Abstract
This study was conducted with the aim to the clustering of provinces in Indonesia of the risk of the COVID-19 pandemic based on coronavirus disease 2019 (COVID-19) data. This clustering was based on the data obtained from the Indonesian COVID-19 Task Force (SATGAS COVID-19) on 19 April 2020. Provinces in Indonesia were grouped based on the data of confirmed, death, and recovered cases of COVID-19. This was performed using the K-Means Clustering method. Clustering generated 3 provincial groups. The results of the provincial clustering are expected to provide input to the government in making policies related to restrictions on community activities or other policies in overcoming the spread of COVID-19. Provincial Clustering based on the COVID-19 cases in Indonesia is an attempt to determine the closeness or similarity of a province based on confirmed, recovered, and death cases. Based on the results of this study, there are 3 clusters of provinces.
Keywords: COVID-19, Clustering, K-means clustering
Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease that is currently circulating around the world (Ahmar and Rusli 2020; Atuahene et al. 2020; Gupta et al. 2020). COVID-19 was first reported in the city of Wuhan, Hubei Province, China in December 2019. COVID-19 is an infectious disease caused by a newly discovered coronavirus—severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—which was first identified in Wuhan (Ahmar and Boj 2020; Azarafza et al. 2021). The first COVID-19 cases in Indonesia were detected on 2 March 2020 in Jakarta. Over time, the pandemic has spread to various provinces in Indonesia. As of 19 April 2020, more than 6575 cases of COVID-19 have been reported in 34 provinces in Indonesia. On April 19, 2020, 6,575 confirmed were cases, 686 recovered while 582 died in Indonesia. Based on COVID-19 data from the Worldometer, last updated: 20 April 2020, 07:53 GMT, Indonesia has the highest confirmed cases of COVID-19 among the Association of Southeast Asian Nations (ASEAN) member states (Worldometer 2020).
Evaluation of the development of COVID-19 cases per province is one of the bases for monitoring the development of COVID-19 cases in Indonesia. However, to date there has been no provincial grouping based on confirmed cases, recoveries, and deaths conducted on this data. The K-means clustering algorithm is a popular unsupervised technique used to identify similarities between objects based on distance vectors suitable for small data sets (Sreedhar et al. 2017). This technique by definition is a kind of cluster algorithm, and has several advantages including briefness, efficiency and celerity (Li and Haiyan 2012). Meanwhile, the purpose of cluster analysis are (1) investigate underlying structure of data, (2) classification: to determine the degree of similarity among data points and (3) compression: a method for organizing and summarizing data into understandable groups (Govender and Sivakumar 2020).
Armstrong, et.al. (2012) said that the K-means algorithm was helpful in segmenting a heterogeneous recovery client population into more homogeneous subgroups and K-means offers a better view of applicant characteristics and needs, which may lead to more targeted rehabilitation options for people in home care. This is in line with Kusrini (2015) that K-means clustering is used since the number of clusters needed for item categorization has already been determined and in addition, Fotouhi & Montazeri-Gh (2013) said that K-means clustering needs less computing than the SAPM process, which benefits the method's capability for accurate traffic grouping. Furthermore, Al-Wakeel and Wu (2016) show that for strongly correlated load profiles, a limited number of clusters is suggested.
By using data mining methods such as the K-means clustering, it is possible to find the main characteristics of each potential province which can be used in an effort to predict future COVID-19 cases based on the provincial data similarity.
Methods and Statistical Analysis
This study was conducted using data obtained on 19 April 2020 from the Indonesian COVID-19 Acceleration Task Force website (https://covid19.go.id/peta-sebaran). Data were analyzed using the K-Means Clustering method as a technique for performing data groupings. Furthermore, the data classification procedure was based on the degree of each component’s membership (Ahmar et.al., 2018). This analysis was performed by using R Software version 3.6.3. as described on the website (https://uc-r.github.io/) and this study, we using R Software version 3.6.3.
The research steps were carried out as follows:
Data on the confirmed, recovered, and death cases were obtained from the Indonesia COVID-19 website (https://covid19.go.id/peta-sebaran).
This data were extracted into 3 parts which include the confirmed, recovered, and death according to the different provinces.
When there is a predominant data compared to others then, that particular set is made into 1 group and excluded from the analysis process.
The following packages were Installed and executed; tidyverse (version 1.3.0), cluster (version 2.1.0), and factoextra (version 1.0.7) of R Software version 3.6.3.
-
Data obtained in stage 2 were further loaded on the R Software.
library("readxl")
data <- read_excel("C:\\datacovid19indonesia.xlsx")
- Data Preparation:
- Rows are observations, columns are variables.
-
Any missing values of the data are deleted or estimated.To remove any missing value that might be present in the data, type this:data <- na.omit(data)
-
The data were standardized (i.e. scaled) in order to make variables comparable.To scale/standardize data using the R function scale:data <- scale(data)head(data)
-
Clustering distances measurement was carried out using Euclidean distances.
euclidean <- get_dist(data)
fviz_dist(euclidean, gradient = list(low = "#00AFBB", mid = "white", high = "#FC4E07"))
- The K-means analysis process can be described as follows:
-
Determinine the number of clusters (k) using optimal clusters. The three (3) most popularly used optimal clusters, include:
-
Elbow methodset.seed(123)fviz_nbclust(data, kmeans, method = "wss")
-
Silhouette methodset.seed(123)fviz_nbclust(data, kmeans, method = "silhouette")
-
Gap statisticset.seed(123)fviz_gap_stat(gap_stat)
The optimal cluster is seen from the fviz_nbclust function of each method. Furthermore, the optimal cluster value in the Elbow Method is the k value which drops drastically on the visualization graph meanwhile, in the Silhoute and Gap statistics, it appears automatically on the graph. -
-
Extracting resultsBased on the optimal cluster method approach in the previous step, the optimal cluster will be obtained. The number of clusters was used to calculate the k-means clustering value.For example, in the previous stage, the value of k = 2 was obtained.set.seed(123)endkmeans <- kmeans(data, 2, nstart = 25)print(endkmeans)Based on these results, k-means clustering results will be obtained. This result can be visualized using the code:fviz_cluster(endkmeans, data = data)
-
Result and Discussion
Based on the descriptive statistical analysis (Table 1) out of the 34 provinces in Indonesia, the maximum confirmed cases were 3032 with 234 recovered and 287 death cases meanwhile, there were provinces without recovered and deaths cases. On average, the number of confirmed cases was 193 with a standard deviation of 528.
Table 1.
Descriptive statistics of COVID-19 in Indonesia
| Variable | Observations | Obs. with missing data | Obs. without missing data | Minimum | Maximum | Mean | Std. deviation |
|---|---|---|---|---|---|---|---|
| Confirmed | 34 | 0 | 34 | 1 | 3032 | 193 | 528 |
| Recovered | 34 | 0 | 34 | 0 | 234 | 20 | 43 |
| Deaths | 34 | 0 | 34 | 0 | 287 | 17 | 50 |
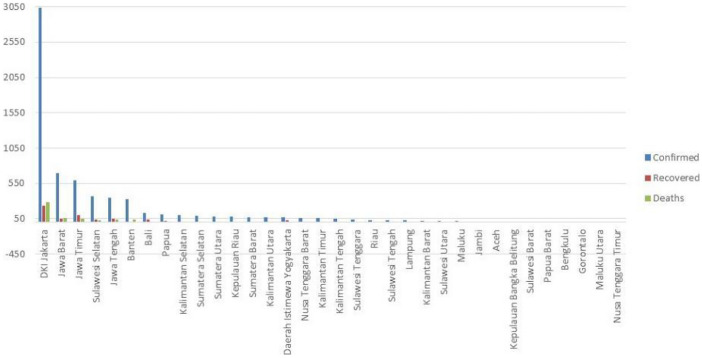
In Fig. 1, Jakarta obviously had more cases hence, the province became the epicenter of data center therefore, Jakarta formed one special group and was not included in the data clustering process (Pamula et al. 2011). Epicentrum is based in Jakarta because it is the capital of the country and the center of the economy in Indonesia.
Fig. 1.
Number of COVID-19 cases each Province in Indonesia
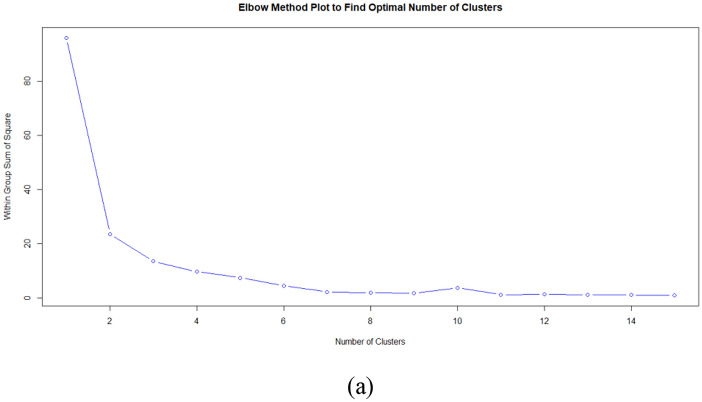
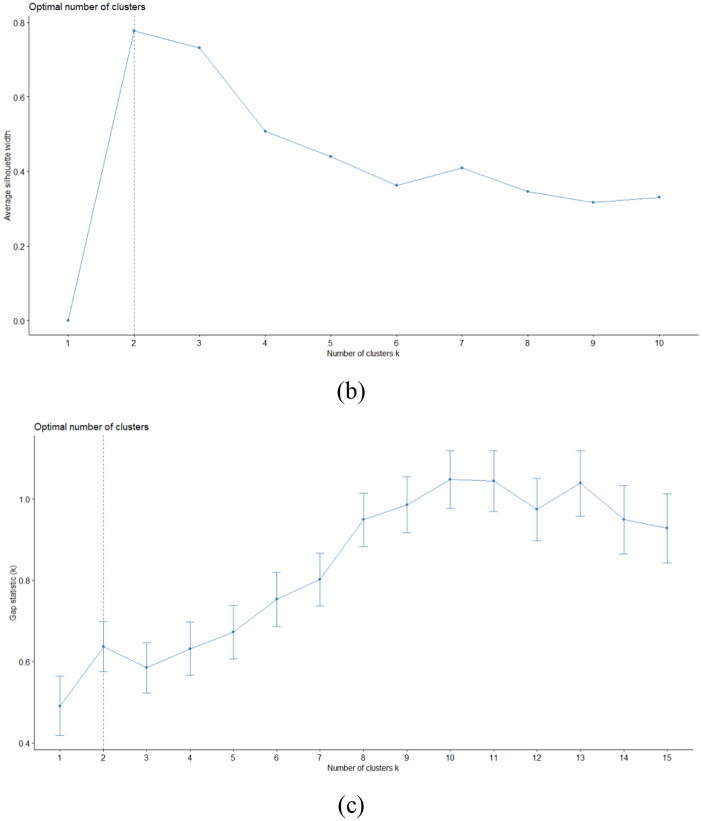
Moreover, the optimal number of k groups was determined using the three(3) most commonly used approaches namely Elbow, Silhouette, and Gap Statistics. The results can be seen in Fig. 2a, b, and c.
Fig. 2.
Result of a Elbow, b Silhouette, and c Gap Statistic to find k optimal
Based on Fig. 2, the Elbow method obtained optimal k at k = 2, the Silhouette method obtained many optimal clusters at k = 2, and the Gap statistics obtained optimal k value to form clusters at k = 2.
Therefore, based on the results from these methods, it can be concluded that the optimal k value to form a cluster is 2. Furthermore, the Clustering analysis results using K-means with k = 2 are presented in Table 2.
Table 2.
Results of provincial clustering in Indonesia with K-Means clustering*
| Province | Cluster |
|---|---|
| Jawa Barat | 1 |
| Jawa Timur | 1 |
| Sulawesi Selatan | 1 |
| Jawa Tengah | 1 |
| Banten | 1 |
| Bali | 2 |
| Papua | 2 |
| Kalimantan Selatan | 2 |
| Sumatera Selatan | 2 |
| Sumatera Utara | 2 |
| Kepulauan Riau | 2 |
| Sumatera Barat | 2 |
| Kalimantan Utara | 2 |
| Daerah Istimewa Yogyakarta | 2 |
| Nusa Tenggara Barat | 2 |
| Kalimantan Timur | 2 |
| Kalimantan Tengah | 2 |
| Sulawesi Tenggara | 2 |
| Riau | 2 |
| Sulawesi Tengah | 2 |
| Lampung | 2 |
| Kalimantan Barat | 2 |
| Sulawesi Utara | 2 |
| Maluku | 2 |
| Jambi | 2 |
| Aceh | 2 |
| Kepulauan Bangka Belitung | 2 |
| Sulawesi Barat | 2 |
| Papua Barat | 2 |
| Bengkulu | 2 |
| Gorontalo | 2 |
| Maluku Utara | 2 |
| Nusa Tenggara Timur | 2 |
*Does not include Province of DKI Jakarta
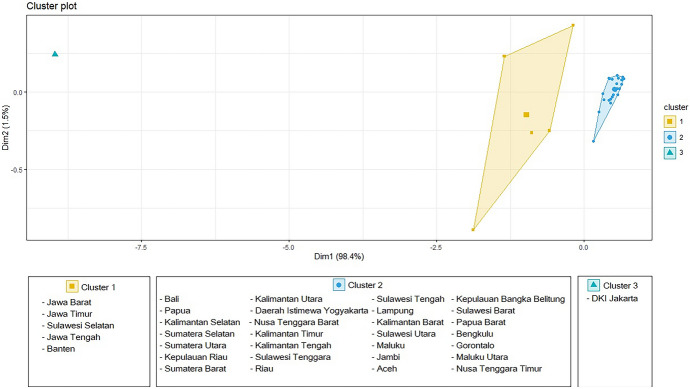
As shown in Table 2, it can be seen that Cluster 1 consists of 5 provinces and Cluster 2 consists of 28 provinces. When combined with the DKI Jakarta Cluster, there will be 3 provincial clusters in Indonesia based on COVID-19 data (Fig. 3).
Fig. 3.
Results of Provincial Clustering in Indonesia with K-Means Clustering
This study is consistent with Zarikas, et.al. (2020), which showed that clustering active cases in a region is useful for drawing conclusions about the disease impact which spreads rapidly in an area. Furthermore, Azarafza et al. (2021) stated that the pattern of transmitting infection between provinces was estimated using the clustering method. Therefore, based on these opinions, it can be concluded that by conducting provincial clusters, one is being provided with an overview of disease spread patterns and solutions related to this distribution pattern.
Conclusion
Provincial grouping/clustering based on the COVID-19 cases in Indonesia is an attempt to determine the closeness or similarity of a province based on confirmed cases, recovered cases, and deaths cases. Based on the results of this study, there are 3 clusters of provinces, each consisting: Clusters 1 (Jawa Barat, Jawa Timur, Sulawesi Selatan, Jawa Tengah); Cluster 2 (Bali,Papua, Kalimantan Selatan, Sumatera Selatan, Sumatera Utara, Kepulauan Riau, Sumatera Barat, Kalimantan Utara, Daerah Istimewa Yogyakarta, Nusa Tenggara Barat, Kalimantan Timur, Kalimantan Tengah, Sulawesi Tenggara, Riau, Sulawesi Tengah, Lampung, Kalimantan Barat, Sulawesi Utara, Maluku, Jambi, Aceh, Kepulauan Bangka Belitung, Sulawesi Barat, Papua Barat, Bengkulu, Gorontalo, Maluku Utara, Nusa Tenggara Timur); and Cluster 3 (DKI Jakarta). The results of the provincial cluster are expected to provide input to the government in making policies related to restrictions on community activities or other policies in overcoming the spread of COVID-19.
Acknowledgements
The authors would like to answer the referees for their helpful suggestions.
Author contributions
Authors contributed to the manuscript equally.
Funding
The author declares that there is no funding for this study.
Data availability
The data in this study can be accessed at: Website of COVID-19 of Indonesia (https://covid19.go.id/peta-sebaran) or Harvard Dataverse (https://doi.org/10.7910/DVN/JUSYYX).
Declarations
Conflict of interest
The author declares that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dahlan Abdullah, Email: dahlan@unimal.ac.id.
Ansari Saleh Ahmar, Email: ansarisaleh@unm.ac.id.
R. Rusli, Email: rusli.siman@unm.ac.id
Rahmat Hidayat, Email: rahmat@pnp.ac.id.
References
- Ahmar AS, Boj E. The date predicted 200.000 cases of covid-19 in Spain. J. App. Sci. Eng. Technol. Educ. 2020;2(2):188–193. doi: 10.35877/454RI.asci22102. [DOI] [Google Scholar]
- Ahmar AS, Rusli R. Will covid-19 cases in the world reach 4 million a forecasting approach using SutteARIMA. JOIV: Int. J. Inform. Vis. 2020;4(3):159–161. doi: 10.30630/joiv.4.3.389. [DOI] [Google Scholar]
- Ahmar A. S., Napitupulu D., Rahim R., Hidayat R., Sonatha Y., Azmi M. Using k-means clustering to cluster provinces in Indonesia. J. Phys: Conf. Ser. 2018;1028:012006. [Google Scholar]
- Al-Wakeel A., Wu J. K-means based cluster analysis of residential smart meter measurements. Energy Procedia. 2016;88:754–760. doi: 10.1016/j.egypro.2016.06.066. [DOI] [Google Scholar]
- Armstrong JJ, Zhu M, Hirdes JP, Stolee P. K-Means cluster analysis of rehabilitation service users in the home health care system of ontario: examining the heterogeneity of a complex geriatric population. Arch. Phys. Med. Rehabil. 2012;93(12):2198–2205. doi: 10.1016/j.apmr.2012.05.026. [DOI] [PubMed] [Google Scholar]
- Atuahene S, Kong Y, Bentum-Micah G. Covid-19 pandemic, economic loses and education sector management. Quant. Econ. Manag. Stud. 2020;1(2):103–109. doi: 10.35877/454RI.qems162. [DOI] [Google Scholar]
- Azarafza M, Azarafza M, Akgun H. Clustering method for spread pattern analysis of coronavirus (covid-19) infection in iran. J. Appl. Sci. Eng. Technol. Educ. 2021;3(1):1–6. doi: 10.35877/454RI.asci31109. [DOI] [Google Scholar]
- Fotouhi A, Montazeri-Gh M. Tehran driving cycle development using the k-means clustering method. Scientia Iranica. 2013;20(2):286–293. [Google Scholar]
- Govender P, Sivakumar V. Application of k-means and hierarchical clustering techniques for analysis of air pollution: A review (1980–2019) Atmos. Pollut. Res. 2020;11(1):40–56. doi: 10.1016/j.apr.2019.09.009. [DOI] [Google Scholar]
- Gupta R. R., Arya R. K., Kumar J., Shubham T. How covid-19 brought indian wedding industry to a halt? JINAV: J. Inform. Vis. 2020;1(2):83–91. [Google Scholar]
- Kusrini K. Grouping of retail items by using K-means clustering. Procedia Comput. Sci. 2015;72:495–502. doi: 10.1016/j.procs.2015.12.131. [DOI] [Google Scholar]
- Li Y, Haiyan Wu. A clustering method based on k-means algorithm. Phys. Procedia. 2012;25:1104–1109. doi: 10.1016/j.phpro.2012.03.206. [DOI] [Google Scholar]
- Pamula, R. Deka, J. K., Nandi, S.: An outlier detection method based on clustering. In 2011 Second International Conference on Emerging Applications of Information Technology, pages 253–256. IEEE, 2011.
- Sreedhar C., Kasiviswanath N, Reddy P. C. Clustering large datasets using k-means modified inter and intra clustering (km-i2c) in hadoop. J. Big Data. 2017;4(1):27. doi: 10.1186/s40537-017-0087-2. [DOI] [Google Scholar]
- Worldometer. COVID-19 coronavirus pandemic, 2020. URL http://worldometers.info/coronavirus. Last accessed 20 April 2020.
- Zarikas V., Poulopoulos S. G., Zoe G., Zervas E. Clustering analysis of countries using the covid-19 cases dataset. Data in Brief. 2020;31:1–8. doi: 10.1016/j.dib.2020.105787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data in this study can be accessed at: Website of COVID-19 of Indonesia (https://covid19.go.id/peta-sebaran) or Harvard Dataverse (https://doi.org/10.7910/DVN/JUSYYX).