Abstract
The last eukaryote common ancestor (LECA) possessed mitochondria and all key traits that make eukaryotic cells more complex than their prokaryotic ancestors, yet the timing of mitochondrial acquisition and the role of mitochondria in the origin of eukaryote complexity remain debated. Here, we report evidence from gene duplications in LECA indicating an early origin of mitochondria. Among 163,545 duplications in 24,571 gene trees spanning 150 sequenced eukaryotic genomes, we identify 713 gene duplication events that occurred in LECA. LECA’s bacterial-derived genes include numerous mitochondrial functions and were duplicated significantly more often than archaeal-derived and eukaryote-specific genes. The surplus of bacterial-derived duplications in LECA most likely reflects the serial copying of genes from the mitochondrial endosymbiont to the archaeal host’s chromosomes. Clustering, phylogenies and likelihood ratio tests for 22.4 million genes from 5,655 prokaryotic and 150 eukaryotic genomes reveal no evidence for lineage-specific gene acquisitions in eukaryotes, except from the plastid in the plant lineage. That finding, and the functions of bacterial genes duplicated in LECA, suggests that the bacterial genes in eukaryotes are acquisitions from the mitochondrion, followed by vertical gene evolution and differential loss across eukaryotic lineages, flanked by concomitant lateral gene transfer among prokaryotes. Overall, the data indicate that recurrent gene transfer via the copying of genes from a resident mitochondrial endosymbiont to archaeal host chromosomes preceded the onset of eukaryotic cellular complexity, favoring mitochondria-early over mitochondria-late hypotheses for eukaryote origin.
Keywords: evolution, paralogy, gene transfer, endosymbiosis, gene duplication, eukaryote origin
Significance
The origin of eukaryotes is one of evolution’s classic unresolved issues. At the center of debate is the relative timing of two canonical eukaryotic traits: cellular complexity and mitochondria. Gene duplications fostered the evolution of novel eukaryotic traits and serve as a rich phylogenetic resource to address the question. By investigating gene duplications that trace to the last eukaryotic common ancestor we found evidence for mitochondria preceding cellular complexity in eukaryote evolution. Our results demonstrate that gene duplications were already rampant in the last eukaryote common ancestor, and we propose that the vast majority of duplications resulted from cumulative rounds of gene transfers from the mitochondrial ancestor to the genome of the archaeal host cell.
Introduction
The last eukaryote common ancestor (LECA) lived about 1.6 Ba (Betts et al. 2018; Javaux and Lepot 2018). It possessed bacterial lipids, nuclei, sex, an endomembrane system, mitochondria, and all other key traits that make eukaryotic cells more complex than their prokaryotic ancestors (Speijer et al. 2015; Gould et al. 2016; Zachar and Szathmáry 2017; Barlow et al. 2018; Betts et al. 2018). The closest known relatives of the host lineage that acquired the mitochondrion are, however, small obligately symbiotic archaea from enrichment cultures that lack any semblance of eukaryotic cell complexity (Imachi et al. 2020). This steep evolutionary grade separating prokaryotes from eukaryotes increasingly implicates mitochondrial symbiosis at eukaryote origin (Gould et al. 2016; Imachi et al. 2020). Yet despite the availability of thousands of genome sequences, and five decades to ponder Margulis (Margulis et al. 2006) resurrection of endosymbiotic theory (Mereschkowsky 1910; Wallin 1925), the timing, and evolutionary significance of mitochondrial origin remains a polarized debate. Gradualist theories contend that eukaryotes arose from archaea by slow accumulation of eukaryotic traits (Cavalier-Smith 2002; Booth and Doolittle 2015; Hampl et al. 2019) with mitochondria arriving late (Pittis and Gabaldón 2016), whereas symbiotic theories have it that mitochondria initiated the onset of eukaryote complexity in a nonnucleated archaeal host (Imachi et al. 2020) by gene transfers from the organelle (Martin and Müller 1998; Lane and Martin 2010; Gould et al. 2016; Martin et al. 2017).
Information from gene duplications can help to resolve this debate. Gene and genome duplications are a genomic proxy for biological complexity and are the hallmark of eukaryotic genome evolution (Ohno 1970). Gene families that were duplicated during the transition from the first eukaryote common ancestor (FECA) to LECA could potentially shed light on the relative timing of mitochondrial acquisition and eukaryote complexity if they could be inferred in a quantitative rather than piecemeal manner. Duplications of individual gene families (Hittinger and Carroll 2007) and whole genomes (Scannell et al. 2006; Van De Peer et al. 2009) have occurred throughout eukaryote evolution. This is in stark contrast to the situation in prokaryotes, where gene duplications are rare at best (Treangen and Rocha 2011) and whole-genome duplications of the kind found in eukaryotes are altogether unknown. In an earlier study, Makarova et al. (2005) used a liberal criterion and attributed any gene present in two major eukaryotic lineages as present in LECA. Their approach overlooks eukaryotic lineage phylogeny, leading to the inference of 4,137 families that might have been duplicated in LECA. More recently, Vosseberg et al. (2021) examined nodes in trees derived from protein domains that could be scored as duplications among the 7,447–21,840 genes that they estimated to have been present in LECA and used branch lengths to estimate the timing of duplication events. However, they did not report integer numbers for duplications because of their approach based on the analyses of very large protein-domain trees instead of discrete protein-coding gene trees. Here, we addressed the problem of which, what kind of, and how many genes were duplicated in LECA and discuss the implications of our findings for the mitochondria-early versus mitochondria-late debate.
Results and Discussion
To ascertain when the process of gene duplication in eukaryote genome evolution commenced and whether mitochondria might have been involved in that process, we inferred all gene duplications among the 1,848,936 protein-coding genes present in 150 sequenced eukaryotic genomes. For this, we first clustered all eukaryotic proteins using a low stringency clustering threshold of 25% global amino acid identity (see Materials and Methods) in order to recover the full spectrum of eukaryotic gene duplications in both highly conserved and poorly conserved gene families. We emphasize that we employed a clustering threshold of 25% amino acid identity because our procedure was designed to allow for the construction of alignments and phylogenetic trees for each cluster. The 25% threshold keeps the alignments and trees out of the “twilight zone” of sequence identity (Jeffroy et al. 2006), where alignment and phylogeny artifacts based on comparisons of nonhomologous amino acid positions arise.
We then identified all genes that were duplicated across 150 sequenced eukaryotic genomes. In principle, genes present only in one copy in any genome could have also undergone duplication, with losses leading to single-copy status. Quantifying duplications in such cases are extremely topology-dependent. We therefore focused our attention on genes for which topology-independent evidence for duplications existed, that is, genes that were present in more than one copy in at least one genome. Eukaryotic gene duplications were found in all six supergroups: Archaeplastida, Opisthokonta, Mycetozoa, Hacrobia, SAR, and Excavata (Adl et al. 2012), whereby 941,268 of all eukaryotic protein-coding genes, or nearly half the total, exist as multiple copies in at least one genome. These are distributed across 239,012 gene families, which we designate as multicopy gene families. However, 89.7% of these gene families harbor only recent gene duplications, restricted to a single eukaryotic genome (inparalogs). The remaining 24,571 families (10.3%) harbor multiple copies in at least two eukaryotic genomes, with variable distribution across the supergroups (fig. 1). Opisthokonts (animals and fungi) together harbor a total of 22,410 multicopy gene families present in at least two genomes. The animal lineage harbors 19,530 multicopy gene families, the largest number of any lineage sampled, followed by the plant lineage (Archaeplastida) with 6,495 multicopy gene families. Of particular importance for the present study, among the 24,571 multicopy gene families, we identified 1,823 that are present as multiple copies in at least one genome from all six supergroups and are thus potential candidates of gene duplications tracing to LECA. In order to distinguish between the possibility of 1) duplications within supergroups after diversification from LECA and 2) duplications giving rise to multiple copies in the genome of LECA, we used phylogenetic trees.
Fig. 1.
Distribution of multicopy genes across 150 eukaryotic genomes. All eukaryotic protein-coding genes were clustered, aligned, and used for phylogenetic inferences. The resulting gene families present as multiple copies in more than one genome are plotted (see Materials and Methods). The figure displays the 24,571 multicopy gene families (horizontal axis) and the colored scale indicates the number of gene copies in each eukaryotic genome (vertical axis). The genomes were sorted according to a reference species tree (supplementary data 7) and taxonomic classifications were taken from NCBI. Animals and fungi together form the opisthokont supergroup.
To infer the relative phylogenetic timing of eukaryotic gene duplication events, we focused our attention on the individual protein alignments and maximum-likelihood trees for all 24,571 gene families with paralogs in at least two eukaryotic genomes. We then assigned gene duplications in each tree to the most recent internal node possible, allowing for multiple gene duplication events and losses as needed (see Materials and Methods) and permitting any branching order of supergroups. This approach minimized the number of inferred duplication events and identified a total of 163,545 gene duplications, 160,676 of which generated paralogs within a single supergroup (inparalogs at the supergroup-level). An additional 2,869 gene duplication events trace to the common ancestor of at least two supergroups (fig. 2a and supplementary table 1). The most notable result however was the identification of 713 gene duplication events distributed in 475 gene trees that generated paralogs in the genome of LECA before eukaryotic supergroups diverged. For these 475 gene trees, the resulting LECA paralogs are retained in at least one genome from all six supergroups, as indicated in red in figure 2a. The sample of 475 genes provides a conservative estimate of genes that duplicated in LECA. Among the 1,823 gene families having multiple copies in members of all six supergroups, note that only in 475 families (26%) do the duplications actually trace to LECA in the trees. These results indicate that most duplications in eukaryotes are lineage specific (figs. 1 and 2), and furthermore raise caveats regarding earlier estimates of duplications in LECA (Makarova et al. 2005; Vosseberg et al. 2021) based on more permissive criteria.
Fig. 2.

Distribution of paralogs descending from gene duplications across six eukaryotic supergroups. (a) The figure shows the distribution of paralogs resulting from gene duplications in eukaryotic-specific genes (E-O) and eukaryotic genes with prokaryotic homologs (E-P) (see Materials and Methods for details). Duplicated genes refer to the numbers of gene trees with at least one duplication event with descendant paralogs across the supergroups (filled circles in the center). Number of duplication events refers to the total number of gene duplications. The red row circles indicate gene duplications with descendant paralogs in species from all six supergroups and, thus, tracing to LECA regardless of the eukaryotic phylogeny. An early study assigned 4,137 duplicated gene families to LECA but attributed all copies present in any two major eukaryotic groups to LECA (Makarova et al. 2005). In the present sample, we find 2,869 gene duplication events that trace to the common ancestor of at least two supergroups. Our stringent criterion requiring paralog presence in all six supergroups leaves 713 duplications in 475 gene families in LECA. (b) Rooted phylogeny of eukaryotic supergroups that maximizes compatibility with gene duplications. Gene duplications mapping to five edges are shown (b1, b2, … , b5). The tree represents almost exactly all edges containing the most duplications, the exception is the branch joining Hacrobia and SAR because the alternative branch joining SAR and Opisthokonta is better supported. However, the resulting subtree ((Opisthokonta, SAR),(Archaeplastida, Hacrobia)) accounts for 249 duplications, fewer than the (Opisthokonta,(Archaeplastida,(SAR, Hacrobia))) subtree shown (262 duplications). The position of the root identifies additional gene duplications tracing to LECA (table 1 and supplementary table 4).
LECA’s Duplications Constrain the Position of the Eukaryotic Root
The six supergroups plus LECA at the root represent a seven-taxon tree with the terminal edges bearing 97% of gene duplication events (fig. 2). Gene duplications that map to internal branches of the rooted supergroup tree can result from duplications in LECA followed by vertical inheritance and differential loss in some supergroups, or they result from more recent duplications following the divergence from LECA. Branches that explain the most duplications are likely to reflect the natural supergroup phylogeny, because support for conflicting branches is generated by random nonphylogenetic patterns of independent gene losses (Van De Peer et al. 2009). There is a strong phylogenetic signal contained within the eukaryotic gene duplication data (fig. 2). Among all possible internal branches, those supported by the most frequent duplications are compatible with the tree in figure 2b, which places the eukaryotic root on the branch separating Excavates from other supergroups, as implicated in previous studies of concatenated protein sequences (Hampl et al. 2009; He et al. 2014). However, massive gene loss in specific supergroups (in excavates, e.g., see fig. 1) could impair identification of the eukaryotic root (Zmasek and Godzik 2011; Ku et al. 2015; Albalat and Cañestro 2016). Indeed, the high frequency of duplications that trace to LECA readily explains why resolution of deep eukaryotic phylogeny or the position of the eukaryotic root with traditional phylogenomic approaches (Ren et al. 2016) is so difficult (see also supplementary table 2): LECA was replete with duplications and paralogy. Paralogy imposes conflicting signals onto phylogenetic systematics, but gene duplications harbor novel phylogenetic information in their own right (fig. 2), as shared gene duplications discriminate between alternative eukaryote supergroup relationships.
Eukaryotic Duplications Are Not Transferred across Supergroups
Like the nucleus, mitochondria, and other eukaryotic traits (Speijer et al. 2015; Gould et al. 2016; Zachar and Szathmáry 2017; Barlow et al. 2018; Betts et al. 2018; Imachi et al. 2020), the lineage-specific accrual of gene and genome duplications distinguish eukaryotes from prokaryotes (Ohno 1917; Scannell 2006; Hittinger and Carroll 2007; Van De Peer et al. 2009; Treangen and Rocha 2011). Nonetheless, one might argue that the distribution of duplications observed here does not reflect lineage-dependent processes at all, but lateral gene transfers (LGTs) among eukaryotes instead (Andersson et al. 2003; Keeling and Palmer 2008; Leger et al. 2018). That is, a duplication could, in theory, originate in one supergroup and one or more gene copies could subsequently be distributed among other supergroups via eukaryote-to-eukaryote LGT. However, were that theoretical possibility true then neither duplications, nor any trait, nor any gene could be traced to LECA because all traits and genes in eukaryotes could, in the extreme, simply reflect 1.6 Byr of lineage-specific invention within one supergroup followed by lateral gene traffic among eukaryotes rather than descent with modification (Andersson et al. 2003; Keeling and Palmer 2008; Leger et al. 2018).
However, the present data themselves exclude the deeply improbable eukaryote-to-eukaryote lateral duplication transfer theory in a subtle but strikingly clear manner. How so? Figures 1 and 2a show that 30,439 gene lineages bearing duplications (93% of the total) are restricted in their distribution to “only one supergroup,” whereas only 2,245 (7% of the total) are shared among two to five supergroups. That is, only 7% of the duplications are shared across supergroups, hence they are the only possible candidates for LGT among supergroups. For the sake of argument, let us entertain the extreme assumption that all 2,245 patterns of shared but nonuniversal duplications involved intersupergroup LGT, recalling that there is no intersupergroup LGT in 93% of the genes (fig. 2 and supplementary table 1). With that generous assumption, the intersupergroup LGT frequency would be maximally 7%. That is an extreme upper bound, though, because the observed 93% frequency for duplicates that are supergroup specific and thus have absolutely no observable intersupergroup LGT should apply equally to the 7% of duplications shared across supergroups. Thus, the more realistic maximum estimate is that 0.49% of duplications (7% of 7%) might have been generated by intersupergroup LGT. This estimate is based solely upon the distribution of the duplicates and the premise that eukaryote supergroups are monophyletic. As it concerns the 475 genes with duplications that trace to LECA (fig. 2 and supplementary table 1), this means that 0.49% out of 475, or about 2.3 genes in our data might have been caused by intersupergroup LGT. That is a very low frequency and is consistent with independent genome-wide phylogenetic tests presented previously (Ku et al. 2015) for the paucity of eukaryote-to-eukaryote LGT. If we count duplication events (fig. 2a, right panel) rather than gene lineages (fig. 2a, left panel), the picture is even more vertical, because 98% of the events are supergroup-specific, hence lacking any patterns that could reflect LGT, meaning that maximally 0.04% (2% of 2%) or 0.19 duplications among 475 (which rounds to zero genes) could be the result of lateral transfer. The supergroup-specific distributions of duplications themselves thus provide very strong evidence that the distribution of duplicated genes in eukaryotes is not the result of eukaryote-to-eukaryote LGT phenomena (Andersson et al. 2003; Keeling and Palmer 2008; Leger et al. 2018) but the result of vertical evolution within supergroups accompanied by gene birth, death (Nei et al. 1997), and differential gene loss (Ku et al. 2015).
LECA’s Duplications Support an Early Mitochondrion
Arguably, the timing of mitochondrial origin is the central so far unresolved issue at the heart of eukaryote origin. Several alternative theories for eukaryogenesis have been proposed (reviewed in Martin et al. 2001; Embley and Martin 2006; Poole and Gribaldo 2014; López-García and Moreira 2015; Eme 2017). Symbiogenic theories posit a causal role for mitochondrial endosymbiosis at the origin of cellular eukaryotic complexity (Lane and Martin 2010) with the host being a garden variety archaeon (Martin and Müller 1998). Gradualist theories posit an autogenous origin of eukaryote cell complexity with little or no contribution of the mitochondrion to eukaryogenesis (Cavalier-Smith 2002; Gray 2014). Intermediate theories posit the existence of endosymbioses prior to the origin of mitochondria. These include an endosymbiotic origin of the nucleus (Lake and Rivera 1994), an endosymbiotic origin of peroxisomes (de Duve 2007), an endosymbiotic origin of flagella (Margulis et al. 2000), the lateral acquisition of the cytoskeleton (Doolittle 1998) or, more liberally, additional symbioses preceding the mitochondrion in unconstrained numbers, as long as each symbiosis “explains the origin of any eukaryotic innovation as a response to an endosymbiotic interaction” (Gabaldón 2018). Most current theories posit an origin of the host from archaea (Martin et al. 2015; Spang et al. 2015; Zaremba-Niedzwiedzka et al. 2017; Imachi 2020), though theories for eukaryote origins from actinobacteria (Cavalier-Smith 2002), and planctomycetes (Cavalier-Smith and Chao 2020) are discussed. Notwithstanding such diversity of views, the main divide among theories for eukaryote origin remains the relative timing of mitochondrial origin, that is did the mitochondrion initiate or culminate eukaryote origin (Martin et al. 2001; Embley and Martin 2006; Poole and Gribaldo 2014; López-García and Moreira 2015; Eme et al. 2017)? Alternative theories for eukaryote origin generate distinct predictions about the nature of gene duplications in LECA.
Gradualist theories entailing an archaeal host (Cavalier-Smith 2002; Booth and Doolittle 2015; Pittis and Gabaldón 2016; Hampl et al. 2019) predict genes of archaeal origin and eukaryote-specific genes to have undergone numerous duplications during the origin of eukaryote complexity, prior to the acquisition of the mitochondrion. In that case, the mitochondrion arose late, hence bacterial-derived genes would have accumulated fewer duplications in LECA than archaeal-derived or eukaryote-specific genes (fig. 3a). Models invoking gradual lateral gene transfers (LGT) from ingested (phagocytosed) food prokaryotes prior to the origin of mitochondria (Doolittle 1998) also predict more duplications in archaeal-derived and eukaryote-specific genes to underpin the origin of phagocytotic feeding, but do not predict duplications specifically among acquired genes (whether from bacterial or archaeal food) because each ingestion contributes genes only once.
Fig. 3.
Alternative models for eukaryote origin generate different predictions with respect to duplications. In each panel, gene duplications during the FECA to LECA transition (boxed in upper portion) are enlarged in the lower portion of the panel. (a) Cellular complexity and genome expansion in an archaeal host predate the origin of mitochondria. (b) Mitochondria enter the eukaryotic lineage early, duplications in mitochondrial-derived, host-derived, and eukaryotic-specific genes occur, genome expansion affects all genes equally. (c) Gene transfers from a resident endosymbiont generate duplications in genes of bacterial origin in an archaeal host. (d) Observed frequencies from gene duplications that trace to LECA (see supplementary table 1). BE refers to eukaryotic genes with bacterial homologs only; AE refers to eukaryotic genes with archaeal homologs only; and Euk refers to eukaryotic genes without prokaryotic homologs. (e) Schematic representation of serial gene transfers from the mitochondrion (white arrows) to the host’s chromosomes.
By contrast, transfers from the endosymbiotic ancestors of organelles continuously generated gene duplications in the host’s chromosomes (Timmis et al. 2004; Allen 2015), a process that continues to the present day in eukaryotic genomes (Timmis et al. 2004; Portugez et al. 2018). Symbiogenic theories posit that the host that acquired the mitochondrion was an archaeon of normal prokaryotic complexity (Martin and Müller 1998; Lane and Martin 2010; Gould et al. 2016; Martin et al. 2017; Imachi et al. 2020) and hence lacked duplications underpinning eukaryote complexity. There are examples known in which bacteria grow in intimate association with archaea (Imachi et al. 2020) and in which prokaryotes become endosymbionts within other prokaryotic cells (Martin et al. 2017). However, there are two different ways in which mitochondria could promote the accumulation of duplications. If energetic constraints (Lane and Martin 2010) were the sole factor permitting genome expansion, duplications would accrue in all genes regardless of their origin, such that gene duplications in the wake of mitochondrial origin should be equally common in genes of bacterial, archaeal, or eukaryote-specific origin, respectively (fig. 3b). If, on the other hand, the role of mitochondria in gene duplications was mechanistic rather than purely energetic, genes of mitochondrial origin should preferentially undergo duplication. This is because the mechanism of gene transfers from resident organelles involve endosymbiont lysis and the “copying” (Allen 2015) of organelle genomes to the host’s chromosomes followed by recombination and mutation (Portugez et al. 2018). Gene transfers from resident endosymbionts specifically generate duplications of endosymbiont genes because new copies of the same genes are recurrently transferred (Timmis et al. 2004; Allen 2015) (fig. 3c).
The duplications in LECA reveal a vast excess of duplications in LECA’s bacterial-derived genes relative to archaeal-derived and eukaryote-specific genes (fig. 3d). Of all gene families tracing to LECA, 26% experienced at least one duplication event during the transition to LECA from FECA. Notably, the excess proportion of duplicates among genes of bacterial origin is significant as judged by the two-tailed binomial test (P=1.3×10−10; proportion of duplicates at 95% CI=[35–44%]; df=1). On the other hand, genes of archaeal origin show significantly fewer duplicates (P=8.4×10−7; proportion of duplicates 95% CI=[8–17%]; df=1) with the proportion of duplicates being similar to eukaryote-specific genes (fig. 3d).
Do Bacterial Genes in LECA Stem from the Mitochondrion?
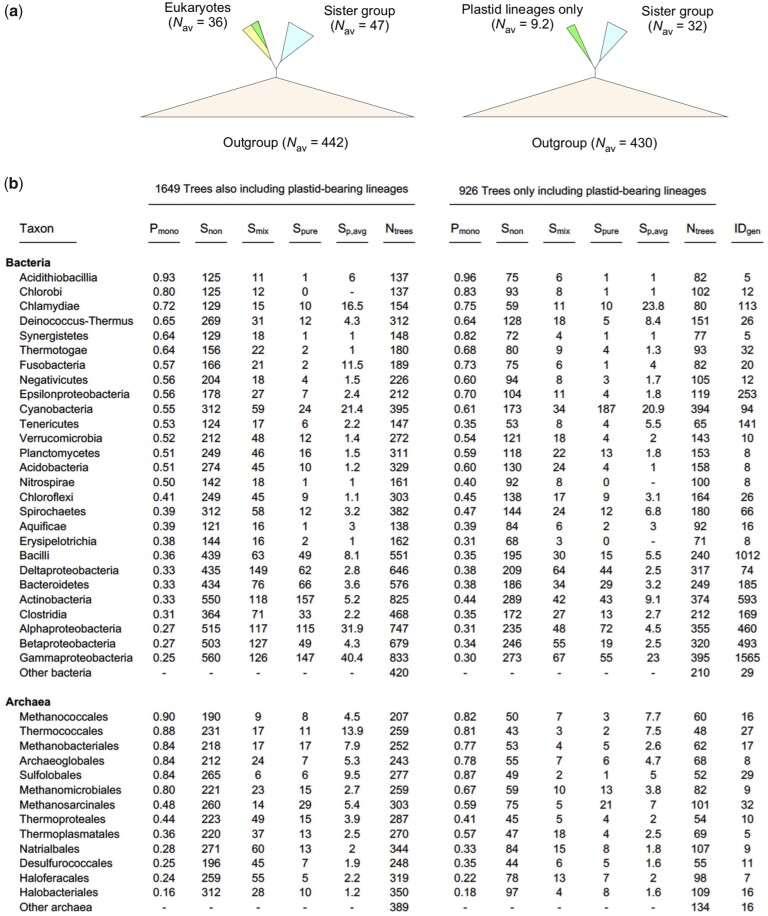
If bacterial genes in LECA stem from the mitochondrion, as opposed to 1) eukaryote-to-eukaryote gene transfers, which were already excluded for >99% of the families with duplications in this data on the basis of their distributions alone, or 2) multiple lineage-specific acquisitions from bacteria via LGT, then the bacterial genes should trace to the eukaryote common ancestor. That is, the eukaryotes should form a monophyletic clade in gene trees that connect prokaryotic and eukaryotic genes. To test this, we generated clusters, alignments, and trees for genes shared by prokaryotes and eukaryotes from 22,471,723 million genes from 5,655 genomes and including 150 eukaryotes (see Materials and Methods). The results from the 2,575 trees that contained at least five prokaryotic and at least two eukaryotic sequences are summarized in figure 4. As with the duplications themselves, eukaryote gene evolution is again vertical. Out of the 2,575 trees only 475 did not recover eukaryotes as monophyletic. However, none of these 475 trees rejected eukaryote monophyly using the Shimodaira–Hasegawa (SH) test (see Materials and Methods) and only 25 trees (1% of the total) rejected eukaryote monophyly using the Kishino–Hasegawa (KH) test. Applying the approximately unbiased (AU) test, only three trees out of 475 rejected eukaryote monophyly. This traces gene origin of ≥1,649 out of the 2,575 genes shared by prokaryotes and eukaryotes to LECA, and the origin of ≤926 genes to the archaeplastidal ancestor because the latter trees contain only photosynthetic eukaryotic lineages (fig. 4a).
Fig. 4.
Identification of prokaryotic sisters in 2,575 eukaryotic–prokaryotic gene trees. (a) The individual trees were rooted on the branch leading to the largest prokaryotic clade deriving the sister group to eukaryotes. The average number of sequences in the eukaryotic clade, sister group, and outgroup are indicated. (b) The list of bacterial (top) and archaeal (bottom) phyla occurring in the trees exclusive to plant lineages (right) and all other trees (left). Archaeal and bacterial phyla with less than five representative species in the data set were collapsed into “other archaea” and “other bacterial” groups. Pmono refers the proportion of trees with a branch (split) separating the species of the phylum from the others; Snon refers to the number of occurrence of the phylum only in the outgroup clade; Smix refers to the number of occurrences of the phylum as a mixed sister (more than one phylum in the clade); Spure refers to the number of occurrences of the phylum as pure sister (as the single phylum); Sp.avg shows the average size of the sister group when the phylum occurs as a pure sister clade. Ntrees show the number of occurrences of the phyla across all trees. IDgen refers to the total number of species in each phylum.
The 1,649 trees that trace prokaryotic gene origins to LECA fall into two classes with regard to the sister group of the eukaryotic gene: 966 in which the prokaryotic sister group to eukaryotes contained members of only one phylum (a “pure” sister, Spure in fig. 4, 59% of the trees) and those in which the sister to the eukaryotes contained members of more than one phylum (a “mixed” sister, 41% of the trees). The only way to obtain a mixed sister topology of prokaryotic sequences for a eukaryotic gene is via LGT among prokaryotes (Ku and Martin 2016). If we exclude the reality of LGT among prokaryotes, and interpret mixed sister topologies at face value, they would suggest that eukaryotes arose before the diversification of the diverse prokaryotic phyla present in our sample, which would be incompatible with accounts of eukaryote age (Parfrey et al. 2011; Betts et al. 2018), and would furthermore have LECA arising at different times, depending on the membership in the sister group. LGT among the prokaryotic reference sequences in the mixed sister cases (Ku and Martin 2016; Nagies et al. 2020) is clearly the simpler explanation. The pure sister was bacterial in 49% of the trees and archaeal in only 9.5% of the trees. Only in 115 trees (7.0%) was the bacterial pure sister clade alphaproteobacterial. These 115 trees are readily explained because they stem from the mitochondrion, even though the alphaproteobacterial-derived genes in eukaryotes do not all reside in the “same” alphaproteobacterial genome as previously observed (Ku et al. 2015; Nagies et al. 2020), requiring LGT among alphaproteobacteria, at least, to account for the topology. Yet, the crucial and previously underinvestigated issue concerns the remaining 695 pure sister bacterial origin cases (86%) that trace to LECA but reside in a genome that does not carry an alphaproteobacterial taxon label (fig. 4), as recently set forth in a study that examined the phylogeny of only the more conserved fraction of genes shared by prokaryotes and eukaryotes (Nagies et al. 2020).
There are two general ways to explain the 86% of nonalphaproteobacterial genes that trace to LECA. The first is to take one specific aspect of the trees—namely, the taxon label of the sister group—at face value and interpret the data as evidence for independent individual contributions to eukaryotes (via LGT or via multiple resident symbionts) by all of the bacterial phyla in the sample. At the level of the taxa listed in figure 4, that would mean 26 different bacterial donors to LECA in addition to the alphaproteobacterial contribution, and donations from 13 different archaeal host taxa. With 39 donor phyla, LECA already looks like a grab bag of genes. At the level of genus, the taxon labels of the trees would mean 794 different bacterial donors to LECA under permissive models (Gabaldón 2018), followed by a particularly ad hoc sudden stop of gene influx to eukaryotes after the FECA to LECA transition, because the eukaryotes are monophyletic in these trees. The suggestion of symbiont acquisition and gene transfers without constraints (Gabaldón 2018) carries a hidden and seldom spelled out corollary (Martin 1999). Namely, it entails the strict condition that all of the nonalphaproteobacterial bacterial genes in question not only resided in the genome of members of the 27 different phylum level bacterial taxa at the time of donation to LECA (fig. 4) but furthermore, and crucially, that those genes evolved “vertically” within the chromosomal confines of those respective phyla during the 1.6 Byr since eukaryotes arose. Such unrestricted donor theories (Gabaldón 2018) assume that the present-day phylum taxon label on the gene accurately identifies the donor phylum at the time of transfer. But that is true “if and only if” the gene has been vertically inherited within that phylum (no interphylum LGT) since its donation to LECA (Martin 1999; Esser et al. 2007).
Such theories of unrestricted LGT to eukaryotes with strictly vertical gene evolution among prokaryotes are unlikely and resoundingly rejected by the data. If we look beyond the mere taxon label of the sister group (fig. 4), we see that the putative 27 bacterial donor lineages themselves do not evolve in a vertical manner. The average level of monophyly for bacterial phyla in the 1,649 trees that trace to LUCA is 47% (Pmono in fig. 4). Alphaprotebacteria were monophyletic in only 27% of the trees in which they occurred, as were generalists with large genomes such as betaproteobacteria (27%) and actinobacteria (33%). Specialists like chlorobi or chlamydia with more restricted pangenomes were more monophyletic (80% and 72%, respectively). Halophilic archaea, which are known to have acquired many genes from bacteria (Nelson-Sathi et al. 2012), are the least monophyletic prokaryotes sampled (halobacteriales, 16%, fig. 4). For the 926 genes that, based on their distribution, trace to the archaeplastidal common ancestor (fig. 4, right panel), the bacterial phyla have a higher proportion of monophyly (P=0.006, V = 67, using two-tailed Wilcoxon signed-rank test) than for those genes that trace to LECA. Plastids are younger than mitochondria, hence the genes from the ancestral plastid genome have had less time to migrate across prokaryotic genomes than genes from the ancestral mitochondrial genome. For the prokaryotic genes and phyla in question, evolution is not a vertical process. The bacterial reference system against which to infer the origin of eukaryotic genes that stem from the mitochondrion (or the plastid) is a system of mosaic (Martin 1999) or fluid (Esser et al. 2007) chromosomes. These findings are fully consistent with a recent larger scale investigation of gene verticality across genomes (Nagies et al. 2020).
If we accept the evidence that LGT in prokaryotes is real and if we accept the evidence that mitochondria were once endosymbiotic bacteria, then the expectation for the phylogeny of a gene that was acquired from the mitochondrion is that it traces to a single origin in LECA, which the genes in this study do, but “not” that it traces to alphaproteobacteria. This is because LGT among prokaryotes preceding and subsequent to the origin of mitochondria generates the illusion of many donors by shuffling the taxon labels attached to genes in mosaic bacterial chromosomes (Martin 1999). Most current studies still equate mitochondrial origin with an alphaproteobacterial sister group relationship (Vosseberg et al. 2021), but if we look at all the data, it is clear that such an interpretation is too strict. For example, Vosseberg et al. (2021) found that about 7% of the eukaryotic protein-domains that they examined branched with alphaproteobacterial homologs. But looking beyond the eukaryotic branch, Nagies et al. (2020) found that only about 35% of alphaproteobacterial genes recover alphaproteobacteria monophyly to begin with, and only 16% of the 220 trees in which alphaproteobacteria appeared as the sole sister of all eukaryotes recovered aphaproteobacteria as monophyletic among prokaryotes. To investigate mitochondrial origin from the standpoint of genes, it is not enough to identify the relationship of eukaryote genes to prokaryotic homologs. One has also to investigate the relationship of prokaryotic homologs to each other, because they are the reference system for comparison.
It is because of LGT among prokaryotes that many different groups are implicated as donors of genes to LECA (fig. 4; see also Nagies et al. 2020). There is no evidence independent of gene phylogenies to suggest or support theories for the participation of spirochaetes (Margulis et al. 2006), actinobacteria (Cavalier-Smith 2002), cyanobacteria (Cavalier-Smith 1975), deltaproteobacteria (López-García and Moreira 1999), planctomycetes (Cavalier-Smith and Chao 2020), or multiple donor lineages (Gabaldón 2018) at eukaryote origin (Embley and Martin 2006). One could of course argue that those conflicting theories for contributions from many different prokaryotic lineages are all simultaneously true, but then theories for eukaryogenesis would no longer be constrained by observations in data, and any assertion about eukaryote origin would be permissible as a line of evidence, an untenable state of affairs. The same sets of considerations apply to the cyanobacterial origin of plastids (fig. 4).
If we let go of the belief that sister group relationships between eukaryotic genes and prokaryotic homologs (fig. 4) identify the prokaryotic lineages that donated genes (Martin 1999; Nagies et al. 2020), and take into account the functions encoded by nuclear genes of bacterial origin that were duplicated in LECA (figs. 2 and 4;table 1), the simplest interpretation of the data in our view is that the bacterial duplicates in LECA were donated by the mitochondrion. Other more complicated interpretations are imaginable, but these interpretations do not simultaneously account for the phylogenetic behavior of the bacterial reference phylogeny set, which we have done here and elsewhere (Nagies et al. 2020). Our data furthermore show that eukaryotic genes are of monophyletic origin. With large genomic samples spanning thousands of reference prokaryotic genomes, eukaryotic gene evolution is clearly vertical, both in terms of lineage-specific distribution of gene duplications (fig. 1) and in terms of likelihood ratio tests (Nagies et al. 2020).
Table 1.
Functional Categories of Genes Duplicated in LECAa
| Categoryb | (n) | Bacterial | Archaeal | Universal | Eukaryotic |
|---|---|---|---|---|---|
| Metabolism | (141) | 64 | 2 | 58 | 17 |
| Protein modification, folding, degradation | (89) | 30 | 8 | 30 | 21 |
| Ubiquitination | 3 | 1 | — | 9 | |
| Proteases | 9 | 1 | 7 | 1 | |
| Kinase/phosphatase/modification | 12 | 6 | 19 | 9 | |
| Folding | 6 | — | 4 | 2 | |
| Novel eukaryotic traits | (61) | 8 | 4 | 12 | 37 |
| Cell cycle | 1 | 1 | 2 | 5 | |
| Cytoskeleton | 4 | — | 1 | 19 | |
| Endomembrane (ER; Golgi; vesicles) | 2 | 2 | 8 | 10 | |
| mRNA splicing | 1 | 1 | 1 | 3 | |
| Mitochondrion | (47) | 29 | — | 9 | 9 |
| Carbon metabolism | (37) | 26 | — | 11 | — |
| Glycolysis | 10 | — | 5 | — | |
| Reserve polysaccharides, other | 16 | — | 6 | — | |
| Cytosolic translation | (36) | 15 | 7 | 10 | 4 |
| Nucleic acids | (55) | 13 | 7 | 15 | 20 |
| Histones | — | — | 2 | 8 | |
| RNA | 8 | 3 | 6 | 4 | |
| DNA | 5 | 4 | 7 | 8 | |
| Membranes (excluding endomembrane) | (46) | 18 | 1 | 12 | 15 |
| Transporters, plasma associated | 8 | 1 | 9 | 14 | |
| Lipid synthesis | 10 | — | 3 | 1 | |
| Redox | (15) | 11 | — | 4 | — |
| Hypothetical | (229) | 81 | 9 | 61 | 78 |
| Total | 295 | 38 | 222 | 201 |
Note.—n, number of duplicated genes in the corresponding category.
About 475 genes duplicated in LECA and present in all six supergroups plus 281 genes with duplications tracing to the common ancestors of excavates and other supergroups. The annotation, source (bacterial, archaeal, present in bacteria and archaea, eukaryote specific), and the numbers of duplications for each cluster are given in supplementary tables 3 and 4. All categories listed had representatives on both the 475 and the 281 list except mRNA splicing, present in the 475 list only.
The categories do not strictly adhere to KEGG or gene ontology classifications, instead they were chosen to reflect the processes that took place during the FECA to LECA transition. The largest number of duplications in LECA for any individual gene was 12, a dynein chain known from previous studies to have undergone duplications in the common ancestor of plants animals and fungi (Kollmar 2016).
Can Positive Selection Explain Excess Bacterial Duplications?
The vast excess of bacterial duplications (fig. 3) and the phylogenies of 2,575 genes that would address the question of gene origin (fig. 4) speak in favor of bacterial acquisition in LECA from a single-resident endosymbiont, the mitochondrion, prior to the origin of eukaryote complexity. Yet one could still imagine numerous individual gene acquisitions in LECA from different donors with a blanket ad hoc hypothesis of “positive selection” increasing the copy number of bacterial-related functions to account for the excess of bacterial-derived duplications (table 1). However, the selection proposal would not explain the excess of bacterial over archaeal or eukaryote-specific genes with the same functional category, as is widely observed in table 1. That is, selection would have to be invoked as a special plea on a bacterial-gene-for-bacterial-gene basis, requiring yet one additional corollary of positive selection for each duplication. Because we observe over 900,000 duplications in the present data, the selection theory to account for duplications carries a burden of too many corollary assumptions.
On the other hand, it is possible that duplications are fundamentally mechanistic in origin, via chromosome mispairing, translocations, genome duplications, or via duplicative transfers from a resident endosymbiont as we argue in this paper. In a context of mosaic, fluid bacterial genomes (Martin 1999; Esser et al. 2007) permitting LGT among prokaryotes (fig. 4) (Nagies et al. 2020), we would require no corollary assumptions of ad hoc selection. The mechanism of transfer from the endosymbiont generates the excess of bacterial duplications and does so across all functional categories (table 1).
The Functions of Bacterial Duplicates Polarize Events at LECA’s Origin
Gene duplications speak to more than phylogeny. Gene duplications are a standard proxy for the evolution of complexity, as diversification of function and form is canonically underpinned by gene family expansion (Ohno 1970). Accordingly, we observe that the morphologically most complex multicellular eukaryotes—plants, animals, and fungi—harbor the largest numbers of duplications (fig. 1). As outlined above, the simplest interpretation of the present data is that complexity started with the mitochondrion. That is not only true for the present data on duplications, is also true from a purely physiological standpoint (Martin et al. 2017) and a bioenergetic standpoint (Lane and Martin 2010).
The functions of genes that were duplicated in LECA help to polarize events in LECA’s evolution. For example, LECA had a mitochondrion. LECA’s gene duplications in 47 genes with mitochondrial functions include pyruvate dehydrogenase complex, enzymes of the citric acid cycle, components involved in electron transport, a presequence cleavage protease, the ATP–ADP carrier, and seven members of the eukaryote-specific mitochondrial carrier family that facilitates metabolite exchange between the mitochondrion and the cytosol (table 1 and supplementary tables 3 and 4). A recent study estimated that some genes for mitochondrial function were probably duplicated in LECA, but interpreted the data as evidence for mitochondria-intermediate hypothesis (Vosseberg et al. 2021). The methodology used in Vosseberg et al. has major limitations because: 1) the timing of gene duplications was inferred using an approach that equates branch-lengths from phylogenetic trees to time, which is expected to be valid “only if” the evolutionary rate is constant across genes (substitutions and gene loss, for example); 2) prokaryotic sequences were arbitrarily removed from gene trees, inflating the estimates of duplications in genes of archaeal origin; 3) the use of trees for which the same gene sequence can be represented simultaneously in multiple trees, biasing the estimates of duplications and their origin; and 4) the use of too liberal thresholds for gene clustering which result in aberrantly large gene families (see supplementary fig. 5, Supplementary Material online), a potential source of tree reconstruction errors. By contrast, we do not infer time from branch lengths, we did not remove sequences that did not fit our expectations, and gene membership in our gene families is always unique.
Our findings clearly indicate that canonical energy metabolic functions of mitochondria were established in LECA, underscored by additional functions performed by mitochondria in diverse eukaryotic lineages: ten genes for enzymes of the lipid biosynthetic pathway (typically mitochondrial in eukaryotes; Gould et al. 2016), the entire glycolytic pathway (mitochondrial among marine algae; Río Bártulos et al. 2018), and 11 genes involved in redox balance are found among bacterial duplicates. The largest category of duplications with annotated functions concerns metabolism and biosynthesis (table 1).
Many products of bacterial-derived genes operate in the eukaryotic cytosol (Martin et al. 1993; Esser et al. 2004). This is because at the outset of gene transfer from the endosymbiont, there was no mitochondrial protein import machinery (Martin and Müller 1998; Dolezal et al. 2006), and no nucleus, such that the products of genes transferred from the endosymbiont were active in the compartment where the genes were cotranscriptionally translated (French et al. 2007). Gene transfers in large, genome sized fragments from the endosymbiont, as they occur today (Timmis et al. 2004; Portugez 2018), furthermore, permitted entire pathways to be transferred, because the unit of biochemical selection is the pathway and its product, not the individual enzyme (Martin 2010). In the absence of upstream and downstream intermediates and activities in a pathway, the product of a lone transferred gene is generally useless for the cell, expression of the gene becomes a burden, and the transferred gene cannot be fixed (Martin 2010).
Bacterial-derived duplications are present in functions that underpinned the origin of cell compartmentation in LECA (table 1). LECA possessed an endomembrane system consisting of bacterial lipids, as symbiogenic models predict (Gould et al. 2016). Bacterial duplicates, not archaeal duplicates, dominate lipid synthesis and membrane biogenesis (table 1). Functions of bacterial duplicates are also involved in mRNA splicing, a selective force at the origin of the nucleus (Garg and Martin 2016; Eme et al. 2017). The origin of protein import into mitochondria was essential to mitochondrial origin (Dolezal et al. 2006) and encompasses many bacteria-derived duplicates (table 1). LECA’s duplicates of bacterial origin are also involved in the origin of eukaryotic-specific traits, including the cell cycle, the cytoskeleton, endomembrane system, and mRNA splicing (table 1). Eukaryote complexity required intracellular molecular movement in the cytosol, which is realized by motor proteins. The protein with the most duplications found in LECA is a light chain dynein with 12 duplications (supplementary table 3), in agreement with previous studies of dynein evolution that document massive dynein gene duplications early in eukaryote evolution (Kollmar 2016).
Notably, ten of the 20 genes encoding cytoskeletal functions that were duplicated in LECA (supplementary tables 3 and 4) encode dynein or kinesin motor proteins (see also Tromer et al. 2019). The bacterial duplicate contribution vastly outnumbers the archaeal contribution to these categories, which are dominated by eukaryote-specific genes, indicating that eukaryotes not only acquired genes, but they also invented new ones as well (Lane and Martin 2010). Duplications in LECA depict bacterial carbon and energy metabolism in an archaeal host supported by genes that were recurrently donated by a resident symbiont, in line with the predictions of symbiotic theories for the nature of the first eukaryote (Martin and Müller 1998; Martin et al. 2017; Imachi et al. 2020). The functions of duplications are consistent with the predictions of symbiogenic theories but contrast with gradualist theories positing eukaryote origin from an archaeal lineage that attained eukaryote-like complexity in the absence of the mitochondrial endosymbiont (Cavalier-Smith 2002; Booth and Doolittle 2015; Pittis and Gabaldón 2016; Hampl et al. 2019).
What Does This Say about the Biology of LECA?
Gene transfers from the mitochondrion can generate duplications of bacterial-derived genes. What mechanisms promoted genome-wide gene duplication at the prokaryote–eukaryote transition? Population genetic parameters such as variation in population size (Zachar and Szathmáry 2017) apply to prokaryotes and eukaryotes equally, hence they would not affect gene duplications specifically in eukaryotes, but recombination processes (Garg and Martin 2016) in a nucleated cell could. Because LECA possessed meiotic recombination (Speijer et al. 2015), it was able to fuse nuclei (karyogamy). Karyogamy in a multinucleate LECA would promote the accumulation of duplications in all gene classes and promote genome expansion to its energetically permissible limits (Lane and Martin 2010) because unequal crossing between imprecisely paired homologous chromosomes following karyogamy generates duplications (Ohno 1970; Scannell et al. 2006; Hittinger and Carroll 2007; Van De Peer 2009). At the origin of meiotic recombination, chromosome pairing and segregation cannot have been perfect from the start; the initial state was likely error-prone, generating nuclei with aberrant gene copies, aberrant chromosomes, and even aberrant chromosome numbers. In cells with a single nucleus, such variants would have been lethal; in multinucleate (syncytial or coenocytic) organisms, defective nuclei can complement each other through mRNA in the cytosol (Garg and Martin 2016). Multinucleate forms are present throughout eukaryotic lineages (fig. 5), and ancestral reconstruction of nuclear organization clearly indicates that LECA itself was multinucleate (fig. 5 and supplementary fig. 1, Supplementary Material online). The multinucleate state enables the accumulation of duplications in the incipient eukaryotic lineage in a mechanistically nonadaptive manner, whereby duplications are implicated in the evolution of complexity (Ohno 1970; Scannell et al. 2006; Hittinger and Carroll 2007; Van De Peer 2009), as observed in the animal lineage (fig. 1). The syncytial state presents a viable intermediate state in the transition from prokaryote to eukaryote genetics.
Fig. 5.
Ancestral state reconstruction for nuclear organization in eukaryotes. Presence and absence of the multinucleate state in members of the respective group are indicated. Resolution of the branches (polytomy vs. dichotomy) does not alter the outcome of the ancestral state reconstruction, nor does position of the root on the branches leading to Amoebozoa, Excavata, or Opisthokonta. LECA was a multinucleate, syncytial cell, not uninucleate (see supplementary fig. 1, Supplementary Material online). Together with mitochondrion and sex, the multinucleate state is ancestral to eukaryotes and fostered accumulation of duplications (see text).
Conclusion
Serial transfers of mitochondrial DNA to the chromosomes of the host are not only a mechanism of gene duplication, they are a form of endosymbiont genome duplication in which an original copy is retained in the organelle and remains functional. Gene duplications in LECA support an early origin of mitochondria and record the onset of the eukaryotic gene duplication process, a hallmark of genome evolution in mitosing cells (Ohno 1970; Scannell et al. 2006; Hittinger and Carroll 2007; Van De Peer 2009; Treangen and Rocha 2011).
Materials and Methods
Protein Clustering and Tree Reconstruction for Gene Duplication Inferences
Protein sequences for 150 eukaryotic genomes were downloaded from NCBI, Ensembl Protists, and JGI (see supplementary data 1 for detailed species composition). To construct gene families, we performed an all-vs-all BLAST (Altschul et al. 1997) of the eukaryotic proteins and selected the reciprocal best BLAST hits with e-value ≤10−10. The protein pairs were aligned with the Needleman–Wunsch algorithm (Rice et al. 2000) and the pairs with global identity values <25% were discarded. The retained global identity pairs were used to construct gene families with the Markov clustering algorithm (Enright et al. 2002) (version 12-068) with default parameters. Because in this study we were interested in gene duplications, we considered only the gene families with multiple gene copies in at least two eukaryotic genomes. Our criteria retained a total of 24,571 multicopy gene families.
Protein-sequence alignments for the individual eukaryotic multicopy gene families were generated using MAFFT (Katoh 2002), with the iterative refinement method that incorporates local pairwise alignment information (L-INS-i, version 7.130). The alignments were used to reconstruct maximum likelihood trees with IQ-tree (Nguyen et al. 2015), using default settings (version 1.6.5), and the trees were rooted with MAD (Tria et al. 2017) (supplementary data 2).
Inference of Gene Duplication
Gene duplications were inferred from gene trees by assigning duplication events to internal nodes in the rooted topologies. Given a rooted gene tree with n leaves, let S be the set of species labels for the leaves. For the case of paralogous gene trees, there is at least one leaf pair, a and b, such that sa=sb. Assigning a gene duplication to the last common ancestor of the pair a and b corresponds to the evolutionary scenario that minimizes paralog losses in the gene tree. For each rooted gene tree, we performed pairwise comparisons of all leaf pairs with identical species labels to infer all the internal nodes corresponding to gene duplications using the minimal loss criterion for each leaf pair. Note that, this approach considers the possibility of multiple gene duplications per gene tree (supplementary fig. 2, Supplementary Material online). We summarized the gene duplication inferences from all gene trees by evaluating the distribution of descendant paralogs across the eukaryotic supergroups for each gene duplication event (fig. 2).
The inferences of gene duplications in the present work are based on trees that were rooted with MAD (Tria et al. 2017). A recent comparison of MAD with other methods showed that MAD performs better than other rooting methods currently in use (Wade et al. 2020).
Inference for the Origin of Eukaryotic Duplicates
For identification of homologs in prokaryotes, we used all protein-coding genes from 5,656 prokaryotic genomes downloaded from RefSeq (Pruitt et al. 2007) (see supplementary data 3) and compared them against eukaryotic protein-coding genes using Diamond (Buchfink et al. 2015) to perform sequence searches with the “more-sensitive” parameter. A eukaryotic gene family was considered to have homologs in prokaryotes if at least one gene of the eukaryotic family had a significant hit against a prokaryotic gene (e-value <10−10 and local identity ≥25%). Gene families with homologs only in archaeal genomes were considered as genes of archaeal origin and similarly for bacteria. Gene families with significant hits in both archaea and bacteria (universal) could have originated from either archaea or bacteria.
We purposefully avoided using trees to inferring the origin of eukaryotic genes because of low levels of sequence conservation entailing a large number of prokaryotic homologs. Note, however, that we reconstructed trees for the subset of eukaryote–prokaryote genes with sufficient sequence conservation (see below). We found that the presence–absence of homologs across prokaryotic taxa remarkably recapitulates the distribution of prokaryotic sisters derived from phylogenetic trees serving, thus, as a validation of our approach (supplementary table 5).
Prokaryote–Eukaryote Protein Clustering and Tree Reconstruction
To assemble a data set of conserved genes for phylogenies linking prokaryotes and eukaryotes, eukaryotic, archaeal, and bacterial protein sequences were first clustered separately before homologous clusters between eukaryotes and prokaryotes were identified as described (Ku et al. 2015). Eukaryotic sequences for the 150 genomes (supplementary data 1) were clustered with MCL (Enright et al. 2002) using global identities from best reciprocal BLAST (Altschul et al. 1997) hits for protein pairs with e-value ≤10−10 and global identity ≥40%. The clusters with genes distributed in more than one eukaryotic genome were retained. Similarly, prokaryotic protein sequences from 5,655 genomes (see supplementary data 3, except for MK-D1 for which the genome was unavailable by the time the data were compiled) were clustered using the best reciprocal BLAST for protein pairs with e-value ≤10−10 and global identity ≥25%, for archaea and bacteria separately. The resulting clusters with gene copies in at least five prokaryotic genomes were retained. The most universally distributed clusters comprise 20–40 proteins, the majority of which are involved in translation (supplementary fig. 4, Supplementary Material online). Eukaryotic and prokaryotic clusters were merged using the reciprocal best cluster procedure. We merged a eukaryotic cluster with a prokaryotic cluster if ≥50% of the eukaryotic sequences in the cluster have their best reciprocal BLAST hit in the same prokaryotic cluster and vice versa (cut-offs: e-value ≤10−10 and local identity ≥30%). We refer to the merged cluster as eukaryotic–prokaryotic cluster (EPC).
Protein-sequence alignments for 2,575 EPCs were generated using MAFFT (Katoh 2002) (L-INS-i, version 7.130). The alignments were used to reconstructed maximum-likelihood trees with IQ-tree (Nguyen et al. 2015) (version 1.6.5) employing default settings (supplementary data 4).
Tests for Eukaryote Monophyly
For 475 gene trees where eukaryotes were not recovered as monophyletic, we conducted the Shimodaira–Hasegawa (Shimodaira and Hasegawa 1999) (SH), Kishino–Hasegawa (Kishino and Hasegawa 1989) (KH), and approximately unbiased (AU) test (Shimodaira 2002) to determine whether the observed nonmonophyly was statistically significant. We reconstructed trees constraining eukaryotic sequences to be monophyletic, but not imposing any other topological constraint, using FastTree (Price et al. 2010) (version 2.1.10 SSE3) and recording all trees explored during the tree search with the “-log” parameter (supplementary data 5). The sample of monophyletic trees was used as input in IQ-tree (Nguyen et al. 2015) (version 2.0.3; parameter: “-zb 100000 –au”) to perform the SH, KH, and AU tests against the unconstrained tree (nonmonophyletic). If the best-constrained tree did not show significant difference relative to the unconstrained tree (P <0.05), then we considered that eukaryotic monophyly cannot be rejected.
Inference of Prokaryotic Sisters
To infer prokaryotes sisters to eukaryotes in the gene trees we used the unconstrained tree if eukaryotes were recovered as monophyletic and the constrained tree if eukaryotes were not recovered as monophyletic, since the SH test did not reject eukaryote monophyly for any gene tree (see main text). Note that in unrooted trees for which eukaryotes are monophyletic, the prokaryotic side of the tree is bisected by one internal node into two prokaryotic subclades, each subclade being the potential sister to eukaryotes (see fig. 4a). We considered the prokaryotic subclade with the smallest number of leaves for our inferences of sister-relations and the prokaryotic phyla present in the sister clade and outgroup clade was recorded for each tree. The sister clades were scored as a “pure” sister when only a single prokaryotic phylum was present in the clade or as “mixed” sister when more than one phylum was present.
Ancestral Reconstruction of Eukaryotic Nuclear Organization
Ancestral state reconstructions were performed on the basis of a morphological character matrix, using maximum parsimony as implemented in Mesquite 3.6 (https://www.mesquiteproject.org/, accessed June 2019). The reference eukaryotic phylogeny includes 106 taxa (ranging from genus to phylum level) to reflect the relations within the eukaryotes and reduce taxonomic redundancy. The phylogeny includes members of six supergroups: Amoebozoa (Mycetozoa), Archaeplastida, Excavata, Hacrobia, Opisthokonta, and SAR, and was constructed by combining branches from previous studies (Burki et al. 2010; Yoon et al. 2010; Adl et al. 2012; Powell and Letcher 2014; Burki et al. 2016; Cavalier-Smith et al. 2016; Derelle et al. 2016; Spatafora et al. 2016; Yang et al. 2016; Archibald et al. 2017; Krabberød et al. 2017; McCarthy and Fitzpatrick 2017; Roger et al. 2017; Spatafora et al. 2017; Bass et al. 2018; Cavalier-Smith et al. 2018; Tedersoo et al. 2018; Irwin et al. 2019). The nuclear organization for each taxon was coded as 0 for nonmultinucleate, 1 for multinucleate or 0/1 if ambiguous according to the literature (Byers 1979; Willumsen et al. 1987; Barthel and Detmer 1990; Daniels and Pappas 1994; Walker et al. 2006; Steiner 2010; Yoon et al. 2010; Adl et al. 2012; Niklas et al. 2013; Maciver 2016; Spatafora et al. 2016; Archibald et al. 2017; Bloomfield et al. 2019) (supplementary data 6). In order to account for uncertainties of lineage relations among eukaryotes, we used a set of phylogenies with alternative root positions (Vossbrinck et al. 1987; Stechmann and Cavalier-Smith 2002; Katz and Grant 2015) (altogether a total of 15 different roots) as well as the consideration of polytomies for debated branches (supplementary data 6). All ancestral state reconstruction rendered LECA as multinucleated, with no ambiguity. Ambiguous reconstructions, however, were observed within supergroups in some topologies but did not pose ambiguity to the reconstructed state in LECA.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the European Research Council (Grant No. 666053), the Volkswagen Foundation (Grant No. 93 046), and the Moore Simons Initiative on the Origin of the Eukaryotic Cell (Grant No. 9743) for financial support. We also thank Damjan Franjević (Department of Biology, University of Zagreb, Croatia) for helpful discussions.
Author Contributions
All authors conceived and designed the study. J.B. and J.S. prepared the data sets with contribution from all the authors. F.D.K.T. performed gene duplication inferences, functional annotation of genes, and the tests for eukaryotic monophyly. J.B. and F. N. performed the analyses of eukaryotic sisters. J.S. compiled the eukaryotic phylogenies and performed ancestral state reconstructions. All authors wrote the paper.
Data Availability
Supplementary tables and data used in this study are available under the link https://doi.org/10.6084/m9.figshare.12249260.
Code Availability
Custom Matlab scripts used to perform data analysis are available upon request.
Literature Cited
- Adl SM, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59(5):429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet. 17(7):379–391. [DOI] [PubMed] [Google Scholar]
- Allen JF. 2015. Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc Natl Acad Sci U S A. 112(33):10231–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Blast and Psi-Blast: protein database search programs. Nucleic Acid Res. 25:2289–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, et al. 2003. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr Biol. 13:94–104. [DOI] [PubMed] [Google Scholar]
- Archibald JM, et al. 2017. Handbook of the protists. Cham: Springer Nature. [Google Scholar]
- Barlow LD, Nývltová E, Aguilar M, Tachezy J, Dacks JB. 2018. A sophisticated, differentiated Golgi in the ancestor of eukaryotes. BMC Biol. 16(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel D, Detmer A. 1990. The spermatogenesis of Halichondria panicea (Porifera, Demospongiae). Zoomorphology 110:9–15. [Google Scholar]
- Bass D, et al. 2018. Clarifying the relationships between microsporidia and cryptomycota. J Eukaryot Microbiol. 65(6):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts HC, et al. 2018. Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat Ecol Evol. 2:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield G, et al. 2019. Triparental inheritance in Dictyostelium. Proc Natl Acad Sci U S A. 116(6):2187–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A, Doolittle WF. 2015. Eukaryogenesis, how special really? Proc Natl Acad Sci U S A. 112(33):10278–10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Burki F, et al. 2010. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol Biol. 10:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, et al. 2016. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc R Soc Lond B. 283:20152802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers TJ. 1979. Growth, reproduction, and differentiation in Acanthamoeba. Int Rev Cytol. 61:283–338. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. 2020. Multidomain ribosomal protein trees and the planctobacterial origin of neomura (eukaryotes, archaebacteria). Protoplasma 257(3):621–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, et al. 2016. 187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol Phylogenet Evol. 99:275–296. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, et al. 2018. Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria. Protoplasma 255(5):1517–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1975. The origin of nuclei and of eukaryotic cells. Nature 256:463–468. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 52(Pt 2):297–354. [DOI] [PubMed] [Google Scholar]
- Daniels EW, Pappas GD. 1994. Reproduction of nuclei in Pelomyxa palustris. Cell Biol Int. 18(8):805–812. [DOI] [PubMed] [Google Scholar]
- de Duve C. 2007. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 8(5):395–403. [DOI] [PubMed] [Google Scholar]
- Derelle R, et al. 2016. Phylogenomic framework to study the diversity and evolution of Stramenopiles (= Heterokonts). Mol Biol Evol. 33(11):2890–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313(5785):314–318. [DOI] [PubMed] [Google Scholar]
- Doolittle FW. 1998. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14(8):307–311. [DOI] [PubMed] [Google Scholar]
- Embley T, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440(7084):623–630. [DOI] [PubMed] [Google Scholar]
- Eme L, et al. 2017. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 15(12):711–723. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, et al. 2004. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 21(9):1643–1660. [DOI] [PubMed] [Google Scholar]
- Esser C, Martin W, Dagan T. 2007. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 22:180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Santangelo TJ, Beyer AL, Reeve JN. 2007. Transcription and translation are coupled in Archaea. Mol Biol Evol. 24(4):893–895. [DOI] [PubMed] [Google Scholar]
- Gabaldón T. 2018. Relative timing of mitochondrial endosymbiosis and the “pre-mitochondrial symbioses” hypothesis. IUBMB Life. 70(12):1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SG, Martin WF. 2016. Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Genome Biol. Evol. 8:1950–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Garg SG, Martin WF. 2016. Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol. 24(7):525–534. [DOI] [PubMed] [Google Scholar]
- Gray MW. 2014. The pre-endosymbiont hypothesis: a new perspective on the origin and evolution of mitochondria. Cold Spring Harb Perspect Biol. 6:a016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Čepička I, Eliáš M. 2019. Was the mitochondrion necessary to start eukaryogenesis? Trends Microbiol. 27(2):96–104. [DOI] [PubMed] [Google Scholar]
- Hampl V, et al. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic ‘supergroups’. Proc Natl Acad Sci U S A. 106(10):3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, et al. 2014. An alternative root for the eukaryote tree of life. Curr Biol. 24(4):465–470. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449(7163):677–681. [DOI] [PubMed] [Google Scholar]
- Imachi H, et al. 2020. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577(7791):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin NA, et al. 2019. Phylogenomics supports the monophyly of the Cercozoa. Mol Phylogenet Evol. 130:416–423. [DOI] [PubMed] [Google Scholar]
- Javaux EJ, Lepot K. 2018. The Paleoproterozoic fossil record: implications for the evolution of the biosphere during Earth’s middle-age. Earth Sci Rev. 176:68–86. [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22(4):225–231. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Grant JR. 2015. Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst Biol. 64(3):406–415. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer LD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 9(8):605–618. [DOI] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. 1989. Evaluation of the maximum-likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 29(2):170–179. [DOI] [PubMed] [Google Scholar]
- Kollmar M. 2016. Fine-tuning motile cilia and flagella: evolution of the dynein motor proteins from plants to humans at high resolution. Mol Biol Evol. 33(12):3249–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabberød AK, et al. 2017. Single cell transcriptomics, mega-phylogeny, and the genetic basis of morphological innovations in Rhizaria. Mol Biol Evol. 34(7):1557–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C, et al. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524(7566):427–432. [DOI] [PubMed] [Google Scholar]
- Ku C, Martin WF. 2016. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70% rule. BMC Biol. 14(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Rivera MC. 1994. Was the nucleus the first endosymbiont? Proc Natl Acad Sci U S A. 91(8):2880–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467(7318):929–934. [DOI] [PubMed] [Google Scholar]
- Leger MM, et al. 2018. Demystifying eukaryote lateral gene transfer. Bioessays 40(5):e1700242. [DOI] [PubMed] [Google Scholar]
- López-García P, Moreira D. 1999. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 24:88–93. [DOI] [PubMed] [Google Scholar]
- López-García P, Moreira G. 2015. Open questions on the origin of eukaryotes. Trends Ecol Evol. 30(11):697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK. 2016. Asexual amoebae escape Muller's ratchet through polyploidy. Trends Parasitol. 32(11):855–862. [DOI] [PubMed] [Google Scholar]
- Makarova KS, et al. 2005. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 33(14):4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Chapman M, Guerrero R, Hall J. 2006. The last eukaryotic common ancestor (LECA): acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proc Natl Acad Sci U S A. 103(35):13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, et al. 2000. The chimeric eukaryote: origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci U S A. 97(13):6954–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. 1999. Mosaic bacterial chromosomes: a chalenge en route to a tree of genomes. Bioessays 21:99–104. [DOI] [PubMed] [Google Scholar]
- Martin W. 2010. Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond B Biol Sci. 365(1541):847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Brinkmann H, Savonna C, Cerff R. 1993. Evidence for a chimeric nature of nuclear genomes: eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci U S A. 90(18):8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, et al. 2001. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 382(11):1521–1539. [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392(6671):37–41. [DOI] [PubMed] [Google Scholar]
- Martin WF, Garg S, Zimorski V. 2015. Endosymbiotic theory for eukaryote origin. Philos Trans R Soc Lond B. 370:20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF, Tielens AGM, Mentel M, Garg SG, Gould SB. 2017. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol. Mol Biol Rev. 81:e00008–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy CG, Fitzpatrick DA. 2017. Multiple approaches to phylogenomic reconstruction of the fungal kingdom. Adv Genet. 100:211–266. [DOI] [PubMed] [Google Scholar]
- Mereschkowsky C. 1905. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol Centralbl. 25:593–604. English translation in Martin W, Kowallik KV. 1999. Annotated English translation of Mereschkowsky’s 1905 paper ‘Über Natur und Ursprung der Chromatophoren im Pflanzenreiche ’. Eur J Phycol., 34:287–295. [Google Scholar]
- Nagies FSP, Brueckner J, Tria FDK, Martin WF. 2020. A spectrum of verticality across genes. PLoS Genet. 16(11):e1009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. 1997. Evolution by birth and death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A. 94(15):7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, et al. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 109(50):20537–20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ, et al. 2013. The evo‐devo of multinucleate cells, tissues, and organisms, and an alternative route to multicellularity. Evol Dev. 15(6):466–474. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Heidelberg (Berlin: ): Springer. [Google Scholar]
- Parfrey LW, et al. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 108:1364–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittis AA, Gabaldón T. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531(7592):101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AM, Gribaldo S. 2014. Eukaryotic origin: how and when was the mitochondrion acquired? Cold Spring Harb Perspect Biol. 6(12):a015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugez S, Martin WF, Hazkani-Covo E. 2018. Mosaic mitochondrial-plastid insertions into the nuclear genome show evidence of both non-homologous end joining and homologous recombination. BMC Evol Biol. 18(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MJ, Letcher PM. 2014. 6 Chytridiomycota, Monoblepharidomycota, and Neocallimastigomycota. In: McLaughlin DJ, Spatafora JW, editors. 2nd ed. The Mycota Part VII A. Systematics and evolution. Heidelberg (Berlin: ): Springer. p. 141–175. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35(Database issue):D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, et al. 2016. Phylogenetic resolution of deep eukaryotic and fungal relationships using highly conserved low-copy nuclear genes. Genome Biol Evol. 8(9):2683–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, et al. 2000. EMBOSS: the European Molecular Biology Open software suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Río Bártulos C, et al. 2018. Mitochondrial glycolysis in a major lineage of eukaryotes. Genome Biol Evol. 10(9):2310–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Muñoz-Gómez SA, Kamikawa R. 2017. The origin and diversification of mitochondria. Curr Biol. 27(21):R1177–R1192. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440(7082):341–345. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3):492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 16:1114–1116. [Google Scholar]
- Spang A, et al. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, et al. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108(5):1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, et al. 2017. The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol Spectr. 5(5):1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer D, Lukeš J, Eliáš M. 2015. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc Natl Acad Sci U S A. 112(29):8827–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. 2002. Rooting the eukaryote tree by using a derived gene fusion. Science 297(5578):89–91. [DOI] [PubMed] [Google Scholar]
- Steiner JM. 2010. Technical notes: growth of Cyanophora paradoxa. J Endoc Cell Res. 20:62–67. [Google Scholar]
- Tedersoo L, et al. 2018. High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Div. 90:135–159. [Google Scholar]
- Timmis JN, Ayliff MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5(2):123–135. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Rocha EPC. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7(1):e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tria FDK, Landan G, Dagan T. 2017. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol. 1:0193. [DOI] [PubMed] [Google Scholar]
- Tromer EC, van Hooff JJE, Kops GJPL, Snel B. 2019. Mosaic origin of the eukaryotic kinetochore. Proc Natl Acad Sci U S A. 116(26):12873–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Peer Y, Maere S, Meyer A. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 10(10):725–732. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, et al. 1987. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326(6111):411–414. [DOI] [PubMed] [Google Scholar]
- Vosseberg J, et al. 2021. Timing the origin of eukaryotic cellular complexity with ancient duplications. Nat Ecol Evol. 5(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade T, et al. 2020. Assessing the accuracy of phylogenetic rooting methods on prokaryotic gene families. PLoS One 15(5):e0232950–e0233022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, et al. 2006. Ultrastructural descripton of Breviata anathema, n. gen., n. sp., the organism previously studied as “Mastigamoeba invertens”. J Eukaryot Microbiol. 53(2):65–78. [DOI] [PubMed] [Google Scholar]
- Wallin IE. 1925. On the nature of mitochondria. IX. Demonstration of the bacterial nature of mitochondria. Am J Anat. 36:131–139. [Google Scholar]
- Willumsen NB, et al. 1987. A multinucleate amoeba, Parachaos zoochlorellae (Willumsen 1982) comb, nov., and a proposed division of the genus Chaos into the Genera Chaos and Parachaos (Gymnamoebia, Amoebidae). Archiv Protist. 134:303–313. [Google Scholar]
- Yang EC, et al. 2016. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci Rep. 6:21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, et al. 2010. Evolutionary history and taxonomy of red algae. In: Seckbach J, Chapman DJ, editors. Red algae in genomic age. Dordrecht: Springer. p. 27–45. [Google Scholar]
- Zachar I, Szathmáry E. 2017. Breath-giving cooperation: critical review of origin of mitochondria hypotheses. Biol Direct. 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541(7637):353–358. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Godzik A. 2011. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 12(1):R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary tables and data used in this study are available under the link https://doi.org/10.6084/m9.figshare.12249260.