Abstract
Objective:
This study performed an integrated analysis of the cellular and transcriptional differences in peripheral immune cells between patients with Systemic Lupus Erythematosus (SLE) and healthy controls (HC).
Methods:
Peripheral blood was analyzed using standardized flow cytometry panels. Transcriptional analysis of CD4+ T cells was performed by microarrays and Nanostring assays.
Results:
SLE CD4+ T cells showed an increased expression of oxidative phosphorylation and immunoregulatory genes. SLE patients presented higher frequencies of activated CD38+HLA-DR+ T cells than HC. Hierarchical clustering identified a group of SLE patients among which African Americans were overrepresented, with highly activated T cells, and higher frequencies of Th1, Tfh, and plasmablast cells. T cell activation was positively correlated with metabolic gene expression in SLE patients but not in HC.
Conclusions:
SLE subjects presenting with activated T cells and a hyperactive metabolic signature may represent an opportunity to correct aberrant immune activation through targeted metabolic inhibitors.
1. Introduction
Systemic Lupus Erythematosus (SLE) is a highly heterogeneous disease that affects predominantly women and has a higher prevalence in African American (AA), Hispanic, and Asian populations in the United States. Many immunological abnormalities affect both innate and adaptive immune cells during SLE pathogenesis and maintenance, including in CD4+ and CD8+ T cells and more specifically, T follicular helper (Tfh), regulatory T cells (Treg), T effector (Teff), as well as T memory subsets [1]. Increased HLA-DR+ [2, 3] and CD38+ [1] expression has been demonstrated on both CD4+ and CD8+ T cell subsets from SLE patients compared to healthy controls (HC). Previous studies have reported that CD8+HLA-DR+ T cell frequency correlated very strongly with disease flares [3]. Canonically, HLA-DR and CD38 expression on CD8+ T cells is associated with an active viral response, especially in human immunodeficiency virus (HIV) infection [4]. Moreover, CD38hiCD8+ T cells, which present with defective cytotoxic activity, are expanded in SLE patients with recurrent infections [5]. It remains unclear what role CD8+ T cells might have in direct SLE pathogenesis.
In parallel with differences in activation, CD4+ T cells from SLE patients have metabolic abnormalities including elevated activity of mammalian target of rapamycin (MTOR) glycolysis, and oxidative metabolism [6]. Chronic T cell stimulation increases oxidative metabolism [7], which may parallel the increased oxidation observed in SLE T cells that are chronically stimulated by autoantigens. Many metabolic alterations have been reported in CD4+ T cells of patients with SLE as well as in mouse models of the disease [6], but how these alterations contribute to lupus pathogenesis and correlate with the expansion of pathogenic T cell subsets during ongoing disease is not yet understood.
Herein, we compared immune phenotypes in peripheral leukocytes, with a particular focus on CD4+ and CD8+ T cells, in parallel with transcriptional programs in CD4+ T cells between SLE patients and HCs, and evaluated whether these variables could be used to define subsets of patients.
2. Materials and methods
2.1. Patients
Female SLE patients fulfilling at least four 1997 American College of Rheumatology (ACR) criteria for SLE were enrolled between 2013–2015 at the University of Florida (UF) Lupus clinic. Informed consent was obtained from 35 SLE patients and 19 female HCs in accordance with an UF Institutional Review Board (IRB) approved protocol. AA, Caucasian American (CA) and Hispanic American (HA) subjects were enrolled to perform three types of assays: gene expression measured with microarrays or NanoString code sets, and immunophenotyping (Table 1). The patients presented with stable disease and low SLE Disease Activity Index (SLEDAI) of 3 – 4, and were treated with conventional treatments. Although attempts were made to match SLE and HCs, the SLE cohort was older than the HC cohort in the immunophenotyping dataset (Table 1).
Table 1.
Demographic characteristics of SLE patients and HC
| Dataset | Microarray | Nanostring | Immunophenotyping | |||
|---|---|---|---|---|---|---|
| Clinical Status | SLE | HC | SLE | HC | SLE | HC |
| Female # | 7 | 8 | 35 | 19 | 39 | 23 |
| Age average (range) | 33.1 (24–54) | 39.0 (27–59) | 43.0 (24–69) | 37.5 (20–56) | 45.8 (26–69)*** | 30.9 (20–52) |
| Race # (%) | ||||||
| African American | 1 (14.3) | 2 (28.6) | 13 (37.1) | 3 (15.8) | 19 (48.7) | 5 (21.7) |
| Caucasian | 5 (71.4) | 5 (71.4) | 14 (40.0)‡ | 16 (84.2) | 15 (38.5)† | 16 (69.6) |
| Hispanic | 1 (14.3) | 1 (14.3) | 8 (22.9) | 0 (0) | 5 (12.8) | 2 (8.7) |
| SLEDAI average (range) | 4 (0 – 18) | 3 (0 – 14) | 3.1 (0–12) | |||
| Drug regimen # (%) | ||||||
| Hydroxychloroquine | 6 (85.7) | 32 (91.4) | 37 (94.8) | |||
| Mycophenolate Mofetil | 3 (42.9) | 17 (48.6) | 17 (43.6) | |||
| Prednisone | 3 (42.9) | 7 (20.0) | 8 (20.5) | |||
| Methotrexate | 1 (14.3) | 2 (5.7) | 2 (5.1) | |||
| Azathioprine | 1 (14.3) | 7 (20.0) | 10 (25.6) | |||
| Tacrolimus | 0 (0) | 1 (2.9) | 0 (0) | |||
| Dapsone | 1 (14.3) | 0 (0) | 0 (0) | |||
Age and race distributions were compared for each data set.
Student’s t test ***p<0.001,
Fisher’s exact test †p<0.05,
p<0.01
2.2. Gene Expression
CD4+ T cells were isolated by negative selection from whole blood using the RosetteSep CD4+ T cell enrichment cocktail (Stemcell Technologies) and Ficoll-Plaque Plus (GE Healthcare) by gravity centrifugation. RNA isolated from eight HC and seven SLE patients (one individual with a pair of samples 6 months apart) using the RNeasy Mini Kit (Qiagen) was prepared for GeneChip™ Human Transcriptome Array 2.0 microarrays (Affymetrix, Thermo Fisher Scientific) with fragmentation, labeling, hybridization and scanning as a single batch. Microarray CEL files were processed by Partek Genomics Suite 6.6 (Partek) using the RMA normalization procedure. Gene set enrichment analysis was carried out with GSEA software [8] on RMA normalized values using the curated KEGG database v6.0 or HALLMARKS v6.0. A custom gene expression panel (Supplemental Table 1) was designed for nCounter® XT Assay (NanoString Technologies). 50 ng of RNA from CD4+ T cell samples were hybridized to cartridges and scanned according to manufacturer instructions. Data was analyzed with nSolver 2.5 with geometric mean normalization to housekeeping genes as well as to positive and negative spike-in controls.
2.3. Hierarchical Clustering
Hierarchical clustering of immunophenotypes or gene expression data was performed using clustermap with the Ward clustering method within the Seaborn library (version 0.9.0) in Python (version 3.6.6). Missing data were imputed as the group mean.
2.4. Immunophenotyping
Peripheral blood collected in EDTA tubes was stained for six flow cytometry panels covering Teff, Treg, Tfh, T memory, dendritic cell (DC)/monocyte/natural killer (NK), and B cell subsets (Supplementary Fig. 1). Panel design has been previously described [9], except the Tfh panel, which was designed in-house to quantify precursor and memory Tfh [10, 11]. Antibodies are listed in Supplemental Table 2. Samples were analyzed on a BD LSRFortessa (FACSdiva software, BD Biosciences) within 24 h of collection. Data were analyzed using FlowJo (FlowJo, Treestar) and graphed in GraphPad Prism software v7 (GraphPad).
2.5. Statistical Analysis
Z-scores of SLE subjects were normalized to the mean and standard deviation (SD) of the HC cohort. Correlations were evaluated by nonparametric Spearman’s rank. Fisher’s exact test was used to evaluate associations for categorical data. Statistical comparisons between groups were assessed by multiple t-tests with a False Discovery Rate (q) set to 5% using the Two-step up method of Benjamini, Krieger, and Yekutieli in GraphPad Prism software version 7.0b.
3. Results
3.1. Transcriptional profiling of SLE CD4+ T cells shows metabolic dysregulation
Microarray analysis of eight SLE and eight HC ex vivo CD4+ T cell samples was performed to provide for a broad transcriptional signature. The top five pathways differentially expressed between SLE and HC CD4+ T cells included three metabolic pathways: OXPHOS, MTORC1 signaling, and MYC targets (Fig. 1A). An enrichment plot for OXPHOS showed increased expression of metabolic genes in SLE samples (Fig. 1B). A heatmap of specific genes in the OXPHOS pathway clearly illustrates their increased expression in SLE CD4+ T cells (Fig. 1C). In addition, increased expression of VDCA1 in SLE CD4+ T cells was also observed (fold change = 1.34, p = 0.020), which is consistent with the mitochondrial membrane hyperpolarization that has been associated with oxidative stress and mitochondrial dysfunction in SLE [12]. To validate these findings, we designed a custom Nanostring array to test a targeted set of metabolic genes selected from the microarray, as well as a set of T cell immune function genes in an independent cohort consisting of 19 HC and 35 SLE patients (Table 1). Three members of the electron transport chain (NDUFB6, encoding for a subunit of complex 1, SDHD for complex II, and COX6A1 for complex IV), as well as SUCLG1, an enzyme that produces succinate in the tricarboxylic acid (TCA) cycle, were expressed at higher levels in SLE CD4+ T cells using this method (Fig. 1D). These results suggest that the characteristic metabolic alterations of SLE CD4+ T cells result from a global alteration of transcriptional programs, and confirm the major contribution of OXPHOS metabolism that has been reported in these cells [13, 14].
Fig. 1.
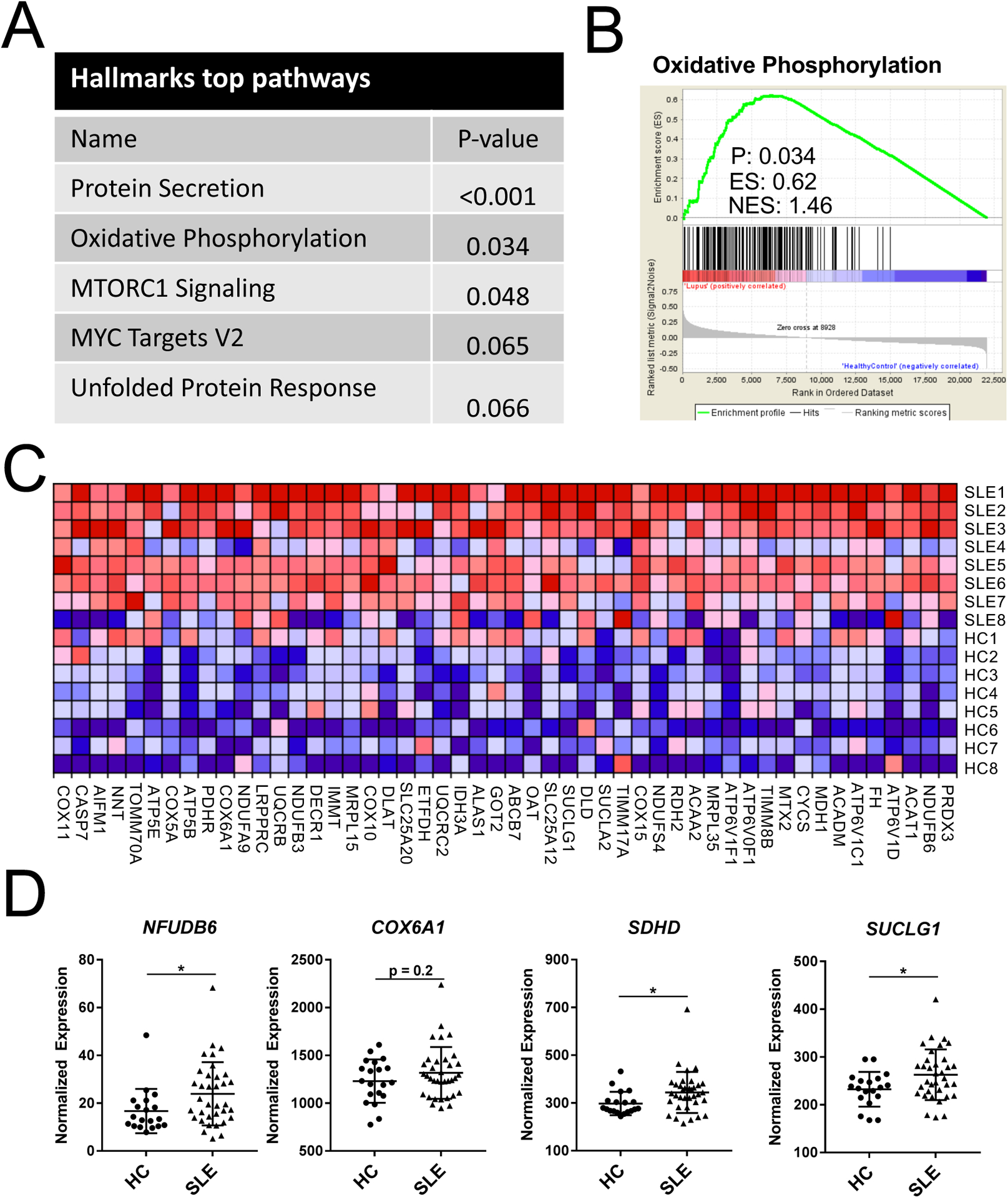
SLE CD4+ T cells have increased oxidative metabolism gene signature when compared with HC. A. Top five HALLMARKS pathway analysis differentially expressed between SLE and HC CD4+ T cells. B. Gene set enrichment plot for OXPHOS. C. Heatmap of microarray data for oxidative metabolism genes. Each row represents a SLE or HC subject. 131 of 193 total OXPHOS pathway genes comprised the core enrichment set. Genes are ranked right to left by enrichment score, and only the top 50 are shown due to space limitation. D. Nanostring nCounter validation of OXPHOS genes overexpressed in SLE CD4 T cells. N = 19 HC and 35 SLE, *p < 0.05. Student’s t-tests.
3.2. Immune and metabolic gene expression is globally upregulated in CD4+ T cells of a subset of SLE patients
We extended our analysis to all 35 genes in the NanoString set, which included both immunoregulatory and metabolic genes, and we identified four differentially expressed genes between HC and SLE subjects (Fig. 2A). Expression of FOXP3, IRF4, TIGIT, and IKZF2, which encodes for HELIOS, was significantly increased in SLE CD4+ T cells (Fig. 2B). We also observed a general trend of increased expression of all 35 genes assessed in SLE CD4+ T cells with an average z-scored increase of 0.60 (inter-quantile range (IQR) 0.25 – 0.84, Fig. 2A). Only three genes (BCL6, CD40L, CXCR5) exhibited mean expressions that trended to be lower in SLE than HC CD4+ T cells. Consistent with this, the first cluster in an unsupervised analysis of this gene expression dataset consisted entirely of SLE patients with broadly higher expression across all probed genes (Supplemental Fig. 2). When limited to all SLE patients, the analysis identified two main clusters (GE1 and GE2), which were again characterized by broad differences in gene expression (Supplemental Fig. 3A). We compared the cumulative gene expression of the two SLE clusters to HC and found that both the immunoregulatory and the metabolic genes were greatly increased for GE2, while GE1 was similar to HC (Fig. 2C and D). We found 20 of the 35 measured genes to be significantly increased in GE2 over GE1, which included the four genes that were increased in all SLE samples over HC (Supplemental Fig. 3B and C). Interestingly, one gene, BCL6, the master transcriptional regulator of the Tfh subset, was expressed at a higher level in the GE1 group (Supplemental Fig. 3C). While age, SLEDAI, and treatments did not differ between clusters (Supplemental Fig. 3D, E and not shown), GE2 SLE patients more frequently reported as AA (Fisher exact test, p < 0.05). Taken together, we have identified a subset of SLE subjects who exhibit globally increased expression of immunoregulatory and metabolic genes in their CD4+ T cells.
Fig. 2.
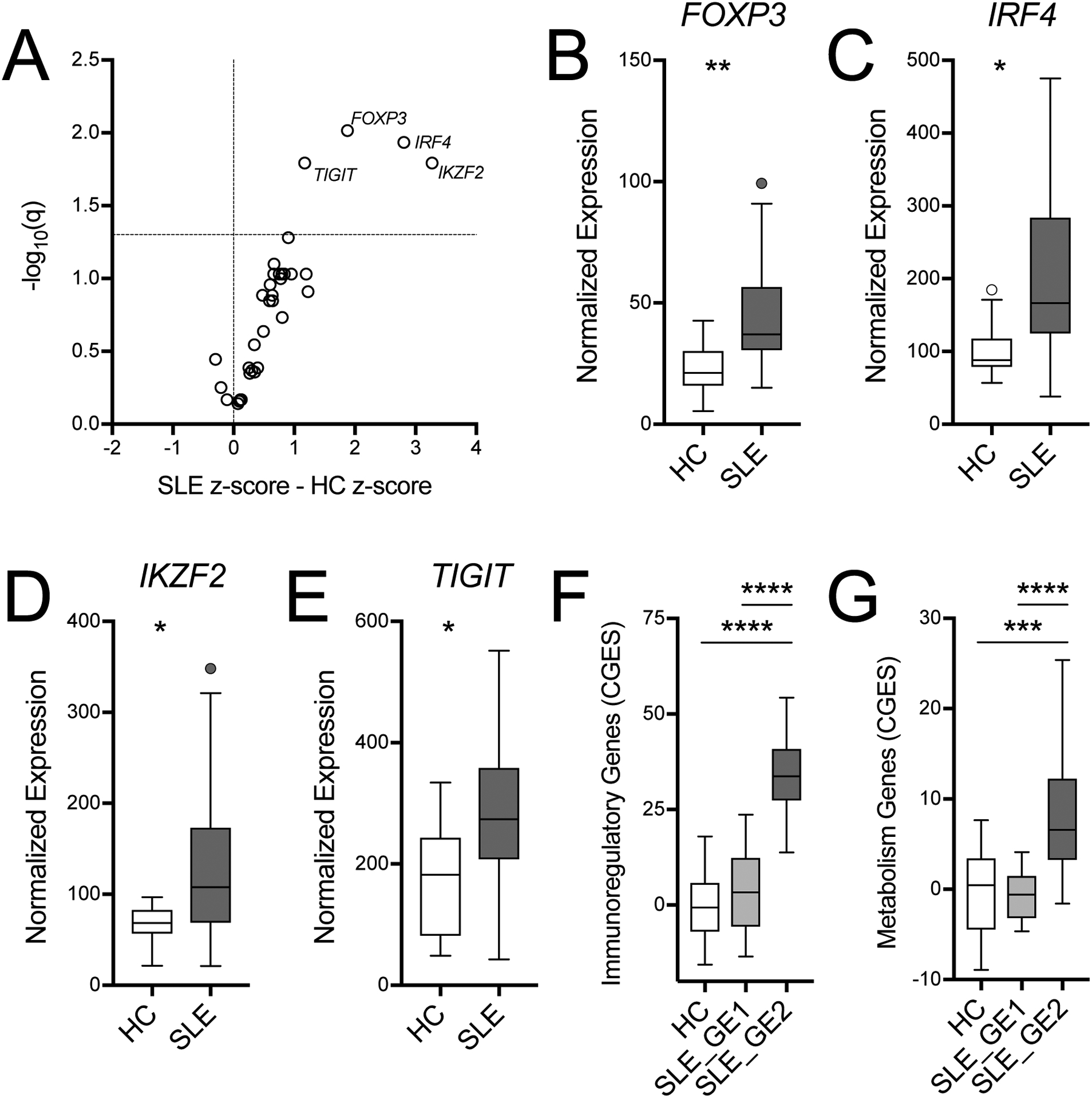
Immune gene expression is altered in SLE CD4+ T cells. A. Volcano plot of gene expression from a Nanostring code set of 35 genes indicates that 4 genes are significantly upregulated in SLE CD4 T cells. B. Normalized expression of individual genes: FOXP3, IRF4, IKZF2, and TIGIT. SLE cluster GE2 had a higher cumulative gene expression score (CGES) for immunoregulatory (C) and metabolic genes (D) than either SLE cluster GE1 or HC. Gene function is defined in Supplemental Table 1. CGES was computed as the sum of z-scored gene expression. Box plots are Tukey box and whiskers. Two-stage step-up method FDR t-test *q < 0.05, **q < 0.01. HC: n = 19, SLE: n = 35, GE1: n = 17; GE2: n = 18.
3.3. SLE patients display activated CD4+ and CD8+ T cell immunophenotypes
An activated immune signature involving both innate and adaptive immune subsets has been previously reported in SLE patients, as well as in mouse models of the disease [15]. The increased expression of immunoregulatory and metabolic genes that we found in CD4+ T cells of SLE patients corroborated these findings in one subgroup of patients. To better understand the functional significance of these findings, we performed an unbiased analysis of peripheral immune cell distributions between SLE patients and HCs using standardized human immunophenotyping panels (Supplemental Fig. 1) [9]. From a total of 62 immunophenotypes examined, we identified seven that were significantly different between patients and HC (Fig. 3A and B). HC subjects exhibited higher frequencies of non-class-switched memory B cells (16.5%, 5.4–21.4%, [median, IQR]) and plasmacytoid DCs (pDCs, 24.0%, 17.5–30.2%) as compared to SLE subjects ([2.3%, 1.0–4.0%] and [9.7%, 5.27–14.0%], respectively). Conversely, within several T cell subsets, SLE subjects had higher frequencies of CD38+HLA-DR+ activated cells. These included CD4+ effector memory (Tem, Fig. 3C) for which the median CD38+HLA-DR+ frequency was 13.6% (IQR 7.2–23.6%) for SLE and 6.24% (4.5–7.3%) for HC, CD8+ Tem (Fig. 3D), for which the median CD38+HLA-DR+ frequency was 33.7% (22.9–45.4%) for SLE and 13.9% (7.4–19.4%) for HC, and CD8+ Temra (Tem that re-express CD45RA) for which the median CD38+HLA-DR+ frequency was 32.6% (25.4–48.9%) for SLE and 17.6% (IQR 9.4–30.8%) for HC. Finally, within CD4+ Teff subsets (Fig. 3E), SLE patients had higher frequencies of activated Th1 (8.0% [4.6–16.1%] in SLE and 4.3% [3.8–7.1%] in HC) and activated Th1/17 (3.7% [2.5–8.6%] in SLE and 2.6% [1.6–3.3%] in HC). In addition to these top seven immunophenotypes, we compared the frequency of Tfh and T peripheral helper (Tph) cells, two populations that have been positively correlated with disease severity [16, 17]. There was a trend for Tfh cells and a significantly higher frequency of Tph cells in SLE patients as compared to HC (Fig. 3F and G). In addition, among SLE patients, the frequency of Tph cells was higher in AA than in CA and HA (Fig. 3H).
Fig. 3.
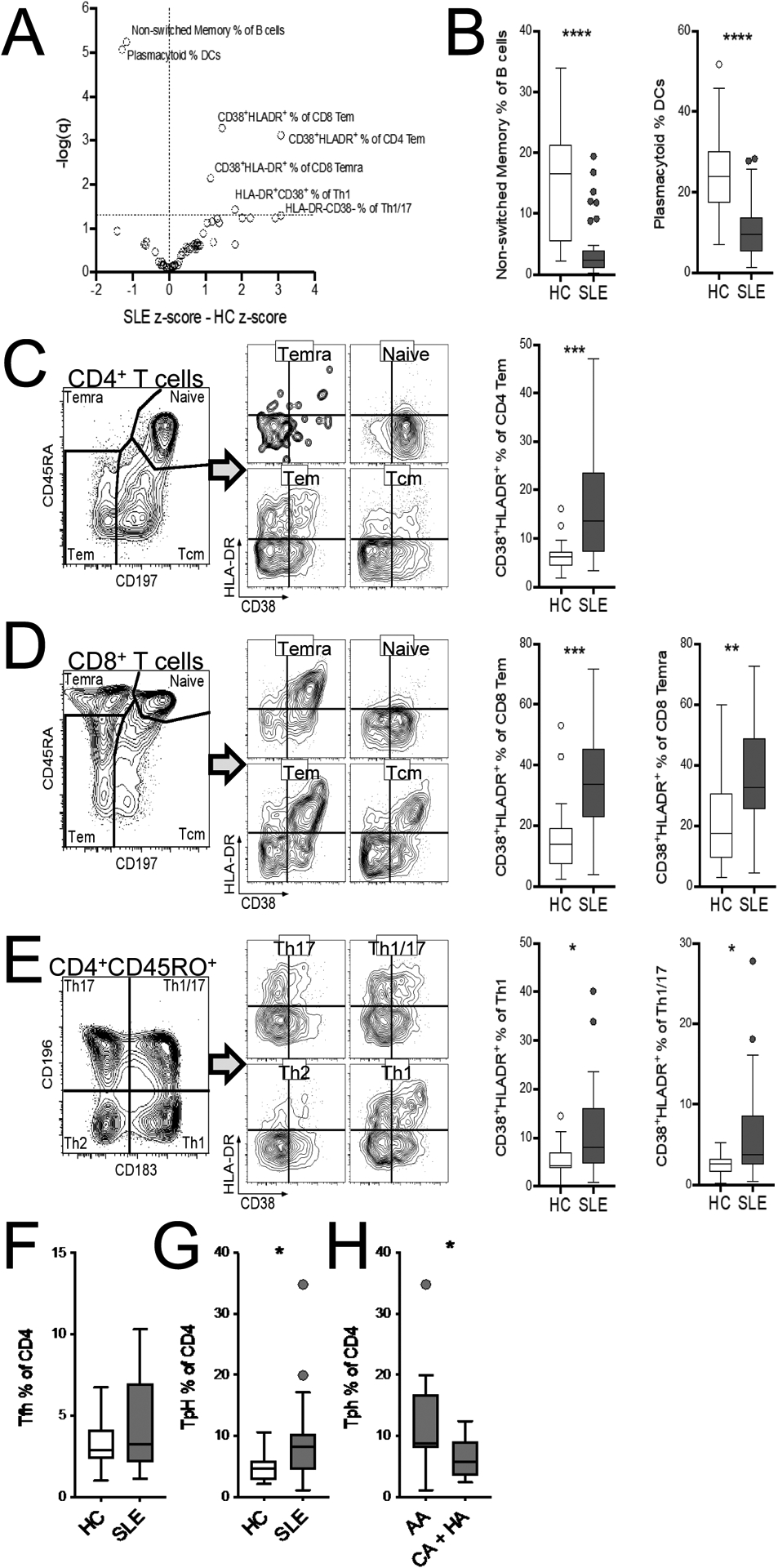
Immunophenotyping identified circulating immune subset differences in SLE subjects (N = 39) versus HC (N = 23). A. Volcano plot of SLE z-scored immunophenotype difference from HC. The horizontal dashed line indicates q = 0.05. B. Comparison of the seven top immunophenotypes between SLE and HC subjects, with representative gatings of activated (CD38+HLA-DR+) naïve and memory CD4+ (C) and CD8+ (D) subsets, as well as activated (CD38+HLA-DR+) CD4+CD45RO+ Teff subsets (E). Comparison of the frequency of Tfh (F) and Tph (G) among CD4+ T cells between SLE and HC subjects. G. Comparison of the frequency of of Tph cells among CD4+ T cells between AA and the combination of CA and HA patients. Box plots are Tukey box and whiskers. Two-stage step-up method FDR T-test * q <0.05, ** q < 0.01, *** q < 0.001, **** q < 0.0001.
We next compared the frequency of the main immunophenotypes between the GE1 and GE2 groups of patients to assess whether the GE2 group with the higher expression of immunoregulatory and metabolic genes presented with specific immunophenotypes. We found no difference in the frequency of Treg cells (Fig. 4A), but the GE2 patients presented a higher frequency of CD45RO+HLA-DR+CD194+ Treg cells (Fig. 4B), which represent effector or memory Treg cells. GE2 patients also presented a decreased expression of CD25 on activated CD4+ T cells (Fig. 4C), increased frequencies of activated Th1, Th1/Th17, Th17, Tfh, Tph cells (Fig. 4D–H), as well as plasmablasts (Fig. 4E). These results showed that a higher expression of immunoregulatory and metabolic genes is associated with the presentation of more activated immunophenotypes in SLE patients.
Fig. 4.
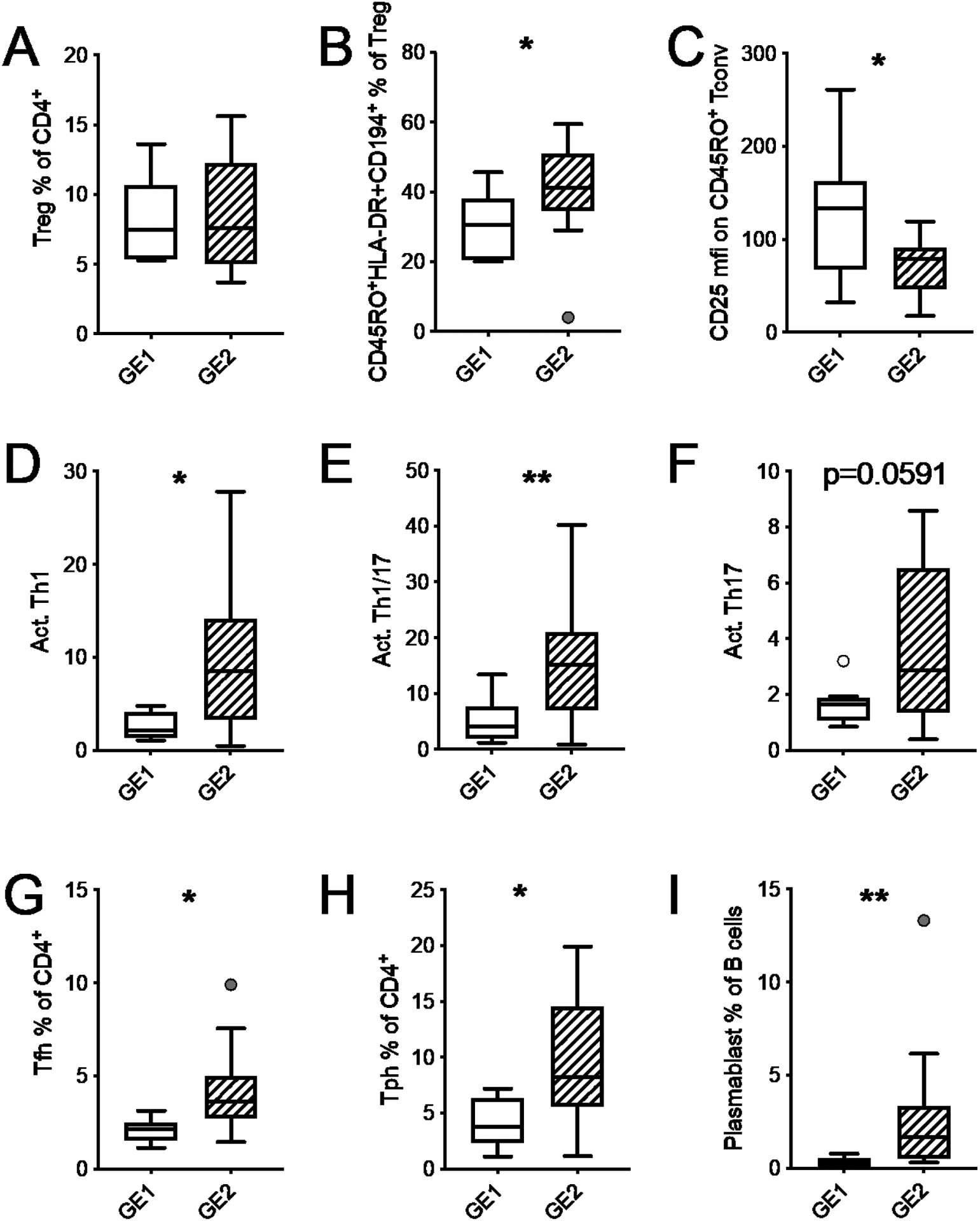
SLE patients from cluster GE2 present activated immunophenotypes. GE1 and GE2 transcriptional clusters were defined in Fig. 2. A. Frequency of Treg cells among CD4+ T cells. B. Frequency of effector cells among Treg cells. C. CD25 expression (mfi) on activated Tconv cells. Frequency of Th1 (D), Th1/Th17 (E), and Th17 (F) among activated CD4+ T cells. Frequency of Tfh (G) and Tph (H) cells among CD4+ T cells. (I) Frequency of plasmablasts among B cells. Box plots are Tukey box and whiskers. Two-stage step-up method FDR T-test * q <0.05, ** q < 0.01.
We next analyzed the correlations between gene expression in total CD4+ T cells and the frequency of immunophenotypes focusing on Tfh and Tph cells, since their expansion strongly correlates with disease activity. No correlation was found between BCL6 and the frequency of either cell type (Fig. 5A and E), but another essential gene that characterizes these subsets, PDCD1 encoding for PD-1, was positively correlated with the frequency of both Tfh and Tph cells (Fig. 5B and F). PD-1 expression on Tfh correlates with disease activity in SLE patients [16]. The expression of IL15R and TIGIT was also correlated with the frequency of Tfh but not Tph cells (Fig. 5C, D, G and H). Not much is known about the expression of IL-15R on Tfh cells, but IL-15 favors the development of Tfh cells [18]. TIGIT+ Tfh cells present a higher B cell helper function than Tfh cells lacking this receptor [19]. Overall, these results suggest that the expansion of Tfh cells in SLE patients correlates with the expression of genes that increase their effector function.
Fig. 5.
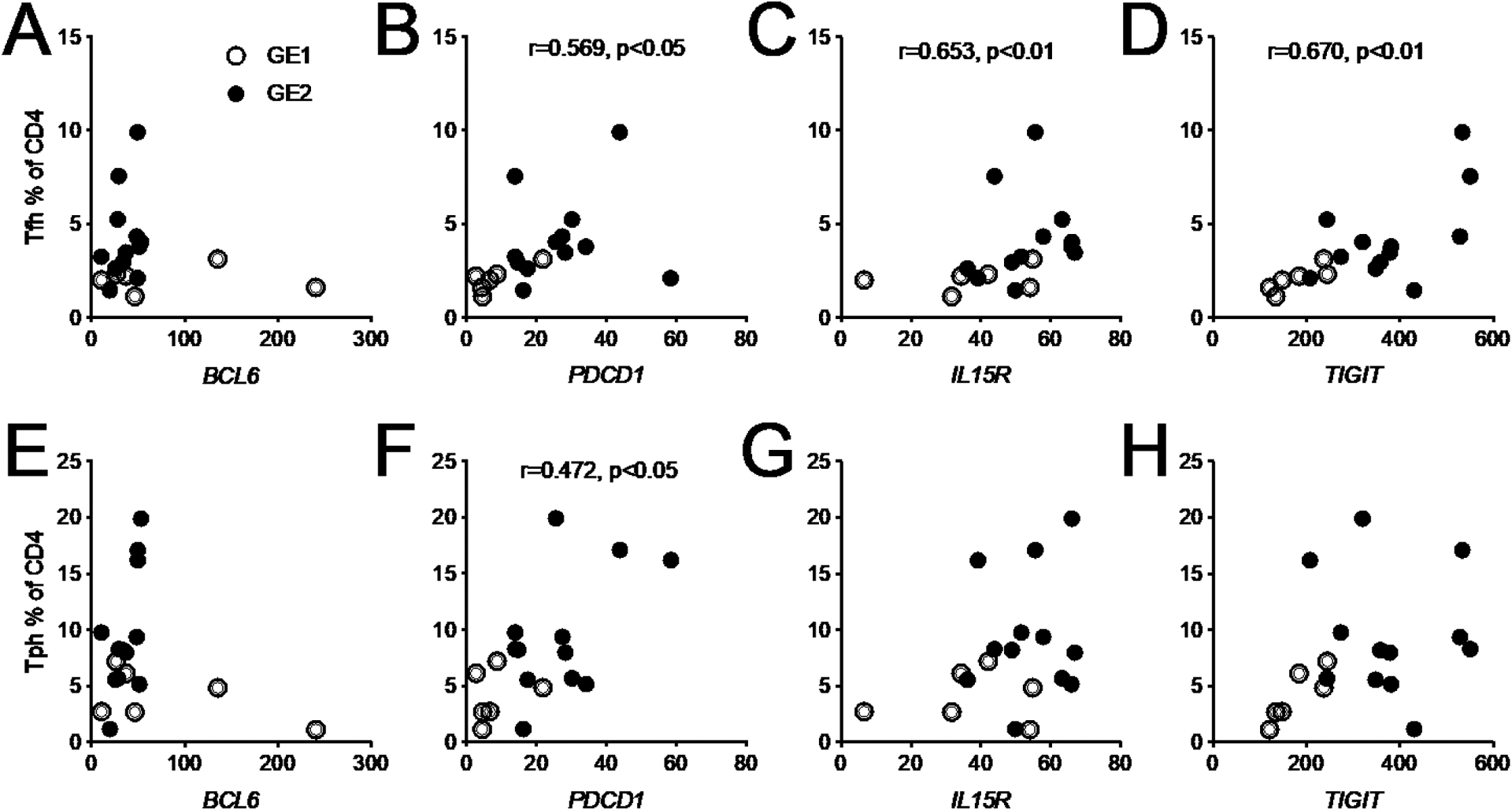
Correlations between the frequency of Tfh (top) or Tph (bottom) cells and the expression in total CD4+ T cells from SLE patients for BCL6, PDCD1, IL15R and TIGIT. GE1 and GE patients are represented by open and closed symbols, respectively. Spearman correlation r values and corresponding p values are shown.
3.4. Hierarchical clustering reveals a distinct group of SLE patients
We next explored the notion that SLE patients may be stratified by their circulating immune profile. When a hierarchical clustering method was used on the entire cohort of SLE patients and HC, all 21 subjects in the top three clusters were SLE patients (clusters 1–3, Supplemental Fig. 4), while only 18 of 41 subjects in the remaining clusters were patients. Subject clustering was driven by three immunophenotype clusters consisting of hyperactivated CD38+HLA-DR+ effector and memory T cells (cluster a), differentiated B cells, NK and monocyte alterations (cluster b), and Tfh subsets (cluster c, Supplemental Fig. 4). Subject cluster 1 was defined by the combination of immunophenotypes cluster a and c, cluster 2 by immunophenotype cluster a only, and cluster 3 by immunophenotype cluster b only. Taken together, we observed profound hyperactivation of T cell subsets, with or without an expansion of Tfh subsets, as well as dysregulation of B cells and innate subsets, that enabled a degree of subject stratification. Moreover, dysregulation in the B cell and innate subsets largely segregated apart from T cell dysregulation.
When applied to SLE patients only, hierarchical clustering of the 62 immunophenotypes revealed two groups defined by 24 immunophenotypes that included activated CD4+ and CD8+ memory and effector T cells, increased proportions of Th1 and Tfh cells, as well as increased B cell differentiation (Fig. 6A and Supplemental Fig. 5A and B). Cluster P1, containing 18 out of 39 SLE patients, displayed increased amounts of these phenotypes relative to P2. Most notably, all CD38+HLA-DR+ activated T cell phenotypes assessed were upregulated, constituting 10 of the 11 most dramatically altered immunophenotypes between P1 and P2 (Supplemental Fig. 5B). Also notable was a B cell differentiation signature in P1 subjects, wherein naïve B cells were decreased while class-switched memory and plasmablasts were increased. Patients in cluster P1 did not differ from P2 in age or SLEDAI (Supplemental Fig. 5C and D). They were, however, significantly enriched for AA patients (14/16 in P1 vs. 5/21 in P2, Fisher’s exact test: p < 0.001). Taken together, these results indicate that a subset of SLE patients present with adaptive immune hyperactivation enriched for activated Th1 and Tfh subsets.
Fig. 6.
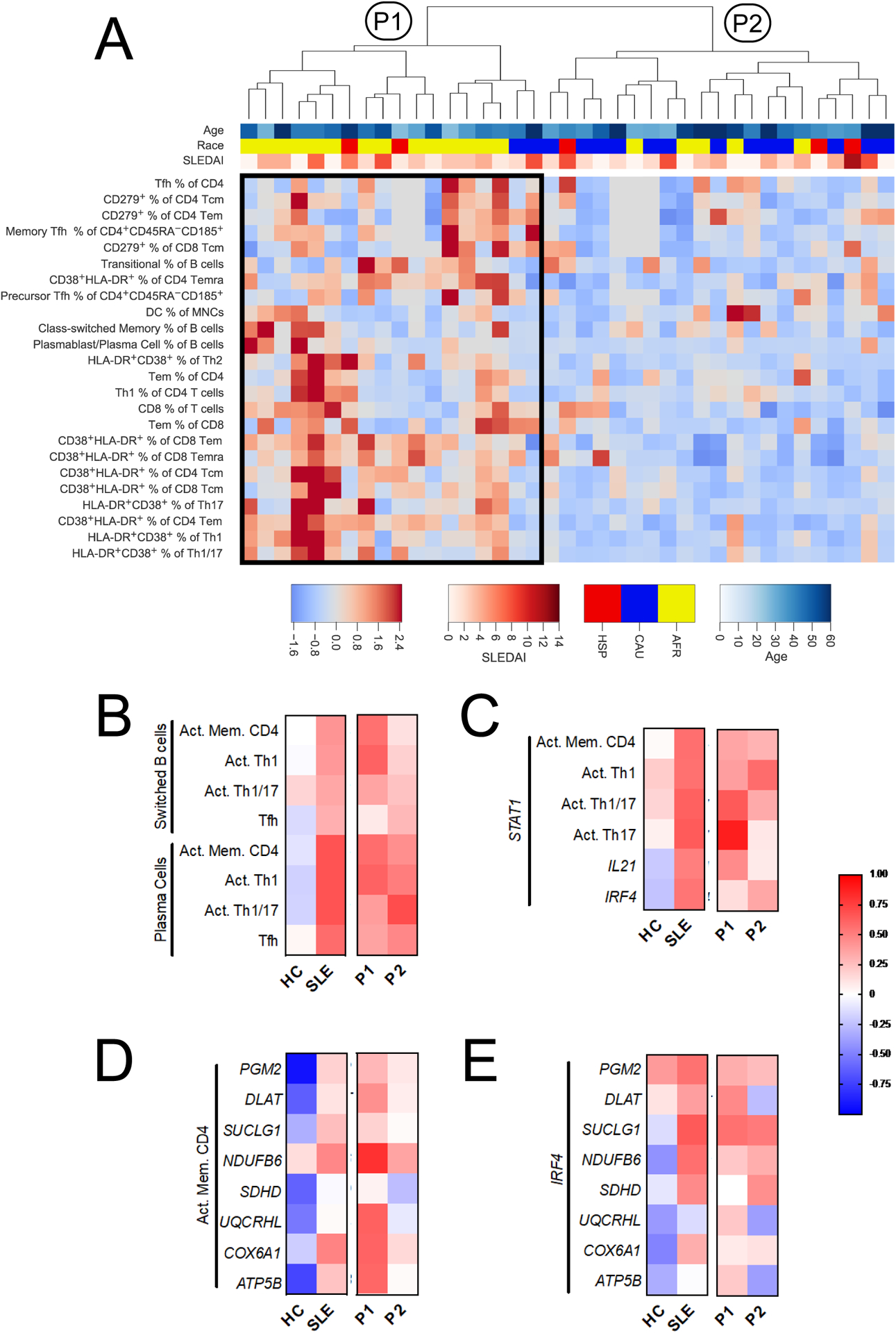
SLE CD4+ T cells present altered patterns of immunophenotype and gene expression correlation. A. Hierarchical clustering of SLE patients by immunophenotypes reveals two major groups, P1 and P2. Cluster P1 featured increased frequencies of the 24 immunophenotypes shown on the heatmap. The color bar legend indicates age (top), ethnicity (middle), and SLEDAI (bottom). Due to space limitations, the phenotype dendrogram is not shown and heatmap excludes all phenotypes that were not in the top phenotype cluster. B – E. Correlations between immunophenotypes and gene expression are shown for HC and SLE, as well as for the SLE immunophenotypes subsets, P1 and P2, defined by hierarchical clustering. B. Correlation between class-switched B cells and plasma cells with CD4+ T cell subsets. C. Correlation between STAT1 gene expression in CD4+ T cells and T cell immunophenotypes, or other genes from the Nanostring code set. D. Correlations between activated memory CD4+ T cells and metabolic gene expression. E. Correlation between the expression of IRF4 and that of metabolic genes. Correlations were evaluated by Spearman r. Activated memory CD4+ T cells (Act. Mem. CD4+, B, C, and D) is the proportion of CD38+HLA-DR+ cells within non-naïve cells (Tcm, Tem, and Temra). For immunophenotype comparisons (B), HC n = 23, SLE n = 39, P1 n = 18, and P2 n = 21. For gene expression and immunophenotype comparisons (C and D), HC n = 9, SLE n = 22, P1 n = 12, and P2 n = 10. For gene expression comparisons (E), HC n = 19 and SLE n = 35, P1 n = 12, and P2 n = 10.
We next assessed whether B cell differentiation phenotypes correlated with activated T cells phenotypes. The frequencies of CD38+HLA-DR+ activated memory CD4+ T cells, Th1, Th1/17, and Tfh cells were each more strongly associated with B cell class switching and plasmablasts differentiation for SLE patients than for HC (Fig. 6B). The frequency of Tph cells was also positively correlated with the frequency of plasmablasts in SLE patients, but the correlation was weaker than for Tfh cells (Tfh: Spearman’s r = 0.572, p = 0.0008, Tph: r = 0.495, p = 0.0046). In addition to confirming that CD4+ T cells contribute to increased class-switching and plasma cell differentiation that characterize SLE, these results implicate the activation of multiple CD4+ T cell subsets in favoring this process. Within SLE patients, the individual associations of class-switched B cells with activated memory and Th1 cells were only significant for P1 subjects (Spearman’s correlation tests, p = 0.027 and p = 0.013, respectively). However, since clusters P1 and P2 were defined these immunophenotypes, this latter observation was predictable.
3.5. CD4+ T cell activation is associated with increased OXPHOS gene expression in a subset of SLE patients
We next sought to explore relationships between phenotypes observed by flow cytometry and gene expression in CD4+ T cells. We therefore examined 9 HC and 22 SLE subjects for which immunophenotyping and CD4+ T cell gene expression data were available. Given the increased percentage and activation level of Th1 cells in SLE P1 subjects relative to P2 subjects (Fig. 6), we examined the expression of STAT1, which mediates the signals for both type 1 and type 2 IFNs in all CD4+ T cell subsets [20]. STAT1 expression correlated more with memory and effector activation, including Th1 effectors, in SLE patients compared to HC (Fig. 6C). STAT1 expression also correlated more with IL21 and IRF4 expression in SLE patients. Among SLE patients, STAT1 correlations with T cell activation were similar in the P1 and P2 groups, except for activated Th17 cells, which were correlated with STAT1 expression in the P1 but not P2 group (Spearman’s correlation tests, p = 0.0003 vs. p = 0.81, respectively).
Next, we explored the relationship between T cell activation with metabolic gene expression. In HC subjects, inverse correlations were observed for the frequency of activated memory CD4+ T cells and expression of seven out of eight metabolic genes, indicative of relative metabolic quiescence (Fig. 6D). Conversely, SLE patients had neutral to positive correlations between the expression of these genes and the frequency of activated memory CD4+ T cells, suggesting a dysregulation of metabolic quiescence in circulating CD4+ T cells. When comparing SLE P1 to P2 groups, we observed much stronger associations between the frequency of activated memory CD4+ T cells and metabolic gene expression, especially for OXPHOS genes (Fig. 6D). Finally, we examined the relationship between metabolic gene expression and IRF4, which is a transcription factor with reported roles in anabolic programming of Th1 cells, glucose uptake, and oxygen consumption [21, 22]. We observed positive correlations between IRF4 expression and that of several OXPHOS genes among SLE subjects, while these relationships were negatively correlated among HC samples (Fig. 6E). Together, these results suggest that circulating CD4+ T cells are phenotypically and metabolically more activated in SLE than in HC. Further, the hyperactivated SLE P1 subset is primed for increased OXPHOS activity relative to SLE P2 patients. Overall the results define a group of predominantly AA SLE patients with activated immunophenotypes and high expression of OXPHOS genes.
4. Discussion
SLE is a clinically heterogeneous disease, which, in addition to implications for patient care, has impeded the implementation and interpretation of clinical trials, and hampered our comprehensive understanding of the disease etiology. Transcriptomic analyses comparing peripheral blood mononuclear cells (PBMCs) between SLE patients and HC have identified increased interferon stimulated gene expression, as well as a granulocyte and a plasmablast signatures across several SLE cohorts [23–25]. These transcriptional signatures are not shared between all SLE patients, and it has been proposed that they can be used to stratify SLE cohorts based on distinct clinical and cellular phenotypic profiles [24].
Mouse models of lupus point to dysregulation of immune metabolism as a key feature of SLE pathogenesis [6], including at the transcriptional level [26], but molecular evidence from patients has been limited. An increased oxidative phosphorylation gene signature has been found in the PBMCs of SLE patients [27], and an increased expression of genes associated with an elevated mitochondrial membrane potential has been reported in the T cells from untreated SLE patients [28]. In addition, the glycolysis and gluconeogenesis pathways are downregulated in CD4+ T cells of AA but not CA patients [29]. However, there is no clear consensus on which genes are most representative of metabolic dysfunction in SLE for use as a “metabolic gene signature”. In this study, we observed two metabolic pathways, MTORC1 signaling and OXPHOS were overexpressed in CD4+ T cells of SLE patients. Moreover, the expression of an OXPHOS gene signature was highly enriched in SLE subjects and these findings were validated in an independent cohort. Several studies have implicated these two pathways as central to the dysfunction of lupus T cells [30], and they have been further supported by the therapeutic effects of rapamycin [31] and N-Acetylcysteine [32], respectively, in SLE patients. We have also shown that inhibiting the electron transport chain decreases the response of CD4+ T cells to type 1 IFN [33], indicating that the OXPHOS metabolic gene signature constitutes a major driver of lupus pathogenesis. The expression of many MYC pathway genes were also elevated in SLE CD4+ T cells, although the pathway as a whole did not reach statistical significance. MYC is a central regulator of responses to T cell receptor (TCR) signaling [34]. High expression of cMYC in PBMCs of SLE patients correlates with disease activity [35] and it is controlled, at least in part, at the transcriptional level [36]. Our results suggest that it also contributes to their CD4+ T cell hyperactivation. Overall, our results demonstrate that these pathways are regulated at the transcriptional level and represent a metabolic signature of lupus CD4+ T cells. Although we have previously reported an increased glycolysis in in-vitro stimulated CD4+ T cells from SLE patients [13], we have not found here evidence for a glycolytic gene signature in ex-vivo SLE CD4+ T cells. We have not found evidence either for an increased activation of the pentose phosphate pathway (data not shown), which dominates the metabolites of SLE patients [37]. This suggests that while SLE CD4+ T cells can become more glycolytic upon activation, their steady-state metabolism relies on OXPHOS, at least in patients with low disease activity.
In addition to metabolic genes, we showed globally increased expression of immune genes in CD4+ T cells of SLE patients. Among them, four stood out as highly expressed, FOXP3, IKZF2 (encoding for HELIOs), IRF4, and TIGIT. HELIOS+FOXP3+ Treg cells are expanded in SLE patients with active disease, mostly likely in response to inflammation [38, 39]. Although our cohort was limited to patients with low disease activity, the high expression of FOXP3 and IKZF2 most likely corresponds to the expansion of this subset. IRF4 is a transcription factor involved in Th17 and Tfh cell differentiation, for which consensus binding sites are overrepresented in the promoters of SLE-susceptibility genes [40]. Our results suggest that overexpression of IRF4 may account for this finding. TIGIT is a negative regulator that is highly expressed on SLE CD4+ T cells in correlation with disease activity, and TIGIT+ T cells are more activated than their TIGIT-negative counterparts [41].
Only three genes tended to be expressed at lower levels in SLE patients relative to HC: BCL6, CXCR5 and CD40L. BCL6 is the master regulator of Tfh and Tph cells, which are both expanded in SLE patients [16, 17]. The Tph population is relatively more abundant than the Tfh population and it is characterized by a lack of CXCR5 expression, as well as a relatively low BCL6 expression [17]. However, there was no difference in BCL6 expression between Tfh and Tph cells in our cohort and it was not correlated with their expansion. There was a trend for a negative correlation between the frequency of Tph cells and the expression of CXCR5 in total CD4+ T cells. A more detailed study comparing gene and protein expression levels in these subsets will be necessary to address the significance of this finding.
An immunophenotypic screen of PBMCs from SLE patients and HC showed that unswitched memory B cells and pDCs were significantly reduced in SLE patients, corroborating previous studies [42–44]. This finding confirms that our cohort is representative and that our results are within expectations. Key differences between SLE patients and HC were driven by higher frequencies of HLA-DR+CD38+ T cell subsets. Active disease and autoantibody production has been associated with higher levels of T cell activation in SLE patients [3, 45], which has been linked to abnormal TCR signaling [46]. Here we show that T cell activation represents a major phenotypic feature in SLE patients, even when they present with stable disease activity. The majority of SLE patients belonged to three phenotypic clusters, two of which were characterized by T cell hyperactivation with the addition of Tfh subsets in one of them, while the third subset being dominated by dysregulation of B cell and innate immune subsets. A recent study identified three immunophenotype clusters in Japanese SLE patients: T cell independent, Treg dominant and Tfh dominant, with a dramatic increase in activated T cell subtypes in the latter cluster [1]. This cohort was ethnically homogeneous and had higher disease activity, representing two key differences from our cohort. However, their Tfh dominant cluster overlaps with our cluster a, and its T-independent cluster overlaps with our cluster b. The Treg subsets did not segregate in our cohort for reasons that are unclear.
The frequency of activated HLA-DR+CD38+ T cells separated SLE patients into two clusters in our study. These hyperactivated phenotypes were not specific to any subset of effector T cells. The expression of metabolic genes was globally higher in the CD4+ T cells from patients in the hyperactivated SLE cluster 1, with a positive correlation between activated memory T cell phenotypes. These results correspond to the functional link between T cell activation, proliferation and metabolism in general [47] and the correlation between increased mitochondrial metabolism and activation in T cells from SLE patients [28]. Disease severity and drug regimens were similar between the two clusters, suggesting that these variables were not involved. The only distinctive feature of cluster 1 with hyperactivated T cell phenotypes was a much higher frequency of AA patients. AA SLE patients tend to have a more severe disease [48], and their genetic risk is higher than CA patients [49]. It has been suggested that disease is more B cell-driven in AA patients, who present higher expression of typical activation markers such as CD80, CD86, and CD40L than CA SLE patients [50]. To our knowledge, our study is the first to report globally increased T cell activation in AA SLE patients. There are indications, however, of increased T cell activation in AA subjects that may precede disease manifestations. Naïve CD4+ T cells present a higher level of DNA demethylation in AA HC and SLE patients than in their CA counterparts [51]. Anti-nuclear autoantibody (ANA)-positive healthy AA but not CA subjects present a higher level of T cell activation than ANA-negative individuals [52].
5. Conclusion
Overall, our results suggest that T cell activation is an important contributor to SLE pathogenesis in AA patients. Future studies in larger cohorts should define the relationship between T cell and B cell activation in this group, and whether it corresponds to specific disease manifestations. Furthermore, our study identified a group of patients in which the hyperactivation of T cells correlated with a high metabolic profile. Metformin and mTOR inhibitors have been shown to normalize the activation and effector functions of lupus CD4+ T cells [13, 31, 33]. Recent clinical trials have shown a beneficial effect of these drugs in SLE patients [31, 53]. Our results suggest that the cluster with hyperactivated T cells may serve as a biomarker to identify a subgroup that might respond more readily to treatment with these metabolic inhibitors.
Supplementary Material
Highlights.
CD4+ T cells from SLE patients express higher levels of metabolic genes, including in the oxidative phosphorylation pathway, as well as immunoregulatory genes
SLE patients with low disease activity present higher frequencies of circulating activated CD38+HLA-DR+ CD4+ and CD8+ T cells across several effector subsets than healthy controls.
Among SLE patients, highly activated T cell subsets defines a group in which African American patients were overrepresented, with higher frequencies of Th1 and Tfh cells, as well as plasmablasts than the low activation group.
A higher expression of immunoregulatory and metabolic genes is associated with the presentation of more activated immunophenotypes in SLE patients.
T cell activation was positively correlated with metabolic gene expression in SLE patients but not in healthy controls.
Acknowledgements
The authors would like to thank Annie Lai Chan for human subject recruitment and phlebotomy, and Amanda Posgai from the UF Diabetes Institute for editorial assistance.
Funding
This study was supported by grants from the NIH: R01AI045050 and R01 AI128901 to LM and PO1 AI42288 and R01 DK106191 to TMB. DJP was partially supported by JDRF 2-PDF-2016-207-A-N and AAT by 5T32DK104721.
Footnotes
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Ethical Approval
Human studies were approved by UF IRB# 201900125
Data Sharing Plan
Microarray data has been deposited under GSE103760 in the Gene Expression Omnibus (GEO) database.
References
- [1].Kubo S, Nakayamada S, Yoshikawa M, Miyazaki Y, Sakata K, Nakano K, Hanami K, Iwata S, Miyagawa I, Saito K, Tanaka Y, Peripheral immunophenotyping identifies three subgroups based on T cell heterogeneity in lupus patients, Arthritis Rheumatol, 69 (2017) 2029–2037. [DOI] [PubMed] [Google Scholar]
- [2].Alcocer-Varela J, Alarcon-Riquelme M, Laffon A, Sanchez-Madrid F, Alarcon-Segovia D, Activation markers on peripheral blood T cells from patients with active or inactive systemic lupus erythematosus. Correlation with proliferative responses and production of IL-2, J Autoimmun, 4 (1991) 935–945. [DOI] [PubMed] [Google Scholar]
- [3].Viallard JF, Bloch-Michel C, Neau-Cransac M, Taupin JL, Garrigue S, Miossec V, Mercie P, Pellegrin JL, Moreau JF, HLA-DR expression on lymphocyte subsets as a marker of disease activity in patients with systemic lupus erythematosus, Clin Exp Immunol, 125 (2001) 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gonzalez SM, Taborda NA, Rugeles MT, Role of different subpopulations of CD8(+) T cells during HIV exposure and infection, Front Immunol, 8 (2017) 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Katsuyama E, Suarez-Fueyo A, Bradley SJ, Mizui M, Marin AV, Mulki L, Krishfield S, Malavasi F, Yoon J, Sui SJH, Kyttaris VC, Tsokos GC, The CD38/NAD/SIRTUIN1/EZH2 axis mitigates cytotoxic CD8+ T cell function and identifies patients with SLE prone to infections, Cell reports, 30 (2020) 112–123.e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Teng X, Brown J, Choi SC, Li W, Morel L, Metabolic determinants of lupus pathogenesis, Immunol Rev, 295 (2020) 167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wahl DR, Petersen B, Warner R, Richardson BC, Glick GD, Opipari AW, Characterization of the metabolic phenotype of chronically activated lymphocytes, Lupus, 19 (2010) 1492–1501. [DOI] [PubMed] [Google Scholar]
- [8].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc Natl Acad Sci USA, 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maecker HT, McCoy JP, Nussenblatt R, Standardizing immunophenotyping for the Human Immunology Project, Nat Rev Immunol, 12 (2012) 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, Zhu L, Wei W, Wang C, Karnowski A, Belz GT, Ghali JR, Cook MC, Riminton DS, Veillette A, Schwartzberg PL, Mackay F, Brink R, Tangye SG, Vinuesa CG, Mackay CR, Li Z, Yu D, Circulating precursor CCR7(lo)PD-1(hi) CXCR5⁺ CD4⁺ T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure, Immunity, 39 (2013) 770–781. [DOI] [PubMed] [Google Scholar]
- [11].Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S, Investigators I.A.V.I.P.C.P., Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses, Immunity, 39 (2013) 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Perl A, Gergely P Jr., Nagy G, Koncz A, Banki K, Mitochondrial hyperpolarization: a checkpoint of T-cell life, death and autoimmunity, Trends Immunol, 25 (2004) 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, Morel L, Normalization of CD4+ T cell metabolism reverses lupus, Sci Transl Med, 7 (2015) 274ra218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Piranavan P, Bhamra M, Perl A, Metabolic targets for treatment of autoimmune diseases, Immunometabolism, 2 (2020) e200012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Choi J, Kim ST, Craft J, The pathogenesis of systemic lupus erythematosus-an update, Curr Opin Immunol, 24 (2012) 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Choi JY, Ho JH, Pasoto SG, Bunin V, Kim ST, Carrasco S, Borba EF, Goncalves CR, Costa PR, Kallas EG, Bonfa E, Craft J, Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity, Arthritis Rheumatol, 67 (2015) 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bocharnikov AV, Keegan J, Wacleche VS, Cao Y, Fonseka CY, Wang G, Muise ES, Zhang KX, Arazi A, Keras G, Li ZJ, Qu Y, Gurish MF, Petri M, Buyon JP, Putterman C, Wofsy D, James JA, Guthridge JM, Diamond B, Anolik JH, Mackey MF, Alves SE, Nigrovic PA, Costenbader KH, Brenner MB, Lederer JA, Rao DA, R.A.S.L.E.N. Accelerating Medicines Partnership, PD-1hiCXCR5- T peripheral helper cells promote B cell responses in lupus via MAF and IL-21, JCI Insight, 4 (2019) e130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cooley ID, Read KA, Oestreich KJ, Trans-presentation of IL-15 modulates STAT5 activation and Bcl-6 expression in TH1 cells, Scientific reports, 5 (2015) 15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Godefroy E, Zhong H, Pham P, Friedman D, Yazdanbakhsh K, TIGIT-positive circulating follicular helper T cells display robust B-cell help functions: potential role in sickle cell alloimmunization, Haematologica, 100 (2015) 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP, Trapani JA, Levy DE, Hertzog PJ, Clarke CJ, Johnstone RW, Functional crosstalk between type I and II interferon through the regulated expression of STAT1, PLoS Biol, 8 (2010) e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kratchmarov R, Nish SA, Lin WW, Adams WC, Chen YH, Yen B, Rothman NJ, Klein U, Reiner SL, IRF4 couples anabolic metabolism to Th1 cell fate determination, Immunohorizons, 1 (2017) 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mahnke J, Schumacher V, Ahrens S, Kading N, Feldhoff LM, Huber M, Rupp J, Raczkowski F, Mittrucker HW, Interferon Regulatory Factor 4 controls TH1 cell effector function and metabolism, Sci Rep, 6 (2016) 35521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baechler E, Batliwalla F, Karypis G, Gaffney P, Ortmann W, Espe K, Shark K, Grande W, Hughes K, Kapur V, Gregersen P, Behrens T, Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus, Proc Natl Acad Sci USA, 100 (2003) 2610– 2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, Cepika AM, Acs P, Turner J, Anguiano E, Vinod P, Kahn S, Obermoser G, Blankenship D, Wakeland E, Nassi L, Gotte A, Punaro M, Liu YJ, Banchereau J, Rossello-Urgell J, Wright T, Pascual V, Personalized immunomonitoring uncovers molecular networks that stratify lupus patients, Cell, 165 (2016) 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bennett L, Palucka A, Arce E, Cantrel V, Borvak J, Banchereau J, Pascual V, Interferon and granulopoiesis signatures in systemic lupus erythematosus blood., The Journal of experimental medicine, 197 (2003) 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choi SC, Titov AA, Abboud G, Seay HR, Brusko TM, Roopenian DC, Salek-Ardakani S, Morel L, Inhibition of glucose metabolism selectively targets autoreactive follicular helper T cells, Nat Commun, 9 (2018) 4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gaffney PM, Moser KL, Baechler EC, Defining a new molecular basis of systemic lupus erythematosus through transcriptional profiling, Expert Rev Clin Immunol, 3 (2007) 913–923. [DOI] [PubMed] [Google Scholar]
- [28].Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, Phillips PE, Crow MK, Oess S, Muller-Esterl W, Perl A, Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation, J Immunol, 182 (2009) 2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sharma S, Jin Z, Rosenzweig E, Rao S, Ko K, Niewold TB, Widely divergent transcriptional patterns between SLE patients of different ancestral backgrounds in sorted immune cell populations, J Autoimmun, 60 (2015) 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Perl A, Oxidative stress in the pathology and treatment of systemic lupus erythematosus, Nat Rev Rheumatol, 9 (2013) 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lai ZW, Kelly R, Winans T, Marchena I, Shadakshari A, Yu J, Dawood M, Garcia R, Tily H, Francis L, Faraone SV, Phillips PE, Perl A, Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial, Lancet, 391 (2018) 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lai Z-W, Hanczko R, Bonilla E, Caza TN, Clair B, Bartos A, Miklossy G, Jimah J, Doherty E, Tily H, Francis L, Garcia R, Dawood M, Yu J, Ramos I, Coman I, Faraone SV, Phillips PE, Perl A, N-acetylcysteine reduces disease activity by blocking mTOR in T cells of lupus patients, Arthritis Rheumat, 64 (2012) 2937–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Titov AA, Baker HV, Brusko TM, Sobel ES, Morel L, Metformin inhibits the type 1 IFN response in human CD4(+) T cells, Journal of immunology, 203 (2019) 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang R, Dillon C-P, Shi L-Z, Milasta S, Carter R, Finkelstein D, McCormick L-L, Fitzgerald P, Chi H, Munger J, Green D-R, The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation, Immunity, 35 (2011) 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Deguchi Y, Negoro S, Kishimoto S, Methylation of c-myc gene changes in human lymphoproliferative diseases, Biosci Rep, 7 (1987) 637–643. [DOI] [PubMed] [Google Scholar]
- [36].Eleftheriades EG, Boumpas JE, Balow JE, Tsokos GC, Transcriptional and post-transcriptional mechanisms are responsible for the increased expression of c-myc protooncogene in lymphocytes from patients with systemic lupus erythematosus, Clin Immunol Immunopathol, 52 (1989) 507–515. [DOI] [PubMed] [Google Scholar]
- [37].Perl A, Hanczko R, Lai ZW, Oaks Z, Kelly R, Borsuk R, Asara JM, Phillips PE, Comprehensive metabolome analyses reveal N-acetylcysteine-responsive accumulation of kynurenine in systemic lupus erythematosus: implications for activation of the mechanistic target of rapamycin, Metabolomics, 11 (2015) 1157–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Alexander T, Sattler A, Templin L, Kohler S, Groß C, Meisel A, Sawitzki B, Burmester G-R, Arnold R, Radbruch A, Thiel A, Hiepe F, Foxp3+ Helios+ regulatory T cells are expanded in active systemic lupus erythematosus, Ann Rheum Dis, 72 (2013) 1549–1558. [DOI] [PubMed] [Google Scholar]
- [39].Golding A, Hasni S, Illei G, Shevach EM, The percentage of FoxP3+Helios+ Treg cells correlates positively with disease activity in systemic lupus erythematosus, Arthritis Rheum, 65 (2013) 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dozmorov MG, Wren JD, Alarcon-Riquelme ME, Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes, Epigenetics, 9 (2014) 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mao L, Hou H, Wu S, Zhou Y, Wang J, Yu J, Wu X, Lu Y, Mao L, Bosco MJ, Wang F, Sun Z, TIGIT signalling pathway negatively regulates CD4(+) T-cell responses in systemic lupus erythematosus, Immunol, 151 (2017) 280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jenks SA, Cashman KS, Zumaquero E, Marigorta UM, Patel AV, Wang X, Tomar D, Woodruff MC, Simon Z, Bugrovsky R, Blalock EL, Scharer CD, Tipton CM, Wei C, Lim SS, Petri M, Niewold TB, Anolik JH, Gibson G, Lee FE, Boss JM, Lund FE, Sanz I, Distinct effector B cells induced by unregulated Toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus, Immunity, 49 (2018) 725–739 e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Psarras A, Alase A, Antanaviciute A, Carr IM, Md Yusof MY, Wittmann M, Emery P, Tsokos GC, Vital EM, Plasmacytoid dendritic cells are functionally exhausted while non-haematopoietic sources of type I interferon dominate human autoimmunity, bioRxiv, (2018) 502047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Robak E, Smolewski P, Woźniacka A, Sysa-Jedrzejowska A, Robak T, Clinical significance of circulating dendritic cells in patients with systemic lupus erythematosus, Mediators Inflamm, 13 (2004) 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Spronk PE, Horst G, Van Der Gun BT, Limburg PC, Kallenberg CG, Anti-dsDNA production coincides with concurrent B and T cell activation during development of active disease in systemic lupus erythematosus (SLE), Clin Exp Immunol, 104 (1996) 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Moulton VR, Tsokos GC, T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity, The Journal of clinical investigation, 125 (2015) 2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].MacIver NJ, Michalek RD, Rathmell JC, Metabolic regulation of T lymphocytes, Annu Rev Immunol, 31 (2013) 259–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Petri M, Purvey S, Fang H, Magder LS, Predictors of organ damage in systemic lupus erythematosus: the Hopkins Lupus Cohort, Arthritis Rheum, 64 (2012) 4021–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gensterblum E, Renauer P, Coit P, Strickland FM, Kilian NC, Miller S, Ognenovski M, Wren JD, Tsou PS, Lewis EE, Maksimowicz-McKinnon K, McCune WJ, Richardson BC, Sawalha AH, CD4+CD28+KIR+CD11a(hi) T cells correlate with disease activity and are characterized by a pro-inflammatory epigenetic and transcriptional profile in lupus patients, J Autoimmun, 86 (2018) 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Menard LC, Habte S, Gonsiorek W, Lee D, Banas D, Holloway DA, Manjarrez-Orduno N, Cunningham M, Stetsko D, Casano F, Kansal S, Davis PM, Carman J, Zhang CK, Abidi F, Furie R, Nadler SG, Suchard SJ, B cells from African American lupus patients exhibit an activated phenotype, JCI Insight, 1 (2016) e87310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH, Renal involvement in lupus is characterized by unique DNA methylation changes in naïve CD4+ T cells, J Autoimmun, 61 (2015) 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Slight-Webb S, Smith M, Bylinska A, Macwana S, Guthridge C, Lu R, Merrill JT, Chakravarty E, Arriens C, Munroe ME, Maecker HT, Utz PJ, Guthridge JM, James JA, Autoantibody-positive healthy individuals with lower lupus risk display a unique immune endotype, J Allergy Clin Immunol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sun F, Wang HJ, Liu Z, Geng S, Wang HT, Wang X, Li T, Morel L, Wan W, Lu L, Teng X, Ye S, Safety and efficacy of metformin in systemic lupus erythematosus: a multicentre, randomised, double-blind, placebo-controlled trial, The Lancet Rheumatol, 2 (2020) e210–e216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


