Abstract
FOXP3 deficiency in mice and IPEX syndrome patients results in fatal autoimmunity by altering T regulatory cells (Treg). CD4+ T cells in IPEX patients and Foxp3-deficient mice were analyzed by single-cell cytometry and RNAseq, revealing heterogeneous Treg-like cells, some very similar to normal Tregs, others more distant. Conventional T cells showed no widespread activation or Th bias, but a monomorphic disease signature affected all CD4+ T cells, cell-extrinsic signature since extinguished in mixed bone marrow chimeric mice and heterozygous mothers of IPEX patients. Normal Tregs exerted dominant suppression, extinguishing the disease signature, and revealing in mutant Treg-like cells a small cluster of genes regulated cell-intrinsically by FOXP3, including key homeostatic regulators. We propose a two-step pathogenesis model: cell-intrinsic downregulation of core genes destabilizes Tregs, de-repressing systemic mediators which imprint the disease signature on all T cells, furthering Treg dysfunction. Accordingly, IL2 treatment improved the Treg-like compartment and survival.
INTRODUCTION
T regulatory cells (Treg) that express the transcription factor FOXP3 are dominant negative regulators of many facets of the immune system, controlling immune responses and enforcing peripheral tolerance to self, symbiotic commensals and fetal antigens 1,2. They also have extra-immunological roles in maintaining tissue homeostasis outside of the purely immunological context 3. Tregs share a common transcriptional signature, genes differentially expressed relative to their conventional CD4+ counterparts (Tconv), in mice and humans 4–6. Much of the Treg signature is controlled by FOXP3, the lineage’s defining transcription factor (TF), although FOXP3 is neither fully necessary nor sufficient to establish the Treg transcriptome, requiring input from transcriptional cofactors 7,8. Consistent with their pleiotropic functions, Tregs show a range of phenotypic variation, differing by activation state, effector capabilities, and tissue localization 3,9. Treg heterogeneity has been further refined by single-cell transcriptomics (scRNAseq) 6,10–12.
IPEX (Immune dysregulation-Polyendocrinopathy-Enteropathy-X-linked) is perhaps the prototype of monogenic autoimmune syndromes 13, resulting from FOXP3 loss of function (LOF) and thus Treg deficiency 1,14. X-linked, it is transmitted by asymptomatic carrier females to their male progeny who usually present early after birth with a constellation of autoimmune manifestations dominated by severe enteropathy, eczematous dermatitis and type-1 diabetes, with some less common manifestations (nephropathy, hypoparathyroidism, antibody-mediated cytopenias) 15–18. There are several causes to the variable severity observed in IPEX patients. First, complete LOF mutations are generally more deleterious than missense mutations; and mutations in the DNA-binding forkhead or dimerization (leucine zipper) domains are generally more severe than N-terminal mutations, in keeping with results from our alanine-scan dissection of FOXP3 8,17. But manifestations and severity can vary between patients with the same mutation 17,19, suggesting that genetic modifiers, environmental influences or immunological history modify the disease course in each patient. Mice of the spontaneous scurfy mutant line, or carrying engineered Foxp3 LOFs, show a similar rapidly lethal phenotype, dominated by enteropathy, dermatitis and lymphoproliferation 1,20; partial or slower disease appears in mice with mild Foxp3 missense mutations 8,21,22.
Also contributing to this variability, the relationship between FOXP3 and Treg cells is now recognized not to be obligate. Several reports have documented the existence of Treg-like cells in the absence of viable Foxp3 in scurfy mice 23–25 and some IPEX patients 26–28. In addition, FOXP3 is not exclusive to Treg cells: it is also expressed, albeit at lower levels than in Tregs, early after Tconv activation 29–32, and scRNAseq showed some FOXP3-positive cells otherwise similar to Tconv cells 6.
The advent of single-cell transcriptomics opened the potential to revisit the alterations of CD4+ T cells in IPEX patients, which were previously difficult to address since the markers potentially used to sort Treg or Tconv cells may be themselves perturbed. We have thus performed a deep profiling analysis in IPEX patients that associates flow cytometry, single-cell (for resolution) and conventional (for depth) RNAseq. Complementary analyses in scurfy mice assessed the generality of the conclusions, eschewing the inevitable confounders of patient material, and permitting experimental manipulations to trace the source of the perturbations. The results paint an unexpected landscape of IPEX T cells, with a complex mix of Treg-like cells, some healthy-like and others more perturbed, a surprisingly narrow intrinsic signature of FOXP3 but a dominant “IPEX signature” that affects both Treg and Tconv cells, and suggests a feed-forward loop in T cell perturbations culminating in clinical disease.
RESULTS
Study Cohorts
This study included a primary and a replication cohort for confirmation and refinement (including some re-sampling of initial patients). Altogether, we analyzed peripheral blood mononuclear cells (PBMC) from 15 IPEX patients and 15 healthy donors (HD) collected at two reference clinical centers (Necker Hospital, Paris and Children’s Hospital, Boston; Supplementary Table 1, Fig. 1a). HD had no significant medical history and were recruited during well-child visits or orthopedic follow-ups. IPEX presentation was typical, first symptoms appearing neonatally for most (1–8 weeks). As in other cohorts 15–18, enteropathy (12/14), dermatitis (10/14), food allergy (8/13) and diabetes (6/14) were most common, with less frequent kidney, neurological or pulmonary involvement. Four patients were untreated at the time of sampling, but most were managed by immunosuppression (mainly mTOR inhibitors) – four of them later received bone marrow transplants. Blood samples were collected at different ages (9 months to 26 years) during routine visits, with no concurrent acute events. Five patients were analyzed at two or three time points (6–12 months apart) to assess the stability of the transcriptional characteristics.
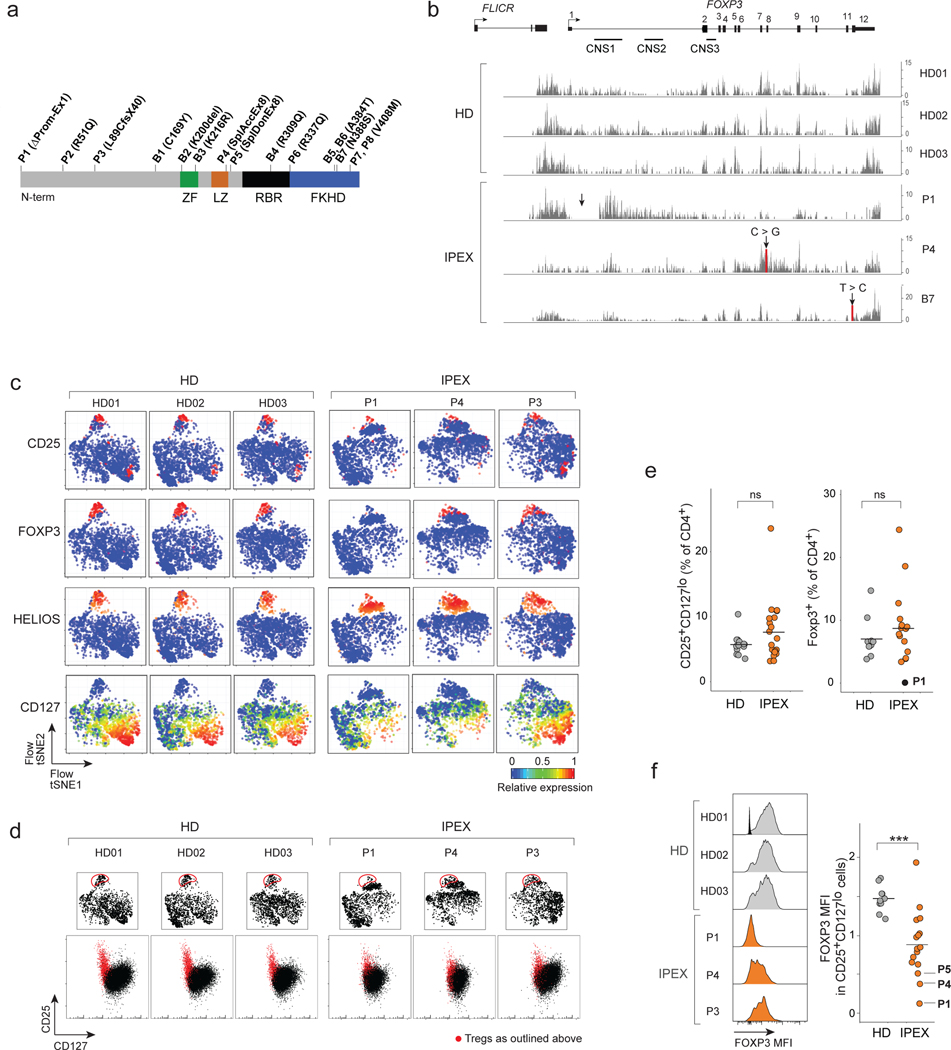
Fig. 1. Identification of Treg-like cells in IPEX by flow cytometry.
a. Position of the FOXP3 mutations in the IPEX cohorts (domains: ZF: zinc-finger; LZ: leucine-zipper; RBR: RUNX1 binding region; FKH: Forkhead). n = 15 IPEX
b. Mapping of RNAseq reads from CD4+CD25+CD127lo cells to the FOXP3 locus in representative samples (see Fig. S1). Arrow indicates mutation.
c. Flow-tSNE plot of CD4+ T cells, from anti-CD3, CD4, CD25, CD127, HELIOS, CD45RA and FOXP3 staining in representative samples (full set in Fig. S3). Color represents scaled expression.
d. Flow cytometric anti-CD25/CD127 plots of CD4+ T cells (bottom row); red: cells gated in the flow-tSNE plot, top row.
e. Proportion of CD25+CD127lo cells and total FOXP3+ cells in HD and IPEX (each dot a sample) (see Fig. S2a), ns non significant, two-sided t.test. n = 15 IPEX, n = 13 HD.
f. FOXP3 expression in CD25+CD127lo HD and IPEX cells. Left: representative cytometry profiles (see Fig. S2b for all other samples; one unstained control overlaid with HD01), quantitation at right (each dot a sample; *** two-sided t.test p < 0.001, n = 15 IPEX, n = 13 HD).
FOXP3 mutations were confirmed by sequencing. Most were missense mutations in different domains, two of them represented twice (V408M and the common A384T). One mutation (P1) was a large deletion spanning the promoter and intron 1; two others affected the exon 8 splice acceptor and donor sites (P4 and P5, respectively). The mutations’ impact was seen in the RNAseq reads at the FOXP3 locus of sorted CD4+CD25hiCD127lo cells (see below). For HD, the traces were very reproducible, reads piling up at the exons, the conserved non-coding sequence (CNS) 1 and the lncRNA FLICR (Fig. 1b, Fig. S1). Profiles from patients were variable: generally normal for the missense mutations; for P1, aberrantly initiated transcripts piled up around CNS1 with essentially no exonic reads; for P4, the splice mutation in exon 8 disturbed later exons and introns. Importantly, though, the FOXP3 locus was active in all IPEX cells, at far greater levels than in Tconv (Fig. S1). FLICR transcripts varied markedly in patient cells, unrelated to the proportion of FOXP3 exonic reads, in line with its independent transcriptional regulation 33.
Identification of Treg-like cells in IPEX patients
Tregs are typically identified by flow cytometry as CD25hiCD127lo, a combination that overlaps well with intracellular FOXP3 expression 34. Such cells have been reported in IPEX patients 26–28, but we first adopted a more global approach to identify Treg-like cells, since expression of these two markers could be perturbed by FOXP3 deficiencies. We analyzed CD4+ PBMCs by multiparameter flow cytometry for a larger panel of Treg markers and intracellular staining for FOXP3 and HELIOS) (Fig. S2). The data from individual staining experiments, including HD and IPEX samples, were then integrated in 2D space by t-Distributed Stochastic Neighbor Embedding (t-SNE; Fig. 1c, Fig. S3). Tregs from HD donors clustered tightly, with characteristic FOXP3, CD25 and HELIOS expression. For IPEX donors, the same region of the plots contained cells that also expressed CD25 and HELIOS, but variable FOXP3 (predictably absent for P1, more normal for P4). Indeed, “gating” Treg-like cells on the tSNE showed that they fell in the expected position of the CD25/CD127 plot (Figs. 1d, Fig. S2a), although with lower CD25 in IPEX samples. The proportion of Treg-like cells, and generally of FOXP3+ cells, were well conserved in IPEX patients (Fig. 1e, Fig. S2b), although the levels of FOXP3 protein were variably reduced (Fig. 1f, Fig. S2b). Other than the low or null levels resulting from promoter or splicing mutations, there was no discernible relationship between FOXP3 intensity and the domains affected by each mutation. These cytometry results confirmed that cells with surface Treg characteristics, but with low or absent FOXP3, could be identified in every one of the IPEX patients.
Widespread perturbations of IPEX CD4+ T cells transcriptomes
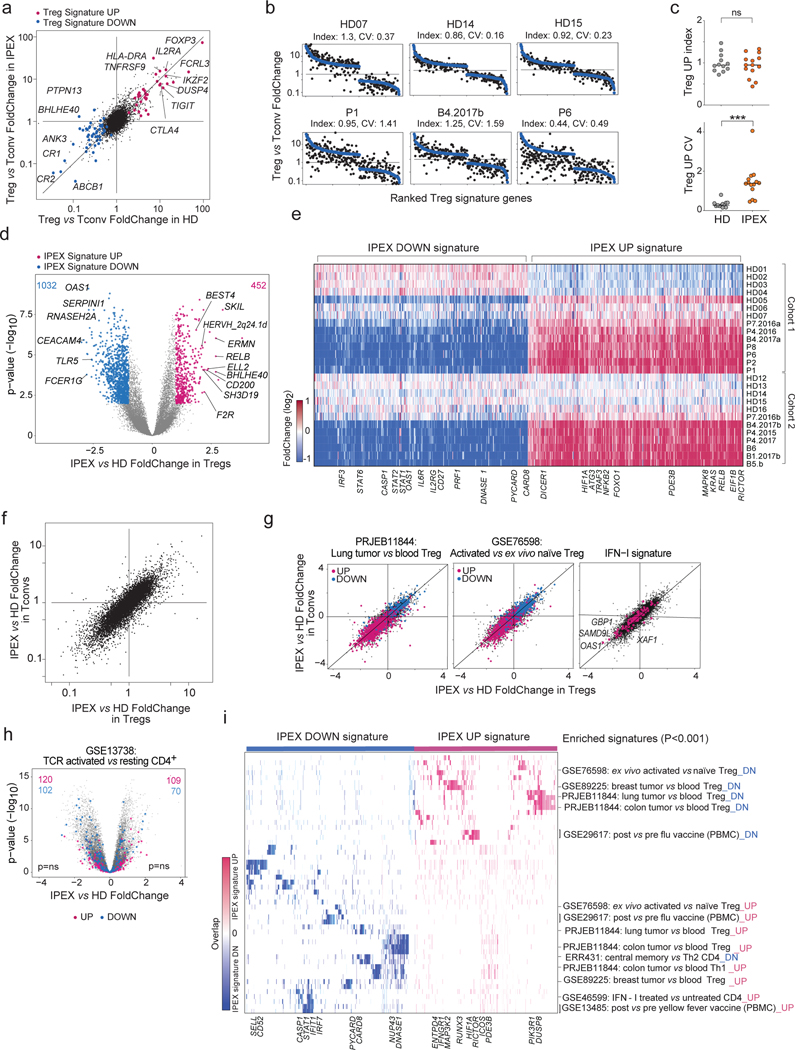
Given the presence of Treg-like CD25hiCD127lo cells in PBMCs from IPEX patients, we performed population low-input RNAseq after sorting these and Tconv (CD25–CD127hi) cells, as a preliminary to single-cell profiling and to help anchor its interpretation (first and replication cohorts profiled independently). Several observations emerged. First, the classic Treg signature was well conserved in Tregs from IPEX donors (Fig. 2a), including prototype transcripts (IL2RA, LRRC32, CTLA4). In ranked FoldChange (FC) plots that display the Treg signature for individual donors, the distribution of signature genes was tight for HD but more variable in IPEX Tregs, ranging from quasi-normal (P7) to markedly affected (P6, P8; Fig. 2b, Fig. S4a) – perhaps surprisingly, P1 with the complete LOF mutation did not show the strongest reduction in Treg signature genes. The Treg signature intensity score seemed better preserved in Tregs from IPEX donors than its coefficient of variation (Fig. 2b,c), suggesting an instability of the Treg signature in the absence of a fully functional FOXP3. These indices did not correlate significantly with clinical outcomes, although patients under immunosuppression trended to a lower index (Fig. S4b); this correlation is confounded, however, as patients with more severe FOXP3 mutations, and thus a lower TregUp index, are clinically more affected and more likely to receive strong treatment. The IPEX mutations did not affect all signature genes equally: some Treg signature genes were actually over-expressed in IPEX Tregs (DUSP4, LRRC32, CTLA4), while most were downregulated as expected (Fig. S4c).
Fig. 2. Transcriptional changes in IPEX Treg and Tconv cells by population RNAseq.
Population RNAseq was performed on sorted Tconv and Treg-like cells from HD (n=12) and IPEX (n = 10) donors. a. The Treg/Tconv FoldChange in HD (x-axis) and IPEX (y-axis); Treg signature 5 genes are highlighted.
b. Ranked FC plots of Treg signature transcripts for individual donors, ranked according to mean FC in all HD (blue dots). FC values for each donor (black dots) computed from the donor’s Treg vs the mean of HD Tconv.
c. Index and coefficient of variation (CV) of Treg Up signature transcripts (each dot is a sample). *** two-sided t.test p < 0.001.
d. FoldChange vs p value (volcano) plot comparing normalized expression in all IPEX to all HD samples. Genes with differential expression (two-sided t.test p<0.01, FC >2) are highlighted (452 up genes and 1032 down genes), and numbers shown.
e. Heatmap of the expression ratio, for IPEX signature genes defined in d, in Tregs from each donor, computed against the mean expression in HD Tregs (each cohort computed against its own HD Treg set).
f. Comparison of mean IPEX effect (all IPEX vs all HD samples) in Treg (x-axis) versus Tconv (y-axis).
g. Same FC/FC plot as in f, but highlighted with representative signatures of tumor-infitrating Tregs, activated Treg vs resting Treg, and IFN-I induced genes.
h. Same volcano plot as in d, but with highlights from a representative CD4+ T activation signature (ns, hypergeometric test).
i. Heatmap, for the IPEX signature genes defined in d, of their overlap with the pathways and signatures that significantly overlap with the IPEX signature (hypergeometric test p<0.001). Present genes are shown by tick marks, color-coded by their IPEX/HD FoldChange.
It is worth mentioning that no patient’s Treg or Tconv cells showed unusual expression of IL4 or IL5, transcripts which have been found to be paradoxically upregulated in response to forced expression of mutant FOXP3 8,22. More generally, there was no specific induction of cytokine genes in Tconv from IPEX patients that might have denoted a loss of Treg control (Fig. S4d).
We then assessed more generally the impact of IPEX mutations on transcriptomes of Treg-like cells. Widespread differences were observed (Fig. 2d), reproducibly in the two cohorts (Fig. S5a, Supplementary Table 2). This “IPEX signature” was consistent in every patient, involving all the same genes, albeit at variable overall intensity (Fig. 2e; one HD did show partial bias, perhaps because of unrecognized pathology). It did not correlate with the Treg signature intensity score (Fig. S5b) and was strikingly similar in Treg and Tconv cells (Fig. 2f), indicating a global impact on CD4+ T cells that transcended the sole effect of FOXP3 in Tregs (only 17 genes of the Treg signature belonged to this IPEX signature, Supplementary Table 2). There was no marked relationship between the main clinical parameters, including current corticosteroid or rapamycin treatment, and the IPEX signature score (Fig. S5c), which was also present in untreated IPEX patients. The IPEX signature was not accompanied by activation of endogenous retroviruses, which might plausibly be reactivated (Fig. S5d). Enrichment analysis showed significant overlap between the IPEX signature and several gene expression signatures of CD4+ T cells (Fig. 2g-i, Supplementary Table 3), but importantly not with the signatures of T cell activation (Fig. 2h), again indicating that the absence of Treg suppression did not result in wholesale T cell activation. If anything, a Treg activation signature 35 was downregulated in IPEX cells (Fig. 2g). Transcripts differentially expressed in tumor-infiltrating Tregs (2 independent studies) were counter-regulated in IPEX CD4+ T cells, as were interferon-stimulated genes (Fig. 2g,i). This broad change was consistent with the up-regulation of major response regulators like RELB, KRAS or HIF1. Overall, our results indicated that the Treg signature was in large part maintained in Tregs from IPEX patients, albeit with a notably high degree of variability. More unexpected was the peculiar transcriptomic footprint shared by IPEX Tregs and Tconvs, which might result from integration of extracellular cues, but not from a generic T cell activation state.
Different types of Tregs in IPEX patients
From these preliminary indications, we proceeded to the crux of the work, applying scRNAseq to identify with unbiased profiling the actual types of FOXP3-deficient Treg-like cells, which might be blurred by the averaging inherent in population profiling, or have escaped recognition because of shifts in the CD25 or CD127 markers. We sorted total CD4+ T cells, to yield granular information on both Treg and Tconv pools, and performed scRNAseq on the two cohorts as above (a total of 52,776 cells passing QC (Supplementary Table 4) from 11 IPEX and 11 HD donors). All results were observed in both the initial and replication cohorts, but are combined below for simplicity. Experimental confounders were minimized by multiplexing IPEX and HD samples in the same scRNAseq libraries, using DNA-coded tags 36. Residual inter-cohort and inter-experiment effects were corrected using the Canonical Correlation Analysis (CCA) and k-nearest neighbor-based integration methodology 37 (Fig. 3a).
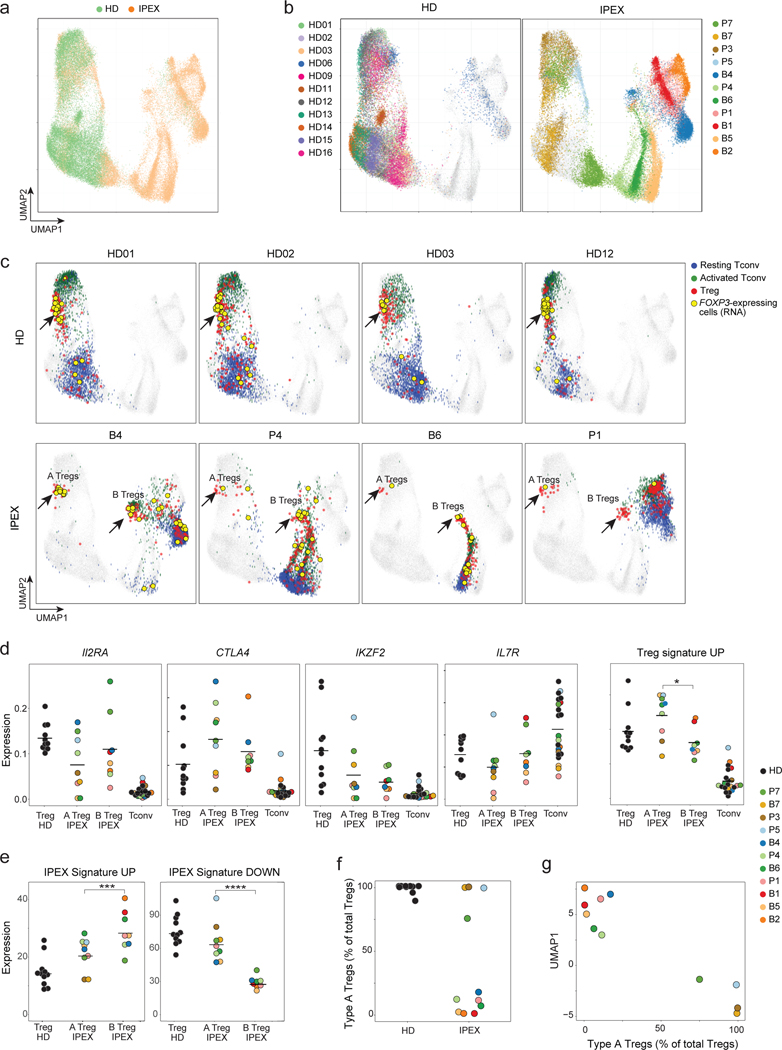
Fig. 3. scRNAseq reveals the heterogeneous effect of FOXP3 deficiency in IPEX Tregs.
scRNAseq was performed on peripheral blood CD4+ T cells from IPEX (n = 11) and HD (n = 11). a. 2D UMAP plot of all CD4+ cells from IPEX and HD samples (52,776 cells altogether, merged with Canonical Correlation Analysis and knn-graph, see Methods)
b. Same UMAP as a, color-coded by individual donor (see also Fig. S6d).
c. Same UMAP as a, with four representative HD and IPEX donors (see Fig. S8 for other donors). Blue, green, and red cells are resting Tconvs, activated Tconvs, and Tregs, respectively. FOXP3-expressing cells (RNA) are yellow. Type A and B Tregs in IPEX donors are indicated by an arrow.
d. Normalized counts expression of IL2RA, CTLA4, IKZF2, IL7R, and the Treg signature in Tregs from HD and IPEX (type A and B) and Tconv cells; each dot is a sample. * t.test p < 0.05
e. Average expression of the IPEX signature in type A and B IPEX Tregs and HD Tregs (normalized counts). *** p < 10–3, **** p < 10–4, two-sided t.test.
f. Proportion of type A Tregs in total Tregs for each sample.
g. Proportion of Type A Tregs plotted against the average UMAP1 dimension for each sample.
In the UMAP projection of the integrated data, CD4+ T cells partitioned sharply according to disease: cells from HD donors clustered together, while those from IPEX patients were much more dispersed (Fig. 3a), each tending to form an island distant to various degrees from the HD group (Fig. 3b). These distances were not batch artefacts (similar HD/IPEX partitions were seen in each batch, Fig. S6a). Replicate samples from 3 patients drawn >1 year apart mapped to the same regions, as did samples from patient P7 collected before and after immunosuppressive treatment (Fig. S6b), indicating that the different locations were patient-specific and not consequences of environmental or treatment variables. Conventional and scRNAseq reflected the same perturbation, as UMAP1 scores axis that partitioned IPEX and HD samples corresponded with the IPEX signature above (Fig. S6c).
We then sought to deconvolute, at single-cell resolution, Tregs among these CD4+ T cells. Tregs were identified in HD and IPEX samples in an unsupervised approach, using community detection in the reciprocal PCA integration network (implemented in Seurat V3 37), also supported by activity at the FOXP3 locus. Three distinct populations of CD4+ T cells could be identified (Fig. S6d): two Tconv populations (resting and activated) and one Treg (confirmed by expression of Treg signature genes FOXP3, IKZF2, CTLA4, IL2RA, TIGIT; Fig. S7a). When Tregs thus identified were displayed onto the UMAP space, HD Tregs formed a tight cluster for all HD samples (Fig. 3c), which co-localized with FOXP3 expression (yellow dots on Fig. 3c). In IPEX samples, Treg-like cells were similarly identified, in proportions equivalent to controls, even for fully FOXP3-deficient P1 (Fig. S7b), but they split into two groups (Fig. 3c, Fig. S8a): some (hereafter referred to as type-A IPEX Tregs) mapped in the same cluster as HD Tregs, while others clustered away, in their respective patient-specific “island” (type-B). Both were truly Tregs, expressing FOXP3 mRNA (Fig. 3c), core Treg transcripts (IL2RA, CTLA4, IKZF2, low IL7R) and the Treg signature (Fig 3d). Type-A and type-B Tregs did differ, with higher levels of Treg signature genes in type-A Tregs, and higher representation in type-B of the IPEX signature (Fig. 3e, Fig. S8b). Finally, proportions varied between IPEX patients (Fig. 3f): type-A Treg dominated in some, almost to HD levels, but were far less abundant in others. This proportion was stable in independent samplings of the same patient, and related to the intensity of the IPEX signature in Treg and Tconv cells: patients with the highest index had the lowest fraction of type-A Tregs (Fig. 3g).
Thus, scRNAseq analysis revealed a subset of Tregs that closely resemble normal Tregs, and another with more extensive perturbations. The origin of these two distinct Treg populations in IPEX patients was unclear. To exclude maternal microchimerism (wild-type Treg cells from the mother could have a competitive advantage in the IPEX offspring), we checked female-specific XIST transcripts: no XIST-positive cells were found among any patient’s Tregs. A striking feature of type-A Tregs was their marked downregulation of the IPEX signature.
IPEX does not affect Tconv phenotypes
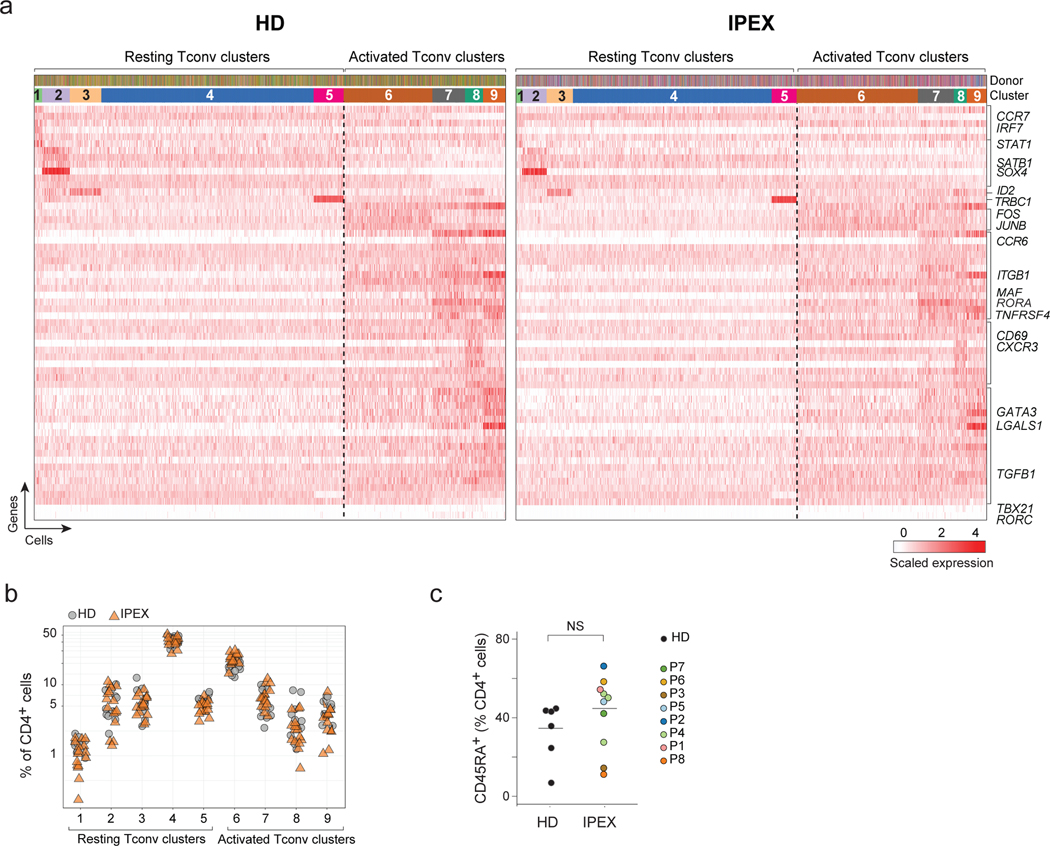
Tregs can affect Tconv polarization in many ways 9, and it is generally assumed that Treg deficiency in IPEX patients leads to excessive Tconv activation and differentiation and an over-representation of their effector states 38,39. Unsupervised clustering of Tconv cells performed in the shared reciprocal PCA space, after regressing out the IPEX effect, distinguished 9 clusters (5 resting and 4 activated/memory types, judging from the expression of characteristic markers and transcription factors like CCR7, SATB1, CD69, TBX21, GATA3) (Fig. 4a). Their distribution, and the expression of defining transcripts, was strikingly similar for HD and IPEX samples (Fig. 4a,b). This observation was confirmed by flow cytometry, which showed similar ranges of CD45RA+ cells among CD4+ T cells in IPEX and HD PBMCs (Fig. 4c). Thus, beyond the shared signature, IPEX disease seemed to have limited impact on other phenotypic aspects of circulating Tconv cells.
Fig. 4. Normal Tconv phenotypes in IPEX patients.
a. From the scRNAseq Tconv datasets (HD and IPEX combined), the IPEX signature was regressed out (see Methods) and biclustering was performed to define clusters in resting and activated Tconv. plotted as a heatmap for expression of the most characteristic genes for each Tconv cluster (each vertical line represents one cell). Top ribbons indicate donor origin and cluster annotations for every cell.
b. Proportion of resting and activated Tconv clusters in total CD4+ cells in HD and IPEX in scRNAseq data.
c. Proportion of CD45RA+ (resting) CD4+ T cells determined by flow cytometry in IPEX and HD. ns, non-significant (two-sided t.test, n= 11 HD, 11 IPEX).
Mixed populations of Foxp3-active cells in Foxp3-deficient mice
The FOXP3 deficiency in IPEX patients thus led to a global change affecting all CD4+ T cells, and to a diversity of Treg-like cells. To assess mutational impacts in a setting devoid of genetic or therapeutic variables, we performed parallel analyses on Foxp3-deficient mice (the previously described Foxp3ΔEGFPcre inactivating reporter line 25, hereafter denoted ΔFoxp3), in which FOXP3 protein is absent, but Foxp3 locus activity can be detected.
We used scRNAseq to analyze the diversity of CD4+ T splenocytes in ΔFoxp3 mice and WT controls (4 mice per group, 18,000 cells altogether, again multiplexing samples in the same libraries, Supplementary Table 4). As in human patients, cells from WT and ΔFoxp3 mice clustered separately on the UMAP projection, here again reflecting a generic ΔFoxp3 signature (Fig. 5a) present in every mutant mouse, and affecting Treg as well as Tconv (Fig. S9a). This cross-cutting “ΔFoxp3 signature” was confirmed by population RNAseq of ΔFoxp3 Tregs. The same perturbation was found in ΔFoxp3 Tregs from spleen and lung, showing that it extended to parenchymal locations, specifically in a site relevant to scurfy disease (Fig. S9b).
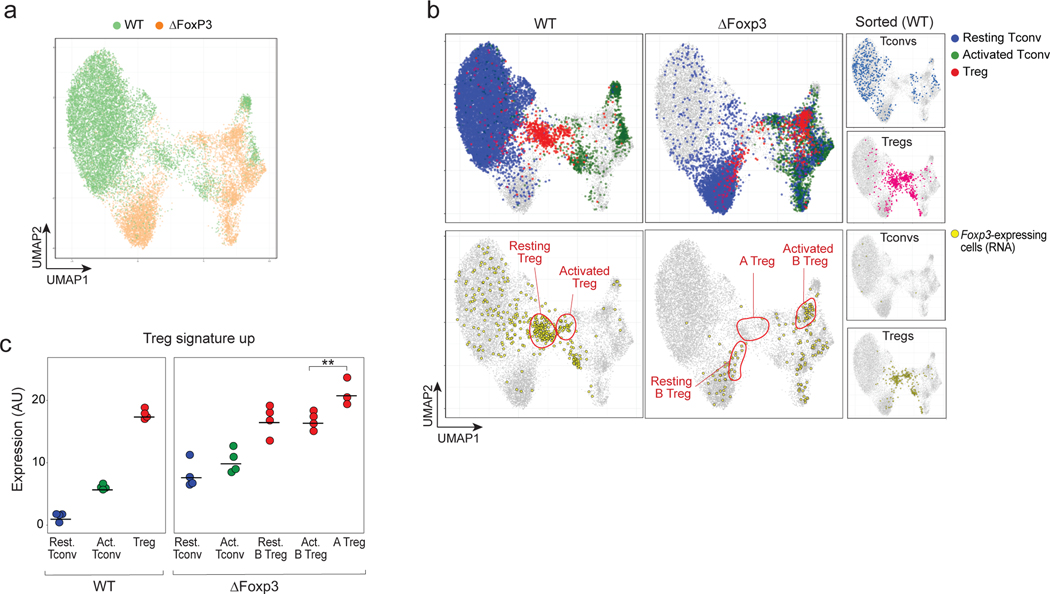
Fig. 5. scRNAseq reveals heterogeneous effects of the Foxp3 ablation in ΔFoxp3 mice.
scRNAseq was performed on CD4+ T cells from WT (n = 4) and ΔFoxp3 (n = 4) mice (18,097 cells altogether).
a. 2D UMAP plot of CD4+ single-cell transcriptomes for WT (green) and ΔFoxp3 (orange)
b. Same UMAP as b. Top row: resting Tconv (blue), activated Tconvs (green), and Tregs (red) (assigned by graph-based clustering on the merged dataset after regressing out the ΔFoxp3 signature, see Methods) are highlighted; bottom: cells with an active Foxp3 locus (GFP or Cre transcripts detected) are colored yellow. The small inserts on the right show sorted control WT Tconvs and WT Tregs, sorted and included as spike-in controls.
c. Treg signature expression in resting Tconvs, activated Tconvs, and Tregs in WT and ΔFoxp3 mice (normalized counts). ** p < 10–2, two-sided t.test.
Unsupervised clustering in the shared CCA space identified, for WT CD4+ cells, resting and activated Tconv and a tight group of Treg cells (blue, green and red, respectively, Fig. 5b top). These assignments were confirmed by the location of sorted Tconv or Treg cells (Fig. 5b, top right), by differential expression of canonical genes (e.g. Ccr7, Sell, Cd44, Tbx21; Fig. S9c, Supplementary Table 5), and at the Foxp3 locus (as Cre and Foxp3 transcripts; Fig. 5b, bottom). Note how Tconv clusters were similarly structured in ΔFoxp3 and WT mice, as they had been in human patients (Fig. S9c).
As for IPEX patients, Treg-like cells of ΔFoxp3 mice were multiform (Fig. 5b): a minor fraction of “A-Tregs” closely resembled normal Tregs, in the same small proportions as in the most complete IPEX deficiencies; a larger proportion of “B-Tregs” that mapped into resting and active areas of the UMAP projection (Fig. 5b), also expressing Foxp3 (Fig. 5b, bottom). Both expressed the Treg signature, highest for Type-A Tregs (Fig. 5c). Together, these Treg-like populations accounted for 7% of total CD4+ T cells, as in WT Tregs (Fig. S9d).
Thus, Treg-like cells in Foxp3-deficient mice showed the same heterogeneity as in human IPEX patients: a mix of healthy and altered Tregs, but with a dominant transcriptional signature that cut across both Treg and Tconv CD4+ T cells.
Cell-intrinsic and extrinsic impact of Foxp3 deficiency.
In humans and mice, FOXP3 deficiency resulted in a heterogenous mix of Treg-like cells, and a strong disease-specific signature unexpectedly shared by Tregs and Tconvs. The latter suggested cell-extrinsic influences on the transcriptomes of CD4+ T cells, which we investigated in two ways.
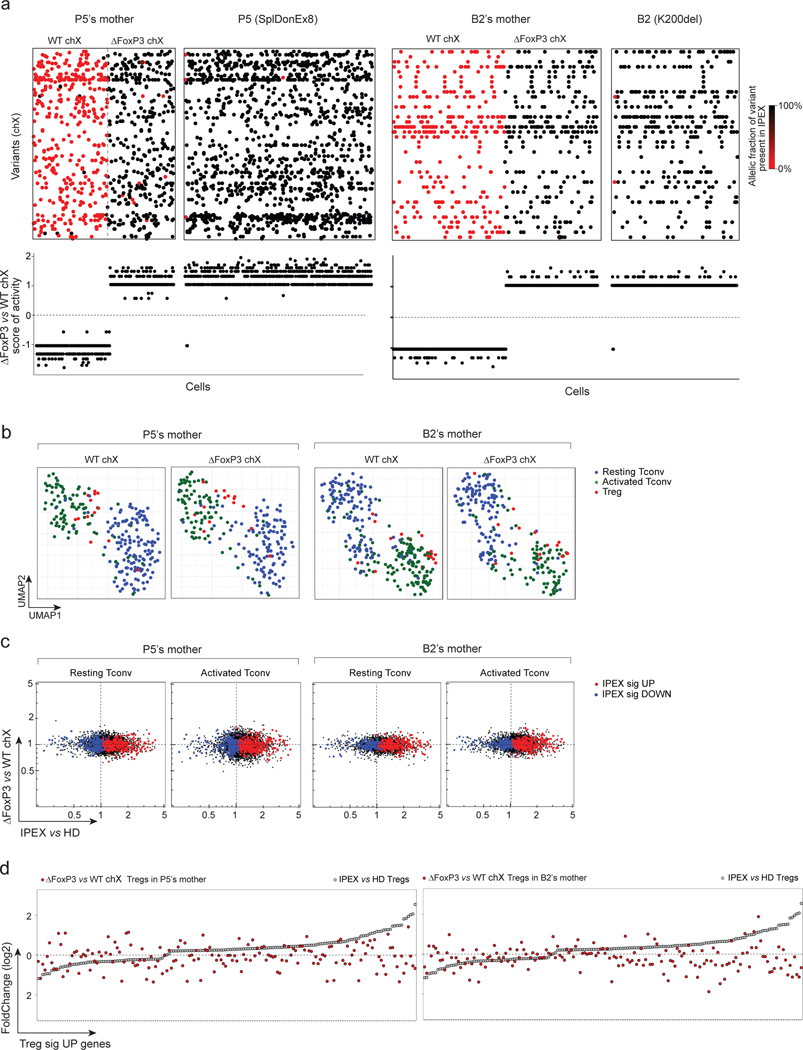
First, we analyzed FOXP3-deficient Tregs present in the same environment as WT cells, in blood CD4+ T cells from two mothers of IPEX patients. X inactivation in females occurs with equal probability, such that half of the T cells in carrier mothers inactivate the mutant X chromosome, and half the WT X chromosome. In the presence of Tregs expressing WT FOXP3, cell-extrinsic signals should be repressed, unmasking the true cell-intrinsic effect of FOXP3 mutations. We performed scRNAseq on blood CD4+ T cells from mothers of patients P5 and B2, analyzing X-chromosome reads that overlapped known SNP variants in order to assign the active X-chromosome of each cell to either the patient’s mutant haplotype or the non-transmitted WT (Fig. 6a, see Methods). Tregs in which the mutant chromosome was active were present in roughly equal proportions to their WT counterparts, as was the case for Tconv. In the UMAP projection, cells with the mutant and WT ChrX active mapped to the same areas, for both Treg and Tconv (Fig. 6b) and all Tregs were type A, irrespective of the active ChrX. The IPEX signature induced in mutant Tconv was fully extinguished (Fig. 6c), with no significant gene expression differences between Tconv with WT or mutant ChrX, highlighting the absence of cell-intrinsic effect of FOXP3 in unchallenged Tconv pools. The Treg UP signature genes that were dysregulated in P5 or B2 Tregs generally normalized in mother’s Tregs expressing the same mutant FoxP3 (Fig. 6d). These results indicate that the signals generating the IPEX signature are cell extrinsic.
Fig. 6. Cell intrinsic and extrinsic effect of FOXP3 deficiency in Tregs and Tconvs of IPEX female carriers (mothers).
scRNAseq was performed on sorted and hashtagged blood CD4+ T cells from two IPEX mothers: P5’s and B2’s mothers (2,880 cells altogether)
a. Because of X inactivation in females a mix of FOXP3-deficient (ΔFOXP3) and -proficient (WT) T cells are present in IPEX mothers (left: P5’s mother, right: B2’s mother). Top: biclustering heatmap showing in each single cell (columns) the allelic fraction of X chromosome variants present in IPEX (P5 and B2) (carrying the FOXP3 mutation). Bottom: FOXP3-deficient vs -proficient X chromosome score of activity for each single cell.
b. 2D UMAP plot of all CD4+ cells from P5’s and B2’s mothers split by active X chromosome (left: FOXP3-proficient, right: FOXP3-deficient). Resting Tconvs, activated Tconvs and Tregs are highlighted in blue, green and red, respectively.
c. scRNAseq data for Tconv cells were collapsed, and expression ratio between FOXP3-deficient and -proficient Tconv (resting or activated) calculated for IPEX (x-axis) and in P5’s and B2’s mothers (y-axis). Up and down IPEX signatures genes in red and blue, respectively.
d. FC plots of FOXP3-deficient vs -proficient Treg in P5’s and B2’s mothers, Treg UP signature genes, ranked according to FC in IPEX vs HD (grey dots).
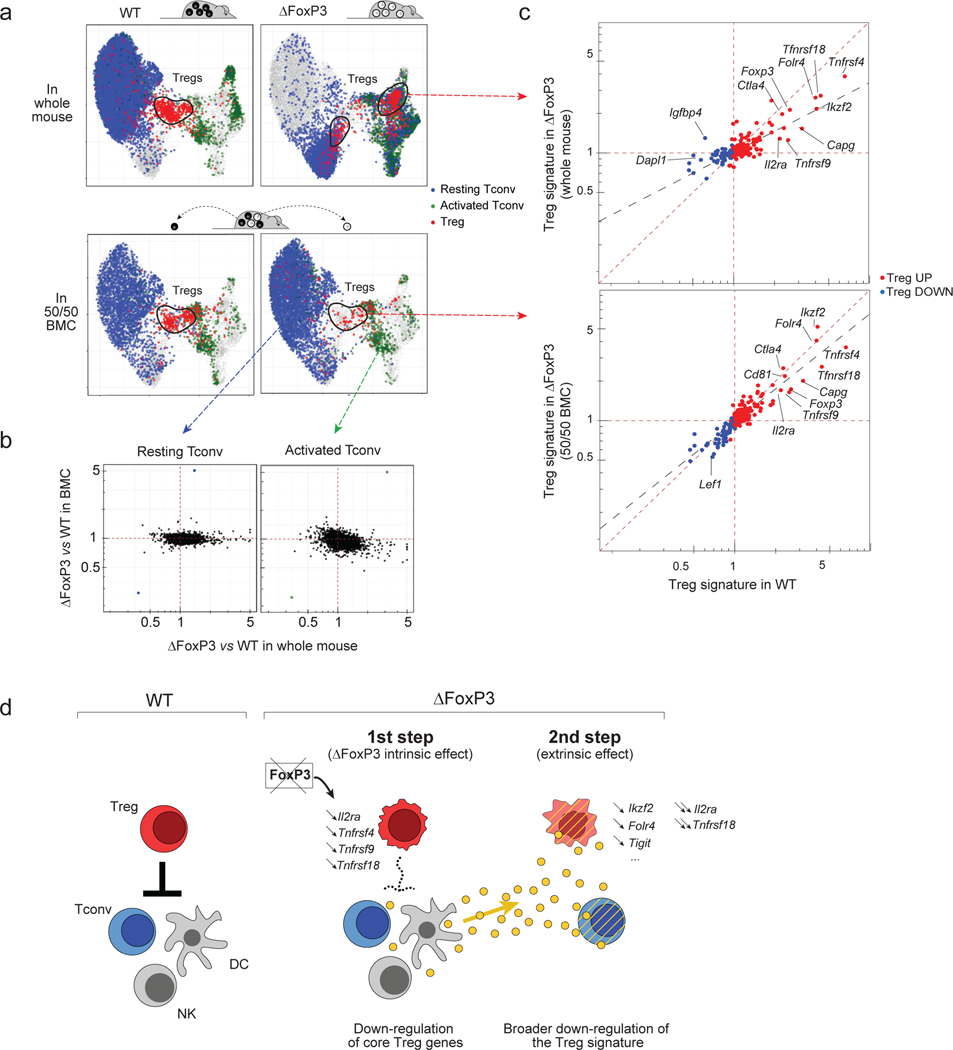
Secondly, we constructed bone marrow chimeras (BMC) by transfer of equal proportions of congenically-marked stem cells from WT and ΔFoxp3 mice, a protocol that prevents the scurfy-like disease that appears after reconstitution with Foxp3-deficient stem cells 40. As in hemizygous human mothers, cell-extrinsic transcriptional hallmarks should be reverted in genetically Foxp3-deficient cells by the presence of WT cells in the same mouse. Ten weeks after reconstitution, 5,556 CD4+ T cells from either ΔFoxp3 or WT compartments of BMC mice were profiled by multiplexed scRNAseq (3 mice/group, data aligned to the same UMAP space as in Fig. 5).
In this mixed setting, many of the characteristics of ΔFoxp3 CD4+ T cells essentially disappeared (Fig. 7a), while WT cells were unchanged. First, on the UMAP plot, resting and activated Tconvs from ΔFoxp3 compartments shifted and overlapped with WT cells from the same mice. Accordingly, the ΔFoxp3 signature was flattened out for mutant cells in the BMC setting (Fig. 7b, Fig. S10a). These results demonstrated the cell extrinsic origin of the ΔFoxp3 signature in Foxp3-deficient Tconv. ΔFoxp3 cells were partially outcompeted by WT Tregs (1:3) in the BMC mice (Fig. S10b), but their phenotypes were also corrected: they were all type-A Tregs that overlapped with WT Tregs on the UMAP (Fig. 7a), and their expression of the Treg signature was almost completely normalized (Fig. 7c, Supplementary Table 5). Only 13 genes remained under-expressed in the ΔFoxp3 Treg compartment, including classic FOXP3 target loci (Il2ra, Tnfrsf4, Tnfrsf9, Tnfrsf18, Capg); no genes were upregulated, confirming that FOXP3 is mainly a transcriptional activator. The other Treg signature genes were otherwise normally expressed, contrasting with their general downregulation in ΔFoxp3 mouse.
Fig. 7. Cell intrinsic and extrinsic effect of Foxp3 deficiency in Tregs and Tconvs in mice.
scRNAseq was performed on sorted and hashtagged CD4+ T cells from 50/50 mixed BMC mice (WT and ΔFoxp3 hematopoietic stem cells, congenically marked) (8,556 cells altogether, n = 3 mice)
a. Same UMAP as 5b. integrating WT and ΔFoxp3 CD4+ cells from whole mice (top) or from chimeras (bottom row). Resting Tconvs (blue), activated Tconvs (green), and Tregs (red) (identified by graph-based clustering on the merged dataset after regressing out the ΔFoxp3 signature, see Methods) are color-coded as in Fig. 5.
b. scRNAseq data for Tconv cells were collapsed, and expression ratio between WT and ΔFoxp3 Tconv calculated for whole mice (x-axis) or for 50/50 BMC (y-axis); for the latter, WT and ΔFoxp3 cells originated from the same host.
c. Treg/Tconv ratio in ΔFoxp3 cells (y-axis) in whole mice (top) and in 50/50 BMC (bottom), both plotted against the same Treg/Tconv expression ratio in WT (x-axis). Treg signature genes are highlighted. Dashed lines (linear regression) represent the slope of the fit.
d. Two-step model of the impact of FOXP3 deficiency in mice and humans. First, the absence of FOXP3 downregulates a few core Treg signature genes in Tregs (cell-intrinsic effect), leading to perturbed control of other immune cells (which might include known Treg targets like Tconv, DCs, NKs, or other), unleashing a secondary inter-cellular response (e.g. cytokines, cell surface modulators) which imprints the dominant IPEX signature on Treg and Tconv, and further affects Treg functionality, amplifying disease pathology.
Complementing FoxP3 deficiency with IL-2
Thus, the dominant suppressive effect of WT Tregs curtailed the ΔFoxp3 signature in Treg and Tconv, revealing a narrower intrinsic effect of Foxp3 on a minor proportion of the Treg signature, highlighting both intrinsic and extrinsic consequences of the Foxp3 deficiency in the disease context. We thus propose a “two-step” model of the IPEX/scurfy disease (Fig. 7d). First, the intrinsic effect of Foxp3 in Tregs dysregulates a few core Treg genes, which unleashes a systemic response. Secondarily, this milieu imprints broad changes on both Treg and Tconv (IPEX/ΔFoxp3 signatures), and further destabilizes Treg signature expression and dampening Treg effector function.
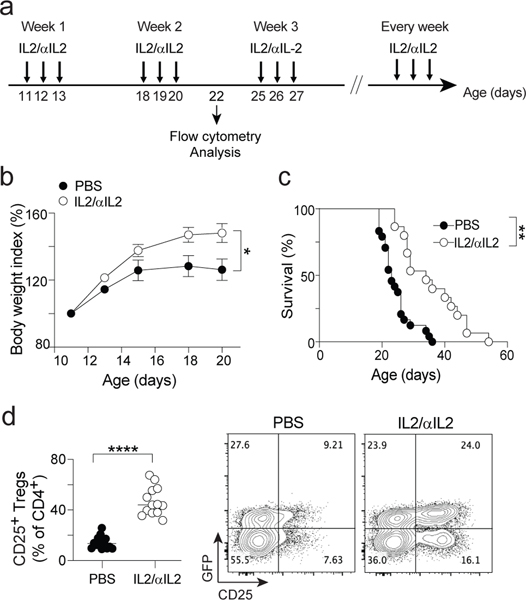
To provide a mechanistic proof-of-concept, we reasoned that enhancing an essential core Treg gene in ΔFoxp3 mice might restore Treg functionality. Il2ra is one of the genes intrinsically regulated by FOXP3 above (see also 8,40,41), and is preferentially expressed in type A Tregs (Fig. 7c). IL2, its ligand, is the key trophic cytokine for Treg, and induces Il2ra expression in a positive feedforward loop 42. IL2 administration might thus complement the deficit due to the missing FoxP3 and, in part, revert the Treg deficiency. Therefore, we evaluated the potential of serial IL2 injections (stabilized in a complex with an anti-IL2 antibody 43) into ΔFoxp3 pups (3 consecutive daily injections per week starting at 11 days of age; Fig. 8a). Compared to the PBS control, IL2-αIL2 complexes partially prevented weight loss (Fig. 8b) and prolonged survival (Fig. 8c) of ΔFoxp3 mice, at least for a period of time. The treatment also expanded the CD25+ fraction within Treg-like cells, thus presumably growing the type A fraction (Fig. 8d). These results show that reverting one of the core deficiencies resulting from the ΔFoxp3 mutation does improve Treg function, at least for some time, and additionally suggest that IL2 therapy might be used to mitigate ΔFoxp3 disease.
Figure 8: IL2 therapy mitigates Foxp3 deficiency disease.
a. IL2 treatment protocol: daily injection of IL2/αIL2 complex for three consecutive days every week starting 11 days after birth.
b. Body weight index growth curves of vehicle (PBS) and IL2 (IL2/αIL2) treated ΔFoxp3 mice (mean +/− SEM). * One-way ANOVA p < 0.05 (b), n = 10 and 19 (PBS and IL2 groups, respectively)
c. Survival curves of vehicle (PBS) and IL2 (IL2/αIL2) treated ΔFoxp3 mice ** Log Rank test p < 10–4, n = 24 and 15 (PBS and IL2 groups, respectively)
d. Proportion of CD25+GFP+ Tregs cells in vehicle (PBS) and IL2 (IL2/αIL2) treated ΔFoxp3 mice (day 22). Right: Representative flow cytometric CD25/GFP plots of CD4+ T cells. **** two-sided t.test p < 10–4, n = 14 and 13 (PBS and IL2 groups, respectively)
DISCUSSION
These results provide unprecedented cellular and genomic resolution of a primary immunodeficiency, and reveal unexpectedly multifarious molecular and cellular consequences of FOXP3 deficiency. There were surprisingly limited cell-intrinsic perturbations in Treg cells, associated with a dominant and monomorphic signature of cell-extrinsic origin, that cuts across the transcriptome of all CD4+ T cells and amplifies Treg perturbations.
We thus propose a two-step model of the disease’s molecular pathogenesis. FOXP3 is actually important for only very few Treg genes: when the milieu was normalized by the presence of WT Tregs the cell-intrinsic impact of the ΔFoxp3 deficiency was exerted on only a handful of genes (Il2ra, Tnfrsf4, Tnfrsf9, Tnfrsf18, Capg, Ikzf2, Ctla4). This surprisingly short list corresponds to a “core set” of genes that are expressed by all Tregs 6, are directly transactivated by and bind FOXP3 8,44. These genes encode the major homeostatic regulator of Treg cells (IL2RA), and several members of the TNFR superfamily, which are also connected to Treg homeostasis and function 45. We propose that their downregulation is the first step (accordingly, IL2RA deficiency also causes an IPEX-like disease). This initiates a systemic reaction which constitutes the second step of the FOXP3 deficiency syndrome: a broadly shared signature that marks both Treg and Tconv cells, further perturbs Treg signature transcripts, and amplifies in a vicious circle the defect in Treg suppressive activity.
What, then, is this IPEX signature, the hundreds of genes that were impacted equivalently in Treg and Tconv cells? It was shared among all IPEX patients, with different intensities rather than qualitative differences, stressing its common etiology. It proved stable over several years in each multiply-tested patient, before and after (and unrelated to) treatment. The genes involved were not simply T cell activation genes, as might have been expected from a loss of suppression (activation-induced transcripts were actually repressed). We hypothesize that it is due to an inductive element(s), delivered via cytokine or cell-cell contact, normally repressed by Tregs but unleashed by the FOXP3 deficiency. This signal might originate from other T cells, or from other immunocytes, much as dendritic and NK cells are the first responders to acute Treg ablation in DTR systems 46,47. The presence of WT Tregs in mixed BMC mice or in heterozygous females would restore the negative feedback, thus reinstating better Treg function, evoking the “infectious tolerance” concept 48, the presence of normal suppressors improving the tone and function of defective ones. Pathway and ontology analysis revealed no clear match, except for effects on a few IFN- and TNF-induced genes. There was, on the other hand, an intriguing anti-correlation for genes over-expressed in tumor-infiltrating Tregs 49), CD4+ T cells from IPEX patients turned off much of the tumor Treg signature.
This principal effect cutting across both Treg and Tconv evokes the debated hypothesis that FOXP3 deficiency has an intrinsic effect in Tconv cells, associated with the transient induction of FOXP3 in activated Tconv 29–32. The mixed chimeras and the mother-son pairs demonstrated that the FOXP3 deficiency affected Tconv cell-extrinsically. However, it remains an open question whether this disease signature contributes to pathology via further dampening of Treg function, or through Tconv defects.
That FOXP3 is not absolutely required for Treg differentiation and homeostasis is now well established, with the existence of Treg-like cells described repeatedly in mice and humans lacking FOXP3 23–28. These “Treg wannabes” were reported to maintain some Treg features (self-reactivity, partial Treg signature, activity at the FOXP3 locus) while acquiring some Tconv characteristics (no in vitro anergy, cytokine expression). The unexpected insight emerging from our single-cell analysis of deficient humans and mice was the wide array of cells with Treg-like characteristics and/or an active FOXP3 locus. Rather than a single population of Treg wannabes, several distinct populations were present: first, a large component of B-Tregs, with many Treg transcriptional characteristics, but perturbed by the IPEX signature. Second, and most mysterious, the subset of A-Tregs that seemed almost unaffected relative to WT Tregs: full Treg signature, active FOXP3, and no IPEX signature, as if these cells had somehow become non-responsive to the systemic influence, generating escape variants, e.g. by dampening receptors or signaling, or because they reside in protected niches. But, then, why only for Tregs?
These results show that it can be misleading to infer the transcriptional footprint of a TF, and its mechanistic causality in disease, solely from the transcriptome of deficient cells, since it can be perturbed as here by dominant cell-extrinsic effects. When the extrinsic effects were blocked in the chimeras, the true core signature of Foxp3 proved much smaller than the first analysis of the deficiency had suggested. IPEX is a rare disease, and our cohorts were not powered to robustly detect clinical correlates of the patients’ cellular and transcriptional characteristics. On the other hand, there was no obvious correlation between clinical severity indicators and the integrated gene expression metrics. This finding is congruent with the notion that molecular severity of the FOXP3 mutations in IPEX patients only loosely correlates with clinical severity 19,50 - here, the null mutation in patient P1 did not determine the most severe disease.
There are some implications of our findings for therapeutic strategies in IPEX. Current management, when bone marrow transplantation is not an option, involves immunologic dampening via immunosuppressants. Our results might suggest harnessing those type A Tregs that are present in the patients by sustaining their homeostasis, or complementing the small set of primary FOXP3 targets identified here. Indeed, the effect of the IL2/αIL2 treatment validates this proposition (it would be interesting to see if combination of IL2 with other activators of the core gene set, e.g. TNF family members, would further improve efficacy). Alternatively, damage might be avoided in patients by blocking the signaling mechanism that imparts the dominant IPEX signature to all T cells.
In conclusion, the new landscape of Treg and Tconv cells revealed in IPEX patients by single-cell analysis and their correction in mixed cell contexts, have opened a new perspective on the disease, and on the role of FOXP3 and Tregs in immune homeostasis.
ONLINE METHODS
Mice
Foxp3IRES-GFP/B6 51, Foxp3ΔEGFPiCrexR26YFP (ΔFoxp3) and Foxp3ΔIRES-GFP/B6 (ΔFoxp3) mice on the C57Bl/6J background were maintained in our colony. Except when specified, 3-week-old male mice were used in this study (Supplementary Table 4). Mice were housed under specific pathogen-free conditions and all experimentation was performed following the animal protocol guidelines of Harvard Medical School and Boston Children’s Hospital (HMS IACUC protocol 02954).
Human cohorts
Male IPEX and healthy donor whole-blood samples were obtained under protocols reviewed and approved by the local Institutional Review Boards at each center (Boston Children’s Hospital 04–09-113R, Necker/Imagine C 15–13_CODECOH_AR, HMS IRB15–0504). Anonymized clinical data included age at onset and at blood sampling, clinical symptoms (enteropathy, diabetes, eczema, other autoimmune diseases and allergy), ancillary testing, treatments and long-term outcomes (Supplementary Table 1). Two independent cohorts of samples were profiled (Supplementary Table 1). Each cohort contains samples from both centers. Cohort 1 samples were processed in two different scRNAseq runs, cohort 2 (replication cohort) in three scRNAseq runs. Samples from each cohort were sorted and sequenced as one batch for population RNA sequencing (Supplementary Table 4).
Peripheral blood mononuclear cells (PBMC) isolation
Whole blood was collected in K2 EDTA tubes and processed within a few hours. An equal volume of room-temperature PBS/2mM EDTA was mixed into 15ml of blood and carefully layered over 14ml Ficoll Hypaque solution (GE Healthcare). After centrifugation for 30 min at 900 g (with no break), at room temperature, the mononuclear cell layer was washed three times with excess HBSS (Gibco) (10min at 400 g) and resuspended in 2ml of HBSS. The pellet was resuspended in 90% FBS-10% DMSO, 20 million cells/mL, 1mL per vial, cooled progressively in isopropyl alcohol (Mr. Frosty™ Freezing Container, Thermo Fisher) for 24h and stored in liquid nitrogen for long term.;
Vials were thawed in 10 mL 10% FBS RPMI, cells washed (500g for 5 min) and resuspended in FACS buffer (phenol red–free RPMI, 2% FBS, 0.1% azide and 10mM HEPES, pH 7.9). After cell counting, the samples were allocated for flow cytometry, population RNA sequencing and scRNAseq.
Flow cytometric profiling of IPEX and HD
Cells were stained for flow cytometry in 100 μL of FACS buffer (phenol red–free DMEM, 2% FBS, 0.1% azide and 10mM HEPES, pH 7.9) for 10 min with 10uL FcBlock (supernatant of clone 2.4G2, ATCC HB-197, hybridoma cultures) and the following antibodies: CD3 BV605 or A700 (2μL, OKT3, Biolegend), CD4 PerCP-Cy5.5 (2 μL, OKT4, Biolegend), CD25 PE-Cy7 (3 μL, BC96, Biolegend), CD127 A488 (3 μL, A019D5, Biolegend), CD45RA PB (3μL, HI100, Biolegend). After permeabilization/fixation for 2 h on ice, FOXP3 (APC, clone PCH101; Biolegend; 2 μL) and anti-HELIOS PE (PE, clone 22F6, Biolegend, 2 μL) staining was performed overnight at 4°C in the dark in accordance with the manufacturer’s instructions (eBioscience™ FOXP3 / Transcription Factor Staining Buffer Set). Data was recorded on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo v10. FOXP3 MFI was normalized to the mean of healthy donors. To generate tSNE plots from the flow cytometry data, compensated, scaled data in the Lymphocyte/Singlet/CD3+/CD4+ gate were exported in .csv format from FlowJo. In R, the matrix containing CD3, CD4, FOXP3, HELIOS, CD25, CD127, CD45RA expression was centered and scaled (by marker) before performing PCA using the prcomp function, and tSNE projections calculated with Rtsne(dims = 2, perplexity = 50, check_duplicates = F, pca = F, max_iter = 500).
Low-input RNA sequencing of human and mouse samples.
RNA-seq was performed with the standard ImmGen low-input protocol (www.immgen.org). Human Tregs and Tconvs were sorted as DAPI– CD4+CD25hiCD127lo and CD4+CD25–CD127hi, respectively on a Moflo Astrios Cell Sorter (Beckman Coulter). Mouse Tregs and Tconvs were sorted as DAPI–TCRβ+CD4+G/YFP+CD25+ and DAPI–TCR+CD4+G/YFP–, respectively. A total of 1,000 cells were double-sorted directly into 5ul of lysis buffer (TCL Buffer (QIAGEN) supplemented with 1% 2-Mercaptoethanol). Smart-seq2 libraries were prepared as previously described 52 with slight modifications. Briefly, total RNA was captured and purified on RNAClean XP beads (Beckman Coulter). Polyadenylated mRNA was then selected using an anchored oligo(dT) primer (50 –AAGCAGTGGTATCAACGCAGAGTACT30VN-30) and converted to cDNA via reverse transcription. First strand cDNA was subjected to limited PCR amplification followed by Tn5 transposon-based fragmentation using the Nextera XT DNA Library Preparation Kit (Illumina). Samples were then PCR amplified for 12 cycles using barcoded primers such that each sample carries a specific combination of eight base Illumina P5 and P7 barcodes for subsequent pooling and sequencing. Paired-end sequencing was performed on an Illumina NextSeq 500 using 2 × 38bp reads with no further trimming.
Reads were aligned to the human genome (GENCODE GRCh38 primary assembly and gene annotations v27) with STAR 2.5.4a (https://github.com/alexdobin/STAR/releases). The ribosomal RNA gene annotations were removed from GTF (General Transfer Format) file. The gene-level quantification was calculated by featureCounts (http://subread.sourceforge.net/). Raw read counts tables were normalized by median of ratios method with DESeq2 package from Bioconductor (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) and then converted to GCT and CLS format. Samples with less than 1 million uniquely mapped reads were excluded from normalization to mitigate the effect of poor quality samples on normalized counts. Genes with a minimum read count of 5 in all replicates of a population (31,448 human genes) were retained. A pseudo count of 1 was added and log2-transformed prior to quantile normalization. Quantile-normalized counts were converted back to a linear scale.
IPEX signature genes were identified by computing the ratio of expression in Tregs or Tconv of all IPEX patients vs all healthy donors (p.value as simple uncorrected t.test). IPEX-up indices for each individual were calculated by selecting the 100 transcripts most over-expressed in IPEX Tregs overall (FC>3), calculating their differential expression in Treg cells of each patient relative to the mean of all HD Tregs, and averaging the log2 of these foldchanges (and similarly for IPEX-down indices from transcripts with IPEX/HD <0.37). The TregUp indices were similarly calculated (expression in Tregs of each individual over mean expression in all healthy donors, average the log2 of these foldchanges).
Geneset enrichment analysis.
Gene signatures were curated from published datasets (references in the signature name) 6. The Human and Mouse Treg signatures have been reported 4,5. Data were downloaded from the Gene Expression Omnnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). Only datasets containing replicates were used. To reduce noise, genes with a coefficient of variation between biological replicates <0.6 in either comparison groups were selected. Up- and downregulated transcripts were defined as having a fold change in gene expression >1.5 or <0.6 and a t-test p-value <0.05. Other signatures were obtained from MSigDB C7 Immunologic signatures collection 53. Geneset enrichment analysis with the IPEX signature was performed using the hypergeometric distribution and type I error was controlled using FDR. Signatures with p-value <0.001 (all with FDR <6%) are reported.
scRNAseq analysis of human PBMC samples
scRNAseq was performed in several batches (different experiment dates in Supplementary Table 1 and Fig. S4): two for cohort 1 and three for cohort 2. Cohort 1 samples were profiled with the 10X Genomics Single Cell 3′ Reagent Kit (V2 chemistry), Cohort 2 samples with the 10X Genomics Single Cell 3′ Reagent Kit (V3 chemistry) and sample barcoding with DNA-tagged antibodies (“hashtagging”) 36. See also Supplementary Table 4.
Cell sorting and pooling using cell hashtagging.
Cells were stained in 100 μL of FACS buffer (phenol red–free RPMI, 2% FBS, 0.1% azide and 10mM HEPES, pH 7.9) for 10 min with 10uL FcBlock (homemade) and the following antibodies: CD3 A700 (2μL, OKT3, Biolegend), CD4 PerCP-Cy5.5 (2 μL, OKT4, Biolegend), CD25 PE-Cy7 (3 μL, BC96, Biolegend), CD127 A488 (3 μL, A019D5, Biolegend), 5 uL (2.5ug) of a unique hashtag antibody (TotalSeq™-A0251 to A0258 anti-human Hashtag 1 to 8 Antibody). 8,000 DAPI–CD3+CD4+ T cells were single sorted using the Moflo Astrios Cell Sorter (70 um nozzle, Beckman Coulter) in 30 ul of PBS-BSA 0.1%. Samples with different hashtag antibodies were sorted in the same tube and the total volume was adjusted to 30 uL.
scRNAseq libraries (10x Genomics).
Cells were encapsulated in one channel per sample (cohort 1 runs) or in 1 channel per pool (cohort 2 samples) of a 10x Chromium instrument, and libraries were constructed with the Single Cell 3′ Reagent Kit (V2 for cohort 1 and V3 chemistry for cohort 2) (https://support.10xgenomics.com/single-cell-gene-expression/libraryprep/). Libraries were sequenced on the NextSeq 500 platform (28/8/0/91, Read1/i7/i5/Read2). Gene counts were obtained by aligning reads to the hg38 transcriptome using CellRanger software (v3.0.2) (10X Genomics) (default parameters).
Hashtag libraries.
Hashtag libraries were made separately as described in Stoeckius et al. 36 (https://citeseq.files.wordpress.com/2019/02/cell_hashing_protocol_190213.pdf). In brief, at the cDNA amplification step in the Single Cell 3′ Reagent Kit protocol, the yield of HTO products was increased using an “additive” primer to cDNA PCR. Hashtag-derived cDNAs (<180bp) and mRNA-derived cDNAs (>300bp) were then separated using 0.6x SPRI bead selection. The supernatant contains the Hashtag-derived cDNA that is purified with two rounds of 2x SPRI beads. The sequencing oligos are added by PCR which also amplifies the Hashtag library. Libraries were sequenced on the NextSeq 500 platform (28/8/0/91, Read1/i7/i5/Read2).
Hashtag count matrices were obtained from CITE-Seq-Count package (https://zenodo.org/record/2590196) (default parameters). Each droplet from the Gene Count matrix was matched to a Hashtag using the HTODemux function from the Seurat v3.1.2 package. Doublets (droplets with 2 hashtags) were excluded and cells were assigned to the max hashtag signal. The Hashtag count data was also analyzed by tSNE for a visual check (clear separated clusters for each hashtag). All single cells from the gene count matrix were matched unambiguously to a single hashtag (and therefore their original donor).
Quality control.
Cells with less than 500 (V2) or 1000 (V3) counts (V2) were excluded. Dead cells with more than 10% of counts mapping to mitochondrial genes were excluded. Doublets were excluded using scrublet (doublet score > 0.3) 54. Finally, cells were annotated using singleR 55 with the BluePrintEncode reference data. Cells that were not annotated as CD4+ cells were excluded.
Batch correction.
Batch correction was performed using the Integration method in Seurat V3 as described in Stuart et al. 37 and only used to visualize the whole dataset with UMAP. Cohort 1 and cohort 2 samples were first normalized independently using the SCTransform function in Seurat V3 with parameters to regress out the following variables: experiment date, percent of mitochondrial gene mapping and the number of counts of each single cell (SCTransform(vars.to.regress = c(“experiment_date”,”percent_mito”, “nCount_RNA”), variable.features.n = 500). Integration of both cohorts together was then performed using the top 500 variable genes in both cohorts. Each cohort was projected into a common CCA (Canonical Correlation Analysis) space. Anchors (robust pairwise cell correspondence between datasets) were found by knn and snn graphs (FindIntegrationAnchors function) and they were used to transform the data in an integrated space. We used this integrated space, reduced to the top 40 PCs by PCA to visualize with UMAP 56. The number of significant principal components (PCs) was determined by comparison to PCA over a randomized matrix, as described by Klein et al. 57. Of note, another batch correction method, scAlign 58 produced similar results.
Clustering.
We found shared clusters across HD and IPEX samples (“regressing out the IPEX effect”) using the Integration method in Seurat V3 as described in Stuart et al. 37. This time, each sample was first normalized independently using the SCTransform function in Seurat V3 with parameters to regress out the following variables: percent of mitochondrial gene mapping and the number of counts of each single cell (SCTransform(vars.to.regress = c(“percent_mito”, “nCount_RNA”), variable.features.n = 500). Integration of samples together was then performed using the top 500 variable genes. Each cohort was projected into a common reciprocal PCA space. Anchors (robust pairwise cell correspondence between samples) were found by knn and snn graphs (FindIntegrationAnchors) and they were used to transform the data in a shared space. We used this shared space and reduced it to the top 32 most significant PCs by PCA for clustering 56. In this space, a shared nearest neighbor graph was constructed from a k-nearest-neighbor graph (k = 20) by pruning cell-cell edges with less than 1/15 neighbor overlap. Community detection using the Louvain algorithm at a resolution of 0.5 found 11 clusters. Automated annotation using singleR with the BluePrintEncode reference data and manual annotation using canonical markers (see Figs. 3, 4, Fig. S7) clearly distinguished resting Tconvs, activated Tconvs, and Tregs. Type A IPEX Tregs were defined as Tregs in IPEX samples that overlapped with the HD Tregs (top left quadrant in UMAP). Type B IPEX Tregs are all the others.
Differential gene expression taking into account technical and biological variables.
We used limma-trend 59, as benchmarked in 60. Briefly, default TMM normalization from edgeR was applied to the SCTransform corrected count matrix. A linear model was fitted to the data using a contrast matrix including confounding variables (lmFit function). In order to find cluster-defining markers shared across HD and IPEX samples (i.e. regressing out the IPEX effect), or markers between type A and B IPEX Tregs, the contrast matrix contained the sample origin as a confounding variable. Empirical Bayes method was then used to estimate the overall trend of gene expression variance and adjust the genewise-wise residual variances towards this global trend (less variance for genes trending high and more for those trending low) (eBayes(trend = TRUE, robust = TRUE)). The TopTable function was then used to extract the differential statistics corresponding to the contrast of interest. Benjamin-Hochberg correction was applied to control type I error. Adjusted p-value < 0.05 were deemed significant.
Analysis of active and inactive X chromosomes carrying FOXP3 mutation in IPEX mothers cells
scRNAseq was performed as described in the previous paragraph of peripheral blood CD4+ cells from two mothers of IPEX patients: P5 and B2’ mother. (see also Supplementary Table 4). 919 cells from P5’s mother and 1,961 cells from B2’s mother were analyzed after QC. Because X inactivation in females occurs randomly during embryogenesis, a mix of FOXP3-deficient (ΔFOXP3) and -proficient (WT) T cells are present in IPEX mothers (female carriers). We sought to identify them using X chromosome variants in IPEX patients (P5 and B2) and their mother. For each sample, we started from the cell ranger bam file (possorted_genome_bam.bam), converted it back to fastq (bamtofastq-1.2.0 --nthreads 32 possorted_genome_bam.bam fastq) and aligned the reads using STAR v020201 ($STAR_PATH/STAR --genomeDir GRCh38_fasta --readFilesIn $READ2 --runThreadN 32 --outSAMtype BAM SortedByCoordinate) to the GRCh38 human genome in order to be able to run bcftools. Variants on the X chromosome were called using bcftools v1.10.2 (bcftools mpileup -Q 30 -A -x -Ou --threads 32 -r X -f $FASTA_REF Aligned.sortedByCoord.out.bam | bcftools call -mv -Ov -o calls.vcf). We then used scAlleleCount (https://github.com/barkasn/scAlleleCount) to obtain for each single cell the reference and alternate allele count for the X chromosome variants (cell x variant matrices). We then constructed the IPEX (ΔFoxp3) X chromosome haplotype (present in sons and carrying the FOXP3 mutation) and the complementary haplotype (present in the mother and carrying the “WT” FOXP3). To do so, we only focused on high confidence variants that were present both in the mother and in the son with a quality >10. We then filtered the uninformative variants (variants with 100% allelic fraction in the mothers, present in both X chromosome). Using the son’s as a reference, we constructed the IPEX (ΔFoxp3) X chromosome haplotype and the complementary WT haplotype. For each single cell the distances to the ΔFoxp3 and the WT haplotypes were computed and we used the ratio as a score to identify whether the active X chromosome harbored WT or mutant FOXP3 (see Fig. 6). There was no evidence of X chromosome recombination in mothers. In IPEX, 3/815 were assigned to an active WT chromosome in P5 and 2/1612 in B2 (misclassification or microchimerism). 469 P5’s mother cells and 513 B2’s mother cells were confidently assigned to a ΔFoxp3 or WT active X chromosome (see Fig. 6).
Bone marrow chimeras
C57BL/6J mice were irradiated with 10 Gy and reconstituted with equal proportions of congenically labeled T cell-depleted bone marrow cells from WT (Foxp3ires–gfpxCD45.1) and ΔFoxp3 (Foxp3ΔEGFPiCrexR26–YFPxCD45.1/CD45.2) mice. Bone marrow cells were harvested from femurs, tibias and hip bones from two WT and two ΔFoxp3 male mice. After red blood cell lysis with ACK for 1 min at 4°C, T cells were depleted: bone marrow single-cell suspensions were incubated with 20 uL of biotinylated anti-CD3e antibodies (OKT3, Biolegend) for 10 min in 2 mL of MACS buffer (PBS, FBS 0.5%, EDTA 2mM), washed, and then incubated with 200 μL of streptavidin beads (Dynabeads Biotin Binder, 11047; Thermo Fisher Scientific) for 20 min in 5 mL of MACS buffer. Isolation of the CD3-negative population was performed after three magnet incubations for 2 min. A total of 4 million cells (2 million WT and 2 million ΔFoxp3) were injected i.v. in each mouse. Mice were treated for 2 weeks with trimethoprim-sulfamethoxazol and analyzed 10 weeks later.
scRNAseq profiling of CD4+ T splenocytes cells in mice and bone marrow chimeras
scRNAseq profiling of mouse samples was performed in two experiments (see also Supplementary Table 4). In the first experiment, 2 WT mice and 2 ΔFoxp3 mice were profiled with the 10X Genomics Single Cell 3′ Reagent Kit (V2 chemistry), one channel per mouse. In the second experiment, CD4+ T cells from 3 bone marrow chimeras, 2 WT mice and 2 ΔFoxp3 mice were pooled with sorted WT Tregs and Tconvs as controls and profiled with the 10X Genomics Single Cell 3′ Reagent Kit (V3 chemistry). Both experiments were analyzed together, after batch correction.
scRNAseq libraries.
Spleens were harvested. After red blood cell lysis with ACK for 1 min at 4°C, 30% of splenocytes (~30 million cells) were stained for sorting by flow cytometry in 200 uL of FACS buffer (PBS, 2% BSA) for 20 min at 4°C in the dark, with the following antibodies (1/100 dilution): 100 uL of FcBlock (homemade), TCRβ PE-Cy7 (H57–597; BioLegend), CD4 PerCPCy5.5 (GK1.5; BioLegend), DAPI, CD45.1 PE-Cy7 (clone A20, Biolegend), CD45.2 A700 (clone 104, Biolegend). Whole ΔFoxp3 and WT mouse CD4+ T cells were sorted as DAPI–TCRβ+CD4+. For BMC mice, ΔFoxp3 CD4+ T cells were sorted as DAPI–TCRβ+CD4+CD45.1+CD45.2+. WT CD4+ T cells were sorted as DAPI–TCRβ+CD4+CD45.1+CD45.2–. 40–50,000 cells were single sorted in 50 uL of PBS-BSA 0.05% and the total volume was adjusted to a concentration of 1,000 cells/uL after cell counting with a hemocytometer. In the first experiment, cells from each tube were encapsulated in one channel of a 10x Chromium instrument, and libraries were constructed with a Single Cell 3′ Reagent Kit (V2 chemistry) (https://support.10xgenomics.com/single-cell-gene-expression/libraryprep/). In the second experiment, in order to pool samples for cell hashtagging, cells were sorted in 50 uL of PBS-BSA 0.05% in two different tubes (5,000 to 10,000 sorted cells for each sample) for sequencing in two 10X lanes, and libraries were constructed with a Single Cell 3′ Reagent Kit (V3 chemistry) (see also Supplementary Table 4).
Libraries were sequenced on the NextSeq 500 platform (28/8/0/91, Read1/i7/i5/Read2). Gene counts were obtained by aligning reads to the mm10 transcriptome using CellRanger software (v3.0.2) (10X Genomics) (default parameters). The mm10 transcriptome was complemented with the transgene sequences of the Ires-Gfp, Yfp, Cre in order to map reads to the Foxp3 locus in Foxp3ires–gfp/B6 and Foxp3ΔGFPiCrexR26–YFP /B6 mice. In Foxp3ires–gfp/B6 mice, Foxp3 locus expression was calculated as the sum of reads mapping to Foxp3 and Ires-Gfp. In Foxp3ΔGFPiCrexR26–YFP /B6 mice, Foxp3 locus expression was calculated as the sum of reads of mapping to Foxp3 and Cre (not GFP because the Read 2 length is not long enough to reach the GFP sequence).
Hashtag libraries.
See similar section in the human analysis (scRNAseq analysis of human PBMC samples)
Quality control.
See similar section in the human analysis (scRNAseq analysis of human PBMC samples). SingleR was used with the Immgen reference data.
Batch correction.
Experiment 1 and 2 were profiled with 10X V2 and V3, respectively. Batch correction was performed using the Integration method in Seurat V3 as described in Stuart et al. 37 and only used to visualize the whole mouse dataset with UMAP. Experiment 1 (10x V2) and experiment 2 (10X V3) samples were first normalized independently using the SCTransform function in Seurat V3 with parameters to regress out the following variables: percent of mitochondrial gene mapping and the number of counts of each single cell (SCTransform(vars.to.regress = c(“percent_mito”, “nCount_RNA”), variable.features.n = 500). Integration of both experiments together was then performed using the top 500 variable genes in both cohorts. Each cohort was projected into a common CCA space. Anchors were found by knn and snn graphs (FindIntegrationAnchors) and they were used to transform the data in an integrated space. We used this integrated space, reduced to the top 50 PCs by PCA to visualize with UMAP. The number of significant principal components (PCs) was determined by comparison to PCA over a randomized matrix, as described by Klein et al. 57.
Clustering.
We found shared clusters across samples using the Integration method in Seurat V3 as described in Stuart et al. 37. This time, each sample was first normalized independently using the SCTransform function in Seurat V3 with parameters to regress out the following variables: percent of mitochondrial gene mapping and the number of counts of each single cell (SCTransform(vars.to.regress = c(“percent_mito”, “nCount_RNA”), variable.features.n = 500). Integration of samples together was then performed using the top 500 variable genes. Each cohort was projected into a common CCA space. Anchors were found by knn and snn graphs (FindIntegrationAnchors) and they were used to transform the data in a shared space. We used this shared space and reduced it to the top 28 most significant PCs by PCA for clustering. In this space, a shared nearest neighbor graph was constructed from a k-nearest-neighbor graph (k = 20) by pruning cell-cell edges with less than 1/15 neighbor overlap. Community detection using the Louvain algorithm at a resolution of 1.5 found 11 clusters. Automated annotation using singleR with the Immgen reference data and manual annotation using canonical markers (see Fig. S9) clearly distinguished resting Tconvs, activated Tconvs, resting Tregs and activated Tregs. Type A IPEX Tregs were defined as Tregs in IPEX samples that overlapped with the HD Tregs. Type B IPEX Tregs are all the others.
Differential gene expression.
See similar section in the human analysis (scRNAseq analysis of human PBMC samples).
Flow cytometry analysis of mouse cells
A single cell suspension was obtained from murine splenocytes after mechanical dissociation through a 40 μm strainer using a syringe plunger. Red blood cell lysis was performed using 1 mL of ACK (Lonza) for 1 min on ice. After careful perfusion with 5 mL of sterile PBS, the right lung was harvested, minced, and enzymatically dissociated in RPMI containing 0.5 mg/mL DNASE I (Sigma), 1.5 mg/mL collagenase IV (Sigma), 10% FBS and 2 mM EDTA with constantly stirring at 37°C for 35 min. Single cell suspensions were then filtered through a 70-μm strainer and washed twice with 10% FBS + 2mM EDTA RPMI buffer. Lung CD4+ T cells were enriched before staining and sorting, using negative magnetic selection (Stem Cell).
After Fc blocking, antibody staining was done in ice-cold buffer (DMEM without phenol red-2% FBS, 1mM EDTA) for 15 min at a dilution of 1/100 using with antibodies against CD45 (30-F11; BioLegend), CD4 (GK1.5; BioLegend), CD25 (PC61; BioLegend), TCRβ (H57–597; BioLegend), CD19 (SA011F11; BioLegend) and DAPI. The stained samples were then analyzed by flow cytometry on a BD Fortessa or Symphony analyzers, and processed using FlowJo Version 10 (Tree Star).
IL2 treatment
IL2/anti-IL2 mAb complex was formed by incubating 1μg of recombinant mouse IL2 (biolegend) and 9 μg of purified anti-mouse IL2 mAb (clone JES6–1A12) (Bio-X-cell) for 20 min at 37°C in sterile PBS. This complex was administered for 3 consecutive days per week starting at day 11 after birth on the basis of 40 ng of IL2 per gram weight of treated mice.
Statistics
Analysis was done using R-3.6.2. Heatmaps were made with Morpheus (https://software.broadinstitute.org/morpheus) or the pheatmap package in R (https://cran.r-project.org/web/packages/pheatmap/index.html). All other plots are made with ggplot2 61. Statistical tests are described in their respective method section.
Data availability
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE166866 (human population RNAseq), GSE166860 (mouse population RNAseq), GSE167976 (human scRNA-seq) and GSE167575 (mouse scRNA-seq).
Supplementary Material
ACKNOWLEDGEMENTS
We thank: Drs. M. Levings and A. Rudensky for insightful discussions; K. Hattori, C. Araneo, K. Seddu, and the Klarman Cell Observatory team, for help with mice, cell sorting and single-cell profiling. This work was funded by grants from the NIH to CB&DM (AI116834, AI125603, AI150686), TAC (AI085090), LMC (AI153174), the Institut National de la Santé et Recherche Médicale, the European Union Seventh Framework (#269037 and #261387) and Horizon 2020 (#693762), the Agence Nationale pour la Recherche (Investissement d’Avenir ANR-10-IAHU-01) to IA, ES, MD, JL, MC, BN, FRL, FR and NCB. JL was supported by an INSERM Poste d’Accueil and an Arthur Sachs scholarship.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
REFERENCES
- 1.Josefowicz SZ, Lu LF, & Rudensky AY Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wing JB, Tanaka A, & Sakaguchi S. Human FOXP3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 50, 302–316 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Panduro M, Benoist C, & Mathis D. Tissue Tregs. Annu. Rev Immunol 34, 609–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill JA et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 27, 786–800 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Ferraro A et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci U S A 111, E1111–E1120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemmour D et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol 19, 291–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono M. Control of regulatory T-cell differentiation and function by T-cell receptor signalling and Foxp3 transcription factor complexes. Immunology 160, 24–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon HK, Chen HM, Mathis D, & Benoist C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat Immunol 18, 1238–1248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell DJ & Koch MA Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol 11, 119–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C et al. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-Treg phenotype. Cell 174, 285–299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dispirito JR et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci Immunol 3, eaat5861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miragaia RJ et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell BR, Buist NR, & Stenzel P. An X-linked syndrome of diarrhea, polyendocrinopathy, and fatal infection in infancy. J Pediatr 100, 731–737 (1982). [DOI] [PubMed] [Google Scholar]
- 14.Ramsdell F & Ziegler SF FOXP3 and scurfy: how it all began. Nat Rev. Immunol 14, 343–349 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Barzaghi F, Passerini L, & Bacchetta R. Immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome: a paradigm of immunodeficiency with autoimmunity. Front Immunol 3, 211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d’Hennezel E, Bin DK, Torgerson T, & Piccirillo CA The immunogenetics of immune dysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet. 49, 291–302 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Duclaux-Loras R et al. Clinical Heterogeneity of Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked Syndrome: A French Multicenter Retrospective Study. Clin. Transl. Gastroenterol 9, 201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambineri E et al. Clinical, immunological, and molecular heterogeneity of 173 patients with the phenotype of Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked (IPEX) syndrome. Front Immunol 9, 2411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barzaghi F et al. Long-term follow-up of IPEX syndrome patients after different therapeutic strategies: An international multicenter retrospective study. J Allergy Clin. Immunol 141, 1036–1049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godfrey VL, Wilkinson JE, & Russell LB X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am. J. Pathol 138, 1379–1387 (1991). [PMC free article] [PubMed] [Google Scholar]
- 21.Wan YY & Flavell RA Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature 445, 766–770 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Van Gool F et al. A mutation in the transcription factor Foxp3 drives T helper 2 effector function in regulatory T cells. Immunity 50, 362–377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W et al. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol 8, 359–368 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Gavin MA et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 445, 771–775 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Charbonnier LM et al. Functional reprogramming of regulatory T cells in the absence of Foxp3. Nat. Immunol 20, 1208–1219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bacchetta R et al. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest 116, 1713–1722 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otsubo K et al. Identification of FOXP3-negative regulatory T-like (CD4+CD25+CD127low) cells in patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome. Clin Immunol. 141, 111–120 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Boldt A et al. Differences in FOXP3 and CD127 expression in Treg-like cells in patients with IPEX syndrome. Clin. Immunol 153, 109–111 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Walker MR et al. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+. J Clin Invest 112, 1437–1443 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavin MA et al. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A 103, 6659–6664 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allan SE et al. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int. Immunol( 2007). [DOI] [PubMed] [Google Scholar]
- 32.McMurchy AN et al. A novel function for FOXP3 in humans: intrinsic regulation of conventional T cells. Blood 121, 1265–1275 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Zemmour D et al. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci U S A 114, E3472–E3480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seddiki N et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J. Exp. Med 203, 1693–1700 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pesenacker AM et al. A Regulatory T-Cell Gene Signature Is a Specific and Sensitive Biomarker to Identify Children With New-Onset Type 1 Diabetes. Diabetes 65, 1031–1039 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Stoeckius M et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuart T et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakke AC, Purtzer MZ, & Wildin RS Prospective immunological profiling in a case of immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX). Clin. Exp. Immunol 137, 373–378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziegler SF FOXP3: of mice and men. Annu Rev. Immunol 24, 209–226 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Fontenot JD, Gavin MA, & Rudensky AY Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Hori S, Nomura T, & Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Malek TR & Ashwell JD Interleukin 2 upregulates expression of its receptor on a T cell clone. J. Exp. Med 161, 1575–1580 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyman O et al. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311, 1924–1927 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Samstein RM et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 151, 153–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Remedios KA et al. The TNFRSF members CD27 and OX40 coordinately limit TH17 differentiation in regulatory T cells. Sci. Immunol 3, (2018). [DOI] [PubMed] [Google Scholar]
- 46.Kim JM, Rasmussen JP, & Rudensky AY Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol 8, 191–197 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Sitrin J et al. Regulatory T cells control NK cells in an insulitic lesion by depriving them of IL-2. J Exp. Med 210, 1153–1165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cobbold S & Waldmann H. Infectious tolerance. Curr. Opin. Immunol 10, 518–524 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Plitas G et al. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 45, 1122–1134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gambineri E et al. Clinical and molecular profile of a new series of patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome: inconsistent correlation between forkhead box protein 3 expression and disease severity. J Allergy Clin. Immunol 122, 1105–1112 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Bettelli E et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Godec J et al. Compendium of Immune Signatures Identifies Conserved and Species-Specific Biology in Response to Inflammation. Immunity 44, 194–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolock SL, Lopez R, & Klein AM Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aran D et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.( 2018). [DOI] [PubMed] [Google Scholar]
- 57.Klein AM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansen N & Quon G. scAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol. 20, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Law CW, Chen Y, Shi W, & Smyth GK Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soneson C & Robinson MD Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 15, 255–261 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Wickham H ggplot2: Elegant Graphics for Data Analysis(Springer-Verlag, New York, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE166866 (human population RNAseq), GSE166860 (mouse population RNAseq), GSE167976 (human scRNA-seq) and GSE167575 (mouse scRNA-seq).