Abstract
Background
Mounting evidence suggests that circulating microRNAs (miRNAs) are critical indicators of cardiovascular disease. However, prospective studies linking circulating miRNAs to incident acute coronary syndrome (ACS) are limited, and the underlying effect of associated miRNA on incident ACS remains unknown.
Methods and Results
Based on a 2‐stage prospective nested case–control design within the Dongfeng‐Tongji cohort, we profiled plasma miRNAs from 23 pairs of incident ACS cases and controls by microarray and validated the candidate miRNAs in 572 incident ACS case–control pairs using quantitative real‐time polymerase chain reaction. We observed that plasma miR‐4286 was associated with higher risk of ACS (adjusted odds ratio according to an interquartile range increase, 1.26 [95% CI, 1.07–1.48]). Further association analysis revealed that triglyceride was positively associated with plasma miR‐4286, and an interquartile range increase in triglyceride was associated with an 11.04% (95% CI, 3.77%–18.83%) increase in plasma miR‐4286. In addition, the Mendelian randomization analysis suggested a potential causal effect of triglyceride on plasma miR‐4286 (β coefficients: 0.27 [95% CI, 0.01–0.53] and 0.27 [95% CI, 0.07–0.47] separately by inverse variance‐weighted and Mendelian randomization‐pleiotropy residual sum and outlier tests). Moreover, the causal mediation analysis indicated that plasma miR‐4286 explained 5.5% (95% CI, 0.7%–17.0%) of the association of triglyceride with incident ACS.
Conclusions
Higher level of plasma miR‐4286 was associated with an increased risk of ACS. The upregulated miR‐4286 in plasma can be attributed to higher triglyceride level and may mediate the effect of triglyceride on incident ACS.
Keywords: acute coronary syndrome, microRNA, miR‐4286, prospective study, triglyceride
Subject Categories: Epidemiology, Acute Coronary Syndromes, Epigenetics
Nonstandard Abbreviations and Acronyms
- FRS
Framingham Risk Score
- IR
insulin resistant
- miRNA
microRNA
- MR
Mendelian randomization
- UA
unstable angina
Clinical Perspective
What Is New?
In our study, we found that plasma miR‐4286 was associated with higher risk of ACS based on a prospective nested case–control design.
Further Mendelian randomization analysis revealed that triglyceride was causally associated with a higher level of plasma miR‐4286.
Plasma miR‐4286 explained 5.5% of the association of triglyceride with incident ACS, suggesting the mediation effect of miR‐4286 in ACS development.
What Are the Clinical Implications?
These findings identified a novel risk contributor to incident ACS at the circulating miRNA level and revealed its mediation potential in ACS development, suggesting a potential target for intervention or prevention of ACS.
Coronary heart disease (CHD) is a major disease burden worldwide. 1 , 2 Acute coronary syndrome (ACS), including acute myocardial infarction (AMI) and unstable angina (UA), is the main fatal subtype of CHD. 3 To date, the precise prevention and management of ACS remains a challenge because of its complicated pathogenic and physiological processes. Previous studies have demonstrated that the development of endothelial dysfunction and subsequent vulnerable plaque rupture or erosion is a major pathophysiological process of ACS. 4 However, the original regulator in ACS progression remains to be elucidated, especially from an epigenetic aspect.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that are involved in posttranscriptional regulation by binding target mRNAs and play important roles in cardiac remodeling, angiogenesis, and vessel development. 5 Given the feature of high stability and sensitivity, circulating miRNAs have been widely characterized as diagnostic and prognostic biomarkers for cardiovascular disease (CVD). 6 , 7 However, prospective clues on circulating miRNAs and risk of ACS are limited and based on Europeans. 8 , 9 Over the past decades, with the change of lifestyles and westernization of the diet, CVD has been the leading cause of death in China, and among cardiovascular deaths, the majority are attributed to ACS. 10 Thereby, prospective associations of circulating miRNAs with incident ACS and its risk factors among the Chinese need to be investigated.
It is now well‐established that hyperlipidemia is a major risk factor for fatal and nonfatal CHD. 11 , 12 With the development of the genome‐wide association study, Mendelian randomization (MR) has become a useful approach to estimating the causal relationship between exposure and outcome. 13 Recent MR studies have demonstrated that abnormal plasma lipid levels were causally associated with CHD, indicating that the genetic variants of lipid traits played a key role in the development of atherosclerosis and vulnerable plaque. 14 , 15 Previous reviews have summarized that liver‐ or vessel‐enriched miRNAs are involved in regulating cholesterol homeostasis. 16 , 17 Additionally, current studies have identified several circulating miRNAs that are related to lipids or are impacted by lipid‐related dietary interventions. 18 , 19 However, there is still uncertainty regarding whether the relationships are causal between aberrant circulating miRNAs and lipid levels, as well as the mediation potential of associated miRNAs in ACS progression.
Herein, we conducted a 2‐stage nested case–control study in the Dongfeng‐Tongji cohort to investigate the associations of plasma miRNAs with incident ACS, and of the associated target miRNA with cardiovascular traits and risk factors. For lipid traits associated with target miRNA, we further evaluated their causal relationships and estimated the mediation effects of target miRNA on the associations between lipids and incident ACS. An overview of the study design is presented in Figure S1.
Methods
Details of methods are provided in Data S1. Data and materials supporting the conclusions of the article are available from the corresponding author on reasonable request.
Study Design and Participants
This 2‐stage prospective nested case–control study was performed in the Dongfeng‐Tongji cohort, 20 which recruited 27 009 retirees of Dongfeng Motor Corporation during September 2008 and June 2010 in Shiyan, China. The first follow‐up was completed in October 2013 and 14 120 individuals were newly recruited. All of the study participants were free of ACS at the baseline of the study (2013). During follow‐up, 595 cases were newly diagnosed ACS during follow‐up through December 31, 2016, including 147 AMI and 448 UA cases. We randomly selected a control who was free of CVD and cancer at the time of the case event to match each incident ACS case using the incidence density sampling method. 21 Each pair was matched on age (±3 years), sex, and blood drawing time (±1 month). All individuals had completed questionnaires and physical examinations, and provided blood samples in 2013. In the discovery stage, we randomly selected 23 pairs of plasma samples to conduct miRNA microarray detection. In the validation stage, the remaining 572 pairs of plasma samples were used for validation by quantitative real‐time polymerase chain reaction. A detailed sampling flowchart is provided in Figure S2A.
The study was approved by the Ethics and Human Subject Committee of Tongji Medical College (2012‐10) and Dongfeng General Hospital. Each participant provided written informed consent.
Discovery and Validation of Candidate miRNAs in Plasma
We used 400 µL plasma to extract RNA by the miRNeasy Serum/Plasma kit (Qiagen, Germany) according to the instruction handbook. Using the Agilent Human miRNA Microarray, Release 21.0, 8x60K (Agilent Technologies Inc, USA), we selected differentially expressed miRNAs by paired t tests in the discovery stage. Candidate miRNAs reaching the significant level (P<0.05 and absolute fold change >1.3, no multiple testing correction) were included for quantitative real‐time polymerase chain reaction validation. Given that the present study was prospective and the cases were relatively healthy status at baseline, and the expression pattern of circulating miRNAs was complex because of its various sources, the miRNA profiles might not be dramatically altered in comparison with controls at baseline. Therefore, we used the uncorrected P value as screening criterion in line with previously published prospective or circulating miRNA‐based studies. 9 , 22 We quantitated candidate miRNAs by TaqMan Advanced miRNA assay according to the protocols of the manufacturer (Applied Biosystems Inc, USA), and the reactions were carried out by QuantStudio 7 Flex Real‐Time PCR System (Applied Biosystems Inc, USA). The miRNA levels were measured by the 2−ΔCt method.
Bidirectional 2‐Sample MR Analyses
We screened 39 and 58 independent loci separately associated with triglyceride and high‐density lipoprotein cholesterol (HDL‐C) with weak linkage disequilibrium (r 2<0.2) as genetic instrumental variables based on the previous reports on Asians (P<5E‐08). For instrumental variables of miR‐4286, we selected 24 single nucleotide polymorphisms of miR‐4286 based on the genome‐wide association study findings from 340 participants in the Dongfeng‐Tongji study. A detailed sampling flowchart is provided in Figure S2B. The proportion of exposure explained by the genetic instruments (adjusted R 2) and F statistics were computed using linear models that fitted the weighted genetic risk score of exposure and adjusted for age and sex. To estimate causal relationships between lipids and miR‐4286, we used the bidirectional 2‐sample MR analyses by the “TwoSampleMR” and “MRPRESSO” packages, which provide inverse variance–weighted, MR‐Egger, weighted median, and MR‐PRESSO methods. 23 , 24
Statistical Analysis
The power estimation for the discovery stage was calculated by the PASS software (version 11.0.10). In the situation of setting 23 pairs of samples, 81% power was obtained when anticipating to achieve minimum significant condition (fold change=1.3, P=0.05) based on the test of eligible 408 miRNAs on the array after data filtering. Baseline characteristics of the case and control groups in 2 stages were compared by t tests or Mann–Whitney U tests for continuous variables, and χ 2 tests or Fisher exact tests for categorical variables.
Data of plasma miRNAs were natural log‐transformed to improve normality. To investigate the association of candidate miRNAs with incident ACS, we categorized the individual miRNAs into tertiles based on their distribution among the controls and fitted conditional logistic regressions to estimate the odds ratio (OR) and 95% CI for incident ACS. Similarly, we also calculated the OR (95% CI) for incident ACS per interquartile range (IQR) increase in the miRNA level. The multivariable model adjusted for established potential confounders including age, body mass index (BMI), smoking status, drinking status, education level, metabolic equivalent, family history of CHD, diabetes mellitus, hypertension, HDL‐C, low‐density lipoprotein cholesterol, triglyceride, and use of lipid‐lowering medication. Missing data of categorical covariates were replaced with an extra category to indicate missingness. Linear trend P value was estimated by assigning the median value of miRNA to each tertile and using this as a continuous variable in the logistic regression model. We defined the statistical significance as a false discovery rate (FDR) <0.1 using the Benjamini‐Hochberg method. 25 We further estimated the risk difference that represented the absolute risk change for ACS attributed to plasma miR‐4286 using generalized linear regression models assuming an identity link function. 26 We also used elastic net regression models with 10‐fold cross‐validation to test the robustness between miRNAs and incident ACS, which combines the Lasso and Ridge penalties to reduce the occurrence of false‐positives. 27
Sensitivity analysis was conducted by excluding ACS events that had been diagnosed within 1 year after collection of blood samples in 2013. In addition, we conducted restricted cubic splines to estimate the associations between target miRNA and incident ACS. We also performed stratified analyses according to age, sex, BMI, current smokers, current drinkers, hypertension, diabetes mellitus, fasting glucose level, and hyperlipidemia. To evaluate the predictive value of ACS‐associated miRNA, we compared the area under the receiver‐operating characteristic curves between the Framingham Risk Score (FRS) for hard CHD and addition of selected miRNA to the FRS, and also calculated the net reclassification improvement and the integrated discrimination improvement to assess the improvement of predictive ability. The net reclassification improvement was calculated according to low (<10%), moderate (10%–30%), and high (>30%) levels of predicted risk of ACS. To estimate the heterogeneity of association between ACS subtypes, we fitted a multinomial logistic regression model with the outcomes (AMI, UA, or non‐ACS) as the dependent variable and miRNA level as independent variable, and adjusted for age, sex, BMI, smoking status, drinking status, education levels, metabolic equivalent, diabetes mellitus, hypertension, family history of CHD, HDL‐C, low‐density lipoprotein cholesterol, triglyceride, and use of lipid‐lowering medication. P value for heterogeneity was obtained from multinomial logistic regression model in the comparison between AMI and UA.
The relationships between lipid traits and target miRNA were estimated by linear regression. Plasma levels of HDL‐C, total cholesterol, triglyceride, and triglyceride/HDL‐C ratio were natural log‐transformed and included in linear models and further analyses. Multivariable models adjusted for age, sex, BMI, smoking status, drinking status, education level, metabolic equivalent, and use of lipid‐lowering drugs. Calculation of linear trend P value, and definition of statistical significance were the same as above. We presented the percent difference in miRNA level when categorizing the participants according to tertiles of each lipid level, which represented the percent change in miRNA in relation to the upper lipid group versus lower lipid group. The percent difference (95% CI) in miRNA level was also estimated in relation to per IQR increase in each lipid level. 28 Furthermore, we used the “mediation” R package, 29 which contained mediator models and outcome models, to estimate the mediation effects of target miRNA between lipids and incident ACS. The mediated proportion reflected the average mediation effect of target miRNA between lipids and incident ACS, estimated by the proportion of indirect effect in total effect.
Analyses were performed with R software (version 3.4.3, R Core Team) or SAS program (version 9.4, SAS Institute).
Results
Characteristics of the Study Participants
The cases were newly diagnosed with ACS within 0.5 to 3.6 years since the 2013 baseline, and the median follow‐up was 1.8 years. Among the participants, mean±SD age was 67.2±7.6 years, and 57.4% were men. In the validation stage, ACS cases were more likely to have a history of hypertension, with higher BMI, fasting glucose, low‐density lipoprotein cholesterol, triglyceride, and lower HDL‐C levels. By contrast, a consistent but weak group difference was detected between ACS cases and controls in the discovery stage (Table). In comparison with respective controls, both AMI and UA cases had higher blood pressure, fasting glucose, and triglyceride levels. In addition, AMI cases were more likely to be less educated, and physically inactive, while UA cases had higher BMI and lower HDL‐C levels (Table S1).
Table 1.
Baseline Characteristics of the 2‐Stage Nested Case–Control Participants
| Variables | Discovery Stage | Validation Stage | ||||
|---|---|---|---|---|---|---|
| ACS Cases (n=23) | Controls (n=23) | P Value | ACS Cases (n=572) | Controls (n=572) | P Value | |
| Age, y | 68.1±6.7 | 68.1±6.7 | 0.96 | 67.2±7.7 | 67.2±7.6 | 0.91 |
| Male, n (%) | 14 (60.9) | 14 (60.9) | 1.00 | 327 (57.2) | 327 (57.2) | 1.00 |
| BMI, kg/m2 | 24.60±3.45 | 23.90±3.58 | 0.50 | 24.71±3.39 | 24.21±3.04 | 0.009 |
| SBP, mm Hg | 146.83±26.00 | 140.22±22.18 | 0.36 | 148.21±24.44 | 142.78±22.37 | <0.001 |
| DBP, mm Hg | 79.04±11.33 | 79.65±12.54 | 0.86 | 83.28±13.26 | 80.67±12.58 | <0.001 |
| Smoking status, n (%) | 1.00 | 0.08 | ||||
| Current smoker | 5 (21.7) | 5 (21.7) | 136 (23.8) | 105 (18.4) | ||
| Former smoker | 4 (17.4) | 4 (17.4) | 119 (20.8) | 110 (19.2) | ||
| Never smoker | 14 (60.9) | 14 (60.9) | 315 (55.1) | 355 (62.1) | ||
| Drinking status, n (%) | 1.00 | 0.11 | ||||
| Current drinker | 9 (39.1) | 10 (43.5) | 158 (27.6) | 185 (32.3) | ||
| Former drinker | 2 (8.7) | 1 (4.3) | 75 (13.1) | 56 (9.8) | ||
| Never drinker | 12 (52.2) | 12 (52.2) | 336 (58.7) | 330 (57.8) | ||
| Education level, n (%) | 0.87 | 0.22 | ||||
| Primary school or below | 7 (30.4) | 8 (34.8) | 175 (30.6) | 154 (26.9) | ||
| Middle school | 10 (43.5) | 8 (34.8) | 218 (38.1) | 212 (37.1) | ||
| High school or beyond | 6 (26.1) | 7 (30.4) | 174 (30.4) | 196 (34.3) | ||
| Physical activity, MET‐h/wk | 21.00 (12.00, 44.00) | 21.00 (14.25, 42.00) | 0.61 | 21.00 (9.25, 42.00) | 23.31 (13.88, 42.00) | 0.05 |
| Family history of CHD, n (%) | 2 (8.7) | 1 (4.3) | 0.61 | 39 (6.8) | 41 (7.2) | 0.91 |
| Lipid‐lowering medication, n (%) | 1 (4.3) | 1 (4.3) | 1.00 | 85 (14.9) | 82 (14.3) | 0.87 |
| Antihypertensive medication, n (%) | 12 (52.2) | 7 (30.4) | 0.23 | 231 (40.4) | 193 (33.7) | 0.02 |
| Antidiabetic medication, n (%) | 4 (17.4) | 3 (13.0) | 1.00 | 76 (13.3) | 57 (10.0) | 0.10 |
| Hyperlipidemia, n (%) | 11 (47.8) | 6 (26.1) | 0.22 | 380 (66.4) | 374 (65.4) | 0.76 |
| Hypertension, n (%) | 16 (69.6) | 15 (65.2) | 1.00 | 449 (78.5) | 409 (71.5) | 0.01 |
| Diabetes mellitus, n (%) | 9 (39.1) | 4 (17.4) | 0.19 | 160 (28.0) | 134 (23.4) | 0.09 |
| FG, mmol/L | 5.90 (5.11, 6.54) | 5.68 (5.50, 6.15) | 0.92 | 5.80 (5.30, 6.50) | 5.69 (5.20, 6.30) | 0.003 |
| HDL‐C, mmol/L | 1.29 (1.17, 1.66) | 1.44 (1.08, 1.60) | 0.84 | 1.31 (1.16, 1.53) | 1.40 (1.19, 1.65) | <0.001 |
| LDL‐C, mmol/L | 2.67±1.02 | 2.60±0.84 | 0.79 | 2.89±0.82 | 2.78±0.86 | 0.03 |
| TC, mmol/L | 4.75 (4.12, 5.39) | 4.34 (3.91, 5.29) | 0.46 | 4.89 (4.29, 5.46) | 4.84 (4.17, 5.45) | 0.15 |
| triglyceride, mmol/L | 1.37 (0.92, 2.19) | 1.36 (1.00, 1.70) | 0.79 | 1.40 (1.04, 2.02) | 1.23 (0.89, 1.68) | <0.001 |
ACS indicates acute coronary syndrome; BMI, body mass index; CHD, coronary heart disease; DBP, diastolic blood pressure; FG, fasting glucose; HDL‐C, high‐density lipoprotein cholesterol; LDL‐C, low‐ density lipoprotein cholesterol; MET, metabolic equivalent; SBP, systolic blood pressure; and TC, total cholesterol.
Selection of Candidate miRNAs in the Discovery Stage
We screened 33 miRNAs to be significantly associated with incident ACS by paired t tests (P<0.05, fold change>1.3). Via comparing the 33 candidate miRNAs to the miRmine database, we removed 11 extremely low abundance miRNAs in plasma. Finally, we selected 18 candidate miRNAs that could be detected in pooled plasma samples for quantitative real‐time polymerase chain reaction validation (Table S2). Hierarchical clustering showed that the cases and controls were grouped distinctly across the heat map of 18 candidate miRNAs (Figure S3).
Identification of Plasma miR‐4286 Associated With Incident ACS in the Validation Stage
Among 14 miRNAs that were selected for further analyses after quality control, most of them were significantly correlated with each other (Spearman’s rank correlation coefficients range from −0.44 to 0.83; Figure S4). After multivariate adjustment, we observed a strong positive association between miR‐4286 and incident ACS; participants with plasma miR‐4286 in the highest tertile of the distribution had a 1.80‐fold increased risk of ACS compared with participants with plasma miR‐4286 in the lowest tertile (OR, 1.80; 95% CI, 1.28–2.53; P‐trend<0.001, FDR=0.01). Corresponding ORs for an IQR increase in plasma miR‐4286 were 1.26 (95% CI, 1.07–1.48; Figure 1). In addition, compared with the lowest tertile, the greatest risk difference was 10.56% (95% CI, 3.46%–17.66%) for the highest tertile of plasma miR‐4286. Meanwhile, an IQR increase in plasma miR‐4286 was associated with an absolute risk increase in incident ACS of 4.43% (95% CI, 0.94%–7.91%; Figure S5). Elastic net model including all 14 candidate miRNAs showed that only miR‐4286 had a nonzero coefficient (β=0.02), indicating a strong association between miR‐4286 and incident ACS.
Figure 1. Associations of candidate miRNAs with incident ACS in the validation stage.
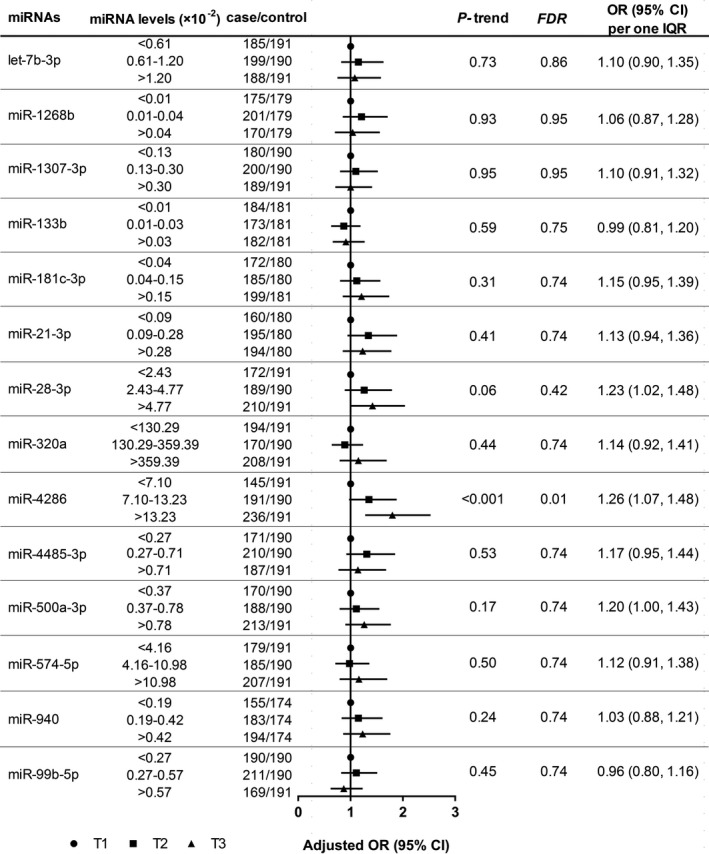
Plasma miRNA levels were normalized to miR‐26b‐5p and were expressed as 2−∆Ct. Adjusted OR (95% CI) for incident ACS was obtained using multivariable conditional logistic regression model with adjustment for age, body mass index, smoking status, drinking status, education levels, metabolic equivalent, diabetes mellitus, hypertension, family history of coronary heart disease, high‐density lipoprotein cholesterol, low‐density lipoprotein cholesterol, triglyceride, and use of lipid‐lowering medication. P‐trend was estimated by assigning the median value of miRNA to each tertile and using this as a continuous variable in the logistic regression model. ACS indicates acute coronary syndrome; FDR, false discovery rate; IQR, interquartile range; miRNAs, microRNAs; and OR, odds ratio.
Of note, the significant result for miR‐4286 was also confirmed by the global mean normalization strategy, and the association remained significant after excluding cases with ACS diagnosed within 1 year (Tables S3 and S4). Cubic spline regression analysis demonstrated a significant nonlinear association between plasma miR‐4286 and incident ACS (nonlinear P=0.002; Figure S6). According to the stratified analyses, no significant interaction was observed between miR‐4286 and ACS‐related risk factors (Table S5). The area under the receiver‐operating characteristic curves for prediction of incident ACS was 0.55 (95% CI, 0.52–0.59, P=0.002), with a sensitivity and specificity being 65.0% and 43.9% at 0.09 of plasma miR‐4286 as a cut‐off value. In comparison with FRS alone, the addition of miR‐4286 increased area under the receiver‐operating characteristic curves from 0.64 (95% CI, 0.61–0.68) to 0.66 (95% CI, 0.63–0.69; P difference=0.07), the net reclassification improvement (95% CI) was 3.2% (0.9%–5.4%, P=0.006), and the integrated discrimination improvement (95% CI) was 1.1% (0.5%–1.8%, P<0.001; Figure S7).
In addition, we observed that miR‐4286 was associated with higher risk of AMI and UA, with no significant heterogeneity observed for AMI and UA (P heterogeneity=0.49; Table S6). It suggested that the associations of plasma miR‐4286 on ACS were consistent across different subtypes. Consequently, we selected miR‐4286 as target miRNA associated with incident ACS to further explore its influencing factors and its role in ACS progression.
Associations of Cardiovascular Traits and Risk Factors With Plasma miR‐4286
We observed a higher miR‐4286 level in females and the significant correlations between lipid traits and plasma miR‐4286 (Tables S7 and S8). Decreased HDL‐C level, increased triglyceride level, and elevated triglyceride/HDL‐C ratio were significantly associated with higher levels of plasma miR‐4286 after multivariable adjustment among all participants and incident ACS cases. According to the results among all participants, comparing the lowest tertiles, the highest tertiles of HDL‐C, triglyceride, and triglyceride/HDL‐C ratio were significantly associated with 18.08% decrease (95% CI, −28.21% to −6.51%; P‐trend=0.003, FDR=0.005), 23.89% increase (95% CI, 8.67%–41.24%; P‐trend=0.001, FDR=0.004), and 23.63% increase (95% CI, 8.37%–41.04%; P‐trend=0.001, FDR=0.004) in plasma miR‐4286, respectively. In addition, an IQR increase in HDL‐C, triglyceride, and triglyceride/HDL‐C ratio was significantly associated with an 11.05% decrease (95% CI, −16.65% to −5.08%), 11.04% increase (95% CI, 3.77%–18.83%), and 15.01% increase (95% CI, 7.16%–to 23.42%) in plasma miR‐4286, respectively (Figure 2). Moreover, we also found the significant associations of triglyceride, HDL‐C, and triglyceride/HDL‐C ratio with incident ACS (Table S9).
Figure 2. Associations of lipid traits with plasma miR‐4286 in the validation stage.
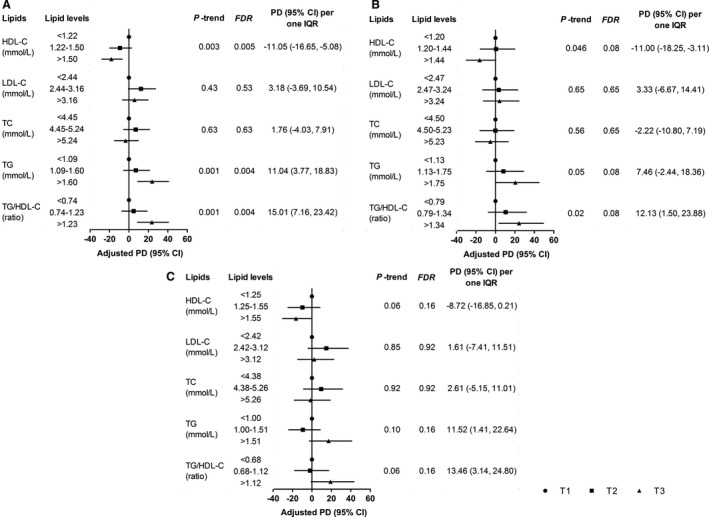
Forest plots of A, B, and C separately show the associations of lipid traits with plasma miR‐4286 among all participants, incident ACS cases, and controls. Adjusted PD (95% CI) for plasma miR‐4286 was obtained using multivariable linear regression model with adjustment for age, sex, body mass index, smoking status, drinking status, education levels, metabolic equivalent, and use of lipid‐lowering medication. P‐trend was estimated by assigning the median value of lipid to each tertile and using this as a continuous variable in the linear regression model. ACS indicates acute coronary syndrome; FDR, false discovery rate; HDL‐C, high‐density lipoprotein cholesterol; IQR, interquartile range; LDL‐C, low‐density lipoprotein cholesterol; PD, percent difference; TC, total cholesterol; and TG, triglyceride.
Causal Relationships Between Lipids and Plasma miR‐4286
The genome‐wide association study results of miR‐4286 are presented in Table S10 and Figure S8, and details of triglyceride and HDL‐C associated instrumental variables information are shown in Tables S11 and S12. The F statistics of triglyceride, HDL‐C, and miR‐4286 were 76.50, 83.65, and 105.41, respectively (Table S13). In the MR analyses, triglyceride was causally associated with an increased level of miR‐4286, and the adjusted β (95% CI) according to per 1‐unit increase was 0.27 (0.01–0.53; P=0.04) by conventional inverse variance–weighted test. Although the significance in MR‐Egger and weighted median tests was marginal, the significant effect was validated by the MR‐PRESSO test (0.27 [0.07–0.47]; P=0.01). The HDL‐C was negatively associated with miR‐4286 in MR‐PRESSO test, the adjusted β (95% CI) according to per 1‐unit increase was −0.20 (−0.39 to −0.01; P=0.04; Figure 3 and Figure S9). Notably, there was no suggestion of a reverse causal effect of miR‐4286 on triglyceride or HDL‐C (Figures S10 and S11).
Figure 3. Causal effects of TG and HDL‐C on plasma miR‐4286.
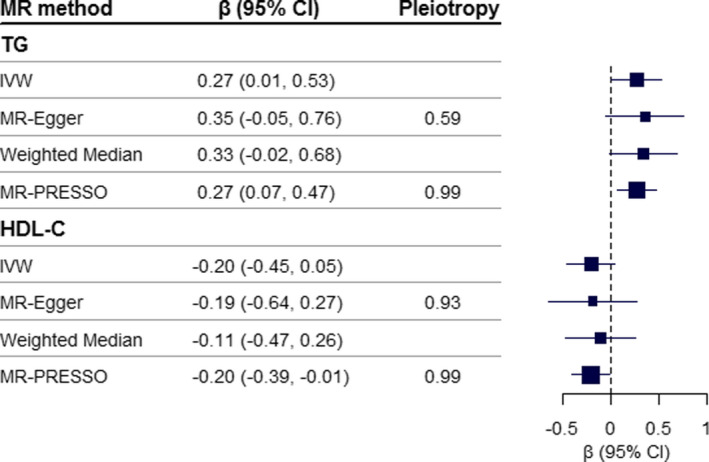
The causal effect (95% CI) of TG and HDL‐C on miR‐4286 was estimated by Mendelian randomization analysis, using 39 TG SNPs and 58 HDL‐C SNPs derived from Asian reports, respectively. Pleiotropy P value derived from the intercept of MR‐Egger test or MR‐PRESSO Global test, a small P value indicates an unbalanced pleiotropy. HDL‐C, high‐density lipoprotein cholesterol; IVW, inverse variance‐weighted; SNPs, single nucleotide polymorphisms; and TG, triglyceride.
Causal Mediation Effects of Plasma miR‐4286 on the Associations of Lipids and Incident ACS
The positive associations of triglyceride and triglyceride/HDL‐C ratio with incident ACS were partly mediated through an increasing level of plasma miR‐4286, and the mediation proportions (95% CI) were 5.5% (0.7%–17.0%; P mediation=0.02) and 6.6% (0.9%–19.5%; P mediation=0.02), respectively. Nevertheless, no significant mediation effect was detected for HDL‐C as exposure, presumably because of the weak association between HDL‐C and incident ACS (Figure 4).
Figure 4. Mediation effects of plasma miR‐4286 on the associations of lipids with incident ACS.
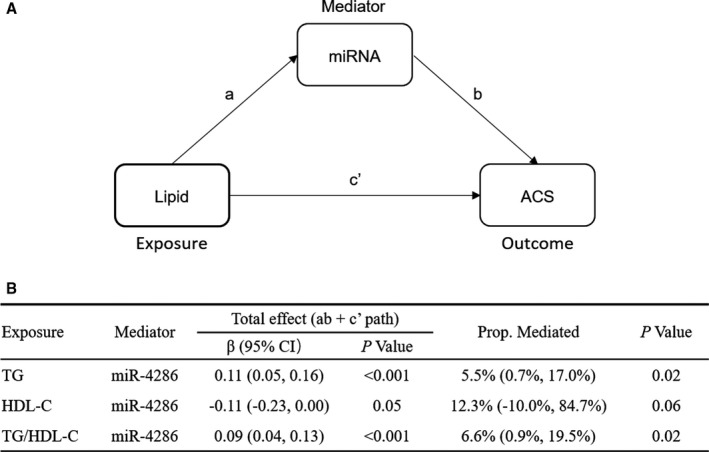
A, Path diagram: Total effect (c)=direct effect (c′)+indirect effect (ab). B, Total effects and the proportion of mediation effects were obtained by quasi‐Bayesian Monte Carlo simulation for 10 000 times in the R package “mediation.” Indirect effects were derived from multiple linear regression for the association of lipid with miRNA level adjusting for age, sex, body mass index, smoking status, drinking status, education levels, metabolic equivalent, and use of lipid‐lowering medication. Direct effects were calculated by logistic regression for the association of lipid with ACS additional adjusting for miRNA level, diabetes mellitus, hypertension, family history of coronary heart disease, and low‐density lipoprotein cholesterol. ACS indicates acute coronary syndrome; HDL‐C, high‐density lipoprotein cholesterol; miRNA, microRNA; and TG, triglyceride.
Discussion
Based on a 2‐stage nested case–control design, we found that plasma miR‐4286 was positively associated with incident ACS and causally related to triglyceride and HDL‐C. Importantly, an increase in plasma miR‐4286 also partially mediated the positive association of triglyceride and triglyceride/HDL‐C ratio with incident ACS. To summarize, our study identified a novel risk contributor of incident ACS and indicated its mediation potential in ACS development.
According to the prospective findings from the Bruneck cohort and HUNT (Nord‐Trøndelag Health Study) cohort, circulating miRNAs associated with risk of AMI were nonoverlapping. 8 , 9 Interestingly, in the present study, we also found none of the predicted biomarkers as abovementioned to be associated with incident ACS. In comparison with the Bruneck cohort including 47 incident AMI cases and 773 controls, we performed the nested case–control design that used age‐ and sex‐matched cases and controls, which enhanced statistical power and reduced heterogeneity. 8 Although the HUNT study also performed the nested case–control design, they have additionally matched on risk factors including BMI, total cholesterol, and HDL‐C. These could be potential explanations for the discrepancy, given that the miRNA identified in the present study was correlated with lipid levels. 9 In addition, the different platform and normalized strategy for miRNA analysis might lead to technical differences. In line with the HUNT study, we also confirmed our result by global mean normalization strategy. Of note, the association between miR‐4286 and incident ACS was moderate in this method. This weaker association could be explained by the fact that the closely associated miRNAs cluster might bias the average of cycle thresholds. 8
As for the predictive potential of plasma miR‐4286 for incident ACS, we observed a slight but significant improvement for net reclassification with the FRS plus miR‐4286, suggesting that miR‐4286 might be a novel ACS risk predictor independent of traditional risk factors. Nevertheless, given that adding plasma miR‐4286 to the FRS model could not significantly increase the area under the receiver‐operating characteristic curves for incident ACS prediction, the optimal combination of biomarkers including miRNAs or clinical traits in ACS risk prediction should be further explored.
Participants with abnormal levels of plasma triglyceride and HDL‐C remain at a high risk of CVD even with normal low‐density lipoprotein cholesterol level. 30 Higher triglyceride and lower HDL‐C levels, as previously reported, were symbolic of dyslipidemia of insulin resistance (IR), which played an important role in the development of CVD. 31 Other studies have demonstrated that the triglyceride/HDL‐C ratio was positively associated with insulin sensitivity, and thus was considered a useful surrogate index to estimate IR. 32 Herein, we identified potentially causal associations of triglyceride and HDL‐C with plasma miR‐4286, as well as a positive association between triglyceride/HDL‐C ratio and miR‐4286. Accordingly, we suggested that an increase in plasma miR‐4286 might be caused by abnormal triglyceride and HDL‐C levels accompanied by impaired insulin sensitivity in the development of ACS. The impact of systematic chronic inflammation caused by impaired insulin sensitivity and abnormal lipid levels might account for the observation, which might stimulate the expression and release of miRNAs involved in the metabolic disturbance and ACS progression. 33
According to pathway enrichment analysis, our result suggested that the highly expressed target genes of miR‐4286 in the cardiovascular system were most enriched in the IR pathway (Figure S12). Previous studies have demonstrated the crucial role of IR in endothelial dysfunction. 34 Indeed, proliferation, migration, and dysfunction of endothelial cells is a key point in the development of atherosclerosis and vulnerable plaque. 35 In this study, miR‐4286 could partially mediate the impact of triglyceride and triglyceride/HDL‐C on incident ACS. It seems possible that endothelium might be a potential target for the mediation effect of miR‐4286 on triglyceride‐associated ACS. Although the mechanism role of miR‐4286 in the IR‐related pathway in endothelial cells was lacking, functional study has found that miR‐4286 played an important part in cell proliferation and migration via regulating the PI3K/AKT pathway in the development of lung cancer. 36 From a clinical perspective, our findings inferred a possible novel target for miRNA‐based therapeutics. 37 Further experimental studies are required to investigate the functional role of miR‐4286 in endothelial cells, especially in insulin‐related pathways.
As mentioned in a review, 38 the pool of circulating miRNAs is the sum of secreted and leaked miRNAs from different cells. MiRNAs derived from active secretion via microvesicles or binding with HDL or Argonaute2 remain stable in the extracellular environment. On the contrary, passively leaked miRNAs from injured cells were susceptible to degradation. According to the EVmiRNA database 39 —an up‐to‐date extracellular vesicle miRNA database including 462 sequencing data sets from 17 tissues—we found that miR‐4286 was undetectable in the secreted exosomes from endothelial cells or epithelial cells. However, it remains to be investigated whether miR‐4286 was passively released from endothelial cells or derived from other cardiovascular cells. In addition, based on the abovementioned database, miR‐4286 was also enriched in exosomes from B lymphoblastoid cell lines and lymph. It suggested that lymphocytes might be one of the sources of plasma miR‐4286.
There are several strengths in the present study. First, our study was a prospective nested case–control design based on a well‐established large prospective cohort, which could provide a relatively strong basis for the associations between plasma miRNAs and incident ACS. Second, we adjusted for traditional risk factors and lipid‐lowering medication used in ACS in all association analyses to reduce potential confounding bias. Third, we further investigated the internal causal relationships of plasma miR‐4286 and lipids through MR analyses and explored the causal mediation effect of miR‐4286 on triglyceride‐incident ACS association.
Several limitations in this study should also be acknowledged. First, compared with previous prospective studies, the follow‐up period of 3 years was relatively short. 8 , 9 However, we have excluded incident ACS cases that were diagnosed within 6 months after blood collections, and our findings were largely unchanged after further excluding cases with a follow‐up period <1 year. Second, the sample size of the microarray exploration data was limited, which may contribute to a relatively high false‐positive rate. Although we used a 1:1 matched strategy to improve the statistical power and verified the association by correcting multiple testing in the validation stage, further long‐term prospective studies with a larger sample size are needed to replicate our results. Third, there is a lack of established endogenous control to normalize the circulating miRNA levels. In response, we chose 3 eligible miRNAs as candidate endogenous controls and used acknowledged NormFinder to estimate the optimal endogenous control and repeated a global mean normalized strategy to check the stability of our results. Fourth, expression patterns and function of miR‐4286 in tissues remain largely unclear. Although there is a lack of evidence for the underlying mechanism, we inferred a potential role of miR‐4286 in regulating insulin sensitivity via functional pathway analysis and provided a basis for further mechanism research. Finally, we could not determine the source of miR‐4286, given that dysregulation of miRNAs in different tissues might be affected by different factors. Further systematic exploration of the source of circulating miR‐4286, as well as its regulatory mechanism, needs to be conducted.
In conclusion, the present study identified a positive association of plasma miR‐4286 with incident ACS in a Chinese population. We indicated the causal effect of triglyceride on plasma miR‐4286, and highlighted a possible mediation role of plasma miR‐4286 in triglyceride‐incident ACS association. Further prospective and functional studies are warranted to systemically elucidate the biological mechanisms behind these observations and translate these results into clinical practice for early and precise prevention and treatment.
Sources of Funding
This work was supported by the National Key Research and Development Program of China (2016YFC0900800), the National Natural Science Foundation of China (81930092), the Fundamental Research Funds for the Central Universities (2019kfyXMBZ015), the 111 Project, and the Program for Changjiang Scholars and Innovative Research Team in University.
Disclosures
None.
Supporting information
Supplemental Methods
Tables S1–S13
Figures S1–S12
References 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58
Acknowledgments
We thank all participants and staffs of Dongfeng‐Tongji cohort for their cooperation.
(J Am Heart Assoc. 2021;10:e018999. DOI: 10.1161/JAHA.120.018999.)
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.120.018999.
For Sources of Funding and Disclosures, see page 11.
References
- 1. GBD . 2017 Causes of Death Collaborators. Global, regional, and national age‐sex‐specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1736–1788. DOI: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilkins JT, Ning H, Berry J, Zhao L, Dyer AR, Lloyd‐Jones DM. Lifetime risk and years lived free of total cardiovascular disease. JAMA. 2012;308:1795–1801. DOI: 10.1001/jama.2012.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Connor RE, Bossaert L, Arntz HR, Brooks SC, Diercks D, Feitosa‐Filho G, Nolan JP, Vanden Hoek TL, Walters DL, Wong A, et al. Part 9: acute coronary syndromes: 2010 international consensus on cardiopulmonary resuscitation and emergency cardiovascular care science with treatment recommendations. Circulation. 2010;122:S422–S465. DOI: 10.1161/CIRCULATIONAHA.110.985549. [DOI] [PubMed] [Google Scholar]
- 4. Crea F, Libby P. Acute coronary syndromes: the way forward from mechanisms to precision treatment. Circulation. 2017;136:1155–1166. DOI: 10.1161/CIRCULATIONAHA.117.029870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. DOI: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res. 2017;120:381–399. DOI: 10.1161/CIRCRESAHA.116.308434. [DOI] [PubMed] [Google Scholar]
- 7. Karakas M, Schulte C, Appelbaum S, Ojeda F, Lackner KJ, Münzel T, Schnabel RB, Blankenberg S, Zeller T. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease‐results from the large AtheroGene study. Eur Heart J. 2017;38:516–523. [DOI] [PubMed] [Google Scholar]
- 8. Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–299. DOI: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 9. Bye A, Røsjø H, Nauman J, Silva GJ, Follestad T, Omland T, Wisløff U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals ‐ the HUNT study. J Mol Cell Cardiol. 2016;97:162–168. DOI: 10.1016/j.yjmcc.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 10. Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, Li X, Wang L, Wang L, Liu Y, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145–1158. DOI: 10.1016/S0140-6736(19)30427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ference BA, Bhatt DL, Catapano AL, Packard CJ, Graham I, Kaptoge S, Ference TB, Guo Q, Laufs U, Ruff CT, et al. Association of genetic variants related to combined exposure to lower low‐density lipoproteins and lower systolic blood pressure with lifetime risk of cardiovascular disease. JAMA. 2019;322:1381–1391. DOI: 10.1001/jama.2019.14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abdullah SM, Defina LF, Leonard D, Barlow CE, Radford NB, Willis BL, Rohatgi A, McGuire DK, de Lemos JA, Grundy SM, et al. Long‐term association of low‐density lipoprotein cholesterol with cardiovascular mortality in individuals at low 10‐year risk of atherosclerotic cardiovascular disease. Circulation. 2018;138:2315–2325. DOI: 10.1161/CIRCULATIONAHA.118.034273. [DOI] [PubMed] [Google Scholar]
- 13. Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. DOI: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 14. Voight BF, Peloso GM, Orho‐Melander M, Frikke‐Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. DOI: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holmes MV, Asselbergs FW, Palmer TM, Drenos F, Lanktree MB, Nelson CP, Dale CE, Padmanabhan S, Finan C, Swerdlow DI, et al. Mendelian randomization of blood lipids for coronary heart disease. Eur Heart J. 2015;36:539–550. DOI: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willeit P, Skroblin P, Kiechl S, Fernández‐Hernando C, Mayr M. Liver microRNAs: potential mediators and biomarkers for metabolic and cardiovascular disease? Eur Heart J. 2016;37:3260–3266. DOI: 10.1093/eurheartj/ehw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feinberg MW, Moore KJ. MicroRNA regulation of atherosclerosis. Circ Res. 2016;118:703–720. DOI: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimura Y, Tamasawa N, Matsumura K, Murakami H, Yamashita M, Matsuki K, Tanabe J, Murakami H, Matsui J, Daimon M. Clinical significance of determining plasma microRNA33b in type 2 diabetic patients with dyslipidemia. J Atheroscler Thromb. 2016;23:1276–1285. DOI: 10.5551/jat.33670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quintanilha BJ, Pinto Ferreira LR, Ferreira FM, Neto EC, Sampaio GR, Rogero MM. Circulating plasma microRNAs dysregulation and metabolic endotoxemia induced by a high‐fat high‐saturated diet. Clin Nutr. 2020;39:554–562. DOI: 10.1016/j.clnu.2019.02.042. [DOI] [PubMed] [Google Scholar]
- 20. Wang F, Zhu J, Yao P, Li X, He M, Liu Y, Yuan J, Chen W, Zhou L, Min X, et al. Cohort profile: the Dongfeng‐Tongji cohort study of retired workers. Int J Epidemiol. 2013;42:731–740. DOI: 10.1093/ije/dys053. [DOI] [PubMed] [Google Scholar]
- 21. Richardson DB. An incidence density sampling program for nested case‐control analyses. Occup Environ Med. 2004;61:e59. DOI: 10.1136/oem.2004.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jernås M, Nookaew I, Wadenvik H, Olsson B. MicroRNA regulate immunological pathways in T‐cells in immune thrombocytopenia (ITP). Blood. 2013;121:2095–2098. DOI: 10.1182/blood-2012-12-471250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR‐Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. DOI: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. DOI: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple hypothesis testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 26. Holmberg MJ, Andersen LW. Estimating risk ratios and risk differences: alternatives to odds ratios. JAMA. 2020;324:1098–1099. DOI: 10.1001/jama.2020.12698. [DOI] [PubMed] [Google Scholar]
- 27. Waldmann P, Mészáros G, Gredler B, Fuerst C, Sölkner J. Evaluation of the lasso and the elastic net in genome‐wide association studies. Front Genet. 2013;4:270. DOI: 10.3389/fgene.2013.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrera‐Gómez J, Basagaña X. Models with transformed variables: interpretation and software. Epidemiology. 2015;26:e16–e17. DOI: 10.1097/EDE.0000000000000247. [DOI] [PubMed] [Google Scholar]
- 29. Tingley D, Yamamoto T, Hirose K, Keele L, Imai K. mediation: R package for causal mediation analysis. J Stat Softw. 2014;59:1–38.26917999 [Google Scholar]
- 30. Chapman MJ, Ginsberg HN, Amarenco P, Andreotti F, Borén J, Catapano AL, Descamps OS, Fisher E, Kovanen PT, Kuivenhoven JA, et al. Triglyceride‐rich lipoproteins and high‐density lipoprotein cholesterol in patients at high risk of cardiovascular disease: evidence and guidance for management. Eur Heart J. 2011;32:1345–1361. DOI: 10.1093/eurheartj/ehr112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–645. DOI: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giannini C, Santoro N, Caprio S, Kim G, Lartaud D, Shaw M, Pierpont B, Weiss R. The triglyceride‐to‐HDL cholesterol ratio: association with insulin resistance in obese youths of different ethnic backgrounds. Diabetes Care. 2011;34:1869–1874. DOI: 10.2337/dc10-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. DOI: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 34. Kim JA, Montagnani M, Koh KK, Quon MJ. Reciprocal relationships between insulin resistance and endothelial dysfunction: molecular and pathophysiological mechanisms. Circulation. 2006;113:1888–1904. DOI: 10.1161/CIRCULATIONAHA.105.563213. [DOI] [PubMed] [Google Scholar]
- 35. Gutiérrez E, Flammer AJ, Lerman LO, Elizaga J, Lerman A, Fernández‐Avilés F. Endothelial dysfunction over the course of coronary artery disease. Eur Heart J. 2013;34:3175–3181. DOI: 10.1093/eurheartj/eht351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ling C, Wang X, Zhu J, Tang H, Du W, Zeng Y, Sun L, Huang JA, Liu Z. MicroRNA‐4286 promotes cell proliferation, migration, and invasion via PTEN regulation of the PI3K/Akt pathway in non‐small cell lung cancer. Cancer Med. 2019;8:3520–3531. DOI: 10.1002/cam4.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. DOI: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 38. Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. DOI: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 39. Liu T, Zhang Q, Zhang J, Li C, Miao YR, Lei Q, Li Q, Guo AY. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47:D89–D93. DOI: 10.1093/nar/gky985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Gara PT, Kushner FG, Ascheim DD, Casey DE Jr, Chung MK, de Lemos JA, Ettinger SM, Fang JC, Fesmire FM, Franklin BA, et al. 2013 ACCF/AHA guideline for the management of ST‐elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;127:e362–e425. [DOI] [PubMed] [Google Scholar]
- 41. Anderson JL, Adams CD, Antman EM, Bridges CR, Califf RM, Casey DE Jr, Chavey WE II, Fesmire FM, Hochman JS, Levin TN, et al. 2012 ACCF/AHA focused update incorporated into the ACCF/AHA 2007 guidelines for the management of patients with unstable angina/non‐ST‐elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;127:e663–e828. [DOI] [PubMed] [Google Scholar]
- 42. Fang Q, Wang Z, Zhan Y, Li D, Zhang K, Zhou T, Yang H, Zhang C, Li X, Min X, et al. Hearing loss is associated with increased CHD risk and unfavorable CHD‐related biomarkers in the Dongfeng‐Tongji cohort. Atherosclerosis. 2018;271:70–76. DOI: 10.1016/j.atherosclerosis.2018.01.048. [DOI] [PubMed] [Google Scholar]
- 43. Xiao Y, Yuan Y, Liu Y, Yu Y, Jia N, Zhou L, Wang H, Huang S, Zhang Y, Yang H, et al. Circulating multiple metals and incident stroke in Chinese adults. Stroke. 2019;50:1661–1668. DOI: 10.1161/STROKEAHA.119.025060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang W, Li Z, Wu L, Cao Z, Liang Y, Yang H, Wang Y, Wu T. Longer habitual afternoon napping is associated with a higher risk for impaired fasting plasma glucose and diabetes mellitus in older adults: results from the Dongfeng‐Tongji cohort of retired workers. Sleep Med. 2013;14:950–954. DOI: 10.1016/j.sleep.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 45. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova‐Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. DOI: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59:S1–S6. DOI: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 47. Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto‐Sietsma SJ. Normalization panels for the reliable quantification of circulating microRNAs by RT‐qPCR. FASEB J. 2015;29:3853–3862. DOI: 10.1096/fj.15-271312. [DOI] [PubMed] [Google Scholar]
- 48. Bargaje R, Hariharan M, Scaria V, Pillai B. Consensus miRNA expression profiles derived from interplatform normalization of microarray data. RNA. 2010;16:16–25. DOI: 10.1261/rna.1688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Navarro‐Quiroz E, Pacheco‐Lugo L, Lorenzi H, Díaz‐Olmos Y, Almendrales L, Rico E, Navarro R, España‐Puccini P, Iglesias A, Egea E, et al. High‐throughput sequencing reveals circulating miRNAs as potential biomarkers of kidney damage in patients with systemic lupus erythematosus. PLoS One. 2016;11:e0166202. DOI: 10.1371/journal.pone.0166202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bhome R, Goh RW, Bullock MD, Pillar N, Thirdborough SM, Mellone M, Mirnezami R, Galea D, Veselkov K, Gu Q, et al. Exosomal microRNAs derived from colorectal cancer‐associated fibroblasts: role in driving cancer progression. Aging (Albany NY). 2017;9:2666–2694. DOI: 10.18632/aging.101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. D'haene B, Mestdagh P, Hellemans J, Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol. 2012;822:261–272. [DOI] [PubMed] [Google Scholar]
- 52. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real‐time quantitative reverse transcription‐PCR data: a model‐based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. DOI: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 53. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. DOI: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang L, Ma L, Guo W, Fang Q, Lai X, Zhang X. Interaction of polymorphisms in APOA4‐APOA5‐ZPR1‐BUD13 gene cluster and sleep duration on 5‐year lipid changes in middle aged and older Chinese. Sleep. 2019;42:1–9. DOI: 10.1093/sleep/zsz115. [DOI] [PubMed] [Google Scholar]
- 55. Wellcome Trust Case Control Consortium . Genome‐wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta‐analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. DOI: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu X, Peloso GM, Liu DJ, Wu Y, Zhang H, Zhou W, Li J, Tang CS, Dorajoo R, Li H, et al. Exome chip meta‐analysis identifies novel loci and East Asian‐specific coding variants that contribute to lipid levels and coronary artery disease. Nat Genet. 2017;49:1722–1730. DOI: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400. DOI: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods
Tables S1–S13
Figures S1–S12
References 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58


