated by autophagic pathways.
Recently, STING1 has been connected to GPX4-mediated lipid peroxidation in the context of ferroptosis161,162. GPX4 depletion increases lipid peroxidation in HSV-1 infected mice, thereby limiting STING1-mediated antiviral immune responses161. One reason is that the lipid product 4-hydroxynonenal (4-HNE) inhibits STING1 activation by the carbonylation of STING1 in mouse primary peritoneal macrophages, suggesting that GPX4 may act as a promoter of STING1-mediated immune response during virus infection. However, the conditional depletion of GPX4 in myeloid cells increases death caused by bacterial infection in mice, suggesting a different role of GPX4 in bacterial innate immunity163. Moreover, mitochondrial or genomic DNA stress activates STING1-dependent autophagy in pancreatic cancer cells, which may cause lipid peroxidation-mediated ferroptosis70,71. Although the mechanism remains obscure, STING1-dependent autophagy may degrade GPX4, promoting ferroptosis71 (Fig. 5). Accordingly, a robust STING1 activation produces excessive lipid peroxidation during ischemia/reperfusion injury, which is associated with cell death in BMDMs164. Thus, the STING1-related ferroptosis pathway is a potential therapeutic target of cancer and tissue damage.
Fig. 5.
STING1 in ferroptosis and ICD. a mtDNA and nDNA activate STING1-dependent autophagy, leading to GPX4 degradation and subsequent lipid peroxidation-mediated ferroptosis, whereas lipid peroxidation negatively regulates STING1 through carbonylation. Ferroptotic damage also activates STING1 in surrounding macrophages by releasing oxidative DNA damage products, such as 8-OHG. b STING1 engages in ICD in two ways: inducing DC-dependent T-cell priming or TAM-mediated T-cell priming. 8-OHG 8-hydroxy-2′-deoxyguanosine, cGAMP cyclic GMP-AMP, CGAS cyclic GMP-AMP synthase, DAMPs damage-associated molecular patterns, DCs dendritic cells, ER endoplasmic reticulum, GPX4 glutathione peroxidase 4, ICD immunogenic cell death, P2RX7 purinergic receptor P2X 7, STING1 stimulator of interferon response cGAMP interactor 1, TAMs tumor-associated macrophages
STING1 plays a dual role in tumor immunity. An acute activation of STING1 is beneficial for antitumor therapy, and chronic activation of STING1 may mediate inflammation that supports tumor growth. Consistent with this idea, ferroptotic damage promotes 8-hydroxy-2′-deoxyguanosine (8-OHG, an oxidized nucleobase product of oxidative DNA damage) release, thereby activating STING1-dependent macrophage polarization162. By maintaining inflammation-related tumorigenesis, this chronic activation of STING1 triggered by ferroptotic DNA damage ultimately promotes KRAS-driven pancreatic ductal adenocarcinoma in mice (Fig. 5), highlighting the dark side of ferroptosis in tumor immunity165. Further research is needed to clarify the role of STING1 in regulating ferroptosis in different immune cells within the tumor microenvironment.
STING1 in mitotic death
Mitotic death is a specific variant of RCD driven by mitotic catastrophe, which suppresses the proliferation of cells undergoing aberrant mitosis60. It usually ends with the formation of large cells with multiple micronuclei and decondensed chromatin166. Ruptured micronuclei can be recognized by CGAS during genome instability to activate an innate immune response167,168. To avoid hyperactivation, CGAS is normally inhibited by nucleosomes169–173.
During mitotic arrest, CGAS triggers the activation of STING1 and IRF3 in response to the increase of micronuclei, which leads to mitotic death/apoptosis by blocking BCL2-like 1, (BCL2L1/BCL-XL)-dependent MOMP inhibition (Fig. 3)174,175. As discussed previously, increased telomeric DNA damage or Burkholderia pseudomallei infection also causes STING1-dependent mitotic death, which is related to the excessive activation of autophagy74,75. These data implicate STING1-mediated mitotic death as an early host defense against tumorigenesis or infection.
STING1 in immunogenic cell death
ICD is implicated in generating antitumor adaptive immunity95. During ICD, dying cells release or expose DAMPs, activate DC-mediated antigen presentation, and ultimately lead to cytotoxic T-cell responses. ICD is associated with multiple cell death modalities (e.g., apoptosis, pyroptosis, necroptosis, and ferroptosis) and can be triggered via a set of therapeutic agents and interventions.
ICD is also induced in some cancer cells by activating the CGAS-STING1 pathway. The effective activation of STING1 by various agonists stimulates ICD-mediated antitumor immunity in colon carcinoma cells176, neuroblastoma cells119, and melanoma cells177 by producing highly immunogenic cancer cell debris or type I IFNs (Fig. 5). Moreover, DNA released from apoptotic MC38 tumor cells can stimulate a STING1-dependent type I IFN response in TAMs (but not DCs) through purinergic receptor P2X 7 (P2RX7) channels, thereby further enhancing antitumor CD8+ T-cell response117 (Fig. 5). Given that CGAS is constitutively active in most tumor cells and results in the production of cGAMP, which can be internalized by bystander cells through gap junctions13, transporter solute carrier family 19 member 1 (SLC19A1)14, or volume-regulated anion channels15,178, targeting the STING1 pathway has the potential to overcome the immunosuppressive tumor microenvironment.
Conclusions and perspectives
In the past 5 years, by combining genetic technology and pharmacological approaches (Table 1), our understanding of the regulation and function of STING1 has rapidly improved (Fig. 6). Accordingly, there is growing interest in the development of natural and synthetic CDN analogs as well as non-CDN small molecule STING1 agonists as clinical drugs for cancer treatment and antiviral therapy (Table 1). However, because tumor-specific T cells can initiate immunosuppressive pathways including CD274 molecule (best known as PD-L1), thereby preventing tumor clearance179, activating the STING1 pathway alone (e.g., using cyclic diadenyl monophosphate) is not sufficient to kill tumors180,181. Therefore, the combined use of STING1 agonists (e.g., using ADU-S100) and immune checkpoint inhibitors may be the best strategy for tumor treatment182,183. In addition, STING1 activation can impair immunotherapy because it is a mediator of IFN-induced cell death of B cells184 or T cells185. Consequently, STING1 inhibitors (Table 1) can alleviate the side effects of STING1 overactivation.
Table 1.
STING1 activators and inhibitors
| Drugs | Targets | Effect | Model/Disease/Cancer Type | Clinical Trial Phase | Clinical Trial ID/Publication Number | References |
|---|---|---|---|---|---|---|
| Natural CDN agonists | ||||||
| c-di-GMP | hSTING1; mSting1 | Antitumor activity | 4T1 and B16 mouse models | 186,187 | ||
| 2′,3′-cGAMP | hSTING1; mSting1 | Antitumor activity | CT26, 4T1, and B16F10 mouse models | 188 | ||
| Synthetic CDN agonists | ||||||
| ML-RR-S2-CDA | hSTING1 mSting1 | Antitumor activity | B16F10, 4T1, and CT26 mouse models | 189 | ||
| ML-RR-S2-CDG | hSTING1 mSting1 | Antitumor activity | B16F10 mouse models | 189 | ||
| 3′,3′-cAIMP | hSTING1 mSting1 | Antiviral activity | HSV2 infection | 190 | ||
| Non-CDN agonists | ||||||
| DMXAA | mSTING1 | Antitumor activity |
Various mouse models |
Failed in phase III clinical trial | 191 | |
| FAA | mSTING1 | Antitumor activity |
Various mouse models |
Failed in phase I clinical trial | 192 | |
| CMA | mSTING1 | Antiviral activity | Murine models | 193 | ||
| α-Mangostin | hSTING1; mSting1 | Antitumor and antiviral activity | 194 | |||
| ABZI | hSTING1; mSting1 | Antitumor activity | CT26 mouse models | 195 | ||
| Benzothiophenes | hSTING1; mSting1 | Antitumor activity | MC38 mouse models | WO2019027858 | ||
| MSA-2 | hSTING1; mSting1 | Antitumor activity | MC38 mouse models | 196 | ||
| SR-717 | hSTING1; mSting1 | Antitumor activity | B16F10 mouse models | 197 | ||
| STING1 agonists currently in clinical trials | ||||||
| ADU-S100 | Synthetic CDN analog | Antitumor activity | B16 mouse models | Failed in phase II clinical trial | 198 | |
| ADU-CL-20 | Synthetic CDN analog | Antitumor activity | Metastatic/recurrent HNSCC | Phase II | NCT03937141 | |
| MK-1454 | Synthetic CDN analog | Antitumor activity | Advanced solid tumors or lymphomas | Phase I | NCT03010176 | |
| MK-2118 | Synthetic CDN analog | Antitumor activity | Advanced solid tumors or lymphomas | Phase I | NCT03249792 | |
| BMS-986301 | Undisclosed | Antitumor activity |
Advanced solid cancers |
Phase I | NCT03956680 | |
| GSK3745417 | diABZI-like | Antitumor activity | Advanced solid tumors | Phase I | NCT03843359 | |
| SB-11285 | CDN analog | Antitumor activity | Advanced solid tumors | Phase I | NCT04096638 | |
| IMSA-101 | cGAMP analog | Antitumor activity | Advanced solid tumors | Phase I/II | NCT04020185 | |
| E7766 | Synthetic CDN analog | Antitumor activity |
Advanced solid tumors; lymphomas |
Phase I | NCT04144140 | |
| STING1 inhibitors | ||||||
| H-151 | Blocks palmitoylation of STING1 | 199 | ||||
| C-178 | Blocks palmitoylation of STING1 | 199 | ||||
| C-176 | Blocks palmitoylation of STING1 | 199 | ||||
| C18 | Blocks cGAMP-induced IFNB production | 200 | ||||
| Astin-C | Blocks recruitment of IRF3 to STING1 | 201 | ||||
Fig. 6.
The network of STING1 in inflammation, immune response, autophagy, and cell death. STING1 can be activated by bacteria-produced CDNs or CGAS-produced cGAMP. The activation of STING1 not only promotes inflammation and immune responses through TBK1-mediated activation of transcription factor, but also contributes to autophagy and various cell death modalities, such as apoptosis, necroptosis, pyroptosis, ferroptosis, mitotic death, immunogenic cell death, and autophagy-dependent cell death. GPX4 glutathione peroxidase 4, CDNs cytosolic cyclic dinucleotides, cGAMP cyclic GMP-AMP, ATG autophagy-related, STING1 stimulator of interferon response cGAMP interactor 1, ER endoplasmic reticulum, IRF3 interferon regulatory factor 3, NLRP3 NLR family pyrin domain containing 3, TBK1 TANK binding kinase 1, CASP caspase, NFKB1 nuclear factor kappa B subunit 1, MLKL mixed-lineage kinase domain-like pseudokinase
In addition to the traditional function of STING1 in mediating inflammation and immune response, emerging evidence has revealed that STING1 is a key regulator of autophagy and cell death after its post-translational modification. For example, palmitoylation at C88/91 residues of STING1 is not only essential for maintaining active STING1 on Golgi23, but also for STING1-mediated T cell death185. K63 or K48-linked STING1 ubiquitination may be involved in SQSTM1-related autophagy76. Generally, HeLa, MEF, HEK-293T, THP-1, and BMDM are widely used cell lines, while STING1−/− mice or xenograft models are widely used animal models to study the function of STING1 in autophagy and cell death. The complex cellular and immune function of STING1 is achieved through its location, modification, and protein–protein interaction. Overall, the activation of STING1 promotes autophagy and mediates many types of cell death, thus highlighting the important role of STING1 in integrating inflammation and immune response under various stresses. Accordingly, the development of STING1 agonists and inhibitors has become a frontier for the treatment of diseases by stimulating or suppressing the immune response. The following questions are worthy of our continued pursuit: Does STING1-dependent control of autophagic signaling contribute to the maintenance of a stress threshold? If STING1 is an autophagy receptor, what is its core substrate? Why does STING1 share a common upstream activation signal during DNA damage but can then lead to different cell death pathways? In addition to the ER, what is the function of STING1 in other subcellular organelles? How do autophagy and cell death pathways combine to control the immune response? How can we evaluate the difference between CGAS-dependent and independent STING1 pathway activation in host defense? Are there any specific biomarkers that can be used to monitor STING1-dependent autophagy and cell death? What are the activities and side effects of different STING1 drugs? How can we develop a STING1-dependent combination drug strategy for tumor treatment?
Competing interests
The authors declare no competing interests.
References
- 1.Tang D, et al. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol. Rev. 2012;249:158–175. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway C., Jr. Innate immunity. N. Engl. J. Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 9.Barber GN. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey B, et al. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 2015;21:401–406. doi: 10.1038/nm.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 13.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie C, et al. SLC19A1 is an importer of the immunotransmitter cGAMP. Mol. Cell. 2019;75:372–381.e375. doi: 10.1016/j.molcel.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahey LJ, et al. LRRC8A:C/E heteromeric channels are ubiquitous transporters of cGAMP. Mol. Cell. 2020;80:578–591. doi: 10.1016/j.molcel.2020.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Torralba D, et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 2018;9:2658. doi: 10.1038/s41467-018-05077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng, L. et al. ALK is a therapeutic target for lethal sepsis. Sci. Transl. Med. 9, eaan5689 (2017). [DOI] [PMC free article] [PubMed]
- 18.Ergun SL, Fernandez D, Weiss TM, Li L. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell. 2019;178:290–301.e210. doi: 10.1016/j.cell.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Zhang BC, et al. STEEP mediates STING ER exit and activation of signaling. Nat. Immunol. 2020;21:868–879. doi: 10.1038/s41590-020-0730-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun MS, et al. TMED2 potentiatescellular IFN responses to DNA viruses by reinforcing MITA dimerization and facilitating its trafficking. Cell Rep. 2018;25:3086–3098.e3083. doi: 10.1016/j.celrep.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 21.Gui X, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobbs N, et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukai K, et al. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567:394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corrales L, et al. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 2017;27:96–108. doi: 10.1038/cr.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonugunta VK, et al. Trafficking-mediated STING degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Rep. 2017;21:3234–3242. doi: 10.1016/j.celrep.2017.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, et al. TMEM173 drives lethal coagulation in sepsis. Cell Host Microbe. 2020;27:556–570.e556. doi: 10.1016/j.chom.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heipertz EL, Harper J, Walker WE. STING and TRIF contribute to mouse sepsis, depending on severity of the disease model. Shock. 2017;47:621–631. doi: 10.1097/SHK.0000000000000771. [DOI] [PubMed] [Google Scholar]
- 31.Zhou B, et al. Extracellular SQSTM1 mediates bacterial septic death in mice through insulin receptor signalling. Nat. Microbiol. 2020;5:1576–1587. doi: 10.1038/s41564-020-00795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019). [DOI] [PubMed]
- 33.Ablasser, A. & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science. 363, eaat8657 (2019). [DOI] [PubMed]
- 34.Cheng Z, et al. The interactions between cGAS-STING pathway and pathogens. Signal Transduct. Target Ther. 2020;5:91. doi: 10.1038/s41392-020-0198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy17, 1–382, (2021). [DOI] [PMC free article] [PubMed]
- 36.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 38.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie Y, et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11:28–45. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Y, Li J, Kang R, Tang D. Interplay between lpid metabolism and autophagy. Front. Cell Dev. Biol. 2020;8:431. doi: 10.3389/fcell.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirawan E, et al. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8:6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- 44.Kim, H. K. et al. TMBIM6 (transmembrane BAX inhibitor motif containing 6) enhances autophagy through regulation of lysosomal calcium. Autophagy17, 1–18, (2020). [DOI] [PMC free article] [PubMed]
- 45.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen SB, et al. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J. Immunol. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins AC, et al. Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saiga H, et al. The recombinant BCG DeltaureC::hly vaccine targets the AIM2 inflammasome to induce autophagy and inflammation. J. Infect. Dis. 2015;211:1831–1841. doi: 10.1093/infdis/jiu675. [DOI] [PubMed] [Google Scholar]
- 50.Watson RO, et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moretti J, et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171:809–823. doi: 10.1016/j.cell.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashiro LH, et al. Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nat. Commun. 2020;11:3382. doi: 10.1038/s41467-020-17156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker ZM, Murphy AA, Leib DA. Role of the DNA sensor STING in protection from lethal infection following corneal and intracerebral challenge with herpes simplex virus 1. J. Virol. 2015;89:11080–11091. doi: 10.1128/JVI.00954-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Q, et al. A synthetic STING agonist inhibits the replication of human parainfluenza virus 3 and rhinovirus 16 through distinct mechanisms. Antivir. Res. 2020;183:104933. doi: 10.1016/j.antiviral.2020.104933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, et al. Inflammation-induced, STING-dependent autophagy restricts Zika virus infection in the Drosophila brain. Cell Host Microbe. 2018;24:57–68. doi: 10.1016/j.chom.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Vaart M, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected] Cell Host Microbe. 2014;15:753–767. doi: 10.1016/j.chom.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Weber MM, et al. Absence of specific Chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Rep. 2017;19:1406–1417. doi: 10.1016/j.celrep.2017.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang AT, et al. IL-26 contributes to host defense against intracellular bacteria. J. Clin. Invest. 2019;129:1926–1939. doi: 10.1172/JCI99550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bialik, S., Dasari, S. K. & Kimchi, A. Autophagy-dependent cell death - where, how and why a cell eats itself to death. J. Cell Sci. 131, jcs215152 (2018). [DOI] [PubMed]
- 60.Galluzzi L, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gump JM, et al. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat. Cell Biol. 2014;16:47–54. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dey A, et al. Inhibition of BMI1 induces autophagy-mediated necroptosis. Autophagy. 2016;12:659–670. doi: 10.1080/15548627.2016.1147670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin SY, et al. Necroptosis promotes autophagy-dependent upregulation of DAMP and results in immunosurveillance. Autophagy. 2018;14:778–795. doi: 10.1080/15548627.2017.1386359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuzawa-Ishimoto Y, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 2017;214:3687–3705. doi: 10.1084/jem.20170558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou W, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu, J. et al. Autophagy-dependent ferroptosis: machinery and regulation. Cell Chem. Biol. 27, 420–435 (2020). [DOI] [PMC free article] [PubMed]
- 67.Chen, X. et al. Ferroptosis: machinery and regulation. Autophagy 1–28 (2020). 10.1080/15548627.2020.1810918. Online ahead of print.
- 68.Li, J. et al. Tumor heterogeneity in autophagy-dependent ferroptosis. Autophagy 1–14 (2021). 10.1080/15548627.2021.1872241. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 69.Zhang, Z. et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy16, 1–24 (2019). [DOI] [PMC free article] [PubMed]
- 70.Li, C. et al. Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy17, 1–13, (2020). [DOI] [PMC free article] [PubMed]
- 71.Kuang F, et al. Cathepsin B is a mediator of organelle-specific initiation of ferroptosis. Biochem. Biophys. Res. Commun. 2020;533:1464–1469. doi: 10.1016/j.bbrc.2020.10.035. [DOI] [PubMed] [Google Scholar]
- 72.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature. 2015;522:492–496. doi: 10.1038/nature14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei W, Sedivy JM. Differentiation between senescence (M1) and crisis (M2) in human fibroblast cultures. Exp. Cell Res. 1999;253:519–522. doi: 10.1006/excr.1999.4665. [DOI] [PubMed] [Google Scholar]
- 74.Nassour J, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ku JWK, et al. Bacterial-induced cell fusion is a danger signal triggering cGAS-STING pathway via micronuclei formation. Proc. Natl Acad. Sci. USA. 2020;117:15923–15934. doi: 10.1073/pnas.2006908117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu D, et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2019;26:1735–1749. doi: 10.1038/s41418-018-0251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Puri C, et al. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prabakaran, T. et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 37, e97858 (2018). [DOI] [PMC free article] [PubMed]
- 79.Srikanth S, et al. The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 2019;20:152–162. doi: 10.1038/s41590-018-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu J, et al. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 2019;216:867–883. doi: 10.1084/jem.20182192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fischer, T. D. et al. STING induces LC3B lipidation onto single-membrane vesicles via the V-ATPase and ATG16L1-WD40 domain. J. Cell Biol. 219, e202009128 (2020). [DOI] [PMC free article] [PubMed]
- 82.McKnight KL, et al. Stimulator of interferon genes (STING) is an essential proviral host factor for human rhinovirus species A and C. Proc. Natl Acad. Sci. USA. 2020;117:27598–27607. doi: 10.1073/pnas.2014940117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho CS, et al. Lipotoxicity induces hepatic protein inclusions through TANK binding kinase 1-mediated p62/sequestosome 1 phosphorylation. Hepatology. 2018;68:1331–1346. doi: 10.1002/hep.29742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamazaki T, et al. Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat. Immunol. 2020;21:1160–1171. doi: 10.1038/s41590-020-0751-0. [DOI] [PubMed] [Google Scholar]
- 85.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Konno H, et al. Pro-inflammation associated with a gain-of-function mutation (R284S) in the innate immune sensor STING. Cell Rep. 2018;23:1112–1123. doi: 10.1016/j.celrep.2018.03.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kemp MG, Lindsey-Boltz LA, Sancar A. UV light potentiates STING (stimulator of interferon genes)-dependent innate immune signaling through deregulation of ULK1 (Unc51-like kinase 1) J. Biol. Chem. 2015;290:12184–12194. doi: 10.1074/jbc.M115.649301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X, et al. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014;5:912–927. doi: 10.1007/s13238-014-0104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu F, et al. Beclin1 haploinsufficiency accentuates second-hand smoke exposure -induced myocardial remodeling and contractile dysfunction through a STING-mediated mechanism. J. Mol. Cell Cardiol. 2020;148:78–88. doi: 10.1016/j.yjmcc.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 90.Decuypere J-P, et al. Ins(1,4,5)P3 receptor-mediated Ca2+ signaling and autophagy induction areinterrelated. Autophagy. 2011;7:1472–1489. doi: 10.4161/auto.7.12.17909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mitzel DN, et al. Age-enhanced endoplasmic reticulum stress contributes to increased Atg9A inhibition of STING-mediated IFN-beta production during Streptococcus pneumoniae infection. J. Immunol. 2014;192:4273–4283. doi: 10.4049/jimmunol.1303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu, J., Kang, R. & Tang, D. ESCRT-III-mediated membrane repair in cell death and tumor resistance. Cancer Gene Ther. 28, 1–4 (2020). [DOI] [PubMed]
- 93.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 94.Broz, P., Pelegrin, P. & Shao, F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2019). [DOI] [PubMed]
- 95.Galluzzi L, et al. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 96.Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020;21:85–100. doi: 10.1038/s41580-019-0173-8. [DOI] [PubMed] [Google Scholar]
- 97.Vanden Berghe T, et al. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 98.Tang D, et al. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galluzzi, L., Bravo-San Pedro, J. M., Kepp, O. & Kroemer, G. Regulated cell death and adaptive stress responses. Cell Mol. Life Sci. 73, 2405–2410 (2016). [DOI] [PMC free article] [PubMed]
- 100.Zindel J, Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu. Rev. Pathol. 2020;15:493–518. doi: 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Q, et al. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy. 2013;9:451–458. doi: 10.4161/auto.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krysko DV, et al. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer. 2012;12:860–875. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- 103.Galluzzi, L. et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer8, e000337 (2020). [DOI] [PMC free article] [PubMed]
- 104.Ramirez MLG, Salvesen GS. A primer on caspase mechanisms. Semin. Cell Dev. Biol. 2018;82:79–85. doi: 10.1016/j.semcdb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Julien O, Wells JA. Caspases and their substrates. Cell Death Differ. 2017;24:1380–1389. doi: 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalkavan H, Green DR. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018;25:46–55. doi: 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 108.Guicciardi ME, Gores GJ. Life and death by death receptors. FASEB J. 2009;23:1625–1637. doi: 10.1096/fj.08-111005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rongvaux A, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McArthur, K. et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science359, eaao6047 (2018). [DOI] [PubMed]
- 112.Riley, J. S. et al. Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J. 37, e99238 (2018). [DOI] [PMC free article] [PubMed]
- 113.Ning X, et al. Apoptotic caspases suppress type I interferon production via the cleavage of cGAS, MAVS, and IRF3. Mol. Cell. 2019;74:19–31. doi: 10.1016/j.molcel.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 114.Chung KW, et al. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 2019;30:784–799. doi: 10.1016/j.cmet.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kato Y, et al. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann. Rheum. Dis. 2018;77:1507–1515. doi: 10.1136/annrheumdis-2018-212988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc. Natl Acad. Sci. USA. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Y, et al. Blockade of the phagocytic receptor MerTK on tumor-associated macrophages enhances P2X7R-dependent STING activation by tumor-derived cGAMP. Immunity. 2020;52:357–373. doi: 10.1016/j.immuni.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 118.Tang CH, et al. Agonist-mediated activation of STING induces apoptosis in malignant B cells. Cancer Res. 2016;76:2137–2152. doi: 10.1158/0008-5472.CAN-15-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang-Bishop, L. et al. Potent STING activation stimulates immunogenic cell death to enhance antitumor immunity in neuroblastoma. J. Immunother. Cancer8, e000282 (2020). [DOI] [PMC free article] [PubMed]
- 120.Gulen MF, et al. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 2017;8:427. doi: 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu Q, et al. STING-mediated intestinal barrier dysfunction contributes to lethal sepsis. EBioMedicine. 2019;41:497–508. doi: 10.1016/j.ebiom.2019.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miao L, et al. Targeting the STING pathway in tumor-associated macrophages regulates innate immune sensing of gastric cancer cells. Theranostics. 2020;10:498–515. doi: 10.7150/thno.37745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Larkin B, et al. Cutting edge: activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 2017;199:397–402. doi: 10.4049/jimmunol.1601999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cui Y, et al. Mycobacterium bovis induces endoplasmic reticulum stress mediated-apoptosis by activating IRF3 in a murine macrophage cell line. Front. Cell Infect. Microbiol. 2016;6:182. doi: 10.3389/fcimb.2016.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Petrasek J, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl Acad. Sci. USA. 2013;110:16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Long, J. et al. Notch signaling protects CD4 T cells from STING-mediated apoptosis during acute systemic inflammation. Sci Adv. 6, eabc5447 (2020). [DOI] [PMC free article] [PubMed]
- 127.Qiao JT, et al. Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism. 2018;81:13–24. doi: 10.1016/j.metabol.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 128.Sze A, et al. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe. 2013;14:422–434. doi: 10.1016/j.chom.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 129.Li N, et al. STING-IRF3 contributes to lipopolysaccharide-induced cardiac dysfunction, inflammation, apoptosis and pyroptosis by activating NLRP3. Redox Biol. 2019;24:101215. doi: 10.1016/j.redox.2019.101215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 131.Luo W, et al. Critical role of cytosolic DNA and its sensing adaptor STING in aortic degeneration, dissection, and rupture. Circulation. 2020;141:42–66. doi: 10.1161/CIRCULATIONAHA.119.041460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McComb S, et al. Type-I interferon signaling through ISGF3 complex is required for sustained Rip3 activation and necroptosis in macrophages. Proc. Natl Acad. Sci. USA. 2014;111:E3206–E3213. doi: 10.1073/pnas.1407068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sarhan J, et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2019;26:332–347. doi: 10.1038/s41418-018-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen D, et al. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc. Natl Acad. Sci. USA. 2018;115:3930–3935. doi: 10.1073/pnas.1717190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schock SN, et al. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ. 2017;24:615–625. doi: 10.1038/cdd.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brault M, et al. Intracellular Nucleic Acid Sensing Triggers Necroptosis through synergistic type I IFN and TNF signaling. J. Immunol. 2018;200:2748–2756. doi: 10.4049/jimmunol.1701492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aden K, et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J. Exp. Med. 2018;215:2868–2886. doi: 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang X, et al. mtDNA-STING pathway promotes necroptosis-dependent enterocyte injury in intestinal ischemia reperfusion. Cell Death Dis. 2020;11:1050. doi: 10.1038/s41419-020-03239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Broz P, Pelegrin P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020;20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 140.Wang Y, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 141.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 142.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 143.Swanson KV, et al. A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med. 2017;214:3611–3626. doi: 10.1084/jem.20171749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim BR, Kim BJ, Kook YH, Kim BJ. Mycobacterium abscessus infection leads to enhanced production of type 1 interferon and NLRP3 inflammasome activation in murine macrophages via mitochondrial oxidative stress. PLoS Pathog. 2020;16:e1008294. doi: 10.1371/journal.ppat.1008294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhong W, et al. Aging aggravated liver ischemia and reperfusion injury by promoting STING-mediated NLRP3 activation in macrophages. Aging Cell. 2020;19:e13186. doi: 10.1111/acel.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang W, et al. STING promotes NLRP3 localization in ER and facilitates NLRP3 deubiquitination to activate the inflammasome upon HSV-1 infection. PLoS Pathog. 2020;16:e1008335. doi: 10.1371/journal.ppat.1008335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gaidt MM, et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell. 2017;171:1110–1124. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ming SL, et al. The human-specific STING agonist G10 activates type I interferon and the NLRP3 inflammasome in porcine cells. Front. Immunol. 2020;11:575818. doi: 10.3389/fimmu.2020.575818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kwon D, Park E, Kang SJ. Stimulator of IFN genes-mediated DNA-sensing pathway is suppressed by NLRP3 agonists and regulated by mitofusin 1 and TBC1D15, mitochondrial dynamics mediators. FASEB J. 2017;31:4866–4878. doi: 10.1096/fj.201700328R. [DOI] [PubMed] [Google Scholar]
- 150.Corrales L, et al. Antagonism of the STING pathway via activation of the AIM2 inflammasome by intracellular DNA. J. Immunol. 2016;196:3191–3198. doi: 10.4049/jimmunol.1502538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tang D, Kroemer G. Ferroptosis. Curr. Biol. 2020;30:R1292–R1297. doi: 10.1016/j.cub.2020.09.068. [DOI] [PubMed] [Google Scholar]
- 152.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen, X., Kang, R., Kroemer, G. & Tang, D. Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 18, 280–296 (2021). [DOI] [PubMed]
- 154.Tang, D., Chen, X., Kang, R. & Kroemer, G. Ferroptosis: molecular mechanisms and health implications. Cell Res.31, 107–125 (2020). [DOI] [PMC free article] [PubMed]
- 155.Stockwell BR, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yan HF, et al. Ferroptosis: mechanisms and links with diseases. Signal Transduct. Target Ther. 2021;6:49. doi: 10.1038/s41392-020-00428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou B, et al. Ferroptosis is a type of autophagy-dependent cell death. Semin. Cancer Biol. 2020;66:89–100. doi: 10.1016/j.semcancer.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 158.Yang M, et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019;5:eaaw2238. doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu Z, et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl Acad. Sci. USA. 2019;116:2996–3005. doi: 10.1073/pnas.1819728116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bai Y, et al. Lipid storage and lipophagy regulates ferroptosis. Biochem. Biophys. Res. Commun. 2019;508:997–1003. doi: 10.1016/j.bbrc.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 161.Jia M, et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 2020;21:727–735. doi: 10.1038/s41590-020-0699-0. [DOI] [PubMed] [Google Scholar]
- 162.Dai E, et al. Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway. Nat. Commun. 2020;11:6339. doi: 10.1038/s41467-020-20154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kang R, et al. BECN1 is a new driver of ferroptosis. Autophagy. 2018;14:2173–2175. doi: 10.1080/15548627.2018.1513758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wu, J. et al. STING-dependent induction of lipid peroxidation mediates intestinal ischemia-reperfusion injury. Free Radic. Biol. Med. 163, 135–140(2020). [DOI] [PubMed]
- 165.Liu J, et al. The dark side of ferroptosis in pancreatic cancer. Oncoimmunology. 2021;10:1868691. doi: 10.1080/2162402X.2020.1868691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Castedo M, et al. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 167.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Harding SM, et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cao D, et al. Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res. 2020;30:1088–1097. doi: 10.1038/s41422-020-00422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kujirai T, et al. Structural basis for the inhibition of cGAS by nucleosomes. Science. 2020;370:455–458. doi: 10.1126/science.abd0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pathare GR, et al. Structural mechanism of cGAS inhibition by the nucleosome. Nature. 2020;587:668–672. doi: 10.1038/s41586-020-2750-6. [DOI] [PubMed] [Google Scholar]
- 172.Michalski S, et al. Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature. 2020;587:678–682. doi: 10.1038/s41586-020-2748-0. [DOI] [PubMed] [Google Scholar]
- 173.Zhao B, et al. The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature. 2020;587:673–677. doi: 10.1038/s41586-020-2749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zierhut C, et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell. 2019;178:302–315. doi: 10.1016/j.cell.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lohard S, et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat. Commun. 2020;11:259. doi: 10.1038/s41467-019-13689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chattopadhyay S, et al. Synthetic immunogenic cell death mediated by intracellular delivery of STING agonist nanoshells enhances anticancer chemo-immunotherapy. Nano Lett. 2020;20:2246–2256. doi: 10.1021/acs.nanolett.9b04094. [DOI] [PubMed] [Google Scholar]
- 177.Klarquist J, et al. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J. Immunol. 2014;193:6124–6134. doi: 10.4049/jimmunol.1401869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou C, et al. Transfer of cGAMP into bystander cells via LRRC8 volume-regulated anion channels augments STING-mediated interferon responses and anti-viral immunity. Immunity. 2020;52:767–781. doi: 10.1016/j.immuni.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 179.Lemos H, et al. STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Res. 2016;76:2076–2081. doi: 10.1158/0008-5472.CAN-15-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Motedayen Aval, L., Pease, J. E., Sharma, R. & Pinato, D. J. Challenges and opportunities in the clinical development of STING agonists for cancer immunotherapy. J. Clin. Med. 9, 3323 (2020). [DOI] [PMC free article] [PubMed]
- 181.Lemos, H. et al. Overcoming resistance to STING agonist therapy to incite durable protective antitumor immunity. J. Immunother. Cancer. 8, e001182 (2020). [DOI] [PMC free article] [PubMed]
- 182.Berger G, Marloye M, Lawler SE. Pharmacological modulation of the STING pathway for cancer immunotherapy. Trends Mol. Med. 2019;25:412–427. doi: 10.1016/j.molmed.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 183.Le Naour J, et al. Trial watch: STING agonists in cancer therapy. Oncoimmunology. 2020;9:1777624. doi: 10.1080/2162402X.2020.1777624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tansakul M, et al. Deficiency of STING promotes collagen-specific antibody production and B cell survival in collagen-induced arthritis. Front. Immunol. 2020;11:1101. doi: 10.3389/fimmu.2020.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wu J, Dobbs N, Yang K, Yan N. Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity. 2020;53:115–126.e115. doi: 10.1016/j.immuni.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang Z, Celis E. STING activator c-di-GMP enhances the anti-tumor effects of peptide vaccines in melanoma-bearing mice. Cancer Immunol. Immunother. 2015;64:1057–1066. doi: 10.1007/s00262-015-1713-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chandra D, et al. STING ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol. Res. 2014;2:901–910. doi: 10.1158/2326-6066.CIR-13-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ohkuri T, et al. Intratumoral administration of cGAMP transiently accumulates potent macrophages for anti-tumor immunity at a mouse tumor site. Cancer Immunol. Immunother. 2017;66:705–716. doi: 10.1007/s00262-017-1975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Corrales L, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Skouboe MK, et al. STING agonists enable antiviral cross-talk between human cells and confer protection against genital herpes in mice. PLoS Pathog. 2018;14:e1006976. doi: 10.1371/journal.ppat.1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lara PN, Jr., et al. Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2011;29:2965–2971. doi: 10.1200/JCO.2011.35.0660. [DOI] [PubMed] [Google Scholar]
- 192.Kerr DJ, Kaye SB. Flavone acetic acid–preclinical and clinical activity. Eur. J. Cancer Clin. Oncol. 1989;25:1271–1272. doi: 10.1016/0277-5379(89)90072-2. [DOI] [PubMed] [Google Scholar]
- 193.Cavlar T, et al. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ovalle-Magallanes B, Eugenio-Perez D, Pedraza-Chaverri J. Medicinal properties of mangosteen (Garcinia mangostana L.): a comprehensive update. Food Chem. Toxicol. 2017;109:102–122. doi: 10.1016/j.fct.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 195.Ramanjulu JM, et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564:439–443. doi: 10.1038/s41586-018-0705-y. [DOI] [PubMed] [Google Scholar]
- 196.Pan, B. S. et al. An orally available non-nucleotide STING agonist with antitumor activity. Science. 369, eaba6098 (2020). [DOI] [PubMed]
- 197.Chin EN, et al. Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science. 2020;369:993–999. doi: 10.1126/science.abb4255. [DOI] [PubMed] [Google Scholar]
- 198.Sivick KE, et al. Magnitude of therapeutic STING activation determines CD8(+) T cell-mediated anti-tumor immunity. Cell Rep. 2018;25:3074–3085. doi: 10.1016/j.celrep.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 199.Haag SM, et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 200.Siu T, et al. Discovery of a novel cGAMP competitive ligand of the inactive form of STING. ACS Med. Chem. Lett. 2019;10:92–97. doi: 10.1021/acsmedchemlett.8b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li S, et al. The cyclopeptide astin C specifically inhibits the innate immune CDN sensor STING. Cell Rep. 2018;25:3405–3421. doi: 10.1016/j.celrep.2018.11.097. [DOI] [PubMed] [Google Scholar]