ABSTRACT
Listeria monocytogenes is a ubiquitous environmental bacterium and intracellular pathogen that responds to stress using predominantly the alternative sigma factor SigB. Stress is sensed by a multiprotein complex, the stressosome, extensively studied in bacteria grown in nutrient media. Following signal perception, the stressosome triggers a phosphorylation cascade that releases SigB from its anti-sigma factor. Whether the stressosome is activated during the intracellular infection is unknown. Here, we analyzed the subcellular distribution of stressosome proteins in L. monocytogenes located inside epithelial cells following their immunodetection in membrane and cytosolic fractions prepared from intracellular bacteria. Unlike bacteria in laboratory media, intracellular bacteria have a large proportion of the core stressosome protein RsbR1 associated with the membrane. However, another core protein, RsbS, is undetectable. Despite the absence of RsbS, a SigB-dependent reporter revealed that SigB activity increases gradually from early (1 h) to late (6 h) postinfection times. We also found that RsbR1 paralogues attenuate the intensity of the SigB response and that the miniprotein Prli42, reported to tether the stressosome to the membrane in response to oxidative stress, plays no role in associating RsbR1 to the membrane of intracellular bacteria. Altogether, these data indicate that, once inside host cells, the L. monocytogenes stressosome may adopt a unique configuration to sense stress and to activate SigB in the intracellular eukaryotic niche.
IMPORTANCE The response to stress mediated by the alternative sigma factor SigB has been extensively characterized in Bacillus subtilis and Listeria monocytogenes. These bacteria sense stress using a supramacromolecular complex, the stressosome, which triggers a cascade that releases SigB from its anti-sigma factor. Despite the fact that many structural data on the complex are available and analyses have been performed in mutants lacking components of the stressosome or the signaling cascade, the integration of the stress signal and the dynamics of stressosome proteins following environmental changes remain poorly understood. Our study provides data at the protein level on essential stressosome components and SigB activity when L. monocytogenes, normally a saprophytic bacterium, adapts to an intracellular lifestyle. Our results support activation of the stressosome complex in intracellular bacteria. The apparent loss of the stressosome core protein RsbS in intracellular L. monocytogenes also challenges current models, favoring the idea of a unique stressosome architecture responding to intracellular host cues.
KEYWORDS: Listeria, stressosome, SigB, intracellular, RsbR1, phosphorylation
INTRODUCTION
Listeria monocytogenes is a ubiquitous Gram-positive bacterium that colonizes a large variety of environmental niches, including soil, decaying plant material, water, and industrial facilities (1, 2). This saprophytic lifestyle is shared with its capacity to infect animal hosts, causing severe foodborne diseases that progress with intracellular infections of phagocytic and nonphagocytic host cells (1, 3, 4). This pathogen withstands extreme stresses, including low pH, high osmolarity, low temperatures, and exposure to bile salts and antimicrobials, among others (5, 6). Such stress responses are fundamental to trigger a successful virulence program upon host contact (7). The stress experienced during this transition is relevant in the early infection stages, including passage through the acidic stomach, the high osmolality of the gut, and the competition with endogenous intestinal microbiota.
In L. monocytogenes and Bacillus subtilis, the alternative sigma factor SigB (σB) mediates the response to stress by modulating the expression of hundreds of genes upon stress perception (6–10). SigB is normally bound to its anti-sigma factor, RsbW, which is itself regulated by the anti-anti-sigma factor RsbV. The association between RsbW and RsbV is favored when the latter is dephosphorylated by the RsbU phosphatase. The ratio between phosphorylated and unphosphorylated RsbV controls the levels of free SigB that associates with the RNA polymerase and, consequently, the intensity of the response (7, 11–13). RsbV phosphorylation is controlled in B. subtilis by two phosphatases, RsbU or RsbP, which respond to either environmental or energy-related stress signals (14–16), whereas in L. monocytogenes the stress signals are integrated in a single RsbU-dependent pathway (17).
Both L. monocytogenes and B. subtilis use a supramacromolecular complex, the stressosome, to perceive and process the external stress signals by yet-unknown mechanisms (18–21). An exception is sensing of blue light, which activates SigB via a stressosome-associated protein named RsbL (7). Although assembled and extensively characterized in vitro using purified recombinant proteins, the architecture of the stressosome in whole cells remains poorly understood. RsbR1 (RsbRA in B. subtilis) and RsbS are core proteins of the stressosome phosphorylated by the kinase RsbT following detection of the stress signal. Until recently, the most widely accepted model predicted that stress stimulates the RsbT kinase to phosphorylate one of the two phosphorylatable threonine residues in RsbR1 and a serine residue in RsbS (22–26). These phosphorylation steps culminate with RsbT release from the stressosome, allowing its interaction with RsbU and the stimulation of the SigB activation cascade. This model was mostly inferred from in vitro studies based on recombinant proteins. Thus, an RsbT variant of B. subtilis lacking its kinase activity does not complex with RsbRA and RsbS (27). Studies performed in L. monocytogenes exposed to oxidative stress identified Prli42, a miniprotein that tethers the stressosome to the membrane by interacting with RsbR1 (18). This work also showed that Prli42 interacts with the RsbR1 paralogues RsbR2 (Lmo0161), RsbL (Lmo0799), and RsbR3 (Lmo1642). A recent study based on subcellular fractionation and analysis of phosphorylation state of stressosome proteins revealed that L. monocytogenes RsbR1 and RsbS are mainly cytosolic and predominantly phosphorylated, regardless of exposure to osmotic stress (28). This work also showed that phosphorylated RbsR1 and its paralogues may play opposing roles in regulating SigB activity (28).
Despite the information obtained in L. monocytogenes exposed to different stresses under laboratory conditions, the role of SigB throughout the infection process remains controversial. Many studies suggest regulatory cross talk between SigB and the virulence regulator PrfA (6); the latter protein is essential for L. monocytogenes invasion and survival within eukaryotic cells (3). The available information assigns a major role to SigB in early events of the infection process mediating bacterial survival in the stomach and the intestinal tract (reviewed in reference 6). The fact that ΔsigB mutants are attenuated by the oral route, although they are fully virulent when injected intravenously (29), provides further support for the apparent dispensability of SigB for virulence. Moreover, ΔsigB mutants proliferate like wild-type bacteria inside eukaryotic cells (30). Nonetheless, other observations suggest that SigB could play a role in the interaction of L. monocytogenes with host cells. First, the SigB-dependent genes opuCA and gadA, which encode a glycine betaine transporter and a glutamate decarboxylase subunit, respectively, are transcribed at higher levels in wild-type bacteria than in a ΔsigB mutant in the cytosol of epithelial cells (30). This observation implies that functions of the SigB regulon known to be required for metabolic readjustments in response to extracellular stresses might also be involved in pathogen survival and/or growth inside host cells. Second, together with PrfA, SigB controls expression of the inlA and inlB genes, encoding surface proteins required for bacterial invasion (31, 32). Third, the P2prfA promoter, which is active in intracellular bacteria, is partially dependent on SigB (30), and, fourth, L. monocytogenes represses motility inside eukaryotic cells, and the flagellar repressor gene mogR has a promoter that depends on SigB (33). This positive regulation agrees with the increased motility displayed by ΔsigB mutants (34).
Taking these observations into account, we investigated L. monocytogenes SigB activity inside eukaryotic cells using approaches that involved monitoring in intracellular bacterial levels, activities, and subcellular distribution of SigB and core stressosome proteins. Consistent with the biochemical data, a SigB-dependent transcriptional reporter fusion confirmed a striking increment of SigB activity as the intracellular infection progressed to reach the pathogen replicative phase in the cytosol of the infected cell.
RESULTS
L. monocytogenes activates SigB and alters localization of stressosome proteins inside epithelial cells.
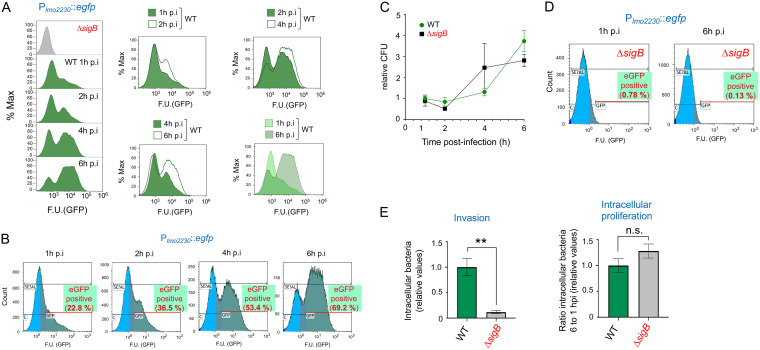
To determine whether L. monocytogenes activates SigB inside host cells, we infected JEG-3 human epithelial cells with a strain bearing a Plmo2230::egfp fluorescent reporter fusion integrated into the chromosome. lmo2230 encodes a predicted arsenate reductase; it forms part of the SigB regulon and is highly dependent on this sigma factor for expression (31, 35, 36). The fluorescence signal derived from the Plmo2230::egfp reporter was monitored by flow cytometry in intracellular bacteria at distinct postinfection times (Fig. 1A and B). Plmo2230::egfp expression increased gradually as intracellular bacteria replicated, from 22.8% of the bacterial population being positive for the reporter at 1 h postinfection (hpi) to 69.2% at 6 hpi (Fig. 1A and B). This postinfection time, 6 h, is coincidental to the maximal number of intracellular bacteria registered. Compared to the number of bacteria at 1 hpi, these numbers accounted for ∼4- and ∼3-fold increases in wild-type and ΔsigB strains, respectively (Fig. 1C). Importantly, no fluorescence signal derived from Plmo2230::egfp was detected in a ΔsigB mutant background at early (1 h) or late (6 h) postinfection times (Fig. 1D). This result demonstrated that the increase in the reporter signal observed in wild-type intracellular bacteria was fully dependent on SigB. In agreement with studies using other epithelial cell lines (13), the ΔsigB mutant replicated intracellularly in JEG-3 cells at similar rates to wild-type bacteria, although it displayed an ∼10-fold-lower capacity to invade these epithelial cells (Fig. 1E).
FIG 1.
L. monocytogenes increases SigB activity inside epithelial cells as infection progresses. Wild-type strain EGD-e (WT) and the isogenic ΔsigB mutant bearing the Plmo2230::egfp reporter fusion were used to infect JEG-3 epithelial cells. (A) Fluorescence intensity registered by flow cytometry at the indicated time points after infection with wild-type bacteria. The right panels show side-to-side comparison in the period from 1 h to 6 h, with a progressive increase in reporter expression. (B) Percentage of wild-type cells positive for expression of the Plmo2230::egfp reporter at the indicated times. Data are representative of the analysis of three biological replicates with one technical replicate for each time point of infection. (C) Intracellular growth of L. monocytogenes wild-type and ΔsigB reporter strains inside JEG-3 epithelial cells. The results are from three independent experiments and are shown as relative values (CFU values from WT and the ΔsigB mutant at 1 hpi were set to 1 and corresponded to 2.7 × 104 and 3.1 × 103 CFU, respectively, in a representative experiment). (D) Signal derived from the Plmo2230::egfp reporter in the ΔsigB mutant at 1 h and 6 h postinfection. (E) Invasion and intracellular proliferation rates of wild-type and ΔsigB reporter strains (CFU from WT at 1 h and 6 h postinfection were set to 1 and corresponded to 2.7 × 104 and 6.7 × 104 CFU, respectively, in a representative experiment). One-tailed P values are indicated by asterisks for comparison between wild type and ΔsigB strains by the t test (**, P < 0.01; n.s., not significant). F.U., fluorescence units.
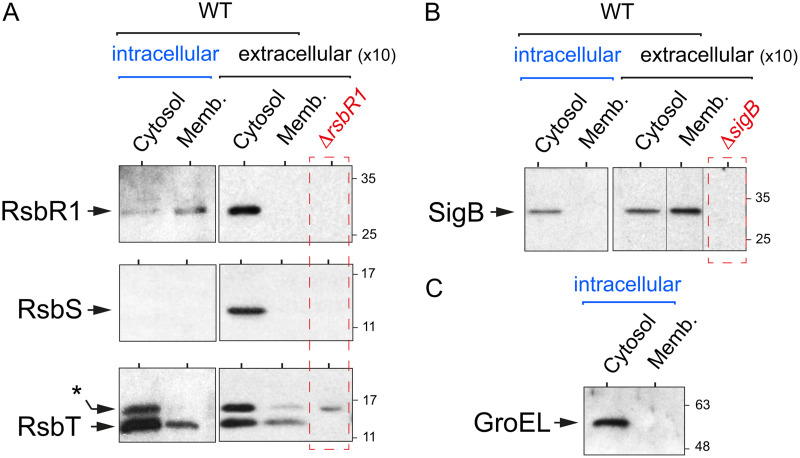
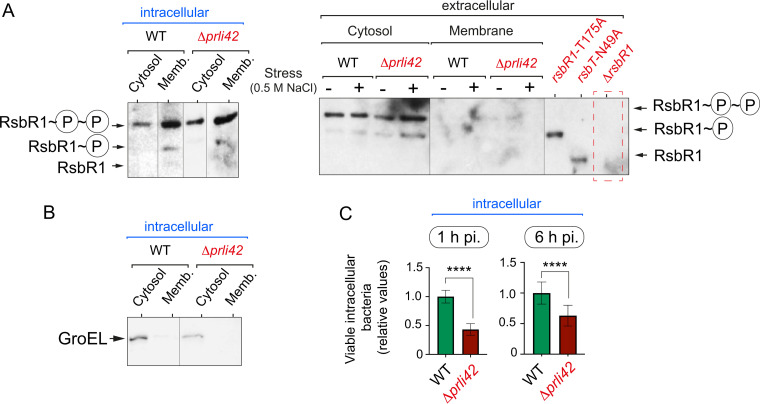
We next sought to determine the levels and distribution of stressosome proteins in bacteria isolated from infected cells. In these bacteria, a large proportion of RsbR1 molecules associate with the membrane, whereas RsbS is undetectable (Fig. 2A). The RsbT kinase, however, does not alter its subcellular distribution in intracellular bacteria, being present in both the membrane and the cytosol but with an increased abundance in the cytosolic location (Fig. 2A). Unlike RsbR1 or RsbT, SigB is detected exclusively in the cytosol of intracellular bacteria (Fig. 2B). The lack of cross-contamination between the distinct subcellular fractions was confirmed in all cases by examining the distribution of the cytosolic chaperone GroEL (Fig. 2C).
FIG 2.
The core stressosome protein RsbR1 is predominantly associated with the membrane in intracellular L. monocytogenes. JEG-3 epithelial cells were infected for 30 min with L. monocytogenes EGD-e wild type (WT) at a multiplicity of infection of 10:1 (bacteria to epithelial cells). Noninternalized bacteria were removed by washing steps, and 10 μg/ml gentamicin was added to the medium. Infected cells were incubated up to 6 h postinfection. At this time, intracellular bacteria were recovered in epithelial cell lysates by high-speed centrifugation. Cytosolic and membrane extracts were prepared from intracellular bacteria (6 hpi) and from extracellular bacteria grown in BHI medium (optical density at 600 nm [OD600] ≈ 0.4), as described elsewhere (28). The extracellular samples corresponded to 10-fold more bacteria. (A) Levels of the stressosome core proteins RsbR1 and RsbS and the kinase RsbT in intra- and extracellular bacteria. *, unspecific band detected in the null ΔrsbR1 polar mutant, as described in reference 28. (B) Levels of the sigma factor SigB. (C) Control assay for the relative enrichment of the cytosolic/membrane fractions based on the detection of the cytosolic chaperonin GroEL. Shown are immunoblots using the respective antibodies raised against each of the indicated proteins. Data correspond to a representative experiment from a minimum of three biological replicates.
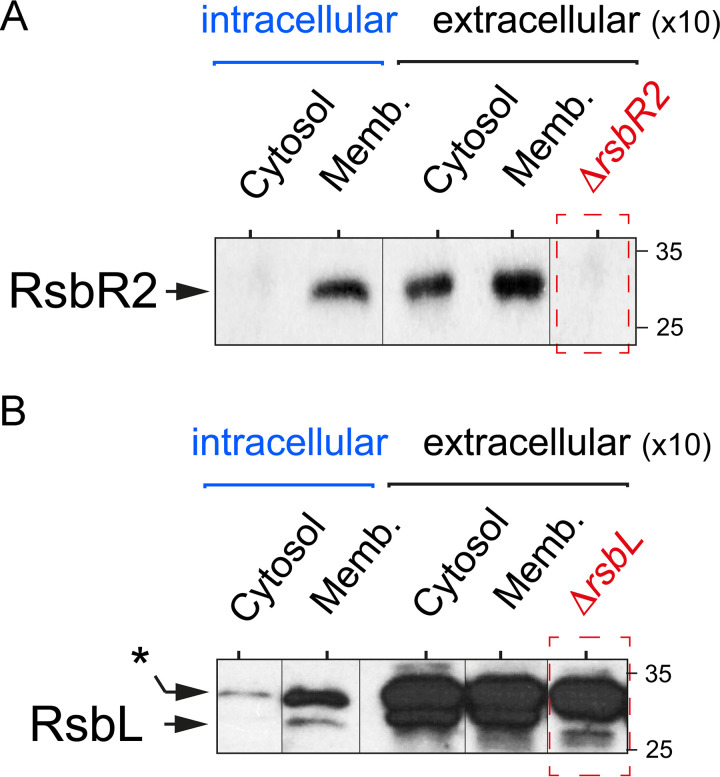
Interestingly, immunoassays with antibodies against the paralogues RsbR2 and RsbL showed that intracellular bacteria have both proteins predominantly associated with the membrane (Fig. 3A and B). Taken together, these results indicated that L. monocytogenes increases SigB activity inside epithelial cells and that the architecture of the stressosome complex may differ in intracellular bacteria compared to bacteria grown in nutritional media.
FIG 3.
The RsbR1 paralogues RsbR2 and RsbL are exclusively associated with the membrane in intracellular L. monocytogenes. Distribution of RsbR2 (A) and RsbL (B) in the cytosol and membrane (Memb.) fractions of intracellular wild-type L. monocytogenes EGD-e after infection of JEG-3 epithelial cells (6 h postinfection). In parallel, samples of extracellular bacteria grown to exponential phase (OD600 ∼ 0.4) in BHI medium corresponding to 10-fold more bacteria were analyzed. Control ΔrsbR2- and ΔrsbL-null mutants were also included for comparison. Shown are representative immunoblots obtained with anti-RsbR2 and anti-RsbL antibodies from a total of two biological replicates. *, unspecific band detected in the ΔrsbL mutant.
RsbR1 is in a fully phosphorylated state in intracellular L. monocytogenes.
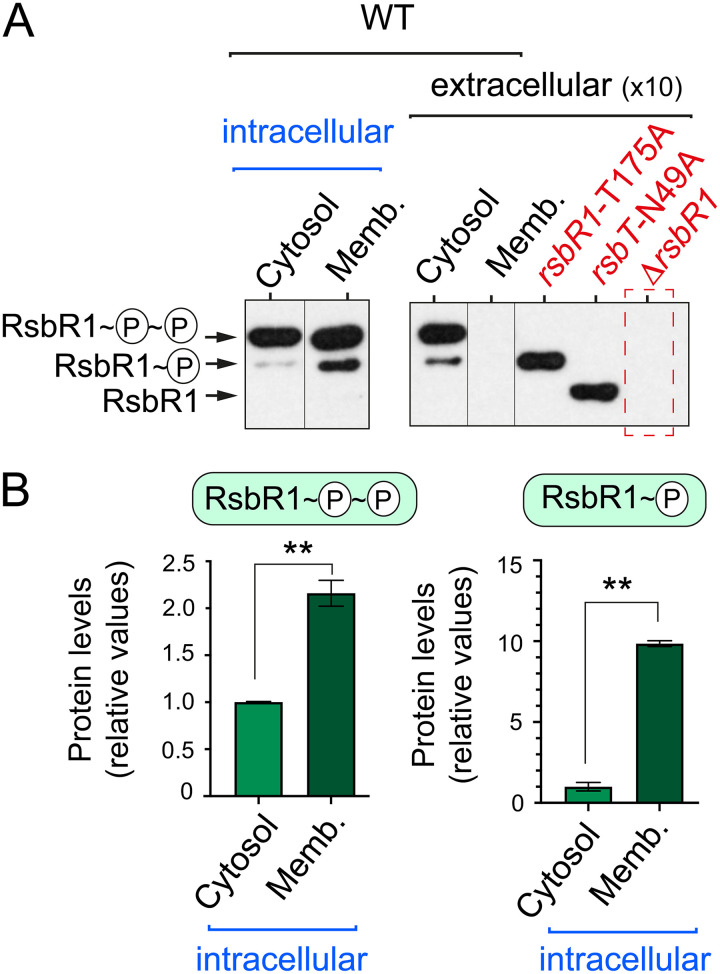
The widely accepted models of stressosome activation include RsbR1 and RsbS phosphorylation by the kinase RsbT following exposure to stress (22–26). L. monocytogenes RsbR1 has two phosphorylatable threonine residues (T175 and T209) with T175 being phosphorylated under basal conditions (37). RsbS has one phosphorylatable serine residue (S56), which in B. subtilis was shown to be phosphorylated only upon stress (22, 26). In contrast to these data, our recent study in L. monocytogenes grown in nutrient medium revealed that RsbR1 is predominantly in a doubly phosphorylated state, regardless of exposure to hyperosmotic stress (28). The analysis in intracellular bacteria also showed predominance of doubly phosphorylated RsbR1, accounting for ∼2-fold enrichment in the membrane relative to the cytosolic pool (Fig. 4A and B). Interestingly, monophosphorylated RsbR1 was highly enriched in the membranes of intracellular bacteria, with ∼10-fold more of this isoform detected in the membrane pool (Fig. 4A and B). Two additional L. monocytogenes mutants, one expressing a kinase-inactive RsbT-N49A variant and the other expressing an RsbR1-T175A variant (the latter not phosphorylatable at residue 175), facilitated the differentiation of the distinct unphosphorylated and phosphorylated isoforms (Fig. 4A). Altogether, these data pointed to RsbR1 in intracellular L. monocytogenes staying in distinct phosphorylated states but with no unphosphorylated forms, a pattern consistent with the increased SigB activity detected under these same infection conditions with the Plmo2230::egfp reporter fusion (Fig. 1).
FIG 4.
RsbR1 is phosphorylated to a large extent by intracellular L. monocytogenes. (A) Phos-tag electrophoresis based on phosphate-Zn2+ ion sequestration and immunoblots were used to analyze RsbR1 phosphorylation in cytosol/membrane extracts prepared from intracellular L. monocytogenes EGD-e wild type (6 h after infection of JEG-3 epithelial cells). In parallel, RsbR1 phospho-isoforms were detected in subcellular fractions from extracellular bacteria grown to exponential phase (OD600 ∼ 0.4) in BHI medium. Control cytosolic samples from rsbR1-T175A, rsbT-N49A, and ΔrsbR1 strains were included. An approximately similar number of bacteria (∼2 × 107) was analyzed in intra- and extracellular extracts. (B) Quantification of the signals obtained for RsbR1 in intracellular bacteria in three independent biological replicates. Data are means and standard deviations. Statistical analysis was performed by t test. **, P < 0.01.
The miniprotein Prli42 is dispensable for RsbR1 association with the membrane of intracellular L. monocytogenes.
Prli42 has been proposed to tether the stressosome to the membrane in L. monocytogenes exposed to oxidative stress (18). RsbR1 levels associated with the membrane were, however, indistinguishable between wild-type and Δprli42 strains following the infection of epithelial cells (Fig. 5A and B). This result ruled out a requirement for Prli42 in the stressosome to associate with the membrane of intracellular bacteria. Nonetheless, the loss of Prli42 had effects on bacterial invasion (ca. 60% less in the absence of Prli42), although no phenotype was discernible in the rate of intracellular replication (Fig. 5C).
FIG 5.
Phosphorylated RsbR1 remains attached to the membrane of intracellular L. monocytogenes in the absence of Prli42. (A) A Phos-tag system was used for the analysis of phosphorylated RsbR1 in subcellular extracts prepared from intracellular L. monocytogenes EGD-e wild-type (WT) and Δprli42 strains growing inside JEG-3 epithelial cells at 6 hpi. In parallel, subcellular fractions from extracellular bacteria subjected to osmotic stress (0.5 M NaCl for 30 min) were analyzed. Control cytosolic samples from extracellular rsbR1-T175, rsbT-N49A, and ΔrsbR1 strains are included. (B) Loading control with anti-GroEL for the samples prepared from intracellular bacteria. (C) Invasion (1 hpi) and proliferation (6 to 1 hpi) rates in JEG-3 epithelial cells of EGD-e wild-type and Δprli42 strains. The results from three independent experiments are shown as ratios of the number of Δprli42 to wild-type bacteria (the number of CFU from the WT at 1 hpi was arbitrarily set to 1 and corresponded to 1.7 × 105 CFU; the number of WT CFU at 6 hpi was 2.5 × 106). One-tailed P values are indicated by asterisks for comparison between wild-type and Δprli42 strains by t test (****, P < 0.0001).
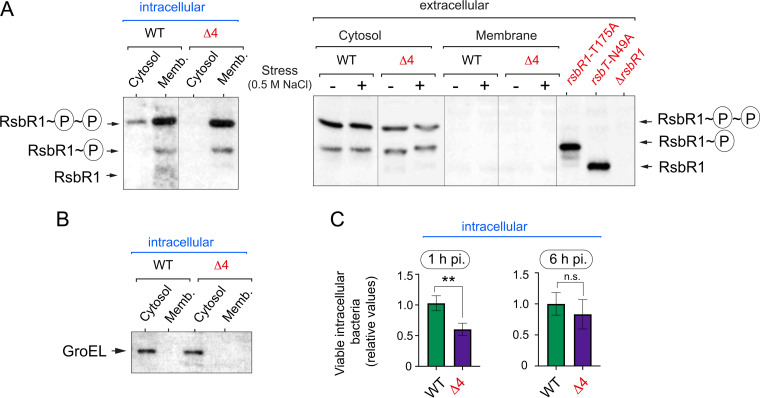
RsbR1 association with the membrane of intracellular L. monocytogenes is modulated negatively by its paralogues.
The RsbR1 paralogues RsbR2, RsbR3, and RsbL were recently shown to be enriched in membrane fractions of L. monocytogenes grown in brain heart infusion (BHI) medium and were proposed to compete with RsbR1 for association with the membrane (28). In that study, a mutant having no functional RsbR1 paralogues (named Δ4) showed higher expression of the Plmo2230::egfp reporter fusion than wild-type bacteria, regardless of stress. This difference, although less than 2-fold for the Δ4 mutant, was statistically significant (28). This increase in SigB activity was correlated with higher association of RsbR1 with the membrane and, as consequence, of the stressosome. Considering that we observed substantial RsbR1 association with the membrane in intracellular L. monocytogenes (Fig. 2A), we infected epithelial cells with the Δ4 mutant lacking functional RsbR1 paralogues. Unlike in wild-type bacteria, RsbR1 was not detected in the cytosol of Δ4 mutant cells (Fig. 6A). Control assays with the chaperone GroEL validated the correct preparation of membrane and cytosolic fractions (Fig. 6B). Interestingly, the Δ4 mutant exhibited an ∼2-fold lower invasion rate than wild-type bacteria, probably due to dysregulation of invasion factors controlled by SigB (Fig. 6C). However, no major differences were seen in the bacterial load at 6 hpi between the two strains (Fig. 6C), indicating that the Δ4 mutant could proliferate at a higher rate than wild-type bacteria inside host cells. Overall, these results reinforced the idea of RsbR1 paralogues fine-tuning the activity of the stressosome.
FIG 6.
Phosphorylated RsbR1 increases association with the membrane of intracellular L. monocytogenes in the absence of its paralogues. (A) A Phos-tag system was used for the analysis of phosphorylated RsbR1 in subcellular extracts prepared from intracellular L. monocytogenes EGD-e wild type (WT) and the Δ4 mutant growing inside JEG-3 epithelial cells at 6 hpi. In parallel, subcellular fractions from extracellular bacteria subjected to osmotic stress (0.5 M NaCl for 30 min) were analyzed. Control cytosolic samples from extracellular rsbR1-T175, rsbT-N49A, and ΔrsbR1 strains are included. (B) Loading control immunoblot with anti-GroEL for the samples prepared from intracellular bacteria. (C) Invasion (1 hpi) and proliferation (6 to 1 hpi) rates in JEG-3 epithelial cells of EGD-e wild-type and Δ4 strains. The results from three independent experiments are shown as ratios of the number of Δ4 to wild-type bacteria (the number of CFU from WT at 1 hpi was arbitrarily set to 1 and corresponded to 1.7 × 105 CFU; the number of CFU from WT at 6 hpi was 2.5 × 106). One-tailed P values are indicated by asterisks for comparison between wild-type and Δ4 bacteria by t test (**, P < 0.01; ns, not significant).
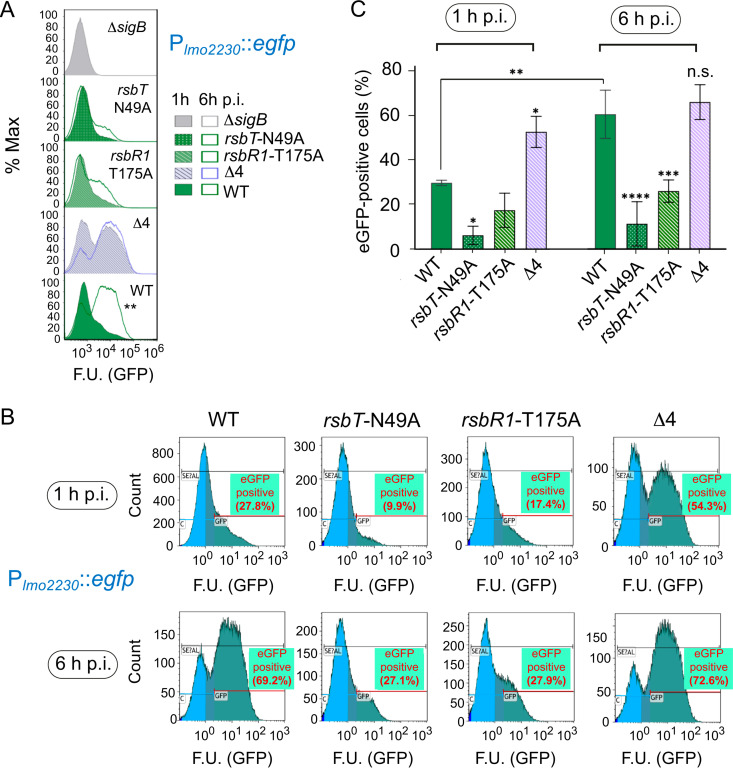
The RsbT kinase and RsbR1 paralogues are essential for adjusting SigB activation in intracellular L. monocytogenes.
The data obtained with the Δ4 mutant suggested that a proper partitioning of RsbR1 between the membrane and cytosol of intracellular L. monocytogenes could modulate the progressive increase of SigB activity observed with the Plmo2230::egfp reporter fusion (Fig. 1). If this hypothesis was true, an increase in SigB activity might be expected at all postinfection times in the absence of RsbR1 paralogues. This was the case for the Δ4 mutant, which showed increased SigB activity even at the earliest time measured, 1 h postinfection, with 54.3% of bacteria being positive for expression of the reporter versus only 27.8% positivity observed for wild-type bacteria (Fig. 7A to C). The rsbT-N49A and rsbR1-T175A mutants, which are defective in RsbT kinase activity and not capable of full phosphorylation of RsbR1, respectively, showed residual SigB activity at 6 hpi that was much lower than that in wild-type or Δ4 bacteria (Fig. 7A to C). These data proved that the SigB activation in intracellular L. monocytogenes requires perception of stress signals by the stressosome, involving RsbR1 phosphorylation at residue T175 by the kinase RsbT and attenuation of the output signal by RsbR1 paralogues.
FIG 7.
The lack of RsbR1 phosphorylation or the absence of RsbR1 paralogues alter the activation pattern of SigB during L. monocytogenes growth inside epithelial cells. Expression of the Plmo2230::egfp reporter fusion in the indicated strains at 1 h and 6 h postinfection of JEG-3 epithelial cells. (A) Side-to-side comparison of SigB activity from 1 h to 6 h of infection for each strain. (B) Representative experiment showing the percentage of eGFP-positive cells for the indicated mutants. (C) Quantification of eGFP-positive cells from three biological replicates. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant (one-way ANOVA with Tukey’s multiple-comparison test). F.U., fluorescence units.
DISCUSSION
The interplay between stress signal perception and virulence is fundamental in most bacterial pathogens. This is especially relevant in L. monocytogenes, a bacterium that inhabits multiple environments. Many studies have shown that two regulators, SigB and PrfA, act as master regulators in the stress and virulence cross talk (reviewed in references 6 and 10). The most accepted view of this interplay suggests opposite activities for these regulators, with SigB being dedicated to coping with “extracellular” stresses while PrfA is responsible for reprogramming gene expression upon perception of host cues, including those derived from the intracellular niche. However, this simple scenario does not reconcile all available information. In some instances, SigB facilitates the expression of PrfA, for example by increasing prfA transcription from the P2prfA promoter (30). Conversely, SigB has also been also proposed to reduce PrfA activity or levels by yet-unknown mechanisms (38). This “attenuation” of PrfA activity has been postulated to occur in the intestinal tract, where the PrfA regulon might be minimally expressed, with SigB predominantly driving the expression of listeriolysin LLO and the InlA and InlB invasins (6).
To our knowledge, no previous study had addressed whether L. monocytogenes exploits SigB to cope with stresses encountered within eukaryotic cells. Stress signals could be sensed within the phagosome due to its acidic environment, nutrient limitation, and/or the presence of reactive oxidants in this compartment. Stress may also occur in the cytosolic phase, in which massive proliferation of the pathogen could lead to bacterial crowding and nutrient starvation at late infection times. It is in this cytosolic phase that L. monocytogenes also spreads to adjacent cells and is subsequently enclosed within a double-membrane vacuole, which is ruptured to initiate a new intracellular growth cycle (3). Despite these potential sources of stress, the fact that the lack of SigB does not affect L. monocytogenes fitness inside host cells is intriguing (30). This lack of a phenotype associated with the loss of SigB in the in vitro infection model has been claimed as proof for the absence of a SigB-mediated response in intracellular L. monocytogenes. In our opinion, the data presented here challenge this view with experimental evidence for a gradual increase of SigB activity in intracellular bacteria.
Our claim is sustained by three pieces of evidence. First, the expression of Plmo2230::egfp reporter increases in the population of intracellular bacteria as the infection progresses, reaching a maximum at 6 h postinfection. This reporter expression is fully dependent on SigB, since it is abrogated in the ΔsigB mutant irrespective of the infection time measured (Fig. 1D). Although an early transcriptomic study identified lmo2230 as a gene that is putatively regulated by PrfA (39), this regulation might be indirect, since there is no PrfA box located in its upstream region. More recent transcriptome studies confirmed a SigB-dependent but PrfA-independent control of lmo2230 expression (40). Second, the SigB protein is detected by Western blotting at high levels in extracts of intracellular bacteria at 6 h postinfection (Fig. 2B); and, third, the core protein RsbR1, essential for stressosome function, is likewise detected in intracellular bacteria associated with the membrane and in a fully phosphorylated state (Fig. 2 and 4). These observations shed light on the frequently discussed interplay between SigB and PrfA in L. monocytogenes upon host contact, opening the possibility of PrfA activity being fine-tuned by SigB not only in extracellular locations, such as the gut, but also inside host cells as the intracellular infection progresses from the phagosomal to the cytosolic phase.
How this cross talk may take place in intracellular bacteria is of much interest for future work. For now, we can only speculate on mechanisms, which are technically challenging to demonstrate considering the scarce amounts of material that can be obtained from intracellular bacteria. For example, future experiments could address at different postinfection times whether there is correlation between the levels of PrfA and SigB produced by intracellular L. monocytogenes and whether the lack of one or the other results in compensatory effects. How PrfA orchestrates the production of the listeriolysin LLO and phospholipases in the phagosomal phase and further arrests such expression for stimulating expression of the actin-polymerizing protein ActA in the cytosolic phase is at present unknown and, based on the findings reported here, the putative contribution of SigB should be tested.
It is also important to note that a study based on transcriptome sequencing (RNA-Seq) and chromatin immunoprecipitation sequencing (ChIP-Seq) analyses showed that CodY, a regulatory protein that becomes active in bacteria starved for branched-chain amino acids (BCAA), binds directly to the upstream sigB region to repress its expression (41). In contrast, CodY was also reported to activate prfA expression from the P2prfA promoter (42), as SigB does (38). CodY has been postulated to sense the cytosolic niche during the intracellular proliferation phase, in which BCAA binding could result in repression of many metabolic and stress genes, including sigB (41, 42). However, a formal demonstration of CodY repressing sigB in intracellular L. monocytogenes is still lacking at both gene expression and protein levels. These apparently disparate data could be better integrated in future studies addressing in intracellular bacteria the dynamics of stressosome proteins and their phosphorylation status, as well as SigB protein levels in the presence/absence of CodY.
We believe that, taken together, our findings challenge the current view of SigB being active exclusively in extracellular L. monocytogenes by demonstrating a predominantly membrane-associated and fully phosphorylated stressosome core protein, RsbR1, that is detected when L. monocytogenes is intracellular. On the other hand, the apparent loss of RsbS in these bacteria is enigmatic, as there is no precedent for any in vivo study sustaining a stressosome complex that could be assembled in the absence of this core protein. We cannot rule out the possibility that the amount of RsbS decreases at such low levels in intracellular bacteria that it became undetectable by standard Western immunoassays. In the latter case, the RsbR1:RsbS stoichiometry, estimated to be 2:1 in stressosome isolated from bacteria (28, 43), should be necessarily compromised by such low levels of RsbS in intracellular L. monocytogenes. To the best of our knowledge, this study represents the first description at the protein level of the stressosome dynamics in L. monocytogenes and SigB activity in the intracellular eukaryotic environment. Future studies involving protein analyses in extracts of bacteria isolated from host cells will be fundamental for dissecting in detail the exact role that SigB plays in modulating virulence functions in the intracellular infection phase.
MATERIALS AND METHODS
Bacterial strains.
All L. monocytogenes strains used in this study are isogenic to the virulent parental wild-type EGD-e strain and are listed in Table 1. Construction and integration of the Plmo2230::egfp transcriptional reporter fusion are described elsewhere (36).
TABLE 1.
L. monocytogenes strains used in this studya
| Strain or genotype | Reference |
|---|---|
| EGD-e (wild type) | 46 |
| ΔsigB | 47 |
| ΔrsbR1 | 28 |
| rsbR1-T175A | 28 |
| rsbT-N49A | 28 |
| Δ4 (rsbL-C56A ΔrsbR2 ΔrsbR3 ΔrsbR4) | 28 |
| ΔrsbL | 48 |
| ΔrsbR2 | This study |
| Δprli42 | 18 |
| EGD-e pKSV7-Plmo2230::egfp | 36 |
| ΔsigB pKSV7-Plmo2230::egfp | 36 |
| rsbR1-T175A pKSV7-Plmo2230::egfp | 28 |
| rsbT-N49A pKSV7-Plmo2230::egfp | 28 |
| Δ4 pKSV7-Plmo2230::egfp | 28 |
All strains are isogenic to EGD-e.
Preparation of intracellular bacterial extracts.
JEG-3 epithelial cells from human placenta (ATCC HTB-36) were propagated in 500-cm2 Nunclon Delta treated square BioAssay dishes until reaching 80% confluence prior to bacterial infection. The cells were infected for 30 min with L. monocytogenes strains previously grown overnight at 37°C in static nonshaking conditions in brain heart infusion (BHI) medium (Becton Dickinson [BD]). The multiplicity of infection (bacteria to epithelial cells) was 10:1. Noninternalized bacteria were removed by three washes with prewarmed phosphate-buffered saline (PBS), pH 7.4, supplemented with 0.9 mM CaCl2 and 0.5 mM MgCl2. The infected cells were then incubated during 30 min in fresh Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 100 μg/ml gentamicin (Sigma-Aldrich). From 1 h postinfection, the cells were maintained in DMEM–10% (vol/vol) FBS containing 10 μg/ml gentamicin. At the required postinfection times, the cells were washed twice in PBS, pH 7.4, and lysed with a scraper in 20 ml of lysis solution (1% phenol, 19% ethanol, 0.4% SDS, 0.1 mg/ml DNase A) per cell culture plate, as described elsewhere (44). The solutions obtained from the lysis were spun down (30,000 × g, 30 min, 4°C), and the supernatant containing epithelial cells debris was discarded. The pellet of intracellular bacteria was washed in 2 ml of PBS, pH 7.4 (30,000 × g, 30 min, 4°C), and kept at −80°C.
Flow cytometry analysis of eGFP-expressing L. monocytogenes.
SigB activity was monitored with the Plmo2230::egfp reporter (28, 36). The analysis of cells expressing enhanced green fluorescent protein (eGFP) at 1 h and 2 h postinfection required the isolation of bacteria from ∼6 × 107 JEG-3 epithelial cells (two 500-cm2 cell culture plates), whereas the analyses at 4 and 6 h postinfection were of ∼1.2 × 108 cells. The pellet of intracellular bacteria was fixed with 4% (wt/vol) paraformaldehyde for 15 min at room temperature. Fixed cells were harvested by centrifugation (30,000 × g, 30 min, 4°C) and resuspended in 300 μl of filtered PBS, pH 7.4. Fluorescence at the single-cell level was quantified by flow cytometry with a Beckman Coulter Gallios analyzer with 488-nm blue laser excitation and 50,000 events recorded for each sample. The collected data were processed with Kaluza software version 2.1 (Beckman Coulter) to plot side and forward scatter values, the percentage of eGFP-positive cells, and the mean fluorescence values.
Bacterial invasion and proliferation rates in JEG-3 cells.
Epithelial JEG-3 cells were cultured in Nunc 24-well plates until 80% confluence (∼4 × 105 cells). At 1 h and 6 h postinfection, the infected cells were lysed in 100 μl of lysis solution (PBS [pH 7.4); 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS) to which 400 μl of PBS, pH 7.4, was added. The number of intracellular bacteria was determined by plating serial dilutions of host cell lysates on BHI agar plates and subsequent colony counting.
Subcellular fractionation and Western blot analysis of stressosome proteins.
The pellet of intracellular bacteria obtained from ∼3 × 108 JEG-3 epithelial cells (five 500-cm2 cell culture plates) was treated with the peptidoglycan hydrolase mutanolysin from Streptomyces globisporus (Sigma-Aldrich), as described previously (28). The protoplasts were resuspended in 100 μl PBS (pH 7.4), 1 μg/ml DNase A, and protease inhibitor cocktail and lysed by sonication. Unbroken cells were removed by centrifugation (20,000 × g, 10 min, 4°C), and the supernatant was subjected to ultracentrifugation (100,000 × g, 1 h, 4°C) to separate cytosol and membrane fractions. The pellet containing membrane proteins was washed with PBS, pH 7.4, by centrifugation (100,000 × g, 1 h, 4°C). The pellet was resuspended in 100 μl PBS, pH 7.4, to adjust the membrane extract to the same number of bacteria as in the cytosolic fraction. Cytosol and membrane fractions from exponential-phase bacteria were obtained as described previously (28, 45).
Identification of phosphorylated RsbR1 with the Phos-tag system.
Phosphorylated RsbR1 was identified in cytosolic and membrane extracts from intracellular bacteria using the phosphate-Zn2+ ion sequestration electrophoresis system with SuperSep Phos-tag precast gels (Fujifilm Wako Chemicals, U.S.A., Richmond, VA). In this system, the migration speed of phosphorylated proteins decreases due to the binding of the metallic ion, and the distinct phosphorylated isoforms are separated as different bands. Samples from exponential-phase bacteria were prepared as previously described (28).
Statistical analyses and densitometry.
Statistical significance was analyzed with GraphPad Prism v8.4.3 software (GraphPad Inc.). The analysis by t test was selected for the comparison of invasion and proliferation rates as well as for evaluating the distribution of the phosphorylated RsbR1 isoforms between cytosolic and membrane extracts. To compare the percentage of positive cells for expression of the Plmo2230::egfp reporter, the statistical analysis was carried out by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison tests. A P value of ≤0.05 was considered significant. Densitometry on bands obtained in immunoblot assays was performed using Image J, available from the National Institutes of Health, USA.
ACKNOWLEDGMENTS
We thank Pascale Cossart (Institut Pasteur, Paris, France) for providing the Δprli42 mutant and Jörgen Johansson (Umeå University, Sweden) for the ΔrsbL mutant. We also thank Henar González for technical assistance.
This work was supported by funds from the European Union’s Horizon 2020 Research-and-Innovation Program under Marie Sklodowska-Curie grant agreement no. 721456 (to F.G.D.P. and C.P.O.) and grant no. PGC2018-096364-B-I00 from the Spanish Ministry of Science and Innovation (to M.G.P.). Charlotte Dessaux and Duarte N. Guerreiro were supported by the Horizon 2020 Research-and-Innovation Program under Marie Sklodowska-Curie grant agreement no. 721456 as Early-Stage Researchers (ESRs).
We declare no competing interests.
Contributor Information
Francisco García-del Portillo, Email: fgportillo@cnb.csic.es.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, González-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NicAogáin K, O'Byrne CP. 2016. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front Microbiol 7:1865. 10.3389/fmicb.2016.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radoshevich L, Cossart P. 2018. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol 16:32–46. 10.1038/nrmicro.2017.126. [DOI] [PubMed] [Google Scholar]
- 4.Johansson J, Freitag NE. 2019. Regulation of Listeria monocytogenes virulence. Microbiol Spectr 7:GPP3-0064-2019. 10.1128/microbiolspec.GPP3-0064-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcari T, Feger M-L, Guerreiro DN, Wu J, O’Byrne CP. 2020. Comparative review of the responses of Listeria monocytogenes and Escherichia coli to low pH stress. Genes (Basel) 11:1330. 10.3390/genes11111330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaballa A, Guariglia-Oropeza V, Wiedmann M, Boor KJ. 2019. Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol Mol Biol Rev 83:e00034-19. 10.1128/MMBR.00034-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerreiro DN, Arcari T, O’Byrne CP. 2020. The σB-mediated general stress response of Listeria monocytogenes: life and death decision making in a pathogen. Front Microbiol 11:1505. 10.3389/fmicb.2020.01505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecker M, Pane-Farre J, Volker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 61:215–236. 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 9.Hecker M, Volker U. 2001. General stress response of Bacillus subtilis and other bacteria. Adv Microb Physiol 44:35–91. 10.1016/s0065-2911(01)44011-2. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Orsi RH, Gaballa A, Wiedmann M, Boor KJ, Guariglia-Oropeza V. 2019. Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Future Microbiol 14:801–828. 10.2217/fmb-2019-0072. [DOI] [PubMed] [Google Scholar]
- 11.Dufour A, Voelker U, Voelker A, Haldenwang WG. 1996. Relative levels and fractionation properties of Bacillus subtilis sigma(B) and its regulators during balanced growth and stress. J Bacteriol 178:3701–3709. 10.1128/jb.178.13.3701-9sigma.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Kang CM, Brody MS, Price CW. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev 10:2265–2275. 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 13.Dorey A, Marinho C, Piveteau P, O'Byrne C. 2019. Role and regulation of the stress activated sigma factor sigma B (σB) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv Appl Microbiol 106:1–48. 10.1016/bs.aambs.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Voelker U, Dufour A, Haldenwang WG. 1995. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of sigma B. J Bacteriol 177:114–122. 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang WG. 1995. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol 177:3771–3780. 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijay K, Brody MS, Fredlund E, Price CW. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol Microbiol 35:180–188. 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 17.Chaturongakul S, Boor KJ. 2006. SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol 72:5197–5203. 10.1128/AEM.03058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Impens F, Rolhion N, Radoshevich L, Becavin C, Duval M, Mellin J, Garcia Del Portillo F, Pucciarelli MG, Williams AH, Cossart P. 2017. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat Microbiol 2:17005. 10.1038/nmicrobiol.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marles-Wright J, Grant T, Delumeau O, van Duinen G, Firbank SJ, Lewis PJ, Murray JW, Newman JA, Quin MB, Race PR, Rohou A, Tichelaar W, van Heel M, Lewis RJ. 2008. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92–96. 10.1126/science.1159572. [DOI] [PubMed] [Google Scholar]
- 20.Pane-Farre J, Quin MB, Lewis RJ, Marles-Wright J. 2017. Structure and function of the stressosome signalling hub. Subcell Biochem 83:1–41. 10.1007/978-3-319-46503-6_1. [DOI] [PubMed] [Google Scholar]
- 21.Quin MB, Berrisford JM, Newman JA, Basle A, Lewis RJ, Marles-Wright J. 2012. The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling. Structure 20:350–363. 10.1016/j.str.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Eymann C, Schulz S, Gronau K, Becher D, Hecker M, Price CW. 2011. In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB. Mol Microbiol 80:798–810. 10.1111/j.1365-2958.2011.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CC, Yudkin MD, Delumeau O. 2004. Phosphorylation and RsbX-dependentdephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J Bacteriol 186:6830–6836. 10.1128/JB.186.20.6830-6836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaidenko TA, Yang X, Lee YM, Price CW. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J Mol Biol 288:29–39. 10.1006/jmbi.1999.2665. [DOI] [PubMed] [Google Scholar]
- 25.Akbar S, Kang CM, Gaidenko TA, Price CW. 1997. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol 24:567–578. 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim TJ, Gaidenko TA, Price CW. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J Bacteriol 186:6124–6132. 10.1128/JB.186.18.6124-6132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodbury RL, Luo T, Grant L, Haldenwang WG. 2004. Mutational analysis of RsbT, an activator of the Bacillus subtilis stress response transcription factor, σB. J Bacteriol 186:2789–2797. 10.1128/jb.186.9.2789-2797.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dessaux C, Guerreiro DN, Pucciarelli MG, O'Byrne CP, García-Del Portillo F. 2020. Impact of osmotic stress on the phosphorylation and subcellular location of Listeria monocytogenes stressosome proteins. Sci Rep 10:20837. 10.1038/s41598-020-77738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garner MR, Njaa BL, Wiedmann M, Boor KJ. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun 74:876–886. 10.1128/IAI.74.2.876-886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazmierczak MJ, Wiedmann M, Boor KJ. 2006. Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology (Reading) 152:1827–1838. 10.1099/mic.0.28758-0. [DOI] [PubMed] [Google Scholar]
- 31.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. 2003. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol 185:5722–5734. 10.1128/jb.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Marquis H, Boor KJ. 2005. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology (Reading) 151:3215–3222. 10.1099/mic.0.28070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori M-A, Soubigou G, Régnault B, Coppée J-Y, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 34.Raengpradub S, Wiedmann M, Boor KJ. 2008. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl Environ Microbiol 74:158–171. 10.1128/AEM.00951-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser KR, Sue D, Wiedmann M, Boor K, O'Byrne CP. 2003. Role of sigmaB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is σB dependent. Appl Environ Microbiol 69:2015–2022. 10.1128/aem.69.4.2015-2022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Utratna M, Cosgrave E, Baustian C, Ceredig R, O'Byrne C. 2012. Development and optimization of an EGFP-based reporter for measuring the general stress response in Listeria monocytogenes. Bioeng Bugs 3:93–103. 10.4161/bbug.19476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misra SK, Milohanic E, Ake F, Mijakovic I, Deutscher J, Monnet V, Henry C. 2011. Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11:4155–4165. 10.1002/pmic.201100259. [DOI] [PubMed] [Google Scholar]
- 38.Ollinger J, Bowen B, Wiedmann M, Boor KJ, Bergholz TM. 2009. Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect Immun 77:2113–2124. 10.1128/IAI.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milohanic E, Glaser P, Coppée J-Y, Frangeul L, Vega Y, Vázquez-Boland JA, Kunst F, Cossart P, Buchrieser C. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47:1613–1625. 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 40.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Bécavin C, Archambaud C, Cossart P, Sorek R. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 8:583. 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobel L, Herskovits AA. 2016. Systems level analyses reveal multiple regulatory activities of CodY controlling metabolism, motility and virulence in Listeria monocytogenes. PLoS Genet 12:e1005870. 10.1371/journal.pgen.1005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobel L, Sigal N, Borovok I, Belitsky BR, Sonenshein AL, Herskovits AA. 2015. The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol Microbiol 95:624–644. 10.1111/mmi.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams AH, Redzej A, Rolhion N, Costa TRD, Rifflet A, Waksman G, Cossart P. 2019. The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation. Nat Commun 10:3005. 10.1038/s41467-019-10782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunez-Hernandez C, Tierrez A, Ortega AD, Pucciarelli MG, Godoy M, Eisman B, Casadesus J, Garcia-del Portillo F. 2013. Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect Immun 81:154–165. 10.1128/IAI.01080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonquieres R, Bierne H, Fiedler F, Gounon P, Cossart P. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol Microbiol 34:902–914. 10.1046/j.1365-2958.1999.01652.x. [DOI] [PubMed] [Google Scholar]
- 46.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 47.Guerreiro DN, Wu J, Dessaux C, Oliveira AH, Tiensuu T, Gudynaite D, Marinho CM, Boyd A, García-Del Portillo F, Johansson J, O’Byrne CP. 2020. Mild stress conditions during laboratory culture promote the proliferation of mutations that negatively affect sigma B activity in Listeria monocytogenes. J Bacteriol 202:e00751-19. 10.1128/JB.00751-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiensuu T, Andersson C, Ryden P, Johansson J. 2013. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol Microbiol 87:909–924. 10.1111/mmi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]