Abstract
We investigated lameness outbreaks at 3 commercial broiler farms in Arkansas. We isolated several distinct bacterial species from Bacterial Chondronecrosis with Osteomyelitis (BCO) lesions from these 3 farms. The results show that BCO-lameness pathogens on particular farms can differ significantly. We characterized genomes for isolates of the 2 most prevalent species, Escherichia coli and Staphylococcus aureus. Genomes assembled for E. coli isolates from all 3 farms were quite different between farms, and most similar to genomes from different geographical locations and hosts. The E. coli phylogenomics suggests frequent host shifts for this species. Genomes for S. aureus isolates from one farm were highly related to those from chicken isolates from Europe. Highly related isolates have also been characterized from chickens in the Arkansas area for at least a decade. Phylogenomics suggest that this S. aureus has been restricted to poultry for more than 40 y. Detailed analysis of genomes from 2 neighboring clades of S. aureus human and chicken isolates, identifies the acquisition of a specific pathogenicity island in the transition from human to chicken pathogen and that pathogenesis for this clade in chickens may depend on this mobile element. Investigation of the evolution of this chicken-restricted clade from 1980 in Ireland, Poland in 2008, Oklahoma in 2010 and Arkansas in 2019, reveals the acquisition of additional virulence determinants including pathogenicity islands. Isolate-specific genome characterizations will help further our understanding of the disease mechanisms of BCO-lameness, a significant animal welfare issue.
Key words: E. coli, S. aureus, lameness, genome, pathogenicity
INTRODUCTION
Lameness poses animal and health welfare issues that result in significant losses in poultry production. Modern broilers selectively bred for rapid growth are particularly prone to leg problems (Wideman, 2016). Bacterial chondronecrosis with osteomyelitis (BCO) is the leading cause of lameness in broiler and broiler breeder flocks (Al-Rubaye et al., 2015; Wideman, 2016; Thøfner et al., 2019). In birds that develop lameness, bacteria translocate into the bloodstream via the integument, respiratory system or gastrointestinal tract (Jiang et al., 2015; Wideman, 2016; Al-Rubaye et al., 2017). These bacteria may have come from the immediate environment, or vertical transfer though the egg (Stalker et al., 2010; Wideman, 2016). Bacteria that survive in the blood may colonize the proximal growth plate of the rapidly growing femorae and tibiae inducing necrosis, leading to BCO-lameness (Wideman and Prisby, 2013; Wideman, 2016; Al-Rubaye et al., 2017). Stressors, or other factors contributing to immunosuppression, can facilitate bacterial colonization and BCO spread in commercial poultry flocks (Mutalib et al., 1983; Andreasen et al., 1993; McNamee et al., 1998; Butterworth, 1999; McNamee and Smyth, 2000; El-Lethey et al., 2003; Wideman, 2016). In our research facility, Staphylococcus agnetis is the primary bacterial isolate from lame broilers induced by growth on raised-wire-flooring (Al-Rubaye et al., 2015; Al-Rubaye et al., 2017).
Genomic analysis of S. agnetis isolates from chickens and dairy cattle demonstrate that the chicken isolates appear to be a clade arising from one branch of the cattle isolates (Shwani et al., 2020). Multiple bacterial species have been historically identified from BCO lesions (Kibenge et al., 1982; Emslie and Nade, 1983; Hocking, 1992; Thorp et al., 1993; Thorp, 1994; McNamee and Smyth, 2000; Butterworth et al., 2001; Tarr et al., 2004; Stalker et al., 2010; Al-Rubaye et al., 2015; Wideman, 2016) but there are few reports on characterization of genetic relatedness within isolates of a species (Butterworth et al., 2001). Genomic analysis of a chicken isolate of Staphylococcus aureus from an outbreak in the United Kingdom suggested a genetic basis for the jump from human to chicken (Lowder et al., 2009). Surveys of 20 broiler flock farms in Australia suggested that avian pathogenic Escherichia coliwere the main BCO isolate (Wijesurendra et al., 2017). Genetic analysis by multilocus sequence type (MLST), pulsed field gel electrophoresis and PCR phylogenetic grouping, of 15 E. coli isolates from 8 flocks in Brazil indicated significant diversity for vertebral osteomyelitis, and arthritis isolates, even within the same flock (Braga et al., 2016). The aim of this study was to characterize the genomes of BCO isolates from 3 different commercial broiler farms in Arkansas.
MATERIALS AND METHODS
Microbiological Sampling and Bacterial Species Identification
Diagnosis and sampling of BCO lesions and blood have been described (Wideman et al., 2013; Wideman and Prisby, 2013; Al-Rubaye et al., 2015; Wideman, 2016). Swabs were streaked onto CHROMagar Orientation (CO; DRG International, Springfield, NJ) plates. Initial characterization of bacterial diversity was based on colony counts according to color, where numerous (>50) colonies of one color were indication of association and low numbers (≤ 10) were considered possible contaminant during sampling. Significant association with BCO was predicated on isolation from multiple sites in the same lame bird based on our prior experiments for inducing BCO in our facilities (Al-Rubaye et al., 2015; Al-Rubaye et al., 2017; Alrubaye et al., 2020b). Further characterization was by restreaking on CHROMagar Staphylococcus (CS; DRG International) (Al-Rubaye et al., 2015; Al-Rubaye et al., 2017). Representative colonies were then diagnosed to species by 16S rRNA gene sequencing (Al-Rubaye et al., 2015; Al-Rubaye et al., 2017).
Air sampling was conducted by waving 3 open CO plates 5 feet above the poultry floor within the building in different locations. Colonies from CO plates were then further evaluated on CS plates, and representative colonies were typed to species as above.
Genomic DNA Isolation and Sequencing
Cultures were preserved in 40% glycerol at -80°C. Working stocks were maintained on tryptic soy agar slants at 4°C. For DNA extraction, staphylococci were grown in tryptic soy broth to log phase and DNA was isolated as described (Shwani et al., 2020). DNA isolation from E. coli used lysozyme treatment, followed by organic extractions (Sambrook et al., 1989). DNA was quantified using a GloMax Multi Jr Detection System (Promega Biosystems Sunnyvale Inc., CA, USA) and purity evaluated with a Nanovue spectrophotometer (Healthcare Biosciences AB Uppsala, Sweden). DNA size was assessed by agarose gel (1.5%) electrophoresis.
Library construction and Illumina MiSeq 2 × 250 sequencing were performed by the Michigan State University Genomics Core Facility. Libraries for Illumina HiSeqX 2 × 125 sequencing were prepared using a RipTide kit (iGenomX, Carlsbad, CA) and sequenced by Admera Health (South Plainfield, NJ). Long reads were generated using Oxford Nanopore-MinION bar-code kit, as described (Shwani et al., 2020).
Genome Assembly and Analysis
De novo genome assemblies from short reads were generated as described (Shwani et al., 2020). For hybrid assemblies the long reads were phase corrected using the Illumina short reads and Ratatosk v0.3 (https://github.com/DecodeGenetics/Ratatosk), before hybrid assembly with Unicycler v0.4.8 (Wick et al., 2017). Unicycler hybrid assembly graphs were further analyzed for contiguity in Bandage 0.8.1 (Wick et al., 2015) to discern and export replicons. The PATRIC (Pathosystems Resource Integration Center) webserver (Davis et al., 2020) was used for some Unicycler assemblies, assembly annotation, and identification of similar genomes. Chromosome-level genomes were obtained from NCBI using genome_updater (https://github.com/pirovc/genome_updater). Genome distances based on SNPs in the core and accessory genomes were computed using PopPUNK (Lees et al., 2019). These were visualized as network clusters, and phylogenetic trees using the Microcreate.org webserver (Argimón et al., 2016). Average Nucleotide Identity (ANI) values were determined using pyANI 0.2.9 (Pritchard et al., 2016). ANI values were subtracted from 1 to generate distance matrices that were submitted to FastME 2.0 (Lefort et al., 2015) to generate Newick trees. Archaeoptryx 0.9928 beta (Han and Zmasek, 2009) was used to transform Newick trees into graphic representations. Assemblies were annotated and compared using the Rapid Annotation using Subsystem Technologies (RAST) and SEED viewer (Aziz et al., 2008; Overbeek et al., 2014). Serotype prediction was using the ECTyper module at GalaxyTrakr.org.
RESULTS AND DISCUSSION
Diagnosis and Microbiological Sampling for Three Broiler Farms
In June of 2016 we surveyed 2 commercial broiler houses on separate farms experiencing outbreaks of BCO-lameness. Both houses had experienced a loss of cooling a week earlier, causing heat stress for several hours. The farms were in rural, central Arkansas separated by 6.3 km, operated by the same integrator, and stocked from the same hatchery. The company veterinarian reported that samples from lame birds had been routinely submitted to a poultry health diagnostics laboratory and were primarily diagnosed as E. coli. For both farms we randomly collected lame birds for necropsy for BCO lesions. Blood and BCO lesion swabs were collected from these birds, and house air was sampled, for bacterial species surveys.
In Farm 1 the birds were 31 d old. We diagnosed and necropsied 6 lame birds (Table 1). KB1 and KB2 were symptomatic of spondylolisthesis/kinky-back (KB). KB1 had BCO lesions in T4, left tibia, and both femora. We obtained thousands (TNTC; too numerous to count) of small green colonies from the T4 sample that were determined to be Enterococcus cecorum. KB2 had BCO of only the left tibia, but no colonies were recovered from sampling from this site. We did recover approximately 50 green colonies from what appeared to be a normal T4 that was E. cecorum. Lame3 and Lame4 both had bilateral BCO of the femora and tibiae. Lame3 had TNTC white colonies from microbiological sampling of T4 that were S. agnetis and 50 purple colonies from the right tibia that were Escherichia coli. Lame5 had bilateral FHN, bilateral tibial dyschondroplasia (TD), and pericarditis. We recovered green colonies (20 from left and TNTC from right) from the TD lesions that were E. cecorum. Due to limited supplies there was no microbiological sampling for Lame4 and Lame6.
Table 1.
Microbiological sampling of bone and blood samples from two commercial broiler farms experiencing BCO outbreaks.
| BCO Diagnoses |
High Colony Counts |
|||||||
|---|---|---|---|---|---|---|---|---|
| Bird | Farm | LT | RT | LF | RF | T4 | Species | Site |
| KB1 | 1 | THN | N | FHN | FHN | KB | Enterococcus cecorum | T4 |
| KB2 | 1 | THN | N | N | N | N | E. cecorum | T4 |
| Lame3 | 1 | THN | THN | FHN | FHN | KB | Escherichia coli | RT |
| Staphylococcus agnetis | T4 | |||||||
| Lame4 | 1 | THN | THN | FHN | FHN | KB | - | - |
| Lame5 | 1 | TD | TD | FHN | FHN | E. cecorum | LT RT | |
| Lame6 | 1 | THN | THN | FHN | FHN | KB | - | - |
| Lame7 | 2 | N | THN | FHN | N | N | E. coli | Blood |
| Lame8 | 2 | THN | THN | FHS | FHN | N | - | |
| Lame9 | 2 | THN | THN | FHN | N | KB | E. coli | blood LT LF |
| Lame10 | 2 | THN | THN | N | FHN | KB | Salmonella enterica | LT RF |
| Lame11 | 2 | THNS | THNC | FHS | FHN | N | E. coli | blood LT LF |
| Lame12 | 2 | THN | THN | FHT | FHT | N | E. coli | blood LT LF |
| Lame13 | 2 | - | - | FHN | FHN | - | - | - |
| Lame14 | 3 | THNS | THNS | FHN | N | - | Staphylococcus aureus | LT LF RT |
| Lame15 | 3 | THN | THNS | FHN | FHN | - | S. aureus | LT LF RT RF |
| Lame16 | 3 | THNS | THNS | FHS | FHN | - | S. aureus | LT |
| Lame17 | 3 | THNS | THNS | N | FHN | - | S. aureus | LT RT RF |
| Lame18 | 3 | THNS | THNS | FHN | FHN | - | E. coli | LT LF RT RF |
| Lame19 | 3 | THNS | THNS | FHN | FHS | - | Staphylococcus epidermidis | LT RF |
| Lame20 | 3 | THNS | THNS | N | N | - | Staphylococcus cohnii | LT RT |
| Lame21 | 3 | THNS | THNS | FHN | FHN | - | S. aureus | LT LF RT RF |
| Lame22 | 3 | THNS | THNS | FHN | FHS | - | Staphylococcus simulans | RF |
| Lame23 | 3 | THNS | THN | FHN | FHN | - | S. aureus | LT LF RT RF |
| Lame24 | 3 | THNS | THN | FHT | FHT | - | S. aureus | LT LF RT RF |
BCO diagnoses are listed along with necropsy comments.
Abbreviations: FHN, femoral head necrosis; FHS, femoral head separation; KB, kinky back; LF, left femur; LT, the left tibia; N, normal; RF, right femur; RT, right tibia; T4, vertebral joint; THN, tibial head necrosis; THNS, THN severe; TD, tibial dyschondroplasia.
High Colony Counts lists the Species diagnosis from 16S rRNA gene sequencing where a large number (>50) of same color colonies were recovered from one or more Site(s) indicating high probability of infection.
For Farm 2 the birds were 41 d of age. We diagnosed and necropsied 7 lame birds (Table 1). Lame7 was diagnosed with BCO lesions of the left femur, and right tibia. We obtained 30 purple colonies that were E. coli from the blood sample. We diagnosed Lame8 with bilateral BCO of tibia and femur. No colonies were obtained from sampling this bird. Lame9 was diagnosed with BCO of bilateral tibia, and the left femur, with evident pericarditis. Approximately 100 purple colonies were produced from sampling of the left femur that were determined to be E. coli. Lame10 was diagnosed with BCO of both tibia and right femur. Lame10 also had pericarditis. We got 40 white colonies from the left tibia, and 70 white colonies from the right femur that were Salmonella enterica. Lame11 had BCO lesions on both tibiae and femora. We got TNTC purple colonies from the left tibia, 3 from the left femur, and 15 from blood that were determined to be E. coli. Lame12 was diagnosed with bilateral BCO of tibia and femur. We recovered approximately 500 purple colonies from blood, 40 purple colonies from the left tibia, and TNTC purple colonies from the left femur that were E. coli. Due to limited supplies there was no microbiological sampling for Lame13.
Average plate counts for air sampling were 80 and 125 for Farm 1 and Farm 2, respectively. The predominant species was Staphylococcus cohnii (∼95%) with 3–4% Staphylococcus lentus and 1–2% E. coli. Air sampling during our broiler experiments with challenges with the BCO pathogen S. agnetis 908 at the University of Arkansas Poultry Research farm typically identify predominantly S. cohnii and 1–3% S. agnetis, but not E. coli (Alrubaye et al., 2020a; Alrubaye et al., 2020b).
In July 2019, we sampled a third commercial broiler farm (Farm 3) that the integrator had identified as experiencing a significant BCO outbreak. This farm was in Northwest Arkansas, more than 88 km from Farms 1 & 2 and operated by a different integrator supplied from a different hatchery. We sampled 11 lame birds at 35 d of age (Table 1). Lame14 was diagnosed with BCO of bilateral tibiae and right femur. We recovered numerous white colonies from the left femur, right tibia, and left tibia that were determined to be Staphylococcus aureus. Lame15 had BCO of all femorae and tibiae. Culture plates had numerous white colonies from all 4 sampled sites that were S. aureus. Lame16 had BCO of all tibiae and femorae. We recovered 10 green colonies from the right femur that were not analyzed, and 10 white colonies from the left tibia that were determined to be S. aureus. We diagnosed Lame17 with BCO of the right femur and both tibiae. We recovered numerous white colonies from all 3 sites that were S. aureus. Lame18 was diagnosed with bilateral BCO of the femorae and the tibiae. We got TNTC purple colonies from both femorae, and 11 total purple colonies from both tibiae, that were E. coli. Lame19 had BCO of all femorae and tibiae. Swabs gave only a few colonies of Staphylococcus epidermidis that we assumed were contaminants during sampling. Lame20 had bilateral BCO of the tibiae. We recovered a few green colonies of S. cohnii which were presumed contaminants during sampling. Lame21 had BCO of all femorae and tibiae. We isolated TNTC white colonies from all 4 sites that were S. aureus. Lame22 was diagnosed with bilateral BCO of femorae and tibiae. Microbial sampling only yielded 10 green colonies from the right femur that were determined to be Staphylococcus simulans. Lame23 was diagnosed with bilateral BCO of femorae and tibiae. Culture plates had only white colonies, TNTC from both femora and tibiae that were S. aureus. Lame24 had BCO of all femorae and tibiae. We recovered more than 100 white colonies from all 4 BCO lesions that were S. aureus.
BCO Genome Assemblies
We characterized genomes for 4 E. coli isolates: 1409 from Farm1, 1413 from Farm 2, and 1512 and 1527 from one bird on Farm 3 (Table 2). A hybrid assembly for 1409 produced 5.05 Mbp in 23 contigs that organized into 4 DNA assembly graphs. We resolved the replicons using the long reads for contiguity analysis of the assembly graphs using Bandage (Wick et al., 2015). The resolved genome contains a 4.84 Mbp chromosome, with episomes of 113.6, 108.7, and 2.3 kbp. The predicted serotype is O16:H32. The hybrid assembly for 1413 produced 5.37 Mbp in 59 contigs and 3 DNA assembly graphs. Unfortunately, the Nanopore reads were not of sufficient quality or length to complete a contiguity analysis of the entire genome, but does identify at least 2 episomes of 98.8 kbp and 2257 bp. The predicted serotype is O78:H4. Draft assemblies were generated for E. coli 1512 and 1527. The assembly of 1512 contained 4.96 Mbp in 152 contigs with a N50 of 150 kbp. The assembly of 1527 was 4.90 Mbp in 179 contigs. The N50 was 97 Kbp with the largest contig of 258 Kbp . Both 1512 and 1527 are predicted to be serotype O78:H17, like 1413. We generated draft assemblies for 14 S. aureus isolates from Farm 3 to examine genome diversity within a farm and within individual birds (Table 2). Two separate colonies from 7 lame bird were used for draft genome assembly (1510 & 1511, 1513 & 1514, 1515 & 1516, 1517 & 1518, 1519 & 1520, 1521 & 1522, 1523 & 1524). The assemblies ranged from 2.79 to 2.82 Mbp in 60 to 96 contigs (excluding contigs < 300 bp). The largest contigs were between 279 and 284 Kbp . N50 values ranged from 58 to 113 kbp. The L50 values ranged from 7 to 14 contigs. Each of the S. aureus assemblies had at least 3 circular contigs (episomes). Table 2 summarizes all BCO isolates and NCBI biosamples, with accessions in Table S1.
Table 2.
Bacterial genome assemblies produced in these analyses are listed by species, isolate designation, host source, assembly genome Status and statistics (Mbp, Contig count), NCBI Biosample.
| Isolate | Source | Status | Mbp | Contigs | Biosample |
|---|---|---|---|---|---|
| Escherichia coli | |||||
| 1409 | RT Lame3 | Finished | 5.063 | 4 | SAMN12285857 |
| 1413 | Blood Lame12 | Finished | 5.375 | 59 | SAMN12285859 |
| 1512 | LF Lame18 | Draft | 4.962 | 152 | SAMN13245724 |
| 1527 | RF Lame18 | Draft | 4.904 | 179 | SAMN13245725 |
| Staphylococcus aureus | |||||
| 1510 | LT Lame14 | Draft | 2.794 | 96 | SAMN13245722 |
| 1511 | RT Lame14 | Draft | 2.804 | 87 | SAMN15589960 |
| 1513 | LF Lame15 | Draft | 2.822 | 78 | SAMN15589961 |
| 1514 | RF Lame15 | Draft | 2.821 | 78 | SAMN15589962 |
| 1515 | RF Lame16 | Draft | 2.820 | 84 | SAMN15589963 |
| 1516 | LT Lame16 | Draft | 2.827 | 79 | SAMN13245723 |
| 1517 | LT Lame17 | Draft | 2.827 | 78 | SAMN15589964 |
| 1518 | RF Lame17 | Draft | 2.817 | 85 | SAMN15589965 |
| 1519 | LT Lame21 | Draft | 2.846 | 60 | SAMN15589966 |
| 1520 | RF Lame21 | Draft | 2.827 | 78 | SAMN15589967 |
| 1521 | RF Lame23 | Draft | 2.820 | 89 | SAMN15589968 |
| 1522 | RT Lame23 | Draft | 2.816 | 89 | SAMN15589969 |
| 1523 | RF Lame24 | Draft | 2.821 | 87 | SAMN15589970 |
| 1524 | LT Lame24 | Draft | 2.820 | 83 | SAMN15589971 |
Abbreviations are as in Table 1.
Phylogenetic Comparisons of E. coliGenomes
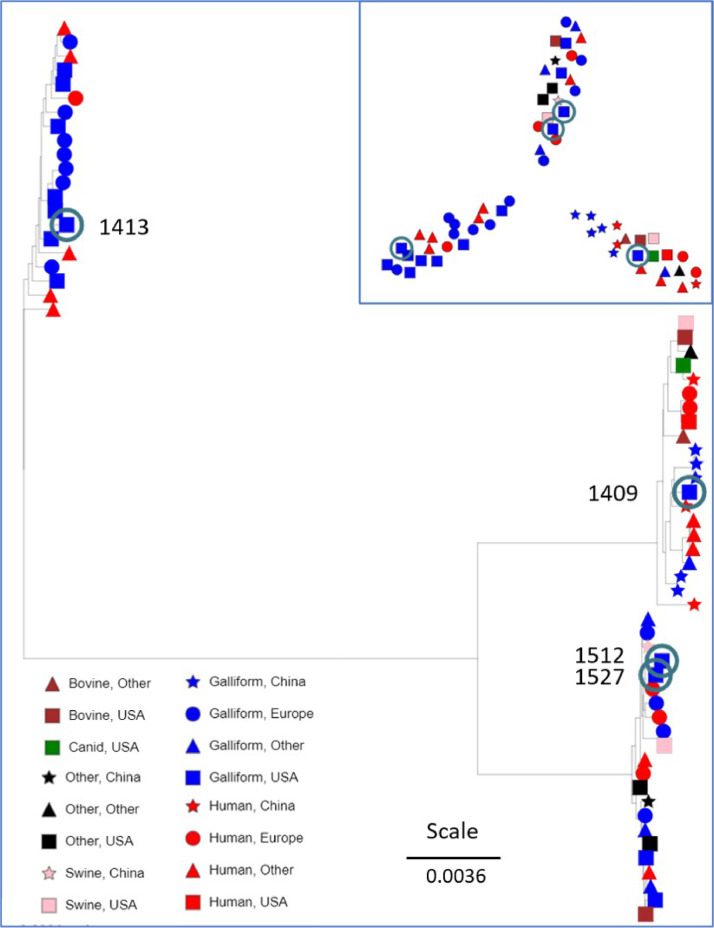
To examine the phylogenetic relationships between E. coli isolates from the 3 farms, we identified the most closely related genomes according to PATRIC for 1409 (Farm 1), 1413 (Farm 2), 1512, and 1527 (Farm 3). We then identified the clades to which the closest related genomes are assigned in the NCBI dendrogram for all E. coli genomes. The E. coli from each farm clearly mapped to very different subgroups in relation to the over 20,000 E. coli genomes in the NCBI dendrogram (data not shown). We downloaded 60 genomes representing the 20 most closely related genomes identified by PATRIC for our 4 new E. coli genomes. Not surprisingly, PATRIC identified the same 20 genomes for 1512 and 1527; as these were from the same lame bird. We used the PopPUNK package (Lees et al., 2019) to generate a neighbor-joining phylogenetic tree for all 64 E. coli genomes based on a lineage model for genome distances computed from SNPs in the shared core genome and accessory genomes. The tree and network (Figure 1) were rendered based on host and geographic sources using the Microreact.org webserver (Argimón et al., 2016). Tree topology was highly similar when we used Average Nucleotide Identity (ANI) to calculate genome distance (Pritchard et al., 2016). The PopPUNK network analysis and phylogenomic tree identified individual clades for 1409, 1413, and the 2 isolates from the same bird from farm 3 (1512 and 1527). Isolate 1409 clustered closely with 3 chicken isolates and a human isolate from China. There were 3 other chicken isolates in this clade, 2 from China and one from Pakistan. The other closely related genomes were from cattle, swine, dog, and human, from China, USA, Mexico, and Europe. Isolate 1413 clustered with isolates from 14 galliform hosts from Europe and USA, with the other 6 genomes from human hosts from South America, Africa and Europe. Isolates 1512 and 1527 are virtually identical, with an ANI of 0.99995, which is not surprising since they were isolated from different sites in the same lame bird. Isolates 1512 and 1527 grouped with a pig isolate from China, a human isolate from France, and one chicken isolate from the UK, with ANI of 0.9996. The other closely related isolates came from human in Japan, sick chickens in Poland, pigs from South Dakota, a FDA water contamination project in Arizona, USA chicken eggs, human clinical samples, a coliseptic turkey in Israel, US citizens afflicted with hemolytic uraemic syndrome during a 2011 outbreak in Germany, Switzerland chicken meat, and US deer feces. Thus, 1409, and 1512/1527 cluster with isolates obtained from a wide variety of hosts, whereas 1413 clusters with isolates primarily from chickens with a lower number of human isolates. The ANI for 1512/5127 relative to 1409 and 1413, is 0.98, while the ANI between 1409 and 1413 is 0.97, so the 3 genomes are equally distant from each other. Note that ANI less than 0.95 is often considered as separate species.
Figure 1.
Neighbor-Joining phylogenetic tree for 64 Escherichia coli genomes based on PopPUNK lineage-model analysis of core genome SNPs as displayed by MicroReact.org. Legend indicates host and region of isolation; scale bar indicates tree distance. Inset is the cluster analysis for network relationships. New isolates from lame broilers are circled in the inset, and circled and labeled in the tree. Additional genome details are provided in Table S1.
Previous genome comparisons between Avian Pathogenic E. coli (APEC) isolates indicated close relationships between two O78 serotypes but marked divergence between the O78 and an O16 serotype (Dziva et al., 2013). We found that 2 E. coli predicted to be O78 serotype, 1413 and 1512/1527, have quite distinct genomes. Molecular diagnosis of E. coli as APEC have relied on presence of particular genes including tsh, iss, cvaC, iroN, iucC sitA, hylF, ompT, and etsA (Rodriguez-Siek et al., 2005; Mellata et al., 2009; Dziva et al., 2013). For isolate χ7122 (chi7122 in Table S1) these genes are clustered on a 103 kbp plasmid (Mellata et al., 2009). We performed tBLASTn queries with the predicted protein products for these 9 genes from χ7122. The results suggest that 1409 is missing tsh, iucC, and etsA, 1413 contains homologs to all 9, and 1512/1527 do as well. However, there was no evidence of clustering in any of these genomes, based on mapping to different contigs. The APEC strains χ7122 (Dziva et al., 2013) and APEC O78 (Mangiamele et al., 2013) are both serotype O78 and were included in the 20 closest genomes for 1512/1527. However, 1413 is also predicted to be serotype O78, and contains the APEC virulence genes, but the phylogenomics places this isolate in a completely distinct E. coli clade.
Phylogenetic Comparisons of S. aureusGenomes
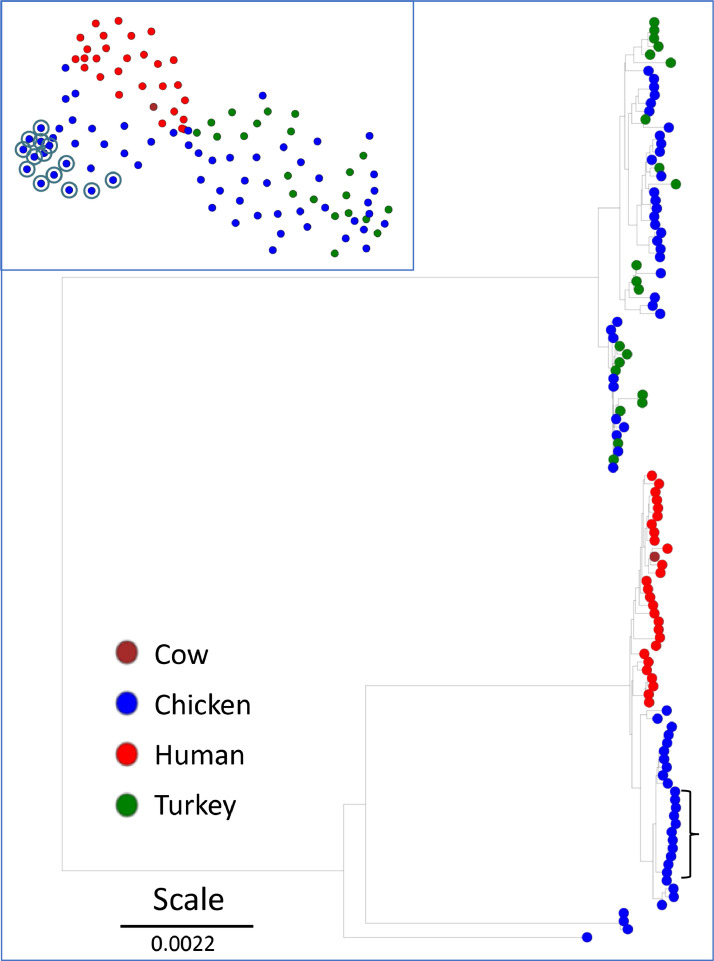
The PATRIC closest-genome report for our new S. aureus chicken assemblies included 11 S. aureus genomes for isolates obtained from chicken hosts. The close relationship of these chicken isolates had been previously demonstrated by using MLST to characterize S. aureus isolates from avian hosts (Lowder et al., 2009). Their analyses suggested a host switch from humans to poultry of the ST-5 sequence type, and a close relationship among S. aureus isolated from the global chicken industry from 1970 to 2000. The genome of isolate ED98 was assembled as the type-strain representing a BCO isolate from S. aureus from 1986 or 1987 in Ireland. We obtained 100 genomes consisting of all S. aureus genomes from NCBI classified with host as chicken (n = 50) or turkey (n = 21). We also included the 29 most closely related S. aureus genomes for which host was known and not chicken, according to the PATRIC report. We generated a PopPUNK network and phylogenomic tree (Figure 2). The cluster and tree results reveal that there are at least 2 distinct populations of S. aureus associated with galliform birds. The cluster that includes our 14 new genomes only contains chicken isolates and has a sister clade of 28 human isolates and one from cow milk. There is a separate network of turkey and chicken isolates that is significantly distinct from the clade containing mammalian and exclusively chicken isolates. ANI analysis of these genomes revealed the same distinct groups.
Figure 2.
Neighbor-Joining phylogenetic tree for 114 Staphylococcus aureus genomes based on PopPUNK lineage-model analysis of core genome SNPs as displayed by MicroReact.org. Genomes were all NCBI genomes where host is turkey (n = 21) or chicken (n = 64; includes the 14 genomes in this report), and the 29 genomes closest to isolate 1516, according to PATRIC, where the host was known, and not-a galliform. Legend indicates host of isolation; scale bar indicates tree distance. Inset is the cluster analysis for network relationships. New isolates from lame broilers are circled in the inset and bracketed in the tree. Additional genome details are provided in Table S1.
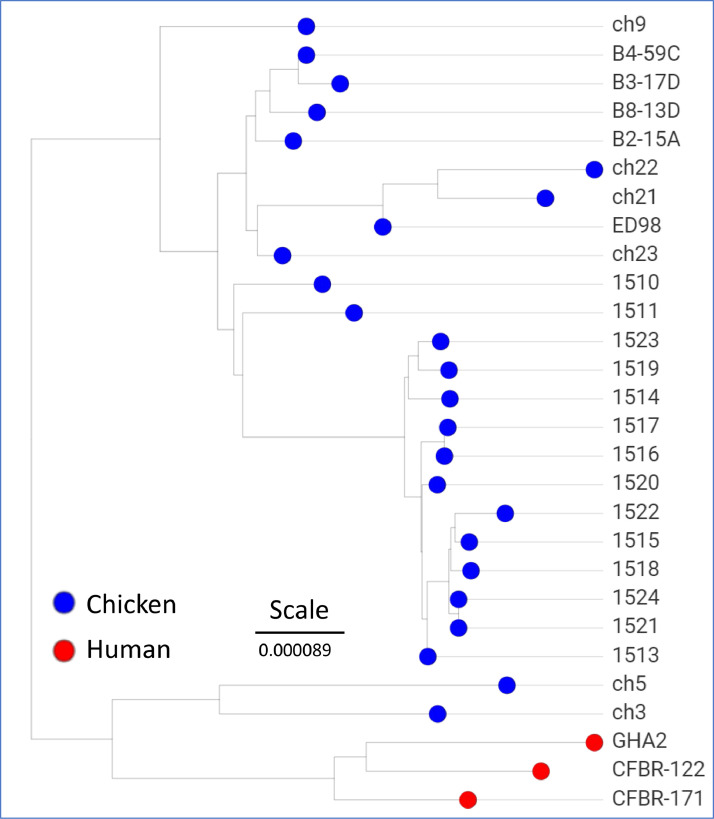
To further resolve the clade of chicken specific isolates we generated a phylgenomic tree (Figure 3) for all 25 chicken isolates in that clade and the 3 closest human isolates (GHA2 from a Ghanian hospital patient, CFBR-122 and CFBR-171 from USA sputum samples). Our 14 new S. aureus chicken BCO isolate genomes cluster together consistent with a clonally-derived population (average ANI of 0.999915). Other chicken isolates in this branch include: 4 isolates from retail chicken meat from 2010 in Tulsa, Oklahoma, USA (B4-59C, B3-17D, B2-15A, B8-13D), 3 isolates from chicken wounds in Poland in 2008 (ch21, ch22, ch23), ED98 (see above) the type strain for the BCO outbreak in Ireland in the 1980s characterized by Lowder et al. (2009). Isolate ch9 was isolated from an infected chicken hock in USA in 1999. Isolates ch3 and ch5 are less related and closely allied with the human isolates. These isolates are recorded as chicken commensals from Belgium in 1976. Additional details on these genomes are provided in Table S1. Our PopPUNK and ANI based phylogenomics are in agreement with the work of Lowder et al. (2009), where they used MLST and mutational analysis of biallelic polymorphisms to demonstrate the tight relationship of poultry isolates of S. aureus. They concluded that the “jump” to chickens likely was associated with the same chicken isolates from Belgium (ch3 and ch5), and likely derived from human isolates in Poland. They suggested that the first instance of association with pathogenesis in chickens is ED98 in 1986 or 1987, and that the host switch represented by ED98 was associated with “acquisition of novel mobile genetic elements from an avian-specific accessory gene pool, and by the inactivation of several proteins important for human disease pathogenesis.” This was evidenced by their demonstration of enhanced resistance to killing by chicken heterophils.
Figure 3.
Neighbor-Joining phylogenetic tree for 28 Staphylococcus aureus genomes based on PopPUNK lineage-model analysis of core genome SNPs as displayed by MicroReact.org. Genomes were the 25 closely related from chicken and the 3 closest related human isolates from Figure 2. Additional details are as for Figure 2.
Genome Evolution of a Chicken-Restricted clade of S. aureus
To further explore the underlying genetics using all available genomes we used the proteome comparison tool that is part of the RAST SEED Viewer to examine the progression from human pathogen (GHA2 and CFBR-171), to either chicken commensal (ch3 and ch5), or to chicken pathogen (ED98, B4-59C, and ch21). First, we used the predicted ED98 proteins as the reference, to identify polypeptides conserved in the other chicken pathogens, and not conserved in the chicken commensals and human pathogens. The cutoffs were to identify polypeptides conserved at >80% in all the chicken pathogens and <80% identity in the commensals and human pathogens. Thus, to identify polypeptides shared by the chicken pathogens but not the chicken commensals or human pathogens. This filtering identified 33 protein encoding genes (PEG) from the ED98 main chromosome and none from the 3 plasmids in ED98 (Table 3 marked with *). There were 2 clusters containing 32 of the 33 PEGs. These 2 clusters were associated with 2 mobile elements: a transposon and a S. aureus pathogenicity island (SaPI). There was also an additional isolated short PEG (PEG 48). To further define any association of the 33 PEGs with the jump from humans to chickens we used tBLASTn to individually query the genomes of 11 S. aureus chicken isolates; 2 chicken commensals and 9 pathogens (indicated in Table S1). We also performed a tBLASTn query of a BLAST database representing the 10 most complete human S. aureus isolates in Figure 2 and an additional 19 representing clades nearby in the NCBI genome tree. The tBLASTn query included all 33 PEGs as well as immediate flanking or intervening PEGs (Table 3). PEG 48 annotated as a hypothetical 56 residue polypeptide and the tBLASTn searches only identify significant matches in the chicken pathogen genomes. Therefore, PEG 48 cannot be discounted as contributing to the jump to chicken pathogenesis. The transposon-related region (PEG 1641-1659) contained no PEG that was highly conserved in all the chicken isolates while also being less conserved in all the human isolates and the chicken commensals. Therefore, the PEGs from the transposon are not supported for significant contribution to the jump from human to chicken pathogen. On the other hand, the SaPI region (PEG 755-773) contained a number of PEGs that appear highly conserved in all the chicken pathogens. Some of these PEGs (i.e., PEG 756–760, 767–771) are highly conserved in some of the human pathogen isolates, but lacking in the chicken commensals. There are an additional 4 hypothetical PEGs (774–778) downstream of the SaPI terminase that appear to be restricted to only the chicken pathogens. Close inspection of the results in Table 3 suggests that isolate ch9 appears to be intermediate between the chicken commensals, ch3 and ch5, and all of the other chicken pathogens in the ED98 clade. This is concordant with our phylogenomic analysis (Figure 3) where ch9 is basal relative to the other chicken pathogens. Thus, ch9 may represent a transitional state between the human pathogens as the bacterial genome acquired increased virulence in chickens. The data are consistent with the S. aureus genome evolving in 2 directions during the host switch from human to chickens: loss of the SaPI to become commensal, and acquisition of new PEGs as part of this SaPI to become chicken-specific. Lowder et al. (2009) had originally identified this SaPI region as associated with the jump to chickens but they only surveyed other avian isolates by endpoint PCR and did not examine at the resolution of individual PEGs.
Table 3.
Percent identity from tBLASTn searches of Staphylococcus aureus genomes using the polypeptide sequences for protein encoding genes from ED98.
| Protein encoding gene |
tBLASTn percent identity for isolate genome(s) |
Count for NCBI S. aureus Desc |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein encoding gene |
29 human |
Commensals |
Pathogens |
X22clade |
||||||||||||||||
| PEG | Polypeptide annotation | Length | ch3 | ch5 | ch9 | ED98 | ch21 | ch22 | ch23 | B2-15A | B3-17D | B4-59C | B8-13D | 1519 | pa3 | ph2 | X22 | ch24 | ||
| 47 | Hypothetical | 638 | 54 | 93 | 93 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 67 | 100 | 100 | 93 | 93 | 0 | 0 | 25 |
| *48 | Hypothetical | 56 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 95 | 100 | 100 | 0 | 0 | 0 | 0 | 28 |
| 49 | Hypothetical | 452 | 81 | 96 | 96 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 93 | 100 | 100 | 96 | 96 | 29 | 29 | 381 |
| 754 | Methionine ABC transporter substrate-binding | 274 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 99 | 99 | 99 | 1012 |
| *755 | Integrase SaPI | 407 | 99 | 45 | 45 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 99 | 100 | 45 | 307 |
| *756 | SAP domain (DNA binding domain) | 463 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 53 |
| *757 | Hypothetical in SaPI | 191 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 98 | 98 | 100 | 30 | 53 |
| *758 | Hypothetical in SaPI | 74 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 53 |
| *759 | Winged helix-turn-helix domain-Containing | 91 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 41 | 394 |
| *760 | Hypothetical in SaPI | 49 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 96 | 368 |
| *761 | Hypothetical in SaPI | 71 | 41 | 0 | 0 | 45 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 42 | 42 | 100 | 0 | 89 |
| *762 | Hypothetical in SaPI | 120 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 100 | 0 | 24 |
| *763 | DUF1474 family | 106 | 84 | 0 | 0 | 49 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 63 | 63 | 100 | 60 | 84 |
| *764 | Primase alpha helix C-terminal Domain-Containing | 290 | 83 | 0 | 0 | 94 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 97 | 97 | 100 | 26 | 200 |
| *765 | DNA helicase | 570 | 21 | 0 | 0 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 99 | 100 | 22 | 200 |
| *766 | Hypothetical in SaPI | 127 | 30 | 0 | 0 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 98 | 98 | 100 | 0 | 199 |
| *767 | Hypothetical in SaPI | 214 | 97 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 95 | 95 | 100 | 92 | 405 |
| *768 | Hypothetical in SaPI | 114 | 97 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 99 | 100 | 98 | 353 |
| *769 | Hypothetical in SaPI | 193 | 98 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 97 | 97 | 100 | 0 | 312 |
| *770 | Capsid morphogenesis B | 43 | 97 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 97 | 97 | 100 | 0 | 312 |
| *771 | Spore coat | 176 | 97 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 98 | 96 | 96 | 100 | 94 | 406 |
| *772 | Hypothetical in SaPI | 114 | 86 | 36 | 36 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 85 | 85 | 100 | 85 | 405 |
| *773 | Terminase SaPI | 190 | 97 | 46 | 46 | 96 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 99 | 100 | 96 | 839 |
| *774 | Hypothetical | 59 | 0 | 0 | 0 | 87 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 42 |
| *775 | Hypothetical | 130 | 0 | 0 | 0 | 95 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 48 |
| *776 | Hypothetical | 403 | 0 | 0 | 0 | 89 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 91 | 91 | 100 | 0 | 48 |
| *777 | Hypothetical | 109 | 44 | 44 | 44 | 52 | 100 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | 100 | 58 | 58 | 100 | 55 | 10 |
| *778 | Hypothetical | 164 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 100 | 0 | 37 |
| 779 | CsbD stress response | 65 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 1011 |
| 1641 | Hypothetical | 121 | 45 | 60 | 60 | 64 | 100 | 100 | 96 | 64 | 64 | 84 | 84 | 64 | 84 | 60 | 60 | 38 | 38 | 7 |
| *1642 | Hypothetical | 198 | 50 | 75 | 75 | 75 | 100 | 99 | 99 | 75 | 75 | 100 | 100 | 75 | 100 | 75 | 75 | 0 | 0 | 7 |
| *1643 | Secretory antigen SsaA-like transposon-related | 343 | 61 | 79 | 79 | 79 | 100 | 100 | 99 | 79 | 79 | 99 | 99 | 79 | 99 | 79 | 79 | 46 | 46 | 7 |
| *1644 | Hypothetical | 644 | 51 | 71 | 71 | 70 | 100 | 100 | 100 | 70 | 70 | 98 | 96 | 70 | 98 | 71 | 71 | 0 | 0 | 7 |
| 1645 | Hypothetical | 78 | 0 | 72 | 82 | 0 | 100 | 100 | 100 | 0 | 0 | 70 | 70 | 0 | 70 | 72 | 72 | 0 | 0 | 3 |
| 1646 | Hypothetical | 56 | 0 | 0 | 0 | 85 | 100 | 100 | 85 | 85 | 85 | 85 | 87 | 85 | 85 | 0 | 0 | 0 | 0 | 2 |
| 1647 | Hypothetical | 454 | 81 | 95 | 95 | 94 | 100 | 100 | 99 | 94 | 94 | 100 | 100 | 94 | 100 | 95 | 95 | 27 | 27 | 27 |
| *1648 | Hypothetical | 111 | 0 | 69 | 69 | 0 | 100 | 100 | 94 | 0 | 0 | 94 | 94 | 0 | 94 | 69 | 69 | 0 | 0 | 7 |
| *1649 | Hypothetical | 77 | 0 | 0 | 0 | 0 | 100 | 100 | 99 | 0 | 0 | 96 | 96 | 0 | 96 | 0 | 0 | 0 | 0 | 4 |
| *1650 | Hypothetical | 73 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 0 | 0 | 94 | 94 | 0 | 94 | 0 | 0 | 0 | 0 | 7 |
| 1651 | Hypothetical | 832 | 85 | 91 | 91 | 90 | 100 | 90 | 100 | 90 | 90 | 100 | 97 | 90 | 100 | 91 | 91 | 0 | 0 | 387 |
| *1652 | Hypothetical transposon-related | 130 | 62 | 78 | 78 | 78 | 100 | 100 | 100 | 78 | 78 | 98 | 93 | 78 | 98 | 78 | 78 | 0 | 0 | 7 |
| 1653 | Hypothetical | 87 | 60 | 72 | 72 | 72 | 100 | 100 | 100 | 72 | 72 | 97 | 82 | 72 | 97 | 72 | 72 | 0 | 0 | 7 |
| 1654 | Hypothetical transposon-related | 359 | 65 | 86 | 86 | 87 | 100 | 96 | 100 | 86 | 86 | 98 | 91 | 86 | 98 | 86 | 86 | 0 | 0 | 6 |
| 1655 | Hypothetical | 359 | 67 | 92 | 92 | 91 | 100 | 100 | 100 | 91 | 91 | 99 | 96 | 91 | 99 | 92 | 91 | 0 | 0 | 26 |
| 1656 | Hypothetical | 101 | 63 | 92 | 92 | 92 | 100 | 100 | 100 | 92 | 92 | 98 | 94 | 92 | 92 | 92 | 92 | 0 | 0 | 27 |
| 1657 | Hypothetical | 112 | 55 | 87 | 87 | 87 | 100 | 100 | 100 | 87 | 87 | 100 | 95 | 87 | 100 | 87 | 87 | 0 | 0 | 27 |
| 1658 | Conserved hypothetical transposon-related | 102 | 26 | 85 | 85 | 85 | 100 | 100 | 100 | 85 | 85 | 100 | 85 | 85 | 100 | 85 | 85 | 0 | 0 | 27 |
| *1659 | Hypothetical protein | 95 | 71 | 78 | 80 | 87 | 100 | 100 | 100 | 87 | 87 | 99 | 91 | 87 | 99 | 78 | 78 | 0 | 0 | 22 |
| 1660 | Acetyl-coenzyme A carboxyl Transferase alpha chain | 315 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 100 | 1014 |
PEG originally identified as possibly chicken pathogen specific are indicated (*). Annotations are derived from SEED augmented by BLASTp queries at NCBI. The 29 human isolates are from the clades surrounding the ED98 clade as described in the text and identified in Table S1. Avian isolates include 2 chicken commensals (ch3 and ch5); 10 chicken pathogens from the ED98 clade; a partridge (pa3), pheasant (ph3) and unknown host (X22) from the X22 clade; and chicken isolate ch24 (not in the ED98 or X22 clade). Count for NCBI S. aureus Desc is the number of descriptions returned (max 5000) with >80% identity for an NCBI tBLASTn search of S. aureus (Taxonomy ID: 1280), which roughly approximates the number of genome entries encoding a highly related polypeptide. Additional genome details are in the text and Table S1.
SaPI are mobile elements that are packaged by “helper” virus assembly systems and integrate at a specific location (Tormo et al., 2008; Dearborn and Dokland, 2012). The SaPI in question is integrated between genes for a Methionine ABC transporter (PEG 754) and a CsbD stress response protein (PEG 779). We used ED98 PEG 755 SaPI integrase as query for a tBLASTn search of the S. aureus genome accessions in NCBI to survey for the number of entries with an integrase homolog (≥80% identity) directly downstream of PEG 754, the Met ABC transporter homolog. Out of 1012 PEG 754 homologs (Table 3) there were 307 entries with a PEG 755 homolog directly downstream (i.e. 307 out of 1012 contained a SaPI at this location). Conversely, there were 568 entries where PEG 779 directly followed PEG 754, consistent with no SaPI inserted in those genomes. Our tBLASTn searches also were consistent with the SaPI mobile element extending from the integrase (PEG 755) through PEG 778, as we never found homologs of PEGs 777, 778, 780 or 781 directly downstream of PEG 754 (data not shown). Further tBLASTn queries using the PEG sequences from 754 to 779 demonstrated variability across the SaPI for whether the ED98 PEG coding sequences are conserved in the genomes of other S. aureus isolates (see Count for NCBI S. aureus Desc column in Table 3). PEG 755, 759, 760, 764-773 appear to be present in many (Count range: 199-839) genomes, consistent with many having defined functions for SaPI mobilization and therefore conserved in many SaPIs. The regions for eleven PEGs 756-758, 761-763, and 774-778 are found in far fewer (Count range: 10-89) genomes.
We then used tBLASTn queries to identify those S. aureus accessions in NCBI that contain homologs (≥80% identity) to these less conserved PEGs. Only 8 accessions contained all 11 of these PEGs: ED98, ch21, ch22, B2-15A, B8-13D, B4-59C, B3-17D and X22 (Table 3). All are chicken pathogens from the same clade as ED98 with the exception of X22, which is a genome deposited by the China Animal Disease Control Center but the host is not specified. Notably, isolates ch3 and ch5, the chicken commensals, have no SaPI downstream of the MetABC transporter (PEG 754). Therefore, this SaPI does not specify chicken colonization, but rather likely significantly contributes to pathogenicity in chickens. Clearly, more work needs to be focused on the actual functions of many of the genes in this SaPI and whether the chicken pathogenicity results from the combination of PEG 48 and this particular SaPI. Some of the PEGs in this SaPI have homologs in SaPIs in human isolates, however, the actual functions of the predicted polypeptides are not well understood (Table 3). Hypothetical PEG 777, in the SaPI appears to be restricted to only the chicken pathogens.
We next explored whether there were signatures of selection in the evolution of the genome of the chicken-restricted clade of pathogens as they progressed from ED98 in the 1980s to the genomes we characterized from 2019. We used the proteome comparison tool in the RAST SEED Viewer to further analyze the evolution of this S. aureus chicken clade (Table S2). We selected our assembly for 1519 to represent the 2019 isolates from Farm 3, as it was the largest assembly with the fewest contigs. The analysis included ED98 representing a 1986-1987 isolate, ch21 from Poland in 2008, and B4-59C from 2010 in Tulsa, Oklahoma retail poultry meat. The SEED Viewer filter was set with the ED98 proteome as reference, to identify predicted proteins absent (<50% identity) in one or more of the other 3 proteomes (Table S2). The analysis suggests that 32 proteins (31 phage and hypothetical proteins, and a efflux pump for Tetracycline resistance) were lost between 1996 and 2008. Eight phage and hypothetical proteins in ED98 and Ch21, were lost between Poland in 2008 and Tulsa in 2010, and only 4 hypothetical proteins in ED98, ch21 and B4-59C, are absent in 1519 in Northwest Arkansas in 2019. We then reversed the analysis with 1519 as the reference to identify new proteins that appeared in the lineage from ED98, through Ch21 to B4-59C to 1519. The analysis identified 35 proteins present in 1519 for which the other 3 genomes lack a protein with 50% or greater identity. Twenty-eight are phage, hypothetical or plasmid-maintenance related. The remaining 7 include a partial coding sequence for phosphoglycerate kinase (PEG 421), an aminoglycoside N6’-acetyltransferase (PEG 1919), a DUF1541 domain-containing protein (PEG 32), and a lead/cadmium/zinc/mercury/copper transporting ATPase (PEG 33).
Two open reading frames (PEG 1916 and 1917) are also novel to the 1519 genome that present overlapping open reading frames (ORFs) and may represent a frame shifted LPXTG cell wall anchor protein with a SdrC adhesin of unknown specificity. The adjacent gene (PEG 1915) is for a SdrD adhesin of unknown specificity. We reexamined this 4512 bp contig for assembly errors and could find none; templated alignments of the Illumina reads for 1519 identified no frame shifts (data not shown). BLASTn queries of S. aureus draft genomes at NCBI found identical regions in several other genomes from human isolates. Therefore, our assembly for PEG 1916 and 1917 is valid, but that does not preclude ribosomal frame-shifting to produce a fusion polypeptide for these 2 ORFs. The Unicycler assembly predicted this 4512 bp contig to be circular, so this contig likely represents a plasmid that encodes one or more adhesin functions. DUF1541 proteins of similar size to that for PEG 32 are found in a wide range of different bacterial species. The divalent cation transporter is found in many different Staphylococcus species. The aminoglycoside-N6’-acetyltransferase (PEG 1919) has no significant BLASTp homologs in any S. aureus genome in NCBI, and the best homologs are 70% identical in isolates of Staphylococcus sciuri, Staphylococcus lentus, and Staphylococcus fleurettii. PEG 1919 is present in a 4357 bp contig that Unicycler did not circularize. However, the contig termini each contain portions of a plasmid recombination MobE mobilization protein that likely could be fused into one ORF using long read sequence data. The other genes in this contig are two hypothetical proteins, a tetracycline resistance predicted region, and an ArsR-family transcriptional regulator. However, this contig appears to possibly contain a mobile element affecting antibiotic resistance with the aminoglycoside transferase and the tetracycline resistance marker.
Further evidence of evolution of this genome is for proteins highly conserved (>80%) in 1519 and B4-59C, but not (<50%) in ED98 and ch21. The isolate genomes from Tulsa 2010 and Arkansas 2019 contain 16 PEGs not found in ED98 and ch21; including a toxic shock syndrome toxin 1 (PEG 327), and a phage associated exotoxin superantigen (PEG 329). Interestingly, this region appears to be a probable SaPI, as inspection of neighboring genes identifies an integrase (PEG 335), terminase (PEG 326) and SaPI associated homologs (PEGs 325 and 330). PEG 327 and 329 are in a 97,219 bp contig predicted as circular that also encodes a number of genes for exotoxins and SaPI functions. The contig only contains 2 phage predicted proteins, so it may be a large plasmid containing many virulence determinants. Additional PEGs found in B4-59C and 1519 genomes but not ED98 and ch21 include a cluster (PEG 2179, 2180, and 2181) of homologs to hypothetical proteins found in SaPIs. However, these PEGs are on a 3446 bp contig in the 1519 assembly. Inspection of the SEED annotation of the B4-59C assembly identifies flanking integrase and terminase homologs and other SaPI associated homologs. So, this region also may represent a functional, mobile SaPI. There were only 3 proteins identified in 1519, B4-59C and Ch21, but not in ED98; 2 are hypothetical (PEG 1197 and 1421) and the other a secretory antigen SsaA-like protein (PEG 49). This secretory antigen has been associated with transposons and also annotates as a CHAP domain protein, or putative cell wall lysis protein.
CONCLUSIONS
The 4 E. coli genomes we characterized from 3 different farms show a wide degree of genomic variation within the broiler industry in Arkansas. Isolates 1512 and 1527 are highly related as they came from different BCO lesions in the same lame bird. We have previously reported that, for individual lame birds, we recover the same species from multiple BCO lesions, and sometimes from the blood (Al-Rubaye et al., 2015; Al-Rubaye et al., 2017). The genomes of E. coli 1409, 1413, and 1512/1527 are very distinct and come from very different clades. Remarkably, 2 neighboring farms (Farm1 and Farm2) supplied by the same hatchery and operated by the same integrator, had very different E. coli (1409 and 1413) involved in BCO lameness outbreaks. Whereas 1409 comes from a clade that seems to be predominantly isolated from poultry, the clades for 1413 and 1512/1527 appear to distribute to a more diverse set of hosts (Figure 1). Reports from Brazil using virulence genes or MLST reported distinct E. coli genotypes within a flock (Braga et al., 2016). However, they did not place their E. coli isolates within the E. coli pangenome. Overall, the E. coli isolates from broiler BCO lesions in Arkansas appear to be highly diverse. These E. coli derive from different clades that contain E. coli closely related to isolates from nonchicken hosts. This is more consistent with the E. coli on each farm originating from other hosts (zoonoses) or each farm could have “evolved” their own E. coli BCO pathogen over many flocks and years.
Conversely, the S. aureus isolates from Farm 3 appear to come from a clade of chicken-specific isolates associated exclusively with chicken hosts for at least 5 decades. The phylogenomics indicate this clade is restricted to, or specialized for, infecting chickens (Figure 2 and 3). Our genomic analyses of the evolution of the genome of this clade of S. aureus that infects chickens demonstrates that the genome continues to evolve from 1986 to 2019. The evidence is for acquisition of additional adhesins and virulence determinants, possibly facilitated by transduction of new virulence determinants in mobile SaPI. One of these SaPI contains a number of genes that appear to be unique to all the chicken pathogens in this clade, but as the genes are all annotated as hypothetical proteins, their role(s) in pathogenesis are not apparent. Further molecular genetics manipulation could help to identify the roles of these hypothetical proteins (e.g., survival in the host, tissue tropism, toxin function, etc.). This clade appears to have been in the Oklahoma, Arkansas region for more than a decade, but how it is transmitted to different farms or flocks is not clear. It could be vertically transmitted from hen to chicks. Alternatively, chicks could be exposed at the hatchery, or workers could spread the bacterium to farms through breakdowns in biosecurity.
ACKNOWLEDGMENTS
Support has been provided in part by grants from Cobb-Vantress, Inc., Zinpro LLC, and the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000. BD was supported by KNOW (Leading National Research Centre), Scientific Consortium “Healthy Animal – Safe Food”, under of Ministry of Science and Higher Education No. 05-1/KNOW2/2015, and Department of Pathology and Veterinary Diagnostics, Institute of Veterinary Medicine, Warsaw University of Life Sciences–SGGW, Poland. Thank you to Dr. Mark Hart and Dr. Karen Christensen for thoughtful comments regarding this manuscript.
DISCLOSURES
The authors state that they have no conflicts or competing interests regarding this work. The funders had no role in the design of this study, the interpretation of the results, or the contents of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2021.101148.
Appendix. Supplementary materials
REFERENCES
- Al-Rubaye A.A.K., Couger M.B., Ojha S., Pummill J.F., Koon J.A., II, Wideman R.F., Jr., Rhoads D.D. Genome analysis of Staphylococcus agnetis, an agent of lameness in broiler chickens. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Rubaye A.A.K., Ekesi N.S., Zaki S., Emami N.K., Wideman R.F., Rhoads D.D. Chondronecrosis with osteomyelitis in broilers: Further defining a bacterial challenge model using the wire flooring model. Poult. Sci. 2017;96:332–340. doi: 10.3382/ps/pew299. [DOI] [PubMed] [Google Scholar]
- Alrubaye A., Ekesi N.S., Hasan A., Koltes D.A., Wideman Jr R., Rhoads D. Chondronecrosis with osteomyelitis in broilers: Further defining a bacterial challenge model using standard litter flooring and protection with probiotics. Poult. Sci. 2020;99:6474–6480. doi: 10.1016/j.psj.2020.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrubaye A.A.K., Ekesi N.S., Hasan A., Elkins E., Ojha S., Zaki S., Dridi S., Wideman R.F., Rebollo M.A., Rhoads D.D. Chondronecrosis with osteomyelitis in broilers: Further defining lameness-inducing models with wire or litter flooring, to evaluate protection with organic trace minerals. Poult. Sci. 2020;99:5422–5429. doi: 10.1016/j.psj.2020.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen J.R., Andreasen C.B., Anwer M., Sonn A.E. Heterophil chemotaxis in chickens with natural Staphylococcal infections. Avian Dis. 1993;37:284–289. [PubMed] [Google Scholar]
- Argimón S., Abudahab K., Goater R.J.E., Fedosejev A., Bhai J., Glasner C., Feil E.J., Holden M.T.G., Yeats C.A., Grundmann H., Spratt B.G., Aanensen D.M. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microbial Genomics. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga J.F.V., Chanteloup N.K., Trotereau A., Baucheron S., Guabiraba R., Ecco R., Schouler C. Diversity of Escherichia coli strains involved in vertebral osteomyelitis and arthritis in broilers in Brazil. BMC Vet. Res. 2016;12:140. doi: 10.1186/s12917-016-0762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth A. Infectious components of broiler lameness: a review. Worlds Poult. Sci. J. 1999;55:327–352. [Google Scholar]
- Butterworth A., Reeves N.A., Harbour D., Werrett G., Kestin S.C. Molecular typing of strains of Staphylococcus aureus isolated from bone and joint lesions in lame broilers by random amplification of polymorphic DNA. Poult. Sci. 2001;80:1339–1343. doi: 10.1093/ps/80.9.1339. [DOI] [PubMed] [Google Scholar]
- Davis J.J., Wattam A.R., Aziz R.K., Brettin T., Butler R., Butler R.M., Chlenski P., Conrad N., Dickerman A., Dietrich E.M., Gabbard J.L., Gerdes S., Guard A., Kenyon R.W., Machi D., Mao C., Murphy-Olson D., Nguyen M., Nordberg E.K., Olsen G.J., Olson R.D., Overbeek J.C., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomas C., VanOeffelen M., Vonstein V., Warren A.S., Xia F., Xie D., Yoo H., Stevens R. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–d612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn A.D., Dokland. T. Mobilization of pathogenicity islands by Staphylococcus aureus strain Newman bacteriophages. Bacteriophage. 2012;2:70–78. doi: 10.4161/bact.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziva F., Hauser H., Connor T.R., van Diemen P.M., Prescott G., Langridge G.C., Eckert S., Chaudhuri R.R., Ewers C., Mellata M., Mukhopadhyay S., Curtiss R., 3rd, Dougan G., Wieler L.H., Thomson N.R., Pickard D.J., Stevens M.P. Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveal the evolution of E. coli lineages pathogenic for poultry via distinct mechanisms. Infect. Immun. 2013;81:838–849. doi: 10.1128/IAI.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lethey H., Huber-Eicher B., Jungi T.W. Exploration of stress-induced immunosuppression in chickens reveals both stress-resistant and stress-susceptible antigen responses. Vet. Immunol. Immunopathol. 2003;95:91–101. doi: 10.1016/s0165-2427(02)00308-2. [DOI] [PubMed] [Google Scholar]
- Emslie K.R., Nade. S. Acute hematogenous Staphylococcal osteomyelitis: a description of the natural history in an avian model. Am. J. Pathol. 1983;110:333–345. [PMC free article] [PubMed] [Google Scholar]
- Han M.V., Zmasek. C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking, P. M. 1992. Musculo-skeletal disease in heavy breeding birds. Pages 297–309 in Bone Biology and Skeletal Disorders in Poultry. C. C. Whitehead, ed. Carfax Publishing Company, Abingdon, UK.
- Jiang T., Mandal R.K., W. R.F., Jr., Khatiwara A., Pevzner I., Kwon Y.M. Molecular survey of bacterial communities associated with bacterial chondronecrosis with osteomyelitis (BCO) in broilers. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibenge F.S.B., Wilcox G.E., Perret D. Staphylococcus aureus isolated from poultry in Australia I. Phage typing and cultural characteristics. Vet. Microbiol. 1982;7:471–483. doi: 10.1016/0378-1135(82)90064-5. [DOI] [PubMed] [Google Scholar]
- Lees J.A., Harris S.R., Tonkin-Hill G., Gladstone R.A., Lo S.W., Weiser J.N., Corander J., Bentley S.D., Croucher N.J. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V., Desper R., Gascuel O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder B.V., Guinane C.M., Ben Zakour N.L., Weinert L.A., Conway-Morris A., Cartwright R.A., Simpson A.J., Rambaut A., Nübel U., Fitzgerald J.R. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiamele P., Nicholson B., Wannemuehler Y., Seemann T., Logue C.M., Li G., Tivendale K.A., Nolan L.K. Complete genome sequence of the avian pathogenic Escherichia coli strain APEC O78. Genome Announc. 2013;1 doi: 10.1128/genomeA.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee P., McCullagh J., Thorp B., Ball H., Graham D., McCullough S., McConaghy D., Smyth J. Study of leg weakness in two commercial broiler flocks. Vet. Rec. 1998;143:131–135. doi: 10.1136/vr.143.5.131. [DOI] [PubMed] [Google Scholar]
- McNamee P.T., Smyth. J.A. Bacterial chondronecrosis with osteomyelitis ('femoral head necrosis') of broiler chickens: a review. Avian Pathol. 2000;29:477–495. doi: 10.1080/030794500750047243. [DOI] [PubMed] [Google Scholar]
- Mellata M., Touchman J.W., Curtiss R., III Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli χ7122 (O78K80H9) PLoS One. 2009;4:e4232. doi: 10.1371/journal.pone.0004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalib A., Riddell C., Osborne A.D. Studies on the pathogenesis of Staphylococcal osteomyelitis in chickens. II. Role of the respiratory tract as a route of infection. Avian Dis. 1983;27:157–160. [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R., Xia F., Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L., Glover R.H., Humphris S., Elphinstone J.G., Toth I.K. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods. 2016;8:12–24. [Google Scholar]
- Rodriguez-Siek K.E., Giddings C.W., Doetkott C., Johnson T.J., Nolan L.K. Characterizing the APEC pathotype. Vet. Res. 2005;36:241–256. doi: 10.1051/vetres:2004057. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Shwani A., Adkins P.R.F., Ekesi N.S., Alrubaye A., Calcutt M.J., Middleton J.R., Rhoads D.D. Whole genome comparisons of Staphylococcus agnetis isolates from cattle and chickens. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.00484-20. e00484-00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker M.J., Brash M.L., Weisz A., Ouckama R.M., Slavic D. Arthritis and osteomyelitis associated with Enterococcus cecorum infection in broiler and broiler breeder chickens in Ontario, Canada. J. Vet. Diagn. Invest. 2010;22:643–645. doi: 10.1177/104063871002200426. [DOI] [PubMed] [Google Scholar]
- Tarr P.E., Sakoulas G., Ganesan A., Smith M.A., Lucey D.R. Hematogenous enterococcal vertebral osteomyelitis: report of 2 cases and review of the literature. J. Infect. 2004;48:354–362. doi: 10.1016/j.jinf.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Thøfner I.C.N., Poulsen L.L., Bisgaard M., Christensen H., Olsen R.H., Christensen J.P. Longitudinal study on causes of mortality in Danish broiler breeders. Avian Dis. 2019;63:400–410. doi: 10.1637/12006-113018-Reg.1. [DOI] [PubMed] [Google Scholar]
- Thorp B.H. Skeletal disorders in the fowl: a review. Avian Pathol. 1994;23:203–236. doi: 10.1080/03079459408418991. [DOI] [PubMed] [Google Scholar]
- Thorp B.H., Whitehead C.C., Dick L., Bradbury J.M., Jones R.C., Wood A. Proximal femoral degeneration in growing broiler fowl. Avian Pathol. 1993;22:325–342. doi: 10.1080/03079459308418924. [DOI] [PubMed] [Google Scholar]
- Tormo M.Á., Ferrer M.D., Maiques E., Úbeda C., Selva L., Lasa Í., Calvete J.J., Novick R.P., Penadés J.R. Staphylococcus aureus pathogenicity Island DNA Is packaged in particles composed of phage proteins. J. Bact. 2008;190:2434. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Schultz M.B., Zobel J., Holt K.E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman R.F. Bacterial chondronecrosis with osteomyelitis and lameness in broilers: a review. Poult. Sci. 2016;95:325–344. doi: 10.3382/ps/pev320. [DOI] [PubMed] [Google Scholar]
- Wideman R.F., Al-Rubaye A., Gilley A., Reynolds D., Lester H., Yoho D., Hughes J.M., Pevzner I. Susceptibility of 4 commercial broiler crosses to lameness attributable to bacterial chondronecrosis with osteomyelitis. Poult. Sci. 2013;92:2311–2325. doi: 10.3382/ps.2013-03150. [DOI] [PubMed] [Google Scholar]
- Wideman R.F., Prisby. R.D. Bone circulatory disturbances in the development of spontaneous bacterial chondronecrosis with osteomyelitis: A translational model for the pathogenesis of femoral head necrosis. Front. Endocrinol. (Lausanne) 2013;3:183. doi: 10.3389/fendo.2012.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesurendra D.S., Chamings A.N., Bushell R.N., Rourke D.O., Stevenson M., Marenda M.S., Noormohammadi A.H., Stent A. Pathological and microbiological investigations into cases of bacterial chondronecrosis and osteomyelitis in broiler poultry. Avian Pathol. 2017;46:683–694. doi: 10.1080/03079457.2017.1349872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.