To,
The Editor
SARS-CoV-2, the novel coronavirus first detected in China in December 2019, has had a far-reaching impact on global health affecting millions worldwide. As the pandemic has evolved over the past year and half, detailed insight has been gained on the multiple variants of this novel coronavirus and their possible role in the resurgence of this pandemic globally. Variants are often defined as a group of viruses that share distinctive mutations inherited over a lineage. If these multiple mutations accumulate in a lineage, these viruses may evolve and develop into newer strains.1 RNA viruses such as SARS-CoV-2 have a high mutation rate which has led to the emergence of multiple variants throughout the globe after the first wave of the pandemic in early 2020. In the United States (US), multiple variants have already been identified till May 2021 including the B.1.1.7, B.1.351, P.1, B.1.427 and B.1.429 variants. All of these have been labelled by the Centers for Disease Control and Prevention (CDC) as “variants of interest”.2 However, recently a variant “B.1.617″ has been classified as a “variant of concern” (VOC) by the World Health organisation (WHO). This is the fourth such variant after B.1.1.7, B.1.351 and P.1 to have been tagged as a “variant of concern” owing to its increased transmission rates and it is suspected to have been a potential contributor to the ongoing massive second wave of pandemic in India.3 In just a few weeks, the B.1.617 variant established itself as the dominant strain in India besides spreading to more than 40 other countries worldwide.
Epidemiology of SARS-CoV-2 variants in India
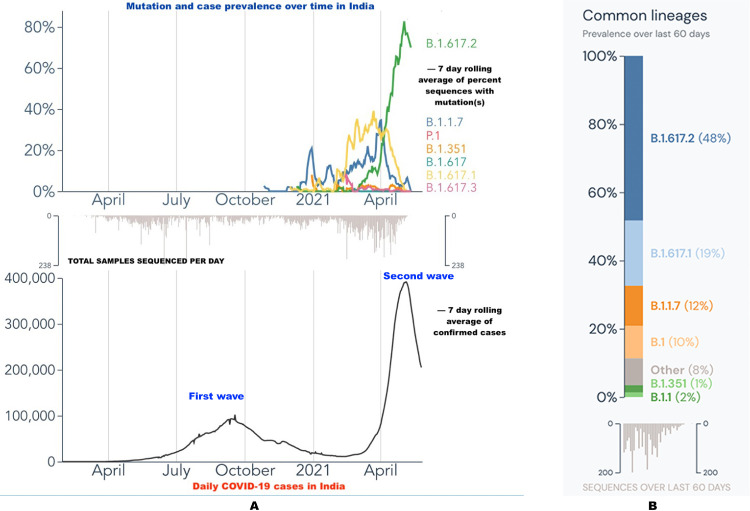
Multiple variants for the novel SARS-CoV-2 including B.1.1.7, B.1.351 and P.1 variant have been classified as “VOC” by the WHO as these variants seem to spread more easily and quickly and have the potential to escape natural/vaccine derived immunity.3 The B.1.617 variant, infamously and wrongly dubbed as the “Indian variant”, was first reported in October 2020 from the state of Maharashtra in India.3 It harbours two important mutations, the E484Q and L452R in the receptor-binding domain (RBD) of the spike protein, leading to increased angiotensin converting enzyme-2 (ACE2) receptor binding. This enhances the transmission capability of this variant leading to greater spread of the disease.4 In addition, these particular mutations in the spike protein also lead to a reduced binding to the selected monoclonal antibodies hence aiding immune escape and conferring increased virulence.4 This particular variant has been reported with three sub-lineages: B.1.617.1, B.1.617.2 and B.1.617.3 with the first two lineages having fueled the second wave of pandemic in India (Fig. 1 A). Data from the Global Initiative on Sharing All Influenza Data (GISAID) database shows that since the beginning of April, both these sub-lineages (B.1.617.1 and B.1.617.2) together account for around 70% of the SARS-CoV-2 genomes sampled in India (Fig. 1B).5., 6., 7. Of these, the B.1.617.2 variant harbouring mutations such as L452R, E484Q, and P681R appears to be more important with its prevalence increasing from just 1% on 1st March 2021 to greater than 70% in the first week of May 2021(Fig. 1A).7 These variants are not only restricted to India and have widely been in circulation among various population groups worldwide since late February 2021.3 The emergence of these novel variants through the accumulation of multiple mutations during the second wave of this COVID-19 pandemic has further iterated the fact that genomic surveillance in the ongoing pandemic is of utmost importance.
Fig. 1.
(A) Line graph showing the mutation and case prevalence over time in India. The novel variant "B.1.617.2" has a sharp rise in April and May 2021 (green line) in the upper half of the figure coinciding with the increasing cases as represented by the second peak in April and May 2021 in the lower half of the figure. (B) Graphical representation of the prevalence of common variants of SARS-CoV-2 over the past 60 days (April and May 2021) in India with “B.1.617.2” being the most dominant variant (48%). (Fig. 1A and 1B source: Outbreak.info; Available online: https://outbreak.info/).7
Impact of these variants in India
RNA viruses including the SARS-CoV-2 owing to their error-prone copying mechanisms can mutate at a faster rate. This can lead to the emergence of multiple variants in a single individual, some of which have a survival advantage in terms of greater affinity to the host receptor, faster replication rates and ability to evade the host immune response.1 Eventually, natural selection among the genetically diverse viral population leads to emergence of newer and more lethal variants of SARS CoV-2.1 There are various problems associated with the emergence of viral variants ranging from diagnostic difficulties, varied symptoms, rapid spread with impact on the disease severity and outcomes. Immunity either natural or following vaccination may not be protective against these novel variants which makes the concept of herd immunity or protective seroprevalence questionable. A major concern regarding the emergence of new variants is whether the available vaccines will be equally efficacious against these variants. COVID-19 variants can notoriously evade the immune response induced by the vaccines leading to infection (asymptomatic/symptomatic) and further bolstering the spread of the disease. This was highlighted in a recent report wherein two fully vaccinated individuals [BNT162b2 (Pfizer–BioNTech) and mRNA-1273(Moderna) respectively] developed breakthrough infection with novel SARS-CoV-2 variants 19 and 36 days following the second dose respectively. This novel variant had considerable similarity to the B.1.1.7 and B.1.526 variants.8 This notion has been further supported through in-vitro studies which reported that plasma from vaccinated individuals were less effective in neutralizing viral variants having E484K-, N501Y- or K417N/E484K/N501 mutations (501Y.V2 and B.1.1.7 variants) in the RBD of S-protein.9 , 10
Real world experience regarding the efficacy of SARS-CoV-2 vaccines against multiple variants have led to mixed results. Oxford/AstraZeneca COVID-19 vaccine (AZD1222) failed to provide protection against mild-to-moderate COVID-19 infection due to the B.1.351 variant.11 However, the vaccine did retain its efficacy against the P.1 variant, B.1.1.7 variant and the recently recognised B.1.617.2 variant. Emergence of a highly transmissible B.1.617.2 variant coinciding with the second wave of pandemic (Fig. 1) did raise concerns regarding the efficacy of vaccines [AZD1222 and BBV152 (Covaxin)] being administered in India. This is of paramount interest in the context of developing countries such as India with limited resources and access to the vaccines. Breakthrough infections two weeks following the second dose of the vaccine have been reported in 13.3% healthcare workers in a small study from India.12 Majority (47.8%) of the breakthrough infections in this study were by the B.1.617.2 variant followed by the B.1 and B.1.1.7 variants.13 A recently released report from Public Health England has stated that both the Pfizer-BioNTech and AZD1222 (two doses) were 87.9% and 59.8% respectively effective against symptomatic disease caused by the B.1.617.2 variant two weeks following the second dose.14 The rapid emergence of these variants can lead to immune evasion in already vaccinated individuals. This highlights the need for a vaccination strategy incorporating multiple variants in a single dose (multivalent vaccines) akin to that of the influenza vaccines based on the prevalent strains in the community. In addition, this also calls for a booster dose in the vaccination regimen for previously vaccinated individuals to cover for potentially significant and lethal SARS-CoV-2 variants in circulation in the community.
Another important aspect with these variants is the impact of the viral genomic mutations on diagnostic accuracy of COVID-19 tests. Molecular tests detect the specific RNA sequences in the viral genomes and mutations within the target sequences can affect the diagnostic performance of these tests leading to false negative results. This is especially significant for tests detecting a single gene target as compared to those incorporating multiple targets. In the US, emergence of the B.1.1.7 variant with deletions positions 69 and 70 on S-gene had an impact on the diagnostic performance of the TaqPath COVID-19 Combo Kit and the Linea COVID-19 Assay Kit leading to false negative results.15 Currently there is no data available regarding the impact of B.1.617 variant on diagnostic accuracy of the molecular tests. There is a need for periodic assessment of the performance of various molecular tests with respect to the circulating variants as highlighted in the recent US food and drug administration (FDA) guidance statement.15
Genomic sequencing - bottlenecks and way ahead
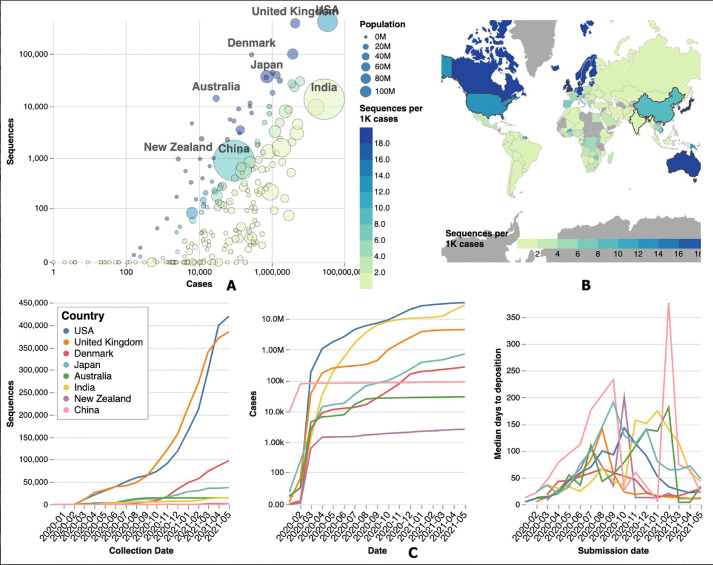
Genomic sequencing plays an important role in identification of novel variants which evolve over a period of time. Variants such as B.1.1.7 and B.1.617 due to their high transmissibility can easily overrun the existing public health infrastructure leading to a catastrophe as experienced in the deadly second wave of the pandemic in UK and India. Emergence of the B.1.1.7 variant and its global spread led to the establishment of the Indian SARS-CoV2 Genomic Consortia (INSACOG) under the Ministry of Health and Family Welfare in late December 2020. This consortium comprises a network of ten national laboratories which is entrusted with monitoring the genomic variations as well as conducting genomic surveillance in India and shares its data with the global databases like GISAID.5 Data from the GISAID database reflects that India trails behind many countries in its quest for genomic sampling of SARS-CoV-2 with just 14,537 (0.05%) of the 27,369,093 cases sequenced [Fig. 2 ].5 This is in contrast to the developed nations such as Australia, UK and the US with sequencing rates of 59.2%, 9.3% and 1.5% respectively. This creates a blind spot for evolution of multiple strains which can then spread far and wide causing multiple waves of the pandemic.
Fig. 2.
(A) Bubble plot comparing the population, actual number of COVID-19 cases and the genome sequencing data (per 1000 cases) deposited in the GISAID database for major countries around the globe. (B) Map comparing the genome sequencing data (per 1000 cases) deposited in the GISAID database across the globe. Developed countries such as United States, United Kingdom and Australia (marked in blue) have the maximum contribution to genomic surveillance while developing nations in Asia and Africa lag well behind (marked in light green). (C) Plot showing the relative contribution of various countries to genomic surveillance, prevalence of disease and the median days to deposition to the GISAID database. India (yellow line) is in the lower part of the curve (far left) in terms of genomic sequencing reflecting poor genomic surveillance however, is ranked second just behind the US (blue line) in terms of absolute number of cases. (Fig. 2A,2B,2C source: COVID CoV Genomics (CG); Available at: covidcg.org) .6
This issue of poor genomic surveillance is not only limited to India but is a problem faced by other resource limited countries. Cost is an important limiting factor especially in developing countries with a need to set-up newer laboratories for genomic sequencing. The problem of poor surveillance is further compounded by lack of technical expertise and limited availability of reagents and other raw materials as most of them need to be imported. In addition, operational issues such as preservation or transport of samples from peripheral laboratories in remote locations further impede genomic surveillance. There is a need for large scale genomic analysis of the variants across the country at multiple points of time. This calls for capacity building, better cooperation among the network labs and involvement of the academic as well as private sector enterprises.3
One of the best strategies to prevent the emergence of newer variants is to prevent the spread of infection at the source. Measures to reduce disease transmission do prevent emergence of newer variants by reducing the disease spread and decreasing chances of viral mutation. This has to be supplemented by vaccination, especially among the high risk groups. However, developing countries such as India have been facing shortage of vaccines coupled by poor rates of vaccination which is further adding to the emergence of these newer variants. Scaling up the manufacturing process and prompt roll out of the vaccines along with equitable distribution among the marginalised population groups could further prevent emergence of novel variants.3 Secondly, in-vitro and real life studies are needed to assess the impact of various mutations on disease transmissibility, severity, diagnostic accuracy of molecular tests and potential for reinfection and failure of vaccination. Vaccines being currently administered or those in pipeline need to be evaluated for protective efficacy against all existing COVID-19 variants in India. And lastly, stringent screening at airports must be conducted regularly to prevent entry of newer variants.
Public health and research communities have regularly emphasized the need for timely tracking of new viral mutations in view of the rapid emergence and spread of new variants of the SARS-CoV-2 virus. A constant vigil to detect and track new COVID-19 variants complimented by large-scale vaccination drive are the need of the hour to curb the potential threat of recurrent waves of the infection, thereby preventing morbidity and mortality.
Financial support and sponsorship
Nil.
Contributions
All the 4 authors contributed to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; & Drafting the work or revising it critically for important intellectual content; & Final approval of the version to be published; & Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Individual contributions
PI, SK and A: Design and drafting of manuscript, acquisition of information. PI, KG and SK: editing, literature review, critical revisions. All authors read and approved the final version of the article.
Declaration of Competing Interest
There are no conflicts of interest to declare
Acknowledgement
We gratefully acknowledge all the Authors from the Originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based.
References
- 1.Fontanet A., Autran B., Lina B., et al. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.About Variants of the Virus that Causes COVID-19. Updated May 20, 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant. (Accessed on 28th May 2021).
- 3.COVID-19 Weekly Epidemiological Update . WHO. 11th May 2021. Available at: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—11-may-2021. (Accessed on 28th May 2021).
- 4.Cherian S, Potdar V, Jadhav S, et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv 2021.04.22.440932. [DOI] [PMC free article] [PubMed]
- 5.Shu Y., McCauley J.GISAID. Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen A.T., Altschuler K., Zhan S.H., et al. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife. 2021;10:e63409. doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullen JL, Tsueng G, Latif AA, et al. and the Center for Viral Systems Biology. Outbreak.info. Available online: https://outbreak.info/(2020). (Accessed on 29th May 2021).
- 8.Hacisuleyman E., Hale C., Saito Y., et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N Engl J Med. 2021 doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z., Schmidt F., Weisblum Y., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier D.A., De Marco A., Ferreira I.A.T.M., et al. Sensitivity of SARS-CoV-2 B1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 11.Madhi S.A., Baillie V., Cutland C.L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyagi K., Ghosh A., Nair D., et al. Breakthrough COVID19 infections after vaccinations in healthcare and other workers in a chronic care medical facility in New Delhi, India. Diabetes Metab Syndr. 2021;15:1007–1008. doi: 10.1016/j.dsx.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baruah S. (2021, May 27). Revealed: B.1.617.2 variant found in healthcare workers of Delhi's Apollo Hospital. The Indian Express. Available at: https://indianexpress.com/article /cities/delhi/revealed-b-1-617-2-variant-found-in-healthcare-workers-of-apollo-hospital-7330960. (Accessed on 28th May 2021).
- 14.Bernal JL, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 variant. medRxiv 2021.05.22.21257658.
- 15.Policy for Evaluating Impact of Viral Mutations on COVID-19 Tests. Available at: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-evaluating-impact-viral-mutations-covid-19-tests. (Accessed on 28th May 2021).