Abstract
Macrophages play crucial and diverse roles in the pathogenesis of inflammatory vascular diseases. Macrophages are the principal innate immune cells recruited to arterial walls to govern vascular homeostasis by modulating the proliferation of vascular smooth muscle cells, the reorganization of extracellular matrix components, the elimination of dead cells, and the restoration of normal blood flow. However, chronic sterile inflammation within the arterial walls draws inflammatory macrophages into intimal/neointimal regions that may contribute to disease pathogenesis. In this context, the accumulation and aberrant activation of macrophages in the neointimal regions govern the progression of inflammatory arterial wall diseases. Herein, we report that myeloid–hypoxia-inducible factor-1α (HIF1α) deficiency attenuates vascular smooth muscle cells and macrophage abundance in stenotic arteries and abrogates carotid neointima formation in vivo. The integrated transcriptomics, Gene Set Enrichment Analysis, metabolomics, and target gene evaluation showed that HIF1α represses oxidative phosphorylation, tricarboxylic acid cycle, fatty acid metabolism, and c-MYC signaling pathways while promoting inflammatory, glycolytic, hypoxia response gene expression in stenotic artery macrophages. At the molecular level, proinflammatory agents utilized STAT3 signaling pathways to elevate HIF1α expression in macrophages. Collectively, this study uncovers that macrophage-HIF1α deficiency restrains the pathogenesis of carotid artery stenosis by rewiring inflammatory and metabolic signaling pathways in macrophages.
Macrophages are the principal innate immune cells that play crucial and diverse roles in the pathogenesis of inflammatory vascular wall diseases.1 Clinical studies have shown that proinflammatory macrophages are associated with symptomatic and high-grade carotid artery stenosis.2,3 Patients with carotid artery stenosis are at an elevated risk for stroke and generally undergo carotid endarterectomy or stenting to restore the normal blood flow to the brain. Many of these patients develop restenosis or in-stent stenosis even after these procedures.4, 5, 6 Current pharmacologic therapies generally target blood pressure regulation and atherothrombotic pathways to treat complications associated with carotid artery stenosis.7, 8, 9 However, there is a lack of information regarding genetic factors or signaling pathways that govern carotid artery stenosis. Thus, the identification of such factors will significantly elevate the current molecular understanding of arterial stenosis and may facilitate the development of novel therapeutic approaches. Inflammatory macrophages that are recruited to stenotic arteries modulate the tissue repair/regeneration through matrix remodeling, migration, and proliferation of vascular smooth muscle cells (VSMCs), production of inflammatory and stenotic factors, and elimination of debris and dead cells.10 In this context, the accumulation and aberrant activation of macrophages within the intimal/neointimal regions govern the progression of inflammatory arterial wall diseases.11 The macrophage-mediated inflammation is an explicitly robust biological response that involves alterations in inflammatory and metabolic gene expression, affecting a significant part of the cellular genome.12 Therefore, transcription factors that govern inflammatory/metabolic gene expression play an essential role in shaping the macrophage phenotypic response.
The hypoxic microenvironment within the diseased arterial walls plays a pivotal role in the development and progression of carotid artery stenosis.13,14 Thus, inflammatory macrophages in these stenotic arteries have to adapt to the hypoxic microenvironment to perform effector functions. Studies over the years have shown that hypoxia-inducible factor-1 (HIF1) mediates adaptive responses to hypoxia/ischemia in metazoan organisms.15 HIF1 is a highly conserved member of the basic helix-loop-helix family of transcription factors that consists of oxygen-sensitive HIF1α and constitutively expressed HIF1β subunits. The HIF1α expression is tightly controlled at the mRNA and protein levels.16 In addition to hypoxia, several cytokines and inflammatory agents are known to elevate HIF1α mRNA expression and protein accumulation, even under normoxic conditions.17,18 Clinical and experimental studies have shown that macrophages within these diseased arterial walls express elevated levels of inflammatory and HIF1α target genes.19,20 Previous studies have shown that HIF1α deficiency attenuates some inflammatory and glycolytic gene expression in macrophages.21 However, whether HIF1α deficiency alters broad inflammatory or metabolic gene programs in macrophages has not been investigated. Moreover, whether myeloid-HIF1α deficiency affects the pathogenesis of carotid artery stenosis has not been examined. This study showed that HIF1α deficiency broadly attenuates proinflammatory gene expression while rewiring metabolic gene programs in macrophages. More importantly, myeloid-HIF1α–deficient mice are highly protected from experimentally induced carotid artery stenosis in vivo.
Materials and Methods
Materials
Anti-HIF1α (14179S), anti-STAT1 (14994S), anti–phosphorylated STAT1-Tyr701 (9167S), anti-STAT3 (4904S), anti–smooth muscle actin (1924S), anti-F480 (70076S), and anti–phosphorylated STAT3-Tyr705 (9145S) antibodies were obtained from Cell Signaling (Danvers, MA). Anti–β-actin (sc-130656) was obtained from Santa Cruz Biotechnology Inc. (Dallas, TX). The horseradish peroxidase–conjugated goat anti-rabbit (A16096) and horseradish peroxidase–conjugated goat anti-mouse (A16066) antibodies were obtained from Thermo Fisher Scientific (Waltham, MA). The wild-type (0664), Lyz2cre (04781), Hif1αfl/fl (7561), and mutant-Stat3 (027952) mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The HRE-luciferase (26731) and pGL2B-HIF1A promoter fragment (40172) were obtained from Addgene (Watertown, MA). The Lipofectamine3000 (L3000-008), TurboFect Transfection Reagent (R0533), bicinchoninic acid assay kit (23227), ECL Western Blotting Substrate (32106), random hexamers (N8080127), oligo-dT primer (18418020), Fast SYBR Green PCR Master Mix (4385612), and custom primers listed in this article were obtained from Thermo Fisher Scientific. The High Pure RNA Isolation Kits (11828665001), protease inhibitors (04693132001), and phosphatase inhibitors (04906837001) were obtained from Roche (Indianapolis, IN). The RAW264.7 cell line (TIB-71) was purchased from ATCC (Manassas, VA). The recombinant mouse macrophage colony-stimulating factor (416-ML) and mouse interferon (IFN)-γ (485-MI-100) were obtained from R&D Systems (Minneapolis, MN). The high-glucose Dulbecco’s modified Eagle’s medium (SH30249.02), phosphate-buffered saline (SH30028.03), and nitrocellulose membranes (10401197) were obtained from GE-Healthcare Life Sciences (Chicago, IL). The radioimmunoprecipitation assay buffer (R0278), bovine serum albumin (A3608), and thioglycollate (108190) were obtained from Millipore-Sigma (St. Louis, MO). The dual-Luciferase reporter assay system (E1960) was obtained from Promega (Madison, WI). The lipopolysaccharide (tlrl-pb5lps) was obtained from InvivoGen (San Diego, CA). The siStat1- and siStat3-specific siRNAs were obtained from GE Dharmacon (Lafayette, CO). All other chemicals and reagents used were of analytical grade and were obtained from commercial sources.
Experimental Animal Studies
All animal procedures were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University (Cleveland, OH) and conformed to guidelines established by the American Association for Accreditation of Laboratory Animal Care. All mice were bred and maintained under pathogen-free conditions, fed standard laboratory chow (2916; ENVIGO, Indianapolis, IN), and kept on a 12-hour light/dark cycle. The control (Lyz2cre/cre on C57BL/6J background) and myeloid-specific Hif1α-deficient (Hif1αfl/fl:Lyz2cre/cre on C57BL/6J background) mice were generating by breeding male and female Hif1αfl/−:Lyz2cre/cre mice. The Hif1αfl/fl:Lyz2cre/cre mice contained two Hif1α floxed and two Lyz2 Cre alleles (C57BL/6J background). Mice with two Cre alleles (Lyz2cre/cre) were used as the control group (C57BL/6J background). The carotid artery ligation model22 was utilized to investigate whether myeloid-HIF1α deficiency affects the pathogenesis of carotid artery stenosis. Accordingly, the left common carotid artery of Hif1αfl/fl:Lyz2cre/cre and Lyz2cre/cre mice was dissected and ligated near the carotid bifurcation to induce arterial stenosis. The sham surgery on the right carotid served as a control. After 28 days of these procedures, study mice were euthanized, and the control/ligated carotid arteries were collected. These carotid arteries were fixed in 10% of buffered formalin and embedded in paraffin. A standardized reference point was set at which location the ligature did not distort the vessel and where elastic laminae were intact. The reference point was situated between 50 and 75 μm from the ligation. The sections at 100, 200, and 300 μm from the reference point were collected for histologic analyses. The images derived from all of the cross-sections were utilized for analyses. The elastin structures within the carotid arteries were visually detected by using a modified Verhoeff Van Gieson elastic stain kit. To determine the VSMCs and macrophage abundance in carotid arteries, serial paraffin sections of carotid arteries were deparaffinized in xylene and rehydrated in graded ethanol series. Samples were subjected to antigen retrieval steps with antigen unmasking solution. Samples were treated with 0.3% H2O2 for 30 minutes at room temperature, and the nonspecific binding was blocked with a blocking buffer. Samples were incubated with rabbit anti–smooth muscle actin or anti-F4/80 antibodies overnight at 4°C. These tissue sections were subsequently incubated with biotin-conjugated goat anti-rabbit IgG for 30 minutes at room temperature. Samples were incubated in ABC reagent, and the immunostaining was visualized using a diaminobenzidine reagent. Images were acquired using a microscope, and ImageJ version 1.53h software (NIH, Bethesda, MD; https://imagej.nih.gov/ij, last accessed February 10, 2021) was utilized for image analyses. The circumferences of the lumen, internal elastic lamina (IEL), and external elastic lamina (EEL) were measured by tracing along the luminal surface, IEL, and EEL, respectively. Under the assumption that the structures were circular, these measurements were used to calculate luminal area, IEL area, and EEL area. The neointimal area was calculated by subtracting the luminal area from the area defined by IEL. The medial area was calculated by subtracting the area within the IEL from the area defined by EEL. The total vessel area was calculated by tracings obtained by the external perimeter of the artery. The quantification of histologic data and statistical analyses were performed by two independent investigators who were blinded to sample genotypes and/or treatments. Furthermore, no experimental animal or data from the experiments were excluded from analyses.
Cell Culture and Metabolic Assays
RAW264.7 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 10 μg/mL streptomycin, and 2 mmol/L glutamine in a humidified incubator (5% CO2 and 37°C). Bone marrow–derived macrophages (BMDMs) were generated by ex vivo differentiation of bone marrow cells. Briefly, bone marrow cells from 8-week–old wild-type, Mut-Stat3, Lyz2cre/cre, and Hif1αfl/fl:Lyz2cre/cre mice were harvested from the femur and tibia. These bone marrow cells were cultured in complete Dulbecco's modified Eagle medium supplemented with recombinant mouse macrophage colony-stimulating factor for 7 days. These BMDMs were collected and utilized for the indicated experiments. The primary macrophages from control and ligated mouse carotid arteries are obtained 4 weeks after the procedure. Briefly, control and ligated carotid arteries were harvested and washed with a copious amount of sterile, ice-cold phosphate-buffered saline to remove blood cells. The carotid tissues were minced with scalpel blades in petri plates under filtered laminar airflow. The minced tissues were incubated with 3 mg/mL collagenase-dispase mixture for 4 hours at room temperature with gentle shaking. The cellular suspension was collected by using a tissue strainer. The anti-F4/80 microbeads (130-110-443; Miltenyi Biotec, Auburn, CA) were used to purify macrophages from these cellular suspensions. The oxygen consumption rate of BMDMs was analyzed by using Seahorse XFe24 Analyzer (Agilent, Santa Clara, CA). Baseline measurements were made to determine basal oxygen consumption rate before the sequential delivery of 1 μmol/L oligomycin, 1 μmol/L fluorocarbonyl cyanide phenylhydrazone, and 1 μmol/L mixture of antimycin A and rotenone through individual injection ports for the measurements of ATP-linked respiration, proton leak, maximum respiration, and spare respiration capacity, respectively. The oxygen consumption rate values were normalized to the protein concentration.
RNA Extraction, Real-Time Quantitative PCR, and Western Blot Analysis
Total RNA was isolated from indicated samples using the High Pure RNA Isolation Kit. The total RNA samples were reverse transcribed using M-MuLV reverse transcriptase in the presence of random hexamers and oligo-dT primers. Real-time quantitative PCR was performed using Universal SYBR Green PCR Master Mix or TaqMan Universal Master Mix on Applied Biosystems (Foster City, CA) Step One Plus real-time PCR system in the presence of gene-specific primers. The list of primers utilized in this study is provided in Table 1. Indicated primary cells and cell lines were lysed in ice-cold radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors. Protein concentration was measured by the bicinchoninic acid protein assay. An equal amount of protein samples was electrophoresed using 8% or 4% to 15% Mini-PROTEAN TGXTM precast gels (Bio-Rad) and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk or 5% bovine serum albumin in Tris-buffered saline with Tween 20 for 1 hour at room temperature. These blots were further incubated with primary antibodies diluted in 5% bovine serum albumin in Tris-buffered saline with Tween 20. After overnight incubation, primary antibodies were removed by washing with Tris-buffered saline with Tween 20. These blots were incubated for 1 hour at room temperature in horseradish peroxidase–conjugated secondary antibodies. Blots were visualized using enhanced chemiluminescence Western Blotting Substrate. The densitometry analyses were performed utilizing ImageJ software. The primary antibodies were used at the following dilutions: HIF1α (1:1000); STAT1, STAT3, phosphorylated STAT1, and phosphorylated STAT3 (1:2500); and β-actin (1:5000).
Table 1.
List of Primers Used for Experiments
| Target gene | Forward primer | Reverse primer |
|---|---|---|
| Glut1 | 5′-GGATCCCAGCAGCAAGAAG-3′ | 5′-CCAGTGTTATAGCCGAACTGC-3′ |
| Hk2 | 5′-TGATCGCCTGCTTATTCACGG-3′ | 5′-AACCGCCTAGAAATCTCCAGA-3′ |
| Pfkfb3 | 5′-CAACTCCCCAACCGTGATTGT-3′ | 5′-TGAGGTAGCGAGTCAGCTTCT-3′ |
| Tpi1 | 5′-AAACCAAGGTCATCGCAGATA-3′ | 5′-CCCGGAGCTTCTCGTGTA-3′ |
| Pgam1 | 5′-GCTGTGGTGTGACCAATGAA-3′ | 5′-TGACTATGCCCCCAGTTACC-3′ |
| Il1α | 5′-GCACCTTACACCTACCAGAGT-3′ | 5′-AAACTTCTGCCTGACGAGCTT-3′ |
| Il1β | 5′-GCAACTGTTCCTGAACTCAACT-3′ | 5′-ATCTTTTGGGGTCCGTCAACT-3′ |
| Icam1 | 5′-CCCACGCTACCTCTGCTC-3′ | 5′-GATGGATACCTGAGCATCACC-3′ |
| Hif1α | 5′-TGAGTTCTGAACGTCGAAAAGA-3′ | 5′-CGGCATCCAGAAGTTTTCTC-3′ |
| Serpine1 | 5′-TTCAGCCCTTGCTTGCCTC-3′ | 5′-ACACTTTTACTCCGAAGTCGGT-3′ |
| Ddx18 | 5′-GAAGCGGAATGCCAAGCTG-3′ | 5′-TCCTGTTTCTTTAGGCACATCTC-3′ |
| Hspe1 | 5′-AGTTTCTTCCGCTCTTTGACAG-3′ | 5′-TGCCACCTTTGGTTACAGTTTC-3′ |
| Xrcc6 | 5′-ATGTCAGAGTGGGAGTCCTAC-3′ | 5′-TCGCTGCTTATGATCTTACTGGT-3′ |
| Ndufs8 | 5′-AGTGGCGGCAACGTACAAG-3′ | 5′-TCGAAAGAGGTAACTTAGGGTCA-3′ |
| Atp6v1f | 5′-GCGGGCAGAGGTAAGCTAATC-3′ | 5′-TTAGGGTGGCGGTTCTTGTTT-3′ |
| Uqcrfs1 | 5′-GAGCCACCTGTTCTGGATGTG-3′ | 5′-GCACGACGATAGTCAGAGAAGTC-3′ |
| Idh2 | 5′-GGAGAAGCCGGTAGTGGAGAT-3′ | 5′-GGTCTGGTCACGGTTTGGAA-3′ |
| Mdh1 | 5′-TTCTGGACGGTGTCCTGATG-3′ | 5′-TTTCACATTGGCTTTCAGTAGGT-3′ |
| Suclg2 | 5′-CCCCGAAGATGGCTGAACC-3′ | 5′-ACCTCCTTTCAAACCGCTATTG-3′ |
| 36B4 | 5′-GCTCCAAGCAGATGCAGCA-3′ | 5′-CCGGATGTGAGGCAGCAG-3′ |
Polar Metabolite Profiling by Liquid Chromatography–Mass Spectrometry
Targeted polar metabolite profiling was performed as described before.23 Briefly, macrophages after indicated treatments were washed with sterilized 0.9% NaCl and incubated with extraction buffer [80% methanol, 20% H2O plus isotopically labeled internal standards (MSK-A2-1.2; Cambridge Isotope Laboratories)]. Cells were harvested by scraping and were subjected to vortexing for 10 minutes at 4°C, and the debris was pelleted by a 10-minute spin at 18,000 × g. The supernatant was then transferred to a new tube and dried under nitrogen. Dried polar samples were resuspended in 100 μL water, and 2 μL was injected into a ZIC-pHILIC 150 × 2.1-mm (5-μm particle size) column. The analysis was conducted on a QExactive benchtop Orbitrap mass spectrometer equipped with an Ion Max source and an HESI II probe, which was coupled to a Dionex UltiMate 3000 UPLC system (Thermo Fisher Scientific). External mass calibration was performed using the standard calibration mixture every 7 days. Chromatographic separation was achieved using the following conditions: buffer A was 20 mmol/L ammonium carbonate and 0.1% ammonium hydroxide; and buffer B was acetonitrile. The column oven and autosampler tray were held at 25°C and 4°C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/minute as follows: 0 to 20 minutes: linear gradient from 80% to 20% B; 20 to 20.5 minutes: linear gradient from 20% to 80% B; 20.5 to 28 minutes: hold at 80% B. The mass spectrometer was operated in full-scan, polarity switching mode with the spray voltage set to 3.0 kV, the heated capillary held at 275°C, and the HESI probe held at 350°C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. The data acquisition was performed over a range of 70 to 1000 m/z, with the resolution set at 70,000, the automatic gain control target at 10e6, and the maximum injection time at 20 milliseconds. Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards. Metabolite levels were normalized to the total protein amount for each condition.
Transient Transfection, Luciferase Assay, and ChIP Analysis
Wild-type mice BMDMs were transfected with indicated siRNA using the Lipofectamine reagent, according to the manufacturer's instructions. These transfected BMDMs were stimulated with cytokines and subjected to specified analyses. RAW264.7 cells were transfected with HIF1α promoter-driven luciferase reporter plasmid in the presence and absence of Stat3-specific siRNA. These cells were stimulated with indicated cytokines, and luciferase reporter activity was measured and normalized according to the manufacturer's instructions. Results are presented as relative luciferase fold change over the control group (cells treated with phosphate-buffered saline).
Chromatin immunoprecipitation (ChIP) analyses were performed using the EZ-Magna ChIP G kit (Millipore-Sigma), according to the manufacturer's instruction. Briefly, BMDMs derived from wild-type mice were separately stimulated with LPS or IFN-γ. ChIP analyses were performed using the anti-STAT3 antibody. Chromatin samples from these experiments were analyzed by real-time quantitative RT-PCR (forward: 5-TGTATGTGTGTGTGAGTGTGTTTG-3; reverse: 5-CAGGTACATGCTGGCTCACT-3). ChIP, performed using isotype IgG, was used as a negative control.
RNAseq Analysis
Quality control of total RNA samples was assessed using Qubit (Thermo Fisher Scientific) for quantification and Agilent 2100 BioAnalyzer (Santa Clara, CA) analysis to assess quality using a cutoff of RNA integrity number >7.0 to select specimens for further analysis. cDNA library for RNA sequencing (RNAseq) was generated from 150 ng of total RNA using the Illumina (San Diego, CA) TruSeq Stranded Total RNA kit with Ribo Zero Gold for rRNA removal, according to the manufacturer's protocol. The resulting purified mRNA was used as input for the Illumina TruSeq kit, in which libraries are tagged with unique adapter indexes. Final libraries were validated on the Agilent 2100 BioAnalyzer, quantified via real-time quantitative PCR, and pooled at equimolar ratios. Pooled libraries were diluted, denatured, and loaded onto the Illumina NextSeq 550 System using a high-output flowcell. The aligned reads were then analyzed with Cufflinks version name 2.2.1 to obtain gene-level expression data using the GENCODE gene annotation for mm10 version GRCm38.p6 and reported as fragments per kilobase per million reads mapped. Differential expression analysis was also performed, and significantly differentially expressed genes were defined using an adjusted P value <0.05 (false discovery rate corrected). Gene expression tables for relevant pairwise comparisons were analyzed for Gene Set Enrichment Analysis (GSEA)24 using GenePattern version v3.9.11-rc.5 b234 (Broad Institute, Cambridge, MA). Pathway data sets that were examined included Kyoto Encyclopedia of Genes and Genomes and Hallmark gene sets. A gene set was considered to be significantly enriched using an FWER cutoff <0.05. Heat maps were generated using ClustVis (latest release December 20, 2018).25 The sequencing data reported in this article have been deposited in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131364).
Quantification and Statistical Analysis
All data, unless indicated, are presented as the means ± SD. The statistical significance of differences between the two groups was analyzed by t-test or two-way analysis of variance with Bonferroni multiple comparison tests.
Results
Myeloid-HIF1α Deficiency Attenuates the Pathogenesis of Carotid Artery Stenosis
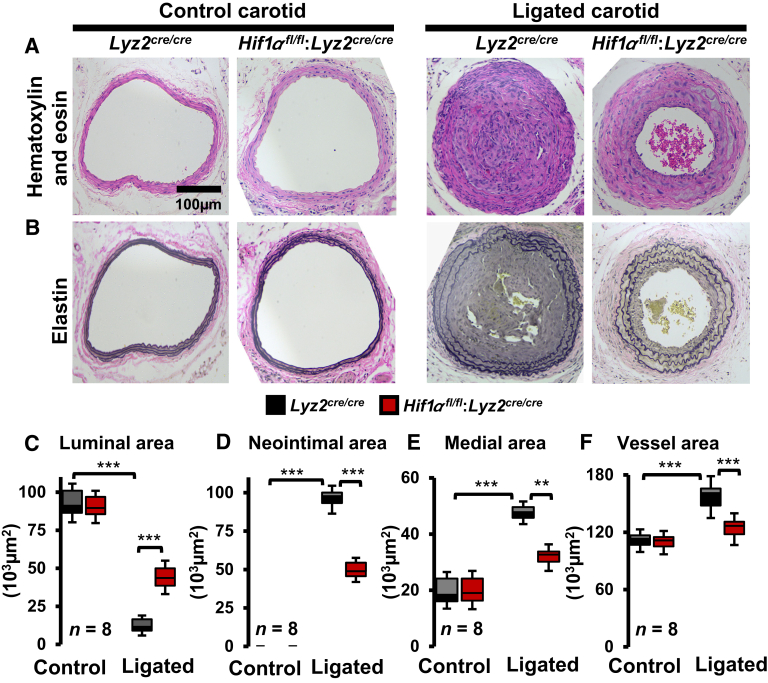
Previous experimental and clinical studies have shown that HIF1α levels are elevated in stenotic arteries.20,26 Therefore, this study was intended to examine whether myeloid-HIF1α deficiency alters experimentally induced carotid artery stenosis in vivo. Accordingly, the carotid artery flow cessation model was utilized to evaluate the effects of myeloid-HIF1α deficiency on intimal hyperplasia and vascular remodeling. The morphometric analyses show that myeloid-HIF1α–deficient mice are highly protected from ligation-induced vascular remodeling (Figure 1, A and B). Interestingly, carotid artery flow cessation dramatically reduced the luminal area in Lyz2cre/cre mice and exhibited extensive carotid artery occlusion (Figure 1C). However, myeloid-HIF1α–deficient mice displayed a modest reduction (approximately 50%) in the luminal area and a partial increase in carotid artery occlusion (Figure 1C). Furthermore, ligation-induced cessation of carotid artery blood flow significantly elevated neointimal formation and medial thickening in Lyz2cre/cre mice (Figure 1, D and E). Surprisingly, myeloid-HIF1α–deficient mice exhibit significant protection from ligation-induced carotid neointimal formation and medial thickening (Figure 1, D and E). Concordantly, Lyz2cre/cre mice displayed a substantial increase in total vessel area compared with myeloid-HIF1α–deficient mice following carotid artery ligation (Figure 1F). Collectively, these analyses show that myeloid-HIF1α deficiency is protective against the pathogenesis of carotid artery stenosis in vivo. Next, studies have examined whether myeloid-HIF1α deficiency alters cellular components of stenotic carotid arteries in vivo. Accordingly, stenotic and control carotid artery transverse sections from Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice were examined for vascular smooth muscle cells and macrophage abundance. The analyses show a profound investment of vascular smooth muscle cells into neointimal regions of stenotic carotid arteries in Lyz2cre/cre mice (Figure 2, A and C). However, myeloid-HIF1α–deficient mice exhibited a dramatic reduction in vascular smooth muscle cell investment into neointimal regions of stenotic carotid arteries (Figure 2, A and C). Interestingly, myeloid-HIF1α deficiency significantly attenuated macrophage abundance in stenotic carotid arteries compared with Lyz2cre/cre mice (Figure 2, B and D). Taken together, these results demonstrate that myeloid-HIF1α deficiency alleviates carotid artery stenosis in vivo.
Figure 1.
Myeloid–hypoxia-inducible factor-1α (HIF1α) deficiency attenuates carotid artery stenosis. A and B: Histologic analyses of control and ligated carotid arteries of Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice. A and B: Representative cross-sections of control and ligated (28 days) carotid arteries with hematoxylin-eosin (A) and elastin (B) staining. C–F: The quantitative analyses of Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice carotid luminal area (C), neointimal area (D), medial area (E), and total vessel area (F) of control and ligated groups. Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (C–F). n = 8 (A–F). ∗∗P < 0.01, ∗∗∗P < 0.001. Scale bar = 100 μm (A and B).
Figure 2.
Myeloid–hypoxia-inducible factor-1α (HIF1α) deficiency reshapes cellular components of stenotic arteries. The Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice control and ligated (28 days) carotid artery cross-sections were stained for vascular smooth muscle cells (A) and macrophages (B) by using anti–smooth muscle actin and anti-F4/80 antibodies, respectively. The vascular smooth muscle cell (C) and macrophage (D) areas were quantified using ImageJ software. Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (C and D). n = 8 (A–D). ∗∗∗P < 0.001. Scale bar = 100 μm (A and B).
HIF1α Deficiency Curtails Inflammatory Genes Involved in Carotid Stenosis
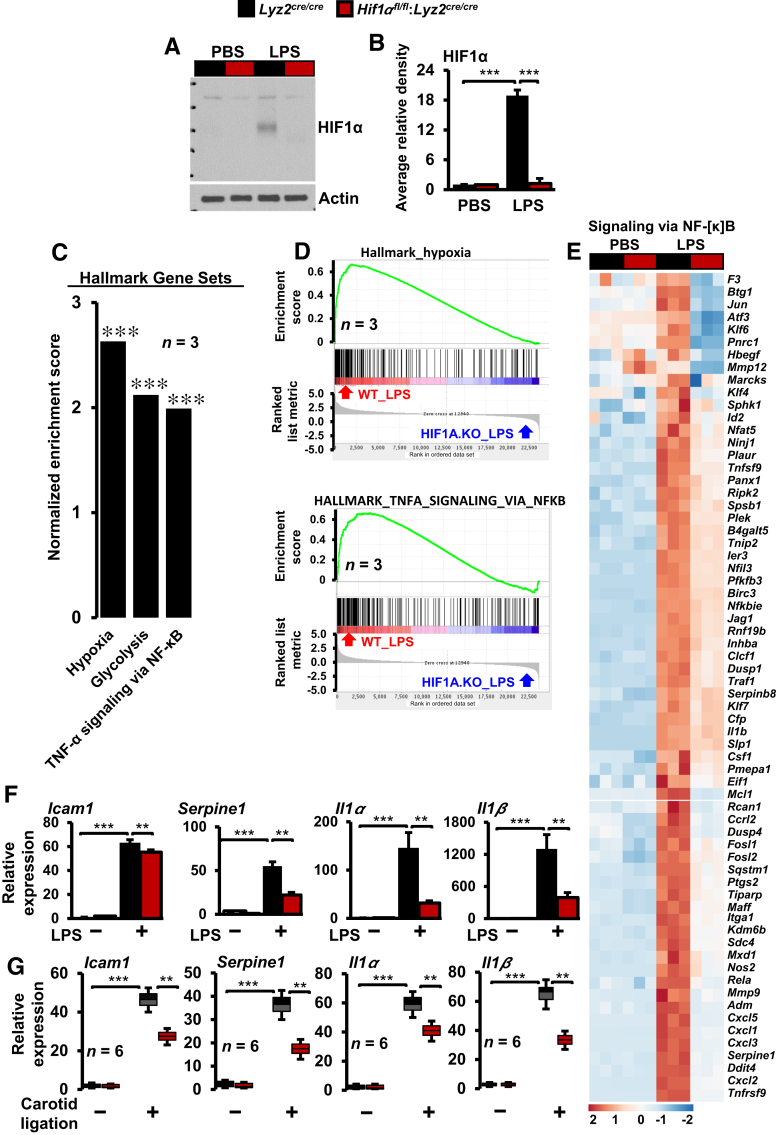
The in vivo analyses demonstrated that myeloid-HIF1α deficiency significantly attenuated macrophage abundance in stenotic arteries and averted substantial vascular remodeling. Therefore, the impact of HIF1α deficiency on inflammatory gene expression was assessed. First, the reduction in HIF1α protein level in Hif1αfl/fl:Lyz2cre/cre mice BMDMs was confirmed by Western blot analysis (Figure 3, A and B). Previous studies have demonstrated that lipopolysaccharides (LPSs) are potent and robust inducers of inflammatory gene expression in macrophages.27 Therefore, LPS was utilized to induce expansive changes in signaling pathways and inflammatory gene expression in macrophages. Accordingly, Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs were stimulated with LPS, and total RNA samples were obtained to perform gene expression profiling studies. To identify signaling pathways that were dysregulated because of HIF1α deficiency, RNAseq data were subjected to GSEA.24 The analyses show that HIF1α deficiency significantly attenuated hypoxia response, glycolysis, and tumor necrosis factor–induced NF-κB–regulated inflammatory gene expression in macrophages (Figure 3, C and D). As shown in Figure 3E, LPS exposure significantly induced a large number of proinflammatory gene targets in Lyz2cre/cre mice BMDMs (Figure 3E). However, the induction of these proinflammatory gene targets was significantly curtailed in HIF1α-deficient macrophages (Figure 3E). Furthermore, these observations were validated on a small subset of inflammatory gene targets by real-time quantitative PCR analyses. As shown in Figure 3F, LPS stimulation robustly induced Icam1, Serpine1, Il1α, and Il1β expression in Lyz2cre/cre mice BMDMs. However, LPS-induced expression of these gene targets was significantly curtailed in HIF1α-deficient BMDMs (Figure 3F). Next, these differentially expressed gene expression profiles were used as a guide to probe dysregulated inflammatory gene targets in stenotic carotid artery macrophages. As shown in Figure 3G, several proinflammatory (Icam1, Serpine1, Il1α, and Il1β) gene targets were substantially attenuated in myeloid-HIF1α–deficient carotid artery macrophages. Collectively, these analyses show that HIF1α deficiency broadly attenuates critical proinflammatory genes involved in carotid artery stenosis.
Figure 3.
Inflammatory gene expression in stenotic artery macrophages. A and B:Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice bone marrow–derived macrophages (BMDMs) were stimulated with 100 ng/mL lipopolysaccharide (LPS) for 4 hours. A and B: The hypoxia-inducible factor-1α (HIF1α) protein levels were evaluated by Western blots (A), and densitometry analysis (B) was performed by utilizing ImageJ software. C and D: Gene Set Enrichment Analysis of RNA-sequencing data (C) and enrichment plots (D) from LPS-treated Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs utilizing Hallmark gene sets. E: Heat map of inflammatory genes that are dysregulated in HIF1α-deficient BMDMs. F: The Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs treated with LPS and total RNA samples were analyzed for expression of Icam1, Serpine1, Il1α, and Il1β by RT-qPCR. G: Total RNA samples from Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice control and stenotic carotid artery macrophages were analyzed for expression of Icam1, Serpine1, Il1α, and Il1β by RT-qPCR. Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (B, C, F, and G). n = 3 (A–E); n = 4 (F); n = 6 (G). ∗∗P < 0.01, ∗∗∗P < 0.001. PBS, phosphate-buffered saline; TNF-α, tumor necrosis factor-α; WT, wild type.
Glycolytic Gene Expression in Stenotic Artery Macrophages
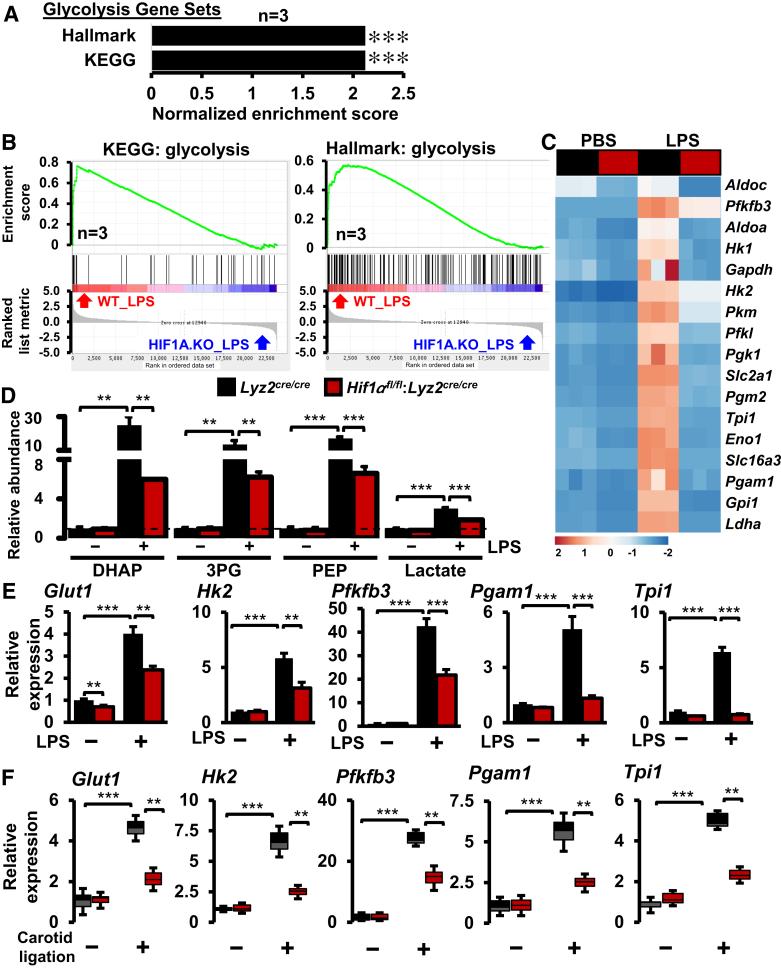
Previous studies have shown that proinflammatory macrophage activation and gene expression are dependent on the processes of glycolysis.28 Therefore, this study intended to examine the glycolytic gene expression changes in stenotic carotid artery macrophages. Interestingly, the GSEA of RNAseq studies utilizing Hallmark and Kyoto Encyclopedia of Genes and Genomes gene sets shows that LPS-induced glycolytic gene expressions were significantly attenuated in HIF1α-deficient BMDMs (Figure 4, A and B). As shown in Figure 4C, LPS exposure vigorously elevated the principle glycolytic gene expression in Lyz2cre/cre mice BMDMs. However, the induction of these glycolytic genes was significantly curtailed in HIF1α-deficient BMDMs (Figure 4C). Concordantly, HIF1α deficiency significantly attenuates LPS-induced augmentation of these glycolytic metabolites in primary macrophages (Figure 4D). Furthermore, real-time quantitative PCR studies show that these critical glycolytic gene (Glut1, Hk2, Pfkfb3, Pgam1, and Tpi1) expression levels were significantly attenuated in LPS-treated HIF1α-deficient BMDMs (Figure 4E). Next, these differentially expressed glycolytic genes were utilized as a lead to probe the dysregulated glycolytic gene targets in stenotic carotid artery macrophages in vivo. As shown in Figure 4F, the expression of these essential glycolytic genes (Glut1, Hk2, Pfkfb3, Pgam1, and Tpi1) was significantly elevated in Lyz2cre/cre mice stenotic artery macrophages. As anticipated, the induction of these glycolytic genes was substantially diminished in Hif1αfl/fl:Lyz2cre/cre mice stenotic carotid artery macrophages (Figure 4F). Collectively, these analyses show that myeloid-HIF1α deficiency attenuates proinflammatory and glycolytic gene expression in stenotic carotid artery macrophages.
Figure 4.
Glycolytic gene expression in stenotic artery macrophages. A and B: Gene Set Enrichment Analysis of RNA-sequencing data (A) and enrichment plots (B) of glycolytic gene sets that are obtained by comparing Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice bone marrow–derived macrophages (BMDMs) treated with lipopolysaccharide (LPS). C: Heat map of select glycolysis genes that are altered in Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs following LPS treatment. D:Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs were stimulated with 100 ng/mL LPS for 6 hours. Indicated glycolytic metabolites levels were quantified by liquid chromatography–mass spectrometry–based targeted metabolomics analyses. E:Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs were stimulated with 100 ng/mL LPS for 6 hours. Total RNA extracts were analyzed for expression of Glut1, Hk2, Pfkfb3, Pgam1, and Tpi1 by RT-qPCR. F: Total RNA samples from Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice control and stenotic carotid artery macrophages were analyzed for expression of Glut1, Hk2, Pfkfb3, Pgam1, and Tpi1 by RT-qPCR. Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (D–F). n = 4 (D and E); n = 6 (F). ∗∗P < 0.01, ∗∗∗P < 0.001. KEGG, Kyoto Encyclopedia of Genes and Genomes; PBS, phosphate-buffered saline; WT, wild type.
HIF1α Deficiency Augments Lipid Metabolism and MYC Pathways
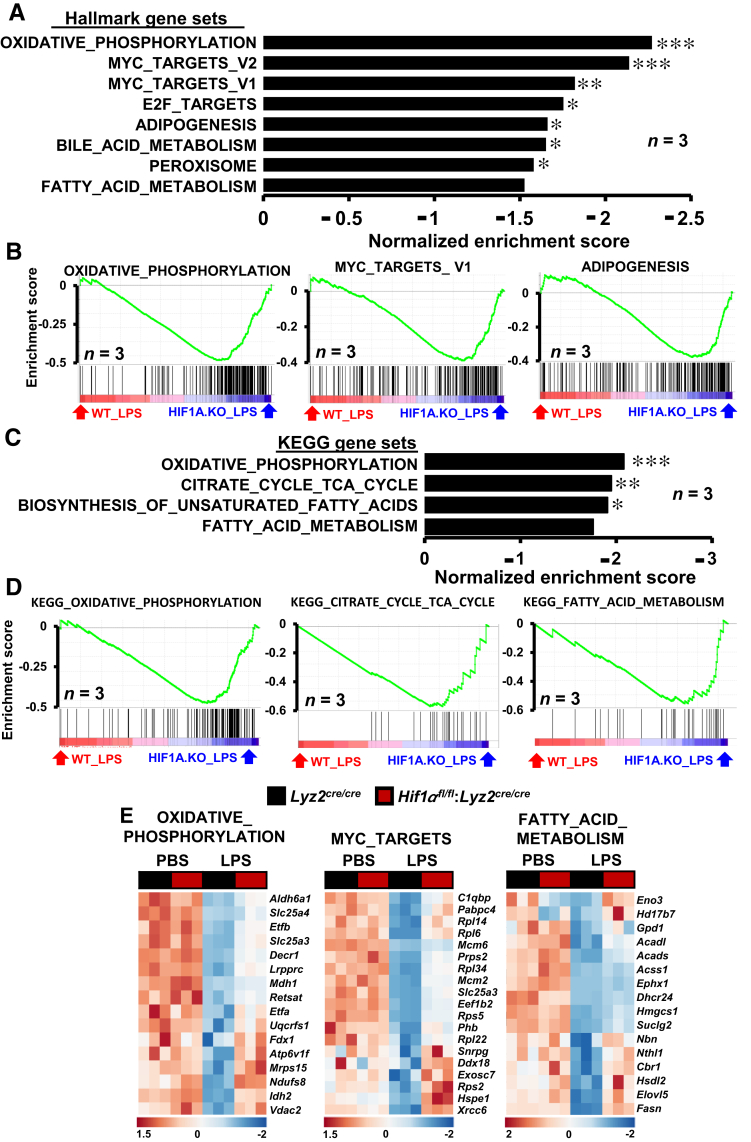
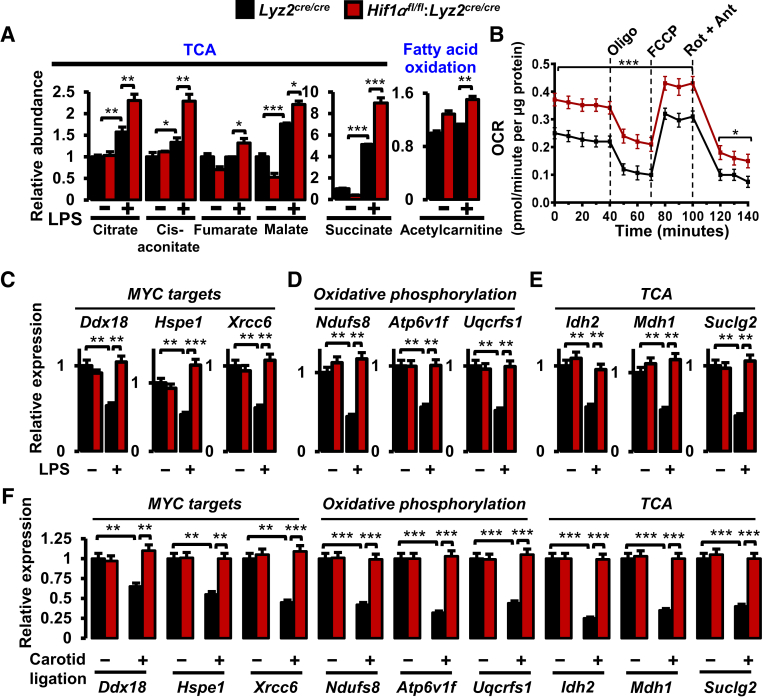
It is conceivable that HIF1α could promote proinflammatory and glycolytic gene expression while repressing anti-inflammatory and lipid metabolism gene expression in stenotic artery macrophages. Therefore, one could hypothesize that HIF1α deficiency might result in heightened signaling and metabolic pathways that support anti-inflammatory macrophage activation. To test this hypothesis, data from RNAseq studies were subjected to GSEA utilizing Hallmark and Kyoto Encyclopedia of Genes and Genomes databases. As shown in Figure 5, A and B, gene targets associated with oxidative phosphorylation, MYC target expression, and fatty acid metabolism are significantly enriched in HIF1α-deficient BMDMs following LPS treatment. More important, concomitant analyses utilizing Kyoto Encyclopedia of Genes and Genomes data sets further corroborated the overrepresentation of oxidative phosphorylation, tricarboxylic acid cycle (TCA), and fatty acid metabolism pathways in HIF1α-deficient BMDMs following LPS exposure (Figure 5, C and D). Furthermore, as shown in Figure 5E, LPS treatment significantly attenuated oxidative phosphorylation, MYC targets, and fatty acid metabolism gene expression in macrophages. Interestingly, LPS-exerted repression of these target genes was substantially abrogated in HIF1α-deficient macrophages (Figure 5E). Moreover, cellular metabolite analyses following LPS exposure show that HIF1α deficiency significantly elevated the critical TCA and fatty acid oxidation metabolite levels in macrophages (Figure 6A). Furthermore, LPS-induced HIF1α-deficient BMDMs exhibited elevated oxygen consumption rates compared with Lyz2cre/cre mice BMDMs treated with LPS (Figure 6B). Moreover, LPS-induced HIF1α-deficient BMDMs also show heightened ATP-linked respiration, proton leak, maximum respiration, and spare respiration capacity (Figure 6B). Subsequently, the GSEA observations (Figure 5) were corroborated on a subset of MYC, oxidative phosphorylation, and TCA gene targets by real-time quantitative PCR analysis. As shown in Figure 6, C–E, HIF1α-deficient BMDMs are highly protected from LPS-exerted repression of MYC targets (Ddx18, Hspe1, and Xrcc6), and genes involved in oxidative phosphorylation (Ndufs8, Atp6v1f, and Uqcrfs1) as well as TCA (Idh2, Mdh1, and Suclg2). Next, GSEA findings (Figure 5) were utilized as a pointer to probe dysregulated oxidative phosphorylation, TCA, and MYC target gene expression in the stenotic carotid artery macrophages. As shown in Figure 6F, expression of MYC targets (Ddx18, Hspe1, and Xrcc6) and genes that facilitate oxidative phosphorylation (Ndufs8, Atp6v1f, and Uqcrfs1) as well as TCA (Idh2, Mdh1, and Suclg2) is significantly attenuated in Lyz2cre/cre mice stenotic artery macrophages. Interestingly, HIF1α deficiency completely abrogates the repression of these MYC targets, oxidative phosphorylation, and TCA gene expression in stenotic artery macrophages. Collectively, these analyses show that HIF1α deficiency results in limited repression of MYC targets, oxidative phosphorylation, and TCA gene expression in the face of inflammatory challenges.
Figure 5.
Gene set enrichments in hypoxia-inducible factor-1α (HIF1α)–deficient macrophages. A and B: RNA-sequencing (RNAseq) data from lipopolysaccharide (LPS)–treated Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice bone marrow–derived macrophages (BMDMs) were subjected to Gene Set Enrichment Analysis (GSEA) utilizing Hallmark gene sets. A and B: The gene sets enriched in Hif1αfl/fl:Lyz2cre/cre mice BMDMs (A) and corresponding enrichment plots (B) are shown. C and D: RNAseq data from LPS-treated Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs were subjected to GSEA utilizing Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets. C and D: The gene sets enriched in HIF1α-deficient mice BMDMs (C) and corresponding enrichment plots (D) are shown. E: Heat map of select oxidative phosphorylation, MYC targets, and fatty acid metabolism genes that are dysregulated in Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs following LPS treatment. FWER P value <0.05 was considered significant. n = 3 (A–D). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (FWER values). PBS, phosphate-buffered saline.
Figure 6.
Impact of hypoxia-inducible factor-1α (HIF1α) deficiency on lipid metabolism and MYC signaling pathways. A:Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice bone marrow–derived macrophages (BMDMs) were stimulated with 100 ng/mL lipopolysaccharide (LPS) for 6 hours. Indicated tricarboxylic acid cycle (TCA) metabolites were quantified by liquid chromatography–mass spectrometry–based targeted metabolomics analyses. B: The oxygen consumption rate (OCR) of LPS-induced Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs. C–E:Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice BMDMs were stimulated with 100 ng/mL LPS for 6 hours. C–E: Total RNA extracts were analyzed for expression of MYC targets (Ddx18, Hspe1, and Xrcc6; C), oxidative phosphorylation (Ndufs8, Atp6v1f, and Uqcrfs1; D), and TCA (Idh2, Mdh1, and Suclg2; E) gene targets by RT-qPCR. F: Total RNA samples from Lyz2cre/cre and Hif1αfl/fl:Lyz2cre/cre mice control and stenotic carotid artery macrophages were analyzed for expression of indicated MYC targets, oxidative phosphorylation, and TCA genes by RT-qPCR. Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (A–F). n = 3 (A and B); n = 4 (C–E); n = 6 (F). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Ant, antimycin A; FCCP, fluorocarbonyl cyanide phenylhydrazone; Oligo, oligomycin; Rot, rotenone.
STAT3 Signaling Supports HIF1α Expression in Macrophages
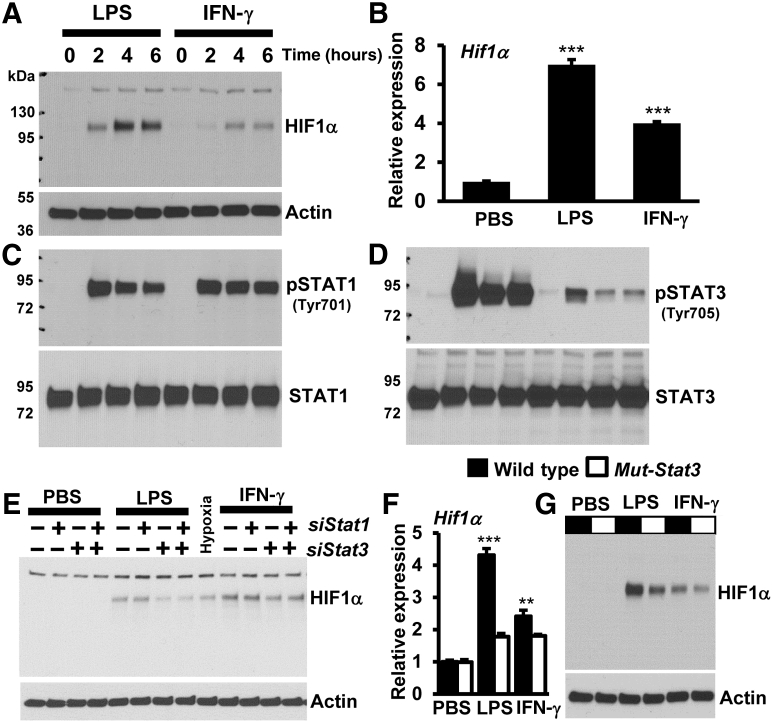
Next, studies were designed to identify signaling events that support HIF1α expression in macrophages. Accordingly, wild-type mice BMDMs were separately stimulated with LPS or IFN-γ, and total protein extracts were subjected to Western blot analyses. Proinflammatory agent exposure elevated HIF1α protein levels in macrophages (Figure 7A). These observations were also recapitulated at Hif1α mRNA expression (Figure 7B). Subsequently, the impact of proinflammatory agent exposure on the activation of STATs was examined in macrophages. The results show that LPS or IFN-γ exposure robustly induced STAT1 and STAT3 phosphorylation in macrophages (Figure 7, C and D). Interestingly, the magnitude of STAT3 phosphorylation level appears to reflect the HIF1α protein expression in macrophages. Therefore, studies were designed to examine whether the silencing of STAT1 or STAT3 expression altered LPS- or IFN-γ–induced HIF1α expression in primary macrophages. As shown in Figure 7E, the silencing of STAT1 expression did not attenuate LPS- or IFN-γ–induced HIF1α expression in wild-type mice BMDMs. However, genetic inhibition of STAT3 substantially attenuated LPS- or IFN-γ–induced HIF1α protein levels in macrophages (Figure 7E). Interestingly, the silencing of both STAT1 and STAT3 did not offer any additional repressive effects on LPS- or IFN-γ–induced HIF1α expression in wild-type mice BMDMs (Figure 7E). More importantly, expression of dominant-negative mutant Stat329 significantly attenuated LPS- or IFN-γ–induced HIF1α mRNA and protein expression in primary macrophages (Figure 7, F and G). Furthermore, analyses show that the silencing of STAT3 expression significantly attenuated LPS- or IFN-γ–induced HIF1α promoter-driven luciferase reporter activity in macrophages (Figure 8A). Next, studies were performed to examine whether STAT3 occupies the Hif1α promoter following proinflammatory cytokine exposure. As shown in Figure 8B, ChIP analysis demonstrated that proinflammatory agent treatment significantly elevated STAT3 accumulation on Hif1α promoter (−1041 to −1051) in wild-type mice BMDMs. Subsequently, studies were designed to examine whether inhibition of STAT3 attenuates HIF1α-driven proinflammatory gene expression in macrophages, and overexpression of HIF1α could reverse this phenotype. Genetic approaches were utilized to silence STAT3 (siStat3) and overexpress the oxygen-stable form of HIF1α (ΔHif1α-P402A/P564A) in macrophages. Accordingly, RAW264.7 macrophages were transfected with a combination of siStat3 siRNA and ΔHif1α plasmid. These cells were stimulated with LPS, and total RNA derived from these experiments was analyzed for proinflammatory gene targets by RT-qPCR analysis. As shown in Figure 8, C–F, inhibition of STAT3 significantly attenuated LPS-induced Il1α, Il1β, Icam1, and Serpine1 expression in macrophages. Surprisingly, overexpression of oxygen-insensitive form of HIF1α completely reversed attenuated Il1α, Il1β, Icam1, and Serpine1 expression in STAT3-deficient macrophages (Figure 8, C–F). Collectively, these studies support a role for STAT3 signaling in the regulation of HIF1α and attendant gene expression in macrophages.
Figure 7.
STAT3 signaling supports hypoxia-inducible factor-1α (HIF1α) expression in macrophages. A–D: Wild-type mice bone marrow–derived macrophages (BMDMs) were treated with 100 ng/mL lipopolysaccharide (LPS) or 10 ng/mL interferon (IFN)-γ for 0 to 6 hours. A and B: HIF1α protein (A) and mRNA (B) expression levels were analyzed by Western blot analysis and RT-qPCR, respectively. C and D: The expression and phosphorylation status of STAT1 (C) and STAT3 (D) were analyzed by Western blots. E: Wild-type mice BMDMs were transfected with siControl, siStat1, or siStat3 siRNA. These cells were stimulated with 100 ng/mL LPS or 10 ng/mL IFN-γ for 4 hours. Total protein extracts were analyzed for HIF1α protein expression by Western blot analysis. F and G: Wild-type and dominant-negative mutant-Stat3 mice BMDMs were stimulated with 100 ng/mL LPS or 10 ng/mL IFN-γ for 4 hours. F and G: HIF1α mRNA (F) and protein (G) expression were analyzed by RT-qPCR and Western blot analysis, respectively. D–F: Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (B and F). n = 4 (C–G). ∗∗P < 0.01, ∗∗∗P < 0.001. PBS, phosphate-buffered saline; pSTAT, phosphorylated STAT.
Figure 8.
STAT3–hypoxia-inducible factor-1α (HIF1α) axis modulates inflammatory gene expression in macrophages. A: RAW264.7 cells were transfected with an HIF1α promoter-driven luciferase reporter construct in the presence of control or Stat3-specific siRNA. These cells were stimulated with lipopolysaccharide (LPS) or interferon (IFN)-γ for 6 hours, and cell lysates were analyzed for luciferase activity. B: Wild-type mice bone marrow–derived macrophages were stimulated with LPS or IFN-γ for 4 hours, and chromatin immunoprecipitation analysis was performed on Hif1α promoter (−1041 to −1051) utilizing anti-STAT3 antibody. C–F: RAW264.7 cells were cotransfected with a combination of Stat3-specific siRNA or oxygen-stable form ΔHIF1α plasmid and stimulated with 100 ng/mL LPS for 5 hours. C–F: Total RNA from these experiments was evaluated for expression of Il1α (C), Il1β (D), Icam1 (E), and Serpine1 (F) by RT-qPCR. Data were analyzed by analysis of variance, followed by Bonferroni post-testing. Values are reported as means ± SD (A–F). n = 3 (A and B); n = 4 (C–F). ∗∗P < 0.01, ∗∗∗P < 0.001. PBS, phosphate-buffered saline.
Discussion
The central finding of this study is that myeloid-HIF1α signaling supports carotid artery stenosis by modulating broad inflammatory and metabolic gene expression in macrophages. The key observations are as follows: i) Myeloid-HIF1α deficiency attenuates carotid neointima formation in vivo. ii) Myeloid-HIF1α deficiency abrogates vascular smooth muscle cells and macrophage abundance in stenotic arteries. iii) HIF1α deficiency broadly restrained proinflammatory and glycolytic gene expression in stenotic artery macrophages. iv) Genes involved in oxidative phosphorylation, TCA, fatty acid metabolism, and MYC target expression are elevated in HIF1α-deficient macrophages ex vivo and in vivo. v) STAT3 signaling supports HIF1α and attendant gene expression in macrophages. Collectively, this study provides evidence that macrophage-HIF1α deficiency curbs carotid artery stenosis by rewiring inflammatory and metabolic signaling pathways in macrophages.
Previous studies have demonstrated that phenotypes of infiltrating macrophages influence carotid artery disease vulnerability in humans.19 In this study, symptomatic patients expressed elevated levels of proinflammatory genes, including chemokine (C-C motif) ligand 2, tumor necrosis factor, IL-6, matrix metalloproteinase 2, and matrix metalloproteinase 9, in carotid artery macrophages.19 Prior studies have also implicated HIF1α as an exclusive transcription factor that supports proinflammatory macrophage activation and functions.21 Besides, HIF1α signaling has been associated with several physiological and pathophysiological processes, including but not limited to angiogenesis, arteriogenesis, vasculogenesis, and vascular homeostasis.30 Interestingly, previous studies have shown that flexible guide wire–induced vascular injury significantly elevates HIF1α mRNA and protein expression in injured arteries.26 Indeed, local inhibition of HIF1α by utilizing siRNA attenuates wire injury–induced VSMC accumulation and neointima formation.26 Likewise, the current study showed that myeloid-HIF1α deficiency attenuated ligation-induced neointima formation and broad vascular remodeling in vivo. Congruent with previous observations, myeloid or local inhibition of HIF1α significantly attenuated angioplasty guide wire–induced neointima formation, VSMC proliferation, and macrophage accumulation in injured femoral arteries.31,32 Local expression of classic HIF1α gene targets is also attenuated in HIF1α-deficient diseased femoral arteries.31,32 Consistent with these observations, myeloid-HIF1α deficiency attenuated neointima formation, medial thickening, and occlusion of carotid arteries in the current study. In addition, carotid ligation altered inflammatory and metabolic gene targets regulated by HIF1α. Concordant with previous studies, inhibition of HIF1α significantly diminished expression of these genes in stenotic artery macrophages. Analyses by Lambert et al33 demonstrate that local inhibition of HIF1α functions by adenoviral-mediated delivery of dominant-negative HIF1α, which significantly curtailed balloon injury–induced carotid artery pathologic remodeling in a rat model. Similarly, Qi et al34 observed that angiotensin II exposure elevated HIF1α mRNA and protein expression in VSMCs. More important, VSMC-specific deficiency of HIF1α significantly attenuated angiotensin II–induced vascular wall remodeling in vivo 34 In this context, the current report shows that myeloid-specific HIF1α deficiency attenuated ligation-induced carotid artery stenosis and arterial wall remodeling in vivo. Furthermore, VSMC-specific deficiency of HIF1α attenuated macrophage recruitment into the vascular wall as well as proinflammatory gene expression in angiotensin II–induced vascular disease model.34 Concordant with these observations, this study shows that myeloid-HIF1α deficiency attenuates macrophage abundance in stenotic carotid artery walls. In addition, this study also indicates that myeloid-HIF1α–deficient mice stenotic carotid artery macrophages expressed lower levels of proinflammatory genes compared with the control group. There is a close relationship between macrophage infiltration and the extent of smooth muscle cell proliferation. Studies by Danenberg et al35 show that macrophage depletion by bisphosphonate-liposomes attenuates neointima formation after carotid balloon injury, indicating that macrophage-mediated inflammation augments VSMC growth. Similarly, macrophage CCR2 depletion abrogates angiotensin II–induced vascular hypertrophy because of the reduction of proliferating VSMCs in the aortic medial layer.36 Furthermore, chemokine (C-C motif) ligand 2–targeted gene therapy inhibits neointima formation after balloon injury,37 stenting,38 and vein graft.39 Thus, further studies are needed to address whether macrophage-HIF1α signaling alters VSMC function by modulating the chemokine (C-C motif) ligand 2–CCR2 axis. Previous studies have implicated STAT3 signaling in VSMC migration and vascular wall remodeling.40,41 In this context, the current study shows that inhibition of STAT3 signaling dwindled cytokine-induced HIF1α and attendant gene expression in macrophages.
In summary, the observations presented herein highlight the importance of HIF1α signaling in amending broad inflammatory and metabolic response in stenotic carotid artery macrophages. The in vivo studies showed that myeloid-HIF1α deficiency significantly alleviates ligation-induced carotid artery remodeling by altering macrophage and VSMC functions. Furthermore, the current study also showed that HIF1α supports proinflammatory and glycolytic gene expression in diseased carotid artery macrophages. More importantly, HIF1α facilitated effective proinflammatory macrophage activation by repressing oxidative phosphorylation, TCA, fatty acid metabolism, and anti-inflammatory c-MYC pathways. At the molecular level, proinflammatory agents utilized STAT3 signaling to elevate HIF1α expression in macrophages. Furthermore, suppression of STAT3 signaling significantly diminished HIF1α and attendant target gene expression in macrophages.
Footnotes
Supported by NIH grants HL126626 and HL141423 and Crohn's and Colitis Foundation Senior Research Award 421904 (G.H.M.).
G.-D.K. and H.P.N. contributed equally to this work.
Disclosures: None declared.
Author Contributions
G.H.M. conceived and designed the study. G.H.M., G.-D.K., and H.P.N. performed experiments. G.H.M., G.-D.K., H.P.N., and E.R.C. analyzed and interpreted the data. G.H.M. wrote the manuscript, which was edited and approved by all authors.
References
- 1.Fayad Z.A., Swirski F.K., Calcagno C., Robbins C.S., Mulder W., Kovacic J.C. Monocyte and macrophage dynamics in the cardiovascular system: JACC macrophage in CVD series (part 3) J Am Coll Cardiol. 2018;72:2198–2212. doi: 10.1016/j.jacc.2018.08.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jander S., Sitzer M., Schumann R., Schroeter M., Siebler M., Steinmetz H., Stoll G. Inflammation in high-grade carotid stenosis: a possible role for macrophages and T cells in plaque destabilization. Stroke. 1998;29:1625–1630. doi: 10.1161/01.str.29.8.1625. [DOI] [PubMed] [Google Scholar]
- 3.Shaikh S., Brittenden J., Lahiri R., Brown P.A., Thies F., Wilson H.M. Macrophage subtypes in symptomatic carotid artery and femoral artery plaques. Eur J Vasc Endovasc Surg. 2012;44:491–497. doi: 10.1016/j.ejvs.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Bonati L.H., Gregson J., Dobson J., McCabe D.J.H., Nederkoorn P.J., van der Worp H.B., de Borst G.J., Richards T., Cleveland T., Muller M.D., Wolff T., Engelter S.T., Lyrer P.A., Brown M.M. Restenosis and risk of stroke after stenting or endarterectomy for symptomatic carotid stenosis in the International Carotid Stenting Study (ICSS): secondary analysis of a randomised trial. Lancet Neurol. 2018;17:587–596. doi: 10.1016/S1474-4422(18)30195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heo S.H., Yoon K.W., Woo S.Y., Park Y.J., Kim Y.W., Kim K.H., Chung C.S., Bang O.Y., Kim D.I. Editor's choice - comparison of early outcomes and restenosis rate between carotid endarterectomy and carotid artery stenting using propensity score matching analysis. Eur J Vasc Endovasc Surg. 2017;54:573–578. doi: 10.1016/j.ejvs.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Martelli E., Patacconi D., DE Vivo G., Ippoliti A. Conventional carotid endarterectomy versus stenting: comparison of restenosis rates in arteries with identical predisposing factors. J Cardiovasc Surg (Toronto) 2016;57:503–509. [PubMed] [Google Scholar]
- 7.Gaba K., Ringleb P.A., Halliday A. Asymptomatic carotid stenosis: intervention or best medical therapy? Curr Neurol Neurosci Rep. 2018;18:80. doi: 10.1007/s11910-018-0888-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aday A.W., Beckman J.A. Medical management of asymptomatic carotid artery stenosis. Prog Cardiovasc Dis. 2017;59:585–590. doi: 10.1016/j.pcad.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Spence J.D., Song H., Cheng G. Appropriate management of asymptomatic carotid stenosis. Stroke Vasc Neurol. 2016;1:64–71. doi: 10.1136/svn-2016-000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nahrendorf M. Myeloid cell contributions to cardiovascular health and disease. Nat Med. 2018;24:711–720. doi: 10.1038/s41591-018-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins P.N. Molecular biology of atherosclerosis. Physiol Rev. 2013;93:1317–1542. doi: 10.1152/physrev.00004.2012. [DOI] [PubMed] [Google Scholar]
- 12.Glass C.K., Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogata A., Kawashima M., Wakamiya T., Nishihara M., Masuoka J., Nakahara Y., Ebashi R., Inoue K., Takase Y., Irie H., Abe T. Carotid artery stenosis with a high-intensity signal plaque on time-of-flight magnetic resonance angiography and association with evidence of intraplaque hypoxia. J Neurosurg. 2017;126:1873–1878. doi: 10.3171/2016.4.JNS16349. [DOI] [PubMed] [Google Scholar]
- 14.Kashiwazaki D., Koh M., Uchino H., Akioka N., Kuwayama N., Noguchi K., Kuroda S. Hypoxia accelerates intraplaque neovascularization derived from endothelial progenitor cells in carotid stenosis. J Neurosurg. 2018;131:884–891. doi: 10.3171/2018.4.JNS172876. [DOI] [PubMed] [Google Scholar]
- 15.Samanta D., Prabhakar N.R., Semenza G.L. Systems biology of oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med. 2017;9 doi: 10.1002/wsbm.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardos J.I., Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta. 2005;1755:107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Blouin C.C., Page E.L., Soucy G.M., Richard D.E. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 18.Palazon A., Goldrath A.W., Nizet V., Johnson R.S. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho K.Y., Miyoshi H., Kuroda S., Yasuda H., Kamiyama K., Nakagawara J., Takigami M., Kondo T., Atsumi T. The phenotype of infiltrating macrophages influences arteriosclerotic plaque vulnerability in the carotid artery. J Stroke Cerebrovasc Dis. 2013;22:910–918. doi: 10.1016/j.jstrokecerebrovasdis.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Guo L., Akahori H., Harari E., Smith S.L., Polavarapu R., Karmali V. CD163+ macrophages promote angiogenesis and vascular permeability accompanied by inflammation in atherosclerosis. J Clin Invest. 2018;128:1106–1124. doi: 10.1172/JCI93025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cramer T., Yamanishi Y., Clausen B.E., Förster I., Pawlinski R., Mackman N., Haase V.H., Jaenisch R., Corr M., Nizet V., Firestein G.S., Gerber H.P., Ferrara N., Johnson R.S. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A., Lindner V. Remodeling with neointima formation in the mouse carotid artery after cessation of blood flow. Arterioscler Thromb Vasc Biol. 1997;17:2238–2244. doi: 10.1161/01.atv.17.10.2238. [DOI] [PubMed] [Google Scholar]
- 23.Kanarek N., Keys H.R., Cantor J.R., Lewis C.A., Chan S.H., Kunchok T., Abu-Remaileh M., Freinkman E., Schweitzer L.D., Sabatini D.M. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559:632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karshovska E., Zernecke A., Sevilmis G., Millet A., Hristov M., Cohen C.D., Schmid H., Krotz F., Sohn H.Y., Klauss V., Weber C., Schober A. Expression of HIF-1alpha in injured arteries controls SDF-1alpha mediated neointima formation in apolipoprotein E deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:2540–2547. doi: 10.1161/ATVBAHA.107.151050. [DOI] [PubMed] [Google Scholar]
- 27.Das A., Yang C.S., Arifuzzaman S., Kim S., Kim S.Y., Jung K.H., Lee Y.S., Chai Y.G. High-resolution mapping and dynamics of the transcriptome, transcription factors, and transcription co-factor networks in classically and alternatively activated macrophages. Front Immunol. 2018;9:22. doi: 10.3389/fimmu.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van den Bossche J., O'Neill L.A., Menon D. Macrophage immunometabolism: where are we (going)? Trends Immunol. 2017;38:395–406. doi: 10.1016/j.it.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Steward-Tharp S.M., Laurence A., Kanno Y., Kotlyar A., Villarino A.V., Sciume G., Kuchen S., Resch W., Wohlfert E.A., Jiang K., Hirahara K., Vahedi G., Sun H.W., Feigenbaum L., Milner J.D., Holland S.M., Casellas R., Powrie F., O'Shea J.J. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood. 2014;123:2978–2987. doi: 10.1182/blood-2013-09-523167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rey S., Semenza G.L. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc Res. 2010;86:236–242. doi: 10.1093/cvr/cvq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christoph M., Ibrahim K., Hesse K., Augstein A., Schmeisser A., Braun-Dullaeus R.C., Simonis G., Wunderlich C., Quick S., Strasser R.H., Poitz D.M. Local inhibition of hypoxia-inducible factor reduces neointima formation after arterial injury in ApoE-/- mice. Atherosclerosis. 2014;233:641–647. doi: 10.1016/j.atherosclerosis.2014.01.048. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama T., Kurobe H., Sugasawa N., Kinoshita H., Higashida M., Matsuoka Y., Yoshida Y., Hirata Y., Sakata M., Maxfield M.W., Shimabukuro M., Takahama Y., Sata M., Tamaki T., Kitagawa T., Tomita S. Role of macrophage-derived hypoxia-inducible factor (HIF)-1α as a mediator of vascular remodelling. Cardiovasc Res. 2013;99:705–715. doi: 10.1093/cvr/cvt146. [DOI] [PubMed] [Google Scholar]
- 33.Lambert C.M., Roy M., Robitaille G.A., Richard D.E., Bonnet S. HIF-1 inhibition decreases systemic vascular remodelling diseases by promoting apoptosis through a hexokinase 2-dependent mechanism. Cardiovasc Res. 2010;88:196–204. doi: 10.1093/cvr/cvq152. [DOI] [PubMed] [Google Scholar]
- 34.Qi D., Wei M., Jiao S., Song Y., Wang X., Xie G., Taranto J., Liu Y., Duan Y., Yu B., Li H., Shah Y.M., Xu Q., Du J., Gonzalez F.J., Qu A. Hypoxia inducible factor 1α in vascular smooth muscle cells promotes angiotensin II-induced vascular remodeling via activation of CCL7-mediated macrophage recruitment. Cell Death Dis. 2019;10:544. doi: 10.1038/s41419-019-1757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danenberg H.D., Fishbein I., Epstein H., Waltenberger J., Moerman E., Mönkkönen J., Gao J., Gathi I., Reichi R., Golomb G. Systemic depletion of macrophages by liposomal bisphosphonates reduces neointimal formation following balloon-injury in the rat carotid artery. J Cardiovasc Pharmacol. 2003;42:671–679. doi: 10.1097/00005344-200311000-00014. [DOI] [PubMed] [Google Scholar]
- 36.Ishibashi M., Hiasa K., Zhao Q., Inoue S., Ohtani K., Kitamoto S., Tsuchihashi M., Sugaya T., Charo I.F., Kura S., Tsuzuki T., Ishibashi T., Takeshita A., Egashira K. Critical role of monocyte chemoattractant protein-1 receptor CCR2 on monocytes in hypertension-induced vascular inflammation and remodeling. Circ Res. 2004;94:1203–1210. doi: 10.1161/01.RES.0000126924.23467.A3. [DOI] [PubMed] [Google Scholar]
- 37.Usui M., Egashira K., Ohtani K., Kataoka C., Ishibashi M., Hiasa K., Katoh M., Zhao Q., Kitamoto S., Takeshita A. Anti-monocyte chemoattractant protein-1 gene therapy inhibits restenotic changes (neointimal hyperplasia) after balloon injury in rats and monkeys. FASEB J. 2002;16:1838–1840. doi: 10.1096/fj.02-0094fje. [DOI] [PubMed] [Google Scholar]
- 38.Egashira K., Nakano K., Ohtani K., Funakoshi K., Zhao G., Ihara Y., Koga J., Kimura S., Tominaga R., Sunagawa K. Local delivery of anti-monocyte chemoattractant protein-1 by gene-eluting stents attenuates in-stent stenosis in rabbits and monkeys. Arterioscler Thromb Vasc Biol. 2007;27:2563–2568. doi: 10.1161/ATVBAHA.107.154609. [DOI] [PubMed] [Google Scholar]
- 39.Tatewaki H., Egashira K., Kimura S., Nishida T., Morita S., Tominaga R. Blockade of monocyte chemoattractant protein-1 by adenoviral gene transfer inhibits experimental vein graft neointimal formation. J Vasc Surg. 2007;45:1236–1243. doi: 10.1016/j.jvs.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 40.Potula H.S., Wang D., Quyen D.V., Singh N.K., Kundumani-Sridharan V., Karpurapu M., Park E.A., Glasgow W.C., Rao G.N. Src-dependent STAT-3-mediated expression of monocyte chemoattractant protein-1 is required for 15(S)-hydroxyeicosatetraenoic acid-induced vascular smooth muscle cell migration. J Biol Chem. 2009;284:31142–31155. doi: 10.1074/jbc.M109.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh N.K., Wang D., Kundumani-Sridharan V., Van Quyen D., Niu J., Rao G.N. 15-Lipoxygenase-1-enhanced Src-Janus kinase 2-signal transducer and activator of transcription 3 stimulation and monocyte chemoattractant protein-1 expression require redox-sensitive activation of epidermal growth factor receptor in vascular wall remodeling. J Biol Chem. 2011;286:22478–22488. doi: 10.1074/jbc.M111.225060. [DOI] [PMC free article] [PubMed] [Google Scholar]