Abstract
Sympatric speciation illustrates how natural and sexual selection may create new species in isolation without geographic barriers. However, recent genomic reanalyses of classic examples of sympatric speciation have revealed complex histories of secondary gene flow from outgroups into the radiation. In contrast, the rich theoretical literature on this process distinguishes among a diverse range of models based on simple genetic histories and different types of reproductive isolating barriers. Thus, there is a need to revisit how to connect theoretical models of sympatric speciation and their predictions to empirical case studies in the face of widespread gene flow. We summarize theoretical differences among different types of sympatric speciation and speciation-with-gene-flow models and propose genomic analyses for distinguishing which models apply to case studies based on the timing and function of adaptive introgression. Investigating whether secondary gene flow contributed to reproductive isolation is necessary to test whether predictions of theory are ultimately borne out in nature.
Keywords: sympatric speciation, genomics, gene flow, introgression, selection, cichlid
What is sympatric speciation?
As Mayr famously quipped, sympatric speciation is like the Lernean Hydra: “which grew two new heads whenever one of its heads was cut off” (p. 451; (Mayr 1963)). The latest incarnation of this phenomenon has occurred over the past decade: sympatric speciation now means two different things to different research groups. We stress that our goal here is not to offer a new definition of sympatric speciation nor grow a new head on the hydra, but only to clarify existing usage so that we can focus on reconciling diverse theoretical models with existing empirical examples of this process.
Subsequent to Mayr’s classic definition based on geography (1947), sympatric speciation was redefined over the past two decades in a population genetic context as the most extreme endpoint on the continuum of divergence with gene flow: panmictic gene flow and no initial divergence at the start of speciation (Bolnick & Fitzpatrick 2007; Fitzpatrick et al. 2008, 2009; Via 2001; Coyne and Orr 2004). In the context of theoretical speciation models, this type of speciation process is the most difficult because the starting conditions involve no pre-existing divergence among loci involved in reproductive isolation. Instead, linkage disequilibrium must build up through time within a population through the action of disruptive natural selection and strong assortative mating by ecotype, despite the countervailing eroding force of recombination (Felsenstein 1981; Dieckmann & Doebeli 1999; Gavrilets 2004; Doebeli et al. 2005).
Recently, the definition of sympatric speciation has been expanded to focus more on biogeographical context in line with Mayr’s original definition (Mallet et al. 2009b), in which the speciation process is defined as sympatric a) as long as diverging populations are within ‘cruising range’ of each other and b) regardless of whether secondary gene flow provided alleles contributing to reproductive isolating barriers in sympatry. Cruising range provides a practical empirical definition of gene flow between diverging sympatric populations, allowing for some geographic or microallopatric population structure (e.g. (Mallet et al. 2009a; Foote 2018)). Allowing for the secondary gene flow of alleles contributing some or all of the reproductive isolation between sympatric populations also expands the definition of sympatric speciation to include both ‘hard’ and ‘easy’ processes under one umbrella. Theoretical models show that speciation is much easier from starting conditions that involve some level of initial divergence and/or restricted gene flow (Otto et al. 2008), for example, if the alleles necessary for reproductive isolation first become physically linked in allopatry.
These two definitions, the population genetic and biogeographic, reflect different perspectives on the value of studying sympatric speciation. The biogeographic definition (for clarity, we will refer to this process as ‘easy sympatric speciation’), with its broader range of starting conditions that are easier to verify in nature, increases, perhaps vastly, the number of empirical speciation events that could be categorized as examples of sympatric speciation. This definition values the frequency of sympatric diverging populations in nature compared to allopatric speciation, as an estimate of the overall importance of “sympatry” in contributing to biodiversity. The population genetic definition (for clarity, we will refer to this more restrictive process as ‘hard sympatric speciation’), with its narrow set of starting conditions that remain challenging to verify in nature, finds value in studying both the easy and hard processes of sympatric speciation defined in theoretical models. Namely it values the theoretical possibility of creating new species solely through the power of divergent selection alone, regardless of whether this process is common in nature. Here we focus on the types of questions that genomic data now allow us to ask to improve the search for examples of both the easy and hard processes of sympatric speciation, and investigate the range of speciation mechanisms found in nature from among those shown to be plausible in theory.
Under the biogeographical definition of sympatric speciation, there is little difference in terms of the speciation mechanisms said to be involved in scenarios that start with initial panmixia (i.e. hard sympatric speciation) versus those that start with some geographic or microallopatric population structure (i.e. easy sympatric speciation (Mallet et al. 2009a; Foote 2018)). However, this contrasts with the theoretical literature which differentiates models of hard sympatric speciation from other models of speciation with gene flow. Indeed, theory teaches us that the hard process of sympatric speciation (without the aid of secondary gene flow contributing to reproductive isolation) is uniquely and notoriously difficult (Coyne & Orr 1989), in part because quite specific conditions of resource availability (e.g., (Dieckmann & Doebeli 1999; Polechová et al. 2005)), mating traits and preferences (e.g., (Weissing et al. 2011; Norvaišas & Kisdi 2012)), and search costs (e.g., (Kopp & Hermisson 2008)) must be met for it to occur. Some argue that the effort to discern the exact geographic scenario and initial conditions of speciation would be better spent on finding loci involved in reproductive isolation (i.e. ‘barrier loci’ (Nosil & Schluter 2011; Ravinet et al. 2017)). This is an important first step and we can glean something about the process of speciation from gene annotations of barrier loci and linkage architecture. However, understanding whether any one particular locus or potential mechanism was necessary for speciation often requires placing genomic discoveries in the context of speciation models that explicitly compare the importance of such factors and mechanisms in driving divergence, models whose outcomes are highly dependent on the initial conditions before sympatric divergence.
Different mechanistic processes underlying divergence in sympatry
Regardless of definition, it is necessary to distinguish among different sympatric divergence processes to understand which classes of speciation models and predictions apply to specific case studies. We here distinguish different scenarios (Fig. 1) that will result in two sister species in sympatry based on whether secondary gene flow aided in population divergence: 1) hard sympatric speciation without gene flow; 2) hard sympatric speciation in the presence of a) neutral secondary gene flow or b) after differential sorting of an ancestral hybrid swarm. In the latter case, we also distinguish whether the ancestral hybrid swarm population achieved panmixia before later divergence (i.e. hard sympatric speciation); otherwise, differential sorting of haplotypes within the hybrid swarm is better described by secondary contact speciation-with-gene-flow models rather than sympatric speciation models. 3) Easy sympatric speciation may be aided by secondary gene flow that a) triggers initial sympatric divergence or b) increases divergence after initial divergence in sympatry becomes stalled, an outcome of many sympatric speciation models without sufficiently strong disruptive selection (Matessi et al. 2001; Bolnick & Doebeli 2003; Bolnick 2006; Bürger & Schneider 2006). Finally, 4) secondary contact after a period of allopatry between two populations can result in coexistence or reinforcement, if there is not collapse into a single admixed population (Turelli et al. 2001; Kirkpatrick & Ravigné 2002; Otto et al. 2008; Garner et al. 2018) or extinction of one or both populations.
Figure 1: Genomic signatures of sympatric speciation and speciation with gene flow.
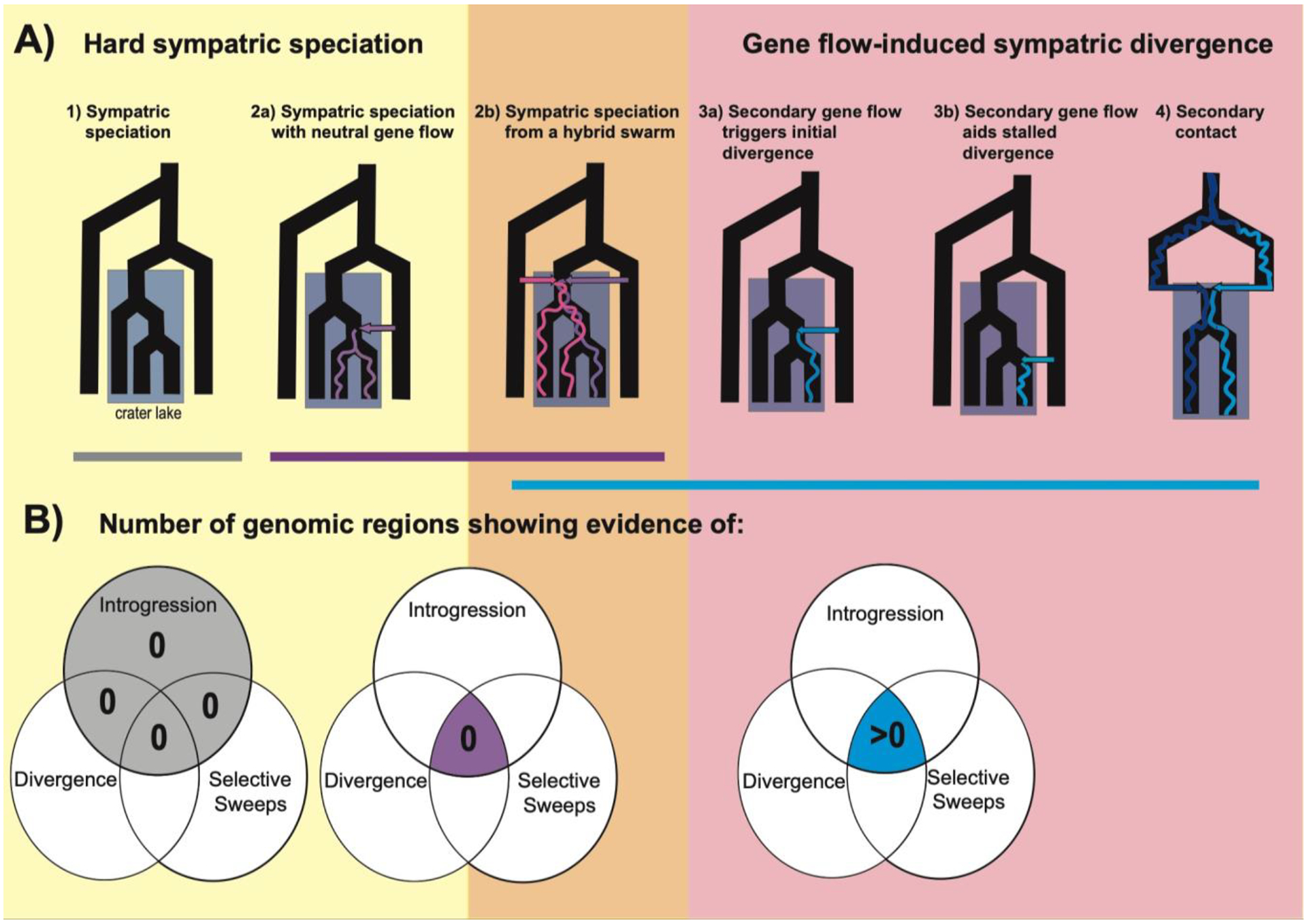
Speciation scenarios are grouped into hard sympatric speciation scenarios (yellow box; harder process in theoretical models) and other divergence scenarios which can also occur in sympatry (red box; easier processes, which we refer to broadly as sympatric divergence here). Speciation from a hybrid swarm (orange box) can fall under either class of scenarios and additional information is necessary to determine what category of speciation models best describe this process. A) The timing of gene flow relative to divergence can be used to distinguish between speciation scenarios. The colored arrows represent gene flow events and the colored lines within the tree are simplified representations of a signature of introgression from that gene flow event into the sympatric species. B) Venn diagrams illustrating the number of genomic windows across the entire genome expected to have overlapping signatures of introgression (e.g. fd outliers), genetic divergence (e.g. Fst and Dxy outliers), and selective sweeps (e.g. SweeD) for each speciation scenario (e.g. see (Richards & Martin 2017)). The highlighted sections of the Venn diagrams indicate the key signature that can be used to distinguish between the scenarios. The scenarios that are expected to leave very similar signatures of overlap are grouped by the bars colored with their respective Venn diagram.
We consider scenarios 1 and 2a to be examples of hard sympatric speciation, whereas scenarios 3 and 4 would be examples of speciation aided by secondary gene flow, a much easier process in theory. Interestingly, hybrid swarm scenarios (2b) exist in a gray area, since substantial initial gene flow from multiple sources may increase ecological or preference variation within a population that is sufficient to trigger later sympatric divergence, even without segregating inversions or genetic incompatibilities (van Doorn & Weissing 2001; Seehausen 2004, 2013). So far, we know of no examples of scenario 1 within any case study of sympatric sister species examined using genomic tools; even long diverged species show some evidence of introgression from outgroups in their past (e.g. (Turissini & Matute 2017)). In contrast, sympatric speciation with neutral gene flow (Scenario 2a, and conditionally Scenario 2b) and speciation aided by gene flow (Scenarios 3 and 4) frequently appear to operate concurrently even within a single sympatric adaptive radiation (e.g. (Wagner & McCune 2009; Martin 2012; Meier et al. 2017)).
It is important to distinguish these scenarios because theoretical models predict that sympatric divergence unaided by any form of secondary gene flow is substantially more difficult than other speciation with gene flow scenarios (Box 1). Gene flow throughout the speciation process allows recombination to break down linkage disequilibrium among alleles associated with ecological divergence and assortative mating. There are also three different types of reproductive isolating traits to consider within sympatric speciation models: the most difficult process involves independently segregating loci for ecotype, female preferences, and male traits within the population, whereas sympatric divergence is much easier if any of these three types of traits are combined (i.e. cannot become disassociated by gene flow and recombination), such as assortative mating based on phenotype matching instead of separate loci for preference and traits (Kopp et al. 2018; Servedio & Boughman 2017) or “magic” traits (such as assortative mating based on microhabitat preference; (Dieckmann & Doebeli 1999; Gavrilets 2004; Servedio et al. 2011)). Sympatric speciation by sexual selection alone is also theoretically possible (albeit considered highly unlikely) if there is substantial preference variation either initially within the population or through secondary gene flow (van Doorn & Weissing 2001; Weissing et al. 2011).
Box 1. Why do we care whether speciation is sympatric?
Inferences from theoretical models predict that, under a scenario of speciation with gene flow (Scenario 3), introgression can make the process of speciation much easier in three ways. First, by introducing additional variation in ecological traits into the population, introgression could potentially facilitate a branching process due to competition for resources (although we are not aware of a model that assesses this precise situation, it can be inferred from the dynamics of (Dieckmann & Doebeli 1999)). Second, introgression of novel alleles for mating preferences may provide a boost in preference variation that could be an important trigger to aid the evolution of assortative mating under a preference/trait mechanism, which requires preference variation to be large ((van Doorn & Weissing 2001; Weissing et al. 2011)). For example, we found evidence of secondary gene flow of olfactory alleles shortly before the rapid divergence of a Cameroon cichlid radiation in Lake Ejagham, which may have boosted preference variation (Poelstra et al. 2018a). Third, secondary gene flow after a period of allopatric isolation may lead to increased linkage disequilibrium between assortative mating and ecological loci or among ecological loci. It seems logical that this might facilitate sympatric speciation as this metric is often described as progress along the speciation continuum. However, initial linkage disequilibrium has been shown not to matter much in at least some scenarios (Felsenstein 1981) because without physical linkage, linkage disequilibrium breaks down quickly. However, physical linkage may enable these alleles to remain in association for a sufficient time for assortative mating to evolve within the population (e.g., (Servedio & Bürger 2018)). Initial linkage disequilibrium may also increase the probability of allelic capture by an inversion or for selection for new mutations within an inversion that may affect both ecology and assortment (Kirkpatrick & Barton 2006). Finally, higher linkage disequilibrium among ecological loci may in some cases increase the probability of sympatric divergence, but this is in effect similar to varying effect sizes of alleles at ecological loci (e.g. many small effect alleles within a region resemble a large-effect locus (Yeaman et al. 2016)). These predictions could also apply to sympatric radiations. For example, some classic sympatric speciation models (e.g. (Dieckmann & Doebeli 1999)) can yield many more than just two species if left to run for more generations (Polechová et al. 2005; Bolnick 2006).
The fundamental difference between sympatric speciation and speciation with gene flow, including secondary contact scenarios, lies in the fact that very often multiple equilibrium states exist in speciation models, such that loss of divergence and maintenance of divergence in the presence of gene flow are both possible outcomes, depending on the starting conditions of a population (this is nicely illustrated for one measure of divergence by (Kirkpatrick & Ravigné 2002), Fig. I). In such cases, speciation is much more easily reached from starting conditions that match those of two populations that have diverged largely in allopatry due to the large amount of allelic variation or pre-existing phenotypic bimodality and assortative mating (e.g., Otto et al. 2008). Even for scenarios of speciation with gene flow that are much easier, such as geographic separation between two incipient species that are undergoing gene flow, differentiation is much more difficult to reach or maintain from an initially homogeneous population than from an initially differentiated one (Cotto & Servedio 2017; Sachdeva & Barton 2017).
Fig I.

Two equilibrium cases exist for the linkage disequilibrium (LD), a proxy for differentiation into two distinct “species” in this proof-of-concept model, that can be maintained between two loci that are under disruptive selection and determine assortative mating. With little initial LD, the one-species equilibrium is likely to be reached even when the intensity of assortment is high. When LD in the traits is initially large, as can be the case if there is initially divergence in allopatry, the two-species equilibrium can be reached instead. Modified from (Kirkpatrick & Ravigné 2002).
Any form of linkage disequilibrium among ecological and mate choice loci formed in allopatry, whether due to physical linkage, selection, or drift, can thus tend to shift the initial starting conditions of panmixia in favor of sympatric divergence (Kirkpatrick & Ravigné 2002). However, linkage disequilibrium without physical linkage subsides within a relatively small number of generations after secondary sympatry and thus may not allow sufficient time for the evolution of assortative mating within the population. In contrast, pre-existing physical linkage among ecological loci has been shown to increase the probability of divergence, especially when it captures already divergent alleles, as is more likely after a period of divergence in allopatry before secondary contact (Kirkpatrick & Barton 2006; Feder & Nosil 2009). Similarly, physical linkage can cause preference and trait alleles to mimic phenotype matching, although even tight linkage can break down over long timescales (shown in a model with population structure: (Servedio & Bürger 2018)). Segregating inversions in the ancestral population are now well-known empirical examples of physical linkage promoting divergence in sympatry (Kirkpatrick & Barton 2006; Fuller et al. 2018). Sympatric divergence is also limited by many other restrictive conditions including the costs of female choosiness and strengths of disruptive selection and assortative mating.
Despite extensive searches for examples of sympatric speciation in the wild, there are few convincing case studies due to the difficulty of ruling out historical allopatric scenarios (see below) and ruling out a role of introgression in speciation. Furthermore, the role of magic traits or matching vs. preference/trait mechanisms is not fully understood in any existing case study. Thus, we still have very limited empirical tests of an extensive theoretical literature and diverse competing models of the notoriously difficult process of sympatric speciation (Gavrilets et al. 2007; Gavrilets 2014; Kopp et al. 2018; Servedio & Boughman 2017).
The classic problem of sympatric speciation
There are four traditional criteria for demonstrating hard sympatric speciation without secondary gene flow (e.g. Scenario 1 in Fig 1A): 1) sister species have to be reproductively isolated, 2) form a monophyletic group, 3) largely overlap in ranges, and 4) have biogeographic and evolutionary histories that make periods of allopatric divergence highly unlikely (Coyne & Orr 2004). Very few case studies have been able to meet these rigorous criteria despite intensive searches (Coyne & Orr 2004; Bolnick & Fitzpatrick 2007). This has led to the prominent status of crater lake cichlids as some of the best examples of sympatric speciation in the wild due to the uniform shape of isolated volcanic lakes which convincingly rule out phases of allopatry due to water level changes (Box 2; (Barluenga et al. 2006)).
Box 2. Evidence for sympatric speciation from crater lake cichlid radiations.
There are relatively few volcanic chains of crater lakes containing fishes in the tropics, notably found in Cameroon, Nicaragua, Tanzania, Uganda, Madagascar, and Papua New Guinea (Sparks 2004; Seehausen 2006; Malinsky et al. 2015a). Although sympatric radiations of endemic fishes are known from other isolated saline, alkali, postglacial, and ancient lakes, only four lineages of cichlids have radiated in the world’s crater lakes (Fig. II). The most diverse radiation is Barombi Mbo, Cameroon with eleven endemic cichlid species, followed by Lake Bermin, Cameroon with nine (Schliewen et al. 1994). Nicaraguan crater lakes reach up to five species (Kautt et al. 2014), the East African craters never exceed two sympatric species (Machado-Schiaffino et al. 2015; Malinsky et al. 2015a), and Madagascar’s crater lakes contain a single endemic cichlid (Sparks 2004). It remains unknown why regional and lineage diversity varies so greatly because there appears to be no relationship between the occurrence of endemic cichlid radiations and crater lake size or age (up to approximately 5 km diameter and 2 million years old) (Wagner et al. 2014).
The evidence for secondary gene flow is remarkably similar across all crater lake cichlid radiations examined with genomic data so far. Admixture proportions with outgroups are frequently detected within the range of 1–4%: 0.6% in Lake Barombi Mbo Sarotherodon (percentage of polyphyletic trees in Saguaro: (Richards et al. 2018)), 1.1% in Lake Massoko Astatotilapia (Patterson’s D: (Malinsky et al. 2015a)), 4.3% in Lake Apoyo Amphilophus (demographic model:(Kautt et al. 2016a)), and 4.4% in Lake Ejagham Coptodon (1,138 fD outliers: (Poelstra et al. 2018b)), although notably these studies all use different metrics of introgression. No case studies have yet found evidence of substantial divergence in allopatry followed by secondary contact (but see Lake Xiloá Amphilophus (Kautt et al. 2016a)). Instead, nearly all studies have concluded sympatric divergence with periodic or continuous gene flow, potentially from an initial hybrid swarm population (i.e. introgression from multiple outgroup populations).
Secondary gene flow may have triggered sympatric divergence in a radiation of three Coptodon cichlids in Lake Ejagham: demographic analyses of whole genomes suggest that this population did not diversify for 8,000 years despite frequent gene flow until an influx of olfactory receptor alleles 1,000 years ago, coinciding with the first sympatric divergence in the lake (Poelstra et al. 2018). Similarly in Lake Victoria, segregating opsin alleles in riverine cichlid populations were differentially sorted among Lake Victorian cichlids and may have triggered their diversification (Meier et al. 2017).
Evidence for hard sympatric speciation in crater lake cichlids without the aid of secondary gene flow remains elusive. Malinsky et al. (Malinsky et al. 2015a) showed that 1.1% introgression occurred long before the divergence of a shallow/deep-water sister species pair of cichlids in Lake Massoko, Tanzania; however, this initial introgression may have aided later sympatric divergence (which admittedly is very difficult to rule out). Very recent sympatric divergence in some crater lakes or the proliferation of many species from a few colonization events may also suggest that divergence occurred in sympatry without the aid of gene flow (Elmer et al. 2010; Kautt et al. 2016b); however, in the former case it remains unclear if incipient divergence will continue to complete reproductive isolation or become stalled as appears to be the case on some species complexes of Cameroon crater lake cichlids (Martin 2012, 2013). Very rare secondary gene flow into the Barombi Mbo cichlid radiation (0.6% introgression) without a clear functional role provides weak evidence of sympatric divergence, but more functional characterization and timing of introgression is needed (Richards et al. 2018). The recent advent of transgenic reporters, CRISPR-Cas9, and in situ hybridization genetic tools within Nicaraguan crater lake cichlids provides much promise for future investigations of the role of introgression in sympatric divergence (Kratochwil et al. 2015, 2017).
Fig. II.

Examples of volcanic crater lakes containing endemic cichlid radiations around the globe: a,d,f) Barombi Mbo, Cameroon and its only outlet stream; b) Lake Apoyo, Nicaragua, c) Lake Massoko, Tanzania, e) Lake Bermin, Cameroon. Satellite images (a-c) from Google Earth; (d-f) by CHM.
The monophyly criterion assumes that monophyly arises only when a single ancestral population underlies the present-day daughter species. This is typically met by inferring a single phylogeny from one or more loci. This single point-estimate view of evolutionary history is problematic because it obscures the presence of non-bifurcating relationships among organisms (e.g. sister species which derived ancestry from multiple source populations due to extensive gene flow or hybrid speciation) and the real variation in evolutionary histories among genes across the genome itself (e.g. (Hahn & Nakhleh 2016)). Few regions of the genome may initially contribute to reproductive isolation resulting in a heterogeneous genomic landscape of differentiation among incipient species (Wu 2001), a pattern now extensively supported across case studies (e.g. (Fontaine et al. 2015; McGirr & Martin 2016; Campbell et al. 2018)). Therefore, monophyletic relationships are consistent with, but not exclusive to a scenario of sympatric speciation. Examining heterogeneous evolutionary histories across regions relevant to speciation is thus crucial for understanding the processes and conditions under which sympatric divergence can occur.
The ‘new’ problem of sympatric speciation
While genomics has increased our ability to resolve evolutionary relationships among organisms, it has also revealed more complex evolutionary histories of multiple colonizations and extensive secondary gene flow in nearly all examples of sympatric speciation that have been examined with genomic data so far ((Barluenga & Meyer 2010; Geiger et al. 2013; Elmer et al. 2014; Igea et al. 2015; Machado-Schiaffino et al. 2015; Malinsky et al. 2015a; Martin et al. 2015; Kautt et al. 2016a; Poelstra et al. 2018a; Richards et al. 2018); e.g. to our knowledge Lord Howe Island palms and indigobirds have not yet been directly examined for secondary gene flow with an outgroup). Indeed, only a handful of genes may directly contribute to the speciation process whereas the rest of the genome is porous to gene flow while reproductive isolation is incomplete (Wu 2001; Wu & Ting 2004). Examples of sympatric speciation without secondary gene flow (Scenario 1) are now even rarer after applying modern genomic tools to search for introgression. Instead, it is still possible that even the hard process of sympatric speciation may occur in the face of secondary gene flow in nearly all these examples (Scenario 2a; (Richards et al. 2018)). Importantly, most evidence of secondary gene flow impacting putative examples of sympatric speciation comes from genome-wide tests of introgression from outgroup lineages that do not look at how that secondary gene flow has impacted reproductive isolating barriers between diverging populations in sympatry (e.g. (Martin et al. 2015; Kautt et al. 2016a)). In case studies of sympatric speciation that involve radiations of species, secondary gene flow may also impact only some of the diverging populations such that some species within a radiation may better represent sympatric speciation scenarios than others. Therefore, introgression detected at the genome-wide level from lineages outside the speciation event tells us that secondary gene flow has occurred, but little about the divergence process among incipient sympatric species and how that gene flow shaped the process of speciation.
The challenge of understanding the hard process of sympatric speciation in the genomic era is establishing or rejecting a functional role for the ubiquitous secondary gene flow present during the speciation process, in effect ruling out scenarios 3 and 4 in favor of scenario 2 (Fig. 1). Even if signatures of secondary gene flow are detected, speciation could still have occurred solely via mechanisms of hard sympatric speciation if that secondary gene flow did not play a causal role in divergence (Scenario 2a and possibly 2b). In contrast, secondary gene flow could play a causal role if it introduced novel genetic variation or physically linked alleles (e.g. a segregating inversion) that promoted divergence, through mechanisms such as inflating variance through the creation of a hybrid swarm (Scenario 2b; ((Seehausen 2004, 2013; Martin 2016; Meier et al. 2017)), adaptive introgression (Scenario 3; (Hedrick 2013; Huerta-Sanchez et al. 2014; Stankowski & Streisfeld 2015; Richards and Martin 2017)), transgressive segregation (Scenarios 2–3; (Rieseberg et al. 1999; Kagawa & Takimoto 2017)), or hybrid speciation ((Schumer et al. 2014)). Here we propose and discuss genomic analyses that may help to establish or reject a functional role of secondary gene flow in the speciation process (Fig. 1). This is necessary to identify putative cases of hard sympatric speciation when gene flow appears to be nearly universal in the wild, particularly among sympatric diverging populations.
Searching for the hard process of sympatric speciation in the genomic era
Although genome-wide analyses of introgression provide a starting point, ultimately consideration of the time of arrival and functional role of each introgressed region within extant sympatric sister species pairs will be necessary to distinguish between hard sympatric speciation in which incidental gene flow does not contribute to reproductive isolating barriers (Scenario 2a) versus easy sympatric speciation in which divergence is aided by secondary gene flow (Scenario 3; e.g. segregating inversions (Feder et al. 2003; Fishman et al. 2013) or balancing selection on regions containing multiple barrier loci (Guerrero & Hahn 2017; Nelson & Cresko 2018)). We suggest four major types of genomic analyses to address questions about the role of secondary gene flow and identify sympatric speciation with gene flow: analyses to 1) estimate the timing of introgression into sympatric sister species relative to their divergence time, 2) infer the presence and timing of selective sweeps within sympatric sister species, 3) annotate candidate adaptive introgression regions for functional elements or trait associations that may be relevant to speciation, and 4) if closely related non-speciating outgroups are available, confirm the lack of selective sweeps of these regions in outgroups. These analyses are by no means trivial, but recently developed methods have made it possible to start addressing such challenging questions.
1). Is the observed secondary gene flow concurrent with divergence times?
Estimating the duration of gene flow and the timing of introgression into a sister species from an outgroup relative to the timing of divergence between sympatric sister species will help distinguish between scenarios of sympatric speciation, speciation with gene flow, and secondary contact. If populations diverged in sympatry independent of any concurrent secondary gene flow (Scenario 2a), we might expect to see weak concordance of the timing of gene flow with divergence times among species, for example discrete gene flow events that date well before or after divergence times among species. In the case of both discrete gene flow events surrounding divergence time estimates or continuous gene flow from the time of colonization to the present, more information about function and selection on regions introgressed near the time of speciation will be needed. Increasingly sophisticated approaches for detecting fine-scale patterns of introgression and inferring the timing and duration of gene flow from genomic data are becoming available (Box 3).
Box 3. Tools for detecting and timing adaptive introgression.
1). Detecting and timing introgression
Although there are a variety of tests to detect gene flow on a local scale or within sliding genomic windows, currently three major types of demographic coalescent modeling approaches can infer the timing of introgression based on different genomic information: 1) the distribution of allele frequencies from genotype data (site frequency spectrum: e.g; (Gutenkunst et al. 2009; Excoffier et al. 2013)), 2) the distribution of haplotype block lengths from phased genomes: e.g; (Loh et al. 2013; Vernot & Akey 2014), and 3) variation in coalescent patterns among gene trees (Gronau et al. 2011).
2). Timing of selective sweeps
Recent methods for estimating the age of a selective sweep exploit different aspects about the pattern of variation surrounding the allele on its haplotypic background. These include heuristic approaches that use point estimates of mean haplotype length or the number of derived mutations within a chosen distance of the site (Hudson 2007; Coop et al. 2008), model-based approaches that use demographic information and summary statistics of allele frequencies and linkage disequilibrium to model a distribution of ages that fit the observed data (Beleza et al. 2013; Nakagome et al. 2016; Ormond et al. 2016), and full sequence approaches that leverage the length of ancestral haplotypes surrounding the beneficial allele and the accumulation of derived mutations (Chen et al. 2015; Smith et al. 2018).
3). Functional analyses of introgressed variants
Functional annotation of introgressed regions minimally involves searching an annotated reference genome for genes with relevant functions known from model organisms. Intergenic regions can be searched for evidence of strong sequence conservation across taxa (Siepel et al. 2005) or potential regulatory elements (reviewed in (Chatterjee & Ahituv 2017)). Additionally, genome wide association studies (GWAS) can identify variants in introgressed regions correlated with reproductive isolating barriers. Functional validation of gene and regulatory element variants through genome-editing experiments is also becoming increasingly tractable for non-model organisms (e.g. (Kratochwil et al. 2017)).
2). Did any of the introgressed regions experience selective sweeps and did the timing of these sweeps align with species divergence time?
We can use information about selective sweeps of introgressed variation to further characterize the role of secondary gene flow in sympatric divergence. When an allele is selectively favored in a population, positive selection may cause it to increase in frequency and form a localized selective sweep of reduced genetic variation surrounding the adaptive variant (Smith & Haigh 1974). Such regions of high differentiation in recently diverged species are often targeted as candidates for speciation genes, although other processes not directly associated with speciation can lead to similar patterns of high heterogeneity in differentiation across a genome (reviewed in (Nachman & Payseur 2012; Cruickshank & Hahn 2014; Ravinet et al. 2017)). If speciation was recent or ongoing, there may be strong signatures of a selective sweep for particular haplotypes in at least one of the sister species for regions involved in the divergence process (e.g. regions containing selective sweeps overlap regions of strong divergence; Fig. 1b). If secondary gene flow was neutral with respect to speciation, we may find no signatures of selective sweeps in those introgressed regions.
Importantly, a sweep of the same introgressed region in both sympatric sister species may be interpreted as adaptation to the same new environment, which may not contribute to reproductive isolation between the pair (dependent on their respective genetic backgrounds; e.g. (Clarkson et al. 2014; Stölting et al. 2015)). However, this pattern is also consistent with the sweep of a region contributing to a ‘one-allele’ mechanism of mate choice (Felsenstein 1981; Kopp et al. 2018; Servedio & Boughman 2017), such as increased female choosiness in both sympatric sister species (e.g. (Ortíz-Barrientos & Noor 2005)), which would contribute to reproductive isolation. Thus, selective sweeps of an introgressed region in both sympatric sister species do not rule out its role in aiding the speciation process.
Alternatively, if selective sweeps are detected, the timing of selective sweeps can give indirect evidence about their role in speciation. If the timing of introgression predates the timing of the selective sweep, it is challenging to infer the importance of an introgressed region for speciation because linkage disequilibrium among loci relevant to speciation may take time to build up. However, the absence of selective sweeps or introgression until long after species divergence would suggest that introgression was not relevant to speciation.
3). Is there support for a causal role of secondary gene flow based on functional genetic analyses of variants in the region?
Another potential source of evidence for the functional importance of gene flow can come from genome-wide association studies (GWAS) between variants in introgressed regions and traits involved in ecological or sexual isolation between sister species. The conservation of sequences within introgressed regions across taxa may also provide strong evidence of a functional role (e.g. PhastCons (Siepel et al. 2005)). However, many complex traits are driven by a large number of variants of small effect and ruling out a functional role for gene flow from gene annotations is difficult (e.g. see the omnigenic model; (Boyle et al. 2017)). Finally, and most powerfully, genome editing and gene expression reporter systems are increasingly tractable in non-model systems (e.g. (Kratochwil et al. 2017; Cleves et al. 2018)). This is ultimately an asymmetric problem: finding evidence that an introgressed region may have contributed to reproductive isolation is far easier than demonstrating that no introgressed regions contributed to reproductive isolation in any way (Richards et al. 2018).
4). Are there similar patterns of selection or divergence in the introgessed regions in closely related outgroup populations?
Thorough investigation of these same regions in outgroups to the sympatric species gives added power to distinguish whether secondary gene flow aided sympatric divergence. If non-diversifying, closely related species exist in similar environments and haven’t diversified in a similar manner but share signatures of selective sweeps in the same regions, then the observed introgression may have been neutral relative to speciation, e.g. due to adaptations to shared changes in climate or pathogens or shared regions of reduced recombination or increased background selection. Similarly, several studies comparing genomic landscapes of differentiation across closely related taxa have found that high differentiation observed in the same genomic regions across taxa reflects the action of linked selection across low-recombination regions rather than selection against gene flow at barrier loci (Van Doren et al. 2017; Vijay et al. 2017; Delmore et al. 2018; Ma et al. 2018).
Concluding Remarks
Sympatric speciation remains among the most controversial evolutionary processes, beloved by theorists and long sought after by empiricists. While evidence of divergence under the biogeographic definition of sympatry is mounting using traditional genetic criteria of monophyly (Bolnick & Fitzpatrick 2007), genomic data has now revealed the ubiquity of secondary gene flow and introgression in many of these examples. Future fine-scale investigations of introgression will likely continue to paint a complex picture of the role of secondary gene flow in speciation. Ruling out a role for secondary gene flow in speciation and discerning which putative cases studies evolved through the hard process of sympatric speciation in the wild will be a formidable task, yet a worthwhile one in its revelation of the sheer power of divergent selection to create species in nature.
Nearly all existing case studies of sympatric speciation involve some form of automatic magic trait, such as assortative mating by habitat (Bush 1975; Feder et al. 2003; Sorenson et al. 2003), along a depth gradient (Malinsky et al. 2015a), or environment-induced phenology shifts (Savolainen et al. 2006). We think an outstanding remaining question is whether the hard process of sympatric speciation occurs in nature without the aid of some form of magic trait, as originally demonstrated to be possible in theory (Dieckmann & Doebeli 1999). The highly polygenic and multi-dimensional nature of adaptation and mate choice suggests that an ‘all-of-the-above’ speciation scenario containing a mix of preference/trait, magic trait, and phenotype matching with each trait affected by a wide distribution of allelic effect sizes with varying times of arrival, will be the norm in nature. In contrast, although numerous and diverse, most speciation models continue to address these mechanisms in a piecemeal fashion with an assumption of large effect alleles. It remains unclear how different mechanisms, effect sizes, and times of arrival will interact and compete within a single model.
Acknowledgements
We thank A. Foote for helpful comments on an early draft and J. Mallet, D. Matute, A. Meyer, J. McGirr, M. St. John, J. Poelstra, B. Reinhard, and J. Hermisson for valuable discussion of content in this manuscript. This work was supported by the National Science Foundation DEB CAREER Grant #1749764 and by the University of North Carolina to CHM.
Footnotes
Data accessibility statement: No new data was used in the production of this manuscript
References
- Barluenga M & Meyer A (2010). Phylogeography, colonization and population history of the Midas cichlid species complex (Amphilophus spp.) in the Nicaraguan crater lakes. BMC Evol. Biol, 10, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barluenga M, Stölting KN, Salzburger W, Muschick M & Meyer A (2006). Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature, 439, 719–723. [DOI] [PubMed] [Google Scholar]
- Beleza S, Santos AM, McEvoy B, Alves I, Martinho C, Cameron E, et al. (2013). The Timing of Pigmentation Lightening in Europeans. Mol. Biol. Evol, 30, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI (2006). Multi-species outcomes in a common model of sympatric speciation. J. Theor. Biol, 241, 734–744. [DOI] [PubMed] [Google Scholar]
- Bolnick DI & Doebeli M (2003). Sexual dimorphism: two sides of the same ecological coin. Evolution (N. Y), 57, 2433–2449. [DOI] [PubMed] [Google Scholar]
- Bolnick DI & Fitzpatrick BM (2007). Sympatric speciation: models and empirical evidence. Annu. Rev. Ecol. Evol. Syst, 38, 459–487. [Google Scholar]
- Boyle EA, Li YI & Pritchard JK (2017). An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell, 169, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R & Schneider KA (2006). Intraspecific Competitive Divergence and Convergence under Assortative Mating. Am. Nat, 167, 190–205. [DOI] [PubMed] [Google Scholar]
- Bush GL (1975). Modes of Animal Speciation. Annu. Rev. Ecol. Syst, 6, 339–364. [Google Scholar]
- Campbell CR, Poelstra JW & Yoder AD (2018). What is Speciation Genomics? The roles of ecology, gene flow, and genomic architecture in the formation of species. Biol. J. Linn. Soc, bly063–bly063. [Google Scholar]
- Chatterjee S & Ahituv N (2017). Gene Regulatory Elements, Major Drivers of Human Disease. Annu. Rev. Genomics Hum. Genet, 18, 45–63. [DOI] [PubMed] [Google Scholar]
- Chen H, Hey J & Slatkin M (2015). A hidden Markov model for investigating recent positive selection through haplotype structure. Theor. Popul. Biol, 99, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, Manske M, et al. (2014). Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat. Commun, 5, 4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Hart JC, Agoglia RM, Jimenez MT, Erickson PA, Gai L, et al. (2018). An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLOS Genet, 14, e1007449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Bullaughey K, Luca F & Przeworski M (2008). The Timing of Selection at the Human FOXP2 Gene. Mol. Biol. Evol, 25, 1257–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto O & Servedio MR (2017). The Roles of Sexual and Viability Selection in the Evolution of Incomplete Reproductive Isolation: From Allopatry to Sympatry. Am. Nat, 190, 680–693. [DOI] [PubMed] [Google Scholar]
- Coyne JA & Orr A (2004). Speciation Sutherland, MA, Sunderland, MA. [Google Scholar]
- Coyne JA & Orr AH (1989). Patterns of speciation in Drosophila. Evolution (N. Y), 43, 362–381. [DOI] [PubMed] [Google Scholar]
- Cruickshank TE & Hahn MW (2014). Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol, 23, 3133–3157. [DOI] [PubMed] [Google Scholar]
- Delmore KE, Lugo Ramos JS, Van Doren BM, Lundberg M, Bensch S, Irwin DE, et al. (2018). Comparative analysis examining patterns of genomic differentiation across multiple episodes of population divergence in birds. Evol. Lett, 2, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann U & Doebeli M (1999). On the origin of species by sympatric speciation. Nature, 400, 354. [DOI] [PubMed] [Google Scholar]
- Doebeli M, Dieckmann U, Metz JA & Tautz D (2005). What we have also learned: adaptive speciation is theoretically plausible. Evolution (N. Y), 59, 691–699. [PubMed] [Google Scholar]
- van Doorn GS & Weissing FJ (2001). Ecological versus Sexual Selection Models of Sympatric Speciation: A Synthesis. Selection, 2, 17–40. [Google Scholar]
- Van Doren BM, Campagna L, Helm B, Llera CJ, Lovette IJ & Liedvogel M (2017). Correlated patterns of genetic diversity and differentiation across an avian family. Mol. Ecol, 26, 3982–3997. [DOI] [PubMed] [Google Scholar]
- Elmer KR, Fan S, Kusche H, Luise Spreitzer M, Kautt AF, Franchini P, et al. (2014). Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat. Commun, 5, 1–8. [DOI] [PubMed] [Google Scholar]
- Elmer KR, Lehtonen TK, Kautt AF, Harrod C & Meyer A (2010). Rapid sympatric ecological differentiation of crater lake cichlid fishes within historic times. BMC Biol, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC & Foll M (2013). Robust Demographic Inference from Genomic and SNP Data. PLOS Genet, 9, e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Berlocher SH, Roethele JB, Dambroski H, Smith JJ, Perry WL, et al. (2003). Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci, 100, 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL & Nosil P (2009). Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution (N. Y), 63, 3061–3075. [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1981). Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution (N. Y), 35, 124–138. [DOI] [PubMed] [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF & Hill JP (2013). Chromosomal rearrangements and the genetics of reproductive barriers in mimulus (monkey flowers). Evolution (N. Y), 67, 2547–2560. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM, Fordyce JA & Gavrilets S (2008). What, if anything, is sympatric speciation? J. Evol. Biol, 21, 1452–9. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM, Fordyce JA & Gavrilets S (2009). Pattern, process and geographic modes of speciation. J. Evol. Biol, 22, 2342–2347. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, et al. (2015). Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science (80-. ), 347, 1258522–1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD (2018). Sympatric Speciation in the Genomic Era. Trends Ecol. Evol, 33, 85–95. [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Leonard CJ, Young RE, Schaeffer SW & Phadnis N (2018). Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLOS Genet, 14, e1007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AG, Goulet BE, Farnitano MC, Molina-Henao YF & Hopkins R (2018). Genomic signatures of reinforcement. Genes (Basel), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S (2004). Fitness landscape and origin of species Princeton University Press, Princeton, NJ. [Google Scholar]
- Gavrilets S (2014). Models of speciation: Where are we now? J. Hered, 105, 743–755. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Vose A, Barluenga M, Salzburger W & Meyer A (2007). Case studies and mathematical models of ecological speciation. 1. Cichlids in a crater lake. Mol. Ecol, 16, 2893–909. [DOI] [PubMed] [Google Scholar]
- Geiger MF, McCrary JK & Schliewen UK (2013). Crater Lake Apoyo revisited: population genetics of an emerging species flock. PLoS One, 8, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronau I, Hubisz MJ, Gulko B, Danko CG & Siepel A (2011). Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet, 43, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF & Hahn MW (2017). Speciation as a sieve for ancestral polymorphism. Mol. Ecol, 26, 5362–5368. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH & Bustamante CD (2009). Inferring the Joint Demographic History of Multiple Populations from Multidimensional SNP Frequency Data. PLOS Genet, 5, e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW & Nakhleh L (2016). Irrational exuberance for resolved species trees. Evolution (N. Y), 70, 7–17. [DOI] [PubMed] [Google Scholar]
- Hedrick PW (2013). Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol, 22, 4606–4618. [DOI] [PubMed] [Google Scholar]
- Hudson RR (2007). The Variance of Coalescent Time Estimates from DNA Sequences. J. Mol. Evol, 64, 702–705. [DOI] [PubMed] [Google Scholar]
- Huerta-Sanchez E, Jin X, Asan, Bianba Z, Peter BM, Vinckenbosch N, et al. (2014). Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature, 512, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igea J, Bogarín D, Papadopulos AST & Savolainen V (2015). A comparative analysis of island floras challenges taxonomy-based biogeographical models of speciation. Evolution (N. Y), 69, 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa K & Takimoto G (2017). Hybridization can promote adaptive radiation by means of transgressive segregation. Ecol. Lett, 21, 264–274. [DOI] [PubMed] [Google Scholar]
- Kautt A, Machado-Schiaffino G, genetics, A.M.-Pl. & 2016, undefined. (n.d.). Multispecies outcomes of sympatric speciation after admixture with the source population in two radiations of Nicaraguan crater lake cichlids journals.plos.org. [DOI] [PMC free article] [PubMed]
- Kautt AF, Machado-Schiaffino G & Meyer A (2016a). Multispecies Outcomes of Sympatric Speciation after Admixture with the Source Population in Two Radiations of Nicaraguan Crater Lake Cichlids. PLoS Genet, 12, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautt AF, Machado-Schiaffino G, Torres-Dowdall J & Meyer A (2016b). Incipient sympatric speciation in Midas cichlid fish from the youngest and one of the smallest crater lakes in Nicaragua due to differential use of the benthic and limnetic habitats? Ecol. Evol, 6, 5342–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M & Barton NH (2006). Chromosome inversions, local adaptation and speciation. Genetics, 173, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M & Ravigné V (2002). Speciation by Natural and Sexual Selection: Models and Experiments. . Am. Nat, 159, S22–S35. [DOI] [PubMed] [Google Scholar]
- Kopp M & Hermisson J (2008). Competitive speciation and costs of choosiness. J. Evol. Biol, 21, 1005–1023. [DOI] [PubMed] [Google Scholar]
- Kopp M, Servedio MR, Mendelson TC, Safran RJ, Rodríguez RL, Hauber ME, et al. (2017). Mechanisms of Assortative Mating in Speciation with Gene Flow: Connecting Theory and Empirical Research. Am. Nat, 191, 1–20. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Sefton MM, Liang Y & Meyer A (2017). Tol2 transposon-mediated transgenesis in the Midas cichlid (Amphilophus citrinellus) — towards understanding gene function and regulatory evolution in an ecological model system for rapid phenotypic diversification. BMC Dev. Biol, 17, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil CF, Sefton MM & Meyer A (2015). Embryonic and larval development in the Midas cichlid fish species flock (Amphilophus spp.): A new evo-devo model for the investigation of adaptive novelties and species differences Evolutionary developmental biology. BMC Dev. Biol, 15, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, et al. (2013). Inferring Admixture Histories of Human Populations Using Linkage Disequilibrium. Genetics, 193, 1233 LP–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wang K, Hu Q, Xi Z, Wan D, Wang Q, et al. (2018). Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proc. Natl. Acad. Sci, 115, E236 LP–E243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Schiaffino G, Kautt AF, Kusche H & Meyer A (2015). Parallel evolution in Ugandan crater lakes: Repeated evolution of limnetic body shapes in haplochromine cichlid fish. BMC Evol. Biol, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, et al. (2015a). Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science (80-. ), 350, 1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, et al. (2015b). Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science (80-. ), 350, 1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Meyer a, Nosil P & Feder JL (2009a). Space, sympatry and speciation. J. Evol. Biol, 22, 2332–41. [DOI] [PubMed] [Google Scholar]
- Mallet J, Meyer A, Nosil P & Feder JL (2009b). Space, sympatry and speciation. J. Evol. Biol, 22, 2332–41. [DOI] [PubMed] [Google Scholar]
- Martin CH (2012). Weak Disruptive Selection and Incomplete Phenotypic Divergence in Two Classic Examples of Sympatric Speciation: Cameroon Crater Lake Cichlids. Am. Nat, 180, E90–E109. [DOI] [PubMed] [Google Scholar]
- Martin CH (2016). The cryptic origins of evolutionary novelty: 1,000-fold-faster trophic diversification rates without increased ecological opportunity or hybrid swarm. Evolution (N. Y), 1–16. [DOI] [PubMed] [Google Scholar]
- Martin CH, Cutler JS, Friel JP, Dening Touokong C, Coop G & Wainwright PC (2015). Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution (N. Y), 69, 1406–1422. [DOI] [PubMed] [Google Scholar]
- Matessi C, Gimelfarb A & Gavrilets S (2001). Long-term Buildup of Reproductive Isolation Promoted by Disruptive Selection: How Far Does it Go? Selection, 2, 41–64. [Google Scholar]
- Mayr E (1963). Animal species and evolution Belknap, Cambridge, MA. [Google Scholar]
- McGirr JA & Martin CH (2016). Novel candidate genes underlying extreme trophic specialization in Caribbean pupfishes. Mol. Biol. Evol, 34, 873–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JI, Marques DA, Mwaiko S, Wagner CE, Excoffier L & Seehausen O (2017). Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun, 8, 14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW & Payseur BA (2012). Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. B Biol. Sci, 367, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagome S, Alkorta-Aranburu G, Amato R, Howie B, Peter BM, Hudson RR, et al. (2016). Estimating the Ages of Selection Signals from Different Epochs in Human History. Mol. Biol. Evol, 33, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TC & Cresko WA (2018). Ancient genomic variation underlies repeated ecological adaptation in young stickleback populations. Evol. Lett, 2, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvaišas P & Kisdi E (2012). Revisiting Santa Rosalia to Unfold a Degeneracy of Classic Models of Speciation. Am. Nat, 180, 388–393. [DOI] [PubMed] [Google Scholar]
- Nosil P & Schluter D (2011). The genes underlying the process of speciation. Trends Ecol. Evol, 26, 160–167. [DOI] [PubMed] [Google Scholar]
- Ormond L, Foll M, Ewing GB, Pfeifer SP & Jensen JD (2016). Inferring the age of a fixed beneficial allele. Mol. Ecol, 25, 157–169. [DOI] [PubMed] [Google Scholar]
- Ortíz-Barrientos D & Noor MAF (2005). Evidence for a One-Allele Assortative Mating Locus. Science (80-. ), 310, 1467 LP–1467. [DOI] [PubMed] [Google Scholar]
- Otto SP, Servedio MR & Nuismer SL (2008). Frequency-dependent selection and the evolution of assortative mating. Genetics, 179, 2091–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelstra J, Richards E & Martin C (2018a). Speciation in sympatry with ongoing secondary gene flow and an olfactory trigger in a radiation of Cameroon cichlids. bioRxiv, 229864. [DOI] [PubMed] [Google Scholar]
- Poelstra JW, Richards EJ & Martin CH (2018b). Speciation in sympatry with ongoing secondary gene flow and a potential olfactory trigger in a radiation of Cameroon cichlids. Mol. Ecol, 27, 1–19. [DOI] [PubMed] [Google Scholar]
- Polechová J, Barton NH & Gavrilefs S (2005). Speciation thrrough competition: a critical review. Evolution (N. Y), 59, 1194–1210. [PubMed] [Google Scholar]
- Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M, et al. (2017). Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J. Evol. Biol, 30, 1450–1477. [DOI] [PubMed] [Google Scholar]
- Richards E, Poelstra J & Martin C (2018). Don’t throw out the sympatric speciation with the crater lake water: fine-scale investigation of introgression provides equivocal support for causal role of secondary gene flow in one of the clearest examples of sympatric speciation. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ & Martin CH (2017). Adaptive introgression from distant Caribbean islands contributed to the diversification of a microendemic adaptive radiation of trophic specialist pupfishes. PLoS Genet, 13, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA & Wayne RK (1999). Transgressive segregation, adaptation and speciation. Heredity (Edinb), 83, 363. [DOI] [PubMed] [Google Scholar]
- Sachdeva H & Barton NH (2017). Divergence and evolution of assortative mating in a polygenic trait model of speciation with gene flow. Evolution (N. Y), 71, 1478–1493. [DOI] [PubMed] [Google Scholar]
- Savolainen V, Anstett M-C, Lexer C, Hutton I, Clarkson JJ, Norup MV, et al. (2006). Sympatric speciation in palms on an oceanic island. Nature, 441, 210–213. [DOI] [PubMed] [Google Scholar]
- Schliewen UK, Tautz D & Paabo S (1994). Sympatric speciation suggested by monophyly of crater lake cichlids. Nature, 368, 629–631. [DOI] [PubMed] [Google Scholar]
- Schumer M, Rosenthal GG & Andolfatto P (2014). How common is homoploid hybrid speciation? Evolution (N. Y), 68, 1553–1560. [DOI] [PubMed] [Google Scholar]
- Seehausen O (2004). Hybridization and adaptive radiation. Trends Ecol. Evol, 19, 198–207. [DOI] [PubMed] [Google Scholar]
- Seehausen O (2006). African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. Biol. Sci, 273, 1987–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O (2013). Conditions when hybridization might predispose populations for adaptive radiation. J. Evol. Biol, 26, 279–281. [DOI] [PubMed] [Google Scholar]
- Servedio M & Bürger R (2018). The Effects on Parapatric Divergence of Linkage between Preference and Trait Loci versus Pleiotropy. Genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servedio MR & Boughman JW (2017). The Role of Sexual Selection in Local Adaptation and Speciation. Annu. Rev. Ecol. Evol. Syst, 48, annurev-ecolsys-110316–022905. [Google Scholar]
- Servedio MR, Doorn GS Van, Kopp M, Frame AM & Nosil P (2011). Magic traits in speciation: ‘magic’ but not rare? Trends Ecol. Evol, 26, 389–397. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res, 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Coop G, Stephens M & Novembre J (2018). Estimating Time to the Common Ancestor for a Beneficial Allele. Mol. Biol. Evol, 35, 1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM & Haigh J (1974). The hitch-hiking effect of a favourable gene. Genet. Res, 23, 23–35. [PubMed] [Google Scholar]
- Sorenson MD, Sefc KM & Payne RB (2003). Speciation by host switch in brood parasitic indigobirds. Nature, 424, 928–931. [DOI] [PubMed] [Google Scholar]
- Sparks JS (2004). Molecular phylogeny and biogeography of the Malagasy and South Asian cichlids (Teleostei: Perciformes: Cichlidae). Mol. Phylogenet. Evol, 30, 599–614. [DOI] [PubMed] [Google Scholar]
- Stankowski S & Streisfeld MA (2015). Introgressive hybridization facilitates adaptive divergence in a recent radiation of monkeyflowers. Proc. R. Soc. London B, 282, 20151666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stölting KN, Paris M, Meier C, Heinze B, Castiglione S, Bartha D, et al. (2015). Genome-wide patterns of differentiation and spatially varying selection between postglacial recolonization lineages of Populus alba (Salicaceae), a widespread forest tree. New Phytol, 207, 723–734. [DOI] [PubMed] [Google Scholar]
- Turelli M, Barton NH & Coyne JA (2001). Theory and speciation. Trends Ecol. Evol, 16, 330–343. [DOI] [PubMed] [Google Scholar]
- Turissini DA & Matute DR (2017). Fine scale mapping of genomic introgressions within the Drosophila yakuba clade. PLOS Genet, 13, e1006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B & Akey JM (2014). Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science (80-. ), 1245938. [DOI] [PubMed] [Google Scholar]
- Vijay N, Weissensteiner M, Burri R, Kawakami T, Ellegren H & Wolf JBW (2017). Genomewide patterns of variation in genetic diversity are shared among populations, species and higher‐order taxa. Mol. Ecol, 26, 4284–4295. [DOI] [PubMed] [Google Scholar]
- Wagner CE, Harmon LJ & Seehausen O (2014). Cichlid species-area relationships are shaped by adaptive radiations that scale with area. Ecol. Lett, 17, 583–592. [DOI] [PubMed] [Google Scholar]
- Wagner CE & McCune AR (2009). Contrasting patterns of spatial genetic structure in sympatric rock-dwelling cichlid fishes. Evolution (N. Y), 63, 1312–1326. [DOI] [PubMed] [Google Scholar]
- Weissing FJ, Edelaar P & van Doorn GS (2011). Adaptive speciation theory: a conceptual review. Behav. Ecol. Sociobiol, 65, 461–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I & Ting C-T (2004). Genes and speciation. Nat. Rev. Genet, 5, 114–122. [DOI] [PubMed] [Google Scholar]
- Wu C-II (2001). The genic view of the process of speciation. J. Evol. Biol, 14, 851–865. [Google Scholar]
- Yeaman S, Aeschbacher S & Bürger R (2016). The evolution of genomic islands by increased establishment probability of linked alleles. Mol. Ecol, 25, 2542–2558. [DOI] [PubMed] [Google Scholar]


