Abstract
Soluble urokinase-type plasminogen activator receptor (suPAR) is a chronic inflammation marker associated with the development of a range of diseases, including cancer and cardiovascular disease. The genetics of suPAR remain unexplored but may shed light on the biology of the marker and its connection to outcomes. We report a heritability estimate of 60% for the variation in suPAR and performed a genome-wide association meta-analysis on suPAR levels measured in Iceland (N = 35,559) and in Denmark (N = 12,177). We identified 13 independently genome-wide significant sequence variants associated with suPAR across 11 distinct loci. Associated variants were found in and around genes encoding uPAR (PLAUR), its ligand uPA (PLAU), the kidney-disease-associated gene PLA2R1 as well as genes with relations to glycosylation, glycoprotein biosynthesis, and the immune response. These findings provide new insight into the causes of variation in suPAR plasma levels, which may clarify suPAR’s potential role in associated diseases, as well as the underlying mechanisms that give suPAR its prognostic value as a unique marker of chronic inflammation.
Subject terms: Genome-wide association studies, Quantitative trait, Genetic markers, Prognostic markers, Inflammatory diseases
Dowsett and colleagues used a genome-wide association approach to investigate the genetic influence on soluble urokinase-type plasminogen activator receptor presence in the plasma of humans. Their findings indicate a 60% heritability factor in British twins, and using a wide sample of Northern European genome samples they identify eleven genetic loci associated with an increase or decrease of this chronic inflammation marker.
Introduction
The plasma protein soluble urokinase-type plasminogen activator receptor (suPAR) is a non-specific biomarker for chronic inflammation (also termed low-grade inflammation) and was recently identified as a key molecule of senescent cells1. It structurally consists of three domains (DI–DIII) and is the soluble form of the membrane-bound receptor uPAR, which is bound to a variety of immune cells, smooth muscle cells, and podocytes by a glycosyl-phosphatidylinositol (GPI) anchor2,3. uPAR is a receptor for urokinase-type plasminogen activator (uPA), an enzyme known for activating plasminogen into plasmin4. Plasmin’s proteolysis of extracellular matrices (ECMs) is essential for fibrin blood clot degradation and clearance4. Other than participating in the plasminogen activator system, uPAR plays a role in various cellular processes including cell adhesion, migration, proliferation, angiogenesis, and chemotaxis2,5. uPAR can be cleaved into its soluble form, suPAR, by several proteases, including uPA, GPI-specific phospholipase D, matrix metalloproteinases (MMPs), cathepsin G, neutrophil elastase, and plasmin6.
In the general population, an elevated plasma suPAR level has been found to predict various health conditions, including incident cancer, cardiovascular disease, diabetes, depression, as well as early mortality7–9. Elevated suPAR levels are also associated with pulmonary diseases including asthma and chronic obstructive pulmonary disease (COPD)10–13. It is also known that increased suPAR levels have strong associations with chronic kidney disease (CKD) across populations14–17, and have been able to independently predict declining eGFR (estimated glomerular filtration rate) and incident CKD18. Mouse models have indicated that suPAR may not only be associated with acute kidney injury, but may be causative in the development of this19. In acute medically ill patients, increased suPAR is associated with readmissions and mortality, independent of clinical presentation, and suPAR is used in clinical routine in some European emergency departments for patient risk assessment20.
Lifestyle factors associated with suPAR have been extensively studied. In particular, smoking has been found to be strongly associated with higher suPAR levels, and suPAR can be lowered by smoking cessation21. Unhealthy diet, inactive lifestyle, and obesity have substantial impacts on suPAR levels in the general population9,22,23. In addition, longitudinal research shows that multiple childhood risk factors (including exposure to adverse experiences, low IQ, and poor self-control) are associated with elevated suPAR in adulthood24.
The genetics of suPAR remain unexplored but may shed light on the biology of the marker and its connection to outcomes. A recent genome-wide association analysis (GWAS) meta-analysis of the well-known inflammatory marker C-reactive protein (CRP) identified 58 associated genetic loci and consequently provided new insight into the genetic etiology of chronic inflammation25. However, CRP and suPAR reflect different aspects of chronic inflammation despite both being used as inflammatory biomarkers26. In addition, unlike CRP, suPAR is a stable biomarker as circadian changes in plasma suPAR are minimal27–29, and suPAR measurements in individuals have been shown to be correlated across five and seven years23,30. We aimed to investigate whether suPAR plasma levels are under the genetic influence and if so, identify associated genetic variants that may facilitate our understanding of suPAR’s biology and its links to associated diseases. Moreover, gaining new insight into suPAR through genetics may potentially improve the marker’s current prognostic capabilities.
Therefore, we performed a heritability analysis in a sample of British twins to estimate the genetic contribution to suPAR levels for the first time. We then performed a GWAS on suPAR levels in a general Icelandic population cohort and in a population of healthy Danish blood donors and combined these in a meta-analysis to identify genetic variants that affect this chronic inflammation marker’s plasma levels. Significant findings were followed-up in two independent cohorts, a sample from Great Britain and another from New Zealand. Furthermore, a pathway-based analysis as well as phenome-wide association studies (pheWASs) were performed to examine the suPAR-associated variants and their predicted genes further.
Results
Heritability of suPAR
We tested if variation in suPAR levels at age 18 years was genetically influenced in the Environmental Risk (E-Risk) Longitudinal Twin Study sample (Great Britain). Within-pair correlations for suPAR levels were r = 0.69 (95% CI: 0.65–0.73) for MZ twin pairs, and r = 0.39 (95% CI: 0.32–0.46) for DZ twin pairs. Using a univariate twin model, we found that additive genetic effects accounted for 60% (95% CI: 38–82%) of the variation in suPAR levels, while shared environmental influences accounted for 10% (95% CI: 0–31%) of the variance and nonshared environmental influences accounted for 30% (95% CI: 26–35%) of the variance in suPAR levels. We additionally calculated the SNP-based heritability based on the general Icelandic population cohort. The SNP heritability estimate was calculated to be 12.5% (SD: 4.8%).
GWAS meta-analysis
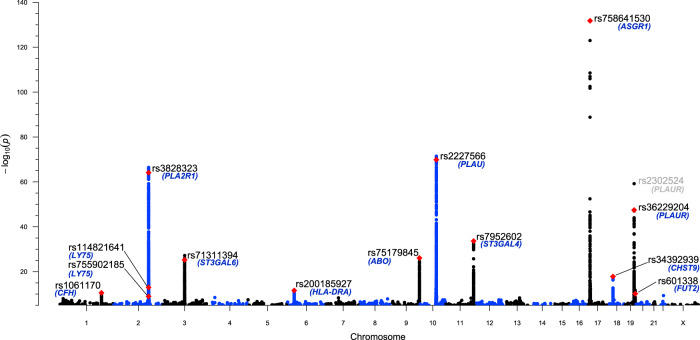
We performed GWASs on plasma suPAR levels in the general Icelandic population cohort (N = 35,559) as well as in the Danish Blood Donor Study (DBDS) (N = 12,177). We performed a meta-analysis of the two GWASs (N = 47,736) and employed a weighted Bonferroni adjustment to determine statistical significance as previously described31. The P-value significance thresholds were 2.0 × 10−7 for high-impact variants (including stop-gained, frameshift, splice-acceptor, or splice-donor variants, N = 11,723), 4.0 × 10−8 for ‘moderate-impact’ variants (including missense, splice-region variants, and in-frame indels, N = 202,336), 3.7 × 10−9 for ‘low-impact’ variants (including upstream and downstream variants, N = 2,896,354), and 6.1 × 10−10 for the ‘lowest-impact’ variants (including intron and intergenic variants, N = 37,239,641). Our GWAS meta-analysis identified 13 independent genome-wide significant genetic variants associated with suPAR across 11 distinct loci in the genome (Fig. 1, Table 1, Supplementary Data 1). The variants were tested for heterogeneity between the two cohorts. Only one variant, the rs71311394 intron variant in ST3GAL6, shows evidence of heterogeneity at P < 0.05. However, the direction of effects for rs71311394 are consistent between the two cohorts and the association with suPAR levels is significant in each cohort (EffectIce = 0.11, PIce = 5.3 × 10−12; EffectDK = 0.21, PDK = 2.1 × 10−18). Two of the 13 genetic variants (rs114821641 and rs755902185 located in the PLA2R1/LY75 locus) were identified via conditional analysis using the Icelandic data exclusively, where linkage disequilibrium (LD) data is available for the same population.
Fig. 1. suPAR GWAS meta-analysis Manhattan plot (N = 47,736), showing the 11 genome-wide significant loci and the 13 independently significant variants associated with suPAR.
The negative log10 transformed P values for variants are plotted by chromosomal location. Y axis begins at P = 1 × 10−5. Red points signify the 13 independently significant variants associated with suPAR, with variant IDs annotated in black and the corresponding gene annotated in blue.
Table 1.
Summary statistics for the 13 independently genome-wide significant variants from the meta-analysis (N = 47,736).
| Variant ID | Chr | Position (Hg38) | Effect allele | Other allele | Effect-allele frequency | Gene | Variant type | Combined effect | 95% CI | Comb. P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1061170 | 1 | 196690107 | C | T | 0.39 | CFH | Missense | 0.048 | 0.034, 0.062 | 2.79 × 10−11 |
| rs3828323 | 2 | 159951564 | C | T | 0.48 | PLA2R1 | Missense | 0.118 | 0.104, 0.132 | 7.50 × 10−65 |
| rs114821641 | 2 | 159858447 | T | C | 0.003 | LY75 | Stop gained | 0.400 | 0.294, 0.506 | 1.08 × 10−13 |
| rs755902185 | 2 | 159864896 | A | AC | 0.0004 | LY75 | Frameshift | 0.445 | 0.302, 0.588 | 1.05 × 10−9 |
| rs71311394 | 3 | 98793766 | G | A | 0.06 | ST3GAL6 | 3 Prime UTR | 0.137 | 0.112, 0.162 | 5.31 × 10−26 |
| rs200185927 | 6 | 32449458 | A | AAAGAAGAAAG | 0.25 | HLA-DRA | Downstream | 0.060 | 0.043, 0.077 | 2.58 × 10−12 |
| rs75179845 | 9 | 133257567 | C | T | 0.10 | ABO | Intron | 0.148 | 0.121, 0.175 | 8.37 × 10−27 |
| rs2227566 | 10 | 73913973 | C | T | 0.46 | PLAU | Splice region | −0.124 | −0.138, −0.110 | 1.36 × 10−70 |
| rs7952602 | 11 | 126363774 | C | G | 0.14 | ST3GAL4 | Intron | −0.131 | −0.152, −0.110 | 2.33 × 10−34 |
| rs758641530 | 17 | 7176936 | C | CCCCCAGCCCCAG | 0.004 | ASGR1 | Intron | 1.089 | 1.002, 1.176 | 1.66 × 10−132 |
| rs34392939 | 18 | 27113190 | GAAA | GAA | 0.30 | CHST9 | Intron | −0.065 | −0.080, −0.050 | 1.58 × 10−18 |
| rs36229204 | 19 | 43671830 | T | C | 0.03 | PLAUR | Upstream | −0.270 | −0.306, −0.234 | 3.55 × 10−48 |
| rs601338 | 19 | 48703417 | G | A | 0.55 | FUT2 | Stop gained | −0.046 | −0.060, −0.032 | 7.21 × 10−11 |
Effect estimate in units of standard deviation per copy increment in the effect allele.
Allele frequencies reported are based on European populations (Supplementary Data 1 reports the cohorts-specific effect allele frequencies).
Table is ordered by genomic locus. Full table available in Supplementary Data 1.
Comparison of genetic variants’ effects for suPAR unadjusted vs adjusted for smoking
As suPAR levels have strong associations with smoking, we investigated whether smoking status would affect the outcome of the suPAR GWAS results. Using the Icelandic cohort, we performed two GWASs; the 30,469 individuals with available information on smoking status, unadjusted for smoking; and the same 30,469 individuals, adjusted for smoking. Smokers had higher suPAR levels than non-smokers (Effect = 0.12 SD, P = 2.2 × 10−23 from t-test). A test of difference in the GWAS results between the two above-mentioned Icelandic GWASs revealed no difference when adjusting for smoking, with heterogeneity p-values ranging from 0.94 to 1.00 (Supplementary Data 2).
Validation of variants from independent cohorts
The lead genetic variants for each suPAR-associated locus were examined in two independent validation cohorts (Table 2 and Supplementary Data 3). We used a sample consisting of 837 individuals of white European-descent non-Maori descent from The Dunedin Multidisciplinary Health and Development Study (Dunedin) cohort, of which eight of the 13 variants were available for replication. A sample of 1444 E-Risk members of white European-descent was also used as a validation cohort, of which six of the 13 variants were available. P-values < 0.05 were considered statistically significant in the validation phase. In the Dunedin cohort, five out of the eight available variants were confirmed and all eight variants had effect estimates in the same direction. In the E-Risk cohort, three out of the six available variants were confirmed and five variants had effect estimates in the same direction.
Table 2.
Results from the validation cohorts of the suPAR-associated variants.
| suPAR GWAS Meta-Analysis Summary Table | Dunedin (New Zealand) | E-Risk (Great Britain) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant ID | Chr | Position (Hg38) | EA | EAF | Gene | Comb. effect | Comb. P-value | ||||||||||
| N | EA | EAF | Effect | p value | N | EA | EAF | Effect | p value | ||||||||
| rs1061170 | 1 | 196690107 | C | 0.39 | CFH | 0.048 | 2.79E−11 | 829 | C | 0.37 | 0.092 | 0.050 | |||||
| rs3828323 | 2 | 159951564 | C | 0.48 | PLA2R1 | 0.118 | 7.50E−65 | 829 | C | 0.49 | 0.032 | 0.481 | 1315 | C | 0.48 | 0.112 | 0.004 |
| rs114821641 | 2 | 159858447 | T | 0.003 | LY75 | 0.400 | 1.08E−13 | ||||||||||
| rs755902185 | 2 | 159864896 | A | 0.0004 | LY75 | 0.445 | 1.05E−09 | ||||||||||
| rs71311394 | 3 | 98793766 | G | 0.06 | ST3GAL6 | 0.137 | 5.31E−26 | 827 | G | 0.08 | 0.253 | 0.003 | 1308 | G | 0.07 | −0.134 | 0.038 |
| rs200185927 | 6 | 32449458 | A | 0.25 | HLA-DRA | 0.060 | 2.58E−12 | ||||||||||
| rs75179845 | 9 | 133257567 | C | 0.1 | ABO | 0.148 | 8.37E−27 | 829 | C | 0.07 | 0.178 | 0.039 | 1413 | C | 0.07 | 0.083 | 0.305 |
| rs2227566 | 10 | 73913973 | C | 0.46 | PLAU | −0.124 | 1.36E−70 | 829 | C | 0.46 | −0.155 | 0.001 | |||||
| rs7952602 | 11 | 126363774 | C | 0.14 | ST3GAL4 | −0.131 | 2.33E−34 | 796 | C | 0.13 | −0.018 | 0.800 | 1273 | C | 0.12 | −0.041 | 0.507 |
| rs754165241 | 17 | 7176936 | C | 0.004 | ASGR1 | 1.089 | 1.66E−132 | ||||||||||
| rs34392939 | 18 | 27113190 | GAAA | 0.3 | CHST9 | −0.065 | 1.58E−18 | ||||||||||
| rs36229204 | 19 | 43671830 | T | 0.03 | PLAUR | −0.270 | 3.55E−48 | 829 | T | 0.02 | −0.299 | 0.035 | 1415 | T | 0.03 | −0.248 | 0.008 |
| rs601338 | 19 | 48703417 | G | 0.55 | FUT2 | −0.046 | 7.21E−11 | 829 | G | 0.49 | −0.001 | 0.978 | 1417 | G | 0.52 | −0.086 | 0.026 |
Results in bold signify effect estimates in the same direction and/or significant P-values.
N number of samples, EA effect allele, EAF effect allele frequency.
Summary of suPAR-associated loci
The following section lists the eleven loci in further detail, describing the variants’ predicted genes based on their position within a given gene or the closest gene, associations from previous studies, and selected annotated gene ontology (GO) biological processes for each gene.
Chr1.q31.3
The missense variant rs1061170-C on chromosome 1 in the gene CFH is associated with an increase in suPAR (effect estimate = 0.048 units of a standard deviation per copy increment in the effect allele; P = 2.79 × 10−11, effect allele frequency (EAF) = 0.39). The variant causes a missense mutation (His > Tyr) in the gene CFH, which encodes the glycoprotein Complement Factor H; a protein that regulates complement activation in an immune response. The variant is known to be associated with age-related macular degeneration32,33. GO terms biological processes associated with the gene include complement activation, regulation of complement-dependent cytotoxicity, and viral process.
Chr2.q24.2
Three independently significant variants at a locus on chromosome 2 were found, resulting in two candidate genes in this locus. The missense variant rs3828323-C in the gene PLA2R1 (phospholipase A2 receptor 1) is associated with an increase in suPAR (effect = 0.118; P = 7.5 × 10−65, EAF = 0.48). Several variants in PLA2R1 have previously been associated to membranous nephropathy, and serum anti-PLA2R1 antibody associates with loss of kidney function34,35. GO terms biological processes associated with the gene include cytokine production, negative regulation of phospholipase A2 activity, and receptor-mediated endocytosis among others.
The remaining two significant variants in this locus are less common and are located in the nearby gene LY75 encoding the protein lymphocyte antigen 75. rs114821641-T causes a stop-gain mutation and an increase in suPAR (effect = 0.400; P = 1.08 × 10−13, EAF = 0.003), and rs755902185 is a deletion which causes a frameshift mutation in LY75 and an increase in suPAR (effect = 0.445; P = 1.05 × 10−9, EAF = 0.0004). Variants in LY75 have previously been associated with Inflammatory Bowel Syndrome and Crohn’s disease36,37. GO terms biological processes associated with the gene include endocytosis, immune response, and inflammatory response.
Chr3.q12.1
The 3 prime untranslated region (UTR) variant rs71311394-G on chromosome 3 in the gene ST3GAL6 (ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 6) is associated with an increase in suPAR (effect = 0.137; P = 5.31 × 10−26). GO terms biological processes associated with the gene include glycolipid biosynthetic processes, cellular response to interleukin-6, and glycosylation among others38.
Chr6.p21.32
The deletion variant rs200185927 on chromosome 6 downstream from HLA-DRA is associated with an increase in suPAR (effect = 0.06; P = 2.58 × 10−12, EAF = 0.25). GO terms biological processes associated with this gene include antigen processing and presentation of exogenous peptide antigen via major histocompatibility complex class II among other immune response processes.
Chr9.q34.2
Intron variant rs75179845-C on chromosome 9 in the gene ABO is associated with an increase in suPAR (effect = 0.148; P = 8.37 × 10−27, EAF = 0.10). The gene determines the blood group of the individual, and GO terms biological processes associated with the gene include lipid and protein glycosylation38.
Chr10.q22.2
Splice-site variant rs2227566-C on chromosome 10 in the gene PLAU is associated with a decrease in suPAR (effect = −0.124; P = 1.36 × 10−70, EAF = 0.46). The variant is located in the splice region of PLAU, the gene encoding the protein uPA, which can bind to uPAR and consequently release the receptor into the bloodstream as suPAR. The protein uPA cleaves plasminogen to form the active form of plasmin and GO terms biological processes associated with this gene include blood coagulation, chemotaxis, fibrinolysis, regulation of smooth muscle cell-matrix adhesion, and migration among others. The lead variant rs2227566-C has been previously associated with asthma and airway hyperresponsiveness39.
Chr11.q24.2
Intron variant rs7952602-C on chromosome 11 in the gene ST3GAL4 (ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 4) is associated with a decrease in suPAR (effect = −0.131; P = 2.33 × 10−34, EAF = 0.14). GO terms biological processes of this gene include glycolipid biosynthetic processes and glycosylation among others38.
Chr17.p13.1
Deletion variant rs758641530 on chromosome 17 in the gene ASGR1 (asialoglycoprotein receptor) is associated with an increase in suPAR (effect = 1.089; P = 1.66 × 10−132, EAF = 0.004). ASGR1 is a lectin that mediates the endocytosis of plasma glycoproteins, and a previous study found that a deletion in this gene is associated with reduced levels of non-HDL cholesterol and a reduced risk of coronary artery disease40.
Chr18.q11.2
Intron variant rs34392939 on chromosome 18 in the gene CHST9 (Carbohydrate Sulfotransferase 9) is associated with a decrease in suPAR (effect = −0.065; P = 1.58 × 10−18, EAF = 0.30). A GO term biological process associated with this gene is proteoglycan biosynthetic process38, and copy number variations of CHST9 associate with hematologic malignancies41.
Chr19.q13.31
Variant rs36229204-T on chromosome 19 in the upstream from PLAUR (encodes uPAR) is associated with a decrease in suPAR (effect = −0.270; P = 3.55 × 10−48, EAF = 0.03). The variant is in high LD with rs4251805 (R2 = 1; D′ = 1) which exists in PLAUR’s 5′ UTR. GO terms biological processes associated with the gene include blood coagulation, chemotaxis, fibrinolysis, and regulation of proteolysis among others.
In addition, we identified PLAUR missense variant rs2302524-C to be independently significant in both the Danish and Icelandic GWASs, but with opposite effect directions (EffectICE = −0.24; PICE = 3.29 × 10−113, EAFICE = 0.18; EffectDK = 0.09, PDK = 4.68 × 10−8, EAFDK = 0.17). Due to the inconsistent direction, it has not been included as a valid 14th suPAR-associated signal and hence is not shown in Table 1. Validation cohorts from New Zealand and Great Britain indicate that rs2302524 is significantly associated with increased suPAR levels as found in the Danish cohort (Supplementary Data 4). The variant has previously been associated with worse baseline lung function (FEV1) in smokers as well as an increased risk of asthma and worse FEV1 in individuals with asthma42,43.
Chr19.q13.33
Stop-gain mutation variant rs601338-G on chromosome 19 in the gene FUT2 (Fucosyltransferase 2) is associated with a decrease in suPAR (effect = −0.046; P = 7.21 × 10−11, EAFICE = 0.39; EAFDK = 0.55). GO terms biological processes associated with the gene include protein glycosylation, L-fucose catabolic process, and regulation of cell adhesion among others.
In summary, we identified 13 genome-wide significant suPAR-associated variants and based on literature searches and GO term annotations, the variants are found in and around 12 genes encoding uPAR/suPAR (PLAUR) and its ligand uPA (PLAU), genes with relations to glycoprotein biosynthesis and glycosylation (ASGR1, ST3GAL4, ST3GAL6, ABO, CHST9, FUT2), genes involved in immune response (LY75, HLA-DRA, CFH), and PLA2R1 (of which variants have been previously associated with membranous nephropathy).
Overrepresentation of biological processes in the suPAR-GWAS-associated set of genes
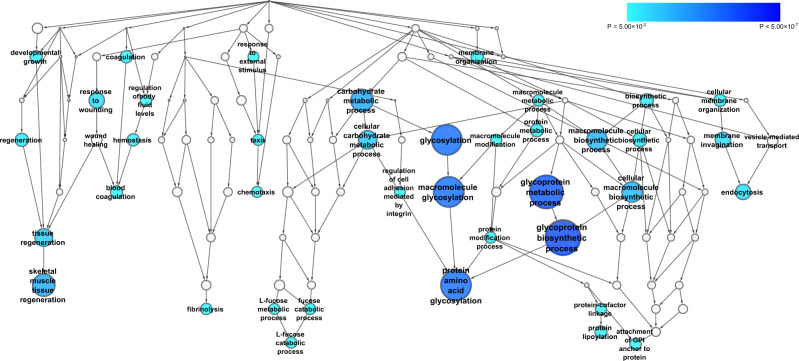
We used the Biological Network Gene Ontology (BiNGO) bioinformatics tool44 to quantitatively assess whether there are GO terms (biological processes) that are statistically overrepresented in our set of 12 suPAR-associated genes. Analysis of the 12 suPAR-associated genes using the BiNGO tool revealed 39 GO term biological processes significantly overrepresented after multiple test corrections. Due to the hierarchical nature of the GO term gene sets, the majority of the 39 biological processes are overlapping and can therefore be grouped into approximately nine biological process branches (Fig. 2). Significant biological processes that are overrepresented include glycoprotein biosynthetic process (P = 8.85 × 10−8), protein amino acid glycosylation (P = 2.14 × 10−6), skeletal muscle tissue regeneration (P = 3.53 × 10−5), endocytosis (P = 7.31 × 10−4), response to wounding (P = 7.86 × 10−4), fibrinolysis (P = 5.86 × 10−3), attachment of GPI anchor to protein (P = 5.86 × 10−3), L-fucose catabolic process (P = 7.52 × 10−3) and chemotaxis (P = 8.47 × 10−3) (Supplementary Data 5).
Fig. 2. Results of the BiNGO pathway over-representation analysis for GO biological processes, using the 12 genes found from the suPAR GWAS meta-analysis.
Significant overrepresented pathway gene sets (P < 0.05 after surviving Benjamini & Hochberg False Discovery Rate correction) are shaded blue with size and darker shades signifying lower P-values as depicted in the legend.
suPAR polygenic risk scores (PRSs) and pheWASs
To investigate whether the combined effect of suPAR-associated genetic variations were associated with specific phenotypes, we performed pheWASs using suPAR PRSs as the exposure. We calculated PRSs for the individuals in the Icelandic population cohort based on the summary statistics from the Danish cohort’s suPAR GWAS. The PRSs explained 0.94% of the suPAR variance in Icelandic individuals.
A total of 14,493 case/control phenotypes and 28,389 quantitative phenotypes in the Icelandic population cohort were tested. After Bonferroni multiple testing correction (P < 7.86 × 10−7), in the case-control pheWAS, we found that suPAR PRSs were associated with type 1 diabetes (effect = 2.212; P = 5.11 × 10−18), autoimmune diseases (effect = 0.354; P = 4.30 × 10−10) and obesity (effect = 0.634; P = 1.04 × 10−7). In the quantitative phenotype pheWAS, we found that suPAR PRSs were associated with increased levels of plasma PLA2R1 (effect = 0.649; P = 1.99 × 10−105), increased levels of B12 (effect = 0.125; P = 7.30 × 10−13), decreased high-density lipoprotein cholesterol (effect = −0.107; P = 3.96 × 10−10), increased fasting plasma glucose (effect = 0.089; P = 6.04 × 10−9), increased alkaline phosphatase (effect = 0.100; P = 1.82 × 10−8), increased potassium (effect = 0.085; P = 2.61 × 10−9), increased BMI (effect = 0.109; P = 1.03 × 10−7) and increased eosinophils (effect = 0.056; P = 7.01 × 10−8) (Supplementary Data 6–7).
We additionally calculated PRSs for the individuals in the UK Biobank based on the summary statistics from the suPAR GWAS meta-analysis (Icelandic cohort + Danish cohort). 15,120 case/control phenotypes and 5,609 quantitative phenotypes were available in the UK Biobank pheWAS. No significant case/control phenotypes associated with suPAR after Bonferroni multiple test correction, but for the quantitative phenotypes, increased mean corpuscular hemoglobin (effect = 0.006; P = 2.40 × 10−11) and increased mean corpuscular volume (effect = 0.005; P = 3.80 × 10−9) were significant (Supplementary Data 8). Single-variant pheWASs were also performed for each of the 13 independently significant variants separately in both the Icelandic and UK Biobank dataset using the same methods and the significant results are available in Supplementary Data 9–12.
Mendelian randomization analyses for suPAR vs. the identified pheWAS findings
We performed Mendelian randomization analyses for suPAR and the identified pheWAS findings using phenotypes available in the Icelandic population cohort. No significant results were found and the removal of outliers detected using MR-PRESSO did not improve the estimates significantly (Supplementary Data 13).
Cis-expression quantitative trait loci (eQTL) analysis of the suPAR-associated variants
We tested if our variants were in high LD (r2 > 0.8) with one more top cis-eQTL based on various tissues and 17 sources including GTEx and Icelandic data. Results are reported in Supplementary Data 14 and sources listed in Supplementary Data 15.
Genetic Correlation between suPAR and CRP
We performed a genetic correlation analysis between suPAR and CRP. The genetic correlation between suPAR and CRP was calculated to be 0.2351 (SD = 0.0533, P = 1.03 × 10−5), using suPAR measurements from the Icelandic/Danish meta-analysis and CRP measurements from UK Biobank. We have additionally checked for CRP variants from the GWAS-catalog for our suPAR-associated variants and their LD-classes (all variants with r2 > 0.80). Only the FUT2 variant (rs601338) was found to be an overlapping variant, where the same variant was reported in Han et al.45. We have additionally searched all the genes that we report to see if they have been reported with CRP, of which the only overlap we find in addition to FUT2 is ABO, where there is a variant at chr9:133266942 (rs643434) reported to associate with CRP in the CRP GWAS meta-analysis by Ligthart et al.25. This variant has r2 = 0.17 with the ABO variant found in our study.
Discussion
Eleven genome-wide significant loci driven by 13 variants were associated with suPAR in our GWAS meta-analysis based on 47,736 individuals. These findings, including our heritability estimate of 60%, provide an updated explanation for the inter-individual variation in suPAR plasma levels.
We identified suPAR-associated variants localized in and near the genes encoding uPAR (PLAUR) and the ligand uPA (PLAU), although how they affect suPAR plasma levels is uncertain. The lead variant rs36229204 at the PLAUR locus is in complete LD with rs4251805 (r2 = 1; D′ = 1) which is located in PLAUR’s 5′ UTR, a region known for its importance in the regulation of a gene’s translation. Similarly, the lead variant rs2227566 at the PLAU locus is located in the splice region of the gene PLAU, but it is also in LD (r2 = 0.44; D′ = 1) with missense variant rs2227564-T (meta-analysis effect = −0.131; P = 1.57 × 10−62, EAF = 0.26) which is the lead variant in the Danish cohort’s suPAR GWAS. This PLAU variant has previously been associated with an increased risk of colorectal cancer, asthma, oral tongue squamous cell carcinoma, and poor coronary collateral circulation in coronary artery disease patients, as well as Alzheimer’s Disease39,46–49. The variant causes a missense mutation in the kringle domain of uPA—the important domain necessary for protein–protein interactions including integrins50. The binding of uPA and suPAR is stabilized by the kringle domain51,52, suggesting that this missense mutation may produce a conformational change that reduces uPA’s ability to bind and/or to cleave uPAR, thereby reducing suPAR levels as seen in the results from this study.
Our results may also provide some insight into suPAR’s possible role in chronic and acute kidney disease18,19. Significant variants were found in and near PLA2R1, where the top variant rs3828323 increases suPAR levels. As a missense variant, the resulting change from non-polar to polar residue may alter the binding of PLA2R1’s ligand PLA2 (secretory phospholipase A2). The residue change is located in the extracellular part of the receptor between two C-type lectin domains, of which one is part of the receptor-binding region for its ligand PLA2. Interestingly, several variants in PLA2R1 have previously been associated to membranous nephropathy, and serum anti-PLA2R1 antibody associates with loss of kidney function34,35. Two studies on membranous nephropathy have identified associated genetic variants, of which one found an association with two variants: rs3828323 and rs3577198234, and the other study identified had rs17831251 as the lead associated variant53. The two variants that are not reported in our study, i.e., rs35771982 and rs17831251 associate with suPAR in our meta-analysis but these associations do not hold up after adjusting for our lead variant (r2 = 0.32 and r2 = 0.19 respectively). This would indicate that the signal reported in these previous membranous nephropathy genetic studies at the PLA2R1 locus is the same as for suPAR in our study. It is well-reported that increased suPAR has strong associations with CKD14–17, though it is unknown how suPAR and PLA2R1 are related to each other in respect to kidney disease. It is possible that when anti-PLA2R1 attaches to podocyte-bound PLA2R1 due to the altered structure of the receptor caused by the genetic variant, it forms immune complexes which consequently activates the immune system. As a result, the inflammatory response would increase suPAR levels and allow suPAR to activate beta3 integrins on the podocytes as shown in the previous studies3,54,55, and this activation would produce the podocyte conformational change that causes membranous nephropathy. It may therefore be hypothesized that it is a variant in PLA2R1 that gives suPAR the appearance of a causal role in the development of membranous nephropathy and other kidney diseases previously shown to have strong associations with suPAR levels. Though rs3828323 in PLA2R1 was the lead variant at this locus in our meta-analysis, it should be noted that two additional independent but rare variants were identified in this locus, located in the gene LY75. The PLA2R1 variant rs3828323 exists in an LD-class which overlaps into both genes. LY75 and PLA2R1 may therefore both be considered candidate genes in this locus. However, from the PRS-based quantitative pheWAS, we found that a higher suPAR PRS was strongly associated with increased PLA2R1 plasma levels (effect = 0.649; P = 1.99 × 10−105), further supporting PLA2R1 as a gene of interest for future studies.
Another noteworthy observation from our study, supported by the results from the BiNGO pathway-based analysis, is that six of the suPAR-associated loci have significant variants in genes encoding proteins that are in some form involved in processes related to glycoprotein biosynthesis or protein glycosylation. These genes include ST3GAL4, ST3GAL6, ABO, CHST9, ASGR1, and FUT2. From the Kyoto Encyclopedia of Genes and Genomes (KEGG) database56 it is also evident that the proteins ST3GAL4, ST3GAL6, ABO and FUT2 participate in the glycosphingolipid biosynthesis (lacto and neolacto series) pathway. Glycosphingolipids are often localized in glycosphingolipid-enriched microdomains called lipid rafts where they have a role in mediating cell–cell interactions and regulating proteins in the same plasma membrane57. Glycosphingolipids may therefore have a regulatory function on suPAR as it has previously been found that uPA-induced uPAR cleavage is strongly accelerated when uPAR is localized in lipid rafts58. ASGR1 is a lectin that mediates the endocytosis of plasma glycoproteins which may also impact uPAR’s role on the plasma membrane. It is well known that uPAR is highly glycosylated59,60 but the function of the glycosylation is not completely understood. However, glycosylation has been found to increase uPAR’s affinity to uPA and enhance suPAR’s solubility60–62. Though the results from our study suggest that uPAR glycosylation may have a molecular function that impacts suPAR plasma levels, the effects of glycosylation and deglycosylation on suPAR detection for the two methods used in this study are not known, and this may affect the results. Nevertheless, the results support previous studies that glycosylation increases uPAR-uPA affinity and suPAR solubility and therefore the genes identified in our study may be considered in future candidate gene studies to investigate their role in affecting the amount of suPAR present in plasma.
Two different suPAR detection methods were used in our study; a proteomics-based assay in the Icelandic population and ELISA in the Danish population. Although a direct comparison between the two different assays using the same samples was not possible, a comparison of the results of the GWAS findings between the Danish and Icelandic population cohorts was performed, of which the results showed high concordance. Only one variant, the rs71311394 variant in ST3GAL6, shows evidence of heterogeneity (P = 0.003). However, the direction of effects for rs71311394 are consistent between the two populations (EffectIce = 0.11 vs. EffectDK = 0.21) and the association with suPAR levels is genome-wide significant in both populations (PIce = 5.3 × 10−12 vs. PDK = 2.1 × 10−18). Given this high degree of similarity in the GWAS findings we believe that a direct comparison between the assays would not add any further genetic insights into our findings.
As our findings indicate that individuals can be genetically predisposed to higher or lower suPAR levels, it may have implications for future precision or personalized medicine practices by potentially improving suPAR’s current prognostic capabilities. It is known that increased suPAR is associated with readmissions and mortality in acute medically ill patients, and suPAR is used in patient risk assessments in some Danish hospitals20. Genetic profiling of patients may contribute to optimized patient treatment by identifying patients genetically predisposed to higher or lower levels of chronic inflammation, thereby enabling risk assessment of a patient at an earlier stage before they reach an acute medically ill stage of their disease progress. However, it must be noted that the SNP-based heritability based on the Icelandic cohort was calculated to be 12.5% (SD: 4.9%). As our univariate twin model estimated that additive genetic effects account for 60% of the variation in suPAR levels, we are unable to account for much of the heritability of suPAR, a situation frequently reported in GWASs of other phenotypes63. Larger studies will likely enable the discovery of more variants that explain some of this missing heritability, as well as future studies focusing on other forms of genetic variation such as copy number variants.
In conclusion, we provide evidence that suPAR plasma levels are under the substantial genetic influence and that 13 independently significant genetic variants at 11 distinct loci influence suPAR levels in Icelandic and Danish individuals. Our data further support genetic links between suPAR-measured chronic inflammation and phenotypes such as diabetes and obesity. Our results indicate that suPAR’s strong associations with chronic kidney disease may be related to a suPAR-associated missense variant in the gene PLA2R1, and that variants in many genes related to glycosylation and glycoprotein biosynthesis pathways affect suPAR levels. Genes identified in this study may be examined as candidate genes in future functional studies to help clarify suPAR’s potential role in the causation of associated diseases, as well as the underlying mechanisms that give suPAR its prognostic value as a unique marker of chronic inflammation.
Methods
Participants
The meta-analysis is based on data from two Northern European population cohorts: a Danish cohort consisting of healthy blood donors, and a general Icelandic population cohort.
Danish cohort
The Danish cohort is based on participants originating from the DBDS, a nationwide research platform utilizing the existing infrastructure in the Danish blood banks64. Participants must be generally healthy and not on medication to be eligible as donors. Upon enrollment, participants gave informed consent, whole blood, plasma, and answered a comprehensive questionnaire. So far, ~110,000 adult DBDS participants have been enrolled with informed consent, whole blood, plasma samples, questionnaire data, and genome-wide genotype data gathered from each65. suPAR was measured in 14,367 consecutive DBDS participants from 1 March 2010 until 10 December 2010, of which 12,177 (84.8%) participated in the GWAS after fulfilling quality control requirements. The project is approved by the Research Ethics Committees by the following three protocols: The DBDS (M-20090237), Genetics of healthy ageing (CVK-1700407), Family study on the genetics of healthy ageing (NVK-1803847). The project is approved by the Danish Data Protection Agency under the combined approval for health care research at The Capital Region of Denmark (P-2019-99).
Icelandic cohort
Plasma samples from 40,004 Icelanders were collected during 2000–2019. Fifty-two percent of the samples were collected as part of the Icelandic Cancer Project (ICP), while the remaining samples (48%) were collected as part of various genetic programs at deCODE genetics, Reykjavík, Iceland. In the ICP, all prevalent and newly diagnosed Icelandic cancer cases and their relatives were invited to participate in a comprehensive study of cancer, along with a control population, randomly selected from the National Registry. The median collection date for samples collected in conjunction with ICP was 1 July 2002, whereas the median collection date for other samples was 15 May 2015. All samples were measured using the SOMAscan platform (SomaLogic), containing 5284 aptamers providing a measurement of relative binding of the plasma sample to each of the aptamers in relative fluorescence units (RFU), corresponding to 4792 proteins, of which suPAR is included. After quality control, unique measurements for N = 35,559 individuals (88.9%) were used for GWAS. All participants who donated samples gave informed consent and the National Bioethics Committee of Iceland approved the study (VSN-15-198) which was conducted in agreement with conditions issued by the Data Protection Authority of Iceland. Personal identities of the participants’ data and biological samples were encrypted by a third-party system (Identity Protection System), approved, and monitored by the Data Protection Authority.
Two independent cohorts agreed to validate the findings from this study: the Environmental Risk Longitudinal (E-Risk) Twin Study from Great Britain, as well as The Dunedin Longitudinal Study from New Zealand, of which the former was additionally used for the twin/heritability analysis.
Environmental risk (E-Risk) longitudinal twin study
Participants were members of the E-Risk longitudinal twin study, which tracks the development of a 1994-95 birth cohort of 2,232 British children66. Briefly, the E-Risk sample was constructed in 1999–2000, when 1116 families (93% of those eligible) with same-sex 5-year-old twins participated in home-visit assessments. This sample comprised 56% monozygotic (MZ) and 44% dizygotic (DZ) twin pairs; sex was evenly distributed within zygosity (49% male). The sample represents socioeconomic conditions in Great Britain, as reflected in the families’ distribution on a neighborhood-level socioeconomic index (ACORN [A Classification of Residential Neighborhoods], developed by CACI Inc. for commercial use): 25.6% of E-Risk families live in “wealthy achiever” neighborhoods compared to 25.3% nationwide; 5.3% vs. 11.6% live in “urban prosperity” neighborhoods; 29.6% vs. 26.9% in “comfortably off” neighborhoods; 13.4% vs. 13.9% in “moderate means” neighborhoods; and 26.1% vs. 20.7% in “hard-pressed” neighborhoods. (E-Risk underrepresents “urban prosperity” neighborhoods because such households are often childless). Home visits were conducted when participants were aged 5, 7, 10, 12, and most recently, 18 years (93% participation). At age 18, each twin was interviewed by a different interviewer. Whole blood was collected from 82% (n = 1700) of the participants. Plasma was available for 1448 participants. The Joint South London and Maudsley and the Institute of Psychiatry Research Ethics Committee approved each phase of the study. Parents gave informed consent and twins gave assent between 5 and 12 years and then informed consent at age 18.
The Dunedin multidisciplinary health and development study
Participants were members of the Dunedin study, a longitudinal investigation of health and behavior in a representative birth cohort. Participants (n = 1037; 91% of eligible births; 52% male) were all individuals born between April 1972 and March 1973 in Dunedin, New Zealand (NZ), who were eligible based on residence in the province and who participated in the first assessment at age 3 years67. The cohort represented the full range of socioeconomic status (SES) in the general population of NZ’s South Island and as adults matched the NZ National Health and Nutrition Survey on key adult health indicators (e.g., body mass index, smoking, GP visits) and the NZ Census of citizens of the same age on educational attainment. The cohort is primarily white (93%), matching South Island demographics67. Assessments were carried out at birth and ages 3, 5, 7, 9, 11, 13, 15, 18, 21, 26, 32, and 38 years. At age 38 years, 95% (n = 961) of the 1007 participants still alive took part. At each assessment, each participant was brought to the research unit for interviews and examinations. Blood from participants of Maori ancestry was not transported to Duke University for cultural reasons, and plasma samples were not available for participants who did not provide blood or due to phlebotomy or defrost cycle problems. The relevant ethics committees approved each phase of the Study and written informed consent was obtained from all participants.
suPAR assessment
Plasma suPAR levels were measured in the DBDS cohort and two validation cohorts (Dunedin and E-Risk) using the CE/IVD-approved suPARnostic AUTO Flex ELISA (ViroGates A/S, Birkerød, Denmark) following the manufacturer’s instructions. The suPARnostic assay utilizes two monoclonal antibodies: a capture antibody directed towards the DIII subunit and a detection antibody against the DII subunit. Full-length suPAR (DIDIIDIII) may be cleaved into DI and DIIDIII, and the assay captures free full-length suPAR (DIDIIDIII) as well as the suPAR fragment (DIIDIII) but not the DI fragment. The DIDIIDIII full-length suPAR molecule can bind urokinase plasminogen activator (uPA) and DIDIIDIII/uPA complexes will not be detected in the suPARnostic assay68. suPAR levels were measured in 14,367 participants in DBDS, 1444 in E-Risk, and 837 in Dunedin. suPAR levels were measured at age 18 in the E-Risk Study, as previously described24 whereas suPAR levels were measured at age 38 in the Dunedin Study, as previously described69.
For the Icelandic cohort, suPAR is one of the plasma proteins measured using the SOMAscan platform as described above.
Genotyping and imputation
Genotyping and imputation of the 110,000 DBDS Genetic Cohort is described in Hansen et al.65. Briefly, DNA purification is performed from the whole blood samples and immediately stored at −20 °C. The samples were genotyped using the Global Screening Array by Illumina, which includes >650,000 variants with custom chip content optimized for comparison with the Illumina Omni Express chip. All genotype data are processed simultaneously for genotype calling, quality control, and imputation. Quality control was conducted in both populations, including using a minimum allele count of 5, the exclusion of individuals or variants with more than 10% missingness, and individuals deviating more than three standard deviations (SDs) from the population heterozygosity (correcting for individuals carrying large copy number variations, >100 Kbp). We performed imputation using a reference panel backbone consisting of (1) UK 1 KG phase 3 and HapMap reference to predict non-genotyped variants with minor allele frequency (MAF) > 1%, and (2) an in-house dataset consisting of N > 6000 Danish whole-genome sequences to improve the prediction of variations with a MAF down to around 0.01%.
The process used to whole-genome sequence the 49,708 Icelanders, as well as the subsequent imputation, has been described in previous publications70,71. Briefly, we sequenced the whole genomes of 49,708 Icelanders using Illumina technology to a mean depth of at least 10× (median 32×). SNPs and indels were identified and their genotypes called using joint calling with Graphtyper72. In total, 166,281 Icelanders were genotyped using Illumina SNP chips and their genotypes were phased using long-range phasing73. All sequenced individuals were also chip-typed and long-range phased, providing information about haplotype sharing that was subsequently used to improve genotype calls. Genotypes of the 32 million high-quality sequence variants were imputed into all chip-typed Icelanders. Using genealogic information, the sequence variants were also imputed into relatives of the chip-typed further increasing the sample size for association analysis and the power to detect associations. All the variants tested had imputation information over 0.8.
For the two validation cohorts, we used Illumina HumanOmni Express BeadChip arrays (Illumina CA, USA) to assay common single nucleotide polymorphism (SNP) variation in the genomes of participants of the E-Risk and Dunedin studies, as previously described74. The resulting database was restricted to SNPs called successfully in >98% of each cohort and in Hardy-Weinberg equilibrium (p > 0.001). Additional SNPs were imputed using the IMPUTE2 software (version 2.3.1, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html)75 and 1000 Genomes version-3 reference panel76. Imputation was conducted on autosomal SNPs appearing in dbSNP (version 140; http://www.ncbi.nlm.nih.gov/SNP/)77 that were called in >98% of each sample. Invariant SNPs were excluded. Prephasing and imputation were conducted using a 50 M base-pair sliding window. The resulting genotype database included genotyped SNPs and SNPs imputed with a 90% probability of a specific genotype among European-descent E-Risk members (n = 1999 children in 1011 families) and among the non-Maori members of the Dunedin cohort (n = 918).
Twin/heritability analysis
To test the genetic contribution to suPAR levels at age 18 in the E-Risk study, we used a univariate twin model comparing correlations between MZ and DZ twins to decompose the phenotypic variation in plasma levels of suPAR into additive genetic, shared environmental, and unique environmental components. We used Mplus Version 7.4 (Muthen & Muthen, Los Angeles, CA) for the analysis. We additionally calculated SNP heritability based on the Icelandic cohort. We estimated the narrow-sense heritability of suPAR with LD score regression78, using an LD score map calculated with high-quality markers from the Icelandic population.
GWAS statistical analysis
The suPAR measurements were each rank-based inverse normal transformed to a standard normal distribution (separately for each sex) and adjusted for age using a generalized additive model. A linear mixed model implemented by BOLT-LMM79, was used to test for association between sequence variants and suPAR levels, assuming an additive genetic model. Thirty-five million variants are tested in Iceland, while 26 million variants are tested in Denmark.
BOLT-LMM accounts for cryptic relatedness and population stratification79, and we additionally used LD score regression to account for distribution inflation in the dataset due to cryptic relatedness and population stratification78. The Danish and Icelandic datasets were combined using a fixed-effect inverse-variance weighted meta-analysis, allowing the populations to have different frequencies for alleles and genotypes but assuming them to have a common effect. Heterogeneity in effect estimates was assessed using a likelihood-ratio test. Effects are given in units of SDs. In total 40 million variants are tested either in Iceland or Denmark, of which 21 million variants are tested in both datasets. Rare variants may therefore be present only in one discovery study.
We accounted for multiple testing by means of a weighted Bonferroni correction, taking into account the higher prior probability of association of certain variant annotations while controlling the family-wise error rate (FWER) at 0.0531. The method has been described previously31 and results in stricter multiple testing correction than the commonly used threshold of 5 × 10−8 (which would not control FWER at 0.05 given that 40 million markers were tested) while being more powerful than simply correcting for 40 million tests using a fixed threshold of 0.05/40,000,000 = 1.25 × 10−9. The resulting significance thresholds were 2.0 × 10−7 for high-impact variants (including stop-gained, frameshift, splice-acceptor, or splice-donor variants, N = 11,723), 4.0 × 10−8 for ‘moderate-impact’ variants (including missense, splice-region variants, and in-frame indels, N = 202,336), 3.7 × 10−9 for ‘low-impact’ variants (including upstream and downstream variants, N = 2,896,354), and 6.1 × 10−10 for the ‘lowest-impact’ variants (including intron and intergenic variants, N = 37,239,641). To identify whether several variants in a single locus are independently associated with suPAR, we performed conditional analysis using Icelandic individual-level data, where LD data are available from the same population. This is in contrast with methods such as GCTA which use summary-level data only. The variants’ predicted genes are based on their actual position within a given gene or the closest gene (from Ensembl variant effect predictor information80).
For previous associations and functions of our suPAR-associated variants and genes, we performed manual searches using PubMed (pubmed.ncbi.nlm.nih.gov). For the variant’s associated gene, we used the UniProt Protein Knowledge Base (UniProtKB) to find the function and annotated GO biological processes listed for each gene (www.uniprot.org/uniprot)81.
For the replication phase, additive genetic association tests between suPAR levels and each of the significant variants were performed using the R package “SNPassoc”. The model suPAR ~variant + sex + PCs1-10 was employed, where variant genotypes were coded as number of risk alleles (0,1,2). For the E-Risk twin study, the analyses employed generalized estimating equation (GEE) linear regression models using the R package ‘gee’, taking into account the clustering of the twins within families.
Comparison of genetic variants’ effects for suPAR unadjusted vs adjusted for smoking
As suPAR levels have strong associations with smoking, we investigated whether smoking status would affect the outcome of the suPAR GWAS results. Smoking status was available for 30,469 of the 35,559 individuals with suPAR measurements in the Icelandic data. This includes 11,093 non-smokers and 19,376 smokers, where “smoker” was defined as “ever smoker”. To assess if the inclusion of smoking as a covariate could have an effect on the GWAS outcomes, we performed two GWASs: (1) the 30,469 individuals with available information on smoking status, unadjusted for smoking; and (2) the same 30,469 individuals, adjusted for smoking. A test of difference (heterogeneity) in the GWAS results was performed.
Biological Network Gene Ontology Analysis
To quantitatively investigate biological processes associated with each gene, we used the Biological Network Gene Ontology (BiNGO) bioinformatics tool44 to assess whether there are GO terms biological processes that are statistically overrepresented in our set of 12 suPAR-associated genes. The BiNGO tool uses GO terms from the Gene Ontology database (www.geneontology.org) and calculates the P values for overrepresented biological processes in our set of genes using the hypergeometric test. This takes into account both the total number of genes from the input dataset and the total number of genes for the specific GO term biological process. A total of 14,306 genes were available in the tool’s reference set. The Benjamini-Hochberg (false discovery rate) correction is calculated to control for multiple testing, where only significantly overrepresented GO term processes with corrected P < 0.05 were considered.
PheWAS statistical analysis
To gain further insights into the possible functional and regulatory role of our newly identified variants, phenotype-wide association analyses (pheWASs) were conducted. pheWASs using suPAR PRSs as the exposure were performed to investigate whether the combined effect of these suPAR-associated genetic variations were associated with specific phenotypes. PRSs were calculated for each individual in the Icelandic population based on the summary statistics of the DBDS suPAR GWAS, and PRSs for suPAR were calculated for each of the 500,000 UK Biobank subjects based on the meta-analysis summary statistics (Denmark + Iceland). Briefly, to generate the suPAR-PRS for the UK Biobank sample we used 630,000 informative variants across the genome and constructed locus allele-specific weightings by applying LDpred to the summary data from the subset meta-analysis GWAS82. Constructing individual weightings, we were able to calculate an aggregated score of suPAR in all included individuals. Subsequently, we assessed the impact of suPAR-PRS on 63,609 traits (binary and quantitative) using a Bonferroni significance threshold of P < 7.86 × 10−7. More specifically, a pheWAS was performed in a comprehensive phenotype dataset within the Icelandic population consisting of 14,493 case/control phenotypes and 28,389 quantitative phenotypes, as well as pheWAS in UK Biobank with 15,120 case/control phenotypes and 5609 quantitative phenotypes83. In addition, single-variant pheWASs were performed in the same datasets using the same Bonferroni significance threshold.
Mendelian randomization analyses for suPAR vs. the identified pheWAS findings
We performed Mendelian randomization analyses for suPAR and the identified pheWAS findings using phenotypes available in the Icelandic population cohort. The analyses were performed using the R Package “MendelianRandomization”84, using the inverse variance weighted (IVW) and MR-Egger methods. The MR-PRESSO global test85 was additionally used to detect possible outliers and remove them. In the instances where outliers were found, the outliers were removed and the IVW method was reutilized. The instrumental variables (IVs) used are the variants we report significant for suPAR, i.e., the lead variants.. Mendelian randomization for suPAR and 10 phenotypes were performed, including type 1 diabetes, autoimmune diseases as a general category, obesity, rheumatoid arthritis, B12, fasting glucose, alkaline phosphatase, and potassium.
Cis-eQTL analysis of the suPAR-associated variants
We tested if our variants were in high LD (r2 > 0.8) with one more top cis-eQTL based on various tissues and 17 sources including GTEx and Icelandic data (See Supplementary Data 15 for list of sources). For the Icelandic data, RNA sequencing and estimation of the association between sequence variants and gene expression have been described in a recent publication86.
Genetic correlation between suPAR and CRP
The genetic correlation between suPAR and CRP was calculated using suPAR from our Iceland/Denmark meta-analysis and CRP from UK biobank. We additionally investigated the overlap of findings between the suPAR-associated variants identified in our study and CRP-associated variants from the GWAS-catalog (https://www.ebi.ac.uk/gwas/). CRP-associated variants reported in the GWAS-catalog were checked for overlap with our suPAR-associated variants + LD-classes (all variants with r2 > 0.80).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank participants in all the included cohorts. Similarly, we thank personnel employed in all blood banks across Denmark for making DBDS inclusion a part of their work routine. This study was supported by the Danish Council for Independent Research (09-069412 and 0602-02634B), Aase og Ejnar Danielsens Fond, AP Møller Fonden, and the Danish Regions (02/2611). The DBDS genetic infrastructure was supported by the Novo Nordic Foundation (NNF17OC0027594). S.B. reports grants from Innovation Fund Denmark, grants from Novo Nordisk Foundation during the conduct of the study; and personal fees from Intomics A/S and Proscion A/S, outside the submitted work. S.B. and K.B. report on behalf of Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, that the following grants supported the study: The Novo Nordisk Foundation (NNF14CC0001 and NNF17OC0027594) and The Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 115881 (RHAPSODY). L.J.H.R. is supported by an international postdoctoral fellowship from the Lundbeck Foundation (grant no. R288-2018-380). The E-Risk Study is funded by the Medical Research Council (UKMRC grant G1002190). Additional support was provided by the National Institute of Child Health and Human Development (grant HD077482) and by the Jacobs Foundation. The authors are grateful to the Study members and their families for their participation. Our thanks CACI, Inc., and to members of the E-Risk team for their dedication, hard work, and insights. We thank the Dunedin Study members, Unit research staff, and Study founder Phil Silva. This research was supported by US-National Institute on Aging grants AG032282 and UK Medical Research Council grant MR/P005918/1. The Dunedin Multidisciplinary Health and Development Research Unit is supported by the New Zealand Health Research Council Programme Grant (16-604), and the New Zealand Ministry of Business, Innovation, and Employment (MBIE).
Author contributions
J.D. contributed to the conception, design, analysis, interpretation of data and drafted the work. E.F., M.K.M., L.J.H.R., and K. Sugden contributed to the design, analysis, interpretation of data and participated in the revision of the first manuscript draft. The DBDS Genomic Consortium contributed to the acquisition of data. G.T., V.T., and M.F. contributed to the acquisition and the analysis of data. S.H.L., L.S., and B.G. contributed to the analysis of data. L.W.T., K.S.B., S.R.O., E.S., C.E., O.B.P., T.F.H, K.B., S.B., R.P., L.A., A.C., T.E.M., and D.G. contributed to the acquisition, interpretation of data and participated in the revision of the first manuscript draft. J.E.O., H.S., K. Stefánsson, and H.U. participated in the acquisition of data, design of the study, interpretation of data, and to the revision of the first manuscript draft. All authors approved the submitted version of the manuscript.
Data availability
The suPAR meta-analysis summary statistics will be made available at https://www.decode.com/summarydata/.
Sequence variants passing GATK filters have been deposited in the European Variation Archive, accession number PRJEB15197.
For information on further access to data included in the meta-analysis, please contact the following authors of the respective cohorts: Hreinn Stefansson for data from the Icelandic cohort (hreinn.stefansson@decode.is) and Sisse Rye Ostrowski for data from the Danish Blood Donor Study (Sisse.Rye.Ostrowski@regionh.dk). The Dunedin Study data and E Risk study data are not publicly available but are available on request by qualified scientists. Requests require a concept paper describing the purpose of data access, ethical approval at the applicant’s institution, and provision for secure data access. Secure access is possible on the Duke University, Otago University, and King’s College London campuses. For UK Biobank please register on https://bbams.ndph.ox.ac.uk/ams/ and apply for the data through there.
Code availability
Variants in the Icelandic and Danish cohorts were imputed using software developed at deCODE genetics based on the IMPUTE HMM model87 as previously described88. A linear mixed model implemented by BOLT-LMM79 was used to test for association between sequence variants and suPAR levels.
We used publicly available software (URLs listed below) in conjunction with the above described algorithms in the sequencing processing pipeline (Whole-genome sequencing, Association testing, RNA-seq mapping, and analysis):
BWA 0.7.10 mem, https://github.com/lh3/bwa
GenomeAnalysisTKLite 2.3.9, https://github.com/broadgsa/gatk/
Picard tools 1.117, https://broadinstitute.github.io/picard/
SAMtools 1.3, http://samtools.github.io/
Bedtools v2.25.0-76-g5e7c696z, https://github.com/arq5x/bedtools2/
Variant Effect Predictor https://github.com/Ensembl/ensembl-vep
BOLT-LMM https://data.broadinstitute.org/alkesgroup/BOLT-LMM/downloads/“
IMPUTE2 v2.3.1 https://mathgen.stats.ox.ac.uk/impute/impute_v2.html
dbSNP v140; http://www.ncbi.nlm.nih.gov/SNP/
LD Score Regression software; https://github.com/bulik/ldsc
BiNGO v3.0.3 https://www.psb.ugent.be/cbd/papers/BiNGO/Download.html
Cytoscape v3.7.1 https://cytoscape.org/download.html
We used R extensively to analyze data and create plots.
Competing interests
The authors E.F., G.T., M.F., M.K.M., V.T., S.H.L., L.S., B.G., D.G., H.S., and K. Stefánsson who are affiliated with deCODE genetics/AMGEN, declare competing interests as employees. J.E.O. is named inventor on patents on suPAR owned by Copenhagen University Hospital Hvidovre, Denmark. J.E.O. is co-founder, shareholder, and board member of ViroGates A/S, Denmark, the company that developed the suPARnostic ELISA. ViroGates A/S had no role in the study design, data collection, analysis, or interpretation of study findings, and/or in the decision to submit the manuscript for publication. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Joseph Dowsett, Email: joseph.dowsett@regionh.dk.
DBDS Genomic Consortium:
Steffen Andersen, Gregor Jemec, Poul Jennum, Rene Kasper Nielsen, Mette Nyegaard, Helene Martina Paarup, Mikkel Petersen, Thomas Werge, Daniel Gudbjartsson, Kari Stefansson, and Unnur Þorsteinsdóttir
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02144-8.
References
- 1.Wagner V, Gil J. T cells engineered to target senescence. Nature. 2020;583:37–38. doi: 10.1038/d41586-020-01759-x. [DOI] [PubMed] [Google Scholar]
- 2.Thuno M, Macho B, Eugen-Olsen J. suPAR: the molecular crystal ball. Dis. Markers. 2009;27:157–172. doi: 10.1155/2009/504294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei C, et al. Modification of kidney barrier function by the urokinase receptor. Nat. Med. 2008;14:55–63. doi: 10.1038/nm1696. [DOI] [PubMed] [Google Scholar]
- 4.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 5.Mahmood, N., Mihalcioiu, C. & Rabbani, S. A. Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front. Oncol.8, 24 (2018). [DOI] [PMC free article] [PubMed]
- 6.Desmedt S, Desmedt V, Delanghe JR, Speeckaert R, Speeckaert MM. The intriguing role of soluble urokinase receptor in inflammatory diseases. Crit. Rev. Clin. Lab. Sci. 2017;54:117–133. doi: 10.1080/10408363.2016.1269310. [DOI] [PubMed] [Google Scholar]
- 7.Eugen-Olsen J, et al. Circulating soluble urokinase plasminogen activator receptor predicts cancer, cardiovascular disease, diabetes and mortality in the general population. J. Intern. Med. 2010;268:296–308. doi: 10.1111/j.1365-2796.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 8.Haastrup E, et al. Soluble urokinase plasminogen activator receptor as a marker for use of antidepressants. PLoS ONE. 2014;9:e110555. doi: 10.1371/journal.pone.0110555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tornkvist PBS, et al. Soluble urokinase plasminogen activator receptor is linearly associated with dietary quality and predicts mortality. Br. J. Nutr. 2019;121:699–708. doi: 10.1017/S0007114518003720. [DOI] [PubMed] [Google Scholar]
- 10.Ivancso, I. et al. Relationship of circulating soluble urokinase plasminogen activator receptor (suPAR) levels to disease control in asthma and asthmatic pregnancy. PLoS ONE8, e60697 (2013). [DOI] [PMC free article] [PubMed]
- 11.Gumus A, et al. Soluble urokinase-type plasminogen activator receptor is a novel biomarker predicting acute exacerbation in COPD. Int. J. Chronic Obstr. Pulm. Dis. 2015;10:357–365. doi: 10.2147/COPD.S77654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Can U, Guzelant A, Yerlikaya FH, Yosunkaya S. The role of serum soluble urokinase-type plasminogen activator receptor in stable chronic obstructive pulmonary disease. J. Investig. Med. 2014;62:938–943. doi: 10.1097/JIM.0000000000000105. [DOI] [PubMed] [Google Scholar]
- 13.Håkansson KEJ, et al. The biomarkers suPAR and blood eosinophils are associated with hospital readmissions and mortality in asthma – a retrospective cohort study. Respir. Res. 2019;20:258. doi: 10.1186/s12931-019-1234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayek SS, et al. Cardiovascular disease biomarkers and suPAR in predicting decline in renal function: a prospective cohort study. Kidney Int. Rep. 2017;2:425–432. doi: 10.1016/j.ekir.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo S, et al. Soluble urokinase-type plasminogen activator receptor in Black Americans with CKD. Clin. J. Am. Soc. Nephrol. 2018;13:1013. doi: 10.2215/CJN.13631217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer F, et al. Association of serum soluble urokinase receptor levels with progression of kidney disease in children. JAMA Pediatr. 2017;171:e172914. doi: 10.1001/jamapediatrics.2017.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz CA, et al. Soluble urokinase-type plasminogen activator receptor (suPAR) and impaired kidney function in the population-based malmo diet and cancer study. Kidney Int. Rep. 2017;2:239–247. doi: 10.1016/j.ekir.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayek SS, et al. Soluble urokinase receptor and chronic kidney disease. N. Engl. J. Med. 2015;373:1916–1925. doi: 10.1056/NEJMoa1506362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayek SS, et al. Soluble urokinase receptor and acute kidney injury. N. Engl. J. Med. 2020;382:416–426. doi: 10.1056/NEJMoa1911481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen LJH, et al. Combining national early warning score with soluble urokinase plasminogen activator receptor (suPAR) improves risk prediction in acute medical patients: a registry-based cohort study. Crit. Care Med. 2018;46:1961–1968. doi: 10.1097/CCM.0000000000003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eugen-Olsen J, Ladelund S, Sorensen LT. Plasma suPAR is lowered by smoking cessation: a randomized controlled study. Eur. J. Clin. Invest. 2016;46:305–311. doi: 10.1111/eci.12593. [DOI] [PubMed] [Google Scholar]
- 22.Haupt TH, et al. Risk factors associated with serum levels of the inflammatory biomarker soluble urokinase plasminogen activator receptor in a general population. Biomark. Insights. 2014;9:BMI.S19876. doi: 10.4137/BMI.S19876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haupt, T. H. et al. Healthy lifestyles reduce suPAR and mortality in a Danish general population study. Immun. Ageing16, 1 (2019). [DOI] [PMC free article] [PubMed]
- 24.Rasmussen, L. J. H. et al. Association of adverse experiences and exposure to violence in childhood and adolescence with inflammatory burden in young people. JAMA Pediatr, 178, 38–47 (2020). [DOI] [PMC free article] [PubMed]
- 25.Ligthart S, et al. Genome analyses of >200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am. J. Hum. Genet. 2018;103:691–706. doi: 10.1016/j.ajhg.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyngbaek S, et al. CRP and suPAR are differently related to anthropometry and subclinical organ damage. Int. J. Cardiol. 2013;167:781–785. doi: 10.1016/j.ijcard.2012.03.040. [DOI] [PubMed] [Google Scholar]
- 27.Andersen O, Eugen-Olsen J, Kofoed K, Iversen J, Haugaard SB. Soluble urokinase plasminogen activator receptor is a marker of dysmetabolism in HIV-infected patients receiving highly active antiretroviral therapy. J. Med. Virol. 2008;80:209–216. doi: 10.1002/jmv.21114. [DOI] [PubMed] [Google Scholar]
- 28.Sier CF, et al. Presence of urokinase-type plasminogen activator receptor in urine of cancer patients and its possible clinical relevance. Lab Invest. 1999;79:717–722. [PubMed] [Google Scholar]
- 29.Thurison T, Christensen IJ, Lund IK, Nielsen HJ, Høyer-Hansen G. Circulating intact and cleaved forms of the urokinase-type plasminogen activator receptor: biological variation, reference intervals and clinical useful cut-points. Clin. Chim. Acta. 2015;439:84–90. doi: 10.1016/j.cca.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen LJH, et al. Association between elevated suPAR, a new biomarker of inflammation, and accelerated aging. J. Gerontol. A Biol. Sci. Med. Sci. 2021;76:318–327. doi: 10.1093/gerona/glaa178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sveinbjornsson G, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat. Genet. 2016;48:314–317. doi: 10.1038/ng.3507. [DOI] [PubMed] [Google Scholar]
- 32.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maugeri A, Barchitta M, Agodi A. The association between complement factor H rs1061170 polymorphism and age-related macular degeneration: a comprehensive meta-analysis stratified by stage of disease and ethnicity. Acta Ophthalmol. 2019;97:e8–e21. doi: 10.1111/aos.13849. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, et al. Single nucleotide polymorphisms in the phospholipase A2 receptor gene are associated with genetic susceptibility to idiopathic membranous nephropathy. Nephron Clin. Pract. 2011;117:c253–c258. doi: 10.1159/000320194. [DOI] [PubMed] [Google Scholar]
- 35.Beck LH, Jr., et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N. Engl. J. Med. 2009;361:11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso A, et al. Identification of risk loci for Crohn’s disease phenotypes using a genome-wide association study. Gastroenterology. 2015;148:794–805. doi: 10.1053/j.gastro.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 37.Hirayama A, et al. Lymphocyte antigen 75 polymorphisms are associated with disease susceptibility and phenotype in Japanese patients with inflammatory bowel disease. Dis. Markers. 2016;2016:6485343. doi: 10.1155/2016/6485343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Begin P, et al. Association of urokinase-type plasminogen activator with asthma and atopy. Am. J. Respir. Crit. Care Med. 2007;175:1109–1116. doi: 10.1164/rccm.200607-1012OC. [DOI] [PubMed] [Google Scholar]
- 40.Nioi P, et al. Variant ASGR1 associated with a reduced risk of coronary artery disease. N. Engl. J. Med. 2016;374:2131–2141. doi: 10.1056/NEJMoa1508419. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X, et al. Examination of copy number variations of CHST9 in multiple types of hematologic malignancies. Cancer Genet. Cytogenet. 2010;203:176–179. doi: 10.1016/j.cancergencyto.2010.07.132. [DOI] [PubMed] [Google Scholar]
- 42.Stewart CE, et al. PLAUR polymorphisms and lung function in UK smokers. BMC Med. Genet. 2009;10:112. doi: 10.1186/1471-2350-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barton SJ, et al. PLAUR polymorphisms are associated with asthma, PLAUR levels, and lung function decline. J. Allergy Clin. Immunol. 2009;123:1391–1400. doi: 10.1016/j.jaci.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Maere S, Heymans K, Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 45.Han X, et al. Using Mendelian randomization to evaluate the causal relationship between serum C-reactive protein levels and age-related macular degeneration. Eur. J. Epidemiol. 2020;35:139–146. doi: 10.1007/s10654-019-00598-z. [DOI] [PubMed] [Google Scholar]
- 46.Przybylowska K, et al. Antigen levels of the urokinase-type plasminogen activator and its gene polymorphisms in colorectal cancer. Cancer Lett. 2002;181:23–30. doi: 10.1016/S0304-3835(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 47.Zhong F, Yang XC, Bu LX, Li NY, Chen WT. Single nucleotide polymorphisms in the u-PA gene are related to susceptibility to oral tongue squamous cell carcinoma in the Northern Chinese Han population. Asian Pac. J. Cancer Prev. 2013;14:781–784. doi: 10.7314/APJCP.2013.14.2.781. [DOI] [PubMed] [Google Scholar]
- 48.Duran J, et al. The PLAU P141L single nucleotide polymorphism is associated with collateral circulation in patients with coronary artery disease. Rev. Espanola De. Cardiologia. 2014;67:552–557. doi: 10.1016/j.recesp.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 49.Wu WL, Jiang H, Wang MY, Zhang DF. Meta-analysis of the association between urokinase-plasminogen activator gene rs2227564 polymorphism and Alzheimer’s disease. Am. J. Alzheimers Dis. Other Dement. 2013;28:517–523. doi: 10.1177/1533317513494450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarui T, et al. Direct interaction of the kringle domain of urokinase-type plasminogen activator (uPA) and integrin alpha v beta 3 induces signal transduction and enhances plasminogen activation. Thromb. Haemost. 2006;95:524–534. doi: 10.1160/TH05-06-0457. [DOI] [PubMed] [Google Scholar]
- 51.Appella E, Blasi F. The growth-factor module of Urokinase is the binding sequence for its receptor. Ann. N. Y. Acad. Sci. 1987;511:192–195. doi: 10.1111/j.1749-6632.1987.tb36247.x. [DOI] [PubMed] [Google Scholar]
- 52.Bdeir K, et al. The kringle stabilizes urokinase binding to the urokinase receptor. Blood. 2003;102:3600–3608. doi: 10.1182/blood-2003-03-0949. [DOI] [PubMed] [Google Scholar]
- 53.Xie J, et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat. Commun. 2020;11:1600. doi: 10.1038/s41467-020-15383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maile LA, et al. Blocking ligand occupancy of the alphaVbeta3 integrin inhibits the development of nephropathy in diabetic pigs. Endocrinology. 2014;155:4665–4675. doi: 10.1210/en.2014-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayek SS, et al. A tripartite complex of suPAR, APOL1 risk variants and alphavbeta3 integrin on podocytes mediates chronic kidney disease. Nat. Med. 2017;23:945–953. doi: 10.1038/nm.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnaar, R. L. & Kinoshita, T. Glycosphingolipids. in Essentials of Glycobiology (eds. rd et al.) 125-135 (Cold Spring Harbor Laboratory Press Copyright 2015–2017 by The Consortium of Glycobiology Editors, La Jolla, California. All rights reserved., Cold Spring Harbor (NY), 2015).
- 58.Cunningham O, et al. Dimerization controls the lipid raft partitioning of uPAR/CD87 and regulates its biological functions. EMBO J. 2003;22:5994–6003. doi: 10.1093/emboj/cdg588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Behrendt N, et al. The human receptor for urokinase plasminogen activator. NH2-terminal amino acid sequence and glycosylation variants. J. Biol. Chem. 1990;265:6453–6460. doi: 10.1016/S0021-9258(19)39348-2. [DOI] [PubMed] [Google Scholar]
- 60.Ploug M, Rahbek-Nielsen H, Nielsen PF, Roepstorff P, Dano K. Glycosylation profile of a recombinant urokinase-type plasminogen activator receptor expressed in Chinese hamster ovary cells. J. Biol. Chem. 1998;273:13933–13943. doi: 10.1074/jbc.273.22.13933. [DOI] [PubMed] [Google Scholar]
- 61.Moller LB, Pollanen J, Ronne E, Pedersen N, Blasi F. N-linked glycosylation of the ligand-binding domain of the human urokinase receptor contributes to the affinity for its ligand. J. Biol. Chem. 1993;268:11152–11159. doi: 10.1016/S0021-9258(18)82104-4. [DOI] [PubMed] [Google Scholar]
- 62.Gardsvoll H, Werner F, Sondergaard L, Dano K, Ploug M. Characterization of low-glycosylated forms of soluble human urokinase receptor expressed in Drosophila Schneider 2 cells after deletion of glycosylation-sites. Protein Expr. Purif. 2004;34:284–295. doi: 10.1016/j.pep.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Blanco-Gómez A, et al. Missing heritability of complex diseases: enlightenment by genetic variants from intermediate phenotypes. BioEssays: N. Rev. Mol., Cell. Dev. Biol. 2016;38:664–673. doi: 10.1002/bies.201600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgdorf KS, et al. Digital questionnaire platform in the Danish Blood Donor Study. Comput. Methods Prog. Biomed. 2016;135:101–104. doi: 10.1016/j.cmpb.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 65.Hansen, T. F. et al. DBDS Genomic Cohort, a prospective and comprehensive resource for integrative and temporal analysis of genetic, environmental and lifestyle factors affecting health of blood donors. Bmj Open9 (2019). [DOI] [PMC free article] [PubMed]
- 66.Moffitt TE. Teen-aged mothers in contemporary Britain. J. Child Psychol. Psychiatry. 2002;43:727–742. doi: 10.1111/1469-7610.00082. [DOI] [PubMed] [Google Scholar]
- 67.Poulton R, Moffitt TE, Silva PA. The Dunedin Multidisciplinary Health and Development Study: overview of the first 40 years, with an eye to the future. Soc. Psychiatry Psychiatr. Epidemiol. 2015;50:679–693. doi: 10.1007/s00127-015-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chew-Harris J, Appleby S, Richards AM, Troughton RW, Pemberton CJ. Analytical, biochemical and clearance considerations of soluble urokinase plasminogen activator receptor (suPAR) in healthy individuals. Clin. Biochem. 2019;69:36–44. doi: 10.1016/j.clinbiochem.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 69.Rasmussen LJH, et al. Cumulative childhood risk is associated with a new measure of chronic inflammation in adulthood. J. Child Psychol. Psychiatry. 2019;60:199–208. doi: 10.1111/jcpp.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–U20. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 71.Jonsson, H. et al. Data Descriptor: Whole genome characterization of sequence diversity of 15,220 Icelanders. Sci. Data4 (2017). [DOI] [PMC free article] [PubMed]
- 72.Eggertsson HP, et al. Graphtyper enables population-scale genotyping using pangenome graphs. Nat. Genet. 2017;49:1654. doi: 10.1038/ng.3964. [DOI] [PubMed] [Google Scholar]
- 73.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belsky DW, et al. Genetic analysis of social-class mobility in five longitudinal studies. Proc. Natl Acad. Sci. USA. 2018;115:E7275–e7284. doi: 10.1073/pnas.1801238115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Consortium, T.U. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2009;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vilhjalmsson BJ, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saevarsdottir S, et al. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature. 2020;584:619–623. doi: 10.1038/s41586-020-2436-0. [DOI] [PubMed] [Google Scholar]
- 87.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 88.Gudbjartsson DF, et al. Sequence variants from whole genome sequencing a large group of Icelanders. Sci. Data. 2015;2:150011. doi: 10.1038/sdata.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The suPAR meta-analysis summary statistics will be made available at https://www.decode.com/summarydata/.
Sequence variants passing GATK filters have been deposited in the European Variation Archive, accession number PRJEB15197.
For information on further access to data included in the meta-analysis, please contact the following authors of the respective cohorts: Hreinn Stefansson for data from the Icelandic cohort (hreinn.stefansson@decode.is) and Sisse Rye Ostrowski for data from the Danish Blood Donor Study (Sisse.Rye.Ostrowski@regionh.dk). The Dunedin Study data and E Risk study data are not publicly available but are available on request by qualified scientists. Requests require a concept paper describing the purpose of data access, ethical approval at the applicant’s institution, and provision for secure data access. Secure access is possible on the Duke University, Otago University, and King’s College London campuses. For UK Biobank please register on https://bbams.ndph.ox.ac.uk/ams/ and apply for the data through there.
Variants in the Icelandic and Danish cohorts were imputed using software developed at deCODE genetics based on the IMPUTE HMM model87 as previously described88. A linear mixed model implemented by BOLT-LMM79 was used to test for association between sequence variants and suPAR levels.
We used publicly available software (URLs listed below) in conjunction with the above described algorithms in the sequencing processing pipeline (Whole-genome sequencing, Association testing, RNA-seq mapping, and analysis):
BWA 0.7.10 mem, https://github.com/lh3/bwa
GenomeAnalysisTKLite 2.3.9, https://github.com/broadgsa/gatk/
Picard tools 1.117, https://broadinstitute.github.io/picard/
SAMtools 1.3, http://samtools.github.io/
Bedtools v2.25.0-76-g5e7c696z, https://github.com/arq5x/bedtools2/
Variant Effect Predictor https://github.com/Ensembl/ensembl-vep
BOLT-LMM https://data.broadinstitute.org/alkesgroup/BOLT-LMM/downloads/“
IMPUTE2 v2.3.1 https://mathgen.stats.ox.ac.uk/impute/impute_v2.html
dbSNP v140; http://www.ncbi.nlm.nih.gov/SNP/
LD Score Regression software; https://github.com/bulik/ldsc
BiNGO v3.0.3 https://www.psb.ugent.be/cbd/papers/BiNGO/Download.html
Cytoscape v3.7.1 https://cytoscape.org/download.html
We used R extensively to analyze data and create plots.