Abstract
As like in mammalian system, the DNA damage responsive cell cycle checkpoint functions play crucial role for maintenance of genome stability in plants through repairing of damages in DNA and induction of programmed cell death or endoreduplication by extensive regulation of progression of cell cycle. ATM and ATR (ATAXIA-TELANGIECTASIA-MUTATED and -RAD3-RELATED) function as sensor kinases and play key role in the transmission of DNA damage signals to the downstream components of cell cycle regulatory network. The plant-specific NAC domain family transcription factor SOG1 (SUPPRESSOR OF GAMMA RESPONSE 1) plays crucial role in transducing signals from both ATM and ATR in presence of double strand breaks (DSBs) in the genome and found to play crucial role in the regulation of key genes involved in cell cycle progression, DNA damage repair, endoreduplication and programmed cell death. Here we report that Arabidopsis exposed to high salinity shows generation of oxidative stress induced DSBs along with the concomitant induction of endoreduplication, displaying increased cell size and DNA ploidy level without any change in chromosome number. These responses were significantly prominent in SOG1 overexpression line than wild-type Arabidopsis, while sog1 mutant lines showed much compromised induction of endoreduplication under salinity stress. We have found that both ATM-SOG1 and ATR-SOG1 pathways are involved in the salinity mediated induction of endoreduplication. SOG1was found to promote G2-M phase arrest in Arabidopsis under salinity stress by downregulating the expression of the key cell cycle regulators, including CDKB1;1, CDKB2;1, and CYCB1;1, while upregulating the expression of WEE1 kinase, CCS52A and E2Fa, which act as important regulators for induction of endoreduplication. Our results suggest that Arabidopsis undergoes endoreduplicative cycle in response to salinity induced DSBs, showcasing an adaptive response in plants under salinity stress.
Subject terms: Plant cell biology, Plant development, Plant molecular biology
Introduction
Among the various environmental stress factors, soil salinity represents one of the major factors, adversely affecting plant growth and development and therefore limiting crop productivity. Worldwide, more than ~ 20% of the cultivated land (~ about 45 hectares) has been shown to be directly affected by salinity and the quantity is increasing day by day1,2. High soil salinity stress has been shown to modulate various physiological and metabolic processes in plants, depending on the severity and duration of the stress2. High soil salinity leads to ionic stress, which results from an excessive accumulation of Na+ and Cl− ions in plant tissues exposed to soils with high salinity and thus creates toxic condition in plant cell3–6. This ion accumulation is complemented with the induction of secondary oxidative stress, and subsequently enhances the production of various reactive oxygen species (ROS), including singlet oxygen, superoxide, hydroxyl radical, and hydrogen peroxide, respectively5,7–9. In contrast to their role as second messenger in mediating salt tolerance, ROS, when produced at enhanced level under stress conditions also cause oxidative damages in various cellular macromolecules, primarily proteins, nucleic acids and lipids, respectively10. Therefore, plants have developed an array of complicated and interlinked mechanisms to adapt and survive in soil with high salt concentrations.
Salinity stress mediated ROS generation serves as one of the crucial factors for the induction of DNA damage3,11. The accumulation of highly reactive free radicals introduces various forms of DNA lesions, including oxidative modification of bases, collapse of replication fork and induction of DNA single and double strand breaks (SSBs and DSBs), thus challenging the cell’s DNA repair machinery12. Among the different types of DNA lesions, double strand breaks (DSBs) in DNA double helix has been considered as one of the most critical forms of damage, resulting in loss of DNA segment and thus, important genetic information. When left unrepaired, DSBs have been found to inhibit DNA replication and transcription, leading to genome instability11,13–15. Thus, soil salinity not only affects the structural, enzymatic and non-enzymatic components of plants but also challenges plant genome stability and thus reduces crop yield. However, information on the mechanism of maintenance of genome stability in plants under salinity stress condition remains mostly elusive.
Various investigations have demonstrated the induction of endoreduplication under salinity stress conditions in plants as part of their salinity tolerance and adaptation strategy16–18. Endoreduplication is the replication of the genome without proceeding into the mitotic phase, resulting in an increase in the cellular ploidy level of an organism over its lifetime18–20 and this condition is termed as ‘endopolyploidy’. The root cortical cells of salt tolerant varieties of Sorghum bicolor (sorghum) display endoreduplication in response to NaCl treatment16. Leaf tissues of Mesembryanthemum crystallinum, an annual halophyte, shows developmental endopolyploidy up to 128C and associated with the increased leaf and root epidermal cell area, suggesting possible role in sequestration or compartmentalization of excess salt18. Another study in Medicago truncatula revealed important link between cellular adaptation under salt stress and enhanced expression of key regulators of endoreduplication, including CCS52 and WEE117. These observations have provided evidence on the adaptive significance of endopolyploidy in plants under salinity stress.
As mentioned earlier, accumulating evidences have indicated that salt stress disrupts genome stability in plants and exposure to increasing salinity increases the incidence of generation of double strand breaks (DSBs) in DNA14,21. Recent studies have indicated that DSBs can induce programmed endoreduplication as a part of DNA damage response (DDR) pathway when the extent of DSB accumulation becomes high22–24. As like mammals, in plants, ATM and ATR (ATAXIA-TELANGIECTASIA-MUTATED and -RAD3-RELATED) function as sensor kinases and play crucial role in the transmission of signals to the downstream cell cycle regulators. The plant-specific NAC domain family transcription factor SOG1 (SUPPRESSOR OF GAMMA RESPONSE 1) has been shown to play crucial role in communicating the signals from both ATM and ATR in presence of DSBs in the genome for the regulation of key genes involved in cell cycle progression, DNA damage repair, endoreduplication and programmed cell death15,23,25–26. In this study, we found that Arabidopsis root tip cells displayed significant increase in cell size and DNA ploidy level without a concomitant change in chromosome number under salinity stress. We have further found that SOG1 promotes G2-M arrest in Arabidopsis under salinity stress, while both ATM-SOG1 and ATR-SOG1 pathways are involved in the salinity mediated induction of endoreduplication.
Results
The loss-of-function mutants of AtSOG1 show less sensitivity to increasing salinity
Previously, several studies have revealed that mutants of AtSOG1 are more resistant to lower concentrations of various DSB inducing agents, including zeocin and gamma irradiation23,25. Furthermore, earlier reports have also demonstrated high salinity mediated induction of DSBs in plants, leading to genome instability14,21. Therefore, based on this information and to understand role of SOG1 in genome stability maintenance mechanism in plants under salinity stress, we have first compared the overall growth responses of the two knockout mutant lines of AtSOG1, sog1-6 (SALK_067631C) and sog1-1 and a transgenic Arabidopsis line overexpressing SOG1 (OE-1) along with the wild-type Arabidopsis in the absence and presence of increasing concentrations of NaCl. Details of NaCl treatment and growth conditions have been described under the “Materials and methods” section. Germination percentage was reduced in all the genotypes under salinity stress, however as compared to wild-type and OE-1 line, the inhibition in seed germination was more prominent in sog1-6 and sog1-1 mutant lines (approximately 53 and 59%, respectively) in presence of increasing concentrations of NaCl (p < 0.01; p < 0.05) (Supplementary Fig. S1A–C). In addition, germination of sog1-6 and sog1-1 mutant lines was significantly compromised at 200 mM concentration of NaCl than wild-type and OE-1 line, respectively (Supplementary Fig. S1C) (p < 0.01).
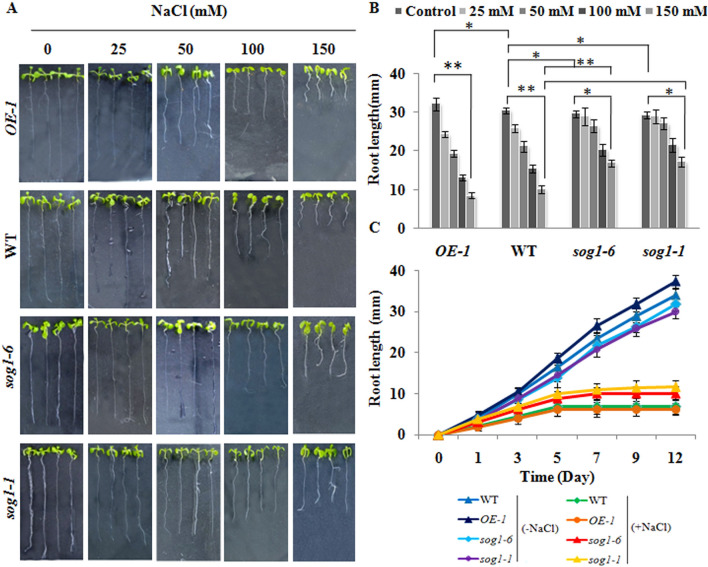
Since roots are immediately exposed to soil salinity, we next specifically analysed the growth response in terms of primary root growth in 12-days old wild-type, OE-1 line, sog1-6 and sog1-1 mutant seedlings grown in absence or presence of different concentrations of NaCl. Under control conditions (without NaCl), the mutants and OE-1 plants did not show any significant difference in primary root growth pattern as compared to the wild-type plants (Fig. 1A,B). On the other hand, in contrast to wild-type plants, root growth in sog1-6 and sog1-1 mutants was not affected under increasing salinity exposure (p < 0.05; p < 0.01) (Fig. 1A,B). Furthermore, when challenged with 150 mM NaCl, primary root growth in wild-type and OE-1 seedlings was appreciably ceased within 3–5 days after germination, whereas, root growth in sog1-6 and sog1-1 mutant seedling was found to be continued up to 5–7 days after germination (Fig. 1C). Quantitative growth response analysis have further indicated significant inhibition of lateral root formation in sog1 mutant seedlings (p < 0.01) than wild-type seedlings under salinity stress (Supplementary Fig. S2A). On the other hand, no significant difference in germination performance and post-germination primary root growth in 12-day old seedlings of wild-type and sog1 mutants could be detected in presence of LiCl (p < 0.05) (Supplementary Fig. S2B,C). Together, these results have provided important clues to indicate the possible functional involvement of SOG1 in regulation of plant response under salinity stress in Arabidopsis.
Figure 1.
Phenotypic responses in presence of increasing salinity. (A,B) Determination of root growth in 12-days old OE-1, wild-type (WT), sog1-6 and sog1-1 mutant lines under untreated condition and in response to increasing salinity. (C) 12-days old seedlings of wild-type, OE-1, sog1-6 and sog1-1 were treated without or with 150 mM NaCl and root growth was measured in 1-day interval. Three independent biological replicates for each genotype, each containing at least 10 plants, were analysed. The mean value of root length in individual experiment was determined and averaged subsequently for the three biological replicates. Error bar in the graphs represents ± SD. Asterisks represent statistically significant differences within a 5 and 1% confidence interval (*p < 0.05, **p < 0.01), respectively based on one-way ANOVA factorial analysis indicating the genotype that differs significantly in primary root growth.
Induction of oxidative damage and DNA double strand breaks in Arabidopsis under salinity stress
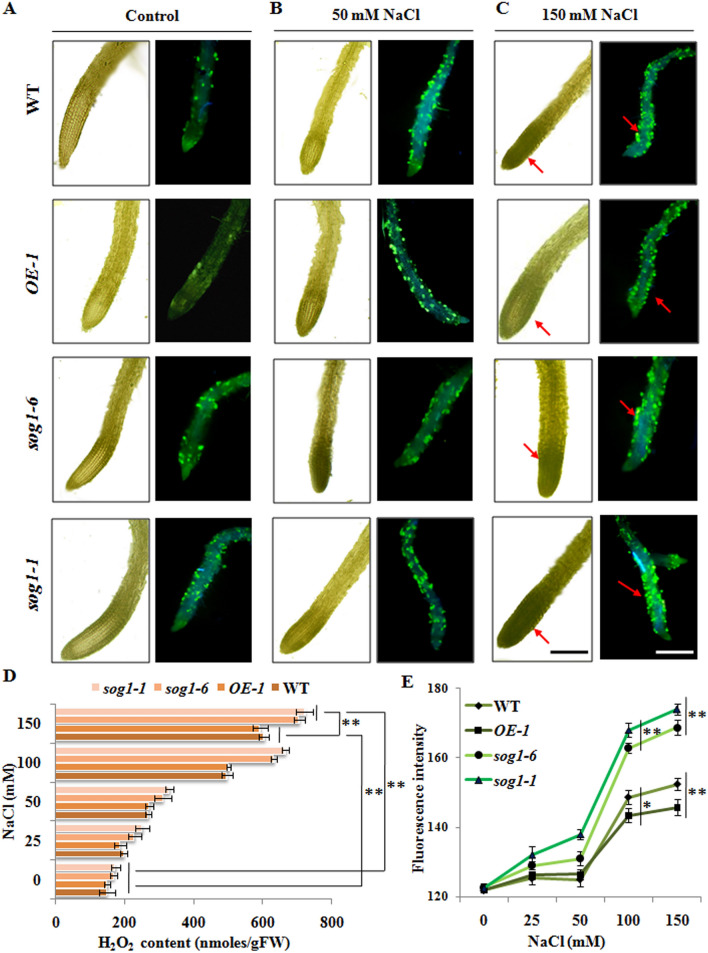
Based on the growth response of sog1 mutants under salinity stress condition, we next investigated the response at the cellular level. Earlier investigations have suggested generation of various reactive oxygen species and induction of oxidative stress in plants under salinity stress27,28. In addition, several previous studies have further demonstrated induction of DSBs in response to high concentration of NaCl treatment in Arabidopsis14,21. Based on this, we next explored whether higher sensitivity of sog1 mutants to NaCl treatment is related to any difference among the wild-type Arabidopsis, OE-1 line and sog1 mutants in terms of induction of oxidative stress response and DNA damage. Qualitative and quantitative analysis of ROS accumulation have revealed prominent level of ROS accumulation, including hydrogen peroxide (H2O2) and superoxide radical generation in the roots of wild-type, sog1-6 and sog1-1 mutant seedlings in presence of increasing concentrations of NaCl (Fig. 2A–C). However, as compared to wild-type Arabidopsis, ROS accumulation signal increased significantly in roots of sog1 mutants under salinity stress, while ROS accumulation signal was appreciably less in OE-1 line plants (p < 0.05; p < 0.01) (Fig. 2D,E). Interestingly, the amount of ROS generated in all the genotypes following salt treatment (150 mM NaCl) was comparable with the level of ROS generated after treatment with H2O2 (20 mM), which has been used as positive control (Supplementary Fig. S3A).Taken together, these results have indicated higher magnitude of induction of oxidative stress in sog1 mutants under salinity stress condition.
Figure 2.
Salinity mediated induction of oxidative stress. (A–C, left panels) Qualitative analysis of H2O2 accumulation levels in the roots of 7-days old wild-type, OE-1, sog1-6and sog1-1 mutant lines in the absence and presence of 50 and 150 mM NaCl, respectively. (A–C, right panels) Localization of superoxide radicals by ROS imaging using CM-H2DCFDA in root tips of all the genotypes in absence and presence of NaCl. Dark regions and green patches indicated by the arrow heads showing H2O2 accumulation and ROS generation, respectively. Representative images from at least three independent experiments with similar results are shown. (D,E) Quantitative estimation of H2O2 content and total ROS in the roots of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant line seedlings, either untreated or treated with increasing concentrations of NaCl. Quantitative data has been represented as the mean value of three independent biological replicates. Error bars represent standard deviation. Asterisks represent statistically significant differences (*p < 0.05, **p < 0.01) relative to controls.
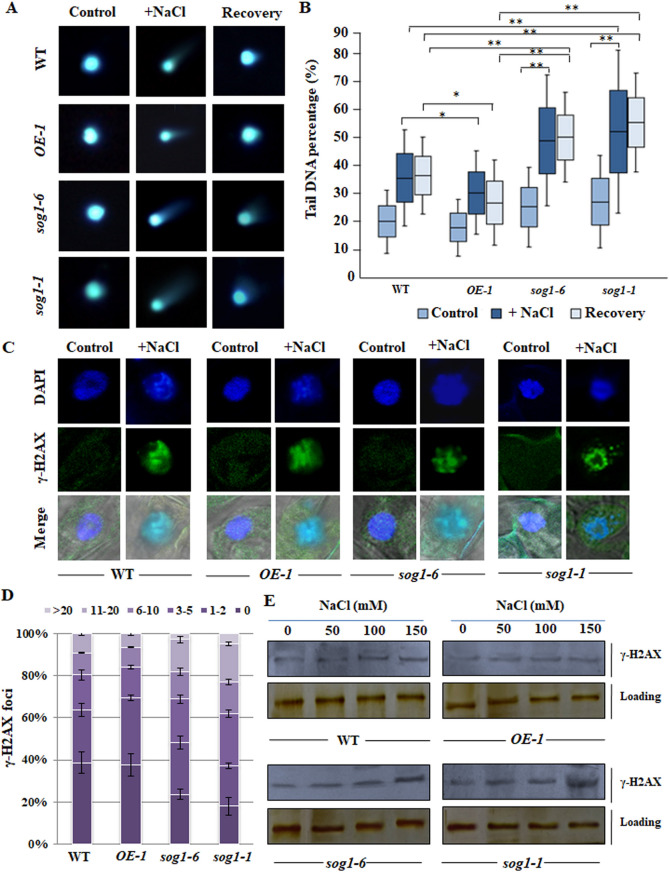
We next investigated the extent of induction of oxidative stress mediated DSBs under salinity stress in wild-type, sog1 mutant and SOG1 overexpressor lines. Comet assay was performed using the nuclei isolated from 7-days old control and salinity stressed seedlings. We have found considerable accumulation level of DSBs in all the genetic backgrounds in the presence of higher concentrations of NaCl than untreated control conditions (Fig. 3A,B). However, consistent with the induction level of oxidative damage, the extent of accumulation of DSBs was significantly high in sog1 mutants as compared to wild-type and OE-1plants (Fig. 3B) (p < 0.05; p < 0.01). In addition, recovery experiments have indicated relatively slower rate of DSB recovery in sog1 mutant lines than wild-type and overexpressor line (OE-1) (Fig. 3A,B) (p < 0.01). Together, these results have indicated that loss of SOG1 function leads to delay in the recovery of DSBs generated under salinity stress and thus increases the extent of accumulation of DSBs, as found in the sog1 mutant lines.
Figure 3.
Salinity induced accumulation of DSBs. (A) Representative Comet images of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant plants incubated without NaCl for 1 h (control) and incubated with 150 mM NaCl for 12 h and then transferred to medium without NaCl for 5 h (recovery) (left, middle and right panel respectively). (B) Box plot of percentages of tail DNA of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant plants. DNA percentage in the comet tails were determined by using TriTek Comet Score software. Box plots are based on analysis of approximately 100 cells per experimental sample from random microscopic fields of three independent biological replicates. Each box represents the interquartile range (IQR) of DNA damage, the line across each box represents the median value and the whiskers represent 5–95 percentile values. Brackets connect box plots of sample groups with significant statistical difference (*p < 0.05, **p < 0.01). (C) Immunostaining of γ-H2AX foci in the root cells of wild-type, overexpressor line and mutant line plants after 12 h of treatment without and with 150 mM NaCl, respectively. (D) Counted numbers of γ-H2AX foci per cell detected after 12 h of treatment with 150 mM NaCl in plants of all the genotypes. According to their number of γ-H2AX foci, approximately 100 nuclei for each sample were grouped into six classes; cell with 0, 1–2, 3–5, 6–10, 11–20 and > 20 foci per nuclei. (E) Detection of γ-H2AX accumulation by immunoblot analysis using the total histone protein isolated from untreated and NaCl treated of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant plants, respectively. The blots were cut prior to incubation with the primary antibody. Replicates of the protein gel blots have been presented in Supplementary Fig. S11.
To further substantiate the salinity mediated induction of DSBs, we have next examined the accumulation level of phosphorylated histone variant H2AX (designated as γ-H2AX), which serves as an important and highly conserved marker for DSB accumulation in plants29,30. In presence of DSBs, the histone variant H2AX becomes phosphorylated at serine 139 at the sites of DNA double strand breaks31. Analysis of γ-H2AX focus formation in the root tip cell nuclei of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings under control and salinity stress conditions have indicated an increased signal of γ-H2AX foci in sog1 mutant lines than wild-type plants after exposure to increasing salinity (Fig. 3C,D). On the other hand, SOG1 overexpressor line plants showed relatively lower number of γ-H2AX foci in comparison to the wild-type plants (Fig. 3C,D). The damage in the mutant plants was particularly predominant at 150 mM concentration of NaCl with about 25–30% of the nuclei possessing more than 10 γ-H2AX foci (Fig. 3D). Although the percentage of nuclei containing a specific number of γ-H2AX foci is slightly different in sog1-1 and sog1-6 mutant lines, the overall trend of foci formation was more or less similar in the two mutants, when compared with wild-type or SOG1overexpressor line plants. In addition, to examine the γ-H2AX accumulation level, protein gel blot analysis was performed using total histone protein isolated from 7-days old wild-type, OE-1, sog1-6 and sog1-1mutant seedlings. Consistent with the results of comet assay, significant increase in γ-H2AX accumulation level was detected in sog1-6 and sog1-1 mutant seedlings in a dose dependent manner under salinity stress as compared to wild-type and OE-1 plants (Fig. 3E; Supplementary Fig. S4A–D).
Previous studies have revealed that several DSB repair genes are upregulated under salinity stress14,21. We have also found increased transcript abundance of various DNA damage response and repair genes, including AtMRE11, AtRAD50, AtNBS1, AtRAD51, AtRAD54 and AtBRCA1, respectively in wild-type Arabidopsis and SOG1 overexpressor line. These genes are mainly involved in homologous recombination (HR) mediated DSB repair pathway15. However, such enhanced expression of the mentioned HR related genes under salinity treatment appeared to be significantly compromised in sog1-1 and sog1-6 mutant background (deficient in SOG1 function). In addition, DNA–protein interaction studies have indicated direct binding of SOG1 to the promoter of some HR related genes, which also showed increased expression under salinity stress in wild-type Arabidopsis (unpublished data). Taken together these observations indicate that SOG1 loss-of-function effectively impairs the HR mediated DSB repair efficiency, thus suggesting involvement of SOG1 in the regulation of repair of DSBs generated in Arabidopsis under salinity stress.
SOG1 participates in the induction of endoreduplication in Arabidopsis root tip cells in presence of increasing salinity
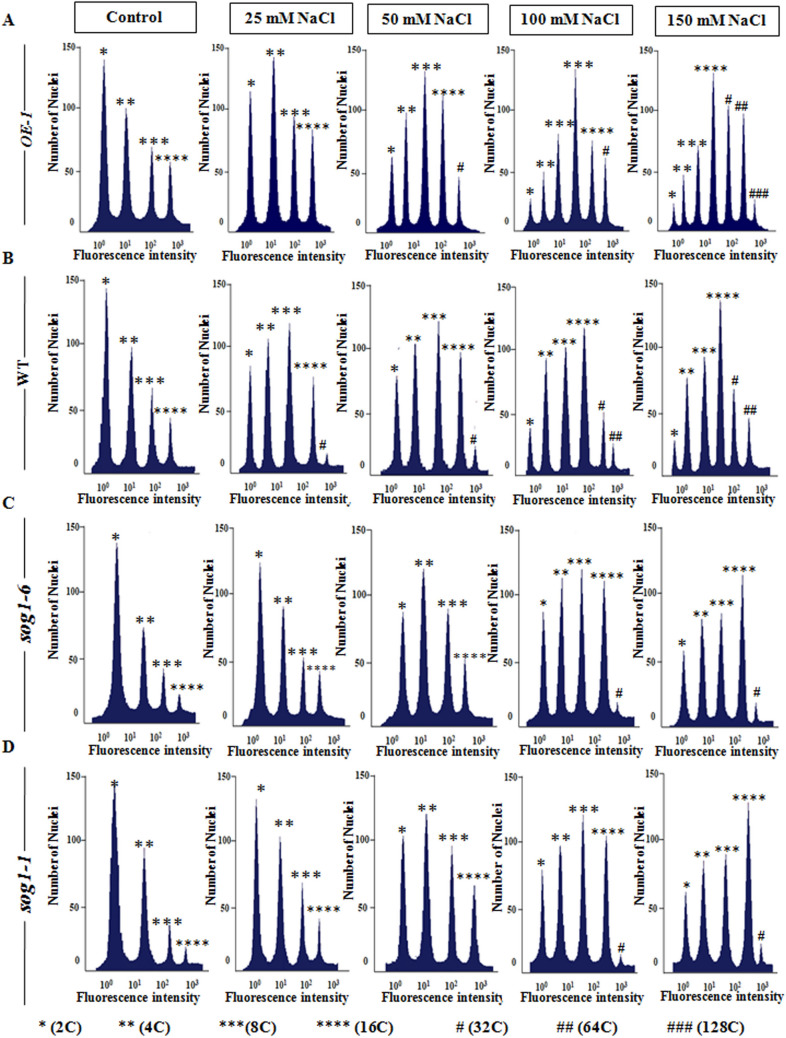
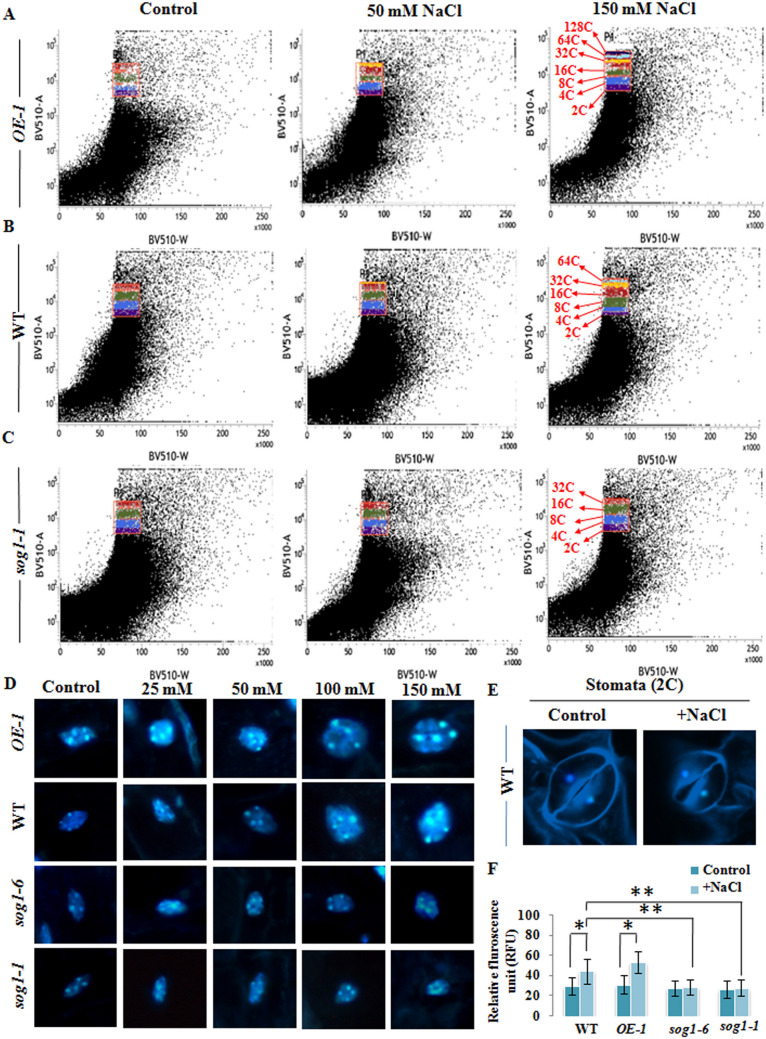
In Arabidopsis thaliana, the epidermal and cortical cells of the developing root tip region have been shown to contain nuclear DNA content up to 16C as a result of developmental endopolyploidy23. Several studies have demonstrated the induction of endoreduplication as part of salt tolerance and adaptation strategy in various plant species including Sorghum bicolor, Mesembryanthemum crystallinum and Medicago truncatula16–18,20,32. Besides, recent studies have revealed that genotoxic stress mediated DSBs promote programmed induction of endoreduplication in Arabidopsis thaliana through ATM-SOG1 and ATR-SOG1 pathways23. Our results have indicated significantly compromised growth response of sog1 mutants under salinity stress, along with higher magnitude of induction of oxidative stress and accumulation of DSBs (Figs. 1, 2, 3). On the other hand, SOG1 overexpressor line, OE-1 showed improved growth performance with relatively reduced level of oxidative stress and DSB induction level as compared to wild-type and sog1 mutant lines, when challenged with increasing concentrations of NaCl (Figs. 1, 2, 3). Based on this, we next investigated whether endoreduplication is involved in modulating the difference in growth response in wild-type, OE-1and sog1 mutant lines under salinity stress and the possible involvement of SOG1 in this regulation. To test this notion, we next examined the salinity induced change in ploidy level in the root cells of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings by performing flowcytometry. Under control conditions, wild-type Arabidopsis and OE-1 seedlings mainly showed presence of 2C (~ 37 and 36%, respectively), 4C (~ 26 and 25%, respectively), 8C (~ 21 and 18%, respectively) and 16C (16.42 and 16.89%, respectively) nuclear populations, while sog1-6 and sog1-1 mutant lines showed approximately 40 and 41.6%, 29.58 and 27.6%, 14.32 and 15.64% and 8.97 and 9.4% of 2C, 4C, 8C and 16C nuclear populations, respectively (Figs. 4A–D, 5A–C; Table 1). At low NaCl concentrations (25–50 mM), although the nuclear populations with 8C and 16C DNA content were increased slightly along with the appearance of prominent 32C peak in wild-type and OE-1 plants, no significant change in nuclear DNA content was detected in sog1-6 and sog1-1 mutant seedlings (Figs. 4A–D, 5A–C, Table 1). On the other hand, at higher concentrations of NaCl (100–150 mM), significant enhancement in polyploid nuclear populations with 16C, 32C and 64C DNA content were detected in wild-type and OE-1 seedlings (~ 27 and 33%, ~ 8 and 16%, ~ 7 and 14.5%, respectively), with the concomitant decrease in 2C populations (Figs. 4A–D, 5A–C, Table 1). Besides 32C and 64C, a small percentage of nuclei with 128C DNA content (~ 4%) was also detected in the OE-1seedlings treated with 150 mM NaCl (Figs. 4A, 5A, Table 1). Together these results have indicated the induction of endoreduplication under salinity stress. On the other hand, both sog1-6 and sog1-1 mutant lines showed only marginal effect of higher concentrations of salt treatment on the root cell nuclear ploidy level, with very low proportion of 32C populations (~ 2 and 3.5%; ~ 1.89 and 2.78%, respectively) could be detected when treated with 100 and 150 mM NaCl, respectively (Figs. 4C,D, 5C; Table 1). Together, these results have demonstrated the induction of DSBs along with the concomitant onset of endoreduplication in Arabidopsis root tip cells under salinity stress. Moreover, SOG1 appears to positively regulate the induction of endoreduplication under salinity stress in Arabidopsis.
Figure 4.
Salinity induced endoreduplication in the root cells. (A–D) Flowcytometric analysis of ploidy level in the DAPI-stained root cell nuclei of 7-days old OE-1, wild-type, sog1-6 and sog1-1 mutant plants treated without or with increasing concentrations of NaCl. A total of 10,000 events were recorded for each run and the data were analysed by FlowJo v. 10.0.6 (Tree Star, Inc.) software. All flow cytometric analyses were repeated with at least three independent biological replicates and representative ploidy distributions of total nuclei are given at the top of each peak.
Figure 5.
Salinity induced endoreduplication and nuclear area expansion. (A–C) Scatter plots showing DNA ploidy distribution in root tip nuclei of 7-days old OE-1, wild-type and sog1-1 mutant plants treated without or with 50 and 150 mM NaCl. (D) Confocal imaging of DAPI-stained nuclei in the root cells of 7-days old OE-1, wild-type, sog1-6 and sog1-1 mutant seedlings (first, second, third and fourth panel, respectively); Scale bar: 10 μm. (E) The nuclei in a pair of wild-type stomata cells represent 2C. Strongly stained dots in each nucleus correspond to centromeres. (F) Measurement of in-situ fluorescence in DAPI-stained root tip nuclei of wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings. Each bar represents the mean value of three independent replicates. Error bar indicates standard deviation. Asterisks indicate significant statistical differences (*p < 0.05, **p < 0.01) from control (without NaCl treatment) using one-way ANOVA factorial analysis.
Table 1.
Percentage of nuclei populations with various DNA content in four genotypes following exposure to increasing concentrations of NaCl.
| Genotype | NaCl treatment (mM) | Percentage of nuclei population with various DNA content | ||||||
|---|---|---|---|---|---|---|---|---|
| 2C | 4C | 8C | 16C | 32C | 64C | 128C | ||
| OE-1 | 0 | 35.62 ± 2.89 | 24.65 ± 1.08 | 23.65 ± 1.25 | 18.89 ± 0.98 | 0.78 ± 0.05 | – | – |
| 25 | 27.43 ± 3.04 | 23.56 ± 1.45 | 24.44 ± 1.11 | 20.78 ± 1.44 | 0.95 ± 0.14 | 0.44 ± 0.09 | – | |
| 50 | 18.77 ± 2.56 | 20.76 ± 0.77 | 24.2 ± 0.96 | 21.44 ± 1.23 | 5.67 ± 0.29 | 1.32 ± 0.06 | – | |
| 100 | 14.67 ± 0.98 | 14.83 ± 0.29 | 16.5 ± 0.33 | 32.44 ± 0.89 | 13.7 ± 0.72 | 10.89 ± 0.98 | 0.87 ± 0.03 | |
| 150 | 7.34 ± 0.23 | 13.5 ± 0.34 | 15.6 ± 0.39 | 30.89 ± 0.17 | 15.87 ± 0.87 | 14.6 ± 1.03 | 3.89 ± 0.55 | |
| WT | 0 | 36.54 ± 3.61 | 26.30 ± 0.55 | 21.17 ± 1.08 | 16.42 ± 0.78 | 0.11 ± 0.09 | – | |
| 25 | 29.65 ± 2.23 | 26.29 ± 1.23 | 22.29 ± 1.32 | 17.81 ± 1.67 | 0.34 ± 0.76 | 0.56 ± 0.32 | ||
| 50 | 22.39 ± 2.96 | 28.26 ± 0.86 | 22.78 ± 2.55 | 18.39 ± 0.65 | 2.71 ± 0.34 | 0.88 ± 0.55 | ||
| 100 | 16.77 ± 1.77 | 19.91 ± 2.45 | 22.17 ± 1.10 | 26.45 ± 2.1 | 9.31 ± 0.19 | 7.03 ± 0.98 | ||
| 150 | 10.58 ± 0.89 | 17.70 ± 3.11 | 18.51 ± 1.07 | 28.09 ± 0.43 | 12.11 ± 0.12 | 10.44 ± 2.55 | ||
| sog1-1 | 0 | 40.90 ± 4.23 | 29.58 ± 3.83 | 14.32 ± 1.22 | 8.97 ± 0.21 | – | – | |
| 25 | 38.54 ± 2.88 | 32.58 ± 3.08 | 15.51 ± 0.84 | 9.45 ± 0.93 | 0.79 ± 0.22 | – | ||
| 50 | 34.04 ± 2.63 | 32.53 ± 1.56 | 16.80 ± 0.89 | 10.41 ± 1.05 | 0.98 ± 0.66 | – | ||
| 100 | 27.27 ± 2.92 | 29.01 ± 3.31 | 15.26 ± 1.74 | 14.71 ± 0.95 | 2.13 ± 0.21 | – | ||
| 150 | 22.30 ± 4.47 | 28.54 ± 3.991 | 13.12 ± 1.54 | 17.78 ± 0.31 | 3.09 ± 0.21 | – | ||
| sog1-6 | 0 | 41.6 ± 2.78 | 27.6 ± 1.98 | 15.64 ± 0.89 | 9.4 ± 0.22 | – | – | |
| 25 | 40.76 ± 3.33 | 28.5 ± 1.44 | 15.92 ± 0.34 | 9.25 ± 0.11 | – | – | ||
| 50 | 36.8 ± 2.25 | 29.4 ± 0.79 | 17.11 ± 1.38 | 9.88 ± 0.17 | 0.55 ± 0.08 | – | ||
| 100 | 29.22 ± 2.55 | 28.7 ± 0.92 | 16.8 ± 1.10 | 15.71 ± 0.68 | 1.89 ± 0.04 | – | ||
| 150 | 23.5 ± 3.01 | 27.4 ± 1.37 | 15.72 ± 0.81 | 18.23 ± 0.48 | 2.78 ± 0.02 | – | ||
To further substantiate the results of flowcytometry, we also examined the root cell nuclei in both the genotypes. Consistent with the altered ploidy distributions, visualisation of DAPI-stained nuclei revealed significant increase in size of nuclei in the root cells of salt treated wild-type and OE-1seedlings with unchanged centromere number (Fig. 5D, first and second panel), indicating the onset of endoreduplication rather than endomitosis. The nuclei in a pair of wild-type stomata cells represent 2C DNA content (Fig. 5E). In addition, prominent increase in DNA content due to salt treatment could be detected in the nuclei of wild-type and SOG1 overexpressor line plants, as evidenced from change in relative fluorescence of nuclei, with an average of 40.5 ± 12.4 versus 29.44 ± 8.4 detected for wild-type and 53.62 ± 10.8 versus 31.22 ± 9.2 detected for OE-1 seedlings (Fig. 5F). On the other hand no significant change in the size of root cell nuclei as well as DNA content could be detected in sog1-6 and sog1-1 mutant seedlings in response to salinity stress (Fig. 5D, third and fourth panel; Fig. 5F). Furthermore, 14-days old untreated and NaCl treated seedlings showed almost similar ploidy distribution pattern like that of the 7-days old seedlings with peaks of 16C and 32C were detected in the roots of salt treated wild-type seedlings. However, the percentage of the polyploid population was decreased as compared to proportions found in 7-days old seedlings (Supplementary Fig. S5A,B). Taken together these results have suggested that induction of endopolyploidy probably serve as an immediate response to salt induced DSBs.
Endoreduplication mediated root epidermal cell area expansion under salinity stress
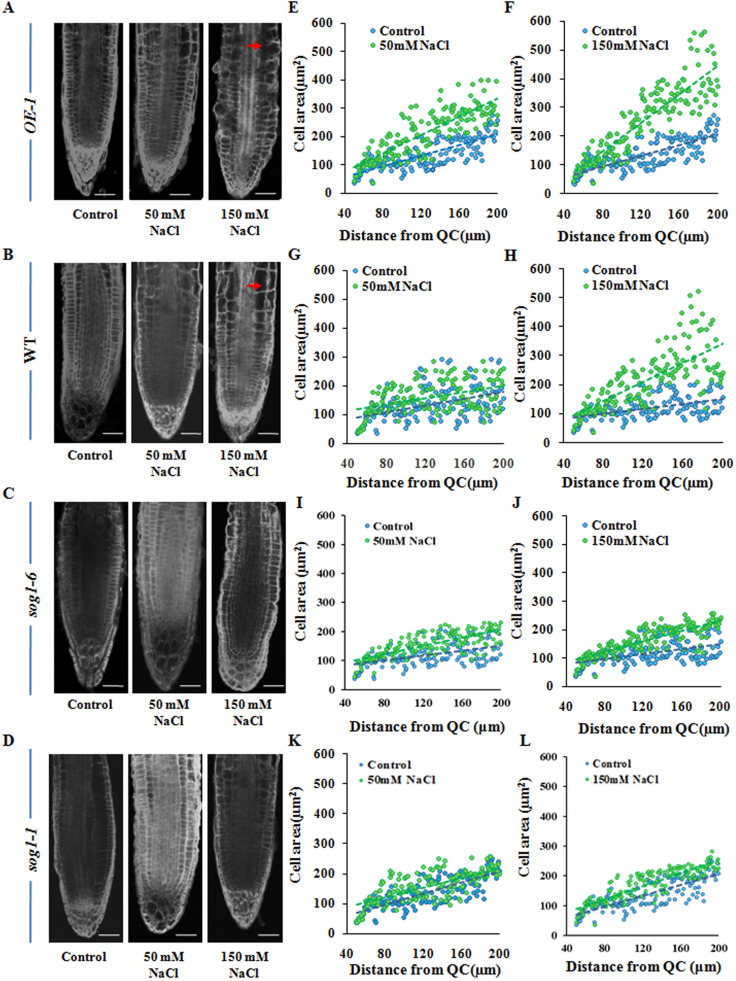
Earlier studies have demonstrated positive correlation between cell area and nuclear DNA content20. Endoreduplication has been shown to be associated with cell area expansion. In Arabidopsis thaliana, endoreduplication has been found to induce cell area expansion of the root tip cells at distances between 160 to 190 μm from the quiescent centre (QC)23,33. Under control condition, it has been shown that in Arabidopsis, endoreduplication generally does not occur in cells located < 200 μm from the QC in root tip epidermal cells. Our flowcytometry data have revealed salinity stress mediated induction of endoreduplication in the root tip cells of wild-type Arabidopsis along with the reduction in 2C DNA containing cell population and an increase in 16C, 32C and 64C cells in a dose dependent manner (Figs. 4B, 5B), indicating entry into the endocycle following salt exposure. This response was significantly compromised in sog1 mutant lines (Figs. 4C,D, 5C), while, as compared to wild-type plants, OE-1 line showed enhanced polyploid nuclear populations under salinity stress conditions (Figs. 4A, 5A). Based on this, we next investigated, whether altered expression of SOG1 and endoreduplication pattern is coupled with the change in cell area in root tip epidermal cells. As shown in Fig. 6, a dose dependent increase in cell area was observed in the root tip epidermal cells of wild-type Arabidopsis seedlings at distances between 160 and 190 μm from the QC following NaCl treatment (Fig. 6B,G,H). The cell expansion was especially prominent in the seedlings treated with 150 mM NaCl (Fig. 6B,H). This response was more prominent in OE-1 line than wild-type plants (Fig. 6A,E,F), while in sog1 mutant seedlings, no significant change in cell area in root tip epidermal cells between 150 and 200 μm from the QC could be detected following NaCl treatment than control condition (Fig. 6C,D,I–L). Taken together, these results have indicated that the onset of endocycle in the root tip epidermal cells of Arabidopsis seedlings under salinity stress is accompanied by cell area expansion and also suggested the involvement of SOG1 in the induction of endoreduplication under salinity stress.
Figure 6.
Salinity induced cell area expansion in the root tip epidermal cells. (A–D) Propidium iodide stained root tip cells of 7-days old OE-1, wild-type, sog1-6 and sog1-1 mutant seedlings respectively, either treated without or with 50 and 150 mM NaCl. Red arrows showing increase in cell size. Scale bar: 50 μm. (E–L) Measurement of area of root tip cells situated between 40 and 200 μm from the quiescent centre (QC) using ImageJ software (NIH). Regression lines included in (E–L); For OE-1 (E,F), R2 = 0.421 (Control), 0.598 (50 mM), 0.787 (150 mM); for wild-type (G,H), R2 = 0.507 (Control), 0.617 (50 mM), 0.713 (150 mM); for sog1-6 (I,J), R2 = 0.228 (Control), 0.236 (50 mM), 0.354 (150 mM) and for sog1-1 (K,L), R2 = 0.181 (Control), 0.191 (50 mM), 0.204 (150 mM). The F value was found to be < 0.001 for all regression analyses.
SOG1 promotes endopolyploidy in first and second pair of leaves under salinity stress along with cell area expansion and trichome branching
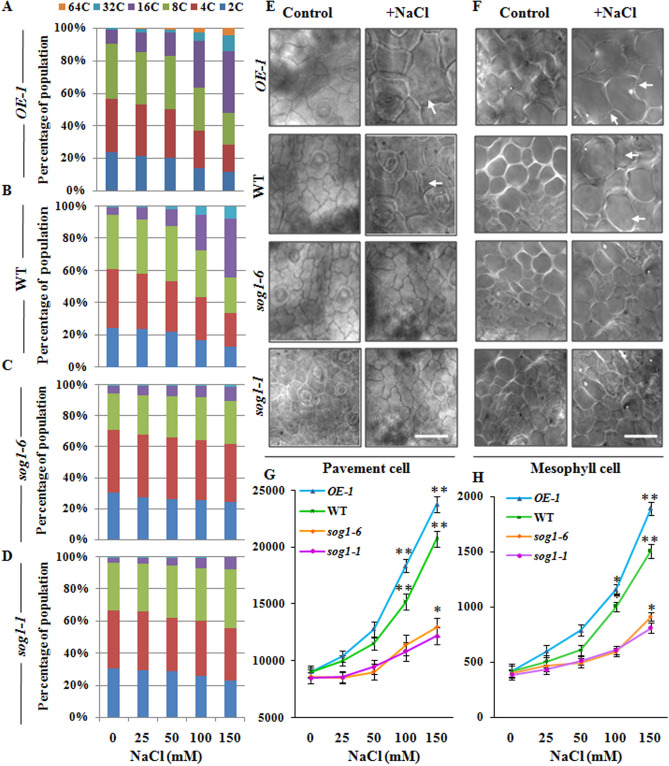
We next investigated whether SOG1 is also involved in the salinity stress mediated induction of endoreduplication in Arabidopsis rosette leaves. Previous studies have indicated presence of up to 16C DNA content in the first and second pair of rosette leaves in 7-days old seedlings, and the ploidy level increased as the leaves grow older34,35. Flowcytometric analysis using the nuclei isolated from first and second pair of leaves of untreated control and NaCl treated 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings have predominantly shown the presence of DNA content up to 16C in all the three genetic backgrounds under control condition (Fig. 7A–D). On the other hand, leaves of wild-type and particularly OE-1 seedlings treated with 150 mM NaCl showed significant increase in 32C nuclei population, which was found to be absent in the leaves of sog1 mutant seedlings under similar conditions (Fig. 7A–D). In addition, analysis of leaf cell area in 7-days old seedlings has clearly indicated expansion in both epidermal pavement cell (Fig. 7E,G) as well as mesophyll cell (Fig. 7F,H) area following NaCl treatment in wild-type Arabidopsis. The response was more prominent in the SOG1 overexpressor line, while only marginal change in leaf cell area under salinity stress was detected in sog1 mutant lines. Together, these results have indicated that apart from root epidermal cells, SOG1 also promotes endoreduplication in leaves of 7-days old Arabidopsis seedlings in presence of increasing concentrations of NaCl.
Figure 7.
Induction of endopolyploidy in leaf epidermal and mesophyll cells. (A–D) Flowcytometric analysis of ploidy level in the nuclei isolated from the first and second pair of leaves of SOG1 overexpressor line OE-1, wild-type, sog1-6 and sog1-1 mutant plants exposed to increasing concentrations of NaCl. (E,F) Change in cell surface area of epidermal pavement cells (Scale bar: 100 μm) and mesophyll cells (Scale bar: 50 μm) of SOG1 overexpressor line OE-1, wild- type, sog1-6 and sog1-1 mutant plants, either in absence or presence of NaCl. Leaf epidermal pavement cells and mesophyll cells in the basal region of primary leaves were photographed. White arrows indicating enlarged pavement and mesophyll cells. (G,H) Measurement of cell surface area of leaf pavement and mesophyll cells in leaves of all the four genotypes exposed to increasing concentrations of NaCl. Leaf surface areas were measured using ImageJ software (NIH). Data of the leaf surface area presented are the mean value of three independent biological replicates. Error bar represents ± SD (n = 40). Asterisks indicate a significant difference between untreated control and treated samples for each genotype based on one-way ANOVA factorial analysis (*p < 0.05, **p < 0.01).
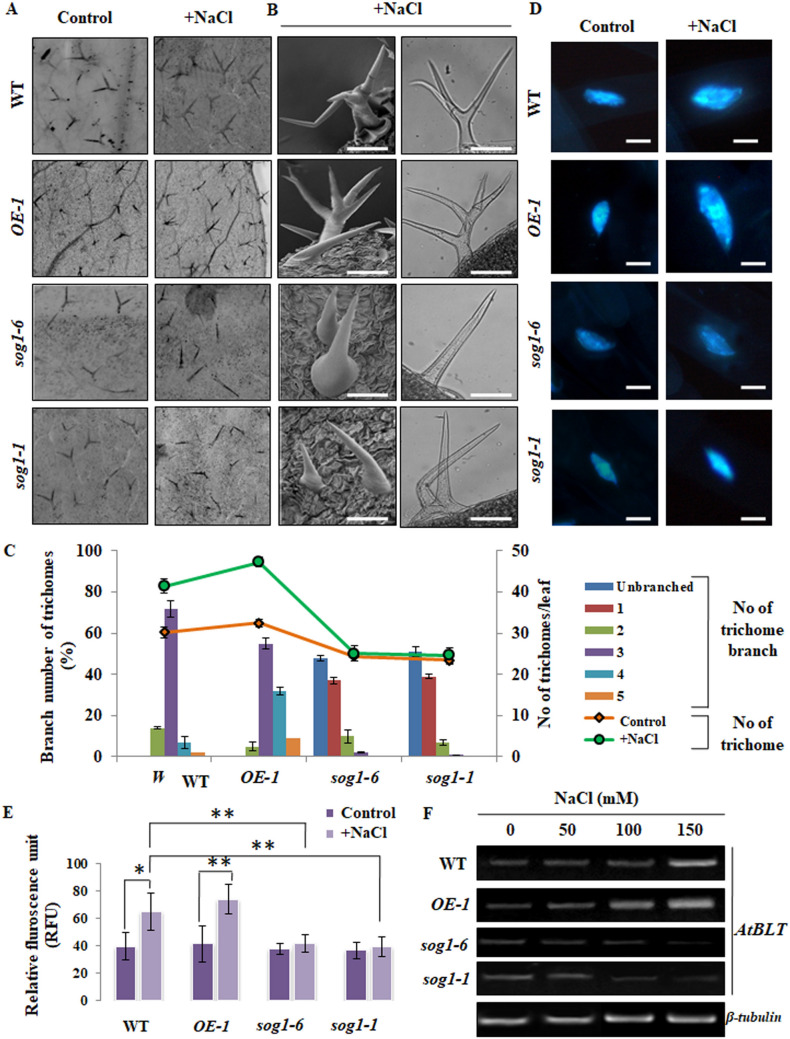
Besides pavement and mesophyll cells, trichome cells in Arabidopsis has been found to undergo several rounds of endoreduplication and reach up to ploidy level of 32C under natural condition36. In addition, branch number of trichomes has been shown to be strongly correlated with the nuclear DNA content36–38, suggesting role of endoreduplication in trichome branch initiation. Based on this, we next investigated whether altered expression level of SOG1 affects trichome density and branching pattern under salinity stress. Trichome density and branching pattern in 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant leaves in the absence and presence of salinity stress were analysed. As shown in Fig. 8A–C, as compared to wild-type plants, 150 mM NaCl treatment increased both density and branching pattern of trichomes in the leaves of OE-1plants, showing trichomes with more than three-branching (Fig. 8A,B, first and second panel; 8C).On the other hand, in sog1-6 and sog1-1 mutant plants, salinity stress reduced both density and branching of trichomes (Fig. 8A,B, third and fourth panel; 8C). Along with the density and branch number, significant change in DNA content in the nuclei of wild-type and OE-1 trichomes was detected in response to salinity stress, with an average of 61.2 ± 12.4 RFU versus 39.22 ± 12.4 RFU for wild-type and 79.4 ± 11.8 RFU versus 41.62 ± 14.5 RFU for OE-1 seedlings (Fig. 8D,E). Together, these results suggest the induction of additional rounds of endoreduplication in the leaf trichomes of wild-type and OE-1 seedlings.
Figure 8.
Change in trichome density, branch number and ploidy level in response to salinity stress. (A) Distribution of leaf trichome in 7-days old wild-type, OE-1, sog1-6 and sog1-1mutant plants, either untreated or treated with 150 mM NaCl. (B) SEM and bright field micrographs of mature trichomes with three branches in wild-type, five branches in OE-1 and unbranched in sog1-6 and sog1-1mutant line plants following NaCl treatment. Scale bar: 100 μm (C) Determination of leaf trichome density and branch number in all the four genotypes. (D) Representative images of DAPI-stained nuclei of 7-days old untreated or NaCl-treated wild-type, OE-1, sog1-6 and sog1-1 mutant plants. Scale bar: 100 μm (E) Measurement of in-situ fluorescence in DAPI-stained trichome nuclei of wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings. (F) Semi quantitative RT-PCR showing expression level of AtBLT gene in all genotypes in response to increasing NaCl concentrations. β-tubulin served as the internal control. Each column represents the mean value of three independent replicates. Images of full-length gels have been presented in Supplementary Fig. S12. Error bars indicate standard deviation. Asterisks indicate significant statistical differences (*p < 0.05, **p < 0.01) from control (without NaCl treatment) using one-way ANOVA factorial analysis.
In Arabidopsis, AtBLT (BRANCHLESS TRICHOMES) gene has been shown to be associated with branching of trichomes and endoreduplication39. Based on nature of trichome branching pattern in OE-1 and sog1 mutant lines under salinity stress, we next investigated whether alteration in the expression level of AtSOG1 affects the expression of AtBLT. Reverse transcription semi quantitative PCR analysis using total RNA from 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings have indicated increased expression level of AtBLT in wild-type and OE-1 line under salinity stress (Fig. 8F). AtBLT expression increased prominently in OE-1 line than wild-type plants under salinity stress (Fig. 8F, first and second panel; Supplementary Fig. S6A). On the other hand, AtBLT expression did not increase significantly in sog1-6 and sog1-1 mutant lines under salinity stress (Fig. 8E, third and fourth panel; Supplementary Fig. S6A). Together, these results indicate that SOG1 upregulates AtBLT expression under salinity stress and could be linked with its effect on endoreduplication and increased trichome branching in presence of higher concentration of NaCl.
Both ATM-SOG1 and ATR-SOG1 pathways participate in salinity mediated induction of endoreduplication in Arabidopsis
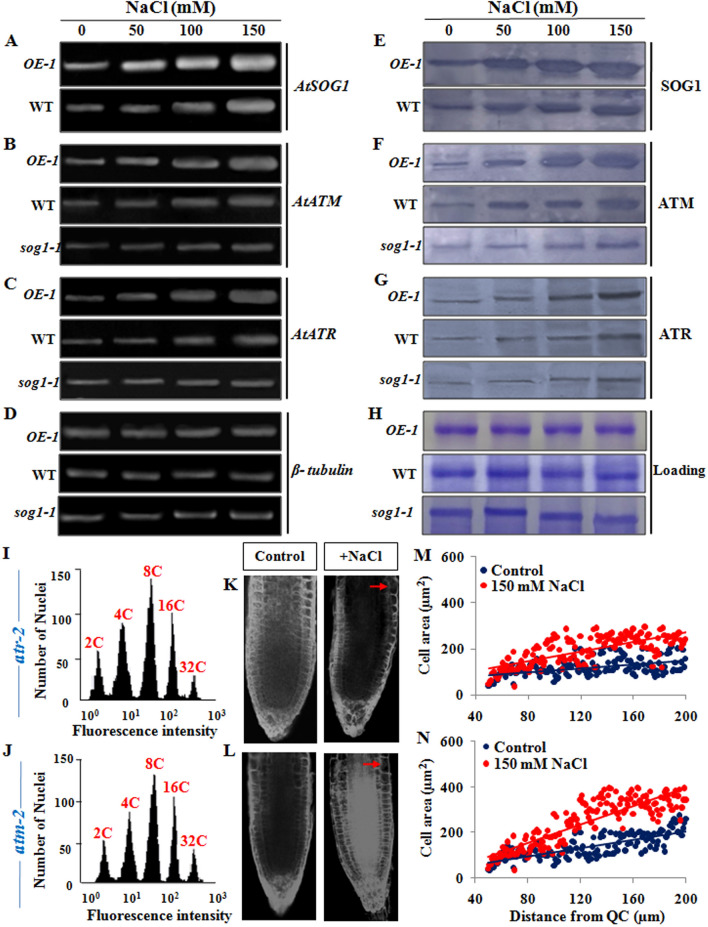
Our results have indicated salinity stress mediated predominant induction of DSBs together with concomitant onset of endopolyploidy in Arabidopsis roots and leaves. Endoreduplication phenotype under salinity stress was more prominent in OE-1 plants than wild-type Arabidopsis, suggesting role of SOG1. Since, SOG1 acts as the central regulator in modulating DNA damage response and found to operate through the ATM-ATR mediated signalling pathway13,15, we next investigated whether SOG1 coupled ATM-ATR pathway is involved in salinity stress mediated induction of endoreduplication. We have analysed the accumulation of ATM and ATR transcripts and protein levels in 7-days old wild-type, OE-1and sog1-1 mutants in the absence or presence of salinity stress (Fig. 9A–C,E–G; Supplementary Fig. S7) Semi quantitative RT-PCR analyses using the total RNA extracted from 7-days old seedlings have revealed that NaCl treatment caused enhanced expression of SOG1 along with ATM and ATR in wild-type and OE-1 seedlings in a dose dependent manner (Fig. 9A–C; Supplementary Fig. S7A–C). This indicates the role of SOG1 in mediating plants’ response under salinity stress. In OE-1 line, transcript abundance of ATM and ATR enhanced approximately 2.5- and 2-folds, respectively as compared to wild-type plants, while only marginal increase in ATM and ATR expression level was found in sog1-1 mutant seedlings in the presence of increasing salinity (Fig. 9B,C, Supplementary Fig. S7B,C), suggesting that loss of SOG1function affects ATM and ATR expression under salinity stress conditions. In each set of samples, transcript quantity was normalized against β-tubulin (Fig. 9D) after background subtraction. Consistent with transcript expression levels, as compared to untreated control, SOG1 protein level increased steadily (p < 0.01) in presence of increasing concentrations of NaCl in wild-type and OE-1 Arabidopsis seedlings (Fig. 9E; Supplementary Fig. S7D). In addition, an enhanced accumulation level of ATM (p < 0.01) and ATR (p < 0.01) proteins could be detected in wild-type and more prominently in OE-1 line than sog1-1 mutant line in presence of increasing concentrations of NaCl (Fig. 9F,G, Supplementary Fig. S7E,F). Protein accumulation was normalized against loading control for each genotype (Fig. 9H). Together, increased transcript abundance of SOG1 in wild-type plant along with enhanced expression level of ATM and ATR following exposure to high salinity have suggested the role of SOG1 in modulating the cellular response in Arabidopsis under salinity stress via the classical ATM-ATR signalling pathway. The accumulation level of ATM and ATR proteins under salinity stress condition in the three different genetic background including wild-type, OE-1 and sog1-1 also supported this notion.
Figure 9.
Salinity induced activation of ATM-SOG1 and ATR-SOG1 pathways. (A) Semi-quantitative reverse transcription-PCR analysis showing expression level of AtSOG1 in OE-1 and wild-type seedlings exposed to increasing concentrations of NaCl for 12 h. (B–D) Transcript accumulation levels of AtATM, AtATR and β-tubulin genes, respectively in OE-1, wild-type and sog1-1 seedlings exposed to increasing concentrations of NaCl for 12 h. β-tubulin served as the housekeeping gene. Quantitative analyses of relative expression of AtSOG1, AtATM and AtATR genes have been presented in Supplementary Fig. S7. Images of full-length gels have been presented in Supplementary Fig. S13. (E) Detection of protein accumulation level of SOG1 in OE-1 and wild-type seedlings exposed to increasing concentrations of NaCl for 12 h. (F–G) Detection of protein accumulation levels of key DNA damage response transducers, including ATM and ATR, respectively in 7-days old OE-1, wild-type and sog1-1 seedlings exposed to increasing concentrations of NaCl. The blots were cut prior to incubation with the respective primary antibodies. (H) Respective loading control of total protein extracts from OE-1, wild-type and sog1-1 seedlings respectively. Immunoblotting was performed by using ~ 50 µg of total protein extracts. Representative gel blot images from at least three independent experiments are shown. Replicates of the protein gel blots have been presented in Supplementary Fig. S14. (I,J) Flowcytometric analysis of ploidy level in the propidium iodide DAPI-stained root cell nuclei of 7-days old atr-2 and atm-2 mutant plants treated without or with 150 mM NaCl. (K,L) Propidium iodide stained root tips of 7-days old atr-2 and atm-2 mutant seedlings respectively, either treated without or with 150 mM NaCl. Red arrows showing enlarged root epidermal cells. Scale bar: 50 μm. (M,N) Measurement of area of root tip cells situated between 40 and 200 μm from the quiescent centre (QC) using ImageJ software (NIH). Regression lines included in (M,N); for atr-2 (M) R2 = 0.224 (Control), 0.506 (150 mM) and for atm-2 (N) R2 = 0.597 (control), 0.744 (150 mM).
To further substantiate the involvement of both ATR and ATM kinases in modulating salinity induced endoreduplication, we next examined the salinity induced endoreduplication and cell expansion in the roots of atr-2 and atm-2 mutants. Endopolyploidy along with cell expansion was observed in both atr-2 and atm-2 mutants following salt treatment (Fig. 9I–N), but both endopolyploidy and expansion was notably lesser than either wild-type or OE-1 line. This trend was similar as shown with zeocin treatment23, but interestingly in our study, salinity induced cell area expansion was found to be more affected in atr-2 mutant as compared to atm-2 mutant, suggesting the crucial involvement of ATR kinase in salinity induced endoreduplication (Fig. 9K–N). Taken together, these results have provided important clue to suggest the induction of replication stress and genotoxicity due to generation of DSBs under salinity stress, resulting in the activation of DNA damage response via SOG1-mediated ATM-ATR pathways.
SOG1 downregulates the M-phase specific signal under salinity stress
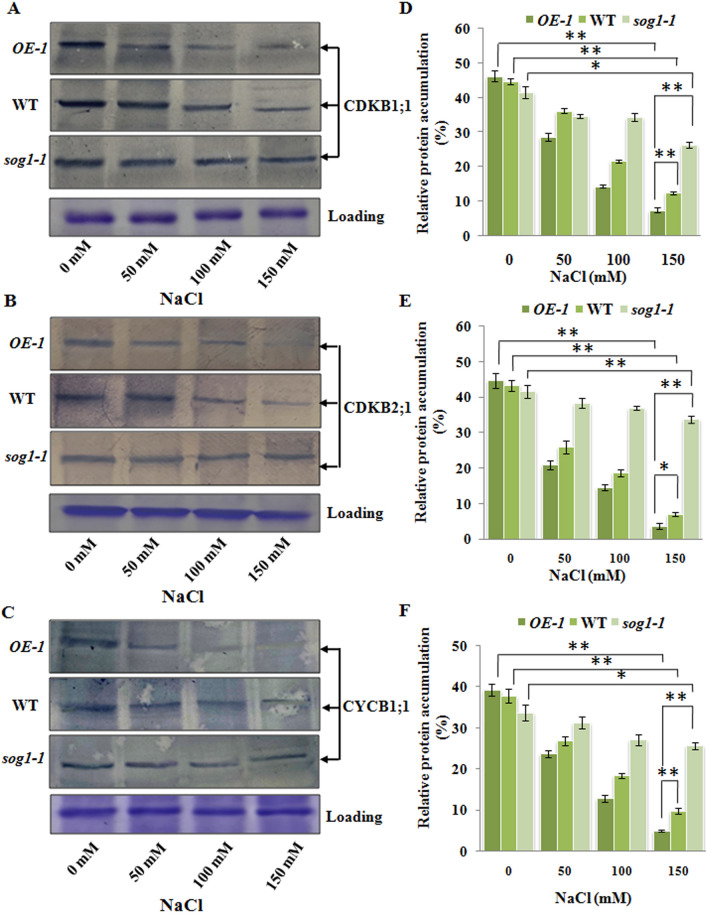
The crucial step for endoreduplication involves the re-entry into the S-phase from G2-phase and simultaneous block of M-phase40. The activities of CDK/cyclin complexes serve as key components regulating the entry of cell from mitotic division into endoreduplication cycle41,42. Downregulation of CDK/cyclin kinase activity has been shown to prevent initiation of mitosis, without inhibiting DNA replication, thus facilitating entry into endoreduplication43,44. As like other eukaryotes, in plants, cell cycle progression is tightly regulated at G1-S and G2-M checkpoints by the activities of specific CDKs-cyclin complexes. G1-S transition has been found to be mainly governed by CDKB1;1, while CYCB1;1/CDKB2;1 complex plays major role in regulating progression of cell cycle through G2-M checkpoint45–48. Moreover, CDKB1;1 activity has been shown to be required for inhibition of endoreduplication49. Our results have provided clue regarding functional involvement of SOG1 in salinity stress mediated induction of endoreduplication. To further investigate this signaling network at the cellular level, we next analysed the accumulation level of some key cell cycle regulators including CDKB1;1, CDKB2;1 and CYCB1;1 in 7-days old wild-type Arabidopsis, SOG1 overexpression line OE-1 and sog1-1 mutant line in the absence and presence of salinity stress. Immunoblot analyses have revealed decreased accumulation levels of CDKB1; 1, CDKB2; 1 and CYCB1; 1 along with the increasing concentrations of NaCl (p < 0.01, p < 0.05). As compared to wild-type seedlings, the reduction of the cell cycle regulatory proteins was relatively higher in OE-1 line under salinity stress (Fig. 10A–C, first and second panel; D–F), suggesting induction of strong signal for inhibition of entry into M phase and thus creating the situation for the onset of endoreduplication. On the other hand, in sog1-1 mutant seedlings only marginal change in the accumulation level of all the three proteins was detected under salinity stress, indicating lack of adequate signal for the onset of endoreduplication due to loss of SOG1 function (Fig. 10A–C, third panel; D–F).
Figure 10.
SOG1 is found to control the entry into M-phase and induce cell cycle arrest under salinity stress. (A–C) Detection of protein accumulation levels of key regulators of cell cycle progression and endoreduplication including CDKB1;1 (A), CDKB2;1 (B), CYCB1;1 (C), respectively in 7-days old OE-1, wild-type and sog1-1 seedlings exposed to increasing concentrations of NaCl for 12 h. Loading control of total protein extracts has been presented in the lowermost panel. Immunoblotting was performed by using ~ 50 µg of total protein extracts. The blots were cut prior to incubation with the respective primary antibody\ies. Representative gel blot images from at least three independent experiments are shown. Replicates of the protein gel blots have been presented in Supplementary Fig. S15. (D–F) Quantification of band intensity to determine the accumulation levels of CDKB1;1 (D), CDKB2;1 (E), CYCB1;1 (F) proteins as detected by immunoblotting. Densitometry analysis was carried out by ImageJ software (NIH). Error bar indicates standard deviation. Asterisks indicate significant statistical differences (*p < 0.05, **p < 0.01) from control (without NaCl treatment) using one-way ANOVA factorial analysis.
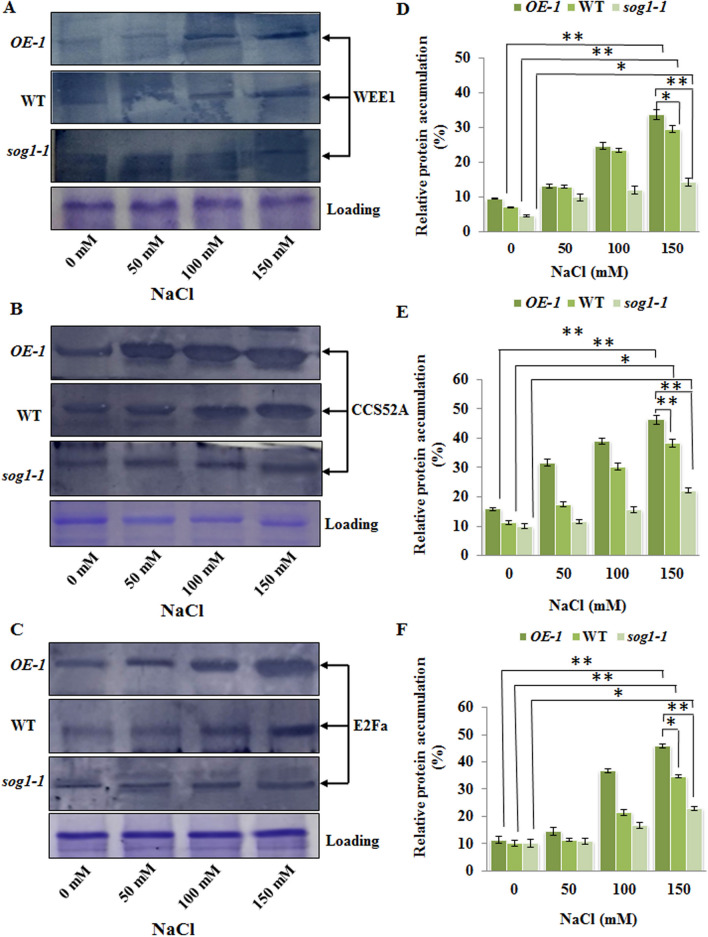
Apart from the CDK-cyclin complex, WEE1 kinase represents another crucial regulatory component of cell cycle progression in plants and acts as potential inhibitor of CDKB activity via phosphorylation, resulting in cell cycle arrest in response to DNA damage50. Earlier studies have indicated that expression of WEE1 is controlled by SOG1 under DNA damaging conditions51. Our results have also shown that accumulation level of WEE1 protein kinase increased appreciably in OE-1 line than wild-type plants under salinity stress (p < 0.01) (Fig. 11A, first and second panel; 11D), while as compared to untreated control condition, WEE1 protein level did not increase significantly in sog1-1 mutant under salinity stress (p < 0.05) (Fig. 11A, third panel; 11D). Taken together, decreased level of CDKB1;1, CDKB2;1 and CYCB1;1 along with increased accumulation of WEE1 in wild-type Arabidopsis and particularly in OE-1 line following exposure to higher NaCl concentrations have indicated role of SOG1 mediated pathway in the downregulation of cellular signal for entry into M-phase of cell cycle and thus generating suitable condition for entry of cell into endoreduplication under salinity stress.
Figure 11.
SOG1 is important in controlling cell cycle progression and entry into endocycle in response to salinity stress. (A–C) Detection of protein accumulation levels of key regulators of cell cycle progression and endoreduplication including WEE1 (A), CCS52A (B) and E2Fa (C), respectively in 7-days old OE-1, wild-type and sog1-1 seedlings exposed to increasing concentrations of NaCl for 12 h. The blots were cut prior to incubation with the respective primary antibody. Loading control of total protein extracts has been presented in the lowermost panel. Representative gel blot images from at least three independent experiments are shown. Replicates of the protein gel blots have been presented in Supplementary Fig. S16. (D–F) Quantification of band intensity for determining the accumulation levels of WEE1 (D) CCS52A (E), E2Fa (F) proteins as detected by immunoblotting. Densitometry analysis was carried out by ImageJ software (NIH). Error bar indicates standard deviation. Asterisks indicate significant statistical differences (*p < 0.05, **p < 0.01) from control (without NaCl treatment) using one-way ANOVA factorial analysis.
SOG1 overexpression enhances FZR/CCS52A and E2Fa accumulation in Arabidopsis under salinity stress
Earlier studies in yeast and Drosophila have indicated that the Fizzy-Related (FZR) family of proteins activate the degradation of A- and B-type cyclins via targeting them to the Anaphase Promoting Complex/ Cyclosome (APC/C) and induce endoreduplication52. In plants, the FZR homologues, Cell-cycle Switch 52A and B (CCS52A and CCS52B) have been shown to act as an activator of the APC/C and promote transition to the endocycle during development53. In Arabidopsis, down-regulation of FZR2/CCS52A1 has been shown to significantly reduce endoreduplication and expansion of trichome and other leaf cells, indicating its crucial role in the induction of endoreduplication54,55. In addition, previous studies in Medicago truncatula have demonstrated that salinity induced endoreduplication is accompanied by enhanced transcript accumulation of CCS52 and WEE1 and maximum ploidy level was observed at 100–150 mM NaCl, which also corresponded to the peak level of expression of CCS52 and WEE117. Furthermore, in plants, the E2Fa transcription factor along with its dimerization partner DPa, act as key regulators, controlling the proliferative status of the cell in Arabidopsis56,57. In endoreduplicating cells, E2Fa-DPa transcription factor induce expression of S-phase genes resulting in additional DNA replication after being released from RETINOBLASTOMA-RELATED (RBR) repression56,58,59. In addition, the activity of CDKB1;1 and E2Fa-DPa has been found to maintain the balance between mitotically dividing cells and endoreduplicating cells in plants49. Our results have shown compromised expression of CDKB2;1-CYCB1;1 along with the enhanced accumulation level of WEE1 protein in OE-1 line, which showed increased population of polyploid nuclei than wild-type plants under salinity stress (Fig. 4; Table 1). Furthermore, protein gel blot analyses have indicated enhanced accumulation of CCS52A protein in OE-1 than wild-type Arabidopsis seedlings in presence of increasing concentrations of NaCl with maximum accumulation detected at 150 mM NaCl (p < 0.01, p < 0.05) (Fig. 11B, first and second panel; E). On the other hand, in sog1-1 mutant line CCS52A accumulation level did not increase significantly under salinity stress (p < 0.05) (Fig. 11B, third panel; E). In addition, in both wild-type and OE-1 line, E2Fa protein accumulation level increased significantly with increasing concentrations of NaCl (p < 0.01, p < 0.05) (Fig. 11C, first and second panel; F), indicating induction of endoreduplication. However, sog1-1 mutant line showed only marginal change in E2Fa protein level under salinity stress (p < 0.05) (Fig. 11C, third panel; F). Together, the enhanced accumulation of CCS52A, WEE1 and E2FA in OE-1 line as compared to wild-type plants have indicated functional involvement of SOG1 in the induction of endoreduplication in Arabidopsis seedlings under salinity stress through the modulation of CDKB1;1, CDKB2;1, CYCB1;1, WEE1, CCS52A and E2Fa protein level and therefore, generation of appropriate signal for the induction of endocycle.
Discussion
Salinity stress impairs plant growth and development via osmotic and hyperionic stress due to reduction in water content and excessive accumulation of Na+ and Cl− ions respectively. In addition, salinity is typically accompanied by oxidative stress due to generation of reactive oxygen species (ROS), which can challenge genome stability and induce DNA lesions7–9. Plants have evolved with several strategies to combat against soil salinity. Multiple studies have demonstrated that salt tolerance and adaptation in plants are governed by various physiological and molecular mechanisms60,61. However, not much information is available about how plants respond to salinity induced genotoxicity and genome instability. Various plants including Sorghum bicolor (sorghum), Mesembryanthemum crystallinum and Medicago truncatula have been found to exhibit additional rounds of endoreduplication in the cells of different organs besides developmental endopolyploidy along with cell area expansion in response to salinity stress16–18. Moreover, recent studies in Arabidopsis thaliana have also revealed that double strand breaks (DSBs) induced by genotoxic agents promote the cells to exit from mitosis and enter into endocycle as a programmed response to DNA damage23. These observations have indicated that plants might switch to endoreduplication cycle as a potential strategy to cope up under stressful conditions.
In this study, we bring further evidence to show that the induction of endoreduplication under salinity stress in Arabidopsis thaliana represents an immediate adaptive response to salinity induced DSBs in the genome. We have further demonstrated the involvement of SOG1 transcription factor in the regulation of onset of endoreduplication in Arabidopsis in response to DSBs generated under salinity stress.
In our study, morphological analyses involving wild-type Arabidopsis, SOG1 overexpressor line (OE-1), sog1-6 and sog1-1 mutant lines have revealed reduced germination performance in both the mutant genotypes as compared to wild-type and overexpressor line in response to increasing NaCl concentrations (Supplementary Fig. S1A–C). Interestingly, root growth in sog1-6 and sog1-1 mutant seedlings was found to be less sensitive in response to increasing NaCl concentrations as compared to wild-type and OE-1 plants (Fig. 1A–C). This indicates maintenance of normal cell cycle, rather than cell cycle arrest in both the mutant lines in absence of functional AtSOG1 gene. On the other hand, wild-type and OE-1 plants with functional AtSOG1 gene displayed controlled root growth in response to increasing salinity. However, at higher NaCl concentration (150 mM), root growth was also drastically affected in sog1 mutants within seven days after germination, indicating cessation of growth (Fig. 1C). This root growth pattern in sog1 mutants under salt stress has been found to be comparable to the response as indicated in the previous report in response to zeocin and aluminium treatment, where mutants of AtSOG1 (sog1-1 and sog1-7, respectively) showed lower sensitivity, as evidenced from less reduction in root growth in sog1 mutants than wild-type plants following zeocin and aluminium treatment23,62. In this study, we also observed relatively less sensitivity of root growth in sog1-6 and sog1-1 mutant as compared to wild-type and OE-1 line under increasing salinity, while germination percentage reduced significantly in both the mutants than the other two genotypes. Previous studies have shown that ROS have been found to play crucial role in dormancy release during seed dry storage through the direct oxidation of a subset of biomolecules63. Moreover, recent studies have also indicated the involvement of SOG1 in oxidative stress response and ROS homeostasis maintenance under abiotic stress conditions64. In sog1 mutants with no functional SOG1 protein, the ROS homeostasis might be disrupted, resulting in induction of oxidative stress and significant damage to the integrity of the genome of seed embryo and thus may lead to delayed germination and reduced germination frequency. Still the reason behind this apparent discrepancy between root growth and germination performance remains puzzling and requires further investigations.
Earlier investigations have demonstrated that salinity stress induces oxidative stress via the excess accumulation of ROS and thus induce genome instability because of oxidative damage of DNA14,21,65. As mentioned above, recent studies have indicated role of SOG1 in oxidative stress response under heavy metal stress conditions78 and regulation of oxidative stress responsive gene OXII, which activates MAPK3 and MAPK6 through phosphorylation following ROS production64, has been shown to be one of the direct targets of SOG151. These observations have suggested involvement of SOG1 in the regulation of oxidative stress response. In the present study, we have found that increasing salt treatment caused significant elevation in ROS generation in the roots of sog1-1 and sog1-6 mutants as compared to wild- type and SOG1 overexpressor line, where ROS generation was found to be relatively less (Fig. 2), indicating role of SOG1 in the regulation of salinity induced oxidative stress response. Furthermore, we have found strong signal for the induction of DNA double strand breaks (DSBs) in Arabidopsis seedlings under salinity stress (Fig. 3A,B). However, as compared to wild-type and OE-1 plants, sog1-6 and sog1-1 mutants showed appreciably higher accumulation level of DSBs along with the slower repair rate (Fig. 3A,B). Together, these results have indicated that accumulation of DSBs in response to salinity possibly depends on the magnitude of oxidative stress and loss of SOG1 function impairs the efficiency of DSB repair in sog1-6 and sog1-1 mutant plants under salinity stress and thus also confirm the crucial role of SOG1 in the regulation of salinity induced DNA damage response and repair in plant genome.
Along with the salinity stress mediated induction of DSBs, we have detected significant population of polyploid nuclei of with up to 64C DNA content in the root tip cells and leaves of wild-type Arabidopsis seedlings in presence of higher concentrations of NaCl, indicating occurrence of additional rounds of endocycles beside the normal developmental polyploidy. Furthermore, transgenic Arabidopsis line overexpressing SOG1(OE-1) showed nuclear DNA ploidy level up to 128C in the root tip cells along with increased nuclei size (Figs. 4A, 5A,D,F). Consistent with nuclear size and DNA content, wild-type and OE-1 plants exhibited prominent increase in root tip epidermal, leaf epidermal (pavement) and mesophyll cell sizes, suggesting the onset of endoreduplication (Figs. 6A,B, 7). On the other hand, the induction of endoreduplication under salinity stress was significantly compromised in sog1-6 and sog1-1 mutant seedlings (Figs. 4C,D, 5C,D,F, 6C,D, 7). Together, these results have suggested function of SOG1 in the induction of endoreduplication in Arabidopsis under salinity stress.
The genotoxic effect of salinity stress appears to be mainly accomplished by the induction of oxidative stress through the generation of reactive oxygen species (ROS)65,66. Excess level of intracellular ROS accumulation induces various forms of mutagenic oxidative lesions in the DNA, including 8-oxo-G and 1,2-dihydro-2-oxoadenine (2-OH-A)67. These lesions are mainly repaired via base excision repair (BER) pathway, but when they occur simultaneously on opposing strands due to prolonged stress conditions, attempted BER may lead to the generation of additional potentially very harmful DNA lesions such as DNA single stand breaks (SSBs) and double strand breaks (DSBs)68. Higher extent of accumulation or slower rate of repair of DSBs often results in structural abnormalities in chromosomes and thus inhibits DNA replication, generating replication stress. Furthermore, ROS oxidize dNTPs to disrupt DNA polymerase activity, leading to reduced replication fork velocity in vitro69. Oxidized bases generated due to ROS activity also acts as physical obstacle to replication forks and causing breakdown of replication forks at fragile sites across the genome and again results in the formation of DSBs70. ATM is predominantly activated due to generation of DSBs in the genome71, while replication stress strongly activates ATR activity. Both these events eventually lead to the activation of DNA damage response and cell cycle arrest via the coordination of SOG115. Therefore, it seems that salinity stress generates both DSBs and replication stress through ROS activity. Our results involving expression studies of ATM, ATR and SOG1 in wild-type Arabidopsis have shown steady state increase in the accumulation of these genes at both transcript and protein level along with increasing concentrations of NaCl (Fig. 9A–C,E–G), suggesting involvement of both ATM and ATR along with SOG1 (Fig. 12). On the other hand, we have found increased expression of ATM and ATR in OE-1 plant under salinity stress, while expression levels of ATM and ATR did not increase significantly in sog1 mutants (Fig. 9A–C,E–G). Another interesting fact is that although ATM mediated phosphorylation of SOG1 is important for regulating the amplitude of DNA damage response in plants26, possible role of SOG1 in regulating the phosphorylation pattern of the DNA damage response related proteins has not been characterized so far. In some of our preliminary experiments using in vitro phosphorylation assays, we failed to detect any phosphorylating activity of SOG1 to some of its well-known targets, including RAD51 and BRCA1 (which are already found to be transcriptionally regulated by SOG151).
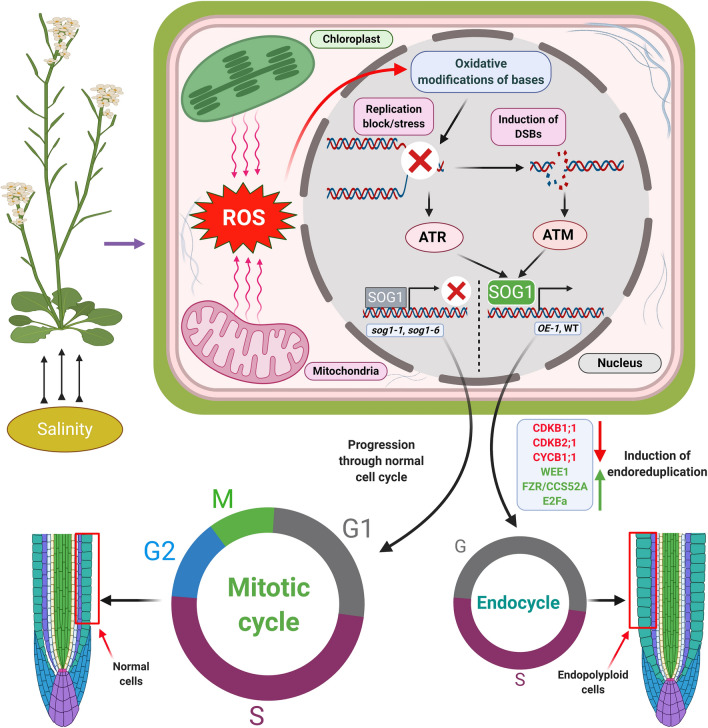
Figure 12.
Salinity induced onset of endoreduplication in Arabidopsis thaliana is mediated by SOG1. Increasing salinity induces replication stress via oxidative modifications of bases, resulting into accumulations of DSBs. ATR-SOG1 and ATM-SOG1 pathways are activated following replication stress and accumulation of DSBs and regulate the expression of various regulators of cell cycle progression and endoreduplication including, CDKB1;1, CDKB2;1, CYCB1;1, CCS52A, WEE1 and E2Fa, resulting in entry into the endocycle. The figure has been created with BioRender.com.
Earlier studies have demonstrated that cells can skip mitosis and enter into endoreduplication by lowering the CDK/cyclin kinase activity and continue to replicate their genomic DNA, resulting in increased DNA content56,72,73. In our study, consistent with the enhanced expression of ATM and ATR, accumulation levels of key cell cycle regulators, including CDKB1;1, CDKB2;1, and CYCB1;1 declined significantly (Fig. 10A–F), while WEE1, E2Fa and CCS52A protein level increased concomitantly in OE-1 plant under salinity stress (Fig. 11A–F). These alterations in the accumulation level of the key cell cycle regulators were appreciably less prominent under sog1 mutant background (Figs. 10A–F, 11A–F). Together, these results have indicated activation of strong signal for cell cycle arrest at G2-M checkpoint in OE-1 line than sog1 mutant lines and therefore, implicating role of SOG1 in this process involving both ATM and ATR. In addition, apart from wild-type plants, although salinity stress has effectively induced DSBs in both OE-1 and sog1 mutants, in OE-1 line, enhanced ATM and ATR level with the concomitant decline in expression of CDKB2;1-CYCB1;1 stimulated cell cycle arrest at G2-M checkpoint, resulting in root growth inhibition, while increased accumulation of E2Fa and CCS52A facilitated effective increase in DNA replication and induction of endoreduplication under salinity stress in OE-1 line more prominently than wild-type Arabidopsis and sog1 mutant lines (Fig. 12).
Previous studies have demonstrated that members of the SIAMESE/SIAMESE-RELATED (SIM/SMR) class of cyclin-dependent kinase inhibitors including SMR5 and SMR7 activate a cell cycle checkpoint in response to ROS-induced DNA damage, resulting in repression of mitosis and onset of endoreduplication73–75. SMR5 and SMR7 are activated through ATM-SOG1 pathway and they are the direct targets of SOG151,64,75. We have also found strong induction of SMR5 and SMR7 expression in wild-type and OE-1 seedlings in response to increasing salinity, but expression was repressed in sog1-1 seedlings, indicating possible involvement of SMR5 and SMR7 in SOG1 mediated salinity induced endoreduplication (Supplementary Fig. S8A,B).
Earlier studies have described that FZR2/CCS52A1 is a key determinant in endoreduplication and cell expansion in Arabidopsis thaliana53. Our results have also shown enhanced accumulation of CCS52A protein level in the presence of functional AtSOG1 gene under increasing salt stress (Fig. 11B). However, no information is available on the transcriptional regulation of both CCS52A1 and CCS52A2 genes by SOG1. Interestingly, we have found some conserved NAC domain binding nucleotide sequences (gaGTCAAca) in the promoters of both CCS52A1 and CCS52A2 genes, approximately 1000 base pairs upstream of the transcriptional start site through in-silico studies. In a separate study, we have detected specific promoter binding of SOG1 protein to this motif both in vitro and in vivo. The binding affinity was found to be increased significantly under salinity stress in vitro. Moreover, we have also found enhanced expression of SOG1 and CCS52A1 in parallel in wild-type Arabidopsis under prolonged salinity (unpublished data). These preliminary observations have suggested that CCS52A1 could be a possible target of SOG1. Consistent with this notion, enhanced expression of CCS52A under SOG1 overexpression line under salinity stress along with the prominent induction of endoreduplication has suggested one possible effective adaptive mechanism in Arabidopsis to survive under salinity stress condition. However, further research is required to understand the additional important interlinked mechanisms.
Materials and methods
Plant material and growth conditions
The wild-type Arabidopsis thaliana and the mutants, including sog1-1, sog1 T-DNA insertion mutant line (SALK_067631C), atm-2 and atr-2 used in the present study are in Col-0 background. The sog1-1, atm-2 and atr-2 mutant seeds were the generous gift from Prof. Anne Britt, University of California. The Arabidopsis sog1‐1 mutant, carrying a missense mutation affecting a highly conserved amino acid in the NAC domain, was originally isolated as a suppressor of the gamma‐sensitive phenotype observed in the DNA repair‐defective xpf mutant and has been characterized previously25,76. The atm-2 and atr-2 are T-DNA knockout mutant lines of AtATM and AtATR respectively and have been characterized previously77. The sog1 T-DNA mutant line (SALK_067631C) was obtained from the T-DNA knockout collections at the Arabidopsis Biological Resource Center (ABRC) and was confirmed for homozygosity by PCR genotyping using specific primers (Supplementary Fig. S9A). Individual plants were examined by PCR using the left border specific primer LBb1.3 (59-ATTTTGCCGATTTCGGAAC-39) and the SALK line specific primers (Supplementary Table 1).The specific position of the T-DNA insertion in AtSOG1 gene was found to be in exon 6 (Supplementary Fig. S9B) and the mutant has been subsequently named as sog1-6. For the generation of SOG1 overexpression lines, the full length CDS of SOG1 (1350 bp) was amplified using specific primers and cloned into the Nco1-Spe1 sites of pCAMBIA1303 binary vector under the control of the constitutive promoter CaMV 35S. The overexpression construct was then transformed into Agrobacterium GV3101 strain. The transgene was then Agro-infiltrated into the wild-type Col-0 plants by floral dip method and the resulting transformants carrying the desired transgene were selected in MS plates containing kanamycin (25 mg/ml). Among the three overexpression lines generated namely OE-1, OE-2 and OE-3, based on the growth performance OE-1was selected for the further study (Supplementary Fig. S10A–C). AtSOG1 transcript and protein expression analyses have indicated that sog1-6 (SALK_067631C) and sog1-1 represent true null mutation lines, while OE-1 transgenic line is a SOG1 overexpressor line (Supplementary Figs. S9C,D, S10D,E). Surface sterilized seeds were grown by essentially following the method described previously14. Briefly, seeds were grown in potting soil (Soilrite) or growth medium supplemented with 0.8% Bactoagar and 1% Sucrose. The seeds in the potting soil or on growth medium-agar plates were cold treated at 4 °C for 3–5 days and then transferred to light chambers and maintained at 22 °C with 16-h-light/8-h-dark periods. To examine the growth response of the SOG1 overexpressor line, OE-1 and sog1 mutants, sog1-1 and sog1-6 along with wild-type (Col-0) Arabidopsis in the presence of increasing concentrations of NaCl during germination and early seedling stage, surface-sterilized seeds were spread on MS agar (containing 0.8% Bactoagar and 1% Sucrose) supplemented without (untreated control) or with different concentrations of NaCl (25, 50, 100 and 150 mM, respectively). After stratification period of 3 days in cold and dark conditions, plates were transferred to growth chambers maintained at 22 °C with a 16-h: 8-h light: dark daily schedule. Germination frequency was calculated three days after germination. One week later, plants were screened for their sensitivity and overall growth response. Growth performance of all the four genotypes was also examined in the presence of LiCl, which served as the negative control.
Root growth assay
Seeds of wild-type Col-0, OE-1, sog1-6 and sog1-1 knockout mutant lines were surface sterilized and placed on ½ MS medium (0.8% Bactoagar and 1% Sucrose) supplemented with different concentrations of NaCl on vertical plates and subjected to cold treatment for 3 days. After the stratification period, the plates were placed in the growth chamber with desired temperature and light conditions. Seedlings were grown under 100 µmol m−2 s−1 light intensity for 12-days. Increase in root length was monitored every day.
H2O2 estimation and localization
Total H2O2 content in untreated or NaCl treated 7-days old wild-type, OE-1, sog1-6 and sog1-1 seedlings was determined following the method described earlier with minor modifications78. Accumulation level of H2O2 in the roots was detected by performing potassium iodide (KI) staining79. Roots of 7-days old seedlings were stained in KI/starch solution [4% (w/v) starch, 0.1 mol/L KI, pH 5.0] for 45 min and then observed using the Zeiss Stemi 508 Stereo microscope.
ROS imaging and quantification
Total ROS was quantitatively estimated from untreated or NaCl treated 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant Arabidopsis seedlings by following the procedure previously described80. For localization of different reactive oxygen species other than H2O2 including various superoxide radicals, ROS imaging was performed using the roots of 7-days old untreated or salt treated OE-1, sog1-6 and sog1-1 mutant seedlings along with the wild-type plants81. Roots were stained with a membrane-permeable fluorescent probe, CM-H2DCFDA [5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate] and imaged in a confocal microscope (Zeiss LSM-510 Meta) using 488 nm excitation and 500–530 nm emission wavelength.
Comet assay
For the evaluation of NaCl induced DNA damages, mainly DSBs, in wild-type, OE-1, sog1-6 and sog1-1 seedlings, comet assay was performed under neutral conditions. For this, nuclear suspension was extracted from 7-days old wild-type and sog1-1 mutant seedlings, either untreated or exposed to various concentrations of NaCl. The comet slides were prepared as described previously82,83 and stained with 2 µg/ml DAPI. Slides were then dried in the dark and examined in a Leica epifluorescence microscope with the excitation and emission wavelengths of 480 nm and 505–550 nm, respectively. Images were taken with the sCMOS fluorescence camera attached to the microscope and subsequently analysed utilizing the TriTek Comet Score software to calculate the tail DNA percentages.
Total histone protein extraction and detection of γ-H2AX
Total histone protein was isolated by acid extraction following previously described method83. Accumulation level of γ-H2AX in wild-type, OE-1 and sog1 mutant seedlings, either untreated or treated with increasing concentrations of NaCl for 12 h, was determined by immunoblotting using γ-H2AX specific primary antibody (Sigma-AldrichH5912) (1:250 dilution) and alkaline phosphatase conjugated goat anti-rabbit IgG secondary antibody (1:1000 dilution)67.
Detection of γ-H2AX focus formation: immunofluorescence staining of root tip nuclei
For the detection of γ-H2AX foci formation, immunostaining of root tip cell nuclei of wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings was performed following the methods described previously with minor modifications84,85. For immunostaining, 7-days old Arabidopsis seedlings were treated with 150 mM NaCl for 12 h. After the incubation period, the seedlings were fixed with 4% paraformaldehyde/PBS for 60 min and then washed in 1× PBS for 5 min at room temperature (25 °C). Washed seedlings were then subjected to digestion in the digestion buffer (containing 0.5% cellulose and 0.025% pectinase in 1× PBS) for 30 min at 37 °C in a waterbath. After the incubation, seedlings were washed thoroughly in 1× PBS for three times, 5 min each. The root tips were then squashed using a cover glass on a glass slide previously coated with poly-l-lysine. The glass slides were then freezed briefly in liquid N2 and the cover glasses were removed subsequently. After air-drying, slides were kept in 0.5% (V/V) Triton-X 100 in 1× PBS for 10 min followed by washing 3 times with 1X PBS at room temperature. 4% BSA (W/V) in 1× PBS solution was dropped on the squashed root tips and kept for about 30 min. The BSA solution was then removed and the primary antibody solution (1:1000) was added to the root tip sample. Rabbit anti-γH2AX (Sigma- Aldrich H5912) was used as the primary antibody. Primary antibody solution was diluted in 1% (W/V) BSA in 1X PBS. Slides were then kept overnight at 4 °C. Next day, the slides were washed in 1× PBS for 5 min. After the washing step, a goat Alexa Flour 488 anti-rabbit (Thermo Fisher A32731) secondary antibody was added in 1:1000 dilution and the slides were again incubated at 37 °C for 1 h in dark. After the incubation period, the slides were washed twice for 5 min in 1× PBS and finally the slides were immersed in 1 μg/ml DAPI (in 1× PBS) for 5 min. Slides were then mounted under 22 × 22 mm cover glass and subsequently visualized in a confocal microscope for imaging of γH2AX foci formation at 63× magnification.
Nuclei isolation and ploidy analysis by flowcytometry
Isolation of nuclei and subsequent analysis of the nuclear DNA content by flowcytometry were performed following the methods described previously with minor modifications34,86. Details of the procedure have been described under Supplementary ‘Materials and Methods’.
Visualization of nuclei by fluorescence microscopy
For the visualization of nuclei in the root tip cells of wild-type, OE-1, sog1-6 and sog1-1mutant lines, 7-days old seedlings were fixed in 4% (v/v) formaldehyde in PEM buffer (25 mm PIPES, 0.5 mm MgSO4, 2.5 mm EGTA, pH 7.2) for about 12 hours87. Roots were then stained with 4,6-diamidino-2-phenylindole (DAPI) and observed under a Leica fluorescence microscope.
Propidium iodide staining
Propidium iodide staining of roots was performed by following previously described method23. Roots of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings were treated without or with different concentrations of NaCl for 12 h and then fixed (50%, V/V methanol and 10% V/V acetic acid). Processed root tips were visualized under confocal microscope (Zeiss LSM-510 Meta). Root tip cell area and distance from the QC were measured using MBF ImageJ software by tracing cell contours (Supplementary ‘Materials and Methods’).
Measurement of leaf cell area expansion
Expansion of Arabidopsis leaf epidermal and mesophyll cells were studied by Differential interference contrast (DIC) microscopy following the method described earlier33,88. First and second pair of rosette leaves from 7-days old wild-type, OE-1, sog1-6 and sog1-1 seedlings were used for the measurements and visualized under a Zeiss Axio ScopeA1 microscope using DIC optics with image captured using Axiocam 503 camera attached with the microscope. Cell areas were measured in ImageJ software. The nature of trichomes were analysed by Scanning electron microscopy89 (Supplementary ‘Materials and Methods’).
Transcript profiling by semi-quantitative PCR
For expression analysis, total RNA was extracted from ~ 100 mg tissue of 7-days old wild-type, OE-1, sog1-6 and sog1-1 mutant seedlings using RNeasy plant mini kit (Qiagen) following manufacturer’s instructions. ~ 1 µg of total RNA was processed further to obtain 1st strand cDNA using Superscript III Reverse Transcriptase (Invitrogen). Semi quantitative reverse transcription PCR was performed using the cDNA samples to examine the transcript accumulation levels of AtATM, AtSOG1, AtATR, AtBLT, AtSMR5 and AtSMR7 genes, respectively using specific expression primers (Supplementary Table 2). The conditions for semi-quantitative reverse transcription-PCR have been described under Supplementary ‘Materials and Methods’.
Total protein extraction and immunoblotting
Total protein was extracted following the procedure described previously14. Details of protein extraction and immunoblotting procedures are provided in Supplementary ‘Materials and Methods’.
Statistical analysis
The quantitative data obtained from all the experiments have been expressed as the mean value of three biological replicates. Error bars indicate ± standard deviation (SD) of three independent replicates. Mean values and standard deviation were calculated using Microsoft Excel 2007 software. Student t-test was performed to test the significance difference between the mean values of the variables. The p-value represents the probability of correlation between two variables. To determine whether the correlation between variables is significant, the p-value is compared to a significance level of 0.05. The p-value of equal or less than 0.05 represents 5% chance that results from the experimental sample occurred due to chance. On the other hand, p-value of equal or less than 0.01 indicates there is only 1% chance. Asterisks represent statistically significant values (p < 0.05; p < 0.01) based on one-way ANOVA factorial analysis using XL Toolbox extension associated with Microsoft Excel 2007 software to assess variance between morphological parameters, quantitative ROS production, trichome branch number and density, transcript and protein accumulation among various genotypes.
Supplementary Information
Acknowledgements
The authors gratefully acknowledge Council of Scientific and Industrial Research, Govt. of India, (Ref. No. 38(1417)/16/EMR-II, dated: 17/05/2016 to SR), UGC, Govt. of India (Start-Up research Grant No.F.30-141/2015 (BSR) and SERB, DST, Govt. of India (Ref. No. ECR/2016/000539 to SR) for providing necessary financial supports. KM is the recipient of SRF fellowship from CSIR, Govt. of India [09/025(0220)/2017-EMRI]. We thank Dr. Swarup Roy Choudhury, IISER Tirupati for the critical review and comments on the manuscript. Authors are also thankful to University Science Instrumentation Centre (USIC), University of Burdwan (Fluorescence microscope), West Bengal, India; Centre for Research in Nanoscience and Nanotechnology (CRNN), Calcutta University, Kolkata, India (BD- Flowcytometer) and, Central Instrument facility (CIF), Bose Institute, Kolkata, India (Confocal microscope) for providing necessary technical support.
Abbreviations
- ANOVA
Analysis of variance
- ATM
ATAXIA-TELANGIECTASIA-MUTATED
- ATR
ATAXIA-TELANGIECTASIA-RAD3-RELATED
- BER
Base excision repair
- BLT
BRANCHLESS TRICHOMES
- CDK
Cyclin dependent kinase
- DAPI
Diamidino-2-phenylindole
- DDR
DNA damage response
- DSBs
Double strand breaks
- PI
Propidium iodide
- ROS
Reactive oxygen species
- SOG1
SUPPRESSOR OF GAMMA RESPONSE 1
- SSBs
Single strand breaks
Author contributions
S.R. conceived the idea and designed the experiments. K.M. performed the experiments. S.R. and K.M. analyzed the results and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41598-024-54831-1
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/21/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41598-024-54831-1
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91293-1.
References
- 1.Flowers TJ. Improving crop salt tolerance. J. Exp. Bot. 2004;55:1–13. doi: 10.1093/jxb/erh003. [DOI] [PubMed] [Google Scholar]
- 2.Gupta B, Huang B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014 doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 4.Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isayenkov S, Maathuis FJM. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019;10:80. doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evelin H, Devi TS, Gupta S, Kapoor R. Mitigation of salinity stress in plants by arbuscular mycorrhizal symbiosis: Current understanding and new challenges. Front. Plant Sci. 2019;10:470. doi: 10.3389/fpls.2019.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsugane K, Kobayashi K, Niwa Y, Ohba Y, Wada K, Kobayashi H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxifcation. Plant Cell. 1999 doi: 10.2307/3870742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez JA, Ferrer MA, Jimenez A, Barcelo AR, Sevilla F. Antioxidant systems and O2-/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001;127:817–831. doi: 10.1104/pp.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isayenkov SV. Physiological and molecular aspects of salt stress in plants. Cytol. Genet. 2012;46:302–318. doi: 10.3103/S0095452712050040. [DOI] [Google Scholar]
- 10.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24(10):453–462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatgilialoglu C, O’Neill P. Free radicals associated with DNA damage. Exp. Gerontol. 2001;36:1459–1471. doi: 10.1016/S0531-5565(01)00132-2. [DOI] [PubMed] [Google Scholar]
- 13.Waterworth WM, Drury GE, Bray CM, West CE. Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 2011;192:805–822. doi: 10.1111/j.1469-8137.2011.03926.x. [DOI] [PubMed] [Google Scholar]
- 14.Roy S, Choudhury SR, Sengupta DN, Das KP. Involvement of AtPolλ in the repair of high salt- and DNA cross-linking agent-induced double strand breaks in Arabidopsis. Plant Physiol. 2013;162:1195–1210. doi: 10.1104/pp.113.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahapatra K, Roy S. An insight into the mechanism of DNA damage response in plants-role of SUPPRESSOR OF GAMMA RESPONSE 1: An overview. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2020 doi: 10.1016/j.mrfmmm.2020.111689. [DOI] [PubMed] [Google Scholar]
- 16.Ceccarelli M, Santantonio E, Marmottini F, Amzallag GN, Cionini PG. Chromosome endoreduplication as a factor of salt adaptation in Sorghum bicolor. Protoplasma. 2006;227:113–118. doi: 10.1007/s00709-005-0144-0. [DOI] [PubMed] [Google Scholar]
- 17.Elmaghrabi AM, et al. Enhanced tolerance to salinity following cellular acclimation to increasing NaCl levels in Medicago truncatula. Plant Cell Tiss. Organ. Cult. 2013;114:61–70. doi: 10.1007/s11240-013-0306-2. [DOI] [Google Scholar]
- 18.Barkla BJ, Rhodes T, Tran KT, Wijesinghege C, Larkin JC, Dassanayake M. Making epidermal bladder cells bigger: Developmental and salinity-induced endopolyploidy in a model halophyte. Plant Physiol. 2018;177:615–632. doi: 10.1104/pp.18.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Storme N, Mason A. Plant speciation through chromosome instability and ploidy change: Cellular mechanisms, molecular factors and evolutionary relevance. Curr. Plant Biol. 2014;1:10–33. doi: 10.1016/j.cpb.2014.09.002. [DOI] [Google Scholar]
- 20.Scholes DR, Paige KN. Plasticity in ploidy: A generalized response to stress. Trends Plant Sci. 2015;20:165–175. doi: 10.1016/j.tplants.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Boyko A, Golubov A, Bilichak A, Kovalchuk I. Chlorine ions but not sodium ions alter genome stability of Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1066–1078. doi: 10.1093/pcp/pcq048. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi N, et al. The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 2008;27(13):1840–1851. doi: 10.1038/emboj.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adachi S, et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Cools T, De Veylder L. Mechanisms used by plants to cope with DNA damage. Annu. Rev. Plant Biol. 2016;67:439–462. doi: 10.1146/annurev-arplant-043015-111902. [DOI] [PubMed] [Google Scholar]
- 25.Yoshiyama KO, Conklin PA, Huefner ND, Britt AB. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12843–12848. doi: 10.1073/pnas.0810304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshiyama KO, et al. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013;14(9):817–822. doi: 10.1038/embor.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez AA, Taleisnik EL. Determination of reactive oxygen species in salt-stressed plant tissues. Methods Mol. Biol. 2012;913:225–236. doi: 10.1007/978-1-61779-986-0_15. [DOI] [PubMed] [Google Scholar]
- 28.Pang CH, Wang BS. Oxidative stress and salt tolerance in plants. In: Lüttge U, Beyschlag W, Murata J, editors. Progress in Botany. Berlin: Springer; 2008. [Google Scholar]
- 29.Friesner JD, Liu B, Culligan K, Britt AB. Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell. 2005;16(5):2566–2576. doi: 10.1091/mbc.e04-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang J, et al. Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol. 2012;194(2):353–363. doi: 10.1111/j.1469-8137.2012.04062.x. [DOI] [PubMed] [Google Scholar]
- 31.Kuo LJ, Yang LX. Gamma-H2AX—A novel biomarker for DNA double-strand breaks. In Vivo. 2008;22(3):305–309. [PubMed] [Google Scholar]
- 32.Leitch IJ, Dodsworth S. Endopolyploidy in Plants. Wiley; 2017. [Google Scholar]
- 33.Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development. 2010;137(1):63–71. doi: 10.1242/dev.035840. [DOI] [PubMed] [Google Scholar]
- 34.Galbraith DW, Harkins KR, Knapp S. Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 1991;96:985–989. doi: 10.1104/pp.96.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li ZY, Li B, Dong AW. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant. 2012;5(1):270–280. doi: 10.1093/mp/ssr086. [DOI] [PubMed] [Google Scholar]
- 36.Hulskamp M, Misra S, Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-X. [DOI] [PubMed] [Google Scholar]
- 37.Guimil S, Dunand C. Cell growth and differentiation in Arabidopsis epidermal cells. J. Exp. Bot. 2007;58:3829–3840. doi: 10.1093/jxb/erm253. [DOI] [PubMed] [Google Scholar]
- 38.Passardi F, Dobias J, Valerio L, Guimil S, Penel C, Dunand C. Morphological and physiological traits of three major Arabidopsis thaliana accessions. J. Plant Physiol. 2007;164:980–992. doi: 10.1016/j.jplph.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Kasili R, et al. BRANCHLESS TRICHOMES links cell shape and cell cycle control in Arabidopsis trichomes. Development. 2011;138:2379–2388. doi: 10.1242/dev.058982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 41.Mironov VV, De Veylder L, Van Montagu M, Inze D. Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell. 1999;11:509–522. doi: 10.1105/tpc.11.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Jager SM, Maughan S, Dewitte W, Scofield S, Murray JA. The developmental context of cell-cycle control in plants. Semin. Cell Dev. Biol. 2005;16:385–396. doi: 10.1016/j.semcdb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 43.De Veylder L, Beeckman T, Inzé D. The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 2007;8(8):655–665. doi: 10.1038/nrm2227. [DOI] [PubMed] [Google Scholar]
- 44.Gutierrez, C. The Arabidopsis cell division cycle. In The Arabidopsis Book (eds. Last, R. et al.) 1–19 (The American Society of Plant Biologists, 2008). [DOI] [PMC free article] [PubMed]
- 45.Meijer M, Murray JA. Cell cycle controls and the development of plant form. Curr. Opin. Plant Biol. 2001;4:44–49. doi: 10.1016/S1369-5266(00)00134-5. [DOI] [PubMed] [Google Scholar]
- 46.Breuer C, Ishida T, Sugimoto K. Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 2010;13:654–660. doi: 10.1016/j.pbi.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Roeder AHK. When and where plant cells divide: A perspective from computational modeling. Curr. Opin. Plant Biol. 2012;15:638–644. doi: 10.1016/j.pbi.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Harashima H, Dissmeyer N, Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013;23:345–356. doi: 10.1016/j.tcb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Boudolf V, et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 2004;16(10):2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cook GS, et al. Plant WEE1 kinase is cell cycle regulated and removed at mitosis via the 26S proteasome machinery. J. Exp. Bot. 2013;64(7):2093–2106. doi: 10.1093/jxb/ert066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogita N, et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018;94(3):439–453. doi: 10.1111/tpj.13866. [DOI] [PubMed] [Google Scholar]
- 52.Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/S0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 53.Cebolla A, et al. The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999;18(16):4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larson-Rabin Z, Li Z, Masson PH, Day CD. FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 2009;149:874–884. doi: 10.1104/pp.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Larson-Rabin Z, Masson PH, Day CD. FZR2/CCS52A1 mediated endoreduplication in Arabidopsis development. Plant Signal Behav. 2009;4:451–453. doi: 10.4161/psb.4.5.8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Veylder L, et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 2002;21(6):1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vlieghe K, et al. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol. 2005;15(1):59–63. doi: 10.1016/j.cub.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 58.Magyar Z, et al. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012;31:1480–1493. doi: 10.1038/emboj.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kállai BM, et al. γ-Tubulin interacts with E2F transcription factors to regulate proliferation and endocycling in Arabidopsis. J. Exp. Bot. 2020;71(4):1265–1277. doi: 10.1093/jxb/erz498. [DOI] [PubMed] [Google Scholar]
- 60.Rajendran K, Tester M, Roy SJ. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 2009;32:237–249. doi: 10.1111/j.1365-3040.2008.01916.x. [DOI] [PubMed] [Google Scholar]
- 61.Roy SJ, Negrão S, Tester M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 62.Sjogren CA, Bolaris SC, Larsen PB. Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell. 2015;27:2501–2515. doi: 10.1105/tpc.15.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailly C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019;476(20):3019–3032. doi: 10.1042/BCJ20190159. [DOI] [PubMed] [Google Scholar]
- 64.Hendrix S, et al. Suppressor of gamma response 1 modulates the DNA damage response and oxidative stress response in leaves of cadmium-exposed Arabidopsis thaliana. Front. Plant Sci. 2020;11:366. doi: 10.3389/fpls.2020.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zvanarou S, et al. Salt stress triggers generation of oxygen free radicals and DNA breaks in Physcomitrella patens protonema. Environ. Exp. Bot. 2020 doi: 10.1016/j.envexpbot.2020.104236. [DOI] [Google Scholar]
- 66.Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidant defense mechanism in plants under stressful conditions. J. Bot. 2012 doi: 10.1155/2012/217037. [DOI] [Google Scholar]
- 67.Markkanen E. Not breathing is not an option: How to deal with oxidative DNA damage. DNA Repair. 2017;59:82–105. doi: 10.1016/j.dnarep.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 68.Zucca E, et al. Silencing of human DNA polymerase lambda causes replication stress and is synthetically lethal with an impaired S phase checkpoint. Nucleic Acids Res. 2013;14:229–241. doi: 10.1093/nar/gks1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graindorge D, et al. Singlet oxygen-mediated oxidation during UVA radiation alters the dynamic of genomic DNA replication. PLoS One. 2015;10(10):e0140645. doi: 10.1371/journal.pone.0140645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sedletska Y, Radicella JP, Sage E. Replication fork collapse is a major cause of the high mutation frequency at three-base lesion clusters. Nucleic Acids Res. 2013;41(20):9339–9348. doi: 10.1093/nar/gkt731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 72.Lee HO, Davidson JM, Duronio RJ. Endoreplication: Polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang K, Ndathe RW, Kumar N, Zeringue EA, Kato N, Larkin JC. The CDK Inhibitor SIAMESE targets both CDKA;1 and CDKB1 complexes to establish endoreplication in trichomes. Plant Physiol. 2020;184(1):165–175. doi: 10.1104/pp.20.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127(18):3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- 75.Yi D, et al. The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell. 2014;26(1):296–309. doi: 10.1105/tpc.113.118943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Preuss SB, Britt AB. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics. 2003;164:323–334. doi: 10.1093/genetics/164.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roy S, Das KP. Homologous recombination defective Arabidopsis mutants exhibit enhanced sensitivity to abscisic acid. PLoS One. 2017;12(1):e0169294. doi: 10.1371/journal.pone.0169294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Velikova V, Yordanov I, Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous poly-amines. Plant Sci. 2000;151:59–66. doi: 10.1016/S0168-9452(99)00197-1. [DOI] [Google Scholar]
- 79.Schutzendbel A, Nikolova P, Rudolf C, Polle A. Cadmium and H2O2-inducedoxidative stress in Populus× canescens roots. Plant Physiol. Biochem. 2002;40:577–584. doi: 10.1016/S0981-9428(02)01411-0. [DOI] [Google Scholar]
- 80.Jambunathan N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. In: Sunkar R, editor. Plant Stress Tolerance, Methods in Molecular Biology. Humana Press; 2010. pp. 291–297. [DOI] [PubMed] [Google Scholar]
- 81.Shin R, Berg RH, Schachtman DP. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 2005;46:1350–1357. doi: 10.1093/pcp/pci145. [DOI] [PubMed] [Google Scholar]
- 82.Kozak J, West CE, White C, da Costa-Nunes JA, Angelis KJ. Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst) 2009;8:413–419. doi: 10.1016/j.dnarep.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 83.Mahapatra K, et al. Assessment of cytotoxic and genotoxic potentials of a mononuclear Fe(II) Schiff base complex with photocatalytic activity in Trigonella. Biochim. Biophys. Acta BBA Gen. Sub. 2019 doi: 10.1016/j.bbagen.2019.129503. [DOI] [PubMed] [Google Scholar]
- 84.Weimer AK, et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016;35(19):2068–2086. doi: 10.15252/embj.201593083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hirakawa T, Hasegawa J, White CI, Matsunaga S. RAD54 forms DNA repair foci in response to DNA damage in living plant cells. Plant J. 2017;90(2):372–382. doi: 10.1111/tpj.13499. [DOI] [PubMed] [Google Scholar]
- 86.Wear EE, et al. Isolation of plant nuclei at defined cell cycle stages using EdU labeling and flow cytometry. Methods Mol. Biol. 2016;1370:69–86. doi: 10.1007/978-1-4939-3142-2_6. [DOI] [PubMed] [Google Scholar]
- 87.Sugimoto K, Williamson RE, Wasteneys GO. New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 2000;124:1493–1506. doi: 10.1104/pp.124.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gegas VC, Wargent JJ, Pesquet E, Granqvist E, Paul ND, Doonan JH. Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J. Exp. Bot. 2014;65:2757–2766. doi: 10.1093/jxb/ert473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen C, Wang S, Huang H. LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis. 2000;26:42–54. doi: 10.1002/(SICI)1526-968X(200001)26:1<42::AID-GENE7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.