Abstract
The Smc5/6 complex facilitates chromosome replication and DNA break repair. Within this complex, a subcomplex composed of Nse1, Nse3 and Nse4 is thought to play multiple roles through DNA binding and regulating ATP-dependent activities of the complex. However, how the Nse1-Nse3-Nse4 subcomplex carries out these multiple functions remain unclear. To address this question, we determine the crystal structure of the Xenopus laevis Nse1-Nse3-Nse4 subcomplex at 1.7 Å resolution and examine how it interacts with DNA. Our structural analyses show that the Nse1-Nse3 dimer adopts a closed conformation and forms three interfaces with a segment of Nse4, forcing it into a Z-shaped conformation. The Nse1-Nse3-Nse4 structure provides an explanation for how the lung disease immunodeficiency and chromosome breakage syndrome-causing mutations could dislodge Nse4 from Nse1-Nse3. Our DNA binding and mutational analyses reveal that the N-terminal and the middle region of Nse4 contribute to DNA interaction and cell viability. Integrating our data with previous crosslink mass spectrometry data, we propose potential roles of the Nse1-Nse3-Nse4 complex in binding DNA within the Smc5/6 complex.
Keywords: The Smc5/6 complex, Nse1-Nse3-Nse4, kleisin-KITE complex, DNA replication and repair, chromosome structure
Introduction
Structural Maintenance of Chromosomes (SMC) family of protein complexes play multiple roles in DNA organization and transaction. They can fold chromatin fibers into higher-order structures, cohere sister chromatids, and facilitate DNA replication and repair. Eukaryotes contain three SMC complexes: cohesin, condensin, and the Smc5/6 complex, and prokaryotes contain the Smc/ScpAB and MukBEF complexes [1]. While the roles of cohesin and condensin in sister chromatid cohesion and chromosome organization have been well established, the Smc5/6 complex appears to exert a range of functions, such as facilitating chromosome replication, regulating homologous recombination, and sumoylating multiple substrates [2–4].
All SMC complexes contain two SMC proteins and a different set of non-SMC subunits. The SMC proteins have similar structures, with a globular hinge domain at one end, an ATPase head domain at the other end, and a long coiled-coil arm that connects the two distal ends [5–6]. Two SMC proteins can associate with each other via their hinge domains and coiled-coil arms and form ATP-mediated interactions between their head domains. A conserved non-SMC subunit, kleisin, can bridge the two SMC proteins by interacting with the head-proximal coiled-coil region (“neck”) of one SMC and the bottom of the head domain of the other SMC [7–10]. The tripartite structure formed by the SMC dimer and kleisin is conserved in most SMC complexes. Several SMC complexes has been suggested to topologically trap DNA molecule(s), providing an explanation for some of the SMC-mediated functions [8, 11–13]. In addition, ATP binding and hydrolysis by some SMC complexes, such as condensin and Smc/ScpAB, enable progressively enlarging DNA loops via loop extrusion [14, 15]. ATP-dependent DNA entrapping and loop extrusion are thought to be the key features of those SMC complexes that can sculpt the genome.
Despite similarities of SMC complexes, three eukaryotic SMC complexes each contains different non-SMC elements. The Smc5/6 complex uniquely contains Nse2 that binds to a coiled-coil region of Smc5 and acts as a SUMO ligase and the Nse5-Nse6 subcomplex that helps to recruit the complex to chromatin [16–21]. In contrast to cohesin and condensin that contain HAWK (HEAT proteins associated with kleisin) subunits, the Smc5/6 complex, like prokaryotic Smc/ScpAB, contains KITE (kleisin-interacting tandem winged-helix elements) subunits [22, 23]. HAWK and KITE are thought to shape the structure of the kleisin subunit, bind to double stranded DNA, and regulate the opening and closing of the SMC head regions [24–28]. In the Smc5/6 complex, the two KITE subunits, namely Nse1 and Nse3, form a dimer and bind to the Nse4 kleisin subunit.
Nse1 and Nse3 each consists of tandem winged helix (WH) domains, and Nse1 further possesses a ring finger domain that could support ubiquitin ligase activity in the human Smc5/6 complex and stability in the fission yeast complex [29, 30]. Mutations disrupting the interaction between Nse1 and Nse3 or between Nse3 and Nse4 cause cell growth defects and hyposensitivity to DNA damaging agents [31]. In human cells, Nse3 is a member of the melanoma-associated antigens (MAGEs) family of proteins that are tumor- and testis-specific antigens and associated with brain development, apoptosis, and differentiation [32, 33]. Two Nse3 mutations have been found to cause a rare lung disease immunodeficiency and chromosome breakage syndrome (LICS) [34].
A recent study examining the interactions among human Nse1, Nse3, Nse4 proteins suggests that the Nse1-Nse3 dimer may shape the Nse4 middle domain to regulate the opening of a gate between Nse4 and the Smc6 neck region (Nse4N gate) [28]. Currently, structural basis of how the Nse1-Nse3 dimer interacts with and affects the Nse4 middle region is unclear. Two different structures of the human Nse1-Nse3 dimer have been reported [29, 35]. One structural model suggests a closed conformation, with Nse1 and Nse3 N-terminal WHs (WHAs) associating with each other in a head-to-head orientation and their C-terminal WHs (WHBs) pointing away from each other [29]. This conformation was based on crystal structure data with limited resolution and uncertainty around the linker between WHA and WHB (WHA-WHB linker) of Nse3. Re-calculation of this data has suggested an open conformation wherein the Nse3’s WHA and WHB domains wrap around the WHA of Nse1 [35].
In this study, we determine the crystal structure of the Xenopus laevis (Xl) Nse4 segment bound to the XlNse1-Nse3 dimer (hereafter referred to as the ‘Nse1-3-4 complex’ or Nse1-3-4). Our structure shows that the Nse1-3 dimer binds to the middle region of Nse4 in a rigid and defined manner via three binding interfaces. The importance of each interface is supported by previous mutagenesis data of human and Schizosaccharomyces pombe (Sp) proteins. We incorporate our structural data with reported crosslink and mass spectrometry (CL-MS) and yeast two-hybrid (Y2H) results to suggest for the spatial orientation of Nse1-3-4 within the Smc5/6 complex [28, 36]. Finally, we show that Nse4 can contribute to DNA binding via positively charged residues located at its disordered N-terminal region and a region C-terminal to the Nse1-3 binding site in vitro, and that these residues are functionally important in cells. Collectively, our data suggests that Nse4 is stiffened upon binding to Nse1-3 and contribute to DNA binding.
Results
Overall structure of the XlNse1-Nse3 dimer binding to XlNse4
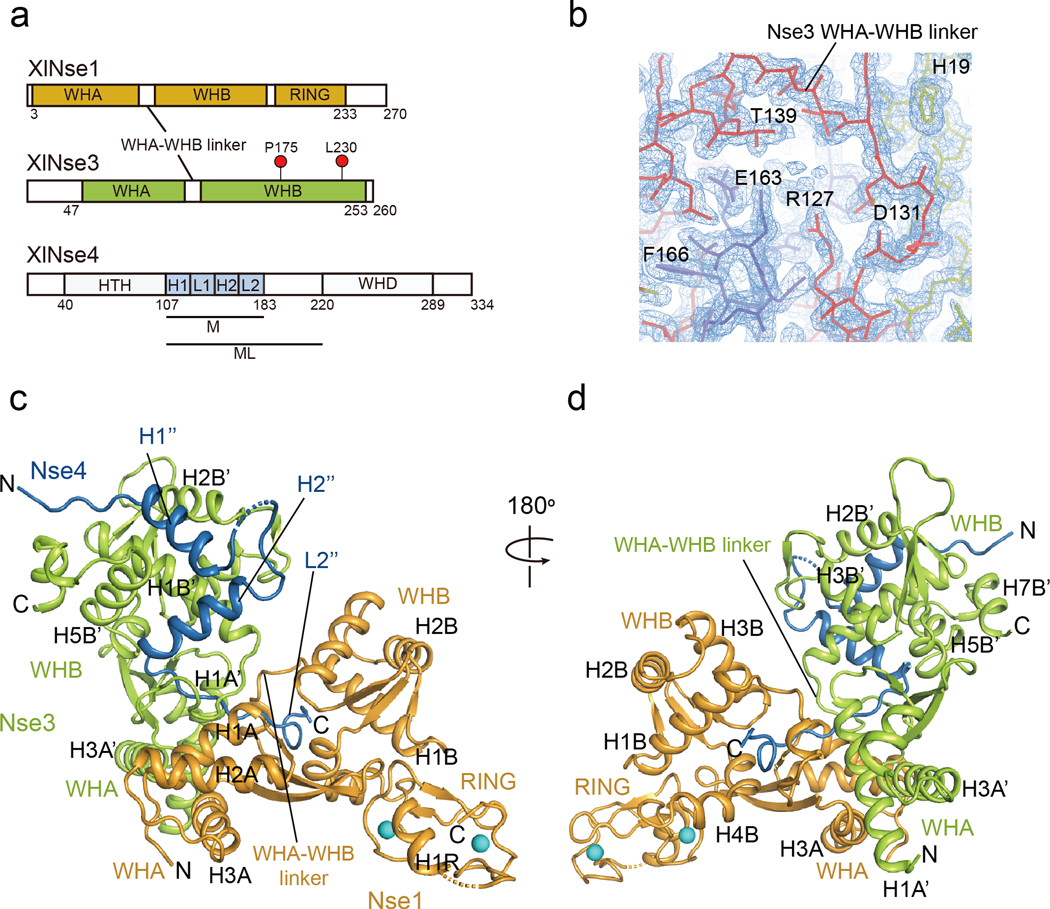
Secondary structure prediction using the PROMALS3D [37] and Psipred [38] programs suggests that XlNse4 is mainly comprised of an N-terminal HTH domain (helix-turn-helix, residues 40 to 107), a middle domain of loops and short helices (residues 108 to 220), and a C-terminal WH domain (residues 221 to 289; Fig. 1a). We obtained crystals that diffracted to a high resolution (1.7 Å) using co-expressed constructs containing the middle domain of XlNse4 (Nse4M; residues 108 to 183) and near full-length XlNse1 (residues 3 to 248) and XlNse3 (residues 45 to 260) (Table 1, Fig. 1a). Figure 1 shows the overall architecture of the Nse1-3 dimer in complex with Nse4M (Fig. 1b–d). The two WH domains of Nse3 (green, 60 × 38 × 48 Å3) are positioned vertically against the two WH domains of Nse1 (orange, 54 × 34 × 21 Å3) (Fig. 1c). The Nse3’s WHA-WHB linker is well-ordered (Fig. 1b and c), unlike the poorly defined linker in the reported human (Homo sapiens or Hs) Nse1-Nse3 structure [29, 35]. The overall arrangement of XlNse1 and XlNse3 in our structure is similar to the closed, but not the open, form of the HsNse1-Nse3 dimer [29, 35] (Fig. S1a). We note that the closed (PDB 3NW0), but not the open conformation (PDB 5HVQ), is consistent with data from a recent CL-MS analysis of the HsNse1-Nse3 complex [36]. For simplicity, we label the secondary structure elements of Nse3 and Nse4 with different superscripts (e.g. H1’ in Nse3 and H1” in Nse4) and the elements in their WHA and WHB domains with A and B, respectively.
Fig. 1.
The overall structure of the XlNse1-3-4 complex.
(a) Diagram of the domain organization of XlNse1, XlNse3, and XlNse4. Nse3 residues corresponding to those mutated in the LICS patients are marked with red circles. Two Nse4 constructs used for structural analyses are marked with M and ML.
(b) 2Fo-Fc map of the Nse3 WHA-WHB linker (red line) showing the connectivity between these two domains. Thick yellow and blue lines represent Nse1 and Nse4, respectively.
(c) Ribbon representation of the structure of the Nse1 (orange)-3 (green)-4 (blue) trimeric complex. The two helices of Nse4 bind to the WHB surface of Nse3, and the extended loop threads through a channel at the interface of Nse1 and Nse3. Two zinc ions at the Nse1 RING finger domain are shown as spheres (cyan).
(d) View of the Nse1-3-4 complex rotated 180° relative to the view shown in (c).
Table 1.
Crystal data collection, refinement and statistics
| Crystal I | Crystal II | Crystal III | Crystal IV | |
|---|---|---|---|---|
| Xl Nse1-3-4 (108–183) | Xl Nse1-3-4 (30–183) | Xl Nse1-3-4 (108–220) | Xl Nse1-3-4 (108–236) | |
| Data collection | ||||
| Wavelength (Å) | 0.9794 | 0.9794 | 1.5418 | 1.5418 |
| Space group | P 212121 | P 212121 | P 212121 | H 3 2 |
| Cell dimensions | ||||
| a, b, c (Å) | 53.5, 66.2, 168.4 | 53.6, 66.5, 169.8 | 53.3, 65.8, 169.3 | 134.8, 134.8, 85.6 |
| Resolution (Å) | 50–1.70(1.73–1.70)1 | 50–1.98(2.01–1.98) | 35.61–2.88(3.05–2.88) | 23.53–2.24(2.31–2.24) |
| Measured reflections | 804796 | 487455 | 95096 | 291938 |
| Unique reflections | 65684 | 40236 | 10702 | 27589 |
| Rsym (%)2 | 10.3(83.2) | 12.2(83.8) | 21.1(75.4) | 6.4(14.8) |
| I/σI | 23.7(2.0) | 24.4(2.6) | 9.1(2.8) | 23.2(8.4) |
| Completeness (%) | 98.7(95.0) | 93.6(93.6) | 75.7(72.4) | 98.9(90.5) |
| Redundancy | 12.3(12.0) | 12.1(12.2) | 8.9(9.2) | 10.2(5.6) |
| Refinement | ||||
| Resolution (Å) | 41.6–1.7 | |||
| No. reflections | 65577 | |||
| Rwork/ Rfree3 | 0.190/0.218 | |||
| No. atoms | ||||
| Protein | 4031 | |||
| Zinc | 2 | |||
| Water | 317 | |||
| Others | 20 | |||
| B-factors (Å2) | ||||
| Protein | 27.3 | |||
| Zinc | 29.6 | |||
| Water | 32.6 | |||
| Others | 40.1 | |||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.006 | |||
| Bond angles (º) | 0.760 | |||
| Ramachandran plot | ||||
| Most favored (%) | 97.72 | |||
| Allowed (%) | 2.28 | |||
| Disallowed (%) | 0 | |||
Values in parentheses are for the highest shell.
Rsym=ΣhΣi |Ih,i−Ih|/ΣhΣi Ih,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h.
R=Σ|Fobs-Fcalc|/ΣFobs, where Fobs = Fpi and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections). R.m.s. deviation in bond lengths and angles are the deviations from ideal values, and the r.m.s. deviation in B factors is calculated between bonded atoms.
In our structure, Nse4M adopts a Z shaped structure composed of two helices (H1” and H2”) followed by a 40 Å long extended L2” loop (Fig. 1c and d). The H1” and H2” helices of Nse4 are arranged in near 90° on the surface groove of WHB of Nse3. The L2” loop of Nse4 extends from the WHA-WHB linker of Nse3 to the interface between Nse3 and Nse1, and exits from a Nse1 channel formed by its S3A strand, three helices (H1B, H3B, and H4B), the L3B loop, and its WHA-WHB linker (Fig. S1b).
We also obtained crystals containing the XlNse1-3 dimer in complex with either the XlNse4’s N-terminal HTH and its middle region (crystal II, residues 30 to 183) or its middle region in a longer form (Nse4ML; crystal III, residues 108 to 220; crystal IV, residues 108 to 236). However, neither the Nse4 N-terminal region (residues 30 to 107) nor the region C-terminal to Nse1-3 binding site (residues 184 to 236) was invisible in the structures derived from these crystals (Table 1).
Structural comparison between the XlNse1-3-4 complex and the HsNse1-Nse3 dimer
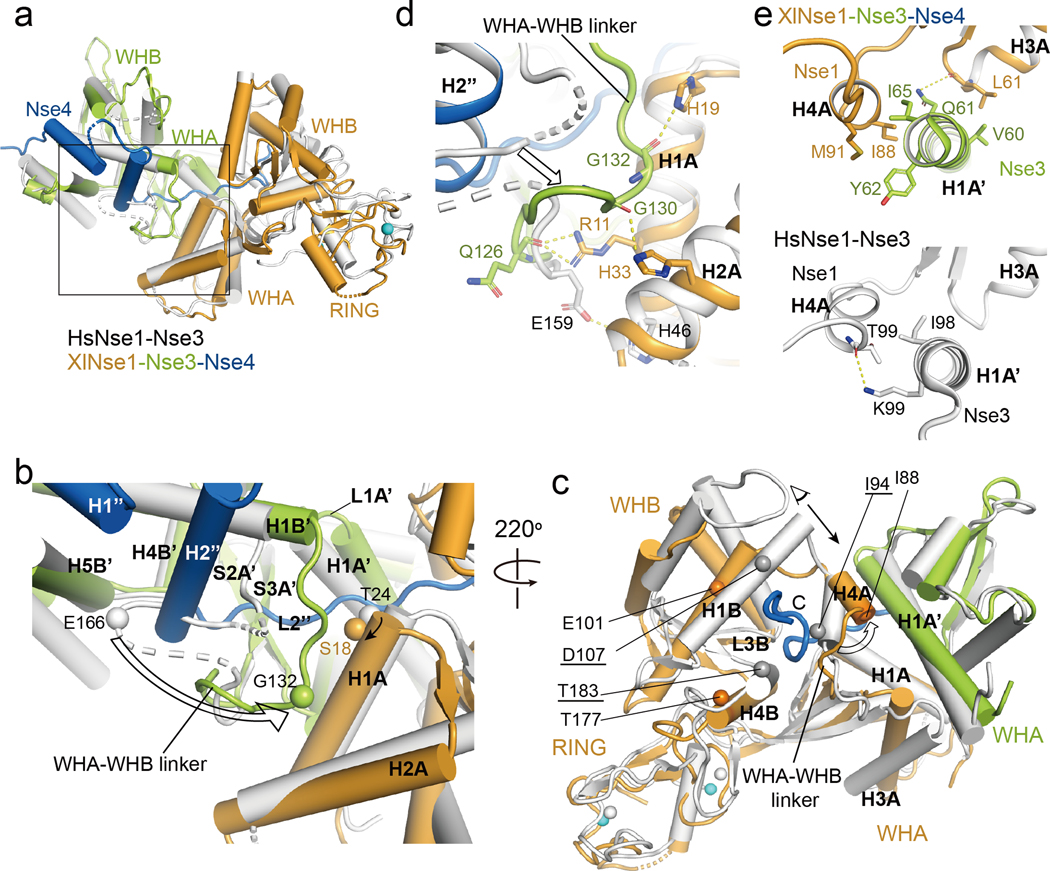
The XlNse1-3-4 complex structure was aligned with the structure of HsNse1-Nse3 dimer (PDB 3NW0) with a root mean square deviation (rmsd) value of 2.1 Å for 428 Cα atoms in Nse1-Nse3 (Fig. 2a). The differences between these structures is partly explained by sequence divergence of Nse1 and Nse3 from the two species, as they share only 48% and 44% sequence identity (Fig. S2, S3). More importantly, Nse4M binding induces the formation of two binding interfaces between XlNse1 and XlNse3 that are not seen in the HsNse1-Nse3 dimer (Fig. 2a–c).
Fig. 2.
Structural comparison of the XlNse1-3-4 and the HsNse1-Nse3 complex.
(a) XlNse3 (green) of the Nse1-3-4 complex is aligned with HsNse3 (white) of the Nse1-Nse3 complex (PDB 3NW0) [29]. XlNse1 and XlNse4 are colored orange and blue, respectively.
(b) Close up view of the box in (a). Binding of the H2” helix and the L2” loop of Nse4 to Nse1-Nse3 results in the shift of the Nse3 WHA-WHB linker toward the H1A helix of Nse1. Ser18 and Gly132 of Nse3 (Thr24 and Glu166 in HsNse3) shown in spheres illustrate the conformational change of the H1A helix of Nse1 and the WHA-WHB linker of Nse3, respectively (indicated by an arrow).
(c) Binding of the extended L2” loop of Nse4 (blue) induces conformational changes in Nse1 (orange). The HsNse1-3 complex is colored white. Selected residues are shown in spheres to highlight the conformational changes of Nse1. Gray and orange spheres represent the residues of Hs and XlNse1, respectively. White and cyan spheres indicate the zinc ions of HsNse1 and XlNse1, respectively. HsNse1 residues corresponding to those of XlNse1 are underlined. Conformational change of the H4A helix is shown with an empty arrow. A black arrow indicates a view for the figure (e).
(d) Close-up view of (b) showing the new interface formed between the WHA-WHB linker of XlNse3 (green) and the H1A and H2A helices of Nse1 (orange). HsNse1-Nse3 complex is colored in white.
(e) Close-up view of (c) in a different angle. The H4A helix of Nse1 shifts toward the H1A’ helix of Nse3, which increases the interface between the H4A helix of Nse1 (orange) and the H1A’ helix of Nse3 (green, top) compared with that of HsNse1-Nse3 (white, bottom).
First, the second helix (H2”) and the L2” loop of Nse4 shape the WHA-WHB linker of Nse3 into a defined structure and shift it towards the WHA of Nse1 by as much as 17.7 Å (Fig. 2b). This change generates a new interface wherein Arg11, His19, and His33 of Nse1 (H1A and H2A) interact with the backbone of the WHA-WHB linker of Nse3 (Fig. 2d). Moreover, these regions collaborate with the L1A’ loop, the S2A’ and S3A’ strands, and the H1B’ and H4B’ helices of Nse3, to form a channel into which the extended loop of Nse4 (L2”) enters (Fig. 2b and S1b).
Second, in order to accommodate the extended L2” loop of Nse4, the H4A helix of Nse1 moves toward the H1A’ helix of Nse3, forming another Nse1 and Nse3 interface (Fig. 2c). The main contacts here involve multiple hydrophobic residues from the H1A’ helix of Nse3 and the H3A and H4A helices of Nse1. Specifically, Leu61 of Nse1 interacts with Val60 of Nse3 and forms a hydrogen bond with Gln61 of Nse3 via its backbone atoms, while Ile88 and Met91 of Nse1 interact with Ile65 and Tyr62 of Nse3, respectively (Fig. 2e). We conclude that Nse4 binding greatly increases the interface between Nse1 and Nse3.
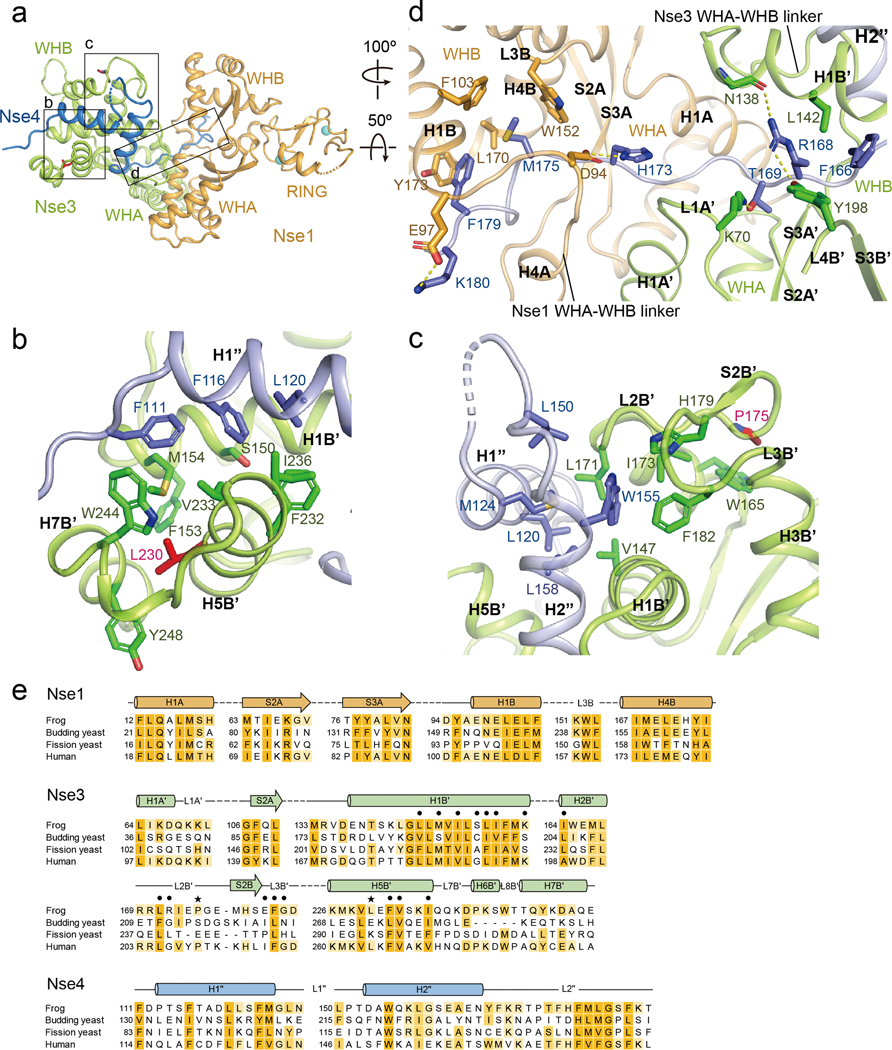
Three interfaces between XlNse4 and the XlNse1-Nse3 dimer
In our structure, the Nse1-Nse3 dimer interacts with Nse4 via three interfaces (Fig. 3a–e). One is formed between the H1” helix of Nse4M and a groove at the WHB of Nse3, with 730 Å buried surface area. Four Nse4M residues, namely Phe111, Phe116, Leu120, and Met124, are directed toward a groove formed by the H1B’, H2B’, H5B’, and H7B’ helices of Nse3, lodging at hydrophobic environment (Fig. 3b and S5a). The importance of this interface is supported by previously reported mutational data for the human and fission yeast proteins. For example, mutating HsNse4 residues equivalent to XlNse4 Phe111, Phe116 and Leu120 reduces binding to HsNse3 in Y2H (Fig. S4) [39]. Similarly, deleting residues corresponding to Ser115-Asp119 of XlNse4 and mutating residues corresponding Gln14 and Met17 of XlNse1 (Fig. S2 and S5c) abrogate the assembly of HsNse1-3-4 complex [28]. Moreover, mutating residues equivalent to the XlNse3 WHB residues that accommodate Phe116 and Leu120 of XlNse4 (Ser150, Met154, Met167, Leu171, Phe232, Val233, or Ile236) reduces SpNse3 and SpNse4 interaction in Y2H and cellular resistance to methanesulfonate (MMS) and hydroxyurea (HU) (Fig. 3e and S3) [31]. Our structural data can also explain how a LICS-causing mutation wherein a phenylalanine replaces Leu264 in HsNse3 can impede HsNse4 binding in Y2H [34]. HsNse3 Leu264 is equivalent to Leu230 of XlNse3 (in H5B’) that is packed against Phe153, Met154, Trp244, and Tyr248 in the WHB groove; Leu230Phe mutation would disrupt the local structure of this groove, thus destabilizing the binding to the H1” helix of Nse4M.
Fig. 3.
The three interfaces between XlNse4 and the XlNse1-Nse3 dimer.
(a) Overall structure of the Nse1 (orange)-3 (green)-4 (blue) complex. Three interfaces are highlighted with boxes.
(b) The first interface, between the first helix of Nse4 (blue) and the WHB of Nse3 (green). Residues in the H1” helix of Nse4 are surrounded by hydrophobic residues in the H1B’, H5B’, and H7B’ helices of Nse3. Leu230, a mutated residue in LICS is shown in a red stick. The interacting residues in Nse3 and Nse4 are also marked in the sequence alignment in Fig. S3 and S4.
(c) The second interface, between the second helix of Nse4 (blue) and the H1B’ and H5B’ helices, S2B’ strand, and L2B’ and L3B’ loops of Nse3 (green). Pro175, a mutated residue in LICS is shown in a red stick. See also Fig. S3 and S4.
(d) The third interface, between Nse4 (the extended loop) and the Nse1-Nse3 complex. The view is 100º and 50º-rotated along the y and the x axis, respectively, relative to that in (a). The L2” loop of Nse4 (blue) interacts with Nse1 (orange) and Nse3 (green) by threading through their interface. The first half of the Nse4 L2” loop is surrounded by Nse3, as well as Nse1 (right). The second half of Nse4 L2” loop is surrounded primarily by Nse1 (left).
(e) Conservation of the residues in the Nse1-3-4 complex in (b)-(d). The point mutations sensitive to DNA damage inducing agents in S. pombe are indicated by dots. LICS related mutations are marked by black stars.
In the second interface, the H2” helix of Nse4M is lodged into a shallow groove of the WHB of Nse3 that is formed by helices H1B’, H5B’, strand S2B’, and loops L2B’ and L3B’ (Fig. 3c). Hydrophobic interactions are dominant here. Six residues in the H2” of Nse4 (Leu150, Trp155, Leu158, Glu161, Tyr165, and Phe166) are directed toward this groove (Fig. 3c, e and S5b). These interactions are augmented by Lys235 of Nse3 (in H5B’) that forms a hydrogen bond with Tyr165 and an ion pair with Glu161 within the H2” helix of Nse4 (Fig. S5b). Further strengthening this interface is achieved by Phe166 of Nse4 that is surrounded by three hydrophobic residues of Nse3 (Leu142, Met146, and Leu149) located within H1B’. The importance of this interface is supported by mutational data from fission yeast studies. Mutating Met214 or Leu248 of SpNse3, equivalent to Met146 or Phe182 of XlNse3, reduces both SpNse4 interaction and DNA damage resistance [31]. Interestingly, a LICS-causing mutation wherein a leucine replaces Pro209 of HsNse3 can impede HsNse4 binding in Y2H [34]. In our structure, the ring of the equivalent Pro175 residue of XlNse3 (in L2B’) is packed against Trp165 and stabilizes the local structure of the L2B’ and L3B’ loops and S2B’ strand, supporting a binding pocket for Trp155 from the H2” of Nse4 (Fig. 3c, e). Replacing Pro175 with leucine in XlNse3 would be expected to interfere this interaction.
The third and most extensive interface is formed between the extended loop (L2”) of Nse4 and the Nse3 and Nse1 interface. The first half of L2” interacts with residues at the channel formed by Nse1 (H1A) and Nse3 (L1A’, L4B’, S2A’, S3A’, H1B’, and WHA-WHB linker) (Fig. 3d, e and S5c, right). This interface is in part supported by hydrogen bonds formed between Arg168 of Nse4 and Tyr198 and Asn138 of Nse3, and between Thr169 of Nse4 and Lys70 of Nse3. This can explain a previous observation that alanine replacement of Tyr264 in SpNse3 (Tyr198 in XlNse3) disrupts the interaction with SpNse1 and SpNse4 in Y2H [31]. The second half of L2” loop of Nse4 threads through the channel formed by Nse1 (S2A, S3A, H1B, H4B, L3B, and WHA-WHB linker) and Nse3 (H1A’) (Fig. 3d and S5c, left). Both hydrophobic interactions and ion-pairs formed between Nse4’s His173 and Nse1’s Asp94 and formed between Nse4’s Lys180 and Nse1’s Glu97 contribute to the stability of this interface. Although the interactions are very extensive, single point mutation affecting this interface is sufficient to reduce SpNse4 binding to the SpNse1-Nse3 dimer and cell viability, suggesting that individual interaction here can be critical for Nse4 binding to Nse1-Nse3 [31]. In conclusion, our structure data delineate three interfaces formed between Nse4M and the Nse1-3 dimer; the importance of these interfaces is well supported by available mutagenesis data. Our data also provide structural explanations for the two disease-causing HsNse3 mutations found in LICS patients.
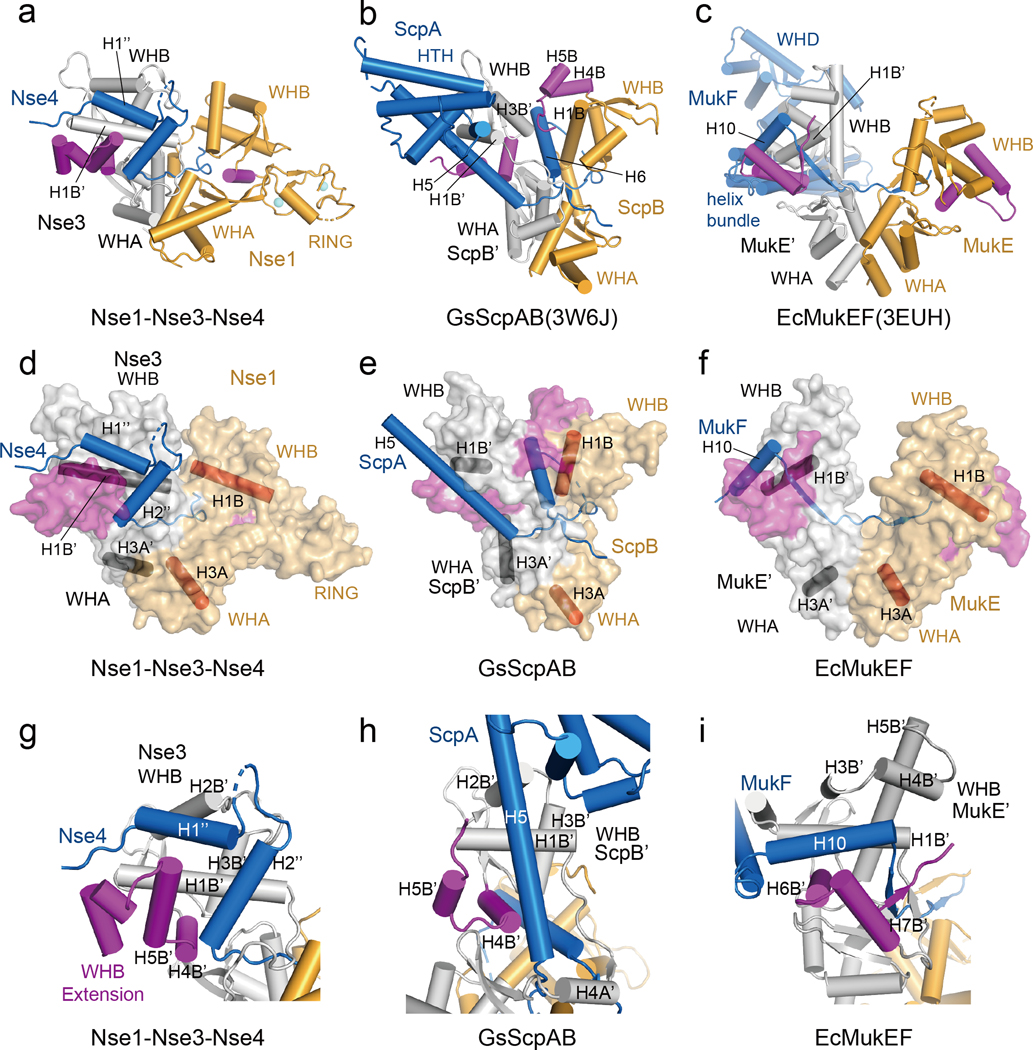
Structural comparison of XlNse1-3-4 and prokaryotic KITE-klesin complexes
The Nse1-3-4 complex shares structural similarities with three prokaryotic KITE-klesin complexes. These include i) ScpAB from Streptococcus pneumoniae (SpScpAB) (PDB: 4I98), ii) ScpAB from Geobacillus stearothermophilus (GsScpAB) (PDB:3W6J), and iii) MukEF from Escherichia coli (EcMukEF) (PDB: 3EUH) [7, 40, 41]. In these complexes, the klesin subunits are ScpA and MukF. The KITE protein ScpB or MukE consists of tandem WH domains and forms a head-to-head homodimer via WHA domains. For convenience, the KITEs binding the N- and C-terminal region of kleisin are referred to as the first and the second KITE, respectively. Figure 4 and S6 show that structures of three KITE-kleisin complexes with the WHA of the second KITE aligning with the WHA of Nse1. Two common features of these complexes include: i) the N-terminal helix (or helices) of the kleisin middle region binds to WHB and WHA of the first KITE or Nse3, and ii) the C-terminal extended loop of kleisin passes through the second KITE or Nse1 (Fig. 4a–c and S6a; the secondary structure elements in the first KITE are marked with superscript). Despite these similarities, there are four major differences (below).
Fig. 4.
Comparison of the Nse1-3-4 complex with ScpAB and MukEF.
(a) Cylinder representation of the XlNse1 (orange)-3 (white)-4 (blue) complex. The WHB C-terminal extensions of Nse1 and Nse3 are colored in magenta.
(b) Cylinder representation of GsScpAB. The WHA of ScpB (orange) is aligned with WHA of XlNse1 in (a). The H6 helix of ScpA (blue) passes through the groove formed by ScpB (orange and magenta) and ScpB’ (white).
(c) Cylinder representation of EcMukEF. The WHA of MukE (orange) is aligned with WHA of XlNse1 in (a). MukF is colored by blue.
(d) Surface representation of the XlNse1 (light orange)-3 (white)-4 (blue) complex in same view with (a). The WHB C-terminal extensions of Nse1 and Nse3 is colored in magenta. The helices from each WH domains of Nse1 and Nse3 are shown in red and black cylinders, respectively, to represent the relative orientations of protomers and domains.
(e) Surface representation of GsScpAB in same view with (b). The helices of ScpB (light orange) and ScpB’ (white) equivalent to the helices of Nse1 and Nse3 are shown in red and black cylinder, respectively. ScpA is colored blue. The WHB C-terminal extensions of both ScpB and ScpB’ are colored magenta.
(f) Surface representation of EcMukEF in same view with (c).
(g) Close-up view of the Nse4 (blue)-Nse3 (white) interface.
(h) Close-up view of the GsScpA (blue)-ScpB’ (white) interface. The structure of the ScpB’ WHB domain is aligned with that of Nse3 in (g).
(i) Close-up view of the EcMukE’ (white)-MukF (blue) interface in same orientation as in (g).
First, the arrangements between WHA and WHB and between the two KITEs differ among complexes (Fig. 4d–f and S6b). Consequently, the complexes show distinct WHA-WHA packaging and WHB domain distances. For example, the two WHBs in MukE are much more open that those in the ScpB homodimer or in Nse1-3.
Second, while Nse4M contains two N-terminal helices, ScpA contains one long helix with 8–10 turns (H4 in SpScpA and H5 in GsScpA) that binds to an elongated groove formed by H1B’, H2B’, H4B’, and H5B’ helices of ScpB’ (Fig. 4g, h and S6c). Similar to ScpA, MukF has a single helix (H10) that engages the grove formed by H1B’, H3B’, H4B’, H6B’ and H7B’ helices of MukE’ (Fig. 4i). One difference between ScpA and MukF is that while the helix H5 of GsScpA is perpendicular to the first helix of the WHB of ScpB’, a corresponding helix of MukF is parallel to the first helix of the WHB of MukE’. The first and second helices of Nse4 are oriented in parallel and perpendicular against the first helix of the WHB of Nse3, respectively. These differences are due to the orientation of helices in first KITE in the complexes as well as the different groove shapes determined in part by the position of the C-terminal extension of WHB. In Nse3 and ScpB’, H4B’ is located to the right side of the WHB C-terminal extension and stabilizes the kleisin (Fig. 4g, h and S6c). However, in MukE’, the equivalent helix (H6B’) is at the left side of the C-terminal extension to interact with the helix H10 of MukF (Fig. 4i).
Third, similar to the extended loop L2” in Nse4, the extended loop of ScpA passes through the channel formed by H1A, H4A, H1B, and H3B (orange) of ScpB, and H1A’ and H4A’ and S3A’ of ScpB’ (white) (Fig. S6d–f). In contrast, the first part of the extended loop of MukF forms a strand and is paired with the C-term extension of WHB while the rest of the loop passes through a channel [41]. Moreover, the extended loop of ScpA exhibits two unique features: i) it makes a sharp turn at the end of the channel, wraps around ScpB, and makes another 90° turn (Fig. S6e and f), and ii) the helix C-terminal to this loop (H5 in SpScpA and H6 in GsScpA) interacts with the groove formed between ScpB’ (H3B’) and ScpB (H1B, H4B, and H5B) (Fig. 4b and S6a, e, f).
Fourth, the C-tail of Nse4ML (residues 184 to 220), which is disordered in our structure, is likely to adapt a different path compared with the corresponding region of ScpA. This is based on structural modeling wherein the WHA domain of Nse3 was aligned with the WHA domain of ScpB’ (Fig. S7a). This model showed a clash between Nse4ML C-tail and the H1B’ and H3B’ of Nse3 (Fig. S7a, green helix). Though this clash could be solved by shifting the Nse4ML C-tail by 18 Å to the cleft formed by H1B and H3B of Nse1, and H1B’and H3B’ and L3B’ loop of Nse3 (Fig. S7a, cyan helix), this revised model disagrees with published CL-MS data [36] (Fig. S7a). Specifically, multiple crosslinked residue pairs detected previously would have Ca-Ca distances larger than permitted by the crosslinker (≤ 30 Å). For example, the model would predict Arg195 of Nse4 is 36 Å away from Glu50 of Nse3; Lys198 of Nse4 and Glu50 of Nse3 are 40 Å away; Lys211 of Nse4 and Lys87 and Glu50 of Nse3 are 50 Å. These analyses suggest that Nse4ML C-tail adapts a different path than the corresponding region of ScpA.
In summary, our data suggest that the XlNse1-3-4 structure has both similarities and differences compared with those of prokaryotic KITE-klesin complexes.
A structural model for an extended Nse4 molecule
Based on the ScpAB structures (PDB 3W6J) [40], we predict that XlNse4 also contains an N-terminal HTH domain (residues 40–107) and an C-terminal WHD domain (residues 221–289) (Fig. S4). CL-MS data derived from full-length HsNse1 and HsNse3 in complex with an HsNse4b fragment (residues 92–212, corresponding to residues 89–215 in XlNse4) (hereafter referred to as CL-MSNSE) [36] provide spatial information to build a model for two regions of XlNse4 that are invisible in our structure, namely its HTH and the Nse4ML C-tail (residues 184–220).
Based on the ScpAB (PDB 3W6J) HTH region (residues 40 to 104, helices NH1-NH3) and CL-MSNSE data, we built a model locating the Nse4’s HTH relative to our Nse1-3-4 structure using the Phyre2 program [42]. This model suggests that Nse4’s HTH is located close to WHB of Nse3, as CL-MSNSE data suggest that Lys100 of XlNse4 can be positioned close to Lys160 and Glu231 of XlNse3 (equivalent residues in HsNSEs are in parenthesis in Fig S7b).
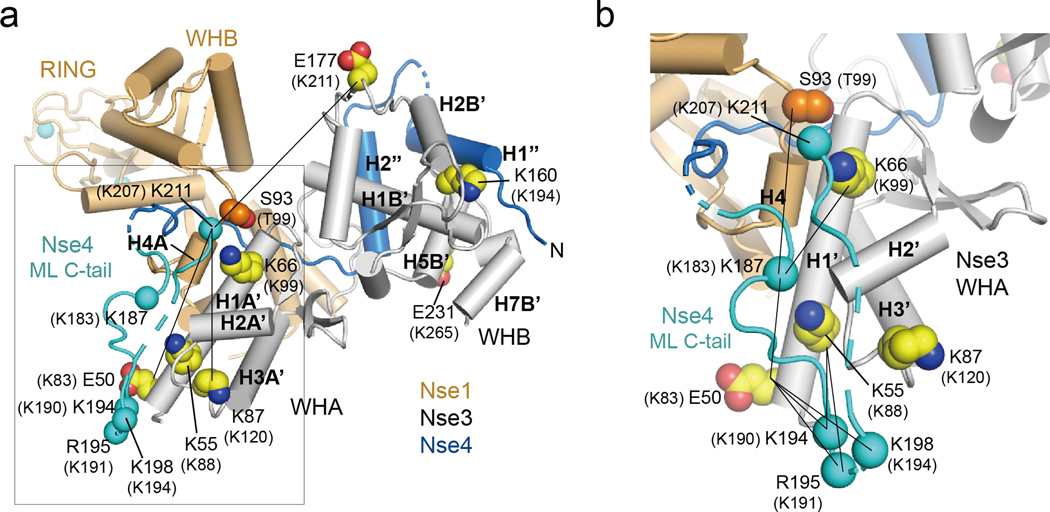
We also modeled Nse4ML C-tail based on three pieces of CL-MSNSE data (Fig. 5a, b). First, CL-MSNSE data suggest that Lys187 of XlNse4 can be in close proximity to Ser93 of XlNse1 and Lys66 and Glu50 of XlNse3. We thus placed the Cα atom of Lys187 of Nse4 within 20 Å of the Cα atoms of these Nse1 and 3 residues (Fig. 5b, solid line). Second, CL-MSNSE data suggest that Lys194, Arg195, and Lys198 of XlNse4 are close in space to Glu50 and Lys55 of XlNse3, indicating that the main-chain between Lys194 to Lys198 of Nse4 is directed toward H1A’ of Nse3. We thus placed Lys194 and Arg195 of Nse4 within 21 Å of Glu50 and Lys55 of Nse3, which results in a 25 Å Cα-Ca distance between Lys198 of Nse4 and Glu50 of Nse3 (Fig. 5b, solid line). Third, CL-MSNSE data suggest that Lys211 of XlNse4 is in close proximity with Glu50, Lys87, and Glu177 of XlNse3. These three XlNse3 residues are located far from each other in the Nse1-3-4 structure: the Cα-Cα distances are 54 Å and 48 Å between Glu50 and Glu177 and between Lys87 and Glu177, respectively. Thus, we placed Lys211 of Nse4 within 30 Å from the Cα atoms of these residues (Fig 5a–b). Taken together, our model suggests that i) one part of the Nse4ML C-tail (residues 187 to 198) is located near the H1A’ helix of Nse3 and aligned in an antiparallel manner with residues 194 to 198 near the N-terminal region of H1A’, and ii) the second part of Nse4ML (residues 199 to 211) is flexible, with Lys211 positioned near the C-terminal of H1A’ and the H4A helix of Nse1 (Fig. 5a, b).
Fig. 5.
Model of the C-terminal region of XlNse4
(a) Modeling a region of the Nse4ML C-tail (residues 187 to 211) based on CL-MSNSE data. Nse1 (orange), Nse3 (white), and Nse4 (blue) are shown in cylinder representation. The modeled region is colored cyan. Solid lines indicate close spacing between Lys211 of Nse4 and Glu50, Lys87 and Glu177 of Nse3. Human Nse1, 3, and 4 residues equivalent to those of Xenopus proteins are in parenthesis.
(b) Close-up view of the Nse4ML C-tail boxed in (a). Residues 187 to 198 of the Nse4ML C-tail (cyan) are placed near the N-terminus of the H1A’ helix (Nse3, white) within ~30 Å of all crosslinked residues. Lys211 is located near Ser93 (Nse1, bright orange) and Lys66 (Nse3). Lines indicate the crosslink pairs between Nse4ML C-tail and Nse1 or Nse3. The equivalent residues in human NSE proteins are in parentheses.
DNA binding by the XlNse1-3-4 complex and XlSmc6
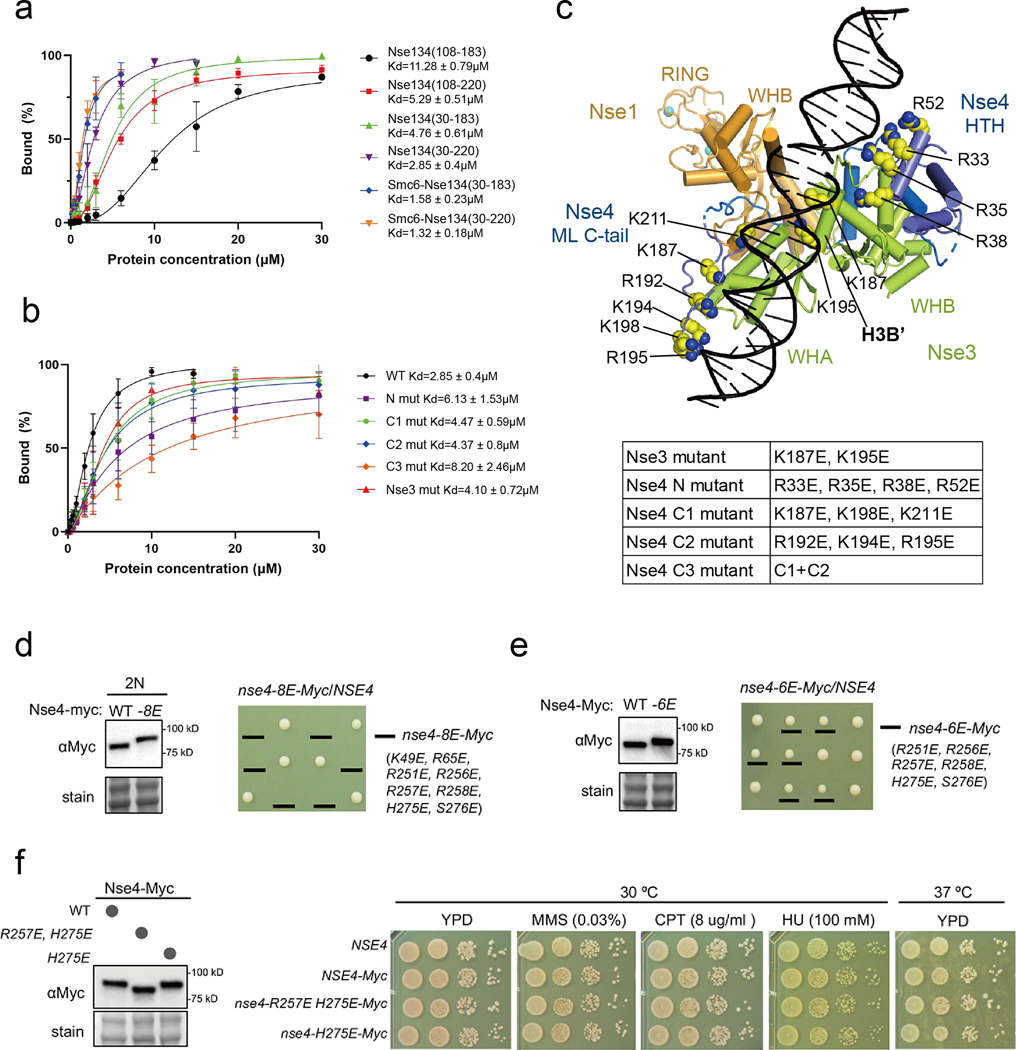
A previous report has suggested that Nse3 is primarily responsible for the Nse1-3-4 complex to bind DNA [27]. To explore whether Nse4 contributes to DNA binding, we analyzed the interaction between Nse1-3-4 complexes containing different Nse4 fragments and a 37 bp double-stranded (ds) DNA with 2 nt overhang at its 5’ end. All examined complexes contained near full length Nse1 (residues 3 to 248) and Nse3 (residues 45 to 260). For the complex also containing Nse4M (residues 108–183), weak DNA binding was detected (Kd ~11.28 μM; Fig. 6a and S8a, lane 2 to 13) and about 50% DNA molecules were shifted in the presence of 75-fold excess protein (Fig S8a, lane 10). The complex containing Nse4ML (residues 108 to 220) increased DNA binding about 2-fold (Kd ~5.29 μM; Fig. 6a, and S8a, lane 15 to 26). The N-terminal extension of Nse4M (residues 30 to 183) also increased the DNA-shifting ability (Kd ~4.76 μM; Fig. 6a and S8a lane 28 to 39). Thus, both the N-terminal region (residues 30 to 107) and the Nse4ML C-tail (residues 184 to 220) appeared to contribute to DNA interaction in our experimental setting. The Nse4 fragment containing bidirectional extensions from Nse4M (residues 30 to 220) showed the strongest DNA shifting ability (Kd ~2.85 μM; Fig. 6a and S8a, lanes 41 to 49). These data suggest that Nse4 may use multiple regions to affect dsDNA interaction in vitro. We note that DNA molecules were shifted to gel wells in most cases suggesting possible DNA-mediated aggregation. Also, the XlNse1-3-4 complex appears to differ from human Nse1-3-4 complex with HsNse4 (residues 92–212) that exhibited robust dsDNA binding ability [27].
Fig. 6.
DNA-binding affinities and cell viability test of various XlNse1-3-4 complex proteins.
(a) Quantification of bound DNA in the Nse1-3-4 complex using various constructs of Nse4 described in Fig. S8a. The amount of bound DNA in the presence of increasing amounts of protein was quantified using ImageJ [54]. Error bars were calculated from three independent experiments.
(b) Quantification of bound DNA in the Nse1-3-4 complex using mutants of Nse3 or Nse4 described in Fig. S8b.
(c) A model of DNA-bound Nse1 (orange)-Nse3 (green)-Nse4 (blue) complex. The N- and C-terminal regions (residues 30 to 104 and 187 to 211) of Nse4 are positioned near the DNA. All residues in Nse3 and Nse4 mutated for EMSA experiments are shown as yellow spheres.
(d) The nse4–8E mutant is inviable. (left) The nse4–8E mutant protein was expressed at wild-type levels. Diploid cells (2N) containing one copy of wild-type untagged Nse4 and another copy of Myc-tagged Nse4, either wild-type (WT) or mutated (−8E), were examined. Protein samples were prepared from asynchronous cultures. Ponceau S stain (Stain) was used for equal loading control. (right) Examples of tetrad analyses from indicated diploid strain. Spore clones were grown at 30 ºC for 2 days and genotyped.
(e) The nse4–6E mutation does not affect protein levels but leads to slow growth on dissection plate. Data are presented in as in panel (d).
(f) The nse4-H275E or –R257E, H275E mutant shows normal growth and genotoxic resistance. (left) Data are presented as the left panel in (d). (right) nse4-H275E or –R257E, H275E cells exhibit wild-type levels of growth at 30 ºC or 37 ºC. Both mutants were resistant to indicated genotoxins. 10-fold serial dilution of cells were spotted on plates and growth was assessed after incubation for 2 days.
Human and fission yeast Nse3 proteins have been reported to bind DNA and this binding relies on two or three basic residues [27]. We thus engineered a charge-inversion mutant of Nse3, in which the corresponding residues, Lys187 and Lys195 (HsNse3 Lys221 and Arg229), were replaced with Glu (Fig. 6b). When in complex with Nse1 and the longest Nse4 fragment tested (residues 30 to 220), this XlNse3 mutant displayed reduced DNA-binding affinity (Kd ~4.10 μM; Fig. 6b and S8b, lane 2 to 13) [27]. However, the mutant complex still behaved better than wild-type Nse1-Nse3 in complex with a shorter Nse4 fragment (residues 108 to 183; Fig. 6b), suggesting that the N-terminal region of Nse4 and Nse4ML C-tail (residues 30 to 107 and 184 to 220, respectively) may compensate for the effects of Nse3 mutations.
To identify residues in the N-terminal region of Nse4 and Nse4ML C-tail that may contribute to DNA binding, we mutated residues that are positively charged and conserved in metazoan. These include Arg33, Arg35, and Arg38 located N-terminal to the HTH region of Nse4. Another conserved residue Arg52 of XlNse4 is equivalent to the human cohesin kleisin residue Lys25, which can be crosslinked with DNA in the nucleotide-free fission yeast cohesin kleisin and is close to DNA (5.2–5.7 Å) in the ATP analog-bound structure (PDB 6YUF and 6WGE) [43, 44]. We thus mutated Arg33, Arg35, Arg38, and Arg52 to Glu (Fig. 6c, Nse4 N). The Nse1-3-4 complex with this quadruple Nse4 mutant exhibited a ~2-fold decrease in dsDNA binding affinity compared with the wild-type complex (Kd ~6.13 μM; Fig. 6b and S8b, lanes 15 to 26). We also mutated the positively charged residues at the Nse4ML C-tail and generated two triple mutants: C1 (Lys187, Lys198, and Lys211 mutated to Glu) and C2 (Arg192, Lys194, and Arg195 mutated to Glu; Fig. 6b–c). Both C1 and C2 mutants slightly decreased affinity to dsDNA, with C2 mutant exhibiting a stronger defect (Fig. 6b and S8b, lanes 28 to 39 and 41 to 52). When the C1 and C2 mutations were combined, there was a greater reduction in DNA-binding affinity, which was ~3-fold lower (Kd ~8.2 μM) than the wild-type complex (Fig. 6b and S8b, lanes 37–40).
Based on DNA-binding analyses, we used the ‘easy interface’ of the HADDOCK program [45] to model dsDNA in the XlNse1-3-4 complex. In our model, the N-terminal region (residues 30 to 104) of Nse4 and the Nse4ML C-tail (residues 187 to 211) locate near the DNA-binding path (Fig. 6c). The mutated residues in the N-terminal region (Arg33, Arg35, Arg38, and Arg52) are located close to dsDNA. All mutated residues located in the Nse4ML C-tail are also positioned near the H1A’ helix of Nse3, and present a positively charged surface together with the positively charged residues from the WHA domain of Nse3 that could provide a DNA-binding surface (Fig. S8c).
We also tested the effects of the Smc6 head domain (residues 85 to 299 and 925 to 1107) (Fig. S8d–e). Addition of Smc6 head region appeared to increase DNA association by the Nse1-3-4 complex containing either Nse4 residues 30–183 or residues 30–220 (Fig. 6a, S8d). The Nse1-3-4 complex containing single point mutations in the Nse4 N-terminal and Nse4ML C-tail did not appear to reduce DNA shifting ability in the presence of Smc6 (Fig. S8e). It is likely that positively charged residues from other parts of the Nse1-3-4 complex and Smc6 can provide sufficiently stable interaction with DNA in these situations. While increased amount of the Smc6-Nse1-3-4 complex reproducibly shifted DNA bands, the band intensity for the protein-bound DNA in the gel wells did not increase accordingly. Similar observations were made in other DNA binding tests described above. It is possible that DNA-protein oligomeric states trapped to the gel wells were not well detected.
In vivo examination of Nse4 residues implicated in DNA binding
We next addressed whether Nse4 residues implicated in DNA binding are important in cells. To this end, we attempted to mutate the corresponding residues in the budding yeast Saccharomyces cerevisiae (Sc) Nse4. Because XlNse4 and ScNse4 share only 18.7% sequence identity, we selected two N-terminal residues and six C-terminal residues among the conserved residues tested above or next to those residues (Fig. S4). We examined these eight residues together as well as in two separate groups.
First, we mutated all eight amino acids into glutamate. The resulting mutant, containing K49E, R65E, R251E, R256E, R257E, R258E, H275E, S276E, is referred to as nse4–8E hereafter. Given that Nse4 DNA binding is likely critical for the protein functions, and thereby could affect cell growth, we replaced only one copy of wild-type NSE4 gene with nse4–8E in a diploid strain. After meiosis, heterozygous diploid strains (nse4–8E/NSE4) gave rise to 2:2 segregation of viable: inviable spore clones. Genotyping of fourteen tetrads showed that all viable clones contained the wild-type copy of NSE4, suggesting that nse4–8E caused cell lethality (Fig. 6d). The lethality was not due to protein level changes as nse4–8E supported wild-type protein levels. We note that the apparent upper shift of the Nse4 protein band in the mutant is likely due to increased numbers of negatively charged residues. This data suggests that the analysed eight residues or a subset of them are essential for ScNse4 functions.
To further analyse the role of the eight residues of ScNse4, we examined the six located at NSe4ML C-tail using a similar strategy as described above. We found that the resulting mutant, referred to as nse4–6E (R251E, R256E, R257E, R258E, H275E, S276E), produced viable, but smaller, spore clones (Fig. 6e). This defect was not caused by protein level changes as the nse4–6E protein level was indistinguishable from that of wild-type Nse4 (Fig. 6e). That nse4-6E led to impaired growth suggests the importance of the tested residues in ScNse4. Given that a more severe defect was associated with nse4–8E compared with nse4–6E, we deduced that the N-terminal residues (Lys49 and Arg65) contribute to the ScNse4 functions. Collectively, our data suggest that the two sets of Nse4 residues located at its N-terminal and its ML C-tail are functionally important.
Further analyses showed that mutations affecting the Nse4 residues Arg257 and His275 did not lead to any noticeable growth defect at 30 ºC or 37 ºC, nor sensitivity to genotoxic agents (Fig. 6f). Thus, either the set of six residues mutated in nse4–6E are collectively important or the other four residues, namely Arg251, Arg256, Arg258, and Ser276, not examined separately, are responsible for the nse6–6E mutant phenotype. Regardless, our genetic data suggest that at least a subset of the ScNse4 residues examined here are critical for its functions in cells. This finding is line with the in vitro finding that the corresponding residues in XlNse4 can be involved in DNA binding.
Discussion
The Smc5/6 complex facilitates chromosome replication and repair. These roles can stem from its DNA binding, ATP-dependent activities, and sumoylation functions. Structural information required to comprehend the Smc5/6 complex’s roles is still lacking. In this work, we present the crystal structure of the XlNse1-3 dimer bound to a XlNse4 middle linker, showing that Nse1-3 stiffens the Nse4 middle linker. Integrating our data and previous CL-MS data, we present a model to explain how an extended Nse4 middle linker can engage with the Nse1-3 dimer. Moreover, our DNA binding and mutagenesis data suggest that two regions of Nse4 could contribute to DNA binding and are functionally important. Integrating our findings and those of other SMC complexes, we discuss possible roles of Nse1-3-4 in Smc5/6 complex interaction with DNA (below).
Our data suggest that Nse4 binding significantly alters the Nse1-Nse3 dimer structure and creates three binding interfaces with the latter. The first two interfaces are formed when two perpendicularly positioned helices at the N-terminal half of Nse4M are docked onto two distinct grooves formed by the Nse3’s WHB. The importance of these grooves is supported by previous mutational data [28, 31, 39]. Significantly, our structure suggests that the XlNse3 residues corresponding to previously identified HsNse3 sites that are mutated in LICS patients contribute to these groove structures, and thus Nse4 binding. Our work provides a structural explanation for how these disease-causing mutations could dislodge Nse4 from Nse1-3 and ultimately interfere with DNA binding by the Smc5/6 complex.
Structural comparison of the XlNse1-3-4 complex with three prokaryotic KITE-klesin complexes reveals that the WHA and WHB domains of KITE proteins are arranged in different orientations in these complexes (Fig. 4d–f and S6b). For instance, the space and geometry between the Nse1’s WHB and Nse3’s WHB domains differ from those of corresponding domains in the prokaryotic KITE subunits (Fig. 4 and S6). Such differences could affect the WHB grooves upon which kleisins bind and thus cause distinct kleisin conformations in the Smc5/6 complex verses prokaryotic SMC complexes. Nevertheless, kleisins in these structures do share common features: in all cases, the N-terminal segment of the kleisin middle region binds to Nse3 or the equivalent prokaryotic KITE proteins, whereas its C-terminal extended loop threads through the WHA and WHB of Nse1 or equivalent prokaryotic KITE subunits. These observations suggest that the structural basis for the KITE-driven compacting of kleisin is largely similar in prokaryotes and eukaryotes.
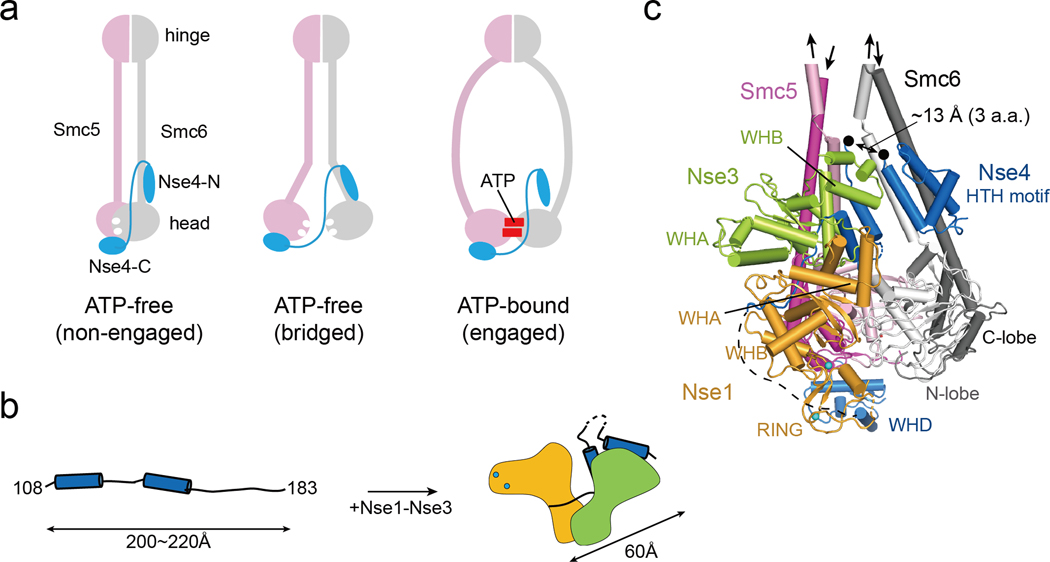
Our structure data can provide a molecular explanation for possible nucleotide-dependent opening of the Nse4N gate (Fig. 7a). Analogous to the proposal that ScpB regulates the binding of ScpA to the SMC neck by constraining ScpA [46], a compact conformation of the middle region of Nse4 seen in our structure suggests that Nse1-3 may also perform a similar role. This idea is consistent with a previous proposal based on studying of Nse4 interaction with Nse1-3 [28]. Our structural data further suggests that Nse4M can fold into the Z-shaped compact form with ~60 Å length, while its extended form could reach a maximum length between 200 and 220 Å (Fig. 7b). If both conformations exist, they could provide plasticity required for potential conformation changes of Smc5 and Smc6 during their ATPase cycles. By integrating our Nse1-3-4 structure, CLMSNSE, and modelling Smc5 and 6 head regions based on condensin head structures [47], we generated a model placing the Nse1-3-4 complex relative to the Smc5/6 heads (Fig 7c). We note that since the Nse5–6 complex is proposed to contact the Smc5 and 6 head regions and the Nse1-3-4 complex, they may affect the overall arrangement of the Smc5/6 complex [36, 48–50]
Fig. 7.
A model of Nse1-3-4 engagement with the Smc5/6 head regions
(a) Three possible configurations for Nse4 engagement with the Smc5/6 head regions according to predominant models of SMC complexes. In the absence of ATP, Smc5/6 head regions can adopt the ‘non-engaged’ or ‘bridged’ state. In both states, the head domains are bridged by Nse4 through two interfaces (Smc6-Nse4-N and Smc5-Nse4-C). In ATP-bound state, the Smc6-Nse4-N interface could open.
(b) A cartoon representation of the folding of Nse4 promoted by the Nse1-Nse3 complex. The Nse4M in the absence of Nse1-Nse3 could adopts an extended, string-like structure with a maximum length of 200−220 Å. Nse1-Nse3 shapes Nse4M to a compact (length ~60 Å) form.
(c) Modeling of the Smc5/6 head regions with Nse1-3-4 (details in Fig. S9). In the ATP-free state, the head domains of Smc5 (pink for the N-lobe and magenta for the C-lobe) and Smc6 (white for the N-lobe and gray for the C-lobe) are not engaged, and the coiled-coils of Smc5 and Smc6 are paired. The Nse4 HTH motif and WH domain interact with the neck of Smc6 and the head of Smc5, respectively. A black arrow links the HTH and the Nse4M. Two black dots indicate residues 104 and 108 of Nse4, and the missing three residues between the two dots are expected to span ~13 Å, if we assume that they are fully extended.
Studies of other types of SMC complexes suggest ATP-dependent topological entrapment of DNA in cells. While the Smc5/6 complex has been shown to topological binding and compact DNA molecules in vitro, how it can topologically entrap DNA remain unclear [13, 48, 49]. Recent structural data suggest that the Smc5/6 complex forms straight rod-shaped conformation, distinct from the arm-bent structure of cohesin and condensin [48, 50]. As arm-bent SMC structure has been suggested to be critical for DNA entrapment, these data raise the possibilities that DNA interactions by the Smc5/6 complex likely have differences from cohesin and condensin.
Our data suggest that Nse4 could contribute to DNA binding using its N-terminal and ML C-tail regions (Fig. 6). The Nse4 N-terminal positively charged residues are partly similar to those in the klesin subunit (Rad21) of cohesin (Fig. S10a). For example, in HsRad21 and SpRad21, Lys10 (similar to Arg38 of XlNse4) and Lys25 (similar to Arg52 in XlNse4) are close to DNA (Fig. S10b, c) [43, 44]. Though deleting N-terminal 17 residues of Rad21 does not affect DNA binding, it impairs topological loading of cohesin onto DNA, suggesting that this region is critical for guiding DNA to into the cohesin structure. It remains to be tested whether the Nse4 N-terminal region plays a similar role. In addition, it is worthwhile to investigate whether DNA could be clamped by the Nse1-3-4 complex on the top and by the Smc5/6 head regions at the bottom. Structural studies for the DNA-bound Smc5/6 complex are required to understand how this complex interact with DNA in various states.
Materials and Methods
Protein expression and purification
Genes encoding XlNse1 (residues 3 to 248), XlNse3 (residues 45 to 260) and XlNse4 (residues 30 to 183, 108 to 183, 108 to 220, and 108 to 236) were amplified by polymerase chain reaction (PCR) and inserted into pCDF-duet (MCS2), pET-duet (MCS2), and pET28a, respectively. The Escherichia coli (Ec) Rosetta (DE3) strain containing the three plasmids was grown in LB media containing 0.4 mM ZnSO4, and overexpression was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18°C overnight. The His-tagged XlNse1-3-4 complex was initially purified by Ni2+-NTA affinity chromatography (GE Healthcare, Chicago, Illinois, U. S. A.). The His-tag was removed by thrombin during dialysis against 25mM Tris-HCl pH 7.4, 200 mM NaCl, 5 mM β-mercaptoethanol, and 5% (v/v) glycerol. The protein complex was further purified using a HiTrapQ ion-exchange column (GE Healthcare) followed by gel filtration on a HiLoad 16/600 Superdex 200 column (GE Healthcare) equilibrated with 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM dithiothreitol (DTT), and 5% (v/v) glycerol. The purified protein was concentrated to 26 mg/ml and used for crystallization.
For electrophoretic mobility shift assay (EMSA), XlSmc6 N- (residues 85 to 299) and C-terminal regions (residues 925 to 1107) were linked by GSGSG and inserted into the pET-duet (MCS1) vector, which contained the gene for XlNse3 (residues 45 to 260) in its MCS2 site. The plasmid containing XlSmc6 and XlNse3 was co-expressed with XlNse1 (residues 3 to 248) and XlNse4 (residues 30 to 183 or 30 to 220) in Ec Rosetta (DE3) cells under the same conditions as the XlNse1-3-4 complex. His-tagged XlSmc6-Nse1-3-4 was first purified by Ni2+-NTA affinity chromatography (GE Healthcare) followed by gel filtration on a HiLoad 16/600 Superdex 200 column (GE Healthcare) equilibrated with 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM DTT, and 5% (v/v) glycerol. The protein complex was concentrated to 2–5 mg/ml and used for EMSA. All mutant proteins were prepared using the same protocol employed for the wild-type protein.
Crystallization and X-ray diffraction data collection
Crystals of the XlNse1 (residues 3 to 248)-Nse3 (residues 45 to 260)-Nse4 (residues 108 to 183) complex were grown from buffer containing 22% (w/v) polyethylene glycol (PEG) 3350, 0.1 M Bis-Tris pH 6.5, and 0.2 M ammonium acetate by the hanging-drop vapor diffusion method at 18°C. Diffraction data were collected at −170°C using crystals frozen in crystallization buffer containing 25% (v/v) glycerol. Diffraction data were collected at the Pohang Accelerator Laboratory (PAL, Beamline 11C), and data integration and scaling were performed using the HKL2000 package [51].
Crystals of both the N- and C-terminally extended XlNse4 (residues 30 to 183, 108 to 220, and 108 to 236) complexed with Nse1 (residues 3 to 248)-Nse3 (residues 45 to 260) were grown in buffer containing 23% (w/v) PEG 3350, 0.1 M Tris-HCl pH 8.0, by the hanging-drop vapor diffusion method at 18°C. Diffraction data were collected from two XlNse1-Nse3-Nse4 (residues 108 to 220 and 108 to 236) crystals using a Rigaku detector (Tokyo, Japan). Data integration and scaling were performed using iMosflm [52]. Diffraction data of XlNse1-Nse3-Nse4 (residues 30 to 183) crystal at −170°C using crystals frozen in crystallization buffer containing 25% (v/v) glycerol. Diffraction data were collected at the PAL (Beamline 11C), and data integration and scaling were performed using the HKL2000 package [51].
Structure determination and refinement
The structure of the XlNse1-3-4 complex was determined by the molecular replacement method. The human Nse1-Nse3 structures (PDB 3NW0, 5HVQ) were used as search models [29, 35]. Human Nse1 was divided into WHA, WHB, and RING domains, and human Nse3 was divided into WHA and WHB domains for searching. We put all 5 ensembles at the same time for phase calculation and initial phases were obtained using the program PHASER [53]. After density modification, an electron density map generated at a resolution of 1.7 Å using the PHENIX program clearly revealed Nse1, Nse3, and Nse4 [53]. Successive rounds of model building using COOT [54] and refinement using PHENIX were performed to build the complete model. The final statistics are summarized in Table 1. For structure determination of XlNse1 (residues 3 to 248) – Nse3 (residues 45 to 260) – Nse4 (residues 108 to 236) complex, we used the XlNse1-Nse3-Nse4 (residues 108 to 183) structure as a search model for the molecular replacement method. The phase calculation and initial phase was obtained using the program PHASER.
Electrophoretic mobility shift assay (EMSA)
A 5’-Fluorescein phosphoramidite (FAM)-labelled 37 bp dsDNA with a 2 nt 5’ overhang was prepared by annealing 5’-FAM-labelled and non-modified oligonucleotides purchased from Bionner (Daejeon, South Korea). For EMSA, the FAM-labelled 37 bp dsDNA was incubated with increasing amounts of proteins in 5 μl reaction buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM DTT, 5% glycerol) on ice for 30 min. Reaction samples were resolved by 10% native polyacrylamide gel electrophoresis (PAGE) in 0.5× TBE buffer. Gels were scanned using a Typhoon-9400 instrument (GE Healthcare) and quantified by ImageJ [55]. Protein-DNA complex ratios were calculated by dividing the intensity of the complex by total DNA. Statistical parameters were derived from triplicate experiments. To determine the dissociation constants (Kd), experimental data were analyzed using specific binding with Hill-slope (non-linear regression) in Prism 9 (GraphPad Software, La Jolla, U. S. A.).
Modelling and docking
For DNA docking, duplex B-DNA was generated from 3DNA [56]. DNA was then docked into the crystal structure of the XlNse1-3-4 complex using the ‘Easy Interface’ of the HADDOCK web docking server [45]. The ‘active residues’ were Lys133 of Nse1, and Lys187 and Lys195 of Nse3, which are DNA-binding residues [27]. The ‘passive residues’ were defined automatically around the active residues. The best structure, with a HADDOCK score of −113.1 +/− 7.6, a cluster size of 29, and a Z-score of −1.3, was aligned to Nse3 of Nse1-Nse3-extended Nse4 model (Fig. 5 and S7b) and shown in Figure 6c and S8c.
Yeast strains and genetic techniques
Standard procedures were used for cell growth and media preparation. Strains used are provided in Table S1 and are isogenic to W1588–4C, a RAD5 derivative of W303 (MATa ade2–1 can1–100 ura3–1 his3–11,15, leu2–3, 112 trp1–1 rad5–535), Replacement wild-type NSE4 allele with nse4 mutant alleles was done using standard PCR-based gene replacement method. All alleles were verified by sequencing. Standard procedures were used for tetrad analyses and spotting assays. At least two biological duplicates were used for each genotype.
Supplementary Material
Highlights.
-
-
The crystal structure of the Nse1-Nse3-Nse4 complex was determined
-
-
The Nse1-Nse3 closed structure forces Nse4 into a Z-shaped conformation
-
-
Nse1-Nse3 drives Nse4 folding, opening the SMC6-Nse4 gate in the engaged state
-
-
The Nse1-Nse3-Nse4 structure explains the lung disease immunodeficiency and chromosome breakage syndrome-causing mutations
-
-
Nse4 and Nse3 interact with DNA, facilitating its entry into the SMC5/6 complex
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korea government (MEST, No. 2018R1A2A1A19021035, 2017M3A9F6029733 and 2017M3A9F6029736), BK21 program (Ministry of Education) to YC and National Institute of General Medical Science (R01GM080670) to XZ. The authors declare that none of the authors have a financial interest related to this work.
Abbreviations
- SMC
structural maintenance of chromosomes
- NSE
Non-SMC elements
- HAWK
HEAT proteins associated with kleisin
- KITE
kleisin-interacting tandem winged-helix elements
- WH
winged helix
- HTH
helix-turn-helix
- MAGEs
melanoma-associated antigens
- LICS
lung disease immunodeficiency and chromosome breakage syndrome
- Nse4M
Nse4 middle domain
- CL-MS
crosslink and mass spectrometry
- Y2H
yeast two-hybrid analyses
Footnotes
Data Availability
Atomic coordinates and the structure factors for the Nse1-3-4 structure has been deposited in the PDB Data Base under accession code 7DG2.
Supplementary Data
Supplementary Data are available at JMB online: Supplementary Figures 1–10 and Supplementary table 1.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Uhlmann F, (2016). SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol, 17, 399–412. [DOI] [PubMed] [Google Scholar]
- 2.Michaelis C, Ciosk R, Nasmyth K, (1997). Cohesin: chromosomal proteins that prevent premature separation of sister chromatids. Cell, 91, 35–45. [DOI] [PubMed] [Google Scholar]
- 3.Strunnikov AV, Hogan E, Koshland D, (1995). SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev., 9, 587–599. [DOI] [PubMed] [Google Scholar]
- 4.Palecek JJ, (2018). SMC5/6: Multifunctional player in replication. Genes (Basel), 10, E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haering CH, Löwe J, Hochwagen A, Nasmyth K, (2002). Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell, 9, 773–788. [DOI] [PubMed] [Google Scholar]
- 6.Melby TE, Ciampaglio CN, Briscoe G, Erickson HP, (1998). The symmetrical structure of structural maintenance of chromosome (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol, 142, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bürmann F, Shin HC, Basquin J, Soh YM, Giménez-Oya V, Kim YG, et al. , (2013). An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat. Struct. Mol. Biol, 20, 371–379. [DOI] [PubMed] [Google Scholar]
- 8.Gligoris TG, Scheinost JC, Bürmann F, Petela N, Chan KL, Uluocak P, et al. , (2014). Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science, 346, 963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J, (2004). Structure and stability of cohesin’s Smc1-kleisin interaction. Mol. Cell, 15, 951–964. [DOI] [PubMed] [Google Scholar]
- 10.Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR, (2006). The SMC5–6 DNA repair complex: bridging of the SMC5–6 heads by the kleisin, NSE4, and non-kleisin subunits. J. Biol. Chem, 281, 36952–36959. [DOI] [PubMed] [Google Scholar]
- 11.Cuylen S, Metz J, Haering CH, (2011). Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol, 18, 894–901. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm L, Bürmann F, Minnen A, Shin HC, Toseland CP, Oh BH, et al. , (2015). SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife. 4, e06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanno T, Berta DG, Sjögren C, (2015). The Smc5/6 complex is an ATP-dependent intermolecular DNA linker. Cell Rep., 12, 1471–1482. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Brandão HB, Le TBK, Laub MT, Rudner DZ, (2017). Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science, 355, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, (2018). Real-time imaging of DNA loop extrusion by condensin. Science, 360, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sergeant J, Taylor E, Palecek J, Fousteri M, Andrews EA, Sweeney S, et al. , (2005). Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5–6) complex. Mol. Cell. Biol, 25, 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H, (2009). Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol. Cell, 35, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Räschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, et al. , (2015). DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science, 348, 1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Blobel G, (2005). A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A, 102, 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan B, Wu J, Meng X, Lei M, Zhao X, (2019). Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107. Mol. cell, 75, 238–251.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oravcová M, Gadaleta MC, Nie M, Reubens MC, Limbo O, Russell P, et al. , (2019). Brc1 Promotes the Focal Accumulation and SUMO Ligase Activity of Smc5-Smc6 during Replication Stress. Mol. Cell. Biol, 39, e00271–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palecek JJ, Gruber S, (2015). Kite proteins: a superfamily of SMC/kleisin partners conserved across bacteria, archaea, and eukaryotes. Structure, 23, 2183–2190. [DOI] [PubMed] [Google Scholar]
- 23.Wells JN, Gligoris TG, Nasmyth KA, Marsh JA, (2017). Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr biol., 27, R17–R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murayama Y, Uhlmann F, (2015). DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell, 163, 1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kschonsak M, Merkel F, Bisht S, Metz J, Rybin V, Hassler M, et al. , (2017). Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell, 171, 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Muir KW, Bowler MW, Metz J, Haering CH, Panne D, (2018). Structural basis for Scc3-dependent cohesin recruitment to chromatin. Elife, 7, e38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zabrady K, Adamus M, Vondrova L, Liao C, Skoupilova H, Novakova M, et al. , (2016). Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res., 44, 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vondrova L, Kolesar P, Adamus M, Nociar M, Oliver AW, Palecek JJ, (2020). A role of the Nse4 kleisin and Nse1/Nse3 KITE subunits in the ATPase cycle of SMC5/6. Sci. Rep, 10, 9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle JM, Gao J, Wang J, Yang M, Potts PR, (2010). MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell, 39, 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pebernard S, Perry JJ, Tainer JA, & Boddy MN, (2008). Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol biol Cell, 19, 4099–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson JJR, Bednarova K, Kozakova L, Liao C, Guerineau M, Colnaghi R, et al. , (2011). Interactions between the Nse3 and Nse4 components of the SMC5–6 complex identify evolutionarily conserved interactions between MAGE and EID families. PLoS One, 6, e17270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker P, Salehi A, (2002). The MAGE proteins: Emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci Res, 67, 705–712. [DOI] [PubMed] [Google Scholar]
- 33.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S, (2001). An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res., 61, 5544–5551. [PubMed] [Google Scholar]
- 34.Van der Crabben SN, Hennus MP, McGregor GA, Ritter DI, Nagamani SC, Wells OS, et al. , (2016). Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease. J. Clin. Investig 126, 2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman JA, Cooper CD, Roos AK, Aitkenhead H, Oppermann UC, Cho HJ, et al. , (2016). Structures of two melanoma-associated antigens suggest allosteric regulation of effector binding. PLoS One, 11, e0148762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adamus M, Lelkes E, Potesil D, Ganji SR, Kolesar P, Zabrady K, et al. , (2020). Molecular insights into the Architecture of the human SMC5/6 complex. J. Mol. Biol, 432, 3820–3837. [DOI] [PubMed] [Google Scholar]
- 37.Pei J, Kim BH, Grishin NV, (2008). PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res., 36, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones DT, (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol, 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 39.Guerineau M, Kriz Z, Kozakova L, Bednarova K, Janos P, Palecek JJ, (2012). Analysis of the Nse3/MAGE-binding domain of the Nse4/EID family proteins. PLoS One, 7, e35813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamada K, Miyata M, Hirano T, (2013). Molecular basis of SMC ATPase activation: role of internal structural changes of the regulatory subcomplex ScpAB. Structure, 21, 581–594. [DOI] [PubMed] [Google Scholar]
- 41.Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, et al. , (2009). Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell, 136, 85–96. [DOI] [PubMed] [Google Scholar]
- 42.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc, 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higashi TL, Eickhoff P, Sousa JS, Locke J, Nans A, Flynn HR, et al. , (2020). A structure-based mechanism for DNA entry into the cohesin ring. Mol. Cell, 79, 917–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Z, Gao H, Bai XC, Yu H, (2020). Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science, 368, 1454–1459. [DOI] [PubMed] [Google Scholar]
- 45.de Vries SJ, van Dijk M, Bonvin AMJJ, (2010). The HADDOCK web server for data-driven biomolecular docking. Nat Protoc., 5, 883–897. [DOI] [PubMed] [Google Scholar]
- 46.Kamada K, Su’etsugu M, Takada H, Miyata M, Hirano T, (2017). Overall shapes of the SMC-ScpAB complex are determined by balance between constraint and relaxation of its structural parts. Structure, 25, 603–616. [DOI] [PubMed] [Google Scholar]
- 47.Lee BG, Merkel F, Allegretti M, Hassler M, Cawood C, Lecomte L, et al. , (2020). Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat. Struct. Mol. Biol, 27, 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serrano D, Cordero G, Kawamura R, Sverzhinsky A, Sarker M, Roy S, et al. , (2020). The Smc5/6 Core Complex Is a Structure-Specific DNA Binding and Compacting Machine. Mol. Cell, 80, 1025–1038.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez-Escribano P, Hormeño S, Madariaga-Marcos J, Solé-Soler R, O’Reilly FJ, Morris K, et al. , (2020). Purified Smc5/6 Complex Exhibits DNA Substrate Recognition and Compaction. Mol. Cell, 80, 1039–1054.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y, Li S, Ser Z, Sanyal T, Choi K, Wan B, et al. , (2020). Integrative analysis reveals unique features of the Smc5/6 complex. bioRxiv, 424863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 52.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG, (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta crystallogr. D Biol. Crystallogr, 67, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. , (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallogr. D Biol. Crystallogr, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta crystallogr. D Biol. Crystallogr, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 55.Schneider CA, Rasband WS, Eliceiri KW, (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng G, Lu XJ, Olson WK, (2009). Web 3DNA– a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res., 37 (Web Server issue), W240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.