ABSTRACT
Acyl coenzyme A (CoA) binding protein (ACBP), also called diazepam-binding inhibitor (DBI), is a phylogenetically conserved protein that is expressed by all eukaryotic species as well as by some bacteria. Since elevated ACBP/DBI levels play a major role in the inhibition of autophagy, increase in appetite, and enhanced lipid storage that accompany obesity, we wondered whether ACBP/DBI produced by the human microbiome might affect host weight. We found that the genomes of bacterial commensals rarely contain ACBP/DBI homologues, which are rather encoded by genomes of some pathogenic or environmental taxa that were not prevalent in human feces. Exhaustive bioinformatic analyses of 1,899 gut samples from healthy individuals refuted the hypothesis that bacterial ACBP/DBI might affect the body mass index (BMI) in a physiological context. Thus, the physiological regulation of BMI is unlikely to be affected by microbial ACBP/DBI-like proteins. However, at the speculative level, it remains possible that ACBP/DBI produced by potential pathogenic bacteria might enhance their virulence by inhibiting autophagy and hence subverting innate immune responses.
IMPORTANCE Acyl coenzyme A (CoA) binding protein (ACBP) can be encoded by several organisms across the domains of life, including microbes, and has shown to play major roles in human metabolic processes. However, little is known about its presence in the human gut microbiome and whether its microbial counterpart could also play a role in human metabolism. In the present study, we found that microbial ACBP/DBI sequences were rarely present in the gut microbiome across multiple metagenomic data sets. Microbes that carried ACBP/DBI in the human gut microbiome included Saccharomyces cerevisiae, Lautropia mirabilis, and Comamonas kerstersii, but these microorganisms were not associated with body mass index, further indicating an unconvincing role for microbial ACBP/DBI in human metabolism.
KEYWORDS: ACBP/DBI, human microbiome
INTRODUCTION
Acyl coenzyme A (CoA) binding protein (ACBP) is also called diazepam-binding inhibitor (DBI). In humans and mice, this small (10 kDa) protein plays a dual role, reflecting its double name. As an intracellular protein, ACBP/DBI binds to medium- and long-chain acyl-CoA esters, reducing their toxicity and facilitating their transport through different subcellular compartments, hence stimulating lipid metabolism (1–3). As an extracellular protein, ACBP/DBI binds to the peripheral benzodiazepine receptor (hence displacing the benzodiazepine diazepam from its binding site), which is the ionotropic gamma-aminobutyric acid type A (GABAA) receptor (GABAAR) possessing another endogenous ligand, γ-aminobutyric acid, the major inhibitory neurotransmitter (4, 5). In the central nervous system, ACBP/DBI can be proteolytically cleaved to yield several neuropeptides, one of which, octadecaneuropeptide (ODN), interacts with a G protein coupled receptor (GPCR) in the central nervous system (6, 7).
ACBP/DBI is ubiquitously expressed and can be released from cells through an unconventional, autophagy-dependent pathway (8). It then acts as a paracrine mediator to inhibit autophagy through an action on GABAAR, which is expressed in many cell types outside the central nervous system (9). Hence, antibody-mediated neutralization of extracellular ACBP/DBI offers the possibility to stimulate autophagy by interrupting a paracrine feedback inhibition loop. In humans, obesity and metabolic syndrome are associated with elevated ACBP/DBI levels in the plasma (10), while anorexia nervosa is characterized by abnormally low concentrations of circulating ACBP/DBI (9, 11). In mice, injection of recombinant ACBP/DBI protein into the peritoneal cavity or the tail vein causes a GABAAR-dependent increase in feeding. This appetite-stimulatory effect of ACBP/DBI is also observed for proteins in which the acyl-CoA binding moiety has been mutated. Conversely, injection of a neutralizing antibody blocks feeding responses and counteracts weight gain or favors weight loss in multiple experimental conditions. These findings suggest that ACBP/DBI is involved in the pathophysiology of human obesity (12).
ACBP/DBI is a phylogenetically conserved protein, as ACBP/DBI homologs have been described in all eukaryotic phyla and even in some bacterial species (13, 14). In the nematode Caenorhabditis elegans and in the insect Drosophila melanogaster, ACBP/DBI orthologs stimulate pharyngeal pumping and mouth hook movement, which are the functional equivalents of mammalian mastication (15). In the yeast Saccharomyces cerevisiae, ACBP/DBI is the only protein known to be released in response to nutrient or oxidative stress (16, 17). Extracellular ACBP/DBI stimulates sporulation of yeast in a GPCR-dependent fashion, hence allowing yeast cells to swarm out to find new food resources (15, 18). Thus, the appetite-stimulatory function of ACBP/DBI appears to be phylogenetically conserved (19–21).
Reportedly, the genomes of some bacteria code for ACBP/DBI orthologs (22, 23). It is well known that human obesity is associated with major shifts in the composition of the intestinal microbiome (24, 25). Moreover, fecal microbial transplantation (FMT) of the stools from obese (but not lean) individuals into mice can transfer features of obesity and metabolic syndrome, establishing cause-effect relationships between alterations in the gut microbiome and the obese phenotype (26, 27).
Intrigued by these observations, we wondered whether specific microbial species in the human gut might encode and express ACBP/DBI-like proteins, thus potentially influencing human metabolism and eating behavior. Here, we report a detailed bioinformatics analysis of ACBP/DBI-like genes within the human gut microbiome and analyze their possible implication in obesity. We found that ACBP/DBI is mostly encoded by eukaryotes, that its presence in bacteria is mostly limited to pathogenic taxa, and that its rare presence in the human gut is not associated with alterations in the body mass index (BMI).
RESULTS
ACBP/DBI-like proteins are rarely encoded in members of the human microbiome.
To assess whether microbial ACBP/DBI ortholog genes could potentially contribute to microbiome-dependent gut metabolism, we first looked for their presence in 99,211 microbial genomes from NCBI as of January 2019. Using an initial set of 1,098 UniRef-annotated orthologous ACBP/DBI sequences (see Materials and Methods) to search these genomes, we found ACBP to be present in 3,635 of them, encompassing 1,668 unique TaxIDs, with the majority belonging to Proteobacteria (89% of genomes). Species with the largest number of genomes encoding ACBP showed it to be part of the core genome of several known pathogens from the Burkholderia genus, as well as those from Saccharomyces cerevisiae and Ralstonia solanacearum (Table 1). While Saccharomyces cerevisiae can be found in the human gut (28, 29), although usually at low abundance, the bacterial taxa in NCBI containing ACBP are at best very rare members of the human microbiome.
TABLE 1.
Top 10 species with the largest number of ACBP/DBI-encoding genomes based on available reference genomes
| Species | No. of genomes with ACBP (% positive from genomes searched) |
|---|---|
| Burkholderia pseudomalleia | 663 (100%) |
| Burkholderia ubonensisa | 291 (100%) |
| Burkholderia cenocepaciaa | 242 (99.1%) |
| Burkholderia multivoransa | 198 (100%) |
| Saccharomyces cerevisiaeb | 109 (93.9%) |
| Burkholderia cepaciaa | 98 (100%) |
| Ralstonia solanacearumc | 80 (100%) |
| Burkholderia stagnalisa | 64 (100%) |
| Burkholderia malleia | 56 (100%) |
| Burkholderia vietnamiensisa | 44 (100%) |
Taxonomy: Bacteria, Proteobacteria, Betaproteobacteria, Burkholderiales, Burkholderiaceae, Burkholderia.
Taxonomy: Eukaryota, Ascomycota, Saccharomycetes, Saccharomycetales, Saccharomycetaceae, Saccharomyces.
Taxonomy: Bacteria, Proteobacteria, Betaproteobacteria, Burkholderiales, Burkholderiaceae, Ralstonia.
Because genomic sequencing captures only a limited fraction of the human microbiome diversity (30–33), we proceeded by searching homologous sequences of known ACBP genes in metagenome-assembled genomes (MAGs). We screened 154,000 MAGs previously recovered from the human microbiome sampled from almost 10,000 individuals spanning diverse geography and lifestyle (Table S1). We found only 129 out of the 154,000 MAGs (0.08%) to encode ACBP, belonging to 14 species-level genome bins (SGBs). One of these SGBs was classified as Deinococcus-Thermus and another as Chitinophagaceae, whereas the remaining 12 all belonged to Proteobacteria, with the closest known taxa being again Burkholderia or taxa linked with sample-processing contamination such as Ralstonia or Acidovorax (34). This exploration of microbial genomes and MAGs thus highlights a lack of ACBP/DBI ortholog genes in microbes of putative relevance in the human microbiome.
Phylogenetic modeling of ACBP/DBI is highly taxonomically consistent.
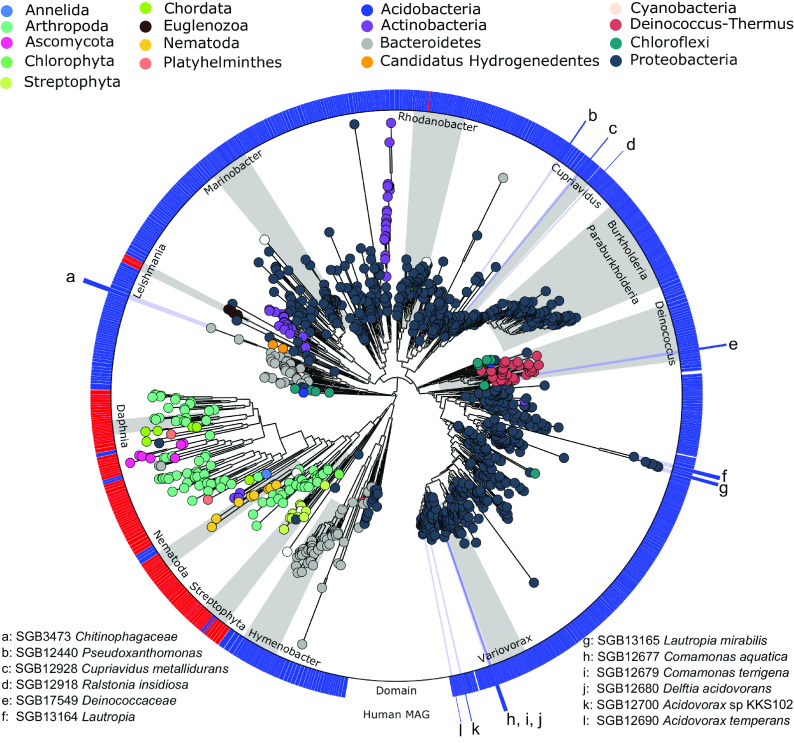
To better assess the sequence diversity of the ACBP/DBI gene, we phylogenetically modeled its sequence variants found in human MAGs and reference genomes from NCBI across different organisms. This analysis revealed very distinct eukaryotic versus microbial ACBP/DBI sequences, despite the relatively short alignment length used for phylogenetic inference (Fig. 1). This distinct pattern between the two domains was also seen when we used pairwise nucleotide identities calculated from multiple sequence alignments (Fig. S1). We found ACBP to be widespread across the domains of life, with ACBP sequences found in eukaryotic phyla including Streptophyta, Arthropoda, Nematoda, Ascomycota, and Chordata and present in 10 different bacterial phyla. Some taxa such as the genera Daphnia and Variovorax exhibited clearly defined clades, while other taxa such as the phyla Arthropoda and Actinobacteria displayed more diverse and paraphyletic phylogenies. The bacterial genera Burkholderia and Paraburkholderia showed a clearly defined subtree. ACBP sequences belonging to MAGs recovered from the human microbiome were widespread across the phylogeny but always maintained a consistent taxonomic structure. This adherence between phylogeny and taxonomy for ACBP/DBI suggests vertical evolutive trajectories for this gene, as a comparison between prokaryotic phylogenies built at the whole-genome level was highly consistent with the phylogenetic tree constructed for the ACBP/DBI gene (Fig. S2), with very limited evidence (if any) of horizontal transfer events and consequently a low likelihood that yet-to-be-characterized taxa not captured by our analysis carry ACBP/DBI ortholog genes.
FIG 1.
Whole phylogeny of ACBP/DBI gene sequences across kingdoms and phyla. The tree was built using 1,223 ACBP/DBI nucleotide sequences retrieved from UniProtKB, reference genomes from NCBI, and human metagenome-assembled genomes (MAGs) belonging to species-level genome bins (SGBs) from reference 30 (see Materials and Methods). Sequences were clustered at 97% identity prior to multiple sequence alignment and the tree was built using 240 nt of aligned positions.
ACBP/DBI is rarely found in human gut microbiomes.
To further investigate whether the few ACBP/DBI-positive genomes and MAGs recovered from the human microbiome could potentially contribute to gut metabolism, we evaluated their prevalence across 7,698 human gut metagenomes present in the curatedMetagenomicData R package (35), spanning different countries, age categories, and health conditions (Fig. 2A; Table S2). We found that the majority of MAGs belonging to these SGBs were very rarely found in samples across different data sets, with two known SGBs classified as Acidovorax sp. 12322_1 (kSGB 12676) and Cupriavidus metallidurans (kSGB 12928) achieving the highest overall prevalence (0.3%).
FIG 2.
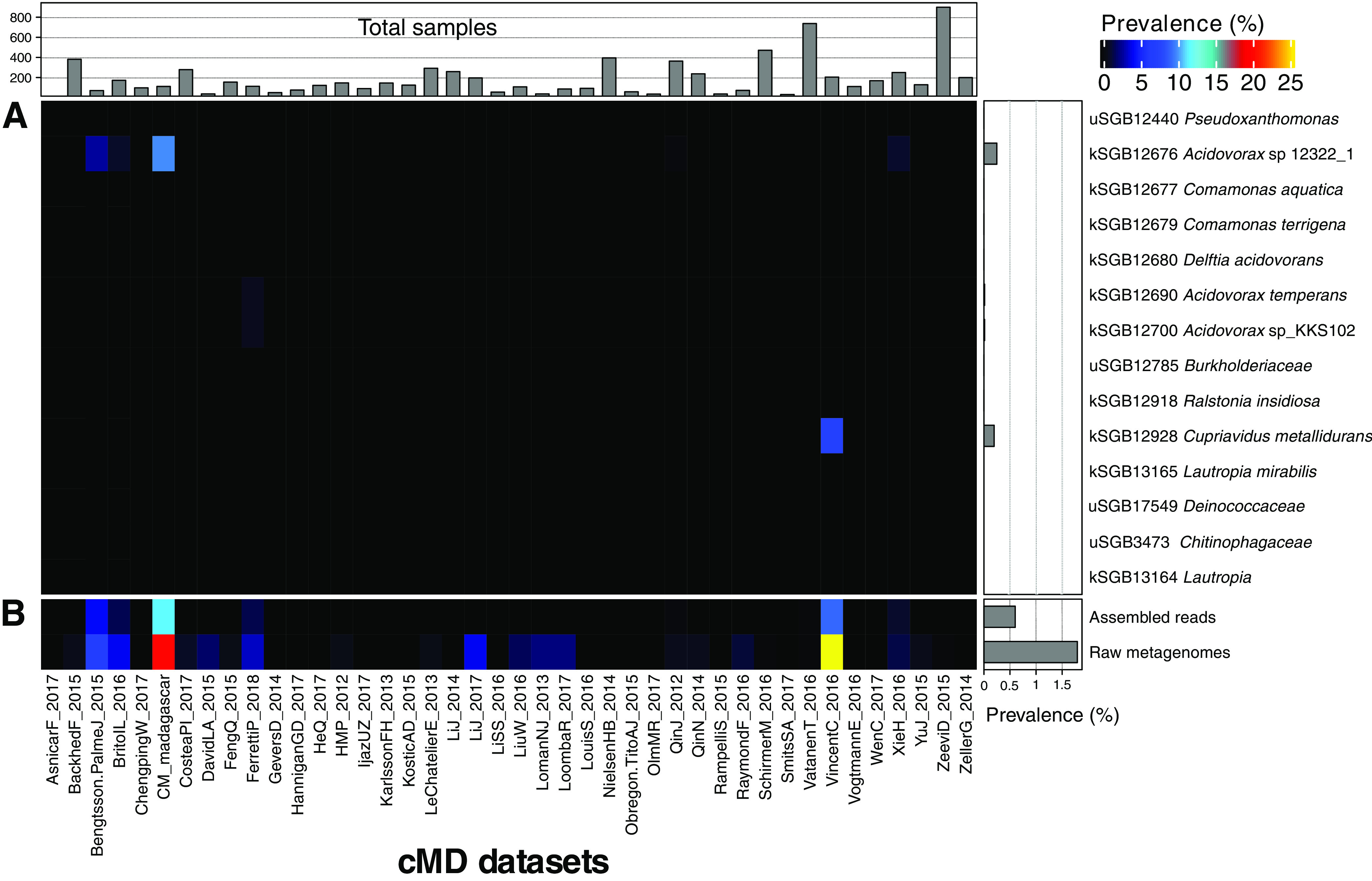
ACBP/DBI is rare in human gut metagenomes. (A) Prevalence of ACBP-encoding SGBs from the human microbiome for all data sets available in curatedMetagenomicData representing 7,698 metagenomic samples from the human gut. (B) The prevalence of assembled reads (contigs) with a significant hit to ACBP sequences and metagenomic reads that map to ACBP sequences with a breadth of coverage of >80% (see Materials and Methods).
Since MAGs rely on the success of metagenomic assembly and binning and thus may miss some low-abundance or hard-to-assemble taxa, we further screened unbinned contigs as well as the raw reads for each sample. The use of unbinned contigs (assembled reads) indeed led to an increase in the overall prevalence of ACBP/DBI-positive samples, but this number remained low (0.6%) (Fig. 2B). When we aligned raw metagenomic reads to the set of retrieved ACBP/DBI sequences, we further observed an increase in the overall relatively low prevalence across samples (1.79%), although we cannot exclude that some of the hits are false positives that inflate the prevalence estimation. Notably, some data sets, such as CM_madagascar from a non-Westernized society (30) and VincentC_2016 comprising fecal microbiome of 98 hospitalized patients treated with antibiotics and that used laxatives (36), showed a higher prevalence of ABCP/DBI in their raw metagenomes compared to others, 19.64% and 25.76%, respectively. On the contrary, 35 data sets (83%) had a prevalence of 0%.
This analysis thus reinforces the very low prevalence of ABCP/DBI-positive taxa and of the ABCP/DBI gene in the human gut microbiome, which appears inconsistent with a hypothesis of a role of this microbial gene variant in human metabolism. Moreover, the taxonomy assignments of the species (from MAGs and NCBI genomes) found to encode ACBP/DBI and occasionally present in some gut microbiome data sets (Fig. 2A) point at sample contamination as a potential source for those taxa. Indeed, Pseudoxanthomonas, Acidovorax, Comamonas, Delftia, Ralstonia, and Cupriavidus have been previously described as common reagent and laboratory contaminants (34).
Lack of correlation between ACBP/DBI-positive species and body mass index.
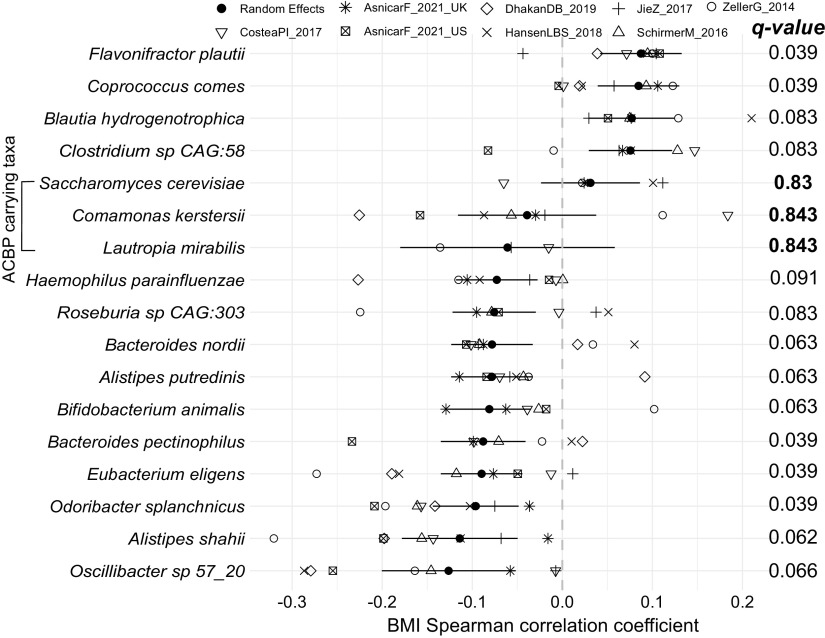
Although we found a low prevalence of ACBP/DBI-encoding members in the human gut microbiome, theoretically there could still be a possibility that low-prevalent low-abundance taxa can somehow contribute to human gut metabolism. To evaluate a possible link between microbial ACBP/DBI ortholog genes and obesity, we performed a meta-analysis of correlations between species-level abundances and BMI as a readout using 1,899 gut samples from healthy individuals curated within the curatedMetagenomicData (35) effort (Fig. 3; Table 2). We found 14 taxa to be significantly associated with BMI (random effects model false-discovery rate [FDR] < 0.1) (Table S3), with species such as Flavonifractor plautii, Coprococcus comes, and Blautia hydrogenotrophica associated with increased BMI, in line with previous reports (37, 38). We also found species associated with decreased BMI, which included Oscillibacter sp. 57_20, Alistipes shahii, and Odoribacter splanchnicus, as previously described (39). However, these 14 species significantly associated with BMI were all ACBP/DBI-negative. Within the limited panel of ACBP/DBI-positive species at least occasionally found in the gut microbiome, only Saccharomyces cerevisiae, Lautropia mirabilis, and Comamonas kerstersii were sufficiently prevalent in these samples to perform the meta-analysis but showed no significant associations (q values > 0.8) (Fig. 3). These results indicate that species found to encode ACBP/DBI in the human gut microbiome do not show associations with BMI.
FIG 3.
ACBP/DBI-carrying taxa present in the human gut show no significant associations with BMI. We performed a meta-analysis of partial correlations (adjusted for age and sex) between species abundances and BMI across 1,899 samples from healthy gut metagenomes using a random effects model. Meta-analysis P values were corrected for multiple hypothesis testing correction using the false discovery rate (q values). ACBP-carrying species are shown, as well as species whose FDR is <10%.
TABLE 2.
Demographic information of gut samples from healthy individuals used in the meta-analysis
| Data set name | n | Age (yrs) |
BMI (kg/m2) |
Sex (n) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | Mean | Min | Max | Female | Male | ||
| AsnicarF_2021_UK | 953 | 45.6 | 18.5 | 65.9 | 25.3 | 18.7 | 40.0 | 686 | 267 |
| AsnicarF_2021_US | 92 | 42.5 | 22.3 | 65.9 | 25.9 | 18.8 | 38.8 | 61 | 31 |
| CosteaPI_2017 | 82 | 50.6 | 29 | 75 | 27.4 | 20.0 | 38.0 | 52 | 30 |
| DhakanDB_2019 | 80 | 35.6 | 19 | 71 | 23.6 | 19.2 | 36.4 | 42 | 38 |
| HansenLBS_2018 | 57 | 48.7 | 22.4 | 65.4 | 28.5 | 21.3 | 35.1 | 30 | 27 |
| JieZ_2017 | 140 | 61.0 | 38 | 107 | 23.7 | 18.8 | 32.1 | 76 | 64 |
| SchirmerM_2016 | 437 | 27.8 | 18 | 75 | 22.9 | 18.8 | 34.4 | 246 | 191 |
| ZellerG_2014 | 58 | 61.0 | 25 | 84 | 24.8 | 20.0 | 34.0 | 31 | 27 |
DISCUSSION
ACBP/DBI plays a major role in the control of appetite and metabolism through a phylogenetically conserved pathway that is conserved in yeast, nematodes, insects, and mammals (15, 20, 21, 40). Intrigued by the observation that ACBP/DBI is a highly conserved protein that is even encoded by some bacterial genes, as well as by the link between human obesity and the gut microbiome, we investigated the prevalence of ACBP/DBI in intestinal commensals and their potential correlation with the body mass index.
The bioinformatic analyses presented in this paper based on extensive available metagenomic data sets suggest that ACBP/DBI-producing bacterial species are rather rare in the human microbiome and are mostly produced by eukaryotic species (as exemplified by the yeast S. cerevisiae) and environmental or potentially pathogenic bacteria (exemplified by Comamonas kerstersii that can cause peritonitis, bacteremia, and sepsis [41–43]), as well as potential sample contaminants. Indeed, the presence of ACBP/DBI-producing species in the human gut appears relatively rare. Moreover, we did not find any correlation between the presence of ACBP/DBI-encoding species and BMI across a large cumulative data set comprising 1,899 samples from healthy gut metagenomes. These results refute the hypothesis that the production of ACBP/DBI by the gut microbiome might affect whole-body metabolism, at least in the context of the normal microbiome.
Despite our findings, it could still be possible that microbes that are strongly associated with the mucosal tissue in the upper intestinal tract (and that hence would be grossly underrepresented in fecal samples) might have some local or systemic effects. It is also noteworthy to mention that the lack of an association between ACBP/DBI gene carriage and obesity found here did not take into account gene expression levels, which could be relevant as they might not mirror gene presence and/or abundance patterns. Moreover, in the context of infections, bacterial ACBP/DBI might exert some physiological effects on the host. However, it is unclear whether prokaryotic ACBP/DBI orthologues possess similar functions as those present in yeast or other eukaryotes, despite previous work showing strong conservation of amino acids at the majority of sites determined to be important for ACBP structure and function across phyla (22). ACBP/DBI inhibits autophagy (9, 19), and autophagy is a potent mechanism to eliminate intracellular bacteria (44), meaning that the subversion of autophagy (also called xenophagy) might contribute to the virulence of pathogenic species. Thus, Streptococcus pneumoniae degrades the essential autophagy protein ATG14 to ensure its survival in host cells (45), while Salmonella enterica serovar Typhimurium targets the V-ATPase-ATG16L1 axis to avoid xenophagy (46), just to mention a few examples. In view of these premises, it might be interesting to generate recombinant bacterial ACBP/DBI proteins and to evaluate them for their autophagy-inhibitory and metabolic effects.
The appetite-stimulatory effects of ACBP/DBI are lost in mice that bear a phenylalanine (F) to isoleucine (I) substitution at position 77 in the N-terminal domain of the gamma2 subunit of GABAAR (10, 47), supporting the contention that this neurotransmitter receptor is responsible for the obesogenic activity of DBI. ACBP/DBI is a GABAAR antagonist, while GABA is a GABAAR agonist. Of note, GABA, the natural agonist of GABAAR, can be produced by a series of bacteria. Reportedly, oral administration of GABA-producing Lactobacillus brevis strains reduces the abundance of mesenteric adipose tissue, enhances insulin secretion following glucose challenge, and improves plasma cholesterol clearance (48). Hence, it is possible that, beyond their documented effects on depression (49, 50), GABA-producing bacteria might affect whole-body metabolism, including appetite control. This hypothesis will be actively investigated by our laboratories.
MATERIALS AND METHODS
Identification of ACBP/DBI sequences and phylogenetic tree reconstruction.
To obtain a more comprehensive set of ACBP/DBI sequences, we downloaded amino acid sequences that matched the keyword “ACBP” from UniProt90 (51), mapped their identifiers to those of the European Molecular Biology Laboratory’s coding sequences using UniParc, and used the resulting DNA sequences to search, using BLASTn (52), all 99,211 microbial genomes available in NCBI, that included the whole set of 17,607 microbial species (16,959 bacteria, 648 archaea) available as of January 2019 and 154,723 metagenome-assembled genomes (MAGs) from reference 30. Matching queries were filtered to include only alignments with >70% identity, alignment length of >100 nt, and an E value of <1 × 10−5. We found no evidence that more permissive minimum alignment lengths lead to increased ACBP/DBI detection.
To build a phylogenetic tree of the known and metagenomically retrieved sequences, we clustered sequences at 97% sequence identity using UCLUST (parameters: “-id 0.97”) (53) and aligned centroid cluster sequences using MAFFT (parameters: “–localpair –maxiterate 1000”) (54). We removed “gappy” regions and ACBP/DBI sequences with insufficient aligned positions from the multiple sequence alignment using Jalview (55), resulting in 240 nucleotides of aligned positions and 1,223 sequences. The tree was built using fastTree (parameters: “-mlacc 2 -slownni -spr 4 -fastest -mlnni 4 -no2nd -nt”) (56) and refined with RAxML (parameters: “-m GTRGAMMA -t”) (57). GraPhlAn (58) was used for tree annotation and visualization.
We used PhyloPhlAn 3 (59) to build a phylogeny on 3,490 reference prokaryotic genomes and 129 MAGs (which we found to contain ACBP/DBI) using the parameters “-diversity high –accurate –force_nucleotides” and the set of up to 400 PhyloPhlAn genome markers. We compared trees built using PhyloPhlAn 3 and ACBP/DBI (with the aforementioned methods) in terms of their normalized pairwise branch lengths and used the tqDist (60) function available in the R quartet package to compare their quartet distances using a random sampling of 477 genomes repeated 1,000 times.
Search of ACBP/DBI sequences in human gut metagenomes.
The prevalence of both known and unknown species-level genome bins (kSGBs and uSGBs) that were found in a repository (https://opendata.lifebit.ai/table/SGB) (30) with ACBP/DBI-encoding MAGs was calculated using 7,698 human gut metagenomes present in the curatedMetagenomicData (cMD) version 1.16.0 R package (35). A given sample was deemed positive if a MAG belonging to the ACBP/DBI-encoding SGB was found.
We used the set of retrieved ACBP/DBI sequences to search, using BLASTn, all contigs assembled from human gut metagenomes available in cMD. Samples were considered to be positive for ACBP if any of their contigs had a significant hit (>70% identity, alignment length of >100 nt, and an E value of <1 × 10−5).
We aligned raw reads from these gut metagenomes to the set of retrieved ACBP/DBI sequences using bowtie2 (61). Resulting BAM files were filtered to keep only alignments with more than 50 nt of matching positions and were used to calculate the breadth of coverage of each sequence using Samtools (62) and VCF utils (63). Samples whose metagenome presented ACBP/DBI sequences with breadth of >80% were considered positive.
Correlations between BMI and species’ abundances.
We used the PREDICT 1 data set comprising 1,001 healthy individuals from the UK and 97 from the US (38), as well as publicly available data sets collected in cMD and profiled with version 3 of MetaPhlAn (64, 65). Of the 57 data sets available, we selected those that had samples with the following characteristics: (i) gut samples collected from healthy adult individuals at first collection (“days_from_first_collection” = 0 or not available [NA]) and (ii) samples with age, sex, and BMI data available. Outlier samples were removed if their BMI value was outside 3.5 and 7.5 times the interquartile range (IQR) of samples meeting the above criteria (IQR = 5.03). Only data sets with at least 50 samples were considered: Asnicar_2020_UK (953 samples out of 1,001), Asnicar_2020_US (92 samples out of 97) (38), CosteaPI_2017 (82 samples out of 279) (66), DhakanDB_2019 (80 samples out of 110) (67), HansenLBS_2018 (57 samples out of 208) (68), JieZ_2017 (140 samples out of 385) (39), SchirmerM_2016 (437 samples out of 471) (69), and ZellerG_2014 (58 samples out of 199) (70).
For each species, Spearman’s correlations with BMI were computed using the pcor.test function from the ppcor R package controlling for age and sex. Resulting correlations were used as input to the metacor function from the meta R package using Fisher’s Z transformation of correlations and the Paule-Mandel estimator of between-study variance in the random effects model. P values from the random-effects model were corrected using false discovery rate (FDR) through the Benjamini-Hochberg procedure, which are reported in the figure as q values. We report q values of ACBP/DBI-carrying taxa found in these data sets, as well as those of species with FDR of <0.1.
ACKNOWLEDGMENTS
This work was supported by the European H2020 program (ONCOBIOME-825410 project) to A.M.T. and N.S. and by the European Research Council (MetaPG-716575 ERC-STG project), the MIUR (“Futuro in Ricerca” RBFR13EWWI_001), the European H2020 program (MASTER-818368 project), a LEO Pharma research award to N.S., the National Cancer Institute of the National Institutes of Health (1U01CA230551 to N.S.), and the Premio Internazionale Lombardia e Ricerca 2019 to G.K. and N.S. G.K. is supported by the Ligue contre le Cancer (équipe labelisée), Agence National de la Recherche (ANR) – Projets blancs, Association pour la recherche sur le cancer (ARC), Association “Ruban Rose”, Cancéropôle Ile-de-France, Fondation pour la Recherche Médicale (FRM), a donation by Elior, Gustave Roussy Odyssea, the European Union Horizon 2020 Project Oncobiome, Fondation Carrefour, High-end Foreign Expert Program in China (GDW20171100085), Institut National du Cancer (INCa), Inserm (HTE), Institut Universitaire de France, LeDucq Foundation, the LabEx Immuno-Oncology (ANR-18-IDEX-0001), the RHU Torino Lumière, the Seerave Foundation, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE), and the SIRIC Cancer Research and Personalized Medicine (CARPEM). This study contributes to the IdEx Université de Paris ANR-18-IDEX-0001.
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Guido Kroemer, Email: kroemer@orange.fr.
Nicola Segata, Email: nicola.segata@unitn.it.
Knut Rudi, Norwegian University of Life Sciences.
REFERENCES
- 1.Mandrup S, Højrup P, Kristiansen K, Knudsen J. 1991. Gene synthesis, expression in Escherichia coli, purification and characterization of the recombinant bovine acyl-CoA-binding protein. Biochem J 276:817–823. 10.1042/bj2760817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duman C, Yaqubi K, Hoffmann A, Acikgöz AA, Korshunov A, Bendszus M, Herold-Mende C, Liu H-K, Alfonso J. 2019. Acyl-CoA-binding protein drives glioblastoma tumorigenesis by sustaining fatty acid oxidation. Cell Metab 30:274–289.e5. 10.1016/j.cmet.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Qiu S, Zeng B. 2020. Advances in understanding the acyl-CoA-binding protein in plants, mammals, yeast, and filamentous fungi. J Fungi (Basel) 6:34. 10.3390/jof6010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudsen J, Mandrup S, Rasmussen JT, Andreasen PH, Poulsen F, Kristiansen K. 1993. The function of acyl-CoA-binding protein (ACBP)/diazepam binding inhibitor (DBI). Mol Cell Biochem 123:129–138. 10.1007/BF01076484. [DOI] [PubMed] [Google Scholar]
- 5.Tonon M-C, Vaudry H, Chuquet J, Guillebaud F, Fan J, Masmoudi-Kouki O, Vaudry D, Lanfray D, Morin F, Prevot V, Papadopoulos V, Troadec J-D, Leprince J. 2020. Endozepines and their receptors: structure, functions and pathophysiological significance. Pharmacol Ther 208:107386. 10.1016/j.pharmthera.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 6.de Mateos-Verchere JG, Leprince J, Tonon MC, Vaudry H, Costentin J. 2001. The octadecaneuropeptide [diazepam-binding inhibitor (33–50)] exerts potent anorexigenic effects in rodents. Eur J Pharmacol 414:225–231. 10.1016/S0014-2999(01)00771-3. [DOI] [PubMed] [Google Scholar]
- 7.Bouyakdan K, Martin H, Liénard F, Budry L, Taib B, Rodaros D, Chrétien C, Biron É, Husson Z, Cota D, Pénicaud L, Fulton S, Fioramonti X, Alquier T. 2019. The gliotransmitter ACBP controls feeding and energy homeostasis via the melanocortin system. J Clin Invest 129:2417–2430. 10.1172/JCI123454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loomis WF, Behrens MM, Williams ME, Anjard C. 2010. Pregnenolone sulfate and cortisol induce secretion of acyl-CoA-binding protein and its conversion into endozepines from astrocytes. J Biol Chem 285:21359–21365. 10.1074/jbc.M110.105858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bravo-San Pedro JM, Sica V, Martins I, Pol J, Loos F, Maiuri MC, Durand S, Bossut N, Aprahamian F, Anagnostopoulos G, Niso-Santano M, Aranda F, Ramírez-Pardo I, Lallement J, Denom J, Boedec E, Gorwood P, Ramoz N, Clément K, Pelloux V, Rohia A, Pattou F, Raverdy V, Caiazzo R, Denis RGP, Boya P, Galluzzi L, Madeo F, Migrenne-Li S, Cruciani-Guglielmacci C, Tavernarakis N, López-Otín C, Magnan C, Kroemer G. 2019. Acyl-CoA-binding protein is a lipogenic factor that triggers food intake and obesity. Cell Metab 30:754–767.e9. 10.1016/j.cmet.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Joseph A, Moriceau S, Sica V, Anagnostopoulos G, Pol J, Martins I, Lafarge A, Maiuri MC, Leboyer M, Loftus J, Bellivier F, Belzeaux R, Berna F, Etain B, Capdevielle D, Courtet P, Dubertret C, Dubreucq J, Thierry DA, Fond G, Gard S, Llorca P-M, Mallet J, Misdrahi D, Olié E, Passerieux C, Polosan M, Roux P, Samalin L, Schürhoff F, Schwan R, Magnan C, Oury F, Bravo-San Pedro JM, Kroemer G. 2020. Metabolic and psychiatric effects of acyl coenzyme A binding protein (ACBP)/diazepam binding inhibitor (DBI). Cell Death Dis 11:502. 10.1038/s41419-020-2716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti E, Tremolizzo L, Bomba M, Uccellini O, Rossi MS, Raggi ME, Neri F, Ferrarese C, Nacinovich R. 2013. Reduced fasting plasma levels of diazepam-binding inhibitor in adolescents with anorexia nervosa. Int J Eat Disord 46:626–629. 10.1002/eat.22129. [DOI] [PubMed] [Google Scholar]
- 12.Sica V, Martins I, Motiño O, Bravo-San Pedro JM, Kroemer G. 2020. Antibody-mediated neutralization of ACBP/DBI has anorexigenic and lipolytic effects. Adipocyte 9:116–119. 10.1080/21623945.2020.1736734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose TM, Schultz ER, Todaro GJ. 1992. Molecular cloning of the gene for the yeast homolog (ACB) of diazepam binding inhibitor/endozepine/acyl-CoA-binding protein. Proc Natl Acad Sci U S A 89:11287–11291. 10.1073/pnas.89.23.11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faergeman NJ, Wadum M, Feddersen S, Burton M, Kragelund BB, Knudsen J. 2007. Acyl-CoA binding proteins; structural and functional conservation over 2000 MYA. Mol Cell Biochem 299:55–65. 10.1007/s11010-005-9040-3. [DOI] [PubMed] [Google Scholar]
- 15.Charmpilas N, Ruckenstuhl C, Sica V, Büttner S, Habernig L, Dichtinger S, Madeo F, Tavernarakis N, Bravo-San Pedro JM, Kroemer G. 2020. Acyl-CoA-binding protein (ACBP): a phylogenetically conserved appetite stimulator. Cell Death Dis 11:7. 10.1038/s41419-019-2205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz-Garcia D, Brouwers N, Malhotra V, Curwin AJ. 2020. Reactive oxygen species triggers unconventional secretion of antioxidants and Acb1. J Cell Biol 219:e201905028. 10.1083/jcb.201905028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188:527–536. 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manjithaya R, Anjard C, Loomis WF, Subramani S. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 188:537–546. 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bravo-San Pedro JM, Sica V, Martins I, Anagnostopoulos G, Maiuri C, Kroemer G. 2019. Cell-autonomous, paracrine and neuroendocrine feedback regulation of autophagy by DBI/ACBP (diazepam binding inhibitor, acyl-CoA binding protein): the obesity factor. Autophagy 15:2036–2038. 10.1080/15548627.2019.1662585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bravo-San Pedro JM, Sica V, Kroemer G. 2019. The elusive “hunger protein”: an appetite-stimulatory factor that is overabundant in human obesity. Mol Cell Oncol 6:e1667193. 10.1080/23723556.2019.1667193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madeo F, Tavernarakis N, Pedro J-S, Kroemer G. 2020. ACBP is an appetite stimulator across phylogenetic barriers. Cell Stress 4:27–29. 10.15698/cst2020.02.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burton M, Rose TM, Faergeman NJ, Knudsen J. 2005. Evolution of the acyl-CoA binding protein (ACBP). Biochem J 392:299–307. 10.1042/BJ20050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raboanatahiry NH, Lu G, Li M. 2015. Computational prediction of acyl-coA binding proteins structure in Brassica napus. PLoS One 10:e0129650. 10.1371/journal.pone.0129650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenburg JL, Bäckhed F. 2016. Diet-microbiota interactions as moderators of human metabolism. Nature 535:56–64. 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Khan MT, Herrema H, Ackermans M, Serlie MJM, de Brauw M, Levels JHM, Sales A, Gerdes VE, Ståhlman M, Schimmel AWM, Dallinga-Thie G, Bergman JJ, Holleman F, Hoekstra JBL, Groen A, Bäckhed F, Nieuwdorp M. 2020. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 69:502–512. 10.1136/gutjnl-2019-318320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF. 2017. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5:153. 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallen-Adams HE, Suhr MJ. 2017. Fungi in the healthy human gastrointestinal tract. Virulence 8:352–358. 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, Collado MC, Rice BL, DuLong C, Morgan XC, Golden CD, Quince C, Huttenhower C, Segata N. 2019. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176:649–662.e20. 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. 2021. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 39:105–114. 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. 2019. A new genomic blueprint of the human gut microbiota. Nature 568:499–504. 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. 2019. New insights from uncultivated genomes of the global human gut microbiome. Nature 568:505–510. 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, Beghini F, Malik F, Ramos M, Dowd JB, Huttenhower C, Morgan M, Segata N, Waldron L. 2017. Accessible, curated metagenomic data through ExperimentHub. Nat Methods 14:1023–1024. 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent C, Miller MA, Edens TJ, Mehrotra S, Dewar K, Manges AR. 2016. Bloom and bust: intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome 4:12. 10.1186/s40168-016-0156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomes AC, Hoffmann C, Mota JF. 2018. The human gut microbiota: metabolism and perspective in obesity. Gut Microbes 9:308–325. 10.1080/19490976.2018.1465157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, Leeming E, Gibson R, Le Roy C, Khatib HA, Francis L, Mazidi M, Mompeo O, Valles-Colomer M, Tett A, Beghini F, Dubois L, Bazzani D, Thomas AM, Mirzayi C, Khleborodova A, Oh S, Hine R, Bonnett C, Capdevila J, Danzanvilliers S, Giordano F, Geistlinger L, Waldron L, Davies R, Hadjigeorgiou G, Wolf J, Ordovás JM, Gardner C, Franks PW, Chan AT, Huttenhower C, Spector TD, Segata N. 2021. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med 27:321–332. 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jie Z, Xia H, Zhong S-L, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen J-Y, Geng Q-S, Zhang Z-W, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K. 2017. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8:845. 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedro J-S, Sica V, Madeo F, Kroemer G. 2019. Acyl-CoA-binding protein (ACBP): the elusive “hunger factor” linking autophagy to food intake. Cell Stress Chaperones 3:312–318. 10.15698/cst2019.10.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaeuffer C, Schramm F, Meyer A, Hansmann Y, Guffroy A, Argemi X. 2018. First case of Comamonas aquatica bacteremia complicated by septic shock. Med Mal Infect 48:540–542. 10.1016/j.medmal.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Tiwari S, Nanda M. 2019. Bacteremia caused by Comamonas testosteroni an unusual pathogen. J Lab Physicians 11:87–90. 10.4103/JLP.JLP_116_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X-J, Qiao X-W, Huang T-M, Li L, Jiang S-P. 2020. Comamonas kerstersii bacteremia. Med Mal Infect 50:288–290. 10.1016/j.medmal.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Levine B, Kroemer G. 2019. Biological functions of autophagy genes: a disease perspective. Cell 176:11–42. 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shizukuishi S, Ogawa M, Ryo A, Ohnishi M. 2020. Streptococcus pneumoniae promotes its own survival via choline-binding protein CbpC-mediateddegradation of ATG14. Autophagy 16:1529–1531. 10.1080/15548627.2020.1776475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp A-K, Li L, Shi X, Zhou Z, Gao W, Li D, He H, Liu X, Ding J, Hottiger MO, Shao F. 2019. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell 178:552–566.e20. 10.1016/j.cell.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Dumitru I, Neitz A, Alfonso J, Monyer H. 2017. Diazepam binding inhibitor promotes stem cell expansion controlling environment-dependent neurogenesis. Neuron 94:125–137.e5. 10.1016/j.neuron.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Patterson E, Ryan PM, Wiley N, Carafa I, Sherwin E, Moloney G, Franciosi E, Mandal R, Wishart DS, Tuohy K, Ross RP, Cryan JF, Dinan TG, Stanton C. 2019. Gamma-aminobutyric acid-producing lactobacilli positively affect metabolism and depressive-like behaviour in a mouse model of metabolic syndrome. Sci Rep 9:16323. 10.1038/s41598-019-51781-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, Lewis K. 2019. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 4:396–403. 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J. 2019. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 4:623–632. 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 51.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh L-SL. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32:D115–D119. 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asnicar F, Thomas AM, Beghini F, Mengoni C, Manara S, Manghi P, Zhu Q, Bolzan M, Cumbo F, May U, Sanders JG, Zolfo M, Kopylova E, Pasolli E, Knight R, Mirarab S, Huttenhower C, Segata N. 2020. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat Commun 11:2500. 10.1038/s41467-020-16366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sand A, Holt MK, Johansen J, Brodal GS, Mailund T, Pedersen CNS. 2014. tqDist: a library for computing the quartet and triplet distances between binary or general trees. Bioinformatics 30:2079–2080. 10.1093/bioinformatics/btu157. [DOI] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 65.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung F-E, Kultima JR, Hayward MR, Coelho LP, Allen-Vercoe E, Bertrand L, Blaut M, Brown JRM, Carton T, Cools-Portier S, Daigneault M, Derrien M, Druesne A, de Vos WM, Finlay BB, Flint HJ, Guarner F, Hattori M, Heilig H, Luna RA, van Hylckama Vlieg J, Junick J, Klymiuk I, Langella P, Le Chatelier E, Mai V, Manichanh C, Martin JC, Mery C, Morita H, O'Toole PW, Orvain C, Patil KR, Penders J, Persson S, Pons N, Popova M, Salonen A, Saulnier D, Scott KP, Singh B, et al. 2017. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 35:1069–1076. 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 67.Dhakan DB, Maji A, Sharma AK, Saxena R, Pulikkan J, Grace T, Gomez A, Scaria J, Amato KR, Sharma VK. 2019. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. Gigascience 8. 10.1093/gigascience/giz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen LBS, Roager HM, Søndertoft NB, Gøbel RJ, Kristensen M, Vallès-Colomer M, Vieira-Silva S, Ibrügger S, Lind MV, Mærkedahl RB, Bahl MI, Madsen ML, Havelund J, Falony G, Tetens I, Nielsen T, Allin KH, Frandsen HL, Hartmann B, Holst JJ, Sparholt MH, Holck J, Blennow A, Moll JM, Meyer AS, Hoppe C, Poulsen JH, Carvalho V, Sagnelli D, Dalgaard MD, Christensen AF, Lydolph MC, Ross AB, Villas-Bôas S, Brix S, Sicheritz-Pontén T, Buschard K, Linneberg A, Rumessen JJ, Ekstrøm CT, Ritz C, Kristiansen K, Nielsen HB, Vestergaard H, Færgeman NJ, Raes J, Frøkiær H, Hansen T, Lauritzen L, Gupta R, Licht TR, et al. 2018. A low-gluten diet induces changes in the intestinal microbiome of healthy Danish adults. Nat Commun 9:4630. 10.1038/s41467-018-07019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Horst RT, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, Fu J, Joosten LAB, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG, Xavier RJ. 2016. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167:1897. 10.1016/j.cell.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 70.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel Doeberitz M, Sobhani I, Bork P. 2014. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 10:766. 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00471-21-s0001.pdf, PDF file, 1.1 MB (2.8MB, pdf)
Supplemental material. Download aem.00471-21-s0002.xlsx, XLSX file, 1.1 MB (173.3KB, xlsx)
Supplemental material. Download aem.00471-21-s0003.xlsx, XLSX file, 1.1 MB (18.3KB, xlsx)
Supplemental material. Download aem.00471-21-s0004.xlsx, XLSX file, 1.1 MB (13.9KB, xlsx)