Summary
This article is based on the address given by the author at the 2020 virtual meeting of the American Society of Human Genetics (ASHG) on October 26, 2020. The video of the original address can be found at the ASHG website.
Main text

It feels very special to be introduced by a former president of ASHG and the very person who introduced me as a fellow to ASHG back in 2003, Professor Cynthia C. Morton. Thank you very much Cynthia for the very kind introduction, which I will come back to later in my talk.
I cannot put in words the tremendous honor it is to win the Curt Stern Award and how much it means to me, my team, my institution, and my family. While it is not my preference to have this lecture taped in my office, I am nonetheless very grateful for the opportunity to give an overview of the work I feel absolutely privileged to undertake on behalf of our entire society to achieve a better understanding of our shared heritage, that is, the human genome.
The power of the software that we call DNA to encode the making of the human body never ceases to amaze me, and even more so considering how deeply this concept has enthralled the general public and the scientific community after the momentous completion of the human genome project. It was the medical implications of the human genome project, however, that brought what once seemed far-fetched to the realm of reality: the exciting possibility that understanding this software will change medicine as we know it.
While it is true that DNA is not your destiny, there are changes in your software that will virtually guarantee an impact on your health, best exemplified by Mendelian phenotypes.
My fascination with Mendelian phenotypes dates back to 1997 when I was still a medical student, and James (Jim) Gusella came to Saudi Arabia to receive King Faisal International Prize in Medicine. He visited my medical school in Riyadh and gave an electrifying talk on how he mapped Huntington disease and eventually cracked its mysterious mutational mechanism. I was fascinated by the simple yet powerful predictability of this Mendelian variant and even more fascinated by how one could literally map a variant onto the human genome even without having a clue as to the gene’s function. I fell in love with Mendelian disorders then and there, and they continue to be my passion.
In the complex world of human genomics, Mendelian disorders epitomize precision and predictability, and that is why they are preventable. They are also, just to add another p, perplexing, because many are so rare, they are not even recognizable by the wider medical community. This is rapidly changing though thanks to the agnostic genome sequencing tools we now have at our disposal. For these tools to work for all clinicians, however, we must first establish the clinical relevance of thousands of genes in the human genome where this remains an open question, and that is one area I try to contribute to as much as possible given the unique patient population I work with.
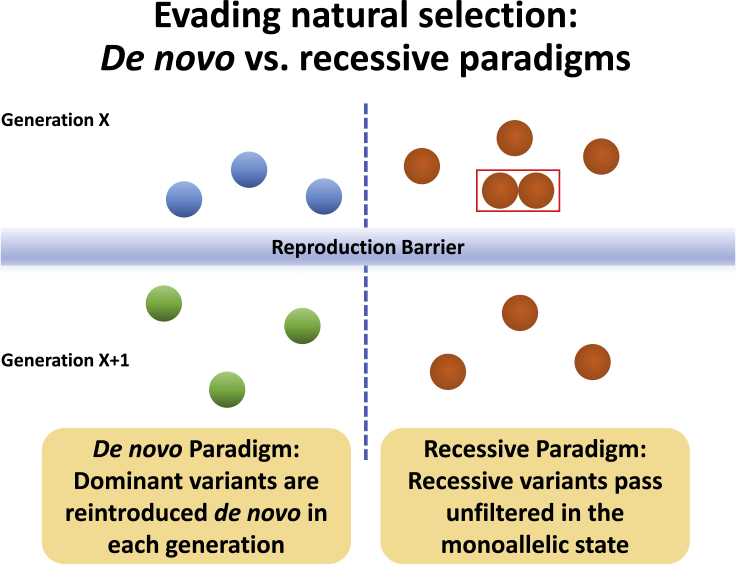
Before I talk about that population, let us first have some background. How do variants that cause Mendelian phenotypes, many of which are very severe, creep into the human gene pool when they are typically not compatible with reproduction, i.e., many have a reproductive fitness of 0? What is their trick to evade natural selection? The one trick that many of us are most familiar with is the de novo mechanism. Just as efficiently, however, recessive variants can also make their way into our gene pool because they can easily pass through the reproduction barrier in the monoallelic but not the bi-allelic state (Figure 1).
Figure 1.
Illustration of the difference between de novo and recessive paradigms in evading natural selection
This illustration assumes a reproductive fitness of 0.
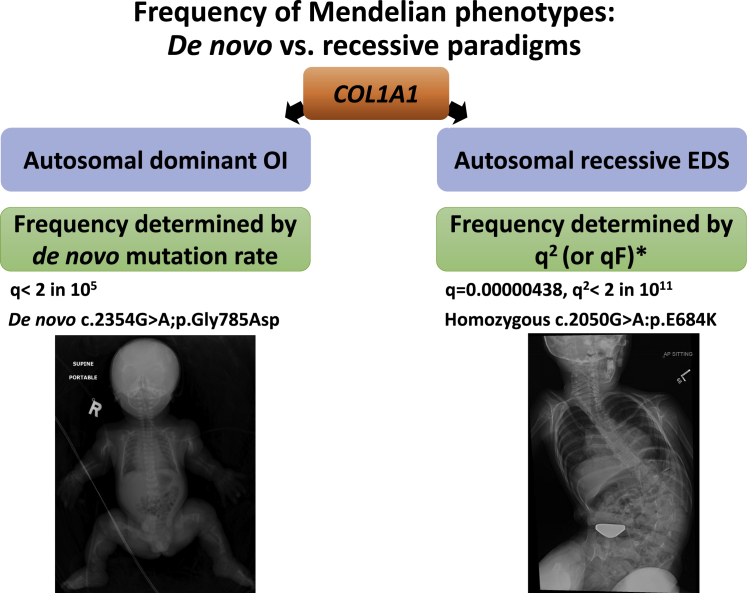
The difference in likelihood, however, between the two scenarios can be vast, as shown in the following example where we can compare the two mechanisms operating on the same gene. The frequency of osteogenesis imperfecta in the baby shown in Figure 2 is the simple function of the de novo mutation rate of COL1A1, which is straightforward, but compare that to the exceedingly rare EDS-like phenotype caused by a recessive COL1A1 variant shown in the same figure.1 It is exceedingly rare because its frequency should be the function of q2, and if q is very small, then q2 is going to be hopelessly rare, that is unless q2 is no longer the determining factor, which is precisely what happens in the consanguineous populations I have been working with for the past 13 years. In a first cousin union, if dad has that very rare recessive COL1A1 variant, then mom’s probability of being a carrier is not 1 in 200,000 but in 1 in 8. In a consanguineous population, therefore, the frequency of recessive disorders is not a function of q2, but rather qF where F is in the inbreeding coefficient.2 This means that there are gene-disease associations that are so rare the only way we can learn about them is through a dedicated effort to study consanguineous populations. Just so you have an idea on how this completely changes the Mendelian diseases landscape in consanguineous versus non-consanguineous populations, I would like you to consider three phenotypes that we know are essentially filtered out by natural selection in a single generation. Take severe intellectual disability as one such an example where the overwhelming majority of causal variants are de novo, which is the complete opposite of what we see in consanguineous populations.3 In stillbirths, you are probably familiar with a recent paper from New York where nearly every variant was dominant, again the complete opposite of the pattern we had observed in our consanguineous population, which is in a way a silver lining because we are able to trace the cause of death in the DNA of the parents when we have no DNA left from the stillbirth (molecular autopsy by proxy).4,5 In male infertility, we have conducted the largest genomic investigation into azoospermia and found that >80% of hits are recessive, and it will be interesting to see what similar studies in outbred populations will show as these become available.6
Figure 2.
Illustration of the difference between de novo and recessive paradigms in shaping the frequency of Mendelian phenotypes
In this example, the two paradigms are compared with the same gene, COL1A1, mutation of which can cause autosomal dominant osteogenesis imperfecta as well as autosomal recessive Ehler-Danlos syndrome. ∗Note that qF is what determines the frequency of affected individuals in inbred populations rather than q2 (see text for explanation).
In consanguineous unions, not only do you get enrichment for even the rarest of autosomal recessive traits, but you also get to map the causal variant readily even when you have no clue what the gene’s function is thanks to the autozygome.7 Combine this with the power of next-generation sequencing (NGS) and you have an exceptional opportunity for a very high throughput novel disease gene discovery pipeline, which will allow us to increase the sensitivity of the clinical genome. Remember, a clinical genome is only as good as our knowledge is about the clinical relevance, or lack thereof, of every single gene. And here too it is salient to contrast the process of novel disease gene discovery under the de novo versus recessive paradigms. For example, in their first major gene discovery paper, the DDD study reported the discovery of 14 genes after analyzing the first few thousands of cases.8 Compare this to a focused effort involving just 143 multiplex consanguineous families enriched for novel gene discovery based on a positional mapping prescreen and this allowed us to identify nearly 70 novel genes.9 Why is it possible to identify far more novel genes with confidence using a far smaller sample size?
In the de novo paradigm, you need multiple mutational events to gain confidence in the gene of interest. Under the recessive paradigm, on the other hand, we are typically dealing with a single mutational event that is rendered homozygous so many times that you can easily cross the threshold for statistical significance by linkage analysis. You do not even require large families when dealing with older founder variants because they get homozygosed in independent consanguinity loops in so many small families that you can still achieve the same result.
Not only do consanguineous populations help us boost the sensitivity of the clinical genome at the gene level, but also at the variant level. Who would have thought that variants in the gene linked to a relatively mild inborn error of metabolism known as serine deficiency can also cause a devastating malformation disorder known as Neu-Laxova syndrome that had been a mystery for decades?10 And who would have thought that variants in the gene linked to microcephaly also cause a perinatally lethal condition, microcephaly-micromelia syndrome, that was described many years ago but remained unexplained until consanguineous families helped us solve this mystery?11 These and so many other surprising allelic series would not have been possible had it not been for the “X-ray” vision enabled by positional mapping that allowed us to reconsider the seemingly unlikely, and it is through these allelic series that we can fully grasp the full phenotypic expression of genes. We have also taken advantage of the power of positional mapping to learn the rules of transcript deleterious variants so we can have a better implementation of RNA-seq to boost the sensitivity of the clinical genome. For example, by analyzing all families that map to single loci, we were able to provide the first unbiased breakdown of coding versus non-coding variants in Mendelian diseases.12 Boosting the sensitivity of the clinical genome need not be limited to rare diseases; we have leveraged the unique structure of consanguineous populations to also identify Mendelian forms of common diseases,13, 14, 15, 16, 17, 18 and there is now a growing realization that a significant minority of patients with all kinds of common diseases have in fact underlying Mendelian variants.
Our endeavor to improve the sensitivity of the clinical genome should not make us lose sight of the importance of improving its specificity. Again, at both the gene and variant levels, the study of consanguineous populations can contribute significantly to minimizing false positives when interpreting the clinical genome. At the gene level, we have taken advantage of naturally occurring “knockout” individuals who are homozygous for loss-of-function variants to challenge gene-disease associations, and we will of course continue doing so as we expand our operation to catalog these knockout events.19,20 At the variant level, I would like you to consider this variant, SLC26A4, GenBank: NM_000441, c.1234G>A (p.Val412Ile), which is very rare both in gnomAD (MAF = 0.0000518) and in our consanguineous population (MAF = 0.00045). One does not know what to make of this variant because even though it is predicted deleterious in silico, it will take more than the entire US population to find a single homozygote (q2) to definitively reveal its phenotypic expression. Not so in our consanguineous population where we can identify not one but two homozygotes despite the much smaller cohort size (3,300 versus 125,411), and because both lack hearing loss, you can have confidence in assigning this a more benign classification. We have done this for thousands of variants, including hundreds of “disease-causing” variants, which we found to cause no phenotypes among homozygotes, and we will continue this effort to contribute as much as we can to improving the specificity of the clinical genome.21
So where does this take us? I always say that while the number of genes is limited, the number of variants is not, so even if we saturate the sensitivity and specificity of the clinical genome at the gene level we will still have work to do at the variant level and these special populations will always be incredibly valuable in this regard. But there is more. While little can be done to prevent a de novo occurrence, there is so much that can be done to prevent recessive alleles from wreaking havoc in the human population. I am not simply referring to prenatal and preimplantation genetic testing but the real possibility of having everyone fully informed of their carrier status of these alleles. This is something we plan to do on a very large scale since we already have a premarital screening program in the country that we can build on. Just to give you an idea of the tremendous opportunity we have here, in a preliminary analysis of 503 couples from Saudi Arabia, we found that the percentage of at-risk couples is >12% and that is only counting a predefined set of pathogenic variants we had detected in that population.22 This is dramatically different from the percentage of at-risk couples in the US, which is 1.2%, so you start to realize how staggering these numbers are. This also showcases the tremendous potential to make a meaningful and lasting impact on the prevalence of these diseases.
In conclusion, I am constantly reminded that achieving a more perfect clinical genome is a goal worth all the effort we and so many of our colleagues around the world make. After all, this is our shared heritage, a heritage that defies boundaries and a heritage that brings us together. A more perfect clinical genome will be a gift to all humanity but only if we all work together to make it happen.
There are so many people to whom I would like to express my most sincere feelings of gratitude so let me start by thanking everyone who contributed to me having this honor whether I remembered to include your name or not. I want to first thank my loving family starting with my parents, who instilled in me the pursuit of excellence, and my wife, Dua, and two sons, Ibrahim and Imen, who not only put up with my tough work schedule but do so with genuine love and encouragement. I want to thank my patients and their families who have always been and will continue to be my source of inspiration and the reason why I do what I do. I have learned from great people throughout my training, but none have impacted me like Professors Cynthia Morton and Richard (Dick) Maas during my Harvard years and beyond. Special thanks to you Cynthia for always being there for me with your advice and guidance. Your very kind introduction clearly shows why I am very lucky to call you a mentor and a friend and for that I am eternally grateful. There are countless people I would like to thank in my institution but most importantly of course my wonderful team members on whose behalf I am receiving this award. I am indebted to the countless collaborators from every continent with whom I have had the honor to collaborate and whom I would love to name one by one, but I simply cannot. I would like to express my sincere gratitude to Cynthia for nominating me, and the other four giants of our field who supported that nomination: Professors James (Jim) Lupski, Jay Shendure, Charles Lee, and Maximilian (Max) Muenke. Special thanks to the Awards Committee and ASHG for this incredible peer-bestowed honor, which I will forever cherish as the highlight of my career.
Finally, I will leave you with a quote by Curt Stern “Genes are involved in all-too-many tragic events when malformations, inborn diseases, or mental deficiencies happen. Sometimes, the involvement of genes brings a smile to one's face.”23 My dream is to change the word “sometimes” to “always” by striving to bring the incredible gift of a more perfect clinical genome to everyone in this world. Thank you.
References
- 1.Alazami A.M., Al-Qattan S.M., Faqeih E., Alhashem A., Alshammari M., Alzahrani F., Al-Dosari M.S., Patel N., Alsagheir A., Binabbas B. Expanding the clinical and genetic heterogeneity of hereditary disorders of connective tissue. Hum. Genet. 2016;135:525–540. doi: 10.1007/s00439-016-1660-z. [DOI] [PubMed] [Google Scholar]
- 2.Abouelhoda M., Sobahy T., El-Kalioby M., Patel N., Shamseldin H., Monies D., Al-Tassan N., Ramzan K., Imtiaz F., Shaheen R., Alkuraya F.S. Clinical genomics can facilitate countrywide estimation of autosomal recessive disease burden. Genet. Med. 2016;18:1244–1249. doi: 10.1038/gim.2016.37. [DOI] [PubMed] [Google Scholar]
- 3.Anazi S., Maddirevula S., Faqeih E., Alsedairy H., Alzahrani F., Shamseldin H.E., Patel N., Hashem M., Ibrahim N., Abdulwahab F. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry. 2017;22:615–624. doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- 4.Stanley K.E., Giordano J., Thorsten V., Buchovecky C., Thomas A., Ganapathi M., Liao J., Dharmadhikari A.V., Revah-Politi A., Ernst M. Causal genetic variants in stillbirth. N. Engl. J. Med. 2020;383:1107–1116. doi: 10.1056/NEJMoa1908753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamseldin H.E., Kurdi W., Almusafri F., Alnemer M., Alkaff A., Babay Z., Alhashem A., Tulbah M., Alsahan N., Khan R. Molecular autopsy in maternal-fetal medicine. Genet. Med. 2018;20:420–427. doi: 10.1038/gim.2017.111. [DOI] [PubMed] [Google Scholar]
- 6.Alhathal N., Maddirevula S., Coskun S., Alali H., Assoum M., Morris T., Deek H.A., Hamed S.A., Alsuhaibani S., Mirdawi A. A genomics approach to male infertility. Genet. Med. 2020;22:1967–1975. doi: 10.1038/s41436-020-0916-0. [DOI] [PubMed] [Google Scholar]
- 7.Alkuraya F.S. Autozygome decoded. Genet. Med. 2010;12:765–771. doi: 10.1097/GIM.0b013e3181fbfcc4. [DOI] [PubMed] [Google Scholar]
- 8.Study D.D.D., McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alazami A.M., Patel N., Shamseldin H.E., Anazi S., Al-Dosari M.S., Alzahrani F., Hijazi H., Alshammari M., Aldahmesh M.A., Salih M.A. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Shaheen R., Rahbeeni Z., Alhashem A., Faqeih E., Zhao Q., Xiong Y., Almoisheer A., Al-Qattan S.M., Almadani H.A., Al-Onazi N. Neu-Laxova syndrome, an inborn error of serine metabolism, is caused by mutations in PHGDH. Am. J. Hum. Genet. 2014;94:898–904. doi: 10.1016/j.ajhg.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds J.J., Bicknell L.S., Carroll P., Higgs M.R., Shaheen R., Murray J.E., Papadopoulos D.K., Leitch A., Murina O., Tarnauskaitė Ž. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat. Genet. 2017;49:537–549. doi: 10.1038/ng.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maddirevula S., Kuwahara H., Ewida N., Shamseldin H.E., Patel N., Alzahrani F., AlSheddi T., AlObeid E., Alenazi M., Alsaif H.S. Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics. Genome Biol. 2020;21:145. doi: 10.1186/s13059-020-02053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Mayouf S.M., Sunker A., Abdwani R., Abrawi S.A., Almurshedi F., Alhashmi N., Al Sonbul A., Sewairi W., Qari A., Abdallah E. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet. 2011;43:1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- 14.Patel N., El Mouzan M.I., Al-Mayouf S.M., Adly N., Mohamed J.Y., Al Mofarreh M.A., Ibrahim N., Xiong Y., Zhao Q., Al-Saleem K.A., Alkuraya F.S. Study of Mendelian forms of Crohn’s disease in Saudi Arabia reveals novel risk loci and alleles. Gut. 2014;63:1831–1832. doi: 10.1136/gutjnl-2014-307859. [DOI] [PubMed] [Google Scholar]
- 15.Maddirevula S., Abanemai M., Alkuraya F.S. Human knockouts of PLA2G4A phenocopy NSAID-induced gastrointestinal and renal toxicity. Gut. 2016;65:1575–1577. doi: 10.1136/gutjnl-2016-312374. [DOI] [PubMed] [Google Scholar]
- 16.Al Mutairi F., Alzahrani F., Ababneh F., Kashgari A.A., Alkuraya F.S. A mendelian form of neural tube defect caused by a de novo null variant in SMARCC1 in an identical twin. Ann. Neurol. 2018;83:433–436. doi: 10.1002/ana.25152. [DOI] [PubMed] [Google Scholar]
- 17.Shaheen R., Alshail E., Alaqeel A., Ansari S., Hindieh F., Alkuraya F.S. T (brachyury) is linked to a Mendelian form of neural tube defects in humans. Hum. Genet. 2015;134:1139–1141. doi: 10.1007/s00439-015-1589-7. [DOI] [PubMed] [Google Scholar]
- 18.Aldahmesh M.A., Khan A.O., Alkuraya H., Adly N., Anazi S., Al-Saleh A.A., Mohamed J.Y., Hijazi H., Prabakaran S., Tacke M. Mutations in LRPAP1 are associated with severe myopia in humans. Am. J. Hum. Genet. 2013;93:313–320. doi: 10.1016/j.ajhg.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alsalem A.B., Halees A.S., Anazi S., Alshamekh S., Alkuraya F.S. Autozygome sequencing expands the horizon of human knockout research and provides novel insights into human phenotypic variation. PLoS Genet. 2013;9:e1004030. doi: 10.1371/journal.pgen.1004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alkuraya F.S. Human knockout research: new horizons and opportunities. Trends Genet. 2015;31:108–115. doi: 10.1016/j.tig.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Abouelhoda M., Faquih T., El-Kalioby M., Alkuraya F.S. Revisiting the morbid genome of Mendelian disorders. Genome Biol. 2016;17:235. doi: 10.1186/s13059-016-1102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stern C. Genes and people. Perspect. Biol. Med. 1967;10:500–523. doi: 10.1353/pbm.1967.0008. [DOI] [PubMed] [Google Scholar]