ABSTRACT
Seagrasses are marine flowering plants that provide critical ecosystem services in coastal environments worldwide. Marine fungi are often overlooked in microbiome and seagrass studies, despite terrestrial fungi having critical functional roles as decomposers, pathogens, or endophytes in global ecosystems. Here, we characterize the distribution of fungi associated with the seagrass Zostera marina, using leaves, roots, and rhizosphere sediment from 16 locations across its full biogeographic range. Using high-throughput sequencing of the ribosomal internal transcribed spacer (ITS) region and 18S rRNA gene, we first measured fungal community composition and diversity. We then tested hypotheses of neutral community assembly theory and the degree to which deviations suggested that amplicon sequence variants (ASVs) were plant selected or dispersal limited. Finally, we identified a core mycobiome and investigated the global distribution of differentially abundant ASVs. We found that the fungal community is significantly different between sites and that the leaf mycobiome follows a weak but significant pattern of distance decay in the Pacific Ocean. Generally, there was evidence for both deterministic and stochastic factors contributing to community assembly of the mycobiome, with most taxa assembling through stochastic processes. The Z. marina core leaf and root mycobiomes were dominated by unclassified Sordariomycetes spp., unclassified Chytridiomycota lineages (including Lobulomycetaceae spp.), unclassified Capnodiales spp., and Saccharomyces sp. It is clear from the many unclassified fungal ASVs and fungal functional guilds that knowledge of marine fungi is still rudimentary. Further studies characterizing seagrass-associated fungi are needed to understand the roles of these microorganisms generally and when associated with seagrasses.
IMPORTANCE Fungi have important functional roles when associated with land plants, yet very little is known about the roles of fungi associated with marine plants, like seagrasses. In this study, we report the results of a global effort to characterize the fungi associated with the seagrass Zostera marina across its full biogeographic range. Although we defined a putative global core fungal community, it is apparent from the many fungal sequences and predicted functional guilds that had no matches to existing databases that general knowledge of seagrass-associated fungi and marine fungi is lacking. This work serves as an important foundational step toward future work investigating the functional ramifications of fungi in the marine ecosystem.
KEYWORDS: seagrasses, Zostera marina, marine fungi, microbial eukaryotes, 18S rRNA, ITS2, eelgrass, mycobiome, core, abundance-occupancy, dispersal limited, plant selected, global distribution
INTRODUCTION
Terrestrial fungi are known to have critical ecological roles as microbial saprotrophs, pathogens, and mutualists (1), and although less is known about fungi in aquatic ecosystems, it is believed that they also have vital ecological roles (e.g., in organic matter degradation, nutrient cycling, and food web dynamics [2–7]). Despite their global importance, the taxonomic, phylogenetic, and functional diversity of marine fungi generally is vastly understudied (8). In comparison to the greater than 120,000 terrestrial fungal species known (9), there are currently only ∼1,900 described species of marine fungi, though estimates of the true diversity of these organisms are much higher (10–12). Recent studies have examined the global distribution of marine planktonic, pelagic, and benthic fungi (13–15), yet the distribution of host-associated fungi in the marine environment is still relatively unknown. Fungi have been reported in association with many marine animals, including sponges (16), corals (17), and other invertebrates (18), with algae and seaweeds (19, 20), and with flowering plants, like seagrasses (21).
Seagrasses are foundation species in coastal ecosystems worldwide and are the only submerged angiosperms (flowering plants) to inhabit the marine environment. One widespread seagrass species, Zostera marina, also known as eelgrass, provides critical ecosystem services in coastal environments throughout much of the Northern Hemisphere (22–24). Previous studies have investigated the composition and structure of the bacterial community associated with Z. marina, including a global survey that was able to identify a core eelgrass root microbiome (25–27). Members of this community are thought to facilitate nitrogen and sulfur cycling for host plant benefit (25, 27–33).
Comparatively, not as much is known about the distribution, diversity, and function of the mycobiome (i.e., the fungal community) associated with Z. marina. Culture-based studies have described a mycobiome composed of taxa in the classes Eurotiomycetes, Dothideomycetes, and Sordariomycetes (34–37). These studies consistently find dominance of a few ubiquitous taxa (e.g., Cladosporium sp.) but also a diverse set of rare taxa that vary among sites and may be endemic to specific locations (e.g., Colletotrichum sp.) (37). One hypothesis for this pattern is that the fungal community is assembled neutrally through stochastic processes.
While culture-independent studies of Z. marina and other seagrass species have more exhaustively characterized the taxonomic diversity of these fungal communities, they have also highlighted how little is known about factors affecting the distribution, function, and community assembly of seagrass-associated fungi (38–42). A common finding among these studies is that taxonomic assignments cannot be made for greater than two-thirds of the fungal sequences associated with seagrasses and that Chytridiomycota lineages are dominant in this ecosystem (39, 40, 42). Culture-independent understanding of the mycobiome of Z. marina has so far focused on a single location in Bodega Bay, California (42). However, site-to-site variation in the mycobiome has now been observed in mycobiome studies from several other seagrass species (38–41). For example, a distance decay relationship was found for the fungal community associated with the seagrass Enhalus acoroides in Singapore and Peninsular Malaysia (40) and for the seagrass Syringodium isoetifolium along Wallace’s Line, a boundary line separating Asian and Australasian taxa (41). Additionally, the global planktonic marine fungal community was found to cluster by ocean (15); therefore, we hypothesize in this study that in addition to a distance decay relationship, we will observe differentiation by ocean basin. Such geographic relationships would support an alternative hypothesis of niche-based community assembly of the mycobiome through deterministic processes, such as environmental filtering and dispersal.
One concept central to the investigation here is the role of stochastic and deterministic drivers in determining the community assembly of the seagrass mycobiome. The Sloan neutral model assumes that random immigrations, births, and deaths determine the relative abundance of taxa in a community (43, 44). The model further assumes that local communities are assembled stochastically from regional pools and that deterministic competitive interactions are not important because species are competitively equivalent (45–47). Stochastic processes supporting the neutral model include priority effects and ecological drift, while deterministic processes include species traits, interspecies interactions (e.g., competition, mutualisms), and environmental conditions (48). Dispersal limitation can be either a stochastic or deterministic process (49). Identifying specific taxa that deviate from the model allows us to predict taxa that are assembled through deterministic processes, including plant selection (50).
Here, we use high-throughput sequencing of marker genes to (i) characterize the fungal community associated with the seagrass Zostera marina on a global scale, (ii) test for a pattern of distance decay within ocean basins, (iii) define a global core mycobiome, (iv) use neutral models to test whether the community is assembled through a stochastic, deterministic, or combination of processes, (v) predict important fungal taxa based on assembly dynamics and global distribution, and (vi) assign functional predictions for the fungal community associated with Z. marina.
RESULTS
Fungal alpha diversity differs between sites, tissues, and oceans.
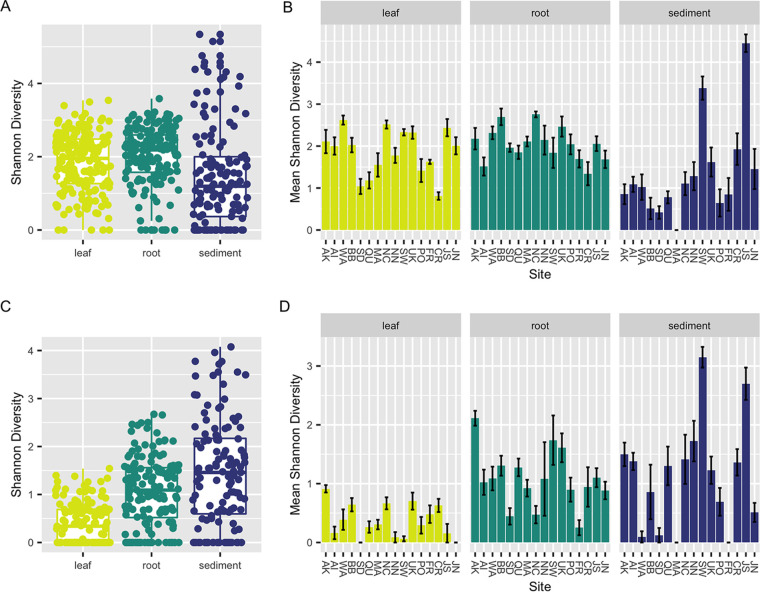
The Shannon index was significantly different between sample types (Kruskal-Wallis [K-W] test, P < 0.001) (Fig. 1) for both the internal transcribed spacer 2 (ITS2) region amplicon and 18S rRNA gene amplicon data sets. Post hoc Dunn tests of both data sets suggest that alpha diversity for leaves was generally lower than that for roots (P < 0.05). However, there were conflicting results for the sediment, with diversity being lower in the sediment than in leaves and roots in the ITS2 region amplicons (P < 0.05) and diversity being higher in the sediment than in leaves and roots in the 18S rRNA gene amplicons (P < 0.05). Alpha diversity for both data sets also was significantly different within each sample type across sites (K-W test, P < 0.001) (Fig. 1). This was driven by diversity being significantly different across some but not all sites (Dunn test, P < 0.05) (see Tables S1 to S6 in the supplemental material). For leaves, this was caused by lower diversity at some sites (SD, CR, QU [see Table 1 for site codes]) in the ITS2 region amplicon data and higher diversity of leaves at one site (AK) in the 18S rRNA gene amplicon data. For roots, this was driven by higher diversity at two sites (NC, BB) in the ITS2 region amplicon data and, for the 18S rRNA gene data, higher diversity at one site (AK) and lower diversity at others (SD, NC, FR). For the sediment for both data sets, observed alpha diversity differences were due to higher diversity at two sites (JS, SW). Alpha diversity for leaves was significantly different across oceans for the ITS2 region amplicon data set (K-W test, P = 0.014) but was not significantly different for roots or sediment between oceans or for leaves, roots, or sediment between oceans for the 18S rRNA gene amplicon data set (P > 0.05).
FIG 1.
Within-sample diversity varies across tissues and sites. Here we depict sample Shannon diversities for the ITS2 region for each sample type (leaf, root, sediment) using box plots (A) and within each sample type at each collection site (see Table 1 for site codes) using bar charts (B). Shannon diversities for the 18S rRNA gene amplicon data are also plotted for each sample type (leaf, root, sediment) using box plots (C) and within each sample type at each collection site (see Table 1 for site codes) using bar charts (D). For panels B and D, the standard error of the mean Shannon diversity at each site for each sample type is represented by an error bar, and bars are colored by sample type. Post hoc Dunn results for pairwise site comparisons for each sample type can be found in Tables S1 to S6 in the supplemental material. For panels A to D, zero values indicate samples that are dominated by a single taxon.
TABLE 1.
Description of collection sitesa
| Site code | Site | Site name | Collection mo | Collection yr | Latitude | Longitude |
|---|---|---|---|---|---|---|
| AK | Alaska—North | Kotzebue | October | 2016 | 64.485428 | −164.76189 |
| AI | Alaska—South | Izembek | October | 2016 | 55.328899 | −162.82121 |
| BB | California—North | Westside Park, Bodega Bay | December | 2016 | 38.319755 | −123.05514 |
| SD | California—South | Shelter Island | November | 2016 | 32.713756 | −117.22547 |
| QU | Canada | Pointe-Lebel | September | 2016 | 49.11237 | −68.17593 |
| CR | Croatia | Posedarje | September | 2016 | 44.21155 | 15.4906946 |
| FR | French Mediterranean | Bouzigues, Etang de Thau | October | 2016 | 43.446971 | 3.661503 |
| JN | Japan—North | Akkeshi-ko estuary | September | 2016 | 43.021167 | 144.903217 |
| JS | Japan—South | Ikunoshima | September | 2016 | 34.297834 | 132.91631 |
| MA | Massachusetts | Dorothy Cove | October | 2016 | 42.42014 | −70.91544 |
| NC | North Carolina | Middle Marsh | April | 2017 | 34.692458 | −76.622589 |
| NN | Norway | Røvika | July | 2016 | 67.2667233 | 15.2560633 |
| PO | Portugal | Culatra | October | 2016 | 37.01427 | −7.493273 |
| SW | Sweden | Torseröd | August | 2016 | 58.3131 | 11.5488 |
| UK | Wales | Porthdinllaen | March | 2017 | 52.990731 | −4.450321 |
| WA | Washington | Willapa Bay | September | 2016 | 46.474 | −124.028 |
Specifics of each collection site, including the site code, site name, month of sample collection, year of sample collection, and site latitude and longitude, are given.
Fungal community structure differs across sites, tissues, and oceans.
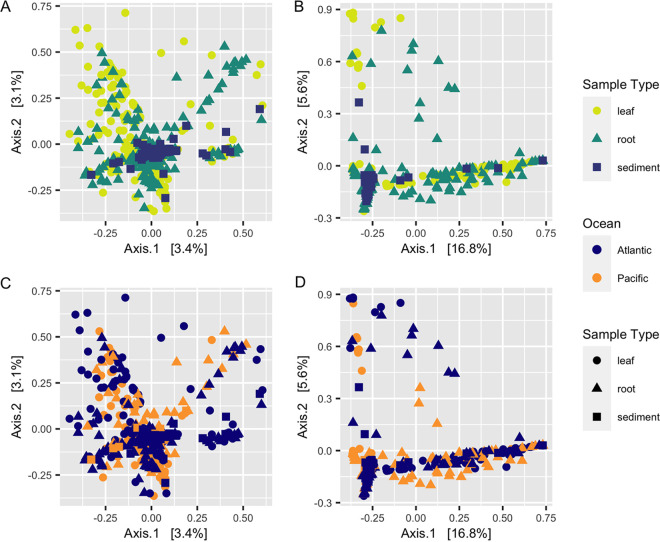
Similar to alpha diversity, fungal beta diversity was significantly different for both data sets using all three ecological metrics (Bray-Curtis dissimilarity, Aitchinson distance, Hellinger distance) across sample types (permutational multivariate analysis of variance [PERMANOVA], P < 0.001) (Fig. 2A and B), across sites (P < 0.001) (Fig. S1), and across oceans (P < 0.001) (Fig. 2C and D). Post hoc pairwise PERMANOVA tests using the ITS2 region amplicon data indicated significant differences in beta diversity across all sample types (P < 0.001) and all sites (P < 0.01). These results were generally consistent with the 18S rRNA gene sequence data, which supported differences in community structure across sample types (P < 0.001) and across the majority of, but not all, collection sites (P < 0.05).
FIG 2.
Community structure varies between tissues and ocean basins. Principal-coordinate analysis (PCoA) visualization of Hellinger distances of fungal communities associated with leaves, roots, and sediment based on ITS2 region amplicon data (A and C) and 18S rRNA gene amplicon data (B and D). (A and B) Points in the ordination are colored and represented by shapes based on sample type: leaf, yellow circles; root, green triangles; sediment, blue squares. (C and D) Points in the ordination are colored by ocean (Pacific, blue; Atlantic, orange) and represented by shapes based on sample type (circles), root (triangles), or sediment (squares).
Within-group variance (i.e., dispersion) also differed significantly for the ITS2 region amplicon data using all three beta diversity metrics across sample types (betadisper, P < 0.01) and sites (P < 0.01) but did not vary across oceans (P > 0.05). Mean dispersion between sites in the 18S rRNA gene data was not significant for two of the ecological metrics (Bray-Curtis, P = 0.79; Hellinger, P = 1), and similarly, the mean dispersion between oceans was not significant for two of the ecological metrics (Bray-Curtis, P = 0.07; Aitchinson, P = 0.26). Mean dispersion was otherwise consistent with the significant results observed in the ITS2 region amplicon data. PERMANOVA results have been shown to confuse dispersion differences and centroid differences when a balanced design is not used. Therefore, these results may indicate that either mean centroids, mean dispersions, or both are differing between sample types and sites here.
Mantel tests suggest weak distance decay relationships within oceans.
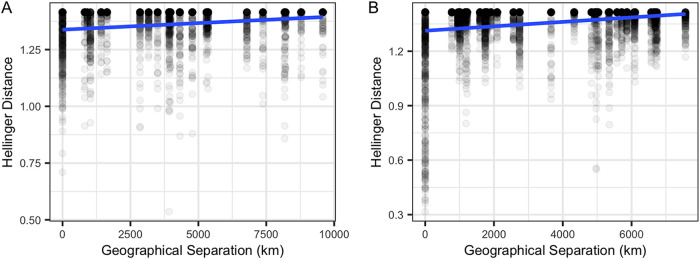
Mantel tests indicated a small, but significant positive relationship between both metrics of community structure (Bray-Curtis, Hellinger) and geographic distance for leaves across the Pacific Ocean for the ITS2 region and 18S rRNA gene amplicon data sets (P < 0.001) (Fig. 3A; Fig. S2A and Table S7). This relationship was also detected for leaves across the Atlantic Ocean (P < 0.001) (Fig. 3B; Fig. S2B and Table S7). Mantel correlograms suggest that this pattern is driven by sites with the closest proximity, such that sites closer together have more similar fungal communities than sites further away (Fig. S3). This pattern is sustained for both data sets for the Pacific Ocean but not for the Atlantic Ocean, when community structure comparisons from the same location are removed (P < 0.05) (Table S7). In the Pacific Ocean, roots had a positive relationship with geographic distance for both the ITS2 region and 18S rRNA gene amplicon data sets (P < 0.001) (Fig. S2C and S4A and Table S7), and an even stronger significant positive relationship was observed for roots in the Atlantic Ocean (P < 0.001) (Fig. S2D and S4B and Table S7). However, upon removal of comparisons from the same location, a pattern of distance decay was observed only in the 18S rRNA gene data set for roots in the Atlantic Ocean (P < 0.001) (Table S7), suggesting that the earlier observed pattern in the Pacific was driven by high similarity in community structure between roots at the same site. Sediment in the Pacific Ocean had a weak relationship with distance, with conflicting significance for the ITS2 region (Bray-Curtis, P < 0.001; Hellinger, P = 0.824), but had strong significant correlations for 18S rRNA gene amplicon data sets (P < 0.001) (Fig. S2E and S4C and Table S7). Upon removal of beta diversity comparisons from the same location, no distance decay pattern was observed in the Pacific (P > 0.05) (Table S7). In contrast, in the Atlantic Ocean, sediment had much more robust positive relationships with geographic distance for both data sets (P < 0.001) (Fig. S2F and S4D and Table S7), which generally remained even when comparisons from the same location were excluded (Table S7). Multiple-regression analyses further confirmed all significant patterns of observed distance decay (P < 0.001).
FIG 3.
Mantel tests suggest a distance decay relationship. Scatterplots depict the weak but significant positive distance-decay relationship between leaf fungal community beta diversity (Hellinger distance) using the ITS2 region amplicon data and geographical distance (km) between sites from the Pacific Ocean (r = 0.1767, P = 0.0001) (A) and Atlantic Ocean (r = 0.1057, P = 0.0001) (B).
Global core leaf, root, and sediment mycobiomes.
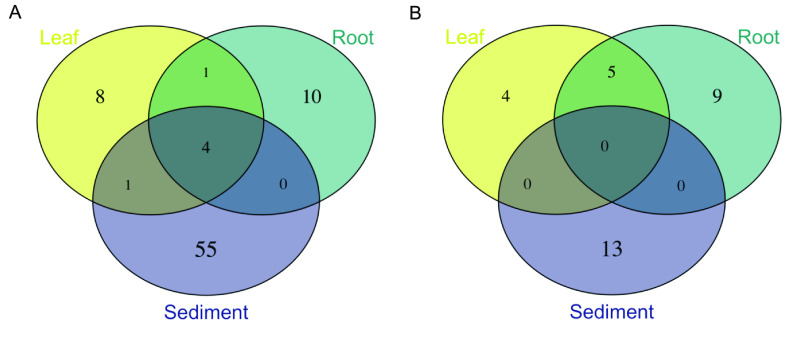
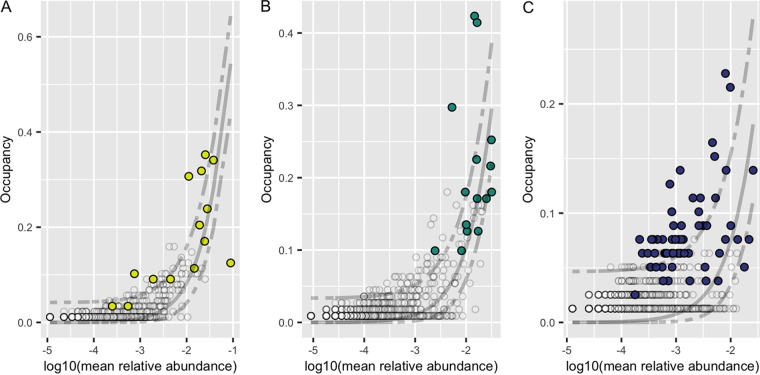
We utilized abundance-occupancy distributions of amplicon sequence variants (ASVs) to infer global Z. marina leaf, root, and rhizosphere sediment core mycobiomes based on ASV rank contributions to beta diversity. A total of 14, 15, and 60 ASVs were predicted as being in the leaf, root, and sediment cores, respectively, based on the ITS2 region amplicon data (Fig. 4A; Table S8). Four ASVs overlapped across all three cores; this included generalist fungi with widespread distributions such as Cladosporium sp. and Malassezia restricta (37, 51). Interestingly, only one ASV was shared between leaf and root cores, Saccharomyces paradoxus (ITS_SV260). The leaf core was dominated by unclassified Capnodiales spp., while the root core was dominated by unclassified Sordariomycetes spp. The sediment core was more diverse but was composed mostly of Ascomycota, particularly members in the Pleosporales and Agaricales.
FIG 4.
Overlap between predicted core mycobiomes of individual Z. marina tissues. Venn diagrams representing shared core ASVs as defined by abundance-occupancy distributions for each sample type (leaf, root, sediment) for ITS2 region amplicon data (A) and 18S rRNA gene amplicon data (B). For comparison, overlap of the entire mycobiome of individual Z. marina tissues is shown in Fig. S5 in the supplemental material.
Smaller core mycobiomes were predicted from the 18S rRNA gene amplicon data with only 9, 14, and 13 ASVs placed in the leaf, root, and sediment cores, and no ASVs overlapped between the three cores (Fig. 4B; Table S9). Generally unclassified Chytridiomycota lineages (including Lobulomycetaceae spp.), Sordariomycetes spp., and Saccharomycetales sp. dominated these core communities.
Neutral models to predict ASV selection.
We applied Sloan neutral models to investigate if ASVs are selected for by Z. marina, assembled through stochastic or deterministic processes (43, 44). ASVs that fall above the neutral model prediction appear in higher occupancy than would be predicted based on their relative abundance and are thus thought to be selected for by the plant environment. ASVs that fall below the neutral model prediction have higher relative abundance than would be predicted based on their occupancy and are thus thought to be either selected against by the plant host or dispersal limited. For the ITS2 region abundance-occupancy distributions, 2.9%, 4.8%, and 3.7% of all ASVs fell above/below the neutral model prediction for leaves, roots, and sediment, respectively (Fig. 5), while for the 18S rRNA gene abundance-occupancy distributions, 7.5%, 6.4%, and 2.2% of all ASVs deviated from the neutral model prediction for leaves, roots, and sediment, respectively (Fig. S6). Further, looking at deviations from the neutral model for ASVs predicted to be in the core mycobiome allows insight into the role of Z. marina in core assembly. We found that of the core leaf, core root, and core sediment ASVs, several were predicted to be plant selected (nleaf = 6, nroot = 7, nsediment = 40), only a few were selected against or dispersal limited (nleaf = 1, nroot = 3, nsediment = 4), and most were neutrally selected (nleaf = 16, nroot = 19, nsediment = 29).
FIG 5.
Abundance-occupancy distributions reveal core mycobiomes. Abundance-occupancy distributions were used to define core members of the leaf (A), root (B), and sediment (C) mycobiomes for the ITS2 region amplicon data. Each point represents an ASV, with predicted core members indicated by a color (leaf = yellow, root = green, sediment = blue) and noncore ASVs in white. Ranked ASVs were predicted to be in the core based on a final percent increase equal to or greater than 10%. A solid line represents the fit of the neutral model, and dashed lines represent 95% confidence around the model prediction. ASVs above the neutral model are predicted to be selected for by the environment (e.g., by the host plant, Z. marina), and those below the model are predicted to be selected against or dispersal limited.
Considering the fit of the neutral model.
Generally, the neutral model had a poor fit for both the ITS2 region (leaf, R2 = 0.31; root, R2 = 0.44; sediment, R2 = −0.76) and 18S rRNA gene (leaf, R2 = 0.49; root, R2 = 0.50; sediment, R2 = 0.08) data sets, with the sediment curves having the worst fit to the neutral model. This could potentially be attributed to the low predicted migration rates (m) for both the ITS2 region (leaf, m = 0.001; root, m = 0.002; sediment, m = 0.001) and 18S rRNA gene (leaf, m = 0.002; root, m = 0.014; sediment, m = 0.014) data sets. These values are consistent with other studies of fungi that used neutral models on abundance-occupancy curves (52) and may be reflective of dispersal limitation playing a stronger role in fungal assembly than bacterial community assembly (53–55).
Global distribution of differentially abundant ASVs.
To investigate variation in fungal community composition at greater taxonomic resolution, we used DESeq2 to identify ASVs whose abundance differed across sample types (Fig. S7 and S8). The greatest number of differentially abundant ASVs was observed between the roots and sediment, with 14 ITS2 region ASVs and four 18S rRNA gene ASVs (Wald test, P < 0.01). This was closely followed by differentially abundant ASVs between leaves and sediment, with 12 ITS2 region ASVs and two 18S rRNA gene ASVs (P < 0.05). The smallest number of differentially abundant ASVs was found between leaves and roots, with three ITS2 region ASVs (P < 0.01). We compared the differentially abundant ASVs to those predicted to be in the leaf, root, and sediment core mycobiomes. We found 14 ASVs that were both differentially abundant between sample types and present in at least one core mycobiome; of those 14, seven were also found to deviate from the neutral model (Table 2). We then examined the global distribution of the 14 ASVs that were both differentially abundant across sample types and predicted to be in the Z. marina core mycobiome. For example, ITS_SV260 (Saccharomyces paradoxus) appears to be globally distributed, neutrally selected, and more abundant on leaves and roots than sediment (P < 0.001) (Fig. S9). In contrast, ITS_SV362 (Lobulomyces sp.) appears to be found only at one site and dispersal limited and is more abundant on leaves than sediment (P < 0.001) (Fig. S10).
TABLE 2.
Predicted differentially abundant core ASVsa
| ASV | Core prediction | Neutral model deviation(s) | Significant DESeq2 comparisons | Taxonomy |
|---|---|---|---|---|
| ITS_SV52 | Leaf, root, sediment | Above, above, none | Root > sediment | Mycosphaerella tassiana |
| ITS_SV60 | Root | Below | Leaf > sediment; root > sediment | Unclassified Sordariomycetes sp. |
| ITS_SV125 | Root | None | Root > sediment | Unclassified Ascomycota sp. |
| ITS_SV234 | Root | None | Leaf > sediment; root > sediment | Unclassified Sordariomycetes sp. |
| ITS_SV260 | Leaf, root | None, none | Leaf > sediment; root > sediment | Saccharomyces paradoxus |
| ITS_SV362 | Leaf | Below | Leaf > sediment; root > sediment | Lobulomyces sp. |
| ITS_SV426 | Leaf, sediment | None, none | Leaf > sediment; root > sediment | Saccharomyces sp. |
| ITS_SV497 | Root | Below | Leaf > sediment; root > sediment | Unclassified Sordariomycetes sp. |
| ITS_SV679 | Sediment | Above | Root > leaf; root > leaf | Pseudeurotium bakeri |
| ITS_SV1045 | Root | Above | Leaf > sediment; root > sediment | Hortaea werneckii |
| 18S_SV756 | Leaf, root | None, none | Root > sediment | Unclassified Chytridiomycetes sp. |
| 18S_SV928 | Leaf, root | Above, above | Leaf > sediment; root > sediment | Saccharomyces sp. |
| 18S_SV968 | Leaf | None | Leaf > sediment; root > sediment | Unclassified Lobulomycetaceae sp. |
| 18S_SV1977 | Root | None | Root > sediment | Chytridium sp. |
ASVs were ranked by abundance-occupancy distributions and then predicted to be in a core based on a final percent increase in beta diversity equal to or greater than 10%. The Sloan neutral model was then applied to the abundance-occupancy distributions to identify ASVs that deviate such that ASVs above the neutral model are predicted to be selected for by the environment (e.g., by the host plant, Z. marina) and those below the model are predicted to be selected against or dispersal limited. Finally, DESeq2 was used to identify ASVs that were differentially abundant between pairwise sample types (leaf, root, sediment). Here, for each predicted core ASV that was also differentially abundant for at least one pairwise comparison, we report the ASV, the core of which it was predicted to be a member (leaf, root or sediment), whether the ASV deviated from the neutral model (above, below, or none), the significant pairwise differential abundance comparisons (e.g., root > sediment means that the ASV was in significantly higher abundance when associated with roots than with sediment), and the taxonomy of the ASV.
Fungi are only a small portion of the Z. marina-associated eukaryotic community.
Fungal sequences made up only a tiny portion of the entire epiphytic eukaryotic community associated with Z. marina, with a mean relative abundance on leaves of 0.50% ± 2.1%, on roots of 0.12% ± 0.4%, and on sediment of 0.23% ± 0.7% in the 18S rRNA gene data set (Fig. S11). The leaf eukaryotic community was generally dominated by diatoms, the root community by both diatoms and Peronosporomycetes (i.e., oomycetes), and the sediment community by both diatoms and dinoflagellates.
Many ASVs have no predicted functional guild.
Although FUNGuild was able to predict the functional guild and trophic mode of 78.6% of ASVs in the ITS2 region amplicon data set, only 10.1% of ASVs had predictions at a confidence of “highly probable.” The most abundant functional guilds assigned at this confidence level included wood saprotroph, ectomycorrhizal, lichenized, endophyte, plant pathogen-wood saprotroph, and fungal parasite (Fig. S12). Comparatively, FUNGuild was able to predict functions for only 35.3% of the ASVs in the 18S rRNA gene data set and only 3.4% of those ASVs had “highly probable” predictions. Generally, the most abundant functional guilds at this confidence level were consistent with those in the ITS2 region data set and included wood saprotroph, ectomycorrhizal, and plant pathogen (Fig. S13). When we further investigated the predicted trophic modes of only the most abundant ASVs (mean relative abundance greater than 0.1%) in both the ITS2 region amplicon and 18S rRNA amplicon data sets, 38.4% and 63.5% of these ASVs, respectively, were unable to be assigned a function, a testament to how little we know about the functional roles of fungi in this system.
DISCUSSION
Overview.
This study is the first to characterize the Zostera marina mycobiome across its full biogeographic distribution using culture-independent methods. We found that the fungal community was different between sites globally and observed a small but significant pattern of distance decay for the Z. marina mycobiome in the Pacific Ocean. We defined a small core mycobiome for leaves, roots, and sediment dominated by Sordariomycetes spp., Chytridiomycota lineages (including Lobulomycetaceae spp.), and Capnodiales spp. Finally, we found the assembly of the mycobiome of Z. marina to be dominated by stochastic processes, although we also observed taxa, including predicted members of the core mycobiome, whose assembly was predicted to be affected by deterministic factors (e.g., environmental filtering, host genetics, dispersal limitation).
Fungal diversity and community structure are generally consistent with previous work.
We observed significant differences in alpha diversity across seagrass tissues, which is consistent with previous seagrass work (38, 40, 42). We also observed differences in alpha diversity across sites, which is consistent with some previous seagrass work (38, 40), while other work found no differences in alpha diversity between sites (39). Additionally, we also observed differences in fungal community structure across tissues and sites. Differences in fungal beta diversity across sites and tissues have been reported previously for seagrasses (26, 38–42). Seasonal differences in fungal colonization of seagrasses have been observed and are likely contributing to the variation observed here between sites, as we were not able to collect from all sites during the same season (56). The global planktonic marine fungal community has been found to cluster by ocean (15), and differences between oceans were observed here as well, suggesting that fungal dispersal is occurring within ocean basins. However, site-to-site variation was a stronger factor than ocean basin in explaining community structure, suggesting that local environmental or host plant filtering may play a critical role in assembling the fungal community associated with Z. marina.
Possible factors driving the distance decay pattern within ocean basins.
It has long been thought that there are few barriers to fungal dispersal (57–60). However, not every fungus is everywhere (61), and there is increasing evidence for rampant environmental filtering and dispersal barriers for host-associated fungi in the marine ecosystem (40, 41, 62). The importance of biogeography for seagrass-associated fungal community structure can be seen in the observation of a small but significant positive distance decay relationship between geographic distance and leaf community structure in the Pacific Ocean and sediment in the Atlantic Ocean. These relationships suggest that sites closer together have more similar fungal communities than sites that are more distant from each other. Previously, similar distance decay relationships were found present in other seagrass-associated fungal communities (40, 41). The observed distance decay relationship is likely driven by a combination of factors, including dispersal limitation, environmental filtering caused by local habitat differences, and priority effects. One limitation of this study is that local environmental conditions (e.g., temperature, salinity, dissolved oxygen) and biogeochemistry (e.g., sediment grain sizes, total organic carbon, carbon/nitrogen ratio) were not measured. Future research should incorporate environmental data to help identify the most important factors driving these patterns.
In addition to local environmental conditions, another factor that might lead to site-specific fungal community composition is host plant genetics. Host plant genotype has been found in other studies to strongly correlate with leaf fungal communities (63–65). The natural dispersal distance of Z. marina is thought to be less than 150 km, and there is some evidence of poor connectivity between locations and rampant inbreeding within locations (66–69). Given the strong population structure and weak dispersal of Z. marina, variation in Z. marina genotypes might be playing a role in structuring the fungal community differences observed here. However, it should be noted that Wainwright et al. failed to find a correlation between S. isoetifolium genetics and fungal community composition in their study (41). Regardless, there is growing evidence that seagrass-associated fungal communities are more similar at closer distances, and future work should look for correlations between environmental factors, Z. marina genetics, Z. marina dispersal, and the fungal community.
Global mycobiome highlights importance of Chytridiomycota lineages.
The global mycobiome of Z. marina was generally composed of taxa previously observed to associate with Z. marina and other seagrass species by using culture-independent methods (see Fig. S14 in the supplemental material) (37–42). This diversity is also in line with cultivation efforts (37, 70, 71). All together, these results are consistent with the seagrass mycobiome being composed of many ASVs, including many Chytridiomycota lineages, for which a specific taxonomic assignment cannot be made based on current data sets that are biased toward terrestrial fungi (39, 40, 42). Likely contributing to this bias, only a few lineages of marine Chytridiomycota have been described using culture-based methods (10, 11), despite their dominance in DNA-based surveys of the marine environment (15, 42, 72). The inability to taxonomically classify fungal sequences is a persistent problem for studies of the marine environment generally and again serves to highlight the need for additional descriptive studies of marine fungi (15, 72–75).
Small core mycobiomes are consistent with work in other seagrass species.
We were able to identify a small “common” core community associated with Z. marina tissues, with only a few ASVs unique to or shared between leaves and roots (Fig. 4). Previously, Trevathan-Tackett et al. (39) were able to identify a small core of eight fungal operational taxonomic units (OTUs) associated with the leaves of the close relative Zostera muelleri, in contrast to Hurtado-McCormick et al. (38), who were unable to identify a core fungal community on Z. muelleri leaves. The Z. marina core leaf and root mycobiomes were dominated by Sordariomycetes spp., Chytridiomycota lineages (including Lobulomycetaceae spp., which have previously been seen to dominate on this species [42]), and Saccharomyces spp. (Table 2; Tables S8 and S9). Sordariomycetes were also found to dominate the core leaf mycobiome in the study by Trevathan-Tackett et al. (39). Only four ASVs overlapped between the core communities for leaves, root, and sediment, and these ASVs were largely assigned to known ubiquitous marine generalists (e.g., Cladosporium [37] and Malassezia [51]).
Considering model fit: are we underestimating the importance of deterministic processes?
The expected shape of a microbial abundance-occupancy distribution is an “S,” with abundant taxa having the highest occupancies and rare taxa having the lowest occupancies (50). However, the abundance-occupancy distributions for the data here do not have this shape. One possible reason for this deviation is an increased incidence of high-abundance but low-occupancy taxa (i.e., “clumping”) (76), which can be suggestive of niche selection (77). Clumping is thought to be affected by spatial variation in habitat quality, localized reproduction, and stochastic immigration-extinction processes (76). Clumping may also be the result of a competitive lottery-based assembly of the mycobiome (i.e., inhibitory priority effects), which means the first species to arrive will take over the entire niche, excluding other group members (78). In addition to not having the expected abundance-occupancy shape, the data had poor fit to the Sloan neutral model, although the fit was generally consistent with other studies of fungi (52) and also of bacteria (44). Thus, the poor fit of the neutral model may indicate that deterministic factors such as competition for niche space, extreme dispersal limitation, and variation in habitat quality may be playing a larger role than expected in assembly dynamics of seagrass-associated fungi.
Saccharomyces sp. is an example of a globally distributed leaf epiphyte.
Both ITS_SV260 (Saccharomyces paradoxus) and 18S_SV928 (Saccharomyces sp.) were globally distributed and more abundant on leaves and roots (P < 0.001) (Fig. S9 and S15). ITS_SV260 was predicted to be neutrally selected, while 18S_SV928 was predicted to be plant selected. S. paradoxus is a wild yeast, the sister species to Saccharomyces cerevisiae, and has been previously observed as a plant endophyte (79, 80). Given the relative abundance of Saccharomyces in both the ITS and 18S rRNA gene data sets on Z. marina leaves and roots, the global distribution of this taxon, and its deviation from the neutral model in the 18S rRNA gene data set (e.g., 18S_SV928), Saccharomyces sp. seems like a good candidate for future work in this system.
Lobulomyces sp. is an example of a dispersal-limited leaf epiphyte.
In comparison, ITS_SV362 (Lobulomyces sp.) was found only at one site, dispersal limited, and is more abundant on leaves (P < 0.001) (Fig. S10). Lobulomycetales have previously been observed in high abundance on and inside Z. marina leaves (42). Members of the Lobulomycetales and marine fungi more generally have been observed to have seasonal dynamics, which may relate to host dynamics and environmental conditions (81–84). Additionally, marine chytrids are known to parasitize seasonal blooms of diatoms (83, 85), and diatoms are the dominant eukaryotes observed on seagrass leaf tissues here (Fig. S11). Future studies should attempt to confirm whether these and other chytrids assigned to the core mycobiome of Z. marina are parasitizing closely associated diatoms or are associated with seagrass leaf tissues directly.
Colletotrichum sp. appear to be endemic to only a few locations.
Previously, a Colletotrichum sp. ASV was found to be an abundant endophyte on and in leaves (42) and was later postulated to be a Z. marina specialist (37). In this study, no ASVs taxonomically assigned as Colletotrichum sp. were defined as part of the global core microbiome. However, one Colletotrichum sp. ASV, ITS_SV219, was found to deviate from the neutral model and was predicted to be dispersal limited. Its global distribution supports a pattern of endemism to only a few locations (Fig. S16). Local adaptation of marine fungi is consistent with patterns of endemism seen in terrestrial fungal studies (86, 87), and Colletotrichum sp. has been seen before as an endemic endophyte in Arabidopsis thaliana (88). One limitation of “core” community analyses generally is that it often underplays the importance of rare microbes which can also be essential for host function (89). Thus, future work should include studies of the functional importance of Colletotrichum sp. and other rare members of the Z. marina mycobiome.
Fungi are not the dominant members of the Z. marina-associated eukaryotic community.
Fungi represented a mean relative abundance of less than 1% in the 18S rRNA amplicon data set. This is generally consistent with the proportion of fungi in other marine eukaryotic studies (e.g., 1.3% fungal sequences in a study by Hassett et al. [15]). Instead, the Z. marina-associated eukaryotic community was generally dominated by diatoms, oomycetes, and dinoflagellates. Diatom dominance was previously observed in a culture-independent study of Z. marina which found that the bacterial and eukaryotic epibiont communities are highly correlated (26). Additionally, oomycetes have been previously cultured in association with Z. marina and are thought to function as opportunistic pathogens or saprotrophs in this system (37, 90, 91).
Functional predictions are consistent with plant-associated lifestyle.
Finally, we used FUNGuild to gain insight into possible functional roles of the mycobiome and found that the seagrass mycobiome is composed of a community of wood saprotrophs, ectomycorrhizal fungi, endophytic fungi, and plant pathogens. This functional distribution fits with what might be expected for a plant-associated community, as well as with what is known of the functional guilds of close relatives of fungal isolates previously isolated from Z. marina (37). However, many dominant members of the fungal community associated with Z. marina were not able to be assigned a functional guild, leaving much functional uncertainty still to be explored in this system. Additional studies characterizing seagrass-associated fungi are needed to understand the taxonomic diversity and functional roles of these fungi in the marine ecosystem generally and in particular when associated with seagrasses.
MATERIALS AND METHODS
Sample collection.
Samples were collected from 16 different globally distributed sites by researchers in the Zostera Experimental Network (ZEN) (Table 1) (92). Samples were collected subtidally at ∼1-m depth using a modified version of the collection protocol previously described by Fahimipour et al. (25). At each of the 16 sites, leaves and roots from individual Z. marina plants and adjacent sediment were collected for 12 individuals, resulting in a total of 576 samples (nleaf = 192, nroot = 192, nsediment = 192).
To obtain Z. marina leaf and root tissues for analysis here, researchers were instructed to (i) gently remove individual Z. marina plants from the sediment, (ii) briefly swish the individual in nearby seawater to remove loosely associated sediment from the roots, (iii) collect ∼5 roots and fully submerge them in a prelabeled 2-ml microcentrifuge tube filled with DNA/RNA Shield (Zymo Research, Inc., Irvine, CA, USA), and (iv) collect a 2-cm section of healthy green leaf tissue and fully submerge it in a prelabeled 2-ml microcentrifuge tube filled with DNA/RNA Shield. A sample of sediment was taken adjacent to each Z. marina individual from 1 cm under the sediment surface using a 6-ml (i.e., 6-cc) syringe. Briefly this was performed by (i) removing the plunger from the syringe, (ii) inserting the barrel of the syringe into the sediment, (iii) inserting the syringe plunger to form an airtight seal, (iv) removing the syringe from the sediment, (v) extruding the sediment until the base of the syringe plunger is at the 3-ml mark, and (vi) using an alcohol-sterilized plastic spatula to transfer ∼0.25 g of sediment into a prelabeled 2-ml microcentrifuge tube filled with DNA/RNA Shield. Samples were preserved in DNA/RNA Shield as it stabilizes DNA/RNA at room temperature. All samples were processed in the field immediately or within 5 h of collection. Samples subsequently were kept at room temperature and mailed to the University of California, Davis, within 2 weeks of sample collection.
Molecular methods.
Samples were shipped from UC Davis to Zymo Research, Inc., for DNA extraction. Samples were transferred to 96-well plate format, with plates including both positive (ZymoBIOMICS microbial community standard) and negative (no-input) controls. DNA was extracted from samples using the ZymoBIOMICS DNA miniprep kit in accordance with the manufacturer’s protocol, with minor modifications as follows. Prior to DNA extraction, samples were heated at 65°C for 5 min to resuspend any white precipitate that had accumulated. Sediment samples were vortexed for 30 s to ensure homogenization and then, by use of a flame-sterilized spatula, transferred into ZR BashingBead lysis tubes until the tubes were two-thirds full. Leaf and root samples were vortexed for 30 s to dissociate any epiphytes, and then all the liquid was transferred into ZR BashingBead lysis tubes. For step 1, ZymoBIOMICS lysis solution was then added to samples such that the final volume was ∼1 ml. For step 2, samples were then subjected to a bead beater on the “homogenize” setting speed for 5 min. For step 4, 600 μl of supernatant was transferred to the filter tube. For step 11, only 50 μl of DNase/RNase free water was used for DNA elution. DNA concentrations for controls and a subset of samples per plate were first quantified with a Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA), and subsequently all samples were quantified using Quant-iT PicoGreen (Thermo Fisher Scientific, Waltham, MA, USA). DNA was then shipped directly to the U.S. Department of Energy Joint Genome Institute (JGI) for amplicon sequencing.
Sequence generation.
The ribosomal internal transcribed spacer 2 (ITS2) region was amplified via PCR using the fungus-specific ITS9F and ITS4R primer set (93, 94), and the 18S rRNA gene was amplified via PCR using the eukaryote-specific 565F and 948R primer set (95). The ITS region is the accepted universal fungal barcoding gene (96) and is made up of two subregions, ITS1 and ITS2. Historically, the ITS1 region has been used to survey fungal communities using short-read sequencing, but it has been shown to have greater length heterogeneity and result in lower phylogenetic richness than the ITS2 region (97–99). Additionally, it should be noted that ITS region primer choice can drastically affect the resulting community analyses (100, 101). The primer sets used here were selected and benchmarked by the JGI (102). Amplicon libraries were prepared according to the JGI’s iTag library construction standard operating protocol (SOP) v.1.0 (103). We briefly summarize the protocol here. Three replicate PCRs for each sample were performed in 96-well plate format with the following conditions: 94°C for 3 min, 35 cycles at 94°C for 25 s, 50°C for 60 s, 72°C for 90 s, and a final extension at 72°C for 10 min. After amplification, replicate PCR products were combined, and then samples were pooled based on DNA quantification of combined PCR replicates. Samples were then pooled at up to 184 samples per sequencing run and sequenced on an Illumina MiSeq (Illumina, Inc., San Diego, CA, USA) in 2 × 300 bp run mode. The resulting sequence data were demultiplexed by the JGI and processed through JGI’s quality-control system, which filters out known contaminant reads using the kmer filter in bbduk and also removes adaptor sequences (104). The quality-controlled sequence read files were downloaded and used for downstream analysis. The JGI iTag standard operating procedure does not include the sequencing of negative controls or blanks. For the ITS2 region, the raw data represented an average read depth of 236,610 with a range of 1 to 1,105,543 reads per sample and, for the 18S rRNA gene amplicon, an average read depth of 177,311 with a range of 3 to 1,095,911 reads per sample.
Sequence processing.
Primers were removed using cutadapt (v. 2.1) (105). The resulting fastq files were analyzed in R (v. 4.0.2) using DADA2 (v. 1.12.1), phyloseq (v. 1.32.0), vegan (v. 2.5-6), microbiome (v. 1.10.0), ecodist (v. 2.0.5), EcoUtils (v. 0.1), DESeq2 (v. 1.28.1), ggplot2 (v. 3.3.2), tidyverse (v. 1.3.0), and many other R packages (106–144). For a detailed walkthrough of the following analysis using R, see the R-markdown summary file (145).
Prior to denoising in DADA2, reads were truncated at the first quality score of 2, and reads with an expected error greater than 2 were removed. Reads were then denoised and merged to generate tables of amplicon sequence variants (ASVs) using DADA2. Prior to downstream analyses, chimeric sequences were identified and removed from tables using removeBimeraDenovo (12.6% of sequences for ITS2 region, 4.5% of sequences for 18S rRNA gene). After chimera removal, samples had an average read depth of 120,270 with a range of 0 to 585,611 reads for the ITS2 region and an average read depth of 109,718 with a range of 0 to 616,884 reads for the 18S rRNA gene amplicon data. Taxonomy was inferred using the RDP Naive Bayesian Classifier algorithm with a modified UNITE (v. 8.2 “all eukaryotes”) database for ITS2 region sequences and the SILVA (v. 138) database for 18S rRNA gene sequences, resulting in 89,754 and 53,084 ASVs, respectively (146–149). The UNITE database was modified to include a representative ITS2 region amplicon sequence for the host plant, Z. marina (GenBank accession no. KM051458.1), as was done previously by Ettinger and Eisen (42). ASVs were then each assigned a unique name by giving each a number preceded by “ITS” or “18S” and then “SV,” which stands for sequence variant (e.g., ITS_SV1, ITS_SV2, etc., and 18S_SV1, 18S_SV2, etc.).
Based on the results of Pauvert et al. (150), ITS-x was not run on the ITS2 region ASVs. However, we removed all ASVs taxonomically assigned as nonfungal at the domain level (e.g., ASVs assigned to the host plant, Z. marina, or other eukaryotic groups or with no domain-level classification) from the ITS2 region ASV table prior to downstream analysis, resulting in a final table of 5,089 ASVs representing 488 samples (nleaf = 179, nroot = 173, nsediment = 136). A total of 88 samples were dropped from the analysis either because they had no sequences after being processed through DADA2 or no remaining sequences after removal of nonfungal ASVs. The remaining samples had an average read depth of 4,990 with a range of 2 to 82,870 reads.
For the 18S rRNA gene ASV table, we generated two different filtered data sets: (i) a fungus-only data set and (ii) a general eukaryotic data set. For data set i, we removed all nonfungal ASVs from the 18S rRNA gene ASV table prior to downstream analysis of the fungi in this data set, resulting in a table of 1,216 fungal ASVs representing 409 samples (nleaf = 146, nroot = 144, nsediment = 119). A total of 167 samples were dropped from the analysis either because they had no sequences after being processed through DADA2 or because they had no remaining sequences after removal of all ASVs classified as nonfungal. For data set ii, we removed ASVs taxonomically classified as noneukaryotic and also as being from embryophytes (e.g., Z. marina) from the 18S rRNA gene ASV table, resulting in a table of 36,582 eukaryotic ASVs representing 556 samples (nleaf = 187, nroot = 187, nsediment = 182). A total of 20 samples were dropped from the analysis either because they had no sequences after being processed through DADA2 or no remaining sequences after filtering ASVs. The remaining samples had an average read depth of 327 with a range of 2 to 8,470 reads.
Sequence analysis and visualization.
To rarefy microbiome data is an ongoing scientific discussion (151–154), and here we use both alternative normalization techniques and rarefaction. For the majority of the analyses, after investigating library sizes and rarefaction curves and to maintain sufficient replication across all sites (Fig. S17), we utilized raw read counts, proportions, centered log-ratio, or Hellinger transformations on the data as appropriate when performing statistical analyses and generating visualizations. Centered log-ratio and Hellinger transformations were performed using the transform function in the microbiome R package. Centered log-ratio (clr) values are scale invariant such that the same ratio is obtained regardless of differences in read counts and thus were suggested as appropriate transformations for microbiome analysis by Gloor et al. (152). We used rarefaction approaches when calculating abundance-occupancy curves to define core taxa. We used rarefy_even_depth in the phyloseq R package to subsample to 1,000 and 100 reads without replacement, respectively, for the ITS2 region and 18S ASV tables (Fig. S17) by following the code described by Shade and Stopnisek (50). These depths were selected after examining rarefaction curves for both data sets to balance maximizing the number of sequences per sample and allowing for curve saturation where possible while also minimizing the number of samples removed for downstream analyses.
To assess alpha (i.e., within-sample) diversity between sample types (leaf, root, and sediment), the Shannon index of samples was calculated in ASV tables containing raw read counts using the estimate_richness function in the phyloseq R package. Raw read counts were used instead of normalizing the data by rarefying, as this kind of subsampling has been shown to be statistically inappropriate (151). To assess alpha diversity across each of the 16 collection sites (Table 1) and across oceans, we first split the data set into different sample types (leaf, root, and sediment), and then for each sample type, we calculated the Shannon index of samples. Kruskal-Wallis tests with 9,999 permutations were used to test for significant differences in alpha diversity across comparisons (sample type, site, or ocean). For comparisons in which the Kruskal-Wallis test resulted in a rejected null hypothesis (P < 0.05), Bonferroni’s corrected post hoc Dunn tests were performed.
To assess beta (i.e., between-sample) diversity, we calculated several ecological metrics (Bray-Curtis dissimilarity, Aitchinson distance, Hellinger distance) by using the ordinate function in phyloseq and visualized them using principal-coordinate analysis. The Bray-Curtis dissimilarity is a widely used ecological metric in microbial analyses which calculates the compositional dissimilarity between samples (155). The Aitchison distance, which is the Euclidean distance of clr-transformed samples, is thought to be better than the Bray-Curtis dissimilarity because it is more stable to data subsampling and is also a true linear distance (152, 156). The Hellinger distance, which is the Euclidean distance of Hellinger distance-transformed data, is based on differences in the proportions of taxa and is thought to be a more ecologically relevant representation of the composition of taxa between samples than the Bray-Curtis dissimilarity, which is biased toward abundant taxa (157, 158).
To test for significant differences in mean centroids between categories of interest (i.e., sample type, site, ocean) for each ecological metric (Bray-Curtis dissimilarity, Aitchinson distance, Hellinger distance), we performed permutational multivariate analyses of variance (PERMANOVAs) with 9,999 permutations, and to account for multiple comparisons, we adjusted P values using the Bonferroni correction (159). We also tested for significant differences in mean dispersions between different categories of interest by using the betadisper and permutest functions from the vegan package in R with 9,999 permutations. The post hoc Tukey’s honestly significant difference (HSD) test was performed on betadisper results that resulted in a rejected null hypothesis (P < 0.05) to identify which categories had mean dispersions that were significantly different.
To test for correlations between the community distances (Bray-Curtis dissimilarity, Hellinger distance) and geographic distances between samples, we first split the data by ocean and sample type and then calculated the geographical distances between samples using the Haversine formula, which accounts for the spherical nature of Earth using the distm function in the geosphere R package. We then performed Mantel tests using 9,999 permutations and generated Mantel correlograms using the mantel and mantel.correlog functions in the vegan R package. Mantel tests were repeated with exclusion of community distances when the geographic distance was zero to assess if patterns persisted in the absence of data from the same site. To further support Mantel test results, we performed multiple regression on distance matrices (MRM) between community distances and geographic distances using 9,999 permutations via the MRM function in the ecodist R package. The code to perform distance decay analyses was adapted from a report by Wainwright et al. (40).
To visualize global fungal community composition across sample types (leaf, root, and sediment), we transformed raw read counts to proportions and collapsed ASVs into taxonomic orders by using the tax_glom function in phyloseq and then removed orders with a mean proportion of less than 1%.
To examine the contribution of specific ASVs to fungal community composition, we used the DESeq2 R package on the raw read counts to examine the log2 fold change (differential abundance) of ASVs across sample types (leaf, root, sediment) in both data sets. We then visualized the global distribution of ASVs found to have significantly different differential abundances (Bonferroni corrected P < 0.05). To do this, we transformed the raw read counts to proportions and then filtered each data set to include only the single ASV of interest by using prune_taxa in the phyloseq R package.
A core microbial community is usually defined as taxa that occur above an arbitrary detection threshold (e.g., greater than 1% relative abundance) and also above an arbitrary occupancy threshold (e.g., from 30% in a report by Ainsworth et al. [160] to 95% in a study by Huse et al. [161]). In an attempt to define “common” core leaf, root, and sediment mycobiomes (“common” as defined by Risely [162]), we used a more standardized approach by building abundance-occupancy curves and then calculating the rank contribution of specific ASVs to beta diversity (Bray-Curtis) to identify putative core ASVs by using code from Shade and Stopnisek (50). ASVs were predicted to be in the core by using the final percent increase in beta diversity described by Shade and Stopnisek (50) with a final percent increase equal to or greater than 10%. We then fit the Sloan neutral model (43) to the abundance-occupancy curves using the code provided by Burns et al. (44) to predict whether core taxa were selected for by the environment (e.g., by the host plant, Z. marina), dispersal limited, or neutrally selected.
To investigate the general composition of the eukaryotic community and assess what proportion of the eukaryotic community is taxonomically classified as fungal, we first transformed raw read counts from the 18S rRNA gene ASV table filtered to include all eukaryotes to proportions and collapsed ASVs into taxonomic phyla using the tax_glom function in phyloseq. For visualization purposes, we then removed phyla with a mean proportion of less than 0.1%. The average relative abundance of eukaryotic phyla was then calculated for each sample type (leaf, root, sediment).
To investigate possible functional roles of seagrass-associated fungi, FUNGuild (v. 1.1) was run on the taxonomic assignments of ASVs from both the ITS2 region and 18S rRNA gene data sets (163). FUNGuild searches the taxonomic assignments at the genus level against an online Guilds database containing taxonomic keywords and functional metadata (e.g., trophic level, guild, etc.) and FUNGuild assignments are given confidence rankings of “highly probable,” “probable,” or “possible.” To assess ecological guilds of high confidence, we first visualized all annotations that were ranked as “highly probable” in either data set. We then investigated functional guilds that were assigned to only highly abundant ASVs in the data. To assess this, we filtered both the ITS2 region and 18S rRNA gene ASV tables to include only ASVs with a mean abundance of greater than 0.1% and then visualized the data in R.
Data availability.
The JGI quality-controlled sequence reads generated for the ITS2 region and the 18S rRNA gene were deposited in GenBank under BioProject ID PRJNA667465 (Sequence Read Archive [SRA] no. SRR12804623 to SRR12805321) and PRJNA667462 (SRA no. SRR12803303 to SRR12804019), respectively. Sequence reads are also available from the JGI Genome Portal (https://genome.jgi.doe.gov/portal/Popandseaspecies/Popandseaspecies.info.html).
ACKNOWLEDGMENTS
We thank the Zostera Experimental Network for helping with sample collection. We are also grateful to Alana J. Firl for assistance in constructing sampling kits, as well as Mikayla Mager and Shuiquan Tang from Zymo Research, Inc., for organizing the DNA extractions. We are very appreciative to Susannah Tringe (ORCID: 0000-0001-6479-8427) from the U.S. Department of Energy Joint Genome Institute for helping organize the sequencing component of this project, Guillaume Jospin (ORCID: 0000-0002-8746-2632) for his assistance downloading the data files from the JGI web server, and John J. Stachowicz and Jeanine L. Olsen for helpful comments and suggestions on the manuscript.
This work was supported in part by a community sequencing proposal from the U.S. Department of Energy Joint Genome Institute, “Population and evolutionary genomics of Zostera marina and its microbiome, and genome diagnostics of other seagrass species” (proposal no. 503251). The work conducted by the U.S. Department of Energy Joint Genome Institute was further supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. This work was also supported in part by grants from the Gordon and Betty Moore Foundation, “Investigating the co-evolutionary relationships between seagrasses and their microbial symbionts” (GBMF333) and “Divergence of Marine Symbiosis After the Rise of the Isthmus of Panama” (GBMF5603).
Footnotes
Supplemental material is available online only.
Contributor Information
Cassandra L. Ettinger, Email: clettinger@ucdavis.edu.
Alfons J. M. Stams, Wageningen University
REFERENCES
- 1.Peay KG, Kennedy PG, Talbot JM. 2016. Dimensions of biodiversity in the Earth mycobiome. Nat Rev Microbiol 14:434–447. 10.1038/nrmicro.2016.59. [DOI] [PubMed] [Google Scholar]
- 2.Grossart H-P, Van den Wyngaert S, Kagami M, Wurzbacher C, Cunliffe M, Rojas-Jimenez K. 2019. Fungi in aquatic ecosystems. Nat Rev Microbiol 17:339–354. 10.1038/s41579-019-0175-8. [DOI] [PubMed] [Google Scholar]
- 3.Grossart H-P, Wurzbacher C, James TY, Kagami M. 2016. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol 19:28–38. 10.1016/j.funeco.2015.06.004. [DOI] [Google Scholar]
- 4.Kagami M, de Bruin A, Ibelings BW, Van Donk E. 2007. Parasitic chytrids: their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 578:113–129. 10.1007/s10750-006-0438-z. [DOI] [Google Scholar]
- 5.Gutiérrez MH, Pantoja S, Tejos E, Quiñones RA. 2011. The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Mar Biol 158:205–219. 10.1007/s00227-010-1552-z. [DOI] [Google Scholar]
- 6.Raghukumar S. 2017. Fungi in coastal and oceanic marine ecosystems, p 17–38. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 7.Orsi W, Biddle JF, Edgcomb V. 2013. Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One 8:e56335. 10.1371/journal.pone.0056335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amend A, Burgaud G, Cunliffe M, Edgcomb VP, Ettinger CL, Gutiérrez MH, Heitman J, Hom EFY, Ianiri G, Jones AC, Kagami M, Picard KT, Quandt CA, Raghukumar S, Riquelme M, Stajich J, Vargas-Muñiz J, Walker AK, Yarden O, Gladfelter AS. 2019. Fungi in the marine environment: open questions and unsolved problems. mBio 10:e01189-18. 10.1128/mBio.01189-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawksworth DL, Lücking R. 2017. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr 5. 10.1128/microbiolspec.FUNK-0052-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones EBG, Gareth Jones EB, Suetrong S, Sakayaroj J, Bahkali AH, Abdel-Wahab MA, Boekhout T, Pang K-L. 2015. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers 73:1–72. 10.1007/s13225-015-0339-4. [DOI] [Google Scholar]
- 11.Jones EBG, Gareth Jones EB, Pang K-L, Abdel-Wahab MA, Scholz B, Hyde KD, Boekhout T, Ebel R, Rateb ME, Henderson L, Sakayaroj J, Suetrong S, Dayarathne MC, Kumar V, Raghukumar S, Sridhar KR, Bahkali AHA, Gleason FH, Norphanphoun C. 2019. An online resource for marine fungi. Fungal Divers 96:347–433. 10.1007/s13225-019-00426-5. [DOI] [Google Scholar]
- 12.Jones EBG. 2011. Are there more marine fungi to be described? Botanica Marina 54. 10.1515/bot.2011.043. [DOI] [Google Scholar]
- 13.Tisthammer KH, Cobian GM, Amend AS. 2016. Global biogeography of marine fungi is shaped by the environment. Fungal Ecol 19:39–46. 10.1016/j.funeco.2015.09.003. [DOI] [Google Scholar]
- 14.Morales SE, Biswas A, Herndl GJ, Baltar F. 2019. Global structuring of phylogenetic and functional diversity of pelagic fungi by depth and temperature. Front Mar Sci 6:131. 10.3389/fmars.2019.00131. [DOI] [Google Scholar]
- 15.Hassett BT, Vonnahme TR, Peng X, Gareth Jones EB, Heuzé C. 2020. Global diversity and geography of planktonic marine fungi. Botanica Marina 63:121–139. 10.1515/bot-2018-0113. [DOI] [Google Scholar]
- 16.Gao Z, Li B, Zheng C, Wang G. 2008. Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl Environ Microbiol 74:6091–6101. 10.1128/AEM.01315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littman R, Willis BL, Bourne DG. 2011. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ Microbiol Rep 3:651–660. 10.1111/j.1758-2229.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 18.Yarden O. 2014. Fungal association with sessile marine invertebrates. Front Microbiol 5:228. 10.3389/fmicb.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gnavi G, Garzoli L, Poli A, Prigione V, Burgaud G, Varese GC. 2017. The culturable mycobiota of Flabellia petiolata: first survey of marine fungi associated to a Mediterranean green alga. PLoS One 12:e0175941. 10.1371/journal.pone.0175941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuccaro A, Schoch CL, Spatafora JW, Kohlmeyer J, Draeger S, Mitchell JI. 2008. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl Environ Microbiol 74:931–941. 10.1128/AEM.01158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borovec O, Vohník M. 2018. Ontogenetic transition from specialized root hairs to specific root-fungus symbiosis in the dominant Mediterranean seagrass Posidonia oceanica. Sci Rep 8:10773. 10.1038/s41598-018-28989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemminga MA, Duarte CM. 2000. Seagrass ecology. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 23.Fourqurean JW, Duarte CM, Kennedy H, Marbà N, Holmer M, Mateo MA, Apostolaki ET, Kendrick GA, Krause-Jensen D, McGlathery KJ, Serrano O. 2012. Seagrass ecosystems as a globally significant carbon stock. Nat Geosci 5:505–509. 10.1038/ngeo1477. [DOI] [Google Scholar]
- 24.Orth RJ, Carruthers TJB, Dennison WC, Duarte CM, Fourqurean JW, Heck KL, Randall Hughes A, Kendrick GA, Judson Kenworthy W, Olyarnik S, Short FT, Waycott M, Williams SL. 2006. A global crisis for seagrass ecosystems. Bioscience 56:987. 10.1641/0006-3568(2006)56[987:AGCFSE]2.0.CO;2. [DOI] [Google Scholar]
- 25.Fahimipour AK, Kardish MR, Lang JM, Green JL, Eisen JA, Stachowicz JJ. 2017. Global-scale structure of the eelgrass microbiome. Appl Environ Microbiol 83:e03391-16. 10.1128/AEM.03391-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bengtsson MM, Bühler A, Brauer A, Dahlke S, Schubert H, Blindow I. 2017. Eelgrass leaf surface microbiomes are locally variable and highly correlated with epibiotic eukaryotes. Front Microbiol 8:1312. 10.3389/fmicb.2017.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ettinger CL, Voerman SE, Lang JM, Stachowicz JJ, Eisen JA. 2017. Microbial communities in sediment from patches, but not the leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 5:e3246. 10.7717/peerj.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capone DG. 1982. Nitrogen fixation (acetylene reduction) by rhizosphere sediments of the eelgrass Zostera marina. Mar Ecol Prog Ser 10:67–75. 10.3354/meps010067. [DOI] [Google Scholar]
- 29.Sun F, Zhang X, Zhang Q, Liu F, Zhang J, Gong J. 2015. Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Appl Environ Microbiol 81:6901–6914. 10.1128/AEM.01382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cúcio C, Engelen AH, Costa R, Muyzer G. 2016. Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front Microbiol 7:440. 10.3389/fmicb.2016.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ettinger CL, Williams SL, Abbott JM, Stachowicz JJ, Eisen JA. 2017. Microbiome succession during ammonification in eelgrass bed sediments. PeerJ 5:e3674. 10.7717/peerj.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crump BC, Wojahn JM, Tomas F, Mueller RS. 2018. Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front Microbiol 9:388. 10.3389/fmicb.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Tomas F, Mueller RS. 2020. Nutrient enrichment increases size of Zostera marina shoots and enriches for sulfur and nitrogen cycling bacteria in root-associated microbiomes. FEMS Microbiol Ecol 96:fiaa129. 10.1093/femsec/fiaa129. [DOI] [PubMed] [Google Scholar]
- 34.Shoemaker G, Wyllie-Echeverria S. 2013. Occurrence of rhizomal endophytes in three temperate northeast pacific seagrasses. Aquat Botany 111:71–73. 10.1016/j.aquabot.2013.05.010. [DOI] [Google Scholar]
- 35.Kirichuk NN, Pivkin MV. 2015. Filamentous fungi associated with the seagrass Zostera marina Linnaeus, 1753 of Rifovaya Bay (Peter the Great Bay, the Sea of Japan). Russ J Mar Biol 41:351–355. 10.1134/S1063074015050053. [DOI] [Google Scholar]
- 36.Petersen L-E, Marner M, Labes A, Tasdemir D. 2019. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the Baltic seagrass Zostera marina. Mar Drugs 17:419. 10.3390/md17070419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ettinger CL, Eisen JA. 2020. Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PLoS One 15:e0236135. 10.1371/journal.pone.0236135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurtado-McCormick V, Kahlke T, Petrou K, Jeffries T, Ralph PJ, Seymour JR. 2019. Regional and microenvironmental scale characterization of the seagrass microbiome. Front Microbiol 10:1011. 10.3389/fmicb.2019.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trevathan-Tackett SM, Allnutt TR, Sherman CDH, Richardson ME, Crowley TM, Macreadie PI. 2020. Spatial variation of bacterial and fungal communities of estuarine seagrass leaf microbiomes. Aquat Microb Ecol 84:59–74. 10.3354/ame01926. [DOI] [Google Scholar]
- 40.Wainwright BJ, Zahn GL, Zushi J, Lee NLY, Ooi JLS, Lee JN, Huang D. 2019. Seagrass‐associated fungal communities show distance decay of similarity that has implications for seagrass management and restoration. Ecol Evol 9:11288–11297. 10.1002/ece3.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wainwright BJ, Zahn GL, Arlyza IS, Amend AS. 2018. Seagrass-associated fungal communities follow Wallace’s line, but host genotype does not structure fungal community. J Biogeogr 45:762–770. 10.1111/jbi.13168. [DOI] [Google Scholar]
- 42.Ettinger CL, Eisen JA. 2019. Characterization of the mycobiome of the seagrass, reveals putative associations with marine chytrids. Front Microbiol 10:2476. 10.3389/fmicb.2019.02476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan WT, Woodcock S, Lunn M, Head IM, Curtis TP. 2007. Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb Ecol 53:443–455. 10.1007/s00248-006-9141-x. [DOI] [PubMed] [Google Scholar]
- 44.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. 2016. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664. 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosindell J, Hubbell SP, Etienne RS. 2011. The unified neutral theory of biodiversity and biogeography at age ten. Trends Ecol Evol 26:340–348. 10.1016/j.tree.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 46.Chave J. 2004. Neutral theory and community ecology. Ecol Lett 7:241–253. 10.1111/j.1461-0248.2003.00566.x. [DOI] [Google Scholar]
- 47.Schmidt VT, Smith KF, Melvin DW, Amaral-Zettler LA. 2015. Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol Ecol 24:2537–2550. 10.1111/mec.13177. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J, Ning D. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe WH, McPeek MA. 2014. Is dispersal neutral? Trends Ecol Evol 29:444–450. 10.1016/j.tree.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Shade A, Stopnisek N. 2019. Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr Opin Microbiol 49:50–58. 10.1016/j.mib.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Amend A. 2014. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog 10:e1004277. 10.1371/journal.ppat.1004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stopnisek N, Shade A. 2020. Prioritizing persistent microbiome members in the common bean rhizosphere: an integrated analysis of space, time, and plant genotype. bioRxiv 10.1101/727461. [DOI] [PMC free article] [PubMed]
- 53.Gumiere T, Durrer A, Bohannan BJM, Andreote FD. 2016. Biogeographical patterns in fungal communities from soils cultivated with sugarcane. J Biogeogr 43:2016–2026. 10.1111/jbi.12775. [DOI] [Google Scholar]
- 54.Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI, Erlandson S, Vilgalys R, Liao H-L, Smith ME, Peay KG. 2014. Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci U S A 111:6341–6346. 10.1073/pnas.1402584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Villarreal Ruiz L, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson K-H, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L-D, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, et al. 2014. Fungal biogeography. Global diversity and geography of soil fungi. Science 346:1256688. 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 56.Mata JL, Cebrián J. 2013. Fungal endophytes of the seagrasses Halodule wrightii and Thalassia testudinum in the north-central Gulf of Mexico. Botanica Marina 56. 10.1515/bot-2013-0047. [DOI] [Google Scholar]
- 57.Finlay BJ. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063. 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 58.Fenchel T, Finlay BJ. 2004. The ubiquity of small species: patterns of local and global diversity. Bioscience 54:777. 10.1641/0006-3568(2004)054[0777:TUOSSP]2.0.CO;2. [DOI] [Google Scholar]
- 59.Hyde KD, Gareth Jones EB, Leaño E, Pointing SB, Poonyth AD, Vrijmoed LLP. 1998. Role of fungi in marine ecosystems. Biodivers Conserv 7:1147–1161. 10.1023/A:1008823515157. [DOI] [Google Scholar]
- 60.Cox F, Newsham KK, Bol R, Dungait JAJ, Robinson CH. 2016. Not poles apart: Antarctic soil fungal communities show similarities to those of the distant Arctic. Ecol Lett 19:528–536. 10.1111/ele.12587. [DOI] [PubMed] [Google Scholar]
- 61.Peay KG, Bidartondo MI, Arnold AE. 2010. Not every fungus is everywhere: scaling to the biogeography of fungal-plant interactions across roots, shoots and ecosystems. New Phytol 185:878–882. 10.1111/j.1469-8137.2009.03158.x. [DOI] [PubMed] [Google Scholar]
- 62.Wainwright BJ, Bauman AG, Zahn GL, Todd PA, Huang D. 2019. Characterization of fungal biodiversity and communities associated with the reef macroalga Sargassum ilicifolium reveals fungal community differentiation according to geographic locality and algal structure. Mar Biodivers 49:2601–2608. 10.1007/s12526-019-00992-6. [DOI] [Google Scholar]
- 63.Bálint M, Tiffin P, Hallström B, O'Hara RB, Olson MS, Fankhauser JD, Piepenbring M, Schmitt I. 2013. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS One 8:e53987. 10.1371/journal.pone.0053987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hunter PJ, Pink DAC, Bending GD. 2015. Cultivar-level genotype differences influence diversity and composition of lettuce (Lactuca sp.) phyllosphere fungal communities. Fungal Ecol 17:183–186. 10.1016/j.funeco.2015.05.007. [DOI] [Google Scholar]
- 65.Sapkota R, Knorr K, Jørgensen LN, O'Hanlon KA, Nicolaisen M. 2015. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol 207:1134–1144. 10.1111/nph.13418. [DOI] [PubMed] [Google Scholar]
- 66.Olsen JL, Stam WT, Coyer JA, Reusch TBH, Billingham M, Boström C, Calvert E, Christie H, Granger S, la Lumière R, Milchakova N, Oudot-Le Secq M-P, Procaccini G, Sanjabi B, Serrao E, Veldsink J, Widdicombe S, Wyllie-Echeverria S. 2004. North Atlantic phylogeography and large-scale population differentiation of the seagrass Zostera marina L. Mol Ecol 13:1923–1941. 10.1111/j.1365-294X.2004.02205.x. [DOI] [PubMed] [Google Scholar]
- 67.Muñiz-Salazar R, Talbot SL, Sage GK, Ward DH, Cabello-Pasini A. 2005. Population genetic structure of annual and perennial populations of Zostera marina L. along the Pacific coast of Baja California and the Gulf of California. Mol Ecol 14:711–722. 10.1111/j.1365-294X.2005.02454.x. [DOI] [PubMed] [Google Scholar]
- 68.Ort BS, Cohen CS, Boyer KE, Wyllie-Echeverria S. 2012. Population structure and genetic diversity among eelgrass (Zostera marina) beds and depths in San Francisco Bay. J Hered 103:533–546. 10.1093/jhered/ess022. [DOI] [PubMed] [Google Scholar]
- 69.Campanella JJ, Bologna PA, Smalley JV, Rosenzweig EB, Smith SM. 2010. Population structure of Zostera marina (eelgrass) on the western Atlantic coast is characterized by poor connectivity and inbreeding. J Hered 101:61–70. 10.1093/jhered/esp103. [DOI] [PubMed] [Google Scholar]
- 70.Sakayaroj J, Preedanon S, Supaphon O, Jones EBG, Phongpaichit S. 2010. Phylogenetic diversity of endophyte assemblages associated with the tropical seagrass Enhalus acoroides in Thailand. Fungal Divers 42:27–45. 10.1007/s13225-009-0013-9. [DOI] [Google Scholar]
- 71.Supaphon P, Phongpaichit S, Sakayaroj J, Rukachaisirikul V, Kobmoo N, Spatafora JW. 2017. Phylogenetic community structure of fungal endophytes in seagrass species. Botanica Marina 60. 10.1515/bot-2016-0089. [DOI] [Google Scholar]
- 72.Hassett BT, Ducluzeau A-LL, Collins RE, Gradinger R. 2017. Spatial distribution of aquatic marine fungi across the western Arctic and sub-arctic. Environ Microbiol 19:475–484. 10.1111/1462-2920.13371. [DOI] [PubMed] [Google Scholar]
- 73.Comeau AM, Vincent WF, Bernier L, Lovejoy C. 2016. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci Rep 6:30120. 10.1038/srep30120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagano Y, Miura T, Nishi S, Lima AO, Nakayama C, Pellizari VH, Fujikura K. 2017. Fungal diversity in deep-sea sediments associated with asphalt seeps at the Sao Paulo Plateau. Deep Sea Res 2 Top Stud Oceanogr 146:59–67. 10.1016/j.dsr2.2017.05.012. [DOI] [Google Scholar]
- 75.Picard KT. 2017. Coastal marine habitats harbor novel early-diverging fungal diversity. Fungal Ecol 25:1–13. 10.1016/j.funeco.2016.10.006. [DOI] [Google Scholar]
- 76.Wright DH. 1991. Correlations between incidence and abundance are expected by chance. J Biogeogr 18:463. 10.2307/2845487. [DOI] [Google Scholar]
- 77.Morella NM, Weng FC-H, Joubert PM, Metcalf CJE, Lindow S, Koskella B. 2020. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc Natl Acad Sci U S A 117:1148–1159. 10.1073/pnas.1908600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verster AJ, Borenstein E. 2018. Competitive lottery-based assembly of selected clades in the human gut microbiome. Microbiome 6:186. 10.1186/s40168-018-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ricks KD, Koide RT. 2019. The role of inoculum dispersal and plant species identity in the assembly of leaf endophytic fungal communities. PLoS One 14:e0219832. 10.1371/journal.pone.0219832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glushakova AM, Ivannikova IV, Naumova ES, Chernov II, Naumov GI. 2007. Massive isolation and identification of Saccharomyces paradoxus yeasts from plant phyllosphere. Mikrobiologiia 76:236–242. [PubMed] [Google Scholar]
- 81.Longcore JE. 1992. Morphology and zoospore ultrastructure of Chytriomyces angularis sp. nov. (Chytridiales). Mycologia 84:442–451. 10.2307/3760197. [DOI] [Google Scholar]
- 82.Seto K, Degawa Y. 2015. Cyclopsomyces plurioperculatus: a new genus and species of Lobulomycetales (Chytridiomycota, Chytridiomycetes) from Japan. Mycologia 107:633–640. 10.3852/14-284. [DOI] [PubMed] [Google Scholar]
- 83.Hassett BT, Gradinger R. 2016. Chytrids dominate arctic marine fungal communities. Environ Microbiol 18:2001–2009. 10.1111/1462-2920.13216. [DOI] [PubMed] [Google Scholar]
- 84.Rojas-Jimenez K, Rieck A, Wurzbacher C, Jürgens K, Labrenz M, Grossart H-P. 2019. A salinity threshold separating fungal communities in the Baltic Sea. Front Microbiol 10:680. 10.3389/fmicb.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor JD, Cunliffe M. 2016. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J 10:2118–2128. 10.1038/ismej.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meiser A, Bálint M, Schmitt I. 2014. Meta-analysis of deep-sequenced fungal communities indicates limited taxon sharing between studies and the presence of biogeographic patterns. New Phytol 201:623–635. 10.1111/nph.12532. [DOI] [PubMed] [Google Scholar]
- 87.Grantham NS, Reich BJ, Pacifici K, Laber EB, Menninger HL, Henley JB, Barberán A, Leff JW, Fierer N, Dunn RR. 2015. Fungi identify the geographic origin of dust samples. PLoS One 10:e0122605. 10.1371/journal.pone.0122605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O'Connell RJ, Schulze-Lefert P. 2016. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165:464–474. 10.1016/j.cell.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Küsel K, Rillig MC, Rivett DW, Salles JF, van der Heijden MGA, Youssef NH, Zhang X, Wei Z, Hol WHG. 2017. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J 11:853–862. 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Man In ’t Veld WA, Man In ’t W, Karin CH, van Rijswick PCJ, Meffert JP, Boer E, Westenberg M, van der Heide T, Govers LL. 2019. Multiple Halophytophthora spp. and Phytophthora spp. including P gemini, P inundata and P chesapeakensis sp. nov. isolated from the seagrass Zostera marina in the Northern hemisphere. Eur J Plant Pathol 153:341–357. 10.1007/s10658-018-1561-1. [DOI] [Google Scholar]
- 91.Man In ’t Veld WA, Man In ’t W, Karin CH, Brouwer H, de Cock AWAM. 2011. Phytophthora gemini sp. nov., a new species isolated from the halophilic plant Zostera marina in the Netherlands. Fungal Biol 115:724–732. 10.1016/j.funbio.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 92.Duffy JE, Reynolds PL, Boström C, Coyer JA, Cusson M, Donadi S, Douglass JG, Eklöf JS, Engelen AH, Eriksson BK, Fredriksen S, Gamfeldt L, Gustafsson C, Hoarau G, Hori M, Hovel K, Iken K, Lefcheck JS, Moksnes P-O, Nakaoka M, O'Connor MI, Olsen JL, Richardson JP, Ruesink JL, Sotka EE, Thormar J, Whalen MA, Stachowicz JJ. 2015. Biodiversity mediates top-down control in eelgrass ecosystems: a global comparative-experimental approach. Ecol Lett 18:696–705. 10.1111/ele.12448. [DOI] [PubMed] [Google Scholar]
- 93.Menkis A, Burokienė D, Gaitnieks T, Uotila A, Johannesson H, Rosling A, Finlay RD, Stenlid J, Vasaitis R. 2012. Occurrence and impact of the root-rot biocontrol agent Phlebiopsis gigantea on soil fungal communities in Picea abies forests of northern Europe. FEMS Microbiol Ecol 81:438–445. 10.1111/j.1574-6941.2012.01366.x. [DOI] [PubMed] [Google Scholar]
- 94.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY. [Google Scholar]
- 95.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner H-W, Richards TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19(Suppl 1):21–31. 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 96.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beeck MOD, Op De Beeck M, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV. 2014. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:e97629. 10.1371/journal.pone.0097629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Liiv I, Kõljalg U, Kisand V, Nilsson H, Hildebrand F, Bork P, Abarenkov K. 2015. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10:1–43. 10.3897/mycokeys.10.4852. [DOI] [Google Scholar]
- 99.Wang X-C, Liu C, Huang L, Bengtsson-Palme J, Chen H, Zhang J-H, Cai D, Li J-Q. 2015. ITS1: a DNA barcode better than ITS2 in eukaryotes? Mol Ecol Resour 15:573–586. 10.1111/1755-0998.12325. [DOI] [PubMed] [Google Scholar]
- 100.De Filippis F, Laiola M, Blaiotta G, Ercolini D. 2017. Different amplicon targets for sequencing-based studies of fungal diversity. Appl Environ Microbiol 83:e00905-17. 10.1128/AEM.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frau A, Kenny JG, Lenzi L, Campbell BJ, Ijaz UZ, Duckworth CA, Burkitt MD, Hall N, Anson J, Darby AC, Probert CSJ. 2019. DNA extraction and amplicon production strategies deeply influence the outcome of gut mycobiome studies. Sci Rep 9:9328. 10.1038/s41598-019-44974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rivers AR. May 2016. iTag amplicon sequencing for taxonomic identification at JGI. https://jgi.doe.gov/wp-content/uploads/2016/06/DOE-JGI-iTagger-methods.pdf.
- 103.Joint Genome Institute. 2017. iTag sample preparation for Illumina sequencing. https://1ofdmq2n8tc36m6i46scovo2e-wpengine.netdna-ssl.com/wp-content/uploads/2019/07/iTag-Sample-Preparation-for-Illumina-Sequencing-SOP-v1.0.pdf.
- 104.Rivers AR. July 2016. iTag amplicon sequencing for taxonomic identification at JGI. https://jgi.doe.gov/wp-content/uploads/2013/05/iTagger-methods.pdf.
- 105.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 106.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie Y. 2014. knitr: a comprehensive tool for reproducible research in R. In Stodden V, Leisch F, Peng RD (ed), Implementing reproducible computational research. Chapman and Hall/CRC, New York, NY. [Google Scholar]
- 108.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 109.Simpson GL. 2019. permute: functions for generating restricted permutations of data. R package version 0.9-5. https://CRAN.R-project.org/package=permute.
- 110.Sarkar D. 2008. Lattice: multivariate data visualization with R. Springer, New York, NY. [Google Scholar]
- 111.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community ecology package. R package.
- 112.Neuwirth E. 2014. RColorBrewer: ColorBrewer palettes. R package.
- 113.Therneau TM. 2020. A package for survival analysis in R. R package.
- 114.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. 2008. Implementing a class of permutation tests: the coin package. J Stat Soft 28:1–23. 10.18637/jss.v028.i08. [DOI] [Google Scholar]
- 115.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. 2006. A Lego system for conditional inference. Am Stat 60:257–263. 10.1198/000313006X118430. [DOI] [Google Scholar]
- 116.Allaire JJ, Xie Y, McPherson J, Luraschi J, Ushey K, Atkins A, Wickham H, Cheng J, Chang W, Iannone R. 2020. rmarkdown: dynamic documents for R. R package.
- 117.Ogle DH, Wheeler P, Dinno A. 2020. FSA: fisheries stock analysis. R package.
- 118.Wickham H. 2007. Reshaping data with the reshape package. J Stat Soft 21:1–20. 10.18637/jss.v021.i12. [DOI] [Google Scholar]
- 119.Baselga A, Orme D, Villeger S, De Bortoli J, Leprieur F. 2018. betapart: partitioning beta diversity into turnover and nestedness components. R package.
- 120.Eddelbuettel D. 2013. Seamless R and C++ integration with Rcpp. 10.1007/978-1-4614-6868-4. [DOI]
- 121.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. 2019. Welcome to the tidyverse. J Open Source Softw 4:1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 123.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan M, Carey V. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R. 2009. ShortRead: a Bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25:2607–2608. 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dray S, Dufour A-B. 2007. The ade4 package: implementing the Duality Diagram for Ecologists. J Stat Soft 22. 10.18637/jss.v022.i04. [DOI] [Google Scholar]
- 126.Hijmans RJ. 2019. geosphere: spherical trigonometry. R package.
- 127.Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Soft 22. 10.18637/jss.v022.i07. [DOI] [Google Scholar]
- 128.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen H. 2018. VennDiagram: generate high-resolution Venn and Euler plots. R package.
- 130.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Becker RA. 2018. maps: draw geographical maps. R package.
- 132.Garnier S. 2018. viridis: default color maps from “matplotlib.” R package.
- 133.Lahti L, Shetty S. 2017. microbiome R package. R package.
- 134.Pedersen TL. 2020. patchwork: the composer of plots. R package.
- 135.Salazar G. 2020. EcolUtils: utilities for community ecology analysis. R package.
- 136.Robinson D, Hayes A. 2020. broom: convert statistical analysis objects into tidy tibbles. R package.
- 137.Bass AJ, Robinson DG, Lianoglou S, Nelson E, Storey JD, from Laurent Gatto WC . 2020. biobroom: turn Bioconductor objects into tidy data frames. R package.
- 138.Wickham H, Seidel D. 2020. scales: scale functions for visualization. R package.
- 139.Yu G. 2020. ggplotify: convert plot to “grob” or “ggplot” object. R package.
- 140.Sprockett D. 2020. reltools: microbiome amplicon analysis and visualization. R package.
- 141.Zeileis A, Croissant Y. 2010. Extended model formulas in R: multiple parts and multiple responses. J Stat Softw 34:1–13. 10.18637/jss.v034.i01. [DOI] [Google Scholar]
- 142.Harrell FE, Jr, et al. 2020. Hmisc: Harrell miscellaneous. R package.
- 143.Elzhov TV, Mullen KM, Spiess A-N, Bolker B. 2016. minpack.lm: R interface to the Levenberg-Marquardt nonlinear least-squares algorithm found in MINPACK, plus support for bounds. R package.
- 144.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ettinger C. 2020. casett/Global_ZM_fungi_amplicons. 10.5281/zenodo.4116550. [DOI]
- 146.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abarenkov K, Zirk A, Piirmann T, Pöhönen R, Ivanov F, Nilsson RH, Kõljalg U. 2020. UNITE general FASTA release for eukaryotes 2. UNITE Community. 10.15156/BIO/786370. [DOI]
- 148.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pauvert C, Buée M, Laval V, Edel-Hermann V, Fauchery L, Gautier A, Lesur I, Vallance J, Vacher C. 2019. Bioinformatics matters: the accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecol 41:23–33. 10.1016/j.funeco.2019.03.005. [DOI] [Google Scholar]
- 151.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Front Microbiol 8:2224. 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cameron ES, Schmidt PJ, Tremblay BJM, Emelko MB. 2020. To rarefy or not to rarefy: Enhancing microbial community analysis through next-generation sequencing. bioRxiv 10.1101/2020.09.09.290049. [DOI]
- 154.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, Knight R. 2017. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monographs 27:325–349. 10.2307/1942268. [DOI] [Google Scholar]
- 156.Aitchison J, Barceló-Vidal C, Martín-Fernández JA, Pawlowsky-Glahn V. 2000. Logratio analysis and compositional distance. Math Geol 32:271–275. 10.1023/A:1007529726302. [DOI] [Google Scholar]
- 157.Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280. 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 158.Rao CR. 1997. An alternative to correspondence analysis using Hellinger distance. 10.21236/ada325255. [DOI]
- 159.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 160.Ainsworth TD, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, Gates RD, Padilla-Gamiño JL, Spalding HL, Smith C, Woolsey ES, Bourne DG, Bongaerts P, Hoegh-Guldberg O, Leggat W. 2015. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9:2261–2274. 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huse SM, Ye Y, Zhou Y, Fodor AA. 2012. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242. 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Risely A. 2020. Applying the core microbiome to understand host-microbe systems. J Anim Ecol 89:1549–1558. 10.1111/1365-2656.13229. [DOI] [PubMed] [Google Scholar]
- 163.Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. 2016. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. 10.1016/j.funeco.2015.06.006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.02795-20-s0001.pdf, PDF file, 1.1 MB (6.9MB, pdf)
Data Availability Statement
The JGI quality-controlled sequence reads generated for the ITS2 region and the 18S rRNA gene were deposited in GenBank under BioProject ID PRJNA667465 (Sequence Read Archive [SRA] no. SRR12804623 to SRR12805321) and PRJNA667462 (SRA no. SRR12803303 to SRR12804019), respectively. Sequence reads are also available from the JGI Genome Portal (https://genome.jgi.doe.gov/portal/Popandseaspecies/Popandseaspecies.info.html).