Abstract
Hermansky–Pudlak syndrome (HPS) is a group of ten autosomal recessive multisystem disorders, each defined by deficiency of a specific gene. HPS-associated genes encode components of four ubiquitously expressed protein complexes: Adaptor Protein (AP)-3 and Biogenesis of Lysosome-related Organelles Complex (BLOC)-1 through −3. All individuals with HPS exhibit albinism and a bleeding diathesis; additional features occur depending on the defective protein complex. Pulmonary fibrosis is associated with AP-3 and BLOC-3 deficiency, immunodeficiency with AP-3 defects, and gastrointestinal symptoms are more prevalent and severe in BLOC-3 deficiency. Therefore, identification of the HPS subtype is valuable for prognosis, clinical management and treatment options. The prevalence of HPS is estimated at 1–9 per 1,000,000. Here we summarize 264 reported and novel variants in ten HPS genes and estimate that ~333 Puerto Rican HPS subjects and ~385 with other ethnicities are reported to date. We provide pathogenicity predictions for missense and splice site variants and list variants with high minor allele frequencies (MAF). Current cellular and clinical aspects of HPS are also summarized. This review can serve as a manifest for molecular diagnostic and genetic counseling aspects of Hermansky-Pudlak syndrome.
Keywords: albinism, biogenesis of lysosome-related organelles, bleeding diathesis, granulomatous colitis, hypopigmentation, pulmonary fibrosis
INTRODUCTION
In 1959, two Czechoslovakian clinicians, Frantisek Hermansky and Paulus Pudlak, described what is now called Hermansky-Pudlak syndrome (HPS) in two unrelated individuals with a bleeding disorder associated with oculocutaneous albinism (Hermansky & Pudlak, 1959). Over the last six decades, the syndrome has expanded to a disorder with 10 distinct genetic causes, with over 715 cases reported worldwide and a vastly improved understanding of clinical symptoms, pathomechanism and therapeutic approaches (Bowman, Bi-Karchin, Le, & Marks, 2019; Gahl et al., 1998; Huizing, Helip-Wooley, Westbroek, Gunay-Aygun, & Gahl, 2008; Huizing, Malicdan, Gochuico, & Gahl, 2017 Oct 26 [Updated 2000 July 24]).
HPS (MIM# 203300) is a genetically heterogeneous autosomal recessive multisystem disorder characterized by oculocutaneous albinism, a bleeding diathesis, and, in some cases, granulomatous colitis, neutropenia, or a fatal pulmonary fibrosis (Gahl et al., 1998; Huizing et al., 2008; Huizing et al, 2017 Oct 26 [Updated 2000 July 24]). These features result from defects in lysosome-related organelles (LROs), such as melanosomes in melanocytes and delta granules in platelets (Bowman et al., 2019; Dell’Angelica, 2004; Marks, Heijnen, & Raposo, 2013; Raposo, Marks, & Cutler, 2007). The ten described human HPS subtypes (HPS-1 through HPS-10), are each associated with a specific gene defect (Table 1). Orthologs of these ten genes also cause HPS in mice and other animal models (Table 1) (Huizing et al., 2008; Li et al., 2004).
Table 1:
Overview of Hermansky-Pudlak Syndrome Subtypes, (Candidate) Genes and Protein Complexes
| HPS Subtype | Gene Name 1 (alternative names) | Protein Complex | Human Locus | mRNA; GeneID 2 (# exons # splice variants) | Protein ID (# amino acids; molecular weight) | Reported Cases | Human Pathogenic Variants | Animal Model 3 |
|---|---|---|---|---|---|---|---|---|
| HPS-1 | HPS1 (BLOC3S1) | BLOC-3 | 10q24.2 |
NM_000195; ID: 3257 (20 exons; 18 variants) |
NP_000186 (700 aa; 79.3 kD) |
~148 4 ~261 (dup16-bp) 5 |
76 4 | pale ear (m) |
| HPS-2 | AP3B1 (ADTB3) | AP-3 | 5q14.1 |
NM_003664; ID: 8546 (27 exons; 2 variants) |
NP_003655 (1094 aa; 121.3 kD) |
~35 | 29 |
pearl (m),
Ap3b1LN (m) |
| HPS-3 | HPS3 (BLOC2S1) | BLOC-2 | 3q24 |
NM_032383; ID: 84343 (17 exons; 2 variants) |
NP_115759 (1004 aa; 113.7 kD) |
~42 6 ~72 (del 3.9-kb) 7 |
37 6 | cocoa (m) |
| HPS-4 | HPS4 (BLOC3S2) | BLOC-3 | 22q12.1 |
NM_022081; ID: 89781 (14 exons; 11 variants) |
NP_071364 (708 aa; 76.9 kD) |
~41 8 | 34 8 | light ear (m) |
| HPS-5 | HPS5 (BLOC2S2) | BLOC-2 | 11p15.1 |
NM_181507; ID: 11234 (23 exons; 3 variants) |
NP_852608 (1129 aa; 127.4 kD) |
~29 | 31 | ruby-eye-2 (m), snow white (z), casper (sb) |
| HPS-6 | HPS6 (BLOC2S3) | BLOC-2 | 10q24.32 |
NM_024747; ID: 79803 (1 exon; 1 variant) |
NP_079023 (775 aa; 83.0 kD) |
~46 9 ~20 (1065insG) 10 |
45 9 |
ruby-eye (m) no privacy (x) |
| HPS-7 | DTNBP1 (BLOC1S8, HPS7) | BLOC-1 | 6p22.3 |
NM_032122; ID: 84062 (10 exons; 5 variants) |
NP_115498 (351 aa; 39.5 kD) |
8 | 4 | sandy (m) |
| HPS-8 | BLOC1S3 (HPS8, RP, BLOS3) | BLOC-1 | 19q13.32 |
NM_212550; ID: 388552 (2 exons; 1 variant) |
NP_997715 (202 aa; 21.3 kD) |
3 6 (448delC) 11 |
4 | reduced pigmentation (m) |
| HPS-9 | BLOC1S6 (HPS9, PLDN, BLOS6) | BLOC-1 | 15q21.1 |
NM_001311255; ID: 26258 (5 exons; 3 variants) |
NP_001298184 (177 aa; 20.3 kD) |
3 | 2 | pallid (m) |
| HPS-10 | AP3D1 (HPS10, ADTD) | AP-3 | 19p13.3 |
NM_001261826; ID: 8943 (32 exons; 2 variants) |
NP_001248755 (1215 aa; 136.7 kD) |
4 | 2 | mocha (m) |
| - | BLOC1S4 (CNO, BLOS4) | BLOC-1 | 4p16.1 |
NM_018366; ID: 55330 (1 exon; 1 variant) |
NP_060836 (217 aa; 23.3 kD) |
- | - | cappuccino (m) |
| - | BLOC1S5, MUTED, BLOS5 | BLOC-1 | 6p24.3 |
NM_201280; ID: 63915 (5 exons; 3 variants) |
NP_958437 (187 aa; 21.6 kD) |
- | - | muted (m) |
| - |
BLOC1S1, BLOS1 |
BLOC-1 (BORC) 12 | 12q13.2 |
NM_001487; ID: 2647 (4 exons; 1 variant) |
NP_001478 (153 aa; 17.2 kD) |
- | - |
Blos1Ell-Cre/loxp
(m) Blos1nestin-Cre/loxp (m) bloc1s1ihb815 (z) |
| - |
BLOC1S2, BLOS2 |
BLOC-1 (BORC) |
10q24.31 |
NM_173809; ID: 282991 (5 exons; 6 variants) |
NP_776170 (142 aa; 16.0 kD) |
- | - |
Bloc1s2−/−
(m) bloc1s2ihb818 (z) |
| - | BLOC1S7, BLOS7 SNAPIN, SNAPAP | BLOC-1 (BORC) 12 |
1q21.3 |
NM_012437; ID: 23557 (4 exons; 1 variant) |
NP_36569 (136 aa; 14.9 kD) |
- | - | snapin−/− (m) |
| - | AP3M1, Mu3A | AP-3 | 10q22.2 |
NM_207012; ID: 26985 (10 exons; 5 variants) |
NP_996895 (418 aa; 46.9 kD) |
- | - | - |
| - | AP3S1, Sigma3A | AP-3 | 5q22.3-q23.1 |
NM_001284; ID: 1176 (6 exons; 6 variants) |
NP_001275 (193 aa; 21.7 kD) |
- | - | - |
| Total: |
385 non-PR
13 333 PR |
264 variants | ||||||
The commonly used HPS-subtype-related gene name is grey highlighted, alternative names are listed in brackets.
Genbank accession numbers of the mRNA encoding the longest isoform (often transcript variant 1), its number of exons, GeneID, and the number of predicted protein-encoding splice variants of each HPS gene. As of November 2019.
Reported vertebrate HPS animal models: m, mouse; sb, stickleback (gasterosteus aculeatus); x, xenopus tropicalis (frog); z, zebrafish (danio rerio), see (Bowman et al., 2019; Huizing et al., 2008) for details and references. See (Bowman et al., 2019) for invertebrate HPS models.
The ~148 reported non-Puerto Rican HPS-1 cases include 11 novel NIH cohort cases. The 76 reported HPS1 variants include 5 novel variants from the NIH HPS cohort.
The ~261 reported HPS-1 cases with the northwest Puerto Rican founder variant c.1472_1487dup16-bp is an estimate and includes 166 cases from the NIH cohort and ~ 95 cases from the literature (Oh et al., 1996; Oh et al., 1998; Santiago Borrero et al., 2006). The number of worldwide cases homozygous for the HPS1 c.1472_1487dup16-bp variant is estimated to be ~ 400 (Santiago Borrero et al., 2006).
The ~42 reported HPS-3 cases include those reported with the Ashkenazi-Jewish founder variants (7 cases) as well as those novel from the NIH cohort (9 cases). The 37 reported HPS3 variants include 11 novel variants from the NIH cohort.
The ~72 HPS-3 cases with the central Puerto Rican founder 3.9-kb del variant (NM_032383.5:c.-2993_217+692del) include 63 cases from the literature (Anikster et al., 2001; Santiago Borrero et al., 2006) and 9 additional cases from the NIH cohort.
The ~41 reported HPS-4 cases include 4 novel cases from the NIH cohort. The 34 reported HPS4 variants include 4 novel NIH cohort variants.
The ~46 reported HPS-6 cases include one novel case from the NIH cohort. The 45 reported HPS6 variants include 2 novel variants from the NIH cohort.
The ~20 reported HPS-6 cases with the c.1065insG variant are part of an extended Israeli Muslim Bedouin family (Schreyer-Shafir et al., 2006).
One Pakistani family with 6 affected HPS-8 cases homozygous for c.448delC was reported (Morgan et al., 2006).
BORC = BLOC-one-related complex (Pu et al., 2015).
PR = Puerto Rican.
HPS is a rare disorder with an estimated worldwide prevalence of 1–9 per 1,000,000 individuals (Christensen, Wagner, Coleman, & Appell, 2017; Huizing et al, 2017 Oct 26 [Updated 2000 July 24]). However, the prevalence per subtype can differ due to founder mutations. HPS-1 is more common in Puerto Rico, particularly in the northwestern part of the island where about 1 in 1,800 people are affected and carry the same homozygous mutation (Witkop, Almadovar, Pineiro, & Nunez Babcock, 1990). HPS-1 has also been reported in a small isolate in a Swiss village (Oh et al., 1998; Schallreuter, Frenk, Wolfe, Witkop, & Wood, 1993) and one in Japan (S. Ito et al., 2005). HPS-3 is common in central Puerto Rico, where about 1 in 4,000 individuals are affected (Anikster et al., 2001; Santiago Borrero et al., 2006). Individuals with HPS have been described in many other regions, including China, India, South America and Western Europe (Arcot Sadagopan et al., 2017; Carmona-Rivera et al., 2011; Hermos, Huizing, Kaiser-Kupfer, & Gahl, 2002; Wei et al., 2016).
The protein products of the HPS genes assemble in four multi-subunit complexes, each involved in distinct steps of membrane trafficking and/or component sorting required for LRO biogenesis (Table 1) (Bowman et al., 2019; Dell’Angelica, 2004; Huizing et al., 2008). The adaptor protein-3 (AP-3) complex consist of 4 subunits and includes the protein products of AP3B1, which is mutated in HPS-2 (Dell’Angelica, Shotelersuk, Aguilar, Gahl, & Bonifacino, 1999), and AP3D1, mutated in HPS-10 (Ammann et al., 2016). Biogenesis of Lysosome-related Organelles Complex (BLOC)-3 consists of the HPS1 and HPS4 proteins (Martina, Moriyama, & Bonifacino, 2003), defective in disease subtypes HPS-1 and HPS-4, respectively (Oh et al., 1998; Suzuki et al., 2002). BLOC-2 consists of HPS3, HPS5 and HPS6 (Di Pietro, Falcon-Perez, & Dell’Angelica, 2004), whose defects cause subtypes HPS-3, HPS-5 and HPS-6 (Anikster et al., 2001; Huizing et al., 2001; Q. Zhang et al., 2003). BLOC-1 consists of 8 subunits (Falcon-Perez, Starcevic, Gautam, & Dell’Angelica, 2002; Starcevic & Dell’Angelica, 2004), including DTNBP1, BLOC1S3, and PLDN, defective in HPS-7, HPS-8 and HPS-9, respectively (Badolato et al., 2012; Li et al., 2003; Morgan et al., 2006). No defects in humans are reported in the other five BLOC-1 or two AP-3 subunits, but some are defective in HPS-like animal models (Table 1) (Bowman et al., 2019); suggesting that these subunits are candidates for additional human HPS subtypes. Of note, BLOC-1 shares 3 subunits (BLOC1S1, BLOC1S2 and SNAPIN) with a BLOC-1 related complex (BORC) (Table 1), which has a distinct function and is likely necessary for life (Pu et al., 2015). Defects in either one of these 3 subunits are lethal or very deleterious in mice and are unlikely to be identified in individuals with HPS.
The HPS clinical spectrum is similar in subjects with defects in genes encoding different subunits of the same AP-3 or BLOC complex. Therefore, HPS clinical features and cell biology are best understood in the context of BLOC‐1, BLOC‐2, BLOC‐3 or AP‐3 disease rather than in the context of each individual gene product (Bowman et al., 2019; Huizing et al., 2008).
Identification of the HPS subtype in each subject is important for several reasons. First, it is clinically valuable for prognosis, clinical management, and consideration of eventual treatment options (Table 2). For example, the fatal pulmonary fibrosis occurs in BLOC-3 and AP-3 deficiency, immunodeficiency is associated with AP-3 defects, while BLOC-2 deficiency results in a relatively milder phenotype without life-threatening features. Second, cell biologists can utilize the association of specific HPS defects in cells and tissues from HPS patients, mice and other animal models to study and understand LRO biology. Third, understanding LRO biology provides insights into the pathomechanism of each HPS subtype that may lead to prospects for development of novel therapies not only for HPS, but also for other LRO disorders.
Table 2:
Hermansky-Pudlak Syndrome Main Clinical Features
| Clinical Feature | Manifestations | LRO Defect (Cell Type) | Deficiency of HPS Complex | Prevention/Therapy |
|---|---|---|---|---|
| Cutaneous Albinism | white/light hair, hypopigmented and sun-sensitive skin 1 | melanosome (skin melanocytes) | AP-3, BLOC-1, -2, -3 |
Preventive care: sun avoidance, sun protection (sunscreen, hat, clothing), periodic skin cancer screening |
| Ocular Albinism | horizontal nystagmus, decreased visual acuity, pale fundus, foveal hypoplasia, iris transillumination 2 | melanosome (retinal pigment epithelial cells) | AP-3, BLOC-1, -2, -3 |
Preventive care: sun avoidance, eye protection (sunglasses, hat) Symptomatic care: vision corrective glasses, ophthalmologic care |
| Bleeding Diathesis | easy bruising, epistaxis, menorrhagia, gingival bleeding, colonic bleeding, prolonged bleeding after trauma or surgery or postpartum 3 | delta granule 3 (platelets) | AP-3, BLOC-1, -2, -3 |
Symptomatic care: local pressure on wounds, topical thrombin, 1-desamino-8D-arginine vasopressin (DDAVP) and other pro-coagulant drugs Therapeutic: platelet transfusion |
| Pulmonary Fibrosis (PF) | nonproductive cough, exertional dyspnea, diffuse rales, hypoxia | lamellar body 4 (type II alveolar epithelial cells) | BLOC-3, AP-3 4 |
Preventive care: avoidance of tobacco products Symptomatic care: supplemental oxygen for hypoxemia, pulmonary rehabilitation Therapeutic: lung transplantation |
| Enterocolitis | abdominal pain, cramps, fever, weight loss, malabsorption, frequent watery and bloody diarrhea. | unknown LRO-membrane formation 5 | BLOC-3, BLOC-2, (BLOC-1) 5 | Therapeutic: corticosteroids, non-steroidal immunomodulator drugs, anti-tumor necrosis factor-alpha drugs (effective for only some subjects) |
| Neutropenia | immunodeficiency | lytic and azurophil granules (neutrophils) | AP-3 (BLOC-1) 6 | Therapeutic: granulocyte colony-stimulating factor (G-CSF) 7 |
| Recurrent Infections | frequent viral and bacterial infections | LRO-related granules (dendritic cells, natural killer cells) | AP-3 | Therapeutic: Not prevented by G-CSF therapy 7 |
Sunburn, photo-aging of the skin, solar keratosis and melanocyte nevi are common in HPS and patients are at risk of developing squamous cell carcinoma, basal cell carcinoma, and melanoma (Toro et al., 1999).
HPS visual acuity is generally stable at 20/200 (legally blind in the United States) or worse. Most HPS patients exhibit nystagmus resulting from abnormal crossing of the optic nerve fibers. Iris transillumination is when a light is shone into the pupil is transmitted back through the iris because of a lack of iris pigmentation (Schneier & Fulton, 2013; Summers et al., 1988).
Absent platelet delta granules (determined by whole mount electron microscopy) is a diagnostic hallmark of HPS. Bleeding tendency varies widely between HPS patients. Due to absent delta granules, a secondary platelet aggregation response cannot occur (Huizing et al, 2017 Oct 26 [Updated 2000 July 24]).
Apart from type II epithelial cell defect, aberrant alveolar macrophage or mast cell function has been suggested to underlie HPS-PF (Kirshenbaum et al., 2016; Mahavadi et al., 2010; Nakatani et al., 2000; Rouhani et al., 2009). Onset of PF is in childhood in AP-3 deficiency (Gochuico et al., 2012) and middle age (30–50 years) in BLOC-3 deficiency (Huizing et al, 2017 Oct 26 [Updated 2000 July 24]). AP-3 related PF has not been described in HPS-10 patients (Ammann et al., 2016; Mohammed et al., 2018). There is no approved medical therapy for HPS PF. Lung transplantation may be considered (El-Chemaly et al., 2018; Gahl et al., 2002; Huizing et al, 2017 Oct 26 [Updated 2000 July 24]; Lederer et al., 2005).
HPS colitis involves intestinal granulomas, erosions and inflammatory cells, and resembles Crohn’s disease. The underlying cause remains unknown. Abnormal endosomal (LRO-related) membrane formation was suggested, leading to ceroid lipofuscin formation, abnormal autophagy and phagocytosis, inflammation (Felipez et al., 2010; Sofia et al., 2017). Some BLOC-2 or BLOC-3 deficient cases develop colitis (Huizing et al, 2017 Oct 26 [Updated 2000 July 24]; Hussain et al., 2006). One BLOC-1 deficient case (HPS-7) developed Crohn’s colitis in adulthood (Lowe et al., 2013). It is unknown if colitis occurs in AP-3 deficiency.
Immunodeficiency was reported in two unrelated individuals with HPS-9 (BLOC-1 deficiency) (Badolato et al., 2012; Okamura et al., 2018) and needs consideration in future BLOC-1 deficient individuals.
G-CSF therapy was only used in HPS-2 patients (AP3B1 deficiency) (Ammann et al., 2016; Fontana et al., 2006). While G-CSF restores neutrophil numbers, it does not prevent recurrent infections caused by defects in innate immunity in HPS-2 (Fontana et al., 2006).
In this report, we provide an overview of the HPS-related genes, their functions and clinical consequences when deficient. We summarize reported human variants in each HPS-associated gene and list unreported variants identified in an HPS patient cohort evaluated at the National Institutes of Health (NIH). This review can serve as a reference for molecular diagnostic aspects of Hermansky-Pudlak syndrome.
HPS SUBTYPE-SPECIFIC MUTATION UPDATE
We searched online literature databases for reported pathogenic variants in 10 HPS-related human genes (as of November 2019). We also list unreported pathogenic gene variants identified in our NIH HPS patient cohort, enrolled in a protocol entitled, “Clinical and Basic Investigations into Hermansky-Pudlak Syndrome” (ClinicalTrials.gov Identifier NTC00001456). Table 1 provides an overview of the HPS subtypes, features of each gene and protein, numbers of reported pathogenic variants, an estimate of reported subjects and reported vertebrate models per subtype. Table 2 lists subtype-specific features, LRO defects, and therapeutic options. Tables 3–8 and Figures 1–7 provide HPS gene-specific pathogenic mRNA and protein variants. Footnotes under each Table describe additional variant-specific information. Pathogenicity predictions of missense and splice site variants are listed in Supp. Tables S1 and S2. Supp. Table S3 lists frequently occurring (mostly missense) variants with a high minor allele frequency (MAF > 0.001) that should be considered as possible polymorphisms when encountered in HPS genetic analyses. Supp. Table S4 lists reported variants in human HPS genes associated with traits other than HPS, which should be considered when these variants are found in future HPS cases.
Table 3:
HPS1 Pathogenic Gene Variants Associated with Hermansky-Pudlak Syndrome Type 1 (HPS-1)
| # | mRNA NM_000195.5 | Amino Acid NP_000186.2 | Exon/Intron | Variant Type 1 | Ethnic Background 2 | References and Footnotes |
|---|---|---|---|---|---|---|
| 1 | del exon 2 3 | - | Exon 2 | Indel | - | (Lasseaux et al., 2018) 4 |
| 2 | c.2T>A | p.Met1Lys 5 | Exon 3 | Start-loss | - | (Lasseaux et al., 2018) 4,5 |
| 3 | c.9delC | p.Cys3Trpfs*26 | Exon 3 | Frameshift | Chinese | (Power et al., 2019) |
| 4 | c.34dupG | p.Glu12Glyfs*12 | Exon 3 | Frameshift | - | (Lasseaux et al., 2018) 4 |
| 5 | c.81delG | p.Leu28* | Exon 3 | Nonsense | Korean | (Sim et al., 2019) |
| 6 | del 121-bp 3,6 | p.Pro41Aspfs*12 | Intron 3/Exon 4 | Indel | Pakistani | (Yousaf et al., 2016) 4,6 |
| 7 | c.97_100delTCAG | p.Ser33Argfs*18 | Exon 3 | Indel | English, Irish, German, Scottish | (Sandrock et al., 2010) 7 |
| 8 | c.166_168delATC | p.Ile56del | Exon 4 | Indel | Afghan | (Oh et al., 1998) |
| 9 | c.212_215delGCTT | p.Cys71Serfs*52 | Exon 4 | Indel | - | (Lasseaux et al., 2018) 4 |
| 10 | c.217delT | p.Ser73Profs*51 | Exon 4 | Frameshift | - | (Lasseaux et al., 2018) 4 |
| 11 | c.255+5G>A | IVS4+5G>A (p.Tyr81Leufs*38) | Intron4 | Splice site | Iranian | (Ghafouri-Fard et al., 2016) |
| 12 | c.288delT | p.Asp97Thrfs*27 | Exon 5 | Frameshift | Japanese | (Ito et al., 2005; Spritz & Oh, 1999) |
| 13 | c.316C>G | p.Arg106Gly 8 | Exon 5 | Missense | Chinese | (Wei et al., 2016) 4 |
| 14 | c.344T>C | p.Leu115Pro | Exon 5 | Missense | Arabic | (Khan et al., 2016) 4 |
| 15 | c.355delC | p.His119Thr*5 | Exon 5 | Frameshift | German, Polish, Russian | (Hermos et al., 2002; Sandrock et al., 2010) |
| 16 | c.391C>T | p.Arg131* | Exon 5 | Nonsense | Caucasian, Chinese, Spanish | (Arcot Sadagopan et al., 2017; Gonzalez-Conejero et al., 2003; Hermos et al., 2002; Wei et al., 2011) |
| 17 | c.397G>T | p.Glu133* | Exon 5 | Nonsense | German, Italian, Ukrainian | (Hermos et al., 2002; Shotelersuk et al., 1998) |
| 18 | c.398+2T>C | IVS5+2T>C | Intron 5 | Splice site | Mexican | novel 4,9,10 |
| 19 | c.398+5G>A | IVS5+5G>A | Intron 5 | Splice site | Chinese, Indian, Japanese | (Furuhashi et al., 2014; Horikawa et al., 2000; Ito et al., 2005; Li et al., 2016; Mai et al., 2019; Natsuga et al., 2005; Oh et al., 1998; Suzuki et al., 2004; Tanaka et al., 2015; Vincent et al., 2009; Wei et al., 2016) 4,11 |
| 20 | c.418delG | p.Ala140Argfs*35 | Exon 6 | Frameshift | European | (Hermos et al., 2002) |
| 21 | c.461G>A | p.Trp154* | Exon 6 | Nonsense | Dutch | (Thielen et al., 2010) |
| 22 | c.467_476del10 | p.Tyr156Cysfs*16 | Exon 6 | Indel | Honduran, Salvadoran | (Carmona-Rivera, Golas, et al., 2011) |
| 23 | c.505G>A | p.Glu169Lys | Exon 6 | Missense- Splice site | Arabic | (Khan et al., 2016) 4,12 |
| 24 | c.507G>A | p.Glu169Glu 13 | Exon 6 | Splice site | African-American | (Merideth et al., 2009) 13 |
| 25 | c.507+1G>A | IVS6+1G>A | Intron 6 | Splice site | Japanese | (Lasseaux et al., 2018; Natsuga et al., 2005) 14 |
| 26 | c.517C>T | p.Arg173* | Exon 7 | Nonsense | Chinese | (Wei et al., 2016) 4 |
| 27 | c.532dupC | p.Gln178Profs*4 | Exon 7 | Frameshift | Japanese | (Ito et al., 2005; Iwakawa et al., 2005) |
| 28 | c.610G>T | p.Glu204* | Exon 7 | Nonsense | Spanish | (Sanchez-Guiu et al., 2014) |
| 29 | c.640delC | p.His214Thrfs*117 | Exon 7 | Frameshift | Chinese | (Wei et al., 2019) 4 |
| 30 | c.695C>T | p.Ala232Val | Exon 8 | Missense | Arabic | (Khan et al., 2016) 4 |
| 31 | c.716T>C | p.Leu239Pro | Exon 8 | Missense | Dutch, German, Irish | (Hermos et al., 2002; Lasseaux et al., 2018; Thielen et al., 2010) 4,15 |
| 32 | c.868–2A>G | IVS9–2A>G | Intron 9 | Splice site | Chinese | (Wei et al., 2019) 4 |
| 33 | c.937G>A | p.Gly313Ser | Exon 10 | Missense- Splice site | Puerto Rican | (Carmona-Rivera, Hess, et al., 2011; Lasseaux et al., 2018) 4,16 |
| 34 | c.956delA | p.Glu319Glyfs*12 | Exon 11 | Frameshift | Chinese | (Wei et al., 2019) 4 |
| 35 | c.962delG | p.Gly321Alafs*10 | Exon 11 | Frameshift | Ukrainian | (Oh et al., 1998) |
| 36 | c.962dupG | p.Thr322Hisfs*131 | Exon 11 | Frameshift | Japanese | (Horikawa et al., 2000) |
| 37 | c.972delC | p.Met325Trpfs*6 | Exon 11 | Frameshift | African-American, Chinese, Japanese, Mexican, Northern European, Puerto Rican | (Carmona-Rivera, Golas, et al., 2011; Carmona-Rivera, Hess, et al., 2011; Hermos et al., 2002; Lasseaux et al., 2018; Merideth et al., 2009; Oh et al., 1996; Oh et al., 1998; Shotelersuk et al., 1998; Wei et al., 2016) 4,17 |
| 38 | c.972dupC | p.Met325Hisfs*128 | Exon 11 | Frameshift | Chinese, Japanese, Northern European, Swiss | (Hermos et al., 2002; Lasseaux et al., 2018; Oh et al., 1996; Oh et al., 1998; Okamura et al., 2019; Wei et al., 2010; Wei et al., 2011; Wei et al., 2019) 4,,17,18 |
| 39 | c.988–1 G>T | IVS11–1G>T | Intron 11 | Splice site | Indian | (Vincent et al., 2009) 19 |
| 40 | del13,966-bp/ins49-bp 3 | p.Gln329fs | Intron 11- Exon 20 |
Indel | Northern European | (Griffin et al., 2005) |
| 41 | c.1080C>G | p.Ser360Arg | Exon 12 | Missense | Canadian, German, Irish, Scottish, Swedish, Ukrainian | novel 4,9,20 |
| 42 | c.1132_1138delATCAACC | p.Ile378Trpfs*4 | Exon 12 | Indel | Chinese | (Wei et al., 2019) 4 |
| 43 | c.1189delC | p.Gln397Serfs*2 | Exon 13 | Frameshift | American, Hispanic, Northern European, Russian, Ukrainian | (Doubkova et al., 2019; Griffin et al., 2005; Hermos et al., 2002; Lasseaux et al., 2018; Oh et al., 1998; Sandrock et al., 2010; Shotelersuk et al., 1998) 4,17,20–24 |
| 44 | c.1228A>T | p.Lys410* | Exon 13 | Nonsense | Ukrainian | novel 9,21 |
| 45 | c.1276_1279dupGGAG | p.Asp427Glyfs*27 | Exon 13 | Indel | Chinese | (Wei et al., 2019) 4 |
| 46 | c.1294_1298delATGGAinsT | p.Met432Serfs*42 | Exon 13 | Indel | Mexican | novel 4,9,10 |
| 47 | c.1323dupA | p.Gln442Thrfs* 11 | Exon 13 | Frameshift | Japanese | (Oh et al., 1996) |
| 48 | c.1342T>C | p.Trp448Arg | Exon 14 | Missense | Pakistani | (Yousaf et al., 2016) 4 |
| 49 | c.[1375delA; c.1388C>A] | p.Ser459Valfs*16 | Exon 14 | Frameshift | Northern European | (Hermos et al., 2002) |
| 50 | c.1423_1428delAAGCGG | p.Lys475_Arg476del | Exon 15 | Indel | - | (Lasseaux et al., 2018) 4 |
| 51 | c.1457_1460dupTTCT | p.Thr488Serfs*95 | Exon 15 | Indel | Chinese | (Wei et al., 2016) 4 |
| 52 | c.1472_1487dup16 24 | p.His497Glnfs*90 | Exon 15 | Indel | NW-Puerto Rican | (Hermos et al., 2002; Oh et al., 1996; Santiago Borrero et al., 2006) 25 |
| 53 | c.1477delA | p.Arg493Glyfs*22 | Exon 15 | Frameshift | Chinese | (Power et al., 2019) |
| 54 | c.1507C>T | p.Gln503* | Exon 15 | Nonsense | Caucasian | (Doubkova et al., 2019) 4 |
| 55 | del exon 15 | deletion | Exon 15 | Indel | Chinese | (Wei et al., 2019) 4 |
| 56 | del exon 15–18 3 | deletion | Ex15–18 | Indel | Chinese | (Wei et al., 2016) 4 |
| 57 | c.1639G>T/c.1645C>T | p.Val547Leu/ p.Arg549Cys | Exon 17 | Missense | Assyrian, English, German, Irish | (Nazarian et al., 2008) 15,26 |
| 58 | c.1691delA | p.Lys564Argfs*22 | Exon 17 | Frameshift | Japanese | (Ito et al., 2005) |
| 59 | c.1744–2A>C | IVS17–2A>C | Intron17 | Splice site | Caucasian, English, German, Irish | (Hermos et al., 2002; Lasseaux et al., 2018; McElvaney et al., 2018; Oetting & King, 1999) 4,22,27 |
| 60 | c.1749G>A | p.Trp583* | Exon 18 | Nonsense | Japanese, Arabic | (Ito et al., 2005) 4,28 |
| 61 | c.1763T>C | p.Leu588Pro | Exon 18 | Missense | Japanese | (Okamura et al., 2019) 4 |
| 62 | c.1787G>T | p.Gly596Val | Exon 18 | Missense | Japanese | (Okamura et al., 2019; Takeuchi et al., 2014) 4 |
| 63 | c.1857+2T>C | IVS18+2T>C | Intron 18 | Splice site | Irish | (McElvaney et al., 2018) 27 |
| 64 | c.1858–1G>A | IVS18–1G>A | Intron18 | Splice site | Dutch, French, German, Irish, Native American | novel 9,29 |
| 65 | c.1887delC | p.Val630Serfs*95 | Exon 19 | Frameshift | Chinese | (Wei et al., 2011) |
| 66 | c.1932delC | p.Tyr645Thrfs*80 | Exon 19 | Frameshift | Chinese | (Wei, Lian, Wang, & Li, 2009; Wei, Zang, Zhang, Yang, & Li, 2015; Wei et al., 2019) 4 |
| 67 | c.1937A>G | p.Tyr646Cys | Exon 19 | Missense | English, Irish, Scottish | novel 9,15,24 |
| 68 | c.1941–2A>G | IVS19–2A>G | Intron 19 | Splice site | Japanese | (Okamura et al., 2019) 4 |
| 69 | c.1996G>A | p.Glu666Lys | Exon 20 | Missense | Korean | (Sim et al., 2019) |
| 70 | c.1996G>C | p.Glu666Gln | Exon 20 | Missense | - | (Lasseaux et al., 2018) 4 |
| 71 | c.1996G>T | p.Glu666* | Exon 20 | Nonsense | Scottish | (Oh et al., 1998) |
| 72 | c.2003T>C | p.Leu668Pro | Exon 20 | Missense | Chinese, Japanese | (Ito et al., 2005; Iwata et al., 2017; Kanazu et al., 2014; Mai et al., 2019; Okamura et al., 2019; Wei et al., 2016) 4 |
| 73 | c.2010_2037del28 | p.His671Trpfs*45 29 | Exon 20 | Indel | - | (Lasseaux et al., 2018) 4,30 |
| 74 | c.2037_2064del28 | p.Leu680Glyfs*36 29 | Exon 20 | Indel | - | (Girot et al., 2019) 30 |
| 75 | c.2037_2068delinsCTGG | p.Leu680Trpfs*36 29 | Exon 20 | Indel | - | (Lasseaux et al., 2018) 4,30 |
| 76 | c.2056C>T | p.Gln686* | Exon 20 | Nonsense | Pakistani | (Yousaf et al., 2016) 4 |
When deletion/insertion is 1 nucleotide it is named Frame shift, when larger it is named Indel.
Extracted from literature reference. ‘-‘ = unreported.
The nomenclature of these HPS1 variants are included in this Table as reported, see reference for each specific variant for more details.
At least one of the reported cases with this variant was identified by next generation sequencing.
This variant likely leads to a loss of protein translation at the start codon of the longest splice variant of HPS1 (NM_000195.5). It is also predicted to affect splicing, as it is located at the exon 2–3 splice junction (Supplemental Table S2).
NC_000010.11:g.10:98435762–98435882 (GRCh38): Genomic 121-bp deletion, including a part of intron 3 and exon 4 (Yousaf et al., 2016).
Two unreported siblings from the NIH HPS cohort with this c.97_100delTCAG variant were of English-Irish-Scottish descent.
Gray highlight: missense variant. See Supplemental Table S1 for pathogenicity predictions.
novel = previously unreported variant detected in the NIH HPS cohort.
This novel HPS1 variant was found heterozygous by next generation sequencing in 2 unreported siblings of Mexican descent from the HPS cohort. They were compound heterozygous for c.398+2T>C and c.1294_1298delATGGAinsT. This splice site variant is predicted to delete the splice junction of exon 18/intron 18 (Supplemental Table S2).
This variant was reported to result in skipping of exon 5 (Suzuki et al., 2004), and is a frequent variant in Japanese HPS patients (Ito et al., 2005).
This variant occurs 3-bp from a splice junction and is predicted to affect the splice site (Supplemental Table S2). No experimental evidence is available (Khan et al., 2016). An alternative intronic splice site, inserting 43-bp of intron 6 sequence may be used as reported for variant c.507G>A occurring in the same codon (Merideth et al., 2009).
This (silent) HPS1 variant p.Glu169Glu results in a splice defect (Merideth et al., 2009).
This variant is reported to result in use of an alternative intronic splice donor site, 44-bp into intron 6, resulting in a frameshift of the coding region (Natsuga et al., 2005).
In vitro studies showed that the HPS1 protein with this missense variant was unstable (Carmona-Rivera et al., 2013).
This (missense) HPS1 variant occurs at the 3’ splice junction of exon 10, resulting in a cryptic intronic splice site and an aberrantly spliced mRNA that includes 144-bp intronic sequence, producing 11 novel amino acids followed by a stop codon (Carmona-Rivera, Hess, et al., 2011).
This HPS1 frameshift variant occurs with a high prevalence in HPS-1 subjects of various ethnic backgrounds.
This variant c.972dupC was reported as an ethnic founder variant in a small isolate in a Swiss village (Oh et al., 1998; Schallreuter et al., 1993)
This variant was reported to result in in-frame skipping of exon 12 and removing 56 amino acids from the protein (Vincent et al., 2009).
This HPS1 variant was identified in one unreported subject of Canadian-German-Irish-Scottish-Swedish-Ukrainian descent from the NIH HPS cohort. This subject is compound heterozygous for c.1080C>G and c.1189delC.
This HPS1 variant was identified in one unreported subject of Ukrainian descent from the NIH HPS cohort. This subject is compound heterozygous for c.1189delC and c.1228A>T.
This HPS1 variant was identified homozygous in one unreported subject of German descent from the NIH HPS cohort.
This HPS1 variant was identified in one unreported subject of German-English-Irish descent in the NIH HPS cohort. This subject is compound heterozygous for c.1189delC and c.1744–2A>C.
This HPS1 variant was found heterozygous in one unreported subject of English-Irish-Scottish in the NIH HPS cohort. This subject is compound heterozygous for c.1189delC and c.1937A>G.
This HPS1 16-bp duplication (c.1472_1487dup16-bp) is originates from a genetic isolate in northwest Puerto Rico (Oh et al., 1996; Santiago Borrero et al., 2006).
These 2 missense variants occur heterozygous on the same allele in two HPS siblings of our NIH cohort, their cells showed aberrant BLOC-3 assembly (Nazarian et al., 2008). Both missense variants are predicted to be deleterious to protein function (Supplemental Table S3). In vitro studies showed that the HPS1 protein with the p.Val547Leu variant was unstable and prevents proper BLOC-3 formation (Carmona-Rivera et al., 2013). No HPS1 coding/splice site variant was detected on the other allele, but this allele appeared to be subject to non-sense mediated mRNA decay (on cDNA analysis), indicating a likely (intronic) gene-truncation variant on this allele.
This HPS1 variant was identified in a subject with of Irish descent with HPS clinical features and accelerated pulmonary fibrosis. He was compound heterozygous for c.1744–2A>C (predicted to cause exon skipping (Oetting & King, 1999)) and c.1857+2T>C (predicted to result in use of alternative intronic splice site 4 base-pairs into intron 18, resulting in a frameshift of the coding region) (Supplemental Table S2) (McElvaney et al., 2018).
This HPS1 variant was identified homozygous by next generation sequencing in one unreported subject of Arabic descent in the NIH HPS cohort.
This HPS1 variant was found homozygous in one unreported subject of Dutch-French-German-Irish-Native American descent in the NIH HPS cohort. This novel splice site variant c.1858–1G>A, is predicted to create an alternative splice site 1-bp into exon 18, resulting in a frameshift of the coding region (Supplemental Table S2).
These Indels occur in the same region and result in a loss of the HPS1 termination codon (codon #701) and extension of the translated HPS1 protein.
Table 8:
HPS6 Pathogenic Gene Variants Associated with Hermansky-Pudlak Syndrome Type 6 (HPS-6)
| No | mRNA NM_024747.5 | Amino Acid NP_079023.2 | Exon/Intron 1 | Variant Type 2 | Ethnic Background 3 | References and Footnotes |
|---|---|---|---|---|---|---|
| 1 | c.62_63insCGGCG | p.Leu22Glyfs*33 | Exon 1 | Indel | - | (Lasseaux et al., 2018) 4 |
| 2 | c.60_64dupGCGGC | p.Leu22Argfs*33 | Exon 1 | Indel | Chinese, Japanese, Portuguese | (Bastida et al., 2019; Okamura et al., 2018; Wei et al., 2016; Wei et al., 2019) 4 |
| 3 | c.87_108dup22 | p.Ser37Leufs*146 | Exon 1 | Indel | Czech, Eastern/Northern European, German, Polish | (Radke et al., 2013; Summers & Schimmenti, 2014) 5 |
| 4 | c.141_143delinsG | p.Pro49Trpfs*126 | Exon 1 | Indel | - | (Lasseaux et al., 2018) 4 |
| 5 | c.155delT | p.Val52Glufs*6 | Exon 1 | Frameshift | Chinese | (Wei et al., 2016) 4 |
| 6 | c.206_210dupGGGCC | p.Trp71Glyfs*158 | Exon 1 | Indel | Chinese | (Wei et al., 2019) 4 |
| 7 | c.223C>T | p.Gln75* | Exon 1 | Nonsense | Italian | (Huizing et al., 2009) |
| 8 | c.233C>G | p.Pro78Arg 6 | Exon 1 | Missense | Japanese | (Okamura et al., 2019) 4 |
| 9 | c.238dupG | p.Asp80Glyfs*96 | Exon 1 | Frameshift | Dutch, German | (Huizing et al., 2009) |
| 10 | c.275T>A | p.Leu92Gln 7 | Exon 1 | Missense | - | (Lasseaux et al., 2018) 4,7 |
| 11 | c.288G>A | p.Trp96* | Exon 1 | Nonsense | Arabic | (Khan et al., 2016) 4 |
| 12 | c.337C>T | p.Arg113Trp | Exon 1 | Missense | - | (Lasseaux et al., 2018) 4 |
| 13 | c.383T>C | p.Val128Ala | Exon 1 | Missense | Caucasian | (Han et al., 2018) 4 |
| 14 | c.448_505dup58 | p.Glu169Glyfs*26 | Exon 1 | Indel | Caucasian | novel 4,8,9 |
| 15 | c.455C>G | p.Ser152* | Exon 1 | Nonsense | - | (Lasseaux et al., 2018) 4 |
| 16 | c.503_504delTG | p.Leu168Argfs*7 | Exon 1 | Indel | Chinese | (Wei et al., 2019) 4 |
| 17 | c.779G>A | p.Gly260Glu | Exon 1 | Missense | Punjabi Afghan | (Hull et al., 2016) 4 |
| 18 | c.815C>T | p.Thr272Ile | Exon 1 | Missense | Dutch, German | (Huizing et al., 2009) |
| 19 | c.823C>T | p.Pro275Ser | Exon 1 | Missense | Pakistani | (Yousaf et al., 2016) 4 |
| 20 | c.877C>T | p.Glu293* | Exon 1 | Nonsense | - | (Shamseldin et al., 2017) 4 |
| 21 | c.895C>T | p.Arg299Trp | Exon 1 | Missense | Chinese | (Wei et al., 2016) 4 |
| 22 | c.896G>C | p.Arg299Pro | Exon 1 | Missense | - | (Lasseaux et al., 2018) 4 |
| 23 | c.905G>A | p.Gly302Asp | Exon 1 | Missense | - | (Lasseaux et al., 2018) 4 |
| 24 | c.902dupT | p.Thr303Hisfs*64 | Exon 1 | Frameshift | Russian-Palestinian | (Hull et al., 2016) 4 |
| 25 | c.913C>T | p.Gln305* | Exon 1 | Nonsense | English, German, Scottish | (Huizing et al., 2009) |
| 26 | c.1065dupG 10 | p.Leu356Alafs*11 | Exon 1 | Frameshift | Israeli Bedouin | (Schreyer-Shafir et al., 2006) 10 |
| 27 | c.1083dupC | p.Gly362Argfs*5 | Exon 1 | Frameshift | Russian-Palestinian | (Hull et al., 2016) 4 |
| 28 | c.1114 C>T | p.Arg372* | Exon 1 | Nonsense | Irish, Native American (Cherokee), Scottish | (O'Brien et al., 2016) |
| 29 | c.1234C>T | p.Gln412* | Exon 1 | Nonsense | Italian | (Huizing et al., 2009) |
| 30 | c.1235_1239dupAGCGG | p.Arg414Serfs*15 | Exon 1 | Indel | Chinese | (Wei et al., 2019) 4 |
| 31 | c.1372delG | p.Glu458Serfs*8 | Exon 1 | Frameshift | Chinese | (Wei et al., 2016) 4 |
| 32 | c.1387C>T | p.Arg463* | Exon 1 | Nonsense | - | (Lasseaux et al., 2018) 4 |
| 33 | c.1513C>T | p.Gln505* | Exon 1 | Nonsense | Chinese | (Wei et al., 2016; Wei et al., 2019) 4 |
| 34 | c.1644delA | p.Gly550Glufs*2 | Exon 1 | Frameshift | Arabic | (Khan et al., 2016) 4 |
| 35 | c.1711_1712insAG | p.Cys571* | Exon 1 | Indel | Czech, Eastern/Northern European, German, Polish | (Radke et al., 2013; Summers & Schimmenti, 2014) 5 |
| 36 | c.1714_1717delCTGT | p.Leu572Alafs*40 | Exon 1 | Indel | Belgian | (Lasseaux et al., 2018; Zhang et al., 2003) 4 |
| 37 | c.1819C>T | p.Arg607* | Exon 1 | Nonsense | Chinese | (Lasseaux et al., 2018; Wei et al., 2019) 4 |
| 38 | c.1865_1866delTG | p.Leu622Argfs*12 | Exon 1 | Indel | German, Irish | (Huizing et al., 2009) |
| 39 | c.1898delC | p.Pro633Leufs*76 | Exon 1 | Frameshift | Japanese | (Miyamichi et al., 2016) 4 |
| 40 | c.1919_1920delTC | p.Val640Glyfs*29 | Exon 1 | Indel | German-Caucasian | (Andres et al., 2017) 4 |
| 41 | c.2038C>T | p.Gln680* | Exon 1 | Nonsense | Japanese | (Miyamichi et al., 2016; Okamura et al., 2018; Okamura et al., 2019) 4, 11 |
| 42 | c.2189dupC | p.Leu731Serfs*28 | Exon 1 | Frameshift | Caucasian | novel 4,8,9 |
| 43 | c.2207T>C | p.Leu736Pro | Exon 1 | Missense | - | (Lasseaux et al., 2018) 4 |
| 44 | del19,972-bp 12 | - | Exon 1 | Indel | English, German, Scottish | (Huizing et al., 2009) |
| 45 | del exon 1 12 | - | Exon 1 | Indel | - | (Lasseaux et al., 2018) 4 |
The HPS6 gene consists of 1 exon.
When deletion/insertion is 1 nucleotide it is named Frameshift, when larger it is named Indel.
Extracted from literature reference. ‘-‘ = unreported.
At least one of the reported individuals with this variant was identified by next generation sequencing.
The subject described in these references was also seen at NIH and is of Eastern/Northern European (Czech, German, Polish) descent. This subject is compound heterozygous for c.87_108dup22-bp and c.1711_1712insAG.
Gray highlight: missense variant. See text and Supplemental Tables for pathogenicity description.
This variant is listed as a variant of uncertain significance with a high MAF in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
novel = previously unreported variant and/or ethnicity, detected in the NIH HPS cohort.
This HPS6 variant was identified by exome sequencing in one unreported subject of Caucasian descent in the NIH HPS cohort. This subject is compound heterozygous for c.448_505dup58-bp and c.2189dupC.
This frameshift variant was previously described as c.1066_1067insG (p.Leu356Argfs*11) (Schreyer-Shafir et al., 2006).
This HPS6 variant c.2038C>T (p.Gln680*) appears to be a Japanese variant, as it occurs in 5 Japanese subjects and is not reported in dbSNP/ExAc databases.
The nomenclature of these HPS6 variants are included in this Table as reported, see reference for each specific variant for more details.
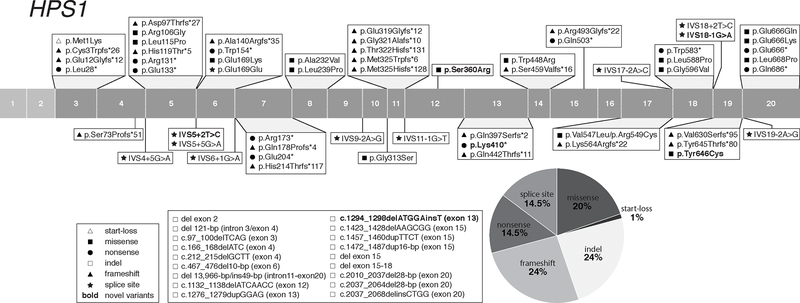
Figure 1:
Distribution of HPS1 Gene Variants
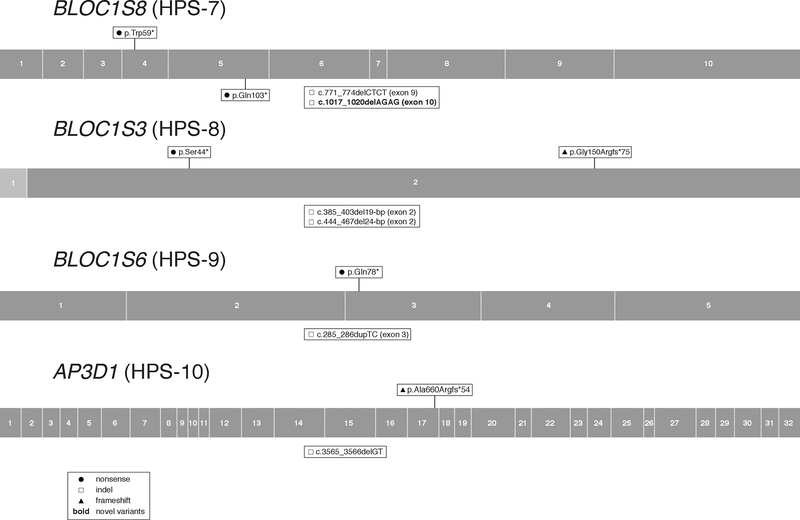
Figure 7:
Distribution of BLOC1S8 (HPS-7), BLOC1S3 (HPS-8), BLOC1S6 (HPS-9), and AP3D1 (HPS-10) Gene Variants
The variant nomenclature in all Tables conforms to human genome variation society (HGVS) recommendations (den Dunnen et al., 2016). The longest mRNA splice variant of each gene is used for variant nomenclature and the GenBank accession number is indicated in each Table. Some previously reported variants are re-named to conform to the current nomenclature convention. Pathogenicity predictions of missense variants (Supp. Table S1) follow the American College of Medical Genetics (ACMG) Standards and Guidelines for interpretation of sequence variants (Richards et al., 2015).
We deposited all unreported variants in the Leiden Open Variation Database 3.0 (http://www.lovd.nl/) (Fokkema et al., 2011) and in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (Landrum et al., 2018). Other databases with variable HPS-related information exist, including the Albinism Database (http://www.ifpcs.org/albinism/), AP3B1base (http://structure.bmc.lu.se/idbase/AP3B1base/), Retina International Mutation Database (http://www.retina-international.org/sci-news/databases/mutation-database) and Oculocutaneous albinism Database (https://ghr.nlm.nih.gov/condition/oculocutaneous-albinism).
HPS-1 (Table 3)
HPS1, also called BLOC3S1, is the first identified HPS-associated gene (Oh et al., 1996), causing HPS Type 1 (HPS-1; MIM# 203300) when defective. HPS1 is located on chromosome 10q24.2; its longest mRNA transcript contains 20 exons and codes for a 700-amino acids protein (~79.3-kD). At least 18 HPS1 protein-coding mRNA transcript variants are predicted; expression patterns and function of these variants remain unknown. The HPS1 protein interacts with HPS4 in the BLOC-3 complex (Carmona-Rivera, Simeonov, Cardillo, Gahl, & Cadilla, 2013; Martina et al., 2003). Pale ear is the murine orthologue of human HPS-1 (Li et al., 2004).
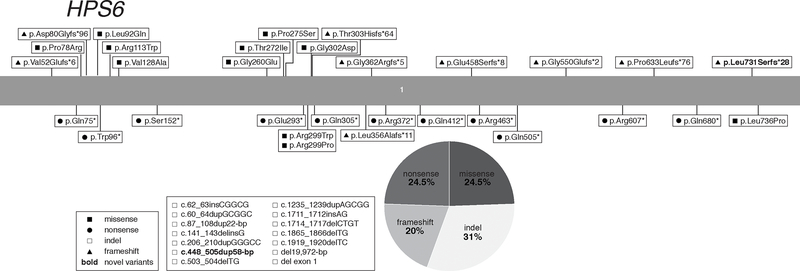
Identification of the HPS1 gene was aided by linkage analysis in northwest Puerto Rican individuals with HPS, who have a homozygous 16-bp duplication (c.1472_1487dup16-bp, p. p.His497Glnfs*90) in HPS1 (Fukai, Oh, Frenk, Almodovar, & Spritz, 1995; Oh et al., 1996). There are currently ~261 Puerto Rican subjects with the HPS1 16-bp duplication founder variant reported in the literature (including the NIH cohort); it is estimated that ~400 such cases exist (Santiago Borrero et al., 2006; Witkop et al., 1990). HPS-1 has also been reported in a small isolate in a Swiss village (c.972dupC, p.Met325Hisfs*128) (Oh et al., 1998; Schallreuter et al., 1993) and one in Japan (c.398+5G>A) (Ito et al., 2005). Apart from the Puerto Rican founder population, there are an additional ~137 HPS-1 cases reported, plus 11 unreported cases from our NIH cohort (Table 1).
We report a total of 76 HPS1 variants associated with the HPS-1 phenotype (Table 3), including 5 novel variants identified in our NIH HPS cohort. The variants are located throughout the entire gene and include 1 (1%) start-loss, 20 (26%) frameshift, 15 (20%) missense, 11 (14.5%) nonsense, 18 (24%) insertions and/or deletions, and 11 (14.5%) splice site variants (Figure 1). All reported HPS1 splice site variants are predicted and/or reported to cause aberrant splicing (Supp. Table S2). Of the 15 reported HPS1 missense variants, 5 occur at an intron/exon boundary and 3 are predicted and/or reported to affect splicing (Supp. Table S2). Of note, one nonsynonymous (silent) variant, p.Glu169Glu (c.507G>A), was reported to result in a splicing defect in two African-American brothers (Merideth et al., 2009). The start-loss variant p.Met1Lys (c.2T>A) likely leads to a loss of protein translation at the start codon of the longest splice variant of HPS1 (NM_000195.5), it is also predicted to affect splicing, as it is located at the exon 2–3 splice site (Supp. Table S2). The MAFs of all HPS1 pathogenic missense variants are very low or not reported, supporting possible pathogenicity (Supp. Table S1). Pathogenicity of some HPS1 missense variants was experimentally assessed (Supp. Table S1). Of the 15 reported HPS1 missense variants, 7 were predicted to be pathogenic (P) or likely pathogenic (LP) by ACMG standards and guidelines (Richards et al., 2015) (Supp. Table S1), while 8 others were classified as variants of uncertain significance (VUS), which should be taken into account when these variants are found in future HPS cases.
Four HPS1 variants (c.1286G>A, p.Arg429His; c.1395G>A, p.Trp465*; c.1888G>A, p.Val630Ile; c.1915G>A, p.Gly639Ser) listed in Supp. Table S4 were found heterozygous in nextgen sequencing studies of probands with other traits than HPS (Stearman et al., 2019; Abouelhoda et al., 2016). Although none of these variants were reported in HPS subjects, they should be taken into account when these variants are found in future HPS cases.
The dbSNP/gnomAd/ClinVar databases list 10 frequently occurring (MAF > 0.001) HPS1 missense variants, with predicted benign or unknown pathogenicity, which should be considered as likely non-pathogenic polymorphisms when encountered in HPS1 genetic analyses (Supp. Table S3).
There are 3 HPS1 frameshift variants that occur with high prevalence in HPS-1 subjects of various ethnic backgrounds and have relatively high MAFs; they are c.972dupC, p.Met325Hisfs*128 (ClinVar MAF 0.000317; frequent in Europeans) and c.972delC, p.Met325Trpfs*6 (ClinVar MAF 0.00002; frequent in Europeans and South Asians) in exon 11 and c.1189delC, p.Gln397Serfs*2 (ClinVar MAF 0.000067; frequent in Europeans) in exon 13. In fact, in our NIH cohort of non-Puerto Rican HPS-1 cases, 19 (39%) of 49 cases carry at least one of these two variants. Hence, analyses of exon 11 and exon 13 of HPS1 could be considered before proceeding to more laborious and costly sequencing techniques in non-Puerto Rican individuals suspected of having HPS-1 disease.
HPS-1 is identified worldwide in individuals with a large spectrum of ethnic backgrounds and is the HPS subtype with the most described cases, even excluding Puerto Rican cases (Table 1). HPS-1 (together with HPS-4) displays the most severe phenotype. In individuals with HPS-1, cutaneous albinism is more profound (higher degree of hypopigmentation of skin and hair) and the ocular findings are more severe than in other subtypes (Huizing et al, 2017 Oct 26 [Updated 2000 July 24]). Of note, Hps1 (and Hps4) variants in mice appear to only have a mild effect on pigmentation and bleeding (Novak, Hui, & Swank, 1984), for unknown reasons. In mice, Hps1 variants appear to impact pigmentation tissue-specific; melanosomes in hair follicles are less affected (i.e. pigmented hair) than in interfollicular melanocytes (i.e. less pigmented skin), perhaps partially explaining the more severe skin pigmentation phenotype in human patients (Nguyen & Wei, 2007). Virtually all HPS-1 subjects develop pulmonary fibrosis by middle age, some develop granulomatous colitis and a majority of female subjects have menorrhagia (Table 2).
HPS-2 (Table 4)
Table 4:
AP3B1 Pathogenic Gene Variants Associated with Hermansky-Pudlak Syndrome Type 2 (HPS-2)
| # | mRNA NM_003664.4 | Amino Acid NP_003655.3 | Exon/Intron | Variant Type 1 | Ethnic Background 2 | References and Footnotes |
|---|---|---|---|---|---|---|
| 1 | c.2T>G | p.Met1Arg 3 | Exon 1 | Start-loss | Australian | (Cetica et al., 2015) |
| 2 | c.62delG | p.Gly21Valfs*20 | Exon 1 | Frameshift | - | (Jessen et al., 2013) |
| 3 | c.155_158delAGAG | p.Glu52Alafs*11 | Exon 2 | Indel | Caucasian, English | (Wenham et al., 2010) |
| 4 | c.177delA | p.Lys59Asnfs*5 | Exon 2 | Frameshift | - | (de Boer et al., 2017) |
| 5 | c.305T>C | p.Leu102Pro 4 | Exon 4 | Missense | - | (Jessen et al., 2013) |
| 6 | c.716G>A | p.Trp239* | Exon 7 | Nonsense | Moroccan | (de Boer et al., 2017) |
| 7 | c.904A>T | p.Arg302* | Exon 8 | Nonsense | - | (Enders et al., 2006) |
| 8 | c.1063_1064delCAinsTATCAATATC | p.Gln355Tyrfs*6 | Exon 10 | Indel | Italian | (Fontana et al., 2006) |
| 9 | c.1095+5G>A | IVS10+5G>A | Intron10 | Splice site | Mexican | (Chiang et al., 2010) |
| 10 | c.1168–1G>C 6 | IVS11–1G>C | Intron11 | Splice site | Dutch | (Dell'Angelica et al., 1999; Gochuico et al., 2012) 5,6 |
| 11 | c.1473+6T>C | IVS14+6T>C | Intron14 | Splice site | - | (Clark et al., 2003) |
| 12 | c.1525C>T | p.Arg509* | Exon 15 | Nonsense | Cajun, Houma Indian | (Huizing et al., 2002) |
| 13 | c.1619dupG | p.Ala541Serfs*25 | Exon 15 | Frameshift | - | (Clark et al., 2003) |
| 14 | g.del8168-bp 7 | del exon 15 | Introns14+15, Exon 15 | Indel | Turkish | (Jung et al., 2006) 7 |
| 15 | g.del1872-bp 7 | del exon 15 | Introns14+15, Exon 15 | Indel | - | (Hengst et al., 2018) 7 |
| 16 | del exon 16 | - | Exon 16 | Indel | - | (Jessen et al., 2013) |
| 17 | c.1739T>G | p.Leu580Arg | Exon 16 | Missense | Dutch | (Dell'Angelica et al., 1999) |
| 18 | c.1754delT | p.Val585Glufs*6 | Exon 16 | Frameshift | Caucasian | (de Boer et al., 2017) |
| 19 | c.1789dupA | p.Ile597Asnfs*12 | Exon 16 | Frameshift | Italian | (Fontana et al., 2006) |
| 20 | c.1839_1842delTAGA | p.Asp613Glufs*38 | Exon 17 | Indel | - | (de Boer et al., 2017; Hengst et al., 2018; Jung et al., 2006) |
| 21 | c.1975G>T | p.Glu659* | Exon 18 | Nonsense | Cajun, Houma Indian | (Huizing et al., 2002) |
| 22 | c.2041G>T | p.Glu681* | Exon 18 | Nonsense | - | (Ammann et al., 2017; Jessen et al., 2013) |
| 23 | g.del624-bp 8 c.del2077_2164 | p.Glu693Valfs*13 | Intron18, Exon 19 | Indel | Maltese | (Wenham et al., 2010) |
| 24 | c.2546T>G | p.Leu849* | Exon 22 | Nonsense | - | (Hengst et al., 2018) |
| 25 | c.2702C>G 9 | p.Ser901Cys | Exon 23 | Missense Splice site | Caucasian | (de Boer et al., 2017) 9 |
| 26 | c.2770delC | p.Leu924Phefs*3 | Exon 23 | Frameshift | - | (Jessen et al., 2013) |
| 27 | c.2944delC | p.Leu982Cysfs*19 | Exon 25 | Frameshift | - | (Hengst et al., 2018) |
| 28 | c.3222_3223delTG 10 | p.Lys1076Asnfs*60 | Exon 27 | Indel-Stop-loss | United Arabic Emirates | (Hengst et al., 2018; Jessen et al., 2013; Kurnik et al., 2013) 10 |
| 29 | inv(5)p15.1-q14.1 11 | - | - | Indel-chrom. inversion | Lebanese | (Jones et al., 2013) 11 |
When deletion/insertion is 1 nucleotide it is named Frame shift, when larger it is named Indel.
Extracted from literature reference. ‘-‘ = unreported.
This variant likely leads to a loss of protein translation in the start codon of the longest splice variant of AP3B1 (NM_003664.4).
Gray highlight: missense variant. See Supplemental Table S1 for pathogenicity predictions.
At least one of the reported cases with this variant was identified by next generation sequencing.
This variant was originally described as del63-bp in the patients’ cDNA (Dell'Angelica et al., 1999), but later found to be due to a gDNA splice site variant, skipping the 63-bp exon 12 (Gochuico et al., 2012).
Described as g.151312_159483del8172-bp (NG_007268) (Hengst et al., 2018). It is possible that del8168-bp reported by (Jung et al., 2006) is the same deletion.
This deletion was reported as: NC_00005.8:g.180242–180866del.
This nucleotide change activates a cryptic donor splice and causes a deletion of 112bp within exon 23 on the mRNA level, resulting in a frame shift and a premature termination codon p.Val900Thrfs*63 (de Boer et al., 2017).
This frameshift in the AP3B1 C-terminal coding region results in a prolonged altered protein, beyond the termination codon, with 42 additional C-terminal amino acids compared to the wild type protein (Kurnik et al., 2013).
Chromosomal inversion breakpoints occur within the AP3B1 gene (Jones et al., 2013).
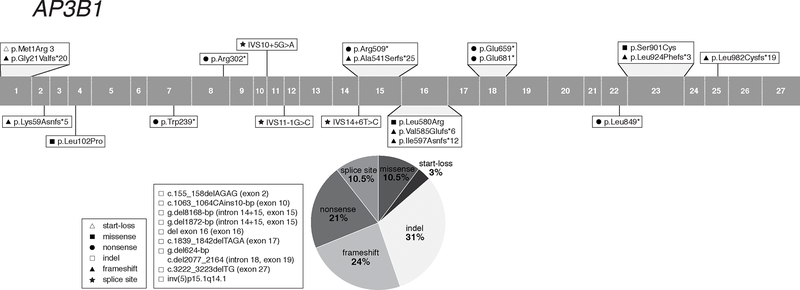
HPS-2 (MIM# 608233) is caused by biallelic pathogenic variants in AP3B1, located on chromosome 5q14.1, and encoding the β3A subunit of AP3 (Dell’Angelica et al., 1999). The longest AP3B1 mRNA transcript contains 27 exons and is translated into the 1094-amino acid protein AP3B1 (AP-3 β3A; ~121-kD). Two AP3B1 protein-coding mRNA transcripts are predicted, varying at the N-terminus, with unexplored expression and functional significance. Vertebrate models of HPS-2 include the pearl and Ap3b1LN mice (Yang et al., 2000; Li et al., 2004). With the recognition that HPS-2 is caused by deficiency of AP-3, HPS became an informative disorder for intracellular vesicle/membrane formation and trafficking (Dell’Angelica et al., 1999). HPS cells and models have since been used extensively for LRO-related cell biology (Huizing et al., 2008).
There are currently ~35 reported HPS-2 cases of various ethnic backgrounds, including Caucasian, Chinese, Lebanese and Mexican. A total of 29 AP3B1 pathogenic variants associated with HPS-2 have been described (Table 4); they are located throughout the gene, including 1 (3%) start-loss, 7 (24%) frameshift, 3 (10.5%) missense, 6 (21%) nonsense, 9 (31%) insertions and/or deletions (including a chromosomal inversion), and 3 (10.5%) splice site variants (Figure 2). There are no known frequently occurring pathogenic variants in AP3B1, nor any apparent founder mutations. All 3 reported AP3B1 splice site variants are predicted and/or reported to cause aberrant splicing (Supp. Table S2). Of the 3 reported pathogenic missense variants, two are likely pathogenic (p.Leu580Arg, p.Ser901Cys), and one is a VUS (p.Leu102Pro).
Figure 2:
Distribution of AP3B1 Gene Variants
Next generation sequencing of different cohorts of subjects with primary immunodeficiency or hemophagocytic lymphohistiocytosis (HLH) reported least 8 AP3B1 variants (Supp. Table S4) in a heterozygous state with or without a variant in another (synergistic) gene. These findings suggest that heterozygous AP3B1 variants may contribute to an immunologic phenotype (Chi et al., 2018; Gallo et al., 2016; Gao, Zhu, Huang, & Zhou, 2015; Miao et al., 2019; Mukda et al., 2017; Tesi et al., 2015; Xu et al., 2017). These variants have not been reported in HPS subjects but were included in this report because they may cause HPS when occurring in a homozygous or compound heterozygous state.
The dbSNP/gnomAd/ClinVar databases list 7 frequently occurring (MAF > 0.001) AP3B1 missense variants and two in-frame 3-bp deletions, with predicted benign or unknown pathogenicity, which should be considered as likely non-pathogenic polymorphisms when encountered in AP3B1 genetic analyses (Supp. Table S3).
Apart from the hypopigmentation and bleeding diathesis of HPS-2, affected individuals are also at risk for developing interstitial lung disease and pulmonary fibrosis in childhood (Gochuico et al., 2012; Hengst et al., 2018); in addition, immunodeficiency associated with neutropenia is the most prevalent clinical feature (Fontana et al., 2006). The immunodeficiency, an impairment of cytotoxic activity, results from T-lymphocyte and/or natural killer cell dysfunction and can present with variable features, from mild recurrent bacterial and viral infections to severe hemophagocytic lymphohistiocytosis (HLH) (Gil-Krzewska et al., 2017; Jessen et al., 2013). These features led to discovery of involvement of AP-3 in different trafficking processes. AP-3 is involved in neutrophil formation (Badolato & Parolini, 2007; Massullo et al., 2005); AP-3 deficient cells mislocalize the neutrophil granule proteins myeloperoxidase and elastase and the lysosomal membrane protein CD63 (de Boer et al., 2017; Di Pietro et al., 2006; Jung et al., 2006; Meng et al., 2010). The AP-3 immunodeficiency also involves defective AP-3 mediated lytic granule exocytosis in natural killer (NK)-cells and cytotoxic T-cells (Clark et al., 2003; Fontana et al., 2006; Gil-Krzewska et al., 2017; Jung et al., 2006). AP-3 deficient dendritic cells showed impaired toll-like receptor recruitment (Mantegazza et al., 2012; Sasai, Linehan, & Iwasaki, 2010), leading to defects in interferon production and antigen presentation in these cells from HPS-2 subjects (Prandini et al., 2016). AP-3-dependent inflammasome positioning and activation was shown in dendritic cells from HPS-2 mice (Mantegazza et al., 2017).
Remarkably, genetic testing in all reported HPS-2 subjects was performed by AP3B1 gene-specific Sanger sequencing. Individuals with HPS-2 that present to an immunologist with (severe) immunodeficiency may escape diagnosis due to emphasis on their immunodeficiency, other mild manifestations (e.g., hypopigmentation, ocular findings, bleeding diathesis) may be overlooked, there may be unfamiliarity with HPS-2, and costs and lack of availability of AP3B1 genetic testing may provide obstacles to diagnosis. However, the recent significant number of heterozygous AP3B1 variants identified by next-generation sequencing in cohorts with immunodeficiency disorders (Chi et al., 2018; Gallo et al., 2016; Gao et al., 2015; Miao et al., 2019; Mukda et al., 2017; Tesi et al., 2015; Xu et al., 2017) emphasizes the importance of including AP3B1 in immunodeficiency-related gene panels and may result in the diagnosis of additional HPS-2 cases.
HPS-3 (Table 5)
Table 5:
HPS3 Gene Variants Associated with Hermansky-Pudlak Syndrome Type 3 (HPS-3)
| No | mRNA NM_032383.5 | Amino Acid NP_115759.2 | Exon/Intron | Variant Type 1 | Ethnic Background 2 | References and Footnotes |
|---|---|---|---|---|---|---|
| 1 | c.-2993_217+692del 3 | - | 5’UTR Intron 1 | Indel | Central Puerto Rican | (Anikster et al., 2001; Torres-Serrant et al., 2010) 3,4 |
| 2 | c.15C>G | p.Tyr5* | Exon 1 | Nonsense | Dutch, German | novel 5,6 |
| 3 | c.87dupG | p.Arg30Alafs*2 | Exon 1 | Frameshift | Japanese | (Saito et al., 2019) |
| 4 | c.319C>T | p.Arg107* | Exon 1 | Nonsense | Japanese | (Okamura et al., 2019) 7 |
| 5 | c.437_439delGAG | p.Gly146del | Exon 1 | Indel | Japanese | (Okamura et al., 2019) 7,8 |
| 6 | c.712+2T>C | IVS2+2 | Intron 2 | Splice site | Chinese | (A. Wei et al., 2016) 7 |
| 7 | c.726_727insTGCCTTACATC | p.Ile243Cysfs*41 | Exon 3 | Indel | Puerto Rican | novel 4,5 |
| 8 | c.728_729insA | p.Ser244Phefs*4 | Exon 3 | Frameshift | Italian, Sicilian | (Boissy et al., 2005) 9 |
| 9 | c.851_852delGA | p.Arg284Lysfs*11 | Exon 3 | Indel | Portuguese | novel 5,11 |
| 10 | c.868C>T | p.Gln290* | Exon 3 | Nonsense | Arabic | (Khan et al., 2016) 7 |
| 11 | c.885–1G>A | IVS3–1G>A | Intron 3 | Splice site | Libyan | (Thielen et al., 2010) |
| 12 | c.1012G>T | p.Glu338* | Exon 5 | Nonsense | Caucasian | novel 5,12 |
| 13 | c.1107_1119del13insC | p.Pro370_Ser373del | Exon 5 | Indel | French-Canadian | novel 5,13,14 |
| 14 | c.1153_1160del8 | p.Val385Lysfs*2 | Exon 5 | Indel | Middle-Eastern | (Trujillano et al., 2017) 7 |
| 15 | c.1163+1G>A | IVS5+1G>A | Intron 5 | Splice site | Ashkenazi Jewish | (Huizing et al., 2001) |
| 16 | c.1189C>T | p.Arg397Trp | Exon 6 | Missense | Canadian, Caucasian, Chinese, German, Japanese, Polish, Russian, Swiss | (Huizing et al., 2001; Nazarian et al., 2008; Okamura et al., 2019; Wei et al., 2016) 7,12,15 |
| 17 | c.1195A>G/ c.1199_1200insATTGC | p.Ser399Gly/ p.Ala401Leufs*16 | Exon 6 | Indel | English, German, Irish, Scottish, Cherokee | novel 5,16 |
| 18 | c.1291delC | p.Leu431Phefs*3 | Exon 7 | Frameshift | Japanese | (Okamura et al., 2019) 7,8 |
| 19 | c.1426dupA | p.Ile476Asnfs*8 | Exon 8 | Frameshift | Japanese | (Saito et al., 2019) |
| 20 | c.1509G>A | p.Met503Ile | Exon 8 | Missense-Splice site | Pakistani | (Yousaf et al., 2016) 7,17 |
| 21 | c.1555_1595dup41 | p.Leu533Phefs*10 | Exon 8 | Indel | Chinese | (Power et al., 2019) |
| 22 | c.1673T>C | p.Leu558Pro | Exon 8 | Missense | - | (Lasseaux et al., 2018) 7 |
| 23 | c.1691+1G>A | IVS9+1G>A | Intron 9 | Splice site | French-Canadian | novel 5,13 |
| 24 | c.1691+2T>G | IVS9+2T>G | Intron 9 | Splice site | Ashkenazi Jewish | (Huizing et al., 2001) |
| 25 | c.1838C>G | p.Ser613* | Exon 10 | Nonsense | Chinese | (Wei et al., 2019) 7 |
| 26 | c.1870G>T | p.Glu624* | Exon 10 | Nonsense | German, Irish | novel 5,18 |
| 27 | c.2208_2209delTC | p.Gln737Alafs*20 | Exon 12 | Indel | Chinese | (Wei et al., 2016) 7 |
| 28 | c.2464C>T | p.Arg822* | Exon 13 | Nonsense | Dutch, German, Portuguese, Spanish | (Bastida et al., 2019) 6,7,11 |
| 29 | c.2482–2A>G | IVS13–2A>G | Intron 13 | Splice site | Irish/German | (Huizing et al., 2001) |
| 30 | c.2589+1G>C | IVS14+1G>C | Intron14 | Splice site | German/Swiss | (Huizing et al., 2001) |
| 31 | c.2589+1G>T | IVS14+1G>T | Intron14 | Splice site | German, Irish | novel 5,18 |
| 32 | c.2628delT | p.Ile877Phefs*25 | Exon 15 | Frameshift | - | (Lasseaux et al., 2018) 7 |
| 33 | c.2733delG | p.Leu912* | Exon 15 | Frameshift | English, German, Irish, Scottish, Cherokee | novel 5,16,19 |
| 34 | c.2739_2742delGAGA | p.Glu913Aspfs*14 | Exon 15 | Indel | - | novel 5,14 |
| 35 | c.2771delA | p.Asn924Ilefs*4 | Exon 15 | Frameshift | Turkish | (Sandrock-Lang et al., 2017) |
| 36 | c.2805G>A | p.Trp935* | Exon 16 | Nonsense | Chinese | (Wei et al., 2016) 7 |
| 37 | c.2888–1612G>A21 | IVS16–1612G>A p.Glu963Alafs*24 | Intron16 | Splice site | English, Irish | (Huizing et al., 2001) 20 |
When deletion/insertion is 1 nucleotide it is named Frame shift, when larger it is named Indel.
Extracted from literature reference. ‘-‘ = unreported.
This HPS3 3.9-kb deletion occurs within two Alu repeats, encompassing exon 1 and originates from a genetic isolate of central Puerto Rico (Anikster et al., 2001). Current nomenclature (as annotated in ClinVar) for this deletion is NM_032383.5(HPS3):c.-2993_217+692del or NC_000003.12:g.149126714_149130632del (GRCh38) or NG_009847.1:g.2131_6049del.
This HPS3 variant was identified in one unreported female subject of Puerto Rican descent in the NIH cohort. This subject was compound heterozygous for c.726delinsTGCCTTACATC and the Central Puerto Rican founder variant g.del3.9-kb.
novel = previously unreported variant detected in the NIH HPS cohort.
This HPS3 variant was identified in one unreported male subject of Dutch-German descent in the NIH HPS cohort. This subject was compound heterozygous for c.15C>G and c.2464C>T.
At least one of the reported cases with this variant was identified by next generation sequencing.
The HPS3 variant c.437_439delGAG occurred compound heterozygous with c.1291delC in a subject of Japanese descent who also had non-segmental vitiligo (Okamura et al., 2019).
This HPS3 variant c.728insA was identified homozygous in one unreported female subject of Sicilian decent in the NIH HPS cohort.
Gray highlight: missense variant. See Supplemental Table S1 for pathogenicity description.
This HPS3 variant was identified in one unreported male subject of Portuguese decent in the NIH HPS cohort. This subject was compound heterozygous for c.851_852delGA and c.2464C>T.
This HPS3 variant was identified in one unreported female subject of Caucasian descent in the NIH HPS cohort. This subject was compound heterozygous for c.1012G>T and c.1189C>T.
This HPS3 variant was identified in one unreported female subject of French-Canadian descent in the NIH HPS cohort. This subject was compound heterozygous for c.1107_1119del13insC and c.1691+1G>A.
This HPS3 variant was identified in one unreported female subject of in the NIH cohort (referred by Dr. Doherty, Carilion Clinic, Roanoke, VA). This subject was compound heterozygous for c.1107_1119del13insC and c.2739_2742delGAGA.
This HPS3 variant c.1189C>T was identified homozygous in one unreported female subject of Canadian-Polish-Russian decent in the NIH cohort. Cells of this patient showed destabilized BLOC-2 assembly, likely due to pathogenicity of this variant (Nazarian et al., 2008).
This HPS3 variant was identified in one unreported female subject of English-German-Irish-Scottish-Cherokee descent in the NIH HPS cohort. This subject was compound heterozygous for c.1195A>G/1199insATTGC and c.2733delG.
This (missense) HPS3 variant c.1509G>A; p.Met503Ile occurs at the exon8/intron 8 splice site junction and may affect splicing. This variant occurs homozygous in 4 subjects of a consanguineous Pakistani family (Yousaf et al., 2016).
This HPS3 variant was identified in one unreported male subject of German-Irish descent. This subject was compound heterozygous for c.1870G>T and c.2589+1G>T.
Subject HPS34 of English-Irish descent in (Huizing et al., 2001) was reported heterozygous for c.2887+2500G>A. We found c.2733delG to be the second HPS3 variant in this subject.
This HPS3 variant was reported in an alternative nomenclature as c.2887+2500G>A. This intronic variant introduces a new consensus splice site that results in insertion of 89-bp (a ‘pseudoexon’) in the patient’s cDNA (Huizing et al., 2001; Vorechovsky, 2010).
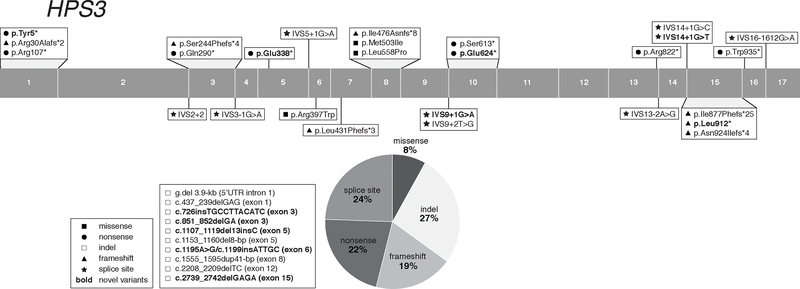
HPS-3 (MIM# 614072) is caused by biallelic pathogenic variants in HPS3, also called BLOC2S1, located on chromosome 3q24. The longest HPS3 mRNA transcript contains 17 exons, encoding a 1004-amino acid protein (~113.7-kD). Two HPS3 protein-coding mRNA variants are predicted, but their expression and functional significance remain unexplored. The HPS3 protein interacts with the HPS5 and HPS6 proteins in BLOC-2 (Di Pietro et al., 2004). The mouse model cocoa is the murine orthologue of human HPS-3 (Suzuki et al., 2001).
The HPS3 gene was identified through homozygosity mapping in a genetic isolate of HPS originating in central Puerto Rico. These individuals are homozygous for a 3.9-kb deletion in HPS3 (NM_032383.5:c.−2993_217+692del or NG_009847.1:g.2131_6049del), detectable with a multiplex PCR amplification assay (Anikster et al., 2001). There are ~63 reported cases homozygous for this deletion (Anikster et al., 2001; Santiago Borrero et al., 2006), and there are an additional 9 such unreported subjects in our NIH cohort. One in 14,000 individuals of central Puerto Rican descent are estimated to be homozygous for this deletion. The carrier frequency in central Puerto-Rico is ~1:32 (Santiago Borrero et al., 2006) and 1:85 in all of Puerto Rico (Torres-Serrant, Ramirez, Cadilla, Ramos-Valencia, & Santiago-Borrero, 2010).
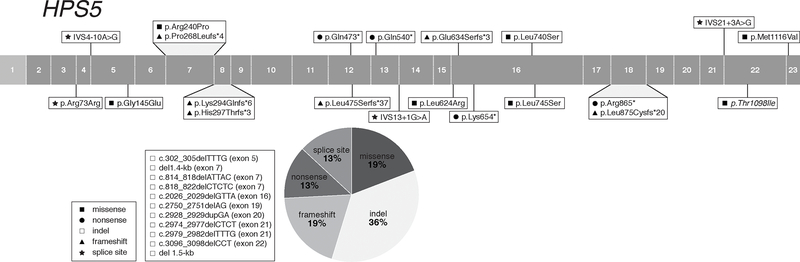
We report 42 non-Puerto Rican HPS-3 subjects, including those described with an Ashkenazi-Jewish founder variant (c.1691+2T>G; 7 cases) (Huizing et al., 2001) and including 9 novel cases from our NIH cohort (Table 5). Apart from central Puerto Rican and Ashkenazi-Jewish cases, HPS-3 subjects with a variety of other ethnic backgrounds are reported, including Arabic, Chinese, northern and southern European, and Pakistani.
We report 37 HPS3 pathogenic variants, including 11 novel variants from our NIH cohort (Table 5). These pathogenic variants are found throughout the gene and include 7 (19%) frameshift, 3 (8 %) missense, 8 (22%) nonsense, 10 (27 %) insertions and/or deletions, and 9 (24%) splice site variants (Figure 3). All HPS3 splice site variants are predicted and/or experimentally demonstrated to cause aberrant splicing (Supp. Table S2). One intronic variant, c.2888–1612G>A (originally reported as c.2887+2500G>A) introduces a new consensus splice site, resulting in cryptic exon activation and insertion of a 89-bp pseudo-exon in the cDNA, leading to a frameshift and premature protein termination (p.Glu963Alafs*24) (Huizing et al., 2001; Vorechovsky, 2010). All 3 reported HPS3 missense variants have a low MAF, one (c.1189C>T, p.Arg397Trp) is predicted likely pathogenic and the other two are predicted VUS (Supp. Table S1).
Figure 3:
Distribution of HPS3 Gene Variants
Next generation sequencing of a cohort of individuals with schizophrenia identified a de novo HPS3 missense variant c.796G>A (p.Glu266Lys) in one proband, suggesting an association of this variant with the schizophrenia phenotype (Fromer et al., 2014) (Supp. Table S4).
This rare and benign variant has not been reported in HPS subjects. In fact, no other variants in any BLOC-2 subunit (HPS3, HPS5, HPS6) have been reported to be associated with a neuronal phenotype in individuals with or without HPS or in animal models. The dbSNP/gnomAd/ClinVar databases list 5 frequently occurring (MAF > 0.001) HPS3 missense variants (Supp. Table S3), with predicted benign or unknown pathogenicity, that should be considered as likely non-pathogenic polymorphisms when encountered in HPS3 genetic analysis.
Subjects with HPS-3 have relatively mild clinical features, including minor hypopigmentation of the hair, skin, and retina. Visual acuity is often only slightly affected, and bleeding tendency is also mild, although some female subjects have significant menorrhagia. Some subjects only appear hypopigmented when compared to their siblings or other family members. Of note, pigmentation and bleeding are also only mildly affected in BLOC-2 (HPS3, HPS5, HPS6) mouse models (Novak et al., 1984). In addition, the bleeding diathesis in BLOC-2-deficient individuals might be exacerbated by disruption not only of platelet dense granules but also of altered maturation of Weibel-Palade bodies in endothelial cells, which are involved in release of von Willebrand factor (Ma et al., 2016).
Pulmonary fibrosis or immunodeficiency has not been reported in HPS-3 (nor in any other BLOC-2 deficient) subjects, and some BLOC-2 deficient individuals develop granulomatous colitis. The diagnosis of HPS-3 may be elusive in some patients due to the mildness of their symptoms.
HPS-4 (Table 6)
Table 6:
HPS4 Pathogenic Gene Variants Associated with Hermansky-Pudlak Syndrome Type 4 (HPS-4)
| No | mRNA NM_022081.5 | Amino Acid NP_071364.4 | Exon/Intron | Variant Type 1 | Ethnic Background 2 | References and Footnotes |
|---|---|---|---|---|---|---|
| 1 | c.45G>A | p.Trp15* | Exon 3 | Nonsense | Uruguayan, Japanese | (Carmona-Rivera, Golas, et al., 2011; Okamura et al., 2018) 3 |
| 2 | c.47delA | p.Asn16Ilefs*11 | Exon 3 | Frameshift | Uruguayan | (Carmona-Rivera, Golas, et al., 2011) |
| 3 | c.57delT | p.Leu20Phefs*7 | Exon 3 | Frameshift | French, German, Irish, Northern European | (Suzuki et al., 2002) 4 |
| 4 | c.123T>A | p.Tyr41* | Exon 4 | Nonsense | Japanese | (Okamura et al., 2018) 3 |
| 5 | c.148C>T | p.Gln50* | Exon 4 | Nonsense | Chinese | (Wei et al., 2019) 3 |
| 6 | c.272T>C | p.Leu91Pro 5 | Exon 4 | Missense | Turkish | (Bastida et al., 2019) 3 |
| 7 | c.276+5G>A | IVS4+5G>A | Intron 4 | Splice site | Pakistani | (Yousaf et al., 2016) 3 |
| 8 | c.357C>G | p.Tyr119* | Exon 5 | Nonsense | Dutch, English, Irish, Polish, Slovak | novel 6,7 |
| 9 | c.412G>T | p.Glu138* | Exon 6 | Nonsense | Indian | (Anderson et al., 2003) |
| 10 | c.416G>A | p.Trp139* | Exon 6 | Nonsense | Chinese | (Power et al., 2019) |
| 11 | c.430G>T | p.Glu144* | Exon 6 | Nonsense | Indian | (Arcot Sadagopan et al., 2017) 3 |
| 12 | c.461A>G | p.His154Arg | Exon 6 | Missense | Caucasian, Japanese | (Anderson et al., 2003; Saito et al., 2013) 8,9 |
| 13 | c.541C>T | p.Gln181* | Exon 7 | Nonsense | Southern Italian | (Suzuki et al., 2002) |
| 14 | c.554G>A | p.Arg185His | Exon 7 | Missense | Indian | (Arcot Sadagopan et al., 2017) 3 |
| 15 | c.596+1G>A | IVS7+1G>A | Intron 7 | Splice site | Japanese | (Okamura et al., 2019) 3 |
| 16 | c.597–2A>T | IVS7–2A>T | Intron 7 | Splice site | - | (Jones et al., 2012) 3 |
| 17 | c.630dupC | p.Ala211Argfs*47 | Exon 8 | Frameshift | Chinese | (Wu et al., 2019) |
| 18 | c.649C>T | p.Arg217* | Exon 8 | Nonsense | Ashkenazi Jewish, English, Polish | (Anderson et al., 2003; Lozynska et al., 2018) |
| 19 | c.664G>T | p.Glu222* | Exon 8 | Nonsense | Indian | (Anderson et al., 2003) |
| 20 | c.706+1G>A | IVS9+1G>A | Intron 9 | Splice site | Dutch, English, Irish, Polish, Slovak | novel 6,7 |
| 21 | c.730C>T | p.Gln244* | Exon 10 | Nonsense | Japanese | (Araki et al., 2014) |
| 22 | c.803G>A 10 | p.Arg268Lys | Exon 10 | Missense-Splice Site | - | (Lasseaux et al., 2018) 3,10 |
| 23 | c.949_972dup24 | p.Ala317_Glu324dup | Exon 11 | Indel | Dutch | (Suzuki et al., 2002) |
| 24 | c.1132C>T | p.Gln378* | Exon 11 | Nonsense | - | novel 11 |
| 25 | c.1318C>T | p.Gln440* | Exon 11 | Nonsense | Turkish | (Sandrock-Lang et al., 2018) |
| 26 | c.1546C>T | p.Gln516* | Exon 11 | Nonsense | Caucasian | novel 6,8 |
| 27 | c.1547_1548delAG | p.Gln516Argfs*42 | Exon 11 | Indel | Indian | (Arcot Sadagopan et al., 2017) 3 |
| 28 | c.1713+5G>C | IVS11+5G>C | Intron 11 | Splicing | Chinese | (Wei et al., 2019) 3 |
| 29 | c.1856C>T | p.Pro619Leu | Exon 13 | Missense | - | (Lasseaux et al., 2018) 3 |
| 30 | c.1858C>T | p.Gln620* | Exon 13 | Nonsense | - | (Sakata et al., 2013) |
| 31 | c.1891C>T | p.Gln631* | Exon 13 | Nonsense | German South Tirol | (Suzuki et al., 2002) |
| 32 | c.1897_1898dupCG | p.Ser634Alafs*3 | Exon 13 | Indel | Japanese | (Okamura et al., 2019) 3 |
| 33 | c.2054delC | p.Pro685Leufs*17 | Exon 14 | Frameshift | Sri Lankan, Spanish | (Bachli et al., 2004; Bastida et al., 2019) 3 |
| 34 | c.2089_2093dupAAGCA | p.Lys699Serfs*5 | Exon 14 | Indel | Austrian, Czech, English, German, Hungaran, Irish, Scandinavian, Swiss | (Anderson et al., 2003; Suzuki et al., 2002) |
When deletion/insertion is 1 nucleotide it is named Frame shift, when larger it is named Indel.
Extracted from literature reference. ‘-‘ = unreported.
At least one of the reported cases with this variant was identified by next generation sequencing.
This HPS4 variant was identified in heterozygous state in one unreported subject of French-German-Irish descent in the NIH HPS cohort.
Gray highlight: missense variant. See Supplemental Table S1 for pathogenicity description.
novel = previously unreported variant detected in the NIH HPS cohort.
This HPS4 variant was identified in one unreported subject of Dutch-English-Irish-Polish-Slovak descent in the NIH HPS cohort. This subject is compound heterozygous for c.357C>G and c.706+1G>A.
This HPS4 variant was identified in one unreported subject of Caucasian descent in the NIH HPS cohort. This individual is compound heterozygous for c.461A>G and c.1546C>T.
This variant was found homozygous in two Japanese siblings with HPS and mental disorder (schizophrenia and major depression). It was suggested that HPS4 gene variants are associated with susceptibility to schizophrenia (Saito et al., 2013) and/or cognitive function (Kuratomi et al., 2013).
This (missense) HPS4 variant c.803G>A; p.Arg268Lys (Lasseaux et al., 2018) occurs at the exon10/intron 11 splice site junction and is predicted to affect splicing (Supplemental Table S2).
This nonsense variant c.1132C>T (p.Gln378*) was found homozygous in one unreported female subject in the NIH cohort (referred by Dr. Everman, Greenwood Genetics Center, Greenville, SC).
HPS-4 (MIM# 614073) is caused by biallelic pathogenic variants in HPS4, also called BLOC3S2, located on chromosome 22q12.1. The longest HPS4 mRNA transcript contains 14 exons, encoding a 708-amino acid protein (~76.9-kD). This major transcript variant is expressed in all tissues tested. A second major mRNA variant contains 12 exons, is alternatively spliced in the 5’ region and is expressed in limited tissues; its function remains unexplored (Anderson, Huizing, Claassen, White, & Gahl, 2003). There are at least 9 additional predicted HPS4 protein-coding mRNA transcripts. The HPS4 protein interacts with the HPS1 protein in BLOC-3 (Carmona-Rivera et al., 2013; Martina et al., 2003).
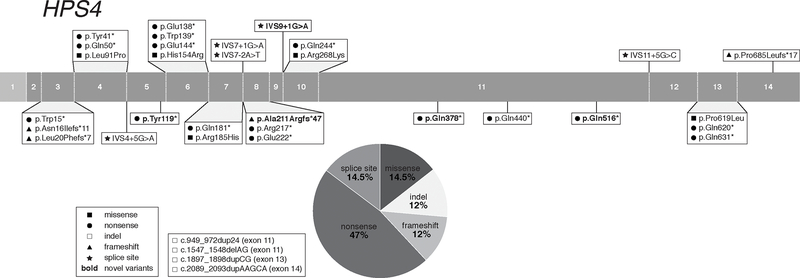
Identification of the human HPS4 gene was initiated based upon Hps4 mutations in the light ear HPS mouse model (Suzuki et al., 2002). Subsequent genetic screening of the human orthologue gene, HPS4, in unclassified HPS subjects identified 7 cases with pathogenic variants (Suzuki et al., 2002). There are currently 37 HPS-4 subjects reported and there are 4 unreported subjects in our NIH cohort. HPS-4 is identified in various populations, including Ashkenazi-Jewish, Chinese, European, Indian, Japanese, Pakistani, Sri Lankan, and Uruguayan (Table 6). We report 34 HPS4 pathogenic variants, including 4 novel variants from our NIH cohort (Table 6). These variants are located throughout the HPS4 gene, with c.2089_2093dupAAGCA (p.Lys699Serfs*5) occurring frequently in individuals of European descent (Anderson et al., 2003; Suzuki et al., 2002). The HPS4 variants include 5 (12 %) frameshift, 5 (14.5%) missense, 16 (47%) nonsense, 4 (12 %) insertions and/or deletions, and 5 (14.5%) splice site variants (Figure 4). The 4 reported HPS4 splice site variants are all predicted to cause aberrant splicing (Supp. Table S2). Of the 5 reported missense variants, one is likely pathogenic (p.His154Arg) and the other 3 are classified as VUS; one of these occurs at a splice site junction and is predicted (but not demonstrated) to cause aberrant splicing (c.803G>A; p.Arg268Lys) (Supp. Table S1). No additional experimental evidence for pathogenicity is available for these missense variants.
Figure 4:
Distribution of HPS4 Gene Variants
Next generation sequencing of cohorts with non-HPS pulmonary fibrosis identified a heterozygous HPS4 frameshift variant (c.1102dupG, p.Asp368Glyfs*4) in a subject with sporadic pulmonary fibrosis (Deng et al., 2018) and a heterozygous HPS4 indel variant (c.1966_1967dupAC, p.Ala657Argfs*46) and a missense variant (c.1396C>T, p.Arg466Cys) in subjects with familial pulmonary fibrosis (Stearman et al., 2019). None of these variants has been reported in HPS subjects but are included in Supp. Table S4 since they may cause HPS when occurring in the homozygous or compound heterozygous state.
The dbSNP/gnomAd/ClinVar databases list 12 frequently occurring (MAF > 0.001) HPS4 missense variants, with predicted benign or unknown pathogenicity (Supp. Table S3); these should be considered as likely non-pathogenic polymorphisms when encountered in HPS4 genetic analysis.
Individuals with HPS-4 have a phenotype similar to that of HPS-1 subjects, including more profound cutaneous and ocular hypopigmentation than in other HPS subtypes, development of pulmonary fibrosis (at middle age) in virtually all subjects, menorrhagia in most female subjects, and occurrence of granulomatous colitis in some subjects (Anderson et al., 2003; Huizing et al., 2008). HPS-4 should be considered in individuals where HPS-1 was suspected based on clinical symptoms (without genetic confirmation). Of note, the HPS4 missense variant p.His154Arg (c.461A>G) was found homozygous in two Japanese siblings with HPS and mental disorder (schizophrenia and major depression). It was suggested that HPS4 gene single nucleotide polymorphisms variants may be associated with susceptibility to schizophrenia (A. Saito et al., 2013) and/or cognitive function (Kuratomi et al., 2013). However, there are no reports of other HPS-related BLOC-3 (HPS1 and HPS4) variants in individuals with HPS to be associated with neurological phenotypes.
HPS-5 (Table 7)
Table 7:
HPS5 Pathogenic Gene Variants Associated with Hermansky-Pudlak Syndrome Type 5 (HPS-5)
| No | mRNA NM_181507.1 | Amino Acid NP_852608.1 | Exon/Intron | Variant Type 1 | Ethnic Background 2 | References and Footnotes |
|---|---|---|---|---|---|---|
| 1 | c.219G>A | p.Arg73Arg 3 | Exon 3 | Splice site | Turkish | (Lasseaux et al., 2018; Michaud et al., 2017) 3,4 |
| 2 | c.285–10A>G | IVS4–10A>G | Intron 4 | Splice site | Turkish | (Stephen et al., 2017) 4 |
| 3 | c.302_305delTTTG | p.Val101Glyfs*3 | Exon 5 | Indel | Cuban, Venezuelan | (Carmona-Rivera, Golas, et al., 2011) |
| 4 | c.434G>A | p.Gly145Glu 5 | Exon 5 | Missense | German, Irish, Welsh | (Nazarian et al., 2008) |
| 5 | c.719G>C | p.Arg240Pro | Exon 7 | Missense | African-French | (Lasseaux et al., 2018; Michaud et al., 2017) 4 |
| 6 | c.803delC 6 | p.Pro268Leufs*4 | Exon 7 | Frameshift | - | (Ringeisen et al., 2013) 6 |
| 7 | del 1.4-kb 7,8 | - | Exon 7 | Indel | African-French | (Michaud et al., 2017) 4,7 |
| 8 | c.814_818delATTAC | p.Ile272Serfs*8 | Exon 7 | Indel | - | (Lasseaux et al., 2018) 4 |
| 9 | c.818_822delCTCTC | p.Thr273Lysfs*7 | Exon 7 | Indel | French | (Michaud et al., 2017) 4 |
| 10 | c.879dupC | p.Lys294Glnfs*6 | Exon 8 | Frameshift | English, Irish | (Huizing et al., 2004) |
| 11 | c.888dupA | p.His297Thrfs*3 | Exon 8 | Frameshift | Turkish | (Korswagen et al., 2008) |
| 12 | c.1417C>T | p.Gln473* | Exon12 | Nonsense | Turkish | (Lasseaux et al., 2018; Michaud et al., 2017) 4 |
| 13 | c.1423delC | p.Leu475Serfs*37 | Exon 12 | Frameshift | Mexican | (Carmona-Rivera, Golas, et al., 2011) |
| 14 | c.1618C>T | p.Gln540* | Exon 13 | Nonsense | Arabic | (Khan et al., 2016) 4 |
| 15 | c.1634+1G>A | IVS13+1G>A | Intron13 | Splice site | Cuban, Venezuelan | (Carmona-Rivera, Golas, et al., 2011) |
| 16 | c.1871T>G | p.Leu624Arg 9 | Exon 16 | Missense | Swiss | (Huizing et al., 2004; Lasseaux et al., 2018; Michaud et al., 2017) 4,9 |
| 17 | c.1900delG | p.Glu634Serfs*3 | Exon 16 | Frameshift | - | (Lasseaux et al., 2018; Michaud et al., 2017) 4 |
| 18 | c.1960A>T | p.Lys654* | Exon 16 | Nonsense | English, German, Scottish | (Botero et al., 2018) 4 |
| 19 | c.2026_2029delGTTA | p.Val676Metfs*8 | Exon 16 | Indel | Turkish | (Zhang et al., 2003) |
| 20 | c.2219T>C | p.Leu740Ser | Exon 16 | Missense | - | (Lasseaux et al., 2018; Michaud et al., 2017) 4 |
| 21 | c.2234T>C | p.Leu745Ser | Exon 16 | Missense | Chinese | (Wei et al., 2016) 4,10 |
| 22 | c.2593C>T | p.Arg865* | Exon 18 | Nonsense | Dutch, English, Irish, Swedish | (Huizing et al., 2004) |
| 23 | c.2624delT | p.Leu875Cysfs*20 | Exon 18 | Frameshift | Dutch, English, Irish, Swedish | (Huizing et al., 2004) |
| 24 | c.2750_2751delAG | p.Glu917Valfs*14 | Exon19 | Indel | French | (Lasseaux et al., 2018; Michaud et al., 2017) 4 |
| 25 | c.2928_2929dupGA | p.Thr977Argfs*15 | Exon 20 | Indel | English, Irish | (Huizing et al., 2004) |
| 26 | c.2974_2977delCTCT | p.Leu992Valfs*17 | Exon 21 | Indel | - | (Lasseaux et al., 2018) 4 |
| 27 | c.2979_2982delTTTG | p.Cys993Trpfs*16 | Exon 21 | Indel | French | (Michaud et al., 2017) 4 |
| 28 | c.3058+3A>G | IVS21+3A>G | Intron 21 | Splice site | Turkish | (Lasseaux et al., 2018; Michaud et al., 2017) 4 |
| 29 | c.3096_3098delCCT | p.Leu1033del | Exon 22 | Indel | French | (Michaud et al., 2017) 4 |
| - 8 | c.3293C>T | p.Thr1098Ile 9 | Exon 22 | Missense | Swiss | (Huizing et al., 2004) 9 |
| 30 | c.3346A>G | p.Met1116Val | Exon 23 | Missense | Chinese | (Wei et al., 2016) 4,10 |
| 31 | del 1.5-kb 8 | - | - | Indel | - | (Lasseaux et al., 2018) 4 |
When deletion/insertion is 1 nucleotide it is named Frame shift, when larger it is named Indel.
Extracted from literature reference. ‘-‘ = unreported.
This (silent) HPS5 variant c.219G>A; p.Arg73Arg occurs at the exon3/intron 3 splice site junction and is predicted to affect splicing (Supplemental Table S2).
At least one of the reported cases with this variant was identified by next generation sequencing.
Gray highlight: missense variant. Gray highlight: missense variant. See Supplemental Table S1 for pathogenicity description.
This variant was previously described as c.1081delC (Ringeisen et al., 2013).
Described as Chr11:18327845 to Chr11:18329253 (Michaud et al., 2017).
The nomenclature of these HPS6 variants are included in this Table as reported, see reference for each specific variant for more details.
Two siblings were homozygous for 2 missense HPS5 variants (hemizygosity was excluded): p.Leu624Arg and p.Thr1098Ile (Huizing et al., 2004). The high MAF and low pathogenicity prediction (Supplemental Table S1) classifies p.Thr1098Ile as a benign SNP. Variant p.Leu624Arg classifies as a likely pathogenic missense and is also identified in other HPS-5 individuals in trans with a pathogenic variant (Lasseaux et al., 2018; Michaud et al., 2017).
These variants were originally reported as c.1892T>C and c.3004A>G, using NM_007216 (HPS5 mRNA Variant 2) nomenclature. For this report, the nomenclature of these variants was converted to NM_181507.1 (HPS5 mRNA Variant 1).
HPS-5 (MIM# 614074) is caused by biallelic pathogenic variants in HPS5, also called BLOC2S2, located on chromosome 11p15.1. The longest HPS5 mRNA transcript contains 23 exons, encoding a 1129-amino acid protein (~127.4-kD). There are 3 HPS5 protein-coding mRNA variants described, each with alternatively spliced 5’ exons (Huizing et al., 2004). The HPS5 protein interacts with HPS3 and HPS6 proteins in BLOC-2 (Di Pietro et al., 2004). Vertebrate models of HPS-5 include the ruby-eye-2 mouse (Zhang et al., 2003), the snow white zebrafish (Daly, Willer, Gregg, & Gross, 2013), and the casper stickleback (Hart & Miller, 2017) (Table 1).
The HPS5 gene was discovered after Hps5 deficiency was identified in the ruby-eye-2 HPS murine model; subsequent sequencing of the human orthologue in unclassified HPS individuals identified one subject with a homozygous 4 base-pair deletion in HPS5 (Zhang et al., 2003). There are now ~29 HPS-5 subjects described worldwide of variable ethnic origins, including Arabic, Chinese, European, Mexican, South-American, and Turkish.
We report 31 HPS5 pathogenic variants (Table 7), including 6 (19%) frameshift, 6 (19%) missense, 4 (13%) nonsense, 11 (36%) insertions and/or deletions, and 4 (13%) splice site variants (Figure 5). The variants are located throughout the HPS5 gene, without an apparent frequently occurring variant. One HPS5 splice variant (c.1634+1G>A) results in skipping of exon 13 (Carmona-Rivera, et al., 2011), another (c.285–10A>G) activates a cryptic splice site leading to an in-frame insertion of 9-bp and reduced HPS5 protein expression (Stephen et al., 2017), and a third variant (c.3058+3A>G) is predicted to cause a splicing defect (Supp. Table S2). A silent HPS5 variant (c.219G>A, p.Arg73Arg; not listed in dbSNP/gnomAd/ClinVar) occurs at the exon 3/intron 3 splice junction and is predicted to weaken the consensus splice site significantly, causing a splice defect (Supp. Tables S1, S2) (Michaud et al., 2017). An additional 6 HPS5 missense variants are reported, three of which are predicted as likely pathogenic (p.Gly145Glu, p.Arg240Pro, p.Leu624Arg) and three as VUS (p.Leu740Ser, p.Leu745Ser, p.Met1116Val) (Supp. Table S1). Of note, two siblings of Swiss origin were reported homozygous for 2 missense HPS5 variants, p.Leu624Arg (c. c.1871T>G) and p.Thr1098Ile (c.3293C>T), and hemizygosity was excluded (Huizing et al., 2004); no pathogenicity predictions were reported at that time. The p.Thr1098Ile variant is a SNP (rs61884288) with a high MAF (0.02362) and low pathogenicity prediction (Supp.l Tables S1, S2, S3). Therefore, this variant should be considered a benign SNP. In contrast, p.Leu624Arg (rs281865102) has no reported allele frequency and is predicted likely pathogenic (Supp. Tables S1); this variant was recently identified in trans with a pathogenic variant in other HPS-5 individuals (Michaud et al., 2017). Therefore, p.Leu624Arg should be considered a pathogenic variant that likely caused the phenotype in the two reported Swiss siblings (Huizing et al., 2004).
Figure 5:
Distribution of HPS5 Gene Variants
There are 10 nonsynonymous HPS5 variants listed in dbSNP with a high ClinVar MAF (>0.001) and low pathogenicity scores (Supp. Table S3). These variants are likely benign SNPs with a small effect on protein function.
Like other BLOC-2 deficient individuals, HPS-5 subjects exhibit a relatively mild phenotype of hypopigmentation and bleeding diathesis. Pulmonary fibrosis or immunodeficiency has not been reported in HPS-5 subjects, and granulomatous colitis occurs some BLOC-2 deficient subjects. The fact that individuals with BLOC-2 deficiency can escape diagnosis or go undiagnosed for decades was illustrated by new diagnoses of HPS-5 in a 92-year-old man, the oldest reported individual with HPS, who had light skin and hair, nystagmus, decreasing visual acuity with age, and a life-long bleeding history (Ringeisen, Schimmenti, White, Schoonveld, & Summers, 2013), and in a 65 year-old man with oculocutaneous albinism and a mild bleeding diathesis (Botero et al., 2018).
HPS-6 (Table 8)
HPS-6 (MIM# 614075) is caused by biallelic pathogenic variants in HPS6, also called BLOC2S3, on chromosome 10q24.32. HPS6 is a one-exon gene, with one mRNA transcript, encoding a 775-amino acid protein (~83.0-kD). HPS6 protein interacts with the HPS3 and HPS5 proteins in BLOC-2 (Di Pietro et al., 2004). Vertebrate models of HPS-6 include the ruby-eye mouse (Zhang et al., 2003) and the no privacy frog (Nakayama et al., 2016).
The HPS6 gene was discovered by finding Hps6 mutated in the ruby-eye HPS mouse model, and subsequent sequencing of unclassified HPS cases identified one individual with a homozygous 4-bp deletion in HPS6 (Zhang et al., 2003). An extended Israeli Muslim Bedouin family had ~ 20 affected individuals homozygous for c.1065insG in HPS6 was reported (Schreyer-Shafir et al., 2006). Another ~45 subjects with HPS6 variants are reported, and there is one unreported case in our NIH cohort; they are from various ethnic backgrounds, including Arabic, Afghan, Caucasian, Chinese, Japanese and Pakistani (Table 8). The HPS6 variant c.2038C>T (p.Gln680*), is reported in ClinVar/gnomAD/dbSNP (rs1131692333) with a very low MAF (0.00001), occurs in 5 Japanese subjects (including 2 sisters) and appears to be a frequent Japanese variant. We report 45 pathogenic HPS6 variants associated with the HPS-6 phenotype (Table 8), including 2 novel variants identified in our NIH HPS cohort. The variants are located throughout the entire coding exon and include 9 (20%) frameshift, 11 (24.5%) missense, 11 (24.5%) nonsense, 14 (31%) insertions and/or deletions, and no splice site variants (Figure 6). Three of 11 reported HPS6 missense variants are predicted to be likely pathogenic; all others are classified as VUS and require experimental pathogenicity evidence and/or familial genetic testing to increase pathogenicity predictions (Supp. Table S1).
Figure 6:
Distribution of HPS6 Gene Variants
Next generation sequencing of a cohort of BRCA1 and BRCA2-negative subjects with breast cancer identified a heterozygous HPS6 stop-loss variant (c.2326T>C, p.*776Arg) in one subject, who also carried heterozygous protein damaging variants in two other genes (Shahi et al., 2019). This variant has not been reported in HPS subjects but is included in Supp. Table S4 because it may cause HPS when occurring in a homozygous or compound heterozygous state.
The dbSNP/gnomAd/ClinVar databases list 4 frequently occurring (MAF > 0.001) HPS6 missense variants, with predicted benign or unknown pathogenicity (Supp. Table S3); these should be considered when encountered in HPS6 genetic analysis.
Like the other two BLOC-2 HPS disorders (HPS-3 and HPS-5 subtypes), HPS-6 subjects exhibit a mild phenotype of hypopigmentation and bleeding diathesis. Pulmonary fibrosis and immunodeficiency have not been reported in HPS-6 and granulomatous colitis occurs in some BLOC-2 deficient subjects. As in other BLOC-2 subtypes, mildness of symptoms may prevent or delay diagnosis.
HPS-7 (Table 9)
Table 9:
Pathogenic Gene Variants Associated with Hermansky-Pudlak Syndrome Types 7 through 10 (HPS-7 - HPS-10)
| # | mRNA | Amino Acid | Exon/Intron | Variant Type 1 | Reference SNP (dbSNP) 2 | Ethnic Background (age, gender) 3 | References and Footnotes |
|---|---|---|---|---|---|---|---|
| HPS-7: DTNBP1 (BLOC1S8, Dysbindin) | |||||||
| NM_032122.4 | NP_115498.2 | ||||||
| 1 | c.177G>A | p.Trp59* | Exon 4 | Nonsense | rs727502866 | Caucasian (77y, F) | (Lowe et al., 2013) 4 |
| 2 | c.307C>T | p.Gln103* | Exon 5 | Nonsense | rs104893945 |
Portuguese (48y, M), Paraguayan (6y, M) Portuguese (26y, M;56y, F) Portuguese (18y, F) Argentinian (M) |
(Li et al., 2003) (Bryan et al., 2017) 5 (Bastida et al., 2019) 5 (Bastida et al., 2019) 5 unreported 5,6 |
| 3 | c.771_774delCTCT | p.Asn257Lysfs*13 | Exon 9 | Indel | - | - (1 case) | (Lasseaux et al., 2018) 5 |
| 4 | c.1017_1020delAGAG | p.Glu340Profs*44 | Exon 10 | Indel | - | Argentinian (M) | unreported 5,6 |
| HPS-8: BLOC1S3 (Reduced Pigmentation) | |||||||
| NM_212550.4 | NP_997715.1 | ||||||
| 1 | c.131C>A | p.Ser44* | Exon 2 | Nonsense | rs281865115 | Iranian (6y, M) | (Cullinane et al., 2012) |
| 2 | c.385_403del19 | p.Ser129Glnfs*90 | Exon 2 | Indel | - | - (1 case) | (Lasseaux et al., 2018) 5 |
| 3 | c.444_467del24 | p.Gln150_Ala157del | Exon 2 | Indel | rs754841982 | - (1 case) | (Lasseaux et al., 2018) 5 |
| 4 | c.448delC | p.Gly150Argfs*75 | Exon 2 | Frameshift | rs281865116 | Pakistani (6 familial cases) | (Morgan et al., 2006) 7 |
| HPS-9: BLOC1S6 (PLDN) | |||||||
| NM_012388.3 8 | NP_036520.1 | ||||||
| 1 | c.232C>T | p.Gln78* | Exon 2 | Nonsense | rs201348482 |
Italian (17y, F), Pakistani (4y, F) Indian (9mo, M) |
(Badolato et al., 2012) 5,9 (Yousaf et al., 2016) 5 (Cullinane et al, NIH unpublished) |
| 2 | c.285_286dupTC | p.His96Leufs*22 | Exon 3 | Indel | - | Japanese (52y, F) | (Okamura et al., 2018) 5,9 |
| HPS-10: AP3D1 | |||||||
| NM_001261826.3 | NP_001248755.1 | ||||||
| 1 | c.1978delG | p.Ala660Argfs*54 | Exon 17 | Frameshift | - | 3 siblings | (Mohammed et al., 2018) 5,10 |
| 2 | c.3565_3566delGT | p.Val1189Leufs*8 | Exon 32 | Indel | rs879255646 | Turkish (3.5y, M) | (Ammann et al., 2017; Ammann et al., 2016) 5,11 |
When deletion/insertion is 1 nucleotide it is named Frameshift, when larger it is named Indel.
Reference SNP numbers for each variant as listed in dbSNP (https://www.ncbi.nlm.nih.gov/snp; searched November 2019), none of the listed SNPs in this Table has a reported allele frequency, suggesting they are rare variants.
Extracted from literature reference. Age (y, years at reporting) and gender (M, male; F, female) of each subject is included for future comparison of additional cases. ‘-‘ = unreported.
This subject also developed granulomatous colitis (Lowe et al., 2013).
The variant in this report was identified by next generation sequencing.
This DTNBP1 variant was identified by exome sequencing in one unreported male subject of Argentinian descent in the NIH HPS cohort referred by Dr. Rosenzweig, NIH Clinical Center, NIH, Bethesda, MD). This subject is compound heterozygous for c.307C>T and c.1017_1020delAGAG.
One extended Pakistani family with 6 affected HPS-8 cases homozygous for c.448delC was reported (Morgan et al., 2006).
Both BLOC1S6 pathogenic variants were reported according to transcript variant 2 (NM_012388.3) nomenclature and are listed as such in this Table to avoid confusion. A longer transcript variant (Variant 1, NM_001311255.1), appeared recently in databases, and future reports may adjust variant nomenclature.
Both the Italian and Japanese subjects had a history of recurrent leucopenia and mild thrombocytopenia, causing immunodeficiency (Badolato et al., 2012; Okamura et al., 2018). The Japanese subject developed schizophrenia in her late forties, a phenotype also associated with DTNBP1 haplotypes.
Next generation sequencing identified this rare AP3D1 frameshift variant c.1978delG homozygous in 3 siblings with seizures, developmental delay, albinism and immunodeficiency. Twin girls died before 6 days of age and their brother died at age 2 years of pneumonia and sepsis (Mohammed et al., 2018).
Next generation sequencing identified this rare AP3D1 variant c.3565_3566delGT in a proband with albinism, neutropenia, immunodeficiency, neurodevelopmental delay, generalized seizures, and impaired hearing. Immunologic investigations excluded HLH in this subject. The proband died at age 3.5 years as result of septic pneumonia (Ammann et al., 2016).
HPS-7 (MIM# 614076) is caused by biallelic pathogenic variants in DTNBP1, also called BLOC1S8 or HPS7, located on chromosome 6p22.3. The longest DTNBP1 mRNA transcript contains 10 exons, encoding a 351-amino acid protein (~39.5-kD) called Dysbindin or HPS7. There are 5 DTNBP1 protein-coding mRNA variants predicted; their expression and functional significance are unknown. Dysbindin is a subunit of BLOC-1 (Li et al., 2003). The sandy mouse is the murine orthologue of human HPS-7 (Li et al., 2003).The DTNBP1 gene was discovered by finding Dtnbp1 mutated in the sandy mouse, and subsequent sequencing of unclassified HPS subjects identified a 48-year old Portuguese female with a homozygous nonsense variant c.307C>T (p.Gln103*) in DTNBP1 (Li et al., 2003). We list 4 DTNBP1 pathogenic variants that cause the HPS-7 phenotype (Figure 7). There are currently seven HPS-7 cases reported, all homozygous for a nonsense or a frameshift DTNBP1 variant, including a Caucasian female diagnosed at age 77 (Lowe et al., 2013) and 4 cases (including 2 siblings) of Portuguese ethnicity homozygous for c.307C>T (p.Gln103*) (Bastida et al., 2019; Bryan et al., 2017; Li et al., 2003) (Table 9). One unreported Argentinian boy in our NIH cohort (referred by Dr. Rosenzweig, NIH Clinical Center, Bethesda, MD), was compound heterozygous for c.307C>T (p.Gln103*) and the novel indel variant c.1017_1020delAGAG (p.Glu340Profs*44).
Next generation has identified heterozygous DTNBP1 variants suggested to contribute to different conditions (Supp. Table S4). Specifically, c.286G>T (p.Glu96*) was reported in a subject with idiopathic pulmonary fibrosis that had no other HPS clinical findings (Deng et al., 2018). The missense variant p.Pro272Ser (c.814C>T) was reported as a low penetrance risk for colorectal cancer (Webb et al., 2006). Several association studies have identified DTNBP1 as a risk allele for schizophrenia, including in European-Americans (Donohoe et al., 2008; Straub et al., 2002; Wang, Xu, Lazarovici, & Zheng, 2017; Zuo et al., 2009). Schizophrenic subjects have reduced hippocampus DTNBP1 mRNA expression (Weickert, Straub, Kleinman, Hyde, & Rothmond, 2006) and DTNBP1 variants are associated with a cognitive response to antipsychotic drug treatment (Scheggia et al., 2018), however, the association of DTNBP1 with schizophrenia has also been challenged (Ghiana & Dell’Angelica, 2011). Notably, no psychiatric illness in HPS-7 subjects has been reported but it is reasonable to consider such features in HPS-7 and BLOC-1 cases.
The dbSNP/gnomAd/ClinVar databases list 4 frequently occurring (MAF > 0.001) DTNBP1 missense variants of predicted benign or unknown pathogenicity (Supp. Table S3), which should be considered as likely non-pathogenic polymorphisms when encountered in DTNBP1 genetic analyses.
Due to the limited number of identified HPS-7 cases, it is difficult to determine whether these individuals are prone to complications other than albinism and a bleeding diathesis. All affected individuals, including two of advanced age, had normal pulmonary function and no signs of immunodeficiency/neutropenia. The 77-year old Caucasian woman had signs of colitis, which was not reported in the other affected subjects.
HPS-8 (Table 9)
HPS-8 (MIM# 614077) is caused by biallelic pathogenic variants in BLOC1S3, also called HPS8, RP or BLOS3, located on chromosome 19q13.32. The longest BLOC1S3 mRNA transcript contains 2 exons, encoding a 202-amino acid protein (~21.3-kD). There exists only one predicted BLOC1S3 mRNA transcript. BLOC1S3/HPS8 is a subunit of BLOC-1 (Starcevic & Dell’Angelica, 2004). Reduced pigmentation is the murine orthologue of human HPS-8 (Gwynn et al., 2004).
HPS-8 was first reported in an extended consanguineous Pakistani family with 6 affected individuals; autozygosity mapping assisted in identifying a homozygous frameshift variant, c.448delC (p.Gly150Argfs*75), in BLOC1S3 in all affected individuals (Morgan et al., 2006). Three additional HPS-8 cases with homozygous pathogenic BLOC1S3 variants have since been reported (Cullinane et al., 2012; Lasseaux et al., 2018) (Table 9, Figure 7).
The dbSNP/gnomAd/ClinVar databases list 3 frequently occurring (MAF > 0.001) BLOC1S3 missense variants, with predicted benign or unknown pathogenicity (Supp. Table S3); these should be considered as likely non-pathogenic polymorphisms when encountered in genetic analyses.
The few reported HPS-8 cases show typical HPS features including hypopigmentation and a bleeding diathesis. None has been reported to exhibit pulmonary fibrosis, granulomatous colitis, immunodeficiency or other complications. Identification of additional HPS-8 cases may confirm or broaden this phenotype.
HPS-9 (Table 9)
HPS-9 (MIM# 614171) is caused by biallelic pathogenic variants in BLOC1S6, also called HPS9, PLDN or BLOS6, located on chromosome 15q21.1. The longest BLOC1S6 mRNA transcript contains 5 exons encoding a 177-amino acid protein (~20.3-kD), called BLOC1S6, HPS9 or Pallidin. There are 3 predicted BLOC1S6 mRNA transcripts with unknown expression patterns and function. The longest variant 1 (NM_001311255.1) only recently appeared in databases, causing all previously described pathogenic variants to be attributed to mRNA splice variant 2 (NM_012388.3). Variant 1 (open reading frame 534-bp) and Variant 2 (open reading frame 519-bp) both have 5 exons and vary in their 5’ UTR and 5’ coding region in exon 1, and each initiate translation at a different start codon; exons 2–5 are identical in both variants. BLOC1S6/HPS9 is a subunit of BLOC-1 (Falcon-Perez et al., 2002; Moriyama & Bonifacino, 2002). Pallid is the murine orthologue of human HPS-9 (Moriyama & Bonifacino, 2002).
All 3 reported HPS-9 cases were identified through exome sequencing. A 17-year-old Italian female (Badolato et al., 2012) and a 4-year-old Pakistani female (Yousaf et al., 2016) were both homozygous for the same HPS9 nonsense variant: c.232C>T, p.Gln78* (NM_012388.3, transcript variant 2). A 52-year-old Japanese female was reported homozygous for c.285_286dupTC, p.H96Lfs*22 (NM_012388.3) (Okamura et al., 2018) (Table 9, Figure 7).
The dbSNP/gnomAd/ClinVar databases list one frequently occurring (MAF > 0.001) BLOC1S6 missense variant (p.Ala12Thr; MAF 0.0030), with predicted benign or unknown pathogenicity (Supp. Table S3); it should be considered as likely non-pathogenic polymorphism when encountered in genetic analyses.
All three HPS-9 subjects exhibited hypopigmentation, visual impairment and a bleeding diathesis. No pulmonary fibrosis or granulomatous colitis was reported in HPS-9 subjects. However, the Italian (17-year-old female) and Japanese (52-year-old female) subjects both had mild thrombocytopenia and recurrent leukopenia, causing immunodeficiency (Badolato et al., 2012; Okamura et al., 2018). In addition, the Japanese subject developed schizophrenia in her late forties, a phenotype previously associated with DTNBP1 haplotypes (Donohoe et al., 2008; Straub et al., 2002; Wang et al., 2017), however, this association appears controversial and needs further consideration(Ghiani & Dell’Angelica, 2011). Both immunodeficiency and schizophrenia should be considered in future evaluations of HPS-9 subjects.
HPS-10 (Table 9)
HPS-10 (MIM# 617050) is caused by biallelic pathogenic variants in AP3D1, also called HPS10 or ADTD, located on chromosome 19p13.3. The longest AP3D1 mRNA transcript contains 32 exons, encoding the 1215-amino acid protein AP3D1 (δ subunit of AP-3; ~136.7-kD). There are 2 predicted protein-coding AP3D1 mRNA transcripts without reported expression or functional data. The AP3D1 protein is a subunit of AP-3 (Ammann et al., 2016; Dell’Angelica et al., 1999). Ap3d1-deficient animal models include the mocha mouse (Kantheti et al., 1998).
There are only 2 AP3D1 pathogenic variants reported to cause the HPS-10 phenotype (Table 9, Figure 7). A homozygous AP3D1 pathogenic indel variant c.3565_3566delGT (p.Val1189Leufs*8) was reported in a Turkish boy, who died at age 3.5 years as result of septic pneumonia (Ammann et al., 2016). The boy had albinism, neutropenia, immunodeficiency, neurodevelopmental delay, generalized seizures, interstitial lung disease and impaired hearing. T cells from the AP3D1-deficient boy showed significantly decreased AP3D1 protein expression compared to healthy control T cells; protein expression levels of other AP-3 subunits (β3A, σ, and μ) were also reduced, consistent with an unstable AP-3 heterotetramer (Ammann et al., 2016). Cytotoxic lymphocytes from the subject exhibited an impaired degranulation response, similar to individuals with pathogenic variants in AP3B1. Immunologic investigations excluded HLH in the subject (Ammann et al., 2016; Enders et al., 2006; Jessen et al., 2013). A homozygous AP3D1 pathogenic frameshift variant, c.1978delG (p.Ala660Argfs*54), was identified in 3 siblings with seizures, developmental delay, albinism and immunodeficiency; twin girls died before 6 days of age and their brother died at age 2 years of pneumonia and sepsis (Mohammed et al., 2018).
Next generation sequencing of cohorts with autism spectrum disorder identified a de novo heterozygous AP3D1 splicing variant (c.273+1G>T) (Takata et al., 2018) and a de novo AP3D1 heterozygous missense variant (p.Gln406Arg) (Iossifov et al., 2014). Next generation sequencing of a schizophrenia cohort identified a de novo heterozygous AP3D1 missense variant (p.Asn605Lys) (Fromer et al., 2014). None of these variants has been reported in HPS subjects but they are included in Supp. Table S4 because they may cause HPS when occurring in a homozygous or compound heterozygous state.
The dbSNP/gnomAd/ClinVar databases list 2 frequently occurring (MAF > 0.001) AP3D1 missense variants, with predicted benign or unknown pathogenicity (Supp. Table S3); these should be considered as likely non-pathogenic polymorphisms when encountered in genetic analyses.
The AP3D1-deficient boys manifested features of albinism and immunodeficiency from birth. These are features characteristic of AP3B1 deficiency (HPS-2) and can likely be attributed to AP-3 deficiency. They did not have an overt tendency for bleeding. They also exhibited neurological findings not previously reported in other HPS subtypes, including microcephaly, severe neurodevelopmental delay, generalized seizures. Hearing impairment occurred in the Turkish boy. The mocha mouse shows a similar HPS-like phenotype with seizures and hearing loss, indicating that these features are likely due to AP3D1 deficiency. A diagnosis of HPS-10 should be considered in individuals with hypopigmentation, immunodeficiency, seizures, hearing loss and possibly other neurologic involvement.
DIAGNOSIS OF HPS
Individuals with HPS can present to different clinical specialties, including dermatology, ophthalmology, pulmonology, hematology, gastroenterology, immunology and neurology. The presence of a combination of hypopigmentation (light hair and skin color), ocular symptoms (nystagmus and decreased visual acuity) (Summers, Knobloch, Witkop, & King, 1988), and a bleeding diathesis (bruising, epistaxis, gingival bleeding, colonic bleeding, prolonged bleeding after minor surgeries) (Gunay-Aygun, Huizing, & Gahl, 2004) leads most physicians to suspect a diagnosis of HPS. The diagnosis of HPS is primarily established by clinical features and platelet phenotyping that show an absence or severe reduction of platelet dense granules, which can be demonstrated by whole mount electron microscopy (EM) (Witkop, Krumwiede, Sedano, & White, 1987). This semi-specialized method is not routinely offered by hematology services. Alternatively, demonstrating an absence of a secondary aggregation response of platelets to exogenous stimuli through platelet aggregation testing also supports the HPS diagnosis (White & Witkop, 1972). Quantification of mepacrine uptake (Billio et al., 2001), and super-resolution immunofluorescence microscopy analyses (Westmoreland et al., 2016) have also been presented as potential alternatives to whole mount EM analysis for diagnosis of dense granule deficiency.
Identification of biallelic variants in one of the 10 HPS-related genes ultimately confirms the HPS diagnosis and the HPS subtype. However, since HPS is a heterogenous genetic disorder, a molecular diagnosis of a particular subtype can be difficult to reach. There is no direct genotype-phenotype association among HPS genes or variants within a gene. When using a single gene testing strategy, we recommend sequence analysis of HPS1 and HPS4 in subjects with more severe clinical manifestations (hypopigmentation, ocular symptoms, bleeding diathesis, pulmonary symptoms) and analysis of HPS3, HPS5 and HPS6 (BLOC-2 subunits), followed by DTNBP1, BLOC1S3 and BLOC1S6 (BLOC-1 subunits) in mildly affected subjects. Subjects with immunological symptoms should be tested for AP3B1 and AP3D1 defects. In this era of next generation sequencing, the genetic diagnosis of HPS is increasingly established by testing all HPS genes simultaneously. This approach can also identify new HPS associated genes, since some subjects with HPS-related symptoms have no apparent pathologic variants in any of the 10 HPS genes. Candidate genes for new HPS subtypes may include genes affected in mouse models of HPS that do not yet have a human counterpart (Table 1), as well as proteins that interact with the BLOC and AP-3 complexes.
The inclusion of HPS-related genes in genetic screening panels of cohorts with clinical features of HPS has resulted in the recent identification of groups of undiagnosed HPS subjects, especially those with milder clinical phenotypes. Targeted sequencing of 990 cases with albinism identified 46 HPS subjects (Lasseaux et al., 2018). A similar study of 21 Arabian individuals with ocular hypopigmentation identified 10 HPS subjects (Khan, Tamimi, Lenzner, & Bolz, 2016), a study of 46 Japanese cases with (OCA-1 and HPS-1 negative) albinism identified 9 HPS subjects (Okamura et al., 2019), and a study of Chinese hypopigmentation cases identified 10 HPS subjects (Wei et al., 2019). Similarly, targeted sequencing of a cohort of 159 cases with bleeding, thrombotic, and platelet disorders identified 6 HPS individuals (Simeoni et al., 2016). These studies suggest that HPS is underdiagnosed, especially when clinical features are mild.
Other recent next generation sequencing approaches identified heterozygous variants in HPS genes that are considered risk alleles for certain conditions (Tables 3–9) these are not classified as HPS-causing pathogenic variants. Heterozygous HPS1, HPS4 and DTNBP1 variants were reported in familial (Stearman et al., 2019) or sporadic (Deng et al., 2018) pulmonary fibrosis cases (Tables 3, 6 and 9). Heterozygous AP3B1 variants have been reported in cases with primary immunodeficiency (Chi et al., 2018; Gallo et al., 2016) or HLH (Gao et al., 2015; Miao et al., 2019; Mukda et al., 2017; Tesi et al., 2015; Xu et al., 2017) (Table 4), and heterozygous variants in AP3D1, HPS3 and HPS4 were reported in cases with autism spectrum disorder or schizophrenia (Fromer et al., 2014; Iossifov et al., 2014; Takata et al., 2018).
Apart from direct sequencing of the exonic regions of HPS genes, alternative molecular methods to establish the HPS type have been used. When a subject’s mRNA has been isolated from whole blood or cultured cells, mRNA expression (i.e., by northern blot or quantitative PCR) and/or cDNA sequencing of each gene can be performed. Another advantage of mRNA availability is that the effects of splicing variants and variants suspected of causing nonsense mediated mRNA decay can be investigated (Anderson et al., 2003; Huizing et al., 2001; Huizing et al., 2004). Immunoblotting of cultured skin fibroblast or platelet rich plasma extracts has also proven helpful to determine or validate the HPS subtype. HPS mouse and human studies have shown that a defect in one HPS protein leads to destabilization of the entire protein complex, i.e., AP-3, BLOC-1, −2, −3 (Ammann et al., 2016; Dell’Angelica et al., 1999; Huizing et al., 2002; Li et al., 2003; Wei et al., 2019). Therefore, the use of immunoblotting with an antibody against one subunit of the complex (AP-3, BLOC-1,−2,−3) allows determination of which complex is defective in unclassified HPS subjects, reducing subsequent sequencing of genes encoding the corresponding subunits (Carmona-Rivera, et al., 2011; Nazarian et al., 2008; Wei et al., 2019).
EPIDEMIOLOGY OF HPS
The worldwide prevalence of HPS is estimated to be 1–9/1,000,000 (Christensen et al., 2017; Huizing et al, 2017 Oct 26 [Updated 2000 July 24]). A few ethnic founder variants occur in HPS genes, in particular in northwest region of Puerto Rico, an estimated 400 individuals are affected (~1/1,800 affected; carrier frequency 1:21) and carry a homozygous 16-bp duplication in HPS1 (c.1472_1487dup16-bp; Table 3) (Santiago Borrero et al., 2006; Witkop et al., 1990), and in Central Puerto Rico a 3.9-kb deletion in HPS3 (NM_032383.5(HPS3):c.−2993_217+692del; Table 5) has an estimated population prevalence of ~1/4000 (carrier frequency 1:32) in (Anikster et al., 2001; Santiago Borrero et al., 2006; Torres-Serrant et al., 2010). Other founder variants without frequency estimates have been reported and are discussed elsewhere in this report for each HPS subtype (Oh et al., 1998; Schallreuter et al., 1993; Ito et al., 2005; Schreyer-Shafir et al., 2006).
Compared to the ~189 reported BLOC-3 deficient individuals (not including ~261 cases with the Puerto Rican HPS1 16-bp duplication variant), there are remarkably fewer reported AP-3 deficient individuals (~35 cases), BLOC-1 deficient individuals (~24 cases) and BLOC-2 deficient individuals (~117 cases; not including ~72 cases with the Puerto Rican HPS3 3.9-kb deletion and ~20 cases with the Israeli Bedouin HPS6 c.1065insG variants) (Table 1).
BLOC-2 deficient cases may escape diagnosis because of their much milder hypopigmentation compared to BLOC-3 deficiency. This is in line with BLOC-2 deficient mice, which inhibit brownish-black eumelanin, but not reddish-yellow pheomelanin production (Hirobe, Ito, & Wakamatsu, 2013), and individuals with HPS have reduced levels of total melanin, but increased pheomelanin production compared to unaffected family members (Okamura et al., 2018), indicating that hypopigmentation might not be detected in HPS cases from fair-skinned families, particularly those with red-blonde hair.
BLOC-2 deficient individuals may get medical attention only for a bleeding diathesis and may be classified as storage pool deficiency, as illustrated by new diagnoses of HPS-5 in 92- and 65-year-old individuals with histories of excessive bleeding (Botero et al., 2018; Ringeisen et al., 2013).
Similarly, AP-3 deficient individuals may avoid an HPS diagnosis or have it delayed because a severe immune disorder dominates medical attention. It is puzzling why so few BLOC-1 deficient individuals are reported. Perhaps BLOC-1 defects are extremely rare or embryonically lethal (although mouse models of BLOC-1 deficiency are viable), or the human phenotypes may have features that are not considered compatible with the HPS phenotype. For example, the absence of platelet delta granules is currently essential for the HPS diagnosis, but a subset of subjects with BLOC-1 may have normal/decreased platelet delta granules and may therefore not be considered for a HPS diagnosis. In addition, reported brain-associated functions of BLOC-1 (A. Ito et al., 2018; Newell-Litwa et al., 2010; Spiegel, Chiu, James, Jentsch, & Karlsgodt, 2015) may underlay a neurologic phenotype of BLOC-1 deficiency. Recognition of these features may facilitate the diagnosis of affected individuals with HPS subtypes associated with BLOC-1 defects. Several association studies have identified DTNBP1 variants as risk alleles for schizophrenia (Donohoe et al., 2008; Straub et al., 2002; Wang et al., 2017; Zuo et al., 2009) and one HPS-9 subject was diagnosed with oculocutaneous albinism and developed schizophrenia in her forties; she was only diagnosed with HPS-9 at age 52 through a whole exome screen (Okamura et al., 2018). However, some reports have challenged brain-associated functions of BLOC-1 and/or a BLOC-1 association with schizophrenia (Ghiani & Dell’Angelica, 2011), suggesting that this issue needs further consideration. With the increased availability of exome/genome sequencing, more HPS subjects with mild or atypical HPS phenotypes will likely be identified and will expand the clinical spectrum of HPS.
CLINICAL MANAGEMENT AND THERAPEUTIC ASPECTS
The myriad of symptoms associated with HPS, some subtype or age specific and some life threatening, require multidisciplinary clinical care (Table 2) (Christensen et al., 2017; Seward & Gahl, 2013).
All individuals with HPS exhibit some degree of albinism, involving hypopigmentation of skin, hair or eyes. The skin is often light- and sun-sensitive and may develop solar keratoses and melanocytic nevi. Subjects are at increased risk for squamous cell carcinoma, basal cell carcinoma, and possibly melanoma. Protection or avoidance from ultraviolet radiation is critical (Toro, Turner, & Gahl, 1999). The hair varies from silvery-white to light brown and hypopigmentation may be evident when affected and unaffected family members are compared. The eyes appear light blue, light green or hazel, but iris color may be darker in mild HPS cases. Iris transillumination is found in most HPS subjects. Due to their albinism, HPS subjects have abnormal crossing of optic nerve fibers (Hoffmann, Lorenz, Morland, & Schmidtborn, 2005) and horizontal nystagmus, the retinal fundus appears pale and visual acuity ranges from 20/60 to 20/400 and can be mildly improved with refractive lenses (Summers et al., 1988). Many subjects are legally blind, i.e., have visual acuity worse than 20/200. BLOC-3 deficient subjects show more severe hypopigmentation than subjects with AP-3 or BLOC-2 defects. The albinism of BLOC-1 individuals has not been well characterized.
The bleeding diathesis of HPS varies in severity in all subtypes, and may include spontaneous bruising, prolonged epistaxis, menorrhagia, pronounced oozing after dental extractions, and excessive surgical blood loss. Topical thrombin, administration of pro-coagulant drugs, or intravenous 1-desamino-8D-arginine vasopressin may ameliorate or prevent bleeding. Platelet transfusion may also be used as prophylaxis or as treatment for bleeding in individuals with HPS (Han et al., 2018; Minkin, Bertetti, Lindsey, & Bovino, 2015; Ozgur & Yilmaz, 2015; Van Avermaete, Muys, & Jacquemyn, 2016). Avoidance of aspirin products and non-steroidal anti-inflammatory drugs is recommended.
A granulomatous colitis involving intestinal granulomas, erosions and inflammatory cells, which resembles Crohn’s disease, occurs in ~10–20% of all BLOC-2 and BLOC-3 subjects (Hussain et al., 2006). One BLOC-1 deficient individual (HPS-7) was diagnosed with Crohn’s colitis in adulthood (Lowe et al., 2013). It is unknown if colitis occurs in AP-3 deficiency. The colitis may respond to corticosteroids or anti-TNF-α drugs; surgical bowel resection is performed in refractory cases (Demirtas, Alahdab, Kani, Atug, & Imeryuz, 2019; Kouklakis et al., 2007; Mora & Wolfsohn, 2011). Abnormal endosomal membrane formation was suggested as an underlying cause for HPS colitis leading to ceroid lipofuscin formation, abnormal autophagy and phagocytosis, and inflammation (Felipez, Gokhale, & Guandalini, 2010; Sofia, Sakuraba, & Rubin, 2017). It was suggested that the presence of risk alleles in Crohn’s disease-associated genes, like NOD2 or ATG16L1, in HPS subjects may contribute to developing colitis (Lozynska et al., 2018). Low vitamin D levels, which may be a factor in HPS subjects avoiding sun exposure, could also contribute to developing colitis (Lozynska et al., 2018).
Ceroid lipofuscin, an amorphous, granular, electron-dense, autofluorescent lipid-protein material, was identified in LROs in HPS cell types, including alveolar macrophages, cells of the gastrointestinal tract, renal tubular cells, bone marrow, lymph nodes, liver, spleen, and heart (Gahl et al., 1998; Harada et al., 2014; Hermansky & Pudlak, 1959; Sparrow et al., 2010; Takahashi & Yokoyama, 1984). Ceroid lipofuscin may accumulate because cells cannot rapidly degrade mistargeted vesicular membranes. Accumulation of ceroid lipofuscin was suggested to underlie the colitis and pulmonary fibrosis in HPS, but this has not been confirmed and other pathogenic mechanisms have been proposed. End stage renal disease attributed to deposition of ceroid lipofuscin has occurred in a few HPS subjects, for which renal transplant is a treatment option (Abdullah, Davis, Quinn, & Mohan, 2018; Gordillo, Del Rio, Thomas, Flynn, & Woroniecki, 2011; Tagboto et al., 2001).
Immunodeficiency and/or neutropenia occurs in AP-3-deficient HPS, resulting in susceptibility to infections (subtypes HPS-2 and HPS-10) (Ammann et al., 2016; de Boer et al., 2017; Fontana et al., 2006; Huizing et al., 2002; Mohammed et al., 2018). No BLOC-2 or BLOC-3 deficient subjects are described with immunodeficiency, but it was reported in two unrelated individuals with HPS-9 (Badolato et al., 2012; Okamura et al., 2018) and therefore needs consideration in future BLOC-1 deficient individuals.
Manifestations of the AP-3 immunodeficiency can vary from mild recurrent viral and bacterial infections to severe hemophagocytic lymphohistiocytosis (HLH) (Dell’Acqua et al., 2019; Enders et al., 2006; Jessen et al., 2013). The neutropenia associated with AP3B1 (HPS-2) deficiency is granulocyte colony-stimulating factor (G-CSF) responsive, but G-CSF therapy has not been used in AP3D1-deficienct subjects, all of whom died before age 3.5 of pneumonia and/or sepsis without signs of HLH. Of note, G-CSF therapy restores the neutrophil numbers, but not the recurrent infections in HPS-2 subjects (Fontana et al., 2006), suggesting defects in innate immunity and bacterial antigen presentation, supported by several studies in AP3-deficient dendritic, natural killer and lymphoblastoid cells from mice and humans (Briken, Jackman, Dasgupta, Hoenig, & Porcelli, 2002; Fontana et al., 2006; Meantegazza et al., 2012; Mantegazza et al., 2017; Sugita et al., 2002; Sasai et al., 2010).
A few AP3B1-deficient subjects developed HLH, which was lethal in 2 subjects (Dell’Acqua et al., 2019; Enders et al., 2006; Fontana et al., 2006). Although the risk of HLH should be considered in AP-3 deficient subjects, preemptive hematopoietic stem cell transplantation (HSCT, therapeutic for HLH but challenging for the subject) has been deemed to not be justified. HSCT could certainly be considered after a severe HLH episode (Dell’Acqua et al., 2019).
Pulmonary fibrosis, a progressive interstitial lung disease with a variable time course, occurs in BLOC-3 (HPS-1 and HPS-4) and AP-3 (HPS-2 and HPS-10) subjects. There are no reports of pulmonary fibrosis in BLOC-1 and BLOC-2 cases. Most BLOC-3 deficient subjects develop pulmonary fibrosis in middle age (30–50 years) and progress to death within a decade (Brantly et al., 2000; Gahl et al., 1998). AP-3 deficiency-related pulmonary fibrosis is reported in a few HPS-2 cases and one HPS-10 case, and symptoms can start as early as childhood (Ammann et al., 2016; Gochuico et al., 2012; Hengst et al., 2018). While the natural history of BLOC-3-related pulmonary fibrosis has been well-documented, that of AP-3 related lung disease needs further elucidation with longitudinal data from more subjects (Gochuico et al., 2012). The exact cause of lung disease in HPS remains unknown; it was suggested that altered LRO formation within alveolar epithelial type II cells may lead to defective formation of lamellar bodies and/or intracellular processing of surfactant proteins, leading to endoplasmic reticulum-stress, apoptosis, and a fibrotic lung phenotype (Guttentag et al., 2005; Mahavadi et al., 2010; Kook et al., 2018). The fibrotic lung phenotype in BLOC-3- or AP-3-deficient mice was shown to be due to a non-hematopoietic cell type and could be averted in Ap3b1-deficient mice by re-expression of Ab3b1 specifically in lung epithelial type 2 cells (Young et al., 2012), strongly supporting the lamellar body defect as causative for lung disease, at least in mice. Abnormal alveolar macrophage or mast cell function was also suggested to underlie HPS-related pulmonary fibrosis (Kirshenbaum et al., 2016; Mahavadi et al., 2010; Nakatani et al., 2000; Rouhani et al., 2009).
No approved medical therapy for or prophylaxis against HPS-related pulmonary fibrosis exists. Maximizing pulmonary function before onset of pulmonary fibrosis by avoidance of cigarette smoke and other lung toxins, treatment of pulmonary infections, influenza and pneumococcal immunization, and regular moderate exercise are recommended. Steroids have no apparent beneficial effect. The anti-fibrotic drug pirfenidone may slow the progression of HPS-related pulmonary fibrosis in some cases (Gahl et al., 2002; O’Brien et al., 2011; O’Brien, Introne, et al., 2018), but it is not an approved therapy for HPS-related pulmonary fibrosis (O’Brien, Gahl, & Gochuico, 2018). Lung transplant is the only known treatment for pulmonary fibrosis, and several individuals with HPS1-related pulmonary fibrosis successfully underwent bilateral or single-lung transplantation (El-Chemaly et al., 2018; Gahl et al., 2002; Lederer et al., 2005).
HPS pathogenesis and therapeutic options continue to be investigated, including through the use of organoids (Korogi et al., 2019; Strikoudis et al., 2019) and explorations of gene therapy (Ikawa et al., 2015; Iyer et al., 2019; Shen et al., 2018).
CONCLUSION
HPS is a rare autosomal recessive disorder characterized by genetic and phenotypic heterogeneity. With the recent rapid evolution of affordable next generation sequencing methods, there is an increased recognition of HPS subjects and new HPS genetic subtypes (Arcot Sadagopan et al., 2017; A. Wei et al., 2016; Yousaf et al., 2016). This is also evidenced by the increased diagnosis of subjects with non-classic HPS phenotypes (i.e. HPS-2, HPS-7, HPS-8, HPS-9, HPS-10) (Ammann et al., 2016; Bryan et al., 2017; Cetica et al., 2015; Iwata et al., 2017; Okamura et al., 2018), as well as diagnosis of ‘unexpected’ HPS in cohorts with albinism (Ito et al., 2005; Khan et al., 2016), immunodeficiency (Badolato et al., 2012), ocular disease (Hull et al., 2016; Miyamichi et al., 2016), or platelet disorders (Jones et al., 2012).
These recent developments have created the need for a current overview of molecular diagnostic and genetic counseling aspects of HPS. We intend this report to serve as a reference for interpretation of molecular data for HPS. The extensive HPS mutational spectrum provides pathogenicity interpretation for future HPS-related variants (such as missense variants currently classified as VUS), phenotype-genotype relationships (in particular atypical symptoms such as autism, schizophrenia and immune deficiency), assistance in genetic counseling for affected individuals (surveillance, management of anticipated symptoms), and tools for cell biologists to elucidate pathways, investigate interactions of HPS-related proteins, and initiate therapeutic efforts.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate the expert laboratory work of Melanie Bryan, Carla Ciccone, Nicholas Cardillo, Carmelo Carmona-Rivera, James Curry, Ricardo Linares, Joseph Roney, Karen L. Simon, and Lisa Vincent. We thank Dr. Emily S. Doherty (Carilion Clinic, Roanoke, VA), Dr. David B. Everman (Greenwood Genetics Center, Greenville, SC) and Dr. Sergio D. Rosenzweig (NIH Clinical Center, NIH, Bethesda, MD) for referring HPS subjects, and Gretchen Golas for excellent patient care. And we thank the Hermansky-Pudlak Syndrome Network for unparalleled support to patients and their families. This work was performed in partial fulfillment of the requirements for a PhD degree of H.P., Sackler Faculty of Medicine, Tel Aviv University, Israel.
Funding:
This study was supported by the Intramural Research Program of the National Human Genome Research Institute (NHGRI; Grant Z01 HG000215), National Institutes of Health, Bethesda, Maryland, United States.
Footnotes
Disclosure statement: The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the the Leiden Open Variation Database (http://www.lovd.nl/) and in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). The data that support the findings of this study are also available from the corresponding author upon reasonable request.
REFERENCES
- Abdullah N, Davis NF, Quinn J, & Mohan P (2018). Living donor renal transplant in a patient with end-stage renal disease due to Hermansky-Pudlak syndrome. BMJ Case Rep, pii:bcr-2017–223376. doi: 10.1136/bcr-2017-223376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abouelhoda M, Sobahy T, El-Kalioby M, Patel N, Shamseldin H, Monies D, … Alkuraya FS (2016). Clinical genomics can facilitate countrywide estimation of autosomal recessive disease burden. Genet Med, 18(12), 1244–1249. doi: 10.1038/gim.2016.37 [DOI] [PubMed] [Google Scholar]
- Ammann S, Lehmberg K, Zur Stadt U, Klemann C, Bode SFN, Speckmann C, … HLH study of the GPOH (2017). Effective Immunological Guidance of Genetic Analyses Including Exome Sequencing in Patients Evaluated for Hemophagocytic Lymphohistiocytosis. J Clin Immunol, 37(8), 770–780. doi: 10.1007/s10875-017-0443-1 [DOI] [PubMed] [Google Scholar]
- Ammann S, Schulz A, Krageloh-Mann I, Dieckmann NM, Niethammer K, Fuchs S, … Ehl S (2016). Mutations in AP3D1 associated with immunodeficiency and seizures define a new type of Hermansky-Pudlak syndrome. Blood, 127(8), 997–1006. doi: 10.1182/blood-2015-09-671636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PD, Huizing M, Claassen DA, White J, & Gahl WA (2003). Hermansky-Pudlak syndrome type 4 (HPS-4): clinical and molecular characteristics. Hum Genet, 113(1), 10–17. doi: 10.1007/s00439-003-0933-5 [DOI] [PubMed] [Google Scholar]
- Andres O, Wiegering V, Konig EM, Schneider AL, Semeniak D, Stritt S, … Schulze H (2017). A novel two-nucleotide deletion in HPS6 affects mepacrine uptake and platelet dense granule secretion in a family with Hermansky-Pudlak syndrome. Pediatr Blood Cancer, 64(5), 1–7. doi: 10.1002/pbc.26320 [DOI] [PubMed] [Google Scholar]
- Anikster Y, Huizing M, White J, Shevchenko YO, Fitzpatrick DL, Touchman JW, … Toro JR (2001). Mutation of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat Genet, 28(4), 376–380. doi: 10.1038/ng576 [DOI] [PubMed] [Google Scholar]
- Araki Y, Ishii Y, Abe Y, Yoshizawa J, Okamoto F, Hozumi Y, & Suzuki T (2014). Hermansky-Pudlak syndrome type 4 with a novel mutation. J Dermatol, 41(2), 186–187. doi: 10.1111/1346-8138.12386 [DOI] [PubMed] [Google Scholar]
- Arcot Sadagopan K, Kathirvel R, Keep RB, Sundaresan P, Huang H, Rolfs A, … Vijayalakshmi P (2017). Cutaneous freckling: Possible new clinical marker for the diagnosis of Hermansky-Pudlak syndrome in Indian Asian patients with oculocutaneous albinism. Ophthalmic Genet, 38(2), 194–196. doi: 10.1080/13816810.2016.1183217 [DOI] [PubMed] [Google Scholar]
- Bachli EB, Brack T, Eppler E, Stallmach T, Trueb RM, Huizing M, & Gahl WA (2004). Hermansky-Pudlak syndrome type 4 in a patient from Sri Lanka with pulmonary fibrosis. Am J Med Genet A, 127A(2), 201–207. doi: 10.1002/ajmg.a.20683 [DOI] [PubMed] [Google Scholar]
- Badolato R, & Parolini S (2007). Novel insights from adaptor protein 3 complex deficiency. J Allergy Clin Immunol, 120(4), 735–741; quiz 742–733. doi: 10.1016/j.jaci.2007.08.039 [DOI] [PubMed] [Google Scholar]
- Badolato R, Prandini A, Caracciolo S, Colombo F, Tabellini G, Giacomelli M, … Kingsmore SF (2012). Exome sequencing reveals a pallidin mutation in a Hermansky-Pudlak-like primary immunodeficiency syndrome. Blood, 119(13), 3185–3187. doi: 10.1182/blood-2012-01-404350 [DOI] [PubMed] [Google Scholar]
- Bastida JM, Morais S, Palma-Barqueros V, Benito R, Bermejo N, Karkucak M, … Rivera J (2019). Identification of novel variants in ten patients with Hermansky-Pudlak syndrome by high-throughput sequencing. Ann Med, 51(2), 141–148. doi: 10.1080/07853890.2019.1587498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billio A, Moeseneder C, Donazzan G, Triani A, Pescosta N, & Coser P (2001). Hermansky-Pudlak syndrome: clinical presentation and confirmation of the value of the mepacrine-based cytofluorimetry test in the diagnosis of delta granule deficiency. Haematologica, 86(2), 220. [PubMed] [Google Scholar]
- Boissy RE, Richmond B, Huizing M, Helip-Wooley A, Zhao Y, Koshoffer A, & Gahl WA (2005). Melanocyte-specific proteins are aberrantly trafficked in melanocytes of Hermansky-Pudlak syndrome-type 3. Am J Pathol, 166(1), 231–240. doi: 10.1016/S0002-9440(10)62247-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero JP, Chen D, Majerus JA, Coon LM, He R, Warad DM, … Nichols WL (2018). Hermansky-Pudlak syndrome subtype 5 (HPS-5) novel mutation in a 65 year-old with oculocutaneous hypopigmentation and mild bleeding diathesis: The importance of recognizing a subtle phenotype. Platelets, 29(1), 91–94. doi: 10.1080/09537104.2017.1361019 [DOI] [PubMed] [Google Scholar]
- Bowman SL, Bi-Karchin J, Le L, & Marks MS (2019). The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic, 20(6), 404–435. doi: 10.1111/tra.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantly M, Avila NA, Shotelersuk V, Lucero C, Huizing M, & Gahl WA (2000). Pulmonary function and high-resolution CT findings in patients with an inherited form of pulmonary fibrosis, Hermansky-Pudlak syndrome, due to mutations in HPS-1. Chest, 117(1), 129–136. doi: 10.1378/chest.117.1.129 [DOI] [PubMed] [Google Scholar]
- Briken V, Jackman RM, Dasgupta S, Hoening S, & Porcelli SA (2002). Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J, 21(4), 825–34. Erratum in: EMBO J, 2002, 21(6), 1504. doi: 10.1093/emboj/21.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan MM, Tolman NJ, Simon KL, Huizing M, Hufnagel RB, Brooks BP, … Gochuico BR (2017). Clinical and molecular phenotyping of a child with Hermansky-Pudlak syndrome-7, an uncommon genetic type of HPS. Mol Genet Metab, 120(4), 378–383. doi: 10.1016/j.ymgme.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Rivera C, Golas G, Hess RA, Cardillo ND, Martin EH, O’Brien K, … Gahl WA (2011). Clinical, molecular, and cellular features of non-Puerto Rican Hermansky-Pudlak syndrome patients of Hispanic descent. J Invest Dermatol, 131(12), 2394–2400. doi: 10.1038/jid.2011.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Rivera C, Hess RA, O’Brien K, Golas G, Tsilou E, White JG, … Huizing M (2011). Novel mutations in the HPS1 gene among Puerto Rican patients. Clin Genet, 79(6), 561–567. doi: 10.1111/j.1399-0004.2010.01491.x [DOI] [PubMed] [Google Scholar]
- Carmona-Rivera C, Simeonov DR, Cardillo ND, Gahl WA, & Cadilla CL (2013). A divalent interaction between HPS1 and HPS4 is required for the formation of the biogenesis of lysosome-related organelle complex-3 (BLOC-3). Biochim Biophys Acta, 1833(3), 468–478. doi: 10.1016/j.bbamcr.2012.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetica V, Hackmann Y, Grieve S, Sieni E, Ciambotti B, Coniglio ML, … Arico M (2015). Patients with Griscelli syndrome and normal pigmentation identify RAB27A mutations that selectively disrupt MUNC13–4 binding. J Allergy Clin Immunol, 135(5), 1310–1318. doi: 10.1016/j.jaci.2014.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli VT, Daniels RW, Godoy R, Hoyle DJ, Kandachar V, Starcevic M, … Dell’Angelica EC (2010). Genetic modifiers of abnormal organelle biogenesis in a Drosophila model of BLOC-1 deficiency. Hum Mol Genet, 19 (5), 861–878. doi: 10.1093/hmg/ddp555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi ZH, Wei W, Bu DF, Li HH, Ding F, & Zhu P (2018). Targeted high-throughput sequencing technique for the molecular diagnosis of primary immunodeficiency disorders. Medicine (Baltimore), 97(40), e12695. doi: 10.1097/MD.0000000000012695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PW, Spector E, Thomas M, & Frei-Jones M (2010). Novel mutation causing Hermansky-Pudlak Syndrome Type 2. Pediatr Blood Cancer, 55(7), 1438. doi: 10.1002/pbc.22793 [DOI] [PubMed] [Google Scholar]
- Christensen S, Wagner L, Coleman MM, & Appell D (2017). The lived experience of having a rare medical disorder: Hermansky-Pudlak syndrome. Chronic Illn, 13(1), 62–72. doi: 10.1177/1742395316655854 [DOI] [PubMed] [Google Scholar]
- Clark RH, Stinchcombe JC, Day A, Blott E, Booth S, Bossi G, … Griffiths GM (2003). Adaptor protein 3-dependent microtubule-mediated movement of lytic granules to the immunological synapse. Nat Immunol, 4(11), 1111–1120. doi: 10.1038/ni1000 [DOI] [PubMed] [Google Scholar]
- Cullinane AR, Curry JA, Golas G, Pan J, Carmona-Rivera C, Hess RA, … Gahl WA (2012). A BLOC-1 mutation screen reveals a novel BLOC1S3 mutation in Hermansky-Pudlak Syndrome type 8. Pigment Cell Melanoma Res, 25(5), 584–591. doi: 10.1111/j.1755-148X.2012.01029.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly CM, Willer J, Gregg R, & Gross JM (2013). snow white, a zebrafish model of Hermansky-Pudlak Syndrome type 5. Genetics, 195(2), 481–494. doi: 10.1534/genetics.113.154898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer M, van Leeuwen K, Geissler J, van Alphen F, de Vries E, van der Kuip M, … Kuijpers TW (2017). Hermansky-Pudlak syndrome type 2: Aberrant pre-mRNA splicing and mislocalization of granule proteins in neutrophils. Hum Mutat, 38(10), 1402–1411. doi: 10.1002/humu.23271 [DOI] [PubMed] [Google Scholar]
- Dell’Acqua F, Saettini F, Castelli I, Badolato R, Notarangelo LD, & Rizzari C (2019). Hermansky-Pudlak syndrome type II and lethal hemophagocytic lymphohistiocytosis: Case description and review of the literature. J Allergy Clin Immunol Pract, 7(7), 2476–2478 e2475. doi: 10.1016/j.jaip.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC (2004). The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol, 16(4), 458–464. doi: 10.1016/j.ceb.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, & Bonifacino JS (1999). Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell, 3(1), 11–21. doi: 10.1016/s1097-2765(00)80170-7 [DOI] [PubMed] [Google Scholar]
- Demirtas CO, Alahdab YO, Kani HT, Atug O, & Imeryuz N (2019). Treatment of Hermansky-Pudlak syndrome Associated granulomatous colitis with anti-TNF agents: case series and review of literature. Eur J Gastroenterol Hepatol, 31(12), 1597–1600. doi: 10.1097/MEG.0000000000001510 [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, … Taschner PE (2016). HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat, 37(6), 564–569. doi: 10.1002/humu.22981 [DOI] [PubMed] [Google Scholar]
- Deng Y, Li Z, Liu J, Wang Z, Cao Y, Mou Y, … Xiong W (2018). Targeted resequencing reveals genetic risks in patients with sporadic idiopathic pulmonary fibrosis. Hum Mutat, 39(9), 1238–1245. doi: 10.1002/humu.23566 [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, & Dell’Angelica EC (2004). Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic, 5(4), 276–283. doi: 10.1111/j.1600-0854.2004.0171.x [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, & Dell’Angelica EC (2006). BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell, 17(9), 4027–4038. doi: 10.1091/mbc.e06-05-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe G, Morris DW, De Sanctis P, Magno E, Montesi JL, Garavan HP, … Foxe JJ (2008). Early visual processing deficits in dysbindin-associated schizophrenia. Biol Psychiatry, 63(5), 484–489. doi: 10.1016/j.biopsych.2007.07.022 [DOI] [PubMed] [Google Scholar]
- Doubková M, Trizuljak J, Vrzalová ZA, Blaháková I, Radová L, Pospíšilová Š, & Doubek M (2019). Novel genetic variant of HPS1 gene in Hermansky-Pudlak syndrome with fulminant progression of pulmonary fibrosis: a case report. BMC Pulm Med, 19(1), 178. doi: 10.1186/s12890-019-0941-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Chemaly S, O’Brien KJ, Nathan SD, Weinhouse GL, Goldberg HJ, Connors JM, … Gochuico BR (2018). Clinical management and outcomes of patients with Hermansky-Pudlak syndrome pulmonary fibrosis evaluated for lung transplantation. PLoS One, 13(3), e0194193. doi: 10.1371/journal.pone.0194193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders A, Zieger B, Schwarz K, Yoshimi A, Speckmann C, Knoepfle EM, … Ehl S (2006). Lethal hemophagocytic lymphohistiocytosis in Hermansky-Pudlak syndrome type II. Blood, 108(1), 81–87. doi: 10.1182/blood-2005-11-4413 [DOI] [PubMed] [Google Scholar]
- Falcon-Perez JM, Starcevic M, Gautam R, & Dell’Angelica EC (2002). BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem, 277(31), 28191–28199. doi: 10.1074/jbc.M204011200 [DOI] [PubMed] [Google Scholar]
- Felipez LM, Gokhale R, & Guandalini S (2010). Hermansky-Pudlak syndrome: severe colitis and good response to infliximab. J Pediatr Gastroenterol Nutr, 51(5), 665–667. doi: 10.1097/MPG.0b013e3181d2dcbb [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, & den Dunnen JT (2011). LOVD v.2.0: the next generation in gene variant databases. Hum Mutat, 32(5): 557–563. doi: 10.1002/humu.21438 [DOI] [PubMed] [Google Scholar]
- Fontana S, Parolini S, Vermi W, Booth S, Gallo F, Donini M, … Badolato R (2006). Innate immunity defects in Hermansky-Pudlak type 2 syndrome. Blood, 107(12), 4857–4864. doi: 10.1182/blood-2005-11-4398 [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, … O’Donovan MC (2014). De novo mutations in schizophrenia implicate synaptic networks. Nature, 506(7487), 179–184. doi: 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai K, Oh J, Frenk E, Almodovar C, & Spritz RA (1995). Linkage disequilibrium mapping of the gene for Hermansky-Pudlak syndrome to chromosome 10q23.1-q23.3. Hum Mol Genet, 4(9), 1665–1669. doi: 10.1093/hmg/4.9.1665 [DOI] [PubMed] [Google Scholar]
- Furuhashi K, Enomoto N, Fujisawa T, Hashimoto D, Inui N, Nakamura Y, & Suda T (2014). Hermansky-Pudlak syndrome with nonspecific interstitial pneumonia. Intern Med, 53(5), 449–453. doi: 10.2169/internalmedicine.53.1311 [DOI] [PubMed] [Google Scholar]
- Gahl WA, Brantly M, Kaiser-Kupfer MI, Iwata F, Hazelwood S, Shotelersuk V, … Bernardini I (1998). Genetic defects and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky-Pudlak syndrome). N Engl J Med, 338(18), 1258–1264. doi: 10.1056/NEJM199804303381803 [DOI] [PubMed] [Google Scholar]
- Gahl WA, Brantly M, Troendle J, Avila NA, Padua A, Montalvo C, … Gochuico B (2002). Effect of pirfenidone on the pulmonary fibrosis of Hermansky-Pudlak syndrome. Mol Genet Metab, 76(3), 234–242. doi: 10.1016/s1096-7192(02)00044-6 [DOI] [PubMed] [Google Scholar]
- Gallo V, Dotta L, Giardino G, Cirillo E, Lougaris V, D’Assante R, … Pignata C (2016). Diagnostics of Primary Immunodeficiencies through Next-Generation Sequencing. Front Immunol, 7, 466. doi: 10.3389/fimmu.2016.00466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhu L, Huang L, & Zhou J (2015). Synergistic defects of UNC13D and AP3B1 leading to adult hemophagocytic lymphohistiocytosis. Int J Hematol, 102(4), 488–492. doi: 10.1007/s12185-015-1807-z [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S, Hashemi-Gorji F, Yassaee VR, Alipour N, & Miryounesi M (2016). A Novel Splice Site Mutation in HPS1 Gene is Associated with Hermansky-Pudlak Syndrome-1 (HPS1) in an Iranian Family. Int J Mol Cell Med, 5(3), 192–195. [PMC free article] [PubMed] [Google Scholar]
- Ghiani CA, & Dell’Angelica EC (2011). Dysbindin-containing complexes and their proposed functions in brain: from zero to (too) many in a decade. ASN Neuro, 3(2), e00058. doi: 10.1042/AN20110010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Krzewska A, Murakami Y, Peruzzi G, O’Brien KJ, Merideth MA, Cullinane AR, … Krzewski K (2017). Natural killer cell activity and dysfunction in Hermansky-Pudlak syndrome. Br J Haematol, 176(1), 118–123. doi: 10.1111/bjh.14390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girot P, Le Berre C, De Maissin A, Freyssinet M, Trang-Poisson C, & Bourreille A (2019). Crohn’s-like acute severe colitis associated with Hermansky-Pudlak syndrome: A case report. World J Gastroenterol, 25(8), 1031–1036. doi: 10.3748/wjg.v25.i8.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gochuico BR, Huizing M, Golas GA, Scher CD, Tsokos M, Denver SD, … Gahl WA (2012). Interstitial lung disease and pulmonary fibrosis in Hermansky-Pudlak syndrome type 2, an adaptor protein-3 complex disease. Mol Med, 18, 56–64. doi: 10.2119/molmed.2011.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Conejero R, Rivera J, Escolar G, Zuazu-Jausoro I, Vicente V, & Corral J (2003). Molecular, ultrastructural and functional characterization of a Spanish family with Hermansky-Pudlak syndrome: role of insC974 in platelet function and clinical relevance. Br J Haematol, 123(1), 132–138. doi: 10.1046/j.1365-2141.2003.04557.x [DOI] [PubMed] [Google Scholar]
- Gordillo R, Del Rio M, Thomas DB, Flynn JT, & Woroniecki RP (2011). Hypertension, chronic kidney disease, and renal pathology in a child with hermansky-pudlak syndrome. Int J Nephrol, 2011, 324916. doi: 10.4061/2011/324916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin AE, Cobb BR, Anderson PD, Claassen DA, Helip-Wooley A, Huizing M, & Gahl WA (2005). Detection of hemizygosity in Hermansky-Pudlak syndrome by quantitative real-time PCR. Clin Genet, 68(1), 23–30. doi: 10.1111/j.1399-0004.2005.00461.x [DOI] [PubMed] [Google Scholar]
- Gunay-Aygun M, Huizing M, & Gahl WA (2004). Molecular defects that affect platelet dense granules. Semin Thromb Hemost, 30(5), 537–547. doi: 10.1055/s-2004-835674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttentag SH, Akhtar A, Tao JQ, Atochina E, Rusiniak ME, Swank RT, & Bates SR (2005). Defective surfactant secretion in a mouse model of Hermansky-Pudlak syndrome. Am J Respir Cell Mol Biol, 33(1), 14–21. doi: 10.1165/rcmb.2004-0293OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwynn B, Martina JA, Bonifacino JS, Sviderskaya EV, Lamoreux ML, Bennett DC, … Peters LL (2004). Reduced pigmentation (rp), a mouse model of Hermansky-Pudlak syndrome, encodes a novel component of the BLOC-1 complex. Blood, 104(10), 3181–3189. doi: 10.1182/blood-2004-04-1538 [DOI] [PubMed] [Google Scholar]
- Han CG, O’Brien KJ, Coon LM, Majerus JA, Huryn LA, Haroutunian SG, … Gochuico BR (2018). Severe bleeding with subclinical oculocutaneous albinism in a patient with a novel HPS6 missense variant. Am J Med Genet A, 176(12), 2819–2823. doi: 10.1002/ajmg.a.40514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Ishimatsu Y, Nakashima S, Miura S, Tomonaga M, Kakugawa T, … Kohno S (2014). An autopsy case of Hermansky-Pudlak syndrome: a case report and review of the literature on treatment. Intern Med, 53(23), 2705–2709. doi: 10.2169/internalmedicine.53.2239 [DOI] [PubMed] [Google Scholar]
- Hart JC, & Miller CT (2017). Sequence-Based Mapping and Genome Editing Reveal Mutations in Stickleback Hps5 Cause Oculocutaneous Albinism and the casper Phenotype. G3 (Bethesda), 7(9), 3123–3131. doi: 10.1534/g3.117.1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst M, Naehrlich L, Mahavadi P, Grosse-Onnebrink J, Terheggen-Lagro S, Skanke LH, … Griese M (2018). Hermansky-Pudlak syndrome type 2 manifests with fibrosing lung disease early in childhood. Orphanet J Rare Dis, 13(1), 42. doi: 10.1186/s13023-018-0780-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansky F, & Pudlak P (1959). Albinism associated with hemorrhagic diathesis and unusual pigmented reticular cells in the bone marrow: report of two cases with histochemical studies. Blood, 14(2), 162–169. [PubMed] [Google Scholar]
- Hermos CR, Huizing M, Kaiser-Kupfer MI, & Gahl WA (2002). Hermansky-Pudlak syndrome type 1: gene organization, novel mutations, and clinical-molecular review of non-Puerto Rican cases. Hum Mutat, 20(6), 482. doi: 10.1002/humu.9097 [DOI] [PubMed] [Google Scholar]
- Hirobe T, Ito S, & Wakamatsu K (2013). The mouse ruby-eye 2(d) (ru2(d) /Hps5(ru2-d) ) allele inhibits eumelanin but not pheomelanin synthesis. Pigment Cell Melanoma Res, 26(5), 723–726. doi: 10.1111/pcmr.12118 [DOI] [PubMed] [Google Scholar]
- Hoffmann MB, Lorenz B, Morland AB, & Schmidtborn LC (2005). Misrouting of the optic nerves in albinism: estimation of the extent with visual evoked potentials. Invest Ophthalmol Vis Sci, 46(10), 3892–3898. doi: 10.1167/iovs.05-0491 [DOI] [PubMed] [Google Scholar]
- Horikawa T, Araki K, Fukai K, Ueda M, Ueda T, Ito S, & Ichihashi M (2000). Heterozygous HPS1 mutations in a case of Hermansky-Pudlak syndrome with giant melanosomes. Br J Dermatol, 143(3), 635–640. doi: 10.1111/j.1365-2133.2000.03725.x [DOI] [PubMed] [Google Scholar]
- Huizing M, Anikster Y, Fitzpatrick DL, Jeong AB, D’Souza M, Rausche M, … Gahl WA (2001). Hermansky-Pudlak syndrome type 3 in Ashkenazi Jews and other non-Puerto Rican patients with hypopigmentation and platelet storage-pool deficiency. Am J Hum Genet, 69(5), 1022–1032. doi: 10.1086/324168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, & Gahl WA (2008). Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet, 9, 359–386. doi: 10.1146/annurev.genom.9.081307.164303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Hess R, Dorward H, Claassen DA, Helip-Wooley A, Kleta R, … Gahl WA (2004). Cellular, molecular and clinical characterization of patients with Hermansky-Pudlak syndrome type 5. Traffic, 5(9), 711–722. doi: 10.1111/j.1600-0854.2004.00208.x [DOI] [PubMed] [Google Scholar]
- Huizing M, Malicdan MCV, Gochuico BR, et al. Hermansky-Pudlak Syndrome. 2017. October 26 [Updated 2000 Jul 24]. In: Adam MP, Ardinger HH, Pagon RA, et al. , editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2021. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1287/ [Google Scholar]
- Huizing M, Pederson B, Hess RA, Griffin A, Helip-Wooley A, Westbroek W, … Gahl WA (2009). Clinical and cellular characterisation of Hermansky-Pudlak syndrome type 6. J Med Genet, 46(12), 803–810. doi: 10.1136/jmg.2008.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Scher CD, Strovel E, Fitzpatrick DL, Hartnell LM, Anikster Y, & Gahl WA (2002). Nonsense mutations in ADTB3A cause complete deficiency of the beta3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr Res, 51(2), 150–158. doi: 10.1203/00006450-200202000-00006 [DOI] [PubMed] [Google Scholar]
- Hull S, Arno G, Holder GE, Plagnol V, Gomez K, Liesner R, … Moore AT (2016). The ophthalmic presentation of Hermansky-Pudlak syndrome 6. Br J Ophthalmol, 100(11), 1521–1524. doi: 10.1136/bjophthalmol-2015-308067 [DOI] [PubMed] [Google Scholar]
- Hussain N, Quezado M, Huizing M, Geho D, White JG, Gahl W, & Mannon P (2006). Intestinal disease in Hermansky-Pudlak syndrome: occurrence of colitis and relation to genotype. Clin Gastroenterol Hepatol, 4(1), 73–80. doi: 10.1016/s1542-3565(05)00858-x [DOI] [PubMed] [Google Scholar]
- Ikawa Y, Hess R, Dorward H, Cullinane AR, Huizing M, Gochuico BR, … Candotti F (2015). In vitro functional correction of Hermansky-Pudlak Syndrome type-1 by lentiviral-mediated gene transfer. Mol Genet Metab, 114(1), 62–65. doi: 10.1016/j.ymgme.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, … Wigler M (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature, 515(7526), 216–221. doi: 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Fukaya M, Saegusa S, Kobayashi E, Sugawara T, Hara Y, … Sakagami H (2018). Pallidin is a novel interacting protein for cytohesin-2 and regulates the early endosomal pathway and dendritic formation in neurons. J Neurochem, 147(2), 153–177. doi: 10.1111/jnc.14579 [DOI] [PubMed] [Google Scholar]
- Ito S, Suzuki T, Inagaki K, Suzuki N, Takamori K, Yamada T, … Tomita Y (2005). High frequency of Hermansky-Pudlak syndrome type 1 (HPS1) among Japanese albinism patients and functional analysis of HPS1 mutant protein. J Invest Dermatol, 125(4), 715–720. doi: 10.1111/j.0022-202X.2005.23884.x [DOI] [PubMed] [Google Scholar]
- Iwakawa J, Matsuyama W, Watanabe M, Yamamoto M, Oonakahara K, Machida K, … Arimura K (2005). Hermansky-Pudlak syndrome with a novel mutation. Intern Med, 44(7), 733–738. [DOI] [PubMed] [Google Scholar]
- Iwata Y, Kobayashi T, Arima M, Numata S, Yagami A, Okamura K, … Matsunaga K (2017). Case of Japanese Hermansky-Pudlak syndrome patient with deeply invasive squamous cell carcinoma and multiple lesions of actinic keratosis on the face and neck. J Dermatol, 44(2), 219–220. doi: 10.1111/1346-8138.13462 [DOI] [PubMed] [Google Scholar]
- Iyer S, Suresh S, Guo D, Daman K, Chen JCJ, Liu P, … Wolfe SA (2019). Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature, 568(7753), 561–565. doi: 10.1038/s41586-019-1076-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen B, Bode SF, Ammann S, Chakravorty S, Davies G, Diestelhorst J, … Ehl S (2013). The risk of hemophagocytic lymphohistiocytosis in Hermansky-Pudlak syndrome type 2. Blood, 121(15), 2943–2951. doi: 10.1182/blood-2012-10-463166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Murden SL, Bem D, Mundell SJ, Gissen P, Daly ME, … group, U. G. s. (2012). Rapid genetic diagnosis of heritable platelet function disorders with next-generation sequencing: proof-of-principle with Hermansky-Pudlak syndrome. J Thromb Haemost, 10(2), 306–309. doi: 10.1111/j.1538-7836.2011.04569.x [DOI] [PubMed] [Google Scholar]
- Jones ML, Murden SL, Brooks C, Maloney V, Manning RA, Gilmour KC, … Mumford AD (2013). Disruption of AP3B1 by a chromosome 5 inversion: a new disease mechanism in Hermansky-Pudlak syndrome type 2. BMC Med Genet, 14, 42. doi: 10.1186/1471-2350-14-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Bohn G, Allroth A, Boztug K, Brandes G, Sandrock I, … Klein C (2006). Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood, 108(1), 362–369. doi: 10.1182/blood-2005-11-4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazu M, Arai T, Sugimoto C, Kitaichi M, Akira M, Abe Y, … Inoue Y (2014). An intractable case of Hermansky-Pudlak syndrome. Intern Med, 53(22), 2629–2634. doi: 10.2169/internalmedicine.53.2446 [DOI] [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, … Burmeister M (1998). Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron, 21(1), 111–122. doi: 10.1016/s0896-6273(00)80519-x [DOI] [PubMed] [Google Scholar]
- Khan AO, Tamimi M, Lenzner S, & Bolz HJ (2016). Hermansky-Pudlak syndrome genes are frequently mutated in patients with albinism from the Arabian Peninsula. Clin Genet, 90(1), 96–98. doi: 10.1111/cge.12715 [DOI] [PubMed] [Google Scholar]
- Kirshenbaum AS, Cruse G, Desai A, Bandara G, Leerkes M, Lee CC, … Metcalfe DD (2016). Immunophenotypic and Ultrastructural Analysis of Mast Cells in Hermansky-Pudlak Syndrome Type-1: A Possible Connection to Pulmonary Fibrosis. PLoS One, 11(7), e0159177. doi: 10.1371/journal.pone.0159177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook S, Qi A, Wang P, Meng S, Gulleman P, Young LR, & Guttentag SH (2018). Gene-edited MLE-15 Cells as a Model for the Hermansky-Pudlak Syndromes. Am J Respir Cell Mol Biol, 58(5), 566–574. doi: 10.1165/rcmb.2017-0324MA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korogi Y, Gotoh S, Ikeo S, Yamamoto Y, Sone N, Tamai K, … Hirai T (2019). In Vitro Disease Modeling of Hermansky-Pudlak Syndrome Type 2 Using Human Induced Pluripotent Stem Cell-Derived Alveolar Organoids. Stem Cell Reports, 13(1), 235. doi: 10.1016/j.stemcr.2019.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen LA, Huizing M, Simsek S, Janssen JJ, & Zweegman S (2008). A novel mutation in a Turkish patient with Hermansky-Pudlak syndrome type 5. Eur J Haematol, 80(4), 356–360. doi: 10.1111/j.1600-0609.2007.01024.x [DOI] [PubMed] [Google Scholar]
- Kouklakis G, Efremidou EI, Papageorgiou MS, Pavlidou E, Manolas KJ, & Liratzopoulos N (2007). Complicated Crohn’s-like colitis, associated with Hermansky-Pudlak syndrome, treated with Infliximab: a case report and brief review of the literature. J Med Case Rep, 1, 176. doi: 10.1186/1752-1947-1-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratomi G, Saito A, Ozeki Y, Watanabe T, Fujii K, Shimoda K, … Akiyama K (2013). Association of the Hermansky-Pudlak syndrome type 4 (HPS4) gene variants with cognitive function in patients with schizophrenia and healthy subjects. BMC Psychiatry, 13, 276. doi: 10.1186/1471-244X-13-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnik K, Bartsch I, Maul-Pavicic A, Ehl S, Sandrock-Lang K, Bidlingmaier C, … Zieger B (2013). Novel mutation in Hermansky-Pudlak syndrome type 2 with mild immunological phenotype. Platelets, 24(7), 538–543. doi: 10.3109/09537104.2012.741275 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, … Maglott DR (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res, 46(D1), D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasseaux E, Plaisant C, Michaud V, Pennamen P, Trimouille A, Gaston L, … Arveiler B (2018). Molecular characterization of a series of 990 index patients with albinism. Pigment Cell Melanoma Res, 31(4), 466–474. doi: 10.1111/pcmr.12688 [DOI] [PubMed] [Google Scholar]
- Lederer DJ, Kawut SM, Sonett JR, Vakiani E, Seward SL Jr., White JG, … Arcasoy SM (2005). Successful bilateral lung transplantation for pulmonary fibrosis associated with the Hermansky-Pudlak syndrome. J Heart Lung Transplant, 24(10), 1697–1699. doi: 10.1016/j.healun.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Li N, Ding YU, Yu T, Li J, Shen Y, Wang X, … Wang J (2016). Causal variants screened by whole exome sequencing in a patient with maternal uniparental isodisomy of chromosome 10 and a complicated phenotype. Exp Ther Med, 11(6), 2247–2253. doi: 10.3892/etm.2016.3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Rusiniak ME, Chintala S, Gautam R, Novak EK, & Swank RT (2004). Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays, 26(6), 616–628. doi: 10.1002/bies.20042 [DOI] [PubMed] [Google Scholar]
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O’Brien EP, … Swank RT (2003). Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet, 35(1), 84–89. doi: 10.1038/ng1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe GC, Sanchez Guiu I, Chapman O, Rivera J, Lordkipanidze M, Dovlatova N, … collaborative, U. G. (2013). Microsatellite markers as a rapid approach for autozygosity mapping in Hermansky-Pudlak syndrome: identification of the second HPS7 mutation in a patient presenting late in life. Thromb Haemost, 109(4), 766–768. doi: 10.1160/TH12-11-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozynska LY, Plawski A, Lozynska MR, Vytvytskyi I, Lozynskyi RY, Prokopchuk N, & Tretiak B (2018). Variant of rare Hermansky - Pudlak syndrome associated with granulomatous colitis: diagnostics, clinical course and treatment. Exp Oncol, 40(1), 73–78. [PubMed] [Google Scholar]
- Ma J, Zhang Z, Yang L, Kriston-Vizi J, Cutler DF, & Li W (2016). BLOC-2 subunit HPS6 deficiency affects the tubulation and secretion of von Willebrand factor from mouse endothelial cells. J Genet Genomics, 43(12), 686–693. doi: 10.1016/j.jgg.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahavadi P, Korfei M, Henneke I, Liebisch G, Schmitz G, Gochuico BR, … Guenther A (2010). Epithelial stress and apoptosis underlie Hermansky-Pudlak syndrome-associated interstitial pneumonia. Am J Respir Crit Care Med, 182(2), 207–219. doi: 10.1164/rccm.200909-1414OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai S, Nishie W, Mai Y, Natsuga K, Nomura T, Suzuki S, … Shimizu H (2019). Speckled lentiginous nevus in a patient with Hermansky-Pudlak syndrome type 1. J Dermatol, 2019 Oct 17. doi: 10.1111/1346-8138.15121 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Mantegazza AR, Guttentag SH, El-Benna J, Sasai M, Iwasaki A, Shen H, … Marks MS (2012). Adaptor protein-3 in dendritic cells facilitates phagosomal toll-like receptor signaling and antigen presentation to CD4(+) T cells. Immunity, 36(5), 782–794. doi: 10.1016/j.immuni.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza AR, Wynosky-Dolfi MA, Casson CN, Lefkovith AJ, Shin S, Brodsky IE, & Marks MS. (2017). Increased autophagic sequestration in adaptor protein-3 deficient dendritic cells limits inflammasome activity and impairs antibacterial immunity. PLoS Pathog, 13(12), e1006785. doi: 10.1371/journal.ppat.1006785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MS, Heijnen HF, & Raposo G (2013). Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol, 25(4), 495–505. doi: 10.1016/j.ceb.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Moriyama K, & Bonifacino JS (2003). BLOC-3, a protein complex containing the Hermansky-Pudlak syndrome gene products HPS1 and HPS4. J Biol Chem, 278(31), 29376–29384. doi: 10.1074/jbc.M301294200 [DOI] [PubMed] [Google Scholar]
- Massullo P, Druhan LJ, Bunnell BA, Hunter MG, Robinson JM, Marsh CB, & Avalos BR (2005). Aberrant subcellular targeting of the G185R neutrophil elastase mutant associated with severe congenital neutropenia induces premature apoptosis of differentiating promyelocytes. Blood, 105(9), 3397–3404. doi: 10.1182/blood-2004-07-2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElvaney OJ, Huizing M, Gahl WA, O’Donovan P, Horan D, Logan PM, … McElvaney NG (2018). Hermansky-Pudlak syndrome with a novel genetic variant in HPS1 and subsequent accelerated pulmonary fibrosis: significance for phenocopy diseases. Thorax, 73(11), 1085–1088. doi: 10.1136/thoraxjnl-2018-211920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merideth MA, Vincent LM, Sparks SE, Hess RA, Manoli I, O’Brien KJ, … Gahl WA (2009). Hermansky-Pudlak syndrome in two African-American brothers. Am J Med Genet A, 149A(5), 987–992. doi: 10.1002/ajmg.a.32757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zhu HY, Qiao C, Xia Y, Kong Y, Zou YX, … Li JY (2019). Pathogenic Gene Mutations or Variants Identified by Targeted Gene Sequencing in Adults With Hemophagocytic Lymphohistiocytosis. Front Immunol, 10, 395. doi: 10.3389/fimmu.2019.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud V, Lasseaux E, Plaisant C, Verloes A, Perdomo-Trujillo Y, Hamel C, … Arveiler B (2017). Clinico-molecular analysis of eleven patients with Hermansky-Pudlak type 5 syndrome, a mild form of HPS. Pigment Cell Melanoma Res, 30(6), 563–570. doi: 10.1111/pcmr.12608 [DOI] [PubMed] [Google Scholar]
- Minkin P, Bertetti R, Lindsey S, & Bovino B (2015). Management of tooth extraction in a patient with a rare bleeding disorder associated with Hermansky-Pudlak syndrome: a case report. J Oral Maxillofac Surg, 73(2), 219–223. doi: 10.1016/j.joms.2014.08.037 [DOI] [PubMed] [Google Scholar]
- Miyamichi D, Asahina M, Nakajima J, Sato M, Hosono K, Nomura T, … Matsumoto N (2016). Novel HPS6 mutations identified by whole-exome sequencing in two Japanese sisters with suspected ocular albinism. J Hum Genet, 61(9), 839–842. doi: 10.1038/jhg.2016.56 [DOI] [PubMed] [Google Scholar]
- Mohammed M, Al-Hashmi N, Al-Rashdi S, Al-Sukaiti N, Al-Adawi K, Al-Riyami M, & Al-Maawali A (2018). Biallelic mutations in AP3D1 cause Hermansky-Pudlak syndrome type 10 associated with immunodeficiency and seizure disorder. Eur J Med Genet. doi: 10.1016/j.ejmg.2018.11.017 [DOI] [PubMed] [Google Scholar]
- Mora AJ, & Wolfsohn DM (2011). The management of gastrointestinal disease in Hermansky-Pudlak syndrome. J Clin Gastroenterol, 45(8), 700–702. doi: 10.1097/MCG.0b013e3181fd2742 [DOI] [PubMed] [Google Scholar]
- Morgan NV, Pasha S, Johnson CA, Ainsworth JR, Eady RA, Dawood B, … Maher ER (2006). A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). Am J Hum Genet, 78(1), 160–166. doi: 10.1086/499338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, & Bonifacino JS (2002). Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic, 3(9), 666–677. doi: 10.1034/j.1600-0854.2002.30908.x [DOI] [PubMed] [Google Scholar]
- Mukda E, Trachoo O, Pasomsub E, Tiyasirichokchai R, Iemwimangsa N, Sosothikul D, … Pakakasama S (2017). Exome sequencing for simultaneous mutation screening in children with hemophagocytic lymphohistiocytosis. Int J Hematol, 106(2), 282–290. doi: 10.1007/s12185-017-2223-3 [DOI] [PubMed] [Google Scholar]
- Mullins C, Hartnell LM, & Bonifacino JS (2000). Distinct requirements for the AP-3 adaptor complex in pigment granule and synaptic vesicle biogenesis in Drosophila melanogaster. Mol Gen Genet, 263(6), 1003–1014. doi: 10.1007/pl00008688 [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Nakamura N, Sano J, Inayama Y, Kawano N, Yamanaka S, … Matsubara O (2000). Interstitial pneumonia in Hermansky-Pudlak syndrome: significance of florid foamy swelling/degeneration (giant lamellar body degeneration) of type-2 pneumocytes. Virchows Arch, 437(3), 304–313. doi: 10.1007/s004280000241 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Nakajima K, Cox A, Fisher M, Howell M, Fish MB, … Grainger RM (2016). no privacy, a Xenopus tropicalis mutant, is a model of human Hermansky-Pudlak Syndrome and allows visualization of internal organogenesis during tadpole development. Dev Biol. doi: 10.1016/j.ydbio.2016.08.020 [DOI] [PubMed] [Google Scholar]
- Natsuga K, Akiyama M, Shimizu T, Suzuki T, Ito S, Tomita Y, … Shimizu H (2005). Ultrastructural features of trafficking defects are pronounced in melanocytic nevus in Hermansky-Pudlak syndrome type 1. J Invest Dermatol, 125(1), 154–158. doi: 10.1111/j.0022-202X.2005.23743.x [DOI] [PubMed] [Google Scholar]
- Nazarian R, Huizing M, Helip-Wooley A, Starcevic M, Gahl WA, & Dell’Angelica EC (2008). An immunoblotting assay to facilitate the molecular diagnosis of Hermansky-Pudlak syndrome. Mol Genet Metab, 93(2), 134–144. doi: 10.1016/j.ymgme.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Chintala S, Jenkins S, Pare JF, McGaha L, Smith Y, & Faundez V (2010). Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J Neurosci, 30(3), 820–831. doi: 10.1523/JNEUROSCI.3400-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TJ, & Wei ML (2007). Hermansky-Pudlak HPS1/pale ear gene regulates epidermal and dermal melanocyte development. J Invest Dermatol, 127(2), 421–428. doi: 10.1038/sj.jid.5700566 [DOI] [PubMed] [Google Scholar]
- Novak EK, Hui SW, & Swank RT (1984). Platelet storage pool deficiency in mouse pigment mutations associated with seven distinct genetic loci. Blood, 63(3), 536–544. [PubMed] [Google Scholar]
- O’Brien K, Troendle J, Gochuico BR, Markello TC, Salas J, Cardona H, … Gahl WA (2011). Pirfenidone for the treatment of Hermansky-Pudlak syndrome pulmonary fibrosis. Mol Genet Metab, 103(2), 128–134. doi: 10.1016/j.ymgme.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KJ, Gahl WA, & Gochuico BR (2018). The curse of idiopathic. J Inherit Metab Dis, 41(1), 3–4. doi: 10.1007/s10545-017-0101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KJ, Introne WJ, Akal O, Akal T, Barbu A, McGowan MP, … Gochuico BR (2018). Prolonged treatment with open-label pirfenidone in Hermansky-Pudlak syndrome pulmonary fibrosis. Mol Genet Metab, 125(1–2), 168–173. doi: 10.1016/j.ymgme.2018.07.012 [DOI] [PubMed] [Google Scholar]
- O’Brien KJ, Lozier J, Cullinane AR, Osorio B, Nghiem K, Speransky V, … Gochuico BR (2016). Identification of a novel mutation in HPS6 in a patient with hemophilia B and oculocutaneous albinism. Mol Genet Metab, 119(3), 284–287. doi: 10.1016/j.ymgme.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting WS, & King RA (1999). Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat, 13(2), 99–115. doi: [DOI] [PubMed] [Google Scholar]
- Oh J, Bailin T, Fukai K, Feng GH, Ho L, Mao JI, … Spritz RA (1996). Positional cloning of a gene for Hermansky-Pudlak syndrome, a disorder of cytoplasmic organelles. Nat Genet, 14(3), 300–306. doi: 10.1038/ng1196-300 [DOI] [PubMed] [Google Scholar]
- Oh J, Ho L, Ala-Mello S, Amato D, Armstrong L, Bellucci S, … Spritz RA (1998). Mutation analysis of patients with Hermansky-Pudlak syndrome: a frameshift hot spot in the HPS gene and apparent locus heterogeneity. Am J Hum Genet, 62(3), 593–598. doi: 10.1086/301757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Abe Y, Araki Y, Wakamatsu K, Seishima M, Umetsu T, … Suzuki T (2018). Characterization of melanosomes and melanin in patients with Hermansky-Pudlak syndrome Types 1,4,6 and 9. Pigment Cell Melanoma Res, 31, 267–276. doi: 10.1111/pcmr.12662 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hayashi M, Abe Y, Kono M, Nakajima K, Aoyama Y, … Suzuki T (2019). NGS-based targeted resequencing identified rare subtypes of albinism: Providing accurate molecular diagnosis for Japanese patients with albinism. Pigment Cell Melanoma Res,32(6), 848–853.. doi: 10.1111/pcmr.12800 [DOI] [PubMed] [Google Scholar]
- Ozgur M, & Yilmaz B (2015). Unexpected intra-operative bleeding due to Hermansky-Pudlak Syndrome. Indian J Anaesth, 59(6), 393–394. doi: 10.4103/0019-5049.158784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power B, Ferreira CR, Chen D, Zein WM, O’Brien KJ, Introne WJ, … Gochuico BR (2019). Hermansky-Pudlak syndrome and oculocutaneous albinism in Chinese children with pigmentation defects and easy bruising. Orphanet J Rare Dis, 14(1), 52. doi: 10.1186/s13023-019-1023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, & Bonifacino JS (2015). BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell, 33(2), 176–188. doi: 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke P, Schimmenti LA, Schoonveld C, Bothun ED, & Summers CG (2013). The unique association of iris heterochromia with Hermansky-Pudlak syndrome. J AAPOS, 17(5), 542–544. doi: 10.1016/j.jaapos.2013.05.012 [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS, & Cutler DF (2007). Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol, 19(4), 394–401. doi: 10.1016/j.ceb.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, … Committee, A. L. Q. A. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeisen AL, Schimmenti LA, White JG, Schoonveld C, & Summers CG (2013). Hermansky-Pudlak syndrome (HPS5) in a nonagenarian. J AAPOS, 17(3), 334–336. doi: 10.1016/j.jaapos.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Rouhani FN, Brantly ML, Markello TC, Helip-Wooley A, O’Brien K, Hess R, … Gochuico BR (2009). Alveolar macrophage dysregulation in Hermansky-Pudlak syndrome type 1. Am J Respir Crit Care Med, 180(11), 1114–1121. doi: 10.1164/rccm.200901-0023OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Kuratomi G, Ito C, Matsuoka H, Suzuki T, Ozeki Y, … Akiyama K (2013). An association study of the Hermansky-Pudlak syndrome type 4 gene in schizophrenic patients. Psychiatr Genet, 23(4), 163–173. doi: 10.1097/YPG.0b013e32836130a9 [DOI] [PubMed] [Google Scholar]
- Saito S, Tanaka R, Sasaki T, Aoki S, Yasuhara R, Nakayama Y, … Kubo A (2019). Subclinical hypopigmentation of the skin and hair in a Japanese patient with Hermansky-Pudlak syndrome type 3. J Dermatol, 2019 Oct 16. doi: 10.1111/1346-8138.15118 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Sakata Y, Kawamura K, Ichikado K, Suga M, & Yoshioka M (2013). Hermansky-Pudlak syndrome type 4 with interstitial pneumonia. Respir Med Case Rep, 9, 38–41. doi: 10.1016/j.rmcr.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Guiu I, Torregrosa JM, Velasco F, Anton AI, Lozano ML, Vicente V, & Rivera J (2014). Hermansky-Pudlak syndrome. Overview of clinical and molecular features and case report of a new HPS-1 variant. Hamostaseologie, 34(4), 301–309. doi: 10.5482/HAMO-14-06-0024 [DOI] [PubMed] [Google Scholar]
- Sandrock-Lang K, Bartsch I, Buechele N, Koehler U, Simon-Gabriel CP, Eckenweiler M, & Zieger B (2017). Novel mutation in two brothers with Hermansky Pudlak syndrome type 3. Blood Cells Mol Dis, 67, 75–80. doi: 10.1016/j.bcmd.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Sandrock-Lang K, Bockelmann D, Eberl W, Schmitt-Kastner S, & Zieger B (2018). A novel nonsense mutation in a patient with Hermansky-Pudlak syndrome type 4. Blood Cells Mol Dis, 69, 113–116. doi: 10.1016/j.bcmd.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Sandrock K, Bartsch I, Rombach N, Schmidt K, Nakamura L, Hainmann I, … Zieger B (2010). Compound heterozygous mutations in 2 siblings with Hermansky-Pudlak syndrome type 1 (HPS1). Klin Padiatr, 222(3), 168–174. doi: 10.1055/s-0030-1249628 [DOI] [PubMed] [Google Scholar]
- Santiago Borrero PJ, Rodriguez-Perez Y, Renta JY, Izquierdo NJ, Del Fierro L, Munoz D, … Cadilla CL (2006). Genetic testing for oculocutaneous albinism type 1 and 2 and Hermansky-Pudlak syndrome type 1 and 3 mutations in Puerto Rico. J Invest Dermatol, 126(1), 85–90. doi: 10.1038/sj.jid.5700034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Linehan MM, & Iwasaki A (2010). Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science, 329(5998), 1530–1534. doi: 10.1126/science.1187029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallreuter KU, Frenk E, Wolfe LS, Witkop CJ, & Wood JM (1993). Hermansky-Pudlak syndrome in a Swiss population. Dermatology, 187(4), 248–256. [DOI] [PubMed] [Google Scholar]
- Scheggia D, Mastrogiacomo R, Mereu M, Sannino S, Straub RE, Armando M, … Papaleo F (2018). Variations in Dysbindin-1 are associated with cognitive response to antipsychotic drug treatment. Nat Commun, 9(1), 2265. doi: 10.1038/s41467-018-04711-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneier AJ, & Fulton AB (2013). The hermansky-pudlak syndrome: clinical features and imperatives from an ophthalmic perspective. Semin Ophthalmol, 28(5–6), 387–391. doi: 10.3109/08820538.2013.825280 [DOI] [PubMed] [Google Scholar]
- Schreyer-Shafir N, Huizing M, Anikster Y, Nusinker Z, Bejarano-Achache I, Maftzir G, … Blumenfeld A (2006). A new genetic isolate with a unique phenotype of syndromic oculocutaneous albinism: clinical, molecular, and cellular characteristics. Hum Mutat, 27(11), 1158. doi: 10.1002/humu.9463 [DOI] [PubMed] [Google Scholar]
- Seward SL Jr., & Gahl WA (2013). Hermansky-Pudlak syndrome: health care throughout life. Pediatrics, 132(1), 153–160. doi: 10.1542/peds.2012-4003 [DOI] [PubMed] [Google Scholar]
- Shahi RB, De Brakeleer S, Caljon B, Pauwels I, Bonduelle M, Joris S, … De Greve J (2019). Identification of candidate cancer predisposing variants by performing whole-exome sequencing on index patients from BRCA1 and BRCA2-negative breast cancer families. BMC Cancer, 19(1), 313. doi: 10.1186/s12885-019-5494-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Maddirevula S, Faqeih E, Ibrahim N, Hashem M, Shaheen R, & Alkuraya FS (2017). Increasing the sensitivity of clinical exome sequencing through improved filtration strategy. Genet Med, 19(5), 593–598. doi: 10.1038/gim.2016.155 [DOI] [PubMed] [Google Scholar]
- Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, … Sherwood RI (2018). Predictable and precise template-free CRISPR editing of pathogenic variants. Nature, 563(7733), 646–651. doi: 10.1038/s41586-018-0686-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotelersuk V, Hazelwood S, Larson D, Iwata F, Kaiser-Kupfer MI, Kuehl E, … Gahl WA (1998). Three new mutations in a gene causing Hermansky-Pudlak syndrome: clinical correlations. Mol Genet Metab, 64(2), 99–107. doi: 10.1006/mgme.1998.2679 [DOI] [PubMed] [Google Scholar]
- Sim W, Kim SY, Han J, Rim TH, Lee JG, Paik HC, & Park MS (2019). Extracorporeal Membrane Oxygenation Bridge to Lung Transplantation in a Patient with Hermansky-Pudlak Syndrome and Progressive Pulmonary Fibrosis. Acute Crit Care, 34(1), 95–98. doi: 10.4266/acc.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeoni I, Stephens JC, Hu F, Deevi SV, Megy K, Bariana TK, … Turro E (2016). A high-throughput sequencing test for diagnosing inherited bleeding, thrombotic, and platelet disorders. Blood, 127(23), 2791–2803. doi: 10.1182/blood-2015-12-688267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia MA, Sakuraba A, & Rubin DT (2017). Two Complex Cases of Hermansky-Pudlak Syndrome Highlight a Potential Biologic Explanation for an Associated Crohn’s Disease Phenotype. ACG Case Rep J, 4, e14. doi: 10.14309/crj.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow JR, Gregory-Roberts E, Yamamoto K, Blonska A, Ghosh SK, Ueda K, & Zhou J (2012). The bisretinoids of retinal pigment epithelium. Prog Retin Eye Res, 31(2), 121–135. doi: 10.1016/j.preteyeres.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel S, Chiu A, James AS, Jentsch JD, & Karlsgodt KH (2015). Recognition deficits in mice carrying mutations of genes encoding BLOC-1 subunits pallidin or dysbindin. Genes Brain Behav, 14(8), 618–624. doi: 10.1111/gbb.12240 [DOI] [PubMed] [Google Scholar]
- Spritz RA, & Oh J (1999). HPS gene mutations in Hermansky-Pudlak syndrome. Am J Hum Genet, 64(2), 658. doi: 10.1086/302257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, & Dell’Angelica EC (2004). Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem, 279(27), 28393–28401. doi: 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Stearman RS, Cornelius AR, Young LR, Conklin DS, Mickler EA, Lu X, … Geraci MW (2019). Familial Pulmonary Fibrosis and Hermansky-Pudlak Syndrome Rare Missense Mutations In Context. Am J Respir Crit Care Med, 200(2), 253–256.. doi: 10.1164/rccm.201902-0457LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen J, Yokoyama T, Tolman NJ, O’Brien KJ, Nicoli ER, Brooks BP, … Malicdan MC (2017). Cellular and molecular defects in a patient with Hermansky-Pudlak syndrome type 5. PLoS One, 12(3), e0173682. doi: 10.1371/journal.pone.0173682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, … Kendler KS (2002). Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet, 71(2), 337–348. doi: 10.1086/341750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikoudis A, Cieslak A, Loffredo L, Chen YW, Patel N, Saqi A, … Snoeck HW (2019). Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep, 27(12), 3709–3723 e3705. doi: 10.1016/j.celrep.2019.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, & Brenner MB (2002). Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity, 16(5), 697–706. doi: 10.1016/s1074-7613(02)00311-4 [DOI] [PubMed] [Google Scholar]
- Summers CG, Knobloch WH, Witkop CJ Jr., & King RA (1988). Hermansky-Pudlak syndrome. Ophthalmic findings. Ophthalmology, 95(4), 545–554. doi: 10.1016/s0161-6420(88)33152-0 [DOI] [PubMed] [Google Scholar]
- Summers CG, & Schimmenti LA (2014). The unique association of iris heterochromia with Hermansky Pudlak syndrome. J AAPOS, 18(2), 209. doi: 10.1016/j.jaapos.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ito S, Inagaki K, Suzuki N, Tomita Y, Yoshino M, & Hashimoto T (2004). Investigation on the IVS5 +5G --> a splice site mutation of HPS1 gene found in Japanese patients with Hermansky-Pudlak syndrome. J Dermatol Sci, 36(2), 106–108. doi: 10.1016/j.jdermsci.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Li W, Zhang Q, Karim A, Novak EK, Sviderskaya EV, … Spritz RA (2002). Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet, 30(3), 321–324. doi: 10.1038/ng835 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Li W, Zhang Q, Novak EK, Sviderskaya EV, Wilson A, … Spritz RA (2001). The gene mutated in cocoa mice, carrying a defect of organelle biogenesis, is a homologue of the human Hermansky-Pudlak syndrome-3 gene. Genomics, 78(1–2), 30–37. doi: 10.1006/geno.2001.6644 [DOI] [PubMed] [Google Scholar]
- Tagboto S, Carr S, Varghese A, Allen A, Feehally J, & Furness P (2001). Iga nephropathy, antineutrophil cytoplasmic antibodies and crescentic glomerulonephritis in a patient with the Hermansky-Pudlak syndrome. Am J Nephrol, 21(1), 58–62. doi: 10.1159/000046221 [DOI] [PubMed] [Google Scholar]
- Takahashi A, & Yokoyama T (1984). Hermansky-Pudlak syndrome with special reference to lysosomal dysfunction. A case report and review of the literature. Virchows Arch A Pathol Anat Histopathol, 402(3), 247–258. doi: 10.1007/bf00695079 [DOI] [PubMed] [Google Scholar]
- Takata A, Miyake N, Tsurusaki Y, Fukai R, Miyatake S, Koshimizu E, … Matsumoto N (2018). Integrative Analyses of De Novo Mutations Provide Deeper Biological Insights into Autism Spectrum Disorder. Cell Rep, 22(3), 734–747. doi: 10.1016/j.celrep.2017.12.074 [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Abe Y, Yamada T, Kawano S, Hozumi Y, Ito S, … Nishigori C (2014). Case of Hermansky-Pudlak syndrome 1 patient with milder symptoms in Japanese. J Dermatol, 41(3), 268–270. doi: 10.1111/1346-8138.12390 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Yang L, Wataya-Kaneda M, Suzuki T, Okamura K, Hozumi Y, … Katayama I (2015). Case of Hermansky-Pudlak syndrome 1 in a Japanese infant. J Dermatol, 42(9), 906–907. doi: 10.1111/1346-8138.12937 [DOI] [PubMed] [Google Scholar]
- Tesi B, Lagerstedt-Robinson K, Chiang SC, Ben Bdira E, Abboud M, Belen B, … Bryceson YT (2015). Targeted high-throughput sequencing for genetic diagnostics of hemophagocytic lymphohistiocytosis. Genome Med, 7, 130. doi: 10.1186/s13073-015-0244-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen N, Huizing M, Krabbe JG, White JG, Jansen TJ, Merle PA, … Zweegman S (2010). Hermansky-Pudlak syndrome: the importance of molecular subtyping. J Thromb Haemost, 8(7), 1643–1645. doi: 10.1111/j.1538-7836.2010.03898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian JH, Wu ZX, Unzicker M, Lu L, Cai Q, Li C, … Sheng ZH (2005). The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J Neurosci, 25(45), 10546–10555. doi: 10.1523/jneurosci.3275-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro J, Turner M, & Gahl WA (1999). Dermatologic manifestations of Hermansky-Pudlak syndrome in patients with and without a 16-base pair duplication in the HPS1 gene. Arch Dermatol, 135(7), 774–780. doi: 10.1001/archderm.135.7.774 [DOI] [PubMed] [Google Scholar]
- Torres-Serrant M, Ramirez SI, Cadilla CL, Ramos-Valencia G, & Santiago-Borrero PJ (2010). Newborn screening for hermansky-pudlak syndrome type 3 in Puerto Rico. J Pediatr Hematol Oncol, 32(6), 448–453. doi: 10.1097/MPH.0b013e3181e5e1f1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Koster J, Marais A, … Abou Jamra R (2017). Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet, 25(2), 176–182. doi: 10.1038/ejhg.2016.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Avermaete F, Muys J, & Jacquemyn Y (2016). Management of Hermansky-Pudlak syndrome in pregnancy and review of literature. BMJ Case Rep, 2016, pii: bcr2016217719. doi: 10.1136/bcr-2016-217719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent LM, Adams D, Hess RA, Ziegler SG, Tsilou E, Golas G, … Gahl WA (2009). Hermansky-Pudlak syndrome type 1 in patients of Indian descent. Mol Genet Metab, 97(3), 227–233. doi: 10.1016/j.ymgme.2009.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorechovsky I (2010). Transposable elements in disease-associated cryptic exons. Hum Genet, 127(2), 135–154. doi: 10.1007/s00439-009-0752-4 [DOI] [PubMed] [Google Scholar]
- Wang H, Xu J, Lazarovici P, & Zheng W (2017). Dysbindin-1 Involvement in the Etiology of Schizophrenia. Int J Mol Sci, 18(10). doi: 10.3390/ijms18102044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb EL, Rudd MF, Sellick GS, El Galta R, Bethke L, Wood W, … Houlston RS (2006). Search for low penetrance alleles for colorectal cancer through a scan of 1467 non-synonymous SNPs in 2575 cases and 2707 controls with validation by kin-cohort analysis of 14 704 first-degree relatives. Hum Mol Genet, 15(21), 3263–3271. doi: 10.1093/hmg/ddl401 [DOI] [PubMed] [Google Scholar]
- Wei A, Lian S, Wang L, & Li W (2009). The first case report of a Chinese Hermansky-Pudlak syndrome patient with a novel mutation on HPS1 gene. J Dermatol Sci, 56(2), 130–132. doi: 10.1016/j.jdermsci.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Wei A, Wang Y, Long Y, Wang Y, Guo X, Zhou Z, … Li W (2010). A comprehensive analysis reveals mutational spectra and common alleles in Chinese patients with oculocutaneous albinism. J Invest Dermatol, 130(3), 716–724. doi: 10.1038/jid.2009.339 [DOI] [PubMed] [Google Scholar]
- Wei A, Yang X, Lian S, & Li W (2011). Implementation of an optimized strategy for genetic testing of the Chinese patients with oculocutaneous albinism. J Dermatol Sci, 62(2), 124–127. doi: 10.1016/j.jdermsci.2011.02.009 [DOI] [PubMed] [Google Scholar]
- Wei A, Yuan Y, Bai D, Ma J, Hao Z, Zhang Y, … Li W (2016). NGS-based 100-gene panel of hypopigmentation identifies mutations in Chinese Hermansky-Pudlak syndrome patients. Pigment Cell Melanoma Res, 29(6), 702–706. doi: 10.1111/pcmr.12534 [DOI] [PubMed] [Google Scholar]
- Wei AH, Zang DJ, Zhang Z, Yang XM, & Li W (2015). Prenatal genotyping of four common oculocutaneous albinism genes in 51 Chinese families. J Genet Genomics, 42(6), 279–286. doi: 10.1016/j.jgg.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Wei A, Yuan Y, Qi Z, Liu T, Bai D, Zhang Y, … Li W (2019). Instability of BLOC-2 and BLOC-3 in Chinese patients with Hermansky-Pudlak syndrome. Pigment Cell Melanoma Res, 32(3), 373–380. doi: 10.1111/pcmr.12748 [DOI] [PubMed] [Google Scholar]
- Weickert CS, Straub R, Kleinman J, Hyde T, & Rothmond D (2006). Reduced dysbindin (DTNBP1) mRNA in hippocampus of patients with schizophrenia. Acta Neuropsychiatr, 18(6), 307–308. doi: 10.1017/S0924270800031719 [DOI] [PubMed] [Google Scholar]
- Wenham M, Grieve S, Cummins M, Jones ML, Booth S, Kilner R, … Mumford AD (2010). Two patients with Hermansky Pudlak syndrome type 2 and novel mutations in AP3B1. Haematologica, 95(2), 333–337. doi: 10.3324/haematol.2009.012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland D, Shaw M, Grimes W, Metcalf DJ, Burden JJ, Gomez K, … Cutler DF (2016). Super-resolution microscopy as a potential approach to diagnosis of platelet granule disorders. J Thromb Haemost, 14(4), 839–849. doi: 10.1111/jth.13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, & Witkop CJ (1972). Effects of normal and aspirin platelets on defective secondary aggregation in the Hermansky-Pudlak syndrome. A test for storage pool deficient platelets. Am J Pathol, 68(1), 57–66. [PMC free article] [PubMed] [Google Scholar]
- Witkop CJ, Almadovar C, Pineiro B, & Nunez Babcock M (1990). Hermansky-Pudlak syndrome (HPS). An epidemiologic study. Ophthalmic Paediatr Genet, 11(3), 245–250. doi: 10.3109/13816819009020986 [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Krumwiede M, Sedano H, & White JG (1987). Reliability of absent platelet dense bodies as a diagnostic criterion for Hermansky-Pudlak syndrome. Am J Hematol, 26(4), 305–311. doi: 10.1002/ajh.2830260403 [DOI] [PubMed] [Google Scholar]
- Wu W, Lin K, Yang Y, Dong Z, Zhang T, Lei W, … Yang Z (2019). A novel mutation causes Hermansky-Pudlak syndrome type 4 with pulmonary fibrosis in 2 siblings from China. Medicine (Baltimore), 98(33), e16899. doi: 10.1097/MD.0000000000016899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XJ, Wang HS, Ju XL, Xiao PF, Xiao Y, Xue HM, … Histiocytosis Study Group of the Chinese Pediatric, S. (2017). Clinical presentation and outcome of pediatric patients with hemophagocytic lymphohistiocytosis in China: A retrospective multicenter study. Pediatr Blood Cancer, 64(4). doi: 10.1002/pbc.26264 [DOI] [PubMed] [Google Scholar]
- Young LR, Gulleman PM, Bridges JP, Weaver TE, Deutsch GH, Blackwell TS, & McCormack FX (2012). The alveolar epithelium determines susceptibility to lung fibrosis in Hermansky-Pudlak syndrome. Am J Respir Crit Care Med, 186(10), 1014–1024. doi: 10.1164/rccm.201207-1206OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf S, Shahzad M, Kausar T, Sheikh SA, Tariq N, Shabbir AS, … Ahmed ZM (2016). Identification and clinical characterization of Hermansky-Pudlak syndrome alleles in the Pakistani population. Pigment Cell Melanoma Res, 29(2), 231–235. doi: 10.1111/pcmr.12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chandrakasan S, Chapman H, Valencia CA, Husami A, Kissell D, … Filipovich AH (2014). Synergistic defects of different molecules in the cytotoxic pathway lead to clinical familial hemophagocytic lymphohistiocytosis. Blood, 124(8), 1331–1334. doi: 10.1182/blood-2014-05-573105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhao B, Li W, Oiso N, Novak EK, Rusiniak ME, … Swank RT (2003). Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet, 33(2), 145–153. doi: 10.1038/ng1087 [DOI] [PubMed] [Google Scholar]
- Zuo L, Luo X, Kranzler HR, Lu L, Rosenheck RA, Cramer J, … Gelernter J (2009). Association study of DTNBP1 with schizophrenia in a US sample. Psychiatr Genet, 19(6), 292–304. doi: 10.1097/ypg.0b013e32832a50bc [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in the the Leiden Open Variation Database (http://www.lovd.nl/) and in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). The data that support the findings of this study are also available from the corresponding author upon reasonable request.