Abstract
The spike protein of SARS-CoV-2 (Severe Acute Respiratory Syndrome coronavirus 2) interacts with the ACE2 receptor in human cells and starts the infection of COVID-19 disease. Given the importance of spike protein's interaction with ACE2 receptor, we selected some antiviral peptides of venom scorpion such as HP1090, meucin-13, and meucin-18 and performed docking and molecular docking analysis of them with the RBD domain of spike protein. The results showed that meucin-18 (FFGHLFKLATKIIPSLFQ) had better interaction with the RBD domain of spike protein than other peptides. We also designed some mutations in meucin-18 and investigated their interactions with the RBD domain. The results revealed that the A9T mutation had more effective interaction with the RBD domain than the meucin-18 and was able to inhibit spike protein's interaction with ACE2 receptor. Hence, peptide “FFGHLFKLTTKIIPSLFQ” can be considered as the potential drug for the treatment of COVID-19 disease.
Keywords: Venom scorpion's peptides, Spike protein, Coronavirus, Docking, Molecular dynamics simulation, Mutation
Abbreviations: RMSD, root mean square deviation; MD simulation, Molecular dynamics simulation; SARS-CoV-2, Severe Acute Respiratory Syndrome coronavirus 2; ACE2, Angiotensin-Converting Enzymes 2; RBD, receptor binding domain
Graphical abstract
Introduction
The worldwide epidemic of the COVID-19 created by the SARS-CoV-2 virus (Severe Acute Respiratory Syndrome caused by coronavirus2) was started in December 2019. The spike protein of the coronavirus plays a major role in bonding to the host cell and is one the most attractive viral protein used for drug design in Covid 19 infection. The spike protein is located on the virus membrane and is a trimeric glycoprotein with two domains, S1 and S2, both critical for the SARS-CoV-2 infection. The S1 domain of spike protein includes one N terminal domain (NTD) and three C terminal domain (CTD 1 to 3). The interactions between the S1-RBD with the PD (peptidase domain) of angiotensin-converting enzymes 2 (ACE2) results in adhering the virus to the host which is followed by the S2 domain and leads to membrane combination and the arrival of viral ribonucleoprotein complex into the host cell. The RBD residues are within the CTD-1 and contain a core structure and the RBM (receptor binding motif). The host recognition by SARS-CoV-2 depends upon the interaction between the hook structured peptidase domain of the ACE2 and the RBM of the RBD region of the S1 domain. An extended loop region of the RBD spans the arch-shaped α1 helix of the ACE2-PD like a bridge; besides, these contacts can be organized into three separate clusters to capture the interacting domain. At the RBD domain Lys 417, Tyr 453, Gln 474, Phe 486, Gln 498, Thr 500 and Asn 501 of the S1-RBD have contacts with Tyr 41, Gln 42, Lys 353, and Arg 357, Asp 30 and His 34 and Gln 24 and Met 82 of ACE2. Besides, two capping loops in the binding domain, which stabilized the interaction with ACE2 by electrostatic interactions, comprised of residues Val 445, Tyr 449, Tyr 473, Gln 474, Ala 475, Glu 484, Gly 485, Phe 486, and Asn 487 [1,2].
The RBD (receptor binding domain) of the S1 domain of the spike protein can use a potential target for the progress of anti-SARS-CoV-2 drugs, which might have the ability to prevent the entrance of SARS-CoV-2 into the host cells. Then, we have considered the interaction of the RBD domain with ACE2. Peptides are potential molecules to be tested against the spike protein. Then some peptides were investigated to inhibit the infection by COVID-19 establishment.
Previous studies showed that scorpion venom might be a significant source of antiviral peptides [3]. Scorpions contain around 2400 species and belong to an olden prosperous group of arachnids that have been characterized in the fossil records coming from about 400 million years ago [3]. Scientists have identified the scorpion venom properties for less than a decade. Scorpion venom has bioactive proteins and peptides that may serve as the leading compounds for designing biotechnological tools and therapeutic drugs [4]. Scorpion venom is a mixture of biologically active compounds, mainly of proteinaceous nature, containing from small peptides to high molecular mass and multi-domain proteins, with complex folding and an extensive amount of post-translational modifications [5,6]. Toxins and neurotoxins are the most studied scorpion venom parts due to their pharmacological action on ion channels. They are disulfide-bridged peptides [DBPs] and can be classified into four main groups containing Na+, K+, Ca2+, and Cl− channels of interacting molecules. These molecules have antimalarial, immunosuppressing, and anticancer functions [7,8]. Examples of the DBPs are chlorotoxin [8,9], BmK CT [10], and BmK AGAP-SYPU2 [11]. Scorpion venom also comprises non-disulfide-bridged peptides [NDBPs] that have structural flexibility and anticancer, antimicrobial, bradykinin-potentiating activity [[12], [13], [14]] such as Bmkn2 [15], Ctry2459 [16], and Mucroporin molecules [[17], [18], [19]]. This class of peptides adopts a random coil configuration in water solution which folds into an α-helical structure in a membrane imitating environment [20]. Meucin-13 and meucin-18 are amphipathic peptides that display widespread cytolytic effects on prokaryotic and eukaryotic cells (bacteria, yeast, erythrocytes, and rat dorsal root ganglion cells) at micromolar concentrations [21]. Experimental studies have discovered that the antimicrobial activity of scorpion venom's mucin-18 was two to four times more effective than meucin-13. Also, some peptides in scorpion venoms have been reported to display antiviral function as well. For example, dermaseptin S4 and caerin 1.1 inhibit HIV-1 virus and viral relocation from dendritic cells to T cells and recombinant scorpion (RScp) inhibits dengue-2 virus replication [22]. Besides, some alpha-helical antimicrobial peptides have the power to destroy several enveloped viruses [22]. The HP1090 from Heterometrus petersii scorpion is an amphipathic peptide that prevents the development of Gram-positive bacteria. Also, the anti-HCV activity of the HP1090 peptide from the scorpion venom on Hepatitis C virus has been described by other investigators [22].
Molecular dynamics (MD) simulation shows the detailed motion of the molecules or atoms during simulation time. This method is usually used for molecular structure improvement and calculation of the system's properties and binding free energy of ligands to proteins [23,24]. Recently, using molecular dynamics for designing a peptide inhibitor to bind to spike protein's RBD domain was accomplish [25]. This study aimed to find a mutation in venom scorpion peptides (HP1090, meucin-13, or meucin-18) for binding to the RBD domain of spike protein virus using docking and molecular dynamics simulation.
Methods
To select a proper peptide for binding to the SARS-CoV-2 spike protein of coronavirus, twenty-three small peptides were obtained from venom scorpion through the literature survey. The name, Uniprot code, and the sequence of all candidate peptides used in this study have been mentioned in Table 1 [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]].
Table 1.
The name and Uniprot code and sequence all peptides of venom scorpion.
| Peptide name | Uniprot code | Peptide SequencesSequences Sequence |
|---|---|---|
| Venom antimicrobial peptide-9 (Meucin-18) | E4VP50 | FFGHLFKLATKIIPSLFQ |
| Venom antimicrobial peptide-6 (Meucin-13) | E4VP07 | IFGAIAGLLKNIF |
| Hp1090 | P0DJ02 | IFKAIWSGIKSLF |
| Mucroporin | B9UIY3 | LFGLIPSLIGGLVSAFK |
| Meucin-25 | P0CH58 | VKLIQIRIWIQYVTVLQMFSMKTKQ |
| Hp1036 | P0DME6 | ILGKIWEGIKSIF |
| Hp1239 | P0DME8 | ILSYLWNGIKSIF |
| Hp1035 | P0DJ03 | IFSAIGGFLKSIF |
| Hp1412 | P0DME9 | IFKAIWSGIKRLC |
| Hp1165 | P0DME7 | ILGEIWKGIKDIL |
| Hp1478 | P0DMF0 | ILGKFCDEIKRIV |
| Potassium channel toxin kappa-KTx 2.7 | P0DJ34 | GNACIEVCLQHTGNPAECDKACD |
| potassium channel toxin kappa-KTx 2.8 | P0DJ35 | GNACIEVCLQHTGNPAECDKPCDK |
| Potassium channel toxin kappa-KTx 3.4 | P0DJ39 | QWINACFNVCMKISSDKKYCKYLCGKS |
| Potassium channel toxin kappa-KTx 3.2 | P0DJ37 | HWINACFNICMKISSDQKYCKSFCG |
| Potassium channel toxin alpha-KTx 5.3 | Q9TVX3 | AVCNLKRCQLSCRSLGLLGKCIGDKCECVKH |
| Bengalin | P0CC11 | GPLTILHINDVHAAFEQFNT |
| Antimicrobial peptide ctriporin | G1FE62 | FLWGLIPGAISAVTSLIKK |
| Antimicrobial peptide AcrAP2 | A0A0A1I6N9 | FLFSLIPNAISGLLSAFK |
| Antimicrobial peptide AcrAP1 | A0A0A1I6E7 | FLFSLIPHAISGLISAFK |
| Ctri9819 | P0DMG0 | NRILPTLIGPL |
| Ctri9677 | P0DMF9 | INWDILIDTIKDKL |
| Ctri9194 | P0DMF5 | YIRDFITRRPPFGNI |
| Ctri10261 | P0DMF8 | FDLGGLIKGVVDLF |
| Ctri10033 | P0DME4 | FLVGILPRMRGFITPFLKKVR |
| Ctri9594 | P0DMF7 | GVVDTLKNLLMGLL |
| Ctri10036 | P0DME3 | FLWSLIPSAISAVTSLIKK |
| Ctri9610 | P0DME2 | FLFNVIPHAINATASLIKK |
| Toxin Acra2 | P0C2A0 | KKDGYIVDSNGCAPECFPTNXGC |
| Toxin b subunit beta | P0C2A3 | ADVPGNYPLNSYGASYYCTI |
| Peptide BmKn2 | Q6JQN2 | FIGAIANLLSKIF |
| Ctry2801 | P0DME5 | FLSLIPGAISAIASLFK |
Among these peptides, the HP1090 with the sequence of IFKAIWSGIKSLF, the meucin-13 with the sequence of IFGAIAGLLKNIF, and the meucin-18 with FFGHLFKLATKIIPSLFQ sequence extracted from Heterometrus Petersii and Meusobuthus Eupeus scorpions had anti-viral and anti-bacterial properties [43] and were able to make helical structures; hence, these structures were selected for extra studies.
Molecular modeling
Because the binding region of ACE2 bounded to the RBD domain has a helical structure, it seems that this structure should be used for peptides. At first, the peptides were subjected to Blast server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the best helical template for HP1090 was recognized as 1T51 and for the meucin-13, the 1T52 and 2L24 PDB codes were distinguished. Then, the 3D structures of these peptides were made using homology modeling; while, homology models were built by applying the Modeller 9.20 software [44]. The Mucin-18 had not appropriate template in the BLAST server and due to this fact, its 3D structure was made via Hyperchem8 software [45]. Five hundred models were produced for each peptide and were arranged by their normalized DOPE (Discrete Optimized Protein Energy) energy [46]. The DOPE energy displays the problematic region of the model structure and the structural model quality. The best homology models with the least DOPE energy in the declared peptides were used for more studies [47]. Then SAVES (https://servicesn.mbi.ucla.edu/SAVES/) [48] and ProSA web servers (https://prosa.services.came.sbg.ac.at/prosa.php) [49] were used for the evaluation of best models.
Molecular dynamics simulation of peptides
Amber 18 package was used for MD simulation [50]. The ff99SB amber force field and TIP3P water model [51] were considered for all simulations. The complexes were placed into truncated octahedral boxes with a buffer distance of 10 Å, and water molecules were added to fill the box. Then the total charge of each system was neutralized by adding Cl− ions. The default protonation of the Amber18 was used for the peptides' titratable residues. Periodic boundary conditions were applied in each system. Long-range electrostatic interactions were calculated using Particle Mesh Ewald (PME) method [52]. All bonds containing H atoms were constrained via the SHAKE algorithm [53]. The non-bonded cutoff distance required for energy minimization was 10 Å. Langevin dynamics were applied to control the systems' temperature. Sander module was used for energy minimization in each system. Ten thousand cycles of minimization were accomplished to delete improper steric interactions and to create better minimum energy via the conjugate gradients and steepest descent method for all parts of the systems. Afterward, the position restraints were executed at the constant volume (NVT) for 100 ps using a restraint force of 10 kcal/mol/Å at the temperature of 100 K. Then, MD simulations were done at the constant pressure (NPT) for 100 ps with a restraint force of one kcal/mol/Å2 at the temperature of 300 K. Next, the MD simulation at NPT ensemble for 100 ps at pressure 1 atm and 310 K with removed restraint force was performed. This step led to equilibrate the system density [54].
Finally, all peptides were separately simulated for 100 ns at the constant temperature and pressure conditions (NPT) at 310 K. During all simulations, the Berendsen thermostat was used to control the temperature, and isotropic pressure scaling was used to control the pressure with a relaxation time of 2 ps. The time step was 2 fs and the coordinates were saved every 1 ps. The Cpptraj module was used for the MD simulation analyses [55]. The ending structures were visualized by and Discovery Studio Visualizer Software These peptides are small and simply lose their initial helical structure in water and biological environments and produce many different conformations. Therefore, one hundred ns MD simulation was performed for each peptide model, and 31 conformations were extracted from the last 80 ns of 100 ns MD simulation.
Docking
The HADDOCK server (http://www.bonvinlab.org/software/haddock2.2) a public docking server that uses experimental and data-driven methods was employed for the peptide-protein docking study [56]. This server mixes the information resulting from biochemical, biophysical, or bioinformatics' methods to improve sampling and scoring schemes. Pervious MD simulation showed that the spike protein has two conformations consisting of “up or open” and “down or close”. In the closed conformational status, the ACE2 recognition motifs (RBDs) were concealed at the spike interface and in the open conformation RBD domain uncovering to spike surface for interaction. The spike protein can interact with the ACE receptor in both closed and open conditions; however, only in the open state, the RBD domain interactions with the PD domain of ACE2 would properly occur and the SARS-CoV-2 can arrive in the cells [57,58]. The crystallographic structure of the RBD domain in combination with the ACE2 receptor (open structure) which exists in the PDB bank was used in our study (PDB code: 6M17, chain E). In this study, we only used the RBD domain and not the full structure of the spike protein. The reason was that the full structure had some defections and also is not the peptides' target. In other words, further parts of the spike protein do not have any impact on our results.
Thirty-one structures achieved from MD simulation for each peptide were separately docked to the RBD domain of the spike protein via the Haddock server [59]. The residues binding to the ACE2 receptor were considered as active residues in the docking method and contained Lys 417, Tyr 453, Gln 474, Phe 486, Gln 498, Thr 500, and Asn 501 [1]. Furthermore, all peptides' resides were considered as active residues. In each complex, the interactions between the peptides and RBD domain were acquired using Ligplot software [60].
Molecular dynamics simulation of the complexes
The best complexes produced by molecular docking studies among 31 docked conformations with the lowest interface energy or HADDOCK docking score were distinctly used for another 50 ns MD simulation of the complexes. Other parameters of the MD simulation were similar to previously mentioned peptides' simulations. The free energy of the binding (ΔG) peptides with RBD domain of the spike protein and per-residue energy decomposition were calculated for each complex using molecular mechanics Generalized Born surface area (MM/GBSA) approaches in Amber 18 [54]. The binding free energies were calculated with MMPBSA script [61] using 100 snapshots extracted from the last ten ns of 50 ns MD simulation using this equation:
| ΔGbinding = Gcomplex-GRBD domain-Gpeptide |
The solvent-accessible surface was obtained from the linear combination of pairwise overlaps [LCPO] model to calculate the non-polar solvation energy [62].
Mutation of Meucin-18
The final structure of complex meucin-18 with RBD domain of the spike protein obtained from 50 ns MD simulation was subjected to BetMuSIC server (http://babylone.ulb.ac.be/beatmusic) [63] to find a mutation with more negative binding energy in complex with the spike protein. The BeAtMuSiC server is a coarse-grained predictor of the changes in binding free energy induced by the point mutations. It depends on a set of statistical potentials resulting from well-known protein structures and merges the mutation effect on interactions' strength at the interface and overall constancy of the complex. This server needs an input structure of the protein-protein complex and gives the probability to assess quickly all probable mutations in a protein chain or at the interface. Then, the first five suggested mutations of BeAtMuSiC server containing A9T, H4Y, A9S, H4F, K7H plus the combination of mutation one and four (A9T + H4F) (in total six mutations) were generated via SPDV Swiss-PDB viewer (https://spdbv.vital-it.ch/) in the final structure of meucin-18 peptide (that was gained from 100 ns MD simulation) distinctly.
MD simulation and docking of mutations
Eventually, the previously mentioned procedure for native peptides (i.e. MD simulation of peptides and docking to RBD domain of spike protein and MD simulation of the complexes) was repeated for six mutated peptides of meucin-18 and finally the best mutation with the lowest MM/GBSA binding energy of peptide in combination with RBD domain of spike protein was chosen.
Results and discussion
Homology modeling results
The best model with the lowest normalized DOPE energy among 5000 models was chosen from HP1090 homology modeling and meucin-13 peptides. The results revealed that these models had proper structures. All residues of the model structures were in favored or allowed regions of the Ramachandran plot (data not displayed), which specifies suitable reliability of the homology model structures. The ProSA website exhibits that Z-scores (overall model quality) achieved for HP1090 and meucin-13 best model, and meucin-18 built model were −0.03, −0.51, and 0.8, respectively; which, emphasizes its suitable quality. Because the meucin-18 had not any template and no homology modeling was accomplished for it, its calculated Z-score was more than other peptides.
Molecular dynamics simulation of peptides
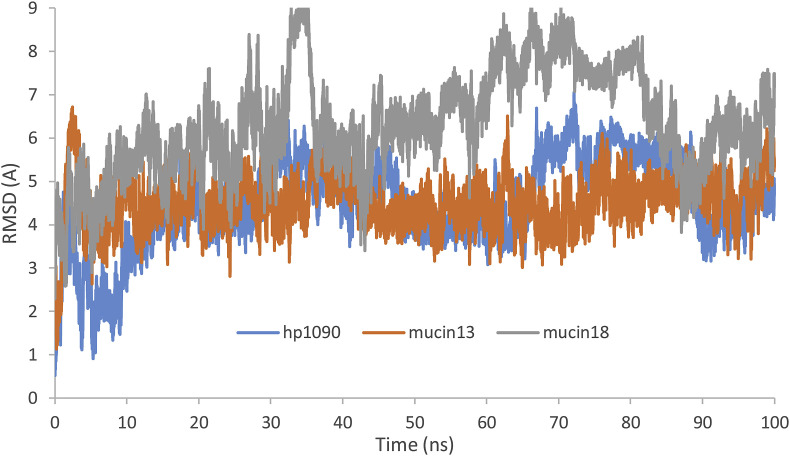
The MD simulations of peptides (three native peptides and six mutations) were carried out. The backbones' root mean square deviations (RMSD) were calculated which are exhibited in Fig. 1 for three native peptides. The RMSD plot is usually used to evaluate the time required for a system to reach structural equilibrium and to estimate the duration of running a simulation. As it is exemplified in Fig. 1, all peptides represented stable dynamics and reached a plateau during the last 10 ns, and then all analyses were performed during the last 10 ns of the simulation. The average of measured backbone's RMSD during the last 10 ns of MD simulation was 4.29 ± 0.45 Å, 4.81 ± 0.54 Å, and 6.16 ± 0.51 Å for HP1090, meucin-13, and meucin-18 peptides, respectively.
Fig. 1.
The backbones' root mean square deviations of all native peptides during 100 ns MD simulation.
The system temperature reached the plateau at about 310 K after 10 ns, and then all systems reached thermal equilibration. The temperature average during the last 10 ns of the simulation for HP1090, meucin-13, and meucin-18 peptides were 309.8 ± 3.2 K, 309.9 ± 2.9 K, and 309.9 ± 2.4 K, respectively. The results of the backbone RMSD, the potential, and temperature showed that the simulation time was adequate in all systems and as a result, these systems were able to reach equilibrium. The RMSD plot and visional examination uncovered that these small peptides make a variable loop in their structure during the simulation. Then, the selection of the final structure was not sufficient for the peptide structure and thirty-one conformations from the last 80 ns of the simulation were extracted from the trajectory with a 3.22 ns interval. These conformations were used in docking simulation.
Docking results
All 31 conformations of each peptide obtained from the peptide's MD simulation were docked to the RBD domain of the spike protein via the Haddock server and 31 docking scores were acquired for each peptide (data not shown). The lowest HADDOCK score, its van der Waals, electrostatic and desolation energy, and Z-score are presented in Table 2 among 31 conformations for each docked peptide. The Haddock score does not have a unit. It can only be expressed in arbitrary units [64]. These complexes were utilized in the next studies.
Table 2.
The best Haddock score and their van der Waals and electrostatic and desolation and Z-score among 31 conformations of each peptide.
| Peptide | HADDOCK score | Cluster size | Van der Waals energy | Electrostatic energy | Desolation energy | Buried Surface Area |
Z-score |
|---|---|---|---|---|---|---|---|
| Meucin-18 | −150.7 ± 4.2 | 134 | −73.5 ± 4.9 | −91.9 ± 30.2 | −61.4 ± 5.7 | 1819.5 ± 56.2 | −1.6 |
| Meucin-13 | −107.2 ± 6.0 | 32 | −55.0 ± 3.8 | −36.1 ± 9.0 | −47.7 ± 5.2 | 1355.7 ± 79.2 | −2.0 |
| Hp1090 | −103.0 ± 8.3 | 15 | −52.8 ± 4.0 | −55.0 ± 15.7 | −42.2 ± 6.2 | 1318.7 ± 87.9 | −2.1 |
| Mut1 | −128.9 ± 5.6 | 66 | −69.5 ± 1.3 | −49.6 ± 25.5 | −54.4 ± 4.1 | 1452.0 ± 52.0 | −2.0 |
| Mut2 | −140.2 ± 6.4 | 68 | −65.3 ± 7.9 | −150.6 ± 8.1 | −50.0 ± 3.9 | 1741.0 ± 86.5 | −2 |
| Mut3 | −139.2 ± 5.9 | 83 | −78.8 ± 1.7 | −90.8 ± 13.3 | −46.9 ± 7.3 | 1965.0 ± 77.9 | −1.8 |
| Mut4 | −119.9 ± 4.2 | 45 | −70.9 ± 4.4 | −53.2 ± 24.4 | −48.3 ± 3.5 | 1545.5 ± 57.8 | −2.3 |
| Mut5 | −131.8 ± 3.9 | 47 | −77.0 ± 2.2 | −65.6 ± 13.2 | −46.7 ± 5.6 | 1718.1 ± 43.0 | −2.2 |
| Mut6 (Mut1+Mut4) | −130.6 ± 28.2 | 6 | −61.0 ± 14.3 | −186.5 ± 28.5 | −35.5 ± 14.1 | 1652.4 ± 225.5 | −2.2 |
Molecular dynamics simulation of the complexes
Best complexes obtained from docking were used for another 50 ns MD simulation. Backbone RMSD of peptides and the spike protein and the temperatures of all systems were calculated and exposed the fact that all peptides of the complexes reach the equilibrium during the last 10 ns of the MD simulation (data not shown). The MM/GBSA binding free energy and also the van der Waals and electrostatic components with standard deviations values were calculated during the last 10 ns of the MD in all peptides were mentioned in Table 3 .
Table 3.
The MM/GBSA binding free energy and electrostatic (ELE) and VDW energies (VDWAALS) components (kcal/mol) of all native and mutated peptides during the last 10 ns of MD simulation.
| Peptide | Generalized Born energy | ELE energy | VDWAALS energy |
|---|---|---|---|
| Hp1090 | −20.78 ± 9.61 | 29.85 ± 28.08 | −34.59 ± 13.88 |
| Meucin-13 | −19.41 ± 8.21 | 19.84 ± 23.39 | −33.77 ± 9.72 |
| Meucin-18 | −40.12 ± 10.08 | −32.90 ± 32.96 | −61.95 ± 13.65 |
| Mut1 | −49.10 ± 7.56 | −149.23 ± 33.53 | −57.97 ± 6.58 |
| Mut2 | −41.66 ± 10.07 | −39.77 ± 51.26 | −61.40 ± 10.46 |
| Mut3 | −47.31 ± 10.32 | 32.20 ± 32.55 | −72.30 ± 10.10 |
| Mut4 | −44.82 ± 5.81 | −6.79 ± 32.48 | −66.77 ± 5.78 |
| Mut5 | −39.34 ± 11.49 | −13.20 ± 23.22 | −71.77 ± 9.79 |
| Mut6 (Mut1+Mut4) | −29.69 ± 8.33 | −26.60 ± 41.28 | −51.30 ± 9.60 |
The results pinpointed that the meucin-18 has better binding power to the RBD domain of the spike protein among other peptides such as HP1090 and meucin-13 and can prevent its binding to the ACE2 receptor. According to MMPBSA results, van der Waals and electrostatic contribution in the meucin-18 were −61.9 and −32.9 kcal/mol, respectively; however, these values for mutation 1 of meucin-18 were −57.9 and −149.2 kcal/mol, respectively (Table 3). Thus, it could be concluded that the mutation in the mucin 18 leads to an increase of electrostatic interaction of the meucin-18 with the RBD domain; because of the alteration in binding conformation of the mutation 1. In other words, in all peptides except mut1 of meucin-18, VDW energy, is more negative and more important than electrostatic energy. In mut1 electrostatics energy is major factor in binding to RBD domain and mutation A9T in meucin-18 lead to enhancement of electrostatic interaction.
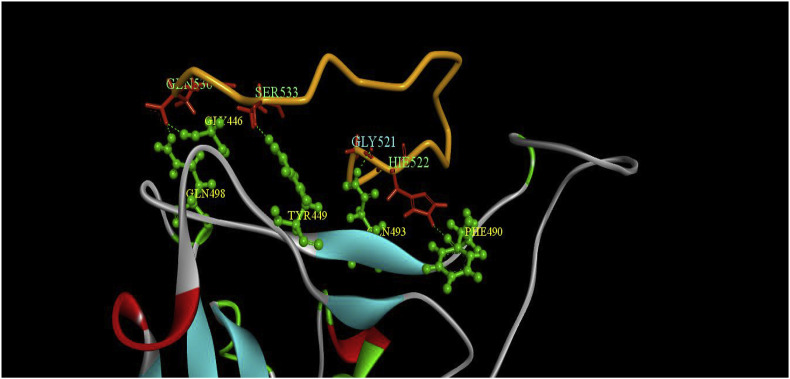
The 3D interaction between the meucin-18 with RBD domain residues after 50 ns MD simulation was obtained via Discovery Studio Visualizer software and presented in Fig. 2 .
Fig. 2.
The 3D representation of the interaction of mucin-18 residues (519–536) (red color in the orange color backbone) with RBD domain residues (green color) of spike after 50 ns MD simulation of the complex. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
It was revealed that Gly 446, Tyr 449, Phe 490, Gln 493, and Gln 498 of the RBD domain have the hydrogen bond with the meucin-18 residues and the Lys 417, Val 445, Tyr 453, Leu 455, Phe 456, Tyr 473, Gln 484, Tyr 489, and Leu 492 of the RBD domain have hydrophobic interaction with the meucin-18 residues.
Among these residues, the Lys 417, Tyr 453, and Gln 498 are the key residues for binding the RBD domain to the ACE2 receptor [1]. Besides, the residues of Tyr 473 and Gln 484 are close to Gln 474 and Phe 486 that are also the key residues in the binding process. Also, energy decomposition of the MM/GBSA binding free energy analysis showed that the Tyr 489, Leu 455, Tyr 449, Phe 490, and Phe 456 had significant interaction with meucin-18 residues. These residues are located at the binding region of the RBD domain with the ACE2 receptor. Besides, Phe 17, Phe 2, His 4, Phe 6, Ile 12, Leu 5, and Lys 11 of meucin-18 had the most interaction with the RBD domain of the spike protein.
Hydrogen bonds number between the meucin-18 and RBD domain were calculated during the simulation time. The Gly 3 (or 521), His 4 (or 522), Ile 12 (or 530), and Ser 15 (or 533) of the mucin-18 had a hydrogen bond with the RBD domain for the duration of more than 10% of the MD simulation time.
The average number of hydrogen bonds between meucin-18 and RBD domain was 2.23 ± 1.5 during the last 20 ns of MD simulation.
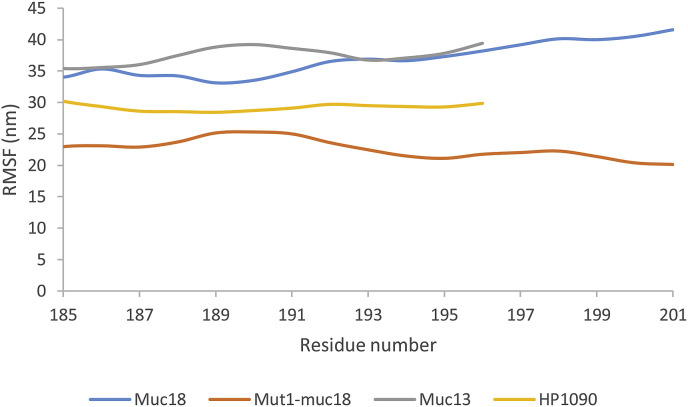
The root mean square fluctuation (RMSF) of all native peptides and mutation 1 of meucin-18 in combination with the RBD domain were calculated (Fig. 3 ). The RMSF all residues of peptides were rather high. It means high rearrangement all peptides during the last 10 ns of MD simulation of peptides in complex with RBD domain. According to the results, the C-terminal resides had more flexibility and RMSF than the N-terminal residues; because they had less interaction with the RBD domain of the spike protein.
Fig. 3.
RMSF of native peptides plus mut1 of mucin 18 in complex with RBD domain during the last 10 ns of MD simulation.
Mutation results
After selection of the meucin-18 as the most proper peptide, BeAtMuSiC server was used for suitable mutations leading to better interaction of the meucin-18 with spike protein. Table 4 shows the differences between binding energy in native and mutated (ΔΔGbinding) structure of the first five mutations in meucin-18 proposed by BetMuSIC server. These results demonstrated that mutation 1 (A9T) causes the greatest decrease in binding free energy and therefore it is the most appropriate mutation for the meucin-18 peptide.
Table 4.
The differences between binding energy between native and mutated (ΔΔGbinding) structures of the first five mutations in meucin-18.
| Name | Mutation | ΔΔGbinding (kcal/mol) |
|---|---|---|
| Mut1 | A9T | −0.22 |
| Mut2 | H4Y | −0.18 |
| Mut3 | A9S | −0.18 |
| Mut4 | H4F | −0.17 |
| Mut5 | K7H | −0.16 |
Docking and MD simulation of the mutated peptide
The structure of the best five mutated peptides of the meucin-18 plus the combination of mutation 1 and 4 (mutation 6) were made and separately simulated for 100 ns and the previous procedure was repeated for these mutations.
Binding free energy of the docking for best conformations of each mutation among total of 31 conformations was implemented in Table 2. Also, the MM/GBSA binding energy of the best conformation in each mutated peptide combined with the RBD domain of the spike protein was measured during the last 10 ns of 50 ns MD simulation and is exhibited in Table 3. According to docking results, mutation 2 (H4Y) of the meucin-18 had a better HADDOCK score with the RBD domain; nevertheless, according to MM/GBSA results, the mutation 1 (A9T) of the meucin-18 had better interaction with the RBD domain. Because MM/GBSA results were obtained from 50 ns MD simulation and were more accurate, we chose mutation 1 or A9T mutation as the most suitable peptide for binding to the RBD domain of the spike protein.
As stressed in Fig. 3, mutation 1 of the meucin-18 leads to a decrease of flexibility or RMSF the meucin-18 residues, then better interaction of the mutation 1 with the RBD domain of the spike protein (in comparison with the meucin-18) occur. The RMSF results are compatible with the MM/GBSA binding free energy results of mutation 1.
The residue of Tyr 449, Glu 484, Gln 498, and Asn 501 of the RBD domain had hydrogen bond and residue of Gly 446, Gly 447, Leu 455, Tyr 473, Gly 485, Phe 486, Tyr 489, Phe 490, Leu 492, Gly 496, Tyr 500, and Tyr 505 of RBD domain had hydrophobic interaction with mutation 1 of the meucin-18.
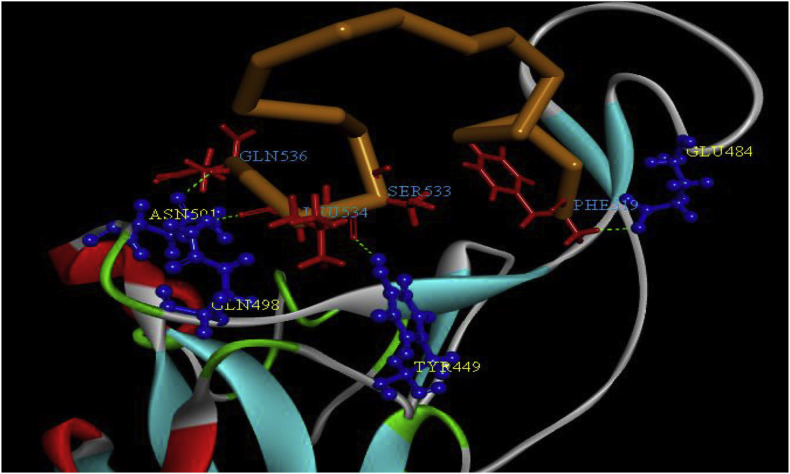
The 3D interaction between the mutation 1 of the meucin-18 with the RBD domain of the spike residues was obtained after 50 ns MD simulation via Discovery Studio Visualizer software and is represented in Fig. 4 .
Fig. 4.
The 3D representation of the interaction of mutation 1 of mucin-18 residues (519–536) (red color in the orange color backbone) with RBD domain residues (blue color) of spike after 50 ns MD simulation of the complex. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
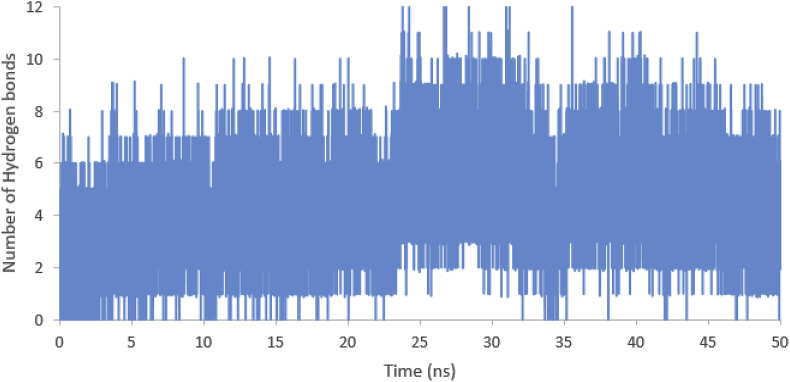
Among these residues, Phe 486, Tyr 500, and Asn 501 were considered as the key residues for binding the RBD domain to ACE2 and the Leu 455 and Gly 496 were close to Tyr 453 and Gln 498 of the RBD domain that were assumed as the key residues for binding RBD to ACE2. Also, the hydrogen bonds between mutation 1 of the meucin-18 and RBD domain were measured during the simulation. The Phe 1 (or 519), Ser 15 (or 533), Leu 16 (or 534), and Gln 18 (or 536) of the mutation 1 of the meucin-18 had hydrogen bond with RBD domain over than 10% of the MD simulation time (Fig. 4). The average number of hydrogen bonds between mutation 1 of mucin-18 with RBD domain during the last 20 ns of 50 ns MD simulation was 4.74 ± 1.6 that is more than the average number of a hydrogen bond between meucin-18 and RBD domain (2.23 ± 1.5). Also, the number of hydrogen bonds during the simulation was shown in Fig. 5 .
Fig. 5.
The number of hydrogen bonds between RBD domain and mutation 1 of mucin-18 during 50 ns MD simulation.
Then binding of mutation 1 to the RBD domain prevents binding of the RBD key residues to the ACE2. Besides, MM/GBSA binding free energy decomposition analysis designated that Tyr 489, Glu 484, Tyr 505, Phe 490, Leu 492, Gln 493, Leu 455, Gln 498, and Asn 501 of RBD domain had considerable interaction with the mutation 1 of meucin-18 residues. These residues are placed at the binding region of the RBD domain to the ACE2 receptor. Besides, Phe 1, Phe 2, Phe 17, Phe 6, Pro 14, and Leu 16 of the mutation 1 of mucin-18 had the highest interaction with the RBD domain of the spike protein.
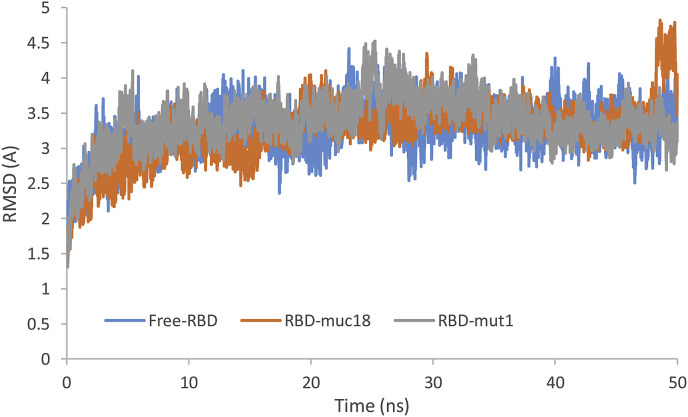
To compare the effect of meucin-18 and mutation 1 of the meucin-18 on the RBD domain structure of the spike protein, the free RBD domain was simulated without any peptide for 50 ns with the same previous conditions. Then, the backbone's RMSD of free RBD domain and in combination with the meucin-18 and mutation 1 of the meucin-18 were calculated (Fig. 6 ).
Fig. 6.
Backbone's RMSD of free RBD domain (free-RBD) and RBD domain in complex with mucin-18 (RBD-muc18) and complex with mutation 1 of mucin-18 (RBD-mut1) during MD simulation.
The average backbone's RMSD of the RBD domain during the last 10 ns of the MD simulation was 3.36 ± 0.25 Å, 3.5 ± 0.42 Å, 3.28 ± 0.17 Å for free RBD and RBD in combination with the meucin-18 and in complex with mutation 1 of meucin-18, respectively. These findings confirm that the mutation 1 peptide can reduce RMSD of the RBD domain of the spike protein and induce a conformational change in the RBD domain which, could result in either eliminating or wrong interaction of the spike protein with the ACE2 receptor.
Conclusion
Nowadays, we are living in a universal health disaster caused by the SARS-CoV-2 virus, which is responsible for the COVID-19 pandemic since the end of 2019. To date, there is not any vaccine against COVID-19 and the main and most significant policy adopted to avoid the spread of this novel coronavirus is social separation.
Souza et al. found two synthetic peptides, comprising Mo-CBP3-PepII and PepKAA interacting with S1 and S2 domains of the spike protein, respectively [55]. Ran Yan et al. using structural analyses showed that HP1090 peptide can inhibit HCV infection before viral entering into cells and quickly kill HCV in vitro. Then HP1090 is a possible anti-HCV leading peptide [22].
Han and Kral identified 15 amino acids from the ACE2 receptor that directly interact with spike protein. Then, they designed two types of peptides including alpha-helix and beta-sheet peptides, and showed that the alpha-helix structures had more potential for binding to spike protein [25].
In this study, we used alpha helix peptides derived from venom scorpion (HP1090, meucin-13, and meucin-18 peptides) that had the suitable potential for binding to spike protein. These peptides had anti-bacterial and anti-viral properties in which their initial structures were obtained via homology modeling or hyprchem8 software. The best models of these peptides were subjected to 100 ns MD simulation via amber 18 package. Because of peptides' small size, helical structures were destroyed during the MD simulation and their structures were transformed to loop conformer. Then, thirty-one conformations of each peptide were extracted from the last 80 ns of MD simulation and each peptides' conformation was docked to the PDB structure of the RBD domain of the spike protein and thereafter the best complex was subjected to another 50 MD simulation for each peptide. Then, the MM/GBSA binding free energy analysis was carried out during the last ten ns of the MD simulation. The results revealed that the meucin-18 peptide (FFGHLFKLATKIIPSLFQ) had better binding free energy comparing other native peptides. Therefore, the final structure of complex meucin-18 with RBD domain acquired from 50 ns MD simulation was subjected to BeAtMuSiC server for mutation proposition in meucin-18 to find a mutation with better binding potency to RBD domain. The 3D structure of the first five proposed mutations plus one additional mutation (a combination of mutations 1 and 4), (in total six mutations) were prepared. The same previous procedure was repeated for six mutations and MM/GBSA binding free energies were calculated. The results showed that mutation 1 (A9T) or (FFGHLFKLTTKIIPSLFQ) had the most negative binding energy and this mutation can prevent interaction RBD domain of spike protein with ACE2 receptor. Also, the energy contribution of the RBD domain's residues interacting with meucin-18 showed that this peptide can interact with key residues of the RBD domain. Furthermore, this peptide can change the native conformation of the RBD domain of spike protein and lead to incorrect interaction of RBD with ACE2 receptor. Mutation 1 (A9T) is even better than native meucin-18 and can be considered as a potential drug for COVID-19 infection; because these peptides can bind to the spike protein and inhibit all downstream processes of the SARS-CoV-2 to enter the cells and create cells' infection.
However, there is an alternative receptor in the human lungs' cells called CD209L, a C-type lectin (also called L-SIGN), which could be used by the coronavirus to cause the infection [58]. The interaction of proposed peptides with CD209L receptor can also be tested by the molecular dynamics simulations in further projects.
A major blockade for using peptides as the drug is their toxicity or ability to lyse eukaryotic cells. There was a significant correlation between amphipathicity and hemolytic activity [65]. The Lys and Arg and Tyr and Trp are amphiphilic amino acids [66], in which these residues can induce hemolytic activity in peptides. Also, previous studies showed that introducing d-amino acids instead of original l-amino acids can significantly reduce the hemolytic activity of helix peptides [67]. Because mutation 1 of meucin-18 (FFGHLFKLTTKIIPSLFQ) has two Lys residues (Lys 7 and Lys 11), to reduce hemolytic properties of this peptide, we propose using D-Lys residue instead of L-Lys in peptide's synthesis over the in vitro experimental studies and as a result, diminishing hemolytic effects. Also, for reducing the hemolytic effect of this peptide, it is suggested that this peptide is used with nanomolar and not with micromolar concentration over the in vivo experiment. In addition, allergenicity's potential of the mentioned peptides were predicted via AllerCatPro server (https://allercatpro.bii.a-star.edu.sg/) [68] and the results showed that none of these peptides had allergenicity (data not shown).
This was the first study reporting scorpion venom peptides as the potential antiviral molecules that can be used to inhibit the SARS-CoV-2 entrance into the cells. The proposed small peptide should be tested both through in vitro and in vivo experiments in further investigations.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to appreciate the supports provided by the University of Shahrekord, Iran for the implementation of this project. The authors also appreciate the attempts of the professional editor, Dr. Mostafa Karbasioun for proofreading and improving the quality of the manuscript.
References
- 1.Prajapat M., Shekhar N., Sarma P., Avti P., Singh S., Kaur H., Bhattacharyya A., Kumar S., Sharma S., Prakash A., Medhi B. Virtual screening and molecular dynamics study of approved drugs as inhibitors of spike protein S1 domain and ACE2 interaction in SARS-CoV-2. J. Mol. Graph. Model. 2020;101:107716. doi: 10.1016/j.jmgm.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robson B. Bioinformatics studies on a function of the SARS-CoV-2 spike glycoprotein as the binding of host sialic acid glycans. Comput. Biol. Med. 2020;122:103849. doi: 10.1016/j.compbiomed.2020.103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Eauclaire M.F., Adi-Bessalem S., Hammoudi-Triki D., Laraba-Djebari F., Bougis P.E. Serotherapy against voltage-gated sodium channel-targeting α-toxins from androctonus scorpion venom. Toxins. 2019;11(2):63. doi: 10.3390/toxins11020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amorim F.G., Cordeiro F.A., Pinheiro-Júnior E.L., Boldrini-França J., Arantes E.C. Microbial production of toxins from the scorpion venom: properties and applications. Appl. Microbiol. Biotechnol. 2018;102(15):6319–6331. doi: 10.1007/s00253-018-9122-2. [DOI] [PubMed] [Google Scholar]
- 5.de la Vega R.C.R., Possani L.D. Overview of scorpion toxins specific for Na+ channels and related peptides: biodiversity, structure–function relationships and evolution. Toxicon. 2005;46(8):831–844. doi: 10.1016/j.toxicon.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Ortiz E., Gurrola G.B., Schwartz E.F., Possani L.D. Scorpion venom components as potential candidates for drug development. Toxicon. 2015;93:125–135. doi: 10.1016/j.toxicon.2014.11.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBin J.A., Maggio J.E., Strichartz G.R. Purification and characterization of chlorotoxin, a chloride channel ligand from the venom of the scorpion. Am. J. Physiol. Cell Physiol. 1993;264(2):C361–C369. doi: 10.1152/ajpcell.1993.264.2.C361. [DOI] [PubMed] [Google Scholar]
- 8.Soroceanu L., Manning T.J., Sontheimer H. Modulation of glioma cell migration and invasion using Cl− and K+ ion channel blockers. J. Neurosci. 1999;19(14):5942–5954. doi: 10.1523/jneurosci.19-14-05942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby D.B., Dyskin E., Yalcin M., Kesavan K., Dahlberg W., Ratliff J., Mousa S.A. Potent pleiotropic anti-angiogenic effects of TM601, a synthetic chlorotoxin peptide. Anticancer Res. 2010;30(1):39–46. [PubMed] [Google Scholar]
- 10.Fan S., Sun Z., Jiang D., Dai C., Ma Y., Zhao Z., Li W. BmKCT toxin inhibits glioma proliferation and tumor metastasis. Canc. Lett. 2010;291(2):158–166. doi: 10.1016/j.canlet.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Shao J.H., Cui Y., Zhao M.Y., Wu C.F., Liu Y.F., Zhang J.H. Purification, characterization, and bioactivity of a new analgesic-antitumor peptide from Chinese scorpion Buthus martensii Karsch. Peptides. 2014;53:89–96. doi: 10.1016/j.peptides.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y., Huang J., Chen Y. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein & cell. 2010;1(2):143–152. doi: 10.1007/s13238-010-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almaaytah A., Tarazi S., Alsheyab F., Al-Balas Q., Mukattash T. Antimicrobial and antibiofilm activity of mauriporin, a multifunctional scorpion venom peptide. Int. J. Pept. Res. Therapeut. 2014;20(4):397–408. doi: 10.1007/s10989-014-9405-0. [DOI] [Google Scholar]
- 14.Tarazi S. Scorpion venom as antimicrobial peptides (AMPs): a review article. Int. Arab. J. Antimicrob. Agent. 2016;5(3) doi: 10.3823/777. [DOI] [Google Scholar]
- 15.Arpornsuwan T., Buasakul B., Jaresitthikunchai J., Roytrakul S. Potent and rapid antigonococcal activity of the venom peptide BmKn2 and its derivatives against different Maldi biotype of multidrug-resistant Neisseria gonorrhoeae. Peptides. 2014;53:315–320. doi: 10.1016/j.peptides.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Hong W., Zhang R., Di Z., He Y., Zhao Z., Hu J., Cao Z. Design of histidine-rich peptides with enhanced bioavailability and inhibitory activity against hepatitis C virus. Biomaterials. 2013;34(13):3511–3522. doi: 10.1016/j.biomaterials.2013.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., Zhao Z., Zhou D., Chen Y., Hong W., Cao L., Wu Y. Virucidal activity of a scorpion venom peptide variant mucroporin-M1 against measles, SARS-CoV and influenza H5N1 viruses. Peptides. 2011;32(7):1518–1525. doi: 10.1016/j.peptides.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akef H.M. Anticancer and antimicrobial activities of scorpion venoms and their peptides. Toxin Rev. 2019;38(1):41–53. doi: 10.1080/15569543.2017.1414847. [DOI] [Google Scholar]
- 19.Boas L.C.P.V., Campos M.L., Berlanda R.L.A., de Carvalho Neves N., Franco O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019;76(18):3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao B., Zhu S. Mesobuthus venom-derived antimicrobial peptides possess intrinsic multifunctionality and differential potential as drugs. Front. Microbiol. 2018;9:320. doi: 10.3389/fmicb.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao B., Sherman P., Luo L., Bowie J., Zhu S. Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. Faseb. J. 2009;23(4):1230–1245. doi: 10.1096/fj.08-122317. [DOI] [PubMed] [Google Scholar]
- 22.Yan R., Zhao Z., He Y., Wu L., Cai D., Hong W., Li W. A new natural α-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides. 2011;32(1):11–19. doi: 10.1016/j.peptides.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Soleimani M., Mahnam K., Mirmohammad-Sadeghi H., Sadeghi-Aliabadi H., Jahanian-Najafabadi A. Theoretical design of a new chimeric protein for the treatment of breast cancer. Res. Pharmaceut. Sci. 2016;11(3):187. doi: 10.4103/2277-9175.180639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghaheh H.S., Ganjalikhany M.R., Yaghmaei P., Pourfarzam M., Sadeghi H.M.M. Improving the solubility, activity, and stability of reteplase using in silico design of new variants. Res. Pharmaceut. Sci. 2019;14(4):359. doi: 10.4103/1735-5362.263560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y., Král P. Computational design of ACE2-Based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., Hong W., Zeng Z., Wu Y., Hu K., Tian X., Cao Z. Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4α in vitro and in vivo. J. Biol. Chem. 2012;287(36):30181–30190. doi: 10.1074/jbc.M112.370312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh A., Roy R., Nandi M., Mukhopadhyay A. Scorpion venom–toxins that aid in drug development: a review. Int. J. Pept. Res. Therapeut. 2019;25(1):27–37. doi: 10.1007/s10989-018-9721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cid-Uribe J.I., Veytia-Bucheli J.I., Romero-Gutierrez T., Ortiz E., Possani L.D. Scorpion venomics: a 2019 overview. Expet Rev. Proteonomics. 2020;17(1):67–83. doi: 10.1080/14789450.2020.1705158. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Y., Luo X., Kuang F., Deng M., Wang M., Zeng X., Liang S. Synthesis and characterization of huwentoxin-IV, a neurotoxin inhibiting central neuronal sodium channels. Toxicon. 2008;51(2):230–239. doi: 10.1016/j.toxicon. [DOI] [PubMed] [Google Scholar]
- 30.Cao Z., Di Z., Wu Y., Li W. Overview of scorpion species from China and their toxins. Toxins. 2014;6(3):796–815. doi: 10.3390/toxins6030796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isbister G.K., Bawaskar H.S. Scorpion envenomation. N. Engl. J. Med. 2014;371(5):457–463. doi: 10.1056/NEJMra1401108. [DOI] [PubMed] [Google Scholar]
- 32.Bergeron Z.L., Bingham J.P. Scorpion toxins specific for potassium (K+) channels: a historical overview of peptide bioengineering. Toxins. 2012;4(11):1082–1119. doi: 10.3390/toxins4111082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janeyuth Chaisakul, et al. Effects of animal venoms and toxins on hallmarks of cancer. J. Canc. 2016;7:11. doi: 10.7150/jca.15309. 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won Y.W., Patel A.N., Bull D.A. Cell surface engineering to enhance mesenchymal stem cell migration toward an SDF-1 gradient. Biomaterials. 2014;35(21):5627–5635. doi: 10.1016/j.biomaterials. [DOI] [PubMed] [Google Scholar]
- 35.Bayatzadeh M.A., Mirakabadi A.Z., Babaei N., Doulah A.H., Doosti A. Characterization, molecular modeling and phylogenetic analysis of a long mammalian neurotoxin from the venom of the ranian scorpion Androctonus crassicauda. Biologia. 2019;1–13 doi: 10.2478/s11756-019-00400-1. [DOI] [Google Scholar]
- 36.Alajmi R., Al-ghamdi S., Barakat I., Mahmoud A., Abdon N., Al-Ahidib M., Abdel-Gaber R. Antimicrobial activity of two novel venoms from Saudi arabian scorpions (Leiurus quinquestriatus and androctonus crassicauda) Int. J. Pept. Res. Therapeut. 2020;26(1):67–74. doi: 10.1007/s10989-019-09816-4. [DOI] [Google Scholar]
- 37.Gao B., Zhu S. Mesobuthus venom-derived antimicrobial peptides possess intrinsic multifunctionality and differential potential as drugs. Front. Microbiol. 2018;9:320. doi: 10.3389/fmicb.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Hu P., Wu W., Wang Y. Peptides with therapeutic potential in the venom of the scorpion Buthus martensii Karsch. Peptides. 2019;115:43–50. doi: 10.1016/j.peptides.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y., Cao L., Zhong M., Zhang Y., Han C., Li Q., Liu F. Anti-HIV-1 activity of a new scorpion venom peptide derivative Kn2-7. PloS One. 2012;7(4) doi: 10.1371/journal.pone.0034947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao L., Dai C., Li Z., Fan Z., Song Y., Wu Y., Li W. Antibacterial activity and mechanism of a scorpion venom peptide derivative in vitro and in vivo. PloS One. 2012;7(7) doi: 10.1371/journal.pone.0040135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baradaran M., Jalali A., Jolodar A., Ghasemian S. New caerin-like antibacterial peptide from the venom gland of the Iranian scorpion Mesobuthus eupeus: cDNA amplification and sequence analysis. Afr. J. Biotechnol. 2012;11(44):10176–10181. doi: 10.5897/AJB11.3373. [DOI] [Google Scholar]
- 42.Oliveira I.S., Ferreira I.G., Alexandre-Silva G.M., Cerni F.A., Cremonez C.M., Arantes E.C., Pucca M.B. Scorpion toxins targeting Kv1. 3 channels: insights into immunosuppression. J. Venom. Anim. Toxins Incl. Trop. Dis. 2019;25 doi: 10.1590/1678–9199–JVATITD–1481–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen P.C., Kuyucak S. Developing a comparative docking protocol for the prediction of peptide selectivity profiles: investigation of potassium channel toxins. Toxins. 2012;4(2):110–138. doi: 10.3390/toxins4020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protocol. Bioinf. 2016;54(1):5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Froimowitz M. HyperChem: a software package for computational chemistry and molecular modeling. Biotechniques. 1993;14(6):1010–1013. [PubMed] [Google Scholar]
- 46.Shen M.Y., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdulazeez S. Molecular simulation studies on B-cell lymphoma/leukaemia 11A (BCL11A) Am. J. Transl. Res. 2019;11(6):3689. [PMC free article] [PubMed] [Google Scholar]
- 48.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26(2):283–291. doi: 10.1107/s0021889892009944. [DOI] [Google Scholar]
- 49.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(suppl_2):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case D.A., et al. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- 51.Tian C., et al. ff19SB: amino-acid-specific protein backbone paramters trained against quantum mechanics energy surfaces in solution. J. Chem. Theor. Comput. 2019;16:528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 52.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 53.Myamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13(8):p952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 54.Farrokhnia M., Mahnam K. Molecular dynamics and docking investigations of several zoanthamine-type marine alkaloids as matrix metaloproteinase-1 inhibitors. Iran. J. Pharm. Res. (IJPR): IJPR. 2017;16(1):173. [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel R.R., Thomas E.C. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9(7):p3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 56.De Vries S.J., Van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5(5):883. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 57.de Oliveira O.V., Rocha G.B., Paluch A.S., Costa L.T. Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1772885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Souza P.F., Lopes F.E., Amaral J.L., Freitas C.D., Oliveira J.T. A molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction with human ACE2 receptor. Int. J. Biol. Macromol. 2020;164:66–76. doi: 10.1016/j.ijbiomac.2020.07.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Bonvin A.M.J.J. The HADDOCK2. 2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 60.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 61.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Holger G., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 62.Weiser J., Shenkin P.S., Still W.C. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J. Comput. Chem. 1999;20(2):217–230. doi: 10.1002/(SICI)1096-987X(19990130)20:2<217::AID-JCC4>3.0.CO;2-A. [DOI] [Google Scholar]
- 63.Dehouck Y., Kwasigroch J.M., Rooman M., Gilis D. BeAtMuSiC: prediction of changes in protein–protein binding affinity on mutations. Nucleic Acids Res. 2013;41(W1):W333–W339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kastritis P., Bonvin A.M.J.J. Are scoring functions in protein-protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark 49. J. Proteome Res. 2010;9:2216–2225. doi: 10.1021/pr9009854. [DOI] [PubMed] [Google Scholar]
- 65.Kondejewski L.H., Jelokhani-Niaraki M., Farmer S.W., Lix B., Kay C.M., Sykes B.D., Hodges R.S. Dissociation of antimicrobial and hemolytic activities in cyclic peptide diastereomers by systematic alterations in amphipathicity. J. Biol. Chem. 1999;274(19):13181–13192. doi: 10.1093/nar/gkt450. [DOI] [PubMed] [Google Scholar]
- 66.Hung A., Mager M., Hembury M., Stellacci F., Stevens M.M., Yarovsky I. Amphiphilic amino acids: a key to adsorbing proteins to nanopatterned surfaces? Chem. Sci. 2013;4(3):928–937. doi: 10.1039/C2SC2163. [DOI] [Google Scholar]
- 67.Huang Y., He L., Li G., Zhai N., Jiang H., Chen Y. Role of helicity of α-helical antimicrobial peptides to improve specificity. Protein & cell. 2014;5(8):631–642. doi: 10.1007/s13238-014-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maurer-Stroh S., Krutz N.L., Kern P.S., Gunalan V., Nguyen M.N., Limviphuvadh V., Eisenhaber F., Gerberick G.F. AllerCatPro-prediction of protein allergenicity potential from the protein sequence. Bioinformatics. 2019;35(17):3020–3027. doi: 10.1093/bioinformatics/btz029. [DOI] [PMC free article] [PubMed] [Google Scholar]