Abstract
Ribosomal L18/L5e (RL18/L5e) is a member of the ribosomal L18/L5e protein family, which has an essential function in translation of mRNA into protein in the large ribosomal subunit. In this study, RL18/L5e was isolated and sequenced from local Pennisetum glaucum (L.) R. Br. cultivar which is known to adapt to environmental stress. The obtained cDNA for PgRL18/L5e was 699 bp in length, with an open reading frame of 564 bp. The deduced protein sequence contained 187 amino acids and comprised an RL18/L5e domain, which shared high sequence identity with orthologous proteins from Viridiplantae. The obtained PgRL18/L5e cDNA contained two exons of 154 and 545 bp, respectively, and an intron of 1398 bp. Secondary and 3D structures of the deduced PgRL18/L5e protein were predicted using in silico tools. Phylogenetic analysis showed close relationships between the PgRL18/L5e protein and its orthologs from monocot species. Multiple sequence alignment showed high identity in the RL18/L5e domain sequence in all orthologous proteins in Viridiplantae. Moreover, all orthologous RL18/L5e proteins shared the same domain architecture and were nearly equal in length. Quantitative real-time PCR indicated a higher transcript abundance of PgRL18/L5e in shoots than in roots of 3-day-old seedlings. Moreover, the expression of PgRL18/L5e in seedlings under cold and drought stress was substantially lower than that in untreated seedlings, whereas the highest expression was shown under heat stress. This study provides insights into the structure and function of the RL18/L5e gene in tolerant crops, which could facilitate the understanding of the role of the various plant ribosomal proteins in adaptation to extreme environments.
Keywords: Ribosomal L18/L5e, Pennisetum glaucum, cDNA, Gene expression, Phylogenetics
1. Introduction
Ribosome assembly is a complex process critical for protein synthesis. In eukaryotic cells, large and small ribosomal subunits comprise four rRNA molecules and approximately 80 ribosomal proteins (RPs) (Lafontaine and Tollervey, 2001). The small subunit includes an 18S rRNA and 33 RPs, whereas the large subunit includes 5S rRNA, 5.8S rRNA, 25S–28S rRNA, and 46 RPs (Warner, 1999, Woolford and Baserga, 2013). Ribosome function in cellular organisms is conserved; similarly, RPs appear to be evolutionarily conserved because many eukaryotic RPs have orthologs in bacteria and archaea (Deshmukh et al., 1995, Lecompte et al., 2002).
In plant cells, accurate regulation of ribosome function in the cytosol and organelles such as chloroplasts and mitochondria is important during plant growth and development (Sormani et al., 2011). Ribosomal proteins in plant organelles are conserved with prokaryotic RPs owing to the prokaryotic origins of chloroplasts and mitochondria (Delwiche and Palmer, 1997, McFadden, 2001).
The cytoplasmic 80S ribosome and chloroplast 70S ribosome from flowering plants have been previously characterized (Stutz and Noll, 1967). Thereafter, many RPs of the cytoplasmic 80S ribosome were identified (Gualerzi and Cammarano, 1970, Barakat et al., 2001, Giavalisco et al., 2005) . One of the cytoplasmic RPs in plants is ribosomal L18/L5e (RL18/L5e) protein, located in the central protuberance of the large subunit (Barakat et al., 2001, Klein et al., 2004). The ortholog of the RL18/L5e protein in bacteria and archaea is known as L18 (Meskauskas and Dinman, 2001). Remarkably, the ortholog of RL18/L5e protein in mammals, MRP-L18, is localized in the mitochondria (Koc et al., 2001). This may be due to translocation of some plant mitochondrial RPs into nuclear DNA during plant evolution (Adams and Palmer, 2003).
The bacterial L18 plays an important role in ribosome assembly through interaction with 5S rRNA, which causes a conformational change that promotes the binding of L5 (the ortholog of eukaryotic L11) to 5S rRNA in the final phase of ribosome assembly. Binding of L18 and L5 to 5S rRNA is essential for the unification of 5S rRNA with 23S rRNA in the large subunit (Steitz et al., 1988, Deshmukh et al., 1995), which contains the peptidyl transferase center where proteins are synthesized (Warner, 1999).
A plant RL18/L5e was first characterized in Arabidopsis (Baima et al., 1995). The RL18/L5e cDNA in Arabidopsis encodes a 187 amino acids (aa) protein that shows sequence homology with orthologous proteins in yeast (Molenaar et al., 1984), African clawed frog (Beccari et al., 1987), and rat (Devi et al., 1988). The role of plant RL18/L5e in translation of mRNA into protein and regulation of protein synthesis during growth and differentiation has been demonstrated in cellular organisms. Additionally, previous studies have reported the involvement of RL18/L5e in plant-pathogen interactions (Leh et al., 2000, Spurgers et al., 2010, de la Cruz et al., 2013, Loutre et al., 2018) (reviewed in (Li, 2019)). Although few reports are available about the role of RL18/L5e in plants under abiotic stress, a previous study has investigated the early transcriptomic response in rice under salinity stress (Kawasaki et al., 2001).
Pearl millet, Pennisetum glaucum (L.) R. Br. ssp. monodii (Maire) Brunken, is a cereal crop that can tolerate abiotic stress and can be grown in harsh environments (D’Andrea et al., 2001). Although the P. glaucum genome has recently been sequenced (Varshney et al., 2017), the molecular mechanisms underlying its growth and stress tolerance remain unclear. Comparative transcriptomics and proteomics of tolerant crops are helpful in understanding the molecular mechanisms that control plant response under stress. The present study aims to characterize the gene encoding the ribosomal L18/L5e family protein RL18/L5e in pearl millet, using cDNA sequencing and gene expression analysis. Obtaining the sequence of RL18/L5e gene is important for analyzing its expression under environmental stress at different developmental stages. These expression data are expected to help in understanding the regulation of RPs under stress in tolerant crops.
2. Experimental procedures
2.1. Database sequence retrieval and primer designing
The sequence of RL18/L5e proteins in Poaceae was retrieved from the UniProt database (http://www.uniprot.org/). The sequence of the Zea mays RL18/L5e protein (accession B4FUY5) was chosen as the query sequence in BLAST to find similar proteins in Setaria italica and Panicum hallii, which are the species closest to P. glaucum. The uncharacterized proteins from Setaria italica and Panicum hallii under accession numbers A0A4U6T2N2 and A0A2T7C9I9, respectively, were selected as queries against the P. glaucum (=Cenchrus americanus) genome in the NCBI database using tBLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). A considerable similarity was found between the query proteins from Setaria italica and chromosome number 2 (accession No. CM007983.2) of P. glaucum in the genomic region from nt 200,453,174 to 200455132. Primers were designed on the basis of the sequence from nt 200,452,995 to 200,455,312 of chromosome 2 of P. glaucum, based on the similarity with RL18/L5e mRNA sequences from Setaria italica and Panicum hallii. Five primer pairs were tested for PgRL18/L5e cDNA amplification. Of these primers, one primer pair (forward: 5′-TTTTTAAGGTCTTGAGCTGTAAT-3′, reverse: 5′- TGTCATGTCCACATCTTCCAG-3′) synthesized a cDNA sequence that included the whole open reading frame (ORF).
2.2. RNA extraction, reverse transcription, and sequencing
The plant material used in this study was a local Saudi pearl millet (Pennisetum glaucum (L.) R. Br. ssp. monodii (Maire) Brunken), cultivar Baydhan, which is grown in Wadi Baydhan in the western region of Saudi Arabia (23.02739°N, 40.41217°E). This local cultivar can tolerate extreme heat which is grown during summer season where the day temperature reaches 44 °C. Pearl millet seeds were surface-sterilized by 5% sodium hypochlorite for 30 min and washed thrice before germinating in Petri dishes at room temperature. Five-day-old seedlings were used to isolate total RNA using TRIzol reagent (Invitrogen); the total RNA was further used to synthesize cDNA with SuperScript® III reverse transcriptase (Invitrogen). The synthesized cDNA was used as a template for PCR using primers designed to amplify PgRL18/L5e cDNA. PCR was performed using 2X Phusion Master Mix with high-fidelity buffer (Thermo Fisher Scientific), 0.4 µmol of each primer, and 2.5 µL of cDNA. PCR cycling was carried out as follows: Initial reaction at 95 °C for 4 min; then 30 cycles of 95 °C for 45 s, 56 °C for 45 s, and 72 °C for 1 min; and finally at 72 °C for 5 min. The PCR product was sequenced by Sanger method (Sanger et al., 1977) using BigDye Terminator v3.1 Cycle Sequencing Kit on ABI genetic analyzer 3730xi (Applied Biosystems). Sequencing results obtained from five replicates were subjected to multiple sequence alignment (MSA) using MUSCLE version 3.8 (https://www.ebi.ac.uk/Tools/msa/muscle/) (Katoh and Standley, 2013). The MSA file was exported to UniGene software (Agarwala et al., 2018). All generated sequences were assembled and the consensus sequence was analyzed using in silico tools.
2.3. In silico sequence analysis and phylogeny
The consensus cDNA sequence was analyzed using UniGene software. The ORF was identified and translated into an amino acid sequence using ExPaSy translation tool (https://web.expasy.org/translate/) (Bairoch, 2005). The deduced protein was identified through HMMER3 on the MyHits from The Swiss Institute of Bioinformatics Web site (http://myhits.isb-sib.ch/) (Pagni et al., 2007). Then, the deduced protein was used as a query in UniProt BLAST to identify orthologous proteins from other plant species. General features of the proteins (isoelectric point and molecular weight) were estimated by ExPASy (https://web.expasy.org/compute_pi/) (Bairoch, 2005). NCBI conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) was used for the prediction of domains in the deduced protein (Marchler-Bauer and Bryant, 2004, Marchler-Bauer et al., 2017). ConSurf was used to estimate the degree of conservation of the PgRL18/L5e protein sequence based on its evolutionary relationships with orthologous proteins (http://consurf.tau.ac.il/) (Glaser et al., 2005, Ashkenazy et al., 2016).
Secondary structure of the deduced protein was predicted using NetSurfP-2.0 (http://www.cbs.dtu.dk/services/NetSurfP/) (Klausen et al., 2019). To predict the tertiary structure of PgRL18/L5e, BLAST and HHBlits search were carried out through the SWISS-MODEL template library (https://swissmodel.expasy.org/) (Guex et al., 2009, Waterhouse et al., 2018). Phosphorylation sites for the PgRL18/L5e protein were predicted using the ScanProsite tool (http://www.expasy.ch/tools/scanprosite/) (de Castro et al., 2006). Protein-protein binding sites were detected using Predict Protein (https://www.predictprotein.org/).
To analyze the phylogenetic relationships between PgRL18/L5e and proteins from other plant species, UniProt BLAST was used to retrieve orthologous RL18/L5e proteins in Viridiplantae. Phylogenetic analysis was performed with 21 selected protein sequences, with a hypothetical protein from Chara braunii as an outgroup. Some of the selected orthologous plant proteins were uncharacterized; therefore, the presence of an RL18/L5e domain in all selected proteins was confirmed using NCBI Batch Web CD-Search tool. Twenty-three RL18/L5e protein sequences, including PgRL18/L5e, were aligned using MUSCLE. The MSA results were imported to MEGA 7.0.26 (Kumar et al., 2016), and a phylogenetic tree was generated using the maximum likelihood method (Felsenstein and Churchill, 1996) using default settings.
2.4. Plant treatments and gene expression measurement
Expression patterns of PgRL18/L5e were examined using quantitative real-time PCR. Three-day-old seedlings were divided into six groups; four groups were treated with different types of stress for 3 days—drought, salt, cold, and heat—in order to analyze the expression of PgRL18/L5e in stressed seedlings. Drought stress was induced by stop irrigation, whereas heat stress was induced by incubating seedlings at 48 °C/38 °C (day/night) with regular irrigation. Cold stress was induced by incubating seedlings at a constant temperature of 12 °C with regular irrigation, whereas salt stress was induced by treating seedlings with 200 mM NaCl. Seedlings in the fifth group were untreated and were used as the control group. To investigate the expression of PgRL18/L5e gene in pearl millet roots, shoots, and seeds; seedlings in the sixth group were grown in normal conditions for 3 days, then were separated into roots, shoots, and seeds tissues.
Total RNA was extracted from untreated seedlings; stressed seedlings (heat, salt, cold, and drought); and untreated roots, shoots, and seeds. Total RNA integrity was assessed using agarose gel electrophoresis; total RNA was then quantified using Qubit® 2 fluorometer (Invitrogen) and Qubit® RNA Assay Kit (Life Technologies). cDNAs were amplified from the eight samples using SuperScript™ VILO™ cDNA Synthesis Kit (Invitrogen). PgRL18/L5e gene-specific primers were designed to span two consecutive exons in cDNA (forward: 5′-CCGGAGGCAATTTCACAGTG-3′, reverse: 5′-CCATGTGGGCGATGGTGCGCTG-3′). The reference gene EF-1α (forward: 5′-GTTACAACCCAGACAAGATTGC-3′ and reverse: 5′-TGGACCTCTCAATCGTGTTG-3′) was used to normalize the PgRL18/L5e expression in all samples. The reactions were performed in three biological replicates. Each reaction consisted of 2X Power SYBR Green RT-PCR mix (Applied Bio Systems), 0.1 µM of each primer, 2 µL of template, and H2O to make up the volume to 12.5 µL. Reaction parameters were as follows: initial reaction at 50 °C for 2 min and then 95 °C for 10 min followed by 55 cycles of 95 °C for 15 s and 60 °C for 1 min.
Cycles to threshold (Ct) values were calculated to compare the expression levels in all tissues and treatments (Livak and Schmittgen, 2001). ΔCt was calculated by normalization of PgRL18/L5e Ct values to EF-1α Ct values. ΔCt values were calibrated using the ΔCt values of PgRL18/L5e for untreated seedlings to obtain ΔΔCt and then 2-ΔΔCt as fold-change for PgRL18/L5e expression in each sample.
3. Results
3.1. PgRL18/L5e cDNA sequencing and sequence analysis
The PgRL18/L5e cDNA sequence was obtained by aligning five sequences. The consensus cDNA sequence was 699 bp in length, with an ORF of 564 bp (Fig. 1). All cDNA sequences were submitted to NCBI GenBank (accession numbers MN624961, MN624962, MN624963 and MN624964). The HMMER3 package from MyHits was used to identify the deduced protein, using the euk database of proteins (some complete proteomes of eukaryotes). The first hit was Zea mays RL18/L5e family protein (UniProt accession B4FUY5) with 90% sequence identity and an E-value of 1.1e-125. Domain prediction using the NCBI Batch Web CD-Search tool revealed that the deduced protein comprised an RL18/L5e superfamily domain (No. cl00379), which also confirmed that the sequenced cDNA corresponded to the PgRL18/L5e gene (Fig. S1.A).
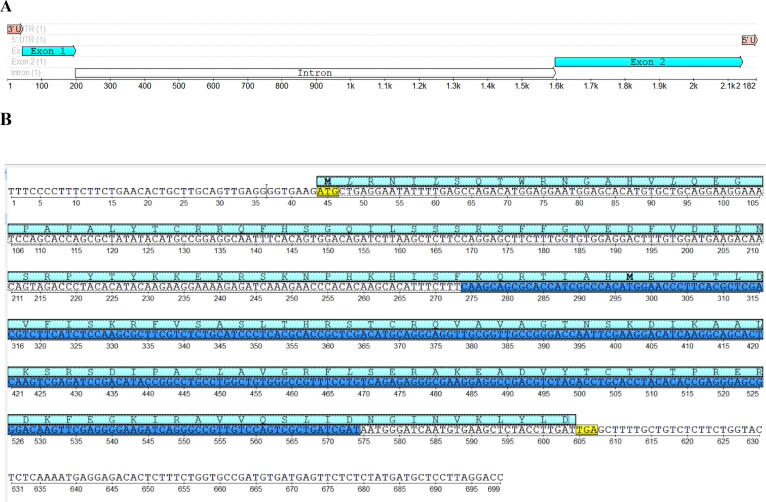
Fig. 1.
The structure of PgRL18/L5e gene. (A): Structure of the PgRL18/L5e gene based on the alignment with the genomic sequence of chromosome 2 of Pennisetum glaucum. The two exons are shown in light blue, the intron is shown as the white arrow, and the 3ʹ and 5ʹ UTR are shown in light pink. (B): The nucleotide sequence and deduced protein sequence for PgRL18/L5e cDNA. The amino acid sequence (light blue) is written above the nucleotide sequence, and the RL18/L5e domain is highlighted in dark blue. The start and stop codons are highlighted in yellow.
The alignment between the PgRL18/L5e cDNA and P. glaucum chromosomes through BLASTn revealed that the constructed PgRL18/L5e cDNA shared 98.71% identity with a 2097 bp genomic region on P. glaucum chromosome 2. Two exons were determined through the alignment of the obtained cDNA and the related genomic region on chromosome 2. The first exon (154 bp) contained the start codon (ATG) starting at nt 44. The second exon (545 bp) contained the end codon (TGA) located on nt 605. The intron between the two exons was 1398 bp long Fig. 1. The ORF was 564 bp long, and the deduced protein obtained by translating this ORF contained 187 aa (NCBI accession numbers; QMU23697, QMU23698 QMU23699 and QMU23700). The NCBI BLASTn search for PgRL18/L5e cDNA against P. glaucum chloroplast genome (NCBI accession KX756179.1) showed no sequence identity. Owing to the unavailability of the complete mitochondrial genome sequence of P. glaucum in NCBI GenBank, PgRL18/L5e cDNA was searched against the complete mitochondrial genome sequences of rice and corn (NCBI accessions BA000029.3 and DQ645536.1, respectively), which also showed no sequence identity.
3.2. In silico characterization of PgRL18/L5e deduced protein
The deduced PgRL18/L5e protein had a molecular mass of 21.328 kDa with a theoretical isoelectric point of 9.78. Protein homology search for PgRL18/L5e protein sequence by NCBI BLASTp showed high identity with RL18/L5e proteins from other Poaceae species. The highest identity (96.79%) was observed with an uncharacterized protein from Setaria italica (Foxtail millet). The second highest identity value (95.19%) was observed with an uncharacterized protein from Panicum hallii. Zea mays RL18/L5e family protein showed 90% identity with PgRL18/L5e (Table S1). However, the NCBI BLASTp search revealed 24.58% to 36.56% sequence identity in the RL18/L5e domain with the orthologous 50S ribosomal L18 proteins from prokaryotes (E-value lower than 0.95; Table S2).
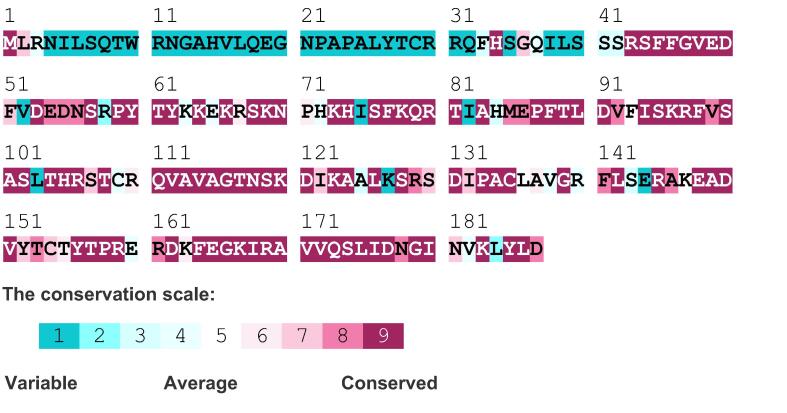
The RL18/L5e superfamily domain spans from aa 78 to 177 (Fig. S1.A). Fig. 2 shows the degree of conservation of the amino acid sequence of the deduced PgRL18/L5e protein as determined by ConSurf. The highly conserved sequence (with a score higher than 6 according to ConSurf) spanned from aa 43 to 187 and included the RL18/L5e domain, which was a highly conserved sequence in all orthologous proteins from Viridiplantae as indicated by MSA (Fig. S1.B). The domain RL18/L5e was highly conserved between all orthologous proteins used in the alignment analysis, with the highest degree of conservation observed with monocot species, especially Setaria italica and Panicum hallii. However, MSA indicated that the PgRL18/L5e protein contained a serine residue at aa 143, which is different from all orthologous proteins from monocots used in the MSA. All analyzed monocots RL18/L5e proteins, except PgRL18/L5e, contained alanine at aa 143, whereas the dicot species contained alanine or serine.
Fig. 2.
Conservation of the PgRL18/L5e protein sequence analyzed by ConSurf. ConSurf scores the most conserved amino acid as 9 and the most variable as 1.
The potential phosphorylation sites in PgRL18/L5e protein sequence were identified using the ScanProsite program. A conserved tyrosine kinase phosphorylation site 1 (PS00007) was located between aa 145 and 152, and an N-myristoylation site (PS00008) was located between aa 36 and 41. Moreover, the PgRL18/L5e protein was found to contain three possible casein kinase II phosphorylation sites (PS00006) and ten protein kinase C phosphorylation sites (PS00005) (Fig. S2.A). The above-mentioned serine residue at aa 143 constituted a protein kinase C phosphorylation site (SER) in an α-helix region of the PgRL18/L5e protein (Fig. S2.B).
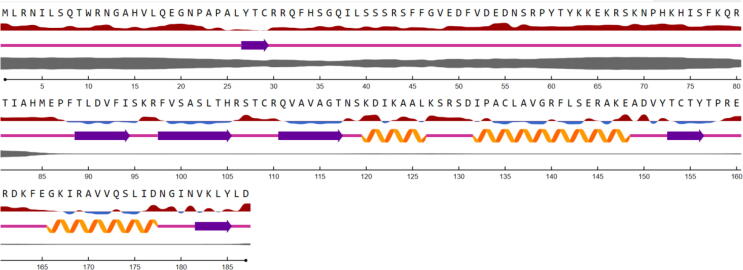
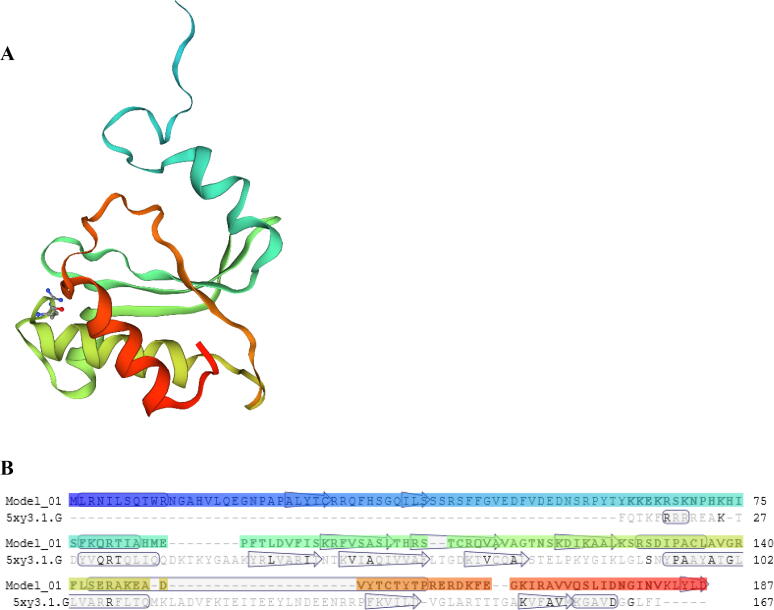
The secondary structure of the deduced PgRL18/L5e protein predicted using NetSurfP-2.0 showed three α-helices and six β-strands, as shown in Fig. 3. SWISS-MODEL was used to develop a tertiary structure model for the deduced PgRL18/L5e protein, based on the known tertiary structures of other homologous proteins. Of the 596 templates in the SWISS-MODEL library, the highest-quality template 5xy3.1.G, which was a ribosomal L5 protein (Li et al., 2017), was selected for 3D structure model building. The alignment of the PgRL18/L5e protein and the 5xy3.1.G template showed 18.49% identity in the region between aa 62 and 182 (Fig. 4.B). On the basis of this alignment, a 3D structure model was built for PgRL18/L5e (Fig. 4.A).
Fig. 3.
Secondary structure of the deduced PgRL18/L5e protein as predicted by NetSurfP-2.0. The relative surface accessibility is shown as  ; the sequences denoted in red are exposed, and those denoted in blue are buried (threshold at 25%). Secondary structure is illustrated as follows:
; the sequences denoted in red are exposed, and those denoted in blue are buried (threshold at 25%). Secondary structure is illustrated as follows:  Helix,
Helix,  Strand,
Strand,  Coil. The thickness
Coil. The thickness  of the line denotes the probability of disordered residues.
of the line denotes the probability of disordered residues.
Fig. 4.
(A): The predicted 3D structure of the sequence from aa 62 to 182 of the deduced PgRL18/L5e protein generated by SWISS-MODEL. The N-terminal is indicated in blue, and the C-terminal is shown in red. (B): The alignment between ribosomal L5 protein (5xy3.1.G) from the SWISS-MODEL template library and PgRL18/L5e protein.
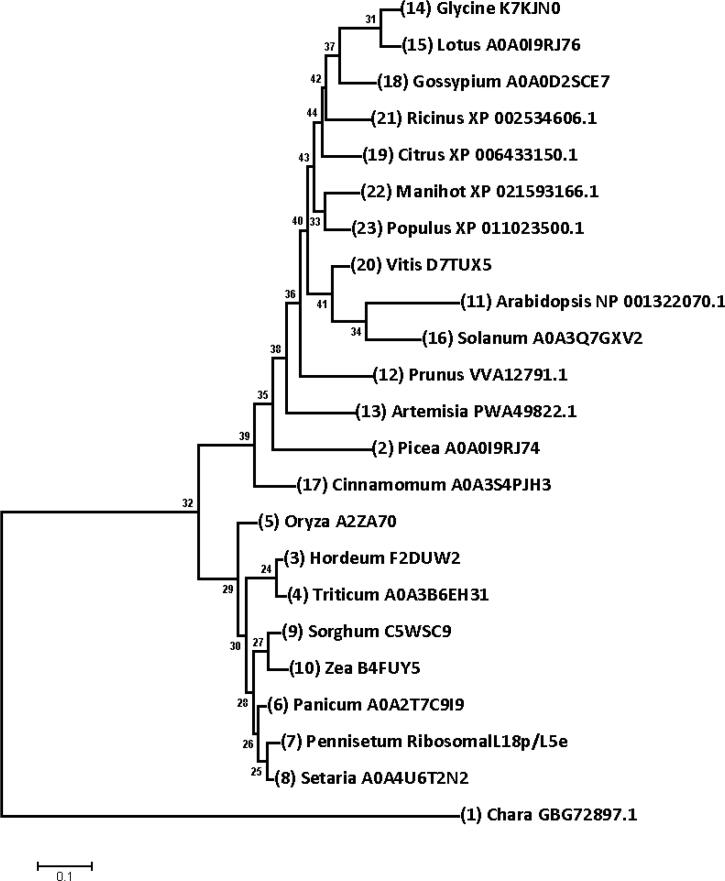
3.3. Phylogenetic relationships
To better understand the relationships between PgRL18/L5e and its orthologous proteins, phylogenetic relationships were analyzed with 22 orthologous proteins from Viridiplantae (Table S3). The constructed maximum likelihood tree (Fig. 5) indicated close relationships between PgRL18/L5e and its orthologous proteins in Poaceae, which clustered together. All orthologous proteins in dicot species were grouped together in the same cluster with RL18 proteins from Cinnamomum micranthum (Magnoliidae) and Picea glauca (Pinidae).
Fig. 5.
Phylogenetic analysis of PgRL18/L5e and homologous proteins. The maximum likelihood tree was built using MEGA 7.0.26. A hypothetical homologous protein from Chara braunii was used as an outgroup.
3.4. Gene expression profiling
The ΔΔCt method was used to analyze PgRL18/L5e expression in stressed pearl millet seedlings and to investigate the tissue distribution of PgRL18/L5e mRNA using expression in untreated seedling tissues as calibrator. PgRL18/L5e was found to be expressed in seedlings under heat and salt stress, but it was expressed at extremely low levels under cold and drought stress (Fig. 6.A). Compared with its expression in untreated seedlings, PgRL18/L5e expression in stressed seedlings was the highest under heat stress (0.92-fold) and the lowest under drought stress (0.00002-fold). Shoot tissues from 3-day-old seedlings showed the highest PgRL18/L5e gene expression (2.2-fold), whereas roots showed the lowest expression levels (0.7-fold; Fig. 6.B).
Fig. 6.
Relative expression of PgRL18/L5e gene in Pennisetum glaucum seedlings analyzed by quantitative real-time PCR. (A): The expression levels of PgRL18/L5e in seedlings treated during germination with four types of stress: heat, salt, cold, and drought. The y-axis corresponds to the relative expression level. (B): The tissue-specific expression levels of PgRL18/L5e in three different tissues— root, seeds, and shoots—from 3-day-old germinated seedlings.
4. Discussion
Pearl millet P. glaucum (L.) is an important crop that widely cultivated in different parts of the world, primarily in Asia and Africa, as a multipurpose cereal (Havilah, 2017, ICRISAT, 1996). In Saudi Arabia, pearl millet is grown in the western and southwestern regions during different seasons with various temperatures and different water needs. In the western region of Saudi Arabia, the local cultivars tolerate extreme heat where the day temperature range from 33 to 46 °C in summer season. In the southwestern region where moderate temperature (range from 22 to 32 °C), other pearl millet cultivars are grown under rainfed or limited irrigation. In the present study, we sequenced and investigated the expression profiles of the RL18/L5e gene from a local pearl millet cultivar, Baydhan, that grown in western of Saudi Arabia during summer season. The RL18/L5e protein is one of the structural constituents of the large ribosomal subunit. This protein has an important role in translation of mRNA into protein. It has been shown in bacteria that in the final steps of the ribosome assembly, the bacterial ortholog L18 binds 5S rRNA and promotes the establishment of the complex of 5S rRNA and 23S rRNA (26S rRNA in plants) (Kuzoff et al., 1998, Woestenenk et al., 2002).
PgRL18/L5e gene encoded a 187 aa ORF. The deduced PgRL18/L5e protein and all the orthologous proteins in Viridiplantae were similar in length and showed similar RL18/L5e superfamily domain architecture. The alignment of the RL18/L5e domain sequence among orthologous RL18/L5e proteins from land plants indicated a high degree of conservation which previously reported of nuclear RPs (Finet et al., 2010). Although the orthologous 50S ribosomal L18 proteins from prokaryotes showed lower degree of conservation with PgRL18/L5e protein, the conserved sequence was in the C-terminal region, indicating the importance of that sequence in 5S rRNA binding (Setterquist et al., 1996). The secondary structure of the conserved sequence from 77 to 187 aa, which comprised the RL18/L5e domain, was shown to have three α-helices and five β-strands. This domain recognizes a specific rRNA motif to bind 5S rRNA (Matthew Michael and Dreyfuss, 1996, Woestenenk et al., 2002). The first 20 N-terminal residues of bacterial L18 are not involved in 5S rRNA binding, but they are required for the binding of L18 with L5 and 23S rRNA (Newberry and Garrett, 1980).
BLAST search showed that PgRL18/L5e is a single-copy gene located on chromosome number 2 in pearl millet genome. Similarly, Arabidopsis RL18/L5e is a single-copy gene that encodes a component of the cytoplasmic ribosome (Baima et al., 1995, Barakat et al., 2001). Although some RP genes are single-copy genes, other RP genes are found in multiple copies as paralogous genes because of gene duplication events during evolution (Sormani et al., 2011).
The PgRL18/L5e protein has potential phosphorylation sites as suggested by the in silico analysis. Previous studies indicated that the orthologous bacterial L18 proteins are modulated by phosphorylation (Bloemink and Moore, 1999, Soung et al., 2008). Bacillus stearothermophilus L18 protein contains a phosphorylation site at a serine residue, and this phosphorylation is critical for protein folding and binding to 5S rRNA, suggesting that phosphorylation of RPs is important for translation or ribosome assembly (Bloemink and Moore, 1999).
On the basis of the available sequenced genomes, orthologous RPs were selected for phylogenetic analysis to represent the main taxa of Viridiplantae. Of the proteins from flowering plants, 7 orthologous proteins were from monocot species, and 12 were from dicots. One orthologous protein from a gymnosperm (Picea glauca) and another from Magnolia (Cinnamomum micranthum) were also selected for the phylogenetic analysis. All selected proteins as well as the outgroup protein from green alga Chara braunii contained an RL18/L5e domain. The phylogenetic relationships observed between PgRL18/L5e and orthologous proteins confirmed the previously reported evolutionary relationships between land plants (Finet et al., 2010). Previous studies on effective phylogenetic markers have suggested that nuclear RPs appeared to be reliable phylogenetic markers because of their conservation in eukaryotes (Philippe et al., 2004, Marlétaz et al., 2006).
PgRL18/L5e was downregulated under all types of environmental stress examined in the present study; the lowest expression levels were observed under cold and drought stress. However, seedling under heat stress showed expression levels similar to those in untreated seedlings, whereas the expression levels under salt stress were intermediate. In contrast, the orthologous RL18/L5e proteins from Arabidopsis and rice were upregulated by salt stress (Kawasaki et al., 2001, Omidbakhshfard et al., 2012). In rice, the upregulation of RL18/L5e proteins in response to salt stress was observed during the first hour after salt treatment; thereafter, this upregulation decreased over time (Kawasaki et al., 2001). The similar expression levels observed between untreated seedlings and seedlings under heat stress may be owing to the tolerance of the local pearl millet cultivar that is grown in the western region of Saudi Arabia during summer when the day/night temperatures reach 44/32 °C. However, to understand the role of PgRL18/L5e in abiotic stress tolerance, further research on expression patterns at different developmental stages under various stresses is required.
The high PgRL18/L5e gene expression in shoots is consistent with the expression patterns of the orthologous RL18/L5e gene in Arabidopsis, which is expressed during growth and differentiation. Arabidopsis RL18/L5e mRNA is expressed in all plant tissues during developmental stages, but the highest transcript levels are observed in differentiated tissues such as young floral buds (Baima et al., 1995). The highest expression levels of Arabidopsis RL18/L5e protein were observed in the cotyledons of 1-day-old seedlings, young leaf laminae, and young flower sepals (Dal Bosco et al., 2004, Ascencio-Ibáñez et al., 2008). The lower expression levels in germinated seeds were consistent with the down-regulated RL18/L5e protein expression in rice embryos at 72 h after germination in comparison with the expression levels during the first hour (Han et al., 2014). These expression profiles signify the crucial function of RL18/L5e in regulation of translation during plant growth and development.
5. Conclusion
The molecular mechanisms underlying the ability of plants to tolerate stress are complex and remain unclear. As reported in previous studies, RL18/L5e gene, a member of the ribosomal L18/L5e family, is regulated by different types of stress. To understand the role of ribosomal protein L18/L5e in the adaptation of crops grown in extreme environmental conditions, we identified the RL18/L5e gene from a traditional pearl millet cultivar. The results of this study are expected to be useful in understanding the role of the cytoplasmic ribosomal protein RL18/L5e in the regulation of synthesis of proteins involved in plant adaptation to stress. Further transcriptomic and proteomic studies are needed for a comprehensive understanding of the role of RPs in response to abiotic stress.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are greatly indebted to the Central Laboratory for Science and Medical Studies at King Saud University for providing sequencing services. We also thank Maha Alrwais for her excellent technical assistance during RNA extraction. This study was financially supported under Project No. 2019/01/9512, from the Deanship of Scientific Research at Prince Sattam bin Abdulaziz University.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2021.03.035.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adams K.L., Palmer J.D. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. https://doi.org/https://doi.org/10.1016/S1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Agarwala R., Barrett T., Beck J., Benson D.A., Bollin C., Bolton E., Bourexis D., Brister J.R., Bryant S.H., Canese K., Cavanaugh M., Charowhas C., Clark K., Dondoshansky I., Feolo M., Fitzpatrick L., Funk K., Geer L.Y., Gorelenkov V., Graeff A., Hlavina W., Holmes B., Johnson M., Kattman B., Khotomlianski V., Kimchi A., Kimelman M., Kimura M., Kitts P., Klimke W., Kotliarov A., Krasnov S., Kuznetsov A., Landrum M.J., Landsman D., Lathrop S., Lee J.M., Leubsdorf C., Lu Z., Madden T.L., Marchler-Bauer A., Malheiro A., Meric P., Karsch-Mizrachi I., Mnev A., Murphy T., Orris R., Ostell J., O’Sullivan C., Palanigobu V., Panchenko A.R., Phan L., Pierov B., Pruitt K.D., Rodarmer K., Sayers E.W., Schneider V., Schoch C.L., Schuler G.D., Sherry S.T., Siyan K., Soboleva A., Soussov V., Starchenko G., Tatusova T.A., Thibaud-Nissen F., Todorov K., Trawick B.W., Vakatov D., Ward M., Yaschenko E., Zasypkin A., Zbicz K. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S., Sessa G., Ruberti I., Morelli G. A cDNA encoding Arabidopsis thaliana cytoplasmic ribosomal protein L18. Gene. 1995;153:171–174. doi: 10.1016/0378-1119(94)00730-G. [DOI] [PubMed] [Google Scholar]
- Bairoch A. Protein identification and analysis tools on the ExPASy server. Proteomics. 2005 doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Barakat A., Szick-miranda K., Chang I., Guyot R., Blanc G., Cooke R., Delseny M., Bailey-serres J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genom. 2001;127:398–415. doi: 10.1104/pp.010265.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari, E., Mazzetti, P., Acidi, C., Cnr, N., Molecolare, B., I, U.R., Moro, P.A., 1987. Volume 15 Number 4 1987 Nucleic Acids Research The nucleotide sequence of the ribosomal protein L14 gene of Xenopus laeYis Nucleic Acids Research 15, 1870–1872. [DOI] [PMC free article] [PubMed]
- Bloemink M.J., Moore P.B. Phosphorylation of Ribosomal Protein L18 Is Required for Its Folding and Binding to 5S rRNA. Biochemistry. 1999;38:13385–13390. doi: 10.1021/bi9914816. [DOI] [PubMed] [Google Scholar]
- Cruz, M.T.S. de la, Adame-García, J., Gregorio-Jorge, J., Jiménez-Jacinto, V., Vega-Alvarado, L., Iglesias-Andreu’, L., Escobar-Hernández, E.E., Luna-Rodríguez, M., 2013. Increase in ribosomal proteins activity: Translational reprogramming in Vanilla planifolia Jacks., against Fusarium infection. J. Chem. Inf. Model. 53, 1689–1699. https://doi.org/10.1017/CBO9781107415324.004
- D’Andrea, a C., Klee, M., Casey, J., 2001. Archaeobotanical evidence for pearl millet (Pennisetum glaucum) in sub-Saharan West Africa. Antiquity 75, 341.
- Dal Bosco C., Lezhneva L., Bieh A., Leister D., Strotmann H., Wanner G., Meurer J. Inactivation of the Chloroplast ATP Synthase γ Subunit Results in High Non-photochemical Fluorescence Quenching and Altered Nuclear Gene Expression in Arabidopsis thaliana. J. Biol. Chem. 2004;279:1060–1069. doi: 10.1074/jbc.M308435200. [DOI] [PubMed] [Google Scholar]
- de Castro E., Sigrist C.J.A., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwiche C.F., Palmer J.D. The origin of plastids and their spread via secondary symbiosis. 1997;86:53–86. doi: 10.1007/978-3-7091-6542-3_3. [DOI] [Google Scholar]
- Deshmukh M., Stark J., Yeh L.C.C., Lee J.C., Woolford J.L. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J. Biol. Chem. 1995;270:30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- Devi K.R.G., Chan Y.-L., Wool I.R.A.G. The Primary Structure of Rat Ribosomal Protein L18. DNA. 1988;7:157–162. doi: 10.1089/dna.1988.7.157. [DOI] [PubMed] [Google Scholar]
- Felsenstein J., Churchill G.A. A hidden markov model approach evolution to variation among sites in rate of evolution. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- Finet C., Timme R.E., Delwiche C.F., Marlétaz F. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr. Biol. 2010;20:2217–2222. doi: 10.1016/j.cub.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Wilson D., Kreitler T., Lehrach H., Klose J., Gobom J., Fucini P. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol. Biol. 2005;57:577–591. doi: 10.1007/s11103-005-0699-3. [DOI] [PubMed] [Google Scholar]
- Glaser F., Rosenberg Y., Kessel A., Pupko T., Ben-Tal N. The ConSurf-HSSP database: The mapping of evolutionary conservation among homologs onto PDB structures. Proteins Struct. Funct. Bioinforma. 2005;58:610–617. doi: 10.1002/prot.20305. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Cammarano P. Species specificity of ribosomal proteins from chloroplast and cytoplasmic ribosomes of higher plants: Electrophoretic studies. Biochim. Biophys. Acta - Nucleic Acids Protein Synth. 1970;199:203–213. doi: 10.1016/0005-2787(70)90709-4. https://doi.org/https://doi.org/10.1016/0005-2787(70)90709-4. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30:S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- Han C., He D., Li M., Yang P. In-depth proteomic analysis of rice embryo reveals its important roles in seed germination. Plant Cell Physiol. 2014;55:1826–1847. doi: 10.1093/pcp/pcu114. [DOI] [PubMed] [Google Scholar]
- Havilah, E.J., 2017. Forages and Pastures: Annual Forage and Pasture Crops—Species and Varieties☆, in: Reference Module in Food Science. Elsevier. https://doi.org/https://doi.org/10.1016/B978-0-08-100596-5.21855-6
- ICRISAT, 1996. Food from thought. Patancheru, India.
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S., Borchert C., Deyholos M., Wang H., Brazille S., Kawai K., Galbraith D., Bohnert H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen M.S., Jespersen M.C., Nielsen H., Jensen K.K., Jurtz V.I., Sønderby C.K., Sommer M.O.A., Winther O., Nielsen M., Petersen B., Marcatili P. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Proteins Struct. Funct. Bioinforma. 2019;87:520–527. doi: 10.1002/prot.25674. [DOI] [PubMed] [Google Scholar]
- Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- Koc E.C., Burkhart W., Blackburn K., Moyer M.B., Schlatzer D.M., Moseley A., Spremulli L.L. The large subunit of the mammalian mitochondrial ribosome: Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger d atasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzoff R.K., Sweere J.A., Soltis D.E., Soltis P.S., Zimmer E.A. The phylogenetic potential of entire 26S rDNA sequences in plants. Mol. Biol. Evol. 1998;15:251–263. doi: 10.1093/oxfordjournals.molbev.a025922. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L.J., Tollervey D. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2001;2:514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- Lecompte O., Ripp R., Thierry J.C., Moras D., Poch O. Comparative analysis of ribosomal proteins in complete genomes: An example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leh V., Yot P., Keller M. The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology. 2000;266:1–7. doi: 10.1006/viro.1999.0073. [DOI] [PubMed] [Google Scholar]
- Li S. Regulation of Ribosomal Proteins on Viral Infection. Cells. 2019;8:508. doi: 10.3390/cells8050508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Guo Q., Zheng L., Ji Y., Xie Y.T., Lai D.H., Lun Z.R., Suo X., Gao N. Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Res. 2017;27:1275–1288. doi: 10.1038/cr.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loutre R., Heckel A.M., Jeandard D., Tarassov I., Entelis N. Anti-replicative recombinant 5S rRNA molecules can modulate the mtDNA heteroplasmy in a glucose-dependent manner. PLoS ONE. 2018;13:1–20. doi: 10.1371/journal.pone.0199258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Geer L.Y., Bryant S.H. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bryant S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004;32:327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlétaz F., Martin E., Perez Y., Papillon D., Caubit X., Lowe C.J., Freeman B., Fasano L., Dossat C., Wincker P., Weissenbach J., Le Parco Y. Chaetognath phylogenomics: a protostome with deuterostome-like development. Curr. Biol. 2006;16:577–578. doi: 10.1016/j.cub.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Matthew Michael W., Dreyfuss G. Distinct domains in ribosomal protein L5 mediate 5 S rRNA binding and nucleolar localization. J. Biol. Chem. 1996;271:11571–11574. doi: 10.1074/jbc.271.19.11571. [DOI] [PubMed] [Google Scholar]
- McFadden G.I. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 2001;37:951–959. doi: 10.1046/j.1529-8817.2001.01126.x. [DOI] [Google Scholar]
- Meskauskas A., Dinman J.D. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001;7:1084–1096. doi: 10.1017/S1355838201001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar C.M.T., Woudt L.P., Jansen A.E.M., Mager W.H., Planta R.J., Donovan D.M., Pearson N.J. Structure and organization of two linked ribosomal protein genes in yeast. Nucleic Acids Res. 1984;12:7345–7358. doi: 10.1093/nar/12.19.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry V., Garrett R.A. The role of the basic N-terminal region of protein l18 in 5S RNA-23S RNA complex formation. Nucleic Acids Res. 1980;8:4131–4142. doi: 10.1093/nar/8.18.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M.A., Omranian N., Ahmadi F.S., Nikoloski Z., Mueller-Roeber B. Effect of salt stress on genes encoding translation-associated proteins in Arabidopsis thaliana. Plant Signal. Behav. 2012;7:1095–1102. doi: 10.4161/psb.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagni M., Ioannidis V., Cerutti L., Zahn-Zabal M., Jongeneel C.V., Hau J., Martin O., Kuznetsov D., Falquet L. MyHits: Improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:433–437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H., Snell E.A., Bapteste E., Lopez P., Holland P.W.H., Casane D. Phylogenomics of eukaryotes: Impact of missing data on large alignments. Mol. Biol. Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setterquist R.A., Smith G.K., Oakley T.H., Lee Y.H., Fox G.E. Sequence, overproduction and purification of Vibrio proteolyticus ribosomal protein L18 for in vitro and in vivo studies. Gene. 1996;183:237–242. doi: 10.1016/S0378-1119(96)00402-7. [DOI] [PubMed] [Google Scholar]
- Sormani R., Masclaux-Daubresse C., Daniele-Vedele F., Chardon F. Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0028070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung G.Y., Miller J.L., Koc H., Koc E.C. Comprehensive Analysis of Phosphorylated Proteins of E. coli Ribosomes. J. Proteome Res. 2008;8:3390–3402. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgers K.B., Alefantis T., Peyser B.D., Ruthel G.T., Bergeron A.A., Costantino J.A., Enterlein S., Kota K.P., Dutch Boltz R.C., Javad Aman M., DelVecchio V.G., Bavari S. Identification of essential filovirion-associated host factors by serial proteomic analysis and RNAi screen. Mol. Cell. Proteomics. 2010;9:2690–2703. doi: 10.1074/mcp.M110.003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J.A., Berg C., Hendrick J.P., La Branche-Chabot H., Metspalu A., Rinke J., Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz E., Noll H. Characterization of Cytoplasmic and Chloroplast Polysomes in Plants: Evidence for Three Classes of Ribosomal Rna in Nature. Proc. Natl. Acad. Sci. 1967;57:774–781. doi: 10.1073/pnas.57.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Shi C., Thudi M., Mariac C., Wallace J., Qi P., Zhang H., Zhao Y., Wang X., Rathore A., Srivastava R.K., Chitikineni A., Fan G., Bajaj P., Punnuri S., Gupta S.K., Wang H., Jiang Y., Couderc M., Katta M.A.V.S.K., Paudel D.R., Mungra K.D., Chen W., Harris-Shultz K.R., Garg V., Desai N., Doddamani D., Kane N.A., Conner J.A., Ghatak A., Chaturvedi P., Subramaniam S., Yadav O.P., Berthouly-Salazar C., Hamidou F., Wang Jianping, Liang X., Clotault J., Upadhyaya H.D., Cubry P., Rhoné B., Gueye M.C., Sunkar R., Dupuy C., Sparvoli F., Cheng S., Mahala R.S., Singh B., Yadav R.S., Lyons E., Datta S.K., Hash C.T., Devos K.M., Buckler E., Bennetzen J.L., Paterson A.H., Ozias-Akins P., Grando S., Wang Jun, Mohapatra T., Weckwerth W., Reif J.C., Liu X., Vigouroux Y., Xu X. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 2017;35:969. doi: 10.1038/nbt.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woestenenk E.A., Gongadze G.M., Shcherbakov D.V., Rak A.V., Garber M.B., Härd T., Berglund H. The solution structure of ribosomal protein L18 from Thermus thermophilus reveals a conserved RNA-binding fold. Biochem. J. 2002;363:553–561. doi: 10.1042/0264-6021:3630553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J.L., Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.