Abstract
In the era of immunotherapy, there lacks of a reliable genomic predictor to identify optimal patient populations in combined radiotherapy and immunotherapy (CRI). The purpose of this study is to investigate whether genomic scores defining radiosensitivity are associated with immune response. Genomic data from Merged Microarray-Acquired dataset (MMD) were established and the Cancer Genome Atlas (TCGA) were obtained. Based on rank-based regression model including 10 genes, radiosensitivity index (RSI) was calculated. A total of 12832 primary tumours across 11 major cancer types were analysed for the association with DNA repair, cellular stemness, macrophage polarisation, and immune subtypes. Additional 585 metastatic tissues were extracted from MET500. RSI was stratified into RSI-Low and RSI-High by a cutpoint of 0.46. Proteomic differential analysis was used to identify significant proteins according to RSI categories. Gene Set Variance Analysis (GSVA) was applied to measure the genomic pathway activity (18 genes for T-cell inflamed activity). Kaplan-Meier analysis was performed for survival analysis. RSI was significantly associated with homologous DNA repair, cancer stemness and immune-related molecular features. Lower RSI was associated with higher fraction of M1 macrophage. Differential proteomic analysis identified significantly higher TAP2 expression in RSI-Low colorectal tumours. In the TCGA cohort, dominant interferon-γ (IFN-γ) response was characterised by low RSI and predicted better response to programmed cell death 1 (PD-1) blockade. In conclusion, in addition to radiation response, our study identified RSI to be associated with various immune-related features and predicted response to PD-1 blockade, thus, highlighting its potential as a candidate biomarker for CRI.
Subject terms: Tumour biomarkers, Cancer genomics
Introduction
Radiotherapy is one of the most effective treatments for uncomplicated locoregional tumours, and around half of all the cancer patients receive this treatment modality during their course of disease management1. With the aid of systems biology, a rank-based radiosensitivity index (RSI) derived from 10 genes (AR, c-JUN [JUN], STAT1, PKC [PRKCB], Rel A [RELA], cABL [ABL1], SUMO1, CDK1, HDAC1, and IRF1), has been generated to predict the survival fraction at 2 Gy (SF2) across 48 cancer cell lines2. The prognostic value of RSI has been validated using several independent data sets, such as those available for the breast and pancreatic cancer, and glioblastoma3–5. Moreover, with the inclusion of a linear-quadratic model, the radiation dose could be further adjusted and personalised6.
Immunotherapy has revolutionized cancer management since the approval of immune checkpoint inhibitors (ICI) for treatment of metastatic melanoma7. Several combinatorial approaches are being investigated to maximize the antitumor immune response, such as radiotherapy combined with immunotherapy (CRI), which possess the potential of inducing durable and synergic effects together. Moreover, over 114 clinical trials involving combined treatment with ICIs and radiotherapy are currently underway8. RSI associates important biological networks regulating signalling in response to radiation9. Here, we studied the robust association of RSI with intratumoral immune landscape. Moreover, we also identified its correlation with immunotherapy, especially the response to PD-1 blockade. By using whole transcriptomic and matched proteomic data from 12832 primary and 585 metastatic tumours, we aimed to improve decision of the combinatorial therapy.
Results
Quality control of RSI across platforms
The output radar plot from sigQC showed that the 10 RSI genes as a signature were characterised by concordant statistical metrics between the Cancer Genome Atlas (TCGA) and Merged Microarray-Acquired dataset (MMD) (Supplementary Fig. 1a). The rank-transformed values for each RSI gene also showed similar distributions (Supplementary Fig. 1b). Taken together, these results suggest that RSI is applicable across these two platforms.
RSI is altered in tumours
A total of 14502 tumour and normal solid tissues were analysed (Supplementary Fig. 2a). Overall, both tumour and normal tissues demonstrated wide distribution of RSI, with more normal tissues displaying low radiosensitivity. This corresponded to a significantly higher mean RSI in normal tissues in both datasets (0.293 [median = 0.289] vs. 0.340 [median = 0.321] in MMD; 0.537 [median = 0.536] vs. 0.629 [median = 0.633] in TCGA, p < 0.001, Supplementary Fig. 2b). Moreover, in different cancer types, we found that the RSI profiles also altered to varied degrees, with significant differential patterns observed in cancer originating in the breast, colon, kidney, and liver (Supplementary Fig. 2c). We also found the differential patterns of RSI gene expression between tumour and normal tissues were specific for these four cancer types (Supplementary Fig. 3a). Further, the RSI is different across cancer types (Supplementary Fig. 3b, ANOVA p < 0.001) and the patterns were similar between the TCGA and MMD, even though most TCGA samples were derived from resected early-stage tumours. Similar to clinical observations, pancreatic cancer, kidney cancer, and melanoma showed higher median RSI, whereas liver cancer showed low median RSI. These findings suggest RSI is altered in tumours and could serve as a biomarker in cancer.
RSI and molecular features associated with immune activation
Studies have shown that defective DNA repair, cancer stemness, and mutational burden are associated with radiation as well as antitumor immune response10,11. To investigate the relationship between RSI and immune response, we used Homologous recombination deficiency (HRD) and RNA stemness score (RNAss), and total mutation burden (TMB) from TCGA and associated them with RSI and other signatures related to radiation response. Signatures derived from RNA-Seq or microarray were compared, as shown in Fig. 1a, most signatures showed low correlation with RSI in TCGA and MMD datasets except for CCL8 (Spearman’s rho [rs] = -0.26, p = 1.12e-8). The negative correlation of CCL8 with RSI was partly supported by the negative association of RSI with a 12-chemokine signature which included CCL89. In addition, we found that RSI was characterised by a generally stronger negative correlation with HRD and RNAss than other signatures (Spearman’s rs = -0.26, p = 2.07e-17; -0.37, p = 2.18e-14), although CCL8 had a higher positive correlation with HRD (Spearman’s rs = 0.28, p = 1.96e-12). Furthermore, our analysis also identified RSI to be negatively correlated with TMB (Spearman’s rs = -0.21, p = 4.52e-6) across most of the epithelial cancer types (Supplementary Fig. 4). The most negative correlation was observed in three gastrointestinal cancers (colon [Spearman’s rs = -0.2697, p = 1.12e-12], pancreas [Spearman’s rs = -0.3512, p = 1.45e-10], and stomach [Spearman’s rs = -0.3372, p = 2.84e-16]).
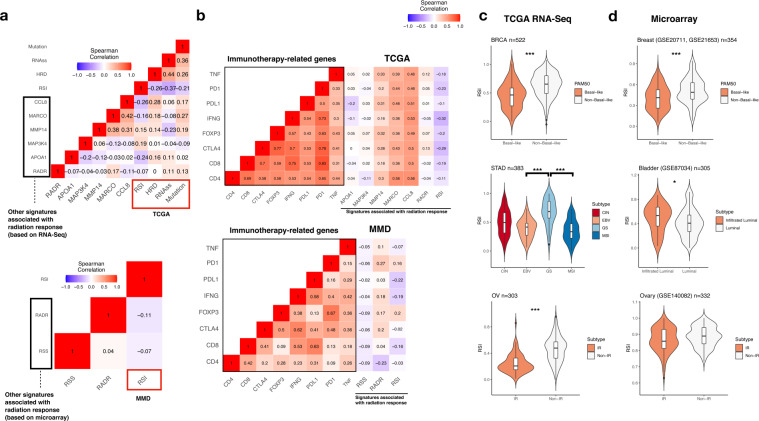
Fig. 1. Association of RSI and other gene signatures with immune-related phenotypes and molecular features.
a Matrix heatmap of Spearman’s correlation among RSI, RADR, RSS, APOA1, MAP3K4, MARCO, CCL8, HRD, RNAss, and TMB in TCGA (upper panel) and MMD (lower panel). Colour bar indicates Spearman’s rho. b Matrix heatmap of Spearman correlation with immunotherapy-related genes (TNF, PD1, PDL1, IFNG, FOXP3, CTLA4, CD8, and CD4) in TCGA (upper panel) and MMD (lower panel). Colour bar indicates Spearman’s rho. c Association of RSI with molecular subtypes in cancer of the BLCA, BRCA, OV, and STAD in TCGA. d Association of RSI with molecular subtypes in cancer of the bladder, breast, ovary, and stomach in MMD. The black horizontal lines represent the median. CIN: chromosomal instability; EBV: Epstein–Barr virus (EBV)-associated; GS: genomically stable; MSI: microsatellite instability; IR: immunoreactive. Mann–Whitney U-test ***p < 0.001, **p < 0.01, *p < 0.05.
We then examined the expression of several immunotherapy-related factors (CD4, CD8A, CTLA4, FOXP3, IFNG, PD1, PDL1, and TNF). Overall, we found that the expression of MMP14, MARCO, and CCL8 signatures was positively correlated with the expression of immunotherapy-related genes. RSI displayed less negative correlations in TCGA, with the strongest negative correlation with IFNG (Spearman’s rs = -0.32, p = 1.02e-11, Fig. 1b). This negative correlation was also observed in MMD (Spearman’s rs = -0.19, p = 3.24e-8). We further identified that the expression of IFNG was negatively correlated with RSI in all cancer sites except for prostate cancer in MMD (Supplementary Fig. 5). Of note, APOA1, MAP3K4, radioresistance score (RADR), and radiosensitivity signature (RSS) correlated poorly with those genes, suggesting that these signatures might have less roles in the crossroads of radiation and immune response.
Using a pre-defined RSI cut-off of 0.46 for tumour stratification, the RSI-Low tumours were found to have significantly higher HRD and RNAss scores than RSI-High tumours (Supplementary Fig. 6). Furthermore, RSI was also significantly associated with different molecular subtypes in cancer (Fig. 1c, d), where tumours with low RSI showed significantly higher proportions of the luminal (bladder), basal-like (breast), immunoreactive (ovary), Epstein–Barr virus (EBV)-associated, and microsatellite instability (MSI) molecular subtypes (stomach) (all p < 0.05). Intriguingly, the RSI-Low tumours displayed both molecular features of MSI and higher TMB in stomach cancer, which were shown to be subgroups with favourable outcomes after immunotherapy12. With higher HRD scores, RSI-Low tumours could be characterised by higher genome instability and subsequently higher mutational burden. Taken together, RSI-Low tumours may represent a special subpopulation and therapeutic target for immunotherapy.
RSI and M1 macrophage polarisation
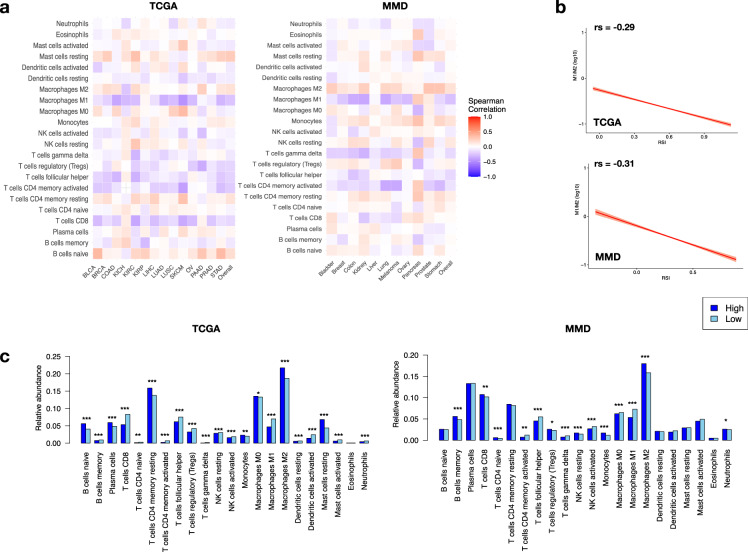
Across the 22 immune cell subtypes, we found the fraction of M1 macrophages to be most negatively correlated with RSI (Spearman’s rs = -0.261, p = 1.07e-10 in MMD, and -0.248, p = 2.16e-18 in TCGA; Fig. 2a). This correlation improved upon examining the relative proportion of M1 and M2 macrophages (M1/2, Spearman’s rs = -0.29, p = 1.16e-10 in TCGA, and -0.31, p = 2.06e-9 in MMD, Fig. 2b). Additionally, as interferon-γ (IFN-γ) has been shown to induce M1 polarisation (M1P)13,14, a strong positive correlation was also observed between intratumoral IFNG expression and log2(M1/M2) (Spearman’s rs = 0.532, p = 1.69e-19 in TCGA; Supplementary Fig. 7). Additionally, RSI-Low tumours harboured significantly higher portion of follicular T helper cells, T cell gamma delta cells, activated NK cells, and M1 macrophages than RSI-High tumours (Fig. 2c).
Fig. 2. RSI and immune cell infiltrates in tumours.
a Heatmaps showing Spearman’s correlation between RSI and the relative fraction of 22 immune cell types obtained from CIBERSORT. b Correlations between RSI and M1/M2 ratio. Relative portion of M1 and M2 macrophages obtained from (M1/M2) was log2 (M1/M2) transformed for plotting. c Distribution of relative abundance of the 22 immune cell subtypes between RSI-High and RSI-Low tumours in TCGA (upper panel) and MMD (lower panel). M1: Macrophage M1; M2: macrophage M2. Mann–Whitney U-test ***p < 0.001, **p < 0.01, *p < 0.05.
The differential expression analysis of 5561 proteins in 95 colorectal adenocarcinoma (COAD) samples indicated high levels of TAP2 (log Fold Change [FC] = -1.33), PARP14 (logFC = -1.25), and GBP4 (logFC = -1.09) in the RSI-Low tumours (Supplementary Fig. 8a and Supplementary Table 1). Concordant with the proteomic analysis, expression of TAP2, PARP14, GBP4, CKB and TRIP6 was significantly high in RSI-Low COAD samples (Supplementary Fig. 8b). TAP1 was included in these differential genes since it forms a heterodimer with TAP2, facilitating antigen presentation to the major histocompatibility complex I (MHCI). Higher expression of these genes was observed in higher M1/M2, especially in cancer of the bladder, breast, colon, melanoma, and stomach (Supplementary Fig. 9). Higher expression of TAP2 has been reported to associate with improved response to anti-PD-1 treatment15. Taken together, RSI-Low tumours were characterised by enhanced antigen presentation machinery and higher M1 proportion, which could lead to pro-inflammatory status and better response to PD-1 blockade.
RSI and immune landscape in cancers
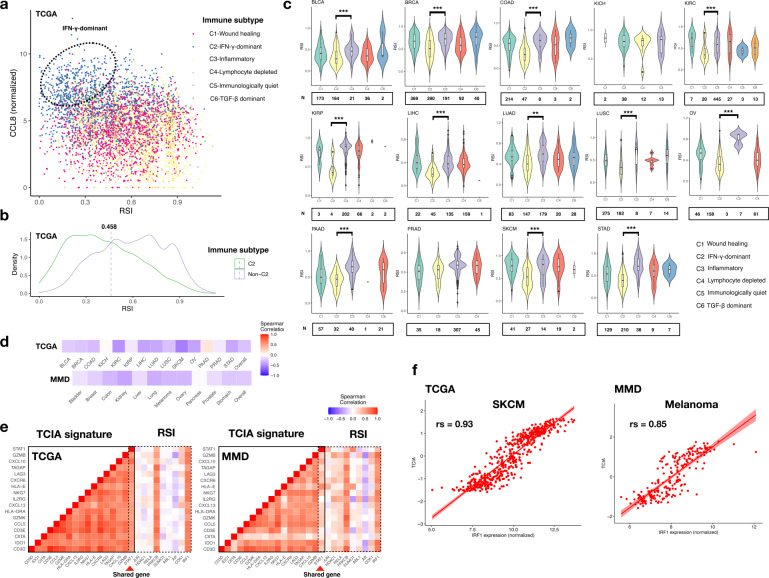
To confirm the role of RSI as a candidate signature associated with immunotherapy, we investigated its distribution across different immune categories. We observed that the related RSI and CCL8 signatures could differentiate C2 (IFN-γ dominant response) from other immune subtypes (Fig. 3a). The density plot also demonstrated a deviated trend of C2 tumours toward lower RSI, with an optimal cut-off value of 0.458, which is very close to the value (0.46) defined previously for RSI stratification (Fig. 3b)2.
Fig. 3. RSI and immune landscape.
a Scatter plot showing distribution of six immune categories along axes of RSI and CCL8. C1, C2, C3, C4, C5, and C6 denote wound healing, IFN-γ-dominant, inflammatory, lymphocyte-depleted, immunologically quiet and TGF-β-dominant pathways, respectively. Dotted circle indicates clustering of the C2 subtype. b Density plot showing distribution of RSI in C2 and non-C2 categories. Dotted line indicates the optimal cut-off value. c Distribution of RSI in TCGA cohorts separated by immune subtypes. The black horizontal lines represent the median. d Spearman’s correlation between RSI and TCIA across the 11 cancer types in TCGA (upper panel) and MMD (lower panel). Colour bar indicates Spearman’s rho. e Matrix heatmap of Spearman’s correlation between the 18 TCIA genes and 10 RSI genes in TCGA (left panel) and MMD (right panel). Red arrowhead indicates the overlapped gene between TCIA and RSI. Colour bar indicates Spearman’s rho. f Correlations between IRF1 expression and TCIA score.
To identify which RSI genes had a higher impact on this difference, we calculated the feature importance for C2 and non-C2 classifications. Among the 10 RSI genes, the mean feature importance was 177.8 (Supplementary Fig. 10a). STAT1 was identified to have the highest feature importance (mean decrease in Gini [MDG] = 283.5), followed by CDK1 (MDG = 236.8), JUN (MDG = 228.5), IRF1 (MDG = 215.4), and ABL1 (MDG = 180.7). The expression of JUN, CDK1, IRF1, and STAT1 was significantly higher in the RSI-Low tumours (Supplementary Fig. 10b). A classifier based on JUN, CDK1, IRF1, STAT1 and ABL1 showed a high classification accuracy of 82% across all malignancies (95% confidence interval [CI] = 0.798–0.841), with an area under the receiver operating characteristic (ROC) curve of 0.8449 (Supplementary Fig. 10c). In the TCGA dataset, C1 (wound healing) and C2 were the main immune subtypes in most cancer sites, except for renal and prostate cancers, which harboured more C3 (inflammatory) subtypes (Fig. 3c). Instead of KICH and PRAD, tumours in the C2 subgroup were generally associated with significantly lower RSI than those in the C3 subgroup (Fig. 4a). These findings suggest that RSI-Low tumours are mainly dominated by IFN-γ-related responses.
Fig. 4. Enrichment of TCIA and survival analysis.
a Heatmap depicting enrichment status of the TCIA defined by the mean z-transformed GSVA scores > 0.32 in the 11 cancer types. Columns have been ordered by increasing the RSI. b KM plots for the OS in patients with cancer of the bladder, breast, colon, ovary, and stomach with enriched TCIA. The RSI-High and RSI-Low tumours have been stratified by the optimal cut-off value for OS in each cancer type.
T cell-inflamed activity (TCIA) is enriched in tumours with low RSI
About 41% tumours in the 11 TCGA cohorts were found to be potentially sensitive to pembrolizumab according to our defined threshold (0.35, Supplementary Fig. 11). We found an overall negative correlation between RSI and TCIA (Spearman’s rs = -0.26, p = 2.38e-15 in TCGA; -0.23, p = 6.87e-13 in MMD, Fig. 3d). The TCIA signature consisted of 18 genes that intersected RSI genes with STAT1 (Fig. 3e). The 18 genes showed high intra-correlation in both TCGA and MMD. Moreover, two RSI genes, PRKCB and IRF1, showed a high positive correlation with the TCIA signature. The highest correlation was found between IRF1 expression and TCIA in melanoma (Spearman’s rs = 0.93, p = 2.12e-15 in TCGA; 0.85, p = 4.43e-15 in MMD, Fig. 3f and Supplementary Table 2). Divided by 0.35 to determine enrichment, TCIA was found to be enriched in most RSI-Low tumours (Fig. 4a). The most pronounced enrichment was observed in colon, kidney, lung, and stomach cancers (Table 1).
Table 1.
Distribution of RSI between TCIA-enrichment and non-TCIA-enrichment.
| TCGA | Low (N) | High (N) | P | MMD | Low (N) | High (N) | P |
|---|---|---|---|---|---|---|---|
| BLCA | <0.001 | Bladder | 0.264 | ||||
| E | 133 | 45 | E | 63 | 14 | ||
| NE | 140 | 108 | NE | 100 | 35 | ||
| BRCA | Breast | ||||||
| E | 176 | 292 | <0.001 | E | 770 | 124 | 0.87 |
| NE | 115 | 625 | NE | 1208 | 200 | ||
| COAD | Colon | ||||||
| E | 69 | 76 | <0.001 | E | 479 | 111 | <0.001 |
| NE | 47 | 132 | NE | 643 | 281 | ||
| KICH | Kidney | ||||||
| E | 2 | 38 | 0.303 | E | 93 | 39 | <0.001 |
| NE | 7 | 44 | NE | 55 | 136 | ||
| KIRC | Liver | ||||||
| E | 132 | 126 | <0.001 | E | 149 | 8 | 0.3978 |
| NE | 54 | 293 | NE | 225 | 19 | ||
| KIRP | Lung | ||||||
| E | 12 | 115 | 0.029 | E | 379 | 25 | <0.001 |
| NE | 16 | 189 | NE | 501 | 92 | ||
| LIHC | Melanoma | ||||||
| E | 154 | 100 | <0.001 | E | 64 | 27 | 0.504 |
| NE | 145 | 22 | NE | 80 | 43 | ||
| LUAD | Ovary | ||||||
| E | 138 | 93 | <0.001 | E | 213 | 45 | 0.068 |
| NE | 94 | 249 | NE | 309 | 96 | ||
| LUSC | Pancreas | ||||||
| E | 142 | 89 | <0.001 | E | 24 | 48 | 0.582 |
| NE | 149 | 172 | NE | 30 | 76 | ||
| SKCM | Prostate | ||||||
| E | 16 | 15 | <0.001 | E | 104 | 0 | 0.075 |
| NE | 4 | 69 | NE | 127 | 6 | ||
| OV | Stomach | ||||||
| E | 97 | 32 | <0.001 | E | 263 | 41 | 0.009 |
| NE | 81 | 93 | NE | 341 | 92 | ||
| PAAD | |||||||
| E | 21 | 53 | 0.586 | ||||
| NE | 36 | 72 | |||||
| PRAD | |||||||
| E | 22 | 203 | 1 | ||||
| NE | 32 | 292 | |||||
| STAD | |||||||
| E | 127 | 62 | <0.001 | ||||
| NE | 91 | 170 |
RSI radiosensitivity index, TCIA T-cell inflamed activity, E Enrichment, NE non-enrichment.
RSI is predictive of survival in several cancer types
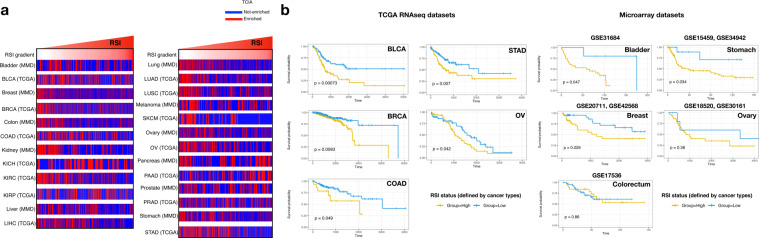
Univariate analysis in the entire TCGA cohort showed that tumours with high RSI and TCIA associated with favourable survival outcomes (Hazard ratio [HR] = 0.29, 95% CI = 0.23–0.36, p < 0.001 for RSI; HR = 0.86, 95% CI = 0.82–0.91, p < 0.001 for TCIA; Supplementary Fig. 12). However, using the optimal cut-off for overall survival (OS) in tumours enriched in TCIA (Supplementary Table 3), cancer of the bladder, breast, and colon with low RSI displayed favourable survival outcomes (Fig. 4b), whereas unfavourable outcomes were observed in cancer of the kidney and prostate (Supplementary Fig. 13).
Metastatic tumours are characterised by low RSI with varied TCIA
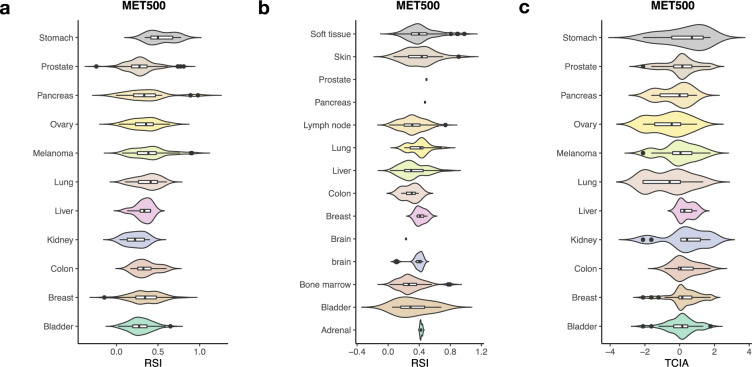
In the MET500 database, the median RSI was found to be less than 0.5, in 10 cancer types (Fig. 5a and Supplementary Table 4). Bone marrow was the most sensitive metastatic site, with higher RSI observed for sites in organs, such as the breast, brain, adrenal gland, and lung (Fig. 5b). For metastases originating from the bladder, breast, and prostate (all N > 50), the median TCIA was above 0 (Fig. 5c). Similar to the primary tumours, even with low RSI, enrichment of TCIA was only achieved in 32.9 % (Bladder, 26/79), 43.4% (Breast, 69/159), and 34.2% (Prostate, 53/155), respectively.
Fig. 5. RSI in the metastatic tissues.
a RSI in metastatic tissues in the MET500 cohort. b RSI of tumours in various metastatic tissues. c TCIA scores in the 11 cancer types. The black horizontal lines represent the median.
Discussion
In the large-scale analysis of RSI across 11 major cancer types, we identified that RSI was related to various immune-relevant genomic and molecular features, and low RSI were associated with dominant IFN-γ signalling response and predicted therapeutic efficacy of PD-1 blockade.
Tumours with defective DNA repair response (DDR) tend to have accumulation of genomic errors, which may trigger increased presentation of tumour-specific neoantigens16. In the context of MHCI, the neoantigens are specifically recognized by T cells, enhancing the antitumor immune response10. In urothelial cancers, mutations in DDR pathways are associated with better efficacy of PD-1/PD-L1 blockade17. Here we demonstrated lower RSI correlated with higher HRD score and higher TMB, suggesting the presence of deficient DNA repair mechanism and potential of responding to immune-based therapies18. HRD score was associated with genes involved in homologous repair, including BRCA1, BRCA2, RAD51B, and RAD51C, and alteration of these genes are linked to radiosensitivity19,20. Additionally, lower RSI also correlated with higher RNAss, which indicated higher degrees of stemness and tumour de-differentiation21. Malta et al. reported higher RNAss is associated with increased PD-L1 protein expression in BLCA, BRCA, COAD, LUAD, KICH, KIRC, KIRP, OV, PAAD, and STAD cohorts in TCGA, suggesting potential effectiveness to ICI for these tumours.
Immune responses are activated in few molecular subtypes, such as the basal-like, immunoreactive, and EBV-associated subtype in cancer of the breast, ovary, and stomach, respectively22–24. These immune-associated subtypes show inter-similarity, and are partly characterised by increased CD8 + T cell infiltrates and up-regulated IFN-γ signalling signatures25–27. Additionally, compared to other molecular features, these subtypes associate with a significantly low RSI. Furthermore, the expression of IFNG, which translates to the actionable IFN-γ response, was negatively correlated with RSI, supporting the up-regulation of IFN-γ signalling activity in these immune-associated subtypes28. Tobin et al. utilized 12 chemokine genes to define the intratumoral immune activation and identified that low RSI significantly associates with high immune activation (using an RSI cut-point of 0.3745)9. Interestingly, CCL8 was listed as one of the chemokine genes, supporting its negative correlation with RSI in the present study. Furthermore, as RSI genes such as STAT1 and IRF1 are downstream of IFN-γ-mediated signalling, RSI has a better correlation with various immune-related molecular features and phenotypes than other genes and gene signatures related to radiation response29.
Tumours with C2 subtype were characterised by the highest M1P, highest CD8 + T cells, and high proliferation rate30. Concordant with this phenotype, here, the RSI-Low tumours showed high percentage of M1 macrophages. Despite an opposite trend observed for the CD8 + T cells in the MMD, RSI-Low tumours also harboured more follicular T helper cells, T cell gamma delta cells, activated NK cells. Of note, these cell types are all capable of IFN-γ secretion, suggesting a dominant role of IFN-γ in RSI-Low tumours31–33. Studies suggest that IFN-γ could transform macrophages to a proinflammatory phenotype and induce M1P34,35. When IFN-γ binds receptor on macrophages, it induces STAT1 homodimerisation, which triggers IFN-γ-dependent signalling36,37. The canonical IRF/STAT signalling is central in modulating the macrophage polarisation. Additionally, IFN-γ induces synthesis of IRF138, which plays an important role in inflammation, immunity, cell proliferation, and apoptosis38,39. Interestingly, STAT1 and IRF1 have been listed in the RSI genes, thus associating radiosensitivity with response to IFN-γ. Further, in the IFN-γ dominant microenvironment, the antigen processing machinery, including MHCI, TAP1, and TAP2 may upregulate40,41. The TAP1 and TAP2 facilitate MHCI antigen presentation42, which subsequently activate the CD8 + T lymphocytes. The IFN-γ released by activated CD8 + T lymphocytes primes the macrophages toward the M1 phenotype43. Here, the intratumoral expression of TAP1 and TAP2 showed higher clustering with increasing M1 population in several cancer types. Therefore, the mechanism regulating this phenomenon may be contributed by common action of local IFN-γ response.
An increased expression of PD-L1 on the surface of tumour cells or immune cells has been associated with an improved response to the PD-1 checkpoint blockade44. However, single measurement of PD-L1 would limit the understanding of the interaction between cancer and immune cells45. Therefore, genomics analyses have been proposed to elucidate the complex tumour microenvironment (TME)46–48. M Ayers et al. utilized patient cohorts from three KEYNOTE trials and confirmed the robustness of a pan-tumour T cell-inflamed gene signature in predicting the response to PD-1 blockade49. The TCIA, defined by the gene signature, is characterised by IFN-γ signalling, cytotoxic effector molecules, antigen presentation, and T cell active cytokines. This “hot” immune-inflamed TME associates with high CD8 + lymphocytes, myeloid cells, monocytic cells, high IFN levels, and stimulation of chemokines, including CXCL9, CLCL10, and CXCL11, which predict benefit from the PD-1 blockade50. Here, we observed enrichment of TCIA in tumours with low RSI, which could be correlated to favourable OS in several cancer types. However, IFN-γ plays a dual role in cancer progression51, and upon prolonged activation of IFN-γ in the TME, the tumour cells may develop resistance, thus activating tumorigenic pathways41. Moreover, upregulation of PD-L1/2 by IFN-γ in cancer, stromal, and myeloid cells would lead to immune evasion52. Furthermore, the C2 subtype was found to associate with a less favourable outcome in the TCGA cohort. In addition to the edited immune response, the absence of survival benefit in several tumours with low RSI, and the corresponding high portion of C2 subtypes could be explained by their aggressiveness and the associated high proliferation30. Furthermore, tumours with re-modelled immune response show loss of genes responsible for antigen presentation, making them less immunogenic30. Therefore, the C2 category is of prognostic value in immunotherapy. The classifier based on five RSI genes, STAT1, CDK1, JUN, and IRF1, and ABL1 sufficiently predicted the presence of the C2 subtype. Taken together, the identification of tumours with the C2 subtype could better characterise the immune status while assessing possible radiation responders.
To enhance the efficacy of PD-1 blockade, selecting tumours with upregulated PD-L1/2 expression is of prognostic significance, especially in tumours with increased exposure to IFN-γ41,53. Additionally, inhibition of the PD-1/PD-L1 axis would prevent exhaustion of CD8 + T cells, and enhance response to immunotherapy54. CRI seems promising, as radiation may help induce immunogenic cell death, elicit innate immune system, and promote antigen presentation to infiltrating CD8 + T cells41,55. Here, the RSI-Low tumours were found to associate with active IFN-γ signalling, M1P, upregulated antigen presentation machinery, response to PD-1 blockade, and response to radiotherapy. Therefore, CRI appears to be the most effective therapy for RSI-Low tumours.
There were some technical limitations in our study. First, we could not obtain the serial RSI change for tumours. Therefore, we could not elucidate the change of radiosensitivity overtime during the radiotherapy. Second, the immune cell infiltration was only the estimation, not the real TME. The TME is complicated might not purely reflect the real fractions of immune cells from RNA sequencing. Third, the metastatic tissues from MET500 dataset were not sufficient for statistical analysis and comparison with the primary tumours, limiting the use of RSI in this situation.
Despite these limitations, with the help of large sample cancer genomics dataset, our study identified strong association between RSI and IFN-γ response and immunotherapy, although future work would be required to elucidate the detailed mechanism and role of RSI in CRI.
Methods
Data source
MMD used in our study have been previously described56. The datasets cover 11 major cancer types (bladder, breast, colon, liver, lung, kidney, melanoma, ovary, pancreas, prostate, and stomach), and comprise of 95 independent Gene Expression Omnibus (GEO) studies (http://www.ncbi.nlm.nih.gov/geo) and a total of 8386 samples, either tumours or relevant normal tissues. Raw data for the 11 cancer types were independently pre-process using author-defined methods or RMA-normalization using the R library affy package57. All the raw data were based on the GPL 570 microarray platform (Affymetrix Human Genome U133 Plus 2.0 Array) and were merged and adjusted by the Combat method using the R library inSilicoMergine package58. Probes annotated with specific genes were collapsed to the maximum expression values, which were adopted for subsequent analyses.
Normalized RNA-Seq data based on Illumina HiSeq platform were extracted for the 11 cancer types and 14 cohorts from the Cancer Genome Atlas (TCGA; abbreviation: BLCA, BRCA, COAD, LIHC, LUAD, LUSC, KICH, KIRC, KIRP, OV, PAAD, PRAD, SKCM, STAD) using the bioinformatics tool Xena browser (https://xenabrowser.net/). Here, rectal adenocarcinoma was included in COAD, which was then used for comparison with colorectal tissues in MMD. Raw RNA-Seq data were quantified using the root square error method (RSEM), and log2 transformed (RSEM + 1). The associated clinical parameters, such as survival and molecular subtypes were obtained for comparative purpose. Finally, 6116 tumour (primary tumour) and normal (solid tissue normal) tissues were retrieved.
Raw RNA-Seq data in MET500 for the 11 epithelial carcinoma were downloaded from the database of Genotypes and Phenotypes, subsequently processed using RSEM59,60, and then normalized using fragments per kilobase of transcript per million mapped reads (FPKM) and log2 transformed (FPKM + 0.001). A total of 585 metastatic tumours from various cancer types and body locations (soft tissue, skin, prostate, pancreas, lymph node, lung, liver, colon, breast, brain, bone marrow, bladder, and adrenal gland) were analysed.
Signature quality control
As RSI was derived from microarray datasets, its use in RNA-Seq platforms has never been elucidated. Prior to proceeding further, we first evaluated the quality of RSI application on TCGA and MMD using sigQC61. sigQC is an R package for gene signature quality control, which encompasses a number of statistical metrics describing the ability of a gene signature to represent a dataset of interest, such as variability of signature genes and co-correlation of signature genes.
Calculation of RSI signature
RSI was calculated using a rank-based linear regression model. 10 RSI genes were indexed from the entire genomics data in the MMD and TCGA, and the expression values were reordered in the respective platforms. Gene with the highest expression value was ranked 10. The RSI was constructed using the following equation:
| 1 |
The RSI value corresponded to the SF2, and therefore a low RSI suggests high radiosensitivity. Furthermore, a previous study used ROC and found a cut-off point of 0.46, which was associated with the best predictive accuracy in pathological response after radiotherapy2. Based on this value, tumours were grouped into RSI-High and RSI-Low, which essentially represent two different types of tumours in terms of response to radiotherapy.
Gene signatures associated with radiation response
To investigate the role of RSI as a promising candidate in linking radiotherapy and immunotherapy, we searched for other gene signatures or genes reported to predict radiation response. A 34-gene RSS was developed and validated by Cui et al. This signature was derived from a microarray platform and was shown to predict the benefit of radiotherapy in breast cancer62. RSS was calculated in MMD according to the author’s method. Another 13-gene RADR was developed by Foy et al. and was shown to predict recurrence after radiotherapy in head and neck squamous cell carcinoma63. It was calculated using a single-sample gene set enrichment analysis tool and was applied well in both RNA-Seq and microarray platforms. In addition to the radiotherapy-associated scores, we searched for genes relevant to the radiation response. In locally advanced breast cancer, APOA1, MAP3K4, and MMP14 were differentially expressed in the responders in the neoadjuvant setting64. MARCO and CCL8, which are associated with immune infiltration and radiation outcome, were also included65. For comparative analyses, RSS and RADR were applied in MMD, whereas RADR, as well as APOA1, MAP3K4, MMP14, MARCO, and CCL8 were applied to the TCGA RNA-Seq platform.
HRD and RNAss
The HRD and RNAss were retrieved from analytic data type in the Xena browser using the TCGA-Pan-Cancer dataset. HRD and RNAss scores were used to represent the degree of HRD and cancer stemness.
Molecular subtypes, TMB and immunotherapy-related genes
To assess the association of RSI with immune-related molecular subtypes in cancers, seven independent datasets with annotated molecular features were obtained (Bladder: accession number = GSE87034 in GEO; Breast: GSE20711 and GSE21653, and BRCA in TCGA; Ovary: GSE140082, and OV in TCGA; Stomach: STAD in TCGA). In cancer originating in the bladder, breast, and ovary, the RSI was dichotomized based on the molecular subtypes associated with immune response. The four molecular subtypes characterised by TCGA in gastric adenocarcinoma22, based on genome stability and infection, were considered for our study to associate with RSI and immune responses. Furthermore, immunotherapy-related factors, including CD4, CD8A, CTLA4, FOXP3, IFNG, PD1, PDL1, and TNF were examined and correlated with RSI. TMB for tumour samples from the TCGA was retrieved from the Genomic Data Commons Data Portal (http://portal.gdc.cancer.gov/projects/). Here, the TMB was defined as the total number of simple somatic mutations observed in each of the TCGA cancer cohorts. Tumours containing at least one mutation were included.
CIBERSORT and polarisation of macrophages
Relative fractions of distinct immune cell types found in the TME were estimated using the beta version of the CIBERSORT (http://cibersort.stanford.edu/). Next, 1000 tumours were randomly selected from cancers originating in the breast, colon, and lung as their gene expression data in the MMD exceeded the 500 Mb file quota. Further, the LM22 signature gene file and 500 permutations were selected as our input parameters. The quantile normalization was disabled for runs with the RNA-Seq data. The LM22 signature gene file contains 547 genes that accurately distinguish 22 types of immune cells, including the T cells, B cells, plasma cells, and NK cells with different activation states, and various subsets of the myeloid lineage, facilitating an overview of immune cells infiltrating the tumours. Moreover, for each immune cell subtypes, the Spearman’s correlation was calculated between the relative levels of immune cells and RSI. Next, the polarisation of macrophages was assessed from the data generated in CIBERSORT. The M1P status was determined using a ratio of the M1 and M2 macrophages, which was obtained based on the fraction of their relative amounts, and was log2 (M1/M2) transformed and correlated with RSI using the Spearman’s rs test.
Proteomic and differential analysis
To investigate the impact of RSI on the protein level, we conducted a proteomic analysis using available data from the NCI Clinical Proteomic Tumor Analysis Consortium (https://cptac-data-portal.georgetown.edu/cptac/s/S016). Relative protein abundance data of 5562 genes for matched colorectal cancer cohort of the TCGA was obtained (TCGA_Colon_VU_Proteome_CDAP_Protein_Report.r2). The cohort comprised of 95 samples (64 colon and 31 rectum tumour tissues). Unshared spectral count values were used to represent the relative protein abundance. The R library edgeR package was used to assess differential protein expression between the RSI-High and RSI-Low tumours66. FC > 1 or FC < − 1 and adjusted p < 0.05 were used to determine the significantly differential proteins. Volcano plot was generated using the R library EnhancedVolcano package.
Gene set variation analysis (GSVA)
TCIA, which associates with response to the PD-1 blockade, was described as an immunotherapy-related pathway by our group49. GSVA scores of this pathway, inclusive of 18 genes, were computed for each tumour sample in the MMD and TCGA datasets. The R/Bioconductor GSVA package was used to calculate these scores, which were then transformed to z-scores67. We applied a cut-off of 0.35 for mean z-score, which was previously identified in melanoma46, to determine whether the samples in our study were enriched or not for TCIA. A heatmap was used for visualization of the correlation between the TCIA enrichment status and RSI.
Determining IFN-γ dominant response
To establish a classifier for C2 subtype, feature importance of the RSI genes in determining IFN-γ dominant response was first quantified using the MDG68. The MDG applies the Random Forest algorithm (RF) and utilizes the Gini impurity to measure the feature importance. An increase in MDG correlates with a decrease in node impurity, and thus enhanced importance. Genes with MDG above mean feature importance were used for building a classifier. Binary classification of the IFN-γ-dominant response subtype (C2) was based on an optimal cut-point determined by the R library cutpointr package. Classifier of the C2 subtype was established using the RF algorithm, and the resultant performance of the model was estimated using the area under the ROC curve with 95% CI. The calculation of MDG and construction of the RF-based classifier were performed using the R library randomForest package.
Survival analysis
Survival data obtained from the TCGA and other independent GEO datasets were analysed using the R library survival package. The RSI allowed stratification into RSI-High and RSI-Low groups based on the optimal cut-point using the surv_cutpoint function in survminer package. Kaplan–Meier (KM) survival curve was derived for OS according to RSI. The HR of RSI and TCIA for OS was calculated and plotted using the hr_plot function in the R library finalfit package. The log-rank test was used to identify survival differences between the groups.
Statistical analysis
All statistical analyses were conducted in R. For comparison of variables under defined conditions, the Mann–Whitney U-test was used to derive the p-value. One-way ANOVA was used for multiple comparisons of RSI across cancer types. For both statistical analyses, p-value < 0.05 was considered statistically significant. The density plot was used to observe the estimated distribution of RSI between the normal and tumour tissues. For comparisons of different datasets (MMD and TCGA), the p values derived for RSI genes for normal vs. tumour tissues were quantified and -log10(p) transformed, and were then visualized by bubble plot.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This study was supported by study project of Tri-Service General Hospital (TSGH-D-110086).
Author contributions
D.Y.H. and H.W.Y. designed the study and wrote the manuscript. D.Y.H., W.Y.F., S.P.C. and L.C.H. performed experiments and analysed the data. L.C.H., Y.J.F., L.C.S., C.H.L. and H.W.Y. helped review the manuscript.
Data availability
All data generated or analysed during this study are available from the corresponding author on reasonable request. Data from TCGA could be downloaded from Xena browser (https://xenabrowser.net/). Data for MET500 could be retrieved from database of Genotypes and Phenotypes (accession number: phs000673.v2.p1). Processed cancer type-specific MMDs are available from ArrayExpress under accession numbers (MTAB-6690, E-MTAB-6691, E-MTAB-6692, E-MTAB-6693, E-MTAB-6694, MTAB-6695, E-MTAB-6696, E-MTAB-6697, E-MTAB-6698, E-MTAB-6699, and E-MTAB-6703). Proteomic data for colorectal cancer are available from the NCI Clinical Proteomic Tumor Analysis Consortium (https://cptac-data-portal.georgetown.edu/cptac/s/S016).
Code availability
The scripts used for the bioinformatics reanalysis are available at https://github.com/YangHongDai/Radiosensitivity_index.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-021-00200-0.
References
- 1.Chen HHW, Kuo MT. Improving radiotherapy in cancer treatment: promises and challenges. Oncotarget. 2017;8:62742–62758. doi: 10.18632/oncotarget.18409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eschrich SA, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J. Radiat. Oncol. Biol. Phys. 2009;75:489–496. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eschrich SA, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clinical cancer research: an official journal of the American Association for. Cancer Res. 2012;18:5134–5143. doi: 10.1158/1078-0432.CCR-12-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed KA, et al. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget. 2015;6:34414–34422. doi: 10.18632/oncotarget.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strom T, et al. Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother. Oncol.: J. Eur. Soc. Therapeutic Radiol. Oncol. 2015;117:159–164. doi: 10.1016/j.radonc.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott JG, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. 2017;18:202–211. doi: 10.1016/S1470-2045(16)30648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat. Rev. Cancer. 2011;11:805–812. doi: 10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J, et al. Trial watch: The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 2018;17:854–855. doi: 10.1038/nrd.2018.210. [DOI] [PubMed] [Google Scholar]
- 9.Strom T, et al. Tumour radiosensitivity is associated with immune activation in solid tumours. Eur. J. Cancer. 2017;84:304–314. doi: 10.1016/j.ejca.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarchoan M, Johnson BA, 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer. 2017;17:209–222. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda A, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl Acad. Sci. USA. 2019;116:9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman AM, Sokol ES, Frampton GM, Lippman SM, Kurzrock R. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol. Res. 2019;7:1570–1573. doi: 10.1158/2326-6066.CIR-19-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 14.Hu X, Chen J, Wang L, Ivashkiv LB. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J. Leukoc. Biol. 2007;82:237–243. doi: 10.1189/jlb.1206763. [DOI] [PubMed] [Google Scholar]
- 15.Li F, et al. In vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in Kras-mutant lung adenocarcinoma. Cancer Disco. 2020;10:270–287. doi: 10.1158/2159-8290.CD-19-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Shih DJH, Lin SY. Role of DNA repair defects in predicting immunotherapy response. Biomark. Res. 2020;8:23. doi: 10.1186/s40364-020-00202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teo MY, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J. Clin. Oncol. 2018;36:1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knijnenburg TA, et al. Genomic and molecular landscape of DNA damage repair deficiency across. The Cancer Genome Atlas. Cell Rep. 2018;23:239–254. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baeyens A, et al. Chromosomal radiosensitivity in BRCA1 and BRCA2 mutation carriers. Int J. Radiat. Biol. 2004;80:745–756. doi: 10.1080/09553000400017937. [DOI] [PubMed] [Google Scholar]
- 20.Zhong X, et al. Rad51 in regulating the radiosensitivity of non-small cell lung cancer with different epidermal growth factor receptor mutation status. Thorac. Cancer. 2016;7:50–60. doi: 10.1111/1759-7714.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malta TM, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen GM, et al. Consensus on molecular subtypes of high-grade serous ovarian carcinoma. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2018;24:5037–5047. doi: 10.1158/1078-0432.CCR-18-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu B, et al. Immune gene expression profiling reveals heterogeneity in luminal breast tumors. Breast Cancer Res. 2019;21:147. doi: 10.1186/s13058-019-1218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konecny, G. E. et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J. Natl Cancer Inst.106, dju249 (2014). [DOI] [PMC free article] [PubMed]
- 26.Gatti-Mays ME, et al. If we build it they will come: targeting the immune response to breast cancer. NPJ Breast Cancer. 2019;5:37. doi: 10.1038/s41523-019-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki S, et al. EBV-associated gastric cancer evades T-cell immunity by PD-1/PD-L1 interactions. Gastric Cancer. 2019;22:486–496. doi: 10.1007/s10120-018-0880-4. [DOI] [PubMed] [Google Scholar]
- 28.Adhikary T, et al. Interferon signaling in ascites-associated macrophages is linked to a favorable clinical outcome in a subgroup of ovarian carcinoma patients. BMC Genomics. 2017;18:243. doi: 10.1186/s12864-017-3630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao L, et al. IRF1 inhibits antitumor immunity through the upregulation of PD-L1 in the Tumor Cell. Cancer Immunol. Res. 2019;7:1258–1266. doi: 10.1158/2326-6066.CIR-18-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, et al. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J. Exp. Med. 2003;198:433–442. doi: 10.1084/jem.20030584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paolini R, Bernardini G, Molfetta R, Santoni A. NK cells and interferons. Cytokine Growth Factor Rev. 2015;26:113–120. doi: 10.1016/j.cytogfr.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoeksema MA, et al. IFN-gamma priming of macrophages represses a part of the inflammatory program and attenuates neutrophil recruitment. J. Immunol. 2015;194:3909–3916. doi: 10.4049/jimmunol.1402077. [DOI] [PubMed] [Google Scholar]
- 36.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 37.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J. Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 40.Cramer LA, Nelson SL, Klemsz MJ. Synergistic induction of the Tap-1 gene by IFN-gamma and lipopolysaccharide in macrophages is regulated by STAT1. J. Immunol. 2000;165:3190–3197. doi: 10.4049/jimmunol.165.6.3190. [DOI] [PubMed] [Google Scholar]
- 41.Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018;9:847. doi: 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Praest P, et al. The influence of TAP1 and TAP2 gene polymorphisms on TAP function and its inhibition by viral immune evasion proteins. Mol. Immunol. 2018;101:55–64. doi: 10.1016/j.molimm.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 43.Bhat P, Leggatt G, Waterhouse N, Frazer IH. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017;8:e2836. doi: 10.1038/cddis.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 46.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conway JR, Kofman E, Mo SS, Elmarakeby H, Van Allen E. Genomics of response to immune checkpoint therapies for cancer: implications for precision medicine. Genome Med. 2018;10:93. doi: 10.1186/s13073-018-0605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayers M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, et al. Application of PD-1 blockade in cancer immunotherapy. Comput Struct. Biotechnol. J. 2019;17:661–674. doi: 10.1016/j.csbj.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandai M, et al. Dual faces of IFNgamma in cancer progression: a role of PD-L1 induction in the determination of pro- and antitumor immunity. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2016;22:2329–2334. doi: 10.1158/1078-0432.CCR-16-0224. [DOI] [PubMed] [Google Scholar]
- 52.Abiko K, et al. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br. J. Cancer. 2015;112:1501–1509. doi: 10.1038/bjc.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Diaz A, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, M. K., Park, B. B. & Uhm, J. Understanding immune evasion and therapeutic targeting associated with PD-1/PD-L1 pathway in diffuse large B-cell lymphoma. Int. J. Mol. Sci.20, 1326 (2019). [DOI] [PMC free article] [PubMed]
- 55.Lhuillier C, Rudqvist NP, Elemento O, Formenti SC, Demaria S. Radiation therapy and anti-tumor immunity: exposing immunogenic mutations to the immune system. Genome Med. 2019;11:40. doi: 10.1186/s13073-019-0653-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bin Lim S, et al. Pan-cancer analysis connects tumor matrisome to immune response. NPJ Precis Oncol. 2019;3:15. doi: 10.1038/s41698-019-0087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 58.Taminau J, et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinf. 2012;13:335. doi: 10.1186/1471-2105-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dhawan A, et al. Guidelines for using sigQC for systematic evaluation of gene signatures. Nat. Protoc. 2019;14:1377–1400. doi: 10.1038/s41596-019-0136-8. [DOI] [PubMed] [Google Scholar]
- 62.Cui Y, Li B, Pollom EL, Horst KC, Li R. Integrating radiosensitivity and immune gene signatures for predicting benefit of radiotherapy in breast cancer. Clin. Cancer Res. 2018;24:4754–4762. doi: 10.1158/1078-0432.CCR-18-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foy JP, et al. A 13-gene expression-based radioresistance score highlights the heterogeneity in the response to radiation therapy across HPV-negative HNSCC molecular subtypes. BMC Med. 2017;15:165. doi: 10.1186/s12916-017-0929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanic M, et al. Molecular signature of response to preoperative radiotherapy in locally advanced breast cancer. Radiat. Oncol. 2018;13:193. doi: 10.1186/s13014-018-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wen, P. et al. Pan-cancer analysis of radiotherapy benefits and immune infiltration in multiple human cancers. Cancers (Basel)12 (2020). [DOI] [PMC free article] [PubMed]
- 66.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Proceedings of the 26th International Conference on Neural Information Processing Systems - Volume 1. (Curran Associates Inc.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are available from the corresponding author on reasonable request. Data from TCGA could be downloaded from Xena browser (https://xenabrowser.net/). Data for MET500 could be retrieved from database of Genotypes and Phenotypes (accession number: phs000673.v2.p1). Processed cancer type-specific MMDs are available from ArrayExpress under accession numbers (MTAB-6690, E-MTAB-6691, E-MTAB-6692, E-MTAB-6693, E-MTAB-6694, MTAB-6695, E-MTAB-6696, E-MTAB-6697, E-MTAB-6698, E-MTAB-6699, and E-MTAB-6703). Proteomic data for colorectal cancer are available from the NCI Clinical Proteomic Tumor Analysis Consortium (https://cptac-data-portal.georgetown.edu/cptac/s/S016).
The scripts used for the bioinformatics reanalysis are available at https://github.com/YangHongDai/Radiosensitivity_index.