Abstract
In recent years, immunotherapies have emerged as effective therapeutic strategies for treating human cancers. However, accumulating evidence has revealed an inconsistency between the response to immune checkpoint inhibitors and programmed death ligand 1 (PD-L1) expression status detected by immunohistochemistry staining. Recent research has revealed that the removal of N-Linked glycosylation significantly enhanced PD-L1 detection, resulting in both more accurate PD-L1 quantification and clinical outcome prediction. In the present study, we evaluated natural and deglycosylated PD-L1 expression in colon cancer using the PD-L1 28–8 antibody. The results of the present study validated the hypothesis that PD-L1 had a higher expression in colon cancer tissues compared with normal tissues. Additionally, colon tumors with defective mismatch repair tended to express higher PD-L1 than those without. Most importantly, the results of the present study indicated that the removal of N-linked glycosylation remarkably enhanced PD-L1 detection. Moreover, the PD-L1 signal intensity of samples with a low natural PD-L1 signal was enhanced more remarkably than that of samples with high signal intensity. Overall, our research provides an improved strategy for patient stratification for anti-PD-1/PD-L1 therapy, which deepens the clinical significance of this established strategy for treatment of colon cancer.
Keywords: biomarkers, DNA mismatch repair, gene expression, immune checkpoint inhibitors, immunotherapy
Introduction
Anti-cancer immunotherapies are currently the most rapidly growing cancer treatments, and have demonstrated encouraging clinical outcomes in the field of oncology. 1,2 Immune checkpoint inhibitors (ICIs) block the interaction between immune checkpoints. One such ICI, programmed cell death 1 (PD-1), and its receptor programmed death ligand 1 (PD-L1) inhibitors, have been confirmed to be instrumental in treating cancer by curtailing immune escape mechanisms (Figure S1). 2,3 Detection of PD-L1 expression by immunohistochemistry (IHC) staining is an important standard by which to guide the utilization of PD-1/PD-L1 ICIs. 4,5 Of note, a growing number of studies have demonstrated an inconsistent correlation between the response to ICIs and the expression status of PD-L1 detected via IHC staining. 6 Accordingly, enhancement of PD-L1 detection may improve patient stratification strategies for anti-cancer immunotherapies.
N-linked glycosylation is a prevalent post-translational modification of PD-L1, and PD-L1 that is heavily glycosylated with N-linked glycan has been found in many types of cancer, exhibiting various patterns on Western blot analyses, whereas the non-glycosylated form of PD-L1 is typically detected at ∼ 33 kDa. 7 A recent study by Lee et al 8 discovered that the removal of N-linked glycosylation could increase the detection of PD-L1 (28-8 clone), more accurately predicting the therapeutic efficacy of PD-1/PD-L1 inhibitors. Their results suggested that the detection of deglycosylated PD-L1 may be a diagnostic biomarker for anti-cancer immunotherapies. However, the efficacy of deglycosylated PD-L1 detection in colon cancer (CC) has not been evaluated in detail.
In the present study, we evaluated the expression of natural and deglycosylated PD-L1 in CC tissues, using the PD-L1 28-8 monoclonal antibody (mAb). As a result, we confirmed that PD-L1 had a much higher expression in CC tissues compared with normal tissues. Additionally, CC with defective mismatch repair (dMMR) tended to have a higher PD-L1 expression than in pMMR tissues. Most importantly, we found that the removal of N-linked glycosylation remarkably enhanced PD-L1 detection, and that the response to deglycosylation was more sensitive in samples with a lower natural PD-L1 signal intensity. Overall, our research further fortifies the clinical significance of deglycosylated PD-L1 detection in CC.
Materials and Methods
PD-L1 antibody (28-8) and the IHC antigen retrieval and detection kit (ab236676) were obtained from Abcam (Cambridge, UK). Recombinant PNGase F (P0708) was obtained from New England Biolabs (Ipswich, MA, USA). The serial high-density CC tissue microarray (TMA) section (HCol-Ade180Sur-06) was obtained from Outdo Biotech (Shanghai, China), which contained 104 CC tissues and 86 paired para-tumor specimens. The TMA contained integrated clinicopathological information and IHC staining for DNA mismatch repair (MMR) genes, including MLH1, MSH2, MSH6, and PMS2. Ethical approval for the study of the TMA slide was granted by the Clinical Research Ethics Committee of Outdo Biotech (Shanghai, China).
Tissue deglycosylation and IHC staining were performed directly on the TMA sections. TMA sections were incubated at 40°C overnight and then at 60°C for 1 hour, deparaffinized with xylene and ethanol, and hydrated in distilled water. Antigen retrieval was performed using a heat-induced epitope retrieval buffer obtained from Abcam (Cambridge, UK). As opposed to the methodology described by Lee et al, 8 we used the thermal denaturation strategy to thoroughly remove the N-linked glycan from PD-L1. Briefly, after washing twice with phosphate-buffered saline (PBS), the TMA sections were incubated with 1 × glycoprotein denaturing buffer at 95°C for 5 minutes, washed 3 times with PBS, treated with or without 5% recombinant PNGase F (P0708, New England Biolabs, Ipswich, MA, USA) dissolved in glycosylated buffer (20% of 10× GlycoBuffer 2, 20% of 10% Nonidet P-40, and 60% H2O, New England Biolabs, Ipswich, MA, USA) at 37°C for 1 hr, and then subjected to IHC staining. 9 Antibody staining was visualized with 3,3’-diaminobenzidine and hematoxylin counterstain. Immunostained sections were scanned using Aperio Digital Pathology Slide Scanners.
The results of the IHC were evaluated using an established semi-quantitative approach to assess the percentage of positive cells in total tumor cells (TPS) as well as the immunoreactivity score (IRS), which equaled the TPS multiplied by staining intensity. The TPS was scored on a scale of 0 to 4, as follows: < 1% = 0, 1%-25% = 1, 25%-50% = 2, 50%-75% = 3, and > 75% = 4. The staining intensity was scored from 0 to 3, as follows: negative = 0; weak = 1; moderate = 2; and strong = 3.
Statistical analyses and visualization were performed using GraphPad Prism 8.0. All error bars denote the standard deviation. The majority of the data between the 2 groups were analyzed by Student’s t-test or Mann-Whitney test. Pearson’s correlation test was used to determine the linear correlation between the 2 variables. The fold change (FC) value before and after deglycosylation was calculated as IRS (after deglycosylation)+0.1 divide IRS (before deglycosylation)+0.1. A log-rank test was performed to assess the difference between the survival curves. A 2-tailed P-value ≤ 0.05 was considered statistically significant.
Results
Comparing PD-L1 Expression in Colon Cancer Tissues and Para-Tumor Tissues Using PD-L1 28-8 Antibody
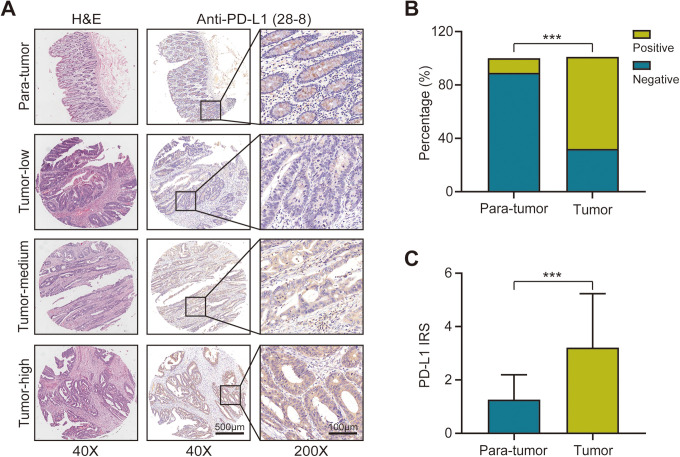
Detection of PD-L1 expression status by IHC assay is the most common method for using patient stratification to guide anti-PD-1/PD-L1 therapy. 10 In the present study, we compared PD-L1 expression status in a variety of CC tissues, including tumor-high, tumor-medium, tumor-low, and para-tumor samples. The representative images exhibited the characteristic PD-L1 staining patterns located in the cytomembrane as well as the cytoplasm (Figure 1A). The positive rate of PD-L1 (IRS ≥ 3) in CC tissues detected using 28-8 mAb was significantly higher than that of para-tumor tissues (Figure 1B). We further analyzed the IRS of PD-L1 using a 28-8 mAb, and found that the IRS of CC tissues was significantly higher than that of para-tumor tissues (Figure 1C). Overall, the expression of PD-L1 was significantly higher in CC tissues than in para-tumor tissues.
Figure 1.
Expression of programmed death ligand 1 (PD-L1) in colon cancer and para-tumor tissues. A, Representative images show samples stained with PD-L1 28-8 monoclonal antibody (mAb). Bar = 500 µm (40x), Bar = 100 µm (200x). B, Bar chart represents expression levels of PD-L1 expression stained by PD-L1 28-8 mAb in tumor and para-tumor tissues, where green is positively stained and blue is negatively stained. P < 0.001. C, Signal intensity of PD-L1 stained by PD-L1 28-8 mAb in tumor and para-tumor tissues. P < 0.001. All data are performed by GraphPad Prism 8.0.
Correlation Between PD-L1 Expression and Different Mismatch Repair Statuses
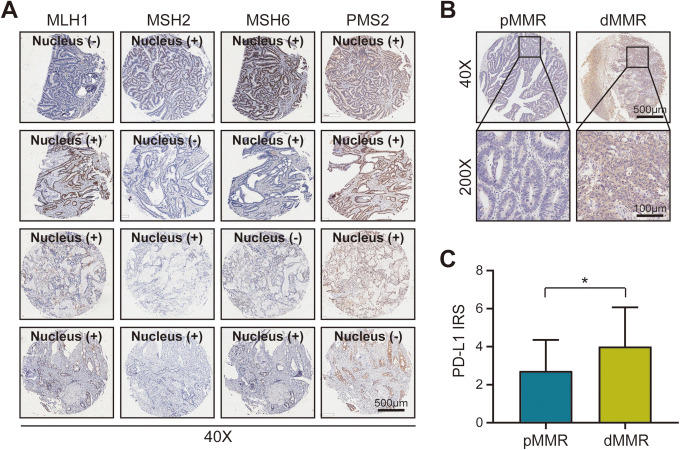
CC patients can be separated into 2 groups based on the mutation patterns involved: defective DNA mismatch repair (dMMR) involves tumors with a high mutation burden, and proficient DNA mismatch repair (pMMR) involves tumors with a much lower mutation burden. dMMR can be detected through a lack of IHC staining in the nuclear proteins MLH1, MSH2, MSH6, and/or PMS2, while pMMR is positive for staining in those 4 proteins. A difference in MMR status was found to be one of the most significant parameters related to PD-L1 expression. 11 Thus, we evaluated the natural PD-L1 expression in various samples with different MMR statuses. The representative images showed that the expression level of PD-L1 was remarkably higher in dMMR CC tissues than in pMMR tissues (Figures 2A and B). Additionally, we further compared the IRS of PD-L1 between dMMR and pMMR CC tissues, and found that there was a statistical difference in PD-L1 expression between the tissues (Figure 2C).
Figure 2.
Signal intensity of programmed death ligand 1 in colon cancer tissues with different mismatch repair status. CC patients can be separated into 2 groups based on the mutation patterns involved: Defective DNA mismatch repair (dMMR) involves tumors with a high mutation burden, and proficient DNA mismatch repair (pMMR) involves tumors with a much lower mutation burden. dMMR can be detected through a lack of IHC staining in the nuclear proteins MLH1, MSH2, MSH6, and/or PMS2, while pMMR is positive for staining in those 4 proteins. Different mismatch repair (MMR) statuses were found to be related to PD-L1 expression. A, Representative images showing dMMR samples stained with the corresponding antibody. Case 1 (line 1): Lack of MLH1; Case 2 (line 2): Lack of MSH2; Case 3 (line 3): Lack of MSH6; Case 4 (line 4): Lack of PMS2. Bar = 500 μm. B, Representative images represent the pMMR and dMMR CC samples stained with PD-L1 28-8 monoclonal antibody (mAb). Bar = 500 µm (40x), Bar = 100 µm (200x). C, Signal intensity of PD-L1 stained by PD-L1 28-8 mAb in pMMR and dMMR CC samples. P = 0.0465. All data are performed by GraphPad Prism 8.0.
Deglycosylation Significantly Enhances PD-L1 Detection in Colon Cancer Tumor Tissues
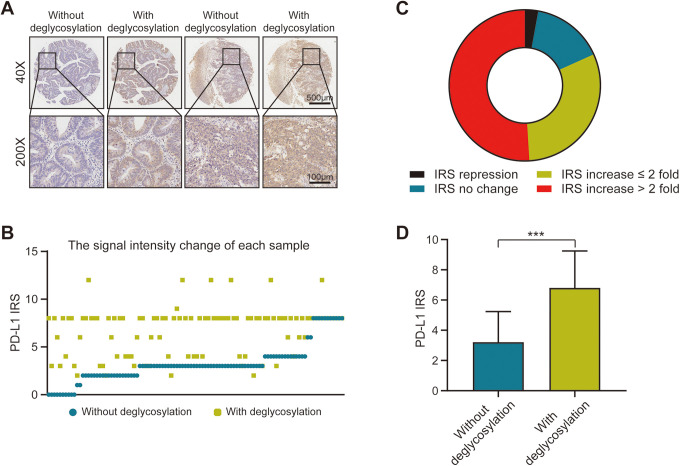
A recent study by Lee et al 8 have demonstrated that antigen retrieval by protein deglycosylation could enhance PD-L1 signal intensity. To determine whether the removal of N-linked glycosylation could increase PD-L1 expression in CC tissues, we evaluated both natural and deglycosylated PD-L1 signal intensities using 28-8 mAb. The representative images show that the removal of N-linked glycosylation remarkably enhanced PD-L1 detection (Figure 3A). We also analyzed the IRS of PD-L1 expression in natural and deglycosylated samples. The results of that analysis showed that the IRS of PD-L1 varied significantly between samples processed with and without deglycosylation (Figures 3B and D). Among these cases, the majority of the samples were categorized as having an increase in IRS. Of note, half of the samples were detected to have more than a 2-fold increase in IRS (Figure 3C). These results revealed that the N-linked glycosylation of PD-L1 critically affects its recognition by the anti-PD-L1 antibody in the clinical stratification of CC patients.
Figure 3.
Signal intensity of PD-L1 in colon cancer tissues after sample deglycosylation. A, Representative images show the samples stained with programmed death ligand 1 (PD-L1) 28-8 monoclonal antibody (mAb), with or without deglycosylation. Bar = 500 µm (40x), Bar = 100 µm (200x). B, Signal intensity of PD-L1 stained by PD-L1 28-8 mAb, before and after deglycosylation. C, Doughnut chart represents the fold change (FC) of PD-L1 immunoreactivity score (IRS) after deglycosylation. Black: IRS repressed after tissue deglycosylation; Green: IRS increased ≤ 2 fold after tissue deglycosylation; Blue: IRS had no change after tissue deglycosylation; Red: IRS increased > 2 fold after tissue deglycosylation. D, Bar chart represents the signal intensity of PD-L1 stained with 28-8 mAb, before and after deglycosylation. Blue: Samples without deglycosylation; Green: Samples with deglycosylation. P < 0.001. All data are performed by GraphPad Prism 8.0.
Correlation Between Response to Deglycosylation and the Immunoreactivity Score in Colon Cancer Tumor Tissues
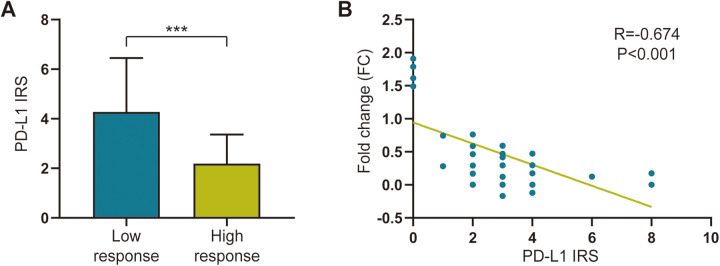
In the present study, we discovered a thought-provoking phenomenon. After removing glycosylation, the lower the expression of PD-L1, the more significant the increase (Figures 4A and B). Therefore, we evaluated the associations between the IRS of natural PD-L1 and the response to deglycosylation. Interestingly, the results of the present study showed that the IRS of natural PD-L1 was negatively related to the deglycosylation response (Figure 4B). Taken together, these results suggest that deglycosylation may preferably reduce false-negative PD-L1 statuses in samples with low natural PD-L1 expression. This novel discovery may be of clinical significance to advanced CC patients with low PD-L1 signal intensity.
Figure 4.
Correlation between response to deglycosylation and immunoreactivity score in colon cancer tumor tissues. A, Bar chart represents signal intensity of programmed death ligand 1 (PD-L1) stained by 28-8 monoclonal antibody (mAb) in samples with high and low response to deglycosylation. Blue: Samples showed low response to deglycosylation; Green: Samples showed high response to deglycosylation. P < 0.001. B, Correlation between signal intensity of PD-L1 stained by 28-8 mAb and fold change (FC), before and after sample deglycosylation. R = -0.674, P < 0.001. All data are performed by GraphPad Prism 8.0.
Discussion
Currently, the signal intensity of PD-L1 is most widely assessed as a biomarker to predict the response to PD-1 or PD-L1 inhibitors. 4,5 However, the role of PD-L1 expression as a predictive biomarker remains inconclusive. 12,13 A recent meta-analysis found that PD-L1 expression alone was not an adequate biomarker for patient selection in anti-PD-1/PD-L1 therapy. 14 However, another meta-analysis suggested that PD-L1 expression was associated with the efficacy of PD-1/PD-L1 blockade therapy. 15 Based on the controversial outcomes of anti-PD-1/PD-L1 therapy, the modulatory mechanisms of PD-L1 are being intensively studied. A growing number of studies have revealed that multiple processes impact the expression status of PD-L1, including gene transcription, genomic alternations, post-transcriptional and post-translational modifications, and exosomal transport. 16 Additionally, several regulatory factors are involved in the regulation of PD-L1 expression, including IFN-γ, IL-10, and miR-34a. 17 Overall, a comprehensive understanding of the regulatory mechanisms of PD-L1 is beneficial for the advanced strategy of PD-L1 detection, and further research is needed to identify the specific mechanisms behind PD-L1 expression.
Recent studies focusing on the various forms of PD-L1 have demonstrated that PD-L1 molecules are located on chromosome 9p24.1, and are broadly distributed in- and outside of cells. 18 Based on the its distribution, PD-L1 can be classified into 5 types: membrane PD-L1, cytoplasmic PD-L1, nuclear PD-L1, soluble PD-L1, and exosomal PD-L1. 18 Membrane PD-L1, located on the cell membrane, has an integrated structure, and suppresses anti-tumor immune responses by binding to its receptor, PD-1. 19 Cytoplasmic PD-L1, located in the cytoplasm, is capable of stimulating cancer properties and protecting cancer cells from death by enhancing the mammalian target of rapamycin complex 1 signal pathway and inhibiting autophagy. 20 Accordingly, researchers have suggested that the knockdown of cytoplasmic PD-L1 with specific RNAs may be a promising immunotherapy for patients with advanced cancer. 20 Nuclear PD-L1 is related to a shorter survival. 21 Soluble PD-L1, which is mainly detected in sera or supernatants, is significantly correlated with PD-L1 expression in mice. 18 However, the mechanism of formation for soluble PD-L1 remains unknown. Current research has revealed 3 possible mechanisms: derivation from membrane PD-L1 via proteolytic enzymes, endogenous translated protein, or splice variant for secretion. 18 Moreover, studies have revealed that the expression of soluble PD-L1 is significantly upregulated in the sera of older individuals, suggesting an association between the expression of soluble PD-L1 and immune status. 22 Of note, exosomal PD-L1 has been reported to inhibit anti-tumor immune responses. 22 Moreover, a recent study revealed that exosomal PD-L1 can predict the potential clinical response to PD-L1 inhibitors early after the initiation of these agents in patients with melanoma. 23 Based on the evidence that different distributions of PD-L1 exhibit different functions, a further understanding of these functions would advance PD-L1 based immunotherapy.
The prognostic value of PD-L1 expression in CC is highly debated. A meta-analysis by Li et al 24 demonstrated that PD-L1 expression was positively correlated with poor overall survival in CC. Moreover, the same study by Li et al 24 revealed that PD-L1 expression was significantly associated with unfavorable clinical prognosis, independent of age, sex, tumor size, tumor stage, and tumor, node, metastasis stage. However, other studies have demonstrated that stromal, rather than epithelial PD-L1 immunostaining, was negatively related to aggressive cancer properties in CC. 25 In the present study, we did not observe a statistical significance in the correlation between patient survival time and PD-L1 expression level in CC patients (Figures S2 and S3). Therefore, comprehensive research with larger clinical samples would be required to validate this in the future.
Although the expression level of PD-L1 has been used to stratify patients who would benefit from ICI therapy, accumulating evidence has revealed an inconsistency between PD-L1 signal intensity and the response to anti-PD-L1/PD-1 therapy. 6 Recent studies have demonstrated that heavy glycosylation of PD-L1 blocks its recognition by PD-L1 antibodies, which could result in an imprecise readout from IHC staining. 8 On the basis of the current research, N-linked glycosylation of PD-L1 blocks its recognition by PD-L1 antibodies in colon cancer. PD-L1 is heavily glycosylated with N-linked glycan and the glycan moiety is crucial for its immunosuppressive mechanism. 7,26 Currently, antibodies are usually generated in the light of synthetic peptide antigens or recombinant protein antigens detected in E. coli or other host organisms. However, post-translational modifications of the antigens are not recognized by the antibodies in order to include those that correspond to the native antigens, thus posing a challenge to recognition of protein glycosylation in higher organisms. 27 -29 Based on the results of the present study, we propose the use of an improved bioassay of PD-L1 detection in CC patients, which removes the glycan moiety of PD-L1 to increase PD-L1 recognition (Figure S4). The results of our study suggest that the deglycosylation of PD-L1 may create a more accurate biomarker with which to predict the clinical responses of PD-L1/PD-1 blockade inhibitors. Moreover, the response to deglycosylation was more sensitive in samples with a lower natural PD-L1 expression, which translates to a reduction in false-negative readouts in clinical practice. Despite the distinct advantages of ICI therapy, this ambitious approach is still limited in the clinical setting due to patient stratification. Therefore, our research presented an optimized method for the detection of PD-L1 by IHC staining for use in the stratification of CC patients to guide anti-PD-1/PD-L1 therapy.
Conclusions
In conclusion, the results of the present study showed that PD-L1 was more highly expressed in CC tissues when compared with normal tissues, especially in tissue samples with dMMR. The most important finding of our study was that the removal of N-linked glycosylation remarkably enhanced PD-L1 detection. Moreover, the PD-L1 signal intensity of samples with a lower natural PD-L1 signal was more remarkably enhanced than that of samples with a higher signal intensity. Overall, our research provides an improved strategy for the stratification of patients with CC for anti-PD-1/PD-L1 therapy, specifically by reducing the number of false-negative readouts detected by current methods.
Supplemental Material
Supplemental Material, sj-docx-1-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-2-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-3-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-4-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-5-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Abbreviations
- CC
colon cancer
- dMMR
defective mismatch repair
- ICIs
immune checkpoint inhibitors
- IHC
immunohistochemistry
- IRS
immunoreactivity score
- FC
fold change
- mAb
monoclonal antibody
- MMR
mismatch repair
- PBS
phosphate-buffered saline
- PD-1
programmed cell death 1
- PD-L1
programmed death ligand 1
- pMMR
proficient mismatch repair
- TMA
tissue microarray
- TPS
tumor proportion score
Footnotes
Authors’ Note: All data generated or analyzed during this study are included in this published article and its additional files. All authors agreed on the manuscript. Ethical approval for the study of tissue microarray slide was granted by the Clinical Research Ethics Committee, Outdo Biotech (No. YBM-05-02).
Author Contributions: DH and JX conceived the study and participated in the study design, performance, coordination and manuscript writing. JX, XY, YM, JM, HW, and JD carried out the assays and analysis. DH revised the manuscript. All authors reviewed and approved the final manuscript. Junying Xu and Xuejing Yang contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Natural Science Foundation of Jiangsu Province of China (BK20171150), Clinical Research Center Fund from Wuxi Science and Technology Bureau (WX18IVJN017) and Key Speciality from Wuxi Health and Family Planning Commission (JZYX04).
ORCID iD: Dong Hua, MD, PhD  https://orcid.org/0000-0001-9906-3252
https://orcid.org/0000-0001-9906-3252
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. [DOI] [PubMed] [Google Scholar]
- 3. Ostrand-Rosenberg S, Horn LA, Haile ST. The programmed death-1 immune-suppressive pathway: barrier to antitumor immunity. J Immunol. 2014;193(8):3835–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodor JN, Boumber Y, Borghaei H. Biomarkers for immune checkpoint inhibition in non-small cell lung cancer (NSCLC). Cancer. 2020;126(2):260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US food and drug administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7(1):278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li CW, Lim SO, Xia W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee HH, Wang YN, Xia W, et al. Removal of N-linked glycosylation enhances PD-L1 detection and predicts anti-PD-1/PD-L1 therapeutic efficacy. Cancer Cell. 2019;36(2):168–178. e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mei J, Xu J, Yang X, et al. A comparability study of natural and deglycosylated PD-L1 levels in lung cancer: evidence from immunohistochemical analysis. Mol Cancer. 2021;20(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsao MS, Kerr KM, Kockx M, et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of blueprint phase 2 project. J Thorac Oncol. 2018;13(9):1302–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luchini C, Bibeau F, Ligtenberg MJL, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. 2019;30(8):1232–1243. [DOI] [PubMed] [Google Scholar]
- 12. Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forde PM, Chaft JE, Smith KN, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med. 2018;378(21):1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen X, Zhao B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: meta-analysis. BMJ. 2018;362: k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X, Guo CY, Tou FF, et al. Association of PD-L1 expression status with the efficacy of PD-1/PD-L1 inhibitors and overall survival in solid tumours: a systematic review and meta-analysis. Int J Cancer. 2020;147(1):116–127. [DOI] [PubMed] [Google Scholar]
- 16. Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76(3):359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen S, Crabill GA, Pritchard TS, et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7(1):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Y, Chen W, Xu ZP, Gu W. PD-L1 distribution and perspective for cancer immunotherapy-blockade, knockdown, or inhibition. Front Immunol. 2019;10:2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hua D, Sun J, Mao Y, Chen LJ, Wu YY, Zhang XG. B7-H1 expression is associated with expansion of regulatory T cells in colorectal carcinoma. World J Gastroenterol. 2012;18(9):971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clark CA, Gupta HB, Curiel TJ. Tumor cell-intrinsic CD274/PD-L1: a novel metabolic balancing act with clinical potential. Autophagy. 2017;13(5):987–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chowdhury S, Veyhl J, Jessa F, et al. Programmed death-ligand 1 overexpression is a prognostic marker for aggressive papillary thyroid cancer and its variants. Oncotarget. 2016;7(22):32318–32328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion. Nat Rev Immunol. 2020;20(4):209–215. [DOI] [PubMed] [Google Scholar]
- 23. Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y, He M, Zhou Y, et al. The prognostic and clinicopathological roles of PD-L1 expression in colorectal cancer: a systematic review and meta-analysis. Front Pharmacol. 2019;10:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wyss J, Dislich B, Koelzer VH, et al. Stromal PD-1/PD-L1 expression predicts outcome in colon cancer patients. Clin Colorectal Cancer. 2019;18(1): e20–e38. [DOI] [PubMed] [Google Scholar]
- 26. Li CW, Lim SO, Chung EM, et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33(2):187–201. e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee BS, Huang JS, Jayathilaka GD, Lateef SS, Gupta S. Production of antipeptide antibodies. Methods Mol Biol. 2010;657:93–108. [DOI] [PubMed] [Google Scholar]
- 28. Rancour DM, Backues SK, Bednarek SY. Protein antigen expression in Escherichia coli for antibody production. Methods Mol Biol. 2010;657:3–20. [DOI] [PubMed] [Google Scholar]
- 29. Spadiut O, Capone S, Krainer F, Glieder A, Herwig C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014;32(1):54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-docx-1-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-2-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-3-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-4-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment
Supplemental Material, sj-docx-5-tct-10.1177_15330338211019442 for Removal of N-Linked Glycosylation Enhances PD-L1 Detection in Colon Cancer: Validation Research Based on Immunohistochemistry Analysis by Junying Xu, Xuejing Yang, Yong Mao, Jie Mei, Huiyu Wang, Junli Ding and Dong Hua in Technology in Cancer Research & Treatment