SUMMARY
Lipids play crucial roles in regulating aging and longevity. In the past few decades, a series of genetic pathways have been discovered to regulate lifespan in model organisms. Interestingly, many of these regulatory pathways are linked to lipid metabolism and lipid signaling. Lipid metabolic enzymes undergo significant changes during aging and are regulated by different longevity pathways. Lipids also actively modulate lifespan and health span as signaling molecules. In this review, we summarize recent insights into the roles of lipid metabolism and lipid signaling in aging and discuss lipid-related interventions in promoting longevity.
INTRODUCTION
Lipids are key biological molecules that contribute to cellular and organismal functions in three principal ways. First, they are fundamental structural elements of cellular membranes, which build a selective barrier separating the cell from the environment and ensuring subcellular compartmentalization. Second, they are key molecules in energy metabolism to fuel the cell. Third, they play active roles in signal transduction by directly acting as signaling molecules, or indirectly by affecting membrane fluidity, providing post-translational modifications or allosteric modulation. Harmful lipid storage and peroxidation have been correlated with aging, but emerging studies also provide evidence to support the active signaling role of lipids in regulating lifespan and longevity. To overview the recent progress in this area, we first illustrate how changes in different classes of lipids are associated with aging and longevity in the context of their functions. We also review changes in brain lipids in association with aging and neurodegenerative diseases. Next, we summarize how well-conserved pro-longevity signaling mechanisms regulate lipid metabolism. In the last section, we focus on a rapidly evolving area of research regarding the longevity regulation by lipid signals, their binding proteins, and receptors.
Age-associated changes in lipid metabolism
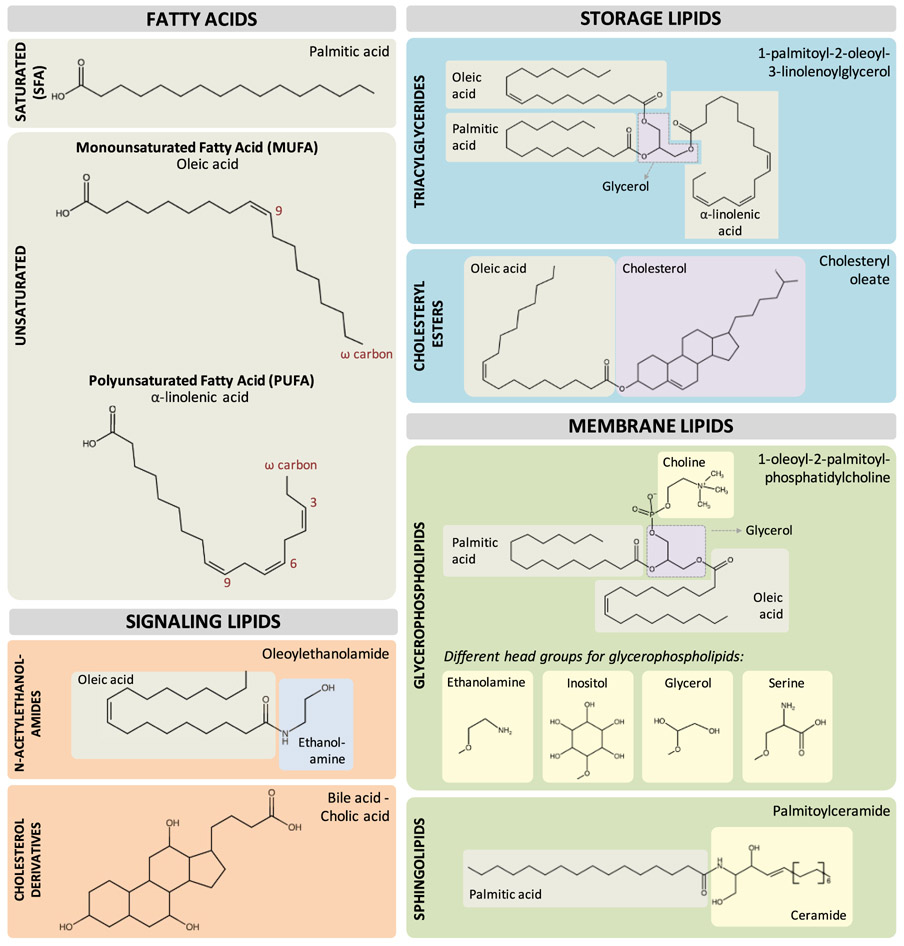
Lipids are broadly defined by their insolubility in water but solubility in organic solvents. To facilitate the discussion in this review, lipids are divided into three categories: fatty acids, phospholipids (glycerophospholipids and sphingolipids), and neutral lipids (triglycerides and cholesteryl esters) (Figure 1; Box 1). Here, we discuss age-associated changes in the metabolism of different lipid species and how direct manipulation of their metabolism modulates longevity.
Figure 1. Different classes of lipid species.
Fatty acids are building blocks of all lipids. Palmitic, oleic, and α-linolenic acid are shown as examples for saturated, monounsaturated, and polyunsaturated FAs, respectively. For storage lipids, triglycerides consist of three FAs attached to a glycerol, whereas cholesterol esters consist of a fatty acid attached to a cholesterol. Glycerophospholipids are lipids with a polar group attached to one of the positions on the glycerol backbone and FAs attached to the other two. For example, phosphatidylcholines have a choline attached to the third position on the glycerol. Other than choline, the head group could be ethanolamine, inositol, serine, and glycerol. Sphingolipids have a sphingoid base attached to one fatty acid. For example, ceramides have sphingosine attached to a fatty acid. For signaling lipids, N-acetylethanolamides (e.g., oleoylethanolamide) and cholesterol-derived bile acids (e.g., cholic acid) are shown as examples.
Box 1. Overview of lipid species.
Fatty acids:
FAs are fundamental building blocks of membrane phospholipids and storage lipids. These carboxylic acids with a hydrocarbon tail can be classified based on their length: short-chain (5 or fewer carbons), medium-chain (6–12 carbons), long-chain (13–21 carbons), and very-long-chain fatty acids (21 or more carbons) and also the number of double bonds in their hydrocarbon tails: saturated (no double bonds, SFA), monounsaturated (a single carbon-carbon double bond, MUFA), and polyunsaturated (more than one carbon-carbon double bond, PUFA). The carbon-carbon double bond is predominantly in cis configuration, which gives unsaturated FAs a “kink” in their shape and makes their biochemical properties distinct from saturated FAs with the same length. Moreover, depending on the location of the first double bond from the terminal methyl group, unsaturated FAs can be grouped into ω-3, ω-6, ω-9, etc. The number and the position of the double bonds strongly impact the physical and physiological properties of unsaturated FAs.
Phospholipids – glycerophospholipids and sphingolipids:
Membrane phospholipids form the structural barrier of the cell and subcellular compartments and also modulate signal transduction by affecting membrane fluidity and curvature, or by generating signaling lipids. The two most abundant membrane phospholipids are glycerophospholipids and sphingolipids, in which FA chains and polar head groups are linked to either a glycerol or sphingosine base, respectively. Glycerophospholipids are derivatives of phosphatidic acid that carry different polar head groups, e.g., phosphatidylethanolamine, phosphatidylcholine, phosphatidylserine, phosphatidylinositol, and cardiolipin. Sphingolipids, on the other hand, have a ceramide base and differ in their polar head groups. Sphingomyelins have a phosphocholine or phosphoethanolamine head group, while cerebrosides and gangliosides carry a single sugar and oligosaccharide residues as a head group, respectively.
Neutral lipids – triacylglycerides and cholesterol esters:
Neutral lipids, comprised triglycerides and sterol esters/cholesterol esters, lack charged groups and are the major form of storage lipids. Triglycerides are glycerolipids that consist of three FAs attached to glycerol. Biosynthesis of triglycerides begin with glycerol-3-phosphate acyltransferase that combines glycerol-3-phosphate, a phosphorylated glycerol, with coenzyme A-bound FAs (acyl-CoA), to generate lysophosphatidic acid. A second acyl-CoA can be added by acyl-CoA:1-acylglycerol-3-phosphate acyltransferase to generate phosphatidic acid, which can be hydrolyzed by lipins to generate diacylglyceride. A third esterification step catalyzed by diacylglyceride acyltransferase (DGAT) produces triglyceride. Cholesterol esters, on the other hand, can be synthesized by two types of enzymes: acyl-CoA:cholesterol acyltransferase and lecithin:cholesterol acyltransferase that uses acyl-CoAs and phospholipids, respectively to esterify cholesterol. Within the cell, neutral lipids are stored in lipid droplets, organelles that play a crucial role in energy metabolism and signal transduction. Neutral lipids are degraded via two different mechanisms: lipolysis and lipophagy. Lipolysis involves metabolization of lipids stored within lipid droplets, via lipid droplet-associated lipases: ATGL toward triglycerides, diacylglycerol lipase (DAGL) and HSL toward diacylglyceride, and monoacylglycerol lipase (MAGL) toward monoglyceride. In addition, diacylglyceride can be metabolized into phosphatidic acids by DGK. Lipophagy also contributes to lipid metabolization from lipid droplets, during which autophagosomes engulf lipid droplets and subsequently fuse with lysosomes, resulting in lipid hydrolysis by lysosomal acid lipases. Ultimately, the end products of lipolysis and lipophagy, free FAs, are subjected to further degradation through mitochondrial or peroxisomal β-oxidation, which cyclically removes 2 carbons from the hydrocarbon tails at a time and generates ATP via the tricarboxylic acid cycle and oxidative phosphorylation. For unsaturated FAs, additional enzymes, such as isomerases and dehydrogenases, are required to process the double bonds before β-oxidation.
Fatty acid composition of phospholipids and aging
Fatty acids (FAs) have been linked with longevity in association with their composition. FA chains can be either saturated (SFA), monounsaturated (MUFA), or polyunsaturated (PUFA) (Figure 1), and the ratio of saturated to unsaturated FAs is usually 40:60 in eukaryotic cells. This ratio not only impacts membrane fluidity, flexibility, and selective permeability but also influences the susceptibility of the membrane to peroxidation. Reactive oxygen species (ROS) are natural byproducts of aerobic metabolism and their accumulation during aging leads to lipid peroxidation that damages membranes and other biological macromolecules. PUFAs in membrane phospholipids are particularly susceptible to ROS attack, and longer-lived species are often found to have a lower membrane PUFA/MUFA ratio than shorter-lived ones. For example, the longest-living metazoan species, the mollusk Arctica islandica, has more MUFAs in their cellular and mitochondrial membrane phospholipids than other shorter-lived mollusk species (Munro and Blier, 2012). The longest-living rodents, naked mole-rats, have very low levels of the PUFA docosahexaenoic acid in phospholipids when compared with mice (Mitchell et al., 2007). Paradoxically, studies on human erythrocyte and lymphocyte membranes found that centenarians have more PUFAs compared with every other age group but decreased susceptibility to peroxidation (Ponnappan et al., 1996; Rabini et al., 2002). Centenarians also possess an increased ratio of ω-3 to ω-6 PUFAs (Rabini et al., 2002). In the Leiden longevity study, which analyzed the plasma lipidome in the middle-aged offspring of nonagenarians and their spouse controls, female offspring show a lower ratio of PUFA/MUFA, but male offspring show no difference (Gonzalez-Covarrubias et al., 2013). Thus, the relationship between FA saturation and longevity is tissue specific, sex linked, and species dependent.
Furthermore, comparative profiling of sphingomyelins between centenarians and control elderly individuals has revealed increased levels of specific FA moieties as biomarkers of healthy aging in humans, including SM41:2, SM36:2, and SM34:1 (Montoliu et al., 2014). A similar overall increase in these sphingomyelins was also detected in another study as a lipidomic signature of familial longevity (Gonzalez-Covarrubias et al., 2013). Sphingomyelinases convert sphingomyelins to ceramides, and their increased activity during aging may lead to the reduction of sphingomyelins and the induction of ceramides seen in the elderly (Sacket et al., 2009). Low serum ceramide levels in elderly women have been associated with reduced risk of dementia and Alzheimer’s disease (Mielke et al., 2012). Ceramides can also be synthesized from sphingosine by ceramide synthases and degraded by ceramidases. In C. elegans, there are three homologs encoding ceramide synthases, hyl-1, hyl-2, and lagr-1. The hyl-2 mutant is short lived, but the hyl-1;lagr-1 double mutant is long lived (Mosbech et al., 2013). It is shown that HYL-1 and HYL-2 are more specific for the formation of ceramide species containing C20–C22 and C26 FA chains, respectively, and the long-lived hyl-1;lagr-1 double mutant has decreased levels of C16–C18 and C26 but increased levels of C22 species (Menuz et al., 2009). In Drosophila, deleting the homolog of the alkaline ceramidase, Dacer, increases the level of C20-C24 ceramides, oxidative stress resistance, and lifespan (Yang et al., 2010). These studies support the importance of specific ceramides and sphingomyelins in regulating longevity, which may be associated with their FA moieties. On the other hand, mutations of mouse Lass1, a ceramide synthase gene, are identified in flincher and toppler mutants (Zhao et al., 2011). These mice are not short lived but exhibit significant weight loss together with neurodegenerative and cellular aging traits (Zhao et al., 2011). A hyperactive LASS1 allele in humans, in combination with specific variants in human Ras1 and apolipoprotein E (APOE), is also associated with exceptional longevity (Jazwinski et al., 2010). The specificity of the mammalian LASS1 toward FA moieties is currently unknown.
Neutral lipid mobilization and longevity
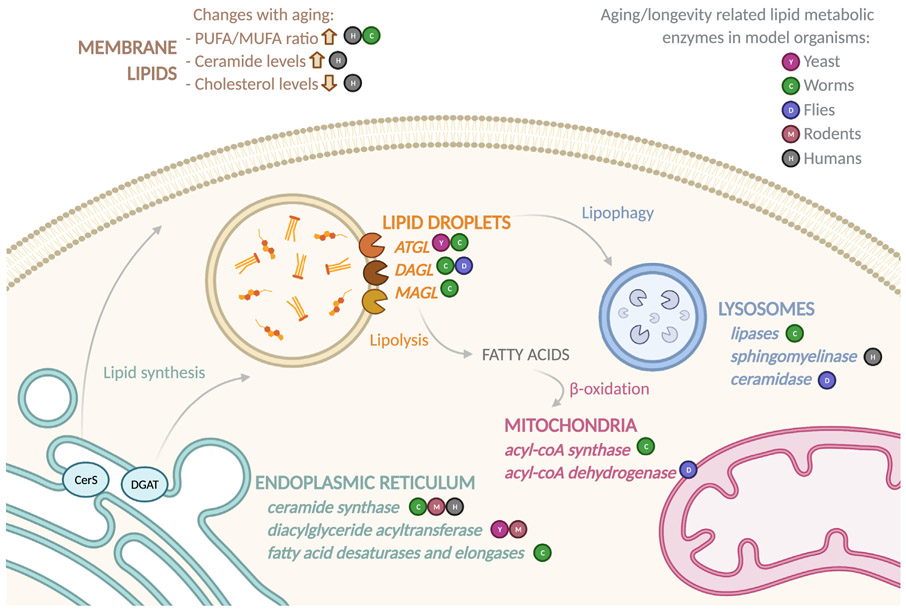
Biosynthesis and mobilization of neutral lipids are catalyzed by a variety of enzymes (Box 1), which have been linked with longevity regulation (Figure 2). In budding yeast, deletion of triglyceride lipases or overexpression of diacylglyceride acyltransferase (DGAT) increases lipid accumulation and lifespan (Handee et al., 2016). In contrast, in female mice, DGAT1 deficiency promotes longevity and leanness without reducing food intake. These DGAT1-deficient mice also show reduced cholesterol biosynthesis and are protected against age-onset obesity and inflammation in white adipose tissue (Streeper et al., 2012). In C. elegans, overexpression of adipose triglyceride lipase (ATGL) or its muscle-specific activation by protein kinase A prolongs lifespan (Schmeisser et al., 2019; Zaarur et al., 2019). Furthermore, in both C. elegans and Drosophila, overexpression of diacylglyceride lipase (DAGL) or knockdown of diacylglycerol kinase (DGK) increases lifespan and oxidative stress tolerance, while the mutation of DAGL shows the opposite effects (Lin et al., 2014). More recently, pharmacological screens have identified a monoacylglyceride lipase (MAGL) inhibitor, JZL184, as a potent pro-longevity inducer in C. elegans (Chen et al., 2019). Interestingly, most of these studies reveal a correlation between increased lipid mobilization and longevity.
Figure 2. Lipid metabolic changes associated with aging.
Specific lipid metabolic enzymes localize to different cellular organelles. The activities of those enzymes are shown to influence lifespan and health span in various model organisms, as well as humans. Age-associated changes are also detected in specific lipid profiles.
Increased lipid mobilization promotes a metabolic shift toward FA β-oxidation, which has been associated with several well-known longevity mechanisms (Ratnappan et al., 2014; Um et al., 2004; Xu et al., 2012). Furthermore, activation of mitochondrial β-oxidation by overexpressing acyl-CoA synthetase 2 (acs-2) in the C. elegans intestine, a major fat storage site, decreases fat storage and promotes longevity (Ramachandran et al., 2019). In Drosophila, female flies heterozygous for the Enigma gene, which encodes the acyl-CoA dehydrogenase, show lifespan extension and decreased triglyceride storage (Mourikis et al., 2006), also supporting increased lipolysis and FA oxidation associated with longevity.
Cholesterol in brain aging
Lipids represent up to 50% of the brain’s dry weight, and the most abundant lipid species in the brain are glycerophospholipids and cholesterol (Figure 1). In the central nervous system, cholesterol is predominantly found in myelin sheaths and in the plasma membrane of neurons and astrocytes. In addition to its canonical role of fine-tuning membrane fluidity and permeability, cholesterol physically interacts with signaling receptors to control their conformation and mobility. During aging, about 20%–40% of cholesterol is lost from several brain regions (Söderberg et al., 1990). The reduction of caveolin-1, a lipid raft resident protein that binds to cholesterol and organizes a multitude of receptors and signaling proteins, is also detected in the aged brain, which negatively impacts synaptic activity and contributes to neurodegeneration (Egawa et al., 2016). Since cholesterol cannot traverse the blood-brain barrier, neurons have to either synthesize it de novo or obtain it from astrocytes. The transfer of cholesterol from astrocytes to neurons is primarily mediated by brain apolipoproteins, which play a crucial role in regulating aging and longevity (Wang and Eckel, 2014). In particular, APOE is the most abundant brain apolipoprotein, and its encoding gene has three major allelic variants referred to as ε2, ε3, and ε4. The ε4 allele has been linked with an increased risk of Alzheimer’s disease, but the ε2 allele is associated with neuroprotective, anti-Alzheimer’s effects and human longevity (Schächter et al., 1994). Cholesterol derivatives, bile acids (Figure 1), are also detected in the brain, and their levels decrease by about 70% during aging in rats (Zheng et al., 2016). It is currently unknown whether the decrease is due to decreased transport of bile acids across the blood-brain barrier or decreased bile acid synthesis in the brain. Further understanding of brain cholesterol regulation will help prevent and treat age-related cognitive decline and neurodegeneration.
Pro-longevity signaling pathways in regulating lipid metabolism
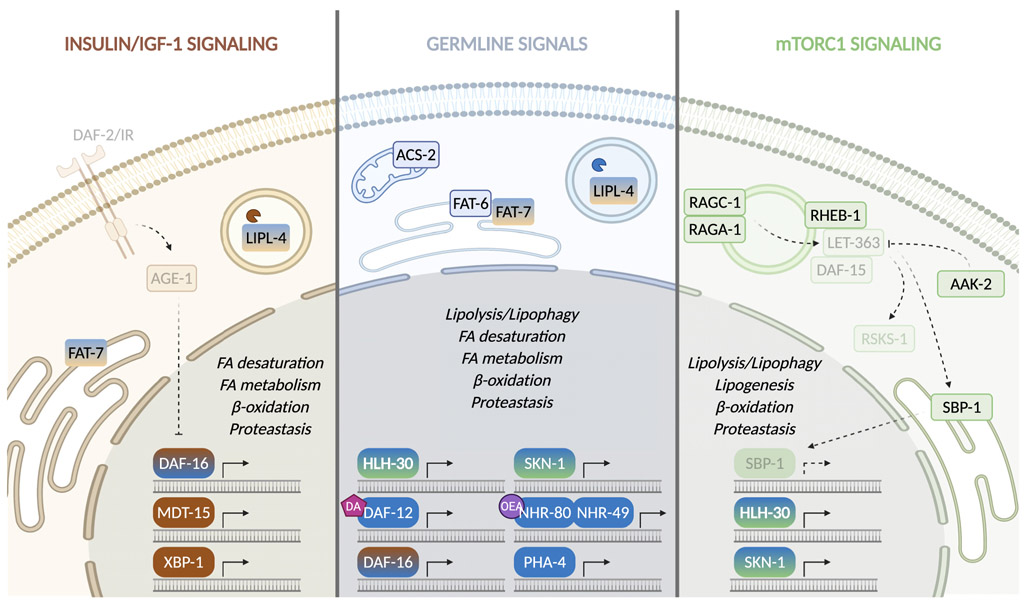
Molecular characterization of pro-longevity signaling pathways has begun to uncover mechanistic links between lipid metabolism and longevity. Here, we focus on three highly conserved signaling mechanisms in regulating longevity: insulin/IGF-1 signaling (IIS), mechanistic target of rapamycin (mTOR) signaling, and germline endocrine signaling (Figure 3). Dietary restriction is another well-known intervention in promoting longevity, and its effects on lipid metabolism are nicely summarized in recent reviews, e.g., Diego et al. (2019).
Figure 3. Pro-longevity signaling pathways regulating lipid metabolism.
Molecular characterization of conserved pro-longevity signaling mechanisms, including IIS, mTOR signaling, and germline endocrine signals, have uncovered interesting mechanistic links between lipid metabolism and longevity. In C. elegans, multiple transcription factors/mediators and nuclear receptors act in these pathways to regulate different lipid metabolic processes.
IIS
The discovery of C. elegans age-1 and daf-2 mutations that could double lifespan opened the era of molecular genetics of aging (Friedman and Johnson, 1988; Kenyon et al., 1993). daf-2 and age-1 encode the insulin/IGF-1 receptor and the downstream phosphoinositide 3-kinase, respectively, and their pro-longevity effects are completely dependent on daf-16, a FOXO transcription factor (Kenyon et al., 1993; Ogg et al., 1997). Later studies confirmed the high conservation of this longevity-regulating pathway in fruit flies, mice, and human (Lapierre and Hansen, 2012). Interestingly, several, but not all, long-lived mutant alleles of daf-2 show increased fat storage, and further analyses revealed that site-specific mutations differentially regulate longevity and fat storage (Perez and Van-Gilst, 2008). Thus, the longevity effect caused by IIS reduction is not simply a result of overall fat storage increase but may be attributed to specific changes in lipid metabolism.
In fact, specific lipid metabolic genes are found to be differentially expressed upon reduced IIS, including FA desaturases and elongases, lipases, and lipid transport proteins (Murphy et al., 2003; Shmookler Reis et al., 2011). FA profiling also revealed that the activity of Δ-9 desaturases and the percentage of MUFAs are positively correlated with increased lifespans in the daf-2 and age-1 mutants, while the percentage of PUFAs displays a negative correlation (Shmookler Reis et al., 2011). Inactivation of a Δ-9 desaturase encoding gene, fat-7, suppresses the longevity of the daf-2 mutant (Murphy et al., 2003), while fat-7 overexpression prolongs lifespan by inducing MUFA accumulation (Han et al., 2017). Furthermore, supplementation of the MUFA oleic acid is sufficient to prolong lifespan in C. elegans (Han et al., 2017). A lysosomal acid lipase, lipl-4, is also upregulated in the daf-2 mutant in a DAF-16/FOXO-dependent manner (Wang et al., 2008). Inactivation of lipl-4 suppresses the lifespan extension of the daf-2 mutant (Wang et al., 2008), while overexpression of this lipase is sufficient to extend lifespan by increasing both lipophagy and mitochondrial β-oxidation (Lapierre et al., 2011; Ramachandran et al., 2019).
Interestingly, despite the ubiquitous expression of the DAF-16/FOXO transcription factor, its activity in the fat storage tissue is specifically responsible for the longevity regulation. In C. elegans, this regulation can be mediated by the DAF-16 target, mdt-15, which encodes a transcriptional mediator subunit (Libina et al., 2003; Zhang et al., 2013). MDT-15 cooperates with nuclear receptors and the sterol regulatory element binding protein (SREBP) to regulate genes involved in lipid metabolism (Arda et al., 2010), which may result in the production of lipid signals from the fat storage tissue to affect distant organs. DAF-16 also collaborates with XBP-1 (X-box-binding protein 1) to control the unfolded protein response of the endoplasmic reticulum in the intestine and lifespan (Henis-Korenblit et al., 2010). Recently, overexpression of the active form of XBP-1 was shown to prolong lifespan and promote lipid metabolic changes, such as the induction of lipophagy, upregulation of the MUFA oleic acid, and decrease in triglycerides (Daniele et al., 2020; Imanikia et al., 2019). Importantly, most of these lipid metabolic changes are necessary for the longevity effect conferred by XBP-1 activation (Imanikia et al., 2019). In Drosophila, activation of the dFOXO transcription factor specifically in the head and peripheral fat body prolongs lifespan (Giannakou et al., 2004; Hwangbo et al., 2004). Moreover, adipose-specific knockout of the insulin receptor in mice reduces fat mass, increases basal metabolic rate, and extends lifespan (Blüher et al., 2003; Katic et al., 2007). Similarly, mice carrying additional copies of PTEN, which reverses IIS activation, show significant lifespan extension along with increased energy expenditure from the induction of mitochondrial uncoupling in the brown adipose tissue (Ortega-Molina et al., 2012). Thus, in response to reduced IIS, lipid metabolic remodeling in the fat storage tissue plays a critical role in the systemic regulation of longevity.
mTOR signaling
Another well-conserved longevity regulatory mechanism involves mTORC1, composed of three core components, Raptor, mLST8, and mTOR kinase. As a key regulator of growth and metabolism, mTORC1 activity is tightly influenced by growth factors, energy levels, and nutrient availability within the cell. The complex regulation of mTORC1 activity is well reviewed in (Liu and Sabatini, 2020). Briefly, growth factors, such as insulin and IGF-1, can stimulate mTORC1 via their responsive kinases; intracellular energy levels block mTORC1 activation via the AMP/ATP sensor, AMPK; and nutrients, such as amino acids, cholesterol, and glucose, regulate mTORC1 by triggering its lysosomal localization and activation by Rheb. Upon activation, mTORC1 can phosphorylate and regulate a series of downstream effectors, such as ribosomal S6 kinase (S6K), TFEB, SREBP, and Nrf2 transcription factors. Reduced mTORC1 signaling was first shown to extend lifespan in C. elegans when let-363/mTOR is knocked down (Vellai et al., 2003). Similar pro-longevity effects occur when reducing multiple components in mTORC1 signaling, including Rag GTPases, raga-1 and ragc-1, rheb-1/Rheb, daf-15/Raptor, and rsks-1/S6K (Blackwell et al., 2019). Overexpression of hlh-30/TFEB or constitutively active skn-1/Nrf2 extends lifespan as well (Blackwell et al., 2019). Importantly, the longevity-promoting effect by mTORC1 reduction is conserved in yeast, Drosophila, and mammals (Liu and Sabatini, 2020). Furthermore, AAK-2/AMPK activation leads to lifespan extension, which is seen in both C. elegans and Drosophila (Apfeld et al., 2004; Stenesen et al., 2013). Besides genetic modification, pharmacological inhibition of mTORC1 by rapamycin or activation of AMPK by metformin is sufficient to prolong lifespan and health span in various organisms ranging from yeast to mice (Liu and Sabatini, 2020), and their longevity-promoting effects are currently being investigated in multiple clinical studies in humans.
mTORC1 signaling plays a crucial role in regulating lipid metabolism, including lipogenesis, lipolysis, lipophagy, and FA β-oxidation. As a master control of anabolism, mTORC1 activation promotes lipid biogenesis by activating the SREBP transcription factor and subsequently upregulating lipogenic genes (Li et al., 2010). mTORC1 also inhibits lipolysis and lipophagy to slow lipid mobilization, which involves the downregulation of ATGL expression (Chakrabarti et al., 2010), hormone-sensitive lipase (HSL) activity (Soliman et al., 2010), and lysosomal and autophagic gene expression (Settembre et al., 2012). Moreover, inhibition of mTORC1 signaling promotes FA β-oxidation by enhancing oxidative gene expression and upregulating mitochondrial activity (Cunningham et al., 2007; Sengupta et al., 2010). Therefore, mTORC1 reduction leads to a metabolic reprogramming toward increased lipid utilization and FA β-oxidation, which may contribute to the longevity effect. In supporting this idea, let-363/mTOR knockdown in C. elegans increases the expression and activity of lipases and lipophagy, which are both required for the lifespan extension (Lapierre et al., 2011, 2013). In mammals, administration of the pro-longevity drug rapamycin decreases lipogenesis (Yecies et al., 2011), increases lipolysis (Chakrabarti et al., 2010) and FA β-oxidation (Sipula et al., 2006), as does the administration of the pro-longevity drug metformin (Pernicova and Korbonits, 2014). How this metabolic switch toward lipid mobilization by mTORC1 reduction contributes to longevity, and whether this increased lipid mobilization in specific tissues generates systemic signals to improve health span at the whole organism level would be interesting topics to be addressed in future studies.
Germline signals
Germline signals were first shown to regulate longevity in C. elegans, where ablation of germline precursor cells leads to a lifespan extension of more than 50% (Hsin and Kenyon, 1999). This longevity effect can also be recapitulated using the loss-of-function mutant of glp-1, which encodes the C. elegans Notch receptor and is required for germline stem cell maintenance (Arantes-Oliveira et al., 2002). In Drosophila, genetic removal of germ cells extends lifespan (Flatt et al., 2008); in mice, transplantation of ovaries from young to old animals leads to lifespan extension in the old recipients (Cargill et al., 2003); and in men, castration has been associated with longevity (Min et al., 2012). Interestingly, further studies in C. elegans have revealed the involvement of lipid metabolism in germline-mediated longevity.
In C. elegans, the pro-longevity effect conferred by germline loss requires nuclear receptors, DAF-12, NHR-80, and NHR-49, along with transcription factors, HLH-30, PHA-4, DAF-16, and SKN-1 (Goudeau et al., 2011; Hsin and Kenyon, 1999; Lapierre et al., 2011,2013; Ratnappan et al., 2014; Steinbaugh et al., 2015). The activation of these nuclear receptors and transcription factors upregulates a group of lipid metabolic genes involved in FA β-oxidation (e.g., acs-2/acyl-CoA synthetase), FA desaturation (e.g., fat-7/Δ-9 desaturase), lipolysis, and lipophagy (e.g., lipl-4/lysosomal acid lipase), which are required for the longevity effect in the glp-1 mutant and are sufficient to prolong lifespan upon overexpression (Goudeau et al., 2011; Ramachandran et al., 2019; Ratnappan et al., 2014; Wang et al., 2008). Paradoxically, although many of these metabolic genes positively regulate lipid mobilization (O’Rourke and Ruvkun, 2013; Ramachandran et al., 2019), the glp-1 mutant shows increased fat storage (O’Rourke et al., 2009). This fat storage increase might be due to a passive accumulation of unused yolk lipids resulted from the lack of oocyte formation in the glp-1 mutant, which can activate the skn-1 transcription factor in the intestine to increase lipid mobilization (Steinbaugh et al., 2015). Whether other transcription factors and nuclear receptors can be activated through similar lipid-related mechanisms remains unknown. Interestingly, both the DAF-12 and NHR-80 nuclear receptors utilize endogenous lipids as ligands (Folick et al., 2015; Motola et al., 2006). Thus, it is possible that these nuclear receptors are also sensitive to lipid accumulation in the glp-1 mutant, and their activation and cooperation with transcription factors may induce lipid mobilization and consequently promote longevity.
Lipid signaling in longevity regulation
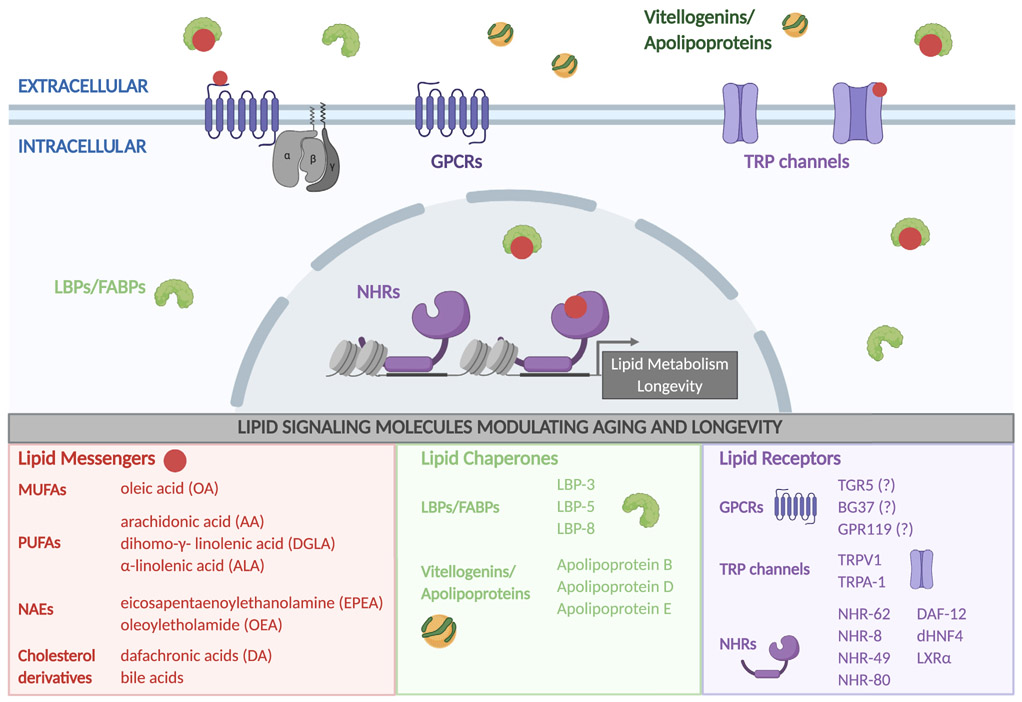
Lipids are crucial signaling molecules that regulate nuclear transcription and cellular communication. Emerging studies have revealed the active role of lipid signals in longevity regulation, the significance of lipid-binding proteins in shuttling hydrophobic lipids in the aqueous environment, as well as the engagement of receptors in recognizing and transducing lipid signals (Figure 4). This section will focus on how specific lipid signaling factors are employed to modulate aging and longevity.
Figure 4. Lipid signaling in longevity regulation.
Lipid messengers linked with the longevity regulation are summarized, which can act through GPCRs and TRP channels on the membrane and nuclear receptors. Lipid chaperones, including FABPs and apolipoproteins (LBPs and vitellogenins in C. elegans), facilitate intracellular and intercellular transportation of lipids and their signaling effects. Although not shown in the figure, changes in the membrane lipid environment and lipid modifications of GPCRs and TRP channels also modulate their activities.
Lipid messengers modulating aging and longevity
FAs and their derivatives have been linked with longevity regulation through distinct signaling mechanisms. As discussed above, increased MUFA biosynthesis has been associated with the longevity effects conferred by IIS reduction and germline loss, and supplementation of specific MUFAs is sufficient to prolong lifespan in C. elegans (Han et al., 2017). This MUFA-induced longevity effect may be mediated by the SKN-1/Nrf2 transcription factor, given that oleic acid supplementation induces its nuclear translocation (Steinbaugh et al., 2015). SKN-1/Nrf2 is a key regulator of oxidative stress responses and has been implicated in longevity from C. elegans to mice (Blackwell et al., 2015). Although the molecular mechanisms by which oleic acid activates SKN-1/Nrf2 remain unclear, a study on the ω-3 PUFA, α-linolenic acid (ALA), provides interesting insights. In C. elegans, ALA supplementation extends lifespan in a SKN-1-dependent manner; however, ALA does not activate SKN-1 directly (Qi et al., 2017). Instead, oxylipins, the peroxidated products of ALA, are responsible for the SKN-1/Nrf2 activation and lifespan extension (Qi et al., 2017). Future studies are necessary to examine whether this lipid-peroxidation-mediated mechanism is also responsible for the activation of SKN-1/Nrf2 by MUFAs and to reveal the molecular basis by which those peroxidated lipids activate SKN-1/Nrf2.
Fatty acyl ethanolamides, commonly referred as N-acylethanolamines (NAEs), are FA-derived lipid messengers (Figure 1). NAEs differ in their FA moieties, and mammalian NAEs, such as arachidonylethanolamide/anandamide (AEA), palmitoylethanolamide (PEA), oleoylethanolamide (OEA), and linoleoylethanolamide (LEA), are best known for their diverse roles in regulating mood, learning and memory, inflammation, and allergies, as well as food intake and energy metabolism (Ezzili et al., 2010). In C. elegans, AEA, PEA, OEA, and LEA have been identified, along with palmitoleoylethanolamide (POEA) and eicosapentaenoylethanolamide (EPEA) (Lucanic et al., 2011). Interestingly, the level of EPEA is decreased by dietary restriction and mTOR inactivation, and supplementation of EPEA abrogates the longevity effect conferred by both mechanisms (Lucanic et al., 2011). In addition, overexpression of the worm NAE degradation enzyme, fatty acid amide hydrolase (FAAH), faah-1, reduces the level of EPEA, POEA, LEA, and AEA, but not OEA or PEA, and extends lifespan under well-fed conditions (Lucanic et al., 2011). Thus, EPEA plays a negative role in regulating longevity, but its associated signaling mechanism is currently unknown. On the other hand, OEA has been identified as a positive regulator of longevity, which is induced in the long-lived transgenic C. elegans overexpressing a lysosomal acid lipase, lipl-4 (Folick et al., 2015). Supplementation of an OEA analog resistant to FAAH hydrolysis sufficiently prolongs lifespan and induces the transcription of genes involved in mitochondrial β-oxidation (Folick et al., 2015). Thus, different NAEs can exert distinct effects on longevity, which may associate with the specificity of their binding/transporting proteins, membrane and nuclear receptors, and/or tissues of action.
Cholesterol derivatives are also emerging as a group of lipid messengers capable of regulating longevity. Once known exclusively for their role in nutrient absorption, cholesterol-derived bile acids are crucial signaling molecules regulating metabolism and inflammation (Chiang, 2009) (Figure 1). In the liver, cholesterol is converted to the primary bile acids, cholic acid (CA), and chenodeoxycholic acid (CDCA), which are further conjugated to glycine and taurine. In the intestine, conjugated CA and CDCA are deconjugated and dehydroxylated into the secondary bile acids, deoxycholic acid and lithocholic acid (LCA), respectively (Chiang, 2009). Bile acids can activate G-protein-coupled receptors (GPCRs) and nuclear receptors, such as G-protein bile acid receptor 1 (GPBAR1/TGR5), farnesoid X receptor (FXR) and liver X receptor (LXR) (Schaap et al., 2014). The involvement of bile acids in longevity regulation was first revealed through the transcriptome and biochemical analysis of the Little mice, which live 20% longer than their wild-type littermates (Amador-Noguez et al., 2004). These long-lived mice have increased levels of bile acids, especially CA, which upregulates xenobiotic detoxification genes in an FXR dependent manner (Amador-Noguez et al., 2007). Increased bile acid levels have also been associated with methionine restriction, which promotes longevity in C. elegans, Drosophila, and mice (Lapierre and Hansen, 2012). CA supplementation extends both lifespan and health span in progeroid mice (Bárcena et al., 2018). Furthermore, LCA was identified from a chemical screen to prolong the lifespan of yeast cells (Arlia-Ciommo et al., 2018) and fruit flies (Staats et al., 2018). In C. elegans, there is another cholesterol-derived lipid messenger, called dafachronic acid (DA) that modulates lifespan through DAF-12, the FXR homolog (Gerisch et al., 2007). NHR-8, the C. elegans LXR homolog, is required for cholesterol and DA metabolism, and its loss results in DA deficiency and reduced lifespan (Magner et al., 2013). Together, these studies support the functional significance of cholesterol derivatives in regulating aging and longevity across different species.
FABPs and lipoproteins in aging regulatory lipid signaling
Being hydrophobic, lipid molecules must be bound to proteins in order to diffuse through the aqueous environment. A key family of such lipid-binding proteins are FA-binding proteins (FABPs), which facilitate the trafficking of FAs and their derivatives, as well as their signaling effects (Hotamisligil and Bernlohr, 2015) (Figure 4). C. elegans has nine FABPs termed LBPs. One of them, lbp-8, is transcriptionally upregulated in the long-lived lipl-4 transgenic strains and mediates the longevity effect (Folick et al., 2015). The LBP-8 protein localizes at the lysosomal surface in the intestine of C. elegans and binds to several PUFAs as well as oleic acid and OEA (Folick et al., 2015; Tillman et al., 2019). Upon induction of LIPL-4, LBP-8 enhances its nuclear translocation and shuttles OEA to activate the nuclear receptor complex NHR-49-NHR-80 (Folick et al., 2015). Moreover, overexpression of LBP-8 sufficiently induces the transcription of mitochondrial β-oxidation genes, activates JUN-1/JUN and SKN-1/Nrf2 anti-oxidant signaling, and in turn promotes longevity (Folick et al., 2015; Ramachandran et al., 2019). Another FABP, LBP-5, localizes to both the cytoplasm and nucleus and binds to a variety of SFAs, MUFAs, and PUFAs (Xu et al., 2011). The loss-of-function mutant of lbp-5 shows increased fat storage as does the nhr-49 mutant and decreased expression of nhr-49 target genes (Van-Gilst et al., 2005; Xu et al., 2011). Interestingly, stearic acid supplementation specifically induces NHR-49 target gene expression, which requires LBP-5 (Xu et al., 2011), but whether stearic acid directly binds to LBP-5 remains unclear. In mammals, FABP4/5-deficient mice display improved metabolic fitness without a change in lifespan (Charles et al., 2017). While these studies demonstrate the importance of FABPs in metabolic health and aging, they implicate that different FABPs can cooperate with specific lipid messengers to exert different effects. Further studies are required to advance our understanding of the signaling roles of FABPs and their specificity in regulating longevity.
Another class of lipid-binding proteins are apolipoproteins that transport lipids through lymphatic and circulatory systems (Figure 4). In addition to APOE mentioned above, another brain apolipoprotein, APOD, is known to be upregulated during human aging and in Alzheimer’s disease patients (Kalman et al., 2000). A meta-analysis of age-related gene expression profiles from vertebrate models and humans has identified APOD as the most significantly upregulated gene with an increasing age (Magalhães et al., 2009). The APOD induction is likely protective because overexpression of human APOD or the Drosophila homolog, Glial lazarillo (Glaz), extends lifespan of fruit flies (Muffat et al., 2008; Walker et al., 2006). In contrast, the absence of Glaz reduces resistance to oxidative stress and starvation, decreases neutral lipid storage and shortens lifespan (Sanchez et al., 2006). Overexpression of another fly APOD homolog, Neural lazarillo (Nlaz), also promotes longevity, and its loss-of-function mutation decreases lifespan (Hull-Thompson et al., 2009). In the nervous system, mitochondrial-defect-induced ROS in neurons leads to lipid droplet accumulation in neighboring glial cells, which is dependent on Glaz and Nlaz (Liu et al., 2015). In the neuron/astrocyte co-culture system, hyperactive neurons use APOE to shuttle excess peroxidated FAs to astrocytes, leading to increased FA β-oxidation there (Ioannou et al., 2019). In mice deficient for ApoD, lipid peroxidation is increased in the brain, alongside reduced tolerance to oxidative stress, and impaired learning and locomotor abilities (Ganfornina et al., 2008). Together, these discoveries reveal that neighboring glial cells endocytose apolipoprotein-FA particles and protect neurons from the toxicity of excessive free FAs and their peroxidation. In C. elegans, lipoproteins, encoded by vit genes, mediate the lipid transfer from the intestine to the gonad to form yolk (Matyash et al., 2001). Upon cessation of reproduction with age, yolk can be seen in the body cavity throughout the organism (Ackerman and Gems, 2012). This ectopic fat deposition in non-adipose tissues is thought to lead to age-associated lipotoxicity and loss-of-tissue functions. Supporting this idea, knocking down the vit genes increases C. elegans lifespan (Murphy et al., 2003; Seah et al., 2016). The mammalian vit ortholog is APOB, and its hepatic levels are decreased in dietary-restricted and long-lived mice (Seah et al., 2016). Interestingly, rare variants of the APOB gene correlate with extreme longevity in centenarian populations (Cash et al., 2014). Future studies on these lipoproteins are expected to reveal the underlying molecular and biochemical mechanisms by which lipoproteins are linked with age-associated diseases and contribute to improved lifespan.
Receptors responding to the longevity-associated lipids
Nuclear receptors work as molecular switches whose transactivation activity is controlled by the presence of lipophilic ligands, with 21 members in Drosophila, 48 in mammals, and 284 in C. elegans (Evans and Mangelsdorf, 2014). In response to specific lipid signals, nuclear receptors are implicated in the regulation of aging and longevity in C. elegans and Drosophila (Figure 4). DAF-12, the first deorphanized C. elegans nuclear receptor, is a DA receptor and mediates the lifespan regulatory effect of DA (Gerisch et al., 2007). NHR-8/LXR also responds to DA signals under dietary-restriction conditions and is required for dietary-restriction-induced longevity (Thondamal et al., 2014). Peroxisome proliferator-activating receptors (PPARs) and hepatocyte nuclear factor 4 (HNF4) receptors are major mediators of FA-related signaling (Evans and Mangelsdorf, 2014). Several of their C. elegans homologs, NHR-49, NHR-80, and NHR-62, play active roles in regulating lifespan. NHR-49 is required for the longevity effects conferred by germline loss, lipl-4 overexpression, and AMPK activation (Burkewitz et al., 2015; Folick et al., 2015), and its mutant exhibits elevated triglyceride levels and shortened lifespan (Van-Gilst et al., 2005). ALA supplementation increases lifespan via NHR-49 (Qi et al., 2017), but whether ALA is a ligand of NHR-49 remains unclear. NHR-80 is required for the longevity induced by germline loss and lipl-4 overexpression (Folick et al., 2015; Goudeau et al., 2011). NHR-80 enhances the expression of fat-6, encoding the desaturase that converts stearic acid to oleic acid (Goudeau et al., 2011), and directly binds to the pro-longevity lipid, OEA (Folick et al., 2015). Interestingly, OEA is a known agonist of mammalian PPARα to produce satiety and reduce body weight, and OEA treatment improves metabolic health in obese mice (Fu et al., 2003). However, whether the OEA treatment exerts a pro-longevity effect in mammals is currently unknown. Moreover, NHR-49 and NHR-80 form a complex and regulate genes involved in FA desaturation and mitochondrial β-oxidation (Pathare et al., 2012). It is possible that different FA signals synergize their effects through the coordination between their responding nuclear receptors. NHR-62 is required for hallmarks of dietary restriction, such as longevity, decreased triglycerides, and enhanced autophagy (Heestand et al., 2013). To date, NHR-62 remains an orphan nuclear receptor, with no known FA ligand. The single Drosophila ortholog for HNF4, dHNF4, regulates genes involved in FA oxidation in response to nutrient deprivation (Palanker et al., 2009). Despite increased fat storage, dHNF4-null mutant larvae are sensitive to starvation, suggesting their incapability to mobilize the stored fat for energy production (Palanker et al., 2009). During adulthood, dHNF4-dependent conversion of free FAs to hydrocarbons is required for waterproof cuticles and desiccation resistance, and hydrocarbon deficiency in the dHNF4-mutant adults leads to lifespan reduction (Storelli et al., 2019). Thus, nuclear receptors play crucial roles in transducing lipid signals to transcriptional responses and linking lipid metabolism and aging regulation.
GPCRs are another key class of receptors that perceive lipid signals and activate intracellular signaling pathways. Most of the GPCR signal transduction occurs at the plasma membrane and is therefore greatly influenced by lipid-protein interactions (Figure 4). First, post-translational lipid modifications of GPCRs and G proteins, such as palmitoylation, myristoylation, and isoprenylation, impact their membrane binding, trafficking, and signaling (Escribá et al., 2007). Second, membrane components, such as phospholipids and sphingolipids, have been reported to affect ligand binding by allosteric modulation, specifically by changing the conformational dynamics of transmembrane domains and loop regions of GPCRs (Sengupta et al., 2018). Altered membrane structure and fluidity due to lipid peroxidation can also lead to instability of GPCRs (Oliveira et al., 2019). Finally, many intercellular lipid messengers directly act on GPCRs as ligands, such as NAEs, prostaglandins, and lysophospholipids (Kostenis, 2004). Changes in the membrane lipid environment of GPCRs modulate their signaling activities and have been implicated in pathologies associated with aging (Alemany et al., 2007). Although several pro-longevity lipid signals act on GPCRs, the role of these GPCRs in aging regulation remains unclear. For example, OEA can bind to GPR119 in addition to PPARα and regulate the secretion of glucagon-like peptide-1 in intestinal L-cells (Lauffer et al., 2009). Glucagon-like peptide-1 exerts protective effects against age-related diseases, such as type II diabetes and neurodegeneration (Cho et al., 2014; Grieco et al., 2019). However, the role of GPR119 in aging remains unknown. Bile acids act through not only nuclear receptors but also GPCRs, such as TGR5 and BG37 (Kawamata et al., 2003; Maruyama et al., 2002). Whether signaling cascades activated by bile acids through these receptors are involved in the regulation of aging are not yet determined.
Lipids also impact the activity of ion channels, including transient receptor potential (TRP) channels (Ciardo and Ferrer-Montiel, 2017)(Figure 4). TRP channels are nonselective cation channels that are activated by thermal, mechanical, or chemical stimuli, including lipids, which modulate TRP channels by direct binding or indirectly by affecting their membrane localization (Ciardo and Ferrer-Montiel, 2017). In the last decade, TRP channels have been implicated in aging regulation. In C. elegans, TRPA-1 was found as a thermosensor to prolong lifespan in a temperature-dependent manner, and its overexpression in neurons leads to lifespan extension (Xiao et al., 2013). Interestingly, selective overexpression of trpa-1 in the intestine is also sufficient to extend lifespan, suggesting a non-neuronal effect of the TRP channel in regulating longevity. Given the connection between low temperature and lipid metabolic adaptation, it would be interesting to know whether TRPA-1 senses lipid metabolic cues in the intestine to regulate lifespan. Furthermore, mutant worms deficient in the TRPV channel, OCR-2, and knockout mice lacking TRPV1 are both long lived (Lee and Ashrafi, 2008; Riera et al., 2014). The TRPV1-knockout mice are also protected from diet-induced obesity by increasing their thermogenic capacity (Motter and Ahern, 2008). Although several NAEs and ω-3 PUFAs modulate TRPV1 activities (Ciardo and Ferrer-Montiel, 2017), whether these lipid signals play a role in TRPV1-mediated longevity remains an interesting open question.
PERSPECTIVE
Although lipid research has been long hindered by the lack of effective profiling and imaging tools, the field has progressed rapidly in recent years owing to the technical advancements in mass spectrometry (MS) and chemical imaging. Lipidomics, the analysis of lipid metabolites by MS, has greatly expanded our knowledge about the lipidome in long-lived species and centenarians. Moreover, lipidomics holds great promise to investigate the lipid profiles of subcellular organelles. These analyses can be achieved by rapid immuno-purification of whole organelles (Abu-Remaileh et al., 2017; Chen et al., 2017) and consequent MS profiling of lipid species. Future applications of these approaches in long-lived species and models will open doors to discover novel organelle-specific lipid signaling in longevity regulation. On the other hand, chemical imaging methods, specifically coherent Raman scattering (CRS) microscopy, enable label-free visualization of lipid molecules in live cells and organisms (Cheng and Xie, 2015). CRS microscopy is employed to visualize the distribution and heterogeneity of lipids across different cells and tissues (Chen et al., 2020; Yu et al., 2017). Furthermore, combined with the use of bioorthogonal (e.g., alkyne) and isotope (e.g., deuterium) tags to minimally label the lipids of interest, CRS microscopy is useful for tracking the incorporation, synthesis, and degradation of lipids in vivo (Lin and Wang, 2017; Mutlu et al., 2020; Wei et al., 2014; Yu et al., 2017). Overall, MS and CRS microscopy make inroads in revealing the chemical specificity and spatiotemporal dynamics of lipids in the regulation of aging and longevity.
Future work will discover more novel health-promoting and lifespan-extending lipid signals and better understand the molecular mechanisms underlying their benefits. Low hanging fruits might be those common lipid signatures shared by multiple pro-longevity pathways. New lipid regulatory mechanisms might also be discovered through investigating lipid signaling components, such as carriers, transporters, and receptors, for their effects on lifespan and health span. To better understand the role of lipid signaling, structural and biochemical characterization of lipid-protein interactions are crucial. Such studies will reveal how lipid signaling can be transduced within the cell and between tissues to systemically modulate health span and lifespan. It would be also important to dissect which tissues/cells/organelles are responsible for these lifespan-extending lipid signals and how they coordinate with each other. These mechanistic studies of anti-aging lipid-protein interactions will likely provide potential druggable sites that could be targeted to promote longevity and healthy aging.
ACKNOWLEDGMENTS
This work has been supported by the NIH grants R01AG045183 (M.C.W.), R01AT009050 (M.C.W.), R01AG062257 (M.C.W.), DP1DK113644 (M.C.W.), and the Welch Foundation Research Grant (M.C.W.). M.C.W is an HHMI investigator. Figures were made using BioRender. We apologize to colleagues whose work we were not able to cite due to space limitations.
REFERENCES
- Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, and Sabatini DM (2017). Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman D, and Gems D (2012). The mystery of C. elegans aging: an emerging role for fat. Distant parallels between C. elegans aging and metabolic syndrome? BioEssays 34, 466–471. [DOI] [PubMed] [Google Scholar]
- Alemany R, Perona JS, Sánchez-Dominguez JM, Montero E, Cañizares J, Bressani R, Escribá PV, and Ruiz-Gutierrez V (2007). G protein-coupled receptor systems and their lipid environment in health disorders during aging. Biochim. Biophys. Acta 1768, 964–975. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Dean A, Huang W, Setchell K, Moore D, and Darlington G (2007). Alterations in xenobiotic metabolism in the long-lived Little mice. Aging Cell 6, 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Noguez D, Yagi K, Venable S, and Darlington G (2004). Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell 3, 423–441. [DOI] [PubMed] [Google Scholar]
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, and Curtis R (2004). The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev 18, 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, and Kenyon C (2002). Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505. [DOI] [PubMed] [Google Scholar]
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Van-Gilst M, Sequerra R, Doucette-Stamm L, Yamamoto KR, and Walhout AJM (2010). Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol. Syst. Biol 6, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlia-Ciommo A, Leonov A, Mohammad K, Beach A, Richard VR, Bourque SD, Burstein MT, Goldberg AA, Kyryakov P, Gomez-Perez A, et al. (2018). Mechanisms through which lithocholic acid delays yeast chronological aging under caloric restriction conditions. Oncotarget 9, 34945–34971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena C, Quirós PM, Durand S, Mayoral P, Rodríguez F, Caravia XM, Mariño G, Garabaya C, Fernández-García MT, Kroemer G, et al. (2018). Methionine restriction extends lifespan in progeroid mice and alters lipid and bile acid metabolism. Cell Rep 24, 2392–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Sewell AK, Wu Z, and Han M (2019). TOR signaling in Caenorhabditis elegans development, metabolism, and aging. Genetics 213, 329–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, and Isik M (2015). SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med 88, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M, Kahn BB, and Kahn CR (2003). Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574. [DOI] [PubMed] [Google Scholar]
- Burkewitz K, Morantte I, Weir HJM, Yeo R, Zhang Y, Huynh FK, Ilkayeva OR, Hirschey MD, Grant AR, and Mair WB (2015). Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 160, 842–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill SL, Carey JR, Müller HG, and Anderson G (2003). Age of ovary determines remaining life expectancy in old ovariectomized mice. Aging Cell 2, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash TP, Pita G, Domínguez O, Alonso MR, Moreno LT, Borrás C, Rodríguez-Mañas L, Santiago C, Garatachea N, Lucia A, et al. (2014). Exome sequencing of three cases of familial exceptional longevity. Aging Cell 13, 1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti P, English T, Shi J, Smas CM, and Kandror KV (2010). Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes 59, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles KN, Li MD, Engin F, Arruda AP, Inouye K, and Hotamisligil GS (2017). Uncoupling of metabolic health from longevity through genetic alteration of adipose tissue lipid-binding proteins. Cell Rep 21, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AL, Lum KM, Lara-Gonzalez P, Ogasawara D, Cognetta AB, To A, Parsons WH, Simon GM, Desai A, Petrascheck M, et al. (2019). Pharmacological convergence reveals a lipid pathway that regulates C. elegans lifespan. Nat. Chem. Biol 15, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, and Sabatini DM (2017). Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat. Protoc 12, 2215–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Lemieux GA, Camp CH, Chang TC, Ashrafi K, and Cicerone MT (2020). Spectroscopic coherent Raman imaging of Caenorhabditis elegans reveals lipid particle diversity. Nat. Chem. Biol 16, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JX, and Xie XS (2015). Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine. Science 350, aaa8870. [DOI] [PubMed] [Google Scholar]
- Chiang JYL (2009). Bile acids: regulation of synthesis. J. Lipid Res 50, 1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YM, Fujita Y, and Kieffer TJ (2014). Glucagon-like peptide-1: glucose homeostasis and beyond. Annu. Rev. Physiol 76, 535–559. [DOI] [PubMed] [Google Scholar]
- Ciardo MG, and Ferrer-Montiel A (2017). Lipids as central modulators of sensory TRP channels. Biochim. Biophys. Acta Biomembr 1859, 1615–1628. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, and Puigserver P (2007). mTOR controls mitochondrial oxidative function through a YY1–PGC-1α transcriptional complex. Nature 450, 736–740. [DOI] [PubMed] [Google Scholar]
- Daniele JR, Higuchi-Sanabria R, Durieux J, Monshietehadi S, Ramachandran V, Tronnes SU, Kelet N, Sanchez M, Metcalf MG, Garcia G, et al. (2020). UPRER promotes lipophagy independent of chaperones to extend life span. Sci. Adv 6, eaaz1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diego I. de, Peleg S, and Fuchs B (2019). The role of lipids in aging-related metabolic changes. Chem. Phys. Lipids 222, 59–69. [DOI] [PubMed] [Google Scholar]
- Egawa J, Pearn ML, Lemkuil BP, Patel PM, and Head BP (2016). Membrane lipid rafts and neurobiology: age-related changes in membrane lipids and loss of neuronal function. J. Physiol 594, 4565–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribá PV, Wedegaertner PB, Goñi FM, and Vögler O (2007). Lipid–protein interactions in GPCR-associated signaling. Biochim. Biophys. Acta 1768, 836–852. [DOI] [PubMed] [Google Scholar]
- Evans RM, and Mangelsdorf DJ (2014). Nuclear receptors, RXR, and the big bang. Cell 157, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzili C, Otrubova K, and Boger DL (2010). Fatty acid amide signaling molecules. Bioorg. Med. Chem. Lett 20, 5959–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D’Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, and Tatar M (2008). Drosophila germ-line modulation of insulin signaling and lifespan. Proc. Natl. Acad. Sci. USA 105, 6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, and Wang MC (2015). Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, and Johnson TE (1988). A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodríguez De Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, and Piomelli D (2003). Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature 425, 90–93. [DOI] [PubMed] [Google Scholar]
- Ganfornina MD, Do Carmo SD, Lora JM, Torres-Schumann S, Vogel M, Allhorn M, Gonzáilez C, Bastiani MJ, Rassart E, and Sanchez D (2008). Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 7, 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, and Antebi A (2007). A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc. Natl. Acad. Sci. USA 104, 5014–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jünger MA, Hafen E, Leevers SJ, and Partridge L (2004). Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305, 361. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I, van der Kloet FM, Houwing-Duistermaat J, Vreeken RJ, Hankemeier T, and Slagboom EP (2013). Lipidomics of familial longevity. Aging Cell 12, 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, and Aguilaniu H (2011). Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol 9, e1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieco M, Giorgi A, Gentile MC, d’Erme M, Morano S, Maras B, and Filardi T (2019). Glucagon-like peptide-1: a focus on neurodegenerative diseases. Front. Neurosci 13, 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Schroeder EA, Silva-García CG, Hebestreit K, Mair WB, and Brunet A (2017). Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handee W, Li X, Hall KW, Deng X, Li P, Benning C, Williams BL, and Kuo MH (2016). An energy-independent pro-longevity function of triacylglycerol in yeast. PLoS Genet 12, e1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, Habermann B, and Antebi A (2013). Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet 9, e1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, and Kenyon C (2010). Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. USA 107, 9730–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, and Bernlohr DA (2015). Metabolic functions of FABPs—mechanisms and therapeutic implications. Nat. Rev. Endocrinol 11, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, and Kenyon C (1999). Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366. [DOI] [PubMed] [Google Scholar]
- Hull-Thompson J, Muffat J, Sanchez D, Walker DW, Benzer S, Ganfornina MD, and Jasper H (2009). Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Genet 5, e1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, and Tatar M (2004). Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562–566. [DOI] [PubMed] [Google Scholar]
- Imanikia S, Sheng M, Castro C, Griffin JL, and Taylor RC (2019). XBP-1 remodels lipid metabolism to extend longevity. Cell Rep 28, 581–589.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou MS, Jackson J, Sheu SH, Chang CL, Weigel AV, Liu H, Pasolli HA, Xu CS, Pang S, Matthies D, et al. (2019). Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell 177, 1522–1535.e14. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM, Kim S, Dai J, Li L, Bi X, Jiang JC, Arnold J, Batzer MA, Walker JA, Welsh DA, et al. (2010). HRAS1 and LASS1 with APOE are associated with human longevity and healthy aging. Aging Cell 9, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman J, McConathy W, Araoz C, Kasa P, and Lacko AG (2000). Apolipoprotein D in the aging brain and in Alzheimer’s dementia. Neurol. Res 22, 330–336. [DOI] [PubMed] [Google Scholar]
- Katic M, Kennedy AR, Leykin I, Norris A, McGettrick A, Gesta S, Russell SJ, Bluher M, Maratos-Flier E, and Kahn CR (2007). Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell 6, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, et al. (2003).A G protein-coupled receptor responsive to bile acids. J. Biol. Chem 278, 9435–9440. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, and Tabtiang R (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- Kostenis E (2004). A glance at G-protein-coupled receptors for lipid mediators: a growing receptor family with remarkably diverse ligands. Pharmacol. Ther 102, 243–257. [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CDDM, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. (2013). The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun 4, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Gelino S, Meléndez A, and Hansen M (2011). Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol 21, 1507–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, and Hansen M (2012). Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab 23, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer LM, Iakoubov R, and Brubaker PL (2009). GPR119 is essential for Oleoylethanolamide-induced glucagon-like peptide-1 secretion from the intestinal enteroendocrine L-cell. Diabetes 58, 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, and Ashrafi K (2008). A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Brown MS, and Goldstein JL (2010). Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. USA 107, 3441–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, and Kenyon C (2003). Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489–502. [DOI] [PubMed] [Google Scholar]
- Lin C-CJ, and Wang MC (2017). Microbial metabolites regulate host lipid metabolism through NR5A–Hedgehog signalling. Nat. Cell Biol 19, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Chen YC, Kao T-Y, Lin Y-C, Hsu T-E, Wu Y-C, Ja WW, Brummel TJ, Kapahi P, Yuh C-H, et al. (2014). Diacylglycerol lipase regulates lifespan and oxidative stress response by inversely modulating TOR signaling in Drosophila and C. elegans. Aging Cell 13, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol 21, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, and Bellen HJ (2015). Glial lipid droplets and ROS Induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Held JM, Vantipalli MC, Klang IM, Graham JB, Gibson BW, Lithgow GJ, and Gill MS (2011). N-acylethanolamine signalling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature 473, 226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães J.P. de, Curado J, and Church GM (2009). Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magner DB, Wollam J, Shen Y, Hoppe C, Li D, Latza C, Rottiers V, Hutter H, and Antebi A (2013). The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab 18, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Nakamura T, Itadani H, and Tanaka K (2002). Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun 298, 714–719. [DOI] [PubMed] [Google Scholar]
- Matyash V, Geier C, Henske A, Mukherjee S, Hirsh D, Thiele C, Grant B, Maxfield FR, and Kurzchalia TV (2001). Distribution and transport of cholesterol in Caenorhabditis elegans. Mol. Biol. Cell 12, 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz V, Howell KS, Gentina S, Epstein S, Riezman I, Fornallaz-Mulhauser M, Hengartner MO, Gomez M, Riezman H, and Martinou JC (2009). Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324, 381–384. [DOI] [PubMed] [Google Scholar]
- Mielke MM, Bandaru VVR, Haughey NJ, Xia J, Fried LP, Yasar S, Albert M, Varma V, Harris G, Schneider EB, et al. (2012). Serum ceramides increase the risk of Alzheimer disease: the women’s health and aging study II. Neurology 79, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KJ, Lee CK, and Park HN (2012). The lifespan of Korean eunuchs. Curr. Biol 22, R792–R793. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Buffenstein R, and Hulbert AJ (2007). Membrane phospholipid composition may contribute to exceptional longevity of the naked molerat (Heterocephalus glaber): a comparative study using shotgun lipidomics. Exp. Gerontol 42, 1053–1062. [DOI] [PubMed] [Google Scholar]
- Montoliu I, Scherer M, Beguelin F, DaSilva L, Mari D, Salvioli S, Martin F-PJ, Capri M, Bucci L, Ostan R, et al. (2014). Serum profiling of healthy aging identifies phospho- and sphingolipid species as markers of human longevity. Aging (Albany, NY: ) 6, 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech MB, Kruse R, Harvald EB, Olsen ASB, Gallego SF, Hannibal-Bach HK, Ejsing CS, and Færgeman NJ (2013). Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PLoS One 8, e70087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, and Mangelsdorf DJ (2006). Identification of ligands for DAF-12 that govern Dauer Formation and reproduction in C. elegans. Cell 124, 1209–1223. [DOI] [PubMed] [Google Scholar]
- Motter AL, and Ahern GP (2008). TRPV1-null mice are protected from diet-induced obesity. FEBS Lett 582, 2257–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourikis P, Hurlbut GD, and Artavanis-Tsakonas S (2006). Enigma, a mitochondrial protein affecting lifespan and oxidative stress response in Drosophila. Proc. Natl. Acad. Sci. USA 103, 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J, Walker DW, and Benzer S (2008). Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proc. Natl. Acad. Sci. USA 105, 7088–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro D, and Blier PU (2012). The extreme longevity of Arctica islandica is associated with increased peroxidation resistance in mitochondrial membranes. Aging Cell 11, 845–855. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, and Kenyon C (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283. [DOI] [PubMed] [Google Scholar]
- Mutlu AS, Gao SM, Zhang H, and Wang MC (2020). Olfactory specificity regulates lipid metabolism through neuroendocrine signaling in Caenorhabditis elegans. Nat. Commun 11, 1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke EJ, and Ruvkun G (2013). MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol 15, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke EJ, Soukas AA, Carr CE, and Ruvkun G (2009). C. elegans Major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab 10, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, and Ruvkun G (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- Oliveira P.G. de, Ramos MLS, Amaro AJ, Dias RA, and Vieira SI (2019). Gi/o-protein coupled receptors in the aging brain. Front. Aging Neurosci. 11, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Efeyan A, Lopez-Guadamillas E, Muñoz-Martin M, Gómez-López G, Cañamero M, Mulero F, Pastor J, Martinez S, Romanos E, et al. (2012). Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab 15, 382–394. [DOI] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, and Thummel CS (2009). Drosophila HNF4 regulates lipid mobilization and β-oxidation. Cell Metab 9, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare PP, Lin A, Bornfeldt KE, Taubert S, and Van-Gilst MR (2012). Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet 8, e1002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CL, and Van-Gilst MR (2008). A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab 8, 266–274. [DOI] [PubMed] [Google Scholar]
- Pernicova I, and Korbonits M (2014). Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol 10, 143–156. [DOI] [PubMed] [Google Scholar]
- Ponnappan U, Holley DH, and Lipschitz DA (1996). Effect of age on the fatty acid composition of phospholipids in human lymphocytes. Exp. Gerontol 31, 125–133. [DOI] [PubMed] [Google Scholar]
- Qi W, Gutierrez GE, Gao X, Dixon H, McDonough JA, Marini AM, and Fisher AL (2017). The ω-3 fatty acid α-linolenic acid extends Caenorhabditis elegans lifespan via NHR-49/PPARα and oxidation to oxylipins. Aging Cell 16, 1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabini RA, Moretti N, Staffolani R, Salvolini E, Nanetti L, Franceschi C, and Mazzanti L (2002). Reduced susceptibility to peroxidation of erythrocyte plasma membranes from centenarians. Exp. Gerontol 37, 657–663. [DOI] [PubMed] [Google Scholar]
- Ramachandran PV, Savini M, Folick AK, Hu K, Masand R, Graham BH, and Wang MC (2019). Lysosomal signaling promotes longevity by adjusting mitochondrial activity. Dev. Cell 48, 685–696.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnappan R, Amrit FRG, Chen SW, Gill H, Holden K, Ward J, Yamamoto KR, Olsen CP, and Ghazi A (2014). Germline signals deploy NHR-49 to modulate fatty-acid β-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet 10, e1004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera CE, Huising MO, Follett P, Leblanc M, Halloran J, Van Andel R, de Magalhaes Filho CD, Merkwirth C, and Dillin A (2014). TRPV1 pain receptors regulate longevity and metabolism by neuropeptide signaling. Cell 157, 1023–1036. [DOI] [PubMed] [Google Scholar]
- Sacket SJ, Chung HY, Okajima F, and Im DS (2009). Increase in sphingolipid catabolic enzyme activity during aging. Acta Pharmacol. Sin 30, 1454–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D, López-Arias B, Torroja L, Canal I, Wang X, Bastiani MJ, and Ganfornina MD (2006). Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Curr. Biol 16, 680–686. [DOI] [PubMed] [Google Scholar]
- Schaap FG, Trauner M, and Jansen PLM (2014). Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol 11, 55–67. [DOI] [PubMed] [Google Scholar]
- Schächter F, Faure-Delanef L, Guénot F, Rouger H, Froguel P, Lesueur-Ginot L, and Cohen D (1994). Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet 6, 29–32. [DOI] [PubMed] [Google Scholar]
- Schmeisser S, Li S, Bouchard B, Ruiz M, Des Rosiers C, and Roy R (2019). Muscle-specific lipid hydrolysis prolongs lifespan through global lipidomic remodeling. Cell Rep 29, 4540–4552.e8. [DOI] [PubMed] [Google Scholar]
- Seah NE, de Magalhaes Filho CD, Petrashen AP, Henderson HR, Laguer J, Gonzalez J, Dillin A, Hansen M, and Lapierre LR (2016). Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy 12, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Prasanna X, Mohole M, and Chattopadhyay A (2018). Exploring GPCR–lipid interactions by molecular dynamics simulations: excitements, challenges, and the way forward. J. Phys. Chem. B 122, 5727–5737. [DOI] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis RJ, Xu L, Lee H, Chae M, Thaden JJ, Bharill P, Tazearslan C, Siegel E, Alla R, Zimniak P, and Ayyadevara S (2011). Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany, NY: ) 3, 125–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipula IJ, Brown NF, and Perdomo G (2006). Rapamycin-mediated inhibition of mammalian target of rapamycin in skeletal muscle cells reduces glucose utilization and increases fatty acid oxidation. Metabolism 55, 1637–1644. [DOI] [PubMed] [Google Scholar]
- Söderberg M, Edlund C, Kristensson K, and Dallner G (1990). Lipid compositions of different regions of the human brain during aging. J. Neurochem 54, 415–423. [DOI] [PubMed] [Google Scholar]
- Soliman GA, Acosta-Jaquez HA, and Fingar DC (2010). mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids 45, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats S, Rimbach G, Kuenstner A, Graspeuntner S, Rupp J, Busch H, Sina C, Ipharraguerre IR, and Wagner AE (2018). Lithocholic acid improves the survival of Drosophila melanogaster. Mol. Nutr. Food Res 62, e1800424. [DOI] [PubMed] [Google Scholar]
- Steinbaugh MJ, Narasimhan SD, Robida-Stubbs S, Moronetti Mazzeo LEM, Dreyfuss JM, Hourihan JM, Raghavan P, Operarña TN, Esmaillie R, and Blackwell TK (2015). Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. eLife 4, e07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenesen D, Suh JM, Seo J, Yu K, Lee KS, Kim JS, Min KJ, and Graff JM (2013). Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab 17, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storelli G, Nam HJ, Simcox J, Villanueva CJ, and Thummel CS (2019). Drosophila HNF4 directs a switch in lipid metabolism that supports the transition to adulthood. Dev. Cell 48, 200–214.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeper RS, Grueter CA, Salomonis N, Cases S, Levin MC, Koliwad SK, Zhou P, Hirschey MD, Verdin E, and Farese RV (2012). Deficiency of the lipid synthesis enzyme, DGAT1, extends longevity in mice. Aging (Albany, NY: ) 4, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thondamal M, Witting M, Schmitt-Kopplin P, and Aguilaniu H (2014). Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat. Commun 5, 4879. [DOI] [PubMed] [Google Scholar]
- Tillman MC, Khadka M, Duffy J, Wang MC, and Ortlund EA (2019). Structural characterization of life-extending Caenorhabditis elegans lipid binding protein 8. Sci. Rep 9, 9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, and Thomas G (2004). Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205. [DOI] [PubMed] [Google Scholar]
- Van-Gilst MR, Hadjivassiliou H, Jolly A, and Yamamoto KR (2005). Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, and Müller F (2003). Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- Walker DW, Muffat J, Rundel C, and Benzer S (2006). Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr. Biol 16, 674–679. [DOI] [PubMed] [Google Scholar]
- Wang H, and Eckel RH (2014). What are lipoproteins doing in the brain? Trends Endocrinol. Metab. 25, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, O’Rourke EJ, and Ruvkun G (2008). Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Hu F, Shen Y, Chen Z, Yu Y, Lin CC, Wang MC, and Min W (2014). Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 11, 410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Zhang B, Dong Y, Gong J, Xu T, Liu J, and Xu XZS (2013). A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell 152, 806–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Joo HJ, and Paik YK (2011). Novel functions of lipid-binding protein 5 in Caenorhabditis elegans fat metabolism. J. Biol. Chem 286, 28111–28118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gopalacharyulu P, Seppänen-Laakso T, Ruskeepää AL, Aye CC, Carson BP, Mora S, Orešič M, and Teleman AA (2012). Insulin signaling regulates fatty acid catabolism at the level of CoA activation. PLoS Genet 8, e1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Gong ZJ, Zhou Y, Yuan JQ, Cheng J, Tian L, Li S, Lin XD, Xu R, Zhu Z-R, and Mao C (2010). Role of Drosophila alkaline ceramidase (Dacer) in Drosophila development and longevity. Cell. Mol. Life Sci 67, 1477–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee C-H, and Manning BD (2011). Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab 14, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Mutlu AS, Liu H, and Wang MC (2017). High-throughput screens using photo-highlighting discover BMP signaling in mitochondrial lipid oxidation. Nat. Commun 8, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaarur N, Desevin K, Mackenzie J, Lord A, Grishok A, and Kandror KV (2019). ATGL-1 mediates the effect of dietary restriction and the insulin/IGF-1 signaling pathway on longevity in C. elegans. Mol. Metab. 27, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee SJ, and Kenyon C (2013). Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab 17, 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Spassieva SD, Jucius TJ, Shultz LD, Shick HE, Macklin WB, Hannun YA, Obeid LM, and Ackerman SL (2011). A deficiency of ceramide biosynthesis causes cerebellar Purkinje cell neurodegeneration and lipofuscin accumulation. PLoS Genet 7, e1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Chen T, Zhao A, Wang X, Xie G, Huang F, Liu J, Zhao Q, Wang S, Wang C, et al. (2016). The brain metabolome of male rats across the lifespan. Sci Rep-Uk 6, 24125. [DOI] [PMC free article] [PubMed] [Google Scholar]