Abstract
Epigenetic dysregulation and disruption of gene enhancer networks are both pervasive in human cancers, and yet, their roles in keratinocyte cancers are poorly understood. Utilizing patient samples, Yao et al. (2020) provide an initial framework for understanding the underlying mechanisms by integrating enhancer and transcriptional alterations that occur during the progression of basal cell and squamous cell carcinomas.
Epigenetic enhancer function: Maintaining the balance in development, differentiation, and cancer
Mutations in epigenetic regulators are ubiquitous in human cancer. An abundance of evidence supports the idea that alterations in the epigenome may contribute to all of the hallmarks of human cancer, driving both tumor initiation and progression (Flavahan et al., 2017). Given the reversibility and targetability of epigenetic changes, gaining deeper insights into these mechanisms is a critical step toward harnessing the potential of these therapies (Mohammad et al., 2019). Indeed, chromatin engineering may offer opportunities to target multiple cancer-driving pathways simultaneously (Baskin and Haynes, 2019), whether it be through drugs that directly target the catalytic activities of chromatin-modifying enzymes or through novel CRISPR-based epigenome editing approaches.
Similar to epigenetic enzymes, gene enhancers have been demonstrated to serve as essential mediators of cell type–specific gene expression during development and differentiation. Therefore, perhaps not surprisingly, enhancer dysregulation has emerged as a major driver of tumorigenesis (Sur and Taipale, 2016). Mutations in chromatin modifiers are highly prevalent in both basal cell carcinoma (BCC) and squamous cell carcinoma (SCC). Work from The Cancer Genome Atlas has demonstrated that SCCs of all tissue types harbor the highest frequency of mutations in epigenetic regulators (Campbell et al., 2018). In cutaneous SCC, mutations in chromatin modifiers have also been associated with poor survival outcomes (Li et al., 2015). Supporting the links between disrupted epigenetics and enhancer dysregulation, mutations in the histone methyltransferases and master enhancer regulators KMT2C (MLL3) and KMT2D (MLL4) are among the top 20 most frequently mutated genes in both BCC and cutaneous SCC according to the Catalogue of Somatic Mutations in Cancer database (https://cancer.sanger.ac.uk/cosmic). However, despite emerging evidence supporting the importance of these enzymes in skin homeostasis (Lin-Shiao et al., 2018), there remains a significant gap in understanding how epigenetic enhancer dysfunction may contribute to keratinocyte (KC) carcinogenesis.
Chromatin immunoprecipitation sequencing meets KC carcinomas
In the Journal of Investigative Dermatology (2020), Yao et al. (2020) began to tackle these outstanding questions by mapping genome-wide enhancer alterations during the progression of both BCC and SCC. The authors collected BCC and SCC tumors from patients who were undergoing Mohs surgery along with normal adjacent tissue to serve as controls. They then performed chromatin immunoprecipitation sequencing (ChIP-seq) for the canonical histone modification associated with active gene enhancers, such as acetylated histone H3 lysine 27 (H3K27ac). Using standard bio-informatic pipelines to define the altered peaks between normal and tumor tissue, the authors found that SCCs displayed significantly more altered H3K27ac sites than BCCs (4,858 vs. 695 differential peaks, respectively). In both cancers, there were more gains than losses of active enhancers in the tumor tissues than in the controls. By identifying the nearest annotated gene to each altered enhancer, the major altered pathways associated with these differential enhancers could be implicated. Enhancers gained in BCC were enriched for numerous developmental pathways (skin, epidermis, skeletal system, and hair follicle development) as well as Wnt signaling. In contrast, enhancers gained in SCC were dominated by immunological processes (Figure 1). Both of these observations are consistent with the known etiopathogenesis of these cancers. BCC is frequently driven by the dysregulation of a major developmental pathway (Hedgehog signaling), and SCCs are very frequent in immunosuppressed individuals. Recent single-cell sequencing studies have further underscored the complex interplay between tumor KCs and the immune system in cutaneous SCC (Ji et al., 2020).
Figure 1. Enhancer chromatin accessibility can determine whether a gene is transcriptionally active or repressed.
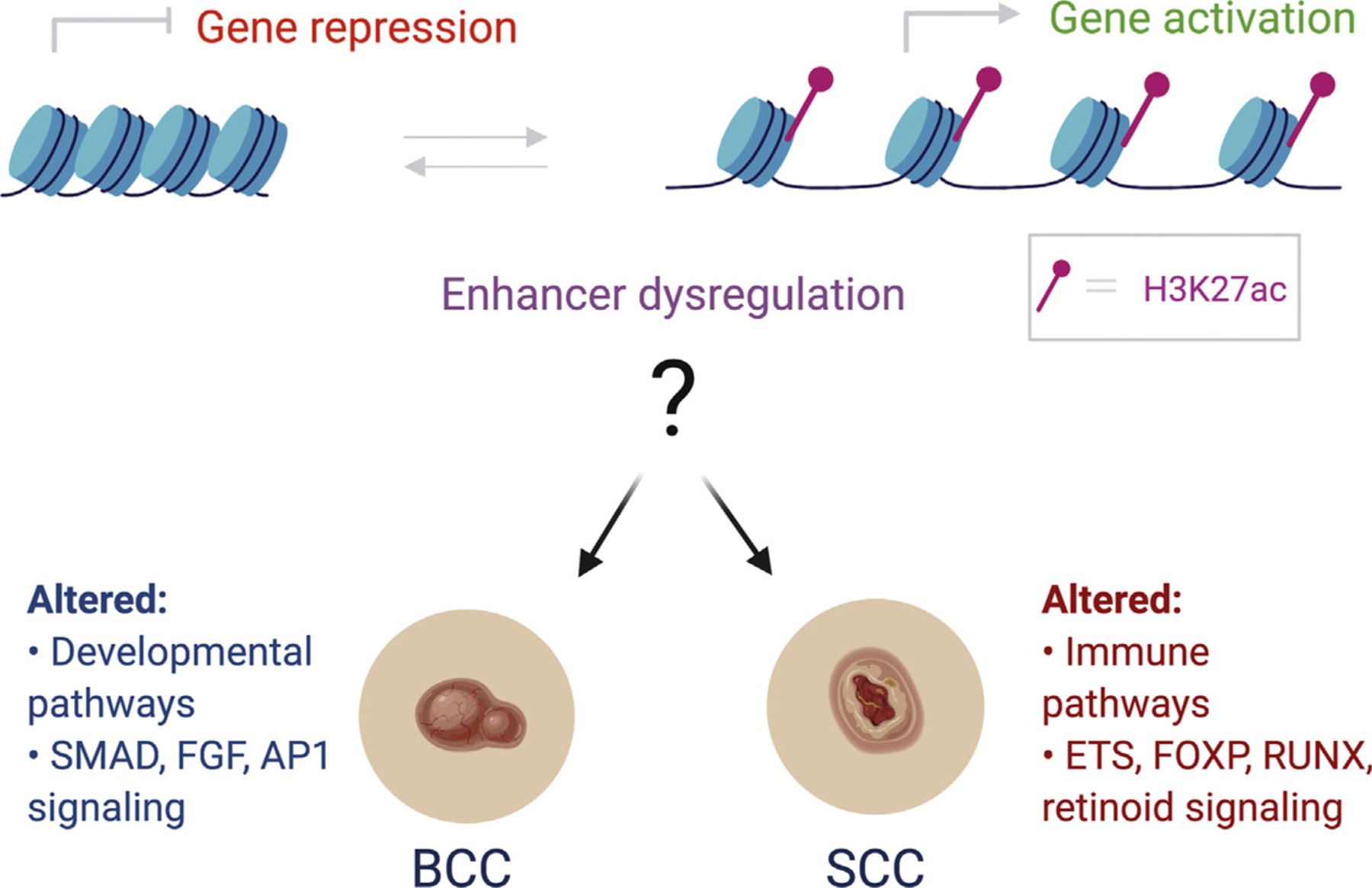
Dysregulation of these processes may be a key driver of KC cancers such as BCC and SCC. The authors mapped H3K27ac, a modification associated with active enhancers, in human patient samples to gain insights into epigenetic contributions to these ubiquitous cancers. In the case of BCC, aberrant enhancer activation is associated with developmental pathways and is enriched for transcription factor motifs such as SMAD3 and/or SMAD4, FGFR2, and FOSL2. In SCC, altered enhancers are enriched for immune-related pathways and display gains in ETS1, FOXP3, RUNX1, and NF-κB/Rel transcription factor sites along with a loss of H3K27ac enrichment and/or enhancer activity at retinoid-related transcription factor motifs. AP-1, activator protein-1; BCC, basal cell carcinoma; FGF, fibroblast GF; H3K27ac, acetylated histone H3 lysine 27; KC, keratinocyte; SCC, squamous cell carcinoma.
The authors then performed multiple analyses to determine whether the gained and lost enhancers were enriched for specific transcription factor–binding sites that might offer clues into the underlying mechanisms. BCCs gained enhancers associated with SMAD2, SMAD3, JUN, and other activator protein-1 factors such as FOSL2, whereas SCC gained enhancers that were enriched for FOXP3, ETS1, and RUNX motifs (Figure 1). Intriguingly, the immune-associated NF-κB/Rel motifs were only observed among the enriched SCC enhancers. With regard to lost enhancers, SCCs were enriched for numerous retinoid-related transcription factor motifs, consistent with evidence suggesting that systemic and topical retinoids may serve as chemo-preventive and/or treatment for SCCs (Harwood et al., 2005).
To ultimately understand the significance of epigenetic and enhancer alterations, it is important to integrate these results with the transcriptional alterations that occur in response to them. Whereas the authors did not perform RNA sequencing (RNA-seq) on the identical samples on which they performed the ChIP-seq, they did take advantage of some recently published RNA-seq data (GSE125285), which captured the same contrast between both normal adjacent skin and BCC and SCC tumors. In this study, the authors focused specifically on two genes potentially unique to the different cancers, FGFR2 in BCC and FOXP1 in SCC, as examples of genes harboring both gains in enhancer H3K27ac signal and in gene expression that may be important in uniquely driving those specific cancers. They highlight WNT5A as a gene with enrichment of H3K27ac in both BCC and SCC and a known role in promoting both cellular proliferation and migration. Collectively, these results offer a framework for testing several novel hypotheses related to the origins of these common cancers.
A look toward the future
Whereas there have been some clinical success stories utilizing epigenetic therapies (Mohammad et al., 2019), much remains to be learned and tested. Frequently, these compounds have demonstrated substantial efficacy both as single agents and in combination with other anticancer therapies, including immunotherapies. The skin may be even more amenable to the potential for epigenome-targeted drugs given that many of the systemic toxicities observed in clinical trials may be avoided by taking advantage of the opportunity to deliver topical agents. Studies such as the one by Yao et al. (2020) are important steps toward gaining insights to test hypotheses that may identify novel approaches.
A major limitation of this work is the limited number of patient samples. Future studies will involve additional samples as well as the mapping of many other histone modifications, integrating patient-matched gene expression data and taking advantage of emerging technologies such as CUT&RUN and CUT&TAG. The latter are ChIP-seq approaches that require very low cell numbers, even down to single cells. Another potential confounding issue to consider is the highly heterogeneous nature of human skin. The samples utilized in this study likely included a number of admixed cell types, ranging from KCs to a variety of immune cells, melanocytes, and fibroblasts. Because the performance of ChIP-seq on human patient tissues is technically challenging at this time, the authors have made a very important contribution that future studies can build on.
As sequencing costs decrease and technologies continue to improve, integrated multiomics analyses will provide an unprecedented view of the underlying mechanistic changes driving these cancers and in turn offer new therapeutic opportunities. For example, recent work by Ji et al. (2020) synthesized both single-cell transcriptome profiling and spatial transcriptomics from human patients with SCC, offering unique insights into both the complex interplay between cells and systems as well as the heterogeneity in these cancers. When these multiomics approaches and cutting-edge machine learning analytical methods are combined and applied on a large scale to patient samples, investigators and clinicians will be in a better place to predict both clinical outcomes and therapeutic responses.
Clinical Implications.
Epigenetic and enhancer dysregulation are key drivers of human cancers.
Both basal and squamous cell carcinomas display unique and common enhancer alterations in comparison with normal skin.
A mechanistic understanding of these changes will offer insights into new potential therapies for these common cancers.
Footnotes
CONFLICT OF INTEREST
The author states no conflict of interest.
REFERENCES
- Baskin NL, Haynes KA. Chromatin engineering offers an opportunity to advance epigenetic cancer therapy. Nat Struct Mol Biol 2019;26:842–5. [DOI] [PubMed] [Google Scholar]
- Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep 2018;23:194–212.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science 2017;357:eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CA, Leedham-Green M, Leigh IM, Proby CM. Low-dose retinoids in the prevention of cutaneous squamous cell carcinomas in organ transplant recipients: a 16-year retrospective study. Arch Dermatol 2005;141: 456–64. [DOI] [PubMed] [Google Scholar]
- Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 2020;182:497–514.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Hanna GJ, Laga AC, Haddad RI, Lorch JH, Hammerman PS. Genomic analysis of meta-static cutaneous squamous cell carcinoma. Clin Cancer Res 2015;21:1447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Shiao E, Lan Y, Coradin M, Anderson A, Donahue G, Simpson CL, et al. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev 2018;32: 181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med 2019;25:403–18. [DOI] [PubMed] [Google Scholar]
- Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer 2016;16:483–93. [DOI] [PubMed] [Google Scholar]
- Yao Q, Epstein CB, Banskota S, Issner R, Kim Y, Bernstein BE, et al. Epigenetic alterations in keratinocyte carcinoma. J Investig Dermatol 2021;141:1207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


