Abstract
Trapa L., an annual floating-leaved herb, is widely distributed in the old world and has important edible and medicinal values. However, the taxonomy and phylogeny of Trapa are unclear. Here, we reported the complete chloroplast genome of a wild species with small nuts, T. incisa. The complete chloroplast genome size of T. incisa was 155, 453 bp, consisting of two inverted repeat (IR) regions (24, 388 bp), one large single copy (LSC) region (88, 398 bp) and one small single copy (SSC) region (18, 279 bp). A total of 129 genes were annotated, including 83 protein-coding genes, 38 tRNA genes and 8 rRNA genes. Among them, 19 genes were duplicated (6 protein-coding genes, 9 tRNA genes and 4 rRNA genes). The phylogenomic analysis suggested a close relationship between T. incisa and T. maximowiczii.
Keywords: Trapa incisa, complete chloroplast genome, Lythraceae, phylogeny
Water chestnut Trapa L. (Lythraceae) is an annual floating-leaved aquatic herb native to the temperate to subtropical regions of Africa, Asia, and Europe (Chen et al. 2007). Besides the important ecological values, the Trapa plants have been commercially cultivated as edible fruits in India, China and Italy (Suriyagoda et al. 2007). However, because of the various morphological traits and shortage of effective identification methods, the taxonomy and phylogeny of the genus are still unclear (Kim et al. 2010; Li et al. 2017). Molecular information is urgently needed to improve the situation of the genus. Previous studies showed that the nut size offered the best diagnostic criteria for the classification of Trapa species (Xiong et al. 1990; Fan et al. 2016). Trapa incisa is a typical species with small nut size. In this study, the chloroplast genome sequence of T. incisa was released, and the phylogenetic relationship was reconstructed within Lythraceae. The basic genetic information is helpful to species identification and systematic relationships construction within Trapa.
An individual of T. incisa was collected from the Wuhan Botanical Garden, Chinese Academy of Sciences, Hubei, China (114.613°E; 30.543°N). The voucher specimen was deposited at the Herbarium of Wuhan Botanical Garden (HIB: Yuanyuan Chen, yychen@wbgcas.cn) under the voucher number yychen20180066. Genome DNA was isolated from 0.5 g fresh leaves using the modified CTAB method (Doyle and Doyle 1987). The purified DNA was used to build a sequencing library with the Illumina NovaSeq 6000 platform. Finally, a total of 5.31 G raw data was obtained for further analysis. The complete chloroplast genome was assembled by GetOrgnelle v1.71 (Jin et al. 2020). The resultant genome was annotated by the genome annotator GeSeq (Tillich et al. 2017) with T. bicornis and T. maximowiczii as references; and the results were manually adjusted by Geneious (Kearse et al. 2012). The complete annotation chloroplast genome of T. incisa was deposited in GenBank with an accession number of MW543307.
The complete chloroplast genome length for T. incisa was 155, 453 bp with the quadripartite structure, including two inverted repeat (IR) regions (24, 388 bp), one large single copy (LSC) region (88, 398 bp) and one small single copy (SSC) region (18, 279 bp). The overall GC content was 36.4%. A total of 129 genes were annotated, consisting of 83 protein-coding genes, 38 tRNA genes and 8 rRNA genes. Among them, 19 genes were duplicated, including 6 protein-coding genes, 9 tRNA genes and 4 rRNA genes.
Phylogenetic analyses
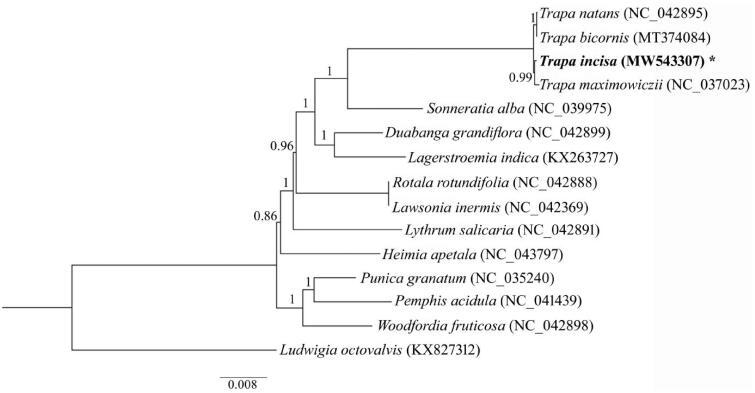
A maximum likelihood (ML) phylogenetic tree was constructed based on the 14 published complete plastomes of Lythraceae, with Ludwigia octovalvis (NC031385) as an out-group. The genome sequences were initially merged by BioEdit (Hall 1999), then aligned using MAFFT (Katoh et al. 2019). The ML tree was computed by PhyML v.3.0 (Stéphane et al. 2010) under the best model (TVM + G + I) and evaluated by Jmodeltest (Darriba et al. 2012). The phylogenetic tree strongly supported a close relationship between T. incisa and T. maximowiczii, which all have small seeds (Figure 1). Additionally, Sonneratia was closely related to Trapa within the family Lythraceae, which was also suggested by previous studies (Graham et al. 2005).
Figure 1.
Phylogenetic tree using maximum-likelihood (ML) based on plastomes of 14 Lythraceae species with Ludwigia octovalvis as an outgroup. Numbers near the nodes represent ML bootstrap values.
Funding Statement
This work was supported by the National Scientific Foundation of China [31100247], Talent Program of Wuhan Botanical Garden of the Chinese Academy of Sciences [Y855291B01] and the High-level Talent Training Program of Tibet University [2018-GSP-018].
Disclosure statement
The authors declare no potential conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MW543307.1/) under the accession no. MW543307. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726367, SRX10752768, and SAMN18928742, respectively.
References
- Chen JR, Ding BY, Funston AM.. 2007. Trapaceae, flora of China. Vol. 13. Beijing: Science Press; St. Louis (MO): Missouri Botanical Garden Press; p. 290–291. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Fan XR, Li Z, Chu HJ, Li W, Liu YL, Chen YY.. 2016. Analysis of morphological plasticity of Trapa L. from China and their taxonomic significance. Plant Sci J. 34:340–351. [Google Scholar]
- Graham SA, Hall J, Sytsma K, Shi SH.. 2005. Phylogenetic analysis of the Lythraceae based on four gene regions and morphology. Int J Plant Sci. 166(6):995–1017. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ.. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Na HR, Choi HK.. 2010. Molecular genotyping of Trapa bispinosa and T. japonica (Trapaceae) based on nuclear AP2 and chloroplast DNA trnL-F region. Am J Bot. 97(12):e149–152. [DOI] [PubMed] [Google Scholar]
- Li XL, Fan XR, Chu HJ, Li W, Chen YY.. 2017. Genetic delimitation and population structure of three Trapa taxa from the Yangtze River, China. Aquat Bot. 136:61–70. [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stéphane G, François DJ, Vincent L, Maria A, Wim H, Olivier G.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Suriyagoda L, Arima S, Suzuki A, Hoque A.. 2007. Variation in growth and yield performance of seventeen water chestnut accessions (Trapa spp.) collected from Asia and Europe. Plant Prod Sci. 10(3):372–379. [Google Scholar]
- Xiong ZT, Huang DS, Wang HQ, Sun XZ.. 1990. Numerical taxonomic studies in Trapa in Hubei III: numerical evaluations of taxonomic characters. Plant Sci J. 8:47–52. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MW543307.1/) under the accession no. MW543307. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726367, SRX10752768, and SAMN18928742, respectively.