ABSTRACT
Ulvan is an important marine polysaccharide. Bacterial ulvan lyases play important roles in ulvan degradation and marine carbon cycling. Until now, only a small number of ulvan lyases have been characterized. Here, a new ulvan lyase, Uly1, belonging to polysaccharide lyase family 24 (PL24) from the marine bacterium Catenovulum maritimum, is characterized. The optimal temperature and pH for Uly1 to degrade ulvan are 40°C and pH 9.0, respectively. Uly1 degrades ulvan polysaccharides in the endolytic manner, mainly producing ΔRha3S, consisting of an unsaturated 4-deoxy-l-threo-hex-4-enopyranosiduronic acid and a 3-O-sulfated α-l-rhamnose. The structure of Uly1 was resolved at a 2.10-Å resolution. Uly1 adopts a seven-bladed β-propeller architecture. Structural and site-directed mutagenesis analyses indicate that four highly conserved residues, H128, H149, Y223, and R239, are essential for catalysis. H128 functions as both the catalytic acid and base, H149 and R239 function as the neutralizers, and Y223 plays a supporting role in catalysis. Structural comparison and sequence alignment suggest that Uly1 and many other PL24 enzymes may directly bind the substrate near the catalytic residues for catalysis, different from the PL24 ulvan lyase LOR_107, which adopts a two-stage substrate binding process. This study provides new insights into ulvan lyases and ulvan degradation.
IMPORTANCE Ulvan is a major cell wall component of green algae of the genus Ulva. Many marine heterotrophic bacteria can produce extracellular ulvan lyases to degrade ulvan for a carbon nutrient. In addition, ulvan has a range of physiological bioactivities based on its specific chemical structure. Ulvan lyase thus plays an important role in marine carbon cycling and has great potential in biotechnological applications. However, only a small number of ulvan lyases have been characterized over the past 10 years. Here, based on biochemical and structural analyses, a new ulvan lyase of polysaccharide lyase family 24 is characterized, and its substrate recognition and catalytic mechanisms are revealed. Moreover, a new substrate binding process adopted by PL24 ulvan lyases is proposed. This study offers a better understanding of bacterial ulvan lyases and is helpful for studying the application potentials of ulvan lyases.
KEYWORDS: ulvan, ulvan lyase, polysaccharide lyase family 24, marine bacterium, substrate recognition, catalytic mechanism
INTRODUCTION
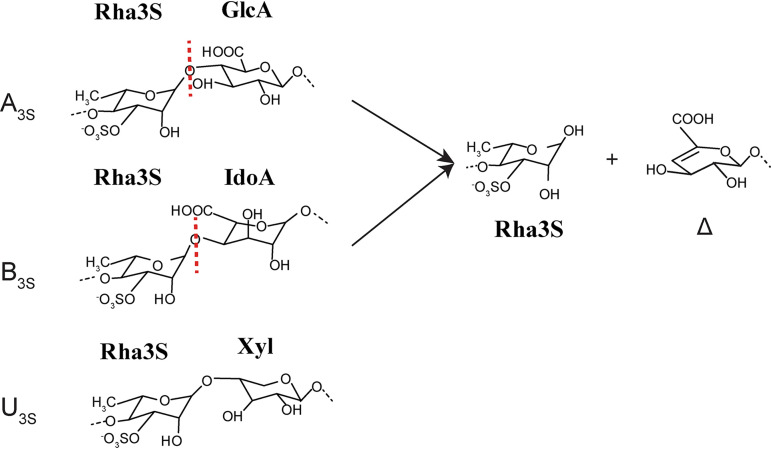
Marine macroalgae are major components of coastal ecosystems. Marine algal polysaccharides contain unique sugars, which are often heavily sulfated and differ substantially from terrestrial plant polysaccharides (1). Ulvan is the major cell wall polysaccharide of green algae of the genus Ulva, accounting for up to 29% of the dry weight (2, 3). Ulvan is a random polymer of 3-O-sulfated α-l-rhamnose (Rha3S), β-d-glucuronic acid (GlcA), α-l-iduronic acid (IdoA; the C-5 epimer of GlcA), and xylose (Xyl) (4). Based on the type of disaccharide unit, ulvan can be classified into one of three groups: that is, A3S, B3S and U3S. A3S consists of Rha3S and GlcA, B3S consists of Rha3S and IdoA, and U3S is composed of Rha3S and Xyl (Fig. 1). A3S and B3S are the common disaccharide repetitive units within ulvan polysaccharides, and U3S occurs in smaller amounts. Because of its bioactivity and chemical versatility, ulvan has application potentials in many areas, including food, medicine, and biomaterials (5–7).
FIG 1.
The disaccharide repetition units encountered in ulvan and the degradation product of ulvan lyases on ulvan. Three disaccharide repetition units, A3S, B3S, and U3S, exist in ulvan. Only A3S and B3S can be decomposed by ulvan lyases, leading to a reducing end on one fragment and an unsaturated ring (Δ, 4-deoxy-l-threo-hex-4-enopyranosiduronic acid) on the nonreducing end of the other fragment.
Ulvan lyases, first identified in 2011, can degrade ulvan, targeting the glycosidic 1,4 O-linkage between Rha3S and GlcA (A3S) or between Rha3S and IdoA (B3S), by a β-elimination mechanism with the formation of an unsaturated 4-deoxy-l-threo-hex-4-enopyranosiduronic acid (Δ) at the nonreducing end (Fig. 1) (8, 9). Until now, 17 ulvan lyases have been characterized (9), which are distributed into the polysaccharide lyase (PL) families 24, 25, 28, 37, and 40 in the Carbohydrate-Active enZYmes (CAZy) database (8–21). Of these, the structures of only three have been solved and characterized, namely, LOR_107 from Alteromonas sp. (the PL24 family) (18), PLSV_3936 from Pseudoalteromonas sp. (the PL25 family) (13), and NLR48 from Nonlabens ulvanivorans (the PL28 family) (19). The ulvan lyases LOR_107 from PL24 and PLSV_3936 from PL25 adopt the β-propeller scaffold, and the ulvan lyase NLR48 from PL28 adopts a β-jelly roll fold. The proposed catalytic mechanisms of these three lyases are significantly different. For LOR_107, the residue His146 is proposed to be both the catalytic base and acid aided by His167 and Tyr243 (18). PLSV_3936 adopts the typical His/Tyr mechanism, in which Tyr188 is most likely the catalytic base and His123 is most likely the catalytic acid (13). NLR48 uses different key catalytic residues, depending on the substrate at the +1 site. With GlcA at the +1 site, Tyr281 acts as the catalytic base and acid; with IdoA at the +1 site, Lys162 acts as the catalytic base and Tyr281 acts as the catalytic acid (19). Ulvan lyases play ecological roles in ulvan degradation in the ocean and have important applications in biotechnological fields, such as preparation of functional oligomers. Therefore, identification of more ulvan lyases and clarification of their characteristics, structures, and catalytic mechanisms will broaden our understanding of the degradation of ulvan polysaccharides.
The PL24 family currently contains four identified ulvan lyases in the CAZy database (LOR_61, LOR_107, PLSV_3875, and PLSV_3925). These are divided into two types: a short type (∼59 kDa) and a long type (∼110 kDa) (10). LOR_107 and PLSV_3875 belong to the short type, and LOR_61 and PLSV_3925 belong to the long type (10). The short-type ulvan lyases only contain the catalytic domain, whereas the long-type lyases comprise a catalytic domain and an unknown domain at the C terminus. Members of the PL24 family are known to specifically cleave the glycosidic bond between GlcA and Rha3S (A3S) by β-elimination, which differs from the other family ulvan lyases that can cleave both the glycosidic bonds of GlcA-Rha3S and IdoA-Rha3S (9). LOR_107 is the only enzyme with its structure reported in this family, adopting the β-propeller scaffold (18).
Of the 49 sequences of the PL24 family available in the CAZy database, 21 are from Catenovulum strains, indicating that Catenovulum is likely an important ulvan-degrading group. However, no ulvan lyases from Catenovulum have been characterized. Catenovulum maritimum Q1T is a marine bacterium isolated from the marine alga Porphyra yezoensis Ueda, collected from coastal areas of Weihai, China (22), which contains a putative PL24 ulvan lyase gene and a putative PL25 ulvan lyase gene. In this study, the putative PL24 ulvan lyase from Catenovulum maritimum Q1T, named Uly1, was characterized. Uly1 is a new ulvan lyase of the PL24 family, which is an endoenzyme with ΔRha3S as its main product. The three-dimensional structure of Uly1 was also solved at a 2.10-Å resolution, and the important residues in Uly1 for substrate binding and catalysis were analyzed. A comparison of the conformation of the catalytic canyon and the residues involved in substrate binding between Uly1 and the PL24 ulvan lyase LOR_107 suggests that Uly1 may adopt a different substrate binding process from that of LOR_107. These results provide new insights into ulvan lyases and ulvan degradation in the ocean.
RESULTS AND DISCUSSION
Sequence analysis of Uly1.
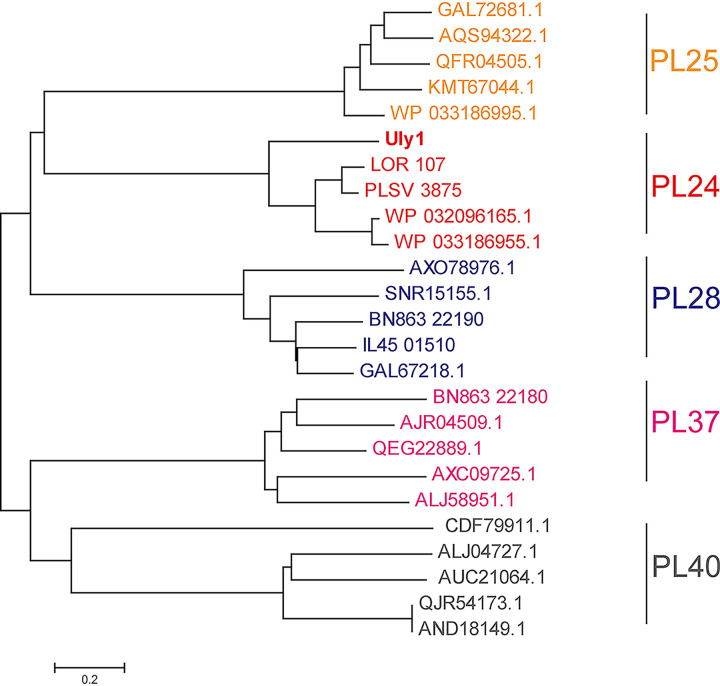
The uly1 gene of the marine bacterium Catenovulum maritimum Q1T is 1,539 bp in length and was predicted to encode a putative PL24 ulvan lyase (Uly1) of 512 amino acid residues. Uly1 contains a 20-residue signal peptide predicted by the SignalP 4.1 server and a catalytic domain (A21 to Q512). Phylogenetic analysis of Uly1 and other reported ulvan lyases suggests that Uly1 is a PL24 ulvan lyase (Fig. 2). Only 4 ulvan lyases have been characterized in the PL24 family. Of these, Uly1 has the highest sequence identity (50%) with the ulvan lyase LOR_107 (covering 94% of the Uly1 sequence) (18). Nevertheless, differences in the remaining 50% of the sequence leave open the possibility that Uly1 may be a new PL24 ulvan lyase with some different properties from the other characterized ulvan lyases. Therefore, we further characterized Uly1.
FIG 2.
Phylogenetic analysis of the ulvan lyases from the PL24, PL25, PL28, PL37 and PL40 families. The sequences are from the CAZy database, including all the characterized ulvan lyase sequences. The unrooted phylogenetic tree was constructed by neighbor joining with a Poisson model.
Expression and characterization of Uly1.
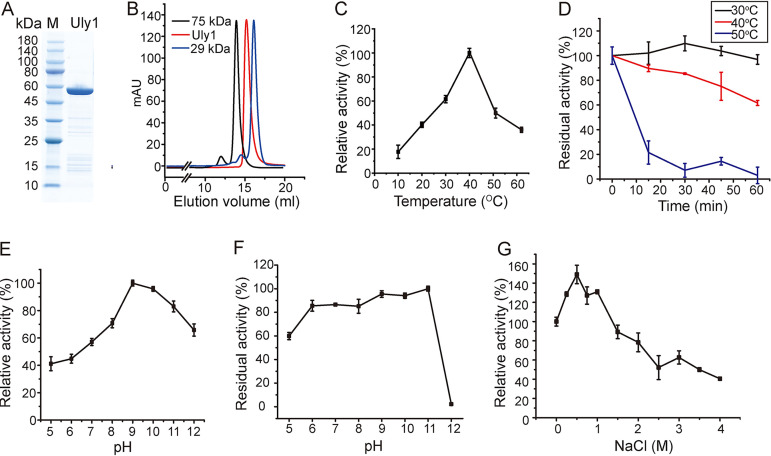
The gene of Uly1, without the signal peptide sequence, was overexpressed in Escherichia coli BL21(DE3). SDS-PAGE analysis showed that the recombinant Uly1 was approximately 55 kDa (Fig. 3A), consistent with its theoretical molecular mass of 55.8 kDa. Gel filtration analysis indicated that Uly1 is present as a monomer in solution (Fig. 3B). Because all the enzymes characterized in the PL24 family are ulvan lyases, the activity of Uly1 toward ulvan was measured along with three other polysaccharides, chondroitin sulfate, heparan sulfate, and alginate, to investigate its substrate specificity. Of these polysaccharides, the recombinant Uly1 was active only toward ulvan, with a specific activity of 20.16 ± 1.40 U/mg at 40°C, demonstrating that Uly1 is an ulvan lyase. The optimal temperature of Uly1 toward ulvan was 40°C (Fig. 3C). The thermostability of Uly1 was further investigated. Approximately 50% of the activity was lost after incubation at 40°C for 1 h, and approximately 70% of its activity was lost after incubation at 50°C for 15 min (Fig. 3D).
FIG 3.
Biochemical characterization of Uly1. (A) SDS-PAGE analysis of the purified recombinant Uly1. M, molecular mass marker. (B) Gel filtration analysis of the form of Uly1 in solution. Conalbumin (75 kDa; GE Healthcare) and carbonic anhydrase (29 kDa; GE Healthcare) were used as protein size markers. The predicted molecular mass of Uly1 without the signal peptide is 55.8 kDa (https://web.expasy.org/protparam/). (C) Effect of temperature on Uly1 activity. The highest activity of Uly1 at 40°C was taken as 100%. (D) Effect of temperature on Uly1 stability. Uly1 was incubated at 30, 40, and 50°C for 0 to 60 min. The residual activity was determined at 40°C and pH 9.0. The activity of Uly1 incubated at 4°C was taken as 100%. (E) Effect of pH on Uly1 activity. Experiments were performed at 40°C in Britton-Robinson buffer ranging from pH 5.0 to 12.0. The highest activity at pH 9.0 was taken as 100%. (F) Effect of pH on Uly1 stability. Uly1 was incubated in Britton-Robinson buffer ranging from pH 5.0 to 12.0 for 1 h. The residual activity was determined at 40°C and pH 9.0. The highest activity of Uly1 incubated at pH 11.0 was taken as 100%. (G) Effect of salinity on Uly1 activity. The activity of Uly1 at 0 M NaCl was taken as 100%. All experiments were repeated three times.
Uly1 exhibited maximal activity at pH 9.0 (Fig. 3E) and retained more than 80% of its activity after incubation at pH 6.0 to 11.0 for 1 h (Fig. 3F). The optimum pH of a majority of the characterized ulvan lyases ranged from 7.5 to 9.0 (9), which reflects their adaptation to the alkaline marine environment. In saline solutions containing 0 to 4 M NaCl, Uly1 exhibited its highest activity at 0.5 M NaCl, which was 1.5-fold higher than that at 0 M NaCl (Fig. 3G), indicating the adaptation of Uly1 to the salt concentration of the seawater. While no metal ions among Mg2+, Li+, K+, Ca2+, and Cu2+ had an enhancing effect on its activity, 1 mM Co2+ and Mn2+ reduced Uly1 activity by 53.40 and 91.44%, respectively (Table 1).
TABLE 1.
Effects of metal ions on Uly1 activity
| Compound | Relative activity (%)a |
|
|---|---|---|
| 0.25 mM | 1 mM | |
| Mg2+ | 102.71 ± 5.91 | 84.89 ± 5.72 |
| Li+ | 103.35 ± 8.53 | 97.18 ± 2.90 |
| K+ | 106.09 ± 9.57 | 91.94 ± 8.55 |
| Mn2+ | 60.68 ± 6.01 | 8.56 ± 9.38 |
| Ca2+ | 106.46 ± 2.16 | 83.41 ± 7.99 |
| Cu2+ | 105.21 ± 14.37 | 78.08 ± 4.72 |
| Co2+ | 99.83 ± 8.84 | 46.60 ± 5.05 |
The enzyme activity of Uly1 at 40°C and pH 9.0 without the addition of metal ion was taken as 100%. All experiments were repeated three times.
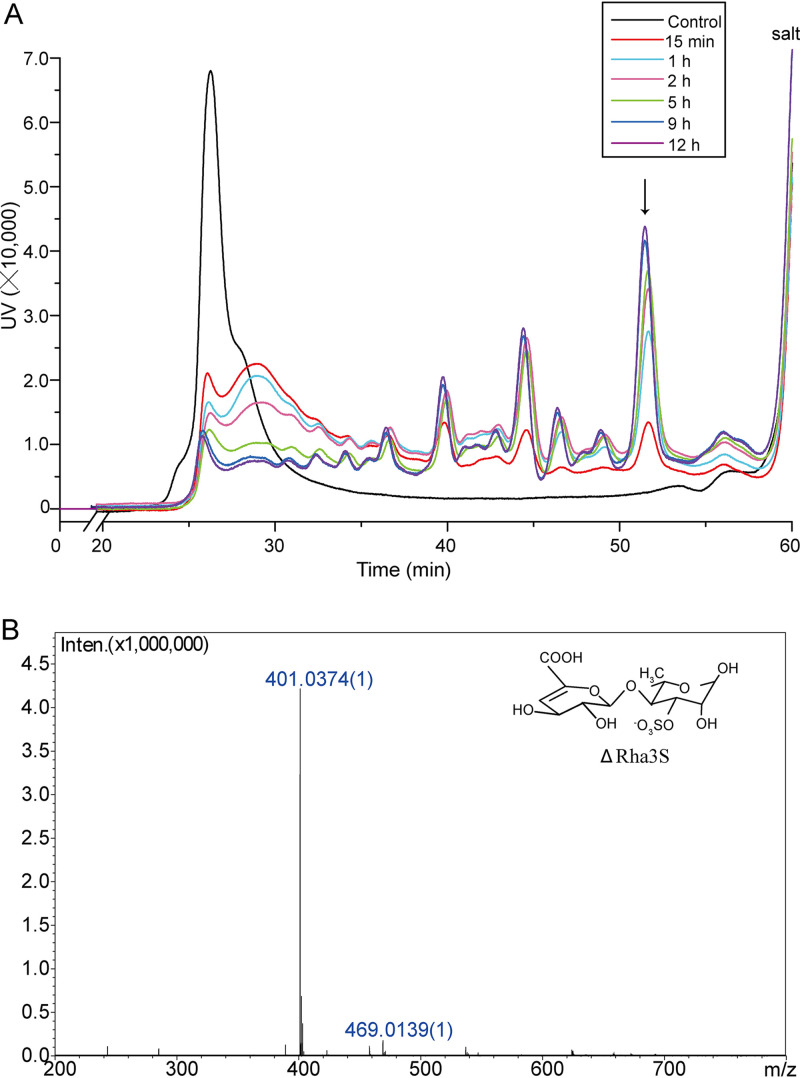
To determine the polysaccharide degradation pattern, the enzymatic products of Uly1 over a time course of 0 to 12 h were separated on a Superdex peptide 10/300 GL column. The products formed five large peaks and some small ones. These peaks displayed changes in amount but consistency in location with the increase in degradation time from 15 min to 12 h (Fig. 4A), indicating that Uly1 has several specific cleavage sites on ulvan and thus is an endolytic lyase. The peaks separated by gel filtration chromatography were further identified by negative electrospray ionization mass spectrometry (ESI-MS). Only the highest peak was successfully identified, with the others failing, probably due to the small amount and/or the impurity of these peaks. (One peak contains several products, and the amount of each is quite low.) The highest peak had an m/z value of 401, which is consistent with the molecular weight of the disaccharide ΔRha3S (Fig. 4B). This indicates that the main product released from ulvan by Uly1 is ΔRha3S.
FIG 4.
The enzymatic products of ulvan generated by Uly1 over a time course of 0 to 12 h. (A) Gel filtration analysis of the enzymatic products over a time course of 0 to 12 h. Gel filtration chromatography analysis was performed using a Superdex peptide 10/300 GL column monitored at a wavelength of 210 nm. Ulvan treated with Uly1 that was preinactivated by boiling was taken as the control. (B) ESI-MS analysis of the major peak indicated by an arrow in panel A. The major peak has an m/z value of 401, consistent with the molecular weight of the disaccharide ΔRha3S.
Kopel et al. reported that the PL24 ulvan lyases LOR_107, LOR_61, PLSV_3875, and PLSV_3925, with different molecular masses or sources of strains, all preferentially cleaved GlcA residues (10). Therefore, as a PL24 ulvan lyase, Uly1 may also preferentially cleave GlcA; however, this needs further confirmation.
Overall structure of Uly1.
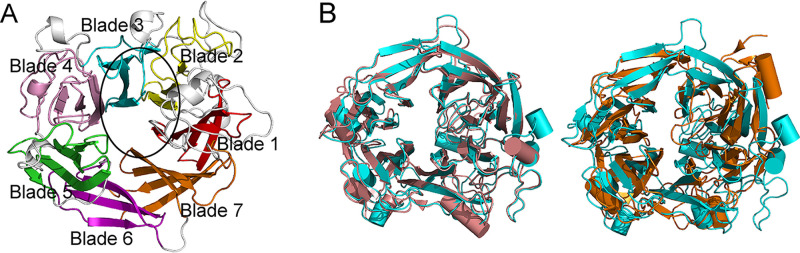
At the time the structure of Uly1 was solved, a similarity search of the Protein Data Bank (PDB) database showed that no suitable structure model could be used for Uly1 structure construction. Therefore, the crystal structure of the wild-type (WT) Uly1 was solved by the single-wavelength anomalous dispersion method using a selenomethionine (SeMet) derivative. Uly1 crystals belong to the P1211 space group, and the structure was solved at a resolution of 2.10 Å. The statistics for refinement are summarized in Table 2. Structure data show that each asymmetric unit contains two Uly1 molecules. Gel filtration analysis indicated that Uly1 is present as a monomer in solution (Fig. 3B), suggesting that the intermolecular contacts observed in the crystal originate from crystal packing. Uly1 adopts the fold of a seven-bladed β-propeller. Each propeller consists of four antiparallel β-strands (Fig. 5A). The catalytic center is located at the canyon on the top of the propeller (Fig. 5A). After the structure of Uly1 was solved, three ulvan lyase structures were successively reported, including those of PLSV_3936 from PL25 (13), LOR_107 from PL24 (18), and NLR48 from PL28 (19). The β-propeller structure of Uly1 is similar to those of PLSV_3936 and LOR_107. The root mean square deviation (RMSD) value (based on all amino acid residues in their structures) between Uly1 and PLSV_3936 is 3.37 Å, and that between Uly1 and LOR_107 is 1.32 Å. As shown in Fig. 5B, PLSV_3936 and Uly1 are significantly different in overall structure, and LOR_107 is the closest structural homologue. It has been reported that two Ca2+ ions are present in each independent molecule of the two LOR_107 molecules in the asymmetric unit at an identical position distant from the conserved catalytic residues, and they appear to play a strictly structural role (18). In the Uly1 structure, the electron density map indicates that there are many metal ions that are identified as Ca2+ based on their coordination geometry. However, these Ca2+ ions do not exist in exactly the same positions in the two molecules of an asymmetric unit. Inductively coupled plasma-optical emission spectrometry (ICP-OES) results showed that each Uly1 monomer contains only approximately 0.1 Ca2+. All these data suggest that the bound Ca2+ ions in the Uly1 crystal structure are most likely obtained from calcium acetate in the crystallization solution.
TABLE 2.
Diffraction data and refinement statistics of Uly1
| Parameter | Value fora: |
|
|---|---|---|
| Uly1 | SeMet Uly1 | |
| Data collection statistics | ||
| Space group | P1211 | P1211 |
| Unit cell dimensions | ||
| a, b, c (Å) | 58.10, 99.44, 95.00 | 57.99, 100.00, 95.30 |
| α, β, γ (°) | 90.00, 90.55, 90.00 | 90.00, 90.94, 90.00 |
| Wavelength (Å) | 0.9791 | 0.9791 |
| Resolution (Å) | 50.00−2.10 (2.14−2.10) | 50.00–1.80 (1.80–1.83) |
| Redundancy | 6.6 (6.5) | 6.8 |
| Completeness (%) | 99.1 (100) | 99.6 |
| Rmergeb | 0.22 (0.34) | 0.11 |
| Refinement statistics | ||
| Resolution range (Å) | 47.50−2.10 | 44.65−1.80 |
| Rwork (%) | 15.90 | 15.70 |
| Rfree (%) | 19.00 | 18.20 |
| B factor (Å2) | ||
| Protein | 26.20 | 17.31 |
| Solvent | 33.99 | 28.96 |
| Ligands | 39.28 | 26.89 |
| RMSD from ideal geometry | ||
| Length (Å) | 0.014 | 0.008 |
| Angles (°) | 1.19 | 0.86 |
| Ramachandran plot (%)c | ||
| Favored | 97.18 | 97.28 |
| Allowed | 2.82 | 2.72 |
Numbers in parentheses refer to data in the highest-resolution shell.
Rmerge=ΣhklΣi|I(hkl)i − 〈I(hkl)〉|/ΣhklΣi 〈I(hkl)i〉.
The Ramachandran plot was calculated by PROCHECK program in the CCP4i program package.
FIG 5.
Overall structure of Uly1. (A) Overall structure of Uly1 monomer. The seven blades of Uly1 structure are shown in different colors. The catalytic canyon is circled. (B) Structural alignment of Uly1 and the ulvan lyases LOR_107 and PLSV_3936. Uly1, LOR_107, and PLSV_3936 are colored in cyan, salmon, and orange, respectively.
Analysis of the important residues involved in catalysis.
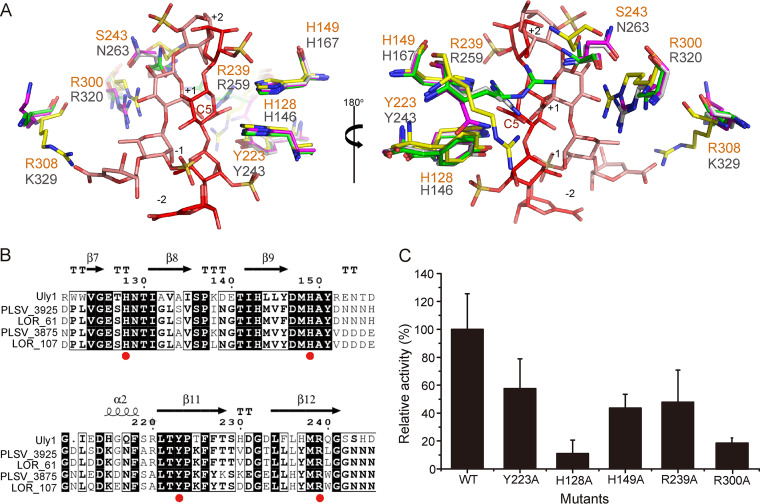
To investigate the crucial amino acid residues in Uly1, we attempted to crystallize different Uly1 mutants with their substrate or products, but this failed. Instead, we made a structural alignment of Uly1 and LOR_107 in complex with the ulvan tetrasaccharide (PDB code 6BYT) (Fig. 6A). The Uly1 residues R239, H149, H128, and Y223 are all close to the tetrasaccharide and are highly conserved in the PL24 ulvan lyases (Fig. 6A and B). The catalytic mechanism of LOR_107 has recently been revealed (18). In LOR_107, H146 plays the roles of both catalytic acid and base, R259 and H167 act as neutralizers of the charge of the GlcA acidic group, and Y243 plays a supporting role in properly orienting the imidazole ring of H146 (18). R239, H149, H128, and Y223 of Uly1 correspond to R259, H167, H146, and Y243 of LOR_107, respectively. To confirm the role of these residues in Uly1 catalysis, these four residues were mutated to alanine, and the activities of the mutants were measured. Based on the structural alignment, the basic amino acid residues H149 and R239 extend toward the carboxylic group of GlcA at the +1 position and can interact with it (Fig. 6A). Mutations on H149 and R239 decreased the activity of Uly1 by approximately 50% (Fig. 6C), indicating that H149 and R239 are likely to act as neutralizers to the negative charge of the +1 carboxyl group. H128 is close to the C-5 atom at subsite +1, and the glucosidic bond (Fig. 6A) and the mutation H128 to alanine almost completely abolished the enzymatic activity of Uly1 (Fig. 6C), indicating that H128 plays a crucial role in catalysis. As with H128, Y223 is also close to the glucosidic bond (Fig. 6A). However, when Y223 was mutated to alanine, the enzyme activity decreased only approximately 40% (Fig. 6C), suggesting that Y223 plays a supporting role in catalysis. Y223 may act as an alternative catalytic acid or help properly orient the imidazole ring of H128, as Y243 does in LOR_107 (Fig. 6A) (18). Therefore, in the catalysis of ulvan, H128 of Uly1 is most likely to be both the catalytic acid and base, H149 and R239 neutralize the charge of the GlcA acidic group, and Y223 plays a supporting role in catalysis.
FIG 6.
Analysis of the important residues in the active site of Uly1. (A) Structural alignment of the important residues of Uly1 and the ulvan lyase LOR_107. The amino acid residues in the structure of Uly1, wild-type LOR_107 structure, and the complex structures of the LOR_107 mutant (R259N) with tetrasaccharide (PDB code 6BYX) and LOR_107 mutant (R320N) with tetrasaccharide (PDB code 6BYT) are colored in yellow, gray, purple, and green, respectively. The bound tetrasaccharides in the complex structures of R259N and R320N occupying the −2 to +2 subsites are shown as salmon and red sticks, respectively. The residues of Uly1 and LOR_107 are labeled in orange and gray letters, respectively. (B) Sequence alignment of Uly1 and characterized PL24 ulvan lyases from the CAZy database. Identical and similar amino acid residues are shaded. Red dots indicate the residues involved in catalysis. The noncatalytic domains of the long ulvan lyases (ulvan lyases LOR_61 and PLSV_3925) are not included. (C) Enzymatic activities of WT Uly1 and its mutants toward ulvan. The activity of WT Uly1 was taken as 100%.
Comparison of the residues involved in the substrate binding of Uly1 with those of LOR_107.
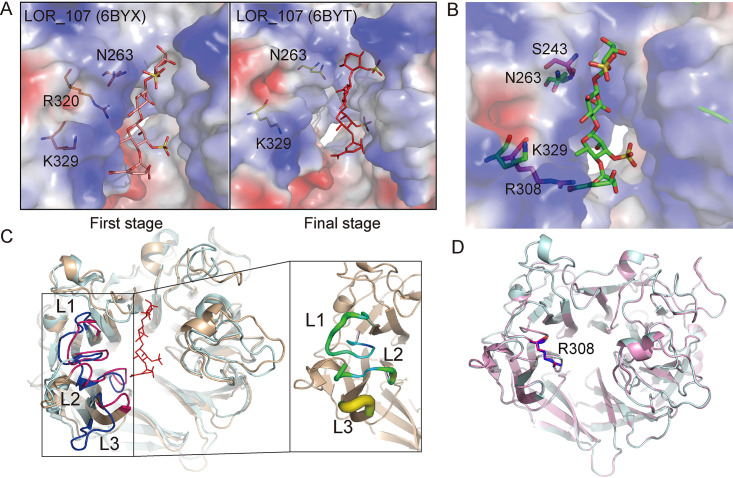
LOR_107 adopts a two-stage substrate binding process containing a substrate sliding event (18). During this process, N263 of LOR_107 plays a crucial role by moving its side chain, making the catalytic canyon narrower and pinning the substrate to the active site (Fig. 7A) (18). However, it was found that in Uly1, the side chain of S243, which spatially corresponds to N263 of LOR_107, is too short to fulfill the same function as N263 of LOR_107 (Fig. 7B). It was also found that the substrate-binding canyon of Uly1 is much narrower than that of LOR_107, mainly caused by three loops on one side of the active center. These loops (L1, residues 300 to 314; L2, residues 361 to 369; and L3, residues 389 to 392) are closer to the other side of the canyon in Uly1 than those in LOR_107 and show low flexibility, with relatively low B factors (Fig. 7C). R308 on the L1 of Uly1 occupies the position of the modeled carboxyl group of the −2 site GlcA in the structural alignment of Uly1 with the LOR_107 mutant in the complex with the ulvan tetrasaccharide (PDB code 6BYX) (Fig. 7B), leading to the inability of Uly1 to bind the saccharide to the bottom left of the catalytic canyon as LOR_107 does in its first binding stage. In addition, the conformations of R308 in the WT structure and the SeMet Uly1 structure, based on the crystals from different crystallization conditions, are alike, suggesting that the conformation of the side chain of R308 is relatively fixed, consistent with its low B factor (Fig. 7D). Based on these analyses, although the possibility that the conformation of the loops (including the conformation of the side chain of R308) changes during substrate binding cannot be excluded, it is less likely that Uly1 first binds the substrate to the left of the catalytic canyon via squeezing the loops, because this needs an uneconomical narrow-enlarge-narrow process during catalysis. Instead, it is more likely that the narrowed substrate-binding canyon allows Uly1 to directly bind the substrate near the catalytic residues, without the need to narrow the catalytic canyon to pin the substrate to the active site as LOR_107 does. Moreover, as shown in the analyses above, Uly1 lacks the key residue (like N263 of LOR_107) related to substrate sliding (Fig. 7B).
FIG 7.
Comparison of the residues involved in the substrate binding in Uly1 with those in LOR_107. (A) The two-stage substrate binding of LOR_107. The figures were generated based on the structures of LOR_107 (PDB codes 6BYX and 6BYT) (18). (B) Structural alignment of Uly1 and the LOR_107 mutant R259N with the tetrasaccharide substrate (PDB code 6BYX). The structure of Uly1 is shown as the electrostatic surface view. The side chains of the amino acid residues of Uly1 are shown as purple sticks. Based on the LOR_107 complex structure, N263, K329, and the bound tetrasaccharide of LOR_107 are modeled into the active center of Uly1 and shown as green sticks. (C) Comparison of the loops on the catalytic canyon of Uly1 with those of LOR_107. The overall structures of Uly1 and LOR_107 are shown in light blue and wheat, respectively. Loops (L1, L2, and L3) of Uly1 and LOR_107 are shown in pink and blue, respectively. In Uly1, L1 includes the residues 300 to 314, L2 includes the residues 361 to 369, and L3 includes the residues 389 to 392. The enlarged view shows the B factor of the three loops of Uly1. (D) Comparison of the side chains of R308 of Uly1 in the WT structure (blue) and the SeMet structure (pink).
In LOR_107, R320 coordinates the initial binding of the substrate (Fig. 7A) (18). When R320 of LOR_107 was mutated to alanine, the mutation resulted in a complete loss of the enzyme activity (18). However, when R300 in Uly1, corresponding to R320 in LOR_107, was mutated to alanine, the activity of the mutant was clearly detectable (remained ∼20%) (Fig. 6C), suggesting that R300 plays an important role in the catalysis of Uly1 but may not play a similar role as R320 does in LOR_107.
Altogether, based on these analyses, it is likely that Uly1 directly binds the substrate near the catalytic residues for catalysis, unlike LOR_107, which undergoes a two-stage substrate binding event in the catalytic process (18).
The proposed substrate binding and catalytic mechanisms of Uly1.
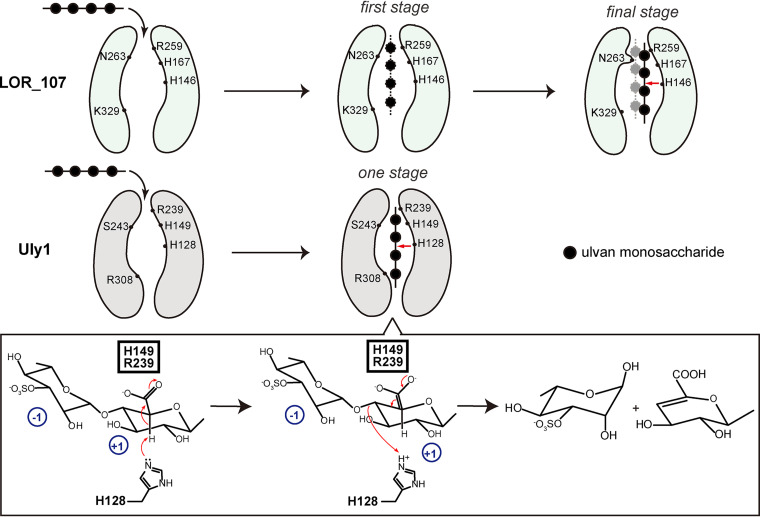
Based on the above structural and biochemical results, the possible substrate binding and catalytic mechanisms of Uly1 for ulvan depolymerization are proposed (Fig. 8). Uly1 possesses a narrower catalytic canyon than that reported for the PL24 ulvan lyase LOR_107. When the substrate approaches the catalytic canyon, the residues involved in substrate binding make the substrate directly bind near the catalytic residues. The key residues H128, H149, R239, and Y223 then mediate the catalytic reaction. H149 and R239 neutralize the negative charge on the carboxylic group of the +1 subsite, H128 acts as both the catalytic acid and base, and Y223 plays a supporting role in catalysis. Although the molecular mechanism of Uly1 catalysis is similar to that of LOR_107, the substrate binding process of Uly1 is likely significantly different from the reported two-stage process of LOR_107 due to differences in the width of the catalytic canyons and the arrangement of amino acids in the catalytic canyons of these two enzymes.
FIG 8.
Schematic diagram of the proposed substrate binding and catalytic mechanisms of Uly1. The reported substrate binding process of LOR_107 is a two-stage process. The catalytic canyon of LOR_107 is wide. The ulvan substrate is first bound to the left of the canyon at the first stage. Then, N263 moves its side chain, narrowing the canyon and locking the substrate to the active site, which is the final stage. However, the catalytic canyon of Uly1 is narrow, and especially, the long side chain of R308 (corresponds to K329 of LOR_107) further narrows one end of the catalytic canyon, resulting in the inability of Uly1 to bind the saccharide to the left of the canyon. Moreover, the side chain of S243 of Uly1, spatially corresponding to N263 of LOR_107, is too short to push the substrate to the other side of the catalytic canyon. Therefore, in Uly1, the substrate is most likely to be directly bound near the catalytic residues and then catalyzed by the catalytic residues. The red arrows indicate the cleavage site of the tetrasaccharide. During the catalytic process, H128 functions as both the catalytic acid and base, and H149 and R239 neutralize the negative charge of the +1 carboxyl. The glucuronic acid at the +1 subsite in the catalytic mechanism diagram is taken as a saccharide example.
Different substrate binding processes in the PL24 family.
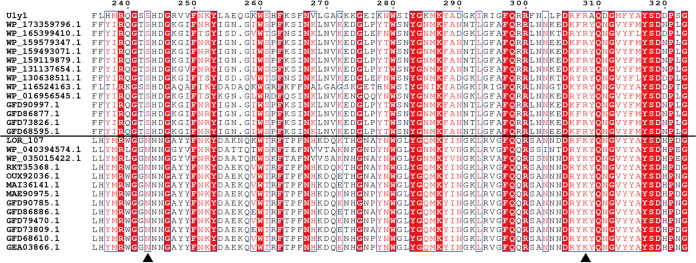
Among the ulvan lyase genes, genes encoding the PL24 family ulvan lyases are the most abundant. The PL24 family includes 50 ulvan lyase sequences, but only 5 (including Uly1) have been characterized. Results presented here suggest that Uly1 likely adopts a different substrate binding process from LOR_107. To investigate the universality of the two substrate binding processes in PL24 ulvan lyases, a sequence alignment of PL24 ulvan lyase sequences was performed. The results show that S243 and R308 of Uly1 are conserved in many PL24 ulvan lyases, and N263 and K329 of LOR_107 are conserved in many other PL24 lyases (Fig. 9). This implies that there are likely two types of substrate binding processes in the PL24 lyases. One type, represented by LOR_107, adopts a two-stage substrate binding process, while the other type, represented by Uly1, may directly bind the substrate near the catalytic amino acid residues for catalysis.
FIG 9.
Sequence alignment of Uly1 and the other PL24 lyases. S243 and R308 of Uly1 and the corresponding residues of the other PL24 lyases are indicated with black triangles.
Conclusions.
Many ulvan lyases secreted by marine bacteria are actively involved in marine ulvan degradation and therefore play important roles in marine carbon cycling. Until now, only a few ulvan lyases have been characterized, which hinders the comprehensive understanding of the degradation of marine ulvan polysaccharides. Herein, a new PL24 ulvan lyase, Uly1, with a single catalytic domain, was characterized in detail. It is an endolytic ulvan lyase with ΔRha3S as the main product. Uly1 adopts the fold of a seven-bladed β-propeller. Compared with the PL24 ulvan lyase LOR_107, which utilizes a two-stage process to bind ulvan for degradation, Uly1 and some other PL24 lyases likely directly bind the substrate near the catalytic amino acid residues for catalysis. The results in this study will provide a better understanding of the PL24 ulvan lyases and ulvan degradation and will be helpful for developing enzymatic tools for the utilization of green alga polysaccharides.
MATERIALS AND METHODS
Gene synthesis, cloning, and mutagenesis.
The 1,539-bp full-length sequence of the Uly1 gene (GenBank accession no. KMT65093.1) from the marine bacterium Catenovulum maritimum Q1T, which is available in the CAZy database, was synthesized by the Beijing Genomics Institute (China). The gene without the signal peptide was then subcloned into the pET-22b vector (TaKaRa, Japan) with a C-terminal His tag. Site-specific mutagenesis was carried out by a QuikChange site-directed mutagenesis kit (Agilent, USA) using the plasmid pET-22b-Uly1 as the template. All the recombinant plasmids were verified by sequencing.
Protein expression and purification.
Recombinant proteins of WT Uly1 and its mutants were overexpressed in E. coli BL21(DE3) (Vazyme, China) and were induced by the addition of 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 15°C for 16 h. Cells were collected and disrupted by sonication in buffer containing 50 mM Tris-HCl buffer (pH 8.0) and 100 mM NaCl. The SeMet derivative of Uly1 was overexpressed in E. coli BL21(DE3) in M9 minimal medium containing 100 mg/liter of lysine, phenylalanine, and threonine, 50 mg/liter of isoleucine, leucine, and valine, 5.2% (wt/vol) glucose, and 0.65% (wt/vol) yeast nitrogen base (YNB), which was then grown at 37°C. When the optical density at 600 nm (OD600) reached 0.6, the culture was cooled to 15°C and 50 mg/liter l-SeMet was added. Fifteen minutes later, the culture was incubated at 15°C at 100 rpm for 16 h under the induction of 0.4 mM IPTG.
The recombinant proteins were first purified by nickel-nitrilotriacetic acid resin (Qiagen, Germany) and then by gel filtration chromatography on a Superdex 200 column (GE Healthcare, USA) in buffer containing 10 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Carbonic anhydrase (29 kDa) and conalbumin (75 kDa) as the protein size standards from GE Healthcare were calibrated in the same buffer in the column.
Biochemical characterization of Uly1.
The concentrations of Uly1 and its mutants were determined by the bicinchoninic acid (BCA) protein assay kit (Thermo, USA), with bovine serum albumin (BSA) as the standard. The metal ions in Uly1 were quantified using ICP-OES (23). The activities of WT Uly1 and its mutants toward ulvan (purchased from Elicityl, France) were measured by the dinitrosalicylic acid (DNS) method (11). The reaction system was composed of 50 mM Tris-HCl buffer (pH 9.0), 10 mg/ml substrate, and 0.5 mg/ml enzyme. The reaction was carried out at 40°C for 30 min at a final volume of 200 μl and terminated by the addition of 100 μl DNS. The reaction mixture was boiled at 100°C for 10 min and immediately cooled on ice for 2 min. Then, the absorbance values were determined at 540 nm. The amount of reducing sugars released into the mixture was determined with rhamnose as the standard. One unit of enzyme activity is defined as the amount of enzyme required to release 1 nmol reducing sugars per min (24).
The optimum temperature for Uly1 activity was determined at a range of 10 to 60°C at pH 9.0. The optimum pH for Uly1 activity was determined at 40°C in Britton-Robinson (B-R) buffer ranging from pH 5.0 to 12.0. B-R buffer was prepared with boric acid, acetic acid, and phosphoric acid, all at a final concentration of 0.04 M in an aqueous solution and was adjusted to different pHs with 0.2 M NaOH. The effect of NaCl on Uly1 activity was determined at NaCl concentrations ranging from 0 to 4.0 M. The effects of selected metal ions on Uly1 activity were examined at pH 9.0 and 40°C at a final concentration of 0.25 or 1 mM.
The action mode and degradation products of Uly1 were analyzed by using ulvan as the substrate. The degradation reaction was carried out at 40°C for different times, from 0 to 12 h. The concentrations of the enzyme and substrate used were both 1 mg/ml. The ulvan degradation products were analyzed by gel filtration chromatography on a Superdex Peptide 10/300 GL column (GE Healthcare, USA) at a flow rate of 0.3 ml/min using 0.2 M ammonium hydrogen carbonate as the running buffer (25, 26). Elution was monitored at 210 nm using a UV detector. LabSolutions software was used for online monitoring and data analysis. The major peak was further identified by ESI-MS on an ion trap time of flight (TOF) hybrid mass spectrometer (LCMS-IT-TOF; Shimadzu, Japan). ESI-MS analysis was set in the negative-ion mode and with the following parameters: source voltage at 3.6 kV, nebulizer nitrogen gas flow rate at 1.5 liter/min, heat block, curved desolvation line temperature at 200°C, and detector voltage at 1.8 kV. The mass acquisition range was set at 200 to 800.
Crystallization and data collection.
The purified Uly1 protein was concentrated to ∼10 mg/ml in the buffer containing 10 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Crystals of WT Uly1 were obtained at 18°C using the sitting drop method in the buffer containing 200 mM calcium acetate hydrate (pH 6.5), 18% (wt/vol) polyethylene glycol 8000 (PEG 8000), and 100 mM sodium cacodylate trihydrate. Crystals of SeMet Uly1 were grown in the buffer containing 200 mM calcium acetate, 100 mM MES (morpholineethanesulfonic acid)-NaOH (pH 6.0), and 20% (wt/vol) PEG 8000. All the X-ray diffraction data were collected on the BL17U1 beamline at the Shanghai Synchrotron Radiation Facility using Area Detector Systems Corporation Quantum 315r. The initial diffraction data sets were processed by the HKL2000 program. Relevant data collection statistics are shown in Table 2.
Structure determination and refinement.
Heavy atoms were searched by SHELXD. The phase problems were solved by the single-wavelength anomalous diffraction (SAD) method using Phenix program Autosol (27). Initial model building was finished by Phenix program AutoBuild (27). Refinement of these structures was performed using Coot and Phenix alternately (27, 28). The quality of the final model is summarized in Table 2. All structure figures were generated using PyMOL software. The RMSD values were calculated using a tool on the PDB database (https://www.rcsb.org/alignment).
Data availability.
The atomic coordinates and structure factors of WT Uly1 (PDB code 7CZH) and SeMet Uly1 (PDB code 7DRQ) have been deposited in the Protein Data Bank.
ACKNOWLEDGMENTS
We thank the staffs of the BL18U1 and BL19U1 beamlines of National Facility for Protein Sciences Shanghai (NFPS) and the Shanghai Synchrotron Radiation Facility for assistance during data collection. We thank Caiyu Sun and Rui Wang from State Key laboratory of Microbial Technology of Shandong University for help and guidance with high-performance liquid chromatography.
F.X., F.D., and X.-H.S. performed all experiments. P.W., Y.-Z.Z., X.-L.C., H.-H.F., and X.-Y.Z. directed the experiments. F.X. and F.D. wrote the manuscript. H.-Y.C., P.W., and C.-Y.L. solved the structures and analyzed the data. Y.-Z.Z. and X.-L.C. designed the research. X.-L.C. and A.M. edited the manuscript.
This work was supported by the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the National Science Foundation of China (grants 91851205, 31630012, U1706207, 31870052, U2006205, and 31800107), the National Key Research and Development Program of China (2018YFC1406700 and 2018YFC0310704), the Program of Shandong for Taishan Scholars (tspd20181203), and the Development of Marine Economy Demonstration City Program during the 13th Five-Year Plan Period—Preparation of Active Marine Protein Feed Additives and Construction of Industrial Chain.
Contributor Information
Peng Wang, Email: wangpeng3331@ouc.edu.cn.
Xiu-Lan Chen, Email: CXL0423@sdu.edu.cn.
Ning-Yi Zhou, Shanghai Jiao Tong University.
REFERENCES
- 1.Zhu Y, Thomas F, Larocque R, Li N, Duffieux D, Cladiere L, Souchaud F, Michel G, McBride MJ. 2017. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ Microbiol 19:2164–2181. 10.1111/1462-2920.13699. [DOI] [PubMed] [Google Scholar]
- 2.Lahaye M, Robic A. 2007. Structure and functional properties of ulvan, a polysaccharide from green seaweeds. Biomacromolecules 8:1765–1774. 10.1021/bm061185q. [DOI] [PubMed] [Google Scholar]
- 3.Robic A, Gaillard C, Sassi JF, Lerat Y, Lahaye M. 2009. Ultrastructure of ulvan: a polysaccharide from green seaweeds. Biopolymers 91:652–664. 10.1002/bip.21195. [DOI] [PubMed] [Google Scholar]
- 4.Collen PN, Jeudy A, Sassi JF, Groisillier A, Czjzek M, Coutinho PM, Helbert W. 2014. A novel unsaturated beta-glucuronyl hydrolase involved in ulvan degradation unveils the versatility of stereochemistry requirements in family GH105. J Biol Chem 289:6199–6211. 10.1074/jbc.M113.537480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bikker P, van Krimpen MM, van Wikselaar P, Houweling-Tan B, Scaccia N, van Hal JW, Huijgen WJJ, Cone JW, Lopez-Contreras AM. 2016. Biorefinery of the green seaweed Ulva lactuca to produce animal feed, chemicals and biofuels. J Appl Phycol 28:3511–3525. 10.1007/s10811-016-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Wal H, Sperber BLHM, Houweling-Tan B, Bakker RRC, Brandenburg W, Lopez-Contreras AM. 2013. Production of acetone, butanol, and ethanol from biomass of the green seaweed Ulva lactuca. Bioresour Technol 128:431–437. 10.1016/j.biortech.2012.10.094. [DOI] [PubMed] [Google Scholar]
- 7.Cunha L, Grenha A. 2016. Sulfated seaweed polysaccharides as multifunctional materials in drug delivery applications. Mar Drugs 14:42. 10.3390/md14030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collen PN, Sassi JF, Rogniaux H, Marfaing H, Helbert W. 2011. Ulvan lyases isolated from the flavobacteria Persicivirga ulvanivorans are the first members of a new polysaccharide lyase family. J Biol Chem 286:42063–42071. 10.1074/jbc.M111.271825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Hu F, Zhu BW, Ni F, Yao Z. 2020. Insights into ulvan lyase: review of source, biochemical characteristics, structure and catalytic mechanism. Crit Rev Biotechnol 40:432–441. 10.1080/07388551.2020.1723486. [DOI] [PubMed] [Google Scholar]
- 10.Kopel M, Helbert W, Belnik Y, Buravenkov V, Herman A, Banin E. 2016. New family of ulvan lyases identified in three isolates from the Alteromonadales order. J Biol Chem 291:5871–5878. 10.1074/jbc.M115.673947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin HM, Xu PP, Guo QQ, Cheng XT, Gao DK, Sun DY, Zhu ZL, Lu FP. 2018. Biochemical characterization of a novel ulvan lyase from Pseudoalteromonas sp. strain PLSV. RSC Adv 8:2610–2615. 10.1039/C7RA12294B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He C, Muramatsu H, Kato S, Ohnishi K. 2017. Characterization of an Alteromonas long-type ulvan lyase involved in the degradation of ulvan extracted from Ulva ohnoi. Biosci Biotechnol Biochem 81:2145–2151. 10.1080/09168451.2017.1379352. [DOI] [PubMed] [Google Scholar]
- 13.Ulaganathan T, Boniecki MT, Foran E, Buravenkov V, Mizrachi N, Banin E, Helbert W, Cygler M. 2017. New ulvan-degrading polysaccharide lyase family: structure and catalytic mechanism suggests convergent evolution of active site architecture. ACS Chem Biol 12:1269–1280. 10.1021/acschembio.7b00126. [DOI] [PubMed] [Google Scholar]
- 14.Foran E, Buravenkov V, Kopel M, Mizrahi N, Shoshani S, Helbert W, Banin E. 2017. Functional characterization of a novel “ulvan utilization loci” found in Alteromonas sp. LOR genome. Algal Res 25:39–46. 10.1016/j.algal.2017.04.036. [DOI] [Google Scholar]
- 15.Konasani VR, Jin CS, Karlsson NG, Albers E. 2018. A novel ulvan lyase family with broad-spectrum activity from the ulvan utilisation loci of Formosa agariphila KMM 3901. Sci Rep 8:14713. 10.1038/s41598-018-32922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The Carbohydrate-Active Enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konasani VR, Jin CS, Karlsson NG, Albers E. 2018. Ulvan lyase from Formosa agariphila and its applicability in depolymerisation of ulvan extracted from three different Ulva species. Algal Res 36:106–114. 10.1016/j.algal.2018.10.016. [DOI] [Google Scholar]
- 18.Ulaganathan T, Helbert W, Kopel M, Banin E, Cygler M. 2018. Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism. J Biol Chem 293:4026–4036. 10.1074/jbc.RA117.001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulaganathan T, Banin E, Helbert W, Cygler M. 2018. Structural and functional characterization of PL28 family ulvan lyase NLR48 from Nonlabens ulvanivorans. J Biol Chem 293:11564–11573. 10.1074/jbc.RA118.003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisky L, Stanetty C, Mihovilovic MD, Schweder T, Hehemann JH, Bornscheuer UT. 2018. Biochemical characterization of an ulvan lyase from the marine flavobacterium Formosa agariphila KMM 3901T. Appl Microbiol Biotechnol 102:6987–6996. 10.1007/s00253-018-9142-y. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, Du CY, Chi YZ, Zuo SQ, Ye H, Wang P. 2019. Cloning, expression, and characterization of a new PL25 family ulvan lyase from marine bacterium Alteromonas sp. A321. Mar Drugs 17:568. 10.3390/md17100568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li DQ, Zhou YX, Liu T, Chen GJ, Du ZJ. 2015. Catenovulum maritimus sp. nov., a novel agarolytic gammaproteobacterium isolated from the marine alga Porphyra yezoensis Ueda (AST58-103), and emended description of the genus Catenovulum. Antonie Van Leeuwenhoek 108:427–434. 10.1007/s10482-015-0495-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang P, Chen XL, Li CY, Gao X, Zhu DY, Xie BB, Qin QL, Zhang XY, Su HN, Zhou BC, Xun LY, Zhang YZ. 2015. Structural and molecular basis for the novel catalytic mechanism and evolution of DddP, an abundant peptidase-like bacterial dimethylsulfoniopropionate lyase: a new enzyme from an old fold. Mol Microbiol 98:289–301. 10.1111/mmi.13119. [DOI] [PubMed] [Google Scholar]
- 24.Miller GL. 1959. Use of dinitrosalicylic acid reagent for detection of reducing sugars. Anal Chem 31:426–428. 10.1021/ac60147a030. [DOI] [Google Scholar]
- 25.Wang YJ, Jiang WX, Zhang YS, Cao HY, Zhang Y, Chen XL, Li CY, Wang P, Zhang YZ, Song XY, Li PY. 2019. Structural insight into chitin degradation and thermostability of a novel endochitinase from the glycoside hydrolase family 18. Front Microbiol 10:2457. 10.3389/fmicb.2019.02457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng YY, Wang DD, Gu JY, Li JG, Liu HH, Li FC, Han WJ. 2017. Biochemical characteristics and variable alginate-degrading modes of a novel bifunctional endolytic alginate lyase. Appl Environ Microbiol 83:e01608-17. 10.1128/AEM.01608-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The atomic coordinates and structure factors of WT Uly1 (PDB code 7CZH) and SeMet Uly1 (PDB code 7DRQ) have been deposited in the Protein Data Bank.