Abstract
CD4+ T helper (Th) cells play a crucial role in the modulation of innate and adaptive immune responses through the differentiation of Th precursor cells into several subsets, including Th1, Th2, Th17, and regulatory T (Treg) cells. Effector Th and Treg cells are distinguished by the production of signature cytokines and are important for eliminating intracellular and extracellular pathogens and maintaining immune homeostasis. Stimulation of naïve Th cells by T cell receptor and specific cytokines activates master transcription factors and induces lineage specification during the differentiation of Th cells. The master transcription factors directly activate the transcription of signature cytokine genes and also undergo post-translational modifications to fine-tune cytokine production and maintain immune balance through cross-regulation with each other. This review highlights the post-translational modifications of master transcription factors that control the differentiation of effector Th and Treg cells and provides additional insights on the immune regulation mediated by protein arginine-modifying enzymes in effector Th cells.
Keywords: arginine-modifying enzyme, CD4 T cell differentiation, effector Th and Treg cell, master regulatory transcription factor, post-translational modifications
INTRODUCTION
The immune system is important in defeating pathogenic antigens and preventing disease outbreaks. In particular, CD4+ T helper (Th) cells play an essential role in immune modulation through timely activation and suppression of innate and adaptive immune responses (Chemin et al., 2019; DuPage and Bluestone, 2016). CD4+ Th cells interact with other immune cells, including CD8+ cytotoxic T cells, macrophages, and B cells, and augment the innate and adaptive immune responses, exerting protective effects against tumor growth and the development of rheumatoid arthritis and atherosclerosis (Ostroumov et al., 2018; Roberts et al., 2015; Tay et al., 2019). Naïve CD4+ Th cells are generated and derived from the thymus and migrate to peripheral organs, including lymph nodes and the spleen, where they are activated by contact with antigens (Yan et al., 2017). Triggering of T cell receptor (TCR) transduces activation signals through the regulation of signaling mediators and induces the differentiation of naïve CD4+ Th cells into various types of effector and regulatory cells (Hwang et al., 2020; Zhu et al., 2010). In addition, environmental cytokines are crucial for lineage specification through the activation of lineage-specific master regulatory transcription factors (Martinez-Sanchez et al., 2018; Pawlak et al., 2020) (Fig. 1A). While stimulation with interleukin (IL)-12 and blockade of IL-4 signaling induce the development of interferon-γ (IFNγ)-producing Th1 cells, inhibition of IFNγ receptor and activation of IL-4-mediated signaling lead to Th2 cell differentiation. Under conditions that block both Th1 and Th2 cell development, stimulation with additional tumor growth factor (TGF)-β induces peripheral regulatory T (Treg) cell development, and the addition of both TGFβ and IL-6 triggers the development of IL-17-producing Th17 cells (Ruterbusch et al., 2020) (Fig. 1B). During lineage commitment of naïve CD4+ Th cells, master regulatory transcription factors such as T-box protein expressed in T cells (T-bet), GATA-binding protein 3 (GATA-3), retinoic acid-related orphan receptor gamma t (RORγt), and forkhead box P3 (FoxP3) are regulated at both the expression and activity level through post-translational modifications (PTMs), thereby producing signature cytokines. Therefore, timely and optimal regulation of the amount and activity of regulatory transcription factors in CD4+ Th cells is critical for defense against pathogenic antigens and immune homeostasis maintenance (Hsu et al., 2020). This review focuses on the understanding of PTMs in master regulatory transcription factors for Th cell lineage specification and provides new insights on the immune function of additional PTMs in Th cells.
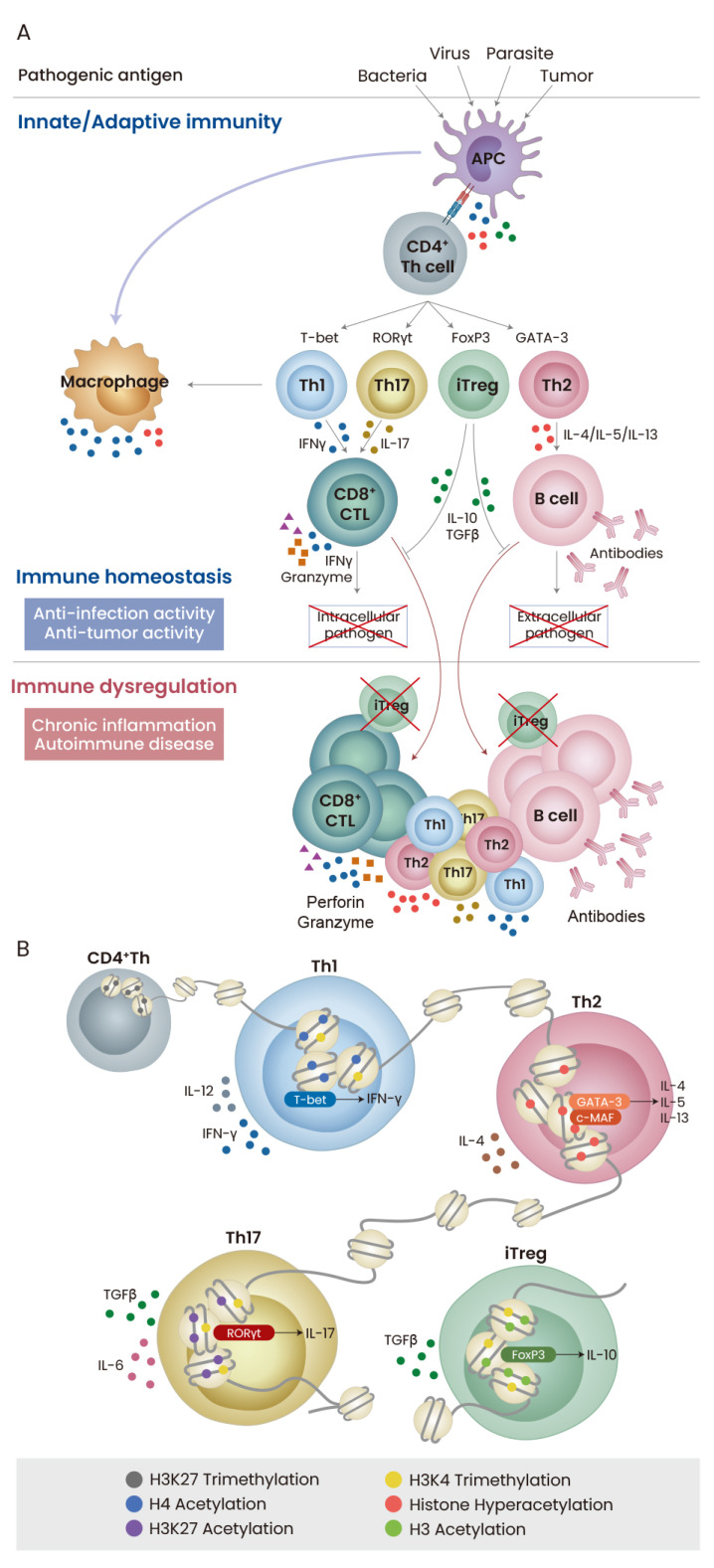
Fig. 1. Immune regulation by Th cell lineage specification.
(A) TCR triggering with pathogenic antigen provided by antigen-presenting cell (APC) induces activation and differentiation of CD4+ Th cells, which subsequently boost the activity of cytotoxic T lymphocytes (CTL) and plasma B cells. Immune response properly coordinated by effector Th and Treg cells is important for eliminating intracellular and extracellular pathogens and maintaining immune homeostasis against disease. (B) Th cell specification is epigenetically and transcriptionally modulated in response to TCR and cytokines stimulation. Despite the importance of histone modifications, the quantitative increase and activation of master transcription factors determine Th cell fate specification.
CURRENT UNDERSTANDING OF GENERAL PTMs
A large number of cellular processes and biological functions are controlled by PTMs that provide protein diversity and complexity through the addition of modifying groups. These modifications include the addition of hydroxyl groups on tyrosine, threonine, and serine, amino groups on arginine and lysine, and carboxyl groups on glutamate and aspartate (Duan and Walther, 2015). Over 200 PTMs have been identified by qualitative and quantitative analyses using mass spectrometry combined with chromatographic and other separation techniques. However, the physiological relevance of many PTMs remains largely unknown (Barber and Rinehart, 2018; Virág et al., 2020). Phosphorylation, acetylation, glycosylation, methylation, SUMOylation, and ubiquitination are the most common PTMs in regulatory proteins. They are mediated by the coordinated actions of kinases, phosphatases, transferases, and ligases and affect subcellular localization, DNA-binding activity, protein stability, and binding to partner proteins (Fig. 2). Phosphorylation is one of the most important PTMs in the regulation of cell cycle, growth, apoptosis, and signal transduction and requires kinases and phosphatases for the reversible modification of serine, threonine, or tyrosine residues. Acetylation of lysine residues, particularly in histones, is essential for the activation of gene transcription and is controlled by histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes (Glozak et al., 2005; Young et al., 2010). Intracellular glycosylation is mediated by O-glycosyltransferase (OGT) (Marth and Grewal, 2008), and protein arginine methyltransferases (PRMTs) and protein lysine methyltransferases (PKMTs) contribute to protein methylation at arginine and lysine, respectively (Guccione and Richard, 2019; Lanouette et al., 2014). Protein SUMOylation and ubiquitination are mediated by the conjugation of small ubiquitin-like modifier (SUMO) and ubiquitin, respectively, to lysine. These modifications are known to mainly mediate protein degradation but also modulate other cellular events such as subcellular localization and transcriptional activity (Wang et al., 2019; Yang et al., 2017). Additional PTMs such as oxidation, reduction, hydroxylation, carboxylation, sulfation, and lipidation have been identified to affect protein properties, but the structural and functional correlations resulting from these PTMs remain to be elucidated (Walsh and Jefferis, 2006).
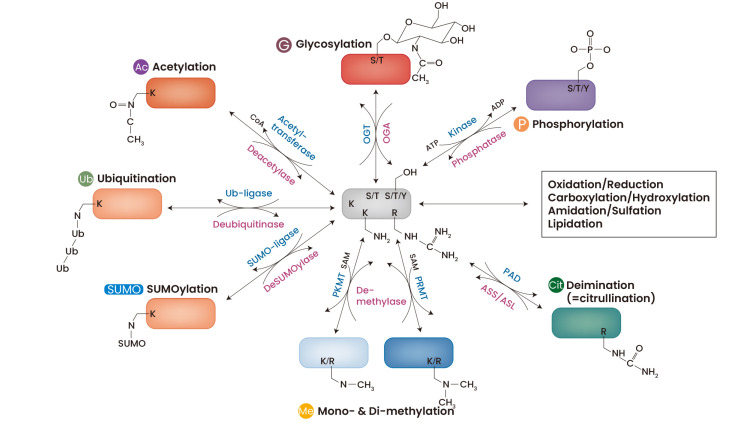
Fig. 2. General PTMs in proteins.
Specific amino acids such as Ser (S), Thr (T), Lys (K), Arg (R), and Tyr (Y) are reversibly changed by various modifying enzymes. Ser, Thr, and Tyr residues are commonly phosphorylated by kinase and dephosphorylated by phosphatase, and OGT glycosylation of Ser and Thr is induced by OGT but is reversibly removed by O-GlcNAcase (OGA). Both Lys and Arg are methylated by transferring methyl group from S-adenosylmethionine (SAM) and selectively acetylated. While Lys is conjugated with ubiquitin (ub) and SUMO by the action of ligases, Arg is converted to citrulline by PADs. Ac, acetylation; Cit, citrullination; G, glycosylation; Me, mono- & di-methylation; SUMO, SUMOylation; P, phosphorylation; ub, ubiquitination.
PTMs OF T-bet AND THEIR EFFECTS ON Th CELL DIFFERENTIATION
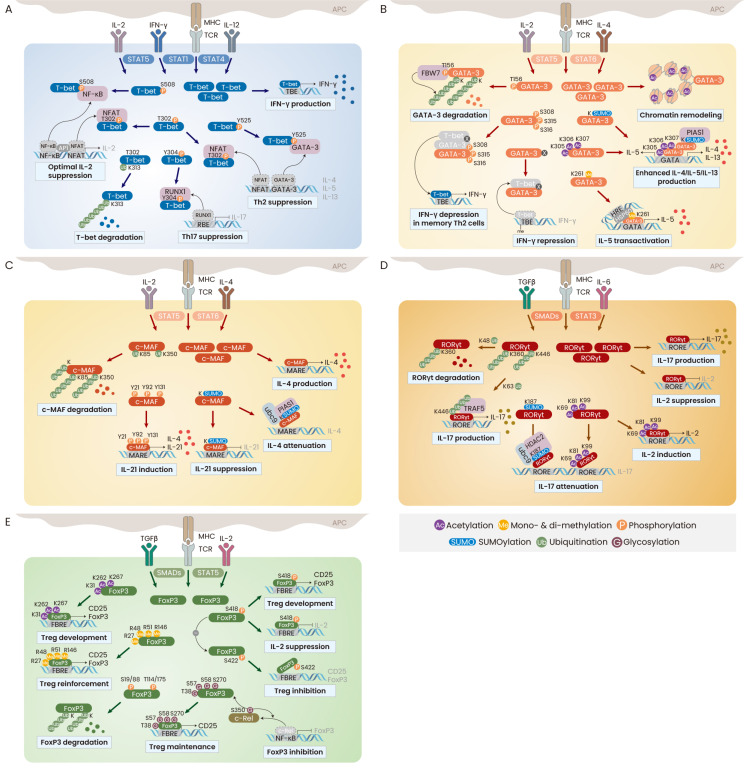
T-bet (also known as TBX21) is a master transcription factor that promotes the differentiation of naive CD4+ Th cells into Th1 cells by directly binding to the IFNγ gene promoter and activating gene transcription (Szabo et al., 2000; 2002). T-bet expression is immediately increased upon TCR stimulation and is further upregulated by treatment with IL-12 and/or IFNγ through activation of signal transducer and activator of transcription (STAT) 1 and STAT4 (Zhu et al., 2012). At the same time, a small amount of the T-bet protein is phosphorylated at Tyr525 through the activation of IL-2-inducible tyrosine kinase (ITK) and interacts with GATA-3, a Th2-specific transcription factor, leading to attenuation of GATA-3 activity and inhibition of Th2 cell development (Hwang et al., 2005b). T-bet also undergoes phosphorylation at Ser508 in Th1 cells and inhibits the transcriptional activity of nuclear factor kappa B (NF-κB) by preventing its DNA-binding to the IL-2 gene promoter. This in turn allows the production of IL-2 to be fine-tuned during Th1 cell differentiation (Hwang et al., 2005a). In addition to modifications at the C-terminal domain of the T-bet protein, PTMs of the T-box domain also regulate the DNA-binding activity and protein stability of T-bet (Jang et al., 2013). Thr302 is an important phosphorylation site, and Thr302 phosphorylation of T-bet specifically and properly restrains the expression of IL-2 and Th2 cytokines through interaction with nuclear factor of activated T cells (NFAT) under conditions of Th1 cell differentiation conditions. Furthermore, Thr302 phosphorylation is closely associated with the control of T-bet stability mediated by polyubiquitination. T-bet undergoes proteasomal degradation through ubiquitination at Lys313, whose mutation enhances T-bet expression in the nucleus, but impairs the upregulation of IFNγ due to a defect in DNA-binding activity. Mutation of Lys313 also leads to failure of Thr302 phosphorylation in T-bet, impairing the ability of T-bet to inhibit IL-2 and Th2 cytokines. In addition, multiple phosphorylation at the Tyr219, Tyr265, and Tyr304 residues in T-bet is mediated by c-Abl tyrosine kinase and is required for T-bet interaction with runt-related transcription factor 1 (RUNX1) and binding to the IFNγ gene promoter (Chen et al., 2011). Tyr304 phosphorylation in T-bet is also essential for the suppression of RUNX1-induced Th17 cell development (Lazarevic et al., 2011) (Fig. 3A). These findings suggest that T-bet plays multiple regulatory roles in the development of not only Th1, but also Th2 and Th17 cells through the fine-tuning of cytokine production, which is mediated by various PTMs. It would be interesting to study the novel PTMs of T-bet in the regulation of FoxP3 expression and Treg cell development.
Fig. 3. PTMs in master transcription factors.
(A) T-bet functions in the Th cell specification. Upon TCR and IL-12 stimulation, T-bet is subjected to PTMs, including phosphorylation at Ser, Thr, and Tyr for inducing optimal Th cell development. T-bet is also ubiquitinated at Lys and undergoes proteasomal degradation. (B) In the presence of IL-4, GATA-3 increases histone acetylation to open the chromatin structure and strongly binds to the promoter of IL-4/IL-5/IL-13 genes as a form with acetylation and SUMOylation. Methylated GATA-3 selectively promotes IL-5 and unknown modification of GATA-3 also represses IFNγ gene transcription. IFNγ repression is inhibited by multiple Ser phosphorylation, which allows partial production of IFNγ in the memory Th2 cells. The GATA-3 protein stability is also regulated by ubiquitination. (C) Stimulation with TCR and IL-4 increases c-MAF expression and enhances c-MAF-mediated IL-4 production. While ubiquitination controls the protein stability of c-MAF, Tyr phosphorylation and SUMOylation are important in context-dependent modulation of IL-21 expression. (D) Blockade of IL-4 and IFNγ signaling and stimulation with TGFβ and IL-6 induces the expression of RORγt. For Th17 cell development, RORγt enhances IL-17 transcription but inhibits IL-2 expression, whereas acetylation reverses these effects. SUMOylation and K63-linked polyubiquitination stabilize the RORγt protein to elevate IL-17 production, while K48-linked polyubiquitination induces its proteasomal degradation. (E) Strong TGFβ stimulation in the absence of IL-4 and IFNγ signaling increases various PTMs in FoxP3. Hyperacetylation, Arg methylation, O-GlcNAcylation, and Ser418 phosphorylation increase the FoxP3 stability to promote the Treg cell development. However, polyubiquitination and Ser422 phosphorylation reduce the amount and activity of FoxP3.
PTMs OF GATA-3 AND c-MAF IN THE REGULATION OF Th CELL DIFFERENTIATION
GATA-3 is highly expressed in a wide variety of cells and tissues, including T cells, neuronal cells, and fetal liver tissues, and is crucial for T cell lineage commitment, neuronal development, and hematopoiesis (Celikkaya et al., 2019; Zaidan and Ottersbach, 2018; Zhu, 2017). It is a key Th2-specific transcription factor that is upregulated by IL-4-dependent activation of STAT6 (Kaplan et al., 1996; Pai et al., 2004). GATA-3 plays important roles in chromatin remodeling of the Th2 cytokine gene locus through the induction of histone lysine acetylation, continuous production of IL-4, IL-5, and IL-13 through KRR acetylation, and timely induction of Th2 cytokines (Jenner et al., 2009; Yamagata et al., 2000; Yamashita et al., 2004). It is also known to directly inhibit IFNγ expression through repressive methylation of histone lysines at the IFNγ locus (Chang and Aune, 2007), suggesting that PTMs of GATA-3 are involved. Indeed, GATA-3 undergoes phosphorylation at Ser308, Thr315, and Ser316 by activated Akt serine/threonine kinase, which results in derepression of IFNγ expression in memory Th2 cells (Hosokawa et al., 2016). Furthermore, Arg261 methylation of GATA3 is selectively required to coordinate IL-5 gene transcription in resting Th2 cells through a specific association with heat shock protein 60 in the IL-5 gene promoter (Hosokawa et al., 2015). Not only the activity of GATA-3 in Th2 cells, but also its protein stability is regulated by PTMs. GATA-3 is ubiquitinated by interacting with F-box and WD repeat domain containing 7 (FBW7) in a phosphorylation-dependent manner at Thr156 (Kitagawa et al., 2014; Song et al., 2018). The protein inhibitor of activated STAT1 (PIAS1) is cloned as a GATA-3-interacing protein and promotes GATA-3-mediated Th2 cytokine production (Zhao et al., 2007). The GATA-3 and PIAS1 complex suggests the SUMOylation of GATA-3, but this has not yet been clarified (Fig. 3B). Identification of additional PTMs in GATA-3 would be helpful for elucidation of its functions in different types of cells, including Th17 and Treg cells and neuronal cells.
Another Th2-specific transcription factor, c-MAF, was originally identified as a cellular counterpart of a viral oncogene and has been shown to regulate cellular differentiation and developmental processes within tissues (Imbratta et al., 2020). The c-MAF protein specifically and strongly binds to the IL-4 gene promoter and induces the upregulation of IL-4 even in Th1 and B cells (Ho et al., 1996). It likely undergoes SUMOylation by directly interacting with ubc9 and PIAS1 and subsequently attenuates c-MAF-mediated IL-4 production (Leavenworth et al., 2009; Lin et al., 2010). Interestingly, SUMOylated c-MAF is essential for the suppression of IL-21 and the pathogenesis of autoimmune diabetes (Hsu et al., 2018). Unlike SUMOylation, phosphorylation of c-MAF at Tyr21, Tyr92, and Tyr131 potentiates both IL-4 production and IL-21 promoter activity, suggesting the potential role of c-MAF in the regulation of Th2- and Th17-mediated immune response (Liu et al., 2015). The protein stability of c-MAF is regulated by general ubiquitin-mediated proteasomal degradation (Zhang et al., 2016) (Fig. 3C).
PTMs OF RORγt FOR Th17 CELL DIFFERENTIATION
Although T-bet and GATA-3 are upregulated upon TCR stimulation, RORγt is increased by additional stimulation with TGFβ and IL-6 in naïve Th cells and induces IL-17 production and subsequent Th17 cell development (Ivanov et al., 2006). RORγt undergoes acetylation at Lys69, Lys81, Lys99, and Lys112 by exogenous p300 expression, but acetylated Lys69, Lys81, and Lys99 residues are deacetylated by histone deacetylase HDAC1 and lysine deacetylase sirtuin 1 (SIRT1) in Th17 cells (Lim et al., 2015; Wu et al., 2015). Deacetylated RORγt is essential for IL-17 production and IL-2 suppression in Th17 cells. The absence of SIRT1 and mutation in RORγt lysines fails to increase IL-17 production and suppress IL-2 production in Th17 cells (Lim et al., 2015). In addition, RORγt undergoes ubiquitination, but the functions of polyubiquitinated RORγt are contradictory. RORγt undergoes proteasomal degradation through Lys48-linked polyubiquitination by the E3 ubiquitin ligase Itch, whereas Itch deficiency results in RORγt accumulation and leads an increase in IL-17 level (Kathania et al., 2016; Rutz and Ouyang, 2016). However, Lys63-linked polyubiquitination stabilizes RORγt through interaction with tumor necrosis factor (TNF) receptor-associated factor 5 (TRAF5), and is essential for promoting IL-17 expression (Wang et al., 2015). Moreover, the deubiquitinases DUBA and USP17 and the ubiquitin ligase UBR5 reciprocally modulate RORγt protein stability by inducing and blocking degradation, respectively, indicating the importance of polyubiquitination of RORγt in the regulation of IL-17 production by Th17 cells (Han et al., 2014; Rutz et al., 2015). It has recently been identified that RORγt is SUMOylated at Lys187 by the function of ubc9, which inhibits IL-17 expression via recruitment of HDAC2 to the IL-17 gene promoter (Singh et al., 2018) (Fig. 3D).
PTMs OF FoxP3 FOR Treg CELL DEVELOPMENT
Th1, Th2, and Th17 effector cells induce an active immune response to eliminate intracellular and extracellular pathogens, while Treg cells are essential to suppress enhanced immune response and maintain immune homeostasis (Asano et al., 1996; Shevyrev and Tereshchenko, 2019). The transcription factor FoxP3 is a dominant regulator that induces Treg cell development and Treg-mediated immune suppression (Hori et al., 2003). Ablation or mutation of FoxP3 leads to the development of dysfunctional Treg cells and an X-linked recessive immune disorder (Bennett et al., 2000; d'Hennezel et al., 2012; Hori et al., 2003). The transcriptional activity of FoxP3 is modulated by its association with several transcription factors, including NF-κB, NFAT, Eos, and RUNX1, which can be mediated by various PTMs of FoxP3 (Bettelli et al., 2005; Ono et al., 2007; Pan et al., 2009; van Loosdregt et al., 2013b; Wu et al., 2006). FoxP3 is phosphorylated at multiple sites of the N-and C-terminal domains by the serine/threonine protein kinases Pim-1, Pim-2, and cyclin-dependent kinase-2 (cdk2). Its phosphorylation at Ser418 upon TCR stimulation allows its DNA-binding activity to be maintained and induces its target gene transcription in Treg cells (Nie et al., 2013). However, under inflammatory conditions, protein phosphatase-1 and Pim-1 kinase induced by TNFα and IL-6, respectively, promote the dephosphorylation of FoxP3 at Ser418. In addition, FoxP3 phosphorylation at Ser422 results in the inhibition of FoxP3 binding to the target gene promoter and the reversal of FoxP3-mediated gene expression (Li et al., 2014; Nie et al., 2013). Moreover, FoxP3 is phosphorylated at multiple sites in the N-terminal domain, including Ser19 and Ser175, by cdk2 and Pim-2, which negatively regulate FoxP3 protein stability and activity (Deng et al., 2015; Morawski et al., 2013). The protein stability of FoxP3 is additionally modulated by acetylation/deacetylation and ubiquitination (Beier et al., 2011; Li et al., 2007; van Loosdregt et al., 2013a). The HAT p300 and the HDAC SIRT1 reciprocally regulate the protein stability of FoxP3 through inhibition and induction of ubiquitin-proteasomal degradation, respectively (Kwon et al., 2012; van Loosdregt et al., 2010; 2011; Wang et al., 2009). FoxP3 undergoes protein degradation through the polyubiquitination of multiple lysines, which is suppressed by the deubiquitinases USP7 and USP21 (Li et al., 2016; van Loosdregt et al., 2013a). Interestingly, FoxP3 undergoes O-linked glycosylation (O-GlcNAcylation) via the addition of N-acetylglucosamine (GlcNAc), which contributes to the FoxP3 protein stability and STAT5 activation in Treg cells (Liu et al., 2019). By contrast, O-GlcNAcylation of c-Rel at Ser350 suppresses FoxP3 gene transcription through repression of its binding to the FoxP3 gene promoter (de Jesus et al., 2021). Simultaneously, O-GlcNAcylation of c-Rel enhances the c-Rel-mediated expression of IL-2 and IFNγ (Ramakrishnan et al., 2013). Further studies are required to identify the effect of O-GlcNAcylation on T cell transcription factors and their functions in immune response (Chang et al., 2020). Furthermore, FoxP3 is arginine mono-and di-methylated at Arg48 and Arg51 by PRMT1 and at Arg27, Arg51, and Arg146 by PRMT5, which are essential for enhancing FoxP3-mediated Treg cell function (Kagoya et al., 2019; Nagai et al., 2019) (Fig. 3E).
NEW INSIGHTS INTO PROTEIN ARGININE MODIFICATIONS DURING Th CELL DIFFERENTIATION
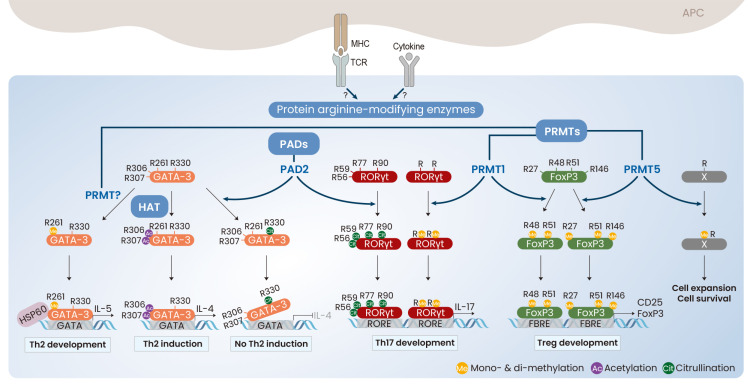
Arginine and lysine are positively charged basic amino acids abundantly found in cellular proteins, and important for controlling protein stability by forming electrostatic interactions on the protein surfaces (Kumar et al., 2000). In particular, the arginine residue provides the protein structure with more stability than lysine due to three asymmetrical nitrogen atoms (Donald et al., 2011). While lysine modifications include acetylation, succinylation, methylation, and ubiquitination, arginine residues in many proteins limitedly undergo citrullination and methylation. Arginine modifications are commonly in histones and important for DNA replication, repair, and transcription. Defects in protein arginine-modifying enzymes and wrongly modified arginine residues in histones are closely associated with the development of arthritis, cancer, heart disease, diabetes, and neurodegenerative diseases (Yang and Bedford, 2013). Arginine modifications in non-histone proteins has only recently emerged, and there is an increasing body of that arginine-modifying enzymes and arginine modifications in non-histone proteins are important for cellular signaling and immune regulation (Boisvert et al., 2005; Curran et al., 2020; Fuhrmann et al., 2015; Swiercz et al., 2007). Protein arginine residues are converted into citrulline via the action of protein arginine deiminases (PADs), which play an important role in the pathogenesis of Th cell-associated autoimmune diseases (Curran et al., 2020; Liu et al., 2018). PAD2 induces the citrullination of both GATA-3 and RORγt. Although the methylation and acetylation of GATA-3 at the arginine residues enhance IL-4 and IL-5 production, citrullination of GATA-3 at Arg330 decreases GATA-3 binding to the promoter of Th2 cytokine genes. However, citrullination of RORγt at 4 arginine residues (Arg56, Arg59, Arg77, and Arg90) strengthens its DNA-binding activity and IL-17 production (Sun et al., 2019). PAD4 expression is also associated with the severity of Th17-related rheumatoid arthritis, but the detailed molecular mechanisms have not yet been elucidated (Harris et al., 2008). Arginine conversion to citrullination seems to activate Th17 cell development but suppress Th2 cell differentiation (Fig. 4). In addition, monomethylation and asymmetric dimethylation of FoxP3, which are caused by PRMT1 at Arg48 and Arg51 enhance FoxP3 expression and promote the immune suppressive activity of Treg cells (Kagoya et al., 2019). PRMT1 directly associates with RORγt and promotes Th17 cell development, whereas PRMT1 inhibition selectively decreases the number of Th17 cells but increases that of FoxP3+ Treg cells through reciprocal recruitment of STAT3 and STAT5 (Sen et al., 2018). In addition, PRMT5 is suggested to be a critical regulator of the expansion and survival of Th cells through monomethylation and asymmetric dimethylation (Snyder et al., 2020; Tanaka et al., 2020; Webb et al., 2017). However, the factors that are methylated by PRMTs during Th cell differentiation remain to be clarified. Therefore, we suggest that protein arginine modifications in transcription factors play positive roles in the activation of Th cell lineage specification and immune response via activation of their DNA-binding and transcriptional activities and suppression of inactivation by other PTMs.
Fig. 4. Protein arginine-modifications in CD4+ Th cells.
Either TCR stimulation or cytokines may induce the expression and activity of protein arginine-modifying enzymes in CD4+ Th cells. PADs induce the conversion of Arg to citrulline of GATA-3 and RORγt and PRMTs integrate monomethyl and dimethyl group into Arg of GATA-3, RORγt, and FoxP3. In addition to the methylation and citrullination, GATA-3 is additionally acetylated by p300 HAT and subsequently promotes the Th2 cell development. Ac, acetylation; Cit, citrullination; G, glycosylation; Me, mono- & di-methylation.
CONCLUSION
In this review, we summarized the current understandings of PTMs of several transcription factors that play crucial roles in the Th cell lineage specification and modulation of immune functions of effector Th (Th1/Th2/Th17) and Treg cells. PTMs, including phosphorylation, acetylation, methylation, O-GlcNAcylation, SUMOylation, and ubiquitination alter the DNA-binding activity of the transcription factors and regulate their interactions with other proteins, affecting subcellular localization, transcriptional activity, and protein stability. We additionally focused on immune regulation by protein arginine-modifying enzymes, particularly PADs and PRMTs, in Th cell differentiation. Despite the importance of PTMs in effector Th and Treg cells, several fundamental questions remain unanswered. Which of TCR stimulation and cytokine signaling induces and activates protein-modifying enzymes in Th cells? How do protein-modifying enzymes select target proteins among various transcription factors? Which protein-modifying enzymes play dominant roles in modifying transcription factors? What percentage of all proteins go through PTMs? It is unclear whether and how one type of PTM controls another in a single protein. Given the functional plasticity of Th cell subsets, further studies on the characterization and functional correlation of additional PTMs should be explored. A profound understanding of PTMs will clarify the correlation between protein structure and function and provide beneficial information for the development of preventive and therapeutic drugs based on the regulation of PTMs.
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation (2018R1A5A2025286 and 2020R1A2C2004679) funded by the Ministry of Education, Science, and Technology.
Footnotes
AUTHOR CONTRIBUTIONS
H.K.K. and M.G.J. summarized Th1-, Th2-, and Th17-specific transcription factors. E.S.H. summarized the outline and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Asano M., Toda M., Sakaguchi N., Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J. Exp. Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber K.W., Rinehart J. The ABCs of PTMs. Nat. Chem. Biol. 2018;14:188–192. doi: 10.1038/nchembio.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier U.H., Akimova T., Liu Y., Wang L., Hancock W.W. Histone/protein deacetylases control Foxp3 expression and the heat shock response of T-regulatory cells. Curr. Opin. Immunol. 2011;23:670–678. doi: 10.1016/j.coi.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., Yoshioka R., Kiyosawa H., Barker D.F., Fain P.R., Shigeoka A.O., Chance P.F. X-Linked syndrome of polyendocrinopathy, immune dysfunction, and diarrhea maps to Xp11.23-Xq13.3. Am. J. Hum. Genet. 2000;66:461–468. doi: 10.1086/302761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Dastrange M., Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., Rhie A., Richard S., Doherty A.J. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005;4:1834–1841. doi: 10.4161/cc.4.12.2250. [DOI] [PubMed] [Google Scholar]
- Celikkaya H., Cosacak M.I., Papadimitriou C., Popova S., Bhattarai P., Biswas S.N., Siddiqui T., Wistorf S., Nevado-Alcalde I., Naumann L., et al. GATA3 promotes the neural progenitor state but not neurogenesis in 3D traumatic injury model of primary human cortical astrocytes. Front. Cell. Neurosci. 2019;13:23. doi: 10.3389/fncel.2019.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Aune T.M. Dynamic changes in histone-methylation 'marks' across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat. Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- Chang Y.H., Weng C.L., Lin K.I. O-GlcNAcylation and its role in the immune system. J. Biomed. Sci. 2020;27:57. doi: 10.1186/s12929-020-00648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin K., Gerstner C., Malmstrom V. Effector functions of CD4+ T cells at the site of local autoimmune inflammation-lessons from rheumatoid arthritis. Front. Immunol. 2019;10:353. doi: 10.3389/fimmu.2019.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Lee S.M., Gao B., Shannon S., Zhu Z., Fang D. c-Abl-mediated tyrosine phosphorylation of the T-bet DNA-binding domain regulates CD4+ T-cell differentiation and allergic lung inflammation. Mol. Cell. Biol. 2011;31:3445–3456. doi: 10.1128/MCB.05383-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran A.M., Naik P., Giles J.T., Darrah E. PAD enzymes in rheumatoid arthritis: pathogenic effectors and autoimmune targets. Nat. Rev. Rheumatol. 2020;16:301–315. doi: 10.1038/s41584-020-0409-1. [DOI] [PubMed] [Google Scholar]
- de Jesus T.J., Tomalka J.A., Centore J.T., Staback Rodriguez F.D., Agarwal R.A., Liu A.R., Kern T.S., Ramakrishnan P. Negative regulation of FOXP3 expression by c-Rel O-GlcNAcylation. Glycobiology. 2021 doi: 10.1093/glycob/cwab001. 2021 Jan 12 [Epub]. https://doi.org/10.1093/glycob/cwab001. [DOI] [PMC free article] [PubMed]
- Deng G., Nagai Y., Xiao Y., Li Z., Dai S., Ohtani T., Banham A., Li B., Wu S.L., Hancock W., et al. Pim-2 kinase influences regulatory T cell function and stability by mediating Foxp3 protein N-terminal phosphorylation. J. Biol. Chem. 2015;290:20211–20220. doi: 10.1074/jbc.M115.638221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Hennezel E., Bin Dhuban K., Torgerson T., Piccirillo C.A. The immunogenetics of immune dysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J. Med. Genet. 2012;49:291–302. doi: 10.1136/jmedgenet-2012-100759. [DOI] [PubMed] [Google Scholar]
- Donald J.E., Kulp D.W., DeGrado W.F. Salt bridges: geometrically specific, designable interactions. Proteins. 2011;79:898–915. doi: 10.1002/prot.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G., Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015;11:e1004049. doi: 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M., Bluestone J.A. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat. Rev. Immunol. 2016;16:149–163. doi: 10.1038/nri.2015.18. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J., Clancy K.W., Thompson P.R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 2015;115:5413–5461. doi: 10.1021/acs.chemrev.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak M.A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Guccione E., Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019;20:642–657. doi: 10.1038/s41580-019-0155-x. [DOI] [PubMed] [Google Scholar]
- Han L., Yang J., Wang X., Wu Q., Yin S., Li Z., Zhang J., Xing Y., Chen Z., Tsun A., et al. The E3 deubiquitinase USP17 is a positive regulator of retinoic acid-related orphan nuclear receptor gammat (RORgammat) in Th17 cells. J. Biol. Chem. 2014;289:25546–25555. doi: 10.1074/jbc.M114.565291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.L., Darrah E., Lam G.K., Bartlett S.J., Giles J.T., Grant A.V., Gao P., Scott W.W., Jr., El-Gabalawy H., Jr., Casciola-Rosen L., Jr., et al. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum. 2008;58:1958–1967. doi: 10.1002/art.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho I.C., Hodge M.R., Rooney J.W., Glimcher L.H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Hosokawa H., Kato M., Tohyama H., Tamaki Y., Endo Y., Kimura M.Y., Tumes D.J., Motohashi S., Matsumoto M., Nakayama K.I., et al. Methylation of Gata3 protein at Arg-261 regulates transactivation of the Il5 gene in T helper 2 cells. J. Biol. Chem. 2015;290:13095–13103. doi: 10.1074/jbc.M114.621524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H., Tanaka T., Endo Y., Kato M., Shinoda K., Suzuki A., Motohashi S., Matsumoto M., Nakayama K.I., Nakayama T. Akt1-mediated Gata3 phosphorylation controls the repression of IFNgamma in memory-type Th2 cells. Nat. Commun. 2016;7:11289. doi: 10.1038/ncomms11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.Y., Fu S.H., Chien M.W., Liu Y.W., Chen S.J., Sytwu H.K. Post-translational modifications of transcription factors harnessing the etiology and pathophysiology in colonic diseases. Int. J. Mol. Sci. 2020;21:3207. doi: 10.3390/ijms21093207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.Y., Yeh L.T., Fu S.H., Chien M.W., Liu Y.W., Miaw S.C., Chang D.M., Sytwu H.K. SUMO-defective c-Maf preferentially transactivates Il21 to exacerbate autoimmune diabetes. J. Clin. Invest. 2018;128:3779–3793. doi: 10.1172/JCI98786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E.S., Hong J.H., Glimcher L.H. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J. Exp. Med. 2005a;202:1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E.S., Szabo S.J., Schwartzberg P.L., Glimcher L.H. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005b;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Hwang J.R., Byeon Y., Kim D., Park S.G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 2020;52:750–761. doi: 10.1038/s12276-020-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbratta C., Hussein H., Andris F., Verdeil G. c-MAF, a Swiss army knife for tolerance in lymphocytes. Front. Immunol. 2020;11:206. doi: 10.3389/fimmu.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jang E.J., Park H.R., Hong J.H., Hwang E.S. Lysine 313 of T-box is crucial for modulation of protein stability, DNA binding, and threonine phosphorylation of T-bet. J. Immunol. 2013;190:5764–5770. doi: 10.4049/jimmunol.1203403. [DOI] [PubMed] [Google Scholar]
- Jenner R.G., Townsend M.J., Jackson I., Sun K., Bouwman R.D., Young R.A., Glimcher L.H., Lord G.M. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoya Y., Saijo H., Matsunaga Y., Guo T., Saso K., Anczurowski M., Wang C.H., Sugata K., Murata K., Butler M.O., et al. Arginine methylation of FOXP3 is crucial for the suppressive function of regulatory T cells. J. Autoimmun. 2019;97:10–21. doi: 10.1016/j.jaut.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Kaplan M.H., Schindler U., Smiley S.T., Grusby M.J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Kathania M., Khare P., Zeng M., Cantarel B., Zhang H., Ueno H., Venuprasad K. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-gammat ubiquitination. Nat. Immunol. 2016;17:997–1004. doi: 10.1038/ni.3488. [DOI] [PubMed] [Google Scholar]
- Kitagawa K., Shibata K., Matsumoto A., Matsumoto M., Ohhata T., Nakayama K.I., Niida H., Kitagawa M. Fbw7 targets GATA3 through cyclin-dependent kinase 2-dependent proteolysis and contributes to regulation of T-cell development. Mol. Cell. Biol. 2014;34:2732–2744. doi: 10.1128/MCB.01549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tsai C.J., Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- Kwon H.S., Lim H.W., Wu J., Schnolzer M., Verdin E., Ott M. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J. Immunol. 2012;188:2712–2721. doi: 10.4049/jimmunol.1100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanouette S., Mongeon V., Figeys D., Couture J.F. The functional diversity of protein lysine methylation. Mol. Syst. Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V., Chen X., Shim J.H., Hwang E.S., Jang E., Bolm A.N., Oukka M., Kuchroo V.K., Glimcher L.H. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat. Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavenworth J.W., Ma X., Mo Y.Y., Pauza M.E. SUMO conjugation contributes to immune deviation in nonobese diabetic mice by suppressing c-Maf transactivation of IL-4. J. Immunol. 2009;183:1110–1119. doi: 10.4049/jimmunol.0803671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Samanta A., Song X., Iacono K.T., Bembas K., Tao R., Basu S., Riley J.L., Hancock W.W., Shen Y., et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu Y., Wang S., Han Z., Zhu F., Ni Y., Liang R., Zhang Y., Leng Q., Wei G., et al. USP21 prevents the generation of T-helper-1-like Treg cells. Nat. Commun. 2016;7:13559. doi: 10.1038/ncomms13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lin F., Zhuo C., Deng G., Chen Z., Yin S., Gao Z., Piccioni M., Tsun A., Cai S., et al. PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J. Biol. Chem. 2014;289:26872–26881. doi: 10.1074/jbc.M114.586651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.W., Kang S.G., Ryu J.K., Schilling B., Fei M., Lee I.S., Kehasse A., Shirakawa K., Yokoyama M., Schnolzer M., et al. SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J. Exp. Med. 2015;212:973. doi: 10.1084/jem.2013237805062015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.S., Tsai P.Y., Hsieh W.Y., Tsao H.W., Liu M.W., Grenningloh R., Wang L.F., Ho I.C., Miaw S.C. SUMOylation attenuates c-Maf-dependent IL-4 expression. Eur. J. Immunol. 2010;40:1174–1184. doi: 10.1002/eji.200939788,. [DOI] [PubMed] [Google Scholar]
- Liu B., Salgado O.C., Singh S., Hippen K.L., Maynard J.C., Burlingame A.L., Ball L.E., Blazar B.R., Farrar M.A., Hogquist K.A., et al. The lineage stability and suppressive program of regulatory T cells require protein O-GlcNAcylation. Nat. Commun. 2019;10:354. doi: 10.1038/s41467-019-08300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.C., Lai C.Y., Yen W.F., Lin Y.H., Chang H.H., Tai T.S., Lu Y.J., Tsao H.W., Ho I.C., Miaw S.C. Reciprocal regulation of C-Maf tyrosine phosphorylation by Tec and Ptpn22. PLoS One. 2015;10:e0127617. doi: 10.1371/journal.pone.0127617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lightfoot Y.L., Seto N., Carmona-Rivera C., Moore E., Goel R., O'Neil L., Mistry P., Hoffmann V., Mondal S., et al. Peptidylarginine deiminases 2 and 4 modulate innate and adaptive immune responses in TLR-7-dependent lupus. JCI Insight. 2018;3:e124729. doi: 10.1172/jci.insight.124729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth J.D., Grewal P.K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 2008;8:874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez M.E., Huerta L., Alvarez-Buylla E.R., Villarreal Lujan C. Role of cytokine combinations on CD4+ T cell differentiation, partial polarization, and plasticity: continuous network modeling approach. Front. Physiol. 2018;9:877. doi: 10.3389/fphys.2018.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski P.A., Mehra P., Chen C., Bhatti T., Wells A.D. Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J. Biol. Chem. 2013;288:24494–24502. doi: 10.1074/jbc.M113.467704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Ji M.Q., Zhu F., Xiao Y., Tanaka Y., Kambayashi T., Fujimoto S., Goldberg M.M., Zhang H., Li B., et al. PRMT5 associates with the FOXP3 homomer and when disabled enhances targeted p185(erbB2/neu) tumor immunotherapy. Front. Immunol. 2019;10:174. doi: 10.3389/fimmu.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H., Zheng Y., Li R., Guo T.B., He D., Fang L., Liu X., Xiao L., Chen X., Wan B., et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-alpha in rheumatoid arthritis. Nat. Med. 2013;19:322–328. doi: 10.1038/nm.3085. [DOI] [PubMed] [Google Scholar]
- Ono M., Yaguchi H., Ohkura N., Kitabayashi I., Nagamura Y., Nomura T., Miyachi Y., Tsukada T., Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- Ostroumov D., Fekete-Drimusz N., Saborowski M., Kuhnel F., Woller N. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell. Mol. Life Sci. 2018;75:689–713. doi: 10.1007/s00018-017-2686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai S.Y., Truitt M.L., Ho I.C. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Yu H., Dang E.V., Barbi J., Pan X., Grosso J.F., Jinasena D., Sharma S.M., McCadden E.M., Getnet D., et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M., Ho A.W., Kuchroo V.K. Cytokines and transcription factors in the differentiation of CD4(+) T helper cell subsets and induction of tissue inflammation and autoimmunity. Curr. Opin. Immunol. 2020;67:57–67. doi: 10.1016/j.coi.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan P., Clark P.M., Mason D.E., Peters E.C., Hsieh-Wilson L.C., Baltimore D. Activation of the transcriptional function of the NF-kappaB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 2013;6:ra75. doi: 10.1126/scisignal.2004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C.A., Dickinson A.K., Taams L.S. The interplay between monocytes/macrophages and CD4(+) T cell subsets in rheumatoid arthritis. Front. Immunol. 2015;6:571. doi: 10.3389/fimmu.2015.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruterbusch M., Pruner K.B., Shehata L., Pepper M. In vivo CD4(+) T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- Rutz S., Kayagaki N., Phung Q.T., Eidenschenk C., Noubade R., Wang X., Lesch J., Lu R., Newton K., Huang O.W., et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 2015;518:417–421. doi: 10.1038/nature13979. [DOI] [PubMed] [Google Scholar]
- Rutz S., Ouyang W. The Itch to degrade ROR-gammat. Nat. Immunol. 2016;17:898–900. doi: 10.1038/ni.3516. [DOI] [PubMed] [Google Scholar]
- Sen S., He Z., Ghosh S., Dery K.J., Yang L., Zhang J., Sun Z. PRMT1 plays a critical role in Th17 differentiation by regulating reciprocal recruitment of STAT3 and STAT5. J. Immunol. 2018;201:440–450. doi: 10.4049/jimmunol.1701654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevyrev D., Tereshchenko V. Treg heterogeneity, function, and homeostasis. Front. Immunol. 2019;10:3100. doi: 10.3389/fimmu.2019.03100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Khare P., Obaid A., Conlon K.P., Basrur V., DePinho R.A., Venuprasad K. SUMOylation of ROR-gammat inhibits IL-17 expression and inflammation via HDAC2. Nat. Commun. 2018;9:4515. doi: 10.1038/s41467-018-06924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder K.J., Zitzer N.C., Gao Y., Choe H.K., Sell N.E., Neidemire-Colley L., Ignaci A., Kale C., Devine R.D., Abad M.G., et al. PRMT5 regulates T cell interferon response and is a target for acute graft-versus-host disease. JCI Insight. 2020;5:e131099. doi: 10.1172/jci.insight.131099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N., Cao C., Tang Y., Bi L., Jiang Y., Zhou Y., Song X., Liu L., Ge W. The ubiquitin ligase SCF(FBXW7alpha) promotes GATA3 degradation. J. Cell. Physiol. 2018;233:2366–2377. doi: 10.1002/jcp.26108. [DOI] [PubMed] [Google Scholar]
- Sun B., Chang H.H., Salinger A., Tomita B., Bawadekar M., Holmes C.L., Shelef M.A., Weerapana E., Thompson P.R., Ho I.C. Reciprocal regulation of Th2 and Th17 cells by PAD2-mediated citrullination. JCI Insight. 2019;4:e129687. doi: 10.1172/jci.insight.129687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz R., Cheng D., Kim D., Bedford M.T. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Sullivan B.M., Stemmann C., Satoskar A.R., Sleckman B.P., Glimcher L.H. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nagai Y., Okumura M., Greene M.I., Kambayashi T. PRMT5 is required for T cell survival and proliferation by maintaining cytokine signaling. Front. Immunol. 2020;11:621. doi: 10.3389/fimmu.2020.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay C., Kanellakis P., Hosseini H., Cao A., Toh B.H., Bobik A., Kyaw T. B cell and CD4 T cell interactions promote development of atherosclerosis. Front. Immunol. 2019;10:3046. doi: 10.3389/fimmu.2019.03046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J., Brunen D., Fleskens V., Pals C.E., Lam E.W., Coffer P.J. Rapid temporal control of Foxp3 protein degradation by sirtuin-1. PLoS One. 2011;6:e19047. doi: 10.1371/journal.pone.0019047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J., Fleskens V., Fu J., Brenkman A.B., Bekker C.P., Pals C.E., Meerding J., Berkers C.R., Barbi J., Grone A., et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity. 2013a;39:259–271. doi: 10.1016/j.immuni.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J., Fleskens V., Tiemessen M.M., Mokry M., van Boxtel R., Meerding J., Pals C.E., Kurek D., Baert M.R., Delemarre E.M., et al. Canonical Wnt signaling negatively modulates regulatory T cell function. Immunity. 2013b;39:298–310. doi: 10.1016/j.immuni.2013.07.019. [DOI] [PubMed] [Google Scholar]
- van Loosdregt J., Vercoulen Y., Guichelaar T., Gent Y.Y., Beekman J.M., van Beekum O., Brenkman A.B., Hijnen D.J., Mutis T., Kalkhoven E., et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- Virág D., Dalmadi-Kiss B., Vékey K., Drahos L., Klebovich I., Antal I., Ludányi K. Current trends in the analysis of post-translational modifications. Chromatographia. 2020;83:1–10. [Google Scholar]
- Walsh G., Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- Wang A., Zhu F., Liang R., Li D., Li B. Regulation of T cell differentiation and function by ubiquitin-specific proteases. Cell. Immunol. 2019;340:103922. doi: 10.1016/j.cellimm.2019.103922. [DOI] [PubMed] [Google Scholar]
- Wang L., de Zoeten E.F., Greene M.I., Hancock W.W. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat. Rev. Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang J., Han L., Zhao K., Wu Q., Bao L., Li Z., Lv L., Li B. TRAF5-mediated Lys-63-linked polyubiquitination plays an essential role in positive regulation of RORgammat in promoting IL-17A expression. J. Biol. Chem. 2015;290:29086–29094. doi: 10.1074/jbc.M115.664573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb L.M., Amici S.A., Jablonski K.A., Savardekar H., Panfil A.R., Li L., Zhou W., Peine K., Karkhanis V., Bachelder E.M., et al. PRMT5-selective inhibitors suppress inflammatory t cell responses and experimental autoimmune encephalomyelitis. J. Immunol. 2017;198:1439–1451. doi: 10.4049/jimmunol.1601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Nie J., Gao Y., Xu P., Sun Q., Yang J., Han L., Chen Z., Wang X., Lv L., et al. Reciprocal regulation of RORgammat acetylation and function by p300 and HDAC1. Sci. Rep. 2015;5:16355. doi: 10.1038/srep16355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Yamagata T., Mitani K., Oda H., Suzuki T., Honda H., Asai T., Maki K., Nakamoto T., Hirai H. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J. 2000;19:4676–4687. doi: 10.1093/emboj/19.17.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Ukai-Tadenuma M., Miyamoto T., Sugaya K., Hosokawa H., Hasegawa A., Kimura M., Taniguchi M., DeGregori J., Nakayama T. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J. Biol. Chem. 2004;279:26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- Yan F., Mo X., Liu J., Ye S., Zeng X., Chen D. Thymic function in the regulation of T cells, and molecular mechanisms underlying the modulation of cytokines and stress signaling (Review) Mol. Med. Rep. 2017;16:7175–7184. doi: 10.3892/mmr.2017.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- Yang Y., He Y., Wang X., Liang Z., He G., Zhang P., Zhu H., Xu N., Liang S. Protein SUMOylation modification and its associations with disease. Open Biol. 2017;7:170167. doi: 10.1098/rsob.170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.L., Page A.J., Cooper N.J., Frisby C.L., Blackshaw L.A. Sensory and motor innervation of the crural diaphragm by the vagus nerves. Gastroenterology. 2010;138:1091–1101.e5. doi: 10.1053/j.gastro.2009.08.053. [DOI] [PubMed] [Google Scholar]
- Zaidan N., Ottersbach K. The multi-faceted role of Gata3 in developmental haematopoiesis. Open Biol. 2018;8:180152. doi: 10.1098/rsob.180152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tong J., Tang X., Juan J., Cao B., Hurren R., Chen G., Taylor P., Xu X., Shi C.X., et al. The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood. 2016;127:1676–1686. doi: 10.1182/blood-2015-07-658203. [DOI] [PubMed] [Google Scholar]
- Zhao X., Zheng B., Huang Y., Yang D., Katzman S., Chang C., Fowell D., Zeng W.P. Interaction between GATA-3 and the transcriptional coregulator Pias1 is important for the regulation of Th2 immune responses. J. Immunol. 2007;179:8297–8304. doi: 10.4049/jimmunol.179.12.8297. [DOI] [PubMed] [Google Scholar]
- Zhu J. GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Front. Immunol. 2017;8:1571. doi: 10.3389/fimmu.2017.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., Paul W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]