Abstract
In this paper, we present a new diffeomorphic registration algorithm for the registration of 3D models to 3D points. A biventricular template is iteratively fitted to the data by a series of implicitly constrained diffeomorphic linear least squares fits with decreasing regularization weights before performing an explicitly constrained diffeomorphic fit. The algorithm has been tested on a set of manual contours from 20 patients with a variety of congenital heart disease. Registration accuracy was assessed by calculating the mean point-to-point distance and the Dice overlap metric. Results showed that the method was able to accurately fit the biventricular model to 3D points and that the deformable model was able to fit all the pathologies while being diffeomorphic. The algorithm took approximately 5 minutes to fit each case, with an average of 52,580 points per case.
I. Introduction
Evaluation of cardiac function is indispensable for the diagnosis and treatment of any cardiovascular disease. Both left ventricular (LV) and right ventricular (RV) function are important to maintaining overall cardiovascular function. Using model-driven or model-based analysis, a prior or template is registered to the patient images by minimizing an objective function defined between the model parameters and the patient image. The segmentation process is therefore constrained by the model, avoiding errors due to the sparse nature of the image data. Both the LV and RV models can be expressed as a small number of parameters allowing simultaneous image processing and shape modelling. One such parameterization is called a subdivision surface introduced by Catmull and Clark [2], which has received increasing attention for cardiac modelling [15, 10, 12].
Subdivision surfaces are a mathematical tool for generating smooth surfaces by means of polygonal meshes. Legget et al. [10] and Sheehan et al. [15] developed a piecewise smooth subdivision surface algorithm to reconstruct both LV and RV geometries from manually traced contours in 3D ultrasound images. Shape-prior and physical constraints were integrated to allow sharp edges, which are commonly found at the mitral annulus and apex [9]. Stebbing et al. [17] have developed a morphable model of the RV using Loop subdivision surfaces. The model was expressed as a linear combination of a set of basis shapes and was able to model the RV by only using a four-parameter statistical model.
A key challenge when fitting a 3D model to a patient’s data, such as magnetic resonance images (MRI), is to satisfy a diffeomorphic constraint where the mapping between the model and the data is invertible, continuous and differentiable. Diffeomorphism preserves the topology of the model and prevents surface folding. A penalty function can be used to avoid surface folding [10, 15, 1], but it does not ensure a diffeomorphic transformation. Large Deformation Diffeomorphic Metric Mapping (LDDMM) [6] and Demons algorithm [18] are two popular diffeomorphic registration algorithms for medical images. In cardiac imaging, Mansi et al. [11] incorporated the information of incompressibility of the myocardium as a constraint within the diffeomorphic demon algorithm. De Craene et al. [4] also proposed a temporal diffeomorphic free form deformation framework incorporating incompressibility of the myocardium. The main drawback of LDDMM and Demon is a high computational time to complete the registration process. Another approach to diffeomorphic registration is based on B-splines control point displacements [3], which was applied to medical images by [14].
We have developed a registration framework by which 3D diffeomorphic displacement fields were generated, ensuring both anatomically realistic results and minimal registration error. This algorithm deforms a finite element model under the combination of implicit and explicit diffeomorphic constraints. Each transformation is guaranteed to be bijective by preventing the determinant of the Jacobian matrix of each element from falling below zero and by limiting the displacement of the coarse mesh within each iteration under explicit constraints. We validated the flexibility and accuracy of this algorithm on 20 patients with cyanotic and acyanotic congenital heart disease (CHD).
II. Methods
A. Template
Our biventricular model was comprised of the LV, the RV and four valves: aortic, mitral, tricuspid and pulmonary (see Fig. 1). At the coarse level, the model consisted of 388 vertices , of which 22 were extraordinary [7]. We modified the model by adding 25 elements using Blender1 to allow more flexibility around the tricuspid corner and the pulmonary valve. Specific knowledge of the final heart shape was incorporated in the coarse model with sharp edges around the valve and at the RV insertion points [9]. The final surface model was obtained after subdividing the coarse mesh twice using the Catmull-Clark subdivision method [2, 5, 16], resulting in 5,806 vertices.
Figure 1.
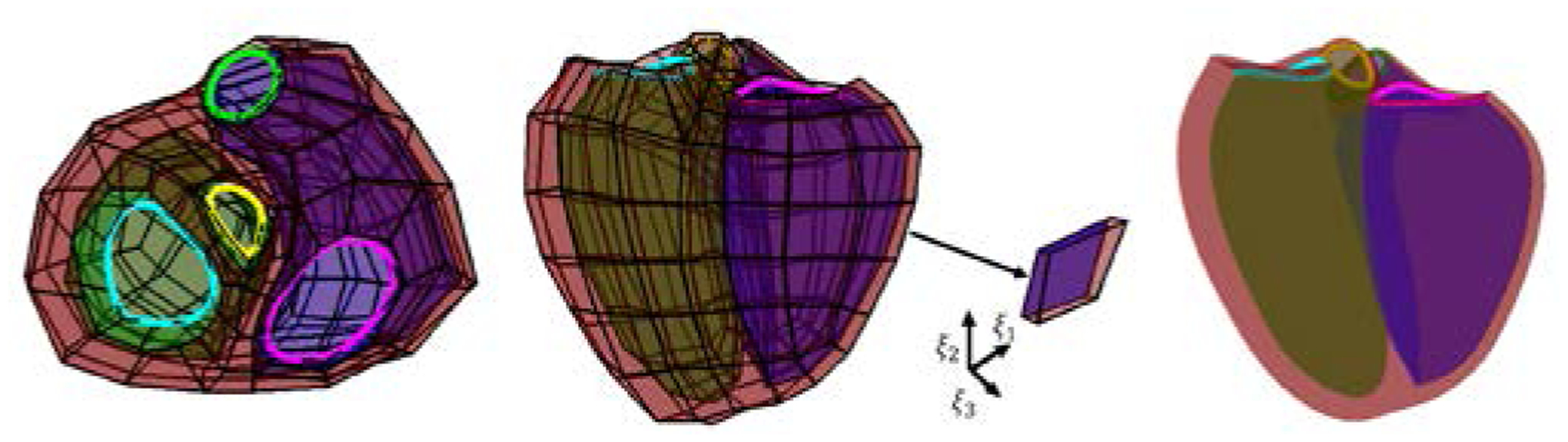
Biventricular coarse mesh (left: basal-apex view and middle: longitudinal view) and its Catmull Clark subdivision (right) with an element extracted showing interpolation directions (ξ1, ξ2, ξ3). Green surface: LV endocardium, blue surface: RV endocardium, red surface: epicardium. The mitral valve is shown in cyan, the aortic valve in yellow, the tricuspid valve in magenta and the pulmonary valve in green.
Catmull-Clark surfaces are a generalization of uniform B-splines and each surface point can be expressed as a linear combination of the coarse mesh, weighted by cubic B-spline coefficients using Stam’s parameterization [16]. Let (u,v) ∈ [0,1] be a location on an element of the coarse mesh. The corresponding surface point is parameterized by
| (1) |
where bi(u,v) is a bicubic B-spline basis function evaluated at (u,v) and are the control points. The coarse points and subdivided points were used to generate a B-splines finite element mesh (FEM). The FEM consisted of 187 elements with the local coordinate system defined by (ξ1, ξ2,ξ3). Each element was defined using Bi-cubic spline interpolation along the circumferential ξ1 and longitudinal direction ξ2, and a linear interpolation along the transmural outward direction ξ3. These interpolation directions are shown in Figure 1. Each surface point with the Cartesian coordinates (x,y,z) was expressed as a linear combination of the coarse points V0 as:
| (2) |
B. Implicitly constrained linear least-squares optimization
Diffeomorphic registration is a time consuming process. To increase the speed of our algorithm, a series of implicitly constrained stiff linear least-square fits were first performed in order to provide a closer solution to the data before performing a constrained diffeomorphic fit. The model was fitted to the data by minimizing the following objective function:
| (3) |
where are the data points, x(ξg) are the surface points at element coordinates ξg corresponding to the closest point on the model. Ereg is a regularization term. In this work, a D-Affine regularization term [8] was used to penalize deviation in strain from affine transformation of the prior and to ensure smooth and physically plausible deformation fields. Hence, Ereg is defined as:
| (4) |
where ||·||F is the Frobenius norm, ηk the smoothing weight in the kth direction and J the Jacobian of the transformation. To discourage self-interaction and folding, the initial smoothing weights used were η1 = η2 = η3 = 1010. To make sure that an iteration does not lead to a nonbijective transformation, implicit constraints based on the determinant of the Jacobian matrix J were added. The sign of the determinant of J gives information about the mapping from the reference template shape (χ1, χ2, χ3) to the patient-shaped model. The deformation, or displacement, from the template’s shape is u = x − χ. Therefore, J is given by:
| (5) |
Where uk is the displacement of the B-spline patch point and can be expressed as a linear combination of the displacement of the coarse mesh and bk, the basis functions. At each iteration, a least-square fit was performed and the determinant of the Jacobian matrix on 4×4×4 Gaussian quadrature points per element was calculated. If all values were positive, the subdivision surface was deformed by updating its control points, x(ξg) were recalculated and ηk were decreased. As long as the deformation is diffeomorphic, the smoothing weights are decreased. If one or more intersections were detected, the model was not updated and explicit diffeomorphic constraints were added.
C. Optimization with explicit diffeomorphic constraints
Minimizing (3) only with D-affine regularization does not lead to diffeomorphic transformations if ηk is too small, as seen Fig. 2. However, in order to fit a large variety of shapes, a highly deformable model and therefore a small ηk is required while keeping D-Affine constraints. Rohde et al. [13] derived sufficient conditions for bijectivity based on the magnitude of the derivatives of displacement. The sufficient condition to guarantee positivity of the Jacobian determinant was derived from Neumann series arguments. They showed that the minimum Jacobian determinant is positive if the magnitude of each gradient is bounded by . Therefore, the following equation assures the bijectivity of the transformation:
| (6) |
where can be pre-calculated for each Gaussian quadrature point for which we want to check det(J). is the scale factor between the template and the deformed model. Equation 3 can be then replaced by a quadratic programming problem with linear constraints. As the maximum displacement was constrained by Equation 6, an iterative process was used. At each iteration the optimization was performed and the model was updated. The scale factor s was then recalculated and the solution found was used as an initial guess for the next iteration. Weights along ξ1, ξ2 and ξ3 were set to 104. The optimization library IBM ILOG CPLEX Optimization Studio2 was used. Fig. 3 presents the overview of the algorithm.
Figure 2.
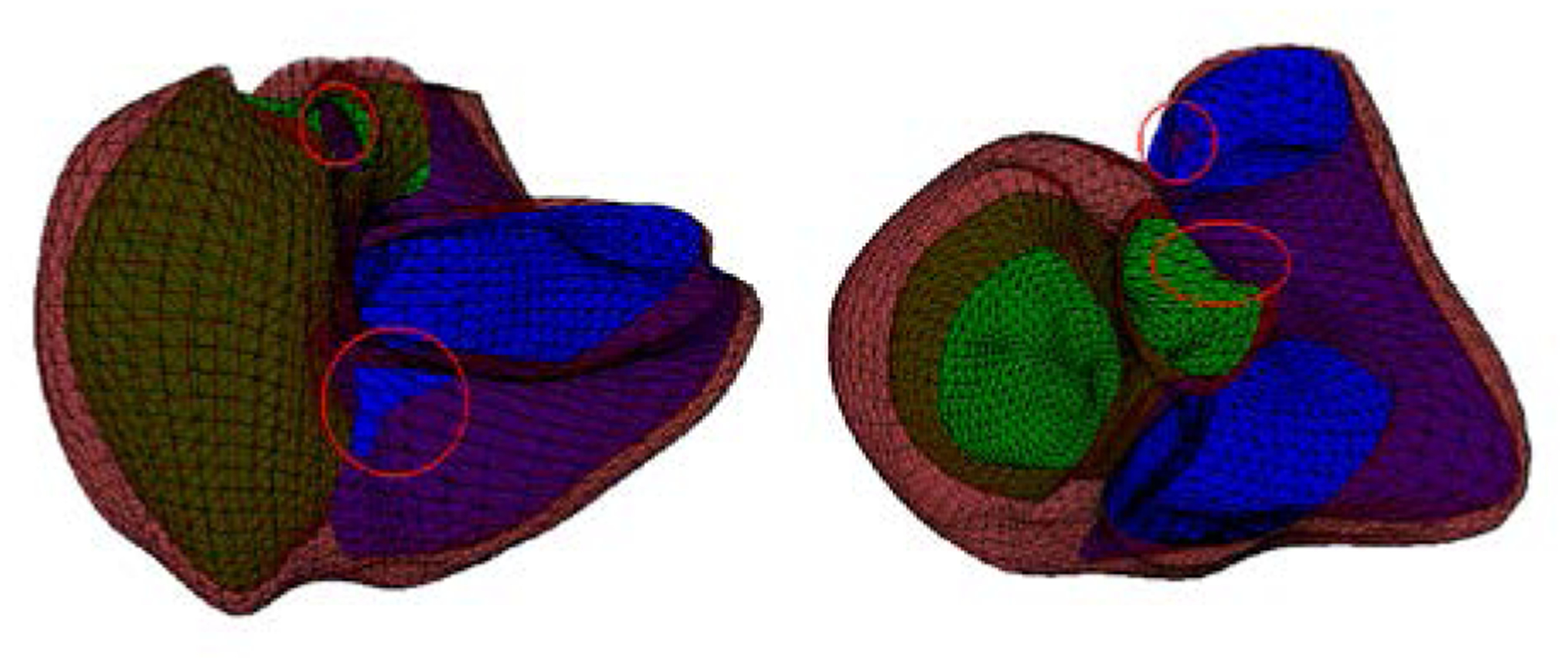
Linear least squares optimization without diffeomorphic constraints for two cases. Surface intersections are circled in red
Figure 3.
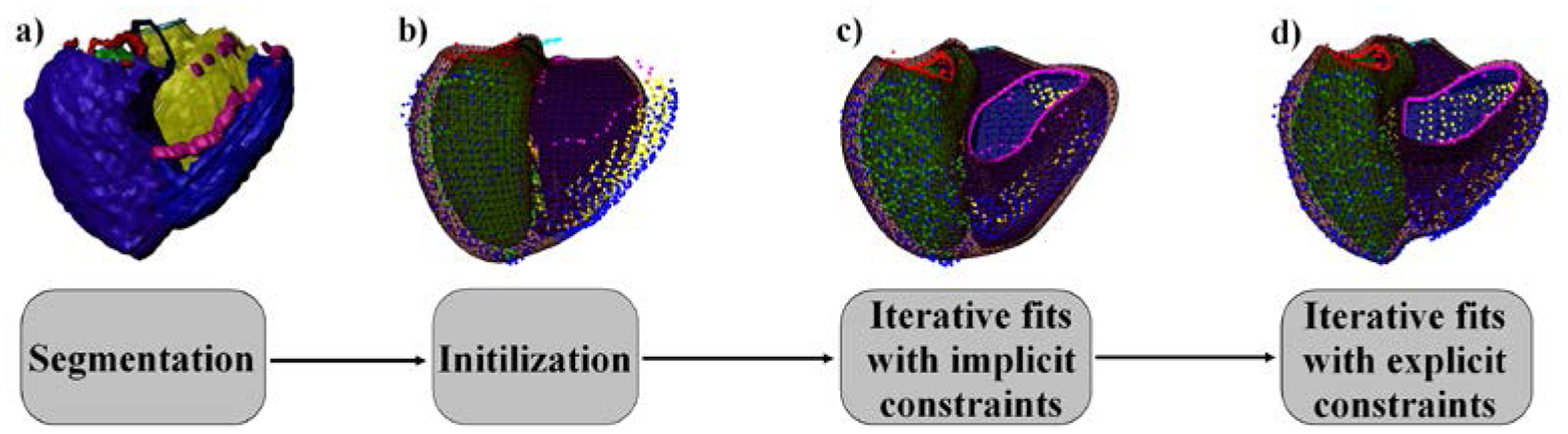
Algorithm overview. LV chamber: green, RV chamber: yellow, epicardium: blue. The mitral valve is shown in red, the aortic valve in black, the tricuspid valve in magenta and the pulmonary valve in cyan
III. Results
A. Data
We validated the proposed algorithm on 3D MRI acquisition of 20 patients with congenital heart disease, including three transposition of the great arteries, five tetralogies of Fallot, one Ebstein’s anomaly, two Marfan’s Syndromes, three bicuspid aortic valves, four coarctations and two double outlet right ventricles. To get an accurate valve orientation, the 3D MR images were re-oriented using Syngo.via (Syngo MR 2004V, Siemens Healthcare, Erlangen, Germany), which produced a range of parallel images to the aortic and the pulmonary valves. Manual segmentation was performed using ITK-Snap [19]. These 3D segmentations were loaded into Blender and separated into surfaces to be projected onto the template. Fig. 4 shows an example of the 3D segmentation image with surface labelling.
Figure 4.
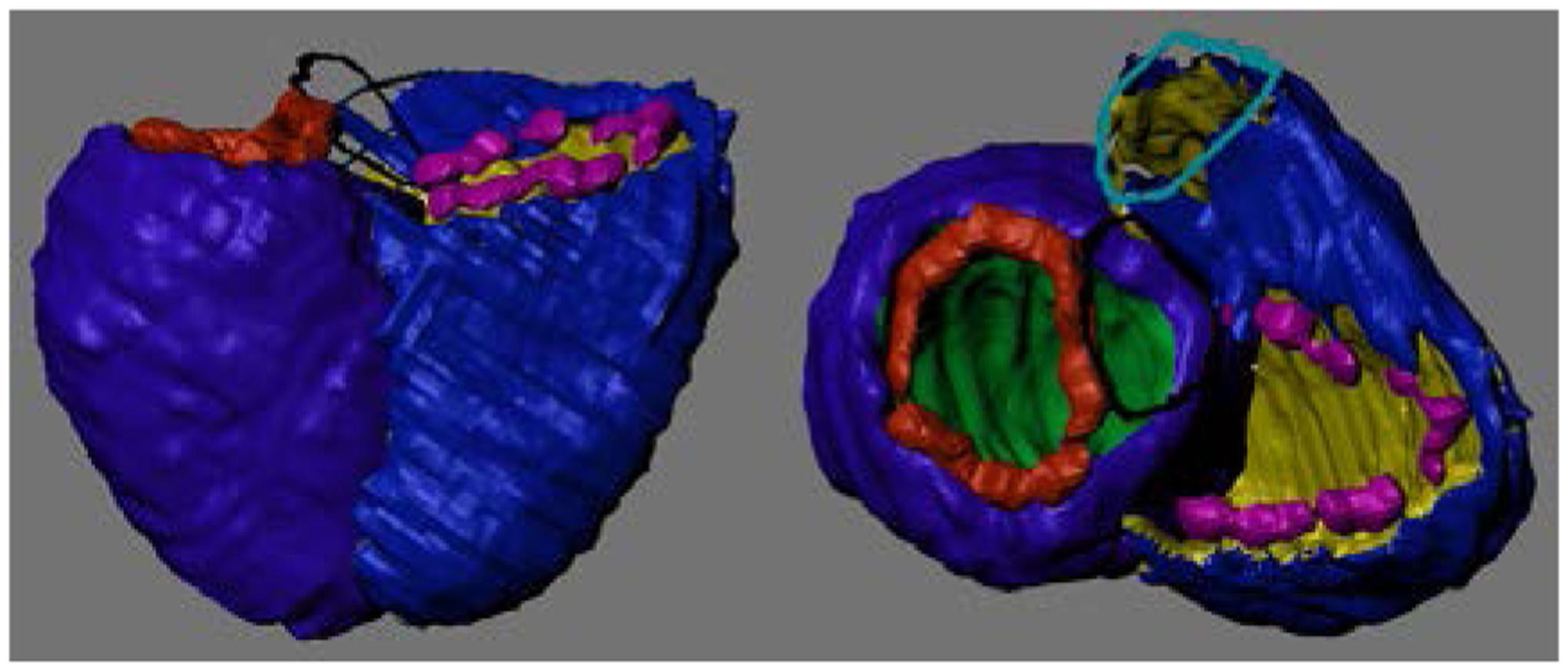
3D segmentation of a heart. The epicardium is shown in blue, RV cavity in yellow, LV cavity in green, pulmonary valve in light blue, tricuspid valve in pink, mitral valve in red and aorta in black
B. Accuracy of registration
Registration accuracy was assessed by using the mean point-to-point distance and the Dice overlap metric. The point-to-point distance was calculated as the distance between a data point and its closest vertex on the surface. Table I lists the mean point-to-point distances and the Dice metric for the RV chamber, the LV chamber and the epicardium. The Dice overlap metric was over 0.88 for each surface, demonstrating a good overlap after fitting. The mean point-to-point distance was smaller than 2 mm, similar to [17]. Fig. 5 shows the results of three different CHD patients.
TABLE I.
Evaluation of the registration accuracy
| Mean point-to-point distance (mm) | Dice metric | |
|---|---|---|
| LV chamber | 1.38 ± 0.18 | 0.95 ± 0.014 |
| RV chamber | 1.55 ± 0.22 | 0.92 ± 0.014 |
| Epicardium | 1.66 ± 0.18 | 0.88 ± 0.024 |
Figure 5.
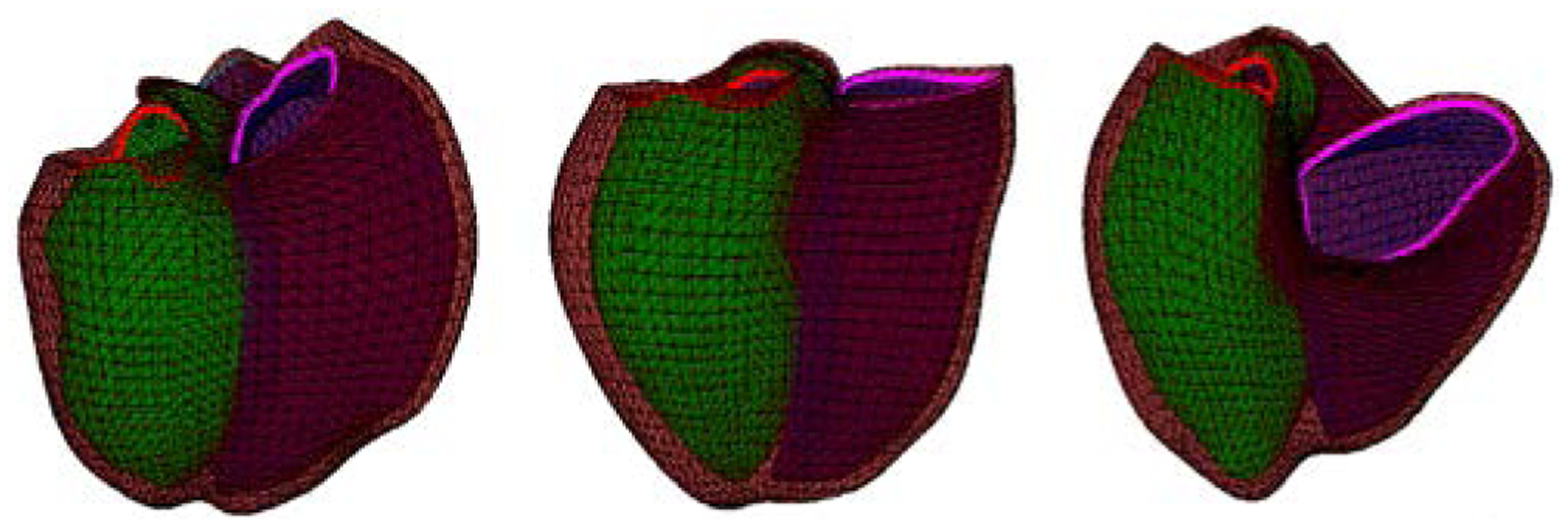
Fit to CHD patient anatomy. Left: Tetralogy of Fallot. Middle: Marfan’s Syndrome, Right: Ebstein’s anomaly
IV. Discussion and conclusion
We have proposed a method for generating diffeomorphic deformation fields. This method is based on the decomposition of deformations, preventing the determinant of the Jacobian matrix falling below zero. Each transformation is guaranteed to be bijective by limiting the displacement of the coarse mesh within each iteration. Topology-preserving registration is a time consuming process. Furthermore, constraining the magnitude of the displacement does not necessarily allow large deformations. Although we added a series of least-square fits compared to the original diffeomorphic fit, the algorithm showed a fast convergence, with an average processing time of 4.8 ± 2.4 minutes (implemented with python programming language) and an average of 52,580 ± 1,463 points to fit. The succession of least-square fits has an important effect on the rate of convergence as it provided a good initialization at a lesser cost. Our results have shown that this technique was able to achieve good registration accuracy while generating diffeomorphic displacement fields. The surface modelling also achieved a good performance as it was able to fit all the CHD pathologies present in our dataset.
Disclosure statement
AM and JO are co-founders of and have an equity interest in Insilicomed, and AM has an equity interest in Vektor Medical. AM and JO serve on the scientific advisory board of Insilicomed, and AM as scientific advisor to both companies. Some of their research grants have been identified for conflict of interest management based on the overall scope of the project and its potential benefit to these companies. The authors are required to disclose this relationship in publications acknowledging the grant support, however the research subject and findings reported here did not involve the companies in any way and have no specific relationship with the business activities or scientific interests of either company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
This work was supported by the New Zealand Heart Foundation Grant 1695 and NHLBI R01HL121754
Footnotes
References
- [1].Cashman Thomas J., and Fitzgibbon Andrew W.. “What shape are dolphins? building 3d morphable models from 2d images.” IEEE transactions on pattern analysis and machine intelligence 35.1 (2013): 232–244. [DOI] [PubMed] [Google Scholar]
- [2].Catmull Edwin, and Clark James. “Recursively generated B-spline surfaces on arbitrary topological meshes.” Computer-aided design 10.6 (1978): 350–355. [Google Scholar]
- [3].Choi Yongchoel, and Lee Seungyong. “Injectivity conditions of 2D and 3D uniform cubic B-spline functions.” Graphical models 62.6 (2000): 411–427. [Google Scholar]
- [4].De Craene Mathieu, et al. “Temporal diffeomorphic free-form deformation: Application to motion and strain estimation from 3D echocardiography.” Medical image analysis 16.2 (2012): 427–450. [DOI] [PubMed] [Google Scholar]
- [5].DeRose Tony, Kass Michael, and Truong Tien. “Subdivision surfaces in character animation.” Proceedings of the 25th annual conference on Computer graphics and interactive techniques. ACM, 1998. [Google Scholar]
- [6].Dupuis Paul, Grenander Ulf, and Miller Michael I.. “Variational problems on flows of diffeomorphisms for image matching.” Quarterly of applied mathematics (1998): 587–600. [Google Scholar]
- [7].Gilbert Kathleen, et al. “Creating shape templates for patient specific biventricular modeling in congenital heart disease.” Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE. IEEE, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gilbert Kathleen, et al. “Rapid d-affine biventricular cardiac function with polar prediction.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hubka Michal, et al. “Three-dimensional echocardiographic measurement of left and right ventricular mass and volume: in vitro validation.” The international journal of cardiovascular imaging 18.2 (2002): 111–118. [DOI] [PubMed] [Google Scholar]
- [10].Legget Malcolm E., et al. “System for quantitative three-dimensional echocardiography of the left ventricle based on a magnetic-field position and orientation sensing system.” IEEE transactions on Biomedical Engineering 45.4 (1998): 494–504. [DOI] [PubMed] [Google Scholar]
- [11].Mansi Tommaso, et al. “Physically-constrained diffeomorphic demons for the estimation of 3D myocardium strain from cine-MRI.” International Conference on Functional Imaging and Modeling of the Heart. Springer, Berlin, Heidelberg, 2009. [Google Scholar]
- [12].Morcos Michael, and Sheehan Florence H.. “Regional right ventricular wall motion in tetralogy of Fallot: a three dimensional analysis.” The international journal of cardiovascular imaging 29.5 (2013): 1051–1058. [DOI] [PubMed] [Google Scholar]
- [13].Rohde Gustavo K., Aldroubi Akram, and Dawant Benoit M.. “The adaptive bases algorithm for intensity-based nonrigid image registration.” IEEE transactions on medical imaging 22.11 (2003): 1470–1479. [DOI] [PubMed] [Google Scholar]
- [14].Rueckert Daniel, et al. “Diffeomorphic registration using B-splines.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Berlin, Heidelberg, 2006. [DOI] [PubMed] [Google Scholar]
- [15].Sheehan Florence H., et al. “Quantitative three dimensional echocardiography: Methodology, validation, and clinical applications.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Berlin, Heidelberg, 1998. [Google Scholar]
- [16].Stam Jos. “Exact evaluation of Catmull-Clark subdivision surfaces at arbitrary parameter values.” Proceedings of the 25th annual conference on Computer graphics and interactive techniques. ACM, 1998. [Google Scholar]
- [17].Stebbing Richard V., et al. “Data-driven shape parameterization for segmentation of the right ventricle from 3D+ t echocardiography.” Medical image analysis 21.1 (2015): 29–39. [DOI] [PubMed] [Google Scholar]
- [18].Thirion J-P. “Image matching as a diffusion process: an analogy with Maxwell’s demons.” Medical image analysis 2.3 (1998): 243–260. [DOI] [PubMed] [Google Scholar]
- [19].Yushkevich Paul A., et al. “User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability.” Neuroimage 31.3 (2006): 1116–1128. [DOI] [PubMed] [Google Scholar]


