Abstract
Purpose of review
In the past decade numerous studies analyzing the genome and transcriptome of large cohorts of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) patients have substantially improved our knowledge of the genetic landscape of these diseases with the identification of heterogeneous constellations of germline and somatic mutations with prognostic and therapeutic relevance. However, inclusion of integrated genetic data into classification schema is still far from a reality. The purpose of this review is to summarize recent insights into the prevalence, pathogenic role, clonal architecture, prognostic impact and therapeutic management of genetic alterations across the spectrum of myeloid malignancies.
Recent findings
Recent multiomic-studies, including analysis of genetic alterations at the single-cell resolution, have revealed a high heterogeneity of lesions in over 200 recurrently mutated genes affecting disease initiation, clonal evolution and clinical outcome. Artificial intelligence and specifically machine learning approaches have been applied to large cohorts of AML and MDS patients to define in an unbiased manner clinically meaningful disease patterns including disease classification, prognostication and therapeutic vulnerability, paving the way for future use in clinical practice.
Summary
Integration of genomic, transcriptomic, epigenomic and clinical data coupled to conventional and machine learning approaches will allow refined leukemia classification and risk prognostication and will identify novel therapeutic targets for these still high risk leukemia subtypes.
Keywords: myeloid, erythroid, mutations, prognosis, single-cells, machine learning
INTRODUCTION
Acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) are a heterogeneous group of diseases characterized by clonal expansion of undifferentiated myeloid precursors, impaired hematopoiesis and bone marrow failure. The threshold of 20% bone marrow blasts distinguishes AML from MDS on morphologic examination [1]. Approximately 30% of MDS cases progress to AML, the incidence of which varies across different MDS subtypes. Progression is generally associated with acquisition of driver mutations leading to clonal outgrowth [2]. In both diseases genetic alterations are progressively acquired overtime in hematopoietic stem cells that first gain a selective advantage (premalignant clone) and then fully transform in malignant clones when secondary mutations are acquired (Fig. 1). According to their biological function and clinical significance, most frequent mutations can be grouped into nine classes including NPM1, myeloid transcription factors, DNA methylation–related genes, epigenetic/chromatin modifiers, tumor suppressor genes, signal transduction pathways, RNA processing and splicing factors, cohesin complex and DNA repair. Mutations in DNA methylation, epigenetic modifiers and splicing machinery genes are typically acquired early, while mutations in transcriptional regulators, signal transduction pathways and chromosomal abnormalities may occur later. In addition to somatic mutations, germline mutations in several genes, such as DDX41 [3], RUNX1 [4,5], GATA2 [6], ANKRD26 [7], ETV6 [8], TP53 [9], CEBPA [10], SAMD9/SAMD9L [11-13], Fanconi anemia genes [14], or telomerase complex genes[15] predispose to AML and MDS in both children and adults [16,17]. Although common targets of mutations are largely similar across age and between AML and MDS, their frequency, type and co-mutational patterns vary by age and disease subtype [18,19]. The main genetic similarities, differences and their impact on prognosis will be discussed in this review.
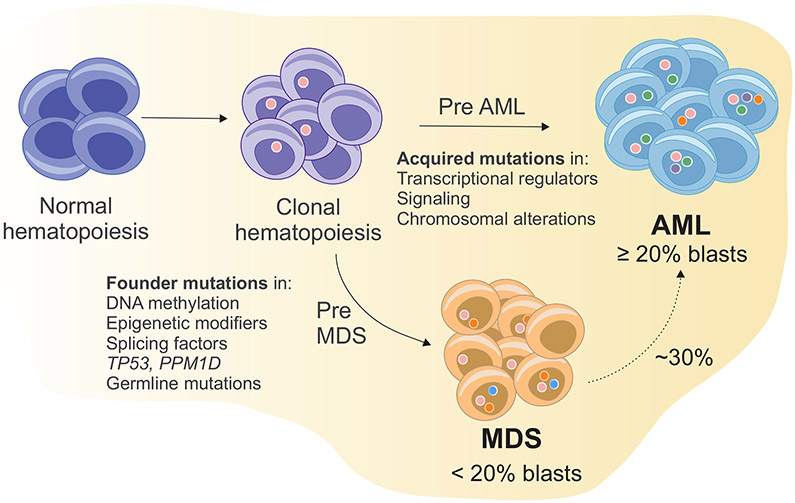
Figure 1. Simplified model of clonal expansion of hematopoietic stem and progenitor cells leading to MDS and AML.
An initiating driver mutation in a hematopoietic stem cell promotes the expansion of mutant stem cells and abnormal hematopoietic progenitor and precursor cells. These cells expand and become dominant. The occurrence of secondary mutations promotes a malignant cell transformation. According to cell morphology features, occurrence of dysplasia and percentage of bone marrow and peripheral blood blasts a diagnosis of AML or MDS is made. In MDS the acquisition of additional driver mutations or the emergence of preexisting ones leads to progression to AML in around 30% of cases.
CONSTELLATIONS OF GENETIC ALTERATIONS
Constellations of genetic alterations in AML
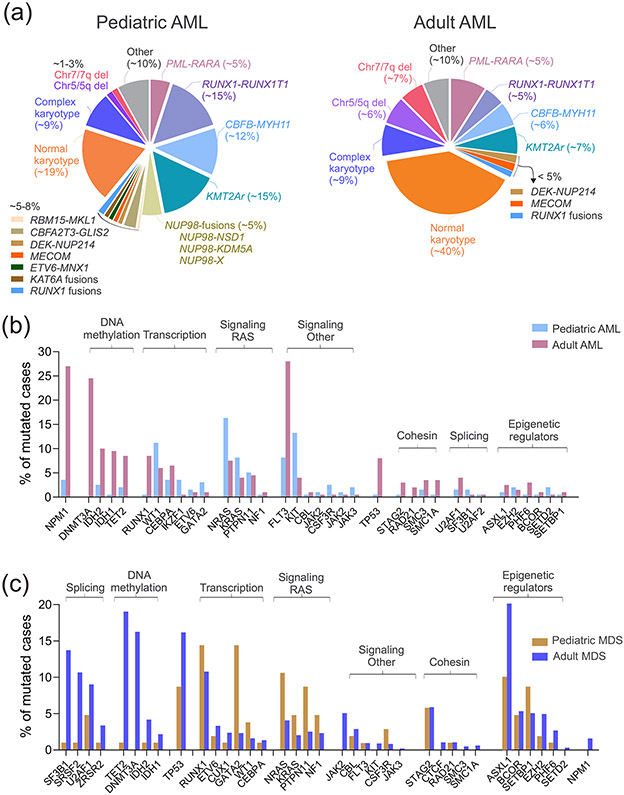
Acute myeloid leukemia includes a clinical and genetic heterogeneous group of hematopoietic malignancies that arise as a result of clonal expansion of undifferentiated myeloid precursors in the bone marrow due to genetic abnormalities that impair self-renewal, proliferation and differentiation. The prevalence of AML increases with age from 20% of all leukemias in childhood (with a peak in infancy) to 80% in older adults [20]. AML can present as either de novo or secondary disease (therapy related or post-antecedent hematologic disorder). Although intensified treatment regimens and risk-adapted patient stratification have improved overall survival, outcome is still dismal with overall 5-year survival of 35%-40% in patients aged <60 years and 5%-15% in older patients [20]. Since the first whole genome sequencing of a single AML patient in 2008 [21], large sequencing studies have considerably increased [19,22,23]. These studies have shown extensive genetic heterogeneity and important differences between the genomic landscapes of pediatric and adult AML. While cytogenetically normal AML represents ~40% of adult AML, it is only 15-20% of pediatric cases (Fig. 2A). In childhood, common gene fusions drive distinct pathogenetic pathways. Among those, KMT2A (also known as mixed lineage leukemia, MLL) rearrangements (KMT2Ar) are common in childhood AML (~15%), especially in infants (~50%), and, although partner dependent, are generally associated with unfavorable outcome [24,25]. The fusion RBM15- MKL1 arising from the t(1;22)(p13;q13) chromosomal rearrangement is a World Health Organization (WHO)-defined subtype of AML that occurs in around 10% of non-Down syndrome AML displaying megakaryoblastic differentiation (AMKL) [26,27], and is associated with intermediate clinical course [28]. Cryptic gene fusions, including NUP98-fusions (4-9% of pediatric AML) [18,29,30], CBFA2T3-GLIS2 (2% of all AML pediatric cases but 20–30% of pediatric AMKL cases) [26,27,31], and MNX1-ETV6 (~ 1% of all pediatric AML cases but 4-30% of AML in children less than 2 years old) [32] are almost exclusively found in pediatric AML and predict poor outcome [33-36]. Similar to MLL, NUP98, encoding a component of the nuclear pore complex, is rearranged to multiple different partner genes in AML. NUP98-KDM5A frequently co-occurs with RB1 gene deletion and is recurrent in acute erythroid leukemia (AEL, ~20% of pediatric cases), while cells with NUP98-NSD1 fusion frequently harbors FLT3 internal tandem duplications (ITD) or WT1 mutations [36-38]. Fusions affecting the core binding factor (CBFB-MYH11), the retinoid acid receptor (PML-RARA and variant RARA rearrangements) or the runt-related transcription factor 1 (RUNX1-RUNX1T1; AML1-ETO) disrupt transcription factors important for myeloid differentiation, occur at any age but with a peak in children and young adults, and are associated with a relatively favorable prognosis in both adults and children [39,40]. Co-occurring genetic alterations are observed and they significantly alter outcome, such as in the case of KIT mutations in core-binding factor leukemias [41-43]. Additional AML-associated cytogenetic abnormalities, more frequently found in adults than pediatric cases, with dismal outcome [44,45] include the DEK proto-oncogene fused to the nucleoporin gene 214 (NUP214) [46] (< 2%) and inv(3) or t(3;3) (1-4%) which is associated with elevated platelet counts, dysplastic megakaryocytes, multilineage dysplasia and MECOM overexpression [44,47-49]. With regard to mutations (Fig. 2B), alterations in transcription factors such as WT1 and GATA2 or in signaling genes (e.g. RAS and KIT) are more prevalent in pediatric cases compared to adult patients. Conversely, mutations in epigenetic modifiers, such as DNMT3A, TET2 and IDH1/2, in NPM1 and TP53 are more frequent in adult AML [19,38,50-53].
Figure 2. Recurrent genetic alterations in AML and MDS.
A) Main cytogenetic abnormalities according to age in pediatric (left panel) and adult (right panel) AML. B) Prevalence of somatic mutations (single nucleotide variations, indels and duplications) in adult versus pediatric AML. Data are from landmark previously published studies [18,19,23,38]. C) Prevalence of somatic mutations (single nucleotide variations, indels and duplications) in adult versus pediatric MDS. Data are from landmark previously published studies [60,62,63]. Genes are grouped according to their biological annotation.
Although the complexity of the AML genetic landscape, the mutation status of only few genes is considered by the current WHO classification model [1]. These include mutations in NPM1, FLT3, CEBPA and a provisional entity represented by RUNX1 mutations. For the remaining subgroups, classification is still based on morphological and immunophenotypic criteria with most entities in this group reminding the old French American British (FAB) classification subtypes [1]. Both recurrent cytogenetic and molecular alterations are also considered for prognostication by two current scoring systems: the European LeukemiaNet[54] and the Medical Research Council [55]. Here, mutations in TP53, ASXL1, DNMT3A and partial tandem duplication (PTD) of KMT2A are added as predictors of adverse outcome. However, except for cases with NPM1 mutations and co-occurring FLT3 ITDs, which are associated with intermediate prognosis compared to the favorable prognosis of NPM1 alone and adverse prognosis of FLT3 ITD alone, these models do not consider: i) functional consequences of distinct hotspot mutations; and ii) concurrent mutations that may dramatically alter the contribution of specific AML disease alleles to clinical outcomes. For example, NPM1 mutations preferentially occur with NRASG12/13 but not NRASQ61 and they generally are associated with favorable outcome [22]. In the case of NPM1-IDH2 mutational co-occurrence, IDH2R140 is significantly associated with NPM1 mutations, while IDH2R172 is mutually exclusive with NPM1 and other class-defining lesions. This hotspot is associated with a distinct gene expression, methylation and metabolic profiles [22]. Moreover, patients who have concurrent mutations of FLT3ITD, DNMT3A, and NPM1 have a very poor clinical outcome, compared to patients with NPM1 and DNMT3A mutations without FLT3ITD. In contrast, in pediatric AML, FLT3ITD and NPM1 mutations co-occur in the absence of DNMT3A mutations in a group of subjects with superior outcomes. However, when FLT3 ITD co-occurs with NUP98-NSD1 or WT1 mutations the prognosis is poor [18].
Morphology-based criteria have led to several changes in leukemia classification. This is notable for AEL, especially the historic FAB M6a (AML, not otherwise specified, NOS, erythroid-subtype) that in the latest revision of the WHO classification [56] was merged to a hybrid group of MDS or AML, NOS non-erythroid subtype. We have recently shown that AEL is a genetically heterogeneous with six defined age-related genomic subgroups:(1) biallelic TP53 mutations, often with concomitant mutations of chromatin regulators, transcription factors and tumor suppressors, 32%; (2) NPM1 mutations, 12%; (3) KMT2A/MLL mutations or rearrangements, 11%; (4)- NUP98-rearrangements, 4%; (5) DDX41 mutations, 3%; and (6) an MDS-like group with mutations in chromatin regulators and splicing factors, 37%; and overall with marked differences in mutation frequency between AEL and non-AEL AML and MDS [36]. For example, AEL has much lower frequency of canonical genes mutated in AML such as FLT3 and NPM1 when compared to non-erythroid AML, but they are more common than in MDS. Conversely, MDS-associated mutations such as SF3B1 and ASXL1 are less frequent in AEL compared to MDS, but more common than in non-erythroid AML. Genomic subgroups but not morphological phenotypes were the strongest predictors of outcome, highlighting the importance of genomic features to properly diagnose and risk-stratify patients [36].
Constellations of genetic alterations in MDS
Myelodysplastic syndromes encompass a spectrum of myeloid neoplasms characterized by ineffective hematopoiesis, cytopenia, abnormal cell morphology and a high propensity of progression to AML in 30% of cases (sAML) [57,58]. MDS is uncommon in children but it increases markedly with age, with a median age of onset of 71-76 years old [59]. It can be de novo or related to prior use of cytotoxic chemotherapy and/or radiation (therapy-related MDS, t-MDS)[1,2]. The latter is commonly associated with monosomies in chromosome 5 or 7, complex cytogenetics and poor outcome [2,60]. Although large DNA-sequencing studies have led to the identification of multiple recurrent mutations [60-65], genetic lesions are not used to define MDS subtypes in the WHO classification of myeloid neoplasms [1] nor in the current traditional scoring systems, including the international prognostic scoring system (IPSS) and the revised IPSS (IPSS-R) [66,67]. The only exceptions are isolated del(5q), which is associated with refractory anemia and normal to increased platelet counts with micromegakaryocytes [68], and SF3B1 mutations which have been included as a diagnostic criterion in MDS with ring sideroblasts (MDS-RS) [1,69-71]. Mutations in SF3B1 occur in 25% of all MDS cases but affect >80% of MDS-RS and are independent predictors of favorable outcomes [71,72]. Mutational targets in MDS overlap those in AML although with a different frequency and prognostic role and include those involved in RNA splicing, epigenetic modification, cohesin complex, transcription, DNA damage response, and signal transduction (Fig. 2C). RNA splicing is the most commonly mutated pathway in MDS, and six genes (SF3B1, TET2, SRSF2, ASXL1, DNMT3A, and RUNX1) are mutated in at least 10% of patients, while the others have a less occurrence and heterogenous patterns. In contrast to adult MDS, Ras/MAPK pathway mutations are common in pediatric MDS (45%), while mutations in RNA splicing genes are rare (2%) [73]. Mutation-interactions are not random, but several specific patterns have been described. For example, splicing mutations are almost mutually exclusive of each other, and, similarly, mutations in genes of cohesin complex, due to synthetic lethality mechanisms. By contrast, splicing mutations are significantly associated with mutations in epigenetic modifiers or transcription factors. For example, SRSF2 mutations are significantly associated with mutations in RUNX1, ASXL1, IDH2, CUX1, TET2, RUNX1 and STAG2. Although gene sequencing results are not included in the current prognostication guidelines, several mutation-prognosis associations have been described. Among those, the most concordant across different studies, include association between mutations in TP53, EZH2, ETV6, RUNX1, ASXL1, and SRSF2 and poor overall survival [61-63,65,74,75]. Notably, somatic mutations can predict overall survival independent of IPSS-R. However, genetic prognostic system may be improved by considering intra-patient co-mutational occurrence, type and number of mutation and variant allele frequency. For example, a recent study in over 3,000 MDS cases showed that MDS patients with monoallelic TP53 mutations did do not differ from TP53 wild-type patients in term of outcome and response to therapy, demonstrating the importance of allelic state in outcome [76**]. Two-thirds of TP53-mutated patients had biallelic targeting including more than one gene mutation, mutation and deletion, mutation and copy neutral loss of heterozygosity. These patients had association with genome instability, treatment resistance, disease progression and dismal outcomes, independently of IPSS-R [76**].
NOVEL GENETIC AND COMPUTATIONAL APPROACHES
Clonal architecture at the single cell resolution
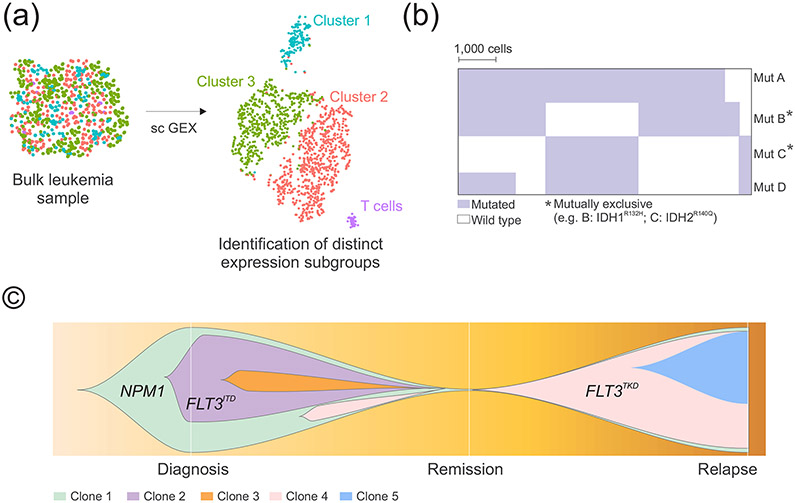
Single-cell sequencing studies can dissect clonal architecture and identify rare populations important for pathogenesis and response to therapy (Fig. 3). Dr. Peter van Galen and colleagues [77] combined single-cell RNA sequencing (Fig. 3A) and genotyping in AML and normal bone marrow cells and identified six malignant cell phenotypes whose abundancy varied between patients and between subclones in the same tumor, as well as across 179 bulk AML profiles from the Cancer Genome Atlas (TCGA) [19] queried with cell-type- specific gene signatures. This analysis yielded seven AML groups with distinct cell-type compositions and associated to characteristic genetic lesions. For example, cells with FLT3 tyrosine kinase domain (TKD) mutations were enriched in AML with differentiation, whereas those with FLT3ITD have higher abundances in primitive stem cells. Interestingly, NPM1 subgroups showed different phenotype according to co-occurrent lesions: a strong stem/progenitor phenotype when co-occurring with FLT3ITD and a more differentiated monocyte- to dendritic cell–like signatures in FLT3ITD-negative cases.
Figure 3. Schematic genomic analysis at the single-cell resolution.
A) Single-cell transcriptome sequencing can detect rare populations with distinct expression patterns and important for leukemogenesis. B) Single cell-DNA sequencing can dissect clonal intra patient genetic heterogeneity and identify co-occurrent or mutually exclusive aberrations. C) Representative fish plot graph showing emergence of mutations responsible for relapse. A schematic example is provided with data from [78**]. Each color represents an individual genetic clone. Abbreviations: sc GEX, single-cell gene expression; mut, mutation.
Single-cell mutation data have the power to unambiguously reveal co- occurrence and mutual exclusivity of driver mutations at the cellular level (Fig. 3B). For example, Dr. Morita and colleagues [78**] described that when multiple signaling pathway genes (e.g. KRAS, NRAS, FLT3) are present in the same patient, at the cellular level they often occur in mutually exclusive clones. A similar mutually exclusive relationship was observed among other functionally redundant mutations such as IDH1 and IDH2 or in the case of TET2 and IDH1. This may have important therapeutic implications since inhibition of one mutation may favor the expansion of the other clone. A notable group of mutations that display statistically significant mutual exclusivity in MDS and AML is that with mutations in splicing factors, although in very rare cases (< 1%) 2 concomitant splicing factor mutations may be present [79**]. By single-cell DNA-sequencing of patient cells with > 1 splicing mutation, Taylor et al [79**] showed that escape from this epistasis occurs when there is i) selection for less common alleles, such as SF3B1non-K700E mutations, that have reduced effects on RNA splicing and/or binding compared with the most common alleles; ii) mutations that occur in cis with preservation of the wild-type allele [79**].
Progress in single-cell technologies allows to simultaneously explore genotype-phenotype correlations by analysis of single-cell DNA and cell surface proteins [78**,80**]. For example, cells with NPM1 or IDH mutations have been described to express lower levels of CD34 and HLA-DR, while cells with a single TP53 mutations have higher levels of CD34 and CD117, but double TP53 mutations have a monocytic immunophenotype [78**,80**].
Moreover, single-cell DNA sequencing studies can dissect the evolution of clonal architecture in response to different therapies (Fig. 3C) [78**,81-83*]. For example, a selection of a small subclone with FLT3 TKD (D835Y) was observed during azacitidine and sorafenib (a FLT3 inhibitor) treatment, which was associated with relapse. Similarly, selection of subclones with NRAS mutation along with the acquisition of PTPN11, FLT3ITD, and IDH1 mutations were observed during treatment with azacytidine and enasidenib (an IDH2 inhibitor) [78**]. Polyclonal emergence of multiple independent kinase activating clones, including FLT3ITD, FLT3TKD, and Ras mutations have been described by single cell analysis also in samples who acquired resistance to venetoclax [83*]. Overall, these findings show that single-cell sequencing studies by dissecting clonal diversity and revealing evolution patterns have important clinical relevance and it is likely that their use will increase in the clinical management of AML.
Machine learning approach in the diagnosis of AML and MDS
The enormous advances in technology and genomics have generated a very large and heterogeneous volume of data from large cohorts of patients. At the same time, advances in hardware and computing have led to an increasing use of machine learning approaches in medicine. “Machine learning” (ML) is an application of artificial intelligence (AI) that defines a data analysis method that automatically learns from data and experience and make decisions without being explicitly programmed [84]. Since both AML and MDS are characterized by high genetic and phenotypic heterogeneity, they represent the good candidates for ML. Warnat-Herresthal et al [85*] used a transcriptomic-based ML approach to predict in an unbiased, entirely data-driven manner, genome-wide predictors of AML in 12,029 transcriptome samples from 105 different studies, including AML, MDS, acute lymphoblastic leukemia and healthy individuals. The results provided evidence that without ancillary data or expert input, the combination of large transcriptomic data with ML allows for the development of robust AML classifiers with>99% accuracy. AI-based image analysis has been successfully used in several studies to classify cells on bone marrow aspirates and predicts diagnosis with sensitivity and specificity > 95% [86,87]. MDS diagnosis strongly relies on morphological interpretation. ML approaches may overcome the limitation due to variable pathologic evaluations. Recently, Nagata et al [88] have used ML to identify patterns of co-occurrence among morphologic features and genomic events in 1079 MDS cases. Novel genotype/ morphology/prognosis associations include STAG2mut and SRSF2mut with myeloid dysplasia and ASXL1mut with megakaryocytic dysplasia. By unsupervised consensus clustering, 5 distinct MDS morphological profiles with unique clinical characteristics were identified, separating patients with different prognoses. Moreover, additional genetic signatures were further classified and associated with specific morphological profiles by Bayesian graphical models and validated in an independent cohort [88]. Overall, these findings demonstrate the power of large data sets and computing in identifying phenotype/genotype associations and assisting in primary diagnosis.
CONCLUSION
The catalogue of mutations affecting pathophysiological and clinical features of myeloid malignancies has exponentially grown in the past decade and showed that pathogenesis is much more complex than that suggested by morphology examination alone. Integrated conventional and molecular approaches are required to comprehensively identify all combinations of mutations, guiding in classification, risk assessment and therapy.
KEY POINTS.
Myeloid leukemia subtypes of prognostic significance are defined by combinatorial mutations, most of which are age dependent.
Single-cell sequencing studies have dissected clonal heterogeneity in AML and MDS and showed distinct correlations between cell-type compositions and genetic lesions, patterns of mutational co-occurrence and exclusivity and clones associated with therapy resistance.
Machine learning has been successfully used to identify phenotype/genotype associations and predict diagnosis of AML and MDS.
Future classification of myeloid malignancies will likely integrate conventional morphological-based, molecular and computational approaches.
Acknowledgments
Financial support and sponsorship
This work was supported by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital and by the National Cancer Institute grant R35CA197695 (C.G.M.)
Footnotes
Conflicts of interest
I.I. has received honoraria from Amgen; C.G.M. received funded research from Abbvie, Loxo Oncology and Pfizer; he received speaking and travel fees from Illumina and Amgen; he holds stock in Amgen.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as: * of special interest or ** of outstanding interest
- 1.Arber DA, Orazi A, Hasserjian R, et al. : The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127:2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Menssen AJ, Walter MJ: Genetics of progression from MDS to secondary leukemia. Blood 2020, 136:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maciejewski JP, Padgett RA, Brown AL, et al. : DDX41-related myeloid neoplasia. Semin Hematol 2017, 54:94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellissimo DC, Speck NA: RUNX1 Mutations in Inherited and Sporadic Leukemia. Front Cell Dev Biol 2017, 5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olofsen PA, Touw IP: RUNX1 Mutations in the Leukemic Progression of Severe Congenital Neutropenia. Mol Cells 2020, 43:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulasekararaj AG, Jiang J, Smith AE, et al. : Somatic mutations identify a subgroup of aplastic anemia patients who progress to myelodysplastic syndrome. Blood 2014, 124:2698–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noris P, Favier R, Alessi MC, et al. : ANKRD26-related thrombocytopenia and myeloid malignancies. Blood 2013, 122:1987–1989. [DOI] [PubMed] [Google Scholar]
- 8.Galera P, Dulau-Florea A, Calvo KR: Inherited thrombocytopenia and platelet disorders with germline predisposition to myeloid neoplasia. Int J Lab Hematol 2019, 41 Suppl 1:131–141. [DOI] [PubMed] [Google Scholar]
- 9.Guha T, Malkin D: Inherited TP53 Mutations and the Li-Fraumeni Syndrome. Cold Spring Harb Perspect Med 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith ML, Cavenagh JD, Lister TA, et al. : Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med 2004, 351:2403–2407. [DOI] [PubMed] [Google Scholar]
- 11.Buonocore F, Kuhnen P, Suntharalingham JP, et al. : Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J Clin Invest 2017, 127:1700–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pastor VB, Sahoo SS, Boklan J, et al. : Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica 2018, 103:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong JC, Bryant V, Lamprecht T, et al. : Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceccaldi R, Sarangi P, D'Andrea AD: The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 2016, 17:337–349. [DOI] [PubMed] [Google Scholar]
- 15.Grill S, Nandakumar J: Molecular mechanisms of telomere biology disorders. J Biol Chem 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy AL, Shimamura A: Genetic predisposition to MDS: clinical features and clonal evolution. Blood 2019, 133:1071–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rafei H, DiNardo CD: Hereditary myeloid malignancies. Best Pract Res Clin Haematol 2019, 32:163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolouri H, Farrar JE, Triche T Jr., et al. : The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018, 24:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research N, Ley TJ, Miller C, et al. : Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013, 368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dohner H, Weisdorf DJ, Bloomfield CD: Acute Myeloid Leukemia. N Engl J Med 2015, 373:1136–1152. [DOI] [PubMed] [Google Scholar]
- 21.Ley TJ, Mardis ER, Ding L, et al. : DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008, 456:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaemmanuil E, Gerstung M, Bullinger L, et al. : Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016, 374:2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyner JW, Tognon CE, Bottomly D, et al. : Functional genomic landscape of acute myeloid leukaemia. Nature 2018, 562:526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winters AC, Bernt KM: MLL-Rearranged Leukemias-An Update on Science and Clinical Approaches. Front Pediatr 2017, 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balgobind BV, Raimondi SC, Harbott J, et al. : Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood 2009, 114:2489–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Rooij JD, Branstetter C, Ma J, et al. : Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet 2017, 49:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masetti R, Pigazzi M, Togni M, et al. : CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood 2013, 121:3469–3472. [DOI] [PubMed] [Google Scholar]
- 28.de Rooij JD, Hollink IH, Arentsen-Peters ST, et al. : NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 2013, 27:2280–2288. [DOI] [PubMed] [Google Scholar]
- 29.Bisio V, Zampini M, Tregnago C, et al. : NUP98-fusion transcripts characterize different biological entities within acute myeloid leukemia: a report from the AIEOP-AML group. Leukemia 2017, 31:974–977. [DOI] [PubMed] [Google Scholar]
- 30.Struski S, Lagarde S, Bories P, et al. : NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia 2017, 31:565–572. [DOI] [PubMed] [Google Scholar]
- 31.Masetti R, Bertuccio SN, Pession A, et al. : CBFA2T3-GLIS2-positive acute myeloid leukaemia. A peculiar paediatric entity. Br J Haematol 2019, 184:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosi S, Mostafa Kamel Y, Owoka T, et al. : Paediatric acute myeloid leukaemia with the t(7;12)(q36;p13) rearrangement: a review of the biological and clinical management aspects. Biomark Res 2015, 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michmerhuizen NL, Klco JM, Mullighan CG: Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood 2020, 136:2275–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruber TA, Larson Gedman A, Zhang J, et al. : An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 2012, 22:683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beverloo HB, Panagopoulos I, Isaksson M, et al. : Fusion of the homeobox gene HLXB9 and the ETV6 gene in infant acute myeloid leukemias with the t(7;12)(q36;p13). Cancer Res 2001, 61:5374–5377. [PubMed] [Google Scholar]
- 36.Iacobucci I, Wen J, Meggendorfer M, et al. : Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet 2019, 51:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Z, Morris SW, Valentine V, et al. : Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 2001, 28:220–221. [DOI] [PubMed] [Google Scholar]
- 38.Shiba N, Yoshida K, Hara Y, et al. : Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood Adv 2019, 3:3157–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel JP, Gonen M, Figueroa ME, et al. : Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012, 366:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pui CH, Carroll WL, Meshinchi S, et al. : Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol 2011, 29:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard JA, Alonzo TA, Gerbing RB, et al. : Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood 2010, 115:2372–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paschka P, Marcucci G, Ruppert AS, et al. : Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol 2006, 24:3904–3911. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa Y, Kawashima N, Atsuta Y, et al. : Prospective evaluation of prognostic impact of KIT mutations on acute myeloid leukemia with RUNX1-RUNX1T1 and CBFB-MYH11. Blood Adv 2020, 4:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamazaki H, Suzuki M, Otsuki A, et al. : A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 2014, 25:415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers HJ, Vardiman JW, Anastasi J, et al. : Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv(3)(q21q26.2)/t(3;3)(q21;q26.2): a Bone Marrow Pathology Group study. Haematologica 2014, 99:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alsabeh R, Brynes RK, Slovak ML, et al. : Acute myeloid leukemia with t(6;9) (p23;q34): association with myelodysplasia, basophilia, and initial CD34 negative immunophenotype. Am J Clin Pathol 1997, 107:430–437. [DOI] [PubMed] [Google Scholar]
- 47.Grimwade D, Hills RK, Moorman AV, et al. : Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116:354–365. [DOI] [PubMed] [Google Scholar]
- 48.Tarlock K, Alonzo TA, Moraleda PP, et al. : Acute myeloid leukaemia (AML) with t(6;9)(p23;q34) is associated with poor outcome in childhood AML regardless of FLT3-ITD status: a report from the Children's Oncology Group. Br J Haematol 2014, 166:254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groschel S, Sanders MA, Hoogenboezem R, et al. : A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014, 157:369–381. [DOI] [PubMed] [Google Scholar]
- 50.Ho PA, Kutny MA, Alonzo TA, et al. : Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: a report from the Children's Oncology Group. Pediatr Blood Cancer 2011, 57:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ley TJ, Ding L, Walter MJ, et al. : DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010, 363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paschka P, Schlenk RF, Gaidzik VI, et al. : IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 2010, 28:3636–3643. [DOI] [PubMed] [Google Scholar]
- 53.Delhommeau F, Dupont S, Della Valle V, et al. : Mutation in TET2 in myeloid cancers. N Engl J Med 2009, 360:2289–2301. [DOI] [PubMed] [Google Scholar]
- 54.Dohner H, Estey E, Grimwade D, et al. : Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129:424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimwade D, Ivey A, Huntly BJ: Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 2016, 127:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arber DA: Revisiting erythroleukemia. Curr Opin Hematol 2017, 24:146–151. [DOI] [PubMed] [Google Scholar]
- 57.Ogawa S: Genetics of MDS. Blood 2019, 133:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cazzola M: Myelodysplastic Syndromes. N Engl J Med 2020, 383:1358–1374. [DOI] [PubMed] [Google Scholar]
- 59.Sekeres MA: Epidemiology, natural history, and practice patterns of patients with myelodysplastic syndromes in 2010. J Natl Compr Canc Netw 2011, 9:57–63. [DOI] [PubMed] [Google Scholar]
- 60.Lindsley RC, Saber W, Mar BG, et al. : Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med 2017, 376:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bejar R, Stevenson K, Abdel-Wahab O, et al. : Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 2011, 364:2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papaemmanuil E, Gerstung M, Malcovati L, et al. : Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122:3616–3627; quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haferlach T, Nagata Y, Grossmann V, et al. : Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28:241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Makishima H, Yoshizato T, Yoshida K, et al. : Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2017, 49:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida K, Sanada M, Shiraishi Y, et al. : Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478:64–69. [DOI] [PubMed] [Google Scholar]
- 66.Greenberg PL, Tuechler H, Schanz J, et al. : Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120:2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Manero G, Shan J, Faderl S, et al. : A prognostic score for patients with lower risk myelodysplastic syndrome. Leukemia 2008, 22:538–543. [DOI] [PubMed] [Google Scholar]
- 68.Venugopal S, Mascarenhas J, Steensma DP: Loss of 5q in myeloid malignancies - A gain in understanding of biological and clinical consequences. Blood Rev 2020:100735. [DOI] [PubMed] [Google Scholar]
- 69.Papaemmanuil E, Cazzola M, Boultwood J, et al. : Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011, 365:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malcovati L, Karimi M, Papaemmanuil E, et al. : SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015, 126:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malcovati L, Stevenson K, Papaemmanuil E, et al. : SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malcovati L, Papaemmanuil E, Bowen DT, et al. : Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 2011, 118:6239–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwartz JR, Ma J, Lamprecht T, et al. : The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun 2017, 8:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cazzola M, Della Porta MG, Malcovati L: The genetic basis of myelodysplasia and its clinical relevance. Blood 2013, 122:4021–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sperling AS, Gibson CJ, Ebert BL: The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer 2017, 17:5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. **.Bernard E, Nannya Y, Hasserjian RP, et al. : Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med 2020, 26:1549–1556.Multi-hit TP53 state in MDS asscoiates with genomic instability, treatment resitance, disease progression and dismal outcome.
- 77.van Galen P, Hovestadt V, Wadsworth Ii MH, et al. : Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 2019, 176:1265–1281 e1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. **.Morita K, Wang F, Jahn K, et al. : Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun 2020, 11:5327.Comprehensive single-cell sequencing study of a large cohort of AML patients.
- 79. **.Taylor J, Mi X, North K, et al. : Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 2020, 136:1477–1486.Elucidation of mechanisms of escape from epistasis of RNA splicing factor mutations.
- 80. **.Miles LA, Bowman RL, Merlinsky TR, et al. : Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020.Dissection of phenotype-genotype association at the single-cell resolution in AML cells.
- 81.McMahon CM, Ferng T, Canaani J, et al. : Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov 2019, 9:1050–1063. [DOI] [PubMed] [Google Scholar]
- 82.Xu L, Durruthy-Durruthy R, Eastburn DJ, et al. : Clonal Evolution and Changes in Two AML Patients Detected with A Novel Single-Cell DNA Sequencing Platform. Sci Rep 2019, 9:11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. *.DiNardo CD, Tiong IS, Quaglieri A, et al. : Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood 2020, 135:791–803.Use of single cell sequencing to to investigate treatment failure to venetoclax combinations.
- 84.Li Y, Wu FX, Ngom A: A review on machine learning principles for multi-view biological data integration. Brief Bioinform 2018, 19:325–340. [DOI] [PubMed] [Google Scholar]
- 85. *.Warnat-Herresthal S, Perrakis K, Taschler B, et al. : Scalable Prediction of Acute Myeloid Leukemia Using High-Dimensional Machine Learning and Blood Transcriptomics. iScience 2020, 23:100780.Use of machine learning approach to predict AML classification.
- 86.Chandradevan R, Aljudi AA, Drumheller BR, et al. : Machine-based detection and classification for bone marrow aspirate differential counts: initial development focusing on nonneoplastic cells. Lab Invest 2020, 100:98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kimura K, Tabe Y, Ai T, et al. : A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Sci Rep 2019, 9:13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. *.Nagata Y, Zhao R, Awada H, et al. : Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes. Blood 2020, 136:2249–2262.Comprehensive implementation of machine learning algorithms to define interdependencies among genetic lesions, morphologies and outcome in MDS.