Abstract
Designing antiviral therapeutics is of great concern per current pandemics caused by novel coronavirus or SARS-CoV-2. The core polymerase enzyme in the viral replication/transcription machinery is generally conserved and serves well for drug target. In this work we briefly review structural biology and computational clues on representative single-subunit viral polymerases that are more or less connected with SARS-CoV-2 RNA dependent RNA polymerase (RdRp), in particular, to elucidate how nucleotide substrates and potential drug analogs are selected in the viral genome synthesis. To do that, we first survey two well studied RdRps from Polio virus and hepatitis C virus in regard to structural motifs and key residues that have been identified for the nucleotide selectivity. Then we focus on related structural and biochemical characteristics discovered for the SARS-CoV-2 RdRp. To further compare, we summarize what we have learned computationally from phage T7 RNA polymerase (RNAP) on its stepwise nucleotide selectivity, and extend discussion to a structurally similar human mitochondria RNAP, which deserves special attention as it cannot be adversely affected by antiviral treatments. We also include viral phi29 DNA polymerase for comparison, which has both helicase and proofreading activities on top of nucleotide selectivity for replication fidelity control. The helicase and proofreading functions are achieved by protein components in addition to RdRp in the coronavirus replication-transcription machine, with the proofreading strategy important for the fidelity control in synthesizing a comparatively large viral genome.
Keywords: RNA dependent RNA polymerase (RdRp), RNA/DNA polymerase (RNAP/DNAP), Fidelity control, Nucleotide selection, Molecular dynamics (MD) simulation, Kinetic modeling
In template-based polymerization or elongation conducted by polymerase enzymes to synthesize RNA or DNA chains of hundreds to thousands of nucleotides, selection of the right (or cognate) nucleotides by the enzyme is highly essential to maintain elongation fidelity over that at equilibrium with Watson-Crick base pairing [1], [2], [3]. To do that, an incoming nucleotide binds to the polymerase into the active site, subjecting to nucleotide selection at multiple stages, and is then added to the 3′-end of the synthesizing chain via phosphoryl-transfer reaction. The nucleotide addition cycle (NAC) indeed proceeds in multiple kinetic steps to allow stepwise nucleotide selectivity [3], [4]. The chemical free energy supports the NAC ensures that the polymerase elongates at non-equilibrium steady state to achieve sufficient speed and accuracy. Although the operation principle appears clear, corresponding structural-function mechanisms of various polymerase systems require elaborated studies for specific understanding, in particular, on i) how an incoming nucleotide is screened upon initial binding to the active site or nearby region, ii) how the nucleotide is subject to selection as it is gradually inserted into the active site to be ready for chemical reaction, and iii) how the nucleotide discrimination is further achieved during chemical reaction, under a two-metal ion (e.g. Mg2+ or Mn2+ ions) catalysis [5], or the incorporated nucleotide can incur additional selectivity even after the chemical addition.
In this work, we briefly review nucleotide selectivity revealed for replication or transcription fidelity control of representative viral RNA dependent RNA polymerases (RdRps) and a couple of structurally similar RNA polymerases (RNAPs), which all have a single-subunit handlike core structure (~500 amino acids, with palm, fingers, and thumb subdomains), with conserved structure motifs on the palm subdomain or additionally on the fingers subdomain to assist NAC and selectivity (see Fig. 1). Two primary conformations (open and closed) of the active site of the polymerases are expected to be involved in the nucleotide binding, insertion, and catalysis reaction [2], [6]. By first reviewing two well-studied RdRps from polio virus (PV) and hepatitis C virus (HCV), we intend to provide structural dynamics clues to support ongoing work on the RdRp from SASR-CoV-2 (or Cov2-RdRp). The RdRps come from positive-strand RNA viruses which represent a large class of human and animal pathogens to cause diseases and pandemics [7], [8]. For further comparison, we summarize our recent findings on nucleotide selectivity in another hand-like viral RNAP from bacteriophage T7 [9], [10], along with a structurally similar RNAP from human mitochondria, which is of constant consideration for antiviral drug side effect investigation [11]. Last, we include a viral phi29 DNAP also bearing a hand-shape structure, which cannot only function as a polymerase and a helicase, but also is furnished with an exonuclease subdomain to support its intrinsic proofreading activity for viral replication fidelity improvements [12], [13]. Note that the fidelity control in virus replication/transcription can generally lead to error rates about 10−3 to 10−6 to balance survival and variations [14], [15], which are primarily achieved by nucleotide selection in polymerase, though proofreading also exists but only for few species such as the coronaviruses (CoVs) [16].
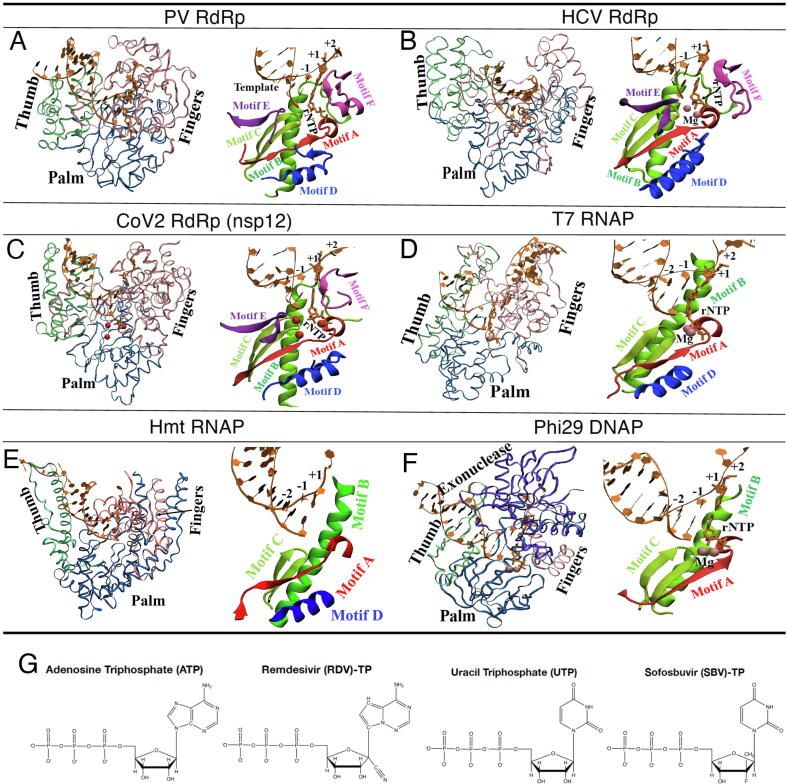
Fig. 1.
The representative single-subunit viral polymerase structures and conserved motifs. (A) The RdRp structure (or 3Dpol) from Polio virus (PV; left; PDB:3OLB) [18], with fingers, palm, thumb subdomains shown in pink, blue, and green, respectively; and with motifs A-F shown (right), colored red for A, green for B, light green for C, blue for D, magenta for E, and purple for F (motif G not shown); (B) The RdRp structure (or NS5B) from hepatitis C virus (HCV; left; PDB:4WTA) [19], and motifs A-F (right); (C) The core structure of SARS-CoV2 RdRp (left; PDB:7BTF; shown for residues S367-F920, no N-terminal part) [20], and motifs A to F (right); (D) The structure of T7 RNAP (left; PDB:1S0V) [21], and the motifs A-D (right). (E) The structure of human mitochondrial RNAP (or POLRMT) (left; PDB:4BOC; similar to T7 RNAP) [22], and the motifs A-D (right); (F) The structure of phi29 DNAP (left; PDB:2PYL) [23], and the motifs A-C (right). (G) The molecular schematics of the triphosphate-form nucleotide analogs from remdesivir (RDV) and sofosbuvir (SBV) along with corresponding natural nucleotide substrates ATP and UTP, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
1. RdRp from Polio virus with a cluster of residues for nucleotide selectivity
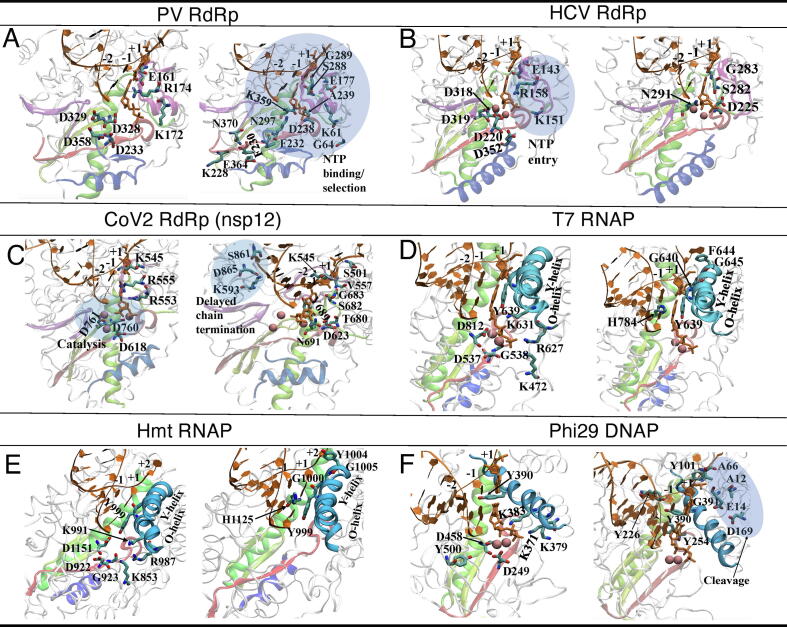
Poliovirus (PV) belongs to the family of Picornaviridae, with a comparatively short genome of size about 7500 nucleotides, and a comparatively high mutation rate [17]. In the core structure of the PV RdRp (or 3Dpol), the finger subdomain has its tip touching the top of thumb subdomain to encircle the active site (see Fig. 1A), as generally for other RdRps. Correspondingly, upon an incoming nucleotide binding and insertion into the active site, the core RdRp structure cannot accommodate large conformational changes on the fingers subdomain. Indeed, only subtle conformational changes have been identified inside the palm domain to allow the active site to transit from open to closed, with the closed conformation featured by extensive interaction with 2′ and 3′ ribose hydroxyl of the nucleotide, prior to the chemical reaction for addition [18], [24]. The nucleotide selection in accompany can be achieved by initial NTP-template base pairing and then the ribose recognition. A cluster of residues (Lys61, Glu177, Ser288, Gly289, Asn297, Asp238; see Fig. 2A) responsible for the ribose hydroxyl recognition is suggested, along with some supporting residues from N-term (Gly64, Ala239), while another cluster (Asp233, Asp328, Asp358) is essentially for two divalent metal ion binding. In particular, motif A and D in the palm subdomain of the RdRp move toward the active site upon its closure; Asp233 on the motif A beta-strand interacts with the two ions for catalysis, and two nearby residues F230 and F232 are in close control of fidelity [18], [25]. For cognate rCTP and non-cognate 2′-dCTP, in vitro biochemical studies show ~117 discrimination factor (i.e. of CTP over that of dCTP) for the wild-type PV RdRp, with mutation G64S, F230I, or F232I slightly improves the fidelity (to ~310, 152 or 200) and lowers the incorporation rate (from ~88 s−1 to ~46, 75 or 56 s−1); mutation K359R on the palm/motif D (see Fig. 2A) significantly improves the fidelity (to ~1300) but further decreases the elongation rate (to ~3 s−1) [25]. Repositioning of Lys359 by the motif-D loop is thus critical for nucleotide selection and incorporation (i.e. via protonating the leaving pyrophosphate group). The involved conformational changes of the motif-D are coordinated by Glu364 that swivels between associating with Lys228 and Asn370 to stabilize the open and closed motif-D loop conformations, respectively [26]. The corresponding mutation K228A slightly increases the polymerization rate and decreases the nucleotide selectivity, while N370A decreases the incorporation rate and increases the selectivity. The above analyses also indicate that the motif A/D induced active site closure can be rate-limiting prior to catalysis (in the presence of Mg2+ but not Mn2+ for PV [27], [28]). In comparison, mutations in the fingers subdomain impact on the elongation rate but little on the fidelity, as the fingers subdomain is likely linked to post-catalytic RNA translocation. All-atom molecular dynamics (MD) had been conducted to reveal correlated motions (within tens of nanoseconds) of a network of residues of the PV RdRp and three others from the picornavirus family [29]. The residues motions are contributed mainly by the structural motifs in regard to the RNA/nucleotide binding and catalysis, as being analyzed via the principal component analyses, which also suggest coevolutionary dynamics in these RdRps. The G64S mutant that improves the fidelity was also examined, which shows notable dynamical impacts to the entire structure in the simulation, enhancing flexibility in the motif D and exterior region of the RdRp but not the active site region [29].
Fig. 2.
The key residues involved in NTP entry, NTP binding or selection, and catalysis in the individual viral polymerases. For clarity, the catalysis/NTP entry related residues are shown (left) separately from those involved in the NTP binding/selection (right). (A) The key residues shown for Polio virus (PV) RdRp (or 3Dpol; PDB:3OLB; the NTP binding/selection residues identified are shaded and detailed in text). (B) The key residues shown for HCV RdRp (NS5B; PDB:4WTA; the NTP entry residues identified are shaded and addressed in text). (C) The key residues shown for SARS-CoV-2 RdRp (PDB:7BTF; the conserved catalytic residues and the residues potentially involved in the delayed chain termination are shaded and addressed in text, along with the key residues to be involved in the NTP-entry, binding/selection). (D) The key residues shown for T7 RNAP, with the O-helix and Y-helix on the fingers subdomain highlighted (PDB:1S0V; the NTP binding/selection residues are addressed in the related work [10]). (E) The key residues shown for human mitochondrial (hmt) RNAP (or POLRMT), with the O-helix and Y-helix highlighted (PDB:4BOC; the residues are shown similarly as in T7 RNAP). (F) The key residues shown for phi29 DNAP, with a comparable helix from the fingers subdomain highlighted [residue 375–390 overlap with the motif B; PDB:2PYL; the proofreading/cleavage involved residues are shaded (with Asp to Ala mutations); other residues are addressed in text].
From the studies on the PV RdRp, one can see that the pre-chemistry open → closed transition of the active site plays a crucial role in balancing the rate of nucleotide incorporation and the selection or fidelity control. The mutations inhibiting the open-closed transition likely improve the fidelity control [14], [15], or vice versa, as the mutations impact on the cluster of residues in the palm subdomain that either facilitate or hinder the active site closure transition. Such a transition is rate limiting before the chemical reaction, which highlights a crucial role of the rate-limiting kinetic step in control of the nucleotide selection, as suggested via our theoretical analyses [30].
2. RdRp from hepatitis C virus with additional nucleotide selectivity
Hepatitis C virus (HCV) belongs to Flaviviridae family of the single-stranded positive-sense RNA virus. NS5B, the C-terminal domain of the HCV polyprotein, functions as the viral RdRp, i.e., the catalytic core of the viral replication complex [31]. With a handlike structure, NS5B also has an extension from the fingers subdomain that encloses the active site and a β-loop originating from the thumb to prevent the binding of duplex RNA (see Fig. 1B). In addition to motif A and D for nucleotide recruitment and selection as being suggested in PV 3Dpol, motif B in NS5B with a loop followed by alpha-helix is also considered for positioning ribose of incoming NTP, using Ser282 and Gly283 [19], [32] (see Fig. 2B). NS5B thus conducts ribonucleotide selectivity using an extensive hydrogen bonding network (motif B-Ser282 and motif A-Asp225 etc) to recognize 2′-hydroxyl of the incoming nucleotide [19]. Mutation S282T provides moderate resistance to 2′-modified nucleotides [33], including sofosbuvir (SBV). The nucleotide prodrug SBV had been successfully designed to target on NS5B for combination treatment of chronic HCV [34]. Indeed, SBV analog can still be allowed to insert by Asn291 and the Watson-Crick paring to form the in-line conformation. In addition, NS5B has been suggested to contain a nucleotide mediated excision mechanism in order to excise the mismatched nucleotide at the 3′-end of the nascent RNA in vitro [32], [35]. Interestingly, motif B can be involved in translocation of the RdRp, which is also linked to fidelity control as mutations in motif B could not only slow down translocation, but also increase the time of residency of a mismatch in the active site to allow it to be excised. Hence, motif B appears to be quite essential in the HCV RdRp function [32]. Motif G, though it mainly interacts with the RNA template, may affect the NTP recognition and selection as it influences the orientation of the template strand/nucleotide or nt [32]. Motif F can also be linked to fidelity control since it guides the nucleotide into the tunnel to be paired with the template nucleotide, as well as to direct the pyrophosphate out of the tunnel after catalysis. Notably, very recent pre-steady state kinetic analyses show that the pyrophosphate release can be rate limiting in NS5B to improve the fidelity [36], which leads to a wide range of error rate from 10−4 to 10−9 for the NS5B elongation complex.
Previous MD simulations had shown apo structure of NS5B under impacts of inhibitors or magnesium ions and a hydrophobic C-terminal regions [37], [38]. Based on a crystal structure of ternary complex of HCV NS5B [19], more recent MD simulations with targeted or steered force and acceleration have been conducted to address the nucleotide entry in the viral RdRp [39]. Two successive checkpoints via motif F to regulate nucleotide traffic into the active site of NS5B have been identified, following a magnesium-bound (MgB) NTP into the entry channel (F1: 144–141; F3: 155–161 [40]) via an entry loop (F2:151–154). In particular, Lys151 forms salt-bridge with motif D-Asp352 to allow the entry loop to open or close (see Fig. 2B); then the salt-bridge between F1-Glu143 and F3-Arg158 coordinates with the NTP passage, which is also affected by arriving of the other magnesium ion (MgA). Correspondingly, the nucleotide seems to be interrogated already prior to insertion into the active site, and such interrogations seem to be comparable to NTP pre-insertion screening or even fingers-closing motions for nucleotide selectivity in some other handlike polymerase (e.g. T7 RNAP addressed below).
3. Probing SARS-CoV2 nucleotide selectivity via nucleotide analogue
SARS-CoV-2 is a newly emerged member of the family Coronaviridae (CoV), which includes SARS (severe acute respiratory syndrome) and MERS (middle east respiratory syndrome) viruses discovered from large-scale epidemics [41], [42]. CoVs are with large genome sizes (~30,000 nucleotides), enveloped, and often employ unusual gene expression strategies. The CoV virus enters the cell via endocytosis, with its ORF1a and OFR1b single-stranded RNA genome translated into proteases and then non-structural proteins (nsp) such the nsp12 (RdRp), nsp13 (helicase), and nsp14 (exonuclease or ExoN) [43]. The CoVs replication/transcription machinery assembles with multiple components. The SARS-CoV-2 RdRp (or nsp12) is the core engine of such a machinery, and its structure in complex with cofactors nsp7 and nsp8 (assisting RNA template binding and the RdRp processivity) has been recently resolved at 2.9-Å resolution from cryo-Electron Microscopy or cryo-EM [44]. A ternary complex structure of nsp12 (at 2.5–2.8 Å resolution; still with nsp7 and nsp8) together with RNA and a prodrug remdesivir (RDV) analog has been obtained soon after also via cryo-EM [20] (see Fig. 1G for schematics of RDV and SBV). The CoV-2 RdRp structure has an N-terminal beta-hairpin (residue 31–50), an extended domain (115–250), adopting a nidovirus RdRp-associated nucleo-tidyltransferase (NiRAN) architecture [45], and an interface region (251–366). The RdRp core structure (S367-F920; shown in Fig. 1C) still has a fingers subdomain (residues L366-A581 and K621-G679), a palm subdomain (residues T582-P620 and T680-Q815), and a thumb subdomain (residues H816-E920). Motifs A-E also locate in the palm subdomain while F and G are in the fingers subdomain. The highly conserved cation-binding Asp618 in motif A (611–626) is comparable to Asp233 in PV 3Dpol and Asp220 in HCV NS5B; the catalytic Asp760-Asp761 in motif C is comparable to Asp328-Asp329 in 3Dpol and Asp318-Asp319 in NS5B; Arg555 in the motif corresponds to 3Dpol Arg174 and NS5B Arg158 (see these residues in Fig. 2C left). The NTP entry channel is formed by hydrophilic residues such as Lys545, Arg555, and Arg553 from motif F (comparable to those in HCV NS5B). Modeling the RDV analog onto Cov2-RdRp following the SBV analog in HCV-NS5B indicates that RDV with an intact ribose can maintain hydrogen bonding network (with Asn691, Thr680 etc) as the native substrate, while the SBV analog interacts with Asn291 in NS5B (corresponding to Asn691 in CoV2-RdRp and Asn297 in 3Dpol) but cannot join the hydrogen bonding network with Ser282 and Asp225 (corresponding to Asp623 in CoV2-RdRp or Asp238 in 3Dpol) due to a fluorine substitution on its sugar moiety. Moreover, the hydrophobic side chain of Val557 in motif F is likely to stack with and stabilize the +1 template RNA uridine base to base pair with the incoming triphosphate form of the nucleotide analog (RDV-TP or RTP). In the ternary complex with the nucleotide analog in its monophosphate form (RDV-MP or RMP) incorporated to the 3′-end of the RNA primer, RMP interacts with side chains form Lys545 and Arg555 (motif F), with magnesium ions nearby and a pyrophosphate at the gate of the NTP entry channel. The motif F and G interact directly with the template RNA strand, with motif F also interacting with Lys545 and Arg555 contacting the +1 base. Hence, it is possible that the motif F can also be involved in the nucleotide binding/selection. The orientation of the RNA template-primer in the active site is indeed similar to that in the PV and HCV RdRp complexes. The RdRp with RMP incorporated in the ternary complex is quite similar overall to the apo RdRp structure (RMSD ~0.7 Å for all Ca atoms). Besides RDV, it has been found via modeling that other nucleotide analogs, e.g., favipiravir, ribavirin, galidesivir, and EIDD-2801 (molnupiravir) can also inhibit SARS-CoV-2 RdRp replication function [46], [47], likely via non-obligate RNA chain termination. For highly potent EIDD-2801 modeled onto the RdRp structure, the N4 hydroxyl group off the cytidine ring forms an extra hydrogen bond with the side chain of Lys545 and cytidine base also forms an extra hydrogen bond with the guanine base from the template strand, as assumed in the model [20]. In additional cryoEM structures obtained for CoV-2 RdRp, a pre-translocation complex with RMV was captured [48], in which Tyr689 (motif B) of the fingers subdomain interacts with the ribose 2′-OH groups of nucleotides at the −2 position from the +1 template nt. A loop region (682–686) in motif B shows interaction with RNA in both pre- and post-translocation; Gly683 on this loop is conserved and likely participates in NTP ribose recognition, active site closure, and mediating translocation. Motif G also interacts with +1 template nt via Ser501 and then Val557; Ser682 and Lys545 jointly stabilize the base of the paired RMP with the +1 template nt (see Fig. 2C right), while motif A-Asp623 stabilizes the ribose group. Another high-resolution cryo-EM structure captures long helical extensions in nsp8 forming positively charged ‘sliding poles’ along exiting RNA to contribute to the RdRp processivity [49]. The above structural studies thus indicate that motif A, B, D along with F and G together can contribute collectively to NTP binding, selection and translocation.
Previous studies showed that RDV inhibits Ebola virus and MERS coronavirus (MERS-CoV) via a delayed chain termination mechanism [50], [51]. The RDV analog thus can evade proofreading from the CoV-ExoN nsp14 [52]. Importantly, recent work on CoV-2 RdRp also clearly indicated that the RDV analog can be readily incorporated (at position i) and then cause the delayed chain termination of the synthesizing RNA, i.e., slightly downstream (at position i +3) [53], highly similar to that being found for SARS-CoV and MERS-CoV. It appears that the incorporated RDV analog, by evading from the nsp14 cleavage, then impacts on the subsequent NTP addition, e.g., by possibly interfering with the RdRp translocation along RNA downstream. In addition, the studies measured RDV-TP with a similar Vmax as ATP and dATP, while its KM is ~ ¼ of that of ATP (KM for dATP is ~103 larger) [53]. It is thus notable that RDV-TP shows even slightly preferred binding/incorporation properties over of its natural nucleotide counterpart ATP, and in this regards it is superior to other broad-spectrum antiviral compounds such as sofosbuvir, favipiravir, and ribavirin, measured together in vitro [53]. Further in vivo studies also demonstrate effectiveness of RDV into human lung cells [54]. Homology modeling based on structure of SARS-CoV RdRp [55] and aligning the modeled CoV-2 RdRp (residues Ser501, Gly590, Gly683, Ser814) with HCV NS5B structure (residues Ala97, Phe193, Gly283, and Ser367) further suggest that steric clash between the incorporated RDV analog and Ser861 would prevent enzyme translocation at i +3 (see Fig. 2C right) [53]. Interestingly, a very recent modeling and all-atom MD simulation study, which was also based on the homology model (i.e. using the SARS-CoV RdRp structure), suggests that the polar 1′-cyano group of RDV at i +3 position can destabilize salt-bridge between Asp865 and Lys593 to hinder the RdRp translocation [56]. In the same study, simulation of RDV interacting with a homology model of ExoN nsp14 shows a steric clash between the RDV 1′-cyano group with Asn104 so that to avoid the nsp14 cleavage. Such a cleavage in general serves as the proofreading mechanism to improve the fidelity in the coronavirus RNA synthesis [52], [57]. Note that a high-resolution crystal structure of the ExoN nsp14 in complex nsp10 from SARS-CoV-2 is then reported very recently [58].
Meanwhile, to illustrate the RDV-TP binding to CoV-2 RdRp, molecular docking and all-atom MD simulations have also been conducted, though these studies were initiated a bit early and still based on homology modeling [59], [60]. In the molecular docking, RTP shows slightly more stabilized binding to RdRp than ATP (binding score −7.5 kcal/mol for RTP vs −7 kcal/mol for ATP) [59]. In comparatively sophisticated MD simulation and alchemical free energy calculation, it is found that RTP is more stabilized (~-2.8 kcal/mol) than ATP at the active site of the modeled RdRp [60]. Note that in that work, ATP/RTP was modeled to the RdRp active site based on structural alignments with the PV RdRp substrate complex, and the modeled complexes were constructed and simulated in the absence of RNA chains [60]. Hence, improved molecular modeling and simulations are expected to illustrate the nucleotide (or analog) binding and selection in the CoV-2 RdRp, in particular, utilizing the newly solved high-solution structures of the CoV-2 RdRp [20], [44], [48], [49]. Our preliminary MD simulation studies using the newly solved CoV-2 RdRp structures confirm that both RTP and ATP can bind much more stably into the closed form of RdRp (PDB:7BV2 [20]) than the open form (modeled from PDB:7BTF [44]); in the open active site of RdRp, RTP seems to be particularly stabilized via base stacking with the template +1 nt, which further facilitates the insertion of RTP into the active site (ms in preparation).
4. Computation analyses of nucleotide selection from pre-insertion to insertion in T7 RNAP
Bacteriophage T7 RNAP and some other single-subunit RNAPs also adopt a hand-shape core structure with palm, fingers, and thumb subdomains [9], [21], [61] (see Fig. 1D). Comparing to the viral RdRps, there is no fingertip counterpart in the RNAPs, so that conformational changes of the fingers subdomain relative to the palm can become significant. In our previous computational work on the T7 RNAP elongation complex (e.g. post-translocation), it is confirmed that the palm-I subdomain (412–553) is most stable while the fingers subdomain (554–784) is highly flexible [62]. In the fingers subdomain, there is a five-helix bundle including the O-helix (627–640; see Fig. 2D); a highly conserved residue Tyr639 located at the C-terminal of the O-helix plays a highly critical role in gating and selecting nucleotides, e.g., by competing with the template +1 nt to interact with the incoming NTP [62]. We later showed via the MD simulation that the O-helix starts opening after the pyrophosphate release [63], e.g. by oscillating to open at pre-translocation (to possibly prevent backtracking), and substantially opens upon the RNAP translocation [64]. Accordingly, Tyr639 also plays a role in the translocation (e.g. assisting the 3′-end of the synthesizing RNA chain to translocate). By aligning the ternary RNAP complex structure with that of the viral RdRP (via the RNA strands near the active site), one cannot however find exact structural correspondence of the O-helix in the viral RdRp. Indeed, only four conserved motifs (A to D) can be found inside the fingers subdomain of T7 RNAP, which are the counterparts of those from the viral RdRps. The O-helix in T7 RNAP, however, is not included in any of these motifs, but locates around the region corresponding to the motif B and F in the viral RdRps. Nevertheless, one can still find that Tyr639 superimposes closely with SARS-CoV-2 R555 (or PV-3Dpol R174 and HCV-NS5B R158).
The structural study of PV RdRp had indicated that the template RNA nt at +1 is fully stacked to the upstream RNA, while other processive polymerases such as T7 RNAP have the template nt at +1 unstacked from the upstream nucleic duplex [18]. Accordingly, such an unstacked template nucleotide provides a pre-insertion site for the incoming NTP. Notably, our simulation studies revealed that a cognate ATP can form hydrogen bonds or base pairing with the template +1 nt in the pre-insertion T7 RNAP complex (though the base pairing was not captured in the crystal structure) [21], [62]. In contrast, a non-cognate NTP (modeled by directly replacing the cognate ATP in the crystal structure by the non-cgonate GTP), would be dragged by Tyr639 closely to be unable to interact with the template nt in the simulation [62]. We then named such a non-cognate NTP positioning (dragged by Tyr639) as a pre-insertion off-path configuration [65]. In comparison, we also modeled non-cognate NTP by performing an alchemical simulation to convert the pre-equilibrated and pre-inserted ATP to the non-cognate GTP, then obtained a pre-insertion configuration of the non-cognate NTP very similar to that of the cognate one, for which we named a pre-insertion on-path configuration [65]. By free energy calculation, we obtained the relative binding free energy of the non-cognate GTP vs the cognate ATP as ~1.8 ± 0.2 kcal/mol at the pre-insertion on-path configuration [65]. Essentially, the structural feature that distinguishes the on-path and off-path of the non-cognate GTP is that the template +1 nt or TTP deviates significantly away from the active site (toward downstream direction) in the off-path configuration; in the on-path GTP/ATP pre-insertion configuration, however, the template TTP stays closely base pairing with GTP or ATP, near the active site. Hence, it appears that via the pre-insertion site of T7 RNAP, nucleotide selection or screening already takes place, leading to two distinct configurations on-path and off-path, for the cognate and non-cognate NTP specie, respectively. Note that the selection is always stochastic, so there is always slight chance that the non-cognate NTP is loaded to the on-path pre-insertion or proceed further to insertion. For the non-cognate NTP at off-path pre-insertion, it would be easy to dissociate from the active site (i.e. almost no dissociation barrier, while the cognate ATP dissociation barrier from the on-path pre-insertion is ~4 kBT [66]).
Our consequent MD simulation studies then focused on revealing nucleotide insertion dynamics and energetics for both cognate and non-cognate NTP species into the active site of T7 RNAP. The nucleotide insertion is a rate-limiting step (~220 s−1) accompanied by the fingers subdomain closing with respect to the palm in T7 RNAP [67]. Our umbrella sampling calculations show that for the cognate ATP, the insertion barrier is ~3 ± 0.6 kBT; for the non-cognate GTP from the on-path pre-insertion, the insertion barrier is similar (~4.0 ± 0.2 kBT); while for the non-cognate GTP from the off-path pre-insertion, the insertion barrier is much larger (~ 7.6 ± 0.6 kBT) [68]. By conducting additional kinetic modeling and chemical master equation approach to quantify non-equilibrium steady state RNAP elongation rates for both the cognate and non-cognate NTP species, we show that with the nucleotide binding and selection energetics from MD simulations, the error rate (for GTP or dATP replacing ATP) right after the initial screening at pre-insertion becomes ~10−2; the error rate is further reduced to ~10−3 (for GTP replacing ATP) upon the nucleotide insertion and selection. After that, the GTP replacing ATP error rate can be further reduced to ~10−4 upon the nucleotide selection during catalysis (i.e. phosphoryl transfer reaction), to be consistent with experimental measurements [69]. It is noted that the error rates related to most non-cognate NTP replacements range around 10−5 ~ 10−6 in T7 RNAP [69]. In comparison, the RNAP selection against dATP over ATP proceeds partially at the initial screening or first checkpoint, via the pre-insertion off-path dissociation, and then partially during the nucleotide insertion. Since dATP replacing ATP error rate was measured ~10−2 [70], our calculation then suggests that catalytic selection does not necessarily happen for the non-cognate dATP over the cognate ATP [68]. Hence, one can see that using both the pre-insertion and insertion as checkpoints for the nucleotide selection in T7 RNAP, the fidelity can be well controlled.
In the theoretical analyses for template-based polymerization or elongation fidelity control in general, we have emphasized two essential properties: (i) The initial screening or the first selection (e.g at pre-insertion) is energetically efficient to possibly support both high speed (or elongation rate) and high fidelity (low error rate) [3]; (ii) The rate-limiting transition (e.g. the nucleotide insertion) is substantial for the nucleotide selection, and modulating the rate of such transition impacts significantly on the fidelity or accuracy (i.e. the error rate) [30]. Indeed, too slow the rate-liming transition would suppress the non-equilibrium driving force, and too fast such transition cannot allow well differentiation for the nucleotide species, and both of which would lead to non-optimal error rates or fidelity control. Hence, a tradeoff to a medium rate of the rate-limiting transition is needed to balance the elongation speed and accuracy [30].
5. Human mitochondrial RNAP to be resistant to antiviral nucleotide analogues
Human mitochondrial transcription also uses a single-subunit RNAP (or POLRMT) as the core engine, which is distantly linked and structurally similar to phage T7 RNAP and family A DNAPs [11]. A similar fingers subdomain and the O-helix on the POLRMT can be well aligned with that in T7 RNAP (see Fig. 1E). Accordingly, the NTP substrate binding and selection mechanisms can be similar in the mitochondrial or mt-RNAP and T7 RNAP. Tyr639 from T7 RNAP has its counterpart as Tyr999 in POLRMT. It was captured that the O-helix (986–1000) binds a sulphate ion with Arg987, Lys991 and Lys853 [71] (corresponding to Arg627, Lys631, and Lys472 in T7 RNAP; see Fig. 2E and D). Asp922 and Asp1151 are the two highly conserved catalytic residues (from motif A and C) for coordinating two magnesium ion activities for catalysis. A clenched conformation (~25° rotation) of the fingers subdomain around the O-helix axis was identified in the initiation complex structure [71]. In the crystal structure of the elongation (e.g. pre-translocation) complex of POLRMT, the active site is widened by rotations of both the palm and fingers subdomains (10° and 15°) [22]. Based on further modeling, it is suggested that NTP 2′-OH may be contacted by Y999 to allow nucleotide selection.
Biochemical studies indicated that the non-cognate GTP replacing ATP error rate for POLRMT is also ~10−4 (dATP replacing ATP ~10−3), similar or slightly better than T7 RNAP, though the kinetics of two systems are quite different (similar KM but kpol much larger in T7 RNAP) [69]. For the NAC, both the pre-chemistry (i.e., the open to close transition; ~80 s−1) and chemistry (~150 s−1) steps of the PLORMT are partially rate-limiting; a post-chemistry step (closed to open) is essentially rate-limiting (15−1); a fast reverse of the pyrophosphate release or rebinding seems to allow pyrophosphorolysis to serve for fidelity control even after the chemistry step [11]. Single molecule experiments had also suggested pausing or backtracking in a mt-RNAP (Rpo41 from yeast) comparable to that of multi-subunit RNAP II [72], with the backtracking usually a link to proofreading activity in the multi-subunit RNAPs. Interestingly, our previous modeling and simulation suggested that mutation of six residues on the O-helix of T7 RNAP to those in the mt-RNAP can lead to less O-helix opening at pre-translocation to possibly allow RNAP backtracking [64]. The mechanism is related to a hypothesis that the O-helix oscillation to opening at pre-translocation would prevent backtracking.
For antiviral drug developments targeting on viral RdRp, it is of high concern about potential drug toxicity or side effects to the human polymerases [73], [74], especially, to the single-subunit hand-shape POLRMT, as it is structurally similar to the viral RdRps, i.e., the antiviral drug target. In particular, the nucleotide analogues as non-obligate chain terminators to viral RdRps may also terminate POLRMT, if being incorporated to the synthesizing RNA. The proved drug RDV and SBV (remdesivir and sofosbuvir) are the exemplary nucleotide analogues, however, causing no obvious side effects to POLRMT. It is therefore important to understand if it is due to the improved fidelity control or particular nucleotide selectivity of POLRMT that prevents the RDV-TP or SBV-TP inhibition. Computational work starts to shed light by predicting drug impacts on the POLRMT via calculating the relative binding free energy upon the nucleotide analog entry to the polymerase [75]. However, as the full nucleotide selectivity comes from multiple checkpoints in the NAC, further investigations seem necessary to reveal systematical fidelity control mechanisms of POLRMT.
6. Phi29 DNA polymerase with exonuclease subdomain for intrinsic proofreading
Many double-stranded DNA viruses that infect either prokaryotes or eukaryotes have the B family polymerase as in eukaryotes [76]. Bacteriophage phi29 DNAP is a hand-shape B-family polymerase with very high processivity along DNA [77], [78]. Phi29 DNAP has an unusual thumb subdomain (small, with 45 residues, 530 to 575; see Fig. 1F), which works as a clamp in concert with other part of the protein to encircle the upstream duplex DNA, i.e., to enhance the processivity. Different from other DNAPs, phi29 DNAP has capability to unwind downstream DNA with intrinsic helicase function, which also assists its high processivity [79]. Essentially, Asp249 and Asp458 are identified as the catalytic carboxylates, Tyr254 as the steric gate to distinguish dNTP and NTP, along with Try390 occupying the NTP insertion site in an apo protein (see Fig. 2F). Starting from a fully open conformation of the polymerase, ~19° rotation of the fingers subdomain of the DNAP leads to the closed conformation, in which Lys371 (pre-B motif), Lys379, and Lys383 (motif B) contact the triphosphate moiety of the incoming dNTP, and Tyr500 (from motif KxY) contacts the priming nucleotide. In contrast with the A-family polymerases with a pre-insertion site (e.g. in T7 RNAP) for initial NTP binding, which is occupied by a conserved tyrosine (e.g Tyr639) to unstack the template nucleotide, the B-family DNAP has been suggested to bind NTP directly into the insertion site, as there is no such tyrosine counterpart to assist the template nucleotide unstacking [23]. In the NTP insertion or substrate state, the NTP together with its template pairing nucleotide stack on the Tyr254, along with Tyr390 nearby [23]. An incoming NTP indeed brings ~14° rotation of the fingers subdomain from the post-translocated state (open or semi-open) to the substrate state (closed) [23]. Re-opening of the fingers subdomain would let Tyr390 to occupy the insertion site again, where Tyr390 and Tyr254 form stable stacking. In addition, rotation of Tyr390 can also break hydrogen bonding with Tyr226 (from motif I/YxGG/A) that is involved in template and primer DNA binding.
Aside from the polymerase region (190–575) with the palm, fingers, and thumb subdomains similar to that of the above RdRps or RNAPs, phi29 DNAP also has an exonuclease subdomain (5–189) at N-terminal region to support its intrinsic proofreading activity (see Fig. 1F). Note that the replicating DNAPs usually have much more stringent fidelity control (i.e., with error rates ranging 10−6 to 10−9) than that of RNAPs or RdRps. The fidelity control of the replicating phi29 DNAP had been examined previously, with nucleotide insertion selectivity, mismatch elongation and proofreading activities well identified [12], [80]. The exonuclease subdomain is structurally conserved across A, B, and C families of polymerases, in which two metal ions are bound with four catalytic carboxylates (Asp12, Glu14, Asp66, and Asp169) [78]. Structural and mutation studies further show that Thr15, Asn62, Phe65, Tyr148, and Leu567 directly contact ssDNA, mutations of Try59, Phe69, Ser122, His61 impact on ssDNA binding, and mutations of Lys143 and Try165 away from the active site also lower the exonuclease activities without changing much the ssDNA affinities.
The proofreading dynamics of Phi29 had also been measured at single molecule level [81], suggesting the DNA primer transferring via two intermediate conformations, one with mechanic force sensitivity and one paused for fidelity check. More recent work shows that Tyr101 and Thr189 particularly coordinate polymerization and exonuclease activities via modulating processivity and fidelity check, respectively [82]. The switching dynamics from polymerization to editing/exonuclease state in a comparable high-fidelity DNAP is also approached computationally lately, with above milliseconds of the switching dynamics and transition path explored [83]. The mechanisms revealed from primer transfer dynamics in the above DNAP system may not directly suggest how comparable events happen in the proofreading of the SARS-CoV2 replication system, as a separate ExoN protein nsp14 is needed to be assembled with nsp12, i.e., the CoV-2 RdRp, to achieve such a function. Nevertheless, lessons learned from Phi29 DNAP as the minimal viral replication machine would help to understand comparatively how fidelity control mechanisms evolve in diverse single-subunit hand-shaped polymerases, which might be evolutionarily connected. Interestingly, a recent work suggests that RNAPs and replicative DNAPs had evolved from a common ancestor that functioned as an RdRp in the RNA-protein world that predated the advent of DNA replication [84].
In a broad context, one can also compare multi-subunit RNAPs from bacteria and eukaryotes with the single-subunit polymerases addressed above. Despite striking differences in the overall molecular shape, crucial structure elements, and regulatory mechanisms between the multi-subunit and single-subunit polymerases, one can still find their essential similarities, e.g., the active sites harboring the metal ions for catalytic addition of NTP, the entry and exit tunnels for NTP recruitment and RNA transcript/primer, respectively [85], [86]. In addition to fidelity control mechanisms addressed previously [2], [4], comparative studies on the nucleo-sugar selection of the multi-subunit RNAPs with respect to the single-subunit species, for example, have been conducted recently [87]. In particular, backtracking related proofreading mechanism plays an important role in the multi-subunit RNAPs for improving fidelity [2], [88]. Notably, comparable backtracking events have also been identified quite recently in the SARS-CoV-2 replication-transcription complex consisting of the RdRp nsp12 and the helicase nsp13 [89], [90], using cryo-EM techniques and unbiased MD simulation methods.
7. Conclusion
The viral polymerases we have discussed here all contain fingers, palm, and thumb subdomains, and these single-subunit enzymes are more or less connected evolutionarily [91], [92], [93]. Among the RdRps, seven conserved motifs have been identified, with A-E in the palm subdomain, F&G in the fingers subdomain. The A-D motifs can be also identified in T7 RNAP and human mt-RNAP or POLRMT, which are similar to A-family polymerases, while phi29 DNAP belongs to B-family polymerases. In the viral RdRps, motif A, B, D are mainly responsible for incoming NTP binding and hence the selection. Additionally, motif F&G can interact closely with template nucleotide to be paring with the incoming NTP. Hence, motif F&G may also participate in the nucleotide selection. Such selectivity is usually conducted upon initial NTP binding, during NTP insertion, and via the catalytic step. A pre-insertion site is located in T7 RNAP and some other A-family polymerases [18], [21], [23], e.g., as a conserved tyrosine assists unstacking the template nucleotide from the upstream DNA. Such a pre-insertion site allows efficient initial selection or screening of the nucleotide species, for example, via on-path (trapped, with dissociation barrier) and off-path (non-trapped, no dissociation barrier) configurations for the cognate and non-cognate nucleotides, respectively, as revealed from our previous modeling and simulations [65], [68]. No such a pre-insertion site exists in the viral RdRps or B-family polymerases, so it is to be determined quantitatively how initial nucleotide screening is possibly conducted in these systems. Further nucleotide selection relies substantially on the nucleotide insertion step, which is accompanied by open to close protein subdomain or active-site conformational transition, pre-chemically, and is often rate-limiting (or partially rate-limiting). In the viral RdRp, only subtle conformational changes around the active site or in the palm subdomain are expected, while A-family polymerases can have large fingers subdomain rotation to accompany with the nucleotide insertion and selection. The corresponding energetics have been revealed computationally for T7 RNAP [68], and can be explored similarly for viral RdRp systems. The catalytic nucleotide selection can be probed from experimental measurements (i.e., on kpol or kmax), yet it is complicated by whether the catalytic step is rate-limiting (or partially rate-limiting); an exact quantification may require quantum chemical calculations. Besides, from knowledges of the viral RdRp systems, post-chemical nucleotide selections such as pyro-phosphorolysis and translocation related chain termination are also feasible, which add further complications to the nucleotide selectivity of these systems. Last, proofreading or editing that serves for fidelity improvements turns out to be critical, in particular, for the large-genome coronavirus replication systems, thus it awaits further investigations for antiviral developments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Current work is supported by National Science Foundation (NSF) Award #2028935. This work also used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. JY has also been supported by the CMCF of UCI via NSF DMS 1763272 and the Simons Foundation grant #594598 and start-up funding from UCI. CL is supported by National Natural Science Foundation of China grant #12005029 and Natural Science Foundation of Chongqing Grant #cstc2020jcyj-msxmX0811 and the Start-up Founding of Chongqing University of Posts and Telecommunication (A2020-029).
References
- 1.Joyce C.M. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sydow J.F., Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Yu J. Efficient fidelity control by stepwise nucleotide selection in polymerase elongation Abstract: Polymerases select nucleotides. Comput Math Methods Med. 2014;2:141–160. [Google Scholar]
- 4.Yuzenkova Y., Bochkareva A., Tadigotla V.R., Roghanian M., Zorov S., Severinov K. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steitz T.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson K.A. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 7.Ahlquist P., Noueiry A.O., Lee W.-M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco-Hernandez R., Jácome R., López Vidal Y. Ponce de León S: Are RNA viruses candidate agents for the next global pandemic? A review. ILAR J. 2017;58:343–358. doi: 10.1093/ilar/ilx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y.W., Steitz T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 10.Long C., Chao E., Da L.-T., Yu J. A viral t7 RNA polymerase ratcheting along DNA with fidelity control. Comput Struct Biotechnol J. 2019;17:638–644. doi: 10.1016/j.csbj.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold J.J., Smidansky E.D., Moustafa I.M., Cameron C.E. Human mitochondrial RNA polymerase: structure–function, mechanism and inhibition. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2012;1819:948–960. doi: 10.1016/j.bbagrm.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Esteban J.A., Salas M., Blanco L. Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 13.Bębenek A., Ziuzia-Graczyk I. Fidelity of DNA replication—a matter of proofreading. Curr Genet. 2018;64:985–996. doi: 10.1007/s00294-018-0820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo E., Holland J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 15.Fitzsimmons W.J., Woods R.J., McCrone J.T., Woodman A., Arnold J.J., Yennawar M. A speed–fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020 doi: 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells V.R., Plotch S.J., DeStefano J.J. Determination of the mutation rate of poliovirus RNA-dependent RNA polymerase. Virus Res. 2001;74:119–132. doi: 10.1016/s0168-1702(00)00256-2. [DOI] [PubMed] [Google Scholar]
- 18.Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 20.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Temiakov D., Patlan V., Anikin M., McAllister W.T., Yokoyama S., Vassylyev D.G. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 22.Schwinghammer K., Cheung A.C., Morozov Y.I., Agaronyan K., Temiakov D., Cramer P. Structure of human mitochondrial RNA polymerase elongation complex. Nat Struct Mol Biol. 2013;20:1298–1303. doi: 10.1038/nsmb.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman A.J., Kamtekar S., Goodman J.L., Lazaro J.M., de Vega M., Blanco L. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. The EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., Peersen O.B. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J Virol. 2015;89:275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J., Perryman J.M., Yang X., Liu X., Musser D.M., Boehr A.K. Rational control of poliovirus RNA-dependent RNA polymerase fidelity by modulating motif-D loop conformational dynamics. Biochemistry. 2019;58:3735–3743. doi: 10.1021/acs.biochem.9b00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Smidansky E.D., Maksimchuk K.R., Lum D., Welch J.L., Arnold J.J. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure. 2012;20:1519–1527. doi: 10.1016/j.str.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehr D.D., Arnold J.J., Moustafa I.M., Cameron C.E. Structure, dynamics, and fidelity of RNA-dependent RNA polymerases. Nucleic Acid Polymerases. Springer. 2014:309–333. [Google Scholar]
- 29.Moustafa I.M., Shen H., Morton B., Colina C.M., Cameron C.E. Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J Mol Biol. 2011;410:159–181. doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long C., Yu J. Balancing Non-Equilibrium driving with nucleotide selectivity at kinetic checkpoints in polymerase fidelity control. Entropy. 2018;20:306. doi: 10.3390/e20040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sesmero E., Thorpe I.F. Using the hepatitis C virus RNA-dependent RNA polymerase as a model to understand viral polymerase structure, function and dynamics. Viruses. 2015;7:3974–3994. doi: 10.3390/v7072808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selisko B., Papageorgiou N., Ferron F., Canard B. Structural and functional basis of the fidelity of nucleotide selection by flavivirus RNA-dependent RNA polymerases. Viruses. 2018;10:59. doi: 10.3390/v10020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutartre H., Bussetta C., Boretto J., Canard B. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob Agents Chemother. 2006;50:4161–4169. doi: 10.1128/AAC.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu S., Doehle B., Rajyaguru S., Han B., Barauskas O., Feng J. In vitro selection of resistance to sofosbuvir in HCV replicons of genotype-1 to-6. Antivir Ther. 2017;22:587–597. doi: 10.3851/IMP3149. [DOI] [PubMed] [Google Scholar]
- 35.Jin Z., Leveque V., Ma H., Johnson K.A., Klumpp K. NTP-mediated nucleotide excision activity of hepatitis C virus RNA-dependent RNA polymerase. Proc Natl Acad Sci. 2013;110:E348–E357. doi: 10.1073/pnas.1214924110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villalba B., Johnson K.A. Rate-limiting pyrophosphate release by hepatitis C virus polymerase NS5B improves fidelity. J Biol Chem. 2020;295:16436–16444. doi: 10.1074/jbc.RA120.015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis B.C., Thorpe I.F. Thumb inhibitor binding eliminates functionally important dynamics in the hepatitis C virus RNA polymerase. Proteins Struct Funct Bioinf. 2013;81:40–52. doi: 10.1002/prot.24154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis B.C., Thorpe I.F. Molecular simulations illuminate the role of regulatory components of the RNA polymerase from the hepatitis C virus in influencing protein structure and dynamics. Biochemistry. 2013;52:4541–4552. doi: 10.1021/bi400251g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouirane K.B., Boulard Y., Bressanelli S. The hepatitis C virus RNA-dependent RNA polymerase directs incoming nucleotides to its active site through magnesium-dependent dynamics within its F motif. J Biol Chem. 2019;294:7573–7587. doi: 10.1074/jbc.RA118.005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruenn J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masters P.S. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discovery. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V, Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020:1–16. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C.-C., Chen M.-Y., Chang Y.-L. Potential therapeutic agents against COVID-19: What we know so far. Journal of the Chinese Medical Association. 2020 doi: 10.1097/JCMA.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(417–428) doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 50.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Zhang D., Wang X., Yuan C., Li Y., Jia X. 1'-Ribose cyano substitution allows Remdesivir to effectively inhibit nucleotide addition and proofreading during SARS-CoV-2 viral RNA replication. Phys Chem Chem Phys. 2021;23:5852–5863. doi: 10.1039/d0cp05948j. [DOI] [PubMed] [Google Scholar]
- 57.Shannon A. Le NT-T, Selisko B, Eydoux C, Alvarez K, Guillemot J-C, Decroly E, Peersen O, Ferron F, Canard B: Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moeller NH, Shi K, Demir Ö, Banerjee S, Yin L, Belica C, Durfee C, Amaro RE, Aihara H: Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. bioRxiv 2021. [DOI] [PMC free article] [PubMed]
- 59.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J Phys Chem B. 2020;124:6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 61.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 62.Duan B., Wu S., Da L.-T., Yu J. A critical residue selectively recruits nucleotides for T7 RNA polymerase transcription fidelity control. Biophys J. 2014;107:2130–2140. doi: 10.1016/j.bpj.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Da L.-T. E C, Duan B, Zhang C, Zhou X, Yu J: A jump-from-cavity pyrophosphate ion release assisted by a key lysine residue in T7 RNA polymerase transcription elongation. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Da L.-T. E C, Shuai Y, Wu S, Su X-D, Yu J: T7 RNA polymerase translocation is facilitated by a helix opening on the fingers domain that may also prevent backtracking. Nucleic Acids Res. 2017;45:7909–7921. doi: 10.1093/nar/gkx495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chao E., Duan B., Yu J. Nucleotide Selectivity at a Preinsertion Checkpoint of T7 RNA Polymerase Transcription Elongation. J Phys Chem B. 2017;121:3777–3786. doi: 10.1021/acs.jpcb.6b11668. [DOI] [PubMed] [Google Scholar]
- 66.Wu S., Wang J., Pu X., Li L., Li Q. T7 RNA polymerase discriminates correct and incorrect nucleoside triphosphates by free energy. Biophys J. 2018;114:1755–1761. doi: 10.1016/j.bpj.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anand V.S., Patel S.S. Transient state kinetics of transcription elongation by T7 RNA polymerase. J Biol Chem. 2006;281:35677–35685. doi: 10.1074/jbc.M608180200. [DOI] [PubMed] [Google Scholar]
- 68.Long C. E C, Da L-T, Yu J: Determining selection free energetics from nucleotide pre-insertion to insertion in viral T7 RNA polymerase transcription fidelity control. Nucleic Acids Res. 2019;47:4721–4735. doi: 10.1093/nar/gkz213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sultana S., Solotchi M., Ramachandran A., Patel S.S. Transcriptional fidelities of human mitochondrial POLRMT, yeast mitochondrial Rpo41, and phage T7 single-subunit RNA polymerases. J Biol Chem. 2017;292:18145–18160. doi: 10.1074/jbc.M117.797480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J., Brieba L.G., Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- 71.Ringel R., Sologub M., Morozov Y.I., Litonin D., Cramer P., Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 72.Zamft B., Bintu L., Ishibashi T., Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci. 2012;109:8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnold J.J., Sharma S.D., Feng J.Y., Ray A.S., Smidansky E.D., Kireeva M.L. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng J.Y., Xu Y., Barauskas O., Perry J.K., Ahmadyar S., Stepan G. Role of mitochondrial RNA polymerase in the toxicity of nucleotide inhibitors of hepatitis C virus. Antimicrob Agents Chemother. 2016;60:806–817. doi: 10.1128/AAC.01922-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freedman H., Winter P., Tuszynski J., Tyrrell D.L., Houghton M. A computational approach for predicting off-target toxicity of antiviral ribonucleoside analogues to mitochondrial RNA polymerase. J Biol Chem. 2018;293:9696–9705. doi: 10.1074/jbc.RA118.002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koonin E.V. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol Direct. 2006;1:39. doi: 10.1186/1745-6150-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salas M., Holguera I., Redrejo-Rodríguez M., de Vega M. DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Φ29 DNA Replication. Front Mol Biosci. 2016;3:37. doi: 10.3389/fmolb.2016.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamtekar S., Berman A.J., Wang J., Lázaro J.M., de Vega M., Blanco L. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage φ29. Mol Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 79.Blanco L., Bernad A., Lázaro J.M., Martín G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 80.Garmendia C., Bernad A., Esteban J.A., Blanco L., Salas M. The bacteriophage phi 29 DNA polymerase, a proofreading enzyme. J Biol Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 81.Ibarra B., Chemla Y.R., Plyasunov S., Smith S.B., Lazaro J.M., Salas M. Proofreading dynamics of a processive DNA polymerase. The EMBO journal. 2009;28:2794–2802. doi: 10.1038/emboj.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Del Prado A., Rodríguez I., Lázaro J.M., Moreno-Morcillo M., de Vega M., Salas M. New insights into the coordination between the polymerization and 3′-5′ exonuclease activities in ϕ29 DNA polymerase. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-018-37513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dodd T., Botto M., Paul F., Fernandez-Leiro R., Lamers M.H., Ivanov I. Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-19165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koonin E.V., Krupovic M., Ishino S., Ishino Y. The replication machinery of LUCA: common origin of DNA replication and transcription. BMC Biol. 2020;18:61. doi: 10.1186/s12915-020-00800-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steitz T.A. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Werner F., Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 87.Mäkinen J.J., Shin Y., Vieras E., Virta P., Metsä-Ketelä M., Murakami K.S. The mechanism of the nucleo-sugar selection by multi-subunit RNA polymerases. Nat Commun. 2021;12:796. doi: 10.1038/s41467-021-21005-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaplan C.D. The architecture of RNA polymerase fidelity. BMC Biol. 2010;8:85. doi: 10.1186/1741-7007-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malone B., Chen J., Wang Q., Llewellyn E., Choi Y.J., Olinares P.D.B. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020;182:1560–1573.e1513. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cermakian N., Ikeda T.M., Miramontes P., Lang B.F., Gray M.W., Cedergren R. On the evolution of the single-subunit RNA polymerases. J Mol Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 92.Filée J., Forterre P., Sen-Lin T., Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 93.de Farias S.T., dos Santos Junior A.P., Rêgo T.G., José M.V. Origin and evolution of RNA-dependent RNA polymerase. Front Genet. 2017;8:125. doi: 10.3389/fgene.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]