Abstract
Nuclear receptor related 1 protein (Nurr1) is an important transcription factor required for differentiation and maintenance of midbrain dopaminergic (DA) neurons. Since decrease in Nurr1 function either due to diminished expression or rare mutation is associated with Parkinson’s disease (PD), upregulation of Nurr1 may be beneficial for PD. However, such mechanisms are poorly understood. This study underlines the importance of peroxisome proliferator-activated receptor (PPAR)α in controlling the transcription of Nurr1. Our mRNA analyses followed by different immunoassays clearly indicated that PPARα agonist gemfibrozil strongly upregulated the expression of Nurr1 in wild-type, but not PPARα−/−, DA neurons. Moreover, identification of conserved PPRE in the promoter of Nurr1 gene followed by chromatin immunoprecipitation analysis, PPRE luciferase assay, and manipulation of Nurr1 gene by viral transduction of different PPARα plasmids confirmed that PPARα was indeed involved in the expression of Nurr1. Finally, oral administration of gemfibrozil increased Nurr1 expression in vivo in nigra of wild-type, but not PPARα−/−, mice identifying PPARα as a novel regulator of Nurr1 expression and associated protection of DA neurons.
Keywords: Parkinson’s disease, Nurr1, Gemfibrozil, PPARα, Dopaminergic neurons
Introduction
PD is the devastating neurodegenerative disease of ventral midbrain, which is characterized by the progressive loss of dopaminergic (DA) neurons [1, 2]. The loss of DA neurons in the substantia nigra pars compacta (SNpc) results in a deficit of dopamine in the striatum. A decrease in striatal level of dopamine impairs the motor activity in PD patients and has also been associated with some non-motor symptoms. Recent studies have identified that DA neurons are associated with the formation of alpha-synuclein aggregates, which is often coupled with significant increase in proinflammatory and oxidative species. While effective, therapeutic interventions utilizing L-dopa and other approved drugs do not meaningfully address the loss of DA neurons in the SNpc and their projections in the striatum [3, 4]. In line with these findings, intriguing targets specific to DA neurons have emerged and may hold significant value for slowing degeneration in at-risk individuals.
One such target is the conserved nuclear orphan receptor family (NR4A) which consists of three members: Nur77 (NR4A1), (NR4A2/nurr1), and NOR1 (NR4A3) [5]. Mutations in the NR4A superfamily of nuclear receptor proteins, including the nurr1 gene, have been identified in subsets in late-onset familial forms of PD [6]. A much broader set of literature links the activity of nurr1 to the development and maintenance of DA neurons [7–9]. Nurr1 protein is highly expressed in mesencephalic DA neurons and appears to be central to their survival [10]. Nurr1 production is critical to the expression of genes associated with the production and storage of DA in DAergic neurons including tyrosine hydroxylase (TH), dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2), and l-aromatic amino acid decarboxylase (AADC) [11–13]. Decreased Nurr1 expression has been reported in postmortem studies investigating the brains of PD patients, especially in neurons containing Lewy bodies [14]. Nurr1 is also expressed in microglia and astrocytes. Its contribution in these cells has been investigated due to the protein’s protective properties that are principally based on its ability to reduce the expression of proinflammatory genes and optimize Lewy body clearance [5, 15, 16]. Nurr1 is also implicated in synaptic plasticity and its positive modulation may be useful for PD patients suffering from comorbid symptoms such as anxiety, sleep disturbances, and general disruptions in the default mode network. Synthetic agonists aimed to increase Nurr1 activity have been also reported to be beneficial in animal models of PD [11, 12, 17, 18].
Gemfibrozil, known as Lopid® in the pharmacy, is an FDA-approved lipid-lowering drug. It has been used safely in humans for the treatment of hypertriglyceridemia for nearly 30 years. It is also an agonist of peroxisome proliferator-activated receptor (PPAR)α. Here, we report that gemfibrozil and another fibrate drug fenofibrate increase Nurr1 expression in DA neurons. Increase in Nurr1 in WT, but not PPARα−/−, DA neurons by gemfibrozil, restoration of gemfibrozil-mediated Nurr1 expression in PPARα−/− DA neurons by over-expression of PPARα, presence of PPRE in the promoter of Nurr1 gene, and recruitment of PPARα to the Nurr1 gene promoter by gemfibrozil treatment suggest that gemfibrozil stimulates the transcription of Nurr1 in DA neurons via PPARα. These results suggest that gemfibrozil and other PPARα activators may be beneficial for DA neurons.
Materials and Methods
Reagents
Dulbecco’s modification of Eagle’s medium (DMEM without L-glutamine) was purchased from Mediatech (Washington, D.C.). Fetal bovine serum (FBS) was obtained from Atlas Biologicals (Fort Collins, CO). Antibiotic-antimycotic solution for cell culture, gemfibrozil, GSK0660 (an antagonist of PPARβ), GW9662 (an antagonist of PPARγ), and 1-methyl-4-phenylpyridinium iodide (MPP+) were obtained from Sigma. All the antibodies used in this study are listed in Table 1.
Table 1.
Antibodies used in this study
| Antibody | Manufacturer | Catalog | Host | Application | Dilution |
|---|---|---|---|---|---|
| Nurr1 | Millipore | ABE1455 | Rabbit | WB&IF | WB—1000 IF—1:250 |
| Tyrosine Hydroxylase | Immunostar | 22941 | Mouse | WB&IF | WB—1:1000 IF—1:250 |
| IgG | Santa Cruz | SC2025 | Mouse | ChIP | 2 μg/106 cells |
| PPAR | Santa Cruz | SC39834 | Mouse | ChIP | 2 μg/106 cells |
| PPAR | Santa Cruz | SC74517 | Mouse | ChIP | 2 μg/106 cells |
| PPAR | Santa Cruz | SC7273 | Mouse | ChIP | 2 |
| RNA polymerase II | Abcam | AB-627 | Mouse | ChIP | 2 μg/106 cells |
| Beta-actin | Abcam | AB-627 | Mouse | WB | WB—1:1000 |
WB western blot, IF immunofluorescence, ChIP chromatin immuno-precipitation
MN9D Cells
MN9D neuronal cell line was obtained from Dr. A. Heller (University of Chicago, Chicago, IL, USA) and maintained in the lab. Cells were grown in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum and allowed to differentiate in neurobasal media containing 2% B27, glutamine, and 1% antibiotic-antimycotic solution. For experiments, prior to gemfibrozil treatment, cells were incubated in neurobasal media containing B27 minus antioxidants.
Isolation of Mouse Primary Dopaminergic Neurons
DA neurons were isolated from mixed glial cultures as described earlier by our lab [19]. Animal maintenance and experimental protocols were approved by the Rush University Animal Care Committee. Briefly, pregnant female C57BL/6 (WT) and PPARα−/− mice were euthanized via cervical dislocation; the embryonic pups (embryonic stage 13.5) removed and then quickly decapitated in serum-free DMEM. Considerable effort was made to isolate only the developing nigra tissue in order to maximize quantities of DA neurons. The isolated nigra tissue was milled three times, and the cells centrifuged at 1000 rpm for 10 min. Pelleted tissue was resuspended in fresh serum-free DMEM. After three rounds of centrifugation and resuspension, the cells were plated in 6-well plates containing coverslips and incubated for 8 days at 37 °C with 5% CO2 in complete DMEM containing 20% FBS and antibiotic-antimycotic prior to their use in subsequent experiments.
Semi-quantitative Reverse Transcriptase-Coupled PCR
Total RNA was isolated from MN9D cells using V Quick-RNA™ MicroPrep kit (Zymogen) following the manufacturer’s protocol. Semi-quantitative reverse transcriptase (RT)-PCR was carried out as described earlier [20, 21] using oligo (dT) as primer and Moloney murine leukemia virus reverse transcriptase (MMLV-RT, Invitrogen) in a 20-μL reaction mixture. The resulting cDNA was appropriately amplified using Promega Master Mix (Madison, WI) and the following were primers (Invitrogen) for murine genes:
Nurr1 sense, 5′-CCGGTCATGGCTTTCCCTAA-3′
Antisense, 5′-AGACAGAGGTAGTTGGGTCGG-3′
Nur77 sense, 5′-AGTTGGGGGAGTGTGCTAGA-3′
Antisense, 5′-TCATAAGTCTGGCTCGGGGA-3′
Nor1 sense, 5′-TGACTCTCCCCCAATCCAGA-3′
Antisense, 5′-GCAGGGCATATCTGGAGGGTA-3′
Gapdh sense, 5′-GGTGAAGGTCGGAGTCAACG-3′
Antisense, 5′-GTGAAGACGCCAGTGGACTC-3′
Amplified products were electrophoresed on 2% agarose (Invitrogen) gels and visualized by ethidium bromide (Invitrogen) staining. Response of the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) genewas usedasa loading control to ascertain that an equivalent amount of cDNA was synthesized from each sample.
Quantitative Real-Time PCR
The mRNA quantification was performed in ABI-Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA) using iTaq™ Fast Supermix With ROX (Bio-Rad) and the primers for nurr1 and gapdh as listed above. The mRNA expression of the targeted genes was normalized to the level of gapdh mRNA, and data were processed by the ABI Sequence Detection System 1.6 software.
Immunostaining
Immunofluorescence was performed as described earlier. Briefly, MN9D cells and/or mouse primary DA neurons were cultured to 70–80% confluence in 8-well chamber slides or 6-well culture dishes containing coverslips, fixed with chilled methanol (Fisher Scientific, Waltham, MA) overnight, followed by two brief rinses with filtered PBS containing Tween 20 (Sigma) and Triton X-100 (Sigma) (PBSt). Samples were blocked with 2% BSA (Fisher Scientific) in PBSt for 30 min and incubated at room temperature under shaking conditions for 24 h in PBSt containing the following anti-rabbit or anti-mouse primary antibodies: Nurr1 (1:250, anti-rabbit; Millipore; AB5778) and TH (1:200 Immunostar, Hudson WI) (Table 1). After three 15-min washes in filtered PBSt, slides were further incubated with Cy2-, Cy3-, or Cy5-labeled secondary antibodies (all 1:200; Jackson ImmunoResearch, West Grove, PA) for 1 h under similar shaking conditions. Following three 15-min washes with filtered PBS, cells were incubated for 4–5 min with 4′,6-diamidino-2-phenylindole (DAPI, 1:10,000; Sigma). The samples were run in an EtOH and xylene (Fisher) gradient, mounted, and observed under an Olympus BX51 fluorescence microscope.
Transduction with a Dominant-Negative Mutant of PPARα (Y464D) and eGFP Lentivirus
Cells were transduced with different lentiviral constructs of PPARα and GFP as following. Briefly, MN9D or primary DA neurons cultured in 6-well pates were transduced with e-GFP-, full-length PPARα, or truncated PPARα Y464D-lentiviral particles at a MOI of 100 in serum-free media containing L-glutamine (Invitrogen) for 48 h. Cells were further incubated for 18 h prior to treatment. Transduction efficiency was measured by comparing relative expression of GFP viewed in the cy2 channel on the Olympus BX51 fluorescence microscope.
Immunoblotting
It was performed as described earlier [22–24]. Briefly, cells were washed 3 times with 1 mL of filtered PBS containing 1:1000 protease/phosphatase inhibitors and centrifuged at 6000 rpm for 5 min. Isolated cell pellets were homogenized in RIPA buffer for 30 min. Total cell lysate was further homogenized and then centrifuged at 15,000 rpm for 15 min. A total of 30 μg protein was loaded and electrophoresed followed by transferring of proteins onto a nitrocellulose membrane (Bio-Rad). The membrane was then blocked in 50% Odyssey Blocking Buffer (Li-COR, Lincoln, NE) followed by blocking with primary antibodies for Nurr1 (1:200, anti-rabbit; Millipore; AB5778) and β-actin (1:800; Abcam, Cambridge, MA). The next day, membranes were washed, incubated in secondary antibodies (all 1:10,000; Li-COR), and visualized under the Odyssey® Infrared Imaging System (Li-COR, Lincoln, NE).
Densitometric Analysis
Protein blots were analyzed using ImageJ (National Institutes of Health, Bethesda). Target bands were normalized to their respective β-actin loading controls. Data are representative of the average fold change with respect to control for three independent experiments.
ChIP Assay
ChIP assay was performed as described earlier [23–25]. Briefly, 2 × 106 MN9D cells were incubated for 2 h with gemfibrozil under serum-free conditions. Cells were fixed by adding paraformaldehyde (4% final concentration), and cross-linked adducts were resuspended and sonicated, resulting in an average chromatin fragment size of 400 bp. ChIP was performed on the cell lysate by overnight incubation at 4 °C with 2 μg of antibodies against PPARα, β, and γ (Santa Cruz Biotechnology). Normal IgG (Santa Cruz Biotechnology) and RNA Poly II (Millipore) were used as a negative and positive control, respectively. The protein DNA complex was next incubated with protein G-agarose (Santa Cruz Biotechnology) for 2 h. To reverse the cross-linking and purify the DNA, precipitates were incubated in a 65 °C incubator overnight and digested with proteinase K. DNA samples were then purified and precipitated, and precipitates were washed with 75% ethanol, air-dried, and resuspended in Tris-EDTA buffer. The following primers were used to amplify fragments flanking PPRE spanning position − 439 and − 461, located on chromosome 2: Set1 sense, 5′-GCTGTTCAGAGAGTCATTAGG-3′, and antisense, 5′-TGGGCAGATAACATACGG-3′; Set2 sense, 5′-GTCATTAGGTTCCTCCCAG-3′, and antisense, 5′-GGCAGCTTAAGCACGGATG-3′. The PCRs were repeated by using varying cycle numbers and different amounts of templates to ensure that results were in the linear range of PCR.
Luciferase Assay
It was performed as described earlier [22, 25]. Briefly, cells plated at 50–60% confluence in 12-well plates were co-transfected with 0.25 μg of tk-PPREx3-Luc, a PPRE-dependent luciferase reporter construct, and 12.5 ng of pRL-TK (a plasmid encoding Renilla luciferase, used as transfection efficiency control; Promega) using Lipofectamine Plus (Invitrogen Life Technologies). After 24 h of transfection, cells were stimulated with different doses of gemfibrozil for 2 h. Firefly and Renilla luciferase activities were analyzed in cell extracts using the Dual Luciferase kit (Promega) in a GloMax 96 microplate luminometer (Promega) and corresponding GloMax 96 microplate luminometer software (Promega) per the manufacturer’s specifications.
Oral Administration of Gemfibrozil and Subsequent Analyses
All animal procedures were conducted in accordance with the Rush University IUCUC protocol (15–056). Six- to eight-week old, litter-matched WT C57BL6J (Jackson Laboratories) and PPARα-null (Jackson Laboratories) were administered either 7.5 mg/kg body weight/14 days of gemfibrozil solubilized in 0.5% methyl cellulose via oral gavage or no treatment (control). On day 15, the mice were heavily anesthetized with a 2:3 ratio of ketamine/xylene and underwent trans-cardiac perfusion with ice cold PBS. The brains were removed and hemisected. Nigral tissue was dissected from one hemisphere and immediately frozen at − 80 °C for western blotting analysis. Nigral sections from other hemisphere were double-labeled for Nurr1 and TH.
Statistics
Values are expressed as means ± S.D. of at least three independent experiments. Statistical analyses for differences were performed via Student’s t test, ANOVA with Tukey post hoc analysis, and two-way ANOVA where applicable. This criterion for statistical significance was p < 0.05. Indicators of significance are described as follows: * = p < 0.03; ** = p < 0.003; *** = p < 0.0003 via Student t tests.
Results
Gemfibrozil and Fenofibrate Increase Nurr1 mRNA and Protein in MN9D Neuronal Cells
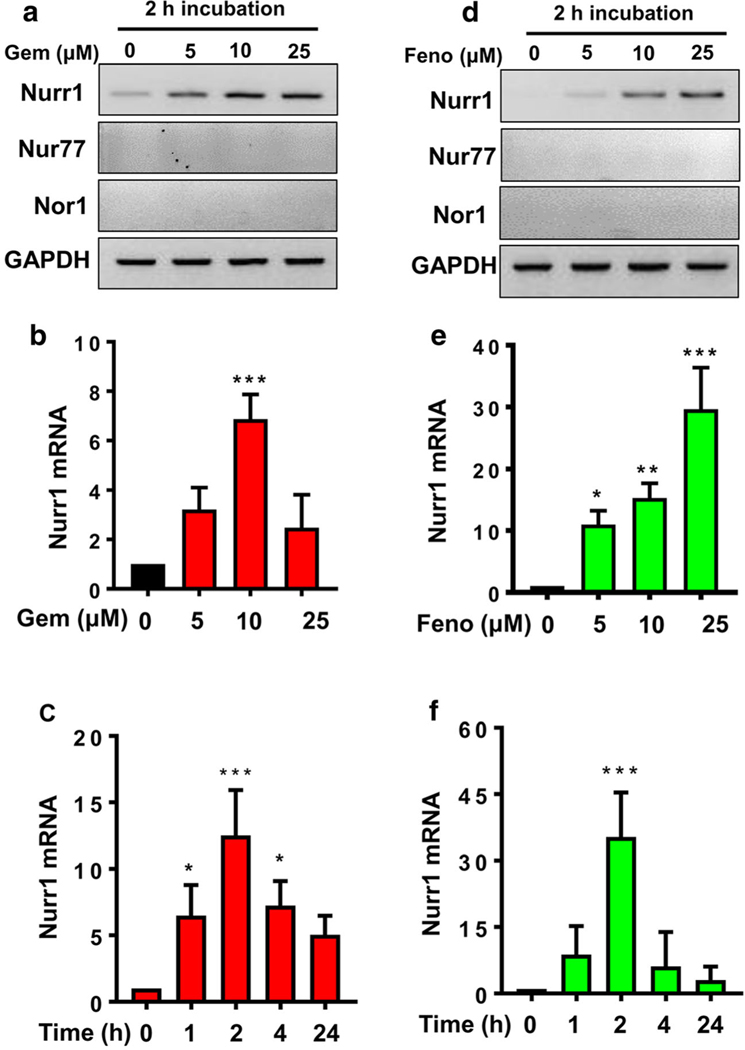
Gemfibrozil and fenofibrate, two common lipid-lowering FDA-approved drugs, are used to improve triglyceride levels in individuals suffering from hyperlipidemia. We [26–28] and others [29] have described that fibrate drugs are also capable of suppressing proinflammatory molecules in glial cells and macrophages. In order to investigate if gemfibrozil could modulate the expression of Nurr1 gene, we performed a series of dose- and time-response studies for Nurr1 gene expression in mouse MN9D dopaminergic neuronal cells. Interestingly, increasing doses of gemfibrozil (Fig. 1a) significantly upregulated the expression of Nurr1 mRNA with maximum at 10 μM concentration. The result was further confirmed by a quantitative real-time PCR (Fig. 1b). Similar to gemfibrozil, fenofibrate was also observed to upregulate the expression of Nurr1 gene in a dose-dependent manner displaying maximum induction at 25 μM concentration (Fig. 1d, e). Moreover, the effect of both gemfibrozil and fenofibrate on the upregulation of Nurr1 gene was specific as these drugs remained unable to alter the expression of Nur77 and Nor1, other isomers of Nurr genes, in MN9D neuronal cells under similar treatment conditions (Fig. 1a, d). Since we could not detect Nur77 and Nor1 in MN9D dopaminergic neuronal cells, to understand whether our PCR conditions were optimum for detecting Nur77 and Nor1, we examined the mRNA expression of Nurr1, Nur77, and Nor1 in liver of normal C57/BL6 mice. As evident from Supplementary Figure 1, mouse liver expressed Nurr1, Nur77, and Nor1 mRNAs, suggesting that our PCR conditions are capable of identifying all three mRNAs. While 10 μM gemfibrozil-mediated increase in Nurr1 was maximum at 2 h (Fig. 1c), 25 μM fenofibrate displayed highest stimulation at 1 h of treatment (Fig. 1f).
Fig. 1.
Both gemfibrozil and fenofibrate increase the expression of Nurr1 mRNA in MN9D neuronal cells. a, b Differentiated MN9D cells were treated with increasing doses of gemfibrozil for 2 h under serum-free condition followed by semi-quantitative and real-time mRNA expression of Nurr1. A one way ANOVA, followed by post hoc Tukey, determined significant differences between the groups [(F(3,8) = 9.59 > Fc = 3.47) *** = p < 0.0003]. c MN9D cells were treated with 10 μM of gemfibrozil for different time points. Significance of the mean was tested by ANOVA followed by post hoc Tukey to determine significant differences between the groups [(F(4,10) = 13.9 > Fc = 3.57) *** = p < 0.0003; * = p < 0.03]. The effect of fenofibrate on the expression of Nurr1 mRNA was evaluated with a dose-dependent study in MN9D cells as shown by d semi-quantitative and e real-time PCR analyses. Significance of the mean was tested by ANOVA followed by post hoc Tukey to determine significant differences between the groups [(F(3,8) = 13.9 > Fc = 3.57); *** = p < 0.0003; **= p < 0.003; * = p < 0.03]. f Time-sensitive expression of Nurr1 mRNA was measured after treating MN9D cells with 25 μM of fenofibrate. Significance of the mean was tested by ANOVA followed by post hoc Tukey to determine significant differences between the groups [(F(4,10) = 13.9 > Fc = 3.7; *** = p < 0.0003]. Results are mean ± SEM of three independent experiments
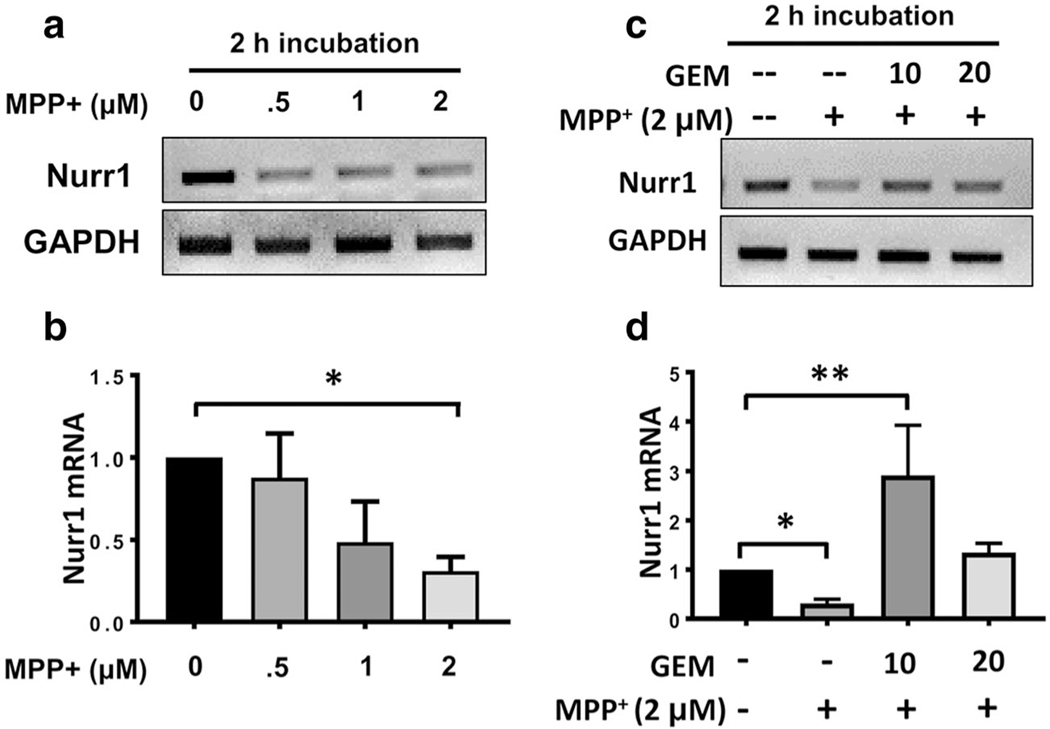
Nurr1 expression has been reported to be downregulated in the postmortem brain tissue isolated from PD patients [14]. Therefore, we examined if a nigral toxin like 1-methyl-4-phenylpyridinium iodide (MPP+) could downregulate the expression of Nurr1 in MN9D cells. Indeed, both semi-quantitative and real-time PCR studies indicate a dose-dependent reduction in Nurr1 expression by MPP+ (Fig. 2a, b). With this finding in mind, our next goal was to test if the addition of gemfibrozil could rescue Nurr1 expression in MPP+-insulted MN9D cells. MN9D cells were pretreated with 2 μM of MPP+ for 2 h for the downregulation of Nurr1 followed by treatment with gemfibrozil for additional 2 h. Interestingly, the addition of 10 μM of gemfibrozil significantly upregulated the Nurr1 expression in MPP+-insulted MN9D cells (Fig. 2c, d). Taken together, these findings indicate that gemfibrozil is capable of upregulating Nurr1 in neuronal cells even in the presence of a nigral toxin.
Fig. 2.
Gemfibrozil rescues Nurr1 mRNA expression in MPP+-insulted MN9D neuronal cells. a Semi-quantitative and b real-time PCR analyses showing a dose-dependent reduction in Nurr1 mRNA expression in MN9D cells upon MPP+ challenge. Significance of the mean was tested by ANOVA followed by a post hoc Tukey to determine differences between the groups [(F(4,10) = 9.06; > Fc = 3.47 * = p < 0.03]. c Semi-quantitative and d real-time PCR studies indicate that the addition of 10 μM of gemfibrozil significantly upregulates Nurr1 mRNA in MN9D cells pre-treated with 2 μM of MPP+. Significance of the mean was tested by ANOVA by a post hoc Tukey [(F(4,10) = 13.7> Fc = 3.47; **= p < 0.003; * = p < 0.03]
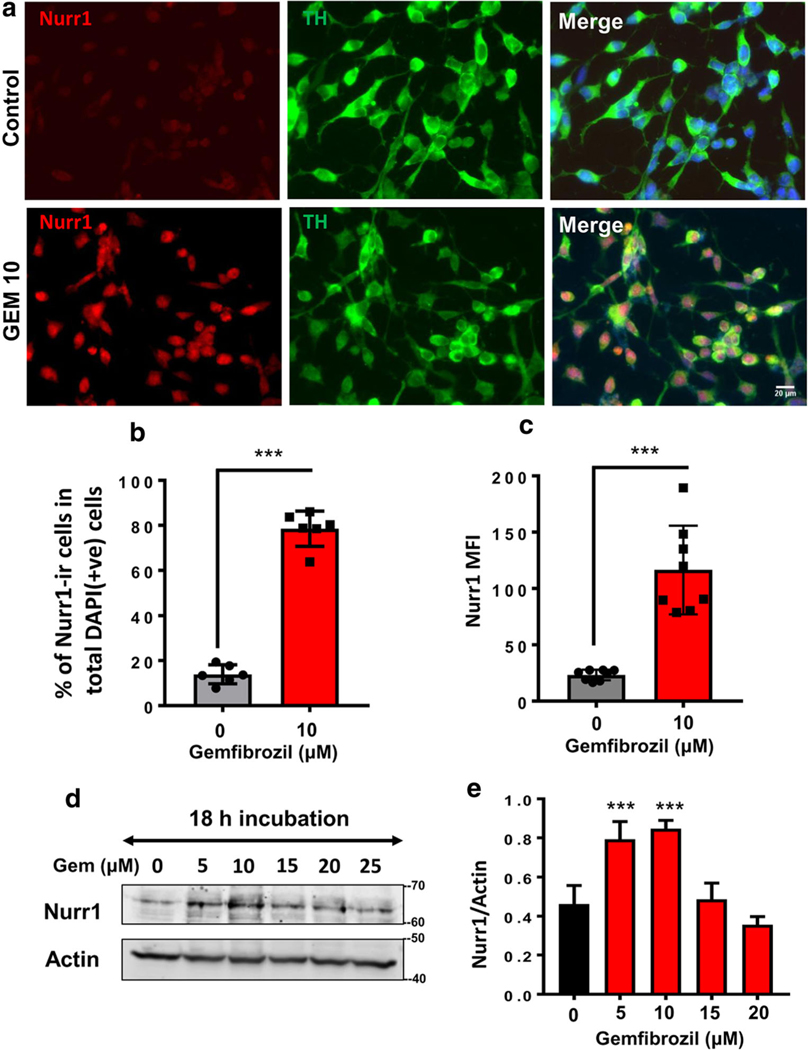
Next, we assessed Nurr1 protein levels in MN9D cells following gemfibrozil stimulation. Both immunofluorescence (Fig. 3a) and immunoblot (Fig. 3d) analyses revealed that 10 μM of gemfibrozil stimulated the expression of Nurr1 protein inMN9D neuronal cells. The result was further confirmed with respective cell counting (Fig. 3b), MFI (Fig. 3c), and densitometric analyses (Fig. 3e). Together, these studies indicate that gemfibrozil has the ability to upregulate both Nurr1 mRNA and protein expression in MN9D neuronal cells.
Fig. 3.
Gemfibrozil upregulates the expression of Nurr1 Protein in MN9D neuronal cells. a Double-labeling immunofluorescence studies were adopted to test the effect of 10 μM gemfibrozil on the expression of Nurr1 (anti-rabbit; red or cy5) in TH–ir MN9D cells (anti-mouse; green or cy2). Results were confirmed after three independent experiments. b Percentage of Nurr1 (red) in total number of cells stained DAPI (blue) was quantified in 10 independent fields. Significance of mean tested with unpaired t test that results in (t = 19.2812; p < 0.0001). c Differentiated MN9D cells were treated with 10 μM of gemfibrozil followed by immunolabeling with Nurr1 (scale bar = 20 μm). Mean fluorescence intensities (MFI) of Nurr1 (cy-5)–ir cells were quantified after normalizing signal with background and total area of the cell. An average of 800 cells were analyzed for MFI per treatment group and averaged to generate resultant MFI. An unpaired t test was done to test the significance of mean (t = 6.6814; p < 0.0001). d Representative immunoblot indicates in the expression of Nurr1 protein with increasing concentrations of gemfibrozil. e Densitometric analysis of three independent blots quantifying Nurr1 expression. Results are mean ± SD of three independent experiments. A one-way ANOVA was adopted to test the significance of mean between groups that results in [F(4,14) = 25.53 > Fc = 3.47; *** = p < 0.0003]
Involvement of PPARα, but Neither PPARβ Nor PPARγ, in Gemfibrozil-Mediated Upregulation of Nurr1 in MN9D Neuronal Cells
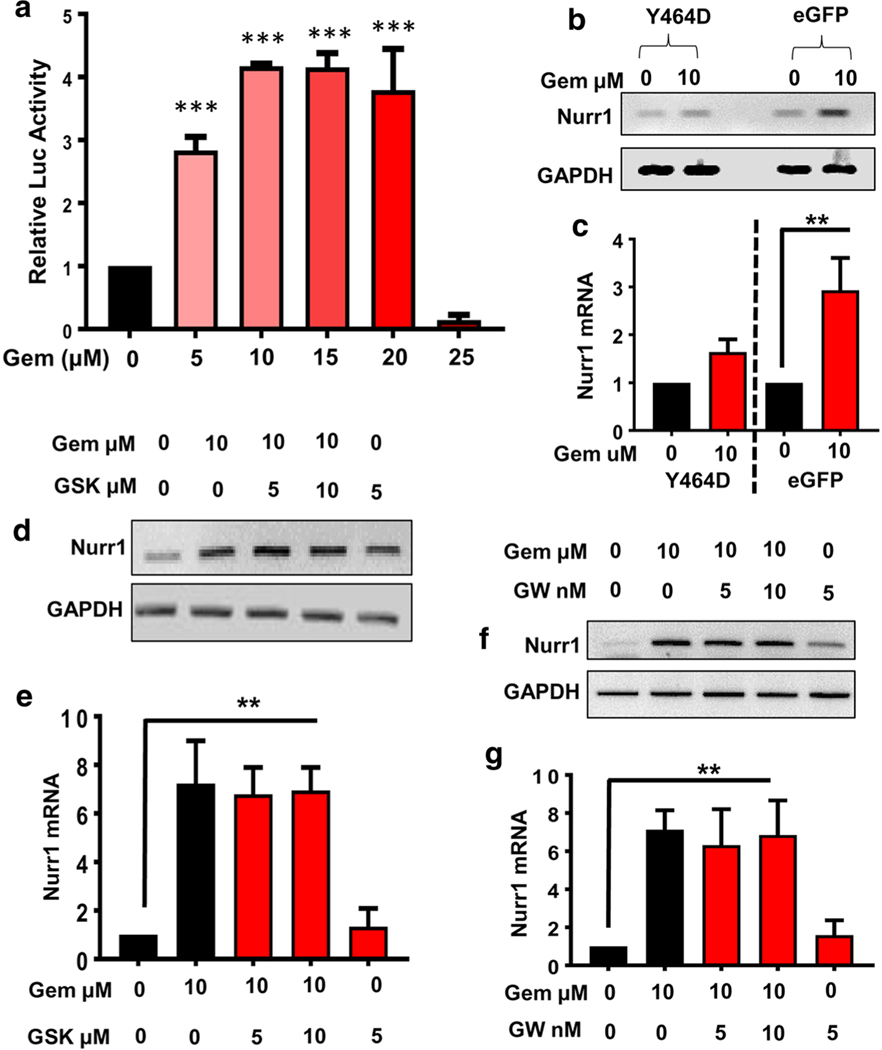
Next, we were interested in identifying the mechanism of gemfibrozil-mediated Nurr1 expression. First, we checked the promoter of Nurr1 gene at the chromosome 2 with the help of MatInspector promoter screening tool of Genomatix Inc. Since gemfibrozil is a well-known pharmacological agonist of PPARα, we checked the Nurr1 promoter for PPARα-responsive element (PPRE). Based on the matrix matching factor and bit score analyses, the Nurr1 promoter harbors a single conserved PPRE site spanning positions − 439 to − 461, suggesting that PPARα might play an important role in the transcription of Nurr1. First, we examined if gemfibrozil was capable of activating PPAR in DA neuronal cells. As evident from Fig. 4a, gemfibrozil dose-dependently induced PPRE-driven luciferase activity in MN9D neuronal cells. Next, we investigated whether PPARα was involved in gemfibrozil-mediated Nurr1 expression in MN9D neuronal cells. Recently, we have observed that disruption of the ligand-binding domain of PPARα by mutating the tyrosine at position 464 to aspartic acid (Y464D) suppresses the function PPARα [22]. Therefore, MN9D cells were transduced with lenti-Y464D-PPARα-e-GFP followed by gemfibrozil treatment. Lenti-e-GFP was used as control and to calculate transduction efficiency. Semi-quantitative RT-PCR (Fig. 4b) and quantitative real-time PCR (Fig. 4c) results clearly indicate that disrupting PPARα LBD by lenti-Y464D-PPARα significantly abrogated gemfibrozil-mediated Nurr1 expression in MN9D cells. On the other hand, lenti-eGFP remained unable to inhibit gemfibrozil-mediated Nurr1 expression (Fig. 4b, c).
Fig. 4.
Involvement of PPARα in gemfibrozil-mediated upregulation of Nurr1 in MN9D neuronal cells. a MN9D cells were transfected with tk-PPREx3-Luc for 24 h followed by the treatment with increasing doses of gemfibrozil (gem) for another 5 h. Then cells were analyzed for PPRE luciferase activity. Results are mean ± SD of three independent experiments with significance of mean tested by one-way ANOVA, which generates [F(5,12) = 101.5 > Fc =3.11; *** = p < 0.0003)]. Semi-quantitative (b) and real-time PCR (c) analyses of Nurr1 gene in MN9D cells transduced with either Y464D-e-GFP (dominant negative mutant of PPARα) or control e-GFP lentiviral construct and treated with 10 μM of gemfibrozil. Results are mean ± SD of three experiments and tested with one-way ANOVA for significance of mean [(F3, 8 = 21.98 > Fc = 4.06); ** = p < 0.003)]. MN9D cells pretreated with different doses of GSK0660 for 1 h were treated with 10 μM of gem for 4 h followed by semi-quantitative (d) and real-time (e) PCR analyses to verify Nurr1 mRNA expression. One-way ANOVA was adopted to test the significance of mean between groups [(F(1,8) = 21.98 > Fc = 4.066); ** = p < 0.003)]. Similar experiments were performed in MN9Dcells with different doses of GW9662. The nurr1 mRNA expression was monitored by semi-quantitative (f) and real-time (g) PCR analyses. Results are mean ± SD of three experiments and tested with one-way ANOVA for significance of mean [F(1,8) = 20.78 > Fc = 3.47); ** = p < 0.003)]
In order to investigate the relative contribution of PPARβ and PPARγ, we performed a series of isoform-specific chemical inhibition studies to block their activity in MN9D cells. As expected, gemfibrozil-treated cells that were not subjected to GSK0060, a specific inhibitor of PPARβ, displayed a significant increase in Nurr1 expression (Fig. 4d, e). However, GSK0060 remained unable to inhibit gemfibrozil-induced Nurr1 expression (Fig. 4d, e). The same experimental setup was replicated to investigate whether gemfibrozil-mediated Nurr1 expression was reduced in the presence of GW9662, a specific inhibitor of PPARγ. Similar to GSK0060, GW9662, at different doses tested, also did not inhibit gemfibrozil-induced Nurr1 expression in MN9D neuronal cells (Fig. 4f, g).
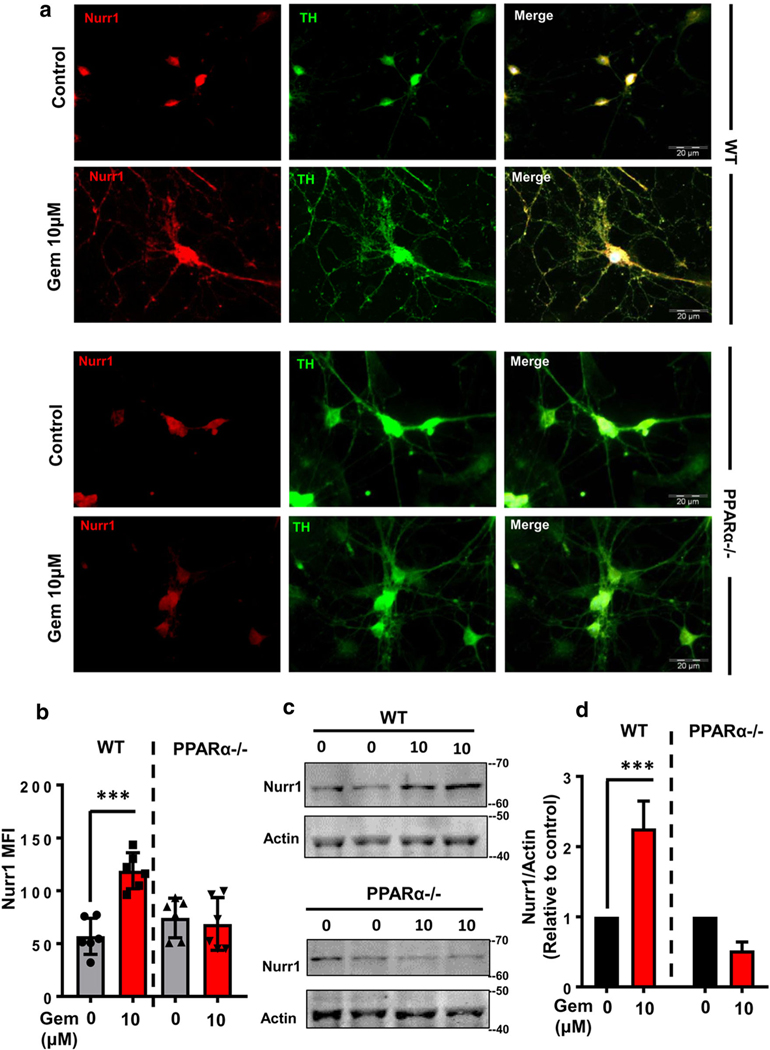
Gemfibrozil Increases Nurr1 in WT, but Not PPARα−/−, Primary Dopaminergic Neurons
To confirm the role of PPARα, next we compared the gemfibrozil-mediated expression of Nurr1 between wild-type and PPARα−/− DA neurons. Briefly, we dissected ventral midbrain tissue of E14.5 fetal brains from both wild-type and PPARα−/− mice, cultured for primary DA neurons and then after 1 week of primary culture, treated with gemfibrozil under serum-free condition. Immunocytochemical analysis of primary DA neurons showed that gemfibrozil markedly increased Nurr1 protein expression in WT neurons (Fig. 5a, b). Western blot analysis of pooled cell lysate complimented our IF studies, and densitometric analysis of immunoblots confirmed a significant increase in Nurr1 protein levels (Fig. 5c, d) by gemfibrozil. Interestingly, Nurr1 expression was not significantly changed when gemfibrozil treatment was carried out in primary DA neurons harvested from PPARα−/− animals (Fig. 5a–d). These results indicate that gemfibrozil increases the level of Nurr1 in DA neurons via PPARα.
Fig. 5.
Gemfibrozil increases Nurr1 expression in primary DA neurons via PPARα. a Mouse primary DA neurons isolated from WT and PPARα−/−mice were treated with 10 μM gemfibrozil for 18 h followed by doublelabeling for Nurr1 (green) and TH (red). Scale bar = 20 μm. b Nurr1 MFI measurements were performed from an average of 800 cells per group [(F(5,15) = 233.6 > Fc = 3.09); *** = p < 0.0003] from two-way ANOVA].c Representative immunoblots display the effect of gemfibrozil on the expression of Nurr1 in mouse primary DA neuron isolates from both WT (top) and PPARα−/− (bottom) fetuses. d Densitometric analyses of Nurr1 were performed from immunoblots, normalized with β-actin and then plotted. Results are mean ± SD of three independent immunoblots. A two-wayANOVA was adopted to justify the significance of mean in Nurr1 expressionbetween WT neurons and PPARα−/− neurons, [F(1,8) = 6.68 > Fc =5.31); *** = p < 0.0003; * = p < 0.03] for treatment; and [F(1,8) = 73.76>Fc = 5.31); *** = p < 0.0003] for genotype
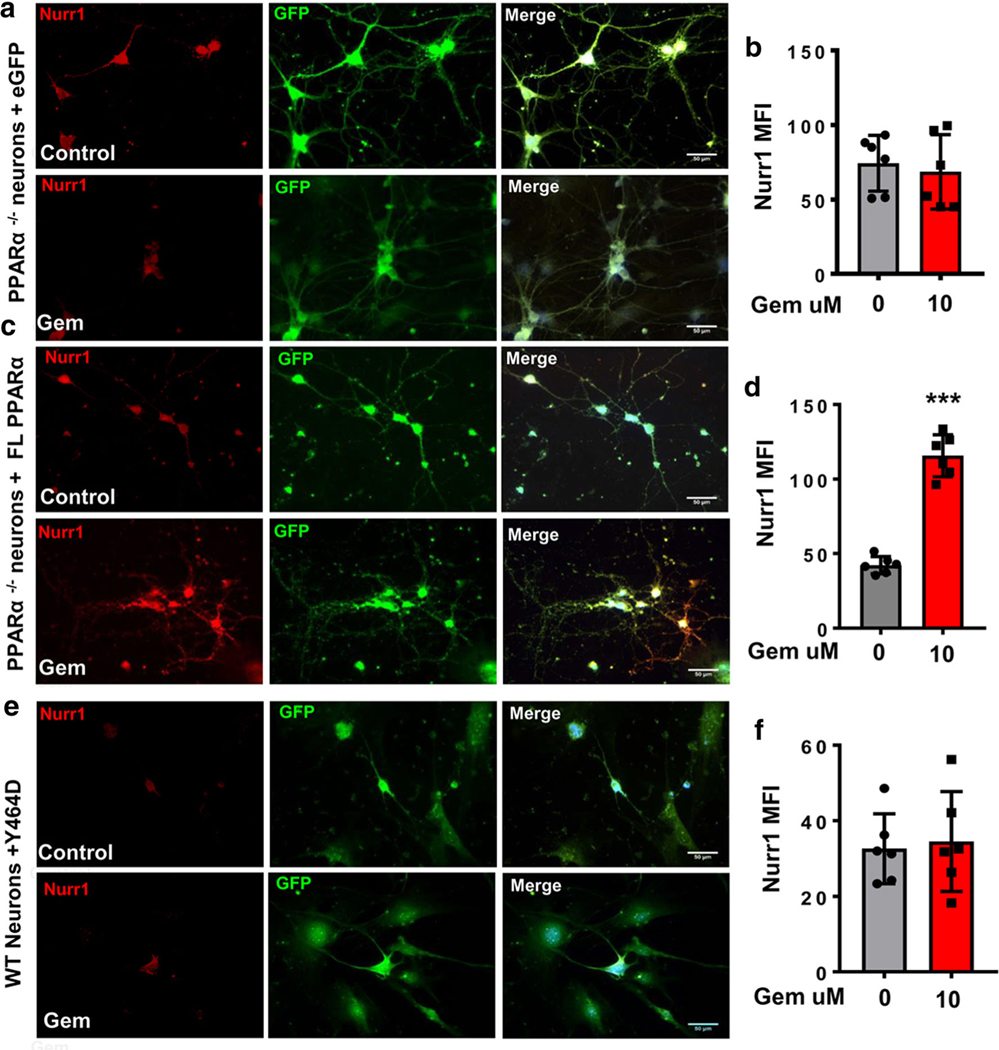
Over-expression of PPARα Restores the Ability of Gemfibrozil in Stimulating the Level of Nurr1 in PPARα−/− DA Neurons
To further confirm the involvement of PPARα in gemfibrozil-mediated Nurr1 expression, we over-expressed full-length PPARα in primary DA neurons isolated from PPARα−/− mice. PPARα−/− DA neurons transduced with lenti-eGFP and treated with 10 μm of gemfibrozil did not display Nurr1 upregulation as compared to untreated cells (Fig. 6a). MFI calculations (Fig. 6b) confirm no significant increase of Nurr1 expression after adjusting for cell size and background fluorescence. Interestingly, however, PPARα−/− DA neurons transduced with lenti-FL-PPARα and treated with 10 μM of gemfibrozil showed a significant increase in Nurr1 expression as compared to untreated cells (Fig. 6c, d). These results indicate that the reinsertion of FL-PPARα re-establishes gemfibrozil-mediated Nurr1 expression in primary DA neurons isolated from PPARα−/− fetuses. Our previous findings indicated that disrupting the LBD of PPARα via the transduction of the Y464D lentiviral construct prevented gemfibrozil-mediated nurr1 expression in MN9D cells. In order to confirm this finding in primary DA neurons, WT DA neurons were transduced with either lenti-Y464D-PPARα for 24 h. Similar to MN9D cells, gemfibrozil treatment remained unable to induce Nurr1 expression in lenti-Y464D-PPARα-transduced WT DA neurons (Fig. 6e, f).
Fig. 6.
Over-expression of PPARα restores Nurr1-upregulating efficacy of gemfibrozil in PPARα−/− DA neurons. Primary DA neurons harvested from PPARα−/− mice were transduced with lenti-empty vector e-GFP (a, b), lenti-full-length PPARα (c, d), or lenti-Y464D PPARα (e, f) for 24 h. Then cells were treated with 10 μM of gemfibrozil for 18 h followed by immunolabeling for Nurr1 (Cy5; red). MFI of Nurr1-ir was quantified for an average of 800 cells per group (b lenti-eGFP; d lenti-FL PPARα; f lenti-Y464D PPARα). A paired t test was utilized to compare significant differences from the means of these two groups resulting in t = 0.448710; p = 0.66 (b). An unpaired t test analysis was performed to show significant differences between the means (t = 11.8110; = p < 0.0003) (d). Unpaired t test shows t = 0.2910; p = 0.77 (f). All results are mean ± SD of three independent experiments
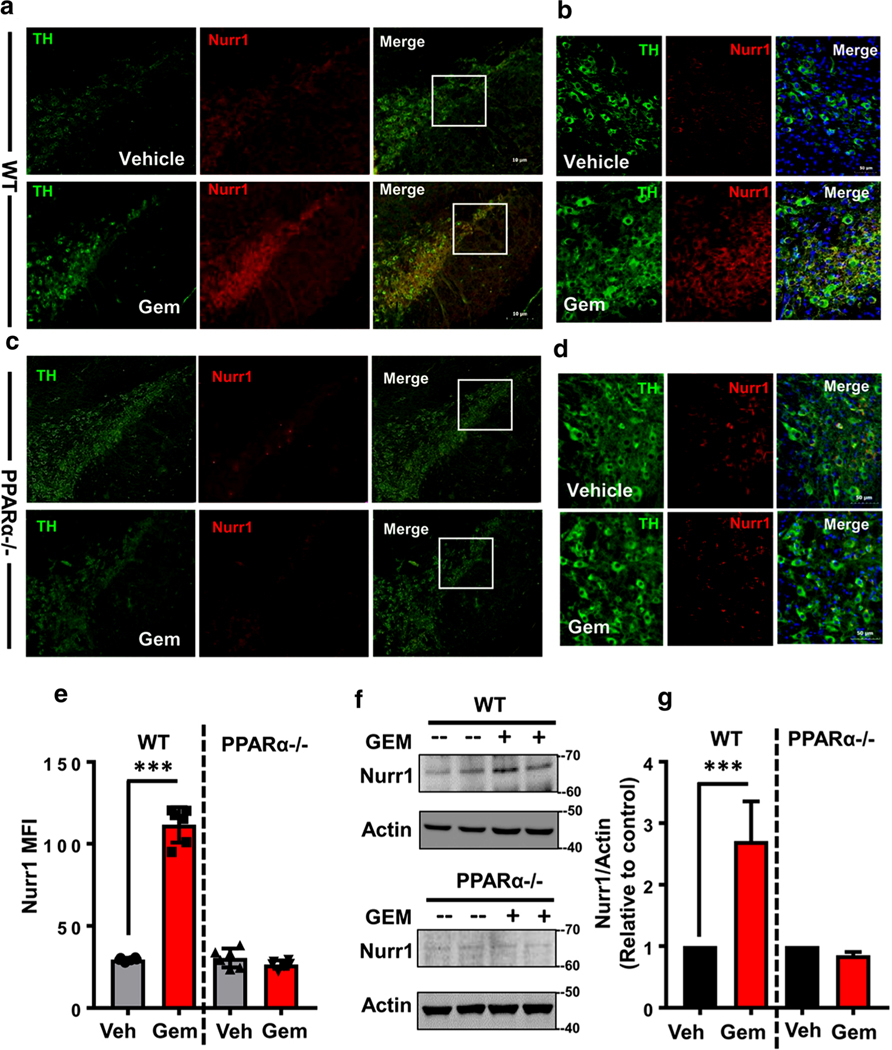
Gemfibrozil Increases Nurr1 Protein In Vivo in the Nigra of WT, but Not PPARα−/−, Mice
Next, we examined whether gemfibrozil was capable of upregulating Nurr1 in vivo in the nigra. Gemfibrozil’s favorable oral bioavailability allowed us to hypothesize that oral gemfibrozil at human equivalent dose (600 mg/adult/day = ~7.5 mg/kg/day) might upregulate Nurr1 expression in vivo in the SNpc. Oral administration of gemfibrozil markedly increased the level of Nurr1 in the nigra compared to vehicle treatment (Fig. 7a, b). MFI calculations indicate a significant increase in Nurr1 signal (Fig. 7e). Increased Nurr1 expression from the WT gemfibrozil-fed group is further represented by the corresponding immunoblot (Fig. 7f) and significance quantified via densitometric analysis (Fig. 7g, left). These results indicate that oral administration of human equivalent dose of gemfibrozil significantly increases nigral Nurr1 expression in mice. However, consistent to our cell culture data, gemfibrozil treatment remained unable to increase Nurr1 in the nigra of PPARα−/− animals as compared to vehicle treatment (Fig. 7c–g).
Fig. 7.
Oral administration of gemfibrozil increases Nurr1 expression in vivo in nigra via PPARα. WT (a, b) and PPARα−/− (c, d) animals were gavage fed 7.5 mg/kg gemfibrozil or 0.5% methyl cellulose (Veh) for 14 days via oral route. After that, nigral sections were double-labeled with anti-mouse TH (cy2) and anti-rabbit Nurr1 (cy5) antibodies. Total n = 5 animals were used per group. e MFI analysis of nigral sections indicates a significant difference between genotypes [(F(3,23) = 233.55> Fc = 3.287); *** = p < 0.0003]. f Representative immunoblots using isolated nigral sections from both WT and PPARα−/− animals in this experimental paradigm. g Densitometric analysis of f indicates a significant increase in Nurr1 protein expression from WT, but not PPARα−/− animals fed GEM as shown by a two-way ANOVA [(F(3, 11) = 20.01 > Fc = 4.757); ***p < 0.0003]
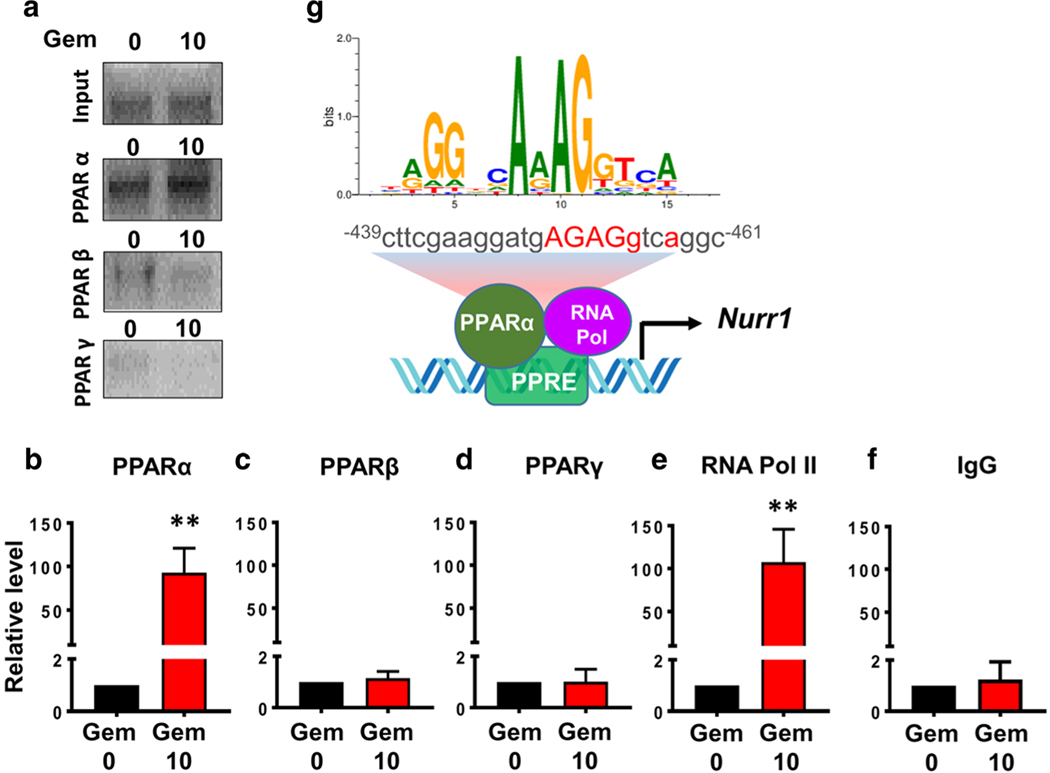
Gemfibrozil Treatment Stimulated the Recruitment of PPARα to the Nurr1 Gene Promoter
Next, to understand whether PPARα was directly involved in the transcription of Nurr1 gene, we examined the recruitment of PPARα to the Nurr1 gene promoter by ChIP assay. The Nurr1 promoter harbors a consensus PPRE spanning positions − 461 to − 439 (Fig. 8g). Cells were treated with 10 μM gemfibrozil for 2 h and after immunoprecipitation of gemfibrozil-treated neuronal chromatin fragments using antibodies against PPARα, we amplified 120-bp fragment from the Nurr1 promoter (Fig. 8a, b). On the other hand, after immunoprecipitation of chromatin fragments with antibodies against PPARβ and PPARγ, we did not see any amplification product (Fig. 8a–d). However, consistent to the recruitment of PPARα to the PPRE, gemfibrozil was able to recruit RNA polymerase to the Nurr1 gene promoter (Fig. 8a, e). These results are specific as no amplification product was observed in immuno-precipitates obtained with control IgG (Fig. 8f). These results demonstrate that gemfibrozil treatment is capable of stimulating the recruitment of PPARα, but neither PPARβ nor PPARγ, to Nurr1 gene promoter in dopaminergic neuronal cells.
Fig. 8.
Gemfibrozil treatment induces the recruitment of PPARα, but neither PPARβ nor PPARγ, to the Nurr1 gene promoter in MN9D neuronal cells. MN9D cells were treated with 10 μM gemfibrozil for 2 h. After that, we performed chromatin immunoprecipitation (ChIP) followed by semi-quantitative (a) and real-time (b–f) PCR of promoter DNA as described in the “Materials and Methods” section. An ANOVA analysis was used to test the significance of the mean differences between groups and shows [(F(9,29) = 23.13 > Fc = 2.40) ** = p < 0.003)]. Post hoc unpaired t tests indicate a significant difference in samples incubated with antibodies to both PPARα (t = 5.654; p = 0.004) and RNA polymerase II (t = 4.794; p = 0.008). g A schema depicts a detailed map of promoter analysis of nurr1 gene. The map reveals a conserved PPAR-responsive element (PPRE) in the promoter of Nurr1 gene at − 439 to − 461 upstream of the Nurr1 start site on chromosome II
Discussion
Nurr1 is a master regulator of DA neuronal development before birth and maintenance throughout life [30, 31]. The dimerization of Nurr1-RXR positively induces the expression of DA genes required for the production of dopamine including tyrosine hydroxylase (TH), dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2), and l-aromatic amino acid decarboxylase (AADC) [5, 10, 32–35]. Mutations in Nurr1 and related genes have been associated with a small minority of late-onset genetic PD patients and neurological disorders such as schizophrenia [36–38]. More broadly, and perhaps more importantly however, Nurr1 protein expression is reported to be decreased in the brains of sporadic PD patients [39–41]. This decreased Nurr1 expression was particularly evident in neurons containing Lewy bodies, which are hallmark pathologies of PD [14]. In animal models, Nurr1 has been reported to be expressed in astrocytes and microglia. Reduced Nurr1 levels in these cells have been reported to increase the expression of their neurodegenerative phenotypes [42, 43]. Expansive literature supports the notion of an increased presence of activated microglia and α-syn aggregation in postmortem PD tissue [44]. Nurr1’s ability to reduce these pathogenic species of activated microglia and astrocytes is thought to be derived through its direct blockade of NF-κB-P65 and induction of the CoREST repressor complex, thereby decreasing IL-6 production from T cells in animal models [45, 46]. Nurr1 has also been shown to a critical regulator of Ret-c signaling expression in DA neurons that has been reported to upregulate GDNF and TH expression in vivo [47]. Trophic factors such as GDNF drive the formation and maintenance of neuronal synapses and, in addition, provide critical support to glutamate absorbing astrocytes, thereby reducing glutamate-mediated excitotoxicity. Animal models of PD such as the A53T overexpressing α-syn derive their pathology from inflammation produced by activated microglia in response to LB production [48]. Nurr1 induction has been shown to decrease the production of proinflammatory CNS-derived immune cells. In addition, it is reported that α-syn aggregates induce the downregulation of Nurr1 which then disrupts GDNF signaling between DA neurons [14]. The induction of Nurr1 achieves a reduction in inflammatory-prone glial populations regardless of insult.
Therefore, agonists of Nurr1 are being considered for clinical trials in PD and other neurodegenerative disorders. Here, we report for the first time that gemfibrozil, a drug approved by the FDA over 30 years ago and prescribed to millions of patients for the treatment of hypertriglyceridemia, increases the expression of Nurr1 in dopaminergic neurons. This result was specific as gemfibrozil did not stimulate the expression of Nur77 and Nor1, other members of the Nurr gene family. Moreover, gemfibrozil was able to upregulate Nurr1 in MPP+-challenged dopaminergic neuronal cells. These studies provide a potentially important mechanism to reduce nigral inflammation and maintain the health of nigral neurons by gemfibrozil.
Mechanisms by which the Nurr1 gene is upregulated are poorly understood. PPARα, a nuclear hormone receptor family transcription factor, is known to control the metabolism of fatty acids in the liver. We [23, 25, 49, 50] and others [51] have demonstrated that PPARα is also present in different parts of the brain. Several lines of evidence presented in this manuscript describe that gemfibrozil upregulates the transcription of Nurr1 via PPARα. First, the promoter of Nurr1 harbors a consensus PPRE. Second, gemfibrozil treatment induced the activation of PPAR in DA neuronal cells. Third, lentiviral over-expression of a dominant-negative mutant (Y464D) of PPARα abrogated gemfibrozil-mediated upregulation of Nurr1 in DA neuronal cells and primary DA neurons. Fourth, gemfibrozil was unable to stimulate the expression of Nurr1 in primary DA neurons isolated from PPARα−/− mice. Fifth, Nurr1 mRNA expression was not affected in gemfibrozil-treated cells upon the inhibition of either PPARβ or PPARγ. Sixth, after lentiviral over-expression of full-length PPARα, gemfibrozil was able to upregulate Nurr1 in PPARα−/− DA neurons. Seventh, similar to cell culture findings, oral administration of gemfibrozil led to significant increase in Nurr1 in the nigra of WT, but not PPARα−/−, mice. Eight, gemfibrozil induced the recruitment of PPARα to the Nurr1 gene promoter in DA neuronal cells. Therefore, PPARα plays a key role in the upregulation of Nurr1 in DA neuronal cells.
It has been shown that gemfibrozil and other fibrate drugs are capable of reducing oxidative stress and lipid peroxidation products [52, 53]. Fibrate drugs are also reported to strengthen the cellular defense machinery by stimulating the activity of anti-oxidant proteins such as superoxide dismutase, catalase, and paraoxonase [54, 55]. It also could be associated with the free radical scavenging ability and metal ion chelation. We have seen while gemfibrozil is capable of upregulating anti-inflammatory molecules like IκBα, IL-1ra, and SOCS3 [24, 27, 56], it also suppresses the expression of proinflammatory molecules [26] in glial cells. Moreover, gemfibrozil is capable of stimulating the production of neurotrophic factors from glial cells [25].Therefore, apart from its lipid-lowering effects, gemfibrozil also possesses anti-inflammatory, immunomodulatory, and anti-oxidative properties. Since PD is associated with increased oxidative stress, elevated neuroinflammation, and decrease in neurotrophic factors [57], treatment with gemfibrozil will not only lead to the upregulation of Nurr1 leading to increased survival of DA neurons but also can be beneficial via its antioxidant, anti-inflammatory, and neurotrophic properties.
Supplementary Material
Acknowledgments
Funding Information This study was supported by a merit award from Veteran Affairs (I01BX003033) and a grant (NS083054) from NIH.
Footnotes
Compliance with Ethical Standards Animal maintenance and experimental protocols were approved by the Rush University Animal Care Committee. All animal procedures were conducted in accordance with the Rush University IUCUC protocol (15–056).
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s12035–019-01649-y) contains supplementary material, which is available to authorized users.
References
- 1.Roy A, Pahan K (2011) Prospects of statins in Parkinson disease. Neuroscientist 17(3):244–255. 10.1177/1073858410385006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanyal J, Chakraborty DP, Sarkar B, Banerjee TK, Mukherjee SC, Ray BC, Rao VR (2010) Environmental and familial risk factors of Parkinsons disease: case-control study. Can J Neurol Sci Le journal canadien des sciences neurologiques 37(5):637–642 [DOI] [PubMed] [Google Scholar]
- 3.Pinto F, Mazza S (1971) Psychic symptoms in levodopa treatment of Parkinsons’ disease. II. Riv Neurobiol 17(2):155–159 [PubMed] [Google Scholar]
- 4.Smith ML, King J, Dent L, Mackey V, Muthian G, Griffin B, Charlton CG (2014) Effects of acute and sub-chronic L-dopa therapy on striatal L-dopa methylation and dopamine oxidation in an MPTP mouse model of Parkinsons disease. Life Sci 110(1):1–7. 10.1016/j.lfs.2014.05.014 [DOI] [PubMed] [Google Scholar]
- 5.Dong J, Li S, Mo JL, Cai HB, Le WD (2016) Nurr1-based therapies for Parkinson’s disease. CNS Neurosci Ther 22(5):351–359. 10.1111/cns.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimes DA, Han F, Panisset M, Racacho L, Xiao F, Zou R, Westaff K, Bulman DE (2006) Translated mutation in the Nurr1 gene as a cause for Parkinson’s disease. Mov Disord 21(7):906–909. 10.1002/mds.20820 [DOI] [PubMed] [Google Scholar]
- 7.Arenas E (2005) Engineering a dopaminergic phenotype in stem/precursor cells: role of Nurr1, glia-derived signals, and Wnts. Ann N Y Acad Sci 1049:51–66. 10.1196/annals.1334.007 [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Traver E, Solis O, Diaz-Guerra E, Ortiz O, Vergano-Vera E, Mendez-Gomez HR, Garcia-Sanz P, Moratalla R et al. (2016) Role of Nurr1 in the generation and differentiation of dopaminergic neurons from stem cells. Neurotox Res 30(1):14–31. 10.1007/s12640-015-9586-0 [DOI] [PubMed] [Google Scholar]
- 9.Alavian KN, Jeddi S, Naghipour SI, Nabili P, Licznerski P, Tierney TS (2014) The lifelong maintenance of mesencephalic dopaminergic neurons by Nurr1 and engrailed. J Biomed Sci 21:27. 10.1186/1423-0127-21-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae EJ, Lee HS, Park CH, Lee SH (2009) Orphan nuclear receptor Nurr1 induces neuron differentiation from embryonic cortical precursor cells via an extrinsic paracrine mechanism. FEBS Lett 583(9):1505–1510. 10.1016/j.febslet.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 11.Kim CH, Han BS, Moon J, Kim DJ, Shin J, Rajan S, Nguyen QT, Sohn M et al. (2015) Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc Natl Acad Sci U S A 112(28):8756–8761. 10.1073/pnas.1509742112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Gao Y, Chang N (2017) Nurr1 overexpression exerts neuroprotective and anti-inflammatory roles via down-regulating CCL2 expression in both in vivo and in vitro Parkinson’s disease models. Biochem Biophys Res Commun 482(4):1312–1319. 10.1016/j.bbrc.2016.12.034 [DOI] [PubMed] [Google Scholar]
- 13.Oh SM, Chang MY, Song JJ, Rhee YH, Joe EH, Lee HS, Yi SH, Lee SH (2016) Combined Nurr1 and Foxa2 roles in the therapy of Parkinson’s disease. EMBO Mol Med 8(2):171. 10.15252/emmm.201506162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decressac M, Kadkhodaei B, Mattsson B, Laguna A, Perlmann T, Bjorklund A (2012) Alpha-synuclein-induced down-regulation of Nurr1 disrupts GDNF signaling in nigral dopamine neurons. Sci Transl Med 4(163):163ra156. 10.1126/scitranslmed.3004676 [DOI] [PubMed] [Google Scholar]
- 15.Hammond SL, Safe S, Tjalkens RB (2015) A novel synthetic activator of Nurr1 induces dopaminergic gene expression and protects against 6-hydroxydopamine neurotoxicity in vitro. Neurosci Lett 607:83–89. 10.1016/j.neulet.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Miranda BR, Popichak KA, Hammond SL, Jorgensen BA, Phillips AT, Safe S, Tjalkens RB (2015) The Nurr1 activator 1,1-bis(3’-indolyl)-1-(p-chlorophenyl)methane blocks inflammatory gene expression in BV-2 microglial cells by inhibiting nuclear factor kappaB. Mol Pharmacol 87(6):1021–1034. 10.1124/mol.114.095398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith GA, Rocha EM, Rooney T, Barneoud P, McLean JR, Beagan J, Osborn T, Coimbra M et al. (2015) A Nurr1 agonist causes neuroprotection in a Parkinson’s disease lesion model primed with the toll-like receptor 3 dsRNA inflammatory stimulant poly(I:C). PLoS One 10(3):e0121072. 10.1371/journal.pone.0121072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh SM, Chang MY, Song JJ, Rhee YH, Joe EH, Lee HS, Yi SH, Lee SH (2015) Combined Nurr1 and Foxa2 roles in the therapy of Parkinson’s disease. EMBO molecular medicine 7(5):510–525. 10.15252/emmm.201404610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy A, Pahan K (2013) Ankyrin repeat and BTB/POZ domain containing protein-2 inhibits the aggregation of alpha-synuclein: implications for Parkinson’s disease. FEBS Lett 587(21):3567–3574. 10.1016/j.febslet.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modi KK, Rangasamy SB, Dasarathi S, Roy A, Pahan K (2016) Cinnamon converts poor learning mice to good learners: implications for memory improvement. Journal of neuroimmune pharmacology : the official journal of the Society on NeuroImmune Pharmacology 11(4):693–707. 10.1007/s1148-1016-9693-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandra G, Kundu M, Rangasamy SB, Dasarathy S, Ghosh S, Watson R, Pahan K (2018) Increase in mitochondrial biogenesis in neuronal cells by RNS60, a physically-modified saline, via phosphatidylinositol 3-kinase-mediated upregulation of PGC1alpha. J Neuroimmune Pharmacol 13(2):143–162. 10.1007/s11481-017-9771-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy A, Kundu M, Jana M, Mishra RK, Yung Y, Luan CH, Gonzalez FJ, Pahan K (2016) Identification and characterization of PPARalpha ligands in the hippocampus. Nat Chem Biol 12(12):1075–1083. 10.1038/nchembio.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbett GT, Gonzalez FJ, Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc Natl Acad Sci U S A 112(27): 8445–8450. 10.1073/pnas.1504890112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh A, Pahan K (2012) Gemfibrozil, a lipid-lowering drug, induces suppressor of cytokine signaling 3 in glial cells: implications for neurodegenerative disorders. J Biol Chem 287(32):27189–27203. 10.1074/jbc.M112.346932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy A, Jana M, Kundu M, Corbett GT, Rangaswamy SB, Mishra RK, Luan CH, Gonzalez FJ et al. (2015) HMG-CoA reductase inhibitors bind to PPARalpha to upregulate neurotrophin expression in the brain and improve memory in mice. Cell Metab 22(2):253–265. 10.1016/j.cmet.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pahan K, Jana M, Liu X, Taylor BS, Wood C, Fischer SM (2002) Gemfibrozil, a lipid-lowering drug, inhibits the induction of nitricoxide synthase in human astrocytes. J Biol Chem 277(48):45984–45991. 10.1074/jbc.M200250200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jana M, Jana A, Liu X, Ghosh S, Pahan K (2007) Involvement of phosphatidylinositol 3-kinase-mediated up-regulation of I kappa B alpha in anti-inflammatory effect of gemfibrozil in microglia. J Immunol (Baltimore, Md :1950) 179(6):4142–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A, Pahan K (2009) Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacol Immunotoxicol 31(3):339–351. 10.1080/08923970902785253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Storer PD, Chavis JA, Racke MK, Drew PD (2005) Agonists for the peroxisome proliferator-activated receptor-alpha and the retinoid X receptor inhibit inflammatory responses of microglia. J Neurosci Res 81(3):403–411. 10.1002/jnr.20518 [DOI] [PubMed] [Google Scholar]
- 30.Terraf P, Babaloo H, Kouhsari SM (2017) Directed differentiation of dopamine-secreting cells from Nurr1/GPX1 expressing murine embryonic stem cells cultured on Matrigel-coated PCL scaffolds. Mol Neurobiol 54(2):1119–1128. 10.1007/s12035-016-9726-4 [DOI] [PubMed] [Google Scholar]
- 31.Kim T, Song JJ, Puspita L, Valiulahi P, Shim JW, Lee SH (2017) In vitro generation of mature midbrain-type dopamine neurons by adjusting exogenous Nurr1 and Foxa2 expressions to their physiologic patterns. Exp Mol Med 49(3):e300. 10.1038/emm.2016.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decressac M, Volakakis N, Bjorklund A, Perlmann T (2013) NURR1 in Parkinson disease—from pathogenesis to therapeutic potential. Nat Rev Neurol 9(11):629–636. 10.1038/nrneurol.2013.209 [DOI] [PubMed] [Google Scholar]
- 33.Bensinger SJ, Tontonoz P (2009) A Nurr1 pathway for neuroprotection. Cell 137(1):26–28. 10.1016/j.cell.2009.03.024 [DOI] [PubMed] [Google Scholar]
- 34.Sacchetti P, Mitchell TR, Granneman JG, Bannon MJ (2001) Nurr1 enhances transcription of the human dopamine transporter gene through a novel mechanism. J Neurochem 76(5):1565–1572 [DOI] [PubMed] [Google Scholar]
- 35.Green AL, Zhan L, Eid A, Zarbl H, Guo GL, Richardson JR (2017) Valproate increases dopamine transporter expression through histone acetylation and enhanced promoter binding of Nurr1. Neuropharmacology 125:189–196. 10.1016/j.neuropharm.2017.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmine A, Buervenich S, Galter D, Jonsson EG, Sedvall GC, Farde L, Gustavsson JP, Bergman H et al. (2003) NURR1 promoter polymorphisms: Parkinson’s disease, schizophrenia, and personality traits. Am J Med Genet B Neuropsychiatr Genet 120B(1):51–57. 10.1002/ajmg.b.20033 [DOI] [PubMed] [Google Scholar]
- 37.Tan EK, Chung H, Chandran VR, Tan C, Shen H, Yew K, Pavanni R, Puvan KA et al. (2004) Nurr1 mutational screen in Parkinson’s disease. Mov Disord 19(12):1503–1505. 10.1002/mds.20246 [DOI] [PubMed] [Google Scholar]
- 38.Buervenich S, Carmine A, Arvidsson M, Xiang F, Zhang Z, Sydow O, Jonsson EG, Sedvall GC et al. (2000) NURR1 mutations in cases of schizophrenia and manic-depressive disorder. Am J Med Genet 96(6):808–813 [DOI] [PubMed] [Google Scholar]
- 39.Chu Y, Le W, Kompoliti K, Jankovic J, Mufson EJ, Kordower JH (2006) Nurr1 in Parkinson’s disease and related disorders. J Comp Neurol 494(3):495–514. 10.1002/cne.20828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobsen KX, MacDonald H, Lemonde S, Daigle M, Grimes DA, Bulman DE, Albert PR (2008) A Nurr1 point mutant, implicated in Parkinson’s disease, uncouples ERK1/2-dependent regulation of tyrosine hydroxylase transcription. Neurobiol Dis 29(1):117–122. 10.1016/j.nbd.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 41.Le W, Pan T, Huang M, Xu P, Xie W, Zhu W, Zhang X, Deng H et al. (2008) Decreased NURR1 gene expression in patients with Parkinson’s disease. J Neurol Sci 273(1–2):29–33. 10.1016/j.jns.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan X, Luo G, Ming M, Pu P, Li L, Yang D, Le W (2009) Nurr1 expression and its modulation in microglia. Neuroimmunomodulation 16(3):162–170. 10.1159/000204229 [DOI] [PubMed] [Google Scholar]
- 43.Lallier SW, Graf AE, Waidyarante GR, Rogers LK (2016) Nurr1 expression is modified by inflammation in microglia. Neuroreport 27 (15):1120–1127. 10.1097/WNR.0000000000000665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang YX, Latchman DS (2008) Nurr1 transcriptionally regulates the expression of alpha-synuclein. Neuroreport 19(8):867–871. 10.1097/WNR.0b013e3282ffda48 [DOI] [PubMed] [Google Scholar]
- 45.Spathis AD, Asvos X, Ziavra D, Karampelas T, Topouzis S, Cournia Z, Qing X, Alexakos P et al. (2017) Nurr1:RXRalpha heterodimer activation as monotherapy for Parkinson’s disease. Proc Natl Acad Sci USA 114(15):3999–4004. 10.1073/pnas.1616874114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK (2009) A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137(1):47–59. 10.1016/j.cell.2009.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galleguillos D, Fuentealba JA, Gomez LM, Saver M, Gomez A, Nash K, Burger C, Gysling K et al. (2010) Nurr1 regulates RET expression in dopamine neurons of adult rat midbrain. J Neurochem 114(4):1158–1167. 10.1111/j.1471-4159.2010.06841.x [DOI] [PubMed] [Google Scholar]
- 48.Wang T, Hay JC (2015) Alpha-synuclein toxicity in the early secretory pathway: how it drives neurodegeneration in Parkinsons disease. Front Neurosci 9:433. 10.3389/fnins.2015.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy A, Pahan K (2015) PPARalpha signaling in the hippocampus: crosstalk between fat and memory. J Neuroimmune Pharmacol 10(1):30–34. 10.1007/s11481-014-9582-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy A, Jana M, Corbett GT, Ramaswamy S, Kordower JH, Gonzalez FJ, Pahan K (2013) Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor alpha. Cell Rep 4(4):724–737. 10.1016/j.celrep.2013.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin C, Deng Y, Liu Y, Gao J, Yan L, Gong Q (2018) Icariside II ameliorates cognitive impairments induced by chronic cerebral hypoperfusion by inhibiting the amyloidogenic pathway: involvement of BDNF/TrkB/CREB signaling and up-regulation of PPARalpha and PPARgamma in rats. Front Pharmacol 9:1211. 10.3389/fphar.2018.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekulic-Jablanovic M, Petkovic V, Wright MB, Kucharava K, Huerzeler N, Levano S, Brand Y, Leitmeyer K et al. (2017) Effects of peroxisome proliferator activated receptors (PPAR)-gamma and -alpha agonists on cochlear protection from oxidative stress. PLoS One 12(11):e0188596. 10.1371/journal.pone.0188596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vazquez M, Merlos M, Adzet T, Laguna JC (1996) Decreased susceptibility to copper-induced oxidation of rat-lipoproteins after fibrate treatment: influence of fatty acid composition. Br J Pharmacol 117(6):1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouedard C, Koum-Besson N, Barouki R, Morel Y (2003) Opposite regulation of the human paraoxonase-1 gene PON-1 by fenofibrate and statins. Mol Pharmacol 63(4):945–956 [DOI] [PubMed] [Google Scholar]
- 55.Mohagheghi F, Khalaj L, Ahmadiani A, Rahmani B (2013) Gemfibrozil pretreatment affecting antioxidant defense system and inflammatory, but not Nrf-2 signaling pathways resulted in female neuroprotection and male neurotoxicity in the rat models of global cerebral ischemia-reperfusion. Neurotox Res 23(3):225–237. 10.1007/s12640-012-9338-3 [DOI] [PubMed] [Google Scholar]
- 56.Corbett GT, Roy A, Pahan K (2012) Gemfibrozil, a lipid-lowering drug, upregulates IL-1 receptor antagonist in mouse cortical neurons: implications for neuronal self-defense. Journal of immunology (Baltimore, Md : 1950) 189(2):1002–1013. 10.4049/jimmunol.1102624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dauer W, Przedborski S (2003) Parkinson’s disease: mechanisms and models. Neuron 39(6):889–909 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.